Abstract
Multispecific drug transporters of the solute carrier and ATP-binding cassette families are highly conserved through evolution, but their true physiologic role remains unclear. Analyses of the organic anion transporter 3 (OAT3; encoded by Slc22a8/Oat3, originally Roct) knockout mouse have confirmed its critical role in the renal handling of common drugs (e.g., antibiotics, antivirals, diuretics) and toxins. Previous targeted metabolomics of the knockout of the closely related Oat1 have demonstrated a central metabolic role, but the same approach with Oat3 failed to reveal a similar set of endogenous substrates. Nevertheless, the Oat3 knockout is the only Oat described so far with a physiologically significant phenotype, suggesting the disturbance of metabolic or signaling pathways. Here we analyzed global gene expression in Oat3 knockout tissue, which implicated OAT3 in phase I and phase II metabolism (drug metabolizing enzymes or DMEs), as well as signaling pathways. Metabolic reconstruction with the recently developed “mouse Recon1” supported the involvement of Oat3 in the aforementioned pathways. Untargeted metabolomics were used to determine whether the predicted metabolic alterations could be confirmed. Many significant changes were observed; several metabolites were tested for direct interaction with mOAT3, whereas others were supported by published data. Oat3 thus appears critical for the handling of phase I (hydroxylation) and phase II (glucuronidation) metabolites. Oat3 also plays a role in bioenergetic pathways (e.g., the tricarboxylic acid cycle), as well as those involving vitamins (e.g., folate), steroids, prostaglandins, gut microbiome products, uremic toxins, cyclic nucleotides, amino acids, glycans, and possibly hyaluronic acid. The data seemingly consistent with the Remote Sensing and Signaling Hypothesis (Ahn and Nigam, 2009; Wu et al., 2011), also suggests that Oat3 is essential for the handling of dietary flavonoids and antioxidants.
Introduction
Oat3 (Slc22a8), originally identified as Roct (Brady et al., 1999), is a multispecific organic anion drug transporter expressed in renal proximal tubule cells, endothelial cells of the brain, and cells of the choroid plexus, where it is believed to comprise part of the “classic” organic anion handling mechanism responsible for the rate-limiting steps in the movement of solutes across the blood–urine, blood–cerebrospinal fluid, and blood–brain barriers. Organic anion transporter (OAT)3 shares several characteristics with OAT1 (Slc22a6) (Lopez-Nieto et al., 1996, 1997) and other related transporters, including the presence of 12 membrane-spanning helices with several consensus extracellular glycolsylation and intracellular protein kinase C sites (Eraly et al., 2004; You, 2004; Klaassen and Aleksunes, 2010; VanWert et al., 2010; Wu et al., 2011); however, OAT3 is phylogenetically and structurally unrelated to other transport proteins, such as the organic anion-transporting polypeptide transporters and the drug-transporting ATP-binding cassette proteins (Hagenbuch and Meier, 2003). Its nearest non-Oat SLC22 relations are the organic cation- and carnitine-transporting OCT and OCTN proteins (Burckhardt and Wolff, 2000; Sweet et al., 2001), as well as the FLIPT proteins (fly-like putative transporters) (Eraly and Nigam, 2002; Enomoto et al., 2003). As with these related transporters, OAT3 is a multispecific transporter (Kusuhara et al., 1999; Sweet et al., 2003), coupling organic anion influx to dicarboxylate efflux (Sweet et al., 1997, 2003; Zhou and You, 2007; Anzai et al., 2012), manifesting the physiologic properties expected for a transporter underlying “classic” organic anion uptake.
OAT3 is a predominant component of the renal organic anion transport apparatus believed to account for the transport of one-third to one-half of the most commonly prescribed drugs (i.e., penicillins, nonsteroidal anti-inflammatory drugs, cephalosporins, angiotensin-converting enzyme inhibitors, diuretics, smallpox and HIV antivirals, methotrexate, and statins) as well as many toxins (Eraly et al., 2003, 2004). In addition, we have analyzed the knockout of Oat3 in mice that are born at expected male/female ratio and are fertile (Sweet et al., 2002). The resulting mutant mice, while free of obvious morphologic abnormalities, manifested a distinct physiologic phenotype characterized by a loss of organic anion transport in the kidney and choroid plexus (Sweet et al., 2002). In addition, basal glomerular filtration rate (determined by 3H-inulin clearance) and p‑aminohippurate clearance were not different between wild-type (WT) controls and Oat3-deficient mice, suggesting that Oat3 is not absolutely required for renal p‑aminohippurate secretion (Sweet et al., 2002). Oat3-deficient mice also displayed systolic blood pressure 10% to 15% lower than WT mice, raising the possibility that OAT3 mediates the specific transport of one more endogenous compounds involved in the regulation of blood pressure (Vallon et al., 2008a). Nevertheless, the endogenous physiologic function(s) of this highly conserved transporter remains to be fully elucidated despite its potential considerable pharmaceutical and pharmacologic worth (Giacomini et al., 2010). In contrast to the Oat1 knockout (Eraly et al., 2006) targeted metabolomics analysis of the Oat3 knockout have revealed surprisingly few changes (Eraly et al., 2008; Vallon et al., 2008a,b, 2012).
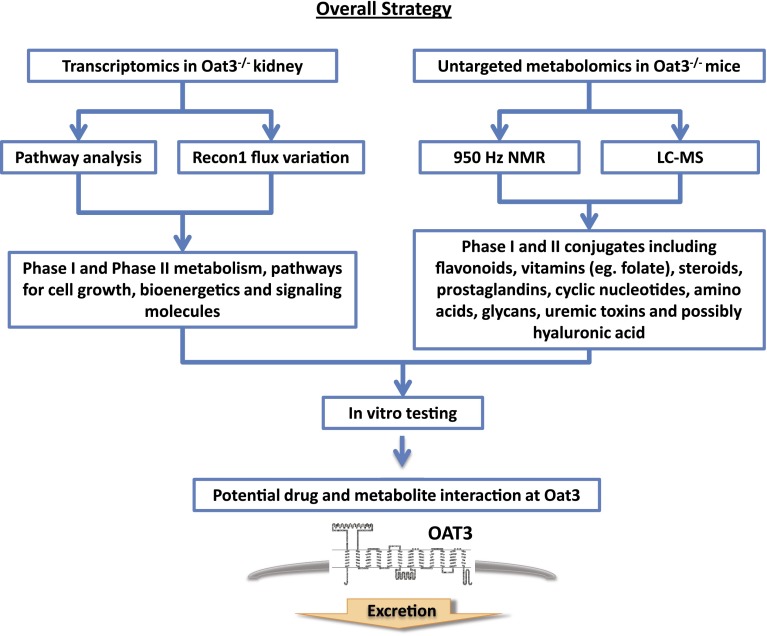
A role for OAT3 and other solute carrier and ATP-binding cassette multispecific drug transporters in remote sensing and signaling during interorgan communication has been proposed (Ahn and Nigam, 2009; Wu et al., 2011). Implicit in this hypothesis is a central role for “drug” transporters like OAT3 in regulating key metabolites and signaling molecules. By using a systems-biology approach that combines computational and wet-laboratory “omics” data obtained from the Oat3-deficient mouse, we observed altered molecular gene networks and concentrations of a battery of cellular metabolites involved in energy production and cell growth. We also found plasma accumulation of a large number of secondary metabolites (phase II) of plant ingredients of dietary origin, particularly those of flavonoid antioxidants. Metabolic reconstruction using transcriptomic data from the knockout and WT was then performed, followed by testing of predictions by two different untargeted metabolomics methods (Fig. 1). In general, the data were consistent with a central role for Oat3 in regulating the cellular metabolism and remote communication.
Fig. 1.
Overall strategy of this study.
Materials and Methods
Chemicals used for in vitro tests were purchased from Sigma-Aldrich (St. Louis, MO) and were analytical grade pure.
Animals.
Oat3/Slc22a8-deficient mice were born with expected sex ratio. Male mice between 12- and 20-weeks-of-age were used in these experiments. For microarray analysis, Oat3/Slc22a8-deficient mice were backcrossed to C57BL/6J for five generations. For metabolomic analysis, Oat3-deficient mice were backcrossed to C57BL/6J for a total of 10 generations, and both control and Oat3-deficient mice were fed the same standard diet. Blood and urine samples were collected, and plasma was isolated and stored at −80°C until analysis was carried out. All animals were handled in accordance with Institutional Animal Care and Use Committee guidelines (http://iacuc.ucsd.edu/index.aspx).
Microarray Analysis.
Total RNA was prepared and purified from WT (n = 3) and knockout kidneys (n = 3), and microarray analyses were performed as previously described (Wikoff et al., 2011). The amplified RNA was labeled by incorporation of biotinylated nucleotides during in vitro transcription and then hybridized to Affymetrix microarrays, washed, and scanned per the standard Affymetrix protocol. Hybridization and scanning were carried out at the UCSD/Department of Veterans Affairs Medical Center GeneChip core laboratory (http://www.vmrf.org/research-websites/gcf), and microarray data analysis was performed as described (Tsigelny et al., 2008).
Recon 1 Analysis.
A variation of Recon 1 (Duarte et al., 2007), a global human metabolic network reconstruction, was used to analyze the differential transcriptomic data from the WT and Oat3-deficient animals. As described previously (Ahn et al., 2011), the National Center for Biotechnology Information Homologene Database (www.ncbi.nlm.nih.gov/homologene) was used to map human Entrez Gene IDs to their mouse homologs. Specific WT and Oat3-deficient models were created using the transcription profiles (binary classification using Affymetrix MAS5.0 presence/absence calls) as data with the GIMME algorithm (http://csbl.bitbucket.org/tiger/doc/tiger/tie/gimme.html) (Becker et al., 2007) and the COBRA toolbox (http://gcrg.ucsd.edu/node/11) (Schellenberger et al., 2011). The mouse biomass pseudo-reaction was used as the objective function. Identical generalized uptake conditions were used for both models and flux spans were calculated following Flux Variability Analysis (Mahadevan and Schilling, 2003; Ahn et al., 2011).
Predicted changes in the metabolic capabilities of the two models were classified into two categories; the active reactions that were shared between the models and the reactions that were not shared between knockout and WT. The most substantive flux changes for the set of shared reactions were identified by rank ordering the reactions and identifying reactions with flux spans that either increased by 2-fold or decreased by 50%. Of note, changes in the flux spans will not necessarily be reflected as increases or decreases in flux, but rather increases or decreases in the range of possible flux for a particular reaction.
Untargeted Metabolomics.
Blood and spontaneous urine samples (at the time of blood collection) from adult male WT control and Oat3-deficient mice (n = 3) were obtained and individual, unpooled samples were subjected to 950 MHz NMR analysis at University of North Carolina metabolomics laboratory.
An untargeted, liquid chromatography/mass spectrometry (LC/MS)-based approach for metabolomics was carried out by the core facility at Scripps Center for Metabolomics and Mass Spectrometry. Plasma samples were obtained from adult male Oat3-deficient and control WT mice (n = 3) and prepared in a manner similar to that previously described for mOAT1 metabolomics (Wikoff et al., 2011). Mass spectrometry data were evaluated using XCMS (https://xcmsonline.scripps.edu/), with nonlinear data aligned with intensity integration. The METLIN database (http://metlin.scripps.edu/) was then used for compound identification. KEGG was used for pathway analysis. Plasma samples from Oat3-deficient mice were compared with plasma samples from wild-types. The samples for metabolomics were then run together and analyzed as a single group. The concentration ratios of revealed metabolites indicate those molecules with altered plasma distribution in the knockout versus WT samples.
Cellular Uptake Assay.
As previously described, confluent monolayers of Chinese hamster ovary (CHO) cells permanently expressing mouse OAT3 (mOAT3-CHO) were grown in 96-well tissue culture plates. The cultured cells were employed in uptake assays using 5-carboxyl fluorescein as a tracer molecule (Ahn et al., 2011).
Statistical Analysis.
The statistical analysis was performed using an unpaired t test. The values are expressed as the means ± S.D.
Results
Altered Expression of Genes Involved in Key Metabolic Pathways in the Oat3-Deficient Kidney.
Oat3 is predominantly expressed in the kidney, where it mediates the uptake of organic anionic solutes in proximal tubular cells (VanWert et al., 2010). Despite the importance of Oat3 in the uptake and elimination of a wide variety of drugs and toxins (Table 1), Oat3-deficient animals are healthy and viable. To determine if a molecular compensatory mechanism exists in the kidney of Oat3-deficient mice, a microarray analysis was carried out comparing renal gene expression in the knockout to that of WT controls. Among the profiled genes, significant expression changes (≥2−fold change in expression either up or down) were observed for more than 100 genes, including Oat3/Slc22a8 (absent in in the Oat3-deficient mice). Detailed annotation of 67 of those gene transcripts that were most significantly elevated (>2.5×) in the Oat3-deficient kidney revealed that at least 23 of them encode gene products related to metabolism, suggesting elevated enzymatic activities (Table 2). For example, expression of the gene related to the human CYP2b10 isoform 1, a phase I drug metabolizing enzyme, was significantly elevated (4.2-fold) in the Oat3-deficient kidney (Table 2). In addition, the expression of Ugt2b5 homolog, a phase II drug-metabolizing enzyme that catalyzes glucuronide conjugation, was also elevated (over 300-fold), whereas elevated expression (3.7-fold) was also observed for 3-oxo-5-alpha-steroid 4-dehydrogenase 2, an enzyme involved in steroid metabolism. Taken together, these expression changes indicate that Oat3 deficiency results in alterations in the expression of phase I and phase II drug-metabolizing enzymes, as well as phase III drug transporters. This finding appears to link Oat3 to the normal physiologic functions of metabolic enzymes and transporters.
TABLE 1.
Known functional alterations in the Oat3-knockout kidney
| Molecule | Class | Reference |
|---|---|---|
| Methotrexate | Drug | VanWert and Sweet, 2008 |
| Penicillin G | Drug | VanWert et al., 2007 |
| Furosemide | Drug | Vallon et al., 2008b |
| Bendroflumethiazide | Drug | Vallon et al., 2008b |
| Ciprofloxacin | Drug | VanWert et al., 2007 |
| Urate | Metabolite | Eraly et al., 2008 |
| Ro 64-0802 (active form of oseltamivir) | Drug | Ose et al., 2009 |
| Aristolochic acid | Toxin | Xue et al., 2011 |
| Zidovudine | Drug | Nagle et al., 2011 |
| Acyclovir | Drug | Nagle et al., 2011 |
| Tenofovir | Drug | Nagle et al., 2011 |
| Lamivudine | Drug | Nagle et al., 2011 |
| Creatinine | Metabolite | Vallon et al., 2012 |
TABLE 2.
Partial list of enzyme transcripts (including some Ests) upregulated in Oat3-knockout kidney
Multiple probes for Phase I (i.e., CYP2b10 isoform 1) and Phase II (i.e., UGT2B5) drug metabolizing enzyme displayed similar levels of expression change. The elevated expression observed for 3-oxo-5-alpha-steroid 4-dehydrogenase 2 suggests enhanced enzymatic metabolism for steroid hormones, many of which are known OAT3 substrates.
| Affy. ID | Gene Name | Description | Fold Change |
|---|---|---|---|
| 1423397_at | Ugt2b5 | UGT2B5 | 371.9 |
| 1440339_at | Enpp1 | Ectonucleotide pyrophosphatase/phosphodiesterase 1 | 120.2 |
| 1459311_at | Est | cAMP-specific 3′,5′-cyclic phosphodiesterase 4D-like | 14.6 |
| 1449486_at | Ces1 | Carboxylesterase 1 | 4.5 |
| 1422257_s_at | Cyp2b10 | CYP2b10 isoform 1 | 4.2 |
| 1440463_at | Est | Similar to peptide N-glycanase (Ngly1) gene | 4.0 |
| 1422960_at | Srd5a2 | 3-oxo-5-alpha-steroid 4-dehydrogenase 2 | 3.7 |
| 1444032_at | Keg1 | Glycine N-acyltransferase-like protein Keg1 | 3.7 |
Utilizing available bioinformatic tools, including GeneSpring, GOBY, and Ingenuity IPA, the genes were further analyzed to investigate the possibility that networks of genes involved in drug metabolism and transport were altered in the knockout animals. These analyses revealed that tricarboxylic acid cycle and energy metabolism, as well as nucleotide and amino acid metabolism, were among some of the most significantly impacted pathways (Table 3). In addition to the core cellular functions implicated, altered functionality was also observed in vitamin and mineral metabolism. Thus, our analyses not only link Oat3 to the normal physiologic functions of metabolic enzymes and transporters, but they also suggest Oat3 may be involved in the handling of nutrients and vitamins.
TABLE 3.
Subsystem prediction of altered metabolic pathways in Oat3 knockout
Transcriptomic analysis was carried out using Affymetrix mouse gene expression array. The most significantly altered sybsystems were listed. It appears that arginine and proline metabolism, blood group biosynthesis, fatty acid metabolism were predicted to be increased in Oat3-deficient kidney, whereas glutathione metabolism, glycine, serine, and threonine metabolism were predicted to be reduced in the Oat3-deficient kidney.
| Up | Down |
|---|---|
| Arginine and proline metabolism | Galactose metabolism |
| Blood group biosynthesis | Glutathione metabolism |
| Chondroitin/heparan sulfate biosynthesis | Glycine, serine, and threonine metabolism |
| Fatty acid metabolism | Glycolysis/gluconeogenesis |
| Galactose metabolism | IMP biosynthesis |
| Glycerophospholipid metabolism | Pentose phosphate pathway |
| Inositol phosphate metabolism | Pyrimidine biosynthesis |
| Nucleotides | Pyruvate metabolism |
| O-glycan biosynthesis | Transport, extracellular |
| Sphingolipid metabolism | Vitamin B12 metabolism |
| Steroid metabolism |
Metabolomic Reconstruction of Potential Physiologic Roles of Oat3 by Recon 1.
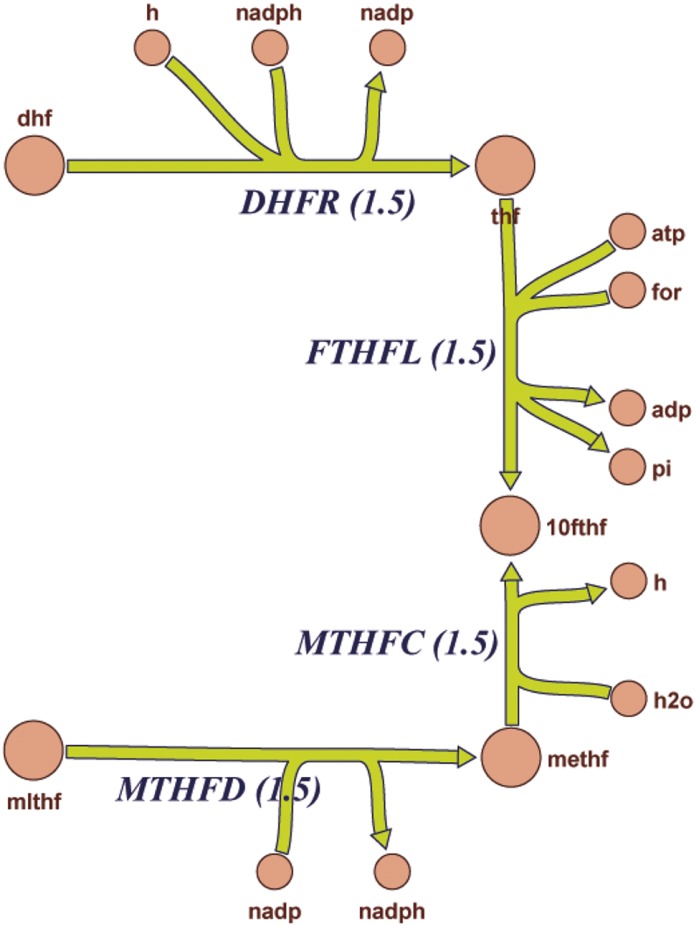
To further investigate this possibility, as well as decipher the cellular response to Oat3 deficiency, global transcriptomic clustering was followed by pathway analysis using mouse Recon 1. This global metabolic network reconstruction is largely based on human Recon 1, which comprises known biochemical and physiologic data (Sigurdsson et al., 2010; Bordbar et al., 2012). These computational analyses revealed alterations in a number of metabolic pathways related to transcriptional regulation, solute handling, and endogenous enzymatic activities (also see Table 2). Among the 285 reactions linked to transcriptomic alteration in Oat3 deficient kidneys by mouse Recon 1, 220 were related to the cell mass growth functionality that includes energy (ATP) production and metabolism of nucleic acids, amino acids, and fatty acids. This raises the possibility that compensatory molecular responses in cell growth and bio-mass occur in the kidneys of Oat3-deficient mice. This genome-wide reconstruction of mouse metabolism based on transcriptomic data also identified 19 additional processes and reactions, including bile acid synthesis, glucuronidation, sulfation, prostaglandin synthesis, and hyaluronan and steroid metabolism, as well as vitamin metabolism, including folate (vitamin B9 derived from dietary sources) (Table 4). As an example, the affected folate metabolism pathway is depicted here, and three reactions involved in the metabolism of folate (DHFR, MTHFD, and MTHFC) are highlighted; on the basis of mouse Recon1 reconstruction, these reactions should be enhanced in the Oat3-deficient kidney (Fig. 2). An overview of the predicted changes in metabolic pathways reactions can be found in Supplemental Fig. 1 and Supplemental Tables 1–4. Thus, comparison of the WT and Oat3-deficient metabolic networks using transcriptomic data highlighted alterations in multiple areas of metabolism, including amino acid, fatty acid, cholesterol, and nucleotide pathways. Moreover, hydroxylation as well as glucuronidation and conjugation reactions (which are usually involved in phase I and phase II drug metabolism) were among the largest differences noted between the WT and Oat3-deficient mouse.
TABLE 4.
Enhanced RECON 1 enzyme reactions in the Oat3 knockout
This is a list of significantly enhanced RECON 1 enzyme reactions (without exchange, transport or biomass-growth related reactions) in the Oat3 knockout.
| Enzyme Reactions | Reaction Name | Subsystem |
|---|---|---|
| P4508B11r | Sterol 12-alpha-hydroxylase | Bile acid biosynthesis |
| P4508B13r | Sterol 12-alpha-hydroxylase (nadh) | Bile acid biosynthesis |
| B3GNT51g | UDP-GlcNAc:bGal b-1,3-N-acetylglucosaminyltransferase 5 | Glycan biosynthesis |
| ST3GAL31g | ST3β-galactoside alpha-2,3-sialyltransferase 3 | Glycan biosynthesis |
| HMGCOASim | Hydroxymethylglutaryl CoA synthase (ir) | Cholesterol metabolism |
| GALT2g | UDP-D-galactose:galactosylxylose galactosyltransferase, Golgi | Chondroitin/heparin sulfate biosynthesis |
| S3T2g | Heparin-glucosamine 3-O-sulfotransferase | Chondroitin/heparin sulfate biosynthesis |
| LTA4H | Leukotriene A-4 hydrolase | Eicosanoid metabolism |
| PGISr | Prostaglandin I2 synthase | Eicosanoid metabolism |
| DHFRa | Dihydrofolate reductase | Folate metabolism |
| MTHFDa | Methylenetetrahydrofolate dehydrogenase (NADP) | Folate metabolism |
| HAS1 | Hyaluronan synthase | Hyaluronan metabolism |
| HAS2 | Hyaluronan synthase | Hyaluronan metabolism |
| S23T3g | β-Galactoside α-2,3-sialyltransferase (complex N-glycan) | Keratan sulfate biosynthesis |
| HSD17B3r | Testicular 17β-hydroxysteroid dehydrogenase | Steroid metabolism |
| PYDXDH | Pyridoxal dehydrogenase | Vitamin B6 metabolism |
| S23T3g | β-Galactoside α-2,3-sialyltransferase (T antigen) | O-glycan biosynthesis |
| MTHFCa | Methenyltetrahydrofolate cyclohydrolase | Folate metabolism |
| 34HPLFM | 3-(4-Hydroxyphenyl-)lactate formation | Ubiquinone biosynthesis |
Enhanced reactions related to folate metabolism (DHFR, MTHFD and MTHFC) are highlighted in Fig. 2.
Fig. 2.
Graphic pathway diagram depicting altered components (enhanced) of folate metabolism in Oat3 knockout kidney generated by Recon 1 analysis. The enhanced pathways of DHFR, MTHFC, and MTHFD were also listed in Table 4. This diagram is a portion of the global overview of predicted global changes in pathways by Recon 1 analysis, which can be found in Supplemental Fig. 1.
Altered Urinary Excretion of Cellular Metabolites in Oat3-Deficient Mice.
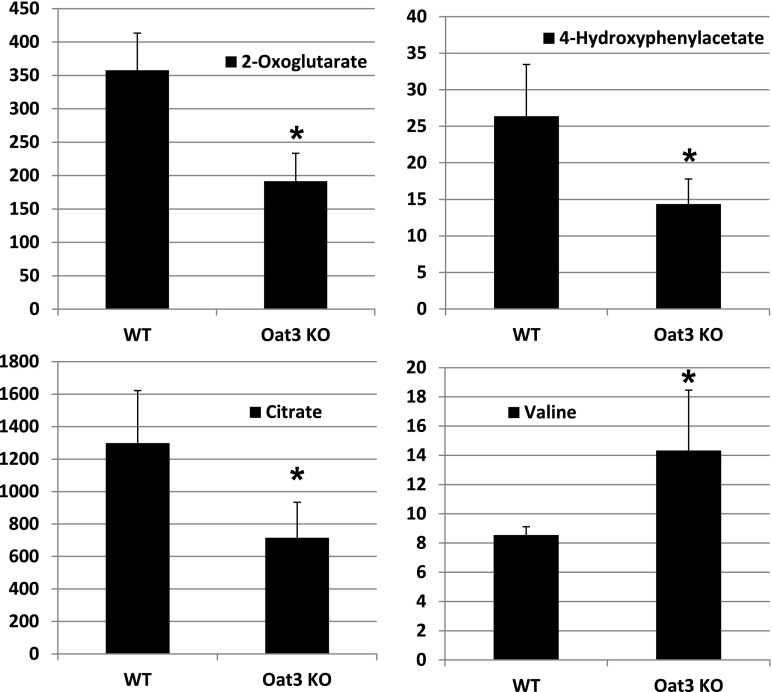
In contrast to findings in the Oat1-deficient animal, targeted metabolomics analysis of the Oat3-deficient animal revealed minimal alterations in a set of about 30 of the most abundant organic anionic endogenous metabolites (Vallon et al., 2008a). Taken together with subsequent analyses that revealed modest alterations in the concentration of urate, thymidine, and flavin mononucleotide in the Oat3-deficient animals (Eraly et al., 2008; Vallon et al., 2008a,b), it seems likely that the metabolic pathways disturbed in the Oat3-deficient mouse are different from those altered in the Oat1-deficient mouse. To confirm and validate the systems biology predictions from the previously described analyses (Table 3), global untargeted metabolomics analyses were performed on serum and urine samples derived from WT and Oat3-deficient mice. After normalization of the concentrations of the detected metabolite to creatinine, 950-MHz NMR-based untargeted metabolomics of urine revealed significant concentration changes in metabolites related to cellular energy metabolism, (e.g., α-ketogluterate, citrate, 4-hydroxyphenylacetate) and growth-related metabolites, such as amino acids (e.g., valine) (Fig. 3); this observation provided support for the Recon 1 predictions in Table 3. However, 950-MHz NMR-based untargeted metabolomics of serum samples did not yield broad concentration changes in the Oat3-deficient animal (data not shown), although nonsignificant reductions in the concentrations of several amino acids, including valine, were detected in the Oat3-deficient animal.
Fig. 3.
Oat3-deficiency results in alterations in the urinary concentration of metabolites. Relative urine concentrations of selected compounds in wild-type and Oat3-deficient mice. Spontaneous urine samples were obtained from adult mice and subjected to untargeted metabolomic analysis using 1H-NMR spectroscopy and multivariate statistical analyses to generate metabolic fingerprints. Urine concentrations of metabolites were normalized to creatinine (n = 3). *P < 0.05.
Global Untargeted Mass Spectrometric Metabolomic Profiling in Oat3-Deficient Mice.
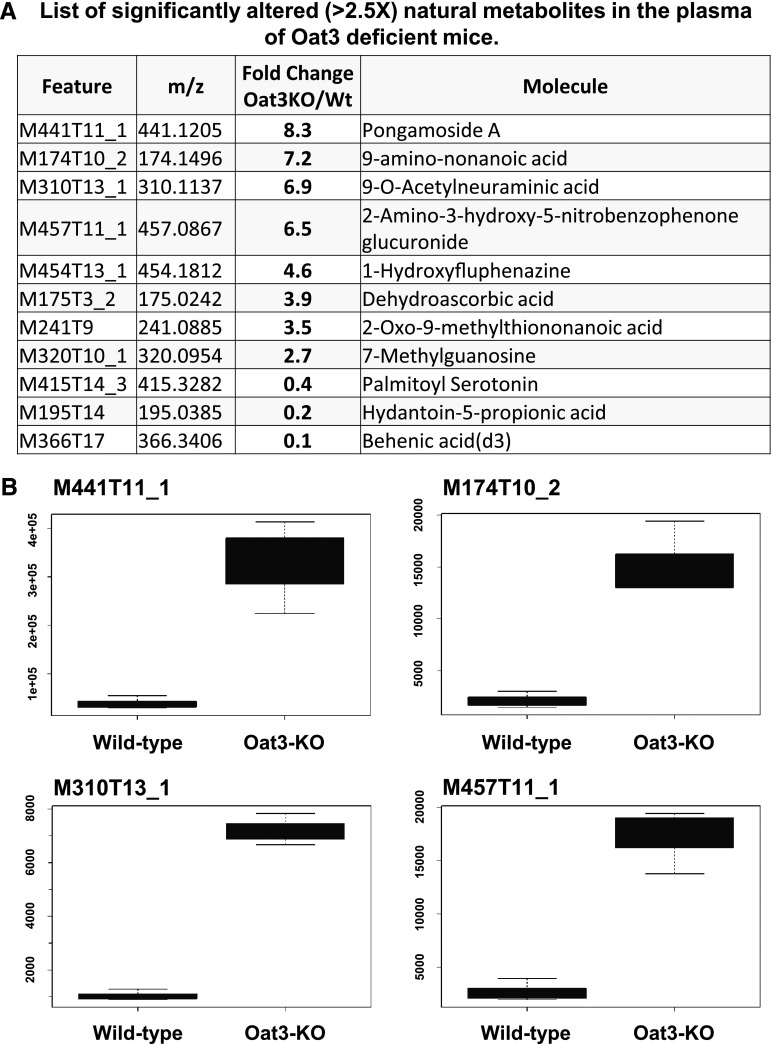
A global untargeted, mass spectrometry-based analysis of blood samples derived from adult Oat3-deficient mice versus WT controls was also performed. Plasma samples from adult C57/BL6 control mice and comparable Oat3-deficient mice were obtained. These samples were subject to LC/MS untargeted metabolomics analysis at Scripps Centers for Metabolomics and Mass Spectrometry; the METLIN metabolomics database was used for compound identification. The plasma concentrations of 1538 features were found to be significantly different between the Oat3-deficient and WT control mice, 982 of which were altered 1.5-fold or higher (either up or down). Of these 982 features, 220 of them were associated with known molecular fragments, of which 30 were clearly identified as known metabolites (Fig. 4; Supplemental Table 5).
Fig. 4.
Oat3-deficiency results in the plasma accumulation of a variety of chemical compounds. (A) List of significantly altered (>2.5×) natural metabolites in the plasma of Oat3-deficient mice by untargeted LC/MS analysis. Additional significantly altered mass-spect features are listed in Supplemental Table 5. (B) Boxplots of relative plasma concentrations for four most highly accumulated compounds in Oat3-deficient plasma. Pongamoside A (P value = 3.3 × 10−2) is a flavonoid metabolite of dietary origin. KO, knockout.
Consistent with metabolic reconstructions, behenic acid (a cholesterol-raising saturated fatty acid in humans), as well as a propionoate (a metabolite linked to carboxylic acid metabolism) and modified serotonin was significantly reduced (11- and 5-fold decrease relative to weight, respectively) in the plasma of Oat3-deficient mice (Fig. 4A). In addition, a glucuronidated molecule (2-amino-3-hydroxy-5-nitrobenzophenone glucuronide) was also among the highest accumulated in the Oat3-deficient mice, consistent with the Recon 1 analysis (Fig. 2; Table 4). Furthermore, methylguanosine, a uremic toxin, was also found to be significantly accumulated in the plasma of Oat3-deficient mice. The two molecules found to be most highly accumulated in Oat3-deficient plasma were pongamoside A and 9-amino-nonanoic acid, both of which are derived from plants (Fig. 4A). Pongamoside A is a plant derivative flavonoid antioxidant, whereas 9-amino-nonanoic acid is a modified nonanoic acid derived from soybeans.
Mass Spectrometry Features of Metabolites That Are Dietary Phenolic Derivatives.
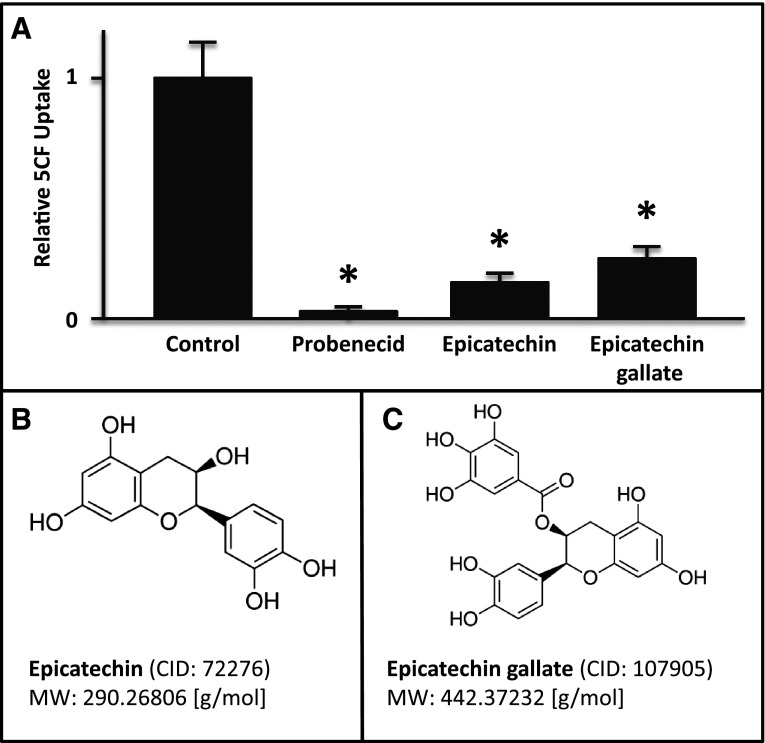
The potential molecules associated with the mass spectrometry fragment features that were most highly accumulated in the Oat3-deficient mice were also examined. Of the 10 features with a minimum of a 5-fold increase in their concentrations in the Oat3-deficient mice (Table 5), at least 7 of them were found to be associated with metabolites of plant origin. For example, feature M457T_1, elevated 13.8-fold in Oat3-deficient plasma, is associated with a group of metabolites of dietary phenolic derivatives. One of these metabolites is epicatechin 3-O-(3-O-methylgallate), also known as an internal metabolite of the ester of epigallocatechin and gallic acid [epigallocatechin gallate (EGCG)]. EGCG is a potent antioxidant found in a number of plants, but it is the most abundant catechin in tea leaves. Interaction of EGCG with OAT3 was confirmed in a cell-based assay that employed mOAT3-expressing CHO cells (mOAT3-CHO). In this assay, EGCG, as well as typical OAT3 substrates (i.e., estrone sulfate) were tested for their ability to inhibit uptake of a preferential OAT3 tracer, 5CF. The uptake of 5CF in the mOAT3-CHO cell was inhibitable by epicatechin (Sigma-Aldrich; 49045-U) and by epicatechin gallate (Sigma-Aldrich; 49060-U) (Fig. 5). Thus, the data support the notion that the broad accumulation of multiple phytophenolic metabolites of dietary origin is most likely the result of a deficiency in OAT3-mediated transport.
TABLE 5.
Mass spectrometry features highly accumulated in the plasma of Oat3-deficient mice
Flavonoid metabolites are in bold.
| Name | Fold Change | P Value | m/z | Retention Time | Putative Associated Molecule |
|---|---|---|---|---|---|
| M301T2_5 | 17.7 | 0.0045 | 301.0746 | 1.59 | Sulfaquinoxaline and others |
| M457T9_1 | 13.8 | 0.0125 | 457.1144 | 9.37 | Epicatechin 3-O-(3-O-methylgallate) and others |
| M573T10 | 9.9 | 0.0075 | 573.1626 | 10.4 | Licuroside and 3 others |
| M441T11_1 | 8.3 | 0.0325 | 441.1205 | 11.27 | Pongamoside |
Fig. 5.
Inhibitory effects of metabolites on uptake of 5CF in Chinese hamster ovary cell permanently expressing mouse OAT3 (mOAT3-CHO). mOAT3-CHO cells were plated in a 96-well plate overnight, and uptake of an OAT3-preferred tracer [5-carboxyl fluorescein (5CF), 20 μM] was carried out for 5 minutes at room temperature. (A) Bar graph illustrating inhibitory effects of epicatechin and epicatechin gallate on OAT3-mediated uptake of 5CF. As expected, probenecid inhibits the uptake in a range that is consistent with previous data. Molecular structures and some chemical properties of epicatechin (B) and epicatechin gallate (C) (n = 3). *P < 0.01.
Discussion
Utilizing a systems biology approach to compare WT and Oat3-deficient mice, the correlation of transcriptomic, computational pathway, and metabolomics analyses (950 MHz NMR plus LC/MS) revealed that the absence of this transporter leads to significant alterations in: 1) several cellular metabolic pathways (e.g., tricarboxylic acid cycle, nucleotide and amino acid metabolism) (Figs. 2 and 3; Supplemental Fig. 1; Tables 2 and 3); 2) the expression of genes encoding DME enzymes with critical roles in phase I and phase II xenobiotic metabolism, as well as phase III drug transport (Tables 2 and 4); 3) pathways involved in the regulation of secondary metabolites, including endogenous signaling molecules (e.g., prostaglandins and steroids), and dietary plant derivatives (e.g., vitamins) (Figs. 2 and 4; Tables 2–4); and 4) pathways involved in the handling of dietary flavonoids (Figs. 4 and 5; Table 5). Among the dietary flavonoids found to accumulate in the plasma of the Oat3-deficient animal, epicatechin is interesting as this molecule has been suggested to impact blood pressure (and have potential cardiovascular benefits), and the Oat3-deficient mice have decreased blood pressure (Vallon et al., 2008a). Nevertheless, taken together, the data help to define the role of this transporter in normal physiology by linking its activity to a variety of pathways regulating levels of key metabolites and signaling molecules.
Oat3 is expressed in the proximal tubules of the kidneys, where it plays an important role in the elimination of numerous organic anions of physiologic, pharmacologic, and toxicologic relevance (Burckhardt and Burckhardt, 2003; Eraly et al., 2004; Nigam et al., 2007; Ahn and Bhatnagar, 2008; Di Giusto et al., 2008; Ahn and Nigam, 2009; Wu et al., 2009, 2011). There is significant overlap in the expression of Oat3 and its close homolog, Oat1, with many proximal tubular cells expressing both of these transporters (Lopez-Nieto et al., 1997; Hwang et al., 2010). Nevertheless, immunocytochemical localization reveals that OAT1 appears to be expressed preferentially in the S1 and S2 segments, whereas OAT3 is found more in the S2 and S3 segments (Hwang et al., 2010); this finding is consistent with their functional localization in knockout animals (Truong et al., 2008). In addition to its renal expression, Oat3 is also expressed in the brain (on the endothelial cell as part of blood–brain barrier) and the choroid plexus, where it is believed to be part of the transport apparatus mediating the elimination of solutes from the brain and central nervous system (Sweet et al., 2002; Vanwert et al., 2007; Truong et al., 2008; Vallon et al., 2008a,b; VanWert and Sweet, 2008; Ose et al., 2009; Nagle et al., 2011; Sweeney et al., 2011; Xue et al., 2011).
While OAT3 is believed to be involved in mediating the disposition, distribution, and elimination of a wide variety of common pharmaceuticals (VanWert et al., 2010; Burckhardt and Burckhardt, 2011), the role of Oat3 in normal physiology has been less clearly defined. In a recent targeted metabolomics study, of the 30 most abundant plasma small organic anion molecules investigated, none were found to accumulate in the plasma of Oat3-deficient mice, which is contrary to the finding from Oat1-deficient mice (Vallon et al., 2008a). The data from this study indicate that Oat3 deficiency leads to changes in the concentration of a variety of small organic metabolites in the blood. Furthermore, transcriptomic and pathway analyses revealed alterations in phase I and phase II xenobiotic metabolism, such as sulfation and glucuronidation; this notion was supported by metabolomics analyses that revealed alterations in the levels of sulfated and glucuronidated small molecules, including components of bio-mass cell growth and flavonoid antioxidants of dietary origin (Table 6). Thus, Oat3 is not only likely key to the handling of endogenous metabolites and signaling molecules with important physiologic functions, but it is also important in the handling of metabolites derived from the gut microbiome. The results seem generally compatible with the proposed role of Oat3 and remote sensing and signaling (Kaler et al., 2006; Ahn and Nigam, 2009; Wu et al., 2011).
TABLE 6.
Known OAT3 metabolites
| Metabolite | Assay | Reference |
|---|---|---|
| Pomgamoside A | Up in Oat3-knockout plasma | This study |
| 9-Amino-nonanoic acid | Up in Oat3-knockout plasma | This study |
| Flavin mononucleotide | Up in Oat3-knockout plasma | Vallon et al., 2008a |
| Thymidine | Up in Oat3-knockout plasma | Vallon et al., 2008a |
| Urate | Down in Oat3-knockout urine | Eraly, 2008 |
| Dideoxycytidine | mOat3, Xenopus. laevis | Truong et al., 2008 |
| 17β-estradiol-d-17β-glucuronide | mOat3, LLC-PK1 | Nagata, 2002 |
| Estrone sulfate | hOAT3, X. laevis | Windass, 2007 |
| Homovanillic acid | rOat3, X. laevis | Mori, 2003 |
| 3-Carboxy-4-methyl-5-propyl-2-furanpropionate | hOAT3, HEK293 | Deguchi, 2004 |
| Indoxyl sulfate | hOAT3, HEK293 | Deguchi, 2004 |
| Prostaglandin E2 | hOAT3, S2 segment | Kimura, 2002 |
| Prostaglandin F2a | hOAT3, S2 segment | Kimura, 2002 |
| Dehydroepiandrosterone sulfate | hOAT3, HEK293 | Ueo, 2005 |
| Genistein-7-O-glucuronide | hOAT3, HEK293 | Wong et al., 2011b |
| Glycitein-7-O-glucuronide | hOAT3, HEK293 | Wong et al., 2011b |
| Quercetin-3′-O-glucuronide | hOAT3, HEK293 | Wong et al., 2011b |
HEK, human embryonic kidney; LLC-PK1, a renal epithelial cell line derived from pig kidneys.
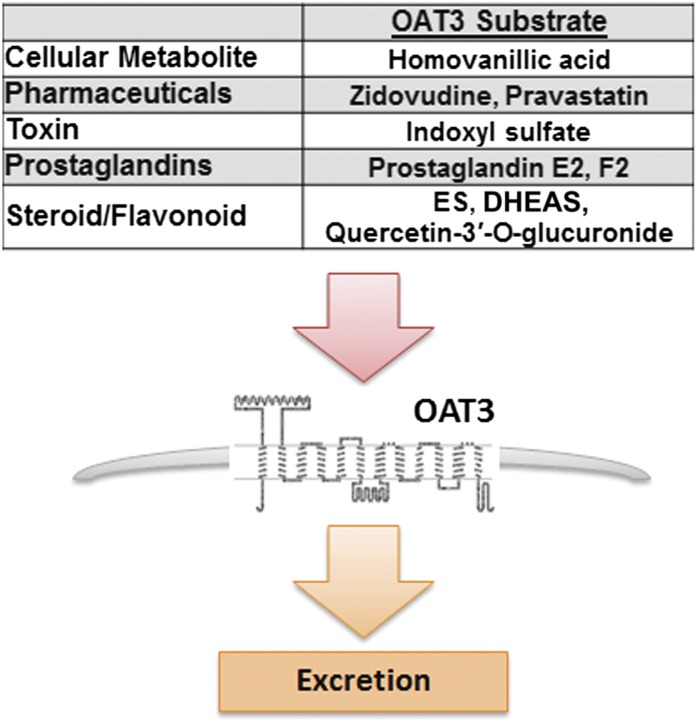
Since this set of metabolites was not found to be altered in a untargeted metabolomics analysis of plasma from Oat1-deficient animals (Ahn et al., 2011; Wikoff et al., 2011), the data support the notion that Oat3 plays a critical and perhaps primary role in the uptake and elimination of these dietary antioxidants. A metabolic reconstruction of the Oat1-deficient animal using similar transcriptomic and metabolomics data revealed alterations in largely different but somewhat overlapping set of pathways (Ahn et al., 2011). Considering the importance of OAT3 in the uptake and elimination of a wide variety of commonly prescribed drugs (e.g., antibiotics, diuretics, nonsteroidal anti-inflammatory drugs) (Giacomini et al., 2010; Klaassen and Aleksunes, 2010; VanWert et al., 2010; Wu et al., 2011) and the increased use of dietary supplements, herbal medicines, botanicals, and probiotics (Bardia et al., 2007; Williamson et al., 2007; Wong et al., 2011a,b), the data raise the possibility of competition between dietary metabolites and drugs for access to OAT3. Moreover, based on the expression of Oat3, such metabolite-drug interactions are likely to affect multiple tissues (Fig. 6).
Fig. 6.
Diagram depicting potential interaction of commonly prescribed pharmaceuticals and endogenous substrates at the site of organic anion transporter 3 for excretion. ES, Estrone sulfate; DHEAS, Dehydroepiandrosterone sulfate.
Supplementary Material
Acknowledgments
The authors thank Bill Webb and Mike McConnell for important contributions to the LC/MS and NMR studies, respectively.
Abbreviations
- CHO
Chinese hamster ovary
- DME
drug metabolizing enzyme
- EGCG
epigallocatechin gallate
- LC/MS
liquid chromatography/mass spectrometry
- OAT1
organic anion transporter 1/Slc22a6
- OAT3
organic anion transporter 3/Slc22a8
- WT
wild-type
Authorship Contributions
Participated in research design: Wu, Jamshidi, Palsson, Eraly, Nigam.
Conducted experiments: Wu, Jamshidi, Eraly, Bush, Liu.
Contributed new reagents or analytic tools: Palsson, Nigam.
Performed data analysis: Wu, Jamshidi, Eraly, Bush, Liu.
Wrote or contributed to the writing of the manuscript: Wu, Jamshidi, Bush, Palsson, Nigam.
Footnotes
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grants GM88824, GM098449 and GM104098] and Eunice Kennedy Shriver National Institute of Child Health and Human Development [Grant HD07160] (to S.K.N.), and National Heart, Lung, and Blood Institute [Grant HL094728] (to S.A.E.)
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Ahn SY, Bhatnagar V. (2008) Update on the molecular physiology of organic anion transporters. Curr Opin Nephrol Hypertens 17:499–505 [DOI] [PubMed] [Google Scholar]
- Ahn SY, Jamshidi N, Mo ML, Wu W, Eraly SA, Dnyanmote A, Bush KT, Gallegos TF, Sweet DH, Palsson BO, et al. (2011) Linkage of organic anion transporter-1 to metabolic pathways through integrated “omics”-driven network and functional analysis. J Biol Chem 286:31522–31531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SY, Nigam SK. (2009) Toward a systems level understanding of organic anion and other multispecific drug transporters: a remote sensing and signaling hypothesis. Mol Pharmacol 76:481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai N, Jutabha P, Amonpatumrat-Takahashi S, Sakurai H. (2012) Recent advances in renal urate transport: characterization of candidate transporters indicated by genome-wide association studies. Clin Exp Nephrol 16:89–95 [DOI] [PubMed] [Google Scholar]
- Bardia A, Nisly NL, Zimmerman MB, Gryzlak BM, Wallace RB. (2007) Use of herbs among adults based on evidence-based indications: findings from the National Health Interview Survey. Mayo Clin Proc 82:561–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker SA, Feist AM, Mo ML, Hannum G, Palsson BO, Herrgard MJ. (2007) Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc 2:727–738 [DOI] [PubMed] [Google Scholar]
- Bordbar A, Mo ML, Nakayasu ES, Schrimpe-Rutledge AC, Kim YM, Metz TO, Jones MB, Frank BC, Smith RD, Peterson SN, et al. (2012) Model-driven multi-omic data analysis elucidates metabolic immunomodulators of macrophage activation. Mol Syst Biol 8:558–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady KP, Dushkin H, Förnzler D, Koike T, Magner F, Her H, Gullans S, Segre GV, Green RM, Beier DR. (1999) A novel putative transporter maps to the osteosclerosis (oc) mutation and is not expressed in the oc mutant mouse. Genomics 56:254–261 [DOI] [PubMed] [Google Scholar]
- Burckhardt BC, Burckhardt G. (2003) Transport of organic anions across the basolateral membrane of proximal tubule cells. Rev Physiol Biochem Pharmacol 146:95–158 [DOI] [PubMed] [Google Scholar]
- Burckhardt G, Burckhardt BC. (2011) In vitro and in vivo evidence of the importance of organic anion transporters (OATs) in drug therapy, in Drug Transporters Handbook of Experimental Pharmacology (Fromm MF, Kim RB. eds), pp 29–104, Springer-Verlag, Berlin: [DOI] [PubMed] [Google Scholar]
- Burckhardt G, Wolff NA. (2000) Structure of renal organic anion and cation transporters. Am J Physiol Renal Physiol 278:F853–F866 [DOI] [PubMed] [Google Scholar]
- Deguchi T, Kusuhara H, Takadate A, Endou H, Otagiri M, Sugiyama Y. (2004) Characterization of uremic toxin transport by organic anion transporters in the kidney. Kidney Int 65:162–174 [DOI] [PubMed] [Google Scholar]
- Di Giusto G, Anzai N, Endou H, Torres AM. (2008) Elimination of organic anions in response to an early stage of renal ischemia-reperfusion in the rat: role of basolateral plasma membrane transporters and cortical renal blood flow. Pharmacology 81:127–136 [DOI] [PubMed] [Google Scholar]
- Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BO. (2007) Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA 104:1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto A, Takeda M, Taki K, Takayama F, Noshiro R, Niwa T, Endou H. (2003) Interactions of human organic anion as well as cation transporters with indoxyl sulfate. Eur J Pharmacol 466:13–20 [DOI] [PubMed] [Google Scholar]
- Eraly SA, Bush KT, Sampogna RV, Bhatnagar V, Nigam SK. (2004) The molecular pharmacology of organic anion transporters: from DNA to FDA? Mol Pharmacol 65:479–487 [DOI] [PubMed] [Google Scholar]
- Eraly SA, Hamilton BA, Nigam SK. (2003) Organic anion and cation transporters occur in pairs of similar and similarly expressed genes. Biochem Biophys Res Commun 300:333–342 [DOI] [PubMed] [Google Scholar]
- Eraly SA, Nigam SK. (2002) Novel human cDNAs homologous to Drosophila Orct and mammalian carnitine transporters. Biochem Biophys Res Commun 297:1159–1166 [DOI] [PubMed] [Google Scholar]
- Eraly SA, Vallon V, Rieg T, Gangoiti JA, Wikoff WR, Siuzdak G, Barshop BA, Nigam SK. (2008) Multiple organic anion transporters contribute to net renal excretion of uric acid. Physiol Genomics 33:180–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eraly SA, Vallon V, Vaughn DA, Gangoiti JA, Richter K, Nagle M, Monte JC, Rieg T, Truong DM, Long JM, et al. (2006) Decreased renal organic anion secretion and plasma accumulation of endogenous organic anions in OAT1 knock-out mice. J Biol Chem 281:5072–5083 [DOI] [PubMed] [Google Scholar]
- Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, Chu X, Dahlin A, Evers R, Fischer V, Hillgren KM, et al. International Transporter Consortium (2010) Membrane transporters in drug development. Nat Rev Drug Discov 9:215–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuch B, Meier PJ. (2003) The superfamily of organic anion transporting polypeptides. Biochim Biophys Acta 1609:1–18 [DOI] [PubMed] [Google Scholar]
- Hwang JS, Park EY, Kim WY, Yang CW, Kim J. (2010) Expression of OAT1 and OAT3 in differentiating proximal tubules of the mouse kidney. Histol Histopathol 25:33–44 [DOI] [PubMed] [Google Scholar]
- Kaler G, Truong DM, Sweeney DE, Logan DW, Nagle M, Wu W, Eraly SA, Nigam SK. (2006) Olfactory mucosa-expressed organic anion transporter, Oat6, manifests high affinity interactions with odorant organic anions. Biochem Biophys Res Commun 351:872–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Takeda M, Narikawa S, Enomoto A, Ichida K, Endou H. (2002) Human organic anion transporters and human organic cation transporters mediate renal transport of prostaglandins. J Pharmacol Exp Ther 301:293–298 [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Aleksunes LM. (2010) Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev 62:1–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuhara H, Sekine T, Utsunomiya-Tate N, Tsuda M, Kojima R, Cha SH, Sugiyama Y, Kanai Y, Endou H. (1999) Molecular cloning and characterization of a new multispecific organic anion transporter from rat brain. J Biol Chem 274:13675–13680 [DOI] [PubMed] [Google Scholar]
- Lopez-Nieto CE, You G, Barros EJG, Beier DR, Nigam SK. (1996) Molecular cloning and characterization of a novel transporter protein with very high expression in the kidney. abstract JASN 7:1301 [Google Scholar]
- Lopez-Nieto CE, You G, Bush KT, Barros EJ, Beier DR, Nigam SK. (1997) Molecular cloning and characterization of NKT, a gene product related to the organic cation transporter family that is almost exclusively expressed in the kidney. J Biol Chem 272:6471–6478 [DOI] [PubMed] [Google Scholar]
- Mahadevan R, Schilling CH. (2003) The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng 5:264–276 [DOI] [PubMed] [Google Scholar]
- Mori S, Takanaga H, Ohtsuki S, Deguchi T, Kang YS, Hosoya K, Terasaki T. (2003) Rat organic anion transporter 3 (rOAT3) is responsible for brain-to-blood efflux of homovanillic acid at the abluminal membrane of brain capillary endothelial cells. J Cereb Blood Flow Metab 23:432–440 [DOI] [PubMed] [Google Scholar]
- Nagata Y, Kusuhara H, Endou H, Sugiyama Y. (2002) Expression and functional characterization of rat organic anion transporter 3 (rOat3) in the choroid plexus. Mol Pharmacol 61:982–988 [DOI] [PubMed] [Google Scholar]
- Nagle MA, Truong DM, Dnyanmote AV, Ahn SY, Eraly SA, Wu W, Nigam SK. (2011) Analysis of three-dimensional systems for developing and mature kidneys clarifies the role of OAT1 and OAT3 in antiviral handling. J Biol Chem 286:243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam SK, Bush KT, Bhatnagar V. (2007) Drug and toxicant handling by the OAT organic anion transporters in the kidney and other tissues. Nat Clin Pract Nephrol 3:443–448 [DOI] [PubMed] [Google Scholar]
- Ose A, Ito M, Kusuhara H, Yamatsugu K, Kanai M, Shibasaki M, Hosokawa M, Schuetz JD, Sugiyama Y. (2009) Limited brain distribution of [3R,4R,5S]-4-acetamido-5-amino-3-(1-ethylpropoxy)-1-cyclohexene-1-carboxylate phosphate (Ro 64-0802), a pharmacologically active form of oseltamivir, by active efflux across the blood-brain barrier mediated by organic anion transporter 3 (Oat3/Slc22a8) and multidrug resistance-associated protein 4 (Mrp4/Abcc4). Drug Metab Dispos 37:315–321 [DOI] [PubMed] [Google Scholar]
- Schellenberger J, Que R, Fleming RM, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S, et al. (2011) Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat Protoc 6:1290–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson MI, Jamshidi N, Steingrimsson E, Thiele I, Palsson BO. (2010) A detailed genome-wide reconstruction of mouse metabolism based on human Recon 1. BMC Syst Biol 4:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney DE, Vallon V, Rieg T, Wu W, Gallegos TF, Nigam SK. (2011) Functional maturation of drug transporters in the developing, neonatal, and postnatal kidney. Mol Pharmacol 80:147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet DH, Bush KT, Nigam SK. (2001) The organic anion transporter family: from physiology to ontogeny and the clinic. Am J Physiol Renal Physiol 281:F197–F205 [DOI] [PubMed] [Google Scholar]
- Sweet DH, Chan LM, Walden R, Yang XP, Miller DS, Pritchard JB. (2003) Organic anion transporter 3 (Slc22a8) is a dicarboxylate exchanger indirectly coupled to the Na+ gradient. Am J Physiol Renal Physiol 284:F763–F769 [DOI] [PubMed] [Google Scholar]
- Sweet DH, Miller DS, Pritchard JB, Fujiwara Y, Beier DR, Nigam SK. (2002) Impaired organic anion transport in kidney and choroid plexus of organic anion transporter 3 (Oat3 (Slc22a8)) knockout mice. J Biol Chem 277:26934–26943 [DOI] [PubMed] [Google Scholar]
- Sweet DH, Wolff NA, Pritchard JB. (1997) Expression cloning and characterization of ROAT1. The basolateral organic anion transporter in rat kidney. J Biol Chem 272:30088–30095 [DOI] [PubMed] [Google Scholar]
- Truong DM, Kaler G, Khandelwal A, Swaan PW, Nigam SK. (2008) Multi-level analysis of organic anion transporters 1, 3, and 6 reveals major differences in structural determinants of antiviral discrimination. J Biol Chem 283:8654–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsigelny IF, Kouznetsova VL, Sweeney DE, Wu W, Bush KT, Nigam SK. (2008) Analysis of metagene portraits reveals distinct transitions during kidney organogenesis. Sci Signal 1:ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueo H, Motohashi H, Katsura T, Inui K. (2005) Human organic anion transporter hOAT3 is a potent transporter of cephalosporin antibiotics, in comparison with hOAT1. Biochemical pharmacology 70:1104–1113 [DOI] [PubMed] [Google Scholar]
- Vallon V, Eraly SA, Rao SR, Gerasimova M, Rose M, Nagle M, Anzai N, Smith T, Sharma K, Nigam SK, et al. (2012) A role for the organic anion transporter OAT3 in renal creatinine secretion in mice. Am J Physiol Renal Physiol 302:F1293–F1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon V, Eraly SA, Wikoff WR, Rieg T, Kaler G, Truong DM, Ahn SY, Mahapatra NR, Mahata SK, Gangoiti JA, et al. (2008a) Organic anion transporter 3 contributes to the regulation of blood pressure. J Am Soc Nephrol 19:1732–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon V, Rieg T, Ahn SY, Wu W, Eraly SA, Nigam SK. (2008b) Overlapping in vitro and in vivo specificities of the organic anion transporters OAT1 and OAT3 for loop and thiazide diuretics. Am J Physiol Renal Physiol 294:F867–F873 [DOI] [PubMed] [Google Scholar]
- VanWert AL, Bailey RM, Sweet DH. (2007) Organic anion transporter 3 (Oat3/Slc22a8) knockout mice exhibit altered clearance and distribution of penicillin G. Am J Physiol Renal Physiol 293:F1332–F1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanWert AL, Gionfriddo MR, Sweet DH. (2010) Organic anion transporters: discovery, pharmacology, regulation and roles in pathophysiology. Biopharm Drug Dispos 31:1–71 [DOI] [PubMed] [Google Scholar]
- VanWert AL, Sweet DH. (2008) Impaired clearance of methotrexate in organic anion transporter 3 (Slc22a8) knockout mice: a gender specific impact of reduced folates. Pharm Res 25:453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Nagle MA, Kouznetsova VL, Tsigelny IF, Nigam SK. (2011) Untargeted metabolomics identifies enterobiome metabolites and putative uremic toxins as substrates of organic anion transporter 1 (Oat1). J Proteome Res 10:2842–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson G, Aeberli I, Miguet L, Zhang Z, Sanchez MB, Crespy V, Barron D, Needs P, Kroon PA, Glavinas H, et al. (2007) Interaction of positional isomers of quercetin glucuronides with the transporter ABCC2 (cMOAT, MRP2). Drug Metab Dispos 35:1262–1268 [DOI] [PubMed] [Google Scholar]
- Windass AS, Lowes S, Wang Y, Brown CD. (2007a) The contribution of organic anion transporters OAT1 and OAT3 to the renal uptake of rosuvastatin. J Pharmacol Exp Ther 322:1221–1227 [DOI] [PubMed] [Google Scholar]
- Wong CC, Barron D, Orfila C, Dionisi F, Krajcsi P, Williamson G. (2011) Interaction of hydroxycinnamic acids and their conjugates with organic anion transporters and ATP-binding cassette transporters. Mol Nutr Food Res 55:979–988 [DOI] [PubMed] [Google Scholar]
- Wong CC, Botting NP, Orfila C, Al-Maharik N, Williamson G. (2011b) Flavonoid conjugates interact with organic anion transporters (OATs) and attenuate cytotoxicity of adefovir mediated by organic anion transporter 1 (OAT1/SLC22A6). Biochem Pharmacol 81:942–949 [DOI] [PubMed] [Google Scholar]
- Wu W, Baker ME, Eraly SA, Bush KT, Nigam SK. (2009) Analysis of a large cluster of SLC22 transporter genes, including novel USTs, reveals species-specific amplification of subsets of family members. Physiol Genomics 38:116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Dnyanmote AV, Nigam SK. (2011) Remote communication through solute carriers and ATP binding cassette drug transporter pathways: an update on the remote sensing and signaling hypothesis. Mol Pharmacol 79:795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Gong LK, Maeda K, Luan Y, Qi XM, Sugiyama Y, Ren J. (2011) Critical role of organic anion transporters 1 and 3 in kidney accumulation and toxicity of aristolochic acid I. Mol Pharm 8:2183–2192 [DOI] [PubMed] [Google Scholar]
- You G. (2004) Towards an understanding of organic anion transporters: structure-function relationships. Med Res Rev 24:762–774 [DOI] [PubMed] [Google Scholar]
- Zhou F, You G. (2007) Molecular insights into the structure-function relationship of organic anion transporters OATs. Pharm Res 24:28–36 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.