Abstract
Increasing evidence suggests a key role for angiopoietin-2 (ANGPT2) in influencing the aggressiveness of chronic lymphocytic leukemia (CLL). In the presence of vascular endothelial growth factor (VEGF), ANGPT2 causes vessel destabilization leading to neoangiogenesis. Accordingly, high expression levels of ANGPT2 and high degree of angiogenesis have consistently been associated with poor prognosis in CLL; however, the molecular mechanisms behind the variability in ANGPT2 expression are still to be discovered. Here, for the first time, we investigated the DNA methylation status of the ANGPT2 promoter in a large CLL cohort (n = 88) using pyrosequencing and correlated methylation data with ANGPT2 expression levels, prognostic factors and outcome. Importantly, methylation levels of the ANGPT2 gene correlated inversely with its mRNA expression levels (p < 0.001). Moreover, low ANGPT2 methylation status was highly associated with adverse prognostic markers, shorter time to first treatment and overall survival. Finally, treatment with methyl inhibitors induced re-expression of ANGPT2 in two B-cell lymphoma cell lines, underscoring the importance of DNA methylation in regulating transcriptional silencing of this gene. In conclusion, we believe that the known variability in ANGPT2 expression among CLL patients could be explained by differential promoter DNA methylation and that low methylation levels of the ANGPT2 promoter have an adverse prognostic impact in CLL.
Keywords: chronic lymphocytic leukemia, prognosis, ANGPT2, gene methylation, pyrosequencing
Introduction
The clinical course of chronic lymphocytic leukemia (CLL) is extremely variable with survival times ranging from less than 1 y to more than 15 y. Hence, while many patients experience an indolent disease without ever needing therapy, others immediately show an aggressive disease and die quickly due to complications related to leukemia or treatment. Moreover, in some patients that seem to be indolent, the disease can acquire an aggressive phenotype that soon requires therapy. Considering this high heterogeneity in CLL, its pathogenesis probably is a complex process involving many different biological pathways. Until today, several genes have been associated to CLL prognosis and a pathogenetic role in CLL has been identified for few of them (e.g., TP53 and NOTCH1).1-4 We and others have recently demonstrated that higher expression of angiopoietin-2 (ANGPT2) in CLL confers poor prognosis and that the ANGPT2 produced by leukemic cells is able to induce an increased angiogenesis, characteristic of CLL patients with an aggressive disease.5-10
ANGPT2 is a secreted glycoprotein able to increase vessel plasticity, by binding to the Tie-2 receptor on endothelial cells and blocking ANGPT1 function. The vessel destabilization induced by ANGPT2 can lead to formation of new vessels or to regression of the existing vessels, depending on the presence or not of vascular endothelial factor (VEGF).11-13 Angiogenesis has always been thought to be a fundamental mechanism in the pathogenesis of solid cancers, but many studies concerning different types of leukemias have also shown the importance of angiogenesis in determining the aggressiveness and progression.14-16 The aberrant ANGPT2 expression found both in cancers and leukemias, together with the evidence that ANGPT2 inhibitors are able to reduce tumor angiogenesis in vivo, strongly suggest a pivotal role of ANGPT2 in this process.17-22
Considering the significance of ANGPT2 in influencing CLL behavior, in particular by acting on angiogenesis, the study of molecular mechanisms responsible for its aberrant and variable expression in CLL could be useful to better elucidate the disease pathogenesis and to identify more efficacious therapies. Many studies on ANGPT2 regulation have been performed on normal endothelial cells but limited data exist on tumor cells. It has been shown that Kaposi’s sarcoma-associated herpes virus (KSHV) promotes ANGPT2 expression by the activation of multiple pathways including ERK, JNK and p38. In particular, ANGPT2 transcription is induced by AP-1 and Ets1, that are activated downstream of these pathways and bind to the ANGPT2 promoter.23 In breast cancer cells, the overexpression of human epidermal growth factor receptor 2 (HER2) upregulates ANGPT2 by activation of the ERK and PI3K/AKT pathways.24 Contrary to this, conflicting data concerning the involvement of ERK and PI3K/AKT in the regulation of ANGPT2 have emerged in studies on pancreatic and colon tumor cell lines, suggesting a cell-type specific control for this gene.25
Recent genome-wide methylation profiling studies performed on CLL patients have identified many significantly different methylated genes between two major, prognostic subgroups of CLL, i.e., poor-prognostic IGHV-unmutated and favorable-prognostic IGHV-unmutated cases.26-28 Of particular interest was the finding of a higher ANGPT2 methylation in the IGHV-mutated subset suggesting a possible epigenetic regulation of this gene.26,27 Since ANGPT2 expression has been demonstrated to vary considerably between these two subgroups of CLL patients,5,6 ANGPT2 gene expression could be directly regulated by DNA methylation. To study this further, we assessed the methylation status of the ANGPT2 gene promoter as well as the ANGPT2 expression levels in a large, well-characterized CLL cohort. For the first time, we here show that ANGPT2 expression is highly dependent on the DNA methylation status, where a lower degree of methylation was associated with particularly poor prognosis in CLL. Furthermore, the direct role of DNA methylation in regulating the ANGPT2 expression levels was re-enforced by treating B-cell lymphoma cell lines with a methyl inhibitor leading to demethylation and re-expression of this gene.
Results
Description of CLL cohort
Overall, the Swedish and Italian CLL materials were very similar regarding the distribution of classical CLL prognostic factors (Table 1). The differences in percentages of treated and dead cases can probably be attributed to a longer follow-up time and a higher proportion of advanced Binet stages (B-C) in the Swedish group. In all 88 CLL patients, the adverse CLL prognostic factors were confirmed to be associated with shorter time to first treatment (TTFT) and overall survival (OS); i.e., advanced Binet stages (p < 0.001 for TTFT and p = 0.003 for OS), IGHV unmutated status (p < 0.001 for TTFT and p < 0.001 for OS), CD38 positivity (p = 0.010 for TTFT and p = 0.011 for OS) and intermediate/high-risk FISH markers (p < 0.001 for TTFT and p < 0.001 for OS) (Table S1).
Table 1. Clinical and molecular characteristics of the present CLL cohort.
| Features | Sweden (n = 40) | Italy (n = 48) | p value | All patients (n = 88) | |||
|---|---|---|---|---|---|---|---|
| Age, years |
|
|
|
||||
| Median |
61 |
65 |
ns |
61 |
|||
| Range |
32–83 |
32–85 |
32–85 |
||||
| Follow up, months |
|
|
|
||||
| Median |
72 |
46 |
0.003 |
59 |
|||
| Range |
11–214 |
0–155 |
0–214 |
||||
| TTFT, months |
|
|
|
||||
| Median |
9 |
35 |
ns |
29 |
|||
| Range |
0–136 |
0–138 |
0–138 |
||||
| |
N° patients |
% |
N° patients |
% |
|
N° patients |
% |
| Sex |
|
|
|
|
|
||
| Male |
30/40 |
75 |
32/48 |
67 |
ns |
62/88 |
71 |
| Female |
10/40 |
25 |
16/48 |
33 |
26/88 |
29 |
|
| Binet stage |
|
|
|
|
|
||
| A |
17/30 |
57 |
33/46 |
72 |
ns |
50/76 |
66 |
| B-C |
13/30 |
43 |
13/46 |
28 |
26/76 |
34 |
|
| IGHV mutational status |
|
|
|
|
|
||
| Mutated (< 98%) |
20/40 |
50 |
28/48 |
58 |
ns |
48/88 |
55 |
| Unmutated (≥ 98%) |
20/40 |
50 |
20/48 |
42 |
40/88 |
45 |
|
| CD38 expression |
|
|
|
|
|
||
| CD38 negative (< 30%) |
21/33 |
64 |
34/43 |
79 |
ns |
55/76 |
72 |
| CD38 positive (≥ 30%) |
12/33 |
36 |
9/43 |
21 |
21/76 |
28 |
|
| FISH stratification |
|
|
|
|
|
||
| Low risk |
22/35 |
63 |
29/39 |
74 |
ns |
51/74 |
69 |
| Intermediate/high risk |
13/35 |
37 |
10/39 |
26 |
23/74 |
31 |
|
| Treatment |
|
|
|
|
|
||
| Yes |
11/14 |
79 |
19/48 |
40 |
0.010 |
30/62 |
48 |
| No |
3/14 |
21 |
29/48 |
60 |
32/62 |
52 |
|
| Death censored |
|
|
|
|
|
||
| Yes |
27/39 |
69 |
10/48 |
21 |
< 0.001 | 37/87 |
42 |
| No | 12/39 | 31 | 38/48 | 79 | 50/87 | 58 | |
Low FISH risk, no abnormalities or del(13q); Intermediate/high FISH risk, del(11q), del(17p) or trisomy 12. FISH, fluorescence in situ hybridization; IGHV, immunoglobulin heavy chain variable genes; TTFT, time to first treatment; ns, not statistically significant.
ANGPT2 expression levels in CLL
ANGPT2 mRNA expression was measured by real-time quantitative PCR (RQ-PCR) in PBMCs from CLL patients observing variable levels with a median value of 2.804×10−6 (range, 0–9.1×10−3). Using ROC analysis and Youden’s index we searched for the best cut-off for ANGPT2 mRNA expression in relation to survival. Based on this cut-off value (2.967×10−6, relative mRNA expression of ANGPT2 to B2M), we divided our CLL cohort in high and low ANGPT2 expressing cases including 40 (45%) and 48 (55%) patients, respectively. High ANGPT2 expressing patients showed significantly shorter TTFT (p = 0.046) and OS (p < 0.001) than patients with low ANGPT2 expression (Fig. 1A and B). Moreover, high ANGPT2 expression was confirmed to be strictly associated with poor-prognostic factors as IGHV unmutated status (p < 0.001) and CD38 positivity (p = 0.037).
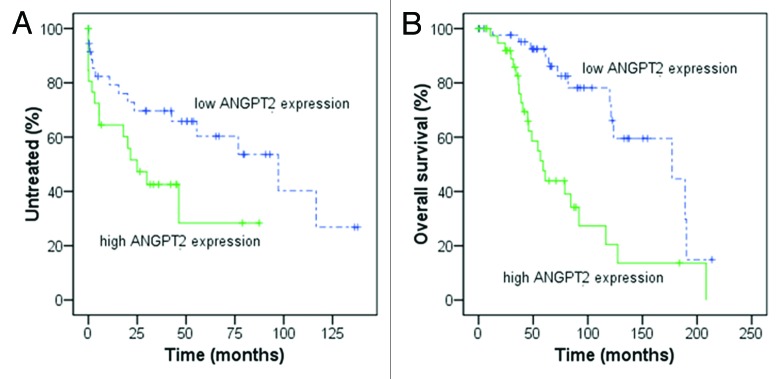
Figure 1. Kaplan-Meier curves for time to first treatment (TTFT) and overall survival (OS) in relation to ANGPT2 mRNA expression. In (A and B), 88 CLL patients were divided according to high and low ANGPT2 expression using the cut-off value 2.967×10−6 (relative mRNA expression of ANGPT2 to B2M). High ANGPT2 expressing cases show significantly shorter TTFT [(A) median 25 vs. 97 mo, p = 0.046; log-rank test) and OS [(B) median 59 vs 177 mo, p < 0.001; log-rank test] compared with low ANGPT2 expressing cases.
Since we used non-purified CLL samples and to exclude any difference between purified/non-purified samples, we sorted the tumor cells in samples from 8/88 patients (Table S2). The ANGPT2 mRNA levels of purified CLL cells (median value 8.800×10−5, range, 0–2.0×10−3) showed a strong correlation with the values detected in the corresponding PBMC samples (r = 0.952, p < 0.001). We also analyzed sorted B cells from two age-matched healthy donors showing low ANGPT2 expression (0–2.2×10−6). Hence, the low expression of ANGPT2 in normal B cells and the similar expression levels in sorted/unsorted CLL samples indeed support the usage of non-sorted samples for this assay. Furthermore, no difference in tumor load was observed between high and low ANGPT2 expressing patients (mean value, 83% and 82%, respectively).
ANGPT2 CpG methylation inversely correlates with mRNA expression
The DNA methylation status of 6 CpG sites located in a CpG island near the transcription start site of the ANGPT2 gene was analyzed by pyrosequencing (Fig. 2). The methylation percentages (%) of each single CpG site and also the average of all 6 CpG sites were very variable among CLL cases (Table 2; Fig. S1), although in each individual patient the six sites showed a similar methylation level. Moreover, the methylation percentage of each single CpG site and of the average of all 6 CpG sites showed significant inverse correlations with ANGPT2 mRNA expression (all sites, p < 0.001) (Fig. S2). In agreement, we observed significantly higher CpG methylation in the ANGPT2 low-expression subset than in the ANGPT2 high-expression cases (average of all 6 CpG sites; median 76% and 50%, respectively, p < 0.001) (Fig. 3A). Similar to the gene expression data, no difference in relation to tumor load was observed between cases with high and low ANGPT2 methylation (mean value, 83% and 82% respectively). Furthermore, although some differences were observed for individual CpG sites, the global methylation level was not statistically different between purified CLL samples (n = 8) and the corresponding unsorted PBMC samples (Table S2). Finally, analysis of normal B-cells from two healthy donors showed high ANGPT2 methylation levels (87% in both samples) as expected.
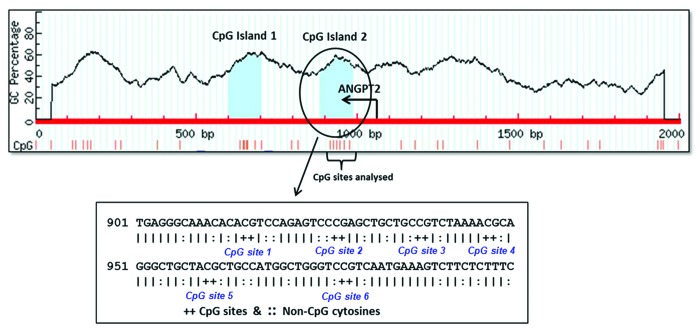
Figure 2. Location of the six CpG sites analyzed by pyrosequencing in relation to transcriptional start site of ANGPT2 gene. The circled region represents the studied CpG island of ANGPT2 gene which was predicted by the site www.urogene.org/methprimer using the following criteria: island size > 100, GC percent > 0.0, obs/exp CpG > 0.6. The start site and the direction of ANGPT2 transcription are indicated by the arrow. The below sequence shows in details the positions of the 6 CpG sites analyzed by pyrosequencing. The CpG site 2 is the same site identified in our previous methylation study.26
Table 2. Methylation percentages of the 6 CpG sites located around the transcription start site of the ANGPT2 gene measured by pyrosequencing in 88 CLL patients.
| Methylation percentage | CpG site 1 |
CpG site 2 |
CpG site 3 |
CpG site 4 |
CpG site 5 |
CpG site 6 |
Average of 6 CpG sites |
|---|---|---|---|---|---|---|---|
| Mean ± SD |
78 ± 19 |
49 ± 20 |
83 ± 25 |
69 ± 21 |
40 ± 19 |
67 ± 15 |
64 ± 17 |
| Median Range |
87 9–100 |
48 6–100 |
100 13–100 |
72 8–100 |
36 0–100 |
73 12–89 |
70 8–94 |
SD, standard deviation.
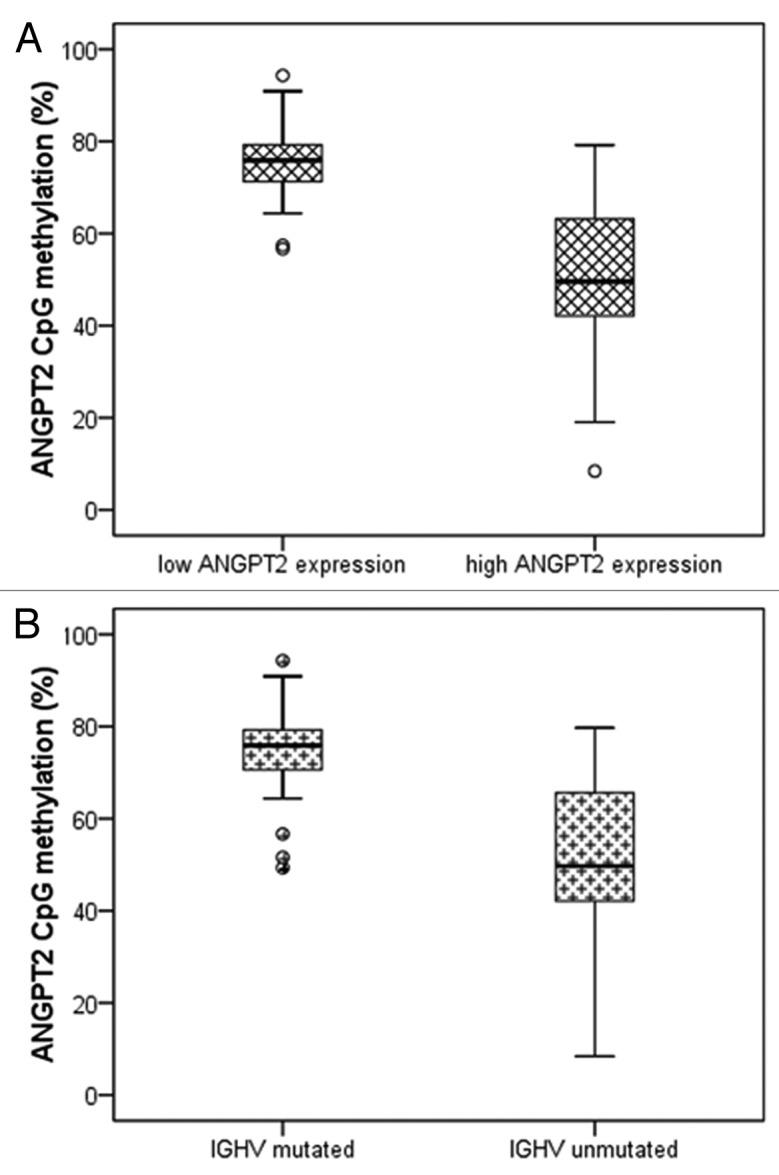
Figure 3. Relations between ANGPT2 CpG methylation, ANGPT2 mRNA expression and IGHV mutational status in 88 CLL patients. The percentage of ANGPT2 methylation (average of all 6 CpG sites) is plotted against ANGPT2 mRNA expression (A) and IGHV mutational status (B). The horizontal lines inside boxes indicate the median values. (A) CLL cases are divided in high and low ANGPT2 expression according to cut off value 2.967×10−6 (relative mRNA expression of ANGPT2 to B2M). High ANGPT2 expressing patients have significantly lower ANGPT2 methylation than low ANGPT2 expressing cases (median 50 vs. 76%; p < 0.001; Mann–Whitney test). (B) IGHV-unmutated patients have significantly lower ANGPT2 methylation than mutated cases (median 50 vs. 76%; p < 0.001; Mann–Whitney test).
Low ANGPT2 CpG methylation is associated with poor prognosis in CLL
By combining with classical CLL prognostic markers, we found a significantly lower ANGPT2 CpG methylation in IGHV-unmutated than in mutated cases (average of all 6 CpG sites; median 50% and 76%, respectively, p < 0.001) (Fig. 3B) and in CD38 positive than in CD38 negative patients (average of all 6 CpG sites, median 60% and 72%, respectively, p = 0.021).
Moreover, we used ROC analysis and Youden’s index to find the best cut-off level of ANGPT2 CpG methylation in relation to survival. The methylation percentage of 73% (average of all 6 CpG sites) was able to divide our CLL cohort in 38 “high ANGPT2 methylation” patients (43%) and 50 “low ANGPT2 methylation” patients (57%). The “low ANGPT2 methylation” subset had a significantly shorter TTFT (median 23 vs. 97 mo, p = 0.022) and OS (median 79 vs. 177 mo, p = 0.018) than the subgroup with high ANGPT2 methylation (Fig. 4A and B). In addition, low ANGPT2 methylation status was confirmed to be significantly associated with high ANGPT2 mRNA expression (p < 0.001) and IGHV unmutated status (p < 0.001) as well as with intermediate/high-risk FISH markers (p = 0.039) (Table 3).
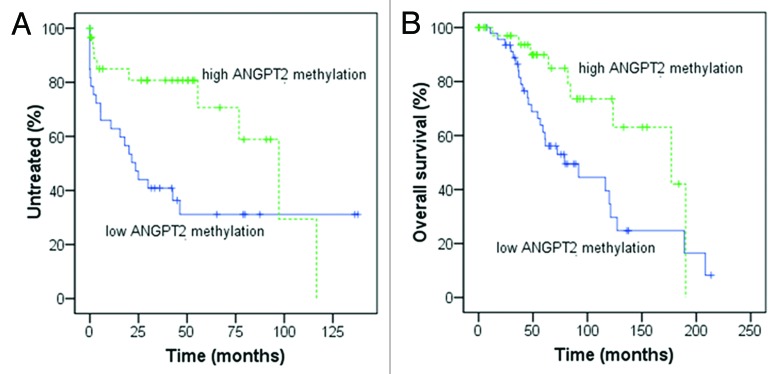
Figure 4. Kaplan-Meier curves for time to first treatment (TTFT) and overall survival (OS) in relation to ANGPT2 CpG methylation. In (A and B), 88 CLL patients were divided according to high and low ANGPT2 methylation using the cut-off value 73% (methylation percentage, average of all 6 CpG sites). Low ANGPT2 methylation cases had significantly shorter TTFT [(A) median 23 vs. 97 mo, p = 0.022; log-rank test] and OS [(B) median 79 vs. 177 mo, p = 0.018; log-rank test] compared with cases with high ANGPT2 methylation.
Table 3. Associations between ANGPT2 methylation status and CLL prognostic factors.
| Low ANGPT2 methylation (< 73%) |
p value | |
|---|---|---|
| |
N° patients (percentage) |
|
| Clinical stage |
|
|
| Binet A |
25/50 (50%) |
ns |
| Binet B-C |
15/26 (58%) |
|
| IGHV mutational status |
|
|
| Mutated (< 98%) |
15/48 (31%) |
< 0.001 |
| Unmutated (≥ 98%) |
35/40 (87%) |
|
| CD38 |
|
|
| CD38 negative (< 30%) |
28/55 (51%) |
ns |
| CD38 positive (≥ 30%) |
15/21 (71%) |
|
| FISH stratification |
|
|
| Low risk |
27/51 (53%) |
0.039 |
| Intermediate/high risk |
18/23 (78%) |
|
| ANGPT2 mRNA expression |
|
|
| Low (< 2.967×10−6) |
15/48 (31%) |
< 0.001 |
| High (≥ 2.967×10−6) | 35/40 (87%) |
Low FISH risk, no abnormalities or del(13q); Intermediate/high FISH risk, del(11q), del(17p) or trisomy 12. FISH, fluorescence in situ hybridization; IGHV, variable region of immunoglobulin heavy chain genes; ns, not statistically significant.
Finally, univariate analyses confirmed that ANGPT2 CpG methylation (cut off < 73%) is a predictor of reduced TTFT (HR 2.407; 95% CI 1.098–5.275, p = 0.028) and OS (HR 2.435; 95% CI 1.137–5.215, p = 0.022), although multivariate analyses including other CLL prognostic factors were not able to show it as an independent prognosticator.
Re-expression of ANGPT2 gene using DNA methyl inhibitor drug
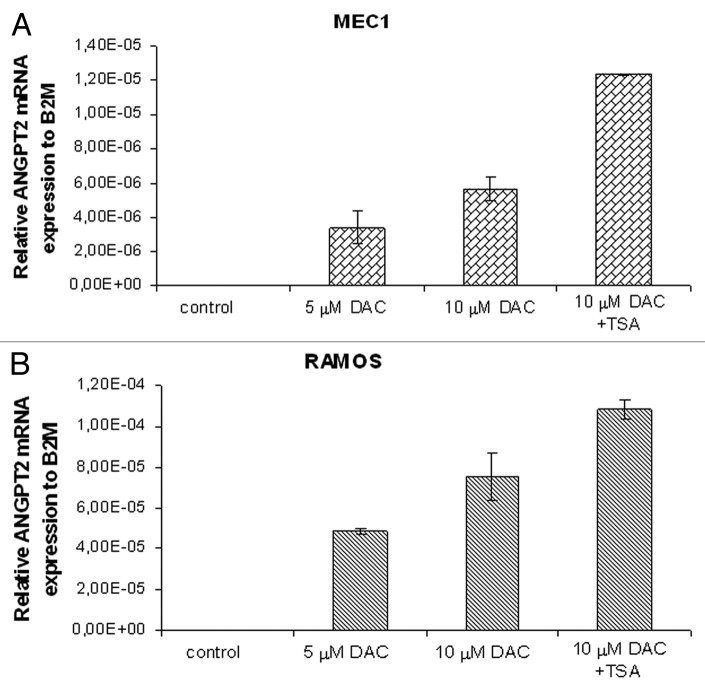
To investigate the role of DNA methylation in transcriptional silencing of the ANGPT2 gene promoter, we examined the effects of increasing concentrations of DNA methyl transferase inhibitor 5-aza-2’-deoxycytidine (DAC), followed by histone deacetylase inhibitor Trichostatin A (TSA) treatment, on ANGPT2 gene expression and promoter methylation in the CLL cell line MEC1 and Burkitt lymphoma cell line RAMOS. Indeed, an increased activation of the ANGPT2 gene was observed in both cell lines after 72 h of incubation with the increasing amounts of DAC. The highest induction of ANGPT2 mRNA expression in the MEC1 and RAMOS cells was observed with DAC treatment followed by overnight incubation of TSA (Fig. 5A and B). Consequently, the methylation percentage of the average of the 6 ANGPT2 CpG sites measured by pyrosequencing was found to proportionally decrease in both cell lines with increasing DAC concentrations (Table S3). Finally, in order to check the percentage of apoptotic cells we performed Annexin V staining for the DAC treated cells on the third day. However, our results showed that except for DAC+TSA cells, there was no significant increase in apoptosis in DAC treated cells compared with untreated cells (Table S3).
Figure 5. The relative expression of ANGPT2 gene to B2M after 72 h of DAC/TSA treatment in the MEC1 and RAMOS cell lines. Real-time PCR analyses showing re-expression of the methylated ANGPT2 gene in the MEC1 and RAMOS cells after treatment with increasing concentrations of the methyl transferase inhibitor 5-aza-2’-deoxycytidine (DAC) are represented in (A and B). In (A), mean ANGPT2 mRNA levels of MEC1 cells are 0 in control (no treatment), 3.39×10−6 with 5 μM DAC and 5.68×10−6 with 10 μM DAC. The last MEC1 sample with highest expression (1.24×10−5, mean ANGPT2 mRNA level) was treated with a combination of DAC (10 μM) and the histone deacetylase inhibitor, Trichostatin A (TSA). In (B), mean ANGPT2 mRNA levels of RAMOS cells are 0 in the control, 4.87×10−5 with 5 μM DAC and 7.54×10−5 with 10 μM DAC. The RAMOS sample with highest expression (1.08×10−4, mean ANGPT2 mRNA level) was obtained with 10 μM DAC plus TSA treatment. All data are represented in graph as mean ± standard deviation of at least 2 independent experiments.
Discussion
In order to get insights into the mechanisms behind the clinical heterogeneity in CLL, many studies have first identified genes with possible prognostic value (e.g., CD38, ZAP70 or CD49d) and then investigated their potential role in CLL pathogenesis. In this way, it has been possible to identify some of the biological pathways that are dysregulated in CLL and that determine its phenotype.4,29,30 In previous studies, we and others have shown that a more aggressive CLL phenotype is characterized by higher levels of ANGPT2, increased angiogenesis and poor prognosis.5-10 Considering the variability in ANGPT2 levels among CLL patients we here investigated the potential relation between ANGPT2 expression and promoter DNA methylation in a well-characterized CLL cohort, in order to understand if epigenetic mechanisms are involved in ANGPT2 regulation.
First, ANGPT2 mRNA levels were measured in 88 CLL patients and the value 2.967×10−6 gave the best cut off to divide CLL cases into high and low ANGPT2 expressing cases. Higher levels of ANGPT2 expression were associated with poor-prognostic factors and with shorter TTFT and OS. This is in agreement with previous studies on the prognostic impact of ANGPT2 in CLL,5-7 supporting the idea that ANGPT2 plays an important role in determining disease aggressiveness. This overexpression leading to increased angiogenesis in CLL8-10 further supports that interactions between leukemic cells and the microenvironment could represent good targets for therapeutic interventions. In fact, the use of drugs able to interfere with these interactions, such as Thalidomide and Lenalidomide, in clinical trials in CLL, is giving preliminary encouraging results,31,32 further underscoring that CLL cells strongly depend on their dialog with microenvironmental elements for their sustainment.
Second, the methylation status of the ANGPT2 gene was analyzed using the quantitative pyrosequencing method and focusing on a CpG island (including 6 CpG sites) located nearby the transcription start site. Importantly, the percentages of methylation of each CpG site and the average of all 6 CpG sites showed negative correlations with ANGPT2 mRNA expression, strongly suggesting that DNA methylation of the ANGPT2 promoter is responsible for transcriptional silencing of this gene. This tight relation between ANGPT2 expression and methylation was also conserved in purified vs. non-purified CLL samples. Considering that high-methylated CLL cases had a more favorable prognosis and the fact that normal B cells showed high levels of ANGPT2 methylation, the apparently aberrant low ANGPT2 methylation levels (and corresponding high ANGPT2 expression) observed in aggressive CLL patients indicate that these latter cases are able to circumvent the normal epigenetic control. Admittedly, the exact mechanisms behind these low levels of methylation in aggressive CLL remains elusive and to be further investigated.
Finally, the involvement of promoter methylation in regulating ANGPT2 expression was confirmed by the induction of ANGPT2 mRNA expression in two B-cell lymphoma cell lines, MEC1 and RAMOS, after treatment with methyl inhibitor drugs. The fact that increasing amounts of ANGPT2 were induced together with reducing levels of CpG methylation in both cell lines by increasing concentrations of DAC, suggests that ANGPT2 transcription is inhibited by DNA methylation in an inversely proportional way. Nevertheless, the highest increase of ANGPT2 expression was obtained with DAC treatment followed by TSA, which is a histone deacetylase inhibitor.
Aberrant gene methylation has previously been demonstrated for other molecules with prognostic and pathogenic value in CLL. For example, ZAP70, that is highly expressed in poor-prognostic CLL patients and is involved in signaling of B cell receptor, was found to be strongly methylated in low expressing patients and vice versa and the same was also observed for TWIST2, LPL and CLLU1.27,33,34 This suggest that DNA methylation plays an important role in CLL pathogenesis28,35 and targeting of such genes using epigenetic drugs could be a useful approach in CLL treatment.36
The pyrosequencing method itself, which was applied here to analyze DNA methylation status of ANGPT2, has many advantages compared with other techniques, such as bisulfite sequencing or combined bisulfite restriction analysis (COBRA) or methyl-specific PCR (MSP). This method is relatively easy to perform, requires less input of DNA and more importantly is highly reproducible and hence could be recommended in routine laboratory for directly analyzing the DNA methylation status of ANGPT2, for example, in order to discriminate low from high ANGPT2 methylation patients and helping to predict their disease course.
In conclusion, we for the first time demonstrate that the expression levels of ANGPT2 are strongly regulated by DNA methylation and that lower DNA methylation levels of the ANGPT2 promoter are associated with poor outcome in CLL. Intriguingly, more aggressive CLL cells somehow appear to “escape” the DNA methylation of the ANGPT2 promoter, potentially by inhibiting the activity of some de novo DNA methyltrasferases,37 resulting in increased expression of ANGPT2. Certainly, the high cut-off value (73%) used in our study to characterize CLL patients with high or low ANGPT2 methylation needs to be investigated further in independent CLL cohorts using similar methodology, aiming to show its significance also in multivariate analyses with other prognostic markers. Nevertheless, our novel data further supports an essential role of ANGPT2 in CLL pathogenesis and points to an important target in this as yet incurable disease.
Materials and Methods
Patients
Peripheral blood mononuclear cells (PBMCs) from 40 CLL cases collected from the Biobank at the Department of Pathology, Uppsala University Hospital and from 48 CLL patients diagnosed at the Hematology Division of Modena were included in this study. All CLL cases were diagnosed according to recently revised criteria38 and tumor samples were collected before treatment with tumor percentage of leukemic cells ≥70%. All patients provided informed consent in accordance with local institutional review board requirements and the Declaration of Helsinki Principles. The clinical characteristics of 88 CLL cases are described in Table 1.
For 8 of these 88 patients and 2 age-matched healthy donors we also collected sorted B cells (Table S2). PBMCs were incubated with CD19-specific Microbeads (Miltenyi Biotech) and separated by AutoMACS (Miltenyi Biotech), obtaining a purity >99% as assessed by flow cytometry using APC-conjugated antibody for CD19 (Miltenyi Biotech).
IGHV mutational analysis
To evaluate the mutational status of the IGHV genes expressed by CLL clones, total RNA was extracted, reverse-transcribed and IGHV-D-J rearrangement was amplified as previously described.39 Alternatively, IGHV-D-J rearrangements were amplified starting from genomic DNA as described elsewhere.40 The tumor-specific IGHV-D-J sequence was aligned to ImMunoGeneTics directories (www.imgt.cines.fr). IGHV gene sequences showing less than 98% identity compared with the corresponding germline sequence were defined as mutated.41
Immunophenotypic analyses of CD38
CD38 expression was analyzed by 3-color immunofluorescence using a FACScalibur flow cytometer (Becton Dickinson), as previously described.42 The 30% cut-off of positive cells was chosen to discriminate CD38 negative from CD38 positive CLL.
Analysis of cytogenetic aberrations
Cytogenetic abnormalities on chromosomes 11, 12, 13 and 17 were detected by interphase FISH using procedures previously described.43 The specific probes LSI-ATM, LSI-D13S319 and LSI-p53 (Vysis), directly labeled with SpectrumGreen (LSI-ATM) or SpectrumOrange (LSI-D13S319 and LSI-p53), were used for chromosomes 11, 13 and 17, respectively. An α satellite DNA probe CEP12 (Vysis), directly labeled with SpectrumGreen, was used to identify aneuploidy of chromosome 12.
ANGPT2 mRNA expression
Total RNA was extracted from PBMCs using RNeasy Mini Kit (Qiagen) and reverse-transcribed with random primers. Five nanograms per reaction of cDNA were analyzed by real-time PCR using TaqMan gene expression assays (Hs00169867_m1, Applied Biosystems) for the ANGPT2 gene, according to manufacturer’s instructions. Beta 2 microglobulin (B2M) was used as internal control (HS99999907_m1, Applied Biosystems). All samples were run in triplicate. The relative expression of ANGPT2 to B2M was calculated using the delta Ct method.
Pyrosequencing analysis of the ANGPT2 promoter
Pyrosequencing was performed to analyze the degree of methylation of 6 CpG sites composing a CpG island located inside the ANGPT2 promoter in all CLL samples. This region includes the CpG site found to have different methylation status between IGHV mutated and unmutated CLL patients in our previous methylation study.26
Genomic DNA was bisulfite converted using the EZ DNA Methylation Kit (Zymo Research) according to manufacturer’s protocol as described previously.26 The PyroMark™ software (Qiagen) was used to design pyrosequencing primers; one forward and one reverse primer for PCR amplification of the desired product (one biotin labeled in the 5′ end) and one sequencing primer. The sequenced target region is from a CpG island located in ANGPT2 promoter containing around 6 CpG sites (Fig. 2). The size of the amplified product containing the target sequence is 340 bp. In brief, bisulfite converted DNA was subjected to PCR amplification using forward primer (5′ - TGTAGGATTTATGTTGGATTTGATATTG - 3′) and reverse primer (5′ - TTCCCACTACAATCTAACAATTTACTACAT - 3′) in a 25 μL total volume. Eighteen to 20 μL of the PCR product was immobilized with 2 μL of Streptavidin Sepharose High Performance (GE Healthcare) followed by annealing with 0.3 μM of sequencing primer (5′ - ATGTTTTTTAGAAAGTTGTTATAGG - 3′) for 2 min at 80 °C. The analysis was performed using the PyroMark™ Q24 pyrosequencer instrument (Qiagen). CpG site methylation analysis was performed with the PyroMark Q24 software, and the CpG site methylation percentage was calculated for each individual CpG site in the target sequence.
DNA methyl inhibitor treatment
CLL cell line MEC144 was cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich), 4 mM glutamine and 1× penicillin/streptomycin (Invitrogen) until confluent. The same culture conditions with 20% FBS were used for the RAMOS cell line.45 Cells were then subdivided to contain approximately 1 million cells/mL per well 12 h before treatment, to allow them to adjust to conditions. Cells were subsequently cultured over 3 d in supplemented RPMI media treated with one of the following treatments: (1) medium containing 5-aza-2’-deoxycytidine (DAC; 5 μM and 10 μM, Sigma-Aldrich) for 72 h whereby medium was changed every 24 h and (2) the above samples followed by the addition of trichostatin A (TSA, 500 nM; Sigma-Aldrich) for overnight. Control cells were cultured in similar way with no drugs added. We performed at least two independent experiments for every treatment condition on both cell lines, analyzing ANGPT2 mRNA expression by RQ-PCR, ANGPT2 CpG methylation by pyrosequencing and apoptotic cell death by flow cytometry using Annexin V Alexa Fluor 488-A (Invitrogen).
Statistical analyses
Data concerning ANGPT2 mRNA expression and CpG methylation in CLL patients were analyzed using SPSS version 19.0 for Windows (SPSS). The cut-off levels were selected according to ROC analysis46 and Youden’s index,47 using survival as state variable. The Pearson Chi-square test was used to evaluate significant differences in categorized variables, whereas the distributions of continuous variables were compared using the Mann–Whitney test. The Spearman test was applied for analyzing correlations between quantitative parameters. TTFT and OS functions were estimated using the Kaplan–Meier method and curves were compared between different groups using the log-rank test. Univariate and multivariate analyses were performed using Cox models. All tests were 2-sided and the statistical significance was reached for p-value ≤ 0.05.
Supplementary Material
Acknowledgments
This work was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC IG10621-R.Mar), Milan, Italy; Programma di Ricerca di Interesse Nazionale (PRIN) 2008, Ministero dell’Università e della Ricerca (MIUR), Rome, Italy; the Swedish Cancer Society; the Swedish Research Council; the Medical Faculty of Uppsala University, Uppsala University Hospital and the Lion's Cancer Research Foundation in Uppsala, Sweden. We thank Prof. Mario Luppi for data and sample collection concerning Italian patients.
R. Marasca and R. Rosenquist supervised the study and revised the manuscript. S. Martinelli and M. Kanduri performed the laboratory research, analyzed data and wrote the paper. R. Maffei, S. Fiorcari and J. Bulgarelli collected biological samples, managed clinical data and analyzed data.
Glossary
Abbreviations:
- ANGPT2
angiopoietin-2
- CLL
chronic lymphocytic leukemia
- VEGF
vascular endothelial growth factor
- IGHV
immunoglobulin heavy chain variable genes
- TTFT
time to first treatment
- OS
overall survival
- DAC
5-aza-2’-deoxycytidine
- TSA
trichostatin A
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
These authors contributed equally as first authors.
These authors contributed equally as last authors.
Previously published online: www.landesbioscience.com/journals/epigenetics/article/24947
References
- 1.Wickremasinghe RG, Prentice AG, Steele AJ. p53 and Notch signaling in chronic lymphocytic leukemia: clues to identifying novel therapeutic strategies. Leukemia. 2011;25:1400–7. doi: 10.1038/leu.2011.103. [DOI] [PubMed] [Google Scholar]
- 2.Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–5. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mansouri L, Cahill N, Gunnarsson R, Smedby KE, Tjönnfjord E, Hjalgrim H, et al. NOTCH1 and SF3B1 mutations can be added to the hierarchical prognostic classification in chronic lymphocytic leukemia. Leukemia. 2013;27:512–4. doi: 10.1038/leu.2012.307. [DOI] [PubMed] [Google Scholar]
- 4.Gunnarsson R, Mansouri L, Rosenquist R. Exploring the genetic landscape in chronic lymphocytic leukemia using high-resolution technologies. Leuk Lymphoma. 2013 doi: 10.3109/10428194.2012.751530. [DOI] [PubMed] [Google Scholar]
- 5.Maffei R, Marasca R, Martinelli S, Castelli I, Santachiara R, Morandi E, et al. Angiopoietin-2 expression in B-cell chronic lymphocytic leukemia: association with clinical outcome and immunoglobulin heavy-chain mutational status. Leukemia. 2007;21:1312–5. doi: 10.1038/sj.leu.2404650. [DOI] [PubMed] [Google Scholar]
- 6.Maffei R, Martinelli S, Santachiara R, Rossi D, Guarnotta C, Sozzi E, et al. Angiopoietin-2 plasma dosage predicts time to first treatment and overall survival in chronic lymphocytic leukemia. Blood. 2010;116:584–92. doi: 10.1182/blood-2009-11-252494. [DOI] [PubMed] [Google Scholar]
- 7.Hüttmann A, Klein-Hitpass L, Thomale J, Deenen R, Carpinteiro A, Nückel H, et al. Gene expression signatures separate B-cell chronic lymphocytic leukaemia prognostic subgroups defined by ZAP-70 and CD38 expression status. Leukemia. 2006;20:1774–82. doi: 10.1038/sj.leu.2404363. [DOI] [PubMed] [Google Scholar]
- 8.Maffei R, Martinelli S, Castelli I, Santachiara R, Zucchini P, Fontana M, et al. Increased angiogenesis induced by chronic lymphocytic leukemia B cells is mediated by leukemia-derived Ang2 and VEGF. Leuk Res. 2010;34:312–21. doi: 10.1016/j.leukres.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 9.Kini AR, Kay NE, Peterson LC. Increased bone marrow angiogenesis in B cell chronic lymphocytic leukemia. Leukemia. 2000;14:1414–8. doi: 10.1038/sj.leu.2401825. [DOI] [PubMed] [Google Scholar]
- 10.Molica S, Vacca A, Ribatti D, Cuneo A, Cavazzini F, Levato D, et al. Prognostic value of enhanced bone marrow angiogenesis in early B-cell chronic lymphocytic leukemia. Blood. 2002;100:3344–51. doi: 10.1182/blood-2002-01-0084. [DOI] [PubMed] [Google Scholar]
- 11.Maisonpierre PC, Suri C, Jones PF, Bartunkova S, Wiegand SJ, Radziejewski C, et al. Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science. 1997;277:55–60. doi: 10.1126/science.277.5322.55. [DOI] [PubMed] [Google Scholar]
- 12.Yancopoulos GD, Davis S, Gale NW, Rudge JS, Wiegand SJ, Holash J. Vascular-specific growth factors and blood vessel formation. Nature. 2000;407:242–8. doi: 10.1038/35025215. [DOI] [PubMed] [Google Scholar]
- 13.Holash J, Maisonpierre PC, Compton D, Boland P, Alexander CR, Zagzag D, et al. Vessel cooption, regression, and growth in tumors mediated by angiopoietins and VEGF. Science. 1999;284:1994–8. doi: 10.1126/science.284.5422.1994. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt T, Carmeliet P. Angiogenesis: a target in solid tumors, also in leukemia? Hematology Am Soc Hematol Educ Program. 2011;2011:1–8. doi: 10.1182/asheducation-2011.1.1. [DOI] [PubMed] [Google Scholar]
- 15.Ribatti D, Scavelli C, Roccaro AM, Crivellato E, Nico B, Vacca A. Hematopoietic cancer and angiogenesis. Stem Cells Dev. 2004;13:484–95. doi: 10.1089/scd.2004.13.484. [DOI] [PubMed] [Google Scholar]
- 16.Moehler TM, Ho AD, Goldschmidt H, Barlogie B. Angiogenesis in hematologic malignancies. Crit Rev Oncol Hematol. 2003;45:227–44. doi: 10.1016/S1040-8428(02)00135-X. [DOI] [PubMed] [Google Scholar]
- 17.Plank MJ, Sleeman BD, Jones PF. The role of the angiopoietins in tumour angiogenesis. Growth Factors. 2004;22:1–11. doi: 10.1080/08977190310001643218. [DOI] [PubMed] [Google Scholar]
- 18.Bach F, Uddin FJ, Burke D. Angiopoietins in malignancy. Eur J Surg Oncol. 2007;33:7–15. doi: 10.1016/j.ejso.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Schliemann C, Bieker R, Thoennissen N, Gerss J, Liersch R, Kessler T, et al. Circulating angiopoietin-2 is a strong prognostic factor in acute myeloid leukemia. Leukemia. 2007;21:1901–6. doi: 10.1038/sj.leu.2404820. [DOI] [PubMed] [Google Scholar]
- 20.Xia Y, Lu RN, Li J. Angiogenic factors in chronic lymphocytic leukemia. Leuk Res. 2012;36:1211–7. doi: 10.1016/j.leukres.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 21.Neal J, Wakelee H. AMG-386, a selective angiopoietin-1/-2-neutralizing peptibody for the potential treatment of cancer. Curr Opin Mol Ther. 2010;12:487–95. [PubMed] [Google Scholar]
- 22.Leow CC, Coffman K, Inigo I, Breen S, Czapiga M, Soukharev S, et al. MEDI3617, a human anti-angiopoietin 2 monoclonal antibody, inhibits angiogenesis and tumor growth in human tumor xenograft models. Int J Oncol. 2012;40:1321–30. doi: 10.3892/ijo.2012.1366. [DOI] [PubMed] [Google Scholar]
- 23.Ye FC, Blackbourn DJ, Mengel M, Xie JP, Qian LW, Greene W, et al. Kaposi’s sarcoma-associated herpesvirus promotes angiogenesis by inducing angiopoietin-2 expression via AP-1 and Ets1. J Virol. 2007;81:3980–91. doi: 10.1128/JVI.02089-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu G, Carter WB. Human epidermal growth factor receptor 2 regulates angiopoietin-2 expression in breast cancer via AKT and mitogen-activated protein kinase pathways. Cancer Res. 2007;67:1487–93. doi: 10.1158/0008-5472.CAN-06-3155. [DOI] [PubMed] [Google Scholar]
- 25.Krug S, Huth J, Göke F, Buchholz M, Gress TM, Göke R, et al. Knock-down of Pdcd4 stimulates angiogenesis via up-regulation of angiopoietin-2. Biochim Biophys Acta. 2012;1823:789–99. doi: 10.1016/j.bbamcr.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Kanduri M, Cahill N, Göransson H, Enström C, Ryan F, Isaksson A, et al. Differential genome-wide array-based methylation profiles in prognostic subsets of chronic lymphocytic leukemia. Blood. 2010;115:296–305. doi: 10.1182/blood-2009-07-232868. [DOI] [PubMed] [Google Scholar]
- 27.Cahill N, Bergh AC, Kanduri M, Göransson-Kultima H, Mansouri L, Isaksson A, et al. 450K-array analysis of chronic lymphocytic leukemia cells reveals global DNA methylation to be relatively stable over time and similar in resting and proliferative compartments. Leukemia. 2013;27:150–8. doi: 10.1038/leu.2012.245. [DOI] [PubMed] [Google Scholar]
- 28.Cahill N, Rosenquist R. Uncovering the DNA methylome in chronic lymphocytic leukemia. Epigenetics. 2013;8:138–48. doi: 10.4161/epi.23439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dal-Bo M, Bertoni F, Forconi F, Zucchetto A, Bomben R, Marasca R, et al. Intrinsic and extrinsic factors influencing the clinical course of B-cell chronic lymphocytic leukemia: prognostic markers with pathogenetic relevance. J Transl Med. 2009;7:76. doi: 10.1186/1479-5876-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenquist R, Cortese D, Bhoi S, Mansouri L, Gunnarsson R. Prognostic markers and their clinical applicability in chronic lymphocytic leukemia: where do we stand? Leuk Lymphoma. 2013 doi: 10.3109/10428194.2013.783913. [DOI] [PubMed] [Google Scholar]
- 31.Chanan-Khan AA, Miller KC, Marshall P, Padmanabhan S, Brady W, Bernstein ZP, et al. Thalidomide (T) in combination with fludarabine (F) as initial therapy for patients (pts) with treatment naive chronic lymphocytic leukemia (CLL): preliminary results of a phase I/II clinical trial. Blood. 2005:106. doi: 10.1182/blood-2005-02-0669. [abstract 2974] [DOI] [PubMed] [Google Scholar]
- 32.Chanan-Khan AA, Miller KC, DiMiceli L, Padmanabhan S, Lawrence D, Bernstein ZP, et al. Results of phase II study of lenalidomide (L) {Revlimid(R)} in patients with relapsed or refractory chronic lymphocytic leukemia (CLL) Blood. 2005:106. [abstract 447] [Google Scholar]
- 33.Amin S, Walsh M, Wilson C, Parker AE, Oscier D, Willmore E, et al. Cross-talk between DNA methylation and active histone modifications regulates aberrant expression of ZAP70 in CLL. J Cell Mol Med. 2012;16:2074–84. doi: 10.1111/j.1582-4934.2011.01503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raval A, Lucas DM, Matkovic JJ, Bennett KL, Liyanarachchi S, Young DC, et al. TWIST2 demonstrates differential methylation in immunoglobulin variable heavy chain mutated and unmutated chronic lymphocytic leukemia. J Clin Oncol. 2005;23:3877–85. doi: 10.1200/JCO.2005.02.196. [DOI] [PubMed] [Google Scholar]
- 35.Rahmatpanah FB, Carstens S, Hooshmand SI, Welsh EC, Sjahputera O, Taylor KH, et al. Large-scale analysis of DNA methylation in chronic lymphocytic leukemia. Epigenomics. 2009;1:39–61. doi: 10.2217/epi.09.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Smet C, Loriot A. DNA hypomethylation and activation of germline-specific genes in cancer. Adv Exp Med Biol. 2013;754:149–66. doi: 10.1007/978-1-4419-9967-2_7. [DOI] [PubMed] [Google Scholar]
- 37.Palamarchuk A, Yan PS, Zanesi N, Wang L, Rodrigues B, Murphy M, et al. Tcl1 protein functions as an inhibitor of de novo DNA methylation in B-cell chronic lymphocytic leukemia (CLL) Proc Natl Acad Sci U S A. 2012;109:2555–60. doi: 10.1073/pnas.1200003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Döhner H, et al. International Workshop on Chronic Lymphocytic Leukemia Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–56. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marasca R, Maffei R, Morselli M, Zucchini P, Castelli I, Martinelli S, et al. Immunoglobulin mutational status detected through single-round amplification of partial V(H) region represents a good prognostic marker for clinical outcome in chronic lymphocytic leukemia. J Mol Diagn. 2005;7:566–74. doi: 10.1016/S1525-1578(10)60589-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ricca I, Rocci A, Drandi D, Francese R, Compagno M, Lobetti Bodoni C, et al. Telomere length identifies two different prognostic subgroups among VH-unmutated B-cell chronic lymphocytic leukemia patients. Leukemia. 2007;21:697–705. doi: 10.1038/sj.leu.2404544. [DOI] [PubMed] [Google Scholar]
- 41.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–54. [PubMed] [Google Scholar]
- 42.Del Poeta G, Maurillo L, Venditti A, Buccisano F, Epiceno AM, Capelli G, et al. Clinical significance of CD38 expression in chronic lymphocytic leukemia. Blood. 2001;98:2633–9. doi: 10.1182/blood.V98.9.2633. [DOI] [PubMed] [Google Scholar]
- 43.Del Principe MI, Del Poeta G, Venditti A, Buccisano F, Maurillo L, Marini R, et al. Clinical significance of soluble p53 protein in B-cell chronic lymphocytic leukemia. Haematologica. 2004;89:1468–75. [PubMed] [Google Scholar]
- 44.Stacchini A, Aragno M, Vallario A, Alfarano A, Circosta P, Gottardi D, et al. MEC1 and MEC2: two new cell lines derived from B-chronic lymphocytic leukaemia in prolymphocytoid transformation. Leuk Res. 1999;23:127–36. doi: 10.1016/S0145-2126(98)00154-4. [DOI] [PubMed] [Google Scholar]
- 45.Klein G, Giovanella B, Westman A, Stehlin JS, Mumford D. An EBV-genome-negative cell line established from an American Burkitt lymphoma; receptor characteristics. EBV infectibility and permanent conversion into EBV-positive sublines by in vitro infection. Intervirology. 1975;5:319–34. doi: 10.1159/000149930. [DOI] [PubMed] [Google Scholar]
- 46.Metz CE. Basic principles of ROC analysis. Semin Nucl Med. 1978;8:283–98. doi: 10.1016/S0001-2998(78)80014-2. [DOI] [PubMed] [Google Scholar]
- 47.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–5. doi: 10.1002/1097-0142(1950)3:1<32::AID-CNCR2820030106>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.