Summary
In blood, transcription factor C/EBPa is essential for myeloid differentiation and has been implicated in regulating self-renewal of fetal liver (FL) hematopoietic stem cells (HSCs). However, its function in adult HSCs has remained unknown. Here, using an inducible knockout model we found that C/EBPa deficient adult HSCs underwent a pronounced expansion with enhanced proliferation, characteristics resembling FL HSCs. Consistently, transcription profiling of C/EBPa deficient HSCs revealed a gene expression programme similar to FL HSCs. Moreover we observed that age-specific C/EBPa expression correlated with its inhibitory effect on HSC cell cycle. Mechanistically we identified N-Myc as C/EBPa downstream target, and loss of C/EBPa resulted in de-repression of N-Myc. Our data establish C/EBPa as a central determinant in the switch from fetal to adult HSCs.
Keywords: Transcription factor, C/EBPa, hematopoietic stem cell, quiescence, proliferation, N-Myc
Introduction
Hematopoiesis is maintained by a small group of multipotent hematopoietic stem cells (HSCs), which can rapidly divide and have an ability to remain quiescence. This ensures appropriate numbers of HSCs to sustain a sufficient supply of mature blood cells throughout lifetime. During early development, fetal HSCs undergo rapid self-renewal divisions, which lead to a massive expansion of the HSC pool to meet the demands of adult hematopoiesis1,2. By contrast, adult HSCs divide infrequently with the majority of cells in a quiescent G0 phase, and are maintained at relatively constant cell numbers3-7. New evidence now shows that the proliferation of HSCs continues until three weeks post-birth, afterwards HSCs undergo an abrupt change in their cycling activity and become quiescent at the end of the 4th week8. This process is also accompanied by alternations in differentiated cell output and self-renewal capacity of HSCs1,9,10. Interestingly, these dramatic and coordinated changes in biological properties correlate with changes in HSC gene expression10,11. It is clear that fetal and adult HSCs are sustained by different transcriptional programs. For instance, transcription factor Sox17 is required for the maintenance of fetal and neonatal, but not adult HSCs11. By contrast, Bmi-112, Gfi-113 and Etv614 are needed for self-renewal of adult, but not fetal HSCs. However, the precise molecular circuitry that actually determines the transition of HSC properties during development remains unknown.
C/EBPa is a member of the basic leucine zipper transcription factor family, which regulates cell-cycle exit and differentiation in various tissues15-17. In hematopoietic system, previous studies have established a critical, non-redundant role of C/EBPa in granulopoiesis. Disruption of C/EBPa blocks the transition from the common myeloid progenitors to the granulocyte/monocyte progenitor, thereby leading to the loss of mature granulocytes18,19. In addition, we have previously reported that C/EBPa negatively regulates fetal HSCs self-renewal, as C/EBPa null FL HSCs exhibit enhanced repopulation activity in competitive transplantation assays, whereas their total number and cell cycle kinetics are not affected19. C/EBPa also appears to be crucial in leukemogenesis, as C/EBPa silencing or mutations leading to reduced activity are frequently found in human acute myeloid leukemia patients20,21. However, despite the significance of C/EBPa in fetal hematopoiesis and human leukemia, the physiological role of C/EBPa in adult hematopoiesis, especially in adult HSC biology, has remained unclear.
In this report, using a C/EBPa conditional knockout mouse model that circumvents the perinatal mortality of the conventional knockout mice, we demonstrate that loss of C/EBPa in adult HSCs confers fetal HSC characteristics, including enhanced proliferation, increased number of functional long term HSCs (LT-HSCs), advanced repopulating ability as well as altered transcriptional profile that resembles fetal HSCs. Furthermore, we identified N-Myc as a downstream target of C/EBPa and showed that transcriptional repression of N-Myc by C/EBPa is at least partially required for acquiring and maintaining adult quiescence of HSCs.
Results
Phenotypic LT-HSCs increase in adult C/EBPa deficient mice
To address whether C/EBPa is involved in adult HSC function, we bred C/EBPaloxP/loxP mice with transgenic Mx1-Cre mice to allow IFN-inducible C/EBPa excision in adult hematopoietic cells. Administration of three doses of poly(inosinic acid) poly(cytidylic acid) [pIpC] in Mx1-Cre+C/EBPaloxP/loxP mice induced the excision of C/EBPa in almost all c-Kit+Sca-1+Lineage- hematopoietic stem and multipotent progenitor cells (HSPCs, also termed as KSLs) four days after the last injection (hereafter referred to as KO, Figure S1a and S1b).
Phenotypic examination of HSCs by flow cytometry showed a ~70-fold increase in the frequency of KSLs in KO bone marrow 5-7 days after pIpC treatment (Figure 1a and 1b). Despite the significant reduction in bone marrow cellularity (Figure 1c), the total number of KSLs increased nearly 12-fold in the bone marrow of KO mice (Figure 1d). KSLs were further analyzed using the combination of CD150 and CD48 (SLAM code) for HSC identification. CD150+CD48-KSLs (hereafter referred to as SLAM+) are enriched for LT-HSCs, while CD48+KSLs contain short-term repopulating HSCs (ST-HSCs) and multipotent progenitors (MPPs)22,23. C/EBPa excision resulted in a decreased percentage of SLAM+ cells within the KSL compartment of KO mice (Figure 1a and 1b); however, the total number of SLAM+KSLs was increased by ~2-fold (Figure 1e). Taken together, loss of C/EBPa led to an expansion of phenotypic LT-HSCs, despite of a disproportional expansion of ST-HSC/MPPs (Figure S1c).
Figure 1.
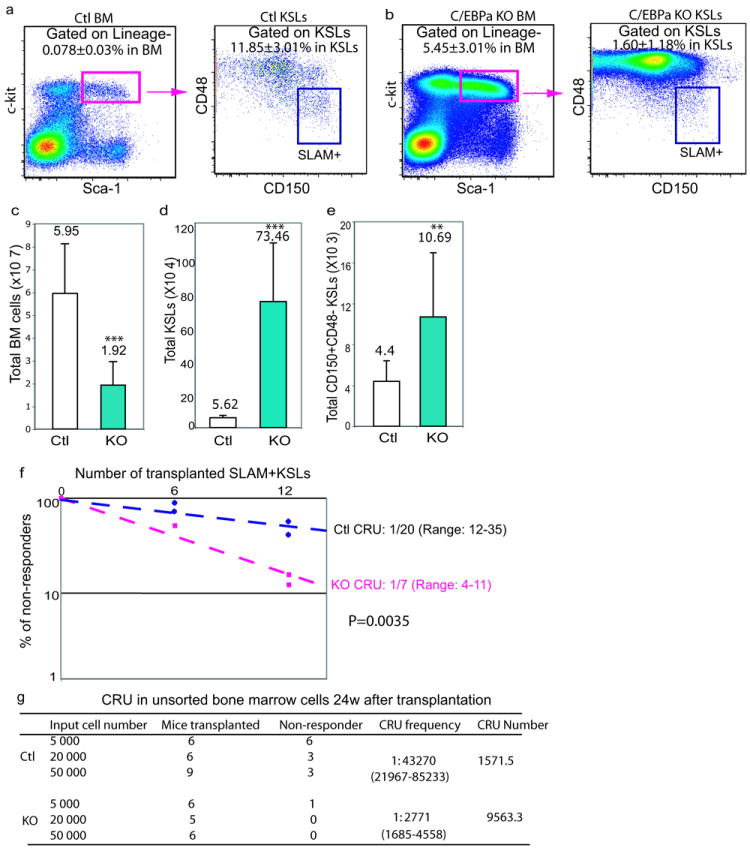
Loss of C/EBPa increases the number of phenotypic and functional hematopoietic stem cells (HSCs)
(a, b) Increased frequency of Lineage-c-kit+Sca-1+ cells (KSLs), but decreased frequency of HSC-enriched CD150+CD48- (SLAM+) in the KSL fraction in C/EBPa conditional knockout (KO) mouse bone marrow. Representative FACS plots analyzing bone marrow cells 5-7 days after last pIpC injections are shown for control (Ctl, a) and KO (b) mice. Numbers in plots indicate average percentages of boxed populations (±SD) among total bone marrow cells or gated populations (p<0.005, data pooled from four independent experiments to obtain n=15 per genotype).
(c-e) Total numbers of bone marrow cells (c), KSLs (d), SLAM+KSLs (e) in the bone marrow (2 femurs, tibias and humerus) of Ctl and KO mice; mean values (±SD) are shown (**p<0.01 and ***p<0.005, data pooled from four independent experiments to obtain n=15 per genotype).
(f, g) Loss of C/EBPa increases the number of functional HSCs in adult mice. Limiting-dilution competitive repopulation analyses using either two doses (6 and 12 cells) of sorted SLAM+KSLs (f) or three doses (5000, 20000, and 50000 cells) of whole bone marrow cells (g). Non-responders are defined as recipients with less than 0.3% donor-derived myeloid and/or lymphoid cells in nucleated peripheral blood cells. The frequency of functional HSCs (competitive repopulation units, CRU) was calculated according to Poisson statistics using L-Calc software (P<0.005). f, plotted is the percentage of non-responders 24 weeks after transplantation versus the number of initial SLAM+KSLs from two independent experiments. g, the frequency and the total number of CRU in bone marrow per mouse were measured and calculated.
See also Figure S1 and table S6 for raw values for figure S1.
Functional LT-HSCs are increased in C/EBPa deficient mice
We next asked whether the increased number of phenotypic SLAM+ LT-HSCs in KO mice correlated with an increase in functional LT-HSCs. Limiting dilution transplantation assays were performed to measure the frequency of competitive repopulation units (CRU)24, which is a reflection of the number of functional HSCs. Analysis of recipients received either control or KO SLAM+KSLs revealed a ~2-fold increase in the frequency of multipotent repopulating cells in KO (Figure 1f) and a 4-5 fold increase in the number of functional LT-HSCs per mouse, when normalized to the total number of SLAM+KLSs (Figure 1e). The increased frequency and total number of functional LT-HSCs in KO mice were further confirmed by transplantation of unsorted whole bone marrow cells. Analysis of recipient animals revealed a 14-fold increase in the frequency of multipotent repopulating cells in KO mice and consequently a ~5-fold increase in the number of CRU per mouse (Figure 1g). The presence of B and T cells, but the lack of donor-derived myeloid cells in bone marrow and peripheral blood, confirmed the reconstitution by KO cells (data not shown). Collectively, these results suggest that loss of C/EBPa leads to a significant expansion of functional LT-HSCs, which are highly enriched in the SLAM+ KSL compartment.
Expansion of HSPC pool is cell intrinsic
To examine whether the expansion of KO HSPCs was hematopoietic cell intrinsic effect, we transplanted bone marrow cells from untreated Mx1-Cre+ C/EBPaloxP/loxP mice into lethally irradiated Mx1-Cre- C/EBPaloxP/loxP mice. In addition, we performed the reciprocal transplants of Mx1-Cre- C/EBPaloxP/loxP bone marrow into Mx1-Cre+ C/EBPaloxP/loxP recipients. C/EBPa excision was induced four months after bone marrow reconstitution (Figure 2a). A significant expansion of KSLs was recapitulated in the recipients with C/EBPa excision exclusively in the hematopoietic compartment, but not in those with C/EBPa excision in the bone marrow environment (Figure 2b, 2c and S2a). Therefore, KSL expansion in KO mice is a hematopoietic cell intrinsic event.
Figure 2.
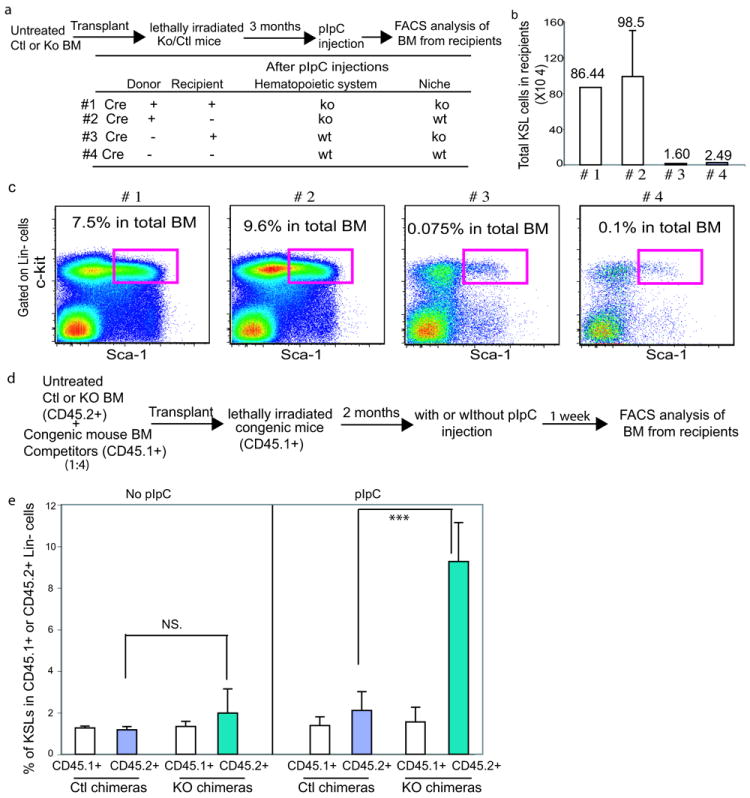
HSC expansion upon loss of C/EBPa is hematopoietic cell intrinsic
(a-c) Bone marrow chimeras. 2×106 total bone marrow cells from either Mx.1-Cre- C/EBPaloxP/loxP or Mx.1-Cre+ C/EBPaloxP/loxP mice were transplanted into lethally irradiated Mx.1-Cre+ C/EBPaloxP/loxP or Mx.1-Cre- C/EBPaloxP/loxP recipients prior to pIpC injections. After stable engraftment (3 months), pIpC was administered to induce deletion. The chimeras were analyzed 2 weeks after the last injection. (a) experimental outline; (b) total number of KSLs in the chimeric mouse bone marrow; mean values (±SD) are shown (n=2 mice for group #1, 3 and 4, and n=3 mice for group #2); and (c) representative FACS analyses of lineage-negative bone marrow cells.
(d, e) Competitive bone marrow chimeras. Untreated Mx.1-Cre- C/EBPaloxP/loxP and Mx.1-Cre+ C/EBPaloxP/loxP bone marrow cells were mixed with CD45.1+ wild type congenic mouse bone marrow at a 1:4 ratio and transplanted into lethally irradiated CD45.1+ recipients. C/EBPa excision was induced 2 months after transplantation by pIpC injection and mice were analyzed 7 days after the last injection. (d) Experiment outline, and (e) Percentages of KSL cells in either CD45.1+ Lin- or CD45.2+ Lin- population in bone marrow chimeras. Values are means (±SD) (n=3 mice for each chimera group, ***p<0.005; NS stands for no statistical significance).
See also Figure S2 and Table S6 for the raw data for panels 2b and 2e.
We also observed that C/EBPa excision led to severe bone marrow cytopenia due to a myeloid differentiation block19. This raises the possibility that the expansion of KO HSPCs is a consequence of either the significant loss of hematopoietic cells or the lack of a myeloid population, an important component of the “hematopoietic environment”. To examine this, lethally irradiated CD45.1+ congenic recipients were reconstituted with undeleted CD45.2+ Mx1-Cre+C/EBPaloxP/loxP or Mx1-Cre- C/EBPaloxP/loxP bone marrow cells mixed with CD45.1+ congenic bone marrow cells at 1:4 ratio. Two months after transplantation, mice were treated with pIpC (Figure 2d). Assessment of the bone marrow one week later revealed less than 10% reduction of bone marrow cellularity and a ~15% decrease in Mac1+Gr1+ myeloid cells in the KO chimera mice (data not shown). Despite these minor changes in cell number, we found that the frequency of donor-derived KSLs in Lin- population consistently displayed a more than 5-fold increase in the KO chimeras, compared to control chimeras (Figure 2e and Figure S2b). Moreover, CD45.1+ wild type congenic competitors in KO chimeras showed little difference in KSL frequency compared to congenic competitors in the control chimera (Figure 2e and S2b). These results demonstrate that the expansion of KSLs observed in KO mice is not the consequence of cell loss or the lack of mature myeloid cells. Collectively, the expansion of KSLs is an intrinsic and HSC specific effect of C/EBPa deletion.
Loss of quiescence of adult HSCs upon C/EBPa excision
C/EBPa has previously been reported as a cell cycle inhibitor15, we therefore tested if loss of C/EBPa increased the number of SLAM+ LT-HSCs via enhancing cell cycle activity. SLAM+KSL cells were stained with a combination of the DNA and RNA dyes Hoechst 33342 and Pyronin Y to distinguish cells in G0, G1, and S-G2/M cell cycle phases. Interestingly, we observed a 2~3-fold increase of cells in G1 and G2-S/M phases 5 days after pIpC treatment (Figure 3a and b). The 5 days interval was chosen to avoid secondary effects of pIpC induced INFa on HSC proliferation, as the transient effect of INFa on HSC proliferation diminishes 4 days after injections25.
Figure 3.
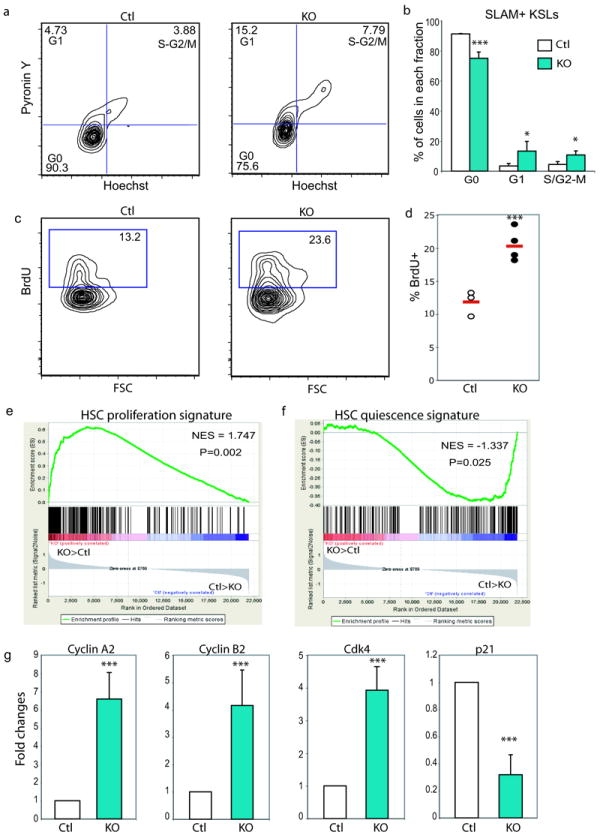
Loss of C/EBPa increases HSC proliferation
(a) Pyronin Y/Hoechst staining showing cell cycle status of SLAM+KSLs in control and KO mice 5 days after pIpC injections.
(b) Distribution of SLAM+KSLs in G0, G1 and S/G2-M phases. Mean values (±SD) are shown (n=3 independent mice for each group, ***p<0.005 and *p<0.05).
(c) Representative FACS plots demonstrating BrdU incorporation in donor derived SLAM+KSLs 20 hours after BrdU injection in reconstituted chimera mouse bone marrow four months after pIpC administration (***p<0.005).
(d) Percentage of BrdU+ cells in donor-derived SLAM+KSLs sorted from control and KO chimeras (***p<0.005).
(e, f) GSEA comparison of KO and control SLAM+KSLs for enrichment/depletion of HSC proliferation-associated gene expression (e) and quiescence associated gene expression (f). The normalized enrichment scores (NES) and p values are indicated in each plot.
(g) qPCR analysis of the expression of selected cell cycle related genes that were differentially expressed by KO versus control SLAM+ KSLs revealed by microarray analysis. The results shown are the relative expression levels and expressed as fold difference compared to the levels (set to 1) detected in control SLAM+ KSLs. The data are the averages ± SD for n=3 independently sorted SLAM+ KSLs per group two weeks after pIpC injections. The average for each sample was calculated from duplicate measurements. (***p<0.005, gapdh normalization).
See also Figure S3 and table S6 for the raw data for panels 3b, 3g, S3b and S3d.
C/EBPa KO mice started to die 10 days following pIpC treatment due to sepsis as a result of granulocytopenia19. To determine the long-term impact of C/EBPa deficiency on HSC proliferation, we created bone marrow chimeric mice as described in Figure 2d. C/EBPa deletion was induced in reconstituted mice 4 month post-transplant and bone marrow cells were analyzed 4 months later. Assessment of cellular proliferation by BrdU incorporation assay in these chimeras revealed persistently increased proliferation of HSCs in the absence of C/EBPa (Figure 3c and d). Of note, the C/EBPa null cell graft was significantly increased after pIpC injection, as ~80% of SLAM+KSLs were derived from KO in KO chimeras, compared to ~20% from control in control chimeras (Figure S3a and b). We further examined the reconstitution ability of KO cells in serial transplantation. The primary transplants as described above were treated with pIpC and two weeks later 2×106 unfractionated whole bone marrow cells were isolated and transplanted into irradiated CD45.1+ recipient mice. Four months after transplantation, we observed a dramatic expansion of KSLs with ~90% of them being KO in mice received the KO chimera bone marrow. In contrast, mice received the control chimera bone marrow retained stable chimerism in the KSL compartment (Figure S3c and d). These results indicate an enhanced competitive repopulation activity of proliferating KO cells.
To further uncover the molecular basis for enhanced HSC proliferation in C/EBPa deficient mice, we performed Affymetrix-based global gene expression analysis on SLAM+KSLs sorted from KO and control mice 7 and 21 days (only 4 out of 13 KO mice survived on day 21) after pIpC injections. In line with the functional cell cycle analysis, gene set enrichment analysis (GSEA) revealed that the expression of proliferation-associated genes was significantly enriched in KO SLAM+KSLs on both day 7 and day 21 (Figure 3e and Table S1), whereas the expression of quiescence-associated genes was dramatically depleted in KOs (Figure 3F and Table S2)26. Quantitative PCR analysis confirmed that C/EBPa KO SLAM+KSLs exhibited significantly increased expression of mitotic cyclin A2, cyclin B2 and cyclin dependent kinase 4 (cdk4), and decreased expression of cell-cycle inhibitor p21, as compared to control (Figure 3g).
In contrast to the cell cycle alterations, no obvious difference in apoptosis was detected between KO and WT cells (Figure S3e), suggesting C/EBPa having a role in maintaining adult quiescence, but not in survival of HSCs.
Loss of C/EBPa leads to transcriptional alterations that resemble fetal liver (FL) HSCs
Although the observed enhanced proliferation, yet expanded HSC pool and advanced reconstitution ability of KO HSCs are unusual phenomena for adult HSCs, they are hallmarks of FL HSCs. This raises a possibility that loss of C/EBPa might restore a fetal transcriptional program in adult HSCs. We then compared the transcriptomes of adult KO, control HSCs, and E15.5 FL HSCs. Unsupervised clustering analysis of differentially expressed genes (DEG) between KO and control showed that KO adult SLAM+KSLs coclustered with FL HSCs, with a distinguishable distance to control adult HSCs (Figure 4a). Among 1551 DEGs between KO and control, 1001 genes overlapped with the 1934 DEGs identified by FL and adult control HSC comparison (1.5-fold change-filtered, P < 0.05). Of note, the vast majority of these overlapped genes (950 out of 1001) over-express or repress accordingly in FL HSCs and KO HSCs, as compared to control HSCs (Figure 4b and Table S3). Moreover, GSEA revealed that the expression of FL HSC associated genes27 was significantly enriched in KO adult HSCs, compared to control adult HSCs (Figure 4c and Table S4). Thus, C/EBPa deletion in adult HSCs leads to the restoration of the gene expression signature of FL HSCs.
Figure 4.
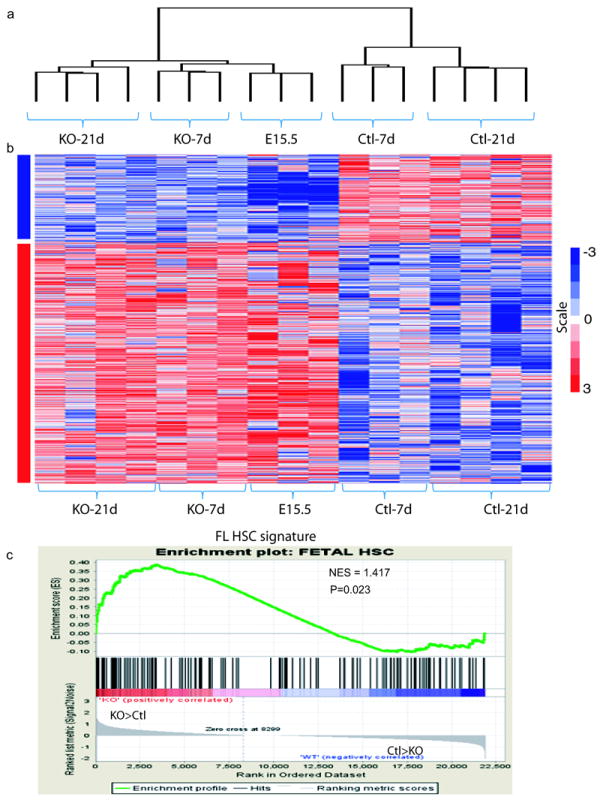
Loss of C/EBPa in adult HSCs results in transcriptional alterations that resemble fetal liver (FL) HSCs
(a) Unsupervised clustering of control, KO bone marrow HSCs (7 days and 21 days after pIpC injection) and E15.5 FL HSCs using differentially expressed genes (DEG) between control and KO.
(b) The heat map represents gene signature shared by KO adult HSCs and E15.5 FL HSCs, but distinct from that of control adult HSCs (1.5 change-filtered, P < 0.05).
(c) GSEA comparison of KO and control adult HSCs for enrichment/depletion of FL HSC associated gene expression. The normalized enrichment scores (NES) and p values are indicated on each plot.
See also Table S1.
Increased levels of C/EBPa correlate with the acquisition of adult quiescence in HSCs
Transition from actively dividing fetal HSCs to quiescent adult HSCs occurs 3-4 weeks after birth8. Since C/EBPa KO HSCs failed to maintain properties of adult HSCs and yet acquired various characteristics of FL HSCs, we then asked whether C/EBPa regulates the developmental transition of HSC proliferation properties. First, we examined C/EBPa expression in HSCs using qPCR and observed that levels of C/EBPa increased during development (Figure 5a). Therefore, C/EBPa expression is inversely correlated with HSC proliferation 8. Moreover, there was a sharp 2-fold increase in C/EBPa expression in HSCs from 4-week old mice, compared to HSCs from 2-week old mice. Notably, this is consistent with the time at which HSCs switch to the non-proliferative state 8. Next, we asked whether levels of C/EBPa in HSCs correlate with inhibition of HSC proliferation. We excised C/EBPa in 1.5-week old and 4.5-week old C/EBPa conditional KO mice, respectively. Assessment of proliferation by BrdU revealed that while there was a nearly 2-fold increase in the percentage of BrdU+ SLAM+KSLs in 4.5-week KO mice (Figure 5c); no significant increase was observed in 1.5-week old mice (Figure 5b). qPCR analysis also confirmed efficient excision of C/EBPa in these cells (data not shown). Last, we asked if over-expression of C/EBPa in fetal HSCs is sufficient to induce the switch from the proliferative to the quiescent state. Mac1low KSLs isolated from FLs were transduced with either MSCV-GFP-C/EBPa (MIG-C/EBPa) or MIG control virus. GFP+c-kit+ cells were isolated 48 hours later and subjected to cell cycle analysis. Strikingly, FL KSLs over-expressing C/EBPa demonstrated a more than 7-fold increase of cells in the quiescent G0 phase, compared to cells infected with control vector, indicating a significantly decreased proliferation (Figure 5d and Figure S4). Together, these data suggest that increased C/EBPa expression contributes to the acquisition of adult HSC quiescence during ontogeny.
Figure 5.
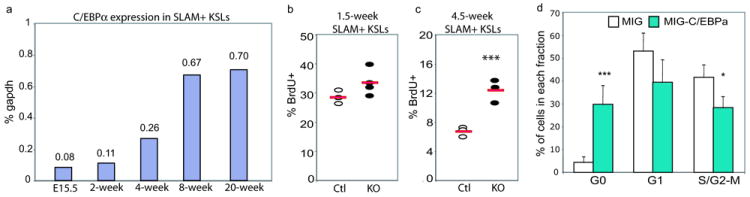
Up-regulation of C/EBPa limits HSC proliferation
(a) qPCR analysis showing levels of C/EBPa in SLAM+KSLs from E15.5 fetal liver (Mac-1low SLAM+KSLs), bone marrow of 2-week old mice (Mac-1low SLAM+KSLs) and 4-, 8- and 20-week old mice (Mac-1- SLAM+KSL), respectively. Mean value of duplicate measurements of C/EBPa levels relative to gapdh from one representative experiment are shown in the plot. Data show result of one of three independent experiments.
(b, c) Percentage of BrdU+ cells in SLAM+ KSLs from 1.5-week old (b) or 4.5-week old control and KO mice (c) 14 hours after Brdu incorporation (***p<0.005). Data are obtained from two independent experiments.
(d) Over-expression of C/EBPa decreased fetal liver KSL proliferation. Percentage of GFP+c-kit+ FL KSLs in G0, G1 and S/G2-M phase 48 hours following infection with MSCV-GFP-C/EBPa (MIG-C/EBPa) or MIG control virus, measured by Pyronin/Hoechest staining. Mean value (±SD) are shown (n=3 of independent FL KSL samples for each group, data collected over two experiments: one sample coming from one experiment and 2 from the other experiment, ***p<0.005, *p<0.05).
See also Figure S4 and table S6 for the raw data for panels 5a and 5d.
N-Myc is a direct target of C/EBPa and mediates its function in the fetal to adult switch of HSC proliferative properties
To identify potential C/EBPa downstream pathways involved, enriched pathway analysis using Ingenuity Systems Pathway Analysis (IPA) was performed on genes that were differentially expressed upon excision of C/EBPa in HSCs. In line with the increased proliferation observed, gene sets and pathways that associate with cell cycle, cytoskeleton organization and HSC self-renewal (such as TGFb and canonical Wnt signaling) were significantly overrepresented in KO HSCs (Figure 6a and Table S5). N-Myc target gene set was among the pathways and gene sets that were most significantly altered. N-Myc is a member of the MYC transcription factor family. Although elimination of either N-Myc or C-Myc in adult mice showed no effect on HSC proliferation, loss of both genes resulted in significantly decreased proliferation, suggesting either redundancy and/or a dosage effect of Myc proteins on HSC proliferation28.
Figure 6.
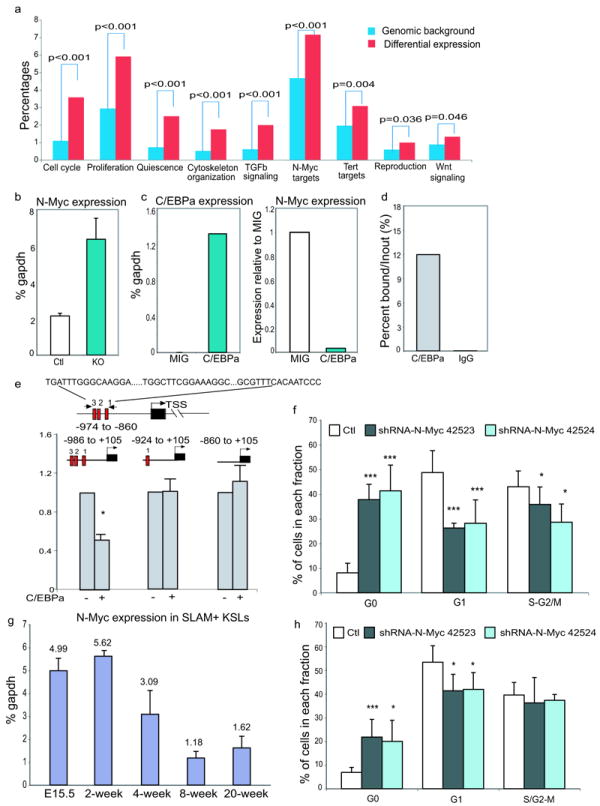
N-Myc is a downstream target of C/EBPa that mediates its regulation of HSC proliferation during fetal to adult transition
(a) Pathway analysis indicating enriched gene sets and pathways downstream of C/EBPa. Percentages in the y axis represent either the percentage of the number of genes in the pathway of interest relative to the total number of genes (Genomic background, blue bars), or the number of differentially expressed genes in the pathway of interest to all differentially expressed genes (Differential expression, red bars).
(b) Up-regulation of N-Myc expression in KO SLAM+KSLs measured by qPCR. Results are shown as mean (±SD) of triplicate measurements of relative mRNA in one representative experiment (normalized to gapdh). Data show results of one of three independent experiments.
(c) Re-introduction of C/EBPa into C/EBPa KO KSLs reduces N-Myc expression. Results from one representative experiment are shown. Left, mean value of duplicate measurements of C/EBPa; right, mean value of the relative N-Myc expression in MIG-C/EBPa infected KO KSLs, with the levels in MIG-infected cells set to 1. Data show results of one of two independent experiments.
(d) ChIP-qPCR analysis confirms specific binding of CEBPa to the N-Myc promoter in hematopoietic stem/progenitor cells (Lineage- c-kit+). Results are shown as mean value of duplicate measurements. Data show results of one of two independent experiments.
(e) Upper, schematic diagram showing three potential C/EBPa binding sites within the N-Myc proximal promoter. Red bars represent consensus C/EBPa binding sites and black bar represents N-Myc gene. lower, reporter assays comparing transcriptional activity of reporter constructs with full length or truncated N-Myc promoters upon the addition of C/EBPa in HEK293 cells. Mean value (±SD) of triplicate measurements of one of two independent experiments are shown.
(f) shRNA-mediated knocking-down of N-Myc in adult C/EBPa-deficient KSLs decreased their proliferation as measured by Pyronin/Hoechst staining. Results are shown as mean value (±SD) (n=7 independent samples for each group, pooled over three experiments).
(g) qPCR of N-Myc in SLAM+KSLs. Results are shown as mean (±SD) of triplicate measurements of N-Myc transcripts in one of two independent experiments.
(h) shRNA-mediated knocking down of N-Myc in FL KSLs decreased their proliferation. Results are shown as mean value (±SD) (n=4 independent samples per group, data pooled over two experiments, ***p<0.005 and *p<0.05).
See also Figure S5 and table S6 for the raw data for panels 6b, 6c, 6d, 6e, 6g, 6h and S5a.
To investigate whether N-Myc mediates the enhanced proliferation of KO HSCs, we first confirmed the increase of N-Myc mRNA in KO SLAM+KSLs by qPCR analysis (Figure 6b). To test whether up-regulation of N-Myc in HSCs is specifically caused by the loss of C/EBPa, we examined N-Myc expression in KO KSLs upon the restoration of C/EBPa expression and observed more than 90% down-regulation of N-Myc expression (Figure 6c). Sequence analysis of the N-Myc promoter revealed three putative C/EBPa binding sites within ~1 kb of the proximal promoter region (Figure 6e). Combined with the rapid down-regulation of N-Myc, this raised the possibility that C/EBPa modulates N-Myc expression through direct transcriptional repression. Chromatin immunoprecipitation (ChIP) in Lin-c-kit+ bone marrow cells clearly showed binding of C/EBPa to endogenous N-Myc promoter (Figure 6d). To further investigate how C/EBPa suppresses N-Myc transcription, we performed luciferase reporter assays with either the wild-type N-Myc promoter or its truncated mutants (Figure 6e). The luciferase activity of the construct containing all three binding sites was repressed by C/EBPa expression. Deletion of the two distal binding sites abolished the repression, suggesting that integrity of these consensus sites in the N-Myc promoter is required for C/EBPa-mediated suppression (Figure 6e). Collectively, we concluded that C/EBPa directly represses N-Myc transcription by binding to the proximal region of the N-Myc promoter.
To determine whether increased expression of N-Myc is responsible for the enhanced proliferation of adult KO HSCs, we knocked down its expression in KO KSLs using lentivirally expressed shRNAs. Among the three shRNAs tested, N-Myc shRNA 42523 and 42524 showed a marked knocking down and were used in subsequent studies (Figure S5a). Knocking down N-Myc by shRNA in KO KSLs led to a 4-fold increase in the frequency of cells in quiescent G0 phase, compared with cells infected with lentivirus carrying control luciferase shRNAs (Figure 6f and Figure S5b). In contrast, knocking-down N-Myc in control adult KSLs showed no such effect on their cell cycle, indicating a specific contribution of N-Myc in C/EBPa regulated HSC proliferation (Figure S5c).
To determine whether the quiescence imposed by increased C/EBPa expression during the transition from fetal to adult hematopoiesis is also modulated through the inhibition of N-Myc expression, we first measured the mRNA levels of N-Myc in sorted SLAM+KSLs from FL and from bone marrow of either newborn or adult mice. Of note, N-Myc showed an inverse expression pattern as compared to C/EBPa (Figure 6g). To examine whether the relatively high levels of N-Myc in FL HSCs mediate their active proliferation, we knocked down N-Myc expression in FL KSLs using shRNA. Reduced levels of N-Myc in FL KSLs resulted in a significant increase of cells in G0 phase (Figure 6h). Together, these data strongly support that C/EBPa regulated fetal to adult switch of HSCs, is at least partially mediated by direct transcriptional repression of N-Myc by C/EBPa.
Discussion
In this study, we demonstrate that deletion of C/EBPa leads to increased proliferation with an expansion of HSC pool. Moreover, the transcriptome of adult C/EBPa KO HSCs resembled that of fetal HSCs. We identified N-Myc as a direct target of C/EBPa and a key mediator of the C/EBPa induced transition from fetal to adult HSCs. Our data establish that a finely orchestrated balance of N-Myc and C/EBPa controls the developmental change of HSC proliferative properties (Figure 7).
Figure 7.
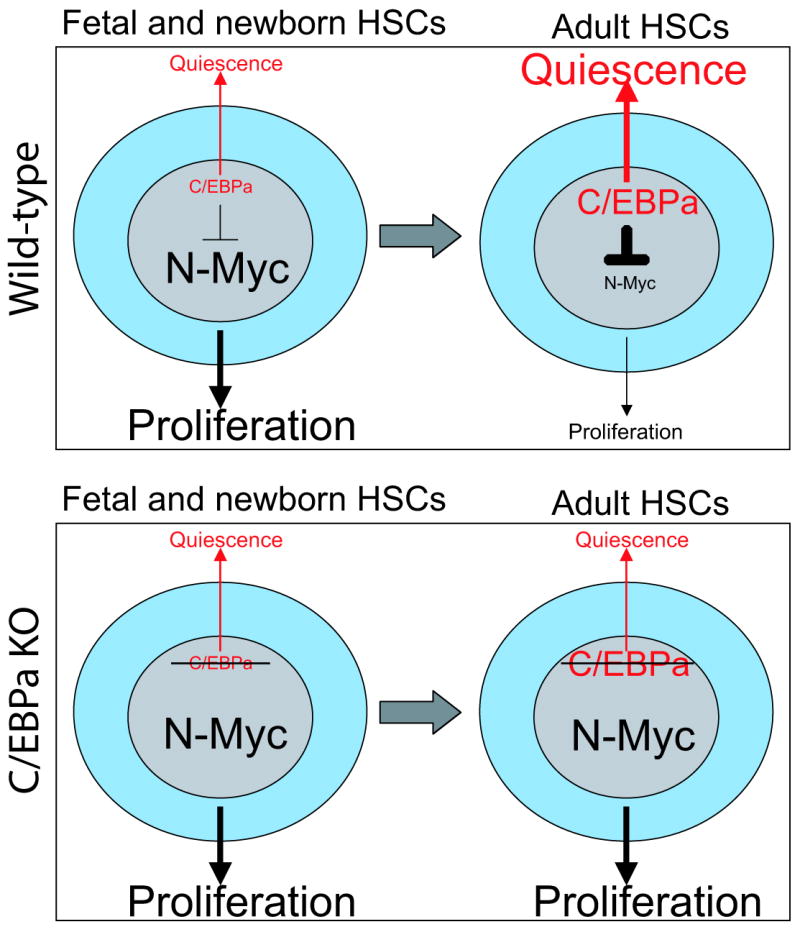
C/EBPa plays a key role in regulating acquisition and maintenance of adult HSC quiescence
Model summarizing the modulation of HSC proliferation properties by C/EBPa during fetal/adult transition. Top panel: Fetal and newborn HSCs express low levels of C/EBPa and high levels of N-Myc, which sustain the active proliferation of fetal HSCs. During the fetal to adult transtition, C/EBPa expression increases in HSCs, which subsequently inhibits N-Myc activity through direct transcriptional repression. As a consequence, HSCs become quiescent. Bottom panel: Inactivation of C/EBPa by conditional ablation of C/EBPa releases its inhibition of N-Myc expression. Elevated N-Myc expression in adult HSCs re-activates their proliferation that resembles the fetal HSC.
One striking feature of HSCs is that actively dividing HSCs become quiescent around 3-4 weeks after birth 8. However, the molecular mechanisms underlying this phenomenon are still under investigation11,27,29. Our data indicate that the fetal to adult switch of HSC is controlled by C/EBPa, probably in a dosage sensitive manner. It is possible that different levels of C/EBPa control different downstream targets in HSCs through either direct transcriptional regulation or indirect mechanisms. For instance, low levels of C/EBPa may modulate genes essential for self-renewal 19,30, whereas increased levels of C/EBPa in adult HSCs may regulate additional sets of factors associated with cell cycle. It is possible that a developmental stage-dependent bone marrow microenvironment triggers the activation of unique genetic programs in HSC. However, the fact that the pace of HSC temporal alternation does not seem to be affected by the microenvironment following transplantation10, rather suggests an intrinsically timed developmental event31,32.
Transcription factor Sox17 has been linked to the transition of fetal to adult HSCs. Sox17 is required for the maintenance of fetal, neonatal but not adult hematopoiesis11,27. Moreover, ectopic expression of Sox17 confers fetal HSC characteristics to adult hematopoietic progenitors27. In C/EBPa KO SLAM+ HSCs, we did not detect the change of Sox17 expression by microarray and qPCR (data not shown), suggesting that C/EBPa might function downstream of Sox17. This hypothesis is supported by the observation that C/EBPa is significantly down-regulated, while its downstream target N-Myc is up-regulated, following the ectopic expression of Sox17 in adult hematopoietic progenitors 27. Intriguingly, although C/EBPa adult KO HSCs exhibit enhanced repopulation ability than the wild-type control as well as enriched expression of fetal HSCs associated signature, they are still unable to fully regain the reconstitution capacity as fetal HSCs, as evidenced by the lower frequency of reconstituted mice and the lower degree of chimerism (Figure S6a and S6b). These data suggested that C/EBPa is a key factor, yet not the only one, that is crucial to FL HSC identity and function. Other factors involving in HSC homing, engraftment and reconstitution might as well be required.
Myc genes are associated with a variety of aspects of cellular physiology including proliferation, apoptosis, differentiation, cellular metabolism, and DNA synthesis 33. Counteraction between C/EBPa and C-Myc has been reported in adipogenesis and myelopoiesis34,35. However, loss of C/EBPa in HSCs did not significantly change levels of c-Myc expression, nor reduced levels of C-Myc by shRNA had an impact on proliferation of KO HSPCs (data not shown). Therefore, it is less likely that elevated protein or enhanced activity of C-Myc contributes significantly to proliferation of KO HSCs. Indeed, in this study we show that enhanced proliferation exhibited in KO HSCs is at least partially mediated by N-Myc. We find that both C/EBPa KO HSCs with characteristics that resemble fetal HSCs and FL HSCs themselves express high levels of N-Myc, and down-regulating N-Myc in either of them diminishes HSC proliferation (Figure 6), indicating its crucial role in maintaining the active state of HSCs. It is likely that N-Myc activates yet to be identified genes modulating cell cycle machinery. As loss of the mouse N-Myc gene results in embryonic lethality between E10.5 and E12.5 probably from neuroectodermal and heart defects36,37, future investigations on HSC behavior and its molecular profiling in mice that the N-Myc gene is specifically disrupted in fetal hematopoiesis would be required to define pathways essential for HSC proliferation. It would be also interesting to know whether N-Myc mediated proliferation also account for the expansion of functional LT-HSCs, which is seen both in C/EBPa deficient mice and wild type FL during early development.
In conclusion, we provide functional and genome-wide evidences that C/EBPa acts as a molecular switch for acquisition and maintenance of adult HSC properties and demonstrate a remarkable capacity of a single transcription factor in determination of HSC identity.
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health grant HL 56745 and the Harvard Stem Cell Institute grant DP-0086-10-00. GA was supported by the Collegio Ghislieri Fellowship Program. EL was supported by FAMRI YCSA and CIA grant. PS was supported by Austrian Research Foundation and European Union.
We thank all members of the Tenen laboratory for helpful discussions; Robert Welner, Christian Bach, Huafeng Xie, Matthias Stadtfeld, Thomas Graf and Xuecui Guo for careful reading of the manuscript and suggestions; Joyce LaVecchio and Girijesh Buruzula from the Harvard Stem Cell Institute/ Joslin Diabetes Center flow cytometry facility for their expertise during cell sorting; Angie Tan Lay Keng and Lee Ming Hui from the NUS-Duke genomic facility in Singapore for their expertise in microarray analysis.
Footnotes
Author Contribution
Contribution: MY and DGT designed the study; MY, HZ, GA, HY, PZ, EL, PS, JZ and MAJ performed research; MY, HZ, HY and PZ analyzed data; MY, HZ and DGT wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
References
- 1.Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL. The purification and characterization of fetal liver hematopoietic stem cells. Proc Natl Acad Sci U S A. 1995;92:10302–6. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ema H, Nakauchi H. Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood. 2000;95:2284–8. [PubMed] [Google Scholar]
- 3.Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–73. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 4.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheshier SH, Morrison SJ, Liao X, Weissman IL. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc Natl Acad Sci U S A. 1999;96:3120–5. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Passegue E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202:1599–611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson A, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–29. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 8.Bowie MB, et al. Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J Clin Invest. 2006;116:2808–16. doi: 10.1172/JCI28310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison DE, Zhong RK, Jordan CT, Lemischka IR, Astle CM. Relative to adult marrow, fetal liver repopulates nearly five times more effectively long-term than short-term. Exp Hematol. 1997;25:293–7. [PubMed] [Google Scholar]
- 10.Bowie MB, et al. Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proc Natl Acad Sci U S A. 2007;104:5878–82. doi: 10.1073/pnas.0700460104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130:470–83. doi: 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park IK, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–5. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 13.Hock H, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–7. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 14.Hock H, et al. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 2004;18:2336–41. doi: 10.1101/gad.1239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson PF. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. J Cell Sci. 2005;118:2545–55. doi: 10.1242/jcs.02459. [DOI] [PubMed] [Google Scholar]
- 16.Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007;17:318–24. doi: 10.1016/j.tcb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 17.McKnight SL. McBindall--a better name for CCAAT/enhancer binding proteins? Cell. 2001;107:259–61. doi: 10.1016/s0092-8674(01)00543-8. [DOI] [PubMed] [Google Scholar]
- 18.Zhang DE, et al. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci U S A. 1997;94:569–74. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang P, et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21:853–63. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Pabst T, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27:263–70. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 21.Wouters BJ, et al. Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1. Blood. 2007;110:3706–14. doi: 10.1182/blood-2007-02-073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–21. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 23.Kim I, He S, Yilmaz OH, Kiel MJ, Morrison SJ. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood. 2006;108:737–44. doi: 10.1182/blood-2005-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szilvassy SJ, Humphries RK, Lansdorp PM, Eaves AC, Eaves CJ. Quantitative assay for totipotent reconstituting hematopoietic stem cells by a competitive repopulation strategy. Proc Natl Acad Sci U S A. 1990;87:8736–40. doi: 10.1073/pnas.87.22.8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Essers MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–8. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 26.Venezia TA, et al. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2004;2:e301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He S, Kim I, Lim MS, Morrison SJ. Sox17 expression confers self-renewal potential and fetal stem cell characteristics upon adult hematopoietic progenitors. Genes Dev. 2011;25:1613–27. doi: 10.1101/gad.2052911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurenti E, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–24. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowie MB, Kent DG, Copley MR, Eaves CJ. Steel factor responsiveness regulates the high self-renewal phenotype of fetal hematopoietic stem cells. Blood. 2007;109:5043–8. doi: 10.1182/blood-2006-08-037770. [DOI] [PubMed] [Google Scholar]
- 30.Iwama A, et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 2004;21:843–51. doi: 10.1016/j.immuni.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Ikuta K, et al. A developmental switch in thymic lymphocyte maturation potential occurs at the level of hematopoietic stem cells. Cell. 1990;62:863–74. doi: 10.1016/0092-8674(90)90262-d. [DOI] [PubMed] [Google Scholar]
- 32.Kikuchi K, Kondo M. Developmental switch of mouse hematopoietic stem cells from fetal to adult type occurs in bone marrow after birth. Proc Natl Acad Sci U S A. 2006;103:17852–7. doi: 10.1073/pnas.0603368103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–90. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 34.Freytag SO, Geddes TJ. Reciprocal regulation of adipogenesis by Myc and C/EBP alpha. Science. 1992;256:379–82. doi: 10.1126/science.256.5055.379. [DOI] [PubMed] [Google Scholar]
- 35.Johansen LM, et al. c-Myc is a critical target for c/EBPalpha in granulopoiesis. Mol Cell Biol. 2001;21:3789–806. doi: 10.1128/MCB.21.11.3789-3806.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawai S, et al. Defects of embryonic organogenesis resulting from targeted disruption of the N-myc gene in the mouse. Development. 1993;117:1445–55. doi: 10.1242/dev.117.4.1445. [DOI] [PubMed] [Google Scholar]
- 37.Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev. 1992;6:2235–47. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- 38.Hirai H, et al. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat Immunol. 2006;7:732–9. doi: 10.1038/ni1354. [DOI] [PubMed] [Google Scholar]
- 39.Santaguida M, et al. JunB protects against myeloid malignancies by limiting hematopoietic stem cell proliferation and differentiation without affecting self-renewal. Cancer Cell. 2009;15:341–52. doi: 10.1016/j.ccr.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irizarry RA, Ooi SL, Wu Z, Boeke JD. Use of mixture models in a microarray-based screening procedure for detecting differentially represented yeast mutants. Stat Appl Genet Mol Biol. 2003;2 doi: 10.2202/1544-6115.1002. Article1. [DOI] [PubMed] [Google Scholar]
- 41.Chua SW, et al. A novel normalization method for effective removal of systematic variation in microarray data. Nucleic Acids Res. 2006;34:e38. doi: 10.1093/nar/gkl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


