Significance
Two types of ubiquitin chain are required to switch on one of the major signaling networks of the innate immune system that triggers the production of inflammatory mediators to combat infection by pathogens. This paper reports the unexpected discovery that both types of ubiquitin chain are attached covalently to one another and that the formation of one type of ubiquitin chain is dependent on the prior formation of the other. One function of these hybrid ubiquitin chains is to permit the colocalization of two different kinase complexes, thereby facilitating the activation of one kinase complex by the other to increase the speed of the response to infection.
Keywords: LUBAC, TNF Receptor-Associated Factor 6, Ubc13, NF-κB
Abstract
Polyubiquitin (pUb) chains formed between the C terminus of ubiquitin and lysine 63 (K63) or methionine 1 (M1) of another ubiquitin have been implicated in the activation of the canonical IκB kinase (IKK) complex. Here, we demonstrate that nearly all of the M1-pUb chains formed in response to interleukin-1, or the Toll-Like Receptors 1/2 agonist Pam3CSK4, are covalently attached to K63-pUb chains either directly as K63-pUb/M1-pUb hybrids or indirectly by attachment to the same protein. Interleukin-1 receptor (IL-1R)-associated kinase (IRAK) 1 is modified first by K63-pUb chains to which M1-pUb linkages are added subsequently, and myeloid differentiation primary response gene 88 (MyD88) and IRAK4 are also modified by both K63-pUb and M1-pUb chains. We show that the heme-oxidized IRP2 ubiquitin ligase 1 interacting protein (HOIP) component of the linear ubiquitin assembly complex catalyzes the formation of M1-pUb chains in response to interleukin-1, that the formation of K63-pUb chains is a prerequisite for the formation of M1-pUb chains, and that HOIP interacts with K63-pUb but not M1-pUb linkages. These findings identify K63-Ub oligomers as a major substrate of HOIP in cells where the MyD88-dependent signaling network is activated. The TGF-beta–activated kinase 1 (TAK1)-binding protein (TAB) 2 and TAB3 components of the TAK1 complex and the NFκB Essential Modifier (NEMO) component of the canonical IKK complex bind to K63-pUb chains and M1-pUb chains, respectively. The formation of K63/M1-pUb hybrids may therefore provide an elegant mechanism for colocalizing both complexes to the same pUb chain, facilitating the TAK1-catalyzed activation of IKKα and IKKβ. Our study may help to resolve the debate about the relative importance of K63-pUb and M1-pUb chains in activating the canonical IKK complex.
Interleukin-1 (IL-1) or ligands that activate Toll-Like Receptors (TLRs) initiate “downstream” signaling events by recruiting the adaptor protein MyD88 (1), IL-1 Receptor (IL-1R)-Associated Kinase 4 (IRAK4), IRAK1, and IRAK2 to form an oligomeric structure termed the Myddosome (2, 3). IRAKs 1 and 2 undergo covalent modification by phosphorylation and ubiquitylation and interact with the E3 ubiquitin ligase TNF Receptor-Associated Factor 6 (TRAF6) (4). TRAF6 can combine with the E2-conjugating complex Ubc13-Uev1a to generate K63-pUb chains in vitro. These K63-Ub chains, not anchored to any other protein, were reported to activate the TAK1 complex in vitro (5–7), probably via conformational changes induced by an interaction with the Npl40 Zinc Finger (NZF) domain of TAB2 and TAB3, which bind to K63-pUb chains specifically in vitro (8, 9). Once activated, TAK1 phosphorylates the canonical IκB kinase (IKK) complex and the mitogen-activated protein (MAP) kinase kinases that activate c-Jun N-terminal kinases (JNKs) and p38 MAP kinases, which then trigger “downstream” signaling events. The IL-1–stimulated activation of the canonical IKK complex and MAP kinases fails to occur in mouse embryonic fibroblasts (MEFs) from TRAF6-deficient mice (10) or TAK1-deficient mice (11) or in MEFs from mice that express an inactive, truncated form of TAK1 (12). Moreover, wild-type TRAF6, but not an E3 ligase-deficient mutant, restores IL-1 signaling to TRAF6-deficient MEFs (10). These observations support the hypothesis that TRAF6-generated K63-pUb chains are required for the IL-1–stimulated activation of TAK1 and that TAK1 expression is essential to activate the IKK complex and MAP kinases in MEFs.
NFκB Essential Modifier (NEMO), an integral component of the canonical IKK complex, was reported to bind K63-pUb chains (13, 14), and NF-κB–dependent gene transcription induced by TNFα or IL-1 was restored to NEMO-deficient MEFs by the reintroduction of wild-type NEMO but not by the pUb-binding–defective mutant NEMO[D311N] (13–15). Moreover, the TNFα-stimulated degradation of IκBα and the TNFα or IL-1–stimulated translocation of the p50/p65 NFκB subunits to the nucleus was impaired in fibroblasts from immune-deficient human patients expressing the NEMO[D311G] mutant. Thus, the activation of the canonical IKK complex is attenuated in cells expressing pUb-binding–defective mutants of NEMO (16).
More recently, NEMO was found to bind linear-Ub oligomers with 100-fold higher affinity than K63-Ub oligomers of equivalent length (17, 18), suggesting that NEMO may bind linear-pUb chains preferentially in vivo. These pUb chains, which are produced by the formation of a peptide bond between the α-amino group of the N-terminal methionine (M) of one ubiquitin and the C-terminal glycine of another ubiquitin, will be termed M1-pUb chains hereafter. M1-Ub linkages can be generated in vitro by the linear ubiquitin assembly complex (LUBAC) (19), which is composed of three components: Heme-Oxidized IRP2 ubiquitin Ligase 1 (HOIL-1), HOIL-1 interacting protein (HOIP), and Sharpin (20–22). Importantly, the IL-1–stimulated activation of the canonical IKK complex was impaired in MEFs from HOIL-1−/− mice (23), suggesting an important role for M1-pUb chains in this process. Moreover, the readdition of LUBAC to HeLa extracts depleted of this E3 ligase restored the activation of the canonical IKK complex in vitro. Furthermore, LUBAC was reported to activate NFκB-dependent gene transcription when overexpressed in wild-type MEFs, but not in NEMO-deficient MEFs (24). It was inferred from these studies that LUBAC-catalyzed M1-pUb chain formation, rather than K63-pUb chain formation, was critical for the activation of the canonical IKK complex.
Here, we report that nearly all of the M1-pUb chains formed when the MyD88 signaling network is activated are attached covalently to K63-pUb chains. We further show that the IL-1–stimulated formation of K63-pUb chains is a prerequisite for the formation of M1-pUb chains and that HOIP binds specifically to K63-pUb chains in vitro. We suggest that the formation of K63-pUb/M1-pUb hybrids permits the recruitment of TAK1 and the canonical IKK complex to the same pUb chains, facilitating the TAK1-catalyzed phosphorylation and activation of the IKKs.
Results
IL-1 Stimulates the Formation of M1-pUb and K63-pUb Chains in IL-1 Receptor Cells.
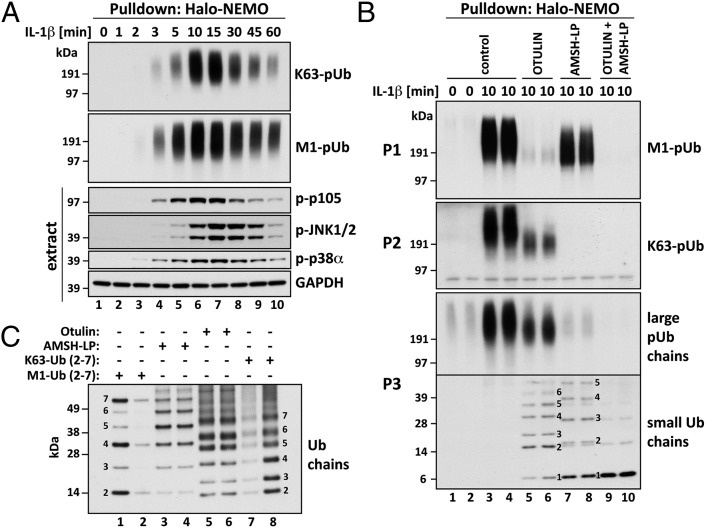
HEK293 cells stably expressing the IL-1 receptor (termed here IL-1R cells) (25) were stimulated with IL-1, and the pUb chains captured from the cell extracts on immobilized Halo-NEMO were analyzed. The Halo-NEMO beads captured M1-pUb and K63-pUb chains from the extracts of IL-1–stimulated cells (Fig. S1A), but did not capture K48-pUb chains or K11-pUb chains, in contrast to Halo–tandem-repeated ubiquitin-binding entities (TUBEs) (26), which captured these types of pUb chain (Fig. S1B). M1-pUb or K63-pUb chains captured by NEMO were not detectable in the absence of IL-1 stimulation, but could be detected within 2 min of exposure to IL-1, the amounts peaking after 10–15 min and declining thereafter (Fig. 1A). The formation of these pUb chains paralleled the phosphorylation of p105, an authenticated physiological substrate of IKKβ (27, 28), whereas the phosphorylation (activation) of JNK and p38α MAPK peaked slightly later (Fig. 1A). The NEMO[D311N] mutant did not capture any pUb chains (Fig. S1A).
Fig. 1.
IL-1 induces the formation of K63-pUb/M1-pUb hybrids. (A) IL-1R cells were stimulated for 10 min with 5 ng/mL IL-1β and lysed with 100 mM iodoacetamide to inhibit deubiquitylases, and the pUb chains and associated proteins were captured from the cell extracts with Halo-NEMO. The captured proteins were identified by immunoblotting with antibodies that recognize K63-pUb or M1-pUb chains specifically (Top two subpanels). The extracts (25 µg protein) were immunoblotted with the antibodies indicated (Bottom four subpanels). (B) As in A, except that the pUb chains captured by Halo-NEMO were incubated in the absence (control) or presence of Otulin, AMSH-LP or both deubiquitylases before SDS/PAGE and immunoblotting with antibodies that recognize M1-pUb chains (P1), K63-pUb chains (P2), or with an antibody that recognizes M1-pUb and K63-pUb chains equally well (P3). In P3, the lower half of the membrane was developed for 10 times longer than the upper half. (C) Authentic M1-pUb oligomers (30 ng, lane 1; 10 ng, lane 2) and K63-Ub oligomers (3 ng, lane 7 and 6 ng, lane 8) were used as markers to demonstrate that the small Ub oligomers formed in B after treatment with AMSH-LP were linked via M1 of ubiquitin (lanes 3 and 4) and that those formed by treatment with Otulin (lanes 5 and 6) were linked via K63.
M1-pUb Chains Captured by NEMO Are Linked to K63-pUb Chains.
Otulin is a deubiquitylase that hydrolyzes M1-pUb chains specifically (29, 30) (Fig. S2A), whereas AMSH-LP cleaves K63-pUb chains specifically (Fig. S2B) (9). As expected, Otulin hydrolyzed nearly all of the M1-pUb chains captured from the cell extracts by immobilized NEMO (Fig. 1B, P1, lanes 5 and 6). However, unexpectedly, Otulin treatment reduced the size of the large K63-pUb chains (Fig. 1B, P2; compare lanes 5 and 6 with lanes 3 and 4) and generated a variety of faster-migrating, small pUb oligomers (Fig. 1B, P3, lanes 5 and 6) that comigrated with K63-pUb chains (Fig. 1C, lanes 5–8) and were hydrolyzed by AMSH-LP (Fig. 1B, P3; compare lanes 9 and 10 with lanes 5 and 6). The generation of these small K63-Ub oligomers explains why the amount of the larger K63-pUb chains was reduced after Otulin treatment. Conversely, treatment with AMSH-LP hydrolyzed all of the K63-pUb chains captured by NEMO (Fig. 1B, P2, lanes 7 and 8) and increased the formation of faster-migrating small Ub oligomers (Fig. 1B, P3, lanes 7 and 8) that comigrated with M1-pUb chains (Fig. 1C, lanes 1–4) and could be hydrolyzed by Otulin (Fig. 1B, P3; compare lanes 9 and 10 with 7 and 8). The reduced size of the large pUb chains after incubation with either Otulin or AMSH-LP could be explained if the M1-pUb chains and K63-pUb chains were attached covalently to different lysine residues on the same protein or if the M1-pUb chains were attached directly to K63-pUb chains as K63-pUb/M1-pUb “hybrid” molecules. However, the generation of small K63-Ub oligomers after incubation with Otulin and small M1-Ub oligomers after incubation with AMSH-LP, which are not attached to any other protein, is compatible only with K63/M1-pUb hybrid formation.
Polyubiquitin Chains Attached to IRAK1 and Captured by NEMO Are K63-pUb/M1-pUb Hybrids.
We identified many proteins by mass spectrometry that were captured by Halo-NEMO from the extracts of IL-1–stimulated IL-1R cells in a ubiquitin-dependent manner because they were not captured by the NEMO[D311N] mutant. Those proteins of relevance to the present study are listed in Table S1. They include many of the proteins known to participate in the MyD88-signaling network (Introduction), namely MyD88 itself, IRAK1, IRAK4, TRAF6, the HOIP, HOIL-1 and Sharpin components of LUBAC and the TAB1, TAB2, TAB3 and TAK1 components of the TAK1 complex (Tables S1 and S2). Proteins captured by Halo-NEMO in a ubiquitin-independent manner included, as expected, IKKα and IKKβ (Table S3), the other components of the canonical IKK complex.
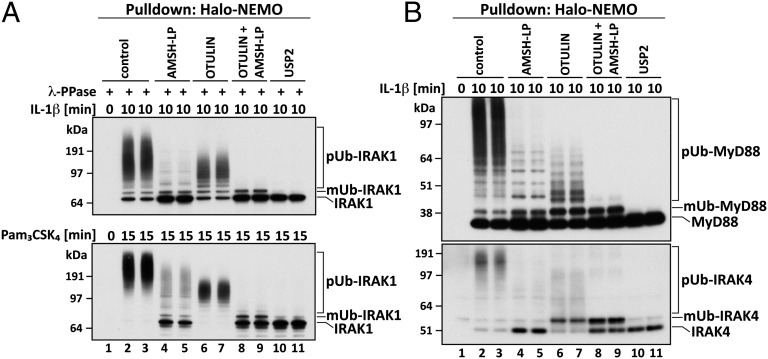
We found that IRAK1 became covalently modified by pUb chains in response to IL-1, explaining why it was captured by Halo-NEMO. Incubation with Otulin reduced the size of the pUb chains attached to IRAK1 (Fig. 2A, Upper, lanes 6 and 7), and these Ub chains could now be hydrolyzed by AMSH-LP, indicating that they were linked via K63 of ubiquitin (Fig. 2A, Upper, lanes 8 and 9). Importantly, none of the IRAK1 was reconverted to either the monoubiquitylated or unmodified forms of the protein after incubation with Otulin (Fig. 2A, Upper; compare lanes 6 and 7 with lanes 2 and 3) even though Otulin hydrolyzed virtually all of the M1-Ub linkages captured by NEMO (Fig. 1B, P1, lanes 5 and 6). These results are not compatible with the M1-pUb chains and K63-pUb chains being attached to different lysine residues in IRAK1. They are also not compatible with IRAK1 being modified initially with M1-Ub linkages and subsequently with K63-Ub linkages. The experiments therefore suggested that IRAK1 was modified by K63-pUb chains initially, to which M1-pUb chains were attached subsequently. These results were not specific to this agonist or cell type because similar results were obtained in human THP1 monocytes stimulated with Pam3CSK4, an activator of the TLR1/2 heterodimer that also signals via MyD88 (Fig. 2A, Lower).
Fig. 2.
Characterization of polyubiquitin chains attached covalently to proteins captured by Halo-NEMO from the extracts of IL-1R cells and THP1 monocytes. (A) IL-1R cells were stimulated with 5 ng/mL IL-1β (Upper) or THP1 monocytes with 1 μg/mL Pam3CSK4 (Lower) and the experiment was then performed as in Fig. 1B, except that the proteins captured by Halo-NEMO were treated for 1 h with λPPase (to dephosphorylate IRAK1) in the absence (control) or presence of the deubiquitylases AMSH-LP, Otulin, AMSH-LP plus Otulin, or USP2. The samples were then subjected to SDS/PAGE and immunoblotted for IRAK1. (B) As in A, except that λPPase was omitted and the PVDF membranes were immunoblotted with antibodies that recognize MyD88 and IRAK4. mUb, monoubiquitylated.
Topology of the pUb Chains Attached to MyD88 and IRAK4.
A significant proportion of the MyD88 and IRAK4 captured by Halo-NEMO from the extracts of IL-1–stimulated IL-1R cells (Fig. 2B) or Pam3Csk4-stimulated THP1 monocytes (Fig. S3A) underwent polyubiquitylation. Similar to IRAK1, treatment with Otulin or AMSH-LP reduced the size of the large pUb chains attached to MyD88 and IRAK4 (Fig. 2B; compare lanes 4–7 with lanes 2 and 3), whereas treatment with Otulin plus AMSH-LP converted MyD88 and IRAK4 to monoubiquitylated and unmodified species. The pUb chains attached to MyD88 and IRAK4 therefore also contain both K63-Ub and M1-Ub linkages. However, in contrast to IRAK1, incubation with Otulin generated some monoubiquitylated MyD88 and IRAK4, suggesting that some of the M1-pUb chains may be attached to these proteins without their prior modification by K63-pUb chains.
Like the nonspecific deubiquitylase USP2 (Fig. 2 A and B, lanes 10 and 11), AMSH-LP not only hydrolyzed K63-pUb chains but also partially reconverted IRAK1 and MyD88, and largely reconverted IRAK4, to the unmodified species (Fig. 2 A and B; compare lanes 4 and 5 with lanes 2 and 3). This indicates that AMSH-LP can cleave the isopeptide bond(s) formed between the C terminus of the first ubiquitin and the ε-amino moiety of a lysine residue(s) on these proteins. However, in the case of IRAK1 and MyD88, this isopeptide bond is hydrolyzed more slowly than K63-Ub linkages by AMSH-LP because increased formation of the monoubiquitylated species is also observed.
Polyubiquitylation of Other Proteins.
The ubiquitylated TRAF6 formed in response to IL-1 was not modified by M1-pUb chains because incubation with Otulin had no effect (Fig. S3B). The ubiquitylated forms of TRAF6 detected after treatment with AMSH-LP plus Otulin may arise from monoubiquitylation of several lysine residues on TRAF6.
Minor amounts of HOIP and HOIL-1 were converted to slower-migrating forms in response to IL-1, which disappeared after USP2 treatment, indicating that they were ubiquitylated species (Fig. S3C). Sharpin was partially converted to a USP2-sensitive monoubiquitylated form in response to IL-1 (Fig. S3C). A small amount of the endogenous NEMO was also converted to a monoubiquitylated form in IL-1–stimulated IL-1R cells (Fig. S3D). Trace amounts of polyubiquitylated NEMO were also detected, which were hydrolyzed by AMSH-LP but not by Otulin (Fig. S3D; compare lanes 3–8), indicating that NEMO is not modified significantly with M1-Ub chains after IL-1 stimulation.
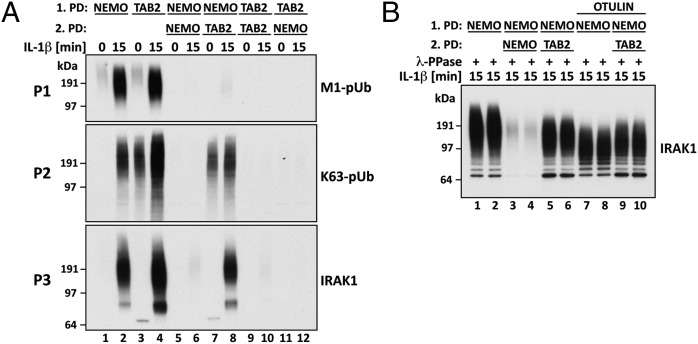
Sequential Capture of pUb Chains by Halo-TAB2 and Halo-NEMO.
The Npl4 Zinc Finger (NZF) domain of the TAB2 component of the TAK1 complex binds specifically to K63-pUb chains (8, 9). However, we found that a tandem repeat of the NZF of TAB2 (termed here Halo-TAB2) also captured all of the large M1-pUb chains present in the extracts of IL-1–stimulated cells (Fig. 3A) because no high-molecular-mass M1-pUb chains were captured by Halo-NEMO from the supernatant obtained after the first Halo-TAB2 pull-down (Fig. 3A, P1; compare lanes 4 and 12). Conversely, no M1-pUb chains were captured from the supernatant of the first Halo-NEMO pull-down by Halo-TAB2 (Fig. 3A, P1; compare lanes 2 and 8). Thus, both Halo-TAB2 and Halo-NEMO deplete the cell extracts of M1-pUb chains. On the other hand, Halo-TAB2 captured K63-pUb chains (Fig. 3A, P2, lanes 7 and 8) and more pUb-IRAK1 (Fig. 3A, P3, lane 8) from the supernatant of the first Halo-NEMO pull-down, whereas Halo-NEMO did not (Fig. 3A, P2 and P3, lanes 5 and 6). The size of pUb-IRAK1 captured by Halo-TAB2 from the supernatant of the first NEMO pull-down was smaller than that captured by Halo-NEMO (Fig. 3B; compare lanes 5 and 6 with 1 and 2). Moreover, unlike the pUb-IRAK1 captured by NEMO (Fig. 3B; compare lanes 7 and 8 with 1 and 2), the pUb-IRAK1 was not reduced in size by treatment with Otulin (Fig. 3B; compare lanes 5 and 6 with 9 and 10). Thus, IL-1 stimulation generates two forms of pUb-IRAK1, one linked to K63/M1-pUb hybrids (captured by Halo-NEMO or Halo-TAB2) and the other linked only to K63-pUb chains (captured by Halo-TAB2 from the supernatant of the first NEMO pull-down).
Fig. 3.
Two distinct forms of pUb-IRAK1 identified by the combined use of Halo-NEMO and Halo-TAB2. (A) The experiment was carried out as in Fig. 2A except that pUb chains and associated proteins were captured with Halo-NEMO or Halo-TAB2 [first pull-down (PD)]. Where indicated, the supernatants from the first PD were subjected to a second PD using either Halo-NEMO or Halo-TAB2 as indicated. Captured pUb chains or IRAK1 were identified by immunoblotting. (B) As in A, except that proteins that were captured by Halo-NEMO or Halo-TAB2 from the supernatant of the first Halo-NEMO pull-down, were incubated with Otulin. The ubiquitylation of IRAK1 was then analyzed by immunoblotting with a specific antibody.
In contrast to Halo-NEMO, Halo-TAB2 captured K63-pUb chains from the extracts of cells not stimulated with IL-1 (Fig. 3A, P2; compare lanes 1 and 3). Taken together, these results establish that NEMO captures M1-pUb chains selectively from the cell extracts, whereas Halo-TAB2 captures only K63-pUb chains. However, both Halo-tagged proteins deplete M1-pUb chains from the extracts of IL-1–stimulated cells because virtually all of the M1-pUb chains are attached covalently to K63-pUb chains.
LUBAC Is the Only E3 Ubiquitin Ligase That Catalyzes the Formation of M1-pUb Chains in IL-1–Stimulated Mouse Embryonic Fibroblasts.
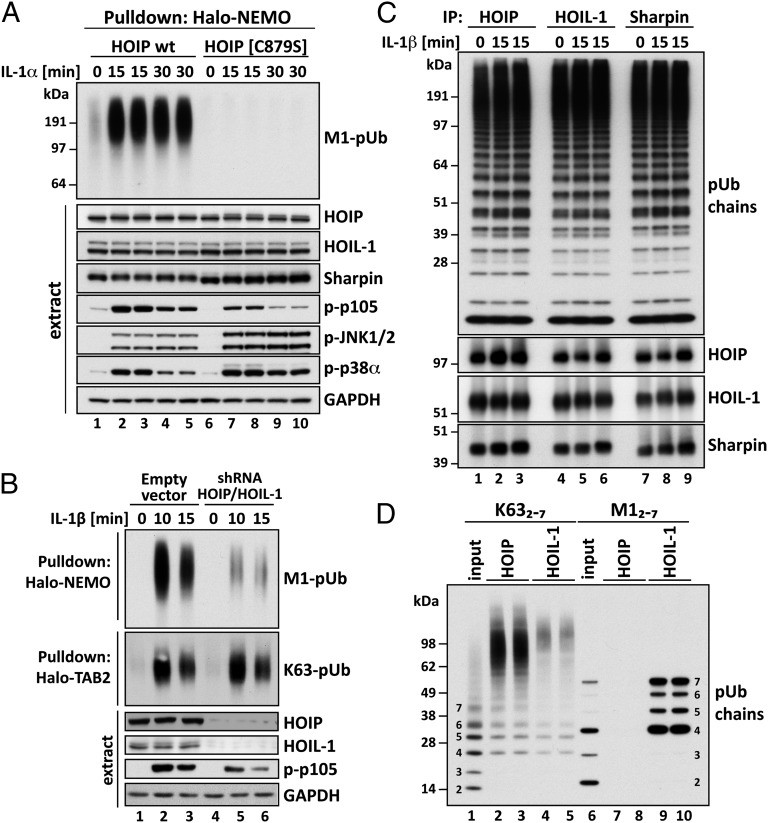
Although LUBAC catalyzes the formation of M1-pUb chains in vitro, it is not established that it is the only E3 ubiquitin ligase that generates these chains in vivo. We therefore generated knock-in mice in which HOIP was replaced by the E3 ligase-inactive HOIP[C879S] mutant, in which the catalytic cysteine was replaced by serine (Fig. S4A). The human HOIP[C885S] mutant, equivalent to Cys879 of murine HOIP, had no detectable E3 ligase activity (Fig. S4B). The HOIP[C879S] mice displayed embryonic lethality, no embryos being detectable from E11 onward; fibroblasts from mouse embryos that were 10.5 d old (MEFs) were therefore used for all subsequent studies. IL-1 did not induce detectable M1-pUb chain formation in HOIP[C879S] MEFs (Fig. 4A), establishing that HOIP is the catalytic subunit of LUBAC and the only E3 ligase that can produce M1-pUb chains in IL-1–stimulated MEFs. The expression of HOIP, HOIL-1, and Sharpin was similar in MEFs from HOIP[C879S] and wild-type mice (Fig. 4A).
Fig. 4.
Studies on the activity, pUb-binding properties, and role of LUBAC. (A) MEFs from mice expressing either wild-type (wt) HOIP or the HOIP[C879S] mutant were stimulated with 5 ng/mL IL-1α for the times indicated. M1-pUb chains captured from the cell extracts with Halo-NEMO were identified by immunoblotting with a specific antibody (Top). Cell extracts (20 μg protein) were immunoblotted with the antibodies indicated (Bottom seven subpanels). (B) IL-1R cells stably expressing shRNAs specific for HOIP and HOIL-1, or an empty control vector, were stimulated with 5 ng/mL IL-1β and pUb chains captured from extracts with Halo-NEMO or Halo-TAB2. The captured M1-pUb (Top) and K63-pUb chains (Top, second subpanel) were identified by immunoblotting with specific antibodies. Aliquots of the extracts (20 μg protein) were also immunoblotted with the antibodies indicated (Bottom four subpanels). (C) IL-1R cells were not stimulated or stimulated for 15 min with 5 ng/mL IL-1β. The cells were lysed without iodoacetamide to prevent the inactivation of LUBAC, which was then immunoprecipitated from the extracts with anti–HOIL-1, anti-Sharpin, or anti-HOIP. The M1-pUb chains formed after 60 min by LUBAC were examined by immunoblotting. The immunoprecipitates were also immunoblotted for HOIP, HOIL-1, and Sharpin. (D) Full-length GST-HOIP and GST-HOIL-1 were mixed with K63-Ub or M1-Ub oligomers, and, after capture on glutathione-Sepharose, the pUb-oligomers bound to each protein were analyzed by immunoblotting with anti-ubiquitin. Lanes 1 and 6 show, respectively, the K63-pUb oligomers and M1-pUb oligomers used. Lanes 2–5 show the K63-pUb chains and lanes 7–10 the M1-pUb chains captured by each protein.
The IL-1–stimulated activation of IKKβ was reduced in MEFs from the HOIP[C879S] mice, as judged by reduced phosphorylation of p105, a well-authenticated physiological substrate of IKKβ (27, 28). Thus, M1-pUb chain formation was required for optimal activation of the canonical IKK complex. In contrast, the IL-1–stimulated activation of JNK and p38α MAP kinase was enhanced in HOIP[C879S] MEFs, demonstrating that M1-pUb chain formation is not required for the activation of these protein kinases (Fig. 4A).
The knock-down of both HOIP and HOIL-1 also reduced the IL-1–stimulated formation of M1-pUb chains and the phosphorylation of p105 in IL-1R cells, without affecting the IL-1–stimulated formation of K63-pUb chains significantly (Fig. 4B).
LUBAC Is Active in Unstimulated Cells.
Because M1-pUb chains were formed only in response to IL-1 in IL-1R cells or to Pam3CSK4 in THP1 cells, we anticipated that LUBAC would be activated by these agonists. However, unexpectedly, LUBAC immunoprecipitated from IL-1R cell extracts with anti-HOIP was already active in serum grown cells not stimulated with IL-1, and activity did not increase in response to IL-1 (Fig. 4C and Fig. S5 A and B). Similar results were obtained in Pam3CSK4-stimulated THP1 cells. It was possible that HOIP had been activated by interaction with the anti-HOIP antibody, but the same results were obtained if LUBAC was immunoprecipitated from the cell extracts with anti–HOIL-1 or anti-Sharpin (Fig. 4C). It is unlikely that antibodies raised against all three components of LUBAC would activate its E3 ligase activity. We therefore conclude that LUBAC does not undergo a stable covalent modification in response to IL-1 that converts it from an inactive to an active form.
The antibodies raised against any one component of LUBAC immunoprecipitated the others, confirming that, as in other cells (31, 32), LUBAC is a hetero-trimeric complex composed of HOIP, HOIL-1, and Sharpin, a complex remaining intact after IL-1 stimulation (Fig. 4C, Lower).
Formation of M1-pUb Chains Requires the Formation of K63-pUb Chains.
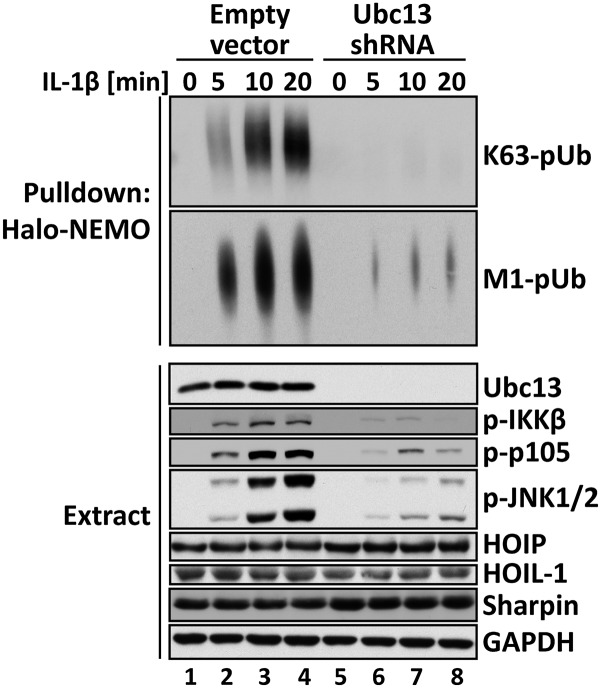
The E2-conjugating complex Ubc13-Uev1a directs the formation of K63-pUb chains in cells. To investigate whether the formation of M1-pUb chains was affected by the formation of K63-pUb chains, we generated IL-1R cells deficient in Ubc13 (Fig. 5). As expected, the IL-1–stimulated formation of K63-pUb chains was almost abolished (Fig. 5, Top) but, interestingly, the IL-1–stimulated formation of M1-pUb chains was also greatly reduced (Fig. 5, Top, second subpanel). Control experiments showed that the expression of HOIP, HOIL-1, and Sharpin was unaltered in the Ubc13-deficient cells (Fig. 5). The phosphorylation of IKKβ, its substrate p105, and JNK was also impaired in the Ubc13-deficient cells (Fig. 5). Importantly, the reexpression of Ubc13 restored the IL-1–stimulated formation of K63-pUb and M1-pUb chains and the activation of the canonical IKK complex and MAP kinases (Fig. S5C). Taken together, these experiments establish that the formation of M1-pUb chains in IL-1R cells requires K63-pUb chain formation.
Fig. 5.
Formation of M1-pUb chains is dependent on the prior formation of K63-pUb chains. IL-1R cells stably expressing shRNA specific for Ubc13 or an empty vector were stimulated for the times indicated with 5 ng/mL IL-1β, and the cells were lysed in the presence of 100 mM iodoacetamide. The pUb chains captured from the cell extracts with Halo-NEMO were subjected to SDS/PAGE and immunoblotting with antibodies that recognize K63-pUb chains (Top) or M1-pUb chains (Top, second subpanel). The cell extracts (20 μg protein) were also subjected to SDS/PAGE and immunoblotted with the antibodies indicated (Bottom eight subpanels).
HOIP Interacts with K63-pUb Chains but Not with M1-pUb Chains.
Because most of the M1-pUb chains were attached covalently to K63-pUb chains, this indicated that K63-pUb oligomers were a major substrate for LUBAC in IL-1–stimulated cells. An N-terminal fragment of HOIP containing its two NZF domains was reported to bind weakly to K48-Ub and M1-Ub oligomers but more strongly to K63-Ub oligomers (33). Here, we found that full-length HOIP captured K63-Ub oligomers (Fig. 4D, lanes 2 and 3), but not M1-Ub oligomers (Fig. 4D, lanes 7 and 8) under the experimental conditions that we used. The NZF of HOIL-1 was reported to interact with M1-Ub dimers (34). We confirmed this finding using full-length HOIL-1 (Fig. 4D, lanes 9 and 10), but also detected weak binding to K63-Ub oligomers (Fig. 4D, lanes 4 and 5). Interestingly, HOIP and HOIL-1 captured longer K63-Ub oligomers preferentially, even though they were minor components in the preparation of mixed K63-Ub oligomers used for these binding studies (Fig. 4D, lane 1). The NZF of Sharpin was reported to interact with M1-Ub and K63-Ub oligomers (20, 22). However, full-length Sharpin failed to interact with M1-Ub, K63-Ub, or K48-Ub oligomers under our experimental conditions, indicating that the binding of Ub oligomers to Sharpin is much weaker than to HOIP or HOIL-1.
Discussion
In this paper we show that most of the M1-pUb chains formed in IL-1–stimulated IL-1R cells or Pam3CSK4-stimulated THP1 monocytes, are attached covalently to K63-pUb chains and that significant formation of M1-pUb linkages cannot take place until K63-pUb chains are produced (Fig. 5 and Fig. S5C). Our finding that HOIP interacts specifically with K63-pUb chains may help to explain why the formation of M1-pUb chains depends on the prior formation of K63-pUb chains (Fig. 4D). However, it cannot be the only reason because substantial amounts of K63-pUb chains were present in serum grown cells not stimulated with IL-1 (Fig. 3A), and yet M1-pUb chains did not accumulate under these conditions, despite LUBAC being active under these conditions. We were also unable to accelerate the LUBAC-catalyzed formation of M1-pUb chains in vitro by adding K63-Ub oligomers to the assays. The recruitment of LUBAC to a MyD88-dependent signaling complex containing the K63-pUb chain-generating E3 ligase TRAF6, and perhaps other K63-pUb chain-generating E3 ligases, would therefore seem to be needed before the M1-pUb chains generated by LUBAC can accumulate and be coupled to K63-pUb chains. Perhaps the interaction of K63-pUb/M1-Ub linkages with one or more proteins in this complex decreases the rate at which they are hydrolyzed by Otulin and/or other deubiquitylases in cells. Alternatively, or in addition, one or more M1-pUb hydrolase(s) may be converted to less active forms when the MyD88-signaling network is activated. The topology of the K63-pUb/M1-pUb hybrids is clearly heterogeneous because the AMSH-LP–catalyzed hydrolysis of K63-pUb chains or the Otulin-catalyzed hydrolysis of M1-pUb chains generated K63-Ub or M1-Ub oligomers, respectively, of varying length (Fig. 1B and Fig. S6). Working out the precise topology of these hybrid pUb chains will be a challenging project.
It was suggested that the activation of the TAK1 complex results from a conformational change induced by the interaction of K63-pUb chains with its TAB2 and TAB3 subunits (6, 7). In contrast, NEMO binds 100-fold more tightly to M1-Ub dimers than to K63-Ub dimers (17, 18) and captures M1-pUb chains specifically from the extracts of IL-1–stimulated cells (Fig. 3). The formation of K63-pUb/M1-pUb hybrids would therefore appear to be a simple device for colocalizing TAK1 and the canonical IKK complex to the same pUb chain, facilitating the TAK1-catalyzed activation of IKKα and IKKβ (Fig. 6). The failure to form M1-pUb linkages in MEFs from HOIP[C879S] knock-in mice (Fig. 4A), and hence K63-pUb/M1-pUb hybrids, may not only reduce the activation of the canonical IKK complex by preventing colocalization with TAK1, but also divert these TAK1 molecules to the activation of MAP kinase kinases, explaining why the IL-1–stimulated activation of JNK and p38α MAP kinase was enhanced in MEFs from the HOIP[C879S] knock-in mice (Fig. 4A). An enhanced activation of JNK was also reported in MEFs from HOIL-1–deficient mice, in which the expression of HOIP was also reduced drastically (23).
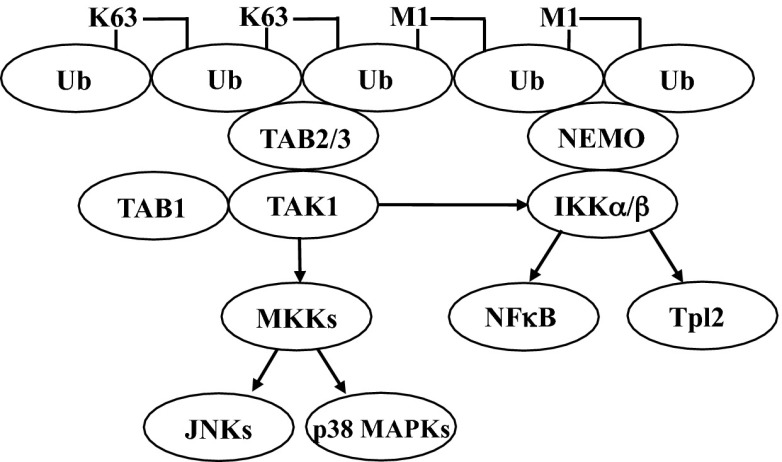
Fig. 6.
Enhanced activation of the canonical IKK complex via the MyD88-dependent formation of K63/M1-linked hybrid ubiquitin chains. The figure shows schematically how the formation of K63/M1-pUb hybrids may permit the corecruitment of the TAK1 and canonical IKK complexes via the interaction of TAB2/3 with K63-pUb chains and NEMO with M1-pUb chains, thereby facilitating the activation of IKKα and IKKβ by TAK1.
It will clearly be of great interest to investigate whether the formation of K63-pUb/M1-pUb hybrids is of more general significance and whether they participate in the regulation of other biological processes dependent on K63-pUb chain formation, such as the cellular response to DNA damage (35).
Materials and Methods
NEMO, the ubiquitin binding-defective mutant NEMO[D311N], a protein expressing two copies of the NZF domain of TAB2, and TUBEs (26) were expressed in Escherichia coli as Halo-tagged proteins. The bacteria were harvested; lysed in 50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 0.1% (vol/vol) 2-mercaptoethanol, 1 mM benzamidine, and 0.2 mM phenylmethylsulfonyl fluoride (PMSF); and sonicated. The bacterial cell lysate was centrifuged to remove debris, and the supernatant was coupled to the HaloLink resin (Promega) by incubation for 5 h at 4 °C as described by the manufacturer. The resin was washed extensively with 50 mM Tris⋅HCl, pH 7.5, 0.5 M NaCl, 0.1 mM EDTA, 270 mM sucrose, 0.03% (wt/vol) Brij 35, 0.1% (vol/vol) 2-mercaptoethanol, 0.2 mM PMSF, and 1 mM benzamidine and stored at 4 °C. To check coupling efficiency, an aliquot of the resin was resuspended in 50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 1 mM DTT containing 0.1 µg/µL TEV protease to release covalently bound NEMO, NEMO[D311N], and the NZF domains of TAB2 or TUBEs followed by SDS/PAGE and staining with Coomassie Blue. All experimental procedures are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Xiaoxia Li for IL-1R cells, the DNA Sequencing Service of the Medical Research Council Protein Phosphorylation and Ubiquitylation Unit (MRC-PPU), and the MRC-PPU’s teams for DNA cloning, protein production, tissue culture, and for outstanding technical support. The work was supported by a Wellcome Trust Senior Investigator Award (to P.C.); by grants from the Medical Research Council, AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck KGaA, Janssen Pharmaceutica, and Pfizer (to P.C.); and by grants from the Medical Research Council (U105192732) and the European Research Council (309756) (to D.K.). This article is dedicated to the late Tony Pawson whose discoveries inspired the research described in this paper.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314715110/-/DCSupplemental.
References
- 1.Martin MU, Wesche H. Summary and comparison of the signaling mechanisms of the Toll/interleukin-1 receptor family. Biochim Biophys Acta. 2002;1592(3):265–280. doi: 10.1016/s0167-4889(02)00320-8. [DOI] [PubMed] [Google Scholar]
- 2.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465(7300):885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Motshwene PG, et al. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem. 2009;284(37):25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye H, et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418(6896):443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 5.Deng L, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103(2):351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 6.Wang C, et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412(6844):346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 7.Xia ZP, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461(7260):114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanayama A, et al. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15(4):535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Komander D, et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10(5):466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh MC, Kim GK, Maurizio PL, Molnar EE, Choi Y. TRAF6 autoubiquitination-independent activation of the NFkappaB and MAPK pathways in response to IL-1 and RANKL. PLoS ONE. 2008;3(12):e4064. doi: 10.1371/journal.pone.0004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shim JH, et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 2005;19(22):2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato S, et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol. 2005;6(11):1087–1095. doi: 10.1038/ni1255. [DOI] [PubMed] [Google Scholar]
- 13.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22(2):245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 14.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat Cell Biol. 2006;8(4):398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 15.Windheim M, Stafford M, Peggie M, Cohen P. Interleukin-1 (IL-1) induces the Lys63-linked polyubiquitination of IL-1 receptor-associated kinase 1 to facilitate NEMO binding and the activation of IkappaBalpha kinase. Mol Cell Biol. 2008;28(5):1783–1791. doi: 10.1128/MCB.02380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubeau M, et al. New mechanism of X-linked anhidrotic ectodermal dysplasia with immunodeficiency: Impairment of ubiquitin binding despite normal folding of NEMO protein. Blood. 2011;118(4):926–935. doi: 10.1182/blood-2010-10-315234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo YC, et al. Structural basis for recognition of diubiquitins by NEMO. Mol Cell. 2009;33(5):602–615. doi: 10.1016/j.molcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahighi S, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136(6):1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Kirisako T, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25(20):4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerlach B, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471(7340):591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 21.Tokunaga F, et al. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature. 2011;471(7340):633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda F, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature. 2011;471(7340):637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11(2):123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 24.Kensche T, et al. Analysis of nuclear factor-κB (NF-κB) essential modulator (NEMO) binding to linear and lysine-linked ubiquitin chains and its role in the activation of NF-κB. J Biol Chem. 2012;287(28):23626–23634. doi: 10.1074/jbc.M112.347195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, et al. Mutant cells that do not respond to interleukin-1 (IL-1) reveal a novel role for IL-1 receptor-associated kinase. Mol Cell Biol. 1999;19(7):4643–4652. doi: 10.1128/mcb.19.7.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hjerpe R, et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10(11):1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lang V, et al. ABIN-2 forms a ternary complex with TPL-2 and NF-kappa B1 p105 and is essential for TPL-2 protein stability. Mol Cell Biol. 2004;24(12):5235–5248. doi: 10.1128/MCB.24.12.5235-5248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waterfield M, Jin W, Reiley W, Zhang M, Sun SC. IkappaB kinase is an essential component of the Tpl2 signaling pathway. Mol Cell Biol. 2004;24(13):6040–6048. doi: 10.1128/MCB.24.13.6040-6048.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keusekotten K, et al. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153(6):1312–1326. doi: 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rivkin E, et al. The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature. 2013;498(7454):318–324. doi: 10.1038/nature12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smit JJ, et al. The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. EMBO J. 2012;31(19):3833–3844. doi: 10.1038/emboj.2012.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walczak H, Iwai K, Dikic I. Generation and physiological roles of linear ubiquitin chains. BMC Biol. 2012;10:23. doi: 10.1186/1741-7007-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haas TL, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36(5):831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 34.Sato Y, et al. Specific recognition of linear ubiquitin chains by the Npl4 zinc finger (NZF) domain of the HOIL-1L subunit of the linear ubiquitin chain assembly complex. Proc Natl Acad Sci USA. 2011;108(51):20520–20525. doi: 10.1073/pnas.1109088108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panier S, Durocher D. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst) 2009;8(4):436–443. doi: 10.1016/j.dnarep.2009.01.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.