Abstract
Exploitation of custom-designed nucleases to induce DNA double-strand breaks (DSBs) at genomic locations of choice has transformed our ability to edit genomes, regardless of their complexity. DSBs can trigger either error-prone repair pathways that induce random mutations at the break sites or precise homology-directed repair pathways that generate specific insertions or deletions guided by exogenously supplied DNA. Prior editing strategies using site-specific nucleases to modify the Caenorhabditis elegans genome achieved only the heritable disruption of endogenous loci through random mutagenesis by error-prone repair. Here we report highly effective strategies using TALE nucleases and RNA-guided CRISPR/Cas9 nucleases to induce error-prone repair and homology-directed repair to create heritable, precise insertion, deletion, or substitution of specific DNA sequences at targeted endogenous loci. Our robust strategies are effective across nematode species diverged by 300 million years, including necromenic nematodes (Pristionchus pacificus), male/female species (Caenorhabditis species 9), and hermaphroditic species (C. elegans). Thus, genome-editing tools now exist to transform nonmodel nematode species into genetically tractable model organisms. We demonstrate the utility of our broadly applicable genome-editing strategies by creating reagents generally useful to the nematode community and reagents specifically designed to explore the mechanism and evolution of X chromosome dosage compensation. By developing an efficient pipeline involving germline injection of nuclease mRNAs and single-stranded DNA templates, we engineered precise, heritable nucleotide changes both close to and far from DSBs to gain or lose genetic function, to tag proteins made from endogenous genes, and to excise entire loci through targeted FLP-FRT recombination.
Keywords: genome editing, CRISPR/Cas9 and TALENs, homology-directed repair, nematode species, dosage compensation
STRATEGIES to engineer heritable, site-directed mutations at endogenous loci have revolutionized our approach toward manipulating and dissecting genome function. Studies of plants and animals alike, whether conducted in whole organisms or cell lines, have benefitted greatly from these genome-editing approaches (Bibikova et al. 2002; Beumer et al. 2006; Doyon et al. 2008; Geurts et al. 2009; Hockemeyer et al. 2009; Holt et al. 2010; Zhang et al. 2010; Hockemeyer et al. 2011; Tesson et al. 2011; Wood et al. 2011; Young et al. 2011; Bedell et al. 2012; Bassett et al. 2013; Cong et al. 2013; Jinek et al. 2013; Mali et al. 2013; Wang et al. 2013; Zu et al. 2013). The most modern tools for modifying complex genomes at single-nucleotide resolution are site-specific nucleases that induce DNA double-strand breaks (DSBs) at specifically designated genomic locations. DSBs trigger repair pathways that can elicit targeted genetic reprogramming, primarily through two mechanisms: error-prone nonhomologous end joining (NHEJ) (Lieber 2010) and precise, homology-directed recombination or repair (HDR) (Chapman et al. 2012). NHEJ rejoins broken ends of chromosomes through an imprecise process that produces nucleotide insertions, deletions, and indels at the DSB site, causing random, heritable null or partial loss-of-function mutations. In contrast, HDR uses homologous DNA as a template to create accurate repairs, guided either by a sister chromosome or by introduced homologous DNA to generate specific deletions or insertions in predictable ways (Chen et al. 2011; Bedell et al. 2012). DSBs made by any site-specific nucleases should possess the capacity to enter the homology-directed repair pathway and generate precisely specified changes in the genome.
Effective nucleases with engineered specificity include the zinc-finger nucleases (ZFNs), the transcription activator-like effector (TALE) nucleases (TALENs), and the RNA-guided CRISPR-associated (Cas9) endonuclease (Urnov et al. 2010; Bogdanove and Voytas 2011; Jinek et al. 2012; Wiedenheft et al. 2012; Gaj et al. 2013; Wei et al. 2013). ZFNs and TALENs contain fusions between the DNA cleavage domain of the FokI endonuclease and a custom-designed DNA-binding domain: either C2H2 zinc-finger motifs for ZFNs or TALE domains for TALENs (Urnov et al. 2010; Carroll 2011; Cermak et al. 2011; Li et al. 2011; Miller et al. 2011; Mussolino et al. 2011). DNA-binding domains of TALEs have a tandem array of repeat units, each 33–35 aa, in which the nucleotide-binding preference of a given repeat is determined by two adjacent amino acids called the repeat variable diresidue (RVD) (Boch et al. 2009; Moscou and Bogdanove 2009). In contrast, the RNA-guided CRISPR-associated Cas9 endonuclease generates DSBs at sites defined by a 20-nt guide sequence contained within an associated CRISPR RNA (crRNA) transcript (Gasiunas et al. 2012; Jinek et al. 2012). Cas9 requires both the guide crRNA and a trans-activating crRNA (tracrRNA), which partially base pairs with the crRNA, to form active enzyme complexes that achieve site-specific DNA recognition and cleavage (Deltcheva et al. 2011; Jinek et al. 2012). Cas9 can also be programmed with a single, chimeric fusion of crRNA and tracrRNA for effective DNA cleavage at a selected site (Jinek et al. 2012; Bassett et al. 2013; Chang et al. 2013; Friedland et al. 2013; Gratz et al. 2013; Shen et al. 2013; Wang et al. 2013; Xiao et al. 2013; Yu et al. 2013).
The model nematode Caenorhabditis elegans (Wood et al. 2011; Friedland et al. 2013) and its relative C. briggsae (Wood et al. 2011), a species diverged by 15–30 MYR (Cutter 2008), have succumbed to genome-editing strategies that yielded site-directed, NHEJ-mediated random mutations, using nucleases with engineered target specificity. However, prior to our current studies, neither species had proved amenable to the site-directed manipulations necessary to introduce precise, HDR-mediated insertions or deletions at endogenous loci without traces of dominant genetic markers or other tools of genetic engineering. Furthermore, the genome-editing approaches had not been extended to male/female free-living species or nematode species having more unusual lifestyles, such as those with a necromenic association with plants or animals. While whole-genome sequencing has facilitated research beyond traditional model nematode species, lack of broadly effective reverse genetic tools has limited cross-species comparisons of gene function needed to explore biological mechanisms.
We report robust strategies, using TALENs and CRISPR/Cas9 for targeted, heritable genome editing across nematode species diverged by 300 MYR (Dieterich et al. 2008), the evolutionary time spanning living reptiles and mammals (Hedges and Kumar 2003). Our protocols proved effective in every species tested: necromenic nematodes (Pristionchus pacificus), male/female species (Caenorhabditis species 9), and hermaphroditic species (e.g., C. elegans). These highly efficient strategies produced precise insertion and deletion of specific DNA sequences at designated endogenous loci. These genome-editing tools will enable nonmodel nematode species to become genetically tractable organisms.
We illustrate the utility of our widely applicable genome-editing strategies by creating tools and reagents of general use to the scientific community and ones specifically needed for analyzing the mechanism and evolution of X chromosome dosage compensation across nematode species. Dosage compensation ensures that males (1X) and females/hermaphrodites (2X) produce equivalent levels of X chromosome products despite having different doses of X chromosomes. In C. elegans, a dosage compensation complex (DCC) is recruited to both X chromosomes of hermaphrodites to repress transcription by half (Pferdehirt et al. 2011; Kruesi et al. 2013) through rex (recruitment element on X) sites that bind the DCC in a DNA-sequence-dependent manner, using DNA motifs enriched on X (MEX) (McDonel et al. 2006; Jans et al. 2009). Pivotal to understanding dosage compensation is the ability to delete combinations of MEX motifs at endogenous rex sites and to insert MEX motifs into new sites on X or autosomes. We achieved those goals and also created reagents to explore the evolution of dosage compensation by deleting from a diverged nematode species the homolog of the C. elegans gene that triggers sex-specific assembly of the DCC onto X.
Materials and Methods
Construction of TALEN vector
To utilize the commercially available Golden Gate TALEN kit for TALEN construction, we modified our previously published TALEN backbone vector [pTY2551 (Wood et al. 2011)] by adding BsmB1 restriction enzyme sites at appropriate positions to insert DNA encoding each TALE. We also modified the vector to include a marker to impose a selection for inserting the new TALE DNA.
The QuikChange Lightning site-directed mutagenesis kit (Agilent no. 210518) was used to modify pTY2551. Primers NTAL6F (5′ ctcaccggggcccccCtgGaGACgaccccagaccaggt 3′) and CTAL6R (5′ cgccaccagatggtcGttGgAGaCGgcagccaacgcggga 3′) were used to introduce the necessary BsmBI sites. Nucleotides in uppercase letters represent modified bases and nucleotides in lowercase letters represent unchanged bases. BsmBI sites are shown in boldface type. The ccdB selection cassette was then introduced using the BsmBI sites.
The ccdB cassette was amplified from the pDEST_R4-R3 gateway plasmid, using primers ccdBTALF (CCC TGG AGA CGA AAC ACA ACA TAT CCA GTC ACT ATG G) and ccdBTALR (CCC TGG AGA CGG AGG ATC CGG CT ACT AAA AGC). This fragment was subcloned into pcr2-1_TOPO vector, using the TOPO TA cloning kit. The TOPO-cloned ccdB cassette was digested with BsmBI, and the 714-bp ccdB fragment was gel extracted for ligation into a BsmBI-digested modified pTY2551 vector to create the final pTY2575 backbone vector. ccdB is a lethal gene that targets DNA gyrase (Bernard et al. 1994). The pTY2575 plasmid can be maintained only in an Escherichia coli strain such as DB3.1, which carries a specific suppressor mutation in the gyrase. After transformation, only DH5α cells carrying plasmids with the desired TALE insert will be viable, because the plasmid will lack the lethal ccdB insert. pTY2575 also includes the T7 promoter necessary for in vitro transcription, the 5′- and 3′-UTRs necessary for germline expression, and the FokI domain required for DNA cleavage.
TALEN DNA construction
TALEs were designed using the TAL Effector Nucleotide Targeter 2.0 website. We used the following online program preferences: for TALEN architecture preference, Miller et al. 2011; for 5′-most nucleotide, “T”. The T is included as part of the TALE design, but is not shown in the construction design, because the T is already encoded in plasmid TY2575 used for TALEN assembly (Supporting Information, Figure S2).
TALENs were constructed using the Golden Gate TALEN kit (Figure S2). We followed the protocol at http://www.addgene.org/TALefector/goldengate/voytas/ with one modification. The plasmids pTAL1, pTAL2, pTAL3, and pTAL4 were replaced with nematode species-specific plasmids having the appropriate 5′- and 3′-UTRs for germline expression: pTY2575 for Caenorhabditis and pTY2634 for Pristionchus. pTY2634 was constructed by replacing the 3′-UTR from pTY2575 with the 3′-UTR from Ppa-rpl-23. The Ppa-rpl-23 3′-UTR sequence is 5′ ATT GTT ATT GTT GAT TGT TAC GCG TGA TGC AAT AAA ATG TGT TAT GCG TTT 3′. The Ppa-rpl-23 3′-UTR and its flanking sequences were excised from pTY2606 with HindIII and StuI and gel purified. The pTY2575 plasmid was digested with HindIII, StuI, and BglII and gel purified to recover only the large HindIII-BglII vector fragment. Vector and 3′-UTR fragments were ligated and transformed into DB3.1 bacteria. Final TALENs were sequenced using primers TALseq2 (5′ TAA CAG CGG TAG AGG CAG TG 3′) and TALseq3 (5′ TCT CCT CCA GCT GCT CGC TCT TC 3′).
TALENs were made using the code in Figure 1, except for the following pairs that utilized the RVD NK as well as NN to recognize Gs:
smo-1 TALEN-L1
HD NI NG HD NN NI NN NK NG HD NG NI HD HD NI NI NN
smo-1 TALEN-R1
NN NG NI NN NN NG NI NN NI NK NN NI HD NG NI NN
smo-1 TALEN-L2
HD NI NG HD NN NI NN NK NG HD NG NI HD HD NI NI
smo-1 TALEN-R2
NI NN NI NN NG NN NG NI NN NN NG NI NN NI NK NN NI HD NG NI
rex-32 TALEN-L1
HD NN HD NI NI NK HD NN NK HD NI NI NN HD NI HD
rex-32 TALEN-R1
NI NG NG NG NG NG HD HD HD NG NG NK NG NG NN HD NK NN NI NN.
NK increases TALEN DNA-binding specificity, but even a limited number of NKs appear to decrease TALEN activity (see Table 1).
Figure 1.
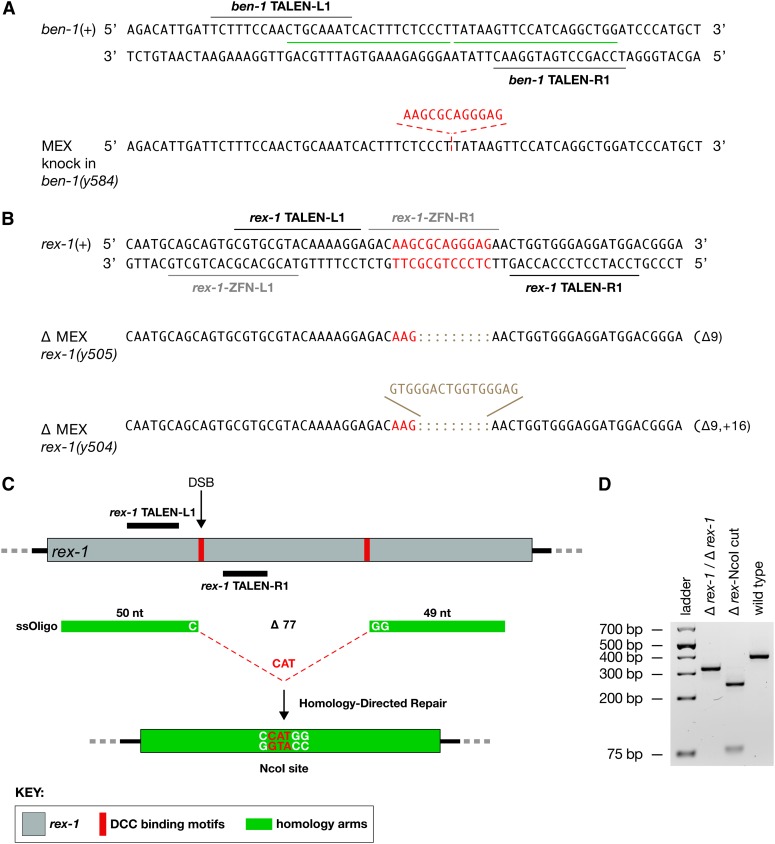
Precise, targeted DNA insertions mediated by homology-directed repair of TALEN-induced DSBs. In a single experiment, animals were injected simultaneously with RNA encoding a ben-1 TALEN pair and three different ssOligos encoding different restriction sites (NcoI, HindIII, and EcoRI) designed to replace the endogenous PsiI site (TTATAA) shown in red. Shown are examples of the HDR-mediated mutations resulting from the single set of injections. Mutations include all three precisely inserted restriction sites and two NHEJ-mediated mutations causing a deletion or an indel. The TALE recognition sequences within ben-1 are shown adjacent to the color-coded repeat variable diresidues (RVDs), which recognize specific nucleotides (nt) of the DNA target site according to the code shown in the key. The locations in ben-1 DNA of the 20-nt homology arms used in each ssOligo are designated by a green line. Each ssOligo is composed of only the 6-nt restriction site to be inserted and the flanking 20-nt homology arms.
Table 1. Summary of genome-editing experiments across nematode species, using TALENs and CRISPR/Cas9.
| Target | DSB-inducing nuclease | Species | Desired change | Knock-in sequence | Animals injected (1 gonad arm) | F1s | All mutations (% of F1s) | Knockouts (%) | Total knock-ins (%) | Precise knock-ins (% of all mutations) |
|---|---|---|---|---|---|---|---|---|---|---|
| ben-1 | TALENs | C. elegans | PsiI → NcoI | CCATGG | 15 | 548 | 22 (4.0) | 10 | 12 | 6 (27) |
| ben-1 | TALENs | C. elegans | PsiI → NcoI | CCATGG | 10 | 440 | 12 (2.7) | 6 | 6 (1.4) | 2 (17) |
| PsiI → EcoRI | GAATTC | 3 (25) | ||||||||
| PsiI → HindIII | AAGCTT | 1 (8) | ||||||||
| ben-1 | TALENs | C. elegans | + MEX | AAGCGCAGGAG | 12 | 382 | 7 (1.8) | 3 | 4 | 4 (57) |
| smo-1 | TALENs | C. elegans | L88K | AAG | 18 | 516 | 1 (0.2) | 0 | 1 | 1 (100) |
| smo-1 | TALENs | C. elegans | L88KA | AAGCGCT | 14 | 729 | 2 (0.3) | 0 | 2 | 2 (100) |
| rex-1 | TALENs | C. elegans | Knockout, NHEJ | 12 | 479 | 12 (2.5) | 12 (2.5) | |||
| rex-1 | TALENs | C. elegans | Knockout, Δ77 bp, homology-directed repair | CAT | 15 | 648 | 85 (13.1) | 61 | 24 | 12 (14) |
| NHEJ | HDR-mediated Δ by insertion | |||||||||
| rex-32 | TALENs | C. elegans | + 5′-FRT | GAAGTTCCTATTCTCTAGAAAGTATAGGAACTTC | 14 | 693 | 5 (0.7) | 3 | 2 | 2 (40) |
| rex-32 | TALENs | C. elegans | + 3′-FRT | Same as 5′-FRT | 12 | 847 | 12 (1.4) | 3 | 9 | 9 (75) |
| rex-32 | FLP-FRT | C. elegans | Knockout by FLP | 18 | 611 | 4 (0.7) | 4 (0.7) | |||
| sdc-2 | TALENs | C. sp. 9 (JU1422) | Knockout, NHEJ | 42 | 42 | 1 (2.4) | 1 (2.4) | |||
| unc-119 | TALENs | P. pacificus (PS312) | Knockout, NHEJ | 16 | 501 | 34 (6.8) | 34 (6.8) | |||
| unc-119 | TALENs | P. pacificus | + HindIII | AAGCTT | 11 | 327 | 18 (5.5) | 12 | 6 | 3 (17) |
| unc-119 | TALENs | P. pacificus | + HA tag | TACCCATACGACGTCCCAGATTACGCT | 15 | 504 | 12 (2.3) | 7 | 5 | 5 (42) |
| gfp | Cas9 | C. elegans | Knockout, NHEJ | 21 | 686 | 8 (1.2) | 8 (1.2) |
Shown are data from all the genome-editing experiments described in the text. The target locus, the DSB-inducing agent, the species, and the types of modifications, including their quantification, are provided. For the knock-in experiments, the inserted sequence is listed, as indicated by “+”. For all experiments, the number of animals injected is provided along with the number of F1s screened from the 8- to 16-hr window postinjection. Because only one of the two gonad arms was injected in each animal, only half the F1s scored had the possibility of receiving a mutation. The frequency was not corrected for this fact. For both knock-in and knockout experiments, the total number of mutations recovered is shown, along with the percentage of total mutations relative to number of F1s screened. For knock-in experiments, three classes of mutations were found: NHEJ-mediated deletions, HDR-mediated imprecise insertions, and HDR-mediated precise insertions. Shown are the total number of knock-ins (precise and imprecise), the number of precise knock-ins, and the number of NHEJ-mediated deletions. The percentage of precise knock-in mutations relative to the total number of mutations is also shown.
TALEN mRNA synthesis and injection
TALEN mRNA was synthesized using the T7 mMessage mMachine kit (Ambion no. A1344), the Poly-A-Tailing kit (Ambion no. A1350), and the Megaclear kit (Ambion no. AM1908) as described in Wood et al. (2011). TALEN mRNA (1000–1500 ng/μl) was injected alone for knockout experiments or with single-stranded DNA nucleotides (ssOligos) (50 ng/μl) for HDR-mediated knock-in experiments.
ssOligo design and synthesis
ssOligos used as repair templates contained the desired insertion sequence flanked on both sides by 20–50 nt of target homology. ssOligos were synthesized by Integrated DNA Technologies (www.idtdna.com).
Mutant screening and isolation
For hermaphroditic species, mutants were screened and isolated according to the protocol (Figure S1) described by Wood et al. (2011), with the exception that the CEL-1 was made using the protocol described in Yang et al. (2000) through the dialysis procedure at the end of step I. For screening, F1 animals laid 8–16 hr postinjection (25°C) were collected and then cloned 2 days postinjection into individual liquid culture wells in a 96-well format. Each liquid well contained 50 μl of S medium and HB101 bacteria (OD = 0.4). Once F2 progeny were present and the wells lacked bacteria, aliquots of animals from each of 4 wells were pooled into a well for the CEL-1 assay. For example, 5 μl from well A1 of plates 1–4 was pooled into plate A, well A1, resulting in 20 μl. Lysis buffer (20 μl) was added to each well, and samples were frozen and then lysed (65°C for 1 hr, followed by 15 min at 95°C). Two microliters from each well was used as a template for PCR in a final reaction volume of 50 μl. S medium is inhibitory to the reaction when present in a ratio >1:25. The CEL-1 assay, which detects and cleaves mismatched DNA, was performed on the PCR products. The process was repeated with the single wells that comprised the positive pooled wells. Individual animals from positive single wells were picked into single wells and their progeny assayed to identify homozygous mutants.
For knock-in experiments that introduced a unique restriction site, PCR fragments were screened with both CEL-1 and the appropriate restriction enzyme. Five microliters of PCR product was digested in a 20-μl reaction for 1–2 hr. Digests were visualized on a 2% agarose gel. The DNA sequence was then obtained from homozygous progeny. For knock-in experiments not introducing a unique restriction site, the DNA sequence of homozygous mutants from CEL-1-positive animals was obtained to determine whether an insertion event had occurred.
For male/female species, mutants were screened and isolated in the following manner (Figure S1). Male/female matings were set up 24 hr prior to injection. Mated females were injected with TALEN mRNA. Progeny were isolated from 8 to 16 hr postinjection and allowed to mature. Matings (one L4 female and one sibling male) were set up in 96-well microtiter plates containing agar rather than liquid medium. Each well had 100 μl of NGM agar and OP50 bacteria. Animals were allowed to mate and produce progeny until the wells lacked bacteria. Animals from starved wells were resuspended in 40 μl of S medium, and 5–20 μl was removed and pooled for screening.
The homozygous C. sp 9 sdc-2(y516) mutant strain derived from this protocol is inviable. The strain is maintained through a cross between sdc-2(y516)/+ XX females and sdc-2(y516) XO males, with frequent monitoring by PCR.
FLP/FRT experiment
FRT sites flanking rex-32 were inserted sequentially. The 5′-FRT was inserted first, using TALENs (pTY2632 and pTY2633) and an ssOligo that contained the FRT sequence and 20 nt of rex-32 homology on either side. The 3′-FRT site was next inserted using TALENs pTY2627 and pTY2628 and an ssOligo containing the FRT site and 40 nt of homology on either side. The length of the homology arms was increased to minimize binding to the previously inserted 5′-FRT site and to optimize binding to the homologous regions flanking the desired insertion site. RNA synthesis, injection, and screening were performed as described above. rex-32 was excised from animals containing both FRT sites, using FLP recombinase, which was introduced into the gonad by injecting FLP-encoding RNA at concentration of 1500 ng/μl. RNA was made by in vitro transcription from a PCR fragment containing the T7 promoter, the FLP recombinase sequence, derived from pGC92 (Voutev and Hubbard 2008), and 5′- and 3′-UTRs appropriate for germline translation of C. elegans mRNAs: 5′-UTR, ATT TAG GTG ACA CTA TAG AAT ACA CGG AAT TCT AGA TGA TCC CCG CGT ACC GAG CT CAGA AAAA; 3′-UTR, GCC TGA GCT CAC GTC GAC CGG GGC CCT GAG ATC TGC TGC AG.
Animals laid 8–16 hr postinjection at 25°C were collected and screened via PCR according to the protocol from Wood et al. (2011) as described above. In a separate experiment, FLP recombinase was supplied by the DNA plasmid pGC92 injected into the gonad.
P. pacificus Western blot
For the Western blot to demonstrate that UNC-119 carried the HA tag, 10 μl of mixed-stage worms were combined with 4× sample buffer [125 mM Tris-HCl (pH 6.8), 20% glycerol, 4% SDS, 10% β-mercaptoethanol, 0.5 mg/ml bromophenol blue] and frozen. Frozen samples were boiled for 5 min, spun at maximum speed in a microcentrifuge for 5 min, and electrophoresed through a 12% acrylamide gel. Standard Western blot protocols were used, and the blot was probed with anti-HA primary antibody (1:1000) (Abcam no. ab1424) and donkey anti-mouse HRP secondary antibody (1:5000) (Jackson ImmunoResearch no. 715.035.151).
Construction of cas9 DNA
The Streptococcus pyogenes cas9 gene, which is codon optimized for eukaryotic expression and carries a C-terminal HA-2xNLS-mCherry-2xNLS fusion tag, was constructed as follows. Oligonucleotides pTY forward (5′-GAC ACG GAA TTC TAG ATA ATC CCC GCG TAC CGA GCT CAG AAA AAA TGG ACA AGA AGT ACA GCA TCG G -3′) and pTY reverse (5′-TCG ATC GTC GAC GTG AGC TCA GGC TCT AGA CTA TTA GTC CAC-3′) were used to PCR amplify the cas9 fusion gene from pDD921 plasmid (a gift from Diane Dickle at Lawrence Berkeley National Laboratory), using Phusion DNA polymerase (NEB). The PCR DNA was digested with EcoRI and SalI (NEB) and ligated into pTY2575 between EcoRI and SalI sites. The resulting cas9 construct (pTY2631) is flanked by a 5′-UTR (5′-GAA TAC ACG GAA TTC TAG ATA ATC CCC GCG TAC CGA GCT CAG AAA AA-3′) and a 3′-UTR (5′-GCC TGA GCT CAC GTC GAC CGG GGC CCT GAG ATC TGC TGC AG-3′) and expressed from the T7 promoter. The plasmid DNA was extracted by the QIAprep Spin Miniprep kit (QIAGEN, Valencia, CA) and DNA sequencing was performed by Quintara Biosciences. Cas9 mRNA was synthesized using the T7 mMessage mMachine kit (Ambion no. A1344), the Poly-A-Tailing kit (Ambion no. A1350), and the Megaclear kit (Ambion no. AM1908), as described in Wood et al. (2011).
Preparation of S. pyogenes Cas9 tracrRNA and crRNA
The tracrRNA and crRNA were synthesized in vitro by T7 transcription, using oligonucleotide templates carrying the T7 promoter sequence. The tracrRNA template was PCR amplified from S. pyogenes M1 GAS genome DNA, using oligonucleotides OLEC1521 (5′-GAA ATT AAT ACG ACT CAC TAT AGA AAA CAG CAT AGC AAG TTA AAA TAA-3′) and OLEC1522 (5′-AAA AAA AGC ACC GAC TCG GTG CCA C-3′) as described by Jinek et al. (2012). The S. pyrogenes bacterium and genomic DNA are available from the American Type Culture Collection (Rockville, MD). The crRNA3 template was annealed from oligonucleotides twGFP T (5′-TAA TAC GAC TCA CTA TAG CTC ACA CAA TGT ATA CAT CAG TTT TAG AGC TAT GCT GTT TTG-3′) and twGFP B (5′-CAA AAC AGC ATA GCT CTA AAA CTG ATG TAT ACA TTG TGT GAG CTA TAG TGA GTC GTA TTA-3′) by heating to 70° and cooling to room temperature. The in vitro T7 transcription reaction consisted of 50 mM tris(hydroxymethyl)aminomethane (pH 8), 5 mM MgCl2, 0.1% Triton X-100, 100 μM spermidine, 1 mM dithiothreitol, 5 mM rNTP, 1 μM DNA template, and 100 μg/ml of purified recombinant T7 polymerase in diethylpyrocarbonate-treated water. The reaction was incubated at 37°C for 5 hr and quenched with 50% formamide containing 10 mM ethylenediaminetetraacetic acid and 0.01% bromophenol blue. The RNA mixture was resolved in a 10% denaturing polyacrylamide gel containing 7 M urea. The RNA band was excised and the RNA was eluted in 10 vol of 0.2 M sodium acetate (pH 5) at 4°C overnight. The RNA was then precipitated with isopropanol, washed with 70% ethanol, and dried under vacuum. The tracrRNA and crRNA were dissolved in EB buffer (QIAGEN) and reannealed by heating to 50°C and cooling on ice.
Cas9 injections
The concentrations for the multiple crRNA:tracrRNA dual-guide RNA injection into strain EG4601 (Ppie-1::gfp::his-33) were as follows: Cas9 mRNA, 459 ng/μl; tracrRNA, 1238 ng/μl; crRNA1, 289 ng/μl; crRNA3, 291 ng/μl; crRNA4, 291 ng/μl; and crRNA6, 291 ng/μl. Prior to injection, the crNRA and tracrRNA were mixed in a tube and allowed to anneal by heating to 50°C for 10 min and cooling to room temperature prior to adding the Cas9 mRNA. Concentrations for the Cas9 mRNA, crRNA3:tracrRNA dual-guide injection into strain EG6171 (Peft-3::gfp) were as follows: Cas9 mRNA, 500 ng/μl; crRNA3, 1248 ng/μl; and tracrRNA, 1248 ng/μl. The concentrations for the single-guide RNA injections into EG6171 were as follows: Cas9 mRNA, 500 ng/μl or 800 ng/μl; and sgRNA3, 2000 ng/μl. RNA injections and progeny screening were carried out using the protocols described above.
Cas9 in vitro DNA cleavage reactions
The Cas9 DNA cleavage reactions with either single-guided (sg)RNA or dual crRNA:tracrRNA were performed in a manner similar to that in Jinek et al. (2012), except the molar ratios for Cas9:guide RNA:target DNA were 0.5:1:0.5 µM (Figure 7D and Figure S4, A and B). The experimental design and concentrations of reagents for the dose-dependent DNA cleavage reactions using sgRNAs are provided in the legend to Figure S5.
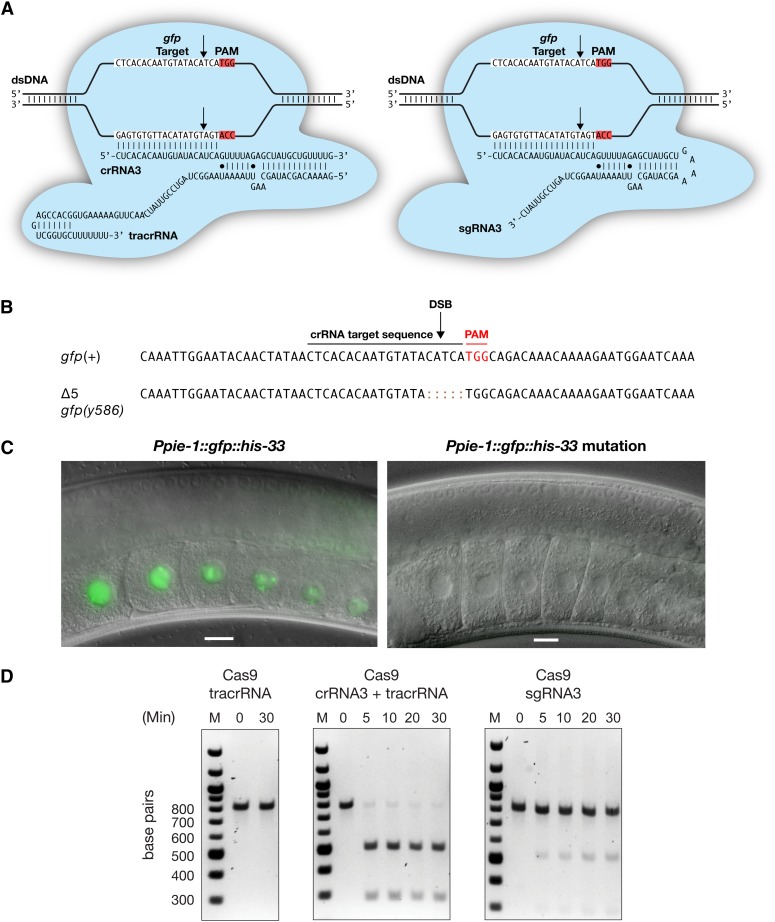
Figure 7.
Heritable, RNA-targeted gene disruption in the C. elegans germline using CRISPR/Cas9. (A) Schematic of Cas9 interacting with its dsDNA target, gfp, and either the dual crRNA:tracrRNA guide RNA (left) or an sgRNA (right), both having the same target DNA. Red highlights the TGG nucleotide protospacer-adjacent motif (PAM), and the arrow designates the DSB site, which occurs in genomic DNA complementary to the crRNA sequence. (B) DNA sequence of the gfp deletion resulting from NHEJ-mediated repair of the Cas9-induced DSB made from the Cas9 complex containing the dual crRNA:tracrRNA guide RNA. (C) Hermaphrodite gonads expressing the wild-type (left) or Cas9-mediated Δ5 mutant (right) version of the Ppie-1::gfp::his-33 transgene, which expresses GFP::histone 2B exclusively in the germline. Bars, 10 µm. (D) In vitro DNA cleavage assays comparing the effectiveness of Cas9-crRNA:tracrRNA and Cas9-sgRNA complexes. Double-stranded DNA cleavage was tested in time-course reactions, using the molar ratio of Cas9:guide RNA:target DNA as 0.5:1:0.5 µM. Reactions were conducted at 37°. The in vitro assays show that the dual RNA guides are more effective at promoting DNA cleavage than the sgRNAs, a finding that recapitulates our results in vivo demonstrating that the dual RNA guides are more effective at promoting mutations than the single RNA guides. M, 100-bp markers.
Results
Precise, heritable DNA insertions can be achieved in C. elegans by homology-directed repair of TALEN-induced DSBs
To establish a strategy in C. elegans for the precise insertion of specific DNA sequences at specifically targeted locations in vivo, we first asked whether exogenously supplied single-stranded DNA could be used as a template for HDR of DSBs induced by TALENs, as shown for zebrafish (Bedell et al. 2012). In the initial experiment, we tested the efficacy of a synthetic ssOligo to guide the replacement of a PsiI restriction site (TTATAA) in the ben-1 locus with an NcoI site (CCATGG) (Figure 1 and Table 1). The donor ssOligo spanned the TALEN cut site near the PsiI site and carried an NcoI site flanked on both sides by 20 nt of DNA homologous to ben-1 sequences. This experimental design allowed us to screen for all TALEN-induced mutations by molecular genotyping, using the CEL-I DNA mismatch assay, and all HDR-mediated NcoI-insertion mutations, using a restriction enzyme digest. Nuclease activity and HDR-mediated insertions were attained in germ cell nuclei by micro-injecting in vitro transcribed TALEN mRNAs and ssOligos into the gonad syncytium of young adult hermaphrodites. This method simultaneously exposes many germ cell nuclei to the nuclease and ssOligo, enabling multiple independent mutagenic events to occur within a single injected parent. Mutagenesis occurs at such high frequency that mutants are readily identified through the CEL-I assay, without reliance on visual phenotype.
From 15 animals injected with the ssOligo and mRNA encoding ben-1 TALENs, 548 F1 progeny were screened, and 22 independent mutations (4.0% frequency) were identified and verified by DNA sequence. Six (27% of all mutations) had precise NcoI substitutions with no other changes, 6 had imprecise insertions containing the NcoI site, and 10 had NHEJ-mediated knockouts (Table 1). Thirty percent of injected animals had mutant progeny, and a single injected animal produced both NHEJ-mediated knockouts and precise HDR-mediated knock-ins. For example, one animal produced six NHEJ events and three precise knock-ins. This proof-of-principle experiment established a robust knock-in protocol for C. elegans (Figure S1) and thereby set the stage for all subsequent cross-species genome-editing experiments in this study.
Targeted insertion of different sequences at a single locus in one experiment
To improve the efficiency of inserting different sequences at a single site, as would be useful for creating an allelic series of mutations in a gene, we asked whether simultaneous introduction of several different ssOligos during a single injection experiment would yield precise insertion of different sequences in different progeny. We introduced mRNA for ben-1 TALENs and ssOligos, encoding three different restriction sites (NcoI, EcoRI, and HindIII) into 10 animals. From 440 F1 progeny, 12 mutations were obtained. Six mutations contained indels from NHEJ events, and 6 mutations had precise insertions: two of the NcoI site, three of the EcoRI site, and one of the HindII site (Figure 1 and Table 1). This experiment demonstrates the ease and efficiency in obtaining multiple different mutations, both NHEJ-mediated deletions and precise HDR-mediated insertions, within a single gene. The strategy permits the recovery of multiple classes of alleles, null, hypomorphic, hypermorphic, and antimorphic, in the same experiment.
Application of the targeted insertion strategy to obtain codon substitutions for the gene encoding SUMO
Many biological processes are regulated by the covalent attachment of a SUMO protein to critical cellular targets (Geiss-Friedlander and Melchior 2007). The C. elegans dosage compensation process is exemplary. Subunits of the DCC are SUMOylated, and SUMOylation is critical for the sex-specific assembly of the DCC onto X chromosomes (Pferdehirt and Meyer 2013). To analyze in detail the role of SUMO in regulating the activities of these proteins, the lysine residues that serve as attachment sites for SUMO modifications must be identified. The amino acid composition of wild-type SUMO makes it difficult to identify the modified residues by the traditional method of proteolysis followed by mass spectrometry. However, protocols requiring expression of a SUMO variant in vivo have been devised for efficient identification of SUMO-modified lysines (Wohlschlegel et al. 2006). We applied our strategy for obtaining precise sequence changes at endogenous loci to create smo-1 gene variants encoding SUMO proteins with either an L88K or an L88KA substitution (Figure 2). L88K improves the efficiency of trypsin cleavage, and L88KA improves cleavage and differentiates between SUMOylated and ubiquitinated substrates. Although the frequency of mutagenesis was relatively low for this experiment (0.3%), all of the mutants recovered carried precise knock-ins of the desired codons (Table 1). The SUMO variant is not only useful for the identification of SUMOylated residues on DCC subunits, but also generally useful to the entire C. elegans community for identifying SUMOylated targets in any biological processes.
Figure 2.
Reprogramming the smo-1 gene by precise DNA insertions to encode SUMO variants that facilitate mass spectrometric identification of SUMO attachment sites within modified proteins. (A) Segment of smo-1 and the two sets of TALE recognition sequences (black lines) within smo-1 for the two TALEN pairs used to induce DSBs for HDR. The ssOligos that serve as templates for HDR are shown below the gene. The smo-1 codon to be replaced is shown in orange, and the replacement sequences are shown in blue and red. The homology arms used in each ssOligo are designated by a green line. (B) Final amino acid sequences of the relevant segment of the SUMO variant.
Targeted HDR to create deletions close to and far from the DSB site
Effective genome-editing strategies must permit the precise deletion of specific sequences in the loci of choice. We developed protocols for this purpose, using the mutagenesis of DCC binding sites as a model. Critical for DCC recruitment to rex sites on X is the 12-bp MEX motif that occurs preferentially at rex sites and is enriched on X chromosomes relative to autosomes (Jans et al. 2009; Pferdehirt et al. 2011). Many rex sites contain multiple MEX motifs. To understand DCC recruitment to rex sites we used our genome-editing strategies to create strains carrying (1) endogenous rex variants with different combinations of MEX deletions and (2) insertions of MEX motifs on autosomes or at new locations on X chromosomes. We first created a MEX insertion strain to determine whether a single MEX motif is sufficient to recruit the DCC to an autosome. For this purpose, we inserted the motif into a region on chromosome III that lacks DCC binding, using the appropriate TALENs and an ssOligo that contained the 12-bp MEX motif flanked by 20 nt of homology on either side (Figure 3A). Seven mutants were isolated from the 382 F1 progeny of 12 injected hermaphrodites (Table 1). Of these mutants, four carried precise 12-bp MEX insertions from HDR, and three carried NHEJ-mediated indels (Figure 3A).
Figure 3.
Targeted HDR and NHEJ to obtain insertions and deletions of DCC binding motifs close to and relatively far from the DSB site. (A) Insertion of a DCC binding motif on an autosome. Shown is a segment of the ben-1 gene on chromosome III and the TALE recognition sequences (black lines) used to target the DSB that induced the insertion of a 12-bp DCC binding motif (MEX) by HDR. The ssOligo contained the DC binding motif (red) flanked by 20-bp homology arms (green). (B) NHEJ-mediated deletion of a DCC binding motif (MEX, in red) within rex-1. Shown is a segment of the rex-1 DCC binding site on X and the TALE recognition sequences (black lines) used to target a DSB. Sequences of representative rex-1 deletion and indel mutations generated by NHEJ are shown. Gray lines show the location of recognition sequences for ZFNs that previously caused numerous rex-1 deletions, none of which knocked out the MEX motif. (C) HDR-mediated deletion of two MEX motifs in rex-1. Schematic diagram of rex-1 (gray) shows the relative location of DCC binding motifs (red). The same TALEN pair used in B was employed to induce a DSB within the first MEX motif. The DSB was repaired by an ssOligo, including homology arms and the sequence CAT, which guided the HDR event to delete both MEX motifs. The precise deletion event was designed to remove 77 bp and insert the nucleotides CAT to create an NcoI site. (D) Agarose gel of PCR products from a rex-1 deletion strain and a wild-type strain verify the deletion. The PCR product from a precise deletion event is 326 bp. NcoI digestion of the Δ rex-1 PCR product yields a 244-bp fragment and a 82-bp fragment. The PCR product from a wild-type strain is 400 bp. The precision of the deletions was confirmed by DNA sequence.
We next targeted rex-1 to obtain a deletion of either one MEX motif or two. This rex site was of particular interest to us because our previous efforts with ZFNs did not result in the series of allelic mutations we desired. All of the 32 ZFN-induced NHEJ deletions occurred to one side of the motif, and only one mutation deleted into the motif, removing only the first 2 bp (Wood et al. 2011). Using the scheme presented in Figure S2, we designed and synthesized a pair of TALENs that positioned the DSB in the middle of the first MEX motif. We obtained 12 NHEJ-mediated mutations from 479 F1s (2.5%), 8 of which disrupted the core of the motif (Figure 3B and Table 1). This result showed that our previous failure to isolate a motif-disrupting mutation was due to limitations of the ZFNs rather than an inherent difficulty in mutagenizing the rex site.
The repetitive nature of the rex-1 locus made it impossible to design TALENs that would create a DSB within the second motif. We therefore adopted an alternative experimental strategy to delete both motifs simultaneously, using the original rex-1 TALEN pair to create a DSB within the first MEX motif and an ssOligo to drive precise HDR-mediated deletion of DNA up to 77 bp from the DSB, removing both MEX motifs (Figure 3C). The ssOligo contained longer homology arms (49 nt and 50 nt) than those of oligos used to insert only short sequences. The ssOligo also carried the sequence CAT between the homology arms to create a unique NcoI site from the desired HDR-mediated event. The experiment was very successful. Of 85 mutations obtained from 648 F1s, 24 had an HDR-mediated deletion, 12 of which were precise. Our genome-editing experiments show that deletions of different sizes can be engineered either immediately adjacent to the DSB by NHEJ or distant from the DSB by HDR, thereby providing flexibility in the choice of DNA sequences targeted by TALENs and hence in TALEN design.
Targeted insertion of FRT sites to obtain large precise deletions through FLP recombinase–FLP recognition target (FLP-FRT) recombination in the germline
Effective genome editing requires the ability to delete large stretches of genomic DNA. We therefore devised a general strategy for obtaining large precise deletions, using the FLP recombinase–FLP recognition target (FLP-FRT) recombination system (Golic and Lindquist 1989), and applied it first to rex sites containing multiple DCC-binding motifs spanning ≤1 kb (Figure 4, A–D). For the purpose of eliminating all DCC-binding activity at such rex sites, removal of each motif individually would be far more labor intensive than simply deleting the entire site. We designed and synthesized two sets of TALENs to insert a 34-bp FRT site at the 5′ and 3′ ends of the rex-32 locus, which contains four strong MEX motifs. Insertion of the 5′-FRT was accomplished with an ssOligo that carried the 34-bp FRT sequence and 20-nt homology arms (Figure 3B). Insertion of the 3′-FRT in the strain carrying the 5′-FRT insertion was then achieved by using an ssOligo with 40-nt homology arms to increase the probability of HDR being driven by homology arm sequences rather than by FRT sequences (Figure 3B). For both FRT insertions, all of the knock-ins were precise, and they represented 40% and 75%, respectively, of all TALEN-mediated mutations for the 5′ and 3′ ends (Table 1).
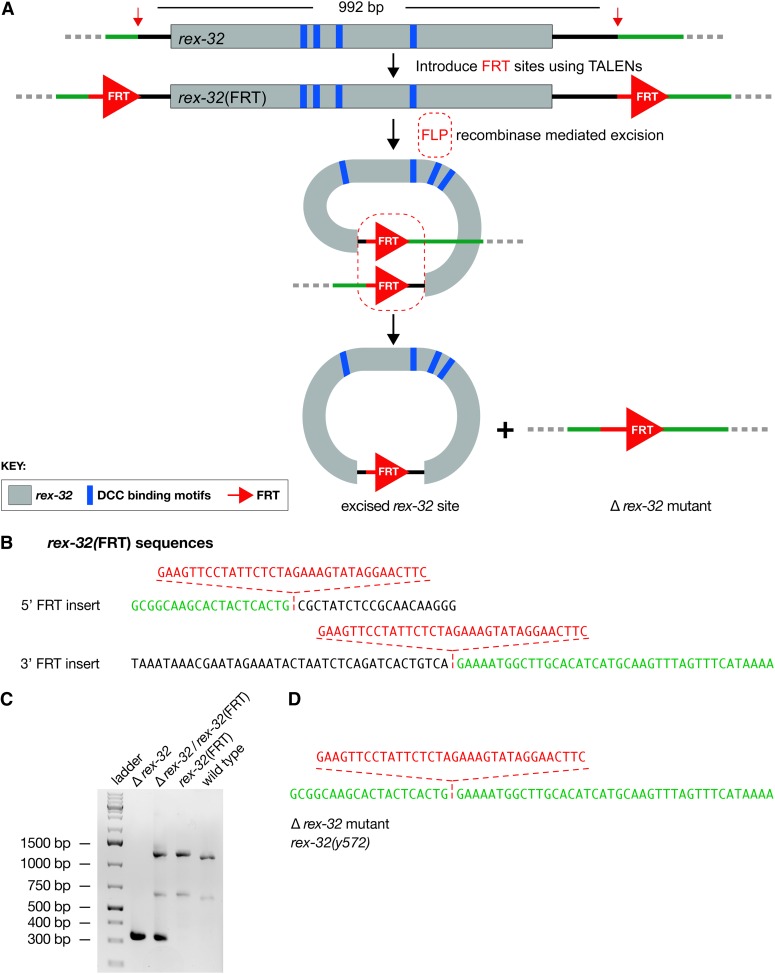
Figure 4.
FLP-FRT-mediated deletion of the rex-32 DCC binding site. (A) Diagram of rex-32 (gray) with its four DCC binding motifs (blue) and the scheme to delete it. FRT sites (red) were inserted at opposite ends of rex-32 via HDR, using the TALENs and ssOligos shown in Figure S3. RNA encoding FLP recombinase was introduced into the germline of the FRT-containing animals, and F2 animals were screened by PCR for the deletion events. (B) DNA sequences of regions carrying the 5′- and 3′-FRT insertions. They are identical to those of the ssOligos used to make the FRT insertions. (C) Agarose gel of PCR products from a homozygous rex-32 deletion strain (360 bp), a heterozygous strain bearing the rex-32 deletion in trans to the FRT-containing rex-32, a homozygous rex-32 FRT strain (1251 bp), and a wild-type strain (1183 bp). The PCR product from the wild-type strain is smaller because it lacks the FRT inserts. The gel verified the deletion, and (D) DNA sequence analysis confirmed that the deletion was precise.
To catalyze the FLP-FRT recombination event that would excise rex-32, RNA encoding the FLP recombinase was injected into the gonads of FRT-containing animals. RNA injections have the advantage over DNA injections in that deletions detected in F1 progeny result from a heritable germline recombination event in the injected animal rather than a nonheritable somatic recombination event in the progeny. From 611 F1 progeny derived from 18 RNA-injected animals, four precise deletions were detected by PCR and verified by DNA sequence analysis (Figure 4, C and D). The RNA protocol is simple and efficient. Furthermore, it is faster and less labor intensive than a DNA-based protocol for delivering FLP recombinase, which we also performed successfully. Only two heritable, precise rex-32-deletion strains were recovered from 1007 F1s, despite having eight positive lines that required tracking for multiple generations. The DNA injections resulted in heritable extrachromosomal arrays that expressed FLP in the progeny, causing somatic, nonheritable recombination events that complicated the analysis. The FLP-FRT system works effectively to delete up to 659 kb in Drosophila (Parks et al. 2004) and should be equally effective in nematodes.
Genome editing in Caenorhabditis species 9, a male/female species
Entire genome sequences are available for numerous species, but lack of reverse genetic tools has hindered cross-species comparisons of gene function. We previously applied our C. elegans nuclease delivery and mutant screening procedures, without modification, to C. briggsae, a hermaphroditic species diverged by 15–30 MYR (Cutter 2008), to explore the evolution of dosage compensation (Wood et al. 2011). We were interested in extending our genome-editing protocols to the male/female strain Caenorhabditis sp. 9 because it forms fertile hybrids with C. briggsae and therefore permits direct cross-species comparisons of gene function in vivo (Figure 5). We changed our general protocol for genome editing to accommodate male/female species by switching the growth conditions to agar-filled microtiter wells, rather than liquid-filled wells, to permit mating (Figure S1). We specifically targeted the dosage compensation gene sdc-2, which triggers sex-specific assembly of the DCC onto X in both C. elegans and C. briggsae, to discover whether its function is conserved in Caenorhabditis (C.) sp. 9 and whether sdc-2 from C. briggsae can function in fertile C. sp. 9/C. briggsae hybrids or vice versa. Although the C. sp. 9 strain JU1422 is not very fertile, we were able to obtain one TALEN-induced, NHEJ-mediated deletion of sdc-2 from 42 F1s that came from 42 injected females. The 14-bp deletion is predicted to cause a truncation of the 350-kDa protein after only 22 aa. The homozygous sdc-2(y516) mutant strain is inviable, yet the hemizygous sdc-2(y516) mutant males appear wild type and sire viable progeny when mated to wild-type females. The XX-specific defects causing inviability of the strain are consistent with a severe disruption in dosage compensation. Successful genome editing in C. sp. 9 opens the way to direct structure–function analysis in C. briggsae/C. sp. 9 hybrids of proteins involved in many biological processes.
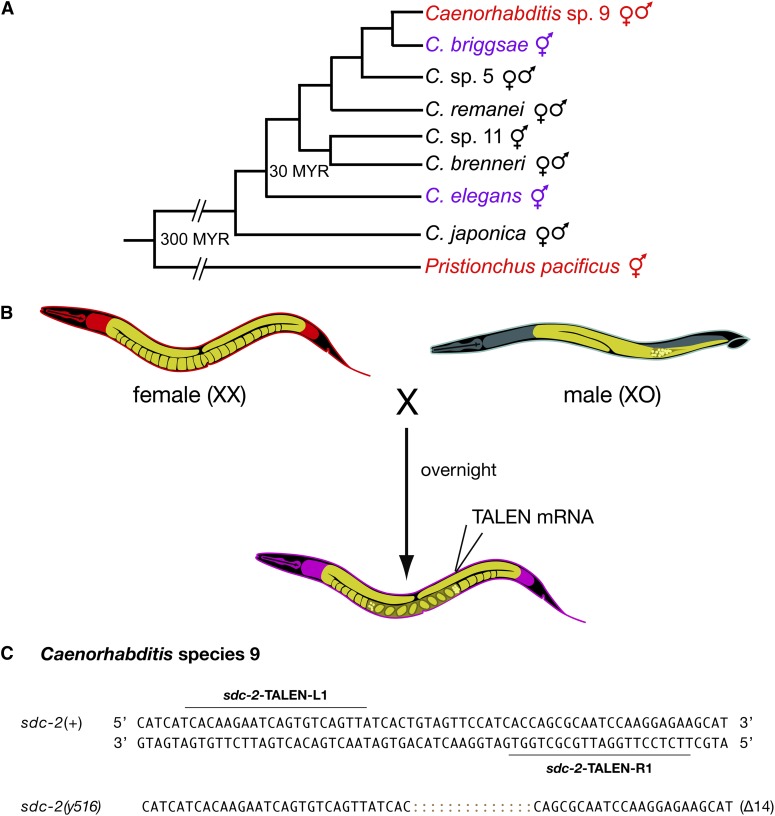
Figure 5.
Genome editing in male/female species. (A) Evolutionary tree for nematode species spanning 300 MYR, from the male/female free-living strain C. sp. 9 to the necromenic nematode strain P. pacificus. Our prior studies described genome editing in C. elegans and C. briggsae (blue), and our current studies describe genome editing for C. sp. 9 and P. pacificus (red). (B) For male/female species, the TALEN mRNA is introduced into females that have been mated overnight. (C) NHEJ-mediated deletion of sdc-2 in C. sp. 9. Shown is a segment of the sdc-2 gene and the TALE recognition sequences (black lines) used to target a DSB. Sequence of the 14-bp deletion in sdc-2 meditated by NHEJ is shown.
Genome editing in the necromenic nematode P. pacificus enabled the production of tagged proteins from endogenous loci
To test the efficacy of our genome-editing strategy across highly diverged species, we applied our protocols to the necromenic nematode P. pacificus, a hermaphroditic species diverged from C. elegans by 300 MYR (Dieterich et al. 2008). P. pacificus lives in close association with scarab beetles and often feeds on the bacteria, fungi, and nematodes growing on dead, rotting beetles. P. pacificus can be cultured in the laboratory on agar plates containing bacteria used for culturing C. elegans. We modified our protocol solely by replacing the 3′-UTR used for translation of TALEN mRNAs in germlines of Caenorhabditis species with the 3′-UTR from the Pristionchus gene rpl-23, shown previously to permit efficient germline RNA translation (Schlager et al. 2009). We initially targeted unc-119 with TALENs to provide the community with a mutant strain enabling unc-119(+) to be used as a co-injection marker for DNA transformation experiments, as in C. elegans. We recovered 34 (6.8%) independent NHEJ-mediated deletions in unc-119 from 501 F1 progeny of 16 injected animals (Table 1). The phenotype is strongly Unc.
We next assessed whether our C. elegans insertion strategy would be effective in P. pacificus. We mutagenized the unc-119 locus, using TALENs and an ssOligo designed to insert a HindIII site. From 327 F1 progeny screened from 11 injected animals, 18 total independent mutations were obtained (5.5%): 12 had NHEJ-mediated deletions and 6 had the HindIII insertion, 3 of which were precise insertions (Table 1).
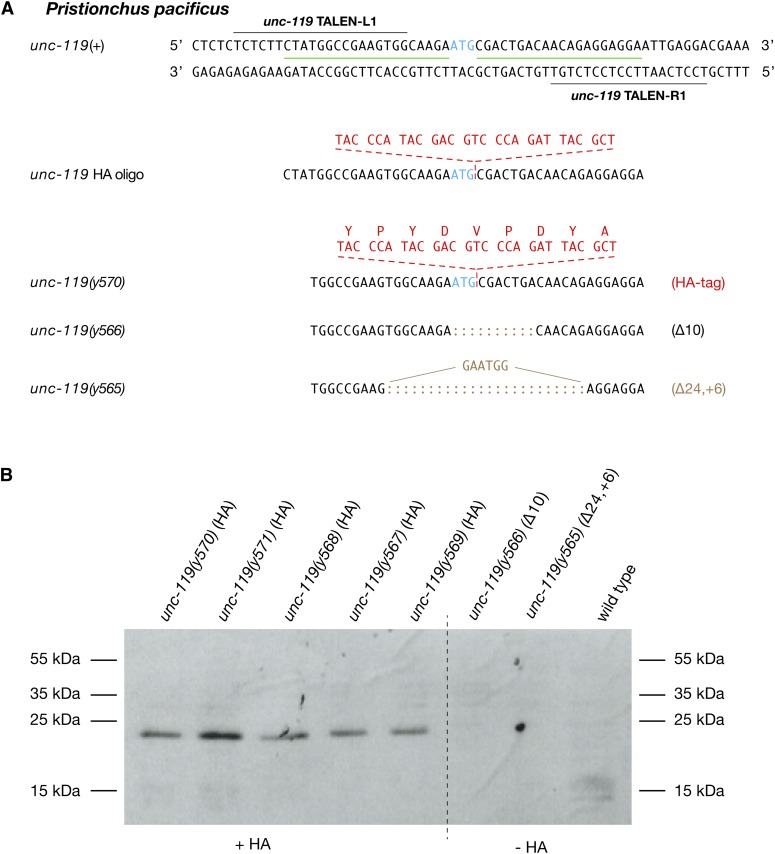
The ability to tag proteins made from an endogenous locus controlled by normal regulatory mechanisms is invaluable for assessing protein function in vivo. Having established the efficacy of the insertion strategy in P. pacificus, we used the strategy to modify the endogenous unc-119 gene to encode an UNC-119 protein with an HA tag at its N terminus (Figure 6A). We inserted a 27-bp sequence encoding the 9-aa HA tag immediately after the ATG translation start codon. Of 504 F1 progeny screened from 15 injected animals, we obtained 12 mutants carrying NHEJ-mediated indels and deletions and 5 mutants carrying HDR-mediated insertions, all of which had precise insertions of the tag (Table 1). None of the insertions caused an Unc phenotype. A Western blot of all five insertion strains using the HA antibody as a probe showed that the UNC-119 protein had indeed been tagged (Figure 6B). None of the controls, neither the wild-type strain (PS312) nor the unc-119 deletion strains from the experiment, were positive for HA antibody staining. These experiments show that our genome-editing protocols are easily portable to species across a broad span of evolutionary time.
Figure 6.
Genome editing in P. pacificus results in an HA-tagged protein made from the endogenous unc-119 locus. (A) Segment of unc-119 spanning the ATG translation start codon (blue) and the TALE recognition sequences within unc-119 (black lines) used to target TALEN-induced DSBs for HDR-mediated insertion of DNA encoding an HA tag. Shown is the DNA sequence of the ssOligo containing the two 20-nt homology arms (green lines) flanking the 27-nt sequence (red) that encodes the 9-aa HA tag. The tag was inserted just after the ATG translation start. Also shown are the sequences of the precise HDR-mediated insertion of the HA tag and the NHEJ-mediated deletions and indels obtained from the same experiment. (B) Western blot of protein extracts from the insertion strains carrying the HA-tagged unc-119 gene, control strains from the same experiment bearing NHEJ-mediated deletions or indels in unc-119, and a wild-type strain. The blot was probed with an antibody directed against the HA tag.
RNA-programmed genome editing using the CRISPR/Cas9 system
TALENs have proved to be highly effective and efficient tools for targeted genetic reprogramming. They have enabled us to establish robust protocols for the precise insertion and deletion of desired sequences at designated loci. However, their use is somewhat constrained by the need to engineer new proteins for each DNA target site. Knowing that protocols designed to deliver site-specific nucleases to the germline and to manipulate nematode genomes through homology-directed DNA repair should be readily transferable to any site-specific nuclease, we directed our efforts toward harnessing the clusters of regularly interspaced short palindromic repeats (CRISPR) CRISPR/Cas9 system, an RNA-guided DNA surveillance system found in bacteria, for use in nematodes.
CRISPR/Cas loci encode RNA-guided adaptive immune systems that identify and destroy foreign DNA (Bhaya et al. 2011; Terns and Terns 2011; Wiedenheft et al. 2012). Unlike the types I and III CRISPR/Cas systems, which are composed of multiprotein surveillance complexes, the type II systems require only a single protein, Cas9, to detect and cleave foreign DNA (Sapranauskas et al. 2011). Cas9 generates blunt DSBs at sites defined by a 20-nt guide sequence contained within an associated crRNA transcript (Gasiunas et al. 2012; Jinek et al. 2012). Cas9 requires both the guide crRNA and a tracrRNA that is partially complementary to the crRNA for site-specific DNA recognition and cleavage (Deltcheva et al. 2011; Jinek et al. 2012). The crRNA:tracrRNA complex can be redesigned as a single chimeric transcript (termed sgRNA) encompassing the features required for both Cas9 binding and DNA target site recognition (Jinek et al. 2012). Using either the sgRNA or separate crRNAs and tracrRNAs, Cas9 can be programmed to cleave double-stranded DNA at any site defined by the guide RNA sequence and the NGG protospacer-adjacent motif (PAM) (Sapranauskas et al. 2011; Jinek et al. 2012). Recent experiments have demonstrated the utility of Cas9-sgRNA complexes for genome engineering in various eukaryotic cell types and whole organisms (Bassett et al. 2013; Chang et al. 2013; Friedland et al. 2013; Gratz et al. 2013; Hwang et al. 2013; Jinek et al. 2013; Mali et al. 2013; Shen et al. 2013; Wang et al. 2013; Xiao et al. 2013; Yu et al. 2013).
Our initial attempts at genome editing using Cas9-sgRNA complexes to target the ben-1 locus with three different sgRNAs in multiple experiments were unsuccessful. We reasoned that dual crRNA:tracrRNA RNA guides might be more effective in vivo for C. elegans than single chimeric sgRNA guides, as observed in some cases for mammalian cells (Cong et al. 2013). We therefore made use of dual RNA guides to target a single-copy, integrated transgene encoding a bifunctional GFP::histone 2B fusion protein expressed solely in the C. elegans germline. We simultaneously introduced tracrRNA, mRNA encoding Cas9, and four different crRNAs to target different sites in gfp. We recovered one gfp mutant carrying a 5-bp deletion that eliminated GFP function (Figure 7, A–C). The general small brood size of the transgenic strain made it difficult to recover the three other independent mutants detected by molecular phenotyping of gfp.
After determining the crRNA that was successful in the dual crRNA:tracrRNA guide experiment from DNA sequence analysis of the mutant, we performed an experiment to compare the efficiency of mutagenesis in vivo of a dual-crRNA:tracrRNA guide vs. a single chimeric sgRNA guide with identical DNA target sequence (Figure 7A). For this experiment, we targeted a different single-copy gfp transgsene, one expressed universally in both the germline and the soma (Peft-3::gfp). In two separate experiments using the sgRNA, we examined 1034 F1 progeny from 30 injected animals and found no mutations. In contrast, the crRNA:tracrRNA complex was again successful for the mutagenesis. From 686 progeny of 21 injected animals, we recovered eight independent NHEJ-mediated mutations (1.2% frequency) (Table 1). Curiously, the frequency of mutagenesis was on par with the frequency from TALENs, but the proportion of insertions, deletions, and indels differed. Deletions and indels are common for TALEN-mediated mutations, but insertions are very rare (<1%). However, in this small Cas9 experiment, the distribution of mutations included a high proportion of insertions: three insertions, two deletions, and three indels.
To understand the basis for the difference in efficacy of the two classes of RNAs in vivo, we performed Cas9-mediated DNA cleavage assays in vitro to compare the dual-crRNA:tracrRNA-Cas9 complex with the chimeric sgRNA-Cas9 complex, using a molar ratio of complexes to target DNA that would reveal a difference in the efficiency of the cleavage reactions, should one exist. We found the dual-crRNA:tracrRNA:Cas9 cleavage reactions to be more efficient than the sgRNA:Cas9 cleavage reactions (Figure 7D). This result was true for four other guide RNAs, two targeting other gfp regions and two targeting a rex site (Figure S4, A and B). The in vitro reactions appear to be good predictors for the success in vivo of RNA-delivered Cas9 experiments.
In a further experiment we assessed DNA cleavage in vitro, using different concentrations of sgRNAs and Cas9 protein (Figure S5). This experiment provides evidence that sgRNAs can be inhibitory under certain conditions and that RNA misfolding is one likely reason for the observed inefficiencies of sgRNAs, although additional factors are probably involved. Together, our Cas9 experiments demonstrate that Cas9-crRNA:tracrRNA complexes function well in C. elegans and provide an efficient and facile tool for RNA-directed reprogramming of genetic information at designated loci.
Discussion
The genetic tools used to manipulate the model organism C. elegans were not previously capable of generating specifically designated nucleotide changes in genomic locations of choice. Not possible, for example, was the ability to modify endogenous loci for the purposes of tagging proteins, perturbing gene function in specific ways, marking chromosomal locations, or changing the position and number of cis-acting regulatory sites. Our genome-editing strategies using CRISPR/Cas9 and TALENs make such custom-designed changes possible not only for C. elegans, but also for nematode species spanning at least 300 MYR of evolutionary divergence, including necromenic nematodes and free-living male/female species. We show that efficient and facile tools now exist for reprogramming genetic information in the germline by the insertion or deletion of specific sequences at any designated locus. Nonmodel nematode species can now be as genetically tractable as model species, thus expanding the repertoire of biological questions to be addressed and the means by which hypotheses can be tested. Precise genome editing provides the opportunity to conduct functional evolutionary studies of diverse biological processes, including one of particular interest to us, dosage compensation.
Two key elements were necessary for advancing the genome-editing technology: (1) the rapid construction and efficient germline delivery of DNA nucleases with engineered target specificity and (2) the ability to exploit DNA damage responses to repair DSBs, using exogenous DNA templates carrying desired sequences. In previous work, we developed an effective mRNA injection strategy to deliver and express ZFNs and TALENs in the germline as well as an efficient strategy to assay putative progeny for NHEJ-mediated mutations. Our current work extended these strategies to achieve RNA-programmed genome editing, using CRISPR-associated Cas9 nuclease. We also expanded the technology to achieve the targeted insertion and deletion of specific DNA sequences, using ssOligos as templates for homology-directed DSB repair. This technology can now be used to achieve homology-directed repair of DSBs made by any engineered nucleases.
Our targeted insertion strategy offers many advantages. It is highly effective. In 7 of 10 experiments, precise insertions represented 100% of all insertions, and in the remaining 3 experiments, they represented 50%. Furthermore, precise insertions comprised a large fraction of all mutations, ranging from 14% to 100%, with a median of 46%. This high frequency makes practical, as demonstrated, the use of several different ssOligos simultaneously to insert different sequences into one locus in a single experiment. This approach permits an allelic series of mutations to be made with ease in one gene in the same experiment.
The insertion strategy is rapid. Four days after injection, molecular genotyping via the CEL-1 mismatch assay revealed the success of the mutagenesis. This speed of analysis is attributable to two factors. First, the frequency of mutagenesis is sufficiently high that identification of mutations requires only a molecular signature, without need for a visual phenotype or genetic selection. Thus, F1 progeny can be scored just after they have reproduced sufficiently to sustain the strain. Second, germline-injected mRNAs used to deliver nucleases and recombinases (TALEN, Cas9, and FLP) are very short lived compared to germline-injected DNAs. Thus, discovery of mutant F1 progeny by the CEL-1 assay indicates the mutations are heritable rather than the consequence of DSBs arising in the somatic tissues of F1s due to enzymes made from inherited DNA arrays. Moreover, the RNA delivery method obviates the need to engineer expression vectors for each different species. Only the 3′-UTR needs to be changed to ensure germline translation in non-Caenorhabditis species.
Another group recently reported the application of CRISPR/Cas9 to induce NHEJ-mediated random mutations at DSB sites in C. elegans, using DNA extrachromosomal arrays to express Cas9 and sgRNAs (Friedland et al. 2013). Success with sgRNAs is likely due to the higher concentration of Cas9 achieved by their expression from DNA arrays. Thus, both Cas9 delivery approaches are successful, as are both chimeric sgRNA guides and dual crRNA:tracrRNA guides, but the Cas9 RNA delivery scheme we used is more successful with dual RNA guides than with sgRNA guides. RNA delivery of Cas9 also has the advantages listed above. Greater efficacy in vivo of dual RNA guides compared to sgRNA guides has been observed previously (Cong et al. 2013) and is likely due to better RNA folding of the dual guides.
Our genome-editing strategy yields precise, clean mutations, without other molecular traces near the endogenous locus. In contrast, a homologous repair event guided by a co-injected DNA plasmid occurs at a lower frequency than repair from an ssOligo template, requiring that both a selection and a counterselection be utilized. That approach leaves a genetic marker near the endogenous locus and is significantly slower than the approach we used, requiring weeks rather than days to recover the insertion (Frokjaer-Jensen et al. 2012).
Because the insertion strategy permits endogenous changes to occur at some distance from the DSB, it provides flexibility in target selection to avoid complications arising from DNA features needed for nuclease target recognition. Both TALENs and CRISPR/Cas9 have specific, although remarkably minimal, constraints for site recognition. TALENs require a 5′ T and CRISPR/Cas9 a 3′ NGG PAM (Miller et al. 2011; Sapranauskas et al. 2011; Jinek et al. 2012). The ability to introduce a specific nucleotide change at a location distant from the DSB permits targets to be chosen that optimize DSB frequency.
For our experiments, the design and construction of TALENs have been rapid and straightforward, and all TALENs yielded the insertions or deletions we sought. However, the CRISPR/Cas9 approach has the ease and simplicity of requiring the synthesis of only a new guide RNA for each new target rather than an entire pair of nucleases. Moreover, experiments in mice indicate that the frequency of mutagenesis using CRISPR/Cas9 is higher than the frequency using TALENs. The frequency is sufficiently high that injection of sgRNAs for two different gene targets into mouse embryos resulted in pups having knockouts of both alleles in both genes. Future experiments will reveal whether the frequency of Cas9 mutagenesis in C. elegans will be comparably high to that in mice (Wang et al. 2013).
Our insertion experiments enabled the first successful utilization of the FLP-FRT system in the C. elegans germline for the purpose of permanently excising an entire single locus from the genome in a precise way, a convenient way to remove multiple cis-acting regulatory motifs in one step. Of course, the insertion of FRT sites in endogenous loci, combined with the temporal or tissue-specific control of FLP expression, will facilitate the more common use of FLP-FRT, to conditionally eliminate gene activity from a desired tissue at a desired time. Engineered FRT sites will also enable the production of insertions, inversions, and larger deletions in the genome.
Our genome-editing strategies have generated many useful reagents and tools for the nematode community and have enabled mechanistic studies of dosage compensation. While future endeavors to improve these strategies will certainly increase their utility, the current genome-editing technology has already transformed the way genetics is conducted in nematode species.
Supplementary Material
Acknowledgments
We thank members of the Meyer and Doudna laboratories for thoughtful discussions and technical support, particularly E. Anderson for construction of rex-1 TALEN plasmids, D. Lapidus for assistance with TALEN experiments, S. Uzawa for microscopy, M. Jinek for initial Cas9 experiments, and D. Stalford for figure graphics. We also thank T. Cline for critical discussions, R. Hong and R. Sommer for advice about P. pacificus, E. J. A. Hubbard for FLP-FRT plasmids, M.-A. Félix for C. sp. 9 strain JU1422, C. Frøkjær-Jensen and E. Jorgensen for gfp strains EG4601 and EG6171, and the Caenorhabditis Genetics Center for other nematode strains. We are grateful to Sangamo BioSciences for providing ZFNs to conduct initial genome-editing experiments in P. pacificus. This research was supported in part by National Institutes of Health grant R0130702 to B.J.M. J.A.D. and B.J.M. are investigators of the Howard Hughes Medical Institute.
Footnotes
Communicating editor: D. I. Greenstein
Literature Cited
- Bassett A. R., Tibbit C., Ponting C. P., Liu J. L., 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4: 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell V. M., Wang Y., Campbell J. M., Poshusta T. L., Starker C. G., et al. , 2012. In vivo genome editing using a high-efficiency TALEN system. Nature 491: 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P., Gabant P., Bahassi E. M., Couturier M., 1994. Positive-selection vectors using the F plasmid ccdB killer gene. Gene 148: 71–74. [DOI] [PubMed] [Google Scholar]
- Beumer K., Bhattacharyya G., Bibikova M., Trautman J. K., Carroll D., 2006. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics 172: 2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D., Davison M., Barrangou R., 2011. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45: 273–297. [DOI] [PubMed] [Google Scholar]
- Bibikova M., Golic M., Golic K. G., Carroll D., 2002. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161: 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., et al. , 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512. [DOI] [PubMed] [Google Scholar]
- Bogdanove A. J., Voytas D. F., 2011. TAL effectors: customizable proteins for DNA targeting. Science 333: 1843–1846. [DOI] [PubMed] [Google Scholar]
- Carroll D., 2011. Genome engineering with zinc-finger nucleases. Genetics 188: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T., Doyle E. L., Christian M., Wang L., Zhang Y., et al. , 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N., Sun C., Gao L., Zhu D., Xu X., et al. , 2013. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23: 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J. R., Taylor M. R., Boulton S. J., 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47: 497–510. [DOI] [PubMed] [Google Scholar]
- Chen F., Pruett-Miller S. M., Huang Y., Gjoka M., Duda K., et al. , 2011. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods 8: 753–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., 2008. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol. Biol. Evol. 25: 778–786. [DOI] [PubMed] [Google Scholar]
- Deltcheva E., Chylinski K., Sharma C. M., Gonzales K., Chao Y., et al. , 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich C., Clifton S. W., Schuster L. N., Chinwalla A., Delehaunty K., et al. , 2008. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat. Genet. 40: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y., McCammon J. M., Miller J. C., Faraji F., Ngo C., et al. , 2008. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 26: 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiacovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C., Davis M. W., Ailion M., Jorgensen E. M., 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9: 117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. A., Barbas C. F., 3rd, 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G., Barrangou R., Horvath P., Siksnys V., 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 109: E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R., Melchior F., 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8: 947–956. [DOI] [PubMed] [Google Scholar]
- Geurts A. M., Cost G. J., Freyvert Y., Zeitler B., Miller J. C., et al. , 2009. Knockout rats via embryo microinjection of zinc-finger nucleases. Science 325: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic K. G., Lindquist S., 1989. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59: 499–509. [DOI] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges S. B., Kumar S., 2003. Genomic clocks and evolutionary timescales. Trends Genet. 19: 200–206. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., et al. , 2009. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 27: 851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D., Wang H., Kiani S., Lai C. S., Gao Q., et al. , 2011. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 29: 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt N., Wang J., Kim K., Friedman G., Wang X., et al. , 2010. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 28: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., et al. , 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans J., Gladden J. M., Ralston E. J., Pickle C. S., Michel A. H., et al. , 2009. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev. 23: 602–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., East A., Cheng A., Lin S., Ma E., et al. , 2013. RNA-programmed genome editing in human cells. Elife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruesi W. S., Core L. J., Waters C. T., Lis J. T., Meyer B. J., 2013. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. Elife 2: e00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Huang S., Zhao X., Wright D. A., Carpenter S., et al. , 2011. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 39: 6315–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M. R., 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79: 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., et al. , 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonel P., Jans J., Peterson B. K., Meyer B. J., 2006. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature 444: 614–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. C., Tan S., Qiao G., Barlow K. A., Wang J., et al. , 2011. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29: 143–148. [DOI] [PubMed] [Google Scholar]
- Moscou M. J., Bogdanove A. J., 2009. A simple cipher governs DNA recognition by TAL effectors. Science 326: 1501. [DOI] [PubMed] [Google Scholar]
- Mussolino C., Morbitzer R., Lutge F., Dannemann N., Lahaye T., et al. , 2011. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 39: 9283–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks A. L., Cook K. R., Belvin M., Dompe N. A., Fawcett R., et al. , 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292. [DOI] [PubMed] [Google Scholar]
- Pferdehirt R. R., Meyer B. J., 2013. SUMOylation is essential for sex-specific assembly and function of the C. elegans dosage compensation complex onto the X chromosomes. Proc. Natl. Acad. Sci. USA (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pferdehirt R. R., Kruesi W. S., Meyer B. J., 2011. An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes Dev. 25: 499–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R., Gasiunas G., Fremaux C., Barrangou R., Horvath P., et al. , 2011. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 39: 9275–9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager B., Wang X., Braach G., Sommer R. J., 2009. Molecular cloning of a dominant roller mutant and establishment of DNA-mediated transformation in the nematode Pristionchus pacificus. Genesis 47: 300–304. [DOI] [PubMed] [Google Scholar]
- Shen B., Zhang J., Wu H., Wang J., Ma K., et al. , 2013. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 23: 720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns M. P., Terns R. M., 2011. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 14: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson L., Usal C., Menoret S., Leung E., Niles B. J., et al. , 2011. Knockout rats generated by embryo microinjection of TALENs. Nat. Biotechnol. 29: 695–696. [DOI] [PubMed] [Google Scholar]
- Urnov F. D., Rebar E. J., Holmes M. C., Zhang H. S., Gregory P. D., 2010. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11: 636–646. [DOI] [PubMed] [Google Scholar]
- Voutev R., Hubbard E. J., 2008. A “FLP-Out” system for controlled gene expression in Caenorhabditis elegans. Genetics 180: 103–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., et al. , 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Liu J., Yu Z., Zhang B., Gao G., et al. , 2013. TALEN or Cas9—rapid, efficient and specific choices for genome modifications. J. Genet. Genomics 40: 281–289. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B., Sternberg S. H., Doudna J. A., 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482: 331–338. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel J. A., Johnson E. S., Reed S. I., Yates J. R., 3rd, 2006. Improved identification of SUMO attachment sites using C-terminal SUMO mutants and tailored protease digestion strategies. J. Proteome Res. 5: 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A. J., Lo T. W., Zeitler B., Pickle C. S., Ralston E. J., et al. , 2011. Targeted genome editing across species using ZFNs and TALENs. Science 333: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A., Wang Z., Hu Y., Wu Y., Luo Z., et al. , 2013. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 41: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Wen X., Kodali N. S., Oleykowski C. A., Miller C. G., et al. , 2000. Purification, cloning, and characterization of the CEL I nuclease. Biochemistry 39: 3533–3541. [DOI] [PubMed] [Google Scholar]
- Young J. J., Cherone J. M., Doyon Y., Ankoudinova I., Faraji F. M., et al. , 2011. Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 108: 7052–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Ren M., Wang Z., Zhang B., Rong Y. S., et al. , 2013. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics 113: 287–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Maeder M. L., Unger-Wallace E., Hoshaw J. P., Reyon D., et al. , 2010. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl. Acad. Sci. USA 107: 12028–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu Y., Tong X., Wang Z., Liu D., Pan R., et al. , 2013. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat. Methods 10: 329–331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.