Abstract
The mitogen-activated protein kinase (MAPK) ERK2 is ubiquitously expressed in mammalian tissues and is involved in a wide range of biological processes. Although MAPKs have been intensely studied, identification of their substrates remains challenging. We have optimized a chemical genetic system using analog-sensitive ERK2,a form of ERK2 engineered to utilize an analog of ATP, to tag and isolate ERK2 substrates in vitro. This approach identified 80 proteins phosphorylated by ERK2, 13 of which are known ERK2 substrates. The 80 substrates are associated with diverse cellular processes, including regulation of transcription and translation, and mRNA processing, as well as regulation of the activity of the Rho-family guanosine triphosphatases. We found that one of the newly identified substrates, ETV3 (a member of the E-twenty six family of transcriptional regulators) was extensively phosphorylated on sites within canonical and non-canonical ERK motifs. Phosphorylation of ETV3 regulated transcription by preventing its binding to DNA at promoters for several thousand genes, including some involved in negative feedback regulation of itself and of upstream signals.
Introduction
Much of the signal processing in eukaryotes involves protein phosphorylation by networks of kinases and phosphatases. Mitogen-activated protein kinases (MAPKs) are serine/threonine kinases that are conserved from yeast to human and represent an integral part of this network (1). In particular, the extracellular-signal regulated kinases 1 and 2 (ERK1/2) are ubiquitously expressed in mammalian cells and involved in many biological processes, including development (2), glucose homeostasis (3), immune function (4), and memory (5). Deregulation of ERK1/2 activity is common in cancer and leads to proliferation, migration, resistance to apoptosis, and loss of differentiated phenotypes (6).
ERK1/2 regulates these various biological processes by phosphorylating hundreds of substrate proteins (7). Substrate recognition is mediated by protein-protein docking sites, and the substrate binding cleft allows for serine or threonine phosphorylation within an amino acid motif with proline strongly preferred at the +1 and common at −2 positions (8). ERK activity and substrate specificity is further regulated by scaffolds and adaptors that assemble the MAPK kinase kinase Raf to MAPK kinase MEK to ERK activation cascade and direct subcellular localization (9). Even though many ERK substrates have been identified, incomplete knowledge of ERK targets remains a hurdle to understanding the myriad biological consequences of ERK activity.
To understand how ERK2 regulates diverse biological responses to cellular stimuli, we sought to identify direct ERK2 substrates and determine quantitative substrate utilization under different biological contexts. However, elucidating enzyme-substrate interactions among signaling molecules is difficult because many substrates are extremely low abundance, interactions are often transient, phosphorylation stoichiometry may be low, and residues are often phosphorylated by several kinases. Moreover, targeted genetic knockdown or chemical inhibition may lead to pleiotropic effects due to complex feedback and crosstalk within signaling networks (10). Despite these concerns, several groups have used global quantitative phosphoproteomics to characterize ERK1/2 signaling by identifying phosphorylation sites that respond to MEK inhibition (10–13).Among these groups, Kosako et al. used a combination of affinity chromatography and 2D gel electrophoresis to identify MEK-dependent phosphorylation sites, including 24 potential ERK targets, many of which were biochemically validated as direct ERK1/2 substrates (13). However, phosphoprotein analysis by itself does not establish direct kinase/substrate relationships and is inherently limited by the dynamic range between abundant phosphorylated proteins and low-abundance substrates. An alternative approach is to selectively tag and enrich direct kinase substrates, allowing more sensitive detection of direct substrates by reducing the background of proteins phosphorylated by other kinases.
Analog-sensitive kinases (AS-kinases) can accommodate unnatural ATP analogs through an expanded ATP-binding pocket; these engineered proteins have been used to identify direct kinase/substrate interactions for several kinases (14). AS-kinases retain their natural substrate specificity. For instance, ERK2 with Q103G substitution (AS-ERK2) uses ATP and ATP analogs efficiently and interacts with known ERK2 substrates (15), and an AS form of cyclin-dependent kinase 2 (AS-CDK2) retains the kinetics and substrate specificity of the wild-type protein (16). Radioactive tagging on γ-phosphate of the ATP analog was originally used for selective labeling and detection of AS-kinase substrates, but purification and identification remained problematic. Two procedures have been reported that use γ-thiol-phosphate ATP analogs to thiophosphorylate substrates of cyclin/CDK complexes in whole cell lysates (17, 18). In both cases, the thiophosphate serves as a nucleophilic “handle” to purify peptides from tryptic digests so that substrates can be identified by tandem mass spectrometry (MS/MS). These experiments rely on the unique ability of the AS-kinase to bind and utilize the ATP analog; any non-specific utilization of the ATP analog by other kinases in the cell will increase the background and may lead to false-positive substrate identification.
AS-ERK2 was used to identify the ubiquitin ligase EDD and nucleoporin TPR as direct substrates but discovery of additional targets has been limited by their low abundance (15). To address this issue, we have optimized the solid-phase capture of thiophosphorylated peptides and added an additional phosphopeptide enrichment to reduce background, thereby enabling low-level substrate identification. Additionally, we have incorporated quantitative mass spectrometry using stable isotope labeling in cell culture (SILAC) (19) to establish statistical thresholds for identification of substrates over background phosphorylation, thereby decreasing false-positive substrate identification. This approach is generally applicable to all AS-kinases.
With this improved methodology, we detected 98 sites directly phosphorylated by ERK2 on 80 proteins from NIH 3T3-L1 fibroblasts. Thirteen of these proteins are known substrates and the rest represent previously unknown kinase/substrate interactions (7).These results reveal a diverse and largely unknown set of substrates, and show that that ERK2 acts on a much broader range of targets than previously recognized. Among the ERK2 substrates, we identified the E-twenty six (ETS) domain-containing protein ETV3. We determined that phosphorylation of this protein by ERK2 was functionally relevant, abrogating the DNA-binding activity of ETV3 at thousands of targets across the genome, thereby providing an additional mechanism for transcriptional regulation downstream of ERK2 activation.
Results
Improved method for solid-phase capture of thiophosphorylated AS-kinase substrates
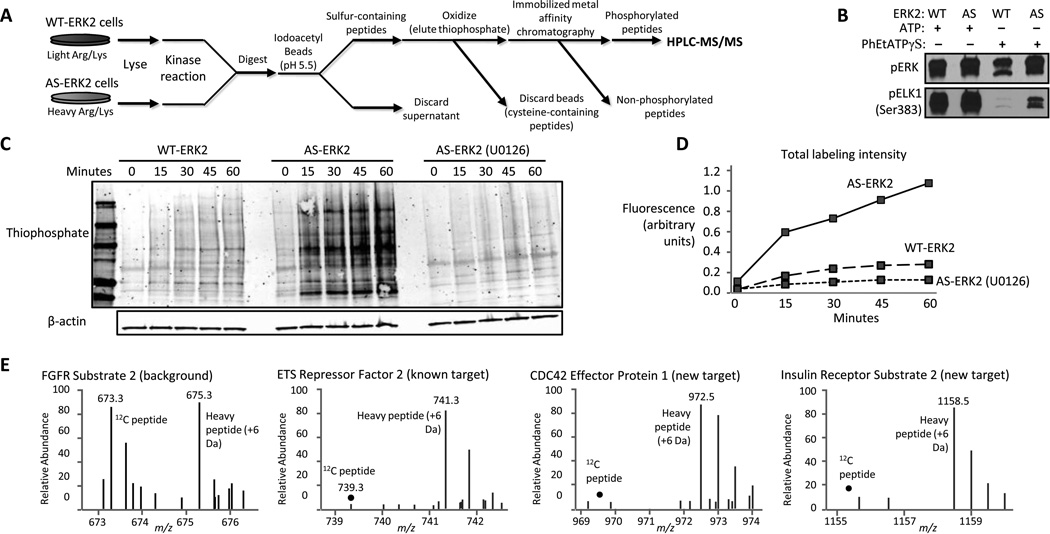
To identify direct substrates of AS-kinases, we began with the method originally developed by Allen et al. (20) and Blethrow et al. (17), in which thiophosphorylated substrate peptides are captured on a solid-phase agarose support functionalized with iodo-acetyl groups, then released by oxidative hydrolysis to yield phosphorylated peptides. We found that oxidative hydrolysis of bead-conjugated thiophosphopeptides released non-specifically adsorbed, non-phosphorylated peptides that can interfere with identification of low-abundance substrates. To reduce the amount of the non-phosphorylated background peptides, we included an additional phosphopeptide enrichment using immobilized metal affinity chromatography (IMAC) (21). We also determined that selective capture of thiophosphorylated peptides was enhanced if the pH of the thiophosphate coupling reaction was decreased to 5.5, from 7.0 used by Blethrow et al, to favor binding of thiophosphate over cysteine. The final protocol is schematically represented in Fig. 1A (a detailed protocol is available in the Supplementary Methods).
Fig. 1. Identification of direct substrates of AS-ERK2.
(A) Substrates of AS-ERK2 were thiophosphorylated in cell lysate, digested, and covalently coupled to iodoacetyl-agarose beads. Phosphopeptides were recovered by oxidative hydrolysis and further enriched by IMAC prior to analysis by LC-MS/MS. (B) Activated wild-type (WT) and AS-ERK2 phosphorylate a known substrate (ELK1). Activated kinases were immunoprecipitated and incubated with ELK1-GST and 50 µM ATP or N6-PhEt-ATPγ Sin kinase buffer for 15 minutes at 30°C. (C) Thiophosphorylation of kinase substrates occurs in lysate of AS-ERK2-expressing cells to a greater extent than occurs in WT-ERK2-expressing cells. 3T3-L1 fibroblasts expressing WT- or AS-ERK2 were stimulated with PMA for 3 minutes or treated with U0126 for 20 minutes followed by PMA and lysed in kinase buffer and thiophosphorylation reactions were carried out for the indicated times. Thiols were alkylated by PNBM followed by Western blot for thiophosphate ester. (D) Quantification of the total signal for each lane from panel C, demonstrating the level and MEK-dependence of the background thiophosphorylation in WT-ERK2 expressing cells. (E) SILAC-labeled peptides provide a quantitative basis to determine whether a given phosphorylation site is a direct ERK2 substrate, with examples of background phosphorylation), a known ERK2 substrate, and two newly identified substrates.
Because the AS-kinase approach relies on the selective affinity of the AS-kinase for ATP analogs, we confirmed the selective utilization of PhEt-ATPγS by AS-ERK2 relative to wild-type ERK2 (WT-ERK2) by an in vitro kinase reaction using PhEt-ATPγS (Fig. 1B). To test specificity of labeling in the context of a whole cell lysate, we expressed matched amounts of AS-ERK2 and WT-ERK2 in NIH 3T3-L1 cells and conducted in vitro reactions in lysate of cells treated with phorbol-12-myristate-13-acetate (PMA). As expected, labeling was strongest in the presence of AS-ERK2, and was decreased by pre-treatment with the MEK inhibitor U0126 (Fig. 1C, D). We also detected labeling in control cells expressing WT-ERK2, indicating utilization of PhEt-ATPγS by wild-type kinases (Fig. 1C, D). The cumulative non-specific labeling by endogenous kinases indicated that a quantitative approach to measuring non-specific labeling was necessary to resolve low abundance substrates from background signals.
Identification of Q103G–ERK2 (AS-ERK2) substrates
To distinguish phosphorylated ERK2 substrates from non-specific background phosphorylation, we used SILAC for quantitative comparison between AS-ERK2 and WT-ERK2 cell lysates. Cells expressing AS-ERK2 were grown in “heavy” L-arginine and L-lysine labeled with 13C or 13C and 15N, whereas WT-ERK2 cells were grown in L-arginine and L-lysine containing entirely 12C and 14N. Samples were combined after in vitro kinase reactions and prior to tryptic digestion. The resulting peptides have distinctive mass signatures: Background peptides have light and heavy ion signals with similar intensity, whereas ERK2 substrate peptides are enriched for heavy ions.
We combined this SILAC quantitative approach with our solid-phase capture protocol to identify thiophosphorylated peptides from six independent labeling reactions with lysates of epidermal growth factor (EGF)-stimulated 3T3-L1 cells expressing WT-ERK2 or AS-ERK2. We used MASCOT (22) to match MS/MS spectra to phosphorylated peptides and examined the results for peptides matching the minimal ERK2 substrate motif: pSer or pThr followed by proline. The MS/MS spectra for all potential ERK2 substrates were manually inspected to ensure proper placement of the phosphorylated residue (Supplementary spectra), and validation with synthetic peptides was used to confirm any low-confidence assignment.
We detected phosphorylated peptides from thirteen reported substrates: Cx43,DYNC1I2, DYNC1LI1, EGFR, ERF, KSR, LMNA, MAPKAP2, NUP153, TPR, STMN1, RPS3, and SORBS3 (7, 13). (A complete annotated list, including the complete names, of all identified ERK2 substrates are provided as table S1.) To differentiate AS-ERK2 substrates from background phosphorylation, we used these thirteen substrates to set a threshold for the quantitative difference in phosphorylation between the AS-ERK2-and WT-ERK2-expressing cells. Signal-over-background ratios for known ERK2 substrates ranged from 6% to 33% (Table 1), whereas non-specific peptides had ratios around 100% with a standard deviation (S.D.) of 19%. Because residual light amino acids in heavy-labeled cells were less than 2%, we concluded that ERK2 substrate sites were subject to varying degrees of non-specific labeling. This result highlights the importance of quantitatively analyzing the negative control to distinguish background from bona fide substrates. To evaluate approximately 200 potential ERK2 substrates, we set a threshold of 3.5 standard deviations below 100% in log-space, corresponding to a light:heavy ratio of 47% and a Bonferroni-corrected familywise error rate of 0.05,assuming that ratios are log-normal distributed under the null-hypothesis.
Table 1.
Previously reported ERK2 substrates and selected previously unknown ERK2 substrates identified as substrates of AS-ERK2. A complete list appears in table S1. Some observations from table S1 are excluded from mean and range calculations because of low signal. Residue numbers are for mouse proteins. Names in parentheses represent alternative designations common in literature.
| Reported ERK1/2 Substrates | Novel ERK2 Substrates | ||||
|---|---|---|---|---|---|
| Protein | Position | WT/AS Ratio† (Range, N) |
Protein | Position | WT/AS Ratio† (Range, N) |
| Cx43 (DJA1) | Ser255 | <26% (NA, 1) | ARHGEF17 | Thr695 | 19% (15–23%, 2) |
| DYNC1I2 | Ser81 | 7% (5–7%, 2) | CDC42EP1 | Ser113 Ter197 |
14% (5–27%, 4) 27% (NA, 1) |
| DYNC1LI2 | Ser516 | <27% (NA, 1) | CDC42EP2 | Ser101 | 8% (5–11%, 4) |
| EGFR | Thr695 | 21% (17–26%, 2) | DOCK1 | Thr1772 | 9% (6–13%, 4) |
| ERF | Thr529 | 6% (2–9%, 6) | ETV3 (PE1) | Ser139 | 16% (16–17%, 2) |
| KSR | Thr260 | 13% (12–14%, 2) | FOX2 (RBM9) |
Thr65 | 41% (NA, 1) |
| LNMA | Ser22 | 11% (7–17%, 3) | GLI2 | Thr885 | 5% (4–4%, 4) |
| MAPKAP2 | Thr320 | 26% (20–32%, 2) | GIGYF2 | Ser30 | 6% (5–8%, 5) |
| NUP153 | Thr610 | 13% (11–15%, 2) | hnRNP H1* | Ser104 | 21% (18–24%, 2) |
| RPS3 | Thr221 | 41% (29–67%, 5) | IRS2 | Thr653 | <21% (19–22%, 2) |
| STMN1 | Ser25 Ser38 |
9% (5–14%, 6) 33% (24–50%, 3) |
STK10 | Thr950 | 9% (6–17%, 4) |
| SORBS3 | Thr648 | 12% (9–15%, 3) | PDXDC1 | Thr691 | 13% (5–26% 5) |
| TPR | Thr2110 Thr2131 Ser2149 |
5% (NA, 1) 6% (5–7%, 5) 7% (4–9%, 4) |
RBPMS | Thr118 | 19% (5–27%, 5) |
The same peptide matches hnRNP H2.
Ratios indicated with “<” are upper-bound values because the light ion peak was undetectable.
A total of 98 peptides from 80 proteins met the criteria for in vitro ERK2 substrates, including 67 previously uncharacterized ERK2 substrates. Fig. 1E shows representative SILAC mass signatures for several phosphorylated peptides. Three phosphopeptides matched the consensus ERK2 motif but had light to heavy ratios close to 100% (ATF7, HIVEP1, G3BP-1), suggesting that these were nonspecific events. In agreement with our data, ATF7 phosphorylation does not respond to MEK inhibition in HeLa cells despite its homology to ATF2 Thr53 (position 71 in human), a well-known ERK2 substrate site (23). Without quantitative data, these peptides could have been mistaken as ERK2 substrates.
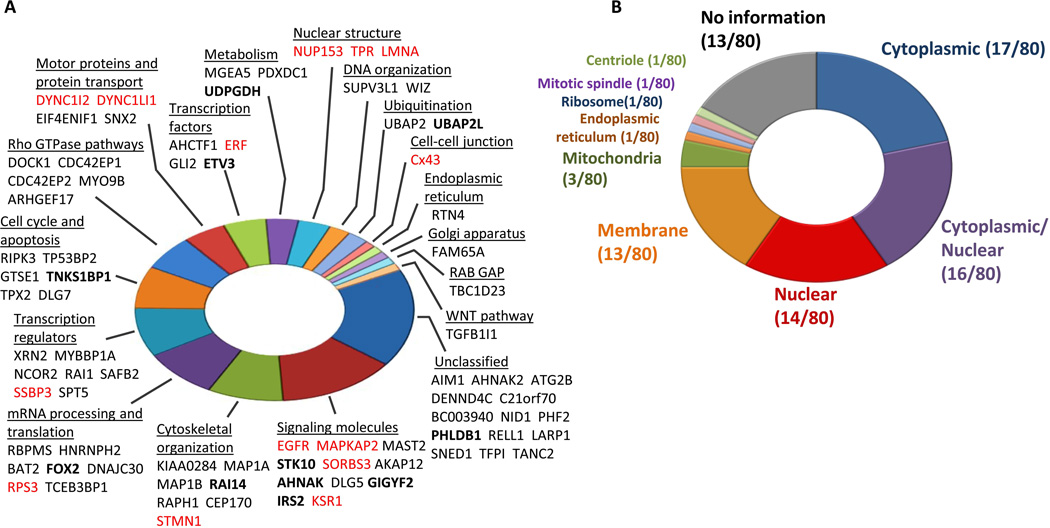
As expected based on the complex circuitry of the ERK2 network, we identified substrates that spanned a range of both upstream and downstream signaling pathways across many biological processes (Fig. 2A). These substrates were associated with multiple cellular compartments, including the cytoplasm, nucleus, and membrane, among other compartments and organelles (Fig. 2B).
Fig. 2. Analysis of AS-ERK2 substrates.
(A) Functional classifications for proteins identified as substrates of AS-ERK2 on the basis of analysis of Gene Ontology (GO)terms, UniProt annotation, and literature review. Proteins in red are previously reported ERK2 substrates, proteins in bold have in vivo evidence of MEK-dependent phosphorylation. (B) AS-ERK2 substrate localization based on “Cellular Component”annotations from GO, giving priority to experimental over automated annotations. Proteins annotated as both nuclear and cytoplasmic are shown as cytoplasmic/nuclear
Validating AS-ERK2 substrate specificity
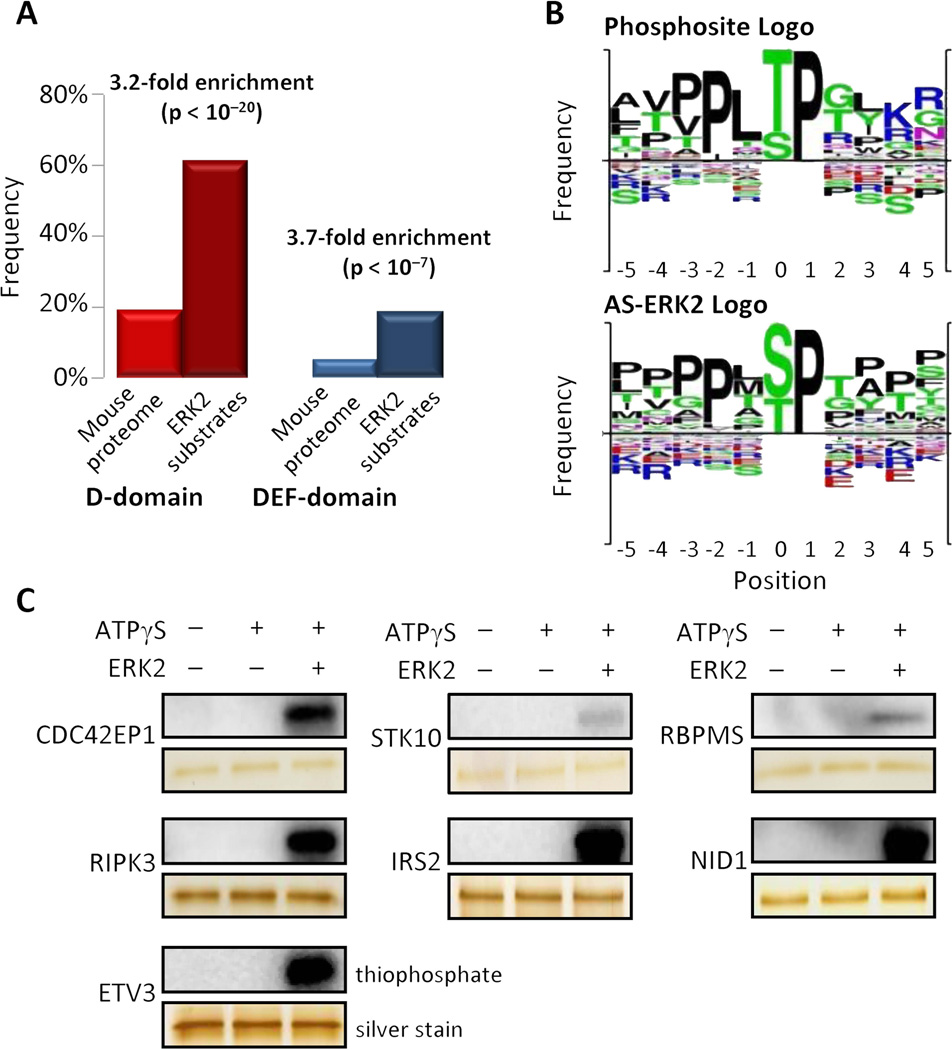
To determine whether the newly identified substrates were consistent with known ERK1/2 binding motifs, we used ScanSite to search the substrates for ERK binding sequences: D-domains (positively charged residues three to five residues ahead of a hydrophobic sequence) and DEF-domains (FXFP, where X is any amino acid and Y may substitute for F) (24). Both motifs were strongly enriched in the AS-ERK2 substrates compared to the SwissProt mouse proteome (Fig. 3A). We also compared phosphorylation site motifs for AS-ERK2 substrates with known ERK2 substrates according to PhosphoSite (www.phosphosite.org) and found that major features of the two motifs were identical (Fig 3B). Although most detected substrates matched the minimal ERK1/2 consensus motif SP/TP, and many matched the full PXSP/PXTP motif, these motifs were neither necessary nor sufficient to identify ERK2 substrates. In fact, one substrate phosphorylation site on the transcriptional repressor ETV3 was found that lacks the adjacent proline in the +1 position.
Fig. 3. Validation of AS-ERK2 substrates.
(A) AS-ERK2 substrates are enriched for ERK1/2-binding motifs. Substrate protein sequences were analyzed by ScanSite (http://scansite.mit.edu) for the presence of ERK 1/2 binding domains with high stringency. The bar plot compares the frequency of high stringency D-domains and DEF-domains between AS-ERK2 substrates and the entire SwissProt mouse proteome., P-values were calculated using a χ 2 -test. (B) Amino acid frequencies around the phosphorylation site are similar for known ERK2 substrates (top plot) and for targets of AS-ERK2 (bottom plot). (C) Newly identified substrates were expressed in E.coli and phosphorylated by wild-type ERK2 through in vitro kinase reactions.
Although the ERK2 Q103G mutation has no effect on substrate specificity (15), we tested several of the substrates identified in this study for phosphorylation by WT-ERK2. Each of the seven recombinant substrate proteins was phosphorylated in vitro by ERK2 to varying degrees (Fig. 3C). We also mapped phosphorylation of recombinant CDC42EP1, IRS2 and ETV3 by mass spectrometry following in vitro kinase reactions using wild-type ERK2 and identified additional ERK2-dependent phosphorylation site son each protein (Fig. 4A, fig. S1).
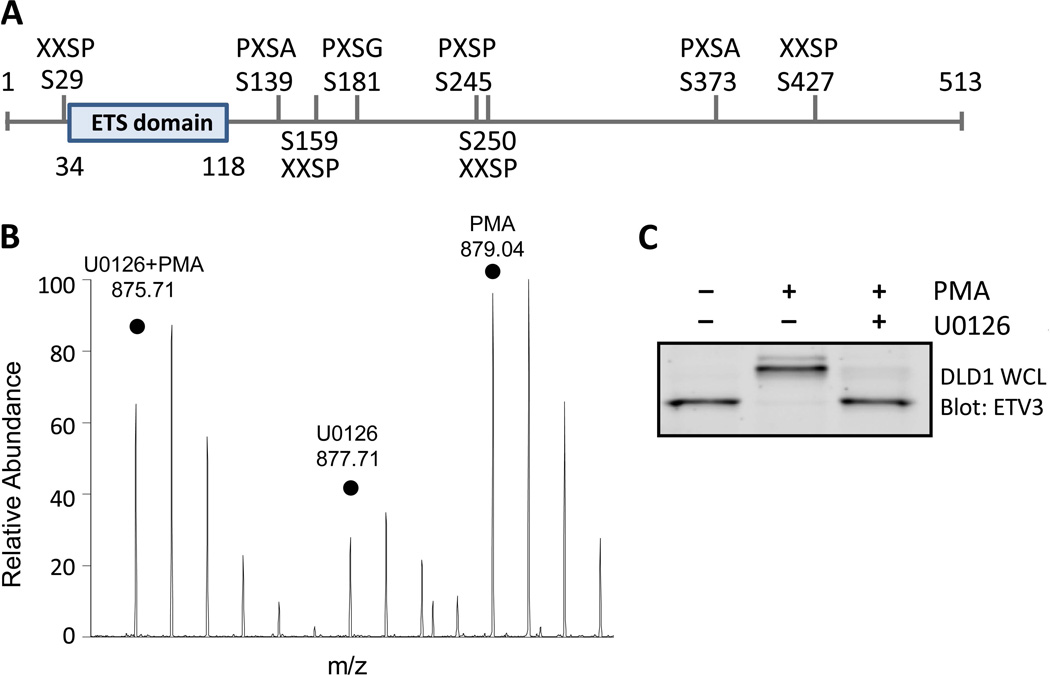
Fig. 4. Phosphorylation of FOX2 in a biological context and wide-spread phosphorylation of ETV3 by ERK1/2.
(A) Phosphorylation sites identified on recombinant ETV3 following in vitro kinase reaction with ERK2, along with the four amino acids surrounding the phosphorylated site to show the similarity of each site to theoptimal ERK1/2 motif (PX[S/T]P). (B) Phosphorylated ERK2 substrate peptide from theFOX2 splicing factor detected by peptide immunoprecipitation in SILAC-labeled 3T3-L1fibroblasts. The three signals represent cells treated with PMA, U0126, or U0126 followed by PMA as indicated. (C) ETV3 exhibits a MEK-dependent gel shift in DLD1cells treated with PMA, U0126, or U0126 pre-treatment followed by PMA.
In vivo MEK dependence of AS-ERK2 substrates
Several of the AS-ERK2 substrates exhibit altered phosphorylation following MEK inhibition. Pan et al. found reduced phosphorylation of AHNAK, ETV3, GIGYF2,PHLDB1, RAI14, TNKS1BP1, and UBAP2L following treatment of EGF-stimulated HeLa cells with U0126 (10). Kosako et al. found MEK-dependent phosphorylation of UDPGDH in 3T3-L1 fibroblasts (13), Old et al. found MEK-dependent phosphorylation of STK10 in the WM115 melanoma cell line (12), and Fritsche et al. found ERK-dependent phosphorylation of IRS2 (25). The combination of our in vitro results and the previously reported cell-based assays supports these proteins as bona fide ERK2 substrates.
To identify additional phosphorylation sites affected by MEK inhibition in an unbiased manner, we treated SILAC-labeled NIH 3T3-L1 fibroblasts with EGF or the phorbol ester PMA with or without U0126 pretreatment (experiments 1 and 2 in table S2). Lysates were digested with trypsin and antibodies recognizing the phosphorylated motifs PXpSP and PXpTP were used to immunoprecipitate peptides containing potential ERK1/2 substrate sites. A number of known ERK2 substrates were identified, including ERF and NUP153, as well as the previously unknown AS-ERK2 substrate FOX2, a splicing factor (Fig. 4B). Table S2 includes a complete list of the peptides identified in this experiment (experiments 1 through 3) and those from a similar experiment in 3T3-L1 adipocytes that also found MEK-dependent phosphorylation of FOX2 following stimulation with insulin (experiment 4).
Regulation of ETV3 by phosphorylation
Ser139 of the transcriptional repressor ETV3 (also called METS and PE-1) was phosphorylated by ERK2 despite lacking proline at the +1 position. Phosphorylation of this site was validated by MS/MS of the synthetic peptide (fig. S2), and an in vitro kinase reaction confirmed that ERK2 phosphorylates ETV3 (Fig. 3C). We detected MEK-dependent ETV3 phosphorylation in cells: Western blot analysis showed a MEK-dependent gel shift in ETV3 in DLD1 colon carcinoma cells and in HEK 293T cell streated with PMA compared with those treated with the MEK inhibitor U0126 (Fig. 4C and fig. S3A).
To determine whether ERK2 phosphorylates other sites on ETV3, we mapped phosphorylation by LC-MS/MS following an in vitro kinase reaction. Eight phosphorylated serine residues were detected, none of which were detected in a negative control (Fig. 4A). Five of the eight sites are followed by proline; whereas two are PXpSAP motifs and one is PXpSG. Although the noncanonical glycine may adopt a proline-like conformation, it seems that proline at −2 and a small amino acid at +1 may be sufficient for recognition by ERK2. Other groups have reported noncanonical sites phosphorylated by various MAPKs (26); therefore, we suspect that these sites may be systematically overlooked because it is uncommon to comprehensively map phosphorylation on putative kinase substrates.
ETV3 is related to ERF, another ETS domain-containing transcriptional repressor. ERF translocates rapidly from nucleus to cytoplasm following phosphorylation by ERK1 or ERK2 but ETV3 has been unresponsive to ERK1/2 activity in reporter gene assays (27). As a result, ETV3 is thought to repress its transcriptional targets regardless of MAPK signaling. Consistent with this previous observation, we detected predominantly nuclear localization of ETV3 in 293T cells transfected with HA-tagged ETV3 regardless of phosphorylation state (fig. S4). We observed the same result in DLD1 colon cancer cells treated with U0126 or stimulated with the PMA and stained for endogenous ETV3(fig. S5).
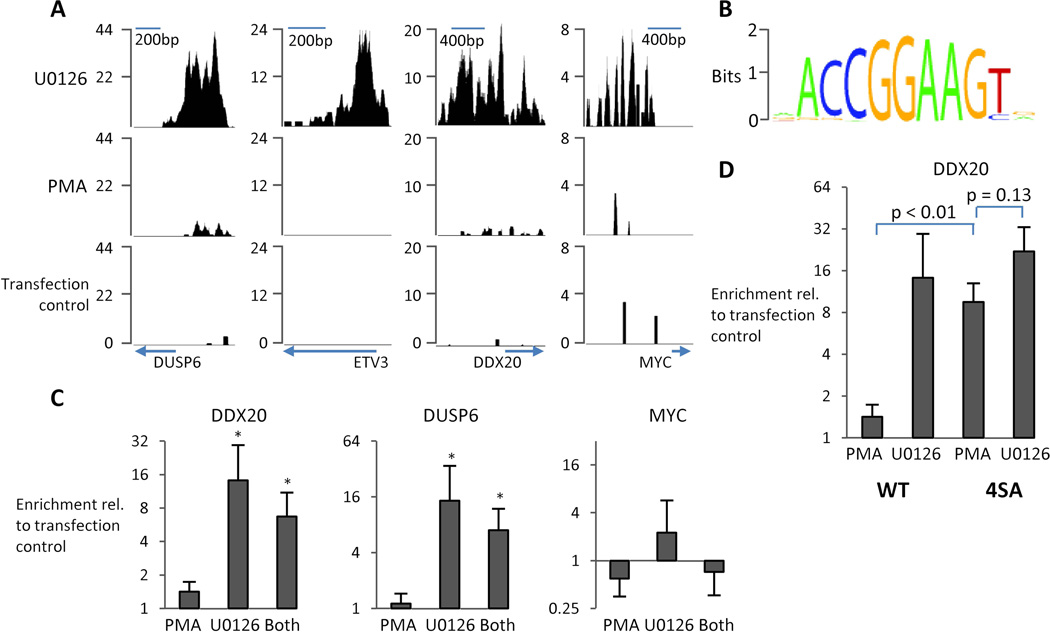
To establish the transcriptional target sites of ETV3 and the effect of phosphorylation by ERK2, we expressed human ETV3 with C-terminal HA-tag in HEK 293T cells and performed ChIP-seq analysis. This experiment revealed several thousand binding sites (tables S3, S4, S5), most of which were within 1 kilobase of known transcription start sites (fig. S6), including previously reported binding near the beginning of the myc gene (Fig. 5A). We also observed binding near the beginning of etv3 itself,etv6 (table S5, also known as tel), and ddx20, a partner required for ETV3 activity, suggesting that ETV3 negatively regulates itself and related ETS factors (Fig. 5A) (28).We used THEME to identify DNA-binding protein motifs near ETV3 binding sites and found that GGAA sequences characteristic of ETS-family proteins were enriched (Fig.5B) (29).
Fig. 5. Phosphorylation by ERK2 prevents DNA binding by ETV3.
(A) Example data from several genes identified by chromatin immunoprecipitation of HA-tagged ETV3followed by high-throughput sequencing. Cells treated with PMA (100 nM) for 20minutes display minimal binding of ETV3 to DNA; cells treated with U0126 (10 µM) for20 minutes yielded several thousand binding sites for ETV3. (B) ETV3 binds a GGAA-containing sequence characteristic of ETS-domain proteins. The most strongly enriched ETS family motif (ELK-1) from TRANSFAC was refined using THEME on high-confidence binding sites (p < 1 × 10−32). (C) ChIP and quantitative PCR for binding sites near myc, ddx20, and dusp6 from ETV3-transfected 293T cells treated as in panel D (N = 6, error bars S.E.M). Greater enrichment of ddx20 and dusp6 is observed in non-phosphorylated and partially phosphorylated conditions (* p <0.01, t-test in log-space).(D) Converting serines 139, 159, 245, and 250 (4SA) to alanine reduces the effect of phosphorylation on DNA binding (N = 4, error bars S.E.M).
Because phosphorylation by ERK eliminates the repressive activity of ERF, we hypothesized that it might have a similar effect on ETV3. Intriguingly, 20 minute stimulation with PMA led to an almost complete loss of DNA binding (Fig. 5A and 5C).
ChIP-PCR was used to confirm binding sites and showed strong phosphorylation-dependent binding near ddx20 and dusp6, which encodes a MAPK phosphatase, and weak binding near myc (Fig. 5C). The weaker binding near myc may reflect the relatively lower signal observed by ChIP-seq. We verified by Western blot that the HA antibody captures ETV3 in these ChIP experiments regardless of phosphorylation state (fig. S3B).Because serines at positions 139, 159, 245, and 250 are homologous to ERK target residues in ERF, we tested the effect of these residues in ETV3 on ERK-responsive DNA binding. Converting those four sites to alanine reduced the PMA-induced gel shift by Western blot (fig. S3C) and reduced the effect of ERK activity on DNA binding (Fig.5D). To test whether phosphorylation impaired DNA binding or simply lead to degradation of ETV3, we compared protein stability in 293T cells treated with cycloheximide and PMA or cycloheximide, PMA, and U0126. ETV3 was constitutively unstable with a half-life of approximately 2 hours, and was unaffected by the state of ERK activity (fig. S7).
The transcriptional regulator STAT3 transiently up-regulates ETV3 mRNA and protein expression in macrophages co-stimulated with interleukin-10 (IL-10) and lipopolysaccharide (30). In the same work, overexpression of ETV3 repressed targets of the p65 (RelA) subunit of the transcription factor NF-κB in COS cells. We identified ETV3 binding sites near many genes encoding proteins involved in NF-κB signaling (nfkb2, nkap, nkiras1, nkiras2, nfkbil1, trib3 and a weak signal near rela) (table S5). This set includes both positive and negative regulators of NF-κB activity, suggesting that inactivation of ETV3 may be an important step in priming the pathway to respond to other signals. ETV3 targets span a wide range of other pathways and processes including ribosomal components and members of the spliceosome (Bonferroni-correct p-value <10−4 using DAVID) (31, 32), and targets involved in p53 signaling and cell cycle control (notably tp53, chek1, cdkn2a, e2f4, ccng2 and mdm4) (table S5).
Discussion
Through an improved solid-phase capture chemical genetics strategy, we identified a large number of known and previously unknown substrates of the AS-ERK2. These additional newly identified substrates reveal a wide range of new connections between ERK2 and other signaling pathways, including adaptors and signaling molecules, regulation and effector proteins of the Rho guanosine triphosphatases (GTPases),transcription factors, splicing regulators, and structural proteins. Validation using wild-type ERK2 showed that these substrates were not artifacts of the Q103G mutation, and cell-based assays indicated that many exhibited MEK-dependent phosphorylation and are, therefore, likely to be physiologically relevant. Only 13 of the 80 substrates have been previously reported, and these represent a small fraction of the approximately 200 reported substrates, suggesting that ERK2 may phosphorylate a much larger number of proteins than previously recognized. Comprehensive identification of these targets will require various complementary technologies and detailed examination of many cell types.
Newly identified substrates expand the already considerable crosstalk between ERK1/2 and other pathways. For example, regulation of cell shape and migration is mediated in part by Rho GTPases, but few ERK1/2 substrates have been reported in Rho signaling pathways (33). Identification of ERK2 substrate sites on the Rho guanosine-nucleotide exchange factors (RhoGEFs) DOCK1 and ARHGEF17, the Rho GTPase activating protein (RhoGAP) MYO9B, and the effector proteins CDC42EP1 and CDC42EP2 suggest that ERK2 regulates these pathways both upstream and downstream from the Rho GTPases. The ERK2 phosphorylation sites on these proteins may provide previously unknown connections between MAPK signaling and regulation of cell morphology and migration. Additional investigation is needed to determine whether these phosphorylation sites function in normal physiological regulation of cytoskeletal structure and cell shape, or in pathological processes, such as cancer metastasis.
Phosphorylation of adaptor proteins may represent feedback loops by which ERK2 regulates activity of the phosphatidylinositol 3-kinase (PI3K) to AKT pathway. Homology between the residues phosphorylated by ERK2 on the adaptor in the insulin signaling pathway IRS2 and well-characterized sites on the related adaptor IRS1 suggests that phosphorylation by ERK2 may modulate the effects of insulin signaling (34). In particular, Ser907 of mouse IRS2 is ERK-responsive in hepatoma cells and liver tissue(25) and may have an functional role. Because ERK is activated by insulin signaling, identification of additional ERK phosphorylation sites raises the possibility that the kinase regulates IRS2 through several modes of feedback. Any effect is likely to be part of normal feedback regulation associated with insulin and other growth factor signaling, and could connect pathological ERK2 activation to insulin resistance caused by inflammation (35). ERK2-mediated phosphorylation of GIGYF2A, a GRB10-associated adaptor protein, is another potential interaction with insulin signaling (36), although this protein is not as well characterized as IRS2.
Another interesting point of crosstalk between ERK2 and other pathways is represented by phosphorylation of the GLI2 transcription factor. Although the ERK2 phosphorylation site has not previously been reported, it is proximal to residues near the C-terminus that are phosphorylated by protein kinase A (PKA) and glycogen synthase kinase 3 (GSK3) to regulate GLI2 stability, and therefore, may underlie the MEK-dependent crosstalk between Hedgehog and EGF receptor signaling (37).
We showed that some of the newly identified substrates, including FOX2 and ETV3, are phosphorylated by ERK2 in a cellular context. In vivo validation is important because kinases are often promiscuous when removed from their biological environment, possibly because of loss of regulation from subcellular localization and protein complexes. Some of the phosphorylation sites identified in vitro have also been observed as MEK-dependent phosphorylation sites in previous phosphoproteomics studies. The combination of in vitro and cell-based assays will continue to be important in elucidating the structure of kinase signaling networks.
ETV3 was the only ERK2 target that we identified with phosphorylation outside the canonical S/TP motif. We found that ETV3 is extensively phosphorylated by ERK2 on at least 8 serine residues, including three in noncanonical motifs. Although ETV3 has previously been reported to be unaffected by ERK activity (27), we found that its DNA-binding activity was rapidly abrogated by phosphorylation of ERK2 target residues. To characterize this effect we used ChIP-SEQ to identify several thousand binding sites for ETV3 across the genome, most of which contain an ETS consensus sequence, and all of which were lost following ERK1/2 activation.
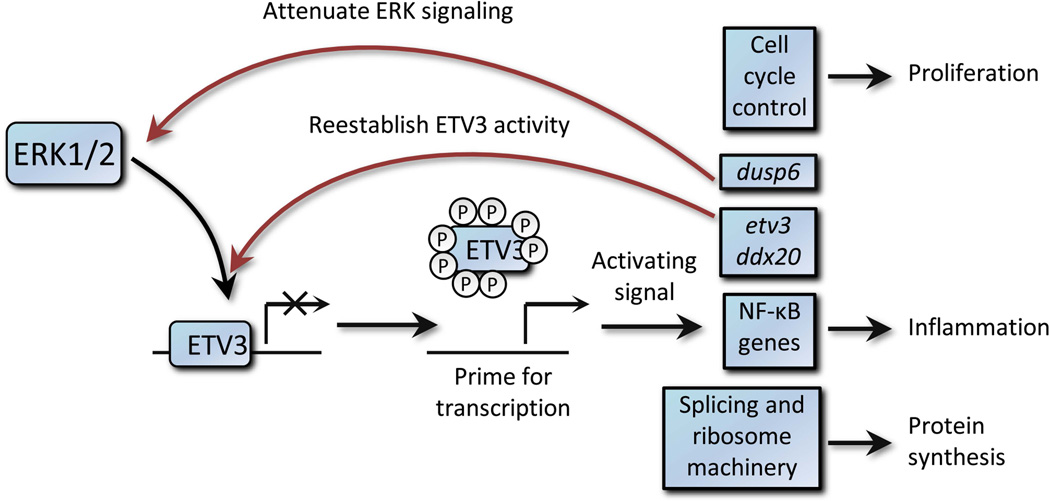
ETV3 target genes including etv3, ddx20, and dusp6 provide negative feedback regulation of ETV3 production and activity. Negative feedback along with constitutive instability may serve to tightly regulate ETV3 abundance. Our date suggest that phosphorylation by ERK2 relieves repression by ETV3, allowing activation of cell cycle control genes including myc, components of the NF-κB pathway, and genes required form RNA processing and translation. This concerted effect may prime cells to respond to additional stimuli, such as IL-10 in bone-marrow derived macrophages (30). Once ERK2 activity ceases, newly translated ETV3 can rapidly repress its target genes (Fig. 6),providing a transient burst of transcriptional activity following ERK activation. We believe that previous work did not observe an interaction between ERK and ETV3 because experiments focused on time-scales of several hours, enough time for repression by ETV3 to be reestablished. Unlike ERF, a related ETS repressor factor, loss of DNA binding by ETV3 does not appear to result from sequestration in the cytoplasm. Although we have not observed phosphorylation of the ETS domain itself, phosphorylation regulates many ETS domain-containing factors through regulation of protein-proteininteractions or auto-inhibitory domains (38, 39). Interestingly, loss of heterozygosity has been associated with the etv3 gene in B-cell chronic lymphocytic leukemia and follicular lymphoma, suggesting that ETV3 functions as a tumor suppressor in these cell types (40).Given the thousands of ETV3-phosphorylation-dependent targets, ETV3 may thus function as a master regulator downstream of ERK activation in some cell types, capable of modulating transcription of genes spanning a broad range of cellular activities.
Fig. 6.
A model for rapid regulation of ETV3 by phosphorylation followed by downstream effects, including negative feedback through increased transcription of etv3,ddx20, and dusp6.
Further development of methods for analysis of thiophosphorylated peptides will continue to expand the range of biological systems amenable to investigation by AS-kinases, and quantitative characterization of background labeling makes it possible to identify substrates even in the presence of WT-kinase through the utilization of PhEt-ATPγS. We expect that comparisons between cell types, growth factor treatments, or genetic perturbations will reveal how MAPK signaling is rewired in different contexts.
Materials and Methods
Antibodies and reagents
Antibodies for phosphorylated ELK1, phosphorylated ERK1/2, phosphorylated motifs, anti-HA Alexa488 conjugate, goat anti-rabbit Alexa488 conjugate, PMA, and recombinant ERK2 were from Cell Signaling Technology. Normal rabbit IgG for immunofluorescence was from Santa Cruz Biotechnology. Monoclonal rabbit antibody against alkylated thiophosphate and p-nitrobenzylmesylate (PNBM) were from Epitomics. N6-(2-Phenylethyl)adenosine-5’-O-(3-thiotriphosphate) (PhEt-ATPγS) was from Biolog (Bremen, Germany). Adenosine-5’-O-(3-thiotriphosphate) (ATPγS), U0126,anti-HA agarose beads, and the antibody recognizing ETV3 were from Sigma-Aldrich. The antibody recognizing HA for ChIP was from Abcam. Media and amino acids for SILAC and iodoacetyl-agarose beads were from Pierce/Thermo Scientific. EGF was from Peprotech. Q103G ERK2 (AS-ERK2) with N-terminal HA tag was a gift from Dr. Roger Davis (University of Massachusetts Medical School). HA-tagged human ETV3 was synthesized by GeneWiz Inc. Genes for mammalian expression were cloned into the pBabe-puro-IRES-EGFP vector (AddGene plasmid 14430)(41).
In vitro substrate labeling
Matched 3T3-L1 cells expressing WT-ERK2 or AS-ERK2 were generated by retroviral transduction followed by puromycin selection, and 3T3-L1 cells were grown in DMEM containing stable isotope labeled L-Lysine and L-Arginine, with 10% dialyzed fetal bovine serum and 230 mg/L L-Proline. Cells were treated as indicated, washed with cold PBS and lysed in 500 µL kinase buffer. Protein amounts were determined by BCA assay and lysates were diluted to uniform concentration. Lysates were immediately supplemented with 1 mM GTP and 50 µM N6-PhEt-ATPγS and incubated at 30°C for 30 to 90 minutes. Time-course reactions were terminated by adding EDTA to 50 mM and placing the reaction on ice. For Western blots, 13.5 µL of lysate was treated with 1.5 µL 25 mM PNBM in 50% DMSO for 2 hours.
Identification of thiophosphorylated peptides
Proteins from kinase reactions were precipitated in methanol/chloroform, suspended in digest buffer by sonication in 100 mM NH4Ac pH 8.9, 1 mM CaCl2, and digested with trypsin (1:100 w/w) overnight with rotation. Digests were acidified with acetic acid (1:6 v/v), desalted on a C18 Sep-Pak (Waters), lyophilized, and resuspended at 20 mg/mL in binding buffer (25 mM HEPES pH 7.0 in 50% ACN) with 1 mM TCEP and 250 µg/mL bovine serum albumin (final pH 5.5). SulfoLink beads (25 µL per mg peptide) weree quilibrated in binding buffer. Peptides were added to beads and rotated overnight in darkness at room temperature. Beads were washed 2x in binding buffer, quenched with DTT, and washed in binding buffer, 5% formic acid, and again in binding buffer. Beads were loaded in a fused silica capillary (530 µm I.D.) and washed briefly with 0.1% acetic acid. Peptides were eluted using 2 mg/mL potassium peroxomonosulfate (Oxone) directly onto a POROS 20 R2 trapping column. The POROS column was washed with 0.2M acetic acid and peptides were eluted with 70% ACN 0.2M acetic acid to a immobilized metal affinity chromatography column (IMAC) for IMAC-LC-MS/MS analysis (21) on an LTQ-Orbitrap XL mass spectrometer (ThermoFisher Scientific). A detailed protocol is available in the Supplementary Methods. Data files were converted to MASCOT generic format (mgf) using DTASuperCharge (version 1.19) and searched using MASCOT(22)(version 2.1) against the NCBI mouse proteome using peptide tolerance of 12 ppm and MS/MS tolerance of 0.7 Da. Peptide identification and phosphorylation site assignments were manually validated to ensure correct identification and quantitation. Any peptide with a low-confidence assignment was confirmed by comparison to a synthetic peptide.
Phosphorylation motif peptide immunoprecipitation
SILAC-labeled fibroblasts were pretreated with either 5 or 10 µM U0126 for 10 minutes and then stimulated with EGF (100 ng/mL) for 20 minutes. Differentiated 3T3-L1 adipocytes were produced as described (42). Adipocytes were pretreated with U0126 at 10 µM for 10 minutes and then with insulin at 150 nM for 15 minutes. Cells were lysed with a solution containing 8 M urea with 1 mM Na3VO4 and processed as described for peptide labeling with 4-plex iTRAQ reagents (AB Sciex). Samples were dissolved (Tris-HCl pH 7.4, 0.03% NP40), immunoprecipitated with antibodies against PXpSP and PXpTP phospho-motifs, and analyzed by IMAC-LC-MS/MS using an LTQ-Orbitrap XL or QStar XL (AB Sciex) mass spectrometer (for fibroblasts or adipocytes respectively) as described (42).
Chromatin immunoprecipitation and high-throughput sequencing (ChIP-Seq)
Three 15-cm plates of HEK 293T cells were transfected with HA-tagged human ETV3 (20 µg plasmid, 60 µL FuGene HD in 0.5 mL PBS) and incubated for two days to a density of 2.5 × 107 cells per plate. Cells were treated for 20 minutes with U0126 (10 µM) with or without PMA (100 nM). A control experiment was conducted with non-transfected cells. The ChIP protocol was modified from methods previously described (43, 44). Briefly, frozen pellets of formaldehyde-fixed cells were resuspended and lysed. The cells were then sonicated using a Misonix Sonicator 3000 at 33W for twelve cycles of a 20-second pulse followed by a 60-second pause. Lysate was centrifuged at maximum speed in a microfuge for 10 minutes at 4°C to remove impurities. The lysate was incubated with 50 µL of Dynal Protein G magnetic beads bound to 10 µg of antibody for 15 hours at 4°C. The purified DNA was prepared for sequencing on a Beckman Coulter SPRI-TE following manufacturer’s instructions. The seq-prepped DNA was PCR amplified using Illumina primers for 18 cycles. Samples were sequenced using Illumina Solexa Genome Analyzers 2.0. Reads were mapped to the hg18 reference genome using the GERALD alignment software package. Uniquely mapping reads were run through the MACS v1.4.0beta peak calling software using default parameters. Reads from 293T cells transfected with ETV3-HA and treated with U0126 were used as the foreground and untransfected 293T cells as the background. Resulting peaks were filtered for p-value < 1e-10. Peaks were mapped to UCSC Known Genes if they fell within a 10kb window around a gene’s transcription start site. ChIP-Seq data has been uploaded to the NCBI Sequence Read Archive.
ChIP-qPCR
8 × 106 293T cells were prepared and processed as described for ChIP-Seq experiments, except that only 106 cells and 5 µg of antibody were used. Samples were analyzed by real-time PCR on a Roche Lightcycler 480 system using Roche SYBR Green I PCR master mix. Primers for ddx20 were 5’ GAGGCGGAGATACGAACT TG 3’ and 5’TACCACATTGGCTGGTGTGT 3’, for dusp6 5’ GCTGGAACAGGTTGTGTTGA 3’and 5’ AAGTGCCCTGGT TTATGTGC 3’, and for myc 5’ CCAACAAATGCAATGGGAGT 3’ and 5’ CCAGAGTCCCAGGGAGAGTG 3’.Mutations in the ETV3-pBabe-IRES-EGFP vector were introduced using Stratagene QuikChange.
Recombinant ERK2 substrates and in vitro kinase reactions
Clones were from OpenBiosystems (clone IDs ETV3: 4918332, RIPK3: 3590770,STK10: 40111177, NID1: 40110859, RBMPS: 3586451), except CDC42EP1 (OriGene catalog #MC205344) and IRS2 (Addgene plasmid 11373). CDC42EP1, ETV3, NID1,RBPMS, and RIPK3 were cloned into pET-16b (Stratagene), and IRS2 and STK10 were inserted into pET-100 (Invitrogen) using directional TOPO cloning. Vectors were transformed into BL21 Star (DE3) E.coli (Invitrogen) and induced with 1 mM IPTG for three hours during log-phase growth of a 2 mL culture. Bacterial pellets were lysed using B-Per bacterial lysis kit with DNAase, lysozyme, and Halt protease inhibitors (Thermo Scientific). Recombinant proteins were recovered from inclusion bodies by solubilization in 8M urea, except RIPK3, which was solubilized with 0.2% sodium dodecyl sulfate in 40 mM Tris-HCl pH 7.5, 100 mM NaCl. Recombinant proteins were stored at 10 ng/µL in 20 mM Tris-HCl pH 7.5, 50 mM NaCl, 50% glycerol.
Kinase reactions used 10–30 ng of substrate, 12.5–25 ng recombinant ERK2, and ATPγS at 50–100 µM in kinase buffer (25 mM Tris-HCl pH 7.5, 5 mM β–glycerolphosphate, 2 mM dithiothreitol, 0.1 mM Na3VO4, 10 mM MgCl2) and were incubated 30 minutes at 30°C. Reactions with RIPK3 were supplemented with 100 mM thiourea to increase solubility. Half of each reaction was silver stained as a loading control; half was treated with PNBM at 2.5 mM for two hours prior to Western blotting for thiophosphate.
Phosphorylation mapping of recombinant proteins by HPLC-MS/MS
Kinase reactions were performed for 1 hour with approximately 100 ng of each substrate and 100 µM ATP. Matched negative control reactions omitted ATP. Each reaction was run by SDS-PAGE and proteins visualized by Coomassie stain or mass-spectrometry compatible silver stain (Thermo Scientific). Matched bands were cut and destained, and processed as described in (45) for overnight digestion in trypsin (Promega, 12.5 ng/uL in 50 mM ammonium bicarbonate pH 8.9) or chymotrypsin (Sigma-Aldrich, 125 ng/uL in 100 mM Tris 10 mM CaCl2 pH 8.0 at 30°C). Reactions were quenched with 5% formic acid in 50% acetonitrile and peptides were eluted by dehydrating the gel band twice with 100% acetonitrile. Peptide samples were dried to about 2 µL in a vacuum centrifuge and resuspended in 30 µL 0.1% acetic acid.
IMAC (21) was used to enrich phosphopeptides from digests of CDC42EP1 and IRS2, and ETV3 was loaded directly onto a C18 pre-column (10 cm, 100 µm internal diameter (I.D.). Pre-columns were placed in-line on an HPLC connected to a C18 column with electrospray tip (10 cm, 50 µm I.D., 1 µm tip flowing at approximately 20–40 nL/min). Peptides were eluted using a piece-wise linear gradient from 0% to 70%acetonitrile in 0.2M acetic acid (4 min: 9.1%, 50 min: 29.4%, 57 min: 42%, 60 min: 70%)and analyzed on an Orbitrap XL hybrid mass spectrometer. The mass spectrometer was running in data-dependent mode where each cycle included an Orbitrap MS scan with 100,000 target resolution followed by isolation and Collision-induced Dissociation of up to six ions (charge state 2–5) for analysis in the LTQ ion trap. MS/MS spectra were extracted using DTASuperCharge (version 1.19) and identified by MASCOT (Matrix Science, version 2.1). Phosphorylated peptides were confirmed by manual inspection of each spectrum and peak annotations are shown in in the Supplementary Spectra.
Immunofluorescence
Approximately 105 per cm2 of DLD1 or HEK 293T cells were thoroughly dispersed and plated overnight on coverslips. HEK 293T cells were then transfected with HA-tagged mouse ETV3 in the pBabe-IRES-EGFP vector (AddGene vector 14430) using FuGene HD (Roche) according to the manufacturer’s instructions and grown for two more days. Cells were treated as indicated in the figure with U0126 (10 µM) in the presence or absence of PMA (100 nM), fixed in 4% formaldehyde for 15 minutes, and washed 3 times with PBS. Cells were blocked for one hour with 5% goat serum (DLD1) or 5%mouse serum (HEK 293T) in PBS with 0.1% Triton-X. DLD1 cells were incubated overnight at 4°C with the antibody recognizing ETV3 at 8 µg/mL in PBS with 1% BSA and 0.1% Triton-X, then washed three times with PBS and incubated for 1 hour with anti-rabbit Alexa488 secondary antibody at 1:1000 dilution with goat anti-rabbit Alexa488 conjugate. Following antibody incubations, slides were washed twice with PBS, treated with DAPI in PBS for 15 minutes, washed once with PBS, treated with phalloidin conjugated to Alexa647 (Invitrogen), washed twice with PBS, and mounted on slides using ProLong Gold (Invitrogen). Images were captured on a DeltaVision Spectris microscope from Applied Precision and processed by software deconvolution.
ERK binding site motif analysis
ScanSite 2.0 (http://scansite.mit.edu) was used to search SwissProt protein sequences for high-stringency ERK1/2 binding sequences (ScanSite motifs “Erk D-domain” and “Erk1 Binding” for D-domain and DEF-domain, respectively) (46) . Where an appropriate protein sequence was not available in SwissProt, protein sequences were uploaded directly from the National Center for Biotechnology Information Entrez protein database. The frequency of each motif in AS-ERK2 substrates was compared to SwissProt mouse proteome (22 Sept 2009 release) using the X2-test.
ETV3 binding motif analysis
To identify an optimal ETV3 binding motif, we searched DNA sequences under ChIP-Seq peaks from the U0126-treated condition for TRANSFAC motifs recognized by ETS family by transcription factors. The motif recognized by ELK-1 was most highly enriched so it was used as the initial hypothesis for motif refinement. The strength of peak signals, as determined by p-value from MACS, was highly correlated with the quality of their match to the ELK-1 binding motif. Peaks with a MACS p-value lower than 1e-25 contained ELK-1 binding motifs with better scores than the median motif score observed in a randomly sampled set of background sequences that matched the distributions of the U0126 peaks in terms of length, guanine-cytosine content, and distance from a transcription start site. We then refined the ELK-1 motif using the sequence of the peaks that passed this threshold as input to THEME (29).
Supplementary Material
Acknowledgements
We thank Kevan Shokat for technical assistance, Brian Joughin for assistance with ScanSite proteome analysis, and Norman Kennedy and Roger Davis for providing antibodies, AS-ERK2DNA, and technical assistance. The Swanson Biotechnology Center at the Koch Institute assisted with DNA sequencing, production of synthetic peptides, and microscopy.
Funding Sources: This work is supported by a grant from Pfizer Inc. and NIH grantsR01DK42816, R01CA118705, U54CA112967 and ES002109. S.M.C. has been supported by a Graduate Research Fellowship from the National Science Foundation, the David H. KochInstitute for Integrative Cancer Research Graduate Fellowship, and the Whitaker Health Science Fellowship. E.F. receives support from the Eugene Bell Career Development Chair.
Footnotes
Supplementary Materials
Supplementary Methods. Detailed protocol for ERK2 substrate labeling and identification References
Figure S1. Additional sites phosphorylated by ERK2 on IRS2, CDC42EP1, and ETV3.
Figure S2. MS/MS spectrum used to identify ETV3 as a direct ERK2 target and the matching spectrum of a synthetic peptide.
Figure S3. HA-tagged ETV3 exhibits MEK-dependent gel shift which is reduced in phosphosite mutants, and an anti-HA antibody captures both the phosphorylated and non-phosphorylated protein in ChIP.
Figure S4. Immunofluorescence for HA-tagged ETV3 expressed in HEK 293T cells showing nuclear localization regardless of ERK activity.
Figure S5. Immunofluorescence for endogenous ETV3 in DLD1 cells showing primarily nuclear localization regardless of ERK activity.
Figure S6. ETV3 binding sites occur near known transcriptional start sites.
Figure S7. Phosphorylation by ERK2 does not affect the stability of ETV3.
Table S1. AS-ERK2 substrates and signal to background ratios.
Table S2. MEK-dependent phosphorylation in peptides isolated by phosphorylation motif peptide immunoprecipitation.
Table S3. Overview statistics of ETV3 ChIP-Seq data.
Table S4. ETV3 binding sites from ChIP-Seq.
Table S5. Gene mappings for ETV3 ChIP-Seq binding sites
Supplementary Spectra. Manually curated spectra used to identify all AS-ERK2 targets.
Author Contributions
S.M.C. designed and performed research and wrote the paper, C.J.L. assisted with research, C.R.C. conducted ChIP experiments, A.L. analyzed ChIP-seq data, K.S. performed adipocyte experiments, E.F. provided support for ChIP experiments and F.M.W. designed and supervised research and wrote the paper.
Competing interests: The authors declare no conflict of interest.
Data availability: ChIP-Seq data has been uploaded to the NCBI Sequence Read Archive (SRP005470). LC-MS/MS data is available from ProteomeCommons (www.proteomecommons.org, group “Identification of ERK2 substrates using chemical genetics”).
References
- 1.Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-Activated Protein Kinase:Conservation of a Three-Kinase Module From Yeast to Human. Physiol. Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 2.Aouadi M, Binetruy B, Caron L, Le Marchand-Brustel Y, Bost F. Role of MAPKs indevelopment and differentiation: lessons from knockout mice. Biochimie. 2006;88:1091–1098. doi: 10.1016/j.biochi.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Bost F, Aouadi M, Caron L, Binétruy B. The role of MAPKs in adipocytedifferentiation and obesity. Biochimie. 2005;87:51–56. doi: 10.1016/j.biochi.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Dong C, Davis RJ, Flavell RA. MAP Kinases in the Immune Response. Ann. Rev.Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- 5.Govindarajan A, Kelleher RJ, Tonegawa S. A clustered plasticity model of long-termmemory engrams : Article : Nature Reviews Neuroscience. Nat. Rev. Neurosci. 2006;7:575–583. doi: 10.1038/nrn1937. [DOI] [PubMed] [Google Scholar]
- 6.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 7.Yoon S, Seger R. The extracellular signal-regulated kinase: multiple substrates regulatediverse cellular functions. Growth Factors. 2006;24:21–44. doi: 10.1080/02699050500284218. [DOI] [PubMed] [Google Scholar]
- 8.Lawrence MC, Jivan A, Shao C, Duan L, Goad D, Zaganjor E, Osborne J, MyGlynn K, Stippec S, Earnest S, Chen W, Cobb MH. The roles of MAPKs in disease. Cell Res. 2008;18:436–442. doi: 10.1038/cr.2008.37. [DOI] [PubMed] [Google Scholar]
- 9.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell. Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 10.Pan C, Olsen JV, Daub H, Mann M. Global effects of kinase inhibitors on signalingnetworks revealed by quantitative phosphoproteomics. Mol. Cell. Proteomics. 2009;8:2796–2808. doi: 10.1074/mcp.M900285-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis TS, Hunt JB, Aveline LD, Jonscher KR, Louie DF, Yeh JM, Nahreini TS, Resing KA, Ahn NG. Identification of novel MAP kinase pathwaysignaling targets by functional proteomics and mass spectrometry. Mol. Cell. 2000;6:1343–1354. doi: 10.1016/s1097-2765(00)00132-5. [DOI] [PubMed] [Google Scholar]
- 12.Old WM, Shabb JB, Houel S, Wang H, Couts KL, Yen CY, Litman ES, Croy CH, Meyer-Arendt K, Miranda JG, Brown RA, Witze ES, Schweppe RE, Resing KA, Ahn NG. Functional proteomics identifies targets of phosphorylation by B-Raf signaling in melanoma. Mol. Cell. 2009;34:115–131. doi: 10.1016/j.molcel.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kosako H, Yamaguchi N, Aranami C, Ushiyama M, Kose S, Imamoto N, Taniguchi H, Nishida E, Hattori S. Phosphoproteomics reveals new ERK MAP kinasetargets and links ERK to nucleoporin-mediated nuclear transport. Nat. Struct. Mol. Biol. 2009;16:1026–1035. doi: 10.1038/nsmb.1656. [DOI] [PubMed] [Google Scholar]
- 14.Bishop AC, Shah K, Liu Y, Witucki L, Kung C, Shokat KM. Design of allele-specific inhibitors to probe protein kinase signaling. Curr. Biol. 1998;8:257–266. doi: 10.1016/s0960-9822(98)70198-8. [DOI] [PubMed] [Google Scholar]
- 15.Eblen ST, Kumar NV, Shah K, Henderson MJ, Watts CK, Shokat KM, J.Weber M. Identification of novel ERK2 substrates through use of an engineered kinase and ATP analogs. J. Biol. Chem. 2003;278:14926–14935. doi: 10.1074/jbc.M300485200. [DOI] [PubMed] [Google Scholar]
- 16.Elphick LM, Lee SE, Child ES, Prasad A, Pignocchi C, Thibaudeau S, Anderson AA, Bonnac L, Gouverneur V, Mann DJ. A quantitative comparison of wild-type and gatekeeper mutant cdk2 for chemical genetic studies with ATP analogues. Chembiochem. 2009;10:1519–1526. doi: 10.1002/cbic.200900052. [DOI] [PubMed] [Google Scholar]
- 17.Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc. Natl. Acad. Sci. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chi Y, Welcker M, Hizli AA, Posakony JJ, Aebersold R, Clurman BE. Identification of CDK2 substrates in human cell lysates. Genome Biol. 2008;9:R149. doi: 10.1186/gb-2008-9-10-r149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ong SE, Mann M. Stable isotope labeling by amino acids in cell culture forquantitative proteomics. Methods Mol. Biol. 2007;359:37–52. doi: 10.1007/978-1-59745-255-7_3. [DOI] [PubMed] [Google Scholar]
- 20.Allen JJ, Li M, Brinkworth CS, Paulson JL, Wang D, Hübner A, Chou WH, Davis RJ, Burlingame AL, Messing RO, Katayama CD, Hedrick SM, Shokat KM. A semisynthetic epitope for kinase substrates. Nat. Methods. 2007;4:511–516. doi: 10.1038/nmeth1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometryand its application to Saccharomyces cerevisiae. Nat. Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 22.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based proteinidentification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Camuzeaux B, Diring J, Hamard PJ, Oulad-Abdelghani M, Donzeau M, Vigneron M, Kedinger C, Chatton B. p38beta2-mediated phosphorylation and sumoylation of ATF7 are mutually exclusive. J. Mol. Biol. 2008;384:980–991. doi: 10.1016/j.jmb.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 24.Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of proteinkinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 2004;68:320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fritsche L, Neukamm SS, Lehmann R, Kremmer E, Hennige AM, Hunder-Gugel A, Schenk M, Häring HU, Schleicher ED, Weigert C. INSULIN-INDUCEDSERINE PHOSPHORYLATION OF IRS-2 VIA ERK1/2 AND mTOR: STUDIES ONTHE FUNCTION OF SER 675 AND SER 907. Am. J. Physiol. Endocrinol. etab. 2011;300:E824–E836. doi: 10.1152/ajpendo.00409.2010. [DOI] [PubMed] [Google Scholar]
- 26.Sluss HK, Davis RJ. H2AX is a target of the JNK signaling pathway that is required for apoptotic DNA fragmentation. Mol. Cell. 2006;23:152–153. doi: 10.1016/j.molcel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Hester KD, Verhelle D, Escoubet-Lozach L, Luna R, Rose DW, Glass CK. Differential repression of c-myc and cdc2 gene expression by ERF and PE-1/METS. Cell Cycle. 2007;6:1594–1604. doi: 10.4161/cc.6.13.4336. [DOI] [PubMed] [Google Scholar]
- 28.Klappacher GW, Lunyak VV, Sykes DB, Sawka-Verhelle D, Sage J, Brard G, Ngo SD, Gangadharan D, Jacks T, Kamps MP, Rose DW, Rosenfeld MG, Glass CK. An induced Ets repressor complex regulates growth arrest during terminalmacrophage differentiation. Cell. 2002;109:169–180. doi: 10.1016/s0092-8674(02)00714-6. [DOI] [PubMed] [Google Scholar]
- 29.Macisaac KD, Gordon DB, Nekludova L, Odom DT, Schreiber J, Gifford DK, Young RA, Fraenkel E. A hypothesis-based approach for identifying the bindingspecificity of regulatory proteins from chromatin immunoprecipitation data. Bioinformatics. 2006;22:423–429. doi: 10.1093/bioinformatics/bti815. [DOI] [PubMed] [Google Scholar]
- 30.El Kasmi KC, Smith AM, Williams L, Neale G, Panopoulos AD, Panopolous A, Watowich SS, Häcker H, Foxwell BM, Murray PJ. Cutting edge: A transcriptionalrepressor and corepressor induced by the STAT3-regulated anti-inflammatory signalingpathway. J. Immunol. 2007;179:7215–7219. doi: 10.4049/jimmunol.179.11.7215. [DOI] [PubMed] [Google Scholar]
- 31.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. GenomeBiol. 2003;4:P3. [PubMed] [Google Scholar]
- 32.Huang dW, Sherman BT, Lempicki RA. Systematic and integrative analysis oflarge gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Pullikuth AK, Catling AD. Scaffold mediated regulation of MAPK signaling andcytoskeletal dynamics: a perspective. Cell. Signal. 2007;19:1621–1632. doi: 10.1016/j.cellsig.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Fea K, Roth RA. Modulation of insulin receptor substrate-1 tyrosinephosphorylation and function by mitogen-activated protein kinase. J. Biol. Chem. 1997;272:31400–31406. doi: 10.1074/jbc.272.50.31400. [DOI] [PubMed] [Google Scholar]
- 35.Luca C, Olefsky JM. Inflammation and insulin resistance. FEBS Lett. 2008;582:97–105. doi: 10.1016/j.febslet.2007.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giovannone B, Lee E, Laviola L, Giorgino F, Cleveland KA, Smith RJ. Two novel proteins that are linked to insulin-like growth factor (IGF-I) receptors by the Grb10 adapter and modulate IGF-I signaling. J. Biol. Chem. 2003;278:31564–31573. doi: 10.1074/jbc.M211572200. [DOI] [PubMed] [Google Scholar]
- 37.Schnidar H, Eberl M, Klingler S, Mangelberger D, Kasper M, Hauser-Kronberger C, Regl G, Kroismayr R, Moriggl R, Sibilia M, Aberger F. Epidermal growth factorreceptor signaling synergizes with Hedgehog/GLI in oncogenic transformation viaactivation of the MEK/ERK/JUN pathway. Cancer Res. 2009;69:1284–1292. doi: 10.1158/0008-5472.CAN-08-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verger A, Duterque-Coquillaud M. When Ets transcription factors meet their partners. Bioessays. 2002;24:362–370. doi: 10.1002/bies.10068. [DOI] [PubMed] [Google Scholar]
- 39.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 40.Green MR, Jardine P, Wood P, Wellwood J, Lea RA, Marlton P, Griffiths LR. A new method to detect loss of heterozygosity using cohort heterozygosity comparisons. BMC Cancer. 2010;10:195. doi: 10.1186/1471-2407-10-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgenstern JP, Land H. series of mammalian expression vectors andcharacterisation of their expression of a reporter gene in stably and transiently transfected cells. Nucleic Acids Res. 1990;18:1068. doi: 10.1093/nar/18.4.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmelzle K, Kane S, Gridley S, Lienhard GE, White FM. Temporal dynamics oftyrosine phosphorylation in insulin signaling. Diabetes. 2006;55:2171–2179. doi: 10.2337/db06-0148. [DOI] [PubMed] [Google Scholar]
- 43.Odom DT, Dowell RD, Jacobsen ES, Nekludova L, Rolfe PA, Danford TW, Gifford DK, Fraenkel E, Bell GI, Young RA. Core transcriptional regulatory circuitry in human hepatocytes. Mol. Syst. Biol. 2006;2:0017. doi: 10.1038/msb4100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brar GA, Kiburz BM, Zhang Y, Kim J, White F, Amon A. Rec8 phosphorylationand recombination promote the step-wise loss of cohesions in meiosis. Nature. 2006;441:532–536. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- 46.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucl. Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.