Significance
In higher organisms, most genes consist of several disconnected regions (exons), which are combined in various ways to produce several different gene transcripts from the same gene. Such alternative exon usage is thought to contribute to the ability of organisms to generate different cell types and tissues from a single genome. However, recent evidence has also suggested that much alternative exon usage might be noise with no particular function. We reconcile these two views by comparing how exons are used in different tissues and for which exons these usage patterns across tissues change or stay similar (are “conserved”) in several primate species. The latter case is an indication that the pattern is of functional importance, and our analysis quantifies how widespread such cases are.
Keywords: alternative isoform regulation, comparative transcriptomics
Abstract
Alternative usage of exons provides genomes with plasticity to produce different transcripts from the same gene, modulating the function, localization, and life cycle of gene products. It affects most human genes. For a limited number of cases, alternative functions and tissue-specific roles are known. However, recent high-throughput sequencing studies have suggested that much alternative isoform usage across tissues is nonconserved, raising the question of the extent of its functional importance. We address this question in a genome-wide manner by analyzing the transcriptomes of five tissues for six primate species, focusing on exons that are 1:1 orthologous in all six species. Our results support a model in which differential usage of exons has two major modes: First, most of the exons show only weak differences, which are dominated by interspecies variability and may reflect neutral drift and noisy splicing. These cases dominate the genome-wide view and explain why conservation appears to be so limited. Second, however, a sizeable minority of exons show strong differences between tissues, which are mostly conserved. We identified a core set of 3,800 exons from 1,643 genes that show conservation of strongly tissue-dependent usage patterns from human at least to macaque. This set is enriched for exons encoding protein-disordered regions and untranslated regions. Our findings support the theory that isoform regulation is an important target of evolution in primates, and our method provides a powerful tool for discovering potentially functional tissue-dependent isoforms.
Alternative exon usage has been observed to affect most multiexon genes in mammals (1–3). As a result, distinct proteins can be produced that differ in their inclusion of regulatory or functional features, including natively unstructured regions (4) and linear motifs (5, 6). These differences have the potential to affect the specificity, efficiency, localization, or life cycle of proteins. Moreover, alternative usage of exons, including those containing untranslated regions (UTRs), can affect the stability, localization, or translation of RNAs. In the literature, a considerable number of examples exist of functional diversity created by the alternative usage of exons (reviewed in ref. 7). For instance, mouse embryonic stem cells express the FOXP1-ES splicing variant of FOXP1; this variant promotes the expression of transcription factors that are necessary to maintain pluripotency (8).
Alternative usage of exons is correlated with organismal complexity, and it is thought that by enhancing proteome diversity, it is essential for the ability of a single genome to generate phenotypically diverse tissues (9). For example, the GAS7 gene encodes multiple tissue-specific isoforms that differ in the inclusion of a region coding for a WW domain that mediates protein interactions (10). Genome-wide, tissue-specific splicing has been found enriched in exons coding for intrinsically disordered regions of proteins and short linear motifs that mediate protein interactions, suggesting a mechanism for the creation of tissue-specific interaction networks (11, 12). Tissue-dependent usage (TDU) of exons also affects noncoding parts of transcripts, such as 3′ UTRs, which often contain binding sites for micro-RNAs and RNA-binding proteins (13). For example, the brain tends to have transcripts with much longer UTRs than other tissues (14).
Despite these observations, which point to the functional importance of exon usage regulation, there is also conflicting evidence: Splicing factor recognition motifs are short and degenerate, exon usage can vary between individuals because of subtle genetic variations in cis-regulatory regions (15, 16), and it has been suggested that a stochastic model of processes in the splicing machinery explains most splicing variation (17). In fact, a comparative analysis of transcriptomes associated with multiple human individuals found a high prevalence for noisy, presumably erroneous, splicing, particularly in low-abundance isoforms (18). Moreover, prominent differences in splicing have been reported between species as close as human and chimpanzee (19, 20). By analyzing transcriptomes of physiologically equivalent organs in mammalian and other vertebrate species, it was recently shown that the splicing variation between species, even between equivalent tissues, exceeds the within-species variation across tissues (21, 22). This finding is in stark contrast to what is observed for overall gene expression levels, in which tissue-dependence patterns show strong conservation (23).
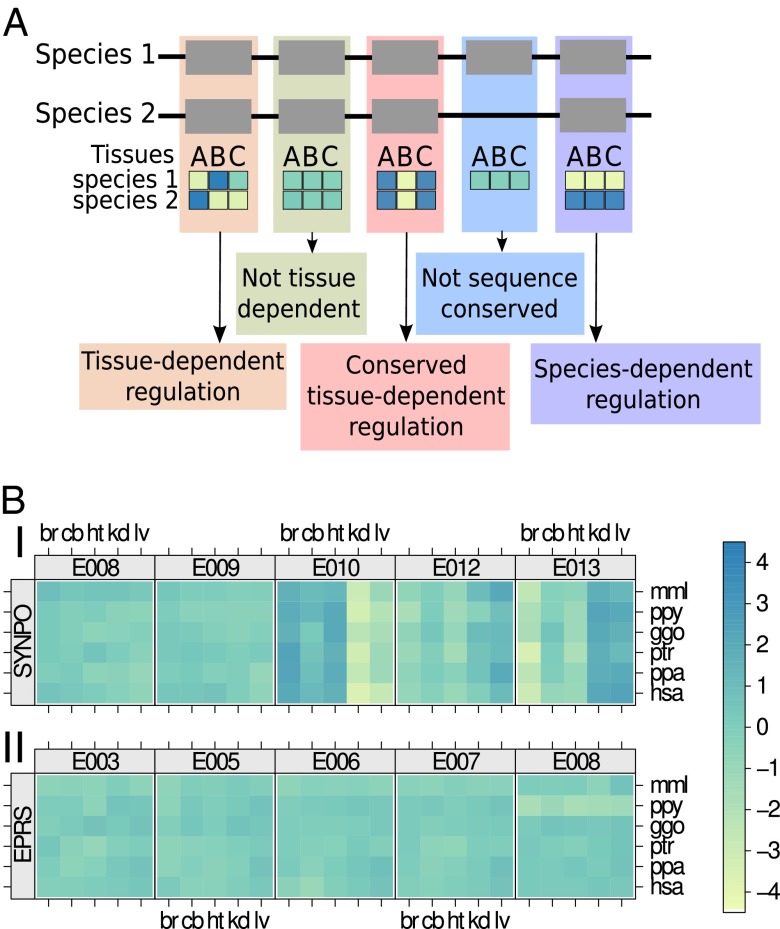
Taken together, the following dilemma arises: alternative exon usage affects almost all human genes, but evidence of the functional consequences of this phenomenon is available for only a relatively small number of genes (7). In many cases, alternative exon usage appears to be a result of “noise” detected only because of sensitive high-throughput sequencing, but is of no phenotypic consequence (24). It is unclear how much of alternative exon usage is noise and how much is biologically relevant. Here we address this question with respect to TDU of exons. Using a sensitive statistical approach to assess the evolutionary conservation of usage patterns, we analyzed transcriptome data of samples of five tissues from six primate species (23). We focus on those exons that are sequence-conserved as one-to-one orthologs across the six species. Thus, our analysis investigates potential conservation of the regulation of exon usage, conditional on the fact that an exon’s sequence features are conserved (25). We define exon usage as the fraction of transcripts originating from a gene that include a specific exon. For each exon, we compute a set of “relative exon usage coefficients” (REUCs), which indicate for each species-tissue combination the relative usage in comparison with the average over all species and tissues (Fig. 1A). Conceptually, this quantity is related to the various measures (all called “percent spliced in”) used by previous analyses (3, 21, 22); however, in contrast to this type of measure, the REUC also allows consideration of boundary exons such as those containing UTRs and includes the effect not only of alternative splicing but also of alternative use of transcription start and polyadenylation sites. In addition, by considering the usage of exons rather than splice junctions, we focus on the effect (relative abundance of RNA segments) rather than the generative mechanisms.
Fig. 1.
Types of exon usage variation across tissues and species. (A) Two hypothetical gene models for two species are depicted, including an example exon that is not sequence-conserved. Also shown are the different relative exon usage patterns considered in this study. (B) Tissue and species dependence of relative exon usage. (Upper) Relative usage patterns of five consecutive exons of gene SYNPO. For each exon, the heat map matrix shows observed changes in the exon’s usage among all transcripts produced from the gene (i.e., the REUC) for all 30 combinations of the six species and the five tissue types [br, brain; cb, cerebellum; ggo, gorilla; hsa, human; ht, heart; kd, kidney; lv, liver; mml, rhesus macaque; ppa, bonobo; ppy, orangutan; ptr, chimpanzee; colors indicate REUC values, i.e., logarithmic fold change (base 2) with respect to the average]. Exon E010 displays a vertical stripe pattern that is indicative of higher relative usage in brain, cerebellum, and heart but is less frequently seen among the genes’ transcripts in liver and kidney. A complementary behavior is observed for exons E012 and E013. (Lower) The relative usage of exon E008 of the gene EPRS showed a strong species effect, as indicated by the horizontal stripe pattern. Across all tissues, this exon is used less frequently in orangutan.
Results
The Data.
We analyzed a subset of the RNA-Seq data presented by Brawand et al. (23), consisting of samples from heart, liver, kidney, brain (whole brain without cerebellum), and cerebellum from human, chimpanzee, bonobo, gorilla, orangutan, and rhesus macaque. For each of the 30 tissue-species combinations, at least 2 individuals were analyzed (in total, 75 samples). Unique, gapped alignments of the cDNA fragments to the genomic reference sequence of the corresponding species were determined. Depending on the sample, between 4,089,237 and 30,765,598 alignments were obtained, summing to a total of 1,356,473,949 alignments (SI Appendix, Table S1).
We generated a mapping graph of one-to-one orthologous exons in the six species, making up a total of 118,695 exons in 10,200 genes (SI Appendix, SI Methods). We only included exons for which the sequences were identical along 90% of their length across all analyzed species. For each sample, we tabulated the number of fragment alignments that were overlapping with each exon. We calculated REUCs by fitting generalized linear models to the numbers of fragments mapped to the exon and the numbers of fragments mapped to the rest of the gene’s exons (SI Appendix, SI Methods). The REUCs indicate the (logarithmic) fold changes of an exon’s usage in each specific tissue-species combination compared with the average usage in all tissues and species. We visualized REUCs in matrix plots such as Fig. 1B.
Exon Usage in Tissues Shows More Interspecies Variation than Gene Expression.
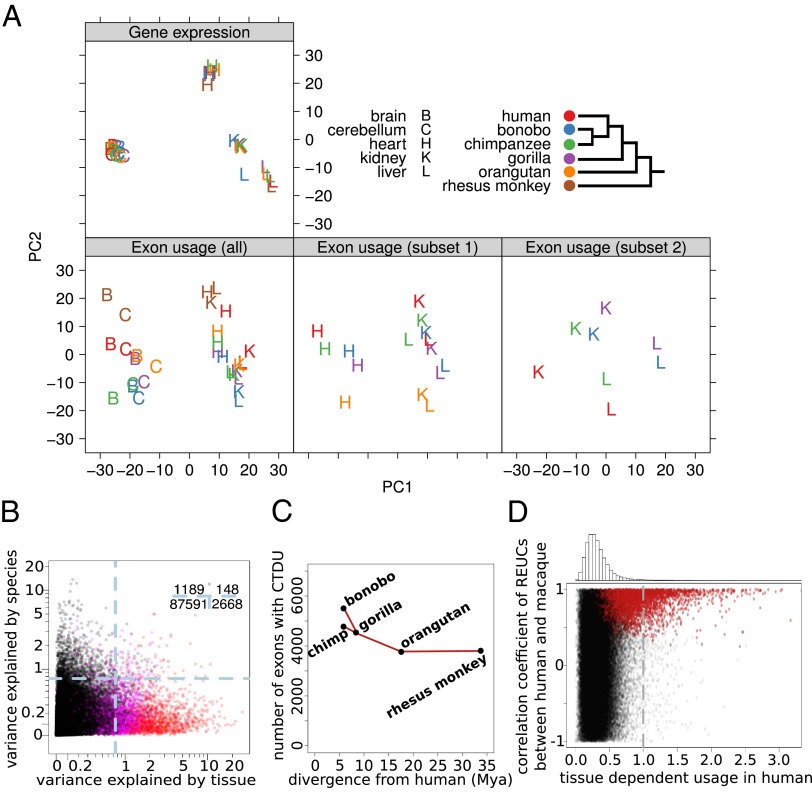
On the level of whole genes, expression patterns across tissues are largely conserved from human to chicken (23). We asked whether a similar principle holds for relative exon usage patterns. To address this question, we performed a principal component analysis (PCA) on the REUCs, as well as on the gene expression levels (Fig. 2A and SI Appendix, SI Methods). Consistent with ref. 21, the PCA for gene expression showed a tight grouping of the gene expression profiles by tissue, whereas species was nearly irrelevant for positioning in the PCA plot. This picture confirms the overall strong conservation of tissue-dependent gene expression. In contrast, in the PCA for the REUCs (i.e., for the individual exons), the conservation signal was much weaker and the distribution more nuanced. We only observed a partial grouping by tissue: brain and cerebellum formed one group along the first principal component, and heart, liver, and kidney formed a second group. However, the second principal component was driven by species; the rhesus macaque, being the most distantly related species, was separated from the rest of the primates.
Fig. 2.
Tissue and species effects on exon usage. (A) Principal component analysis shows that gene expression (Upper) groups tightly by tissue, irrespective of species, whereas exon usage (Lower) shows prominent species-to-species variability. The interplay between species and tissue effects is explored further in the image (Middle, Right), showing PCA analyses of selected subsets. (B) Variance in the REUCs explained by tissue and by species, respectively. The numbers in the top right corner indicate how many exons, represented by dots, are in the four quadrants delineated by the dashed lines. Exons with CTDU between human and macaque are shown in magenta; exons with CTDU between all species pairs (“strictly CTDU exons”) are shown in red. (C) Number of exons whose TDU pattern shows significant conservation between humans and the other primate species, plotted against the time since phylogenetic separation of the species from human, in million years (26). (D) Pearson correlation coefficient of REUCs across tissues between human and macaque versus TDU strength in human. Exons with conserved TDU between human and macaque are plotted in red. The plot shows that if an exon’s usage pattern shows strong differences across tissues in one species (here: human), this pattern tends to be the same in other species (here: macaque), indicating conservation of regulation and suggesting functional importance. However, as the histogram of TDU strengths on shows (Upper), these exons represent only a small fraction of all exons.
We further explored the second group of tissues by PCA on the subset of the data containing only heart, liver, and kidney of the great apes (i.e., without macaque). Again, the distances between species were on the same order as those between tissues, and the most phylogenetically distant species (orangutan) was separated from the others. Even when restricting the PCA to kidney and liver of the four Homininae (i.e., excluding orangutan), pronounced species-related differences were seen.
Together, these results indicate that exon usage has drastically more interspecies variability than gene expression. Although exon usage variability decreases with evolutionary distance, the variation among human, bonobo, and chimpanzee is still of comparable extent to the variation between different tissues. These results contrast with what is observed for gene expression, in which for the majority of genes, expression variation is dominantly associated with tissue, consistent with stabilizing selection that maintains tissue-dependent expression patterns of functional importance. The fact that such widespread conservation is not seen for exon usage raises the question of to what extent differential exon usage across tissues is functional.
Species-Associated Changes of Exon Usage Tend to Be Small and Numerous; Tissue-Associated Changes Tend to Be Large and Less Frequent.
A more detailed picture emerged when we asked for each exon separately whether its usage changes more strongly between tissues or between species. To quantify this, we fitted an analysis-of-variance model to each exon’s REUC values, with tissue and species as explanatory variables (SI Appendix, SI Methods). Again, species dominated over tissues: For 60% of the exons (54,879/91,596), exon usage showed stronger dependence on species than on tissues. For most exons, however, usage variations across tissues or species were small overall. The balance changed once we focused on exons with large effects, taking the 4,005 exons (4.3%) for which the REUC variance explained by either of the two factors exceeded the value 0.75; 69% of these exons varied more strongly between tissues than between species (Fig. 2B).
These results indicate that although interspecies differences dominate the majority of exons with generally small variability in their usage, the minority of exons with high variance are mostly driven by tissue-related differences (SI Appendix, Figs. S2 and S3).
Identification of Conserved Tissue-Dependent Exon Usage.
We aimed to systematically identify exons that showed conserved TDU (CTDU), as exemplified in the matrix plots in Fig. 1 A and B. For each exon and each pair of species, we calculated the covariance between their REUC profiles across the five tissues. (A large covariance value indicates that the rows corresponding to the two species in the exon’s matrix plot show patterns that are strong and similar to each other.) We devised a test to identify exons with statistically significant CTDU. Specifically, we considered the TDU pattern of an exon conserved between two species if its REUCs had a covariance exceeding 0.1. This threshold was chosen such that the estimated false-discovery rate was below 10% (SI Appendix, SI Methods). The number of exons with significant CTDU was higher for closely related species and decreased with phylogenetic distance: we detected significant conservation for 4,760 exons in 2,023 genes between human and chimpanzee and for 3,800 exons in 1,643 genes between human and macaque (Fig. 2C). The curve in Fig. 2C levels off with increasing distance, suggesting the existence of a core set of several thousand exons whose tissue-dependent regulation is conserved beyond the primate clade.
Strong Patterns of Tissue-Dependent Exon Usage Are Frequently Conserved.
Next, we quantified the strength of tissue dependence of exon usage (TDU); we define the TDU strength for an exon in a species as the maximum of absolute differences between the REUCs of the five tissues and their average. Strikingly, we found that high TDU strength in one species is predictive of CTDU across species. Specifically, exons with high TDU strength in human had a strong tendency to show the same pattern of TDU in macaque, as measured by the correlation coefficient between the respective REUCs (Fig. 2D). Thus, of the 1,379 exons (1.5% of the 91,596 exons for which we have REUC values) in which the TDU strength in humans exceeded a value of 1 (indicating that in at least one tissue, the REUC differed by more than twofold from that in the other tissues), 80% showed statistically significant conservation with macaque, and 70% had a correlation coefficient of REUCs between human and macaque larger than 0.7 (Fig. 2D).
This result indicates that whenever an exon’s usage differs strongly between tissues, these differences are frequently conserved, which is consistent with the potential functional importance of such exon usage patterns.
CTDU Occurs Frequently in Disordered Regions of Proteins and in Transcript UTRs.
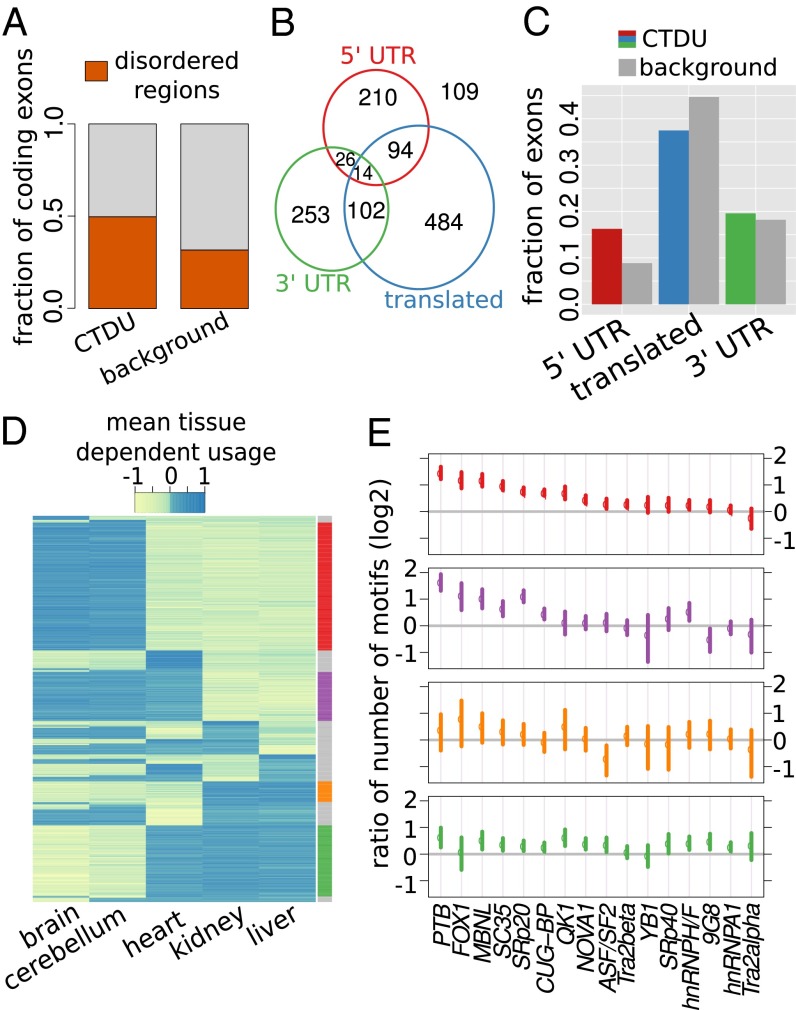
To gain insight into potential functions of CTDU exons, we focused on the 1,292 exons whose tissue-dependent regulation showed significant conservation in all 15 species pairs (referred to in the following as “strictly CTDU exons”). As a reference for subsequent enrichment analyses, we identified two background sets of exons whose distributions of expression strength, exon length, and variance across replicates were matched to our set of strictly CTDU exons. Background set 1 included coding exons only, whereas background set 2 had no such restriction (SI Appendix, SI Methods).
It has been reported that exons with TDU in human (irrespective of conservation) were statistically enriched in exons coding for protein-disordered regions that mediate protein interactions (27), thus enabling interaction networks to rearrange in tissue-specific ways (11, 12). In analogy to that finding, we found an enrichment of exons coding for protein-disordered regions (as predicted by IUPred; ref. 28) among strictly CTDU exons compared with background set 1 (P = 1.3 × 10−12, Fisher’s exact test; Fig. 3A; SI Appendix, SI Methods). Thus, by adding the angle of comparisons across species, our results further support the existence of widespread, conserved, functional roles of alternative usage of protein-disordered regions regulated through alternative exon usage.
Fig. 3.
Features and tissue patterns of strictly CTDU exons. (A) Bar chart showing the fraction of exons of the strictly CTDU and background set of exons that overlap with regions predicted to code for natively disordered protein regions. (B) Venn diagram depicting a nonexclusive categorization of the strictly CTDU exons into translated, 3′-UTRs, and 5′-UTRs. (C) Bar chart indicating how the proportions of these categories compare between the strictly CTDU exons and background 2. (D) Heat map representation of the per tissue REUCs, obtained by averaging across species. Rows are ordered according to usage patterns. (E) Splicing factor motif enrichment in four exon clusters; the colors correspond to the clusters indicated in the right margin of D. The points indicate the mean value of the ratio to the background set of exons, the bars correspond to 95% confidence intervals estimated by bootstrapping.
Next, we classified the strictly CTDU exons into translated exons, 5′- and 3′-untranslated exons, based on human transcript annotations (Fig. 3B). The distribution of CTDU exons in these categories was significantly different from background 2 (P = 3 × 10−14, Fisher’s exact test; Fig. 3C). In particular, 5′ untranslated exons were subject to CTDU more than twice as often as expected from background 2. UTRs often contain regulatory elements, such as protein or miRNA binding sites, that are able to regulate translation and transcript localization (29). These results suggest that differential usage of UTRs has widespread effects, presumably in the posttranscriptional regulation of transcripts in a tissue-dependent manner.
TDU Patterns Are Associated with Splicing Factor Binding Motifs and Suggest a Conserved cis-Regulatory Code.
How is tissue-dependent exon usage regulated? To start addressing this question, we compared the degree of sequence conservation in introns flanking strictly CTDU exons with that in introns next to exons from background 2. We observed a small but highly significant elevation in sequence conservation, as measured by the amount of single-nucleotide variation across species in these introns (P = 1.9 × 10−15 and P = 5.7 × 10−9, Wilcoxon rank sum test; SI Appendix, Fig. S6). This result is consistent with the notion that the need to maintain splicing-related cis-regulatory elements involved in tissue-dependent exon usage results in purifying selection of sequences within these introns. Next, we classified our set of strictly CTDU exons into major usage patterns. For each exon, we summarized its tissue preference by the mean of REUCs across species. Thus, a positive value (e.g., in heart) means the exon’s usage is consistently higher in heart than in the average of all tissues. We considered all 25 − 2 = 30 possible patterns of positive and negative signs across the five tissues. Remarkably, the CTDU exons showed strong preferences for only a small subset of the possible patterns: The four largest classes alone contained 70% (899/1,292) of the exons (Fig. 3D). This categorization also revealed three groups of tissues with largely similar exon abundance profiles within each group; namely, brain and cerebellum, liver and kidney, and heart. The same tissue grouping was seen in a PCA of REUC profiles using only the strictly CTDU exons (SI Appendix, Fig. S4).
Next, we used SFmap (30) to count the number of predicted binding motifs for 18 splicing factors within 200 base pairs up- and downstream of each strictly CTDU exon. Overall, we found higher numbers of motifs compared with the background sets 2 (P < 2.2 × 10−16, Wilcoxon sum rank test). We then used the data to associate particular splicing factors with each of the TDU patterns (Fig. 3E). The four major exon classes were associated with different distributions of splicing factor binding motifs. Consistent with previous results (31), NOVA1 was particularly enriched in the exon classes that showed differential usage of exons in brain and cerebellum compared with rest of the tissues. In some cases, the distributions of splicing factor binding motifs were similar across the exon classes. For instance, SC35, SRp20, MBNL, PTB, and CUG-BP were particularly enriched around those exons that had higher relative usage in brain and cerebellum compared with kidney and liver. Taken together, these results corroborate the existence of a conserved cis-acting code, which contributes to modulating exon usage in tissue-dependent patterns, presumably through the differential (tissue-dependent) activity of transacting factors.
A Resource for Discovering Tissue-Dependently Regulated Exons and Hypothesis Formation.
We created a browsable resource to aid the exploration of our genome-wide analysis. A Web site, provided at www-huber.embl.de/pub/DEUprimates, shows for each gene, matrix plots as in Fig. 1B, alongside the gene and transcript models and annotation of protein domains provided by the Smart (32, 33) and Pfam (34) databases (displayed with the Dalliance browser; ref. 35). The matrix plots for all strictly CTDU exons are also reproduced in the Dataset S1.
Exploration of our set of CTDU exons in this resource corroborated several instances of previously described tissue-specific isoform regulation. For example, we observed that heart tissue mainly expresses the shorter isoforms of the gene ANK1 that differ in their 5′ start site compared with the other tissues (www-huber.embl.de/pub/DEUprimates/allPages/ENSG00000029534.html). Accordingly, we found reports in the literature of shorter, muscle-specific isoforms of this gene (36). In a similar manner, we observed a conserved cerebellum-specific pattern of exon usage of the gene GAS7 consistent with a previous report (10) (www-huber.embl.de/pub/DEUprimates/allPages/ENSG00000007237.html).
In addition to supporting known cases, the data revealed numerous further instances of tissue-dependent isoform regulation. For example, we observed an interesting pattern in the gene COBL (cordon bleu), a gene first characterized to be involved in neural tube formation (37) and then found to catalyze actin nucleation (38). Our data show that the relative usage of the exons at the gene’s 3′ end is strongly and conservedly reduced in heart compared with the other tissues (www-huber.embl.de/pub/DEUprimates/allPages/ENSG00000106078.html). This is intriguing, as this end contains the gene’s three WH2 domains, which allow the protein to bind to actin monomers. Comparison with the Illumina Bodymap 2.0 (a resource of RNA-Seq data across many human tissues, available on Ensembl) confirmed these usage differences and showed that skeletal muscle behaves similar to heart. These observations suggest the COBL gene, whose function has so far been chiefly studied for neuronal tissues (reviewed in ref. 39), might fulfill a role in heart and skeletal muscle tissue that is different from its well-established function in initiating actin polymerization.
Methods
We realigned the data by Brawand et al. (23), tallied read counts for all orthologous exons, calculated REUCs, and performed downstream analyses, as outlined above. Details on the bioinformatics analysis and on the mathematical methods are given in the SI Appendix, SI Methods. A complete, extensively commented transcript of the R session used to analyze the data is provided as Dataset S1.
Discussion
TDU of Exons Is Widespread, but Mostly of Low Amplitude and Not Conserved; a Minority of Exons Shows Strong Tissue Patterns, Which Are Largely Conserved.
Our findings provide an answer to the controversy about conservation of exon usage regulation. They reconcile the two opposing views outlined in the Introduction: the prevalence of apparently nonconserved or noisy splicing seen by recent high-throughput sequencing studies and the existence of well-studied examples, at the single-gene level, of tissue-dependent isoforms with different functions. We find that both views are justified, albeit with important qualifications. Variations in exon usage are prevalent, but they are, with surprisingly few exceptions, of low amplitude, and the lack of conservation of tissue-dependent variation is consistent with the view that most of them have negligible consequences for the phenotypic variation across tissues. The cases of conserved TDU form a minority, yet they are associated with large amplitudes of the between-tissue differences and they are still numerous, totalling several thousand exons in our analysis. These cases are plausible candidates for functional tissue-dependent regulation of exon usage.
Our findings are consistent with previous reports, such as those by Merkin et al. and Barbosa-Morais et al. (21, 22) and recapitulated in Fig. 2A, that exon usage profiles have more interspecies variability than gene expression. It appears that organisms can buffer the minor variations in abundance that are observed for many exons. Such robustness is also consistent with the findings of Pickrell et al. (18), who saw a large variety of isoforms of low abundance in human (HapMap) samples, many of which appear to be physiologically inconsequential. We suggest that small variations in exon usage that are driven by local sequence variations are an important raw material for evolution’s “tinkering.” Once a TDU event turns out to be beneficial, it can be accentuated by evolutionary feedback and, eventually, may become fixed and conserved.
Strength of TDU Can Be Used as a Predictor of Conservation.
Evolutionary conservation is often used as an indicator of functional importance. For this purpose, one would like to have a data set including samples from many different tissues, each taken from several individuals of a suitable range of species. However, it is unlikely that such a resource will soon be available that covers as many tissues as established resources focusing on human only, such as Illumina’s BodyMap 2.0 data set. We demonstrated that strong TDU is highly predictive of conservation (Fig. 2D). Hence, even in the absence of multispecies data, strong TDU seen in one species could already provide sufficient motivation for in-depth study (e.g., by biochemical or genetic means).
The Higher Primates Offer a Good Vantage Point to Analyze Conservation of Tissue-Dependent Exon Usage.
For this study, we chose to focus our analysis on exons with high sequence conservation; we set aside exons affected by larger structural rearrangements or major sequence divergence. The motivation for this choice was the aim of studying exon usage regulation at the level of small cis-regulatory elements and transacting factors. The choice imposes a trade-off between the width of the clade considered and the size of the set of exons available for study. By considering species that diverged within ∼35 million years, we were able to study 118,695 exons in 10,200 genes, therefore reaching genome-wide coverage. Three thousand eight hundred exons in 1,643 genes showed significant CTDU between human and macaque, and even with the more strict criterion of significant evidence for conservation in all 15 species pairs, we detected 1,292 exons. These numbers provide a conservative lower limit, and they can be expected to increase when more tissues or more replicates per tissue-species combination are considered.
Two recent studies focused on the evolution of splicing profiles of several tissues spanning more than 300 million years of evolution (21, 22). Because of the trade-off between the size of the clade and the number of orthologous exons, these studies considered much smaller sets of exons, and their results mainly report on the potential conservation of the qualitative attribute of whether an exon is constitutive or alternative, whereas we were able to use a more sensitive, quantitative measure of relative exon usage.
Is the phylogenetic distance between the species that we considered large enough for neutral drift to occur, and therefore to allow us to detect conservation? This question is answered by the results of Fig. 2 A and B: For the majority of exons, interspecies differences dominate, and those exons with evidence of conservation are highly distinct from what is expected within the null distribution.
Differential Exon Usage Provides Versatile Mechanisms to Achieve Transcriptional Diversity Across Tissues.
Differential exon usage provides a layer of regulation that appears to be essential for the morphological complexity of animals. By integrating information from the genome and the epigenome, this layer of regulation has the flexibility to “rewire” biological networks in the course of the evolution of species in a tissue-specific manner. We have strengthened and extended recent findings (11, 12) that demonstrate the role of alternative exon usage in rearranging protein interaction networks in tissue-specific ways; in particular, we have demonstrated evolutionary conservation of such mechanisms by showing that CTDU exons are enriched for protein-disordered regions, which are frequently involved in mediating protein interactions.
A specific finding of our analysis is the prevalence of UTRs in CTDU exons: 38% of our set of strictly CTDU exons are UTRs of transcripts, and it is plausible to hypothesize that many of them are involved in posttranscriptional regulation. Other recent findings also support the view that UTRs may play a previously underappreciated role in establishing specific functions of tissues and organs: Throughout the evolution of animals, UTR length has increased along with morphological complexity (40). The brain, arguably the most complex tissue in humans, has much longer UTRs than other tissues (14). The use of emerging technologies that allow precise mapping of transcript start and end sites (41) is expected to provide more insight into this largely unexplored terrain.
Supplementary Material
Acknowledgments
We thank John Marioni and Michael Love for helpful discussions and suggestions on the analysis. We also thank Ignacio Schor and Giorgia Guglielmi for critical reading of the manuscript. Funding was provided by the European Commission through the Seventh Framework Programme Health project Radiant (to S.A. and W.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.B.E. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307202110/-/DCSupplemental.
References
- 1.Xu Q, Modrek B, Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30(17):3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5(10):R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hegyi H, Kalmar L, Horvath T, Tompa P. Verification of alternative splicing variants based on domain integrity, truncation length and intrinsic protein disorder. Nucleic Acids Res. 2011;39(4):1208–1219. doi: 10.1093/nar/gkq843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weatheritt RJ, Gibson TJ. Linear motifs: lost in (pre)translation. Trends Biochem Sci. 2012;37(8):333–341. doi: 10.1016/j.tibs.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Weatheritt RJ, Davey NE, Gibson TJ. Linear motifs confer functional diversity onto splice variants. Nucleic Acids Res. 2012;40(15):7123–7131. doi: 10.1093/nar/gks442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelemen O, et al. Function of alternative splicing. Gene. 2013;514(1):1–30. doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabut M, et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147(1):132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 9.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463(7280):457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.You JJ, Lin-Chao S. Gas7 functions with N-WASP to regulate the neurite outgrowth of hippocampal neurons. J Biol Chem. 2010;285(15):11652–11666. doi: 10.1074/jbc.M109.051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buljan M, et al. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol Cell. 2012;46(6):871–883. doi: 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis JD, et al. Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol Cell. 2012;46(6):884–892. doi: 10.1016/j.molcel.2012.05.037. [DOI] [PubMed] [Google Scholar]
- 13.Smibert P, et al. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep. 2012;1(3):277–289. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC (2013). Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome Res 23(5):812–825. [DOI] [PMC free article] [PubMed]
- 15.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464(7289):768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lalonde E, et al. RNA sequencing reveals the role of splicing polymorphisms in regulating human gene expression. Genome Res. 2011;21(4):545–554. doi: 10.1101/gr.111211.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melamud E, Moult J. Stochastic noise in splicing machinery. Nucleic Acids Res. 2009;37(14):4873–4886. doi: 10.1093/nar/gkp471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickrell JK, Pai AA, Gilad Y, Pritchard JK. Noisy splicing drives mRNA isoform diversity in human cells. PLoS Genet. 2010;6(12):e1001236. doi: 10.1371/journal.pgen.1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calarco JA, et al. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev. 2007;21(22):2963–2975. doi: 10.1101/gad.1606907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010;20(2):180–189. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338(6114):1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbosa-Morais NL, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338(6114):1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 23.Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478(7369):343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 24.Kornblihtt AR, et al. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14(3):153–165. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- 25.Irimia M, Rukov JL, Roy SW, Vinther J, Garcia-Fernandez J. Quantitative regulation of alternative splicing in evolution and development. Bioessays. 2009;31(1):40–50. doi: 10.1002/bies.080092. [DOI] [PubMed] [Google Scholar]
- 26.Israfil H, Zehr SM, Mootnick AR, Ruvolo M, Steiper ME. Unresolved molecular phylogenies of gibbons and siamangs (Family: Hylobatidae) based on mitochondrial, Y-linked, and X-linked loci indicate a rapid Miocene radiation or sudden vicariance event. Mol Phylogenet Evol. 2011;58(3):447–455. doi: 10.1016/j.ympev.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero PR, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci USA. 2006;103(22):8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21(16):3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 29.Pichon X, et al. RNA binding protein/RNA element interactions and the control of translation. Curr Protein Pept Sci. 2012;13(4):294–304. doi: 10.2174/138920312801619475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paz I, Akerman M, Dror I, Kosti I, Mandel-Gutfreund Y. SFmap: a web server for motif analysis and prediction of splicing factor binding sites. Nucleic Acids Res. 2010;38(Web Server issue) suppl 2:W281-5. doi: 10.1093/nar/gkq444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302(5648):1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 32.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95(11):5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40(Database issue) D1:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue) D1:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Down TA, Piipari M, Hubbard TJP. Dalliance: interactive genome viewing on the web. Bioinformatics. 2011;27(6):889–890. doi: 10.1093/bioinformatics/btr020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallagher PG, Forget BG. An alternate promoter directs expression of a truncated, muscle-specific isoform of the human ankyrin 1 gene. J Biol Chem. 1998;273(3):1339–1348. doi: 10.1074/jbc.273.3.1339. [DOI] [PubMed] [Google Scholar]
- 37.Carroll EA, et al. Cordon-bleu is a conserved gene involved in neural tube formation. Dev Biol. 2003;262(1):16–31. doi: 10.1016/s0012-1606(03)00323-3. [DOI] [PubMed] [Google Scholar]
- 38.Ahuja R, et al. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131(2):337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kessels MM, Schwintzer L, Schlobinski D, Qualmann B. Controlling actin cytoskeletal organization and dynamics during neuronal morphogenesis. Eur J Cell Biol. 2011;90(11):926–933. doi: 10.1016/j.ejcb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 40.Chen CY, Chen ST, Juan HF, Huang HC. Lengthening of 3’UTR increases with morphological complexity in animal evolution. Bioinformatics. 2012;28(24):3178–3181. doi: 10.1093/bioinformatics/bts623. [DOI] [PubMed] [Google Scholar]
- 41.Pelechano V, Wei W, Steinmetz LM. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497(7447):127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.