Abstract
Members of the tumor necrosis factor receptor superfamily play key roles in innate and adaptive immunity. Here, we review recent structural studies in the intracellular signal transduction of these receptors. A central theme revealed from these structural studies is that upon ligand binding, multiple intracellular proteins form higher-order signaling machines to transduce and amplify receptor activation information to different cellular fates, including NF-κB activation, apoptosis, and programmed necrosis. These studies open a new vista for understanding the biophysical principles in these signaling cascades.
1. INTRODUCTION
The tumor necrosis factor receptor (TNFR) superfamily consists of 29 transmembrane receptors. Members of TNFRs contain an extracellular domain responsible for ligand binding and an intracellular domain that mediates activation of signaling pathway (Aggarwal, 2003; Bodmer, Schneider, & Tschopp, 2002; Locksley, Killeen, & Lenardo, 2001). TNFRs may be divided into two groups: activating receptors and death receptors (DRs). Most TNFRs are activating receptors, such as CD40 and TNFR2, which can activate nuclear factor κB (NF-κB) and mitogen-activated protein kinase (MAPK) pathways. DRs include eight members, such as TNFR1 and Fas, which have a protein interaction module called the death domain (DD) in the intracellular region that mediates extrinsic signal-induced cell death (Wu & Hymowitz, 2009). TNFR1 is a pleiotropic receptor and is able to induce both activating and death signaling pathways to effect cell metabolism, differentiation, and proliferation (Moquin & Chan, 2010; Schröfelbauer & Hoffmann, 2011). It is activated by the ligand TNFα, which is the founding member of the TNF superfamily.
The ligand/receptor interaction at the extracellular domain has been first revealed by the crystal structure of the trimeric TNFβ-bound symmetrically to the extracellular region of three TNFR1 molecules (Banner et al., 1993). Each TNFR1 chain contacts the interfaces between two protomers of a TNF trimer (Wu & Hymowitz, 2009). A number of subsequent structures of ligand/receptor complexes further confirmed the 3:3 symmetrical interactions at the extracellular region. In this review, we focus on the intra-cellular events in TNFR signaling. Inparticular, we illustrate the structural basis for the induction of NF-κB activation, apoptosis, and programmed necrosis.
2. NF-κB ACTIVATION
Members of the TNFR superfamily activate NF-κB in two alternatively pathways, exemplified by TNFR1 and CD40, respectively. Upon binding with TNFα, the intracellular DD of TNFR1 recruits TNF receptor-associated DD protein (TRADD), which in turn recruits receptor-interacting protein kinase 1 (RIP1), cellular inhibitor of apoptosis proteins 1 and 2 (cIAP1 and 2), and TNF receptor-associated factor 2 (TRAF2; Fig. 5.1). TRADD is important for the TNF-induced NF-κB signaling pathway, as in TRADD-deficient MEFs, IκB phosphorylation and degradation are completely abolished (Chen et al., 2008). The N-terminal region of TRADD interacts with the trimeric TRAF domain of TRAF2 in a 3:3 stoichiometry, whereas the C-terminal DD-containing region of TRADD interacts with many other DD-containing proteins, such as FADD and RIP1 (Park et al., 2000).
Figure 5.1.
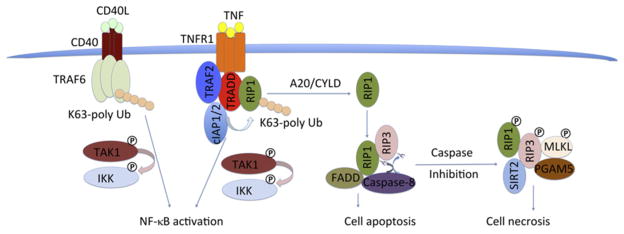
Overview of signaling pathways in the TNF receptor superfamily with TNFR1 and CD40 as prototypes.
The inhibitor of apoptosis proteins cIAP1 and cIAP2 acts as an E3 ligase to form K63 polyubiquitin chains on RIP1 and itself, providing a platform for recruitment of NEMO, the regulatory subunit of the IKK complex (Mahoney et al., 2008). Meanwhile, cIAP1 together with E2 UbcH5 can generate K11 polyubiquitin chains on RIP1 within the endogenous TNFR1 complex and activate NF-κB (Dynek et al., 2010). cIAPs consist of two parts: the N-terminal three baculoviral IAP repeats (BIRs) and CARD and RING domains at the C-terminal region. The structures of BIR1/3 domains, CARD, and RING domains have been determined (Lopez et al., 2011; Mace et al., 2008; Zheng, Kabaleeswaran, Wang, Cheng, & Wu, 2010).
RIP1 is a key factor in mediating TNF-induced signal pathways. In RIP1-deficient T and B cells, TNF-induced NF-κB activation was totally abolished (Feltham et al., 2010). When the E3 ligases TRAF2/cIAP and linear ubiquitin chain assembly complex (LUBAC) ubiquitinate RIP1 in the TNFR1 signaling complex, polyubiquitinated RIP1 engages downstream adaptors such as TGF beta-activated kinase 1 (TAK1) and NEMO to activate IKK, promoting NF-κB transcriptional activity, and leading to cell survival, proliferation, and differentiation (Walczak, 2011).
Besides K63 polyubiquitination, RIP1 and NEMO can also be modified with linear polyubiquitin chain, which is executed by LUBAC, consisting of HOIL-1, HOIP, and SHARPIN (Gerlach et al., 2011; Ikeda et al., 2011). LUBAC can increase the recruitment of cIAP1/2, TRAF2, RIP1, and TAK1 among the TNFR signaling complex, and the depletion of any LUBAC component decreases NF-κB and MAPK activation (Haas et al., 2009).
In the CD40-mediated NF-κB pathway, TRAF6 directly interacts with the intracellular region of the receptor and acts as the ubiquitin ligase to induced K63-linked polyubiquitination (Deng et al., 2000; Fig. 5.1). Similar to the TNFR1 pathway, the polyubiquitin chains engage downstream signaling proteins such as TAK1 and NEMO to activate IKK, leading to IκB phosphorylation, nuclear translocation of NF-κB, and transcription of NF-κB-controlled genes for cell survival, proliferation, and differentiation.
2.1. Structures of TRAFs and TAK1 complex
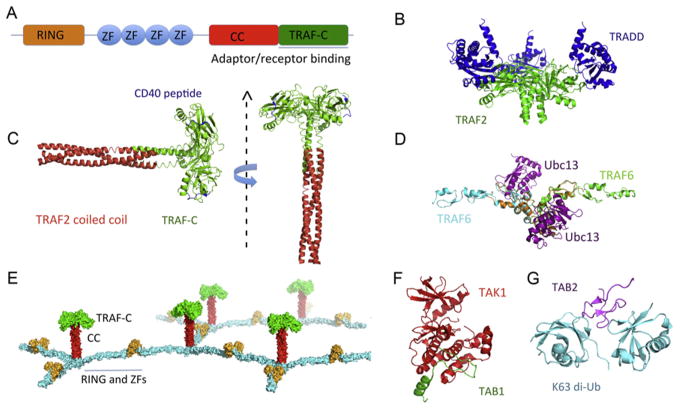
TRAF proteins consist of two parts: an N-terminal RING/zinc-finger domain and a C-terminal coiled-coil/TRAF-C domain (Fig. 5.2A). The N-terminal region of TRAF6 functions as an ubiquitin E3 ligase for K63-linked polyubiquitination (Yin, Lamothe, Darnay, & Wu, 2009; Yin, Lin, et al., 2009). In the TNFR1 pathway, cIAP1 and cIAP2 are the analogous E3 ligases. The coiled-coil/TRAF-C domain mediates interactions with receptors and adaptor proteins, such as TNFR2, CD30, CD40, and TRADD, and is responsible for the specificity and diversity of TRAF recruitment (Park, Burkitt, Villa, Tong, & Wu, 1999; Park et al., 2000; Ye, Cirilli, & Wu, 2002; Ye, Park, Kreishman, Kieff, & Wu, 1999; Ye & Wu, 2000; Zheng et al., 2010). TRAF2 can be recruited to TNFRs either via direct interactions or via intermediate adapter proteins such as TRADD, and TRAF6 is recruited directly to CD40. The interaction between TRADD and TRAF2 (Fig. 5.2B) is much stronger than that in receptor-TRAF2, which ensures the downstream cIAPs recruitment for the direct inhibition of caspase activation in the signaling complex (Park et al., 2000). The trimeric coiled-coil domain of TRAF2 forms a complex with one cIAP2 via direct interaction from two TRAF2 chains (Zheng et al., 2010; Fig. 5.2C).
Figure 5.2.
TRAFs and TAK1 complex structures. (A) Domain organizations of TRAF2 and TRAF6. ZF, zinc finger; CC, coiled coil. (B) Structure of the trimeric TRADD–TRAF2 complex (PDB: 1F3V). (C) Composite C-terminal region structure of TRAF2 based on the structure of TRAF2 CC in complex with cIAP2 BIR1 and TRAF2 CC+TRAF-C in complex with the CD40 peptide, shown in two orientations (PDB: 1CZZ and 3M0A). (D) Ribbon diagram of the TRAF6 RING/ZFs/Ubc13 complex, modeled by superposition of the TRAF6 RING/ZF1/Ubc13 complex with the structure of TRAF6 RING/ZF1–3 (PDB: 3HCS and 3HCT). (E) Model of a 2-dimensional TRAF lattice assembly through trimerization of TRAF domain and dimerization of the N-terminal RING/ZF domains. (F) Structure of TAK1 in complex with an activating TAB1 peptide (PDB: 2EVA). (G) Structure of TAB2 with K63-linked di-Ub complex (PDB: 2WWZ).
Unexpectedly, the N-terminal RING/ZF domains of TRAF6 form a dimer in solution and in the crystal (Yin, Lamothe, et al., 2009; Yin, Lin, et al., 2009; Fig. 5.2D). Dimerization of TRAF6 is important for its E3 ligase activity, which promotes the assembly of polyubiquitination and IκB phosphorylation (Yin, Lamothe, et al., 2009; Yin, Lin, et al., 2009). Based on the symmetry mismatch between the dimeric N-terminal region and the trimeric C-terminal region, a 2D lattice model was proposed to elucidate the infinite oligomerization of TRAF6 and other TRAFs (Fig. 5.2E).
The kinase complex TAK1, also called MAP3K7 and MEKK7 (Yamaguchi et al., 1995), is composed of TAK1 and TAK1-binding proteins (TAB1/2/3; Ishitani et al., 2003). In the TAK1/TAB1 complex structure, the C-terminal lobe of TAK1 kinase domain (KD) forms an extensive interface with an α helix of TAB1 (Brown et al., 2005; Fig. 5.2F). This interaction promotes TAK1 autophosphorylation, most likely through an allosteric mechanism (Ono et al., 2001; Sakurai, Miyoshi, Mizukami, & Sugita, 2000). TAB2 and TAB3 facilitate TAK1 activation via recruiting the K63-linked polyubiquitin chains. The N-terminal zinc-finger domain of TAB2/TAB3 binds diubiquitin identically in crystal structures, and prefers the K63-linked polyubiquitin chains to K48-linked ones as the conformational constraint does not favor the linear linkage (Ono et al., 2001; Sakurai et al., 2000; Fig. 5.2G). The TRAF-polyubiquitin chains act as a scaffold to bring the activated TAK1 to the proximal space of IKK and, in turn, phosphorylate and activate IKK complex.
2.2. Structure of the IKK complex
The IKK complex consists of two catalytic subunits IKKα/IKKβ and a regulatory subunit NEMO (IKKg; Chen, Parent, & Maniatis, 1996; Mercurio et al., 1997). The recently solved IKKβ structure reveals a trimodular architecture (Xu et al., 2011; Fig. 5.3A and B), which is composed of a KD, an ubiquitin-like domain (ULD), and an elongated α-helical scaffold/dimerization domain (SDD). IKKβ is dimeric in solution and in crystal lattice (Xu et al., 2011); however, IKKβ may form a high-order oligomerization during activation as the dimeric conformation does not facilitate the intra-dimeric trans-autophosphorylation. NEMO contains a UBAN (ubiquitin binding in ABIN and NEMO) and a zinc-finger (ZF) domain at its C-terminal end, which mediates the interaction between the IKK complex and the polyubiquitin chains (Rothwarf, Zandi, Natoli, & Karin, 1998; Yamaoka et al., 1998). The UBAN domain prefers linear poly-ubiquitin chains, evidenced in the crystal structures that UBAN binds to both ubiquitins in linear diubiquitin but does not make simultaneous contacts with both ubiquitins in K63-linked diubiquitin (Lo et al., 2009; Rahighi et al., 2009; Yoshikawa et al., 2009; Fig. 5.3C). Most likely, linear and K63-linked polyubiquitin chains play roles in response to different stimuli. NEMO binds to the C-terminal NEMO-binding domain (NBD) of IKKα and IKKβ (Marienfeld, Palkowitsch, & Ghosh, 2006) and forms a parallel four-helix bundled heterotetramer with two molecules of each protein (Rushe et al., 2008). Based on the structural information available for IKKβ and NEMO, a model for the IKK complex was proposed (Ferrao, Li, Bergamin, & Wu, 2012; Zheng, Yin, & Wu, 2011; Fig. 5.3C).
Figure 5.3.
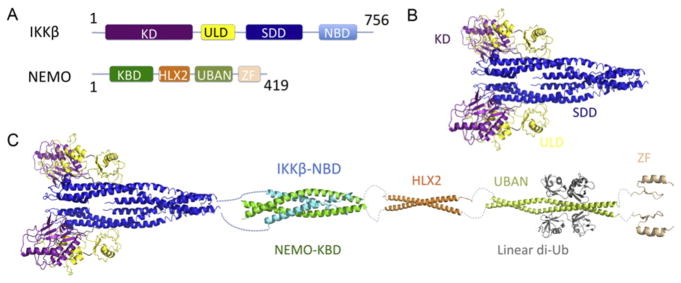
The IKKβ complex. (A) Domain schematics of human IKKβ and NEMO. (B) The dimeric structure of IKKβ, consisting of the KD domain (purple), ULD domain (yellow), and SDD domain (blue). (C) Model of the full-length IKKβ–NEMO complex. The IKKβ NBD (light blue) interacts with the N-terminal kinase-binding domain (KBD) of NEMO (green). The linear representation of NEMO is based on the domain organization, with a HLX2 domain (orange), a UBAN domain (lemon), ubiquitins (gray), and a C-terminal zinc-finger (ZF) domain (light orange).
3. TNFR1 AND FAS-INDUCED APOPTOSIS
TNFR1 and Fas belong to the DR family. Fas induces cell apoptotic death. When the ligand FasL binds to the extracellular region of Fas, the cytosolic region of Fas recruits the adaptor protein FADD via the DD interaction. FADD consists of a DD and a death effector domain (DED), through which FADD recruits caspase-8 and -10 via the interactions with the tandem DEDs in the prodomain of the caspases (Carrington et al., 2006; Strasser, Jost, & Nagata, 2009; Fig. 5.4A). The ternary complex, composed with Fas, FADD, and caspase-8/10, has been traditionally named the death-inducing signaling complex (DISC; Kischkel et al., 1995), which brings the catalytic domains of the caspases into proximity for dimerization and autoprocessing. DDs and DEDs belong to the DD-fold superfamily (Park et al., 2007). Proteins containing these domains form oligomeric complexes through homotypic interactions, which play central roles in different apoptotic and inflammatory pathways (Ferrao & Wu, 2012).
Figure 5.4.
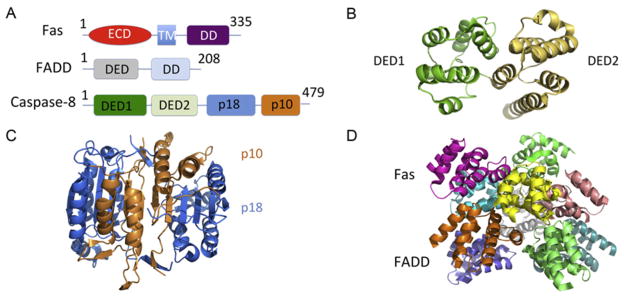
Structures of the death-inducing signaling complex (DISC). (A) Domain schematics of human Fas, FADD, and procaspase-8. The Fas consists of extracellular domain (ECD, red), transmembrane domain (TM, light blue), and intracellular death domain (DD, purple). The FADD consists of DED (gray) and DD (wheat). The caspase-8 consists of N-terminal DED1 (green), DED2 (lime), and C-terminal p18 (blue) and p10 (orange) subunits. (B) The DED1/2 domain structure of vFLIP, which is a viral caspase-8/10 inhibitory protein (PDB: 2BBR). (C) The catalytic domain structures (p18 and p10) of Caspase-8 (PDB: 2Y1L). (D) The Fas DD:FADD DD complex forms a 5:5 asymmetric structure (PDB: 3OQ9).
Upon ligand binding, the cytosolic region of TNFR1 forms a large complex, which includes TRADD, TRAF2/5, cIAP1/2, and RIP1. When K63-linked polyubiquitin chain of RIP1 is removed by the deubiquitinases CYLD (cylindromatosis) or A20 (Sun, 2010; Wilson, Dixit, & Ashkenazi, 2009), or blocked by removal of the E3 ligases cIAP1 and cIAP2 through genetic ablation, RNAi knockdown, or IAP antagonists, RIP1 and its family member RIP3 are recruited to a second complex containing TRADD, FADD, and caspase-8 (Feoktistova et al., 2011; Tenev et al., 2011). In either the TRADD/FADD/caspase-8 or the RIP1/FADD/caspase-8 complex, caspase-8 is activated, which in turn cleaves RIP1 and RIP3, and induces cell apoptosis (Bertrand et al., 2008; Wang, Du, & Wang, 2008).
3.1. Structures of individual proteins in the DISC
The NMR structure of Fas DD reveals an antiparallel six-helical bundle architecture, which is common among the entire DD superfamily (Huang, Eberstadt, Olejniczak, Meadows, & Fesik, 1996). The DD, DED, and full-length (FL) FADD structures have been determined by NMR method (Carrington et al., 2006; Eberstadt et al., 1998; Jeong et al., 1999). The structure of the tandem DED domain of caspase-8 or caspase-10 has not been resolved. However, the structure of the tandem DED domain of a viral caspase-8/10 and FLICE/caspase-8 inhibitory protein (FLIP) from poxvirus Molluscum contagiosum virus has been determined, revealing a dumbbell-shaped arrangement common to all tandem DEDs (Li, Jeffrey, Yu, & Shi, 2006; Yang et al., 2005; Fig. 5.4B). The structure of the catalytic domain of caspase-8 reveals a dimeric structure common to all caspases (Blanchard et al., 1999; Watt et al., 1999; Fig. 5.4C).
3.2. Structure of the Fas DD:FADD DD complex in the DISC
The structure of the Fas DD:FADD DD complex has been elucidated (Wang et al., 2010; Fig. 5.4D). Similar to the structure of PIDD DD: RAIDD DD complex (Park et al., 2007), the class projection averages of negatively stained Fas DD:FADD DD complex shows an asymmetric oligomeric structure. Moreover, it was shown that the Fas DD:FADD DD complex contains a mixture of 5 Fas:5 FADD, 6 Fas:5 FADD, and 7 Fas:5 FADD complexes evidenced by the nanoflow electrospray ionization and tandem mass spectrometry method. Based on the layered structure of the PIDD DD:RAIDD DD complex, a 5:5 core Fas DD:FADD DD complex was built and used to solve a low-resolution crystal structure of the complex (Wang et al., 2010). Mutations on the interfaces between Fas and FADD affected the complex formation and directly explained the dominant-negative effects from Fas mutations that are associated with autoimmune lymphoproliferative syndrome in humans (Wang et al., 2010). The structure shows the similar helical assembly architecture as the death domain complex in Toll-like receptor signaling (Lin et al., 2010).
3.3. DED chains in the DISC
The formation of the DISC is essential for Fas-mediated apoptosis. Recent findings identified the stoichiometry of the Fas DISC (Dickens et al., 2012; Schleich et al., 2012). Among the DISC, the amount of DED proteins procaspase-8/10 and c-FLIP exceeds that of FADD by seven- to ninefold with quantitative western blots, mass spectrometry, and mathematical modeling methods (Dickens et al., 2012; Schleich et al., 2012). One proposed model showed that procaspase-8/10 and c-FLIP could form a caspase-activating chain via their DED domains. Mutations of some key interacting residues in procaspase-8 DED2 abrogate DED chain formation in cells (Dickens et al., 2012; Schleich et al., 2012). Moreover, the DED of FADD and the DED2 of procaspase-8 form filaments in Hela and Jurkat Tag cells, which can be blocked by coexpression of viral antiapoptotic DED-containing proteins (MC159 and E8), but not by bcl-2 family proteins (Siegel et al., 1998). The DED chain assembly in DISC may drive caspase-8 dimerization and activation, leading to cell apoptosis.
4. TNFR1-INDUCED PROGRAMMED NECROSIS
Cell necrosis is distinct from cell apoptosis with swelling and membrane rupture, resulting in the loss of membrane integrity and cytoplasmic leakage (Yuan & Kroemer, 2010). Recent studies showed that programmed necrosis is an alternative route to cell death that is distinct from apoptosis in the immune system (Galluzzi et al., 2012; Han, Zhong, & Zhang, 2011; Kaiser et al., 2011; Mocarski, Upton, & Kaiser, 2011).
In the TNFR1 pathway, active caspase-8 cleaves and inactivates RIP1 (Chan et al., 2003; Lin, Devin, Rodriguez, & Liu, 1999) and RIP3 (Feng et al., 2007). When caspases are inhibited by pharmacological inhibitors or under certain physiological conditions such as viral infections, RIP1 and RIP3 form the necrosome to initiate programmed necrosis or necroptosis (Cho et al., 2009; He et al., 2009; Zhang et al., 2009). The RIP1/RIP3 complex is the core of the necrosome, which also contains other components, such as mixed lineage kinase domain like protein (MLKL).
RIP1 and RIP3 are important in host defense against bacterial and viral infections (Robinson et al., 2012; Upton, Kaiser, & Mocarski, 2010, 2012). Dysregulation of the pathway appears to be involved in many human diseases, such as lymphoproliferative diseases (Ch’en, Tsau, Molkentin, Komatsu, & Hedrick, 2011; Kaiser et al., 2011), atherosclerosis development (Lin et al., 2013), Crohn’s disease (Welz et al., 2011), acute liver injury (Liedtke et al., 2011), ischemic brain injury (Degterev et al., 2005; Northington et al., 2011), myocardial ischemia–reperfusion injury (Oerlemans et al., 2012), and skin inflammation (Bonnet et al., 2011). In addition, RIP3 is responsible for the embryonic lethality of caspase-8−/− mice as the caspase-8−/− /RIP3−/− double knockout mice are viable (Kaiser et al., 2011). Similarly, RIP1 deficiency can rescue the lethality of FADD−/− mice (Zhang et al., 2011), suggesting that necroptosis plays key roles in cell development.
4.1. RIP1/RIP3 forms a functional amyloid signaling complex
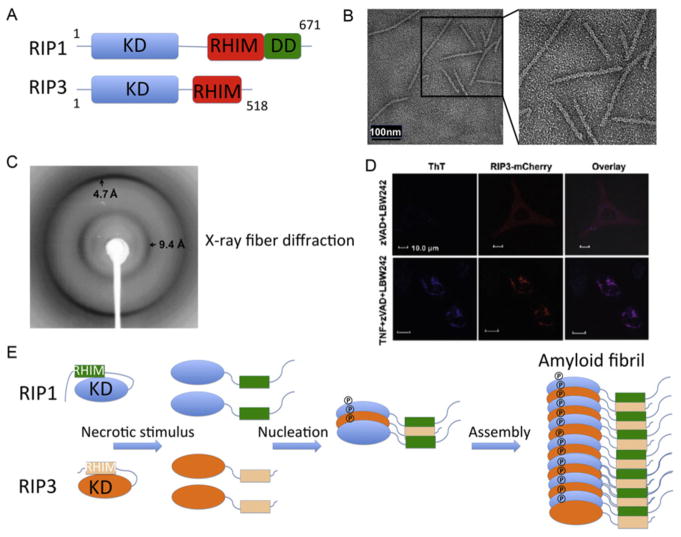
Both RIP1 and RIP3 share a KD and a RIP homotypic interaction motif (RHIM), whereas RIP1 has one more DD at its C-terminal end (Fig. 5.5A). RIP1 and RIP3 form a necrotic signaling complex via the RHIM (Stanger, Leder, Lee, Kim, & Seed, 1995; Sun et al., 1999; Sun, Yin, Starovasnik, Fairbrother, & Dixit, 2002). The RHIM contains a high conserved (I/V/L)Q(I/V/L)G motif. Recombinant RIP1/RIP3-RHIM and RIP1/RIP3 FL complexes eluted from around the void position in gel filtration chromatography, much larger than the expected molecular weight of a heterodimer (Li et al., 2012). The complexes were identified as filamentous structures under electron microscopy (EM; Fig. 5.5B). Moreover, the RIP1/RIP3-FL and endogenous RIP1/RIP3-FL complexes purified with anti-RIP1 antibody from HT-29 cells showed the similar filamentous core structures upon limited proteolysis to remove flanking domains and proteins.
Figure 5.5.
RIP1/RIP3 forms a filamentous structure during TNF-induced programmed necrosis. (A) Domain schematics of human RIP1 and RIP3. (B) EM images of the RIP1/ RIP3–RHIM complex. (C) An X-ray diffraction image of partially aligned RIP1/RIP3 fibrils. (D) Colocalization of ThT with RIP3 puncta in necrotic HeLa cells. (E) A proposed model for RIP1/RIP3 fibril assembly. Phosphorylation and necrosome formation may be mutually reinforcing.
Amyloids are fibrous protein aggregates composed of cross-β cores (Chiti & Dobson, 2006). The RIP1/RIP3 complex showed classical characteristics of β-amyloid fibrils with specific binding to amyloid-interacting dyes (Thioflavin T and Congo Red) and in Fourier transform infrared spectra (Li et al., 2012). In addition, the X-ray fiber diffraction clearly showed equatorial and meridional reflections at 9.4 and 4.7 Å resolutions, respectively, which correspond to inter- and intra-β-sheet spacings in cross-β amyloid structures (Fig. 5.5C). Additionally, in RIP3−/− cells reconstituted with RIP3-mCherry, necrosis induction resulted in formation of Thioflavin T (ThT)-positive clusters (Fig. 5.5D), demonstrating the physiological relevance of the RIP1/RIP3 amyloid structure (Li et al., 2012).
The recombinant and endogenous RIP1/RIP3 complex is ultrastable, consistent with the generally recognized stability of amyloidal structures (Balbirnie, Grothe, & Eisenberg, 2001). Mutagenesis in the RHIM domain and ThT staining experiments showed the core regions of RHIMs are crucial for cluster formation, kinase activity, and programmed necrosis (Li et al., 2012). In addition, both the RHIM domains and kinase activity of RIP1/RIP3 are required for TNF-induced programmed necrosis (Cho et al., 2009; Li et al., 2012). Amyloid-β amyloidogenesis occurs via a nucleated polymerization mechanism (Eisenberg & Jucker, 2012). Similarly, we propose that RIP1 and RIP3 follow a feed-forward nucleation model in which RIP1 and RIP3 kinase activation and the RIP1/RIP3 amyloid scaffold formation are mutually reinforcing (Fig. 5.5E). The amyloid scaffold may function as a crucial platform for recruiting other components, such as MLKL, and trigger the downstream execution mechanisms of necroptosis (Li et al., 2012).
4.2. New components of TNFR-induced cell necrosis
MLKL was recently identified as the downstream substrate of RIP3 (Sun et al., 2012; Zhao et al., 2012). Phosphorylated RIP3 can interact with MLKL and phosphorylate MLKL at sites Thr357 and Ser358. The phosphorylation on the two amino acid residues is necessary but not sufficient for necroptosis. Knockdown of MLKL can abolish TNF-induced cell necrosis. The phosphorylated MLKL may induce downstream JNK activation and reactive oxygen species (ROS) generation, eventually triggering cell death.
Another new component of the necrosome, called PGAM5 (phosphoglycerate mutase/protein phosphatase), was identified by coimmunoprecipitation withRIP3inHela cells (Wang, Jiang, Chen, Du, & Wang, 2012). Knockdown of either types of PGAM5 (long- or short-form variant) led to attenuation in TNF-induced necrosis as well as in generation of ROS and calcium ionophore in Hela cells. In contrast, knocking down RIP3 and MLKL only resulted in blockage of necrosis. The fact that PGAM5 can dephosphorylate mitochondrial fission factor Drp1 and activate its GTPase activity shows PGAM5 functions in multiple necrosis pathways (Wang et al., 2012). In addition, PGAM5L is required for NLRP3 inflammasome activation besides necrotic death (Kang, Yang, Toth, Kovalenko, & Wallach, 2013).
Interestingly, recent studies found that programmed necrosis has relationships with the NAD-dependent deacetylase SIRT2 (Narayan et al., 2012). SIRT2 binds constitutively to the C-terminal RHIM domain of RIP3. However, it is not clear whether the RHIM domain is sufficient for the interaction between the two proteins. Deletion or siRNA knockdown of SIRT2 can block the formation of the RIP1/RIP3 complex in mice. SIRT2 regulates RIP1 acetylation via deacetylation at Lys530 of RIP1, which promotes RIP1/RIP3 complex formation in TNF-induced necrosis. When SIRT2 is inhibited by a specific pharmacological inhibitor AGK2, ischemic injury in the heart and the brain is reduced in mice (Narayan et al., 2012).
5. CONCLUSIONS
Members of the TNF receptor superfamily induce a diverse array of cell fates, including NF-κB activation, apoptosis, and programmed necrosis. Structural studies on the intracellular signaling complexes in these pathways have revealed that higher-order signaling machines are the central entities responsible for transmission of receptor activation information to cellular processes. Because many protein families are shared between the TNF receptor signaling pathway and other aspects of the innate and adaptive immunity system, we propose that higher-order supramolecular assemblies represent a conserved central theme in host defense. These higher-order signaling machines implicate new molecular mechanisms in proximity-driven enzyme activation, threshold behavior, and time delay of activation (Wu, 2013) that may be crucial for mounting an immune response only when the danger is sufficient and persistent to avoid accidental tissue injury associated with inflammation. Future structural and functional studies of these multiprotein assemblies may unveil the molecular basis for oligomerization-induced allosteric changes in enzyme activation in these pathways and provide new and promising avenues for therapeutic intervention.
Acknowledgments
We apologize for incomplete citations due to space limitations and thank funding support from the National Institute of Health (to H. W.).
References
- Aggarwal B. Signaling pathways of the TNF superfamily: A double-edged sword. Nature Reviews Immunology. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- Balbirnie M, Grothe R, Eisenberg DS. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated beta-sheet structure for amyloid. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:2375–2380. doi: 10.1073/pnas.041617698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banner DW, D’Arcy A, Janes W, Gentz R, Schoenfeld HJ, Broger C, et al. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Molecular Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Blanchard H, Kodandapani L, Mittl PR, Marco SD, Krebs JF, Wu JC, et al. The three-dimensional structure of caspase-8: An initiator enzyme in apoptosis. Structure. 1999;7:1125–1133. doi: 10.1016/s0969-2126(99)80179-8. [DOI] [PubMed] [Google Scholar]
- Bodmer JL, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends in Biochemical Sciences. 2002;27:19–26. doi: 10.1016/s0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- Bonnet MC, Preukschat D, Welz PS, van Loo G, Ermolaeva MA, Bloch W, et al. The adaptor protein FADD protects epidermal keratinocytes from necroptosis in vivo and prevents skin inflammation. Immunity. 2011;35:572–582. doi: 10.1016/j.immuni.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Brown K, Vial SC, Dedi N, Long JM, Dunster NJ, Cheetham GMT. Structural basis for 0074he interaction of TAK1 kinase with its activating protein TAB1. Journal of Molecular Biology. 2005;354:1013–1020. doi: 10.1016/j.jmb.2005.09.098. [DOI] [PubMed] [Google Scholar]
- Carrington PE, Sandu C, Wei Y, Hill JM, Morisawa G, Huang T, et al. The structure of FADD and its mode of interaction with procaspase-8. Molecular Cell. 2006;22:599–610. doi: 10.1016/j.molcel.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Chan FK, Shisler J, Bixby JG, Felices M, Zheng L, Appel M, et al. A role for tumor necrosis factor receptor-2 and receptor-interacting protein in programmed necrosis and antiviral responses. Journal of Biological Chemistry. 2003;278:51613–51621. doi: 10.1074/jbc.M305633200. [DOI] [PubMed] [Google Scholar]
- Chen NJ, Chio II, Lin WJ, Duncan G, Chau H, Katz D, et al. Beyond tumor necrosis factor receptor: TRADD signaling in toll-like receptors. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12429–12434. doi: 10.1073/pnas.0806585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- Ch’en IL, Tsau JS, Molkentin JD, Komatsu M, Hedrick SM. Mechanisms of necroptosis in T cells. The Journal of Experimental Medicine. 2011;208:633–641. doi: 10.1084/jem.20110251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annual Review of Biochemistry. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nature Chemical Biology. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, et al. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- Dickens LS, Boyd RS, Jukes-Jones R, Hughes MA, Robinson GL, Fairall L, et al. A death effector domain chain DISC model reveals a crucial role for caspase-8 chain assembly in mediating apoptotic cell death. Molecular Cell. 2012;47:291–305. doi: 10.1016/j.molcel.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO Journal. 2010;29:4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberstadt M, Huang B, Chen Z, Meadows RP, Ng SC, Zheng L, et al. NMR structure and mutagenesis of the FADD (Mort1) death-effector domain. Nature. 1998;392:941–945. doi: 10.1038/31972. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltham R, Moulin M, Vince JE, Mace PD, Wong WW, Anderton H, et al. Tumor necrosis factor (TNF) signaling, but not TWEAK (TNF-like weak inducer of apoptosis)-triggered cIAP1 (cellular inhibitor of apoptosis protein 1) degradation, requires cIAP1 RING dimerization and E2 binding. Journal of Biological Chemistry. 2010;285:17525–17536. doi: 10.1074/jbc.M109.087635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Yang Y, Mei Y, Ma L, Zhu DE, Hoti N, et al. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cellular Signalling. 2007;19:2056–2067. doi: 10.1016/j.cellsig.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M, et al. cIAPs block ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Molecular Cell. 2011;43:449–463. doi: 10.1016/j.molcel.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrao R, Li J, Bergamin E, Wu H. Structural insights into the assembly of large oligomeric signalosomes in the toll-like receptor-interleukin-1 receptor superfamily. Science Signaling. 2012;5(226):re3. doi: 10.1126/scisignal.2003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Current Opinion in Structural Biology. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, et al. Molecular definitions of cell death subroutines: Recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death and Differentiation. 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Molecular Cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Han J, Zhong CQ, Zhang DW. Programmed necrosis: Backup to and competitor with apoptosis in the immune system. Nature Immunology. 2011;12:1143–1149. doi: 10.1038/ni.2159. [DOI] [PubMed] [Google Scholar]
- He S, Wang L, Miao L, Wang T, Du F, Zhao L, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Deribe YL, Skånland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani T, Takaesu G, Ninomiya-Tsuji J, Shibuya H, Gaynor RB, Matsumoto K. Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO Journal. 2003;22:6277–6288. doi: 10.1093/emboj/cdg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong EJ, Bang S, Lee TH, Park YI, Sim WS, Kim KS. The solution structure of FADD death domain. Structural basis of death domain interactions of Fas and FADD. Journal of Biological Chemistry. 1999;274:16337–16342. doi: 10.1074/jbc.274.23.16337. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R, et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 2011;471:368–372. doi: 10.1038/nature09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang TB, Yang SH, Toth B, Kovalenko A, Wallach D. Caspase-8 blocks kinase RIPK3-mediated activation of the NLRP3 inflammasome. Immunity. 2013;24:27–40. doi: 10.1016/j.immuni.2012.09.015. [DOI] [PubMed] [Google Scholar]
- Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, et al. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO Journal. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FY, Jeffrey PD, Yu JW, Shi Y. Crystal structure of a viral FLIP: Insights into FLIP-mediated inhibition of death receptor signaling. Journal of Biological Chemistry. 2006;281:2960–2968. doi: 10.1074/jbc.M511074200. [DOI] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke C, Bangen JM, Freimuth J, Beraza N, Lambertz D, Cubero FJ, et al. Loss of caspase-8 protects mice against inflammation-related hepatocarcinogenesis but induces non-apoptotic liver injury. Gastroenterology. 2011;141:2176–2187. doi: 10.1053/j.gastro.2011.08.037. [DOI] [PubMed] [Google Scholar]
- Lin Y, Devin A, Rodriguez Y, Liu ZG. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes & Development. 1999;13:2514–2526. doi: 10.1101/gad.13.19.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Li H, Yang M, Ren J, Huang Z, Han F, et al. A role of RIP3-mediated macrophage necrosis in atherosclerosis development. Cell Reports. 2013;3:200–210. doi: 10.1016/j.celrep.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signaling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YC, Lin SC, Rospigliosi CC, Conze DB, Wu CJ, Ashwell JD, et al. Structural basis for recognition of diubiquitins by NEMO. Molecular Cell. 2009;33:602–615. doi: 10.1016/j.molcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: Integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Lopez J, John SW, Tenev T, Rautureau GJ, Hinds MG, Francalanci F, et al. CARD-mediated autoinhibition of cIAP1’s E3 ligase activity suppresses cell proliferation and migration. Molecular Cell. 2011;42:569–583. doi: 10.1016/j.molcel.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Mace PD, Linke K, Feltham R, Schumacher FR, Smith CA, Vaux DL, et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. Journal of Biological Chemistry. 2008;283:31633–31640. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- Mahoney DJ, Cheung HH, Mrad RL, Plenchette S, Simard C, Enwere E, et al. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11778–11783. doi: 10.1073/pnas.0711122105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marienfeld RB, Palkowitsch L, Ghosh S. Dimerization of the I kappa B kinase-binding domain of NEMO is required for tumor necrosis factor alpha-induced NF-kappa B activity. Molecular and Cellular Biology. 2006;26:9209–9219. doi: 10.1128/MCB.00478-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, et al. IKK-1 and IKK-2: Cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- Mocarski ES, Upton JW, Kaiser WJ. Viral infection and the evolution of caspase 8-regulated apoptotic and necrotic death pathways. Nature Reviews Immunology. 2011;12:79–88. doi: 10.1038/nri3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moquin D, Chan FK. The molecular regulation of programmed necrotic cell injury. Trends in Biochemical Sciences. 2010;35:434–441. doi: 10.1016/j.tibs.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan N, Lee IH, Borenstein R, Sun J, Wong R, Tong G, et al. The NAD-dependent deacetylase SIRT2 is required for programmed necrosis. Nature. 2012;492:199–204. doi: 10.1038/nature11700. [DOI] [PubMed] [Google Scholar]
- Northington FJ, Chavez-Valdez R, Graham EM, Razdan S, Gauda EB, Martin LJ. Necrostatin decreases oxidative damage, inflammation, and injury after neonatal HI. Journal of Cerebral Blood Flow and Metabolism. 2011;31:178–189. doi: 10.1038/jcbfm.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oerlemans MI, Liu J, Arslan F, den Ouden K, van Middelaar BJ, Doevendans PA, et al. Inhibition of RIP1-dependent necrosis prevents adverse cardiac remodeling after myocardial ischemia-reperfusion in vivo. Basic Research in Cardiology. 2012;107:270. doi: 10.1007/s00395-012-0270-8. [DOI] [PubMed] [Google Scholar]
- Ono K, Ohtomo T, Sato S, Sugamata Y, Suzuki M, Hisamoto N, et al. An evolutionarily conserved motif in the TAB1 C-terminal region is necessary for interaction with and activation of TAK1 MAPKKK. Journal of Biological Chemistry. 2001;276:24396–24400. doi: 10.1074/jbc.M102631200. [DOI] [PubMed] [Google Scholar]
- Park YC, Burkitt V, Villa AR, Tong L, Wu H. Structural basis for self-association and receptor recognition of human TRAF2. Nature. 1999;398:533–538. doi: 10.1038/19110. [DOI] [PubMed] [Google Scholar]
- Park HH, Logette E, Raunser S, Cuenin S, Walz T, Tschopp J, et al. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YC, Ye H, Hsia C, Segal D, Rich RL, Liou HC, et al. A novel mechanism of TRAF signaling revealed by structural and functional analyses of the TRADD–TRAF2 interaction. Cell. 2000;101:777–787. doi: 10.1016/s0092-8674(00)80889-2. [DOI] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Robinson N, McComb S, Mulligan R, Dudani R, Krishnan L, Sad S. Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nature Immunology. 2012;13:954–962. doi: 10.1038/ni.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwarf DM, Zandi E, Natoli G, Karin K. IKK-gamma is an essential regulatory subunit of the IkappaB kinase complex. Nature. 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- Rushe M, Silvian L, Bixler S, Chen LL, Cheung A, Bowes S, et al. Structure of a NEMO/IKK-associating domain reveals architecture of the interaction site. Structure. 2008;16:798–808. doi: 10.1016/j.str.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Sakurai H, Miyoshi H, Mizukami J, Sugita T. Phosphorylation-dependent activation of TAK1 mitogen-activated protein kinase kinase kinase by TAB1. FEBS Letters. 2000;474:141–145. doi: 10.1016/s0014-5793(00)01588-x. [DOI] [PubMed] [Google Scholar]
- Schleich K, Warnken U, Fricker N, Oztürk S, Richter P, Kammerer K, et al. Stoichiometry of the CD95 death-inducing signaling complex: Experimental and modeling evidence for a death effector domain chain model. Molecular Cell. 2012;47:306–319. doi: 10.1016/j.molcel.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Schröfelbauer B, Hoffmann A. How do pleiotropic kinase hubs mediate specific signaling by TNFR superfamily members? Immunological Reviews. 2011;244:29–43. doi: 10.1111/j.1600-065X.2011.01060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RM, Martin DA, Zheng L, Ng SY, Bertin J, Cohen J, et al. Death-effector filaments: Novel cytoplasmic structures that recruit caspases and trigger apoptosis. The Journal of Cell Biology. 1998;141:1243–1253. doi: 10.1083/jcb.141.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger BZ, Leder P, Lee TH, Kim E, Seed B. RIP: A novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–192. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC. CYLD: A tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death and Differentiation. 2010;17:25–34. doi: 10.1038/cdd.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Lee J, Navas T, Baldwin DT, Stewart TA, Dixit VM. RIP3, a novel apoptosis-inducing kinase. Journal of Biological Chemistry. 1999;274:16871–16875. doi: 10.1074/jbc.274.24.16871. [DOI] [PubMed] [Google Scholar]
- Sun L, Wang H, Wang Z, He S, Chen S, Liao D, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- Sun X, Yin J, Starovasnik MA, Fairbrother WJ, Dixit VM. Identification of a novel homotypic interaction motif required for the phosphorylation of receptor-interacting protein (RIP) by RIP3. Journal of Biological Chemistry. 2002;277:9505–9511. doi: 10.1074/jbc.M109488200. [DOI] [PubMed] [Google Scholar]
- Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, et al. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Molecular Cell. 2011;43:432–448. doi: 10.1016/j.molcel.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Upton JW, Kaiser WJ, Mocarski ES. Virus inhibition of RIP3-dependent necrosis. Cell Host & Microbe. 2010;7:302–313. doi: 10.1016/j.chom.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton JW, Kaiser WJ, Mocarski ES. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cyto-megalovirus vIRA. Cell Host & Microbe. 2012;11:290–297. doi: 10.1016/j.chom.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak H. TNF and ubiquitin at the crossroads of gene activation, cell death, inflammation, and cancer. Immunology Reviews. 2011;244:9–28. doi: 10.1111/j.1600-065X.2011.01066.x. [DOI] [PubMed] [Google Scholar]
- Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148:228–243. doi: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- Wang L, Yang JK, Kabaleeswaran V, Rice AJ, Cruz AC, Park AY, et al. The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nature Structural and Molecular Biology. 2010;17:1324–1329. doi: 10.1038/nsmb.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt W, Koeplinger KA, Mildner AM, Heinrikson RL, Tomasselli AG, Watenpaugh KD. The atomic-resolution structure of human caspase-8, a key activator of apoptosis. Structure. 1999;7:1135–1143. doi: 10.1016/s0969-2126(99)80180-4. [DOI] [PubMed] [Google Scholar]
- Welz PS, Wullaert A, Vlantis K, Kondylis V, Fernandez-Majada V, Ermolaeva M, et al. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 2011;477:330–334. doi: 10.1038/nature10273. [DOI] [PubMed] [Google Scholar]
- Wilson NS, Dixit V, Ashkenazi A. Death receptor signal transducers: Nodes of coordination in immune signaling networks. Nature Immunology. 2009;10:348–355. doi: 10.1038/ni.1714. [DOI] [PubMed] [Google Scholar]
- Wu H. Higher order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Hymowitz SG. Structure and function of tumor necrosis factor (TNF) at the cell surface. In: Bradshaw Ralph A, Dennis Edward A., editors. Handbook of cell signaling. Oxford: Academic Press; 2009. pp. 265–275. [Google Scholar]
- Xu G, Lo Y, Li Q, Napolitano G, Wu X, Jiang X, et al. Crystal structure of inhibitor of κB kinase. Nature. 2011;472:325–330. doi: 10.1038/nature09853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, et al. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science. 1995;270:2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- Yamaoka S, Courtois G, Bessia C, Whiteside ST, Weil R, Agou F, et al. Complementation cloning of NEMO, a component of the IκB kinase complex essential for NF-κB activation. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- Yang JK, Wang L, Zheng L, Wan F, Ahmed M, Lenardo MJ, et al. Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition. Molecular Cell. 2005;20:939–949. doi: 10.1016/j.molcel.2005.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Cirilli M, Wu H. The use of construct variation and diffraction data analysis in the crystallization of the TRAF domain of human tumor necrosis factor receptor associated factor 6. Acta Crystallographica Section D: Biological Crystallography. 2002;58:1886–1888. doi: 10.1107/s0907444902013318. [DOI] [PubMed] [Google Scholar]
- Ye H, Park YC, Kreishman M, Kieff E, Wu H. The structural basis for the recognition of diverse receptor sequences by TRAF2. Molecular Cell. 1999;4:321–330. doi: 10.1016/s1097-2765(00)80334-2. [DOI] [PubMed] [Google Scholar]
- Ye H, Wu H. Thermodynamic characterization of the interaction between TRAF2 and receptor peptides by isothermal titration calorimetry. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:8961–8966. doi: 10.1073/pnas.160241997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Lamothe B, Darnay BG, Wu H. Structural basis for the lack of E2 interaction in the RING domain of TRAF2. Biochemistry. 2009;48:10558–10567. doi: 10.1021/bi901462e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nature Structural and Molecular Biology. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa A, Sato Y, Yamashita M, Mimura H, Yamagata A, Fukai S. Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63-linked di-ubiquitin. FEBS Letters. 2009;583:3317–3322. doi: 10.1016/j.febslet.2009.09.028. [DOI] [PubMed] [Google Scholar]
- Yuan J, Kroemer G. Alternative cell death mechanisms in development and beyond. Genes & Development. 2010;24:2592–2602. doi: 10.1101/gad.1984410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhou X, McQuade T, Li J, Chan FK, Zhang J. Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature. 2011;471:373–376. doi: 10.1038/nature09878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q, Luo J, et al. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5322–5327. doi: 10.1073/pnas.1200012109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Kabaleeswaran V, Wang Y, Cheng G, Wu H. Crystal structures of the TRAF2:cIAP2 and the TRAF1:TRAF2:cIAP2 complexes: Affinity, specificity, and regulation. Molecular Cell. 2010;38:101–113. doi: 10.1016/j.molcel.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Yin Q, Wu H. Structural studies of NF-κB signaling. Cell Research. 2011;21:183–195. doi: 10.1038/cr.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]