SUMMARY
Spontaneous nucleation of actin is very inefficient in cells. To overcome this barrier, cells have evolved a set of actin filament nucleators to promote rapid nucleation and polymerization in response to specific stimuli. However, the molecular mechanism of actin nucleation remains poorly understood. This is hindered largely by the fact that actin nucleus, once formed, rapidly polymerizes into filament, thus making it impossible to capture stable multisubunit actin nucleus. Here, we report an effective double-mutant strategy to stabilize actin nucleus by preventing further polymerization. Employing this strategy, we solved the crystal structure of AMPPNP-actin in complex with the first two tandem W domains of Cordon-bleu (Cobl), a potent actin filament nucleator. Further sequence comparison and functional studies suggest that the nucleation mechanism of Cobl is probably shared by the p53 cofactor JMY, but not Spire. Moreover, the double-mutant strategy opens the way for atomic mechanistic study of actin nucleation and polymerization.
INTRODUCTION
Actin is involved in a wide range of cellular processes through tight spatial and temporal control of actin polymerization and disassembly. Formation of the actin nucleus is the rate-limiting step of spontaneous actin polymerization in cells. To cope with this, cells have evolved a set of actin nucleators to promote rapid actin nucleation in response to specific stimuli (Campellone and Welch, 2010; Chesarone and Goode, 2009; Chhabra and Higgs, 2007; Dominguez and Holmes, 2011; Pollard, 2007; Qualmann and Kessels, 2009; Reisler and Egelman, 2007; Robertson et al., 2009). The recent few years have witnessed a fast expansion of the list of actin nucleators with roles in development, genetic disorders, and pathogenic processes. In addition to the well-known actin-related proteins 2/3 (Arp2/3) complex together with various nucleation-promoting factors and formins (Campellone and Welch, 2010; Chesarone et al., 2010; Pollard, 2007), the newest class of actin nucleators is exemplified by Spire (Quinlan et al., 2005), Cobl (Ahuja et al., 2007; Carroll et al., 2003; Gasca et al., 1995; Schwintzer et al., 2011), and JMY (Zuchero et al., 2009) that contain three to four tandem Wiskott-Aldrich syndrome protein-homology domains 2 (W domains).
The W domains are small but ubiquitous actin-binding motifs of ~35 residues (Dominguez, 2004, 2007; Paunola et al., 2002) composed of an N-terminal amphipathic α helix and a C-terminal LKKT-related motif. The N-terminal α helix binds to the conserved hydrophobic cleft between actin subdomains 1 and 3 at the barbed end, while the C-terminal LKKT-related motif extends toward the nucleotide-binding site at the pointed end. Tandem W domains are expected to “stitch” together multiple actin subunits to overcome the largest energy barriers in actin nucleation, the formation of actin dimers and trimers, thus accelerating actin polymerization. Despite various efforts in the past (Rebowski et al., 2008, 2010), this process remains poorly understood, owing to the inability to obtain high-resolution structures of native multisubunit actin nucleus, which, once formed, rapidly proceeds to long and dynamic actin filament.
Here, we report an effective strategy to prevent actin polymerization, thus allowing for the capture of stable actin nucleus for structural study. Using this strategy, we solved the crystal structure of actin, bound with nonhydrolyzable ATP analog adenylyl imidodiphosphate (AMPPNP), in complex with the first two tandem W domains of Cobl to 2.91 Å resolution. The structure guided further biochemical and functional investigations that together support an elegant model for Cobl-mediated actin nucleation. In addition, the structure revealed a hydrophilic binding cleft between actin subdomains 3 and 4 that binds to the hydrophilic side of the N-terminal α helix of the second W domain in Cobl and JMY. The simultaneous binding of two actin subunits to the N-terminal α helix of a W domain facilitates the formation of a compact nucleus for actin polymerization.
Cobl is an evolutionarily conserved protein involved in neural tube closure and is required for development of motile cilia in establishing left-right asymmetry (Ahuja et al., 2007; Carlier et al., 2011; Carroll et al., 2003; Ravanelli and Klingensmith, 2011). JMY was originally characterized as a binding partner of p300 to co-activate many transcription factors including p53 (Shikama et al., 1999), but later also found to activate Arp2/3 complex and de novo nucleate actin filaments (Zuchero et al., 2009). It is involved in key processes such as asymmetric division and cytokinesis in mouse oocytes (Sun et al., 2011), neuritogenesis (Firat-Karalar et al., 2011), and hypoxia-driven cell motility (Coutts et al., 2011). The structural and functional studies reported here for Cobl and JMY shed critical light on how they control actin nucleation for cellular functions and dynamics. Furthermore, the nonpolymerizable actin mutants used in this study provide a valuable tool for studying actin nucleation and oligomeric nuclear actin (de Lanerolle and Serebryannyy, 2011).
RESULTS
Use of Nonpolymerizable Actin Mutants to Obtain a Stable Actin Nucleus
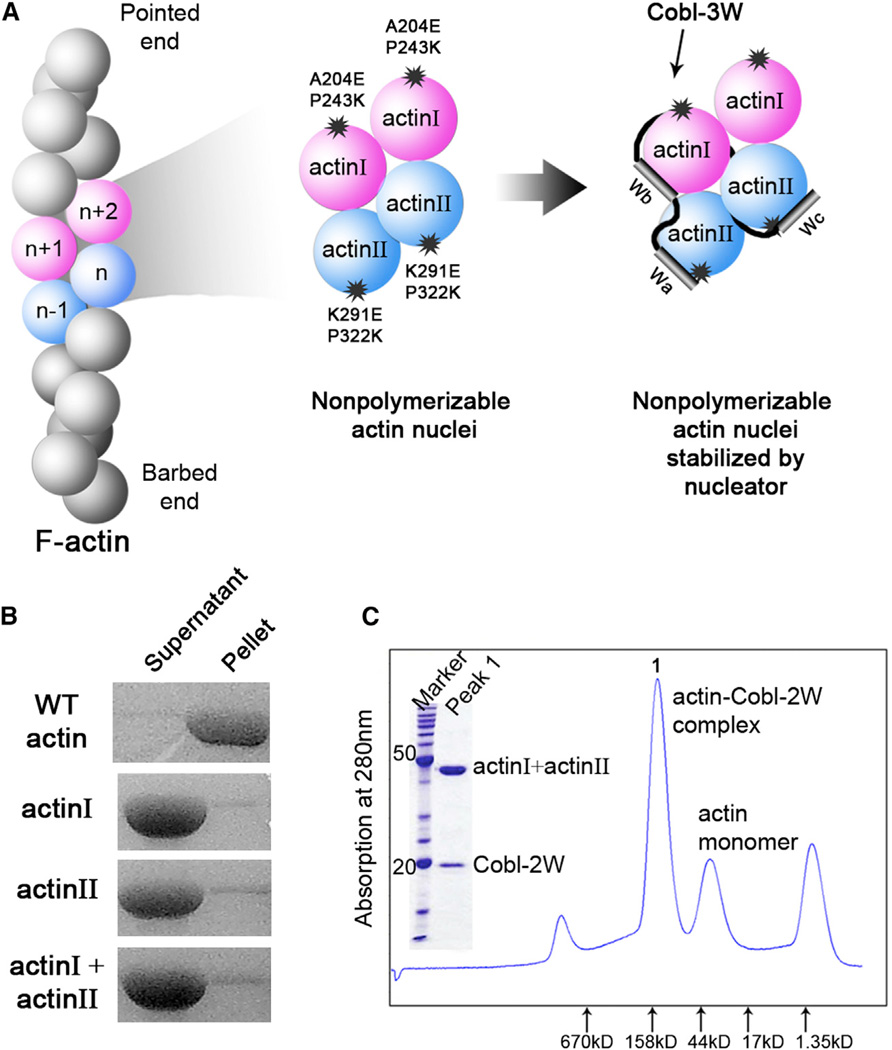
The double-mutant strategy (Figure 1) takes advantage of the fact that actin filament can be viewed as two right-handed long-pitch helices of head-to-tail bound actin subunits through the conserved interactions of the pointed-end surface of actin subunit n −1 and the barbed-end surface of actin subunit n + 1 (Fujii et al., 2010; Holmes et al., 1990; Murakami et al., 2010; Oda et al., 2009) (Figure 1A, left panel). Therefore, actin subunits can be rendered nonpolymerizable if proper mutations are introduced at the pointed- or barbed-end surface. When these two types of nonpolymerizable actin mutants are mixed together, a nucleus comprising two to four actin subunits should form, but without the ability to polymerize (Figure 1A, middle panel). The addition of an actin filament nucleator will stabilize such an actin nucleus for structural studies (Figure 1A, right panel).
Figure 1. The Double-Mutant Strategy for Capturing Stable Actin Nucleus.
(A) Schematic illustration of the strategy. Double mutations introduced at the pointed- or barbed-end interface of an actin subunit make them practically nonpolymerizable. Their mixture, however, gives rise to an actin nucleus that can be further stabilized by an actin filament nucleator.
(B) Under the same condition where WT actin forms filament, neither actin I, II, nor their mixture polymerizes.
(C) The presence of Cobl-2W yields a stable actin-Cobl-2W complex suitable for crystallization.
Two nonpolymerizable actin mutants have been previously created by introducing mutations at the pointed-end surface (Joel et al., 2004; Noguchi et al., 2007; Rould et al., 2006). Of them, the AP-actin containing two surface-accessible substitutions (A204E and P243K) in subdomain 4 has been shown indistinguishable from wild-type (WT) actin in ATP hydrolysis, nucleotide exchange, and protease digestion (Rould et al., 2006). The structures of AP-actin (Ducka et al., 2010; Rould et al., 2006) and WT actin (Aguda et al., 2006; Chereau et al., 2005; Hertzog et al., 2004; Irobi et al., 2004; Kabsch et al., 1990; Otterbein et al., 2001) were highly similar when alone or bound to various individual W domains. This mutant is termed as “actin I” in our double-mutant strategy.
Based on the filamentous actin (F-actin) structures obtained by fiber diffraction and cryo-electron microscopy (Fujii et al., 2010; Holmes et al., 1990; Murakami et al., 2010; Oda et al., 2009), we designed a second actin mutant, termed as “actin II,” that is impaired at the barbed-end surface by two solvent-accessible mutations, K291E and P322K, in subdomain 3 (Figure 1A, middle panel). Actin II was similar to WT actin in nucleotide exchange (Figure S1). WT actin and actin I and II exhibited a comparable binding affinity for individual Cobl-W domains (Table S1). However, their polymerization activities were almost completely abolished when present individually or mixed together (Figure 1B). Therefore, the surface mutations introduced into actin I and II render them nonpolymerizable without affecting their binding to Cobl-W domains.
We then purified Cobl-2W, the first two tandem W domains (Wa and Wb) of Cobl with the highest binding affinity for actin (Ahuja et al., 2007). Cobl-2W is functionally active in actin nucleation and polymerization (Husson et al., 2011). The mixture of Cobl-2W and actin I and II in the presence of AMPPNP gave rise to a well-defined single species of stable actin nuclei, AMPPNP-actin-Cobl-2W, that survived Superdex 200 size exclusion chromatography (Figure 1C). We obtained plate-like crystals of AMPPNP-actin-Cobl-2W that diffracted to 2.91 Å. The crystals were in P1 space group containing two AMPPNP-actin-Cobl-2W complexes in an asymmetric unit (Table 1). Each AMPPNP-actin-Cobl-2W complex consists of one Cobl-2W connecting two actin subunits, actin(Wa) and actin(Wb) (Figure 2A).
Table 1.
Statistics of Data Collection and Structural Refinement
| Data Collection Statistics | |
|---|---|
| Ligand | AMPPNP |
| Space group | P1 |
| Unit cell | |
| a, b, c (Å) | 53.45, 99.80, 118.27 |
| α, β, γ (°) | 65.41, 90.03, 77.77 |
| Resolution (Å)a | 45.02–2.91 (3.07–2.91) |
| Number of unique reflectionsa | 46,313 (6,735) |
| Multiplicitya | 2.7 (2.7) |
| Completeness (%)a | 97.7 (97.3) |
| Rmerge (%)a | 9.0 (49.1) |
| I/σ(I)a | 9.2 (2.2) |
| Structural Refinement Statistics | |
| Resolution (Å) | 45.02–2.91 |
| Number of atoms | 13,002 |
| Rwork / Rfree (%)b | 20.04/25.51 |
| Rmsd bond length (Å)c | 0.008 |
| Rmsd bond angle (°)c | 1.132 |
| Mean B-value (Å2)d | 66.7 |
Values in parentheses are for the highest resolution shell.
The Rfree was calculated by using 5.1% data that were omitted from structural refinement.
The rmsd of bond lengths and angles from ideal geometry for the final model.
The mean temperature factor for all the atoms of the polypeptide chains in the asymmetric unit.
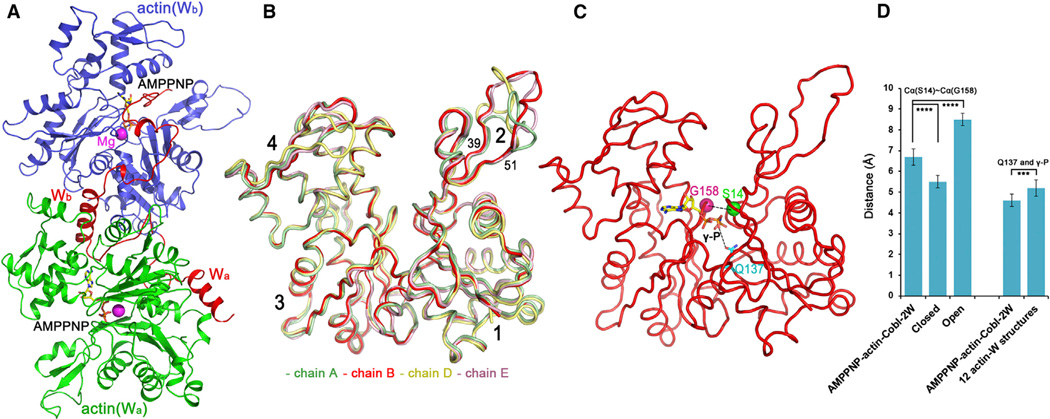
Figure 2. Structure of Actin-Cobl-2W.
(A) AMPPNP-actin-Cobl-2W structure. Cobl is in red with the two tandem W domains labeled as Wa and Wb, while their interacting actin subunits, labeled as actin(Wa) and actin(Wb), are in green and blue, respectively. AMPPNP is shown as ball-and-stick models, and the Mg2+ ions are shown as purple spheres.
(B) Comparison of all four actin subunits in an asymmetric unit of AMPPNP-actin-Cobl-2W structure superimposed on subdomains 3 and 4.
(C) Schematic illustration of the distances of Cα(S14)~Cα(G158) and Q137~γ-phosphate (γ-P) used to characterize the “open” or “closed” conformations of actin, using AMPPNP-actin-Cobl-2W structure (chain B) as an example.
(D) Statistic comparison of actin subunits in the AMPPNP-actin-Cobl-2W structure with “open” and “closed” actin structures. (****p< 0.001; ***p< 0.005 in one-tailed Student’s t test).
The data are represented as mean ± SD. See also Table S2.
The Actin Subunits in AMPPNP-Actin-Cobl-2W Structure Are in “Open” State
Although we did not impose noncrystallographic symmetry in structural refinement, all four actin subunits in an asymmetric unit were very similar (Figure 2B). The only exception was for residues 40–50 within the DNase I binding loop (D-loop) and a surface loop in subdomain 4 in chain D, presumably due to their involvement in different crystal packing. The high structural similarity in the remaining portion of actin should reflect its true conformational state.
All known crystal structures of actin can be classified into two groups according to the “openness” of the nucleotide-binding site (Sablin et al., 2002): the “open” group is represented by a profilin-β-actin structure (Chik et al., 1996) and the structures of nucleotide-free or ADP-bound Arp3 (Nolen et al., 2004; Robinson et al., 2001), and the “closed” group is composed of all other available actin structures. Two conserved residues, S14 and G158, located on the opposite sides of the nucleotide-binding site, are of a shorter distance in the “closed” conformation than in the “open” conformation (Sablin et al., 2002). Here, we used as a criterion the distance between Cα-atoms of S14 and G158 to compare the “openness” of known actin structures (Figure 2C). The averaged Cα(S14)~Cα(G158) distance was 5.5 ± 0.3 Å for a total of 74 “closed” actin crystal structures (including 13 complex structures between actin and individual W domains) and 8.5 ± 0.3 Å for the “open” group (Table S2). For our AMPPNP-actin-Cobl-2W structure, the Cα(S14)~Cα(G158) distance was averaged at 6.7 ± 0.4 Å, which is significantly larger (p < 0.0001) than that of the “closed” group, but smaller (p < 0.0005) than that of the “open” group (Figure 2D). The structures with a larger Cα(S14)~Cα(G158) distance have a larger cleft between actin subdomains 2 and 4.
Q137 was proposed to play a critical role in the ATPase activity of actin (Vorobiev et al., 2003): the Q137A mutation cleaved the γ-phosphate group of bound ATP four times more slowly than WT actin (Iwasa et al., 2008). The flattening of actin subunits upon binding to growing actin filaments was suggested to bring Q137 and the γ-phosphate group closer to allow efficient ATP hydrolysis (Fujii et al., 2010; Oda et al., 2009; Vorobiev et al., 2003). We compared the distances between Q137 and the γ-phosphate group of bound ATP/AMPPNP (Figure 2C) of our AMPPNP-actin-Cobl-2W structure and 13 known complex structures of actin with various individual W domains (Aguda et al., 2006; Chen et al., 2012; Chereau et al., 2005; Didry et al., 2012; Ducka et al., 2010; Hertzog et al., 2004; Irobi et al., 2004). Of the 13 actin-W complex structures, one has an ADP as the ligand and was excluded from this analysis. The actin subunits in our AMPPNP-actin-Cobl-2W structure had an average distance of 4.6 ± 0.3 Å between Q137 and the γ-phosphate group of bound AMPPNP, which was significantly shorter than the average distance of 5.2 ± 0.4 Å in 12 complex structures of actin and individual W domains (p < 0.005) (Table S2; Figure 2D).
In conclusion, judging from the distances of Cα(S14)~ Cα(G158) and Q137~γ-phosphate and the cleft between subdomains 2 and 4, the actin subunits in AMPPNP-actin-Cobl-2W are in a more “open” conformation ready for efficient ATP hydrolysis. This likely resulted from the simultaneous binding of the first two tandem W domains of Cobl, because the binding of individual W domains did not “open” the actin structures (Table S2).
The Actin-Cobl-2W Interfaces
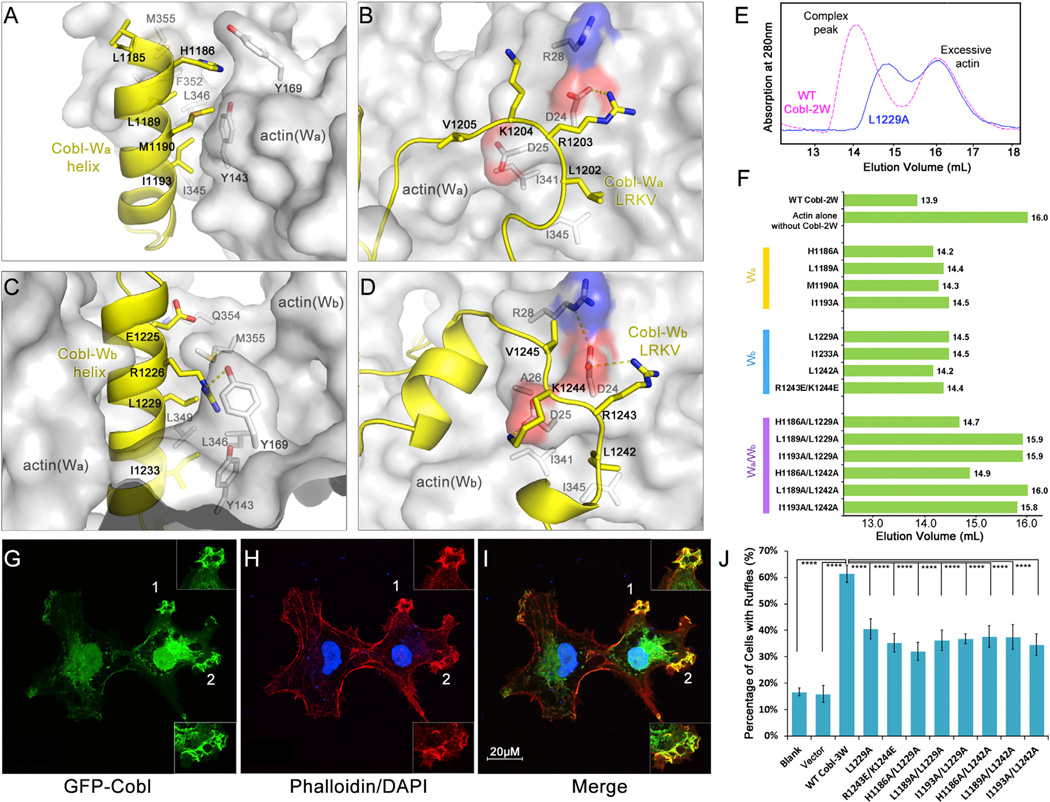
Cobl-2W binds to actin via the N-terminal α helices and C-terminal LRKV motifs of both Wa (Figures 3A and 3B) and Wb (Figures 3C and 3D) domains. The N-terminal α helices bind to the conserved cleft between actin subdomains 1 and 3 at the barbed end (Figures 3A and 3C) that is lined by hydrophobic residues Y143 and Y169 from subdomain 1 and I345, L346, L349, F352, and M355 from subdomain 3. The residues of the N-terminal α helices that are in contact with actin are predominantly hydrophobic, including residues L1185, L1189, M1190, and I1193 on Cobl-Wa helix (Figure 3A) and L1229 and I1233 on Wb helix (Figure 3C). However, hydrophilic residues such as H1186onWa and E1225 and R1226 on Wb also contribute to binding (Figures 3A and 3C). On the other hand, the hydrophobic residues in the LRKV motifs in both Cobl-Wa and Wb bind to the hydrophobic pockets on the surface of actin, while R1203 in Wa and R1243 in Wb each forms a salt bridge with the conserved residue D24 on actin (Figures 3B and 3D).
Figure 3. Actin-Cobl-2W Interfaces.
(A–D) The interface between (A) actin(Wa) and Cobl-Wa, (B) actin(Wa) and Cobl-Wa LRKV, (C) actin(Wb) and Cobl-Wb, and (D) actin(Wb) and Cobl-Wb LRKV.
(E) Comparison of elution profiles on Superdex 200 size exclusion chromatography, using WT and L1229A Cobl-2W as an example.
(F) Mutations of Cobl-2W at the interfaces with actin disrupt the formation of actin-Cobl-2W complex as judged by size exclusion chromatography.
(G–I) Cobl-3W induces the formation of membrane ruffles enriched for F-actin in COS-7 cells. EGFP-Cobl (green) (G) and F-actin stained by phalloidin (red) (H) colocalize in ruffles (I). The nuclei are stained in blue.
(J) Mutations of Cobl-3W at the interfaces with actin impair ruffle induction. Data are presented as mean ± SD from three to five independent experiments (****p < 0.001 in two-tailed Student’s t test).
See also Figure S2.
To investigate the contributions of Cobl-2W residues to the formation of actin-Cobl-2W complex and to the nucleation of actin filaments, we created a series of single and double mutations on key residues of Cobl-2W. The effects of these mutations were first assessed by comparing the elution profiles of actin I and II with WT or mutant Cobl-2W on size exclusion chromatography (Figures 3E and 3F). All the tested single mutations caused varying degrees of right shift of the eluted complex peak (indicating smaller molecular sizes) on size exclusion chromatography (Figures 3E and 3F), presumably due to substantial weakening or complete disruption of actin-Cobl-2W interaction on a particular interface. However, given the presence of multiple interfaces between actin and Cobl-W domains, even a complete disruption of a particular actin-Cobl-W interface (for instance, actin-Cobl-Wa) by a single mutation would still leave the other interface (for instance, actin-Cobl-Wb) intact, thus resulting in relatively small changes in elution volumes for single mutations (Figure 3F). In contrast, mutations on both Wa and Wb domains had much more profound changes. In some cases, they completely distabilized the complex and resulted in elution profiles the same as actin alone without Cobl-2W (Figure 3F). A pyrene-based actin polymerization assay showed that despite the small changes in elution profiles, the actin polymerization activities of single mutations such as I1193A and L1229A were severely compromised or completely lost (Figure S2). The double mutation I1193A/L1229A completely lost its actin polymerization activity.
We further gauged the impacts of these mutations on the ability to promote the formation of actin-rich ruffles in COS-7 cells (Figures 3G–3I). Clearly, all the tested single and double mutations caused significant decreases in actin-rich ruffle induction compared to the WT (p < 0.001) (Figure 3J), confirming that the actin-Cobl interfaces observed in the AMPPNP-actin-Cobl-2W structure are genuine and important for Cobl-mediated filament nucleation in cells. The significant decreases in ruffle induction by these mutants, instead of complete abolishment, could be explained by the fact that actin filament nucleation is a highly dynamic process where even a transient, weak stabilization of actin nuclei could result in accelerated formation of actin filament, albeit at a much lower rate than a fully functional nucleator.
The Two Actin Subunits Adopt a Non-Filament-Like Conformation in the Actin Nucleus
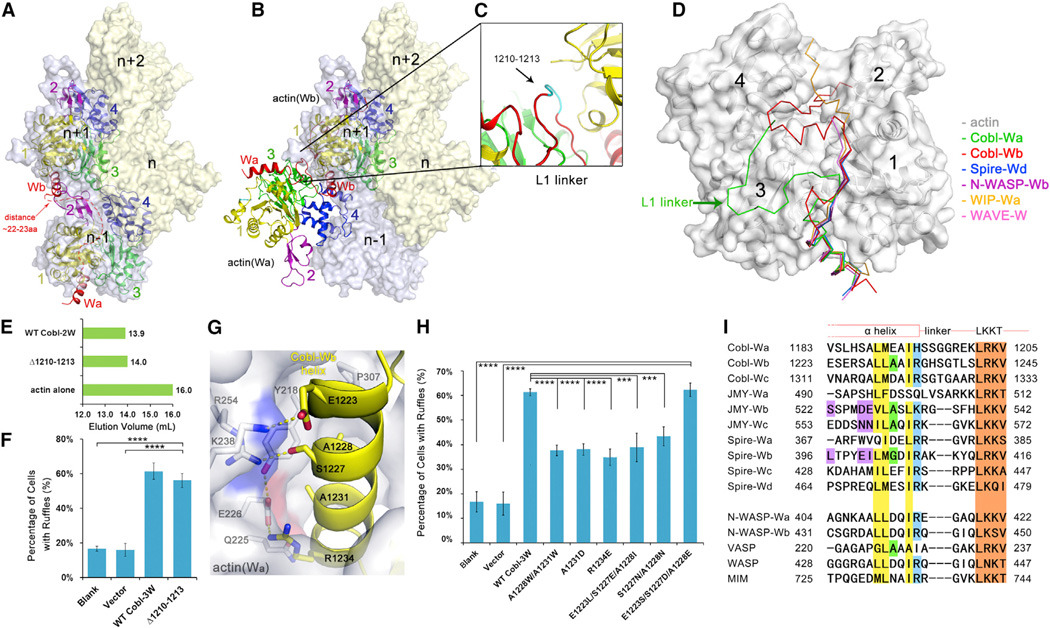
Distinctly different from the head-to-tail conformation in F-actin filament (Figure 4A), the actin(Wa) subunit in actin-Cobl-2W structures is at an angle to actin(Wb). To allow a direct comparison of filament growth based on the existing actin nucleus formed by Cobl with known F-actin structures, we superimposed actin(Wb) onto F-actin n+1 subunit. This superposition resulted in actin(Wa) lying on the outside of F-actin and partially overlapping with actin n−1 subunit (Figure 4B).
Figure 4. The Observed Conformation of Actin-Cobl-2W Is Biologically Relevant.
(A) F-actin structure from fiber diffraction, with two neighboring longitudinal actin subunits highlighted. The number of residues required to span the W-binding sites on two neighboring longitudinal actin subunits was estimated using the structure of actin-WIP-Wa (PDB 2A41) as a reference and shown as a red dashed line.
(B) Superposition of actin-Cobl-2W structure onto F-actin.
(C) The extensively curled L1 linker.
(D) Comparison of known actin-W structures. The PDB codes are Spire-Wd (3MN5), N-WASP-Wb (2A3Z), WIP-Wa (2A41), and WAVE-W (2A40).
(E) Elution profiles of Δ1210–1213.
(F) The mutant Δ1210–1213 is fully active in membrane ruffle induction activity. Data are presented as mean ± SD from three to five independent experiments (****p < 0.001 in two-tailed Student’s t test).
(G) The actin(Wa)-Wb interface.
(H) Comparison of ruffle induction activities for designed mutations. Data are presented as mean ± SD from three to five independent experiments (****p < 0.001; ***p < 0.005 in two-tailed Student’s t test).
(I) Sequence alignment of W domains from tandem W-domain-containing actin filament nucleators and from nucleation-promoting factors. GenBank sequence accession numbers are AAP74341 (Cobl), Q9U4F1 (Spire), AAI30625 (JMY), O00401 (N-WASP), P50552 (VASP), P42768 (WASP), and O43312 (MIM). The LKKT-related motifs are colored in brown. Residues corresponding to the hydrophobic side of the N-terminal α helix, A1231, and R1234 in Cobl-Wb are in yellow, green, and blue, respectively. The key sequence differences on the hydrophilic side of the N-terminal α helix in Spire-Wb, JMY-Wb, and Wc are highlighted in purple.
See also Figures S3, S4, S5, and S6.
The unexpected conformation of actin(Wa) in relation to actin(Wb) urged us to investigate whether it is a biologically meaningful state. To address this, we found that in order to adopt the observed conformation, the linker between Wa and Wb, denoted as L1, was extensively curled (Figure 4C), which is very obvious when various structures of actin subunits with single W domains were aligned (Chereau et al., 2005; Ducka et al., 2010) (Figure 4D). Indeed, if the Wa and Wb domains were destined to bind to two longitudinally neighboring actin subunits n −1 and n + 1, they needed to span a distance of 22–23 residues in extended conformation (shown as a red dashed line in Figure 4A). This would be possible for the total of 22 residues between the two N-terminal α helices of Wa and Wb (residues 1199–1220). However, if this region is shortened by four residues, the remaining 18 residues would not be able to span this distance but would still be sufficient for exhibiting the conformation observed in our structure. In other words, a four residue deletion mutant of Cobl-2W would validate the unexpected conformation of AMPPNP-actin-Cobl-2W as biologically relevant if it does not severely impair actin nucleation and polymerization. We made a four residue deletion at residues 1210–1213 of Cobl (Figure 4C in cyan), denoted as Δ1210–1213, to ensure that the remaining residues 1209 and 1214 could be easily connected without significant structural rearrangement. Impressively, Δ1210–1213 behaved almost the same as WT Cobl in size exclusion chromatography (Figure 4E), pyrene-based polymerization assay (Figure S3) and ruffle induction assay (Figure 4F). Therefore, the non-filament-like conformation of actin-Cobl-2W nucleus revealed in our structure is biologically relevant. In fact, crosslinking and small-angle X-ray scattering studies of tandem W domains in Spire concluded that the actin-Spire-4W complex is also not strictly filament like (Chen et al., 2012; Ducka et al., 2010; Sitar et al., 2011).
Next, to investigate the molecular signatures on Cobl-2W responsible for the unexpected conformation of actin-Cobl-2W, we found that the interface between actin(Wa) and Cobl-Wb might hold the key (Figure 4G). This interface has well-designed complementarities: the actin(Wa) surface presents positively (K238 and R254) and negatively (E226) charged surfaces to which the residues of opposite charges, E1223 at the N terminus and R1234 at the C terminus of Wb, interact. In addition, conserved small polar residues such as S1227 and small hydrophobic residues A1228 and A1231 on Cobl also appear to be important for the interactions.
To test the contributions of these Cobl-Wb residues to the interaction with the actin(Wa) subunit, we performed actin-rich ruffle induction assays on mutant Cobl-3W containing small-to-large mutations A1228W/A1231W, or A1231D, and positive-to-negative mutation R1234E. All of these mutations caused significant loss of ruffle induction activity compared to WT (p < 0.001) (Figure 4H). Pyrene-based polymerization assay on A1231D and R1234E agreed well with the ruffle assay results (Figure S4). Therefore, the interactions between actin and Cobl-2W at this interface play important roles in the nucleation function of Cobl in cells.
Cobl Is Released from Actin Filaments upon ATP Hydrolysis
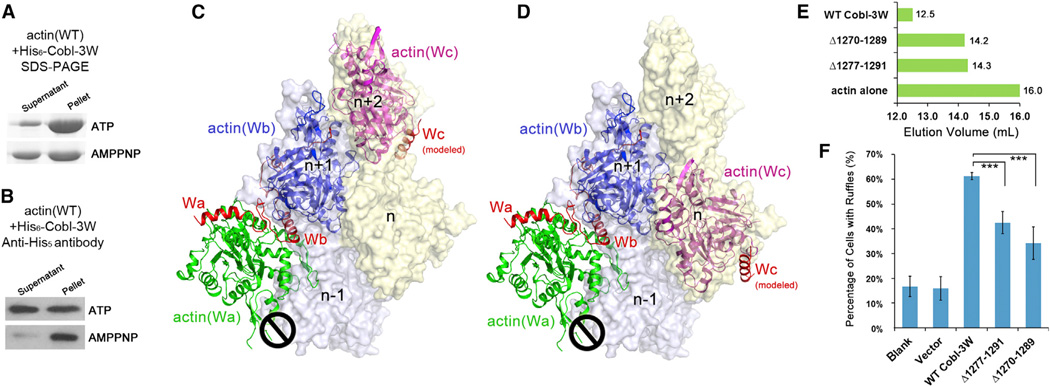
To investigate whether and how Cobl is released from the pointed end of growing filament, we compared the amounts of actin and Cobl-3W in the soluble supernatant fractions versus those in the F-actin pellet fractions in the presence of ATP or AMPPNP. The amounts of actin in supernatant and pellet in different nucleotides were compared on SDS-PAGE (Figure 5A), whereas those of Cobl-3W were monitored by western blot using pentahistidine antibody that recognized the N-terminal his6-tag of Cobl-3W (Figure 5B). Clearly, comparing with the mixture with ATP, AMPPNP resulted in a smaller amount of actin incorporated into the filaments (Figure 5A) and fewer Cobl-3W molecules observed in the supernatant fraction (Figure 5B). This was probably due to the inability to release Cobl-3W from actin filaments in the absence of ATP hydrolysis; thus, fewer actin nuclei could be initiated. Thus, in normal cellular functions, Cobl is likely released from the pointed end of growing filaments and ATP hydrolysis is required for this release.
Figure 5. ATP Hydrolysis Is Required for Cobl Functions and Deletions in the L2 Linker Impair Cobl Function.
(A) SDS-PAGE and (B) Westernwestern blot of the supernatant and pellet fractions of actin polymerization reaction in the presence of ATP or AMPPNP.
(C and D) Models of actin-Cobl-3W superimposed on F-actin in two different modes.
(E) Elution profiles of two Cobl-3W deletions, Δ1270–1289 and Δ1277–1291.
(F) The ruffle induction activities of Δ1270–1289 and Δ1277–1291.
Data are presented as mean ± SD from three to five independent experiments (***p < 0.005 in two-tailed Student’s t test).
Deletions in the Linker L2 Impair Cobl Function
A previous study found that the 65 residue length of the L2 linker, but not the sequence, is required for normal Cobl functions, possibly by allowing Cobl-Wc to bind to the actin subunit on a different protofilament (Ahuja et al., 2007). Based on our AMPPNP-actin-Cobl-2W structure, Wc and actin(Wc) have two most likely locations on actin filaments (Figures 5C and 5D). To probe the impact of shorter deletions on Cobl functions, we removed 20 and 15 residues from L2 of Cobl-3W, termed as Δ1270–1289 and Δ1277–1291, respectively. For both mutants, the complex elution profiles changed (Figure 5E) and the ruffle induction activities were substantially impaired compared to WT (p < 0.005) (Figure 5F). These data suggest that actin(Wc) probably take the longer route as shown in Figure 5D.
JMY Shares a Similar Nucleation Mechanism as Cobl
Once we had identified the key characteristics that bestow Cobl-Wb the ability to bind to two actin subunits simultaneously with a unique conformation, we wondered whether this ability is shared by other W domains. We performed a sequence alignment for the W domains from tandem W-domain nucleators such as Cobl (Wa~Wc), JMY (Wa~Wc), and Spire (Wa~Wd) and from several nucleation-promoting factors such as WASP and N-WASP (Figure 4I). Clearly, the hydrophobic side of the N-terminal a helix that interacts with the hydrophobic cleft between actin subdomains 1 and 3, and the LKKT-related motif, are highly conserved. However, a higher degree of variation exists on the hydrophilic side of the N-terminal α helix. In particular, residues other than positively charged ones such as I and Q are observed on the equivalent position to R1234, and larger residues such as D, E, and N are observed on the equivalent position to A1231. According to the results in Figure 4H, residues that disrupt the ionic interactions between Cobl R1234 and actin E226 or larger residues at A1231 would lead to significant deficiency in actin nucleation functions. Thus, only Spire-Wb, JMY-Wb, and Wc might be able to interact with actin subunits in a similar manner as Cobl-Wb. To further examine these candidates, we tested E1223L/S1227E/A1228I, E1223S/S1227D/A1228E, and S1227N/A1228N, the combinations that are observed in Spire-Wb, JMY-Wb, and Wc, respectively. Only the combination from JMY-Wb (E1223S/S1227D/A1228E) resulted in the same level of activities as WT Cobl in pyrene-based actin polymerization (Figure S5) and cellular ruffle induction assays (p <0.001 when compared with blank or vector) (Figure 4H). The interface of actin-Cobl-Wb can easily accommodate E1223S/S1227D/A1228E from JMY-Wb by forming favorable interactions between the introduced residues E1223S and A1228E on JMY-Wb and K238 and R254 on actin(Wa) (Figure S6). Therefore, JMY-Wb likely employs a similar mechanism to Cobl-Wb in binding to actin (Figure 4B).
DISCUSSION
Models of Actin Filament Nucleation and Polymerization Mediated by Cobl and JMY
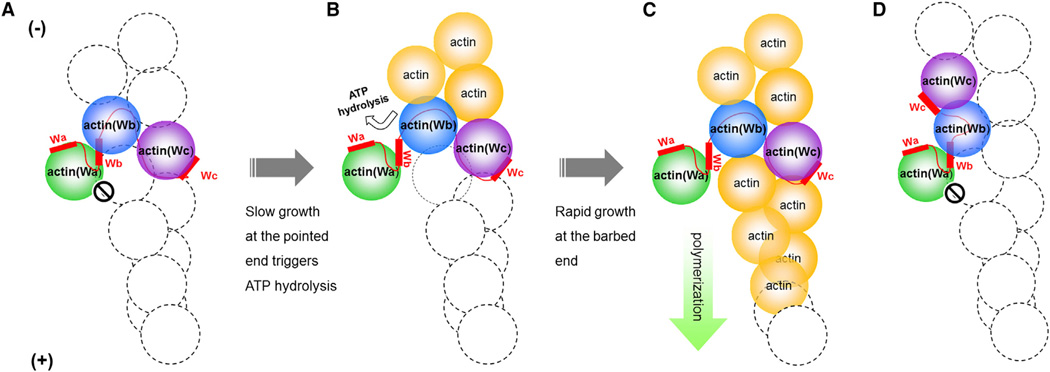
Taken together, we propose a model for the action of Cobl in promoting actin filament nucleation and polymerization (Figures 6A–6C). The AMPPNP-actin-Cobl-2W structure presented here represents the starting state of actin-Cobl nucleus. Because the position of actin(Wa) in ATP (or AMPPNP)-bound state would passively block the growth of actin filament at the barbed end (Figure 6A), Cobl-mediated actin polymerization would start from slow growth at the pointed end. This is similar to the model proposed for formin-mediated nucleation (Otomo et al., 2005). The elongation of actin at the pointed end could induce ATP hydrolysis in Cobl-bound actin subunits, especially in actin(Wb), given that the actin-Cobl nucleus is in a more “open” conformation poised for ATP hydrolysis. The hydrolysis of ATP and subsequent release of inorganic phosphate could induce a conformational change in actin (Murakami et al., 2010) to open the Cobl-binding site between subdomain 1 and 3 in actin(Wb), and the lower affinity of the W domains with ADP-actin compared to ATP-actin would discharge the bound Cobl-Wb and its interacting actin(Wa) (Hertzog et al., 2004) (Figure 6B). The moved-away actin(Wa) would release the steric clash it imposes on the initial nucleus, thus allowing rapid actin filament polymerization at the barbed end (Figure 6C). The slow disassembly at the pointed end would eventually release Cobl to the solution where it could be recycled for another round of actin nucleation.
Figure 6. Proposed Models of Actin Filament Nucleation and Polymerization Mediated by Tandem W Domains.
(A–C) Proposed model of Cobl-mediated actin nucleation. The actin-Cobl nucleus allows the pointed-end but not the barbed-end growth (A). Slow growth at the pointed end triggers ATP hydrolysis in actin(Wb). The bound Cobl-Wb is expulsed from actin(Wb), and the steric hinderance imposed by actin(Wa) is released (B). Rapid growth of actin filament proceeds at the barbed end, and slow depolymerization at the pointed end eventually releases the bound Cobl into solution (C).
(D) Proposed nucleus of actin-JMY.
Although the first two W domains of JMY are expected to bind to actin subunits similarly as observed for Cobl-2W, the actin nucleus formed by JMY is different from that of Cobl in terms of the location of actin(Wc): the shorter L2 linker in JMY could place actin(Wc) on the same protofilament as actin(Wb) (Figure 6D), in opposite to Cobl where actin(Wc) is placed on a different protofilament (Ahuja et al., 2007) (Figure 6A).
The Function of the Hydrophilic Face of N-Terminal α Helix on W Domains
The W domains are small but versatile actin-binding motifs with multifunctionality such as actin monomer sequestration, actin filament nucleation, elongation, and severing (Campellone and Welch, 2010; Carlier et al., 2011; Chesarone and Goode, 2009; Dominguez, 2007; Husson et al., 2010; Paunola et al., 2002; Qualmann and Kessels, 2009). Due to the inability to structurally study actin oligomers in the past, previous studies have focused on the hydrophobic side of the N-terminal amphipathic α helix that binds to the highly conserved hydrophobic cleft located between actin subdomains 1 and 3. Our structure of AMPPNP-actin-Cobl-2W revealed the importance of the hydrophilic side of the N-terminal α helix in the functionality of the W domains, which should be expanded to other W domains in future studies. This is particularly relevant to understanding the molecular mechanisms of W-domain-containing actin-binding proteins of pathogen origins. The more unique features exhibited by the hydrophilic binding face of the W domains may allow specific targeting against pathogens with minimal cytotoxicity. In addition, an in-depth understanding of the sequence-structure-function relationship of the hydrophilic binding face would ultimately assist rational design and engineering of W domains with novel functions.
A General Tool for Structural Study of Actin Nucleation
Unlike previous studies, the AMPPNP-actin-Cobl-2W structure reported in this study represents an atomic structure of native actin nucleus formed by a natural actin filament nucleator. The unexpected conformation of actin subunits in this structure underscores the significance of high-resolution structural studies of this kind. The lack of structural insights into actin nucleation by three classes of specific actin nucleators (Campellone and Welch, 2010; Chesarone et al., 2010; Chesarone and Goode, 2009; Dominguez and Holmes, 2011; Pollard, 2007; Reisler and Egelman, 2007) was largely due to the inability to control the rapid polymerization of actin from a nucleus. This also limited high-resolution structural study of interactions between F-actin and F-actin binding proteins. Therefore, the double-mutant strategy reported in this study provides a valuable tool to overcome these obstacles.
EXPERIMENTAL PROCEDURES
Molecular Cloning
The gene of full-length Drosophila 5C actin was kindly provided by Dr. Loy Volkman (Volkman et al., 1996) and subcloned into pFastBac-Dual vector (Invitrogen) with an N-terminal his6-tag. The gene encompassing the C-terminal residues (1176–1337) of mouse Cobl (GI:32251014)(Cobl-3W) was cloned into pET-45b vector (Clontech Laboratories) with an N-terminal his6-tag. The gene of Cobl-3W was cloned into pEGFP-C1 (Clontech) to allow expression of EGFP-fused Cobl-3W protein. The genes of Cobl-Wa (residues 1176–1224) and Cobl-Wb (residues 1206–1276) were cloned into pGEX-6p1 (Clontech) to generate glutathione S-transferase (GST) fusion proteins. Actin I and II and various Cobl mutants were introduced by site-directed mutagenesis using Quikchange kit (Agilent Technologies) and verified by DNA sequencing.
Expression and Purification of Recombinant Proteins
Expression of actin I and II using baculovirus/insect cell system and lysis of infected insect cells were as previously described (Joel et al., 2004; Rould et al.,2006). Cleared cell lysate was incubated with Ni-NTA agarose (QIAGEN) at 4°C for binding and followed by extensive washes to remove unbound proteins. Bound actin mutants were eluted with 400 mM imidazole and further purified using Source 15Q 4.6/100 PE (GE Healthcare).
Expression of Cobl constructs used E. coli Rosetta(DE3) strains. For GST fused Cobl-W domains, the clarified supernatant was incubated with GST resin (BD Biosciences) at 4°C. Bound proteins were then eluted with 50 mM reduced glutathione and further purified by Superdex 200 10/300 (GE Healthcare). For Cobl-2W, the clarified supernatant was incubated with Ni-NTA agarose (QIAGEN) at 4°C. Bound proteins were eluted in 500 mM imidazole and further purified by Source 15S 4.6/100 PE and Superdex 200 10/300 (GE Healthcare). Purified protein was analyzed by mass spectrometry, revealing an actual molecular weight of 14.33 kDa with an intact N terminus. Thus, the purified protein contains residues 1176–1301 encompassing Wa, L1, Wb, and most of L2, denoted as Cobl-2W.
Binding Affinities between Actin and Individual Cobl-W Domains
WT actin was purified rabbit skeletal muscle as previously described (Spudich and Watt, 1971). The actin-Cobl-W-binding affinities were measured using Octet RED96 (ForteBio) in which GST-Cobl-Wa or GST-Cobl-Wb was loaded onto biosensors coated with GST antibodies (ForteBio, 18–5096), and association and dissociation curves were generated by incubating biosensors with WT actin, actin I, or actin II. The binding affinities were represented as equilibrium dissociation constant KD calculated from Kon and Koff that were measured from the slope of the association and dissociation curves, respectively. Different actin concentrations were used to obtain a full range of binding curves.
Actin Precipitation Assay
Freshly made WT actin, actin I, actin II, or the mixture of actin I and II was in G-buffer (2 mM Tris-HCl [pH 8.0], 0.2 mM CaCl2, 0.5 mM ATP, and 1 mM DTT). Actin polymerization was triggered by bringing the sample to a final concentration of 100 mM KCl and 1 mM MgCl2, which was then incubated for 30 min at room temperature before centrifugation at 90,000 rpm at 4°C to separate filamentous actin in the pellet and soluble actin in the supernatant. The two fractions were then analyzed by SDS-PAGE.
Purification of AMPPNP-Actin-Cobl-2W Complex
Purified actin I, actin II and Cobl-2W were mixed together in 1:1:1 molar ratio in F-buffer (10 mM HEPES [pH 7.6], 100 mM KCl, 1 mM MgCl2, 0.2 mM AMPPNP, and 1 mM DTT). The mixture was then loaded onto Superdex 200 10/300 column (GE Healthcare) to separate AMPPNP-actin-Cobl-2W complex from any extra monomers according to their sizes. The elution volume of actin-Cobl complex was also used to assess the compactness of the complex.
Crystallization, Data Collection, and Structural Refinement
Crystals of AMPPNP-actin-Cobl-2W complex were obtained using vapor diffusion hanging drop method in the solution of 0.1 M Pipes (pH 7.5), 0.16–0.19 M NaCl, and 11% PEG 3350 through serial microseeding. The crystals were frozen in the mother liquid supplied with 18% glycerol. X-ray diffraction data were collected at 100 K at the 21ID-D beamline at Advanced Photon Source. The data were processed using MOSFLM (Battye et al., 2011; Powell, 1999). The structure of an isolated monomeric actin (Protein Data Bank ID code [PDB] 1NWK) (Graceffa and Dominguez, 2003) was used as the search model for molecular replacement by PHENIX (Adams et al., 2010). The model was refined by simulated annealing using PHENIX and then subjected to iterative cycles of positional and B-factor refinement using REFMAC5 (Murshudov et al., 1997) in CCP4 (Collaborative Computational Project, Number 4, 1994) and manual model building in O (Jones et al., 1991) guided by composite omit 2Fo-Fc maps. Additionally, normal-mode-based X-ray crystallographic refinement method (Chen et al., 2007; Poon et al., 2007) was used at the final stage of structural refinement to provide better descriptions of B-factors, resulting in new densities in flexible regions in both Cobl and actin. Three rounds of normal-mode anisotropic B-factor refinement and manual adjustment were applied until the structure was converged.
Pyrene-Based Actin Polymerization Assay
The pyrene-actin polymerization assay was performed as previously described (Ahuja et al., 2007; Husson et al., 2011). Freshly thawed pyrene-labeled rabbit skeletal muscle actin (Cytoskeleton) was incubated on ice for 1 hr in G-buffer to depolymerize actin oligomers. Labeled and unlabeled G-actin was mixed and centrifuged at 90,000 rpm for 30 min at 4°C to remove residual nucleating centers. Actin polymerization was initiated by adding 4 ml actin mixture of 45 mM unlabeled G-actin and 5 mM pyrene-labeled G-actin (final 2 mM actin) into 96 µl reaction mixtures containing tested proteins in F-buffer. The rate of actin polymerization was measured by monitoring the change in fluorescence intensity over time with excitation wavelength 355 nm and emission wavelength 410 nm using FLUOstar Omega (BMG Labtech).
F-Actin-Rich Ruffle Induction Activity Assay of Cobl-3W
COS-7 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, 100 mg/ml streptomycin, and 2 mM L-glutamine (Invitrogen). Transient transfection was carried out using Lipofectamine 2000 Transfection Reagent (Invitrogen) following the manufacturer’s recommendations. At 48 hr posttransfection, the cells were stained with phalloidin and DAPI (Sigma) and analyzed using Zeiss LSM 510 confocal Laser Scanning Microscope. Quantification of ruffles was presented as mean ± SD from three to five independent experiments, each examining 120–300 cells. The statistic analysis was performed using two-tailed Student’s t test.
ATP Hydrolysis and Cobl-3W Release
Freshly made WT actin was diluted to 20 µM and split into two halves. One half of the sample was exchanged into G-buffer containing ATP, while the other half was into AMPPNP. The actin samples with ATP or AMPPNP were separately mixed with 0.5 µM purified Cobl-3W, and actin polymerization for 5 min and precipitation by centrifugation were performed as in “Actin precipitation assay.” The two fractions were analyzed by SDS-PAGE followed by western blot using pentahistidine antibody (Sigma) to detect the Cobl-3W with an N-terminal his6-tag.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Loy Volkman for the gift of the full-length Drosophila 5C actin gene and Dr. B.V. Venkatar Prasad for comments on the manuscript. Q.W. thanks the National Institutes of Health (R01-AI067839) and the Gill-son-Longenbaugh Foundation for support. J.M. acknowledges support from the National Institutes of Health (R01-GM067801), the National Science Foundation (MCB-0818353), and the Welch Foundation (Q-1512). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (grant 085P1000817). J.M. and Q.W. conceived and supervised the project. X.T. initiated the expression of actin, X.C. purified all the proteins, crystallized the complex, refined the structure, and performed all the functional studies. E.K. collected the diffraction data, and F.N. solved the structure. J.M. and Q.W. analyzed the structure and wrote the paper. All authors agreed on the final manuscript.
Footnotes
ACCESSION NUMBERS
The coordinates and structure factors of AMPPNP-actin-Cobl-2W structure reported in this study have been deposited in the Protein Data Bank under ID code 4JHD.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.04.028.
LICENSING INFORMATION
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguda AH, Xue B, Irobi E, Préat T, Robinson RC. The structural basis of actin interaction with multiple WH2/beta-thymosin motif-containing proteins. Structure. 2006;14:469–476. doi: 10.1016/j.str.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM, Qualmann B. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Husson C, Renault L, Didry D. Control of actin assembly by the WH2 domains and their multifunctional tandem repeats in Spire and Cordon-Bleu. Int. Rev. Cell Mol. Biol. 2011;290:55–85. doi: 10.1016/B978-0-12-386037-8.00005-3. [DOI] [PubMed] [Google Scholar]
- Carroll EA, Gerrelli D, Gasca S, Berg E, Beier DR, Copp AJ, Klin-gensmith J. Cordon-bleu is a conserved gene involved in neural tube formation. Dev. Biol. 2003;262:16–31. doi: 10.1016/s0012-1606(03)00323-3. [DOI] [PubMed] [Google Scholar]
- Chen X, Poon BK, Dousis A, Wang Q, Ma J. Normal-mode refinement of anisotropic thermal parameters for potassium channel KcsA at 3.2 A crystallographic resolution. Structure. 2007;15:955–962. doi: 10.1016/j.str.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Sawaya MR, Phillips ML, Reisler E, Quinlan ME. Multiple forms of Spire-actin complexes and their functional consequences. J. Biol. Chem. 2012;287:10684–10692. doi: 10.1074/jbc.M111.317792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, Goode BL. Actin nucleation and elongation factors: mechanisms and interplay. Curr. Opin. Cell Biol. 2009;21:28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat. Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Chik JK, Lindberg U, Schutt CE. The structure of an open state of beta-actin at 2.65 A resolution. J. Mol. Biol. 1996;263:607–623. doi: 10.1006/jmbi.1996.0602. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Coutts AS, Pires IM, Weston L, Buffa FM, Milani M, Li JL, Harris AL, Hammond EM, La Thangue NB. Hypoxia-driven cell motility reflects the interplay between JMY and HIF-1α. Oncogene. 2011;30:4835–4842. doi: 10.1038/onc.2011.188. [DOI] [PubMed] [Google Scholar]
- de Lanerolle P, Serebryannyy L. Nuclear actin and myosins: life without filaments. Nat. Cell Biol. 2011;13:1282–1288. doi: 10.1038/ncb2364. [DOI] [PubMed] [Google Scholar]
- Didry D, Cantrelle FX, Husson C, Roblin P, Moorthy AM, Perez J, Le Clainche C, Hertzog M, Guittet E, Carlier MF, et al. How a single residue in individual β-thymosin/WH2 domains controls their functions in actin assembly. EMBO J. 2012;31:1000–1013. doi: 10.1038/emboj.2011.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R. Actin-binding proteins-a unifying hypothesis. Trends Biochem. Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Dominguez R. The beta-thymosin/WH2 fold: multifunctionality and structure. Ann. N Y Acad. Sci. 2007;1112:86–94. doi: 10.1196/annals.1415.011. [DOI] [PubMed] [Google Scholar]
- Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducka AM, Joel P, Popowicz GM, Trybus KM, Schleicher M, Noegel AA, Huber R, Holak TA, Sitar T. Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation. Proc. Natl. Acad. Sci. USA. 2010;107:11757–11762. doi: 10.1073/pnas.1005347107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Hsiue PP, Welch MD. The actin nucleation factor JMY is a negative regulator of neuritogenesis. Mol. Biol. Cell. 2011;22:4563–4574. doi: 10.1091/mbc.E11-06-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- Gasca S, Hill DP, Klingensmith J, Rossant J. Characterization of a gene trap insertion into a novel gene, cordon-bleu, expressed in axial structures of the gastrulating mouse embryo. Dev. Genet. 1995;17:141–154. doi: 10.1002/dvg.1020170206. [DOI] [PubMed] [Google Scholar]
- Graceffa P, Dominguez R. Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotide-dependent actin dynamics. J. Biol. Chem. 2003;278:34172–34180. doi: 10.1074/jbc.M303689200. [DOI] [PubMed] [Google Scholar]
- Hertzog M, van Heijenoort C, Didry D, Gaudier M, Coutant J, Gigant B, Didelot G, Préat T, Knossow M, Guittet E, Carlier MF. The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell. 2004;117:611–623. doi: 10.1016/s0092-8674(04)00403-9. [DOI] [PubMed] [Google Scholar]
- Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- Husson C, Cantrelle FX, Roblin P, Didry D, Le KH, Perez J, Guittet E, Van Heijenoort C, Renault L, Carlier MF. Multifunctionality of the beta-thymosin/WH2 module: G-actin sequestration, actin filament growth, nucleation, and severing. Ann. N Y Acad. Sci. 2010;1194:44–52. doi: 10.1111/j.1749-6632.2010.05473.x. [DOI] [PubMed] [Google Scholar]
- Husson C, Renault L, Didry D, Pantaloni D, Carlier MF. Cordon-Bleu uses WH2 domains as multifunctional dynamizers of actin filament assembly. Mol. Cell. 2011;43:464–477. doi: 10.1016/j.molcel.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Irobi E, Aguda AH, Larsson M, Guerin C, Yin HL, Burtnick LD, Blanchoin L, Robinson RC. Structural basis of actin sequestration by thymosin-beta4: implications for WH2 proteins. EMBO J. 2004;23:3599–3608. doi: 10.1038/sj.emboj.7600372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa M, Maeda K, Narita A, Maéda Y, Oda T. Dual roles of Gln137 of actin revealed by recombinant human cardiac muscle alpha-actin mutants. J. Biol. Chem. 2008;283:21045–21053. doi: 10.1074/jbc.M800570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel PB, Fagnant PM, Trybus KM. Expression of a nonpolymerizable actin mutant in Sf9 cells. Biochemistry. 2004;43:11554–11559. doi: 10.1021/bi048899a. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Murakami K, Yasunaga T, Noguchi TQ, Gomibuchi Y, Ngo KX, Uyeda TQ, Wakabayashi T. Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release. Cell. 2010;143:275–287. doi: 10.1016/j.cell.2010.09.034. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macro-molecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Noguchi TQ, Kanzaki N, Ueno H, Hirose K, Uyeda TQ. A novel system for expressing toxic actin mutants in Dictyostelium and purification and characterization of a dominant lethal yeast actin mutant. J. Biol. Chem. 2007;282:27721–27727. doi: 10.1074/jbc.M703165200. [DOI] [PubMed] [Google Scholar]
- Nolen BJ, Littlefield RS, Pollard TD. Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP. Proc. Natl. Acad. Sci. USA. 2004;101:15627–15632. doi: 10.1073/pnas.0407149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Iwasa M, Aihara T, Maéda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- Otterbein LR, Graceffa P, Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- Paunola E, Mattila PK, Lappalainen P. WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett. 2002;513:92–97. doi: 10.1016/s0014-5793(01)03242-2. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Poon BK, Chen X, Lu M, Vyas NK, Quiocho FA, Wang Q, Ma J. Normal mode refinement of anisotropic thermal parameters for a supramolecular complex at 3.42-A crystallographic resolution. Proc. Natl. Acad. Sci. USA. 2007;104:7869–7874. doi: 10.1073/pnas.0701204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell HR. The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1690–1695. doi: 10.1107/s0907444999009506. [DOI] [PubMed] [Google Scholar]
- Qualmann B, Kessels MM. New players in actin polymerization-WH2-domain-containing actin nucleators. Trends Cell Biol. 2009;19:276–285. doi: 10.1016/j.tcb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- Ravanelli AM, Klingensmith J. The actin nucleator Cordon-bleu is required for development of motile cilia in zebrafish. Dev. Biol. 2011;350:101–111. doi: 10.1016/j.ydbio.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebowski G, Boczkowska M, Hayes DB, Guo L, Irving TC, Dominguez R. X-ray scattering study of actin polymerization nuclei assembled by tandem W domains. Proc. Natl. Acad. Sci. USA. 2008;105:10785–10790. doi: 10.1073/pnas.0801650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebowski G, Namgoong S, Boczkowska M, Leavis PC, Navaza J, Dominguez R. Structure of a longitudinal actin dimer assembled by tandem w domains: implications for actin filament nucleation. J. Mol. Biol. 2010;403:11–23. doi: 10.1016/j.jmb.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisler E, Egelman EH. Actin structure and function: what we still do not understand. J. Biol. Chem. 2007;282:36133–36137. doi: 10.1074/jbc.R700030200. [DOI] [PubMed] [Google Scholar]
- Robertson AS, Smythe E, Ayscough KR. Functions of actin in endocytosis. Cell. Mol. Life Sci. 2009;66:2049–2065. doi: 10.1007/s00018-009-0001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- Rould MA, Wan Q, Joel PB, Lowey S, Trybus KM. Crystal structures of expressed non-polymerizable monomeric actin in the ADP and ATP states. J. Biol. Chem. 2006;281:31909–31919. doi: 10.1074/jbc.M601973200. [DOI] [PubMed] [Google Scholar]
- Sablin EP, Dawson JF, VanLoock MS, Spudich JA, Egelman EH, Fletterick RJ. How does ATP hydrolysis control actin’s associations? Proc. Natl. Acad. Sci. USA. 2002;99:10945–10947. doi: 10.1073/pnas.152329899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwintzer L, Koch N, Ahuja R, Grimm J, Kessels MM, Qualmann B. The functions of the actin nucleator Cobl in cellular morphogenesis critically depend on syndapin I. EMBO J. 2011;30:3147–3159. doi: 10.1038/emboj.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikama N, Lee CW, France S, Delavaine L, Lyon J, Krstic-Demonacos M, La Thangue NB. A novel cofactor for p300 that regulates the p53 response. Mol. Cell. 1999;4:365–376. doi: 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- Sitar T, Gallinger J, Ducka AM, Ikonen TP, Wohlhoefler M, Schmoller KM, Bausch AR, Joel P, Trybus KM, Noegel AA, et al. Molecular architecture of the Spire-actin nucleus and its implication for actin filament assembly. Proc. Natl. Acad. Sci. USA. 2011;108:19575–19580. doi: 10.1073/pnas.1115465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosintroponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Sun SC, Sun QY, Kim NH. JMY is required for asymmetric division and cytokinesis in mouse oocytes. Mol. Hum. Reprod. 2011;17:296–304. doi: 10.1093/molehr/gar006. [DOI] [PubMed] [Google Scholar]
- Volkman L, Storm K, Aivazachvili V, Oppenheimer D. Overexpression of actin in AcMNPV-infected cells interferes with polyhedrin synthesis and polyhedra formation. Virology. 1996;225:369–376. doi: 10.1006/viro.1996.0611. [DOI] [PubMed] [Google Scholar]
- Vorobiev S, Strokopytov B, Drubin DG, Frieden C, Ono S, Condeelis J, Rubenstein PA, Almo SC. The structure of nonvertebrate actin: implications for the ATP hydrolytic mechanism. Proc. Natl. Acad. Sci. USA. 2003;100:5760–5765. doi: 10.1073/pnas.0832273100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat. Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.