Significance
Transcription of protein-coding genes requires transient binding of many different factors to RNA polymerase II. Thus far, crystal structures of only two such factors in complex with RNA polymerase II are known. Here we report crystal structures of a third polymerase-binding protein, bypass of Ess1 (Bye1), in complex with RNA polymerase II in different functional states. We also show that Bye1 binds histone tails with posttranslational modifications that mark active chromatin and discuss models for Bye1 function in a chromatin context.
Keywords: gene transcription, chromatin modification
Abstract
Bypass of Ess1 (Bye1) is a nuclear protein with a domain resembling the central domain in the transcription elongation factor TFIIS. Here we show that Bye1 binds with its TFIIS-like domain (TLD) to RNA polymerase (Pol) II, and report crystal structures of the Bye1 TLD bound to Pol II and three different Pol II–nucleic acid complexes. Like TFIIS, Bye1 binds with its TLD to the Pol II jaw and funnel. In contrast to TFIIS, however, it neither alters the conformation nor the in vitro functions of Pol II. In vivo, Bye1 is recruited to chromatin via its TLD and occupies the 5′-region of active genes. A plant homeo domain (PHD) in Bye1 binds histone H3 tails with trimethylated lysine 4, and this interaction is enhanced by the presence of neighboring posttranslational modifications (PTMs) that mark active transcription and conversely is impaired by repressive PTMs. We identify putative human homologs of Bye1, the proteins PHD finger protein 3 and death-inducer obliterator, which are both implicated in cancer. These results establish Bye1 as the founding member of a unique family of chromatin transcription factors that link histones with active PTMs to transcribing Pol II.
For transcription of eukaryotic protein-coding genes, RNA polymerase (Pol) II associates transiently with dozens of transcription factors. Different Pol II–associated factors are required for transcription initiation, RNA chain elongation through chromatin, pre-mRNA 5′-capping, splicing, 3′-RNA processing of the nascent transcript, and transcription termination (1–3). To understand how these factors cooperate with Pol II and achieve their functions, structural information on Pol II in complex with transcription factors is required. Thus far, X-ray crystallographic structural information on such complexes is limited to two transcription factors: the initiation factor TFIIB (4–7), and the elongation factor TFIIS (8–11). TFIIS contains three domains, a mobile N-terminal domain, a central domain that binds directly to the Pol II jaw and funnel domains, and a C-terminal zinc ribbon domain that inserts into the polymerase pore (also called the secondary channel) and reaches the Pol II active site (9), to stimulate cleavage of backtracked RNA during transcriptional proofreading and escape from arrest (12).
In the yeast Saccharomyces cerevisiae, there is only a single protein that contains a domain that is distantly homologous to the central, Pol II–associated domain of TFIIS. This protein, bypass of Ess1 (Bye1), has been identified as a multicopy suppressor of Ess1 (13), a peptidyl-prolyl cis-trans isomerase involved in proline isomerization of the C-terminal domain (CTD) of Pol II (14, 15). In Bye1, the central TFIIS-like domain (TLD, residues 232–365) is flanked by an N-terminal plant homeo domain (PHD) (residues 74–134) and a C-terminal Spen paralogue and orthologue C-terminal (SPOC) domain (residues 447–547; Fig. 1A). PHD domains are mostly found in proteins involved in chromatin-mediated gene regulation (16). Consistent with this, the Bye1 PHD domain binds to a histone H3 tail peptide containing trimethylated lysine 4 (H3K4me3) (17). The function of SPOC domains in yeast is unclear, but in higher eukaryotes, SPOC domains are implicated in developmental signaling (18). Bye1 localizes to the nucleus (19), consistent with harboring putative nuclear localization signals in the N-terminal protein region. Based on yeast genetics, it was suggested that Bye1 plays an inhibitory role during transcription elongation (20). It is unknown whether Bye1 binds to Pol II directly, and what the consequences of such binding are for polymerase structure and function.
Fig. 1.
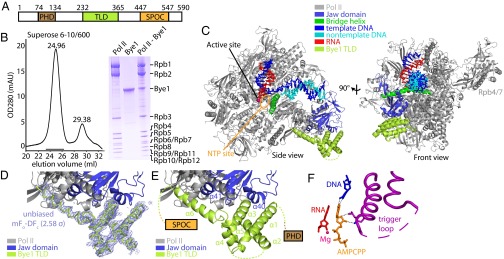
Structure of the Pol II–Bye1 elongation complex. (A) Bye1 domain organization. Bordering residue numbers are indicated. (B) Pol II and Bye1 form a stable complex. Pol II was incubated with an excess of Bye1 and subjected to size-exclusion chromatography. The elution profile (Left) reveals a stable Pol II–Bye1 complex (elution volume, 24.96 mL) and excess Bye1 (29.38 mL). SDS/PAGE analysis of fractions indicated with a black horizontal line confirmed the presence of a Pol II–Bye1 complex (right lane of the Coomassie-stained SDS/PAGE gel shown on the Right). For comparison, pure Pol II and recombinant Bye1 have been included in SDS/PAGE analysis. (C) Ribbon model of the Pol II–Bye1 elongation complex crystal structure. The views correspond to the side and front views of Pol II (46) and are related by a 90° rotation around a vertical axis. (D) Sigma-A–weighted difference electron density (blue mesh, contoured at 2.58σ) for the Bye1 TLD (green). The Fourier coefficients used for map calculation were mFo − DFc (Fo = observed structure factor amplitude, Fc = structure factor amplitude calculated from the Pol II structure alone, m = figure of merit, D = Sigma-A weighting factor). (E) Close-up view of the Pol II–Bye1 interaction. Mobile loops are indicated by dashed lines. (F) Contacts of AMPCPP with the closed trigger loop in the AMPCPP-containing Pol II–Bye1 elongation complex structure. Residues involved in hydrogen bond formation are shown as sticks; hydrogen bonds are indicated by dashed lines. A few residues of the trigger loop that face away from the AMPCPP are mobile (dashed line).
Here we show that Bye1 binds directly to the core of Pol II and report four crystal structures of different Pol II functional complexes bound by Bye1. The structures reveal similarities and differences to the Pol II–TFIIS complex. Together with functional data, our results indicate that Bye1 binds to early Pol II elongation complexes at the beginning of transcribed regions of active genes without changing polymerase structure or function. The polymerase interaction recruits Bye1 to chromatin, where it directly contacts histone H3 tails marked with posttranslational modifications (PTMs) of active transcription.
Results
Bye1 Interacts with Pol II.
To test whether Pol II binds directly to Bye1 in vitro, we incubated pure yeast Pol II with recombinant Bye1 and subjected the sample to size-exclusion chromatography (Materials and Methods). A stable and apparently stoichiometric Pol II–Bye1 complex was obtained (Fig. 1B). To characterize the Pol II–Bye1 interaction, we used surface plasmon resonance. We immobilized Pol II on a Biacore sensor chip and determined Bye1 association and dissociation rates. The ratio of these rates provided a dissociation constant of KD = 3.8 ± 2.2 µM (Fig. S1).
Structure of Bye1-Bound Pol II Elongation Complex.
Cocrystallization of Pol II with full-length Bye1 yielded crystals diffracting to 4.8-Å resolution (Table S1). Structure solution by molecular replacement with free Pol II (21) revealed positive difference density for the Bye1 TLD on the Rpb1 surface, but no density for the two other Bye1 domains. To obtain better diffraction, the Bye1 TLD was expressed in isolation and soaked into preformed Pol II elongation complex crystals containing a DNA–RNA scaffold. Diffraction data to 3.15-Å resolution were obtained (Table S1). Phasing with the Pol II structure (21) revealed positive difference density at the same location observed with full-length Bye1 (Fig. 1 C and D). The Bye1 TLD structure was built with the aid of sequence markers obtained with selenomethionine-labeled protein, and the complex structure was refined to a free R-factor of 21.19% (Table S1).
Bye1 Binds the Polymerase Jaw.
The Bye1 TFIIS-like domain (TLD) fold comprises an N-terminal three-helix bundle (helices α1–α3) followed by two short helices (α4 and α5) that link to an extended C-terminal helix α6 (Fig. 1E). This fold resembles that of TFIIS domain II (helices α1–α5) (9), and helix α6 corresponds to the linker between TFIIS domains II and III (Fig. 2A). The Bye1 TLD binds the Rpb1 jaw domain at the location where TFIIS domain II binds the polymerase (Fig. S2A). Despite this overall similarity, the Pol II contacts by the Bye1 TLD and TFIIS differ. The Bye1 helix α3 binds loop β30–β31 and helix α40 of the Rpb1 jaw domain and induces ordering of loop α40–β29. Helix α6 extends from the jaw into the Pol II funnel, contacting the Rpb1 loops α20–α21 and β29–α41, and strand β32 of the Rpb1 funnel domain. The Bye1 loop α2–α3 contacts the N terminus of Rpb5 (Fig. 2 B–D).
Fig. 2.
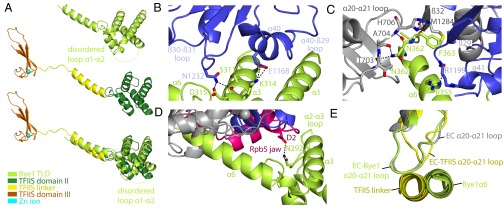
Pol II–Bye1 interaction and comparison with TFIIS. (A) Side view of the Bye1 TLD bound to Pol II (Top), TFIIS (Middle) in its Pol II-bound state (11) with its central domain II (green), linker helix (yellow), and the C-terminal zinc ribbon domain III (orange), and superposition of the two structures (Bottom). (B) Details of the interaction of the Bye1 TLD with the Pol II Rpb1 jaw domain (blue). The view is from the side. Residues involved in hydrogen bond formation or salt bridges (dashed lines) are shown as sticks. (C) Side view of the Pol II Rpb1 funnel and jaw domain–Bye1 TLD interaction. (D) Details of the interaction of the Bye1 TLD with the Pol II Rpb5 jaw domain (magenta). (E) In the Pol II–TFIIS complex structure, conformational changes in Pol II are induced by movements of the Pol II Rpb1 loop α20–α21, which results in opening of a crevice in the polymerase funnel. Loop movements are observed for TFIIS-bound Pol II (yellow) (11), but not for Bye1-bound (this study, green) or unbound Pol II (silver) (8).
Bye1 Does Not Change Pol II Conformation.
TFIIS binding to Pol II induces three major conformational changes in the polymerase elongation complex. It repositions the jaw-lobe module, traps the trigger loop in a locked conformation (9), and realigns the RNA in the active site (8). Although Bye1 resembles part of TFIIS and binds to a similar position on Pol II, it does not induce conformational changes (Fig. 2E). This lack of conformational changes was observed in structures of Pol II complexes with the Bye1 TLD, but also with full-length Bye1. These observations predicted that Bye1 does not impair nucleoside triphosphate (NTP) binding to Pol II, which requires closure of the trigger loop. Indeed, we were able to show closure of the trigger loop and binding of an NTP substrate analogon in the presence of Bye1 by crystallizing an additional complex of Bye1 TLD bound to the Pol II elongation complex with α,β-Methyleneadenosine 5′-triphosphate (AMPCPP) (Fig. 1F; Fig. S2B; Table S1). Furthermore, Bye1 TLD binding also did not prevent backtracking of RNA into the Pol II pore, as seen in another structure of Bye1 bound to arrested Pol II with backtracked RNA (Fig. S2C; Table S1).
Bye1 Does Not Influence Basic Pol II Functions.
These observations suggested that Bye1 had no functional influence on basal transcription. Indeed nuclear extracts prepared from yeast cells lacking the gene encoding Bye1 were active in promoter-dependent in vitro transcription assays, and addition of purified Bye1 to WT nuclear extracts did not alter their activity (Fig. S3). In contrast to TFIIS, Bye1 did not induce Pol II backtracking and RNA cleavage on DNA–RNA scaffolds, but allowed for unperturbed elongation activity in RNA extension assays (Fig. S4). Collectively, these data indicate that Bye1 neither induces structural changes in Pol II functional complexes nor influences their function in vitro.
Bye1 Associates with Chromatin via Its TLD Domain.
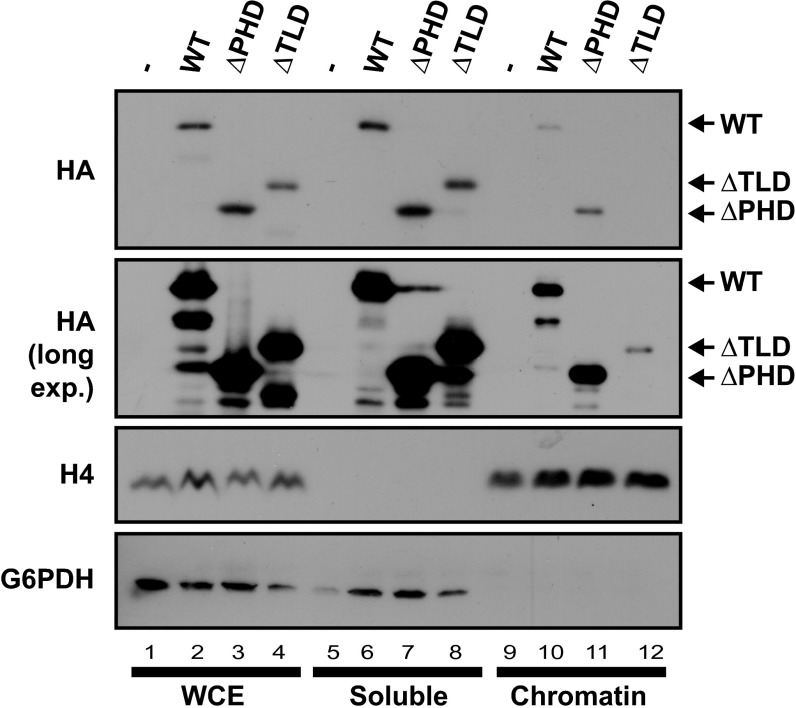
The above results suggested that Bye1 functions in a chromatin context. To investigate whether Bye1 associates with chromatin in vivo and whether its Pol II–binding TLD is required for this, we biochemically separated cell extracts into an insoluble fraction, containing chromatin and associated proteins, and a soluble fraction, containing free proteins in the cytoplasm and nucleoplasm. We used strains harboring plasmids containing hemagglutinin (HA)-tagged full-length Bye1 (WT) or variants lacking either the PHD domain (∆PHD) or the TLD domain (∆TLD) (20). All variants of Bye1 were present at the same level in unfractionated whole cell extract (Fig. 3, lanes 1–4). WT Bye1 and its ∆PHD variant were present in the chromatin and soluble fractions, but the ∆TLD variant associated with chromatin only very weakly (Fig. 3, lanes 9–12). These results demonstrate that a fraction of the Bye1 protein present in cells associates with chromatin and that the TLD of Bye1 is important for the association.
Fig. 3.
Bye1 associates with chromatin via its TLD domain. Immunoblot of whole-cell extract (WCE), chromatin-bound, and soluble cell fraction to hemagglutinin tag (HA), histone 4 (H4), and glucose-6-phosphate-1-dehydrogenase (G6DH). For details, see Materials and Methods.
Bye1 Binds Active Histone Marks via Its PHD Domain.
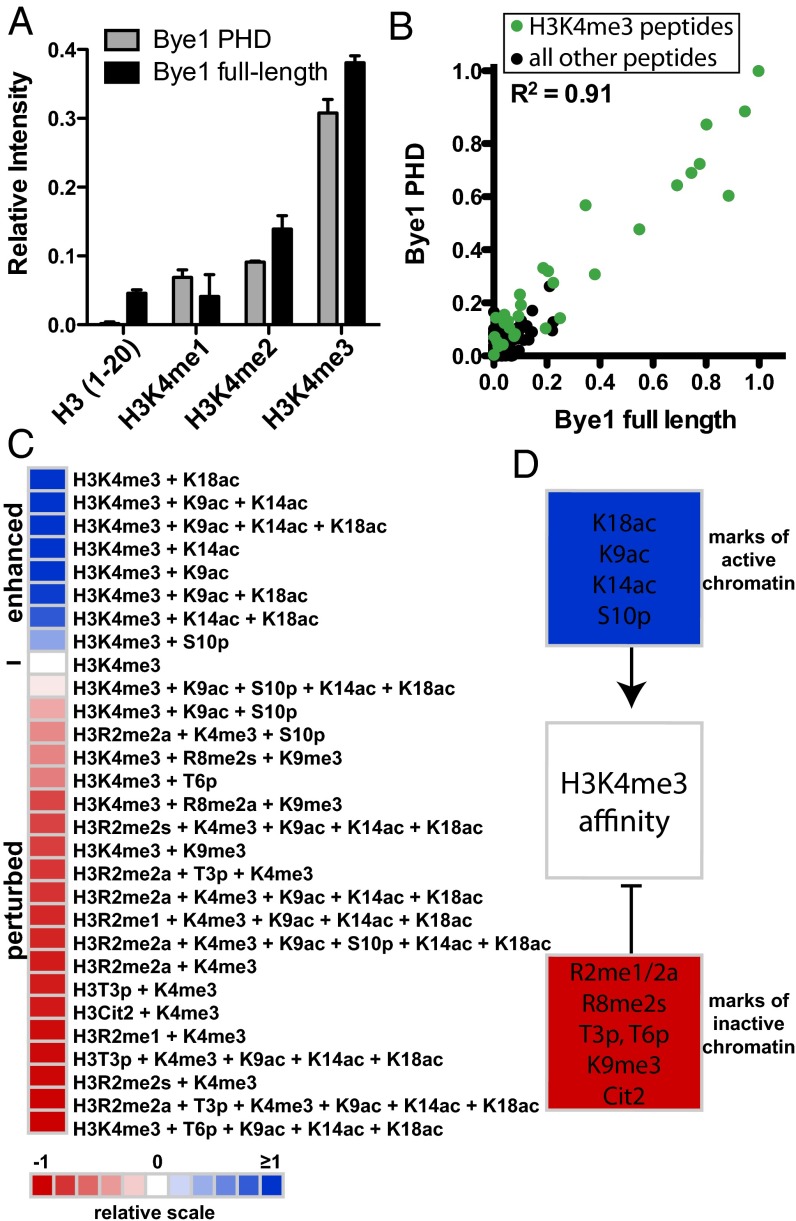
In addition to the observations described above, it has been reported that the Bye1 PHD domain contributes to chromatin association by binding trimethylated H3K4 peptides (17). We therefore investigated binding of the Bye1 PHD domain to roughly 200 differentially modified histone peptides on a microarray (22). Microarrays were spotted at high density (∼4,000 individual features) with histone peptides that encompass known single and combinatorial PTMs on the core and tail domains of the four histone proteins H3, H4, H2A, and H2B and their variants (Table S2). The Bye1 PHD domain and full-length Bye1 bound specifically to H3K4me3 peptides (Fig. 4A). The high correlation between arrays probed with full-length Bye1 and the isolated PHD domain indicated that the histone-binding potential of Bye1 is harbored within its PHD domain (Fig. 4B). The interaction of Bye1 with H3K4me3 was strongly influenced by neighboring PTMs (Fig. 4C). In particular, marks of active transcription (H3K9ac, H3K14ac, H3K18ac, and H3S10p) (23–25) enhanced Bye1 affinity to H3K4me3, whereas marks of transcriptional repression (H3R2 and H3R8 methylation, Cit2, T3 and T6 phosphorylation, and H3K9me3) (23, 26–29) perturbed the interaction with H3K4me3 (Fig. 4D).
Fig. 4.
Bye1 preferentially binds histone peptides carrying PTMs of active transcription. (A) Peptide array binding analysis reveals that Bye1 preferentially associates with H3K4me3 peptides, and its PHD domain is sufficient for this interaction. Results of two independent arrays consisting of 24 individual spots for each peptide (Table S2) are presented as relative mean intensity measurements on a scale from 0 to 1, with 1 being the most significant peptide interaction. (B) Scatter plot correlating relative mean intensity measurements of all peptide interactions (Table S2) from arrays probed with full-length Bye1 and the Bye1 PHD domain. H3K4me3-containing peptides are shown as green dots. All other peptides on the array are shown as black dots. The correlation coefficient was calculated by linear regression analysis using GraphPad Prims v5. (C) Heat map depicting the effects of combinatorial PTMs on the binding of Bye1 to H3K4me3-containing peptides. Average binding intensities are represented relative to the H3K4me3 peptide (0, white). Enhanced (>0, blue) and perturbed (<0, red) interactions are depicted. (D) Summary of modifications enhancing (blue) and perturbing (red) the Bye1 interaction with H3K4me3 peptides.
Bye1 Occupies the 5′-Region of Active Genes.
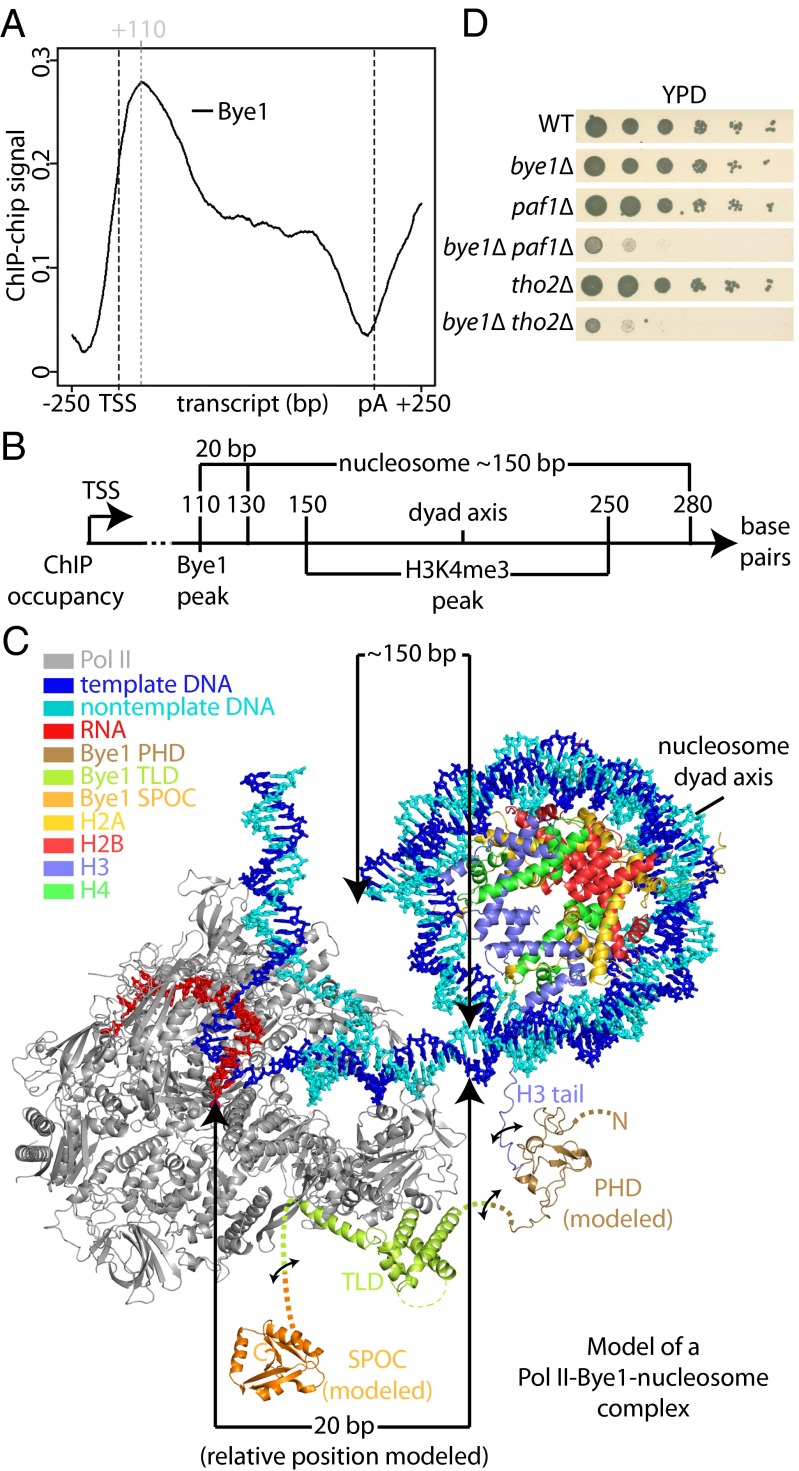
To test whether Bye1 is recruited to actively transcribed genes in vivo, we carried out genomic occupancy profiling with the use of ChIP (SI Materials and Methods). Metagene analysis by averaging occupancy profiles of genes of similar length revealed a Bye1 occupancy peak 110 nucleotides downstream of the transcription start site (TSS) (Fig. 5A). No significant signals were observed in promoter regions and at the polyadenylation (pA) site. Bye1 was found on all active genes, and its occupancy level correlated with those for bona fide Pol II elongation factors such as Spt5 (Fig. S5A). Published ChIP data for H3K4me3 shows a peak at a similar location downstream of the TSS, although the peak is broader (Fig. S5B) (30). These results indicated that Bye1 is recruited to the 5′-region of active genes in vivo, and suggest that H3K4me3 contributes to Bye1 recruitment.
Fig. 5.
Bye1 associates with active genes in front of the +2 nucleosome. (A) Gene-averaged Bye1 ChIP occupancy profile for the median gene length class (1,238 ± 300 nt, 339 genes). (B) Scheme showing occupancies of Bye1 and H3K4me4 (30) derived from ChIP data and nucleosome position derived from microarray and high-throughput DNA sequencing data (31). (C) Model of a Pol II–nucleosome-Bye1 complex based on crystal structures and ChIP occupancy peak positions. Distances in base pairs (bp) are indicated between the Pol II active center and the nucleosome, as well as for the nucleosomal DNA. The model is based on the structure of the nucleosome core particle (1aoi) (47). Modeling was performed with Coot (48). Bye1 PHD and SPOC domains were modeled using Modeler (49). The PHD domain model is based on structures 3kqi, 1wem, 1wew, 2lv9, and 1wep, which were identified by HHpred (50) to be most similar to Bye1 PHD. Binding of the PHD domain to H3K4me3 was modeled based on structure 2jmj. The SPOC domain model is based on structure 1ow1. (D) Bye1 genetically interacts with Paf1 and Tho2. Serial dilutions of strains bye1∆, paf1∆, bye1∆paf1∆, tho2∆, bye1∆tho2∆, and an isogenic WT control strain were placed on yeast extract peptone dextrose (YPD) plates and incubated at 30 °C for 3 d.
To interpret the ChIP data, we generated a 3D topological model of the Bye1-bound Pol II elongation complex approaching the +2 nucleosome of an active yeast gene (Fig. 5C). For the modeling we assumed that Bye1 cross-links to DNA via Pol II in ChIP experiments and set the Pol II active center to nucleotide position +110 downstream of the TSS. We positioned the +2 nucleosome based on its experimentally defined average position (31) (Fig. 5B). We also included models of the flexible Bye1 SPOC and PHD domains, with the latter positioned on the H3 tail emerging from the core nucleosome particle (Fig. 5C). Although the trajectory of the H3 tail is unclear and although the linkers between the Bye1 domains are flexible, the resulting model explained the position of the ChIP peak with high H3K4me3 occupancy. The model also suggests that it is structurally possible for Bye1 to interact simultaneously with the Pol II core and the trimethylated H3 tail in the 5′-region of active genes.
Bye1 Genetically Interacts with Paf1 and Tho2.
To further elucidate Bye1 function, we aimed at identifying genes that interact functionally with the gene encoding Bye1. The yeast bye1∆ strain does not show any obvious growth phenotype (20), suggesting a nonessential function. However, it has been shown for the transcription factor TFIIS that mutation of its functional residues, but not its deletion, confers lethality (32). This finding illustrates that nonessential genes can have very important functions. Screening a yeast deletion strain collection (33, 34) for synthetic growth defects with bye1∆ revealed two candidate genes: paf1 (RNA polymerase II associated protein 1) and tho2 (THO complex subunit 2). Generating bye1∆paf1∆ and bye1∆tho2∆ double mutants in a different genetic background confirmed the synthetic interaction between these genes (Fig. 5D). The genes paf1 and tho2 encode for subunits of two bona fide elongation factor complexes. Paf1 belongs to the five-subunit Paf complex that recruits the histone methyltransferase Set1 to transcribed genes (35). Set1 in turn is responsible for H3K4 trimethylation during transcription (36). The interaction of Bye1 PHD with H3K4me3 is therefore a plausible link between Paf1 and Bye1. Tho2 resides in the four-subunit THO complex that is required for efficient transcription elongation (37). These results strongly support an involvement of Bye1 in transcription elongation through chromatin.
PHF3 and DIDO Are Human Homologs of Bye1.
No homologs in higher eukaryotes have been reported for Bye1. We performed a bioinformatics search based on the Pfam database (38) to identify potential homologs with the same domain organization. We found two human proteins, PHD finger protein 3 (PHF3) and death-inducer obliterator (DIDO), which show the same domain organization as Bye1 (Fig. 6B). Both proteins contain an N-terminal PHD domain, a central TLD domain, and a C-terminal SPOC domain, with linkers of varying lengths in between these domains. Homology for both proteins could not be inferred based on sequence homology [E-value: 2e−04 (PHF3)/5e−04 (DIDO)]. PHF3 has been associated with glioma development because its expression is significantly reduced or lost in glioblastomas (39). DIDO is a potential tumor suppressor showing abnormal expression patterns in patients with myelodysplastic and myeloproliferative diseases (40). Specific binding of the DIDO PHD domain to H3K4me3 has been reported recently (41).
Fig. 6.
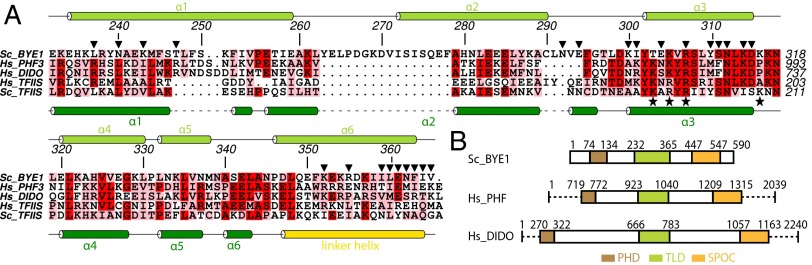
Conservation of Pol II–binding residues in Bye1 human homologs. (A) Amino acid sequence alignment of S. cerevisiae Bye1, H. sapiens PHF3, H. sapiens DIDO, H. sapiens TFIIS, and S. cerevisiae TFIIS. Secondary structure elements are indicated as arrows (β-strands) or rods (α-helices). Loops are indicated with solid lines. Residues that are part of the Pol II–Bye1 interface are marked with black triangles. Residues essential for the Pol II–TFIIS interaction (51) are marked with black asterisks. (B) Domain organization of S. cerevisiae Bye1, H. sapiens PHF3, and H. sapiens DIDO. Numbers for bordering residues are indicated.
To corroborate the homology of PHF3 and DIDO with Bye1, we analyzed the conservation of the Pol II–TLD interface. Both yeast Pol II and Bye1 TLD surfaces forming the interface are well conserved in human Pol II and PHF3/DIDO, respectively (Fig. 6A; Fig. S6). In particular, a salt bridge between yeast Bye1 residue K314 and E1168 in the largest Pol II subunit Rpb1 is conserved in the predicted human PHF3/DIDO–Pol II complexes. Similarly, many hydrogen bonds observed between the Bye1 TLD and Rpb1 (Bye1 residues N292, S311, D315, R355, N362, and F363) are predicted to be conserved in the homologous human complexes. These results indicate that PHF3 and DIDO contain Pol II–binding TLD domains and are human homologs of Bye1, and our structural, biochemical, and genetic results provide a starting point for elucidating the function of these proteins.
Discussion
Here we show that the nuclear protein Bye1 binds to Pol II and report crystal structures of the central TLD domain of Bye1 bound to free Pol II, a Pol II elongation complex with DNA template and RNA transcript, an elongation complex with an NTP analog, and an arrested elongation complex with backtracked RNA. These studies represent only the third high-resolution structural analysis of a transcription factor complex with the polymerase core. Whereas the previously studied factors TFIIB (4–7, 42) and TFIIS (8–12) alter Pol II function by directly affecting catalytic events, Bye1 does not alter basic Pol II functions in vitro. Consistent with this, Bye1 binding to Pol II does not alter Pol II conformation in the structures. Additional functional data in vitro and in vivo indicate that Bye1 occupies active genes in their 5′-region and can bind to histone H3 tails with PTMs of active transcription using its PHD domain. A chromatin-related function of Bye1 may explain published genetic evidence for a negative role of Bye1 in transcription elongation (20).
What could be the function of Bye1 in chromatin transcription? Because the TLD of Bye1 is required for chromatin association of Bye1, it is unlikely that Bye1 first recognizes active chromatin marks and then recruits Pol II to active chromatin regions. We speculate that instead Bye1 binds directly to Pol II during early transcription elongation and tethers surrounding histones containing active PTMs, perhaps to prevent loss of histones during polymerase passage through chromatin.
Materials and Methods
Proteins were prepared either from the natural source (Pol II) or expressed recombinantly in Escherichia coli (Bye1). For surface plasmon resonance analysis, Pol II was immobilized on a biosensor chip (Biacore), and time-resolved affinity measurements of Bye1 dilution series were carried out. Pol II–Bye1 complexes were formed with a 10× molar excess of Bye1 and cocrystallized. For Pol II–Bye1 TLD complexes containing nucleic acids, Pol II and nucleic acids were cocrystallized and Bye1 TLD was soaked into preformed crystals. Diffraction data were collected at the Swiss Light Source, and structures were solved by molecular replacement. For chromatin fractionation, plasmids containing HA-tagged full-length Bye1, Bye1 ∆PHD (∆1–177), and Bye1 ∆TLD (∆177–354) [obtained from S. D. Hanes, Division of Infectious Disease, Wadsworth Center, New York State Department of Health, Albany, New York (20)] were transformed into WT yeast. Chromatin fractionation was performed using a combination of previously described methods (43, 44). Peptide synthesis and validation, microarray fabrication, effector protein hybridization and detection, and data analysis of histone peptide microarrays were performed essentially as described previously (22). Synthetic genetic array analysis was performed as described previously (33, 34). For details, see SI Material and Methods.
Note Added in Proof.
After our paper was submitted for publication, it was reported that the human DNA helicase RECQL5 uses a TLD domain that is homologous to the Bye1 TLD and binds the same Pol II region (45). This finding is consistent with our proposal that human proteins PHF3 and DIDO are homologs of Bye1. Bye1 may thus be the founding member of a new family of transcription factors that link early transcribing Pol II to histones in yeast and human cells.
Supplementary Material
Acknowledgments
We thank Alan Cheung, Stefanie Etzold, Tobias Koschubs, Kristin Leike, and other members of the Cramer laboratory. We thank Stefan Jentsch, Jochen Rech, and Boris Pfander for materials and help with genetic screens. We thank the crystallization facility at the Max-Planck-Institute for Biochemistry in Martinsried. Part of this work was performed at the Swiss Light Source at the Paul Scherrer Institute, Villigen, Switzerland. K.K. was supported by a Boehringer Ingelheim fellowship and the International Max Planck Research School. P.C. was supported by the Deutsche Forschungsgemeinschaft [SFB646, TR5, GraKo1721, SFB960, Center for Integrated Protein Science Munich (CIPSM), Nanosystems Initiative Munich (NIM)] an Advanced Grant of the European Research Council, the LMUinnovativ project Bioimaging Network, the Jung-Stiftung, and the Vallee Foundation. S.B.R. was supported by a Postdoctoral Fellowship from the American Cancer Society (PF-13-085-01-DMC). This work was supported in part by National Institutes of Health Grant GM068088 (to B.D.S.). B.D.S. is a cofounder of EpiCypher, Inc.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data bank, www.pdb.org (PDB ID codes 4bxz, 4by7, 4by1, and 4bxx).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311010110/-/DCSupplemental.
References
- 1.Hahn S, Young ET. Transcriptional regulation in Saccharomyces cerevisiae: Transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011;189(3):705–736. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perales R, Bentley D. “Cotranscriptionality”: The transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36(2):178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mischo HE, Proudfoot NJ. Disengaging polymerase: Terminating RNA polymerase II transcription in budding yeast. Biochim Biophys Acta. 2013;1829(1):174–185. doi: 10.1016/j.bbagrm.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: An RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science. 2004;303(5660):983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 5.Kostrewa D, et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462(7271):323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010;327(5962):206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sainsbury S, Niesser J, Cramer P. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature. 2013;493(7432):437–440. doi: 10.1038/nature11715. [DOI] [PubMed] [Google Scholar]
- 8.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16(6):955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 9.Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114(3):347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 10.Wang D, et al. Structural basis of transcription: Backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324(5931):1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471(7337):249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 12. Wind M, Reines D (2000) Transcription elongation factor SII. BioEssays 22(4):327–336. [DOI] [PMC free article] [PubMed]
- 13.Wu X, et al. The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. EMBO J. 2000;19(14):3727–3738. doi: 10.1093/emboj/19.14.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris DP, Phatnani HP, Greenleaf AL. Phospho-carboxyl-terminal domain binding and the role of a prolyl isomerase in pre-mRNA 3′-End formation. J Biol Chem. 1999;274(44):31583–31587. doi: 10.1074/jbc.274.44.31583. [DOI] [PubMed] [Google Scholar]
- 15.Hani J, Stumpf G, Domdey H. PTF1 encodes an essential protein in Saccharomyces cerevisiae, which shows strong homology with a new putative family of PPIases. FEBS Lett. 1995;365(2–3):198–202. doi: 10.1016/0014-5793(95)00471-k. [DOI] [PubMed] [Google Scholar]
- 16.Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20(2):56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 17.Shi X, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007;282(4):2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ariyoshi M, Schwabe JW. A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev. 2003;17(15):1909–1920. doi: 10.1101/gad.266203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A, et al. TRIPLES: A database of gene function in Saccharomyces cerevisiae. Nucleic Acids Res. 2000;28(1):81–84. doi: 10.1093/nar/28.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X, Rossettini A, Hanes SD. The ESS1 prolyl isomerase and its suppressor BYE1 interact with RNA pol II to inhibit transcription elongation in Saccharomyces cerevisiae. Genetics. 2003;165(4):1687–1702. doi: 10.1093/genetics/165.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armache KJ, Mitterweger S, Meinhart A, Cramer P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J Biol Chem. 2005;280(8):7131–7134. doi: 10.1074/jbc.M413038200. [DOI] [PubMed] [Google Scholar]
- 22.Rothbart SB, Krajewski K, Strahl BD, Fuchs SM. Peptide microarrays to interrogate the “histone code”. Methods Enzymol. 2012;512:107–135. doi: 10.1016/B978-0-12-391940-3.00006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliver SS, Denu JM. Dynamic interplay between histone H3 modifications and protein interpreters: Emerging evidence for a “histone language”. ChemBioChem. 2011;12(2):299–307. doi: 10.1002/cbic.201000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakanishi S, et al. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol. 2008;15(8):881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 2003;4(4):276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 26.Pal S, et al. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007;26(15):3558–3569. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24(21):9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen D, et al. Regulation of transcription by a protein methyltransferase. Science. 1999;284(5423):2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 29.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Schulze JM, et al. Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev. 2011;25(21):2242–2247. doi: 10.1101/gad.177220.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang C, Pugh BF. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009;10(10):R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sigurdsson S, Dirac-Svejstrup AB, Svejstrup JQ. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol Cell. 2010;38(2):202–210. doi: 10.1016/j.molcel.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294(5550):2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 34.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303(5659):808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 35.Krogan NJ, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol Cell. 2003;11(3):721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 36.Roguev A, et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20(24):7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rondón AG, Jimeno S, García-Rubio M, Aguilera A. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J Biol Chem. 2003;278(40):39037–39043. doi: 10.1074/jbc.M305718200. [DOI] [PubMed] [Google Scholar]
- 38.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischer U, et al. PHF3 expression is frequently reduced in glioma. Cytogenet Cell Genet. 2001;94(3–4):131–136. doi: 10.1159/000048804. [DOI] [PubMed] [Google Scholar]
- 40.Fütterer A, et al. Dido gene expression alterations are implicated in the induction of hematological myeloid neoplasms. J Clin Invest. 2005;115(9):2351–2362. doi: 10.1172/JCI24177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gatchalian J, et al. (2013) Dido3 PHD modulates cell differentiation and division. Cell Reports 2013 4(1):148–158. [DOI] [PMC free article] [PubMed]
- 42.Deng W, Roberts SG. TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma. 2007;116(5):417–429. doi: 10.1007/s00412-007-0113-9. [DOI] [PubMed] [Google Scholar]
- 43.Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94(11):5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keogh MC, et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439(7075):497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 45.Kassube SA, Jinek M, Fang J, Tsutakawa S, Nogales E. Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5. Nat Struct Mol Biol. 2013;20(7):892–899. doi: 10.1038/nsmb.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292(5523):1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 47.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sánchez R, Sali A. Evaluation of comparative protein structure modeling by MODELLER-3. Proteins. 1997;(Suppl 1):50–58. doi: 10.1002/(sici)1097-0134(1997)1+<50::aid-prot8>3.3.co;2-w. [DOI] [PubMed] [Google Scholar]
- 50. Soding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33(Web Server issue):W244–W248. [DOI] [PMC free article] [PubMed]
- 51.Awrey DE, et al. Yeast transcript elongation factor (TFIIS), structure and function. II: RNA polymerase binding, transcript cleavage, and read-through. J Biol Chem. 1998;273(35):22595–22605. doi: 10.1074/jbc.273.35.22595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.