Abstract
Antifolates, folate analogs that inhibit vitamin B9 (folic acid)-using cellular enzymes, have been used over several decades for the treatment of cancer and inflammatory diseases. Cellular uptake of the antifolates in clinical use occurs primarily via widely expressed facilitative membrane transporters. More recently, human folate receptors (FRs), high affinity receptors that transport folate via endocytosis, have been proposed as targets for the specific delivery of new classes of antifolates or folate conjugates to tumors or sites of inflammation. The development of specific, FR-targeted antifolates would be accelerated if additional biophysical data, particularly structural models of the receptors, were available. Here we describe six distinct crystallographic models that provide insight into biological trafficking of FRs and distinct binding modes of folate and antifolates to these receptors. From comparison of the structures, we delineate discrete structural conformations representative of key stages in the endocytic trafficking of FRs and propose models for pH-dependent conformational changes. Additionally, we describe the molecular details of human FR in complex with three clinically prevalent antifolates, pemetrexed (also Alimta), aminopterin, and methotrexate. On the whole, our data form the basis for rapid design and implementation of unique, FR-targeted, folate-based drugs for the treatment of cancer and inflammatory diseases.
Keywords: isothermal titration calorimetry, targeted drug delivery
Folic acid, an essential vitamin developed in the 1940s for the treatment of anemia, is converted to functional, naturally occurring metabolites via the action of dihydrofolate reductase and hence can act as a dietary supplement for folates (1). Folates are necessary in eukaryotic cells for single carbon transfer reactions, notably the conversion of homocysteine to methionine and a multitude of steps in de novo nucleotide synthesis (2–5). Folic acid and its reduced derivatives, including 5-methyltetrahydrofolate and 10-formyltetrahydrofolate, are transported via two widely expressed facilitative transporters, the reduced folate carrier (RFC) and the proton-coupled folate transporter (PCFT), and via a family of glycosyl-phosphatidylinositol (GPI)-anchored receptors with limited expression profiles generally described as folate receptors (FRs) (3–7). For >60 y, folate analogs that inhibit intracellular folate-using enzymes, termed antifolates, have been developed to treat a variety of cancer types and inflammatory diseases (8–14). More recently, the emphasis in folate-mediated drug therapy has shifted to obtain a better understanding of transport mechanisms of antifolates because dose-limiting toxicities arise from their transport via RFC, and possibly PCFT, into normal cells (6, 11, 15–17). To date, antifolates approved for clinical use are transported primarily via RFC, although one of the most recently developed antifolates, pemetrexed (PMX), may also transit through PCFT (6, 11, 18–21). Given the toxicities associated with transport via facilitative transporters, the development of therapeutic molecules specifically transported via the FRs has been explored over the last two decades (22–24).
In humans, three genes encoding functional folate receptors termed hFRα, hFRβ, and hFRγ (also FOLR1, FOLR2, and FOLR3, respectively) have been identified (Fig. 1) (2, 3, 25–29). hFRα and hFRβ are anchored at the plasma membrane via a GPI anchor, whereas FRγ is secreted due to the lack of a signal sequence for GPI anchor attachment (30–32). FRα is generally displayed on the apical surface of polarized epithelial cells, particularly in the proximal tubule cells of the kidney and the choroid plexus (2, 33–35). hFRβ is expressed in latter stages of normal myelopoiesis and in the placenta, spleen, and thymus (36, 37). In the normal development of the myelomonocytic lineage, hFRβ can be seen as a differentiation marker coexpressed with cluster of differentiation (CD) 14 at relatively low levels in monocytes but not in CD34+ normal hematopoietic progenitors (38, 39). hFRγ is secreted at low levels from lymphoid cells in the spleen, thymus, and bone marrow. Whereas hFRα has been implicated in folate transcytosis in the kidney and delivery into the central nervous system, the biological functions of hFRβ and hFRγ when expressed normally are unclear (29, 37–43). hFRβ, however, has the capacity to deliver folate and folate-derived molecules into activated macrophages or certain leukemic cells (37, 38, 43). Given the restricted expression of hFRα and hFRβ, the transcytosis mechanism normally used by hFRα, and the generally low expression levels of hFRβ, FR-targeted therapeutics are predicted to show limited toxicity in normal tissues (28).
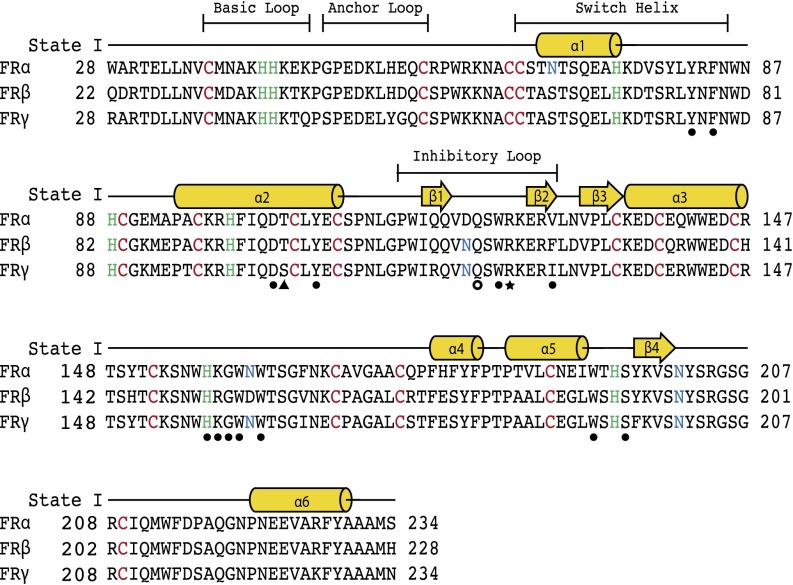
Fig. 1.
Sequence alignment of human folate receptors α, β, and γ. Conserved cysteine and histidine residues are shown in red and green, respectively, and sites of N-glycan attachment are shown in blue. Secondary structure elements of the model representing the folate receptor in state I are presented above the sequences as cylinders for α-helices and arrows for β-strands. Regions that change conformations during proposed trafficking states are labeled with boundaries indicated above the secondary structures. Filled circles denote residues that interact directly with folate and all antifolates examined in this study. Residues that interact with a subset of ligands are shown as follows: PMX-specific, triangle; AMT- and MTX-specific, open circle; and FOL- and MTX-specific, star.
Furthermore, FR-targeted therapies are expected to be effective in the treatment of many types of cancer and inflammatory disease owing to high expression levels of hFRα or hFRβ in disease-causing cells (24, 33, 38). Specifically, hFRα is consistently overexpressed in nonmucinous adenocarcinomas of the ovary, uterus, breast, cervix, kidney, and colon, as well as testicular choriocarcinoma, ependymal brain tumors, malignant pleural mesothelioma, and nonfunctioning pituitary adenocarcinoma (35, 44). hFRβ expression is increased in certain leukemias, most consistently seen in chronic myelogenous leukemia and acute myelogenous leukemia (43, 45). hFRβ expression is also increased in activated synovial macrophages, cells involved in the pathogenesis of rheumatoid arthritis and other inflammatory conditions including psoriasis and Crohn’s disease (37, 39, 46, 47).
As the FRs serve as markers for diseased cells in cancers and inflammatory disease, the development of three distinct types of FR-targeted therapeutics based on antibodies, folate-conjugates, and antifolates are being pursued (22–24). Monoclonal antibodies against hFRα and hFRβ could promote clearance of FR-positive cells by the immune system, folate-conjugates aim to deliver cytotoxic cargo or imaging agents to FR-positive cells, and FR-targeted antifolates would potentially eliminate cytotoxic side effects of current antifolates delivered to normal cells via RFC (12, 16, 45, 48–52). Because these therapies have enormous potential for the treatment of cancer and inflammatory disease, we performed a detailed molecular study of hFRs to understand biological trafficking and principles of ligand binding and release of the folate receptors. This work stands to enhance and accelerate efforts in ongoing FR-targeted drug development.
We present herein a series of six unique hFR structures that outline three distinct states relevant to the endocytic trafficking of the receptor and also provide four structures in complex with folate or antifolate ligands (2, 42). Five of the six unique structures were determined for hFRβ at pH 7.4–8.2, whereas the sixth structure resulted from hFRα crystallized at pH 5.5 (Tables S1 and S2). From comparison of three of these structures, apo-hFRβ, hFRβ in complex with folate, and apo-hFRα at acidic pH, we delineate discrete structural conformations representative of key stages in the endocytic trafficking of FRs and propose models for pH-dependent conformational changes. Our comparison of hFRα and hFRβ structures, although not straightforward, is validated by the high sequence identity (82%) and similarity (92%), of these two receptors (Fig. 1). For example, only one residue that makes a polar contact with ligands in the binding pocket differs between hFRα and hFRβ (K158α/R152β). Our complex structures of hFRβ illustrate the key determinants in ligand binding while also demonstrating the propensity of hFRs to bind similar ligands in distinct, unpredictable modes. Additionally, we examined the pH dependence of hFRs binding to folate and antifolates to test a paradigm in receptor trafficking that proposes release of ligands from the receptor is based on subtle acidification in the endosome (2, 6, 53). On the whole, our data offer a comprehensive understanding of the molecular behavior of folate receptors and provide a basis for unique strategies in FR-mediated drug discovery.
Results
Structures of the hFR in Distinct Biological Trafficking States.
After processing in the secretory pathway, mature hFRs consist of ∼205 residues with posttranslational modifications including the attachment of N-linked glycans and the formation of eight disulfide bonds between 16 conserved cysteines (Fig. 1; Fig. S1) (25, 26, 54). Two human folate receptors, hFRα and hFRβ, have been examined extensively as potential vectors for the delivery of folate-based drugs (6, 55). These receptors are displayed on the cell surface via a GPI anchor and are capable of transporting folate via a cell division cycle 42 protein-dependent endocytic mechanism (30, 53, 56). To understand the molecular details of folate uptake, we purified proteins via heterologous expression in eukaryotic host cells and subsequently used these properly folded glycoproteins to solve structures of hFRs in three states relevant to endocytic transport (Fig. 2).
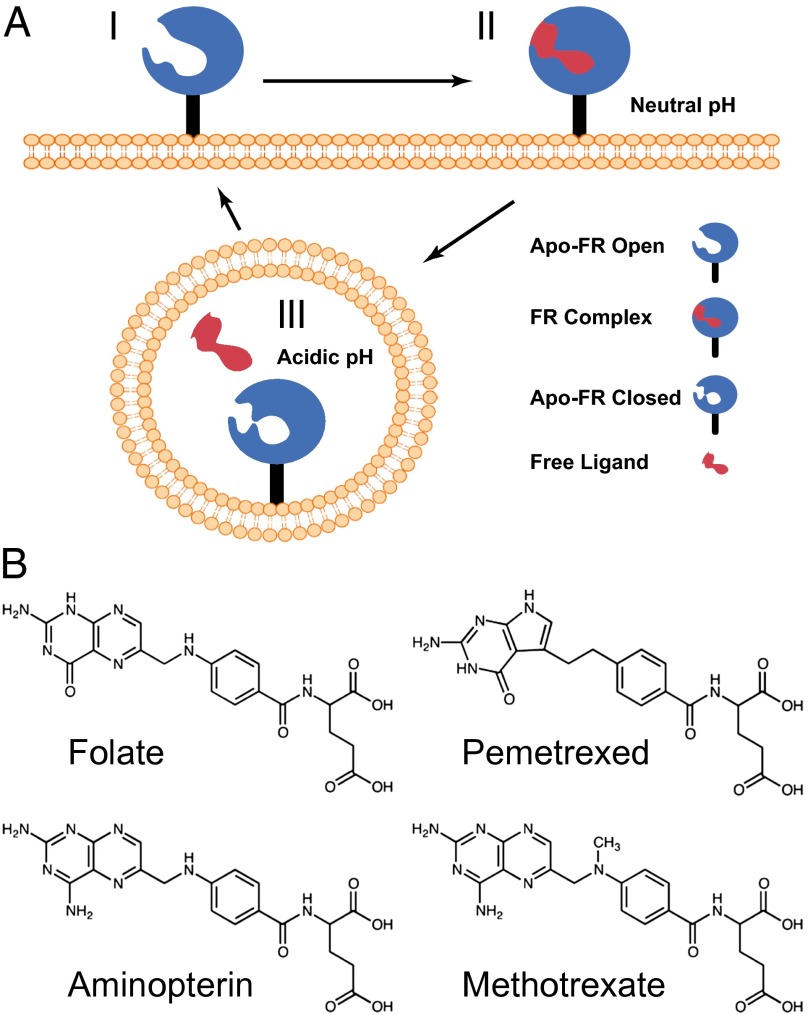
Fig. 2.
Model for trafficking of human folate receptors. (A) Schematic of ligand transport via endocytosis of hFR is depicted with three biological trafficking states. At the cell surface, the receptor at neutral to slightly basic pH is in an apo-FR conformation competent to bind ligand (state I). On ligand binding, structural transitions occur and lead to complex formation (state II). After endocytosis, ligand release occurs in the mildly acidic microenvironment of the recycling endosome. After ligand release and under acidic conditions, the receptor likely adopts a third distinct conformation (state III) before recycling to the cell surface. (B) Folic acid and the clinically prevalent anitfolate drugs examined in this study are shown.
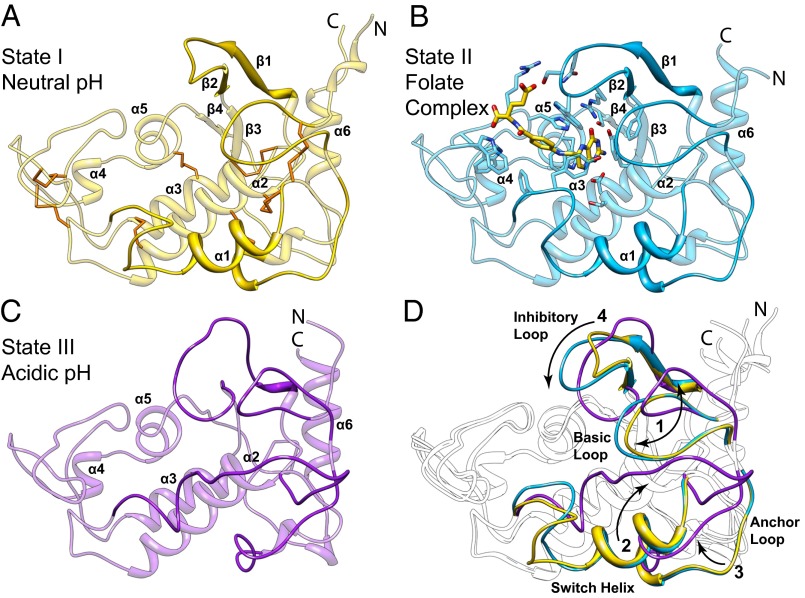
The first structure, hereafter referred to as state I, was determined to a resolution of 1.8 Å (Rfree = 19.3%) for apo-hFRβ crystallized at slightly basic conditions (pH 7.4) and represents the conformation of the receptor at the cell surface before association with folate (Figs. 3A and 4A). After examination of available structures, hFR was found to share structural homology only with chicken riboflavin binding protein (57–59). The majority of apo-hFRβ lacks defined secondary structure, with ∼30% of residues in six α-helices and less than 8% in four short β-strands (Figs. 1 and 3A; Fig. S1). Eight disulfide bonds stabilize the fold by tethering loops and secondary elements. The most prominent feature of apo-hFRβ is a ∼10 × 15 Å cleft with a depth of ∼20 Å. Based solely on the state I structure, we proposed that this cleft is the binding site for ligands and that the receptor is in an open conformation poised to interact with folate. These hypotheses are supported by our additional hFR structures discussed below.
Fig. 3.
Structures of folate receptors depicting states of biological trafficking. Cartoon models representing proposed states I, II, and III (A–C) of folate transport are represented by apo-hFRβ and the hFRβ/FOL complex at near neutral pH, and apo-hFRα at acidic pH, respectively. (A) Cartoon model of the apo-hFRβ structure is shown with conserved disulfides colored orange. (B) A cartoon depiction of the hFRβ/FOL complex shows the position of the ligand binding pocket with residues that interact with folate shown as sticks. (C) The apo-hFRα model illustrates global conformational differences in the structure of apo-hFR at pH 5.5 relative to the same at near neutral pH (cf. A and C). (D) Conformational differences between the three trafficking states are highlighted. Four regions of the folate receptors that undergo significant conformational changes are numbered with arrows to indicate the general direction of movements and colored as seen in A–C. Variable regions in each individual model are emphasized with darker shading of the same color.
Fig. 4.
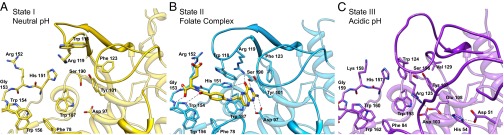
Conformational changes in the hFR ligand binding pocket during states of biological trafficking. Residues that interact with folate in the complex structure are modeled to highlight the movements of conformationally variable loops (darker shading) between the open state at neutral pH (A), the folate complex (B), and the closed state at acidic pH (C). Polar interactions in the folate complex are designated with dashed lines. Tyr-76, which forms hydrophobic interactions benzoyl moiety of the folate ligand, is removed for clarity. Detailed interaction maps can be seen in Figs. S2 and S4–S6.
To elucidate structural determinants for ligand binding and to deduce conformational changes in the receptor that occur when folate (FOL) interacts, a structure of the hFRβ/FOL, termed state II hereafter, was determined to 2.3 Å (Rfree = 26.0%). The hFRβ/FOL complex crystallized at neutral to basic pH values (7.0–8.5), and thus the hFRβ/FOL complex represents the general conformation of the receptor at the cell surface after binding to a ligand (Figs. 2 and 3B). Folate is oriented such that the pterin ring is positioned deep within the aforementioned cleft, the 4-aminobenozyl linker extends through the central region of the cleft, and the glutamyl tail is partially exposed to solvent with atoms leading to the γ-carboxylate protruding from the binding cleft.
To accommodate folate, the conformations of state I and state II differ globally only in two loop regions, one connecting the β1 and β2 strands and another following the α1 helix (cf. Fig. 3 A and B; Movie S1). The movements of these two loops allow interactions of Y76, W118, and R119 with folate. Additional contacts with folate are provided by atoms that do not move significantly between state I and state II (e.g., F78, D97, Y101, G153, W154, W156, W187) or are facilitated by changes in side chain rotamer conformations (e.g., H151, R152, S190). Additional details of all contacts between folate and hFRβ are provided in Fig. S2. The image that emerges from a detailed analysis of folate binding is the following: (i) the pterin ring stacks between Y101 and W187, residues conserved throughout the FR superfamily, and is further stabilized by several polar contacts with side chain atoms (31, 57); (ii) the central 4-aminobenzoyl moiety interacts only via hydrophobic interactions, but not through direct π-π stacking, with aromatic residues Y76, F78, and W118; and (iii) the glutamyl portion is positioned in a pocket wherein main chain atoms, three conserved tryptophans, and a single basic residue (R152) form polar contacts with the carboxylates.
To transport folate, hFRs are encapsulated into an endosome wherein folate release occurs. This release has been proposed to be due to a pH-dependent switch in the hFR/FOL complex as the microenviroment of the endosome acidifies, although recycling endosomes that traffic folate receptors have been reported to only become mildly acidic within a pH range of 5.6–7.2 with an average value of 6.5 (2, 6, 42, 53). Due to the potential for structural changes in the receptor at acidic pH, we determined the structure of apo-hFRα at pH 5.5 in three distinct crystal forms from three protein constructs to resolutions of 1.55, 1.8, and 2.2 Å, with Rfree values of 19.9, 26.9, and 19.9%, respectively. With the exception of changes in the conformation of one loop due to a crystal contact and the degree of order in another, all four molecules in the three crystal forms are statistically identical (Fig. S3). This conformation, termed state III, represents the endosomal conformation of the receptor after folate has been released. On acidification, the receptor undergoes several conformational changes to loops that surround the binding cleft (Fig. 3 C and D).
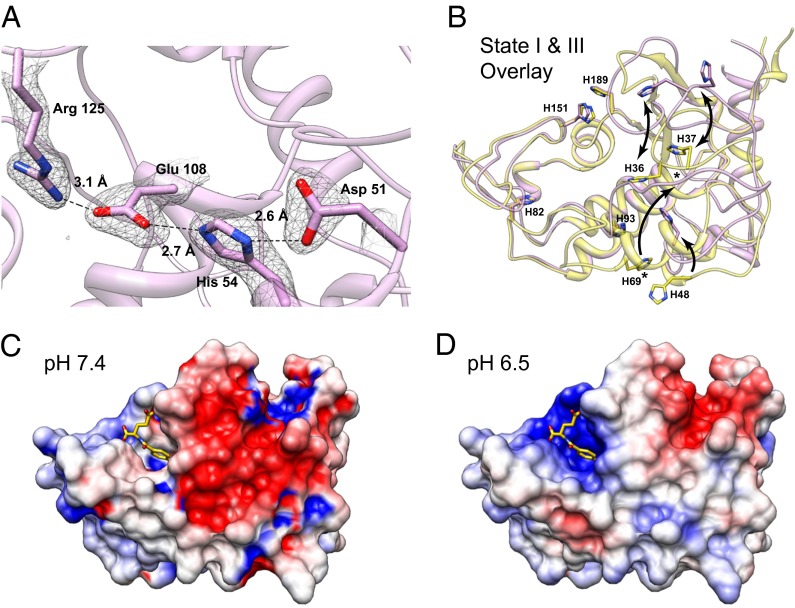
Our three biological trafficking models of hFRs show structural variations in four regions, which are described using the following names: basic loop, switch helix, anchor loop, and inhibitory loop (Figs. 1 and 3D). The basic loop follows the first cysteine residue and includes a conserved tetrapeptide of basic residues: KHHK. In state III, the histidines in the central region of the basic loop have moved with a Cα displacement of ∼9 Å relative to the position in state I or II. The KHHK residues in the basic loop are placed near the C-terminal end of the α2 helix in states I and II, and the loop rotates away from the α2 helix, and thus the folate binding site, in state III at acidic pH, placing the basic loop in a position wherein the side chains of the KHHK motif are largely solvent exposed. Movement of the basic loop allows for the displacement of α1, the switch helix, which undergoes a transition from helix in states I and II to an extended loop in the acidic state III. The movement of the switch helix allows a shift in the position of the anchor loop, which provides two residues, D51 and H54 (D45/H48 in hFRβ), for a series of ionic interactions including E108 and R125 (E102/R119 in hFRβ; Figs. 4 and 5A). As a result, the inhibitory loop is anchored in the folate binding cleft via R125 (R119 in hFRβ), a conserved residue that interacts with folate in state II. In state III, the arginine sidechain is positioned to prevent the pterin ring of folate from entering the binding cleft (Movie S2). Relative to changes between state I and II, the transformations to state III are of much greater magnitude.
Fig. 5.
Histidine residues may promote or stabilize pH-dependent global conformational changes in hFR structures. (A) Conserved residues of hFRα and hFRβ form ionic contacts at low pH. A series of interactions facilitated by D51 and H54, residues from the anchor loop, which are solvent exposed at neutral pH, result in stabilization of Arg125 in a position that occludes the folate binding site. Electron density for the final 2m|Fo-DFc| map contoured at 1.0 σ is shown with residue numbering based on hFRα. (B) Conserved histidines in hFRα and hFRβ may promote ligand dissociation and global changes in conformation based on changes in pH when moving from neutral (states I and II) to acidic (state III) conditions during trafficking. Residue numbers are indicated for hFRβ; equivalent numbering for hFRα would be incremented +6 relative to hFRβ (Fig. 1). Arrows serve as guides for the direction of movement for individual histidines. An asterisk indicates the H69 equivalent position in state III. This residue was not shown explicitly or modeled due to lack of electron density for the sidechain atoms. (C and D) The adaptive Poisson-Boltzmann solver (APBS) software was used to calculate electrostatic surface potentials for the hFRβ folate complex at pH 7.4 and 6.5 to assess possible differences in electrostatic potential experienced by hFRβ/FOL complex during trafficking.
Based on our finding that hFRs have significantly different structures at near neutral and mildly acidic pH values, we examined whether histidines, the most likely residues to be ionized during this pH change, serve as facilitators of global conformational changes in the receptor. Initially, we compared the positions of the eight histidines conserved in hFRα and hFRβ and identified residues that undergo movement between apo-hFR states I and III (Fig. 5B). Four residues, H82, H93, H151, and H189, showed no positional changes between the two apo-hFR models, likely indicating no role for these residues in global pH-dependent conformational changes of the receptor (Movie S3). However, H151 may still perturb folate binding via pH-dependent ionization as it undergoes a rotamer shift to interact with folate at neutral pH (state II). The remaining histidines, H36, H37, H48, and H69, have Cα displacements of 9.3, 8.9, 4.8, and 16.0 Å, respectively. H36 and H37 reside in the basic loop flanked by two conserved lysines and may change conformation due to increased electropositive potential at acidic pH. The movement of H48 (H54 in hFRα) in the anchor loop, which is dependent on movement of both the basic loop and the switch helix, ultimately positions R119 (R125 in hFRα) in the folate binding pocket. The final histidine, H69, is solvent exposed with no contacts that implicate the residue in specific stabilization of state I or state III. On the basis of this structural analysis, we propose that three histidines, H36, H37, and H48, facilitate global structural rearrangements via destabilization of state I or stabilization of state III due to ionization at acidic pH.
In addition to our positional analysis of conserved histidine residues, we also examined the electrostatic surface potential of our hFRβ/FOL complex, crystallized at pH 8, to understand whether surface charge density is a factor in ligand release at acidic pH. Electrostatic potential surfaces were computed at pH values of 7.4, representing a pH value at which folates should bind the hFRs or at pH 6.5, a condition under which the receptor should be capable of folate release (Fig. 5 C and D) (60–62). Strikingly, comparison of these surfaces illustrated a significant increase in electropositive surface potential of the folate binding cleft at pH 6.5. Thus, we conclude that the structure of the hFRβ/folate complex at pH 6.5, which we have been unable to determine experimentally, may be destabilized due to increases in electropositive charge density. Furthermore, these changes in charge density may in part trigger ligand release and could be considered in the development of unique antifolates with desired release properties.
From our structures representing three biological trafficking states of hFRs, we conclude that the receptors are poised in an open conformation at the cell surface and undergo minor rearrangements to make specific contacts with folate. After ligand release under acidic conditions, hFRs undergo a major reorganization wherein the movements of four surface loops ultimately occlude the folate binding site. A key residue, R125, occupies a position in the binding site that is stabilized by a series of ionic interactions mediated by a H54, blocking the association of folate with the receptor, whereas additional histidines may play a role in global conformational changes.
pH Dependence of Ligand Binding to the hFRs.
Because ligand release by the folate receptors is considered a pH-dependent event, we determined the binding affinities of folate and three clinically prevalent antifolates, pemetrexed (PMX), aminopterin (AMT), and methotrexate (MTX) (Fig. 2), for hFRα or hFRβ via isothermal titration calorimetry at pH 7.4 and pH 6.5. A general expectation given the proposed pH dependence model for ligand release is that a lower affinity for folate would be seen at pH 6.5, the average pH of the recycling endosome (Table 1; Table S3) (2, 42, 53, 63). Isothermal titration calorimetry (ITC) measurements demonstrate that hFRα has an extremely high affinity for folate with a dissociation constant (Kd) of ∼10 pM at pH 7.4. In agreement with a pH-dependent release mechanism, the affinity of hFRα for folate at pH 6.5 decreases more than 2,000-fold to a Kd of ∼21 nM. Among the antifolates tested, PMX showed the highest affinity for hFRα at pH 7.4, whereas AMT and MTX bound at an order of magnitude lower affinity than PMX. However, no significant pH dependence was seen in the binding of PMX, AMT, or MTX to hFRα at pH 6.5, indicating that these antifolates may not release effectively from this receptor.
Table 1.
hFR affinities for folate and antifolates measured by ITC
| Ligand | hFRα (Kd ± SD, nM) |
hFRβ (Kd ± SD, nM) |
||
| pH 7.4 | pH 6.5 | pH 7.4 | pH 6.5 | |
| Folic acid | 0.0103 ± 0.0015 | 21 ± 9 | 2.7 ± 1.5 | 23 ± 5 |
| Methotrexate | 65 ± 5 | 88 ± 5 | 40 ± 7 | 332 ± 64 |
| Aminopterin | 65 ± 6 | 71 ± 7 | 144 ± 5* | ND |
| Pemetrexed | 4.5 ± 1.9 | 11 ± 2.5 | 54 ± 17 | 323† |
Unless noted, at least three measurements were made for each data point. ND, not determined.
Two measurements.
Single measurement.
We similarly characterized the pH-dependent binding profiles for hFRβ. Although the binding sites of hFRα and hFRβ are highly conserved with only two divergent residues (V129α/F123β and K158α/R152β), hFRβ exhibited a lesser pH dependence in binding to folate, with Kds of 2.7 and 23 nM at pH 7.4 and 6.5, respectively. However, the Kd of folate to hFRβ represents an upper limit as the combination of the high affinity binding and limitations in ITC based on poor signals prevented an accurate measurement via direct titration and competition experiments similar to those conducted for hFRα yielded poor results based on the same limitations. Thus, the binding of folate for hFRβ is likely tighter than the measured Kd; however, by comparison with our hFRα data, we expect an affinity less than that of hFRα with folate (10 pM). Contrary to the data obtained for hFRα, hFRβ did show pH-dependent decreases in affinity for the antifolates tested (six- to eightfold), supporting the notion that these drugs could be efficiently released by hFRβ. Through examination of sequence variation at the ligand binding site and in conformationally variable loops in hFRβ and hFRα, no simple, sequence-based explanation presents itself to explain the difference in behavior between the two receptors. On the whole, our data indicate that pH plays a role in the release of PMX and MTX from hFRβ, and both hFRα and hFRβ exhibit a pH-dependent decrease in affinity for folate on acidification.
Folate and Antifolates Interact with hFRs in Distinct Binding Modes.
Antifolates have undergone continued development for more than six decades (6, 11). Initially, much of antifolate development relied on empirical structure activity relationship studies involving either inhibition of target enzymes or cytotoxicity in cancer or model cell lines. Later development depended in part on newly determined structural models of target enzymes including dihydrofolate reductase, thymidylate synthase, and glycinamide ribonucleotide formyltransferase. Recent advances in folate-mediated therapies have focused on molecules that are targeted specifically via the folate receptors to tumors or sites of inflammation, but structural models of hFRs to aid in drug design efforts have been lacking. To gain a robust understanding of the molecular determinants of and variability in ligand binding and concurrently provide atomic resolution models to facilitate drug development, we determined crystallographic structures of hFRβ at pH 8–8.2 in complex with the antifolates PMX, AMT, and MTX to resolutions of 2.6, 2.3, and 2.1 Å, respectively (Rfree = 24.4, 23.5, and 23.1%).
The ligand-binding modes of PMX, AMT, and MTX share many characteristics with that of FOL (Fig. 6; Figs. S2 and S4–S7). For all ligands, the bicyclic aromatic moiety is stacked between Y101 and W187, and the α-carboxylate of the glutamyl tail makes similar contacts with G153, W154, and W156. Subtle variations exist in the interactions of the γ-carboxylate with hFRβ. Specifically, the γ-carboxylates of FOL and PMX make contacts with only W118 and R152, whereas the γ-carboxylates of AMT and MTX make additional contacts with Q116. The central regions of each ligand consisting of variable linker atoms flanking a central benzyl ring make nonspecific interactions with hydrophobic residues, and the exact path of these atoms through the binding cleft varies (Fig. 6; Fig. S7).
Fig. 6.
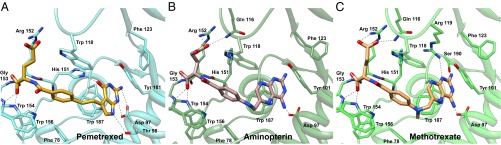
Binding site interactions of antifolates with hFRβ. Interacting residues are highlighted to compare similarities and differences in the amino acid contacts in hFRβ complex structures with (A) pemetrexed, (B) aminopterin, and (C) methotrexate.
The most evident variations in ligand-binding modes for folate and antifolates reside in the position of pterin, pyrollo-pyrimidine, or diaminopteridine moieties. The pterin in FOL is oriented in a fashion most similar to the pyrollo-pyrimidine of PMX. Using O4 of the pterin ring of FOL as a reference, the pyrollo-pyrimidine of PMX is rotated 20°, and the exocyclic oxygen is displaced by 2.4 Å. Rotation of the pyrollo-pyrimidine still affords polar contacts with D97 and H151 equivalent to those seen in FOL. However, PMX does not interact with R119 or S190 and rather forms a new contact with T98. Due to the molecular differences between FOL and PMX, this level of variation seen in the binding modes is not unexpected. In contrast, comparison of the hFRβ/FOL and hFRβ/AMT complexes did yield some unexpected differences in the interactions of the two ligands. AMT is the first antifolate and is based on a single change of the O4 oxygen of FOL to an amine (8). To accommodate this seemingly subtle change on binding, the diaminopteridine of AMT docks such that it is flipped 180° in the binding cleft (Fig. S7). As a consequence, none of the polar contacts between hFRβ and the pterin of FOL are preserved with equivalent atoms for the diaminopteridine ring in AMT. Rather, hFRβ uses the same residues that make contacts with the pterin of FOL, with the exception of H151, to make a series of new contacts with the diaminopteridine. We also determined a structure of hFRβ in complex with MTX, an antifolate that varies from AMT via the addition of a methyl at the N10 position, and we see little distinction in the binding modes of these two diaminopteridine-containing antifolates (9). Taken together, our structural models of four hFR complexes suggest that folate receptors are able to recognize molecules that consist of an aromatic system capable of stacking between Y101 and W187, a largely hydrophobic linker with length matched to the central portion of the binding cleft, and a glutamyl moiety that makes specific polar contacts at the exit of the binding pocket. Additionally, our models show that pterins and 5-substituted pyrollo-pyrimidines are positioned in the binding cleft of hFRs in a similar fashion that is distinct from that seen for diaminopteridines.
Discussion
We determined six unique structures of hFRs that enhance our understanding of conformational changes occurring during biological trafficking of folate receptors and delineate molecular details of multiple binding modes for folate and antifolates. Specifically, we have shown hFRs in three states mimicking the apo-receptor and folate bound receptor at the cell surface and the apo-receptor within the endosome (Figs. 2–4). These results strongly suggest that the pH of the receptor environment plays a major role in structural rearrangements with the folate binding cleft poised in an open conformation at neutral pH and occluded at the acidic pH experienced in the endosome (2, 53).
Perhaps the most anticipated structure determined is the hFR/FOL complex (state II), whereas the most unexpected results are those of apo-hFR crystallized under acidic conditions (state III). The binding mode for folate in state II rationalizes the linkage position for nearly all folate-conjugates, as the cargo for these multifunctional molecules are almost exclusively coupled at or near the γ-carboxylate of the glutamate (28, 48–50, 64). The apo-hFR state III model highlights dramatic structural changes that occur in hFRs as a result of changes in pH. State III is informative as a model of the receptor after ligand release and clearly indicates a mechanism to prevent reassociation of folate via the insertion of an inhibitory loop into the binding cleft of the receptor.
Our trafficking state models provide a foundation for further study, with further analyses of conserved histidines and surface electrostatics a likely focus (Fig. 5). The possibility exists that additional, intermediate states of the receptor could be discovered at the pH values between that of states II and III, but none of our crystallization trials yielded hits in this pH range. Perhaps the receptor undergoes conformational exchange between states under these conditions, preventing crystal formation. We also attempted to capture a state of the receptor in complex with any ligand at acidic pH without success. Through thousands of trials, samples that were crystallized in the presence of ligand at low pH lacked any density for ligand and were consistently found in the apo-hFR state III conformation.
Our three antifolate complex structures establish how folate receptors accommodate these clinically used molecules (Fig. 6). Additionally, we characterized the pH dependence for the binding of folate and select antifolates to both hFRα and hFRβ. These data indicate pH-dependent changes in affinity for folate and lesser changes for antifolates (Table 1). Although we see a decrease in affinity of hFRs for folate at pH values consistent with recycling endosomes, it remains to determine whether the decrease in affinity to ∼20 nM is sufficient to effectively drive release of folates. Additional studies remain to determine whether intrinsic factors such as kinetics of ligand binding and release or extrinsic factors, perhaps other proteins within the endosome, are needed for efficient release and transport of folates. Our data nonetheless provide a more comprehensive view than has been available to date regarding the structural underpinnings of the transport of and molecular interactions with folate receptors. These findings may be extended to the design of folate-conjugate and immunological agents that are highly hFR specific. Furthermore, these designs could be targeted toward either hFRα or hFRβ for specification in disease treatment.
With a series of hFR/ligand complexes in hand (Fig. 6), reasonable predictions can now be made regarding the binding poses for many folate analogs (6, 11). At this stage, we showed two distinct binding modes in the four complexes. FOL and PMX bind similarly and serve as the best models to design new molecules or predict binding modes for additional analogs with either pterin and 5-substituted pyrollo-pyrimidine aromatic systems. Similarly, AMT and MTX bind in the same mode and are useful for studies of additional diaminopteridine analogs. A number of additional folate metabolites or analogs should be pursued for structural study to increase the diversity of available model complexes, including reduced folates and leucovorin (19), as well as the newly developed ONX801 (13) and 6-substituted thieno-pyrimidines, analogs that show varying degrees of specificity for the folate receptors over the reduced folate carrier (51, 52).
Our work suggests at least two additional avenues for potential exploration in antifolate development. The basis for FR-targeted therapy is the notion that cytotoxic drugs transported into cells via the folate receptors, rather than through facilitative transporters widely expressed in normal tissues, will not harm normal cells. FR specificity over RFC and PCFT transporters could be gained by the engineering of molecules that take advantage of the many solvent-filled cavities that surround ligands in the binding cleft. The binding pocket of the receptor is not filled by any of the ligands we examined. Moreover, the ligands use different regions of the binding cleft to accommodate the variations in the bicyclic aromatic regions. One antifolate that may take advantage of additional pockets within the binding cleft, and for which the mode of binding must await structural analysis, is the aforementioned ONX801, a molecule that contains a decorated tricyclic fused ring system (12, 13, 65).
The second possibility for drug development could exploit the pH changes during FR trafficking. For example, the presence of a histidine in the binding pocket could be leveraged in antifolate design to improve ligand release properties. We propose that antifolates that contain an ionizable group with an appropriate pKa, for example an imidazolium moiety, would release efficiency due to the increased local positive charge.
Major outcomes anticipated from our structural studies on hFRs are the development of FR-specific antifolates and folate-conjugates with reduced toxicity profiles in normal tissue owing to lack of transport via RFC. Our FR structures provide a platform for rational drug design and also provide new perspectives in a mature field, both of which will accelerate the development of unique folate-based therapeutics. Having molecular details in place, the focus can now shift to making targeted molecules that bind with high affinity and specificity to a given hFR at extracellular pH (∼7.0–7.4) and are released efficiently at endosomal pH (∼5.0–6.5). The potential of FR-targeted therapeutics in treatment of cancer and inflammatory disease is being explored in detail with lead molecules based on antifolate or folate-conjugate designs in clinical trials, and we expect that our structures will be at the forefront of the development of the next generation of FR-targeted therapeutics.
Materials and Methods
Expression and Purification of hFRs.
DNAs encoding the mature FRs, lacking residues for the N-terminal signal sequence and C-terminal GPI anchor attachment, were amplified by PCR and subcloned as XbaI/EcoRI or BamHI/EcoRI restriction endonuclease fragments into pSGHV0 (66). From the N to C terminus, resultant proteins consist of human growth hormone (hGH), an octahistidine tag, the tobacco etch virus protease site, and either hFR α or β (hGH-hFRα or hGH-hFRβ; Fig. S1). To generate cell lines stably expressing hGH-hFRs, 50 μg plasmid DNA was cotransfected with 5 μg dhfr-containing plasmid into dhfr- Chinese Hamster Ovary (CHO duk-) cells via electroporation at settings of 174 V and 400 μF in a 2-mm gap cuvette. Cells were plated in nucleoside-free selection medium [αMEM, 10% (vol/vol) dialyzed FBS] and incubated at 37 °C under a 5% CO2 atmosphere for approximately 2 wk before clonal selection. Clones were expanded, and expression of secreted hGH-hFRα and hGH-hFRβ was assayed by anti-hGH ELISA. Clones with suitable protein expression levels were subjected to gene amplification in the presence of MTX as described in Leahy et al. (66). After amplification, cell lines expressing hGH-hFR were expanded for growth in roller bottles or hollow fiber bioreactors (FiberCell Systems) containing high glucose DMEM/F12 medium supplemented with 1% (vol/vol) FBS (Life Technologies).
As folate is present in the growth medium, standard purification of both hGH-hFRα and hGH-hFRβ yielded complexes of FRs with folic acid. In a typical purification, 4 L conditioned medium from roller bottles or 240 mL from hollow fiber bioreactors, respectively, was concentrated and exchanged into buffer containing 50 mM Tris, pH 8.0, 500 mM NaCl, 10% (vol/vol) glycerol, and 5 mM imidazole using a Labscale TFF concentrator (Millipore) against a 50-kDa nominal MWCO tangential flow filter. Target hGH-hFR fusion proteins were purified via immobilized metal affinity chromatography (IMAC) with Ni-nitrilotriacetic acid Sepharose resin (Qiagen). During gradient purification, hGH-hFR elutes in a broad peak from 20 to 40 mM imidazole. hGH-hFR target proteins were exchanged into 10 mM Tris, pH 7.5, and 50 mM NaCl before proteolysis with tobacco etch virus (TEV) protease at 4 °C for 16 h. A second passive Ni-nitrilotriacetic acid purification afforded cleaved hFR in the flowthrough and 20 mM imidazole wash fractions. hFR fractions were pooled, concentrated, and subjected to size exclusion chromatography on a Superdex 75 (16/60) column (GE Healthcare) equilibrated in 10 mM Tris, pH 7.5, and 50 mM NaCl buffer. Final clones used in this work expressed 5–8 mg hFR in complex with folate per liter of medium.
For apo-hFR purification, folate was dissociated from the receptor at pH 3.5 and absorbed to activated charcoal. After concentration and buffer exchange to 25 mM Tris, pH 8.0, and 500 mM NaCl, 5% (vol/vol) glycerol, 5 mM imidazole, FBS, and Triton X-100 were added to 30% (vol/vol) and 1% (vol/vol), respectively, to reduce adsorption of hGH-hFR to charcoal. The pH of this solution was adjusted to pH 3.5 with 1 M HCl. One volume equivalent of 80 mg/mL charcoal in 25 mM sodium acetate, pH 3.5, with 1% (vol/vol) Triton X-100 was added to the protein sample, and the resulting slurry was stirred for ∼20 min at 4 °C. After centrifugation and filtration, the protein solution is adjusted to pH 7.4 with 1 M NaOH. The crude protein solution was loaded onto a folate Sepharose affinity column equilibrated in PBS, pH 7.5, and 1% (vol/vol) Triton X-100 (PBS-T) (67, 68). The column was washed with PBS-T, PBS-T supplemented with 500 mM NaCl, and PBS. hGH-hFR was eluted in 10 mM sodium acetate, pH 3.5, and eluent fractions were immediately neutralized with 0.1 volume equivalents of 0.5 M Tris, pH 7.5. The subsequent purification and proteolysis steps were identical to the methods used for the hFR-folate complex.
ITC Analysis of hFRβ with Folate and Antifolates.
For ITC analyses, apo-hFR samples were exchanged into 20 mM sodium citrate phosphate and 100 mM NaCl at pH values of 7.4 or 6.5 via size exclusion chromatography on a Superdex 75 10/300 column (GE Healthcare). Ligands were dissolved in the same buffer, and titrations of hFR proteins into ligands were conducted using a NanoITC low volume calorimeter (TA Instruments). In a typical titration experiment, 300 μL 5–30 μM ligand was placed in the sample cell of 174 μL working volume, whereas 50 μL 100–500 μM hFR was loaded in the titration syringe. Volume per injection varied from 0.18 to 0.8 μL hFR with 150-s intervals between injections. To measure the high affinity binding of hFRα to folate, a competition experiment was necessary wherein 300 µM MTX was included as a competitor in both the sample cell with folate and titration syringe with hFRα. The data were fit to an observed association constant, which was used with the measured affinity of MTX to hFRα to calculate the association constant of hFRα to folate. Enthalpic parameters (∆H) of the hFRα/folate interaction were obtained via a standard titrations. After initial trials and parameter optimization, at least three titrations were carried out for each protein-ligand pair (Table 1; Table S3) (69).
Crystallization of hFRs.
Crystallization trials were carried out using a combination of sitting and hanging drop vapor diffusion techniques. In the early stages of our work, two crystal forms of apo-hFRα were obtained from proteins produced in Sf9 cells (70, 71). All other crystals of hFRα and hFRβ proteins were obtained from proteins secreted from CHO cells (Fig. S1) (66). Additional details regarding manipulation of glycan types for crystallization trials are discussed in SI Materials and Methods. For hFR complexes, ligands were added to apo-hFR in a 1.5:1 molar ratio, and excess ligand was removed via size exclusion chromatography in 10 mM Tris, pH 7.5, and 50 mM NaCl. For all crystallization experiments, equal volumes (150 nL or 1.0 μL) of protein or protein complex and each crystallant solution were combined. Conditions for crystallization were as follows: hFRα-P212121 crystal form, 12 mg/mL protein copurified with MTX in 0.1 M sodium citrate, 35% (wt/vol) jeffamine ED-2001 (MTX not seen in the structure); hFRα-P65 crystal form, 10 mg/mL protein in 0.1 M citrate, pH 4.5, 0.1 M lithium chloride, 17.5% (wt/vol) PEG 8000; hFRα-P1 crystal form, 10 mg/mL protein in 0.1 M citrate, pH 4.5, 0.1 M lithium chloride, 17.5% (wt/vol) PEG 8000, 40% (vol/vol) 2,2,2-trifluoroethanol; hFRβ, 50 mg/mL protein in 1.0 M ammonium citrate tribasic pH 7.0, 0.1 M Bis-Tris propane pH 7.0; hFRβ/FOL, 50 mg/mL protein in 2.4 M ammonium phosphate dibasic, 0.1 M Hepes pH 7.5; hFRβ/PMX, 12 mg/mL protein in 1.5 M ammonium phosphate dibasic, 0.1 M Tris pH 8.5; and both hFRβ/AMT and hFRβ/MTX, 10 mg/mL protein in 20% (wt/vol) PEG 3350, 0.2 M lithium sulfate, 0.1 M Tris-HCl pH 8.0.
Data Collection and Structure Determination.
Crystal parameters, data reduction, and model refinement statistics are presented in Tables S1 and S2. Data were collected on crystals flash frozen in liquid nitrogen via the oscillation method and were reduced using the HKL2000 program suite (72). Phases were calculated via molecular replacement (MR) using Phaser as implemented in AutoMR, a component of the PHENIX software package (45, 73–75). For our initial structure of apo-hFRα (P65 form), model coordinates for chicken riboflavin binding protein (chRfBP) were kindly provided by H. Monaco (University of Verona, Verona, Italy), and a truncated form of the cRfBP model was used as a MR search model in Phaser (57). Our apo-hFRα (P65) model was subsequently used as a search model to obtain phases for two additional apo-hFRα datasets as well as the hFRβ/FOL complex data. In the latter case, the hFRα search model was truncated to the core of the protein as determined via structural alignment with chRfBP. The remaining hFRβ complex structures were phased using the hFRβ/FOL structure coordinates as the search model. Refinement and model building proceeded with phenix.refine and Coot programs, respectively (45, 74, 76, 77). Translation libration screw refinement was implemented in the final stages of refinement (78).
Supplementary Material
Acknowledgments
We thank H. L. Monaco (University of Verona, Verona, Italy) for providing model coordinates of chicken riboflavin binding protein, E. E. Carlson for scientific discussion, and S. M. Deis, D. P. Giedroc, and M. G. Oakley for comments on the manuscript. We gratefully acknowledge support from National Institutes of Health Grants GM094472 and CA166711 (to C.E.D.) and the Indiana University College of Arts and Sciences. All crystallization experiments were carried out in the Indiana University METACyt Crystallization Automation Facility. Results shown in this report for the P65 and P1 crystal forms of apo-hFRα are derived from work performed at beamline 19BM at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the US Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357. Crystallographic data for the remaining six hFR structures were collected with remote assistance provided by Dr. Jay Nix on beamline 4.2.2 at the Advanced Light Source at the Lawrence Berkeley National Laboratory, part of the Molecular Biology Consortium supported by the US Department of Energy and Indiana University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4KM6, 4KM7, 4KMX, 4KMY, 4KMZ, 4KN0, 4KN1, 4KN2).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308827110/-/DCSupplemental.
References
- 1.Hoffbrand AV, Weir DG. The history of folic acid. Br J Haematol. 2001;113(3):579–589. doi: 10.1046/j.1365-2141.2001.02822.x. [DOI] [PubMed] [Google Scholar]
- 2.Kamen BA, Smith AK. A review of folate receptor alpha cycling and 5-methyltetrahydrofolate accumulation with an emphasis on cell models in vitro. Adv Drug Deliv Rev. 2004;56(8):1085–1097. doi: 10.1016/j.addr.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Kamen BA, Wang MT, Streckfuss AJ, Peryea X, Anderson RG. Delivery of folates to the cytoplasm of MA104 cells is mediated by a surface membrane receptor that recycles. J Biol Chem. 1988;263(27):13602–13609. [PubMed] [Google Scholar]
- 4.Matherly LH, Goldman DI. Membrane transport of folates. Vitam Horm. 2003;66:403–456. doi: 10.1016/s0083-6729(03)01012-4. [DOI] [PubMed] [Google Scholar]
- 5.Zhao R, Matherly LH, Goldman ID. Membrane transporters and folate homeostasis: Intestinal absorption and transport into systemic compartments and tissues. Expert Rev Mol Med. 2009;11:e4. doi: 10.1017/S1462399409000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldman ID, Chattopadhyay S, Zhao R, Moran R. The antifolates: Evolution, new agents in the clinic, and how targeting delivery via specific membrane transporters is driving the development of a next generation of folate analogs. Curr Opin Investig Drugs. 2010;11(12):1409–1423. [PubMed] [Google Scholar]
- 7.Matherly LH, Hou Z. Structure and function of the reduced folate carrier a paradigm of a major facilitator superfamily mammalian nutrient transporter. Vitam Horm. 2008;79:145–184. doi: 10.1016/S0083-6729(08)00405-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farber S, Diamond LK. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N Engl J Med. 1948;238(23):787–793. doi: 10.1056/NEJM194806032382301. [DOI] [PubMed] [Google Scholar]
- 9.Burchenal JH, et al. The effects of the folic acid antagonists and 2,6-diaminopurine on neoplastic disease, with special reference to acute leukemia. Cancer. 1951;4(3):549–569. doi: 10.1002/1097-0142(195105)4:3<549::aid-cncr2820040308>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 10.Huennekens FM. The methotrexate story: A paradigm for development of cancer chemotherapeutic agents. Adv Enzyme Regul. 1994;34:397–419. doi: 10.1016/0065-2571(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 11.Visentin M, Zhao R, Goldman ID. The antifolates. Hematol Oncol Clin North Am. 2012;26(3):629–648, ix. doi: 10.1016/j.hoc.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackman AL, Theti DS, Gibbs DD. Antifolates targeted specifically to the folate receptor. Adv Drug Deliv Rev. 2004;56(8):1111–1125. doi: 10.1016/j.addr.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Gibbs DD, et al. BGC 945, a novel tumor-selective thymidylate synthase inhibitor targeted to alpha-folate receptor-overexpressing tumors. Cancer Res. 2005;65(24):11721–11728. doi: 10.1158/0008-5472.CAN-05-2034. [DOI] [PubMed] [Google Scholar]
- 14.Cronstein BN. Low-dose methotrexate: A mainstay in the treatment of rheumatoid arthritis. Pharmacol Rev. 2005;57(2):163–172. doi: 10.1124/pr.57.2.3. [DOI] [PubMed] [Google Scholar]
- 15.Qiu A, et al. Identification of an intestinal folate transporter and the molecular basis for hereditary folate malabsorption. Cell. 2006;127(5):917–928. doi: 10.1016/j.cell.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 16.Liu M, et al. Structure and regulation of the murine reduced folate carrier gene: Identification of four noncoding exons and promoters and regulation by dietary folates. J Biol Chem. 2005;280(7):5588–5597. doi: 10.1074/jbc.M412662200. [DOI] [PubMed] [Google Scholar]
- 17.Qiu A, et al. Rodent intestinal folate transporters (SLC46A1): Secondary structure, functional properties, and response to dietary folate restriction. Am J Physiol Cell Physiol. 2007;293(5):C1669–C1678. doi: 10.1152/ajpcell.00202.2007. [DOI] [PubMed] [Google Scholar]
- 18.Sirotnak FM, Donsbach RC. Comparative studies on the transport of aminopterin, methotrexate, and methasquin by the L1210 leukemia cell. Cancer Res. 1972;32(10):2120–2126. [PubMed] [Google Scholar]
- 19.Min SH, et al. The clinical course and genetic defect in the PCFT gene in a 27-year-old woman with hereditary folate malabsorption. J Pediatr. 2008;153(3):435–437. doi: 10.1016/j.jpeds.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao R, Hanscom M, Chattopadhyay S, Goldman ID. Selective preservation of pemetrexed pharmacological activity in HeLa cells lacking the reduced folate carrier: Association with the presence of a secondary transport pathway. Cancer Res. 2004;64(9):3313–3319. doi: 10.1158/0008-5472.can-03-3953. [DOI] [PubMed] [Google Scholar]
- 21.Chattopadhyay S, Moran RG, Goldman ID. Pemetrexed: Biochemical and cellular pharmacology, mechanisms, and clinical applications. Mol Cancer Ther. 2007;6(2):404–417. doi: 10.1158/1535-7163.MCT-06-0343. [DOI] [PubMed] [Google Scholar]
- 22.Xia W, Low PS. Folate-targeted therapies for cancer. J Med Chem. 2010;53(19):6811–6824. doi: 10.1021/jm100509v. [DOI] [PubMed] [Google Scholar]
- 23.Leamon CP, Jackman AL. Exploitation of the folate receptor in the management of cancer and inflammatory disease. Vitam Horm. 2008;79:203–233. doi: 10.1016/S0083-6729(08)00407-X. [DOI] [PubMed] [Google Scholar]
- 24.Elnakat H, Ratnam M. Distribution, functionality and gene regulation of folate receptor isoforms: Implications in targeted therapy. Adv Drug Deliv Rev. 2004;56(8):1067–1084. doi: 10.1016/j.addr.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Antony AC, Utley C, Van Horne KC, Kolhouse JF. Isolation and characterization of a folate receptor from human placenta. J Biol Chem. 1981;256(18):9684–9692. [PubMed] [Google Scholar]
- 26.Ratnam M, Marquardt H, Duhring JL, Freisheim JH. Homologous membrane folate binding proteins in human placenta: Cloning and sequence of a cDNA. Biochemistry. 1989;28(20):8249–8254. doi: 10.1021/bi00446a042. [DOI] [PubMed] [Google Scholar]
- 27.Antony AC. Folate receptors: Reflections on a personal odyssey and a perspective on unfolding truth. Adv Drug Deliv Rev. 2004;56(8):1059–1066. doi: 10.1016/j.addr.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Parker N, et al. Folate receptor expression in carcinomas and normal tissues determined by a quantitative radioligand binding assay. Anal Biochem. 2005;338(2):284–293. doi: 10.1016/j.ab.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 29.Shen F, Ross JF, Wang X, Ratnam M. Identification of a novel folate receptor, a truncated receptor, and receptor type beta in hematopoietic cells: cDNA cloning, expression, immunoreactivity, and tissue specificity. Biochemistry. 1994;33(5):1209–1215. doi: 10.1021/bi00171a021. [DOI] [PubMed] [Google Scholar]
- 30.Lacey SW, Sanders JM, Rothberg KG, Anderson RG, Kamen BA. Complementary DNA for the folate binding protein correctly predicts anchoring to the membrane by glycosyl-phosphatidylinositol. J Clin Invest. 1989;84(2):715–720. doi: 10.1172/JCI114220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maziarz KM, Monaco HL, Shen F, Ratnam M. Complete mapping of divergent amino acids responsible for differential ligand binding of folate receptors alpha and beta. J Biol Chem. 1999;274(16):11086–11091. doi: 10.1074/jbc.274.16.11086. [DOI] [PubMed] [Google Scholar]
- 32.Wu M, Fan J, Gunning W, Ratnam M. Clustering of GPI-anchored folate receptor independent of both cross-linking and association with caveolin. J Membr Biol. 1997;159(2):137–147. doi: 10.1007/s002329900277. [DOI] [PubMed] [Google Scholar]
- 33.Weitman SD, et al. Cellular localization of the folate receptor: Potential role in drug toxicity and folate homeostasis. Cancer Res. 1992;52(23):6708–6711. [PubMed] [Google Scholar]
- 34.Wu M, Gunning W, Ratnam M. Expression of folate receptor type alpha in relation to cell type, malignancy, and differentiation in ovary, uterus, and cervix. Cancer Epidemiol Biomarkers Prev. 1999;8(9):775–782. [PubMed] [Google Scholar]
- 35.Chancy CD, et al. Expression and differential polarization of the reduced-folate transporter-1 and the folate receptor alpha in mammalian retinal pigment epithelium. J Biol Chem. 2000;275(27):20676–20684. doi: 10.1074/jbc.M002328200. [DOI] [PubMed] [Google Scholar]
- 36.Salazar MD, Ratnam M. The folate receptor: What does it promise in tissue-targeted therapeutics? Cancer Metastasis Rev. 2007;26(1):141–152. doi: 10.1007/s10555-007-9048-0. [DOI] [PubMed] [Google Scholar]
- 37.Puig-Kröger A, et al. Folate receptor beta is expressed by tumor-associated macrophages and constitutes a marker for M2 anti-inflammatory/regulatory macrophages. Cancer Res. 2009;69(24):9395–9403. doi: 10.1158/0008-5472.CAN-09-2050. [DOI] [PubMed] [Google Scholar]
- 38.Ross JF, Chaudhuri PK, Ratnam M. Differential regulation of folate receptor isoforms in normal and malignant tissues in vivo and in established cell lines. Physiologic and clinical implications. Cancer. 1994;73(9):2432–2443. doi: 10.1002/1097-0142(19940501)73:9<2432::aid-cncr2820730929>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 39.Reddy JA, et al. Expression and functional characterization of the beta-isoform of the folate receptor on CD34(+) cells. Blood. 1999;93(11):3940–3948. [PubMed] [Google Scholar]
- 40.Shen F, Wu M, Ross JF, Miller D, Ratnam M. Folate receptor type gamma is primarily a secretory protein due to lack of an efficient signal for glycosylphosphatidylinositol modification: Protein characterization and cell type specificity. Biochemistry. 1995;34(16):5660–5665. doi: 10.1021/bi00016a042. [DOI] [PubMed] [Google Scholar]
- 41.Rijnboutt S, et al. Endocytosis of GPI-linked membrane folate receptor-alpha. J Cell Biol. 1996;132(1-2):35–47. doi: 10.1083/jcb.132.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabharanjak S, Mayor S. Folate receptor endocytosis and trafficking. Adv Drug Deliv Rev. 2004;56(8):1099–1109. doi: 10.1016/j.addr.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Ross JF, et al. Folate receptor type beta is a neutrophilic lineage marker and is differentially expressed in myeloid leukemia. Cancer. 1999;85(2):348–357. doi: 10.1002/(sici)1097-0142(19990115)85:2<348::aid-cncr12>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 44.Bueno R, Appasani K, Mercer H, Lester S, Sugarbaker D. The alpha folate receptor is highly activated in malignant pleural mesothelioma. J Thorac Cardiovasc Surg. 2001;121(2):225–233. doi: 10.1067/mtc.2001.111176. [DOI] [PubMed] [Google Scholar]
- 45.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 46.Nakashima-Matsushita N, et al. Selective expression of folate receptor beta and its possible role in methotrexate transport in synovial macrophages from patients with rheumatoid arthritis. Arthritis Rheum. 1999;42(8):1609–1616. doi: 10.1002/1529-0131(199908)42:8<1609::AID-ANR7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 47.Weinblatt ME, et al. Long-term prospective study of methotrexate in the treatment of rheumatoid arthritis. 84-month update. Arthritis Rheum. 1992;35(2):129–137. doi: 10.1002/art.1780350202. [DOI] [PubMed] [Google Scholar]
- 48.Zhao XB, Lee RJ. Tumor-selective targeted delivery of genes and antisense oligodeoxyribonucleotides via the folate receptor. Adv Drug Deliv Rev. 2004;56(8):1193–1204. doi: 10.1016/j.addr.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 49.Kim SH, Jeong JH, Chun KW, Park TG. Target-specific cellular uptake of PLGA nanoparticles coated with poly(L-lysine)-poly(ethylene glycol)-folate conjugate. Langmuir. 2005;21(19):8852–8857. doi: 10.1021/la0502084. [DOI] [PubMed] [Google Scholar]
- 50.van Dam GM, et al. Intraoperative tumor-specific fluorescence imaging in ovarian cancer by folate receptor-α targeting: First in-human results. Nat Med. 2011;17(10):1315–1319. doi: 10.1038/nm.2472. [DOI] [PubMed] [Google Scholar]
- 51.Deng Y, et al. Synthesis and biological activity of a novel series of 6-substituted thieno[2,3-d]pyrimidine antifolate inhibitors of purine biosynthesis with selectivity for high affinity folate receptors over the reduced folate carrier and proton-coupled folate transporter for cellular entry. J Med Chem. 2009;52(9):2940–2951. doi: 10.1021/jm8011323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L, et al. Synthesis, biological, and antitumor activity of a highly potent 6-substituted pyrrolo[2,3-d]pyrimidine thienoyl antifolate inhibitor with proton-coupled folate transporter and folate receptor selectivity over the reduced folate carrier that inhibits β-glycinamide ribonucleotide formyltransferase. J Med Chem. 2011;54(20):7150–7164. doi: 10.1021/jm200739e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, Chen H, Vlahov IR, Cheng JX, Low PS. Characterization of the pH of folate receptor-containing endosomes and the rate of hydrolysis of internalized acid-labile folate-drug conjugates. J Pharmacol Exp Ther. 2007;321(2):462–468. doi: 10.1124/jpet.106.117648. [DOI] [PubMed] [Google Scholar]
- 54.Antony AC, Kincade RS, Verma RS, Krishnan SR. Identification of high affinity folate binding proteins in human erythrocyte membranes. J Clin Invest. 1987;80(3):711–723. doi: 10.1172/JCI113126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Low PS, Antony AC. Folate receptor-targeted drugs for cancer and inflammatory diseases. Adv Drug Deliv Rev. 2004;56(8):1055–1058. doi: 10.1016/j.addr.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 56.Sabharanjak S, Sharma P, Parton RG, Mayor S. GPI-anchored proteins are delivered to recycling endosomes via a distinct cdc42-regulated, clathrin-independent pinocytic pathway. Dev Cell. 2002;2(4):411–423. doi: 10.1016/s1534-5807(02)00145-4. [DOI] [PubMed] [Google Scholar]
- 57.Monaco HL. Crystal structure of chicken riboflavin-binding protein. EMBO J. 1997;16(7):1475–1483. doi: 10.1093/emboj/16.7.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24(23):2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hasegawa H, Holm L. Advances and pitfalls of protein structural alignment. Curr Opin Struct Biol. 2009;19(3):341–348. doi: 10.1016/j.sbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 60.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 62.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157(1):281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 63.Leamon CP, Low PS. Delivery of macromolecules into living cells: A method that exploits folate receptor endocytosis. Proc Natl Acad Sci USA. 1991;88(13):5572–5576. doi: 10.1073/pnas.88.13.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reddy JA, et al. Folate receptor-specific antitumor activity of EC131, a folate-maytansinoid conjugate. Cancer Res. 2007;67(13):6376–6382. doi: 10.1158/0008-5472.CAN-06-3894. [DOI] [PubMed] [Google Scholar]
- 65.Theti DS, et al. Selective delivery of CB300638, a cyclopenta[g]quinazoline-based thymidylate synthase inhibitor into human tumor cell lines overexpressing the alpha-isoform of the folate receptor. Cancer Res. 2003;63(13):3612–3618. [PubMed] [Google Scholar]
- 66.Leahy DJ, Dann CE, 3rd, Longo P, Perman B, Ramyar KX. A mammalian expression vector for expression and purification of secreted proteins for structural studies. Protein Expr Purif. 2000;20(3):500–506. doi: 10.1006/prep.2000.1331. [DOI] [PubMed] [Google Scholar]
- 67.Fischer C, Rothenberg SP, da Costa M. Preparation of a stable folate-sepharose complex for affinity chromatography. Anal Biochem. 1978;85(1):15–19. doi: 10.1016/0003-2697(78)90267-1. [DOI] [PubMed] [Google Scholar]
- 68.Nishino T, Nishino T, Tsushima K. Purification of highly active milk xanthine oxidase by affinity chromatography on Sepharose 4B/folate gel. FEBS Lett. 1981;131(2):369–372. doi: 10.1016/0014-5793(81)80406-1. [DOI] [PubMed] [Google Scholar]
- 69.Pierce MM, Raman CS, Nall BT. Isothermal titration calorimetry of protein-protein interactions. Methods. 1999;19(2):213–221. doi: 10.1006/meth.1999.0852. [DOI] [PubMed] [Google Scholar]
- 70.Jarvis DL. Developing baculovirus-insect cell expression systems for humanized recombinant glycoprotein production. Virology. 2003;310(1):1–7. doi: 10.1016/s0042-6822(03)00120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hitchman RB, Locanto E, Possee RD, King LA. Optimizing the baculovirus expression vector system. Methods. 2011;55(1):52–57. doi: 10.1016/j.ymeth.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 72.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 73.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adams PD, et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat. 2004;11(Pt 1):53–55. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 75.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 1):61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 77.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 78.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 1):122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.