Toxoplasma gondii secretes a novel dense granule protein, GRA24, that traffics from the vacuole to the host cell nucleus where it prolongs p38a activation and correlates with proinflammatory cytokine production.
Abstract
Toxoplasma gondii, the causative agent of toxoplasmosis, is an obligate intracellular protozoan parasite that resides inside a parasitophorous vacuole. During infection, Toxoplasma actively remodels the transcriptome of its hosting cells with profound and coupled impact on the host immune response. We report that Toxoplasma secretes GRA24, a novel dense granule protein which traffics from the vacuole to the host cell nucleus. Once released into the host cell, GRA24 has the unique ability to trigger prolonged autophosphorylation and nuclear translocation of the host cell p38α MAP kinase. This noncanonical kinetics of p38α activation correlates with the up-regulation of the transcription factors Egr-1 and c-Fos and the correlated synthesis of key proinflammatory cytokines, including interleukin-12 and the chemokine MCP-1, both known to control early parasite replication in vivo. Remarkably, the GRA24–p38α complex is defined by peculiar structural features and uncovers a new regulatory signaling path distinct from the MAPK signaling cascade and otherwise commonly activated by stress-related stimuli or various intracellular microbes.
Toxoplasma gondii is a widespread obligate intracellular protozoan parasite causing toxoplasmosis, a potentially severe disease in immunocompromised or congenitally infected humans. Once it has invaded its hosting cell, the tachyzoite grows inside a parasitophorous vacuole (PV) in any type of nucleated cell, where it directs profound changes in the host cell transcriptome (Martin et al., 2007; Plattner and Soldati-Favre, 2008), which is known to tightly precede the host cell proteome (Nelson et al., 2008). In this regard, when the hosting cells are of mouse origin, IL-12 and IFN-γ were recognized as a pivotal immune axis contributing not only to the control of the tachyzoite population size (Sher et al., 2003; Mashayekhi et al., 2011; Goldszmid et al., 2012) but also to the T. gondii tachyzoite-bradyzoite developmental transition. Interestingly, once delivered within the host cell cytoplasm, lineage-specific polymorphic tachyzoite molecules have been shown to counteract this IL-12p70–IFN-γ axis, either acting directly—e.g., disruption of the autonomous host cell protecting immune processes (Howard et al., 2011)—or indirectly—e.g., subversion of the transcriptome machinery (Hunter and Sibley, 2012). The majority of these tachyzoite molecules identified so far are transiently released early on during host cell invasion from the rhoptry secretory organelles (Hunter and Sibley, 2012). Chief among them, the polymorphic rhoptry protein ROP16 directs tyrosine phosphorylation of STAT3 and STAT6 (Saeij et al., 2007; Yamamoto et al., 2009; Butcher et al., 2011). Once enclosed in the PV, additional effectors, including GRA15 (Rosowski et al., 2011) and GRA16 (Bougdour et al., 2013), are secreted from dense granules (DGs) and traffic to the host cell. Notably, GRA16 once exported beyond the PV membrane (PVM) reaches the host cell nucleus, where it positively modulates genes involved in cell cycle progression and the p53 tumor suppressor pathway (Bougdour et al., 2013).
Considerable effort is currently being made to unravel the intricacy of the parasite and host interplay that leads, for the medically relevant type II and III strains, to the control of acute parasite multiplication and dissemination yet allows persistence of cryptic parasites in deep tissues. What is known is the need for a strong and persistent Th1 response and the correlated and marked proinflammatory cytokine IL-12 production by innate immune cells, including macrophages (MØ), CD8α+ dendritic cells, and neutrophils (Sher et al., 2003; Mashayekhi et al., 2011; Goldszmid et al., 2012). IL-12, in turn, activates natural killer and T cells to produce IFN-γ, which triggers parasite killing through the activation of IRGs (IFN-regulated GTPases; Howard et al., 2011). In mice lacking IL-12 or IFN-γ, parasite expansion throughout the host is not controlled, causing host death (Gazzinelli et al., 1994; Scharton-Kersten et al., 1996; Yap et al., 2006). Although IFN-γ is the main mediator of resistance to Toxoplasma infection, IL-12 synthesis appears as the key cytokine to be tightly regulated in a strain-specific manner by Toxoplasma. In this regard, macrophages are classically (or M1) activated by a type II strain while they are alternatively (or M2) activated by hypervirulent type I and avirulent type III strains (Jensen et al., 2011). While GRA15 type II allele contributes to M1 activation by activating NF-κB and downstream proinflammatory cytokines, in particular IL-12p70 (Jensen et al., 2011; Rosowski et al., 2011), ROP16 triggers a STAT3/6-dependent M2 response that prevents IL-12p70 from being synthesized (Saeij et al., 2007; Butcher et al., 2011; Jensen et al., 2011).
Sustained IL-12p70 production in tissues hosting T. gondii is not exclusively controlled by NF-κB but could also require the MAPK pathways (Kim et al., 2005; Masek et al., 2006). MAPK pathways encompass a series of kinases that activate each other through an orchestrated cascade (MAP4K, then MAP3K, then MAP2K) that ends with the phosphorylation of a specific MAPK that positively or negatively regulates the expression of suites of genes by activating effector proteins, in particular transcription factors. A wide range of stimuli activates MAPKs, including inflammatory cytokines, osmotic stress, LPS, growth factors, osmotic shock, and UV light (Krishna and Narang, 2008; Cuadrado and Nebreda, 2010). Although it has been proposed that Toxoplasma bypasses the MKK-dependent MAPK phosphorylation cascade to induce autophosphorylation of the p38α MAPK and to promote IL-12p70 production (Kim et al., 2005), the tachyzoite molecule subverting the host p38α MAPK pathway remains unknown.
Within this context, we characterize a novel DG-like protein, hereafter referred to as GRA24, that is exported beyond the PV to the host cell nucleus. We discovered that GRA24 directly interacts with p38α, leading to an unusually persistent autophosphorylation and activation of the host kinase. 3D modeling of the complex predicts unusual structural features of the complex that highlight a novel pathway for p38α extended activation. The GRA24-dependent p38α activation was documented in both human stromal cells and mouse macrophages, the latter up-regulating the expression of multiple proinflammatory cytokines (i.e., IL-12p40 and MCP-1/CCL2) in a strain-specific manner. Our results demonstrate a substantial role for GRA24 in the control of parasite burden in the gut of infected mice. Finally, as a link between the cytokine response and GRA24-p38α activation, we identified several transcription factors up-regulated in a GRA24-dependent fashion, among which are Egr-1 and c-Fos. Collectively, our findings identify a GRA24-dependent sustained p38α kinase activation pathway that could contribute shaping and modulating immune response—at least the one driven by the T. gondii tachyzoite developmental stage.
RESULTS
GRA24 is a novel DG-like protein exported to the host cell nucleus
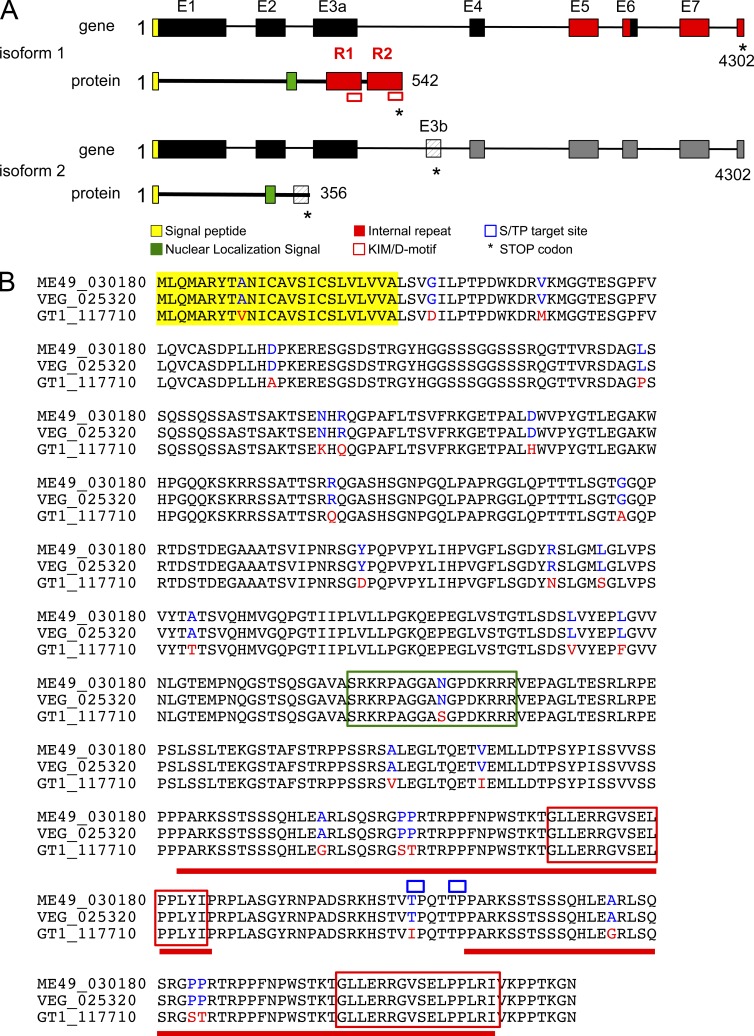
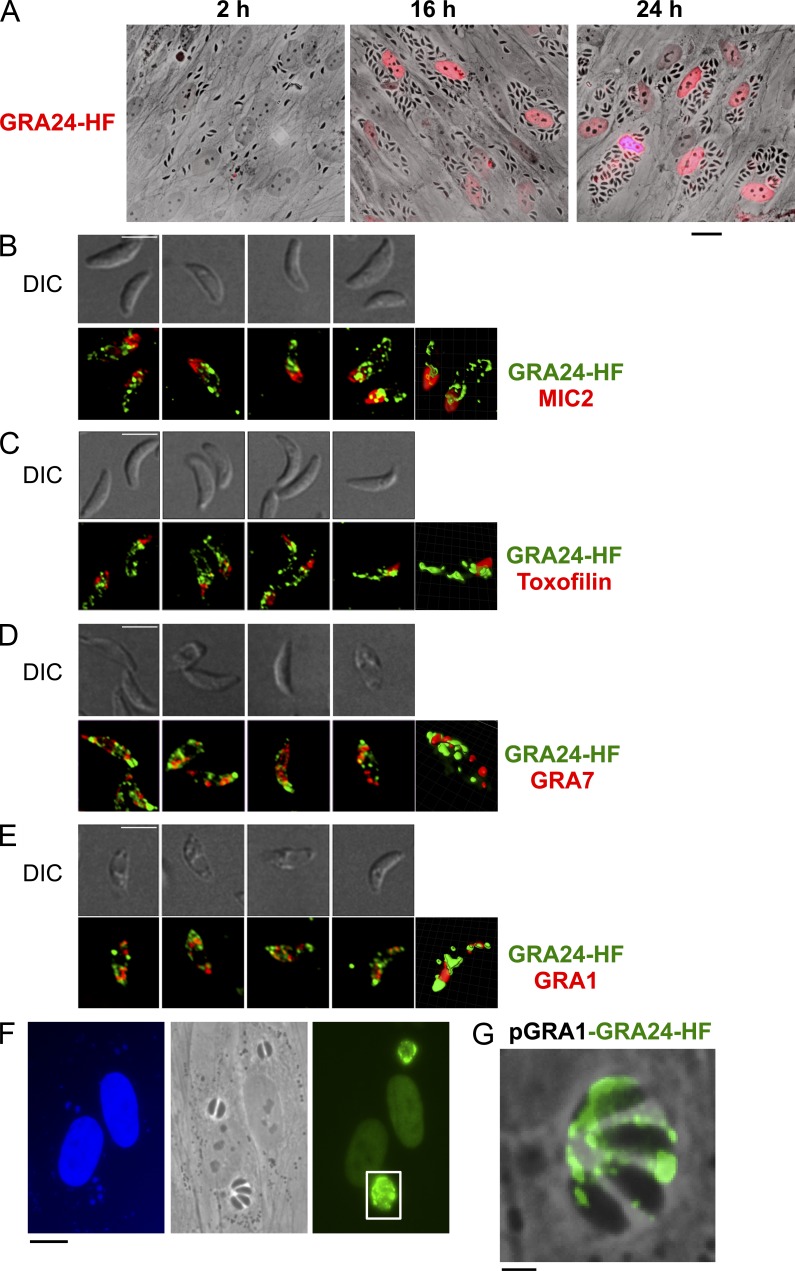
The gene TGME49_030180 encoding GRA24 was originally identified in silico together with GRA16 (Bougdour et al., 2013) as strong parasite candidate genes with attributes to both target the parasite secretory pathway and to reach the nuclei of T. gondii tachyzoite-hosting cells. GRA24 encodes a 542-aa protein with a predicted N-terminal signal peptide, an internal putative bipartite nuclear localization sequence, and two repeats, R1 and R2, at the C-terminal end (Fig. 1 A). GRA24 protein is unique because it has no significant homology with any proteins, not even with the close relative Neospora caninum proteins. In Toxoplasma, GRA24 harbors significant polymorphism between the allele shared by types II and III clonal lineages and the allele typifying type I clonal lineage (Fig. 1 B). In fibroblasts hosting tachyzoites expressing a carboxy-terminal HAFlag-tagged version of GRA24, the protein is distinctly detected in the host cell nucleoplasm at 16 h after invasion and later, whereas there is no detectable protein left in the intracellular tachyzoites (Fig. 2 A). In contrast, in free tachyzoites, GRA24-HAFlag delineates punctuate structures throughout the parasite cytoplasm which occasionally overlap with the canonical DG proteins GRA1 and GRA7 (Fig. 2, D and E) while being excluded from the two typical apical secretory organelles using toxofilin and MIC2 as markers for rhoptry and microneme, respectively (Fig. 2, B and C). Interestingly, when overexpressed under the strong constitutive GRA1 promoter, a significant fraction of GRA24 is seen in the PV space similarly to GRA1, thus suggesting that GRA24 export across the PVM is a limiting step (Fig. 2, F and G). We concluded from these data that GRA24 is a novel DG-like protein that crosses the PVM of maturing vacuoles to reach the host cell nucleus, thereby belonging to the recently defined GRA exported protein subfamily typified by GRA16 (Bougdour et al., 2013).
Figure 1.
The gene GRA24 generates splice variants encoding two distinct proteins. (A) Schematic representation of the GRA24 alternative splice variants and their corresponding GRA24 protein isoforms. The putative signal peptide is shown in yellow (the cleavage site is predicted by SignalP). A putative nuclear localization signal is in green (as predicted by PSORT). The KIM/D-motif (in red square) and the internal repeats (colored in red) are shown. (B) GRA24 amino acid alignment. The predicted amino acid sequence for the primary translation product of the GRA24 gene is shown for type I (TGGT1_117710), II (TGME49_030180), and III (TGVEG_025320) parasites. Single amino acid polymorphisms are indicated for type II and III (in blue) versus type I (in red). The alignment was done by ClustalW2 (EMBL-EBI).
Figure 2.
GRA24 is a novel DG protein exported beyond the PV into the host cell nucleus. (A) Time course of GRA24 secretion and export to the host cell nucleus. Human fibroblasts (HFF) were infected with parasites expressing an HAFlag (HF)-tagged copy of GRA24 (RHku80 GRA24-HF), fixed, and stained with anti-HA antibodies (red). Images are representative of at least four experiments. Bar, 20 µm. (B) GRA24-HAFlag is contained in cytoplasmic organelles distinct from the apical micronemes and rhoptries and partially colocalizing with DG proteins. (B–E) Free parasites were fixed, permeabilized, and successively stained with antibodies raised against HA to label GRA24 (green) and then with antibodies (red) against the micronemal protein MIC2 (B), the rhoptry protein toxofilin (C), and the DG proteins GRA7 (D) and GRA1 (E). The top panels show the DIC images corresponding to the IFA (bottom panels), and the right frames show 3D images processed using MetaMorph and Imaris software. Images are representative of at least two experiments. Bars, 5 µm. (F) IFA of GRA24-HF expressed by the strong promoter of GRA1. HFF monolayers were infected with RHku80 parasites expressing ectopically pGRA1-GRA24-HF. 18 h after infection, cells were fixed and secretion of GRA24 was monitored using anti-HA antibodies (green). Images are representative of at least two experiments. Bar, 10 µm. (G) Higher magnification of a vacuole showing GRA24-HF signal in the vacuolar space. Bar, 2.5 µm.
GRA24 forms a stable dimeric complex with host p38α kinase
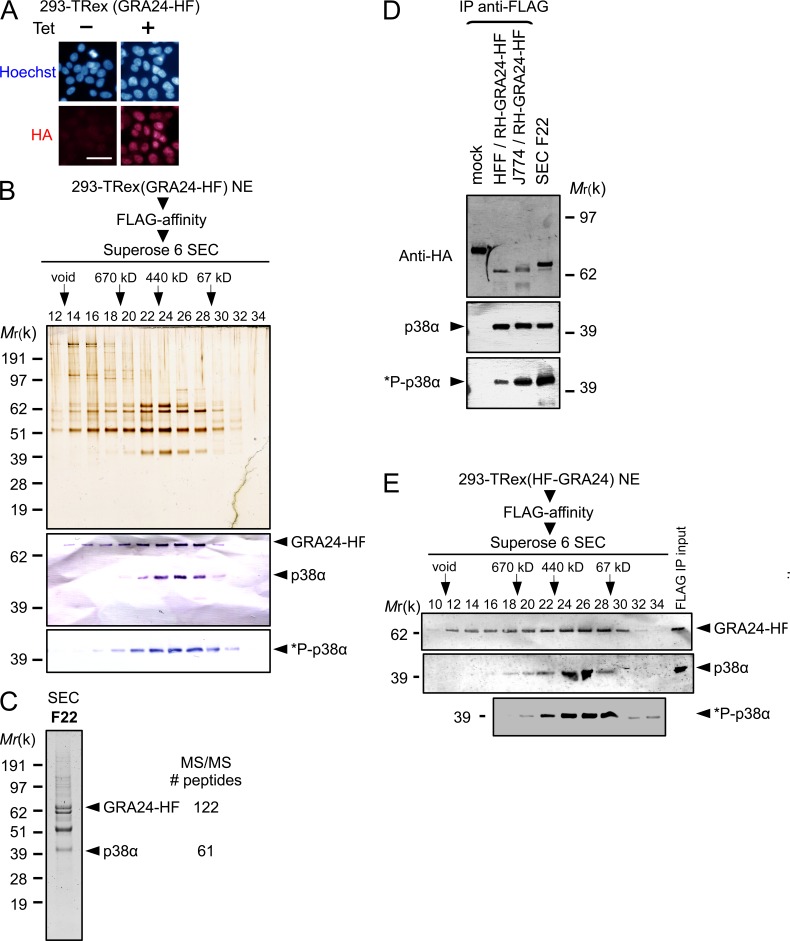
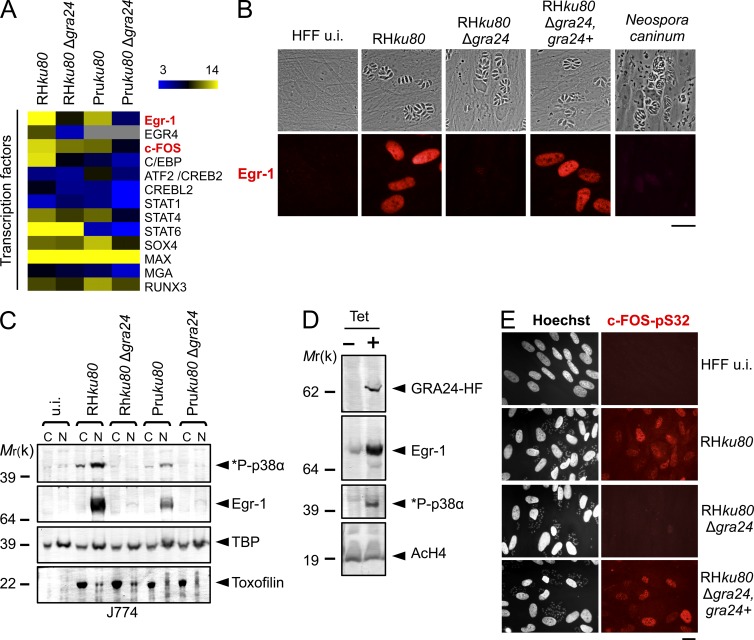
Because GRA24 operates as a secreted DG that crosses the PVM and traffics to the host cell nucleus, we next sought partners of GRA24 by applying chromatography and ion-trap mass spectrometry to nuclear extracts that were prepared from inducible GRA24-HAFlag–expressing 293 cell lines. After induction, the protein localizes in the nucleus (Fig. 3 A), and regardless of the tag position (N- or C-terminal fusion), GRA24 is embedded in a remarkably robust molecular mass complex (400 kD, fractions 22–28; Fig. 3, B and E), which contains the CSBP2/SAPK2α isoform, also named p38α, as the only GRA24 relevant partner (Fig. 3 C). Western blotting confirmed the partnership between GRA24 and p38α (Fig. 3, B and E). The GRA24–p38α complex was also isolated from primary human foreskin fibroblasts (HFFs) and mouse J774 MØ line hosting GRA24-HAFlag–expressing tachyzoites, demonstrating that the complex forms not only in stromal cells but also in macrophages hosting cell-cycling tachyzoites (Fig. 3 D). Using the dual phosphorylation of Thr180/Tyr182 as a hallmark of p38 activation (Coulthard et al., 2009), we confirmed by immunoblotting that p38α in complex with GRA24 is activated (i.e., phospho Thr180/Tyr182 positive) when purified from inducible GRA24-HAFlag–expressing 293 cells (Fig. 3, B and E) or from infected murine and human cells (Fig. 3 D).
Figure 3.
GRA24 forms a dimeric complex with the p38α MAPK. (A) Immunofluorescence analysis (IFA) of GRA24-HAFlag ectopically and stably expressed in 293-TRex cell line. Cells were either left untreated (−) or treated with 1 µg/ml tetracycline for 20 h before fixation and staining with anti-HA antibodies (red) and Hoechst DNA-specific dye (blue). Bar, 2.5 µm. (B) GRA24-associated polypeptides were purified from nuclear extracts of 293-TRex cells tetracycline-induced to express GRA24-HF. Size exclusion chromatography (SEC) of GRA24-containing complexes after Flag affinity selection. Fractions were analyzed by silver staining and immunoblotting to detect GRA24-HF (anti-HA), total p38α, and Thr180/Tyr182 phosphorylated *P-p38α. Data are representative of two experiments. (C) Mass spectrometry analysis of SEC fraction 22. Identity of the proteins with their respective number of peptides is indicated on the right. (D) GRA24-p38α association detected in HFF and J774 MØ infected by RHku80 GRA24-HF (18 h). Tagged proteins were Flag-immunoprecipitated from extracts and eluates were analyzed by immunoblotting. Data are representative of two experiments. (E) GRA24-associated polypeptides were purified from nuclear extracts of 293-TRex cells induced to express an N-terminal HAFlag-tagged GRA24. SEC Fractions were analyzed by immunoblotting to detect HAFlag-GRA24 (anti-HA), total p38α, and Thr180/Tyr182 phosphorylated *P-p38α. Data are representative of two experiments.
Deletion of GRA24 is sufficient to abolish host cell p38α activation by Toxoplasma
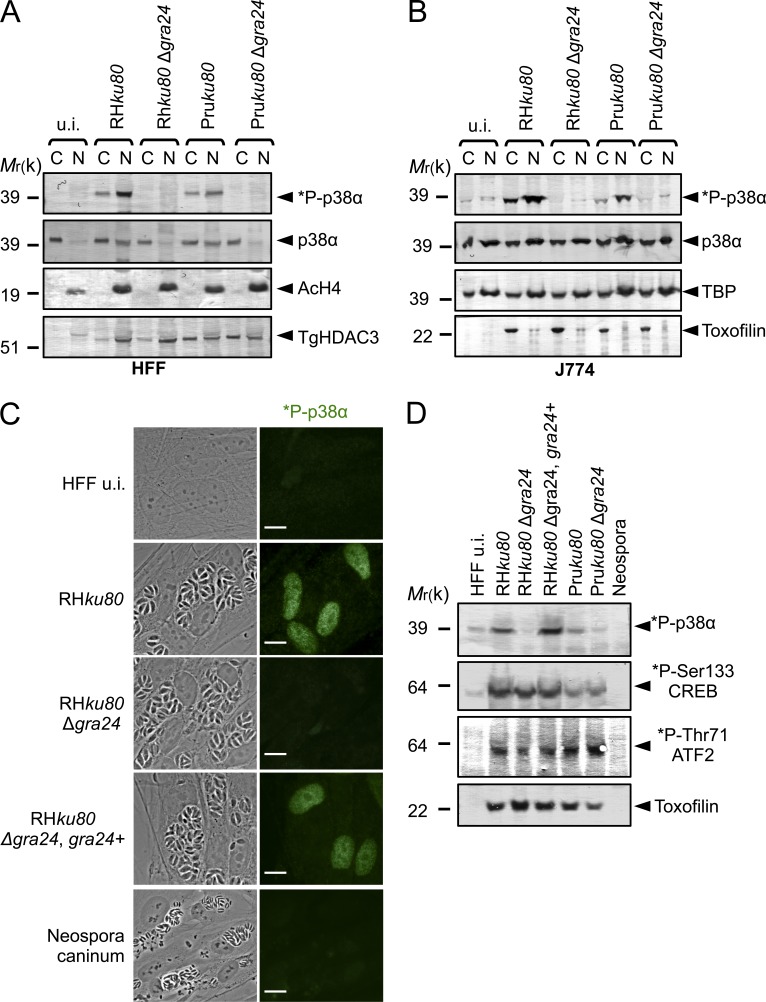
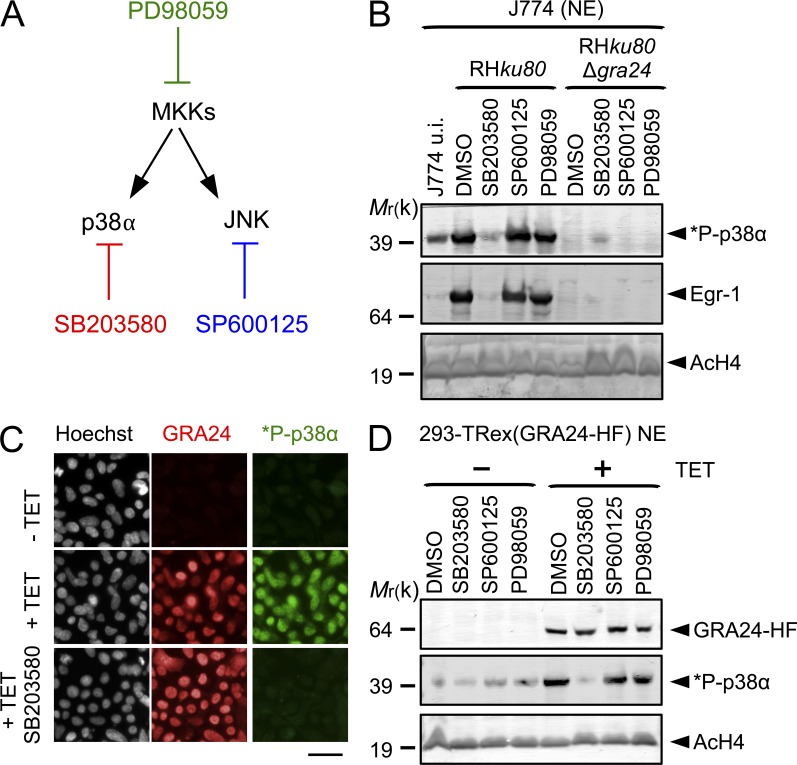
To investigate the role of GRA24 on host cell p38α activation during infection, WT, GRA24 knockout, or complemented tachyzoites were generated. The GRA24 gene was disrupted by homologous recombination (Δgra24) in both type I (RHku80) and type II (Pruku80) strains while the mutant was complemented with a single copy of a GRA24 functional allele driven by its endogenous promoter (Fig. S1 and Table S1). All strains behave similarly in vitro, indicating that GRA24 is not essential for either invasion or intracellular survival (unpublished data). Consistent with our previous data, no phospho-p38α population is ever detected in HFF and J774 MØ cells hosting GRA24 knockout tachyzoites in contrast to cells hosting GRA24 WT virulent type I or cystogenic type II parasite strains (Fig. 4, A and B). Importantly, reintroduction of the GRA24 gene in the mutant restores a parasite’s ability to induce p38α phosphorylation (Fig. 4, C and D), whereas p38α was never found to be activated by Neospora caninum regardless of the multiplicity of infection (MOI) or the duration of infection (Fig. 4, C and D). Remarkably, although p38α activation usually occurs transiently and within minutes in response to most stimuli (Ferreiro et al., 2010), GRA24-dependent p38α phosphorylation is, in contrast, impressively sustained as late as 24 h after the swift tachyzoite invasion step (Fig. 4), a trait which fits well with the unusual long-lasting presence of GRA24 into the host cell (Fig. 2 A) and suggests a direct control on the p38α kinase activity.
Figure 4.
GRA24 elicits phosphorylation and relocation of host p38α in the nucleus. (A and B) Immunoblotting detection of *P-p38α and p38α in HFF (A) and J774 MØ (B) uninfected (u.i.) or infected (24 h) with RHku80, RHku80 Δgra24, Pruku80, and Pruku80 Δgra24 strains. Cytosolic (C) and nuclear (N) cell lysates were probed with the indicated antibodies. Histone H4 acetylated (K5-K8-K12-K16) (nuclear), TBP (host-specific), and TgHDAC3 (parasite-specific) levels are shown as loading controls. (C) Subcellular in situ detection of *P-p38α (green) in HFF uninfected (u.i.) or infected (24 h) with RHku80, RHku80 Δgra24, RHku80 Δgra24, GRA24+, and Neospora caninum strains. Data are representative of at least three experiments. Bars, 10 µm. (D) Immunoblotting detection of *P-p38α, *P-Ser133-CREB, and *P-Thr71-ATF2 in nuclear fraction and Toxofilin (parasite-specific) in cytosolic fraction from HFF uninfected (u.i.) or infected (24 h) with the aforementioned strains and Neospora caninum. Data are representative of two experiments.
Once present in the hosting cell, GRA24 allows sustained p38α autophosphorylation, independently of the prototypic kinase cascade
The prototypic p38α activation is achieved by upstream MAPK kinase (MKK3/6; Ashwell, 2006) or, alternatively, through TAB1-mediated p38α autophosphorylation (Ge et al., 2002). Another pathway, restricted so far to T cells upon T cell receptor activation, includes phosphorylation of Tyr323 by the ZAP-70 tyrosine kinase that leads to p38 autophosphorylation but solely on Thr180 (Salvador et al., 2005). The host p38α autophosphorylation, observed when tachyzoites are nested in their PV, was proposed to reflect a TAB1-dependent mechanism, thus a mechanism bypassing the MKK-dependent MAPK phosphorylation (Kim et al., 2005). In T. gondii–hosting J774 MØ cells exposed to SB203580, a competitive inhibitor of ATP binding blocking p38α autophosphorylation but not trans-phosphorylation by MKKs (Gum et al., 1998), there is a complete abrogation of GRA24-induced p38α phosphorylation, a phenotype which was not observed when cells were treated with PD98059 and SP600125, which have been shown to block the MEK1/2 and the JNK pathways, respectively (Fig. 5, A and B). In addition, expression of GRA24 is sufficient to trigger p38α autophosphorylation because SB203580 inhibits p38α activation in 293 cells induced for GRA24 synthesis, whereas MEK1/2 and JNK inhibitors do not (Fig. 5, C and D). GRA24 is the first molecular switch originating from a eukaryotic parasite that directly activates p38α in a sustained way independently of the prototypic kinase cascade in both human stromal cells and mouse macrophages.
Figure 5.
GRA24 promotes p38α autophosphorylation. (A) Diagram showing the enzymes and their respective inhibitor. (B) Specific pharmacological inhibition of GRA24-dependent p38α phosphorylation in J774 MØ. Cells were incubated for 1 h with p38 (SB203580, 15 µM), SAPK/JNK (SP600125, 25 µM), and MKK (PD98059, 15 µM) inhibitors or DMSO vehicle before infection in presence of the drug with RHku80 and RHku80 Δgra24 strains (18 h). Nuclear fractions were immunoblotted with the indicated antibodies. Data are representative of two experiments. (C) In situ *P-p38α (green) detection in 293-TRex cells before (−Tet) or after (+Tet) conditional GRA24-HF expression (18 h; red). SB203580 impedes p38α phosphorylation. Images are representative of at least three experiments. Bar, 2.5 µm. (D) p38α phosphorylation status in 293-TRex cells pretreated by inhibitors (see B) and induced (+Tet) or not (−Tet) to express GRA24-HF. Nuclear fractions were immunoblotted with the indicated antibodies. Data are representative of two experiments.
GRA24 interacts with p38α through a singular kinase interacting motif (KIM)
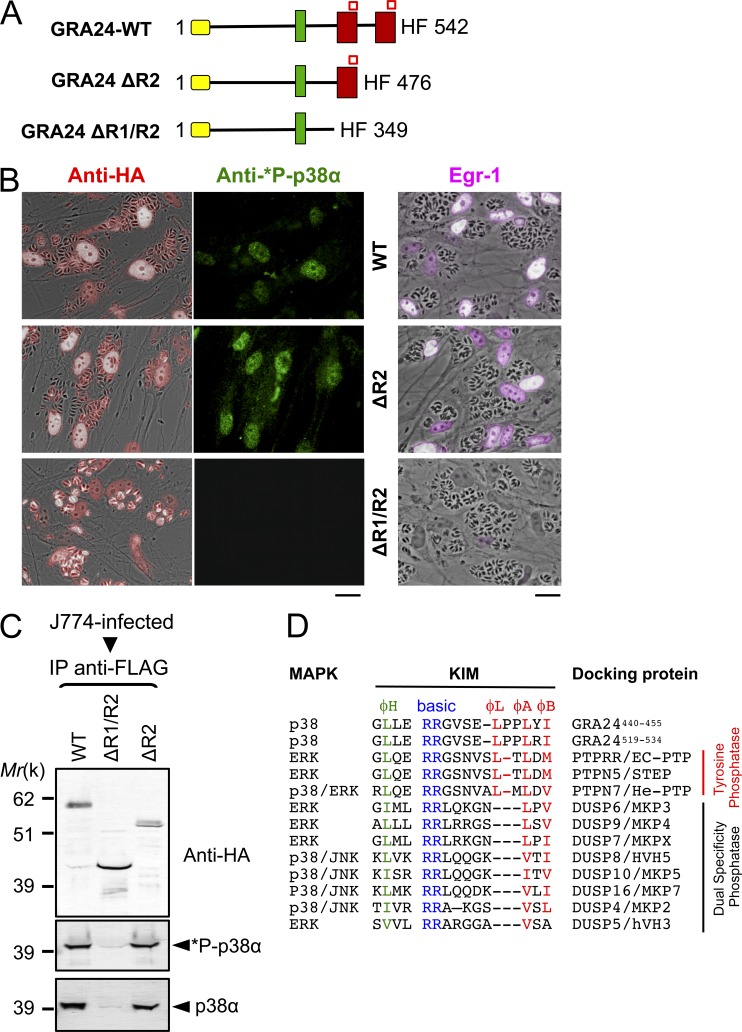
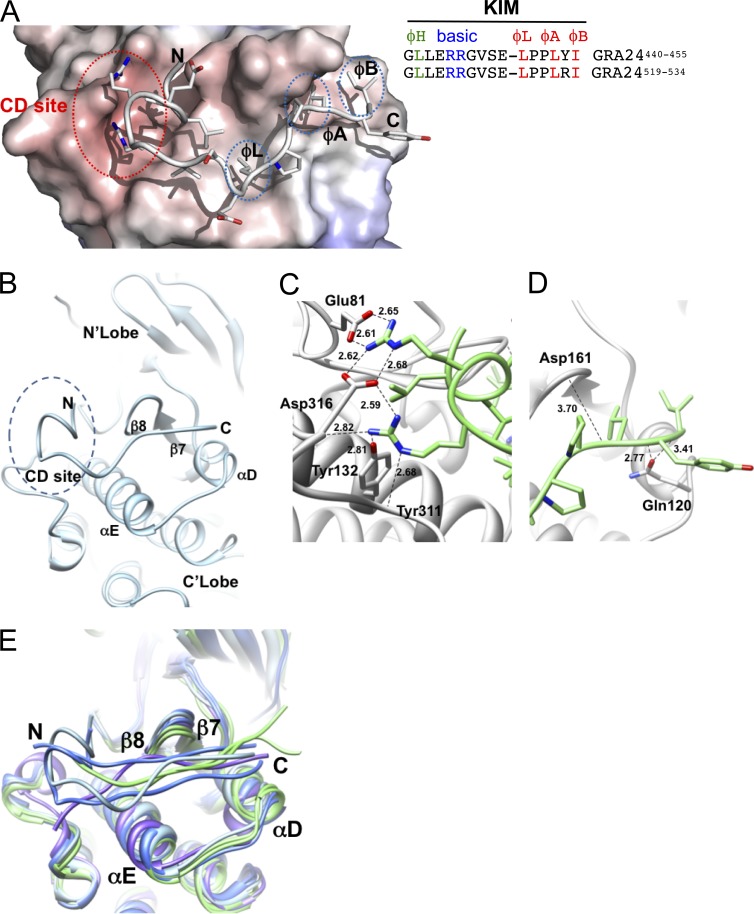
To detail further how GRA24 binding to p38α promotes activation of the kinase, we mapped the GRA24 region interacting with the enzyme. To this end, HAFlag epitope-tagged carboxy-terminal truncated forms of GRA24 were stably expressed in RHku80 parasites (Fig. 6 A). The two chimeric proteins, namely GRA24 ΔR2 and GRA24 ΔR1/R2, migrate at their expected molecular weights (Fig. 6 C) and localize in the host cell nuclei after infection (Fig. 6, B and C), thereby indicating that the removed domains do not affect the export and the transit into the host cell of the chimeric proteins. However, although deleting only repeat R2 has no effect on p38α binding and activation, double deletion of R1 and R2 abolishes GRA24 binding property to p38α (Fig. 6 C), preventing any p38α phosphorylation (Fig. 6 B), thus pointing to the required contribution of GRA24 R1 domain to p38α activation. These observations prompted us to look for the molecular determinants within the repeat that drive the interaction with/activation of p38α. We identified a perfect replicate of the canonical KIM (also known as D motif, Fig. 6 D) defined as a key MAPK-interacting structural element that, in addition, provides specificity toward the MAP kinase partners (Reményi et al., 2005; Francis et al., 2011). The motif is typified by a cluster of basic residues at the N terminus and hydrophobic amino acids near the C terminus (Fig. 6 D). The GRA24-KIM clearly belongs to the tyrosine phosphatase PTP-KIM class (defined in Garai et al., 2012; Fig. 6 D) but displays an unusual insertion of a proline (Pro-451) between hydrophobic canonical residues ϕL and ϕA that, in tandem with Pro-452, confers local rigidity to the linear peptide embedded in the KIM binding site (Fig. 6 D) and, most likely, adds specificity toward p38α (Garai et al., 2012). Our docking models validate the proper binding of GRA24-KIMs to p38α (Fig. 7). More specifically, the two N-terminal arginine residues (Arg-444 and Arg-445) share salt bridges with CD-site Asp-316 and Glu-81 of p38α, whereas both Leu-450 (ϕL) and Leu-453 (ϕA) insert optimally into the shallow hydrophobic pockets (Fig. 7, A and B). Finally, Ile-455 (ϕB) reinforces the hydrophobic interactions between the two partners, as previously reported for the He-PTP Val-31 residue (ϕB; Francis et al., 2011). Collectively, peculiar GRA24-KIM structural features empower GRA24 ability to bind to p38α, thereby accounting for the remarkable resistance to the highly stringent conditions used during the GRA24–p38α complex purification (Fig. 3, B and E). Such binding properties are expected to potentiate p38α autophosphorylation, as KIM-mediated interaction with p38α has been shown to trigger local (Fig. 7 E) as well as long-range conformational changes, in particular within the activation loop that becomes disordered, therefore exposing the phosphorylatable Thr-180 and Tyr-182 residues (Chang et al., 2002; Zhou et al., 2006; Zhang et al., 2011). Of note, the KIM repeat itself, i.e., within R1 and R2 (Fig. 6 A), might favor p38α trans-autophosphorylation via the simultaneous binding of two p38α monomers (Diskin et al., 2007).
Figure 6.
A singular KIM motif in GRA24 repeats is required for p38α autophosphorylation. (A) Diagram of GRA24 C-terminal truncations. Signal peptide (yellow), nuclear localization signal (green), internal repeats (red), and KIM (red square) sequences are shown. (B) IFA of GRA24 chimeric proteins (red), subcellular localization, and p38α phosphorylation status (green) in HFF infected with parasites expressing HF-tagged GRA24 full-length or truncates (see A). Data are representative of three experiments. Bars, 20 µm. (C) J774 MØ were infected (24 h) with parasites expressing HF-tagged GRA24 full-length or truncates. Tagged proteins were Flag-immunoprecipitated and eluates were analyzed by immunoblotting. Data are representative of two experiments. (D) Sequence alignment of KIMs from GRA24 and phosphatases. The N-terminal hydrophobic residue (φH), positively charged residues (basic), and φL-X-φA-X-φB motif are indicated.
Figure 7.
Structural features of the GRA24-KIM1 peptide main chain docked into p38α. (A and B) GRA24-KIM1 peptide docked into p38α (1LEW). The N-terminal side binds to the charged CD site while C-terminal hydrophobic residues insert into hydrophobic pockets ϕL, ϕA, and ϕB. The hydrophobic pocket is formed by the loop between β7 and β8 and the loop joining αD and αE helices. (C and D) GRA24-KIM1 peptide docked and energy minimized (see Materials and methods) into p38 KIM-binding domain. (C) Hydrogen bond network of the two arginines at the CD domain. (D) Peptide hydrophobic C terminus flanked by loops 159–163 and 118–125. (E) Cα-backbone KIM binding site area superposition of p38α, ERK2, and JNK models demonstrating local displacement upon binding. p38α peptides (MEF2A-1lew and MKK3b-1lez green), ERK2 peptides (MKP3-2fys and HePTP-2gph, blue), and JNK (pepJIP1-1ukh, purple) and p38α-GRA24/KIM1 docked peptide (light blue) are shown.
GRA24 activates the expression of proinflammatory genes in macrophages
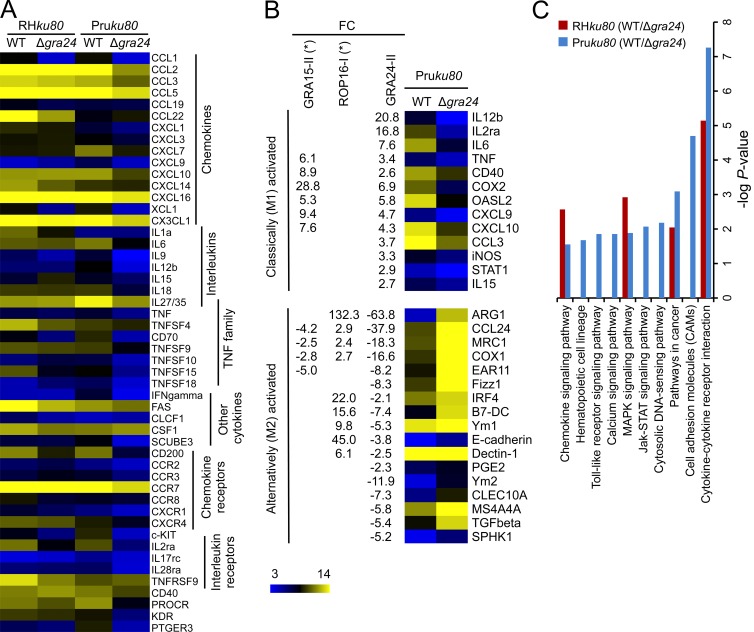
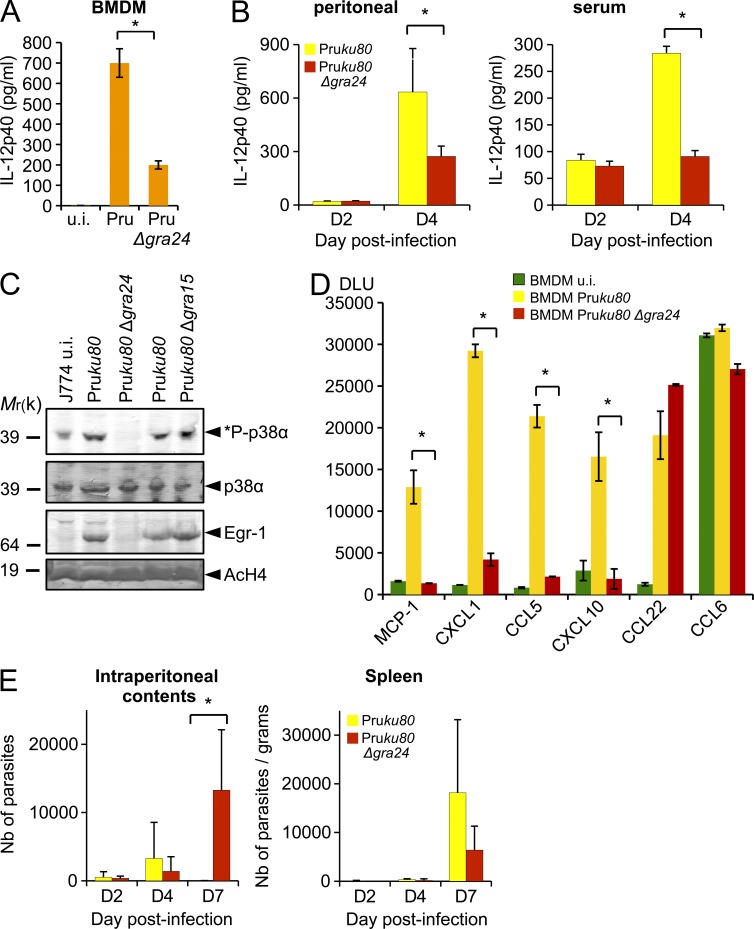
The unique features of the unraveled interaction between GRA24 and p38α and the consequences on p38α activation status prompted us to analyze whether the GRA24–p38α complex contributes to the typical changes in gene expression observed in either stromal or phagocytic cells infected with tachyzoites (Blader et al., 2001; Saeij et al., 2007; Jensen et al., 2011). To address this possibility, we performed a genome-wide expression profiling of mouse BM-derived macrophages (BMDMs) hosting parental versus Δgra24 parasites from both type I and type II strains (Fig. 8 A). We focused our analysis on genes that were altered with more than a twofold change when comparing each Δgra24 mutant with their respective parental strains. KEGG analysis highlights 10 pathways as significantly and selectively induced (P < 0.05) in a GRA24-dependent fashion, a majority of which regulates proinflammatory cytokines (e.g., IL-6, IL-12b, and TNF) and chemokines (e.g., MCP-1, CCL5, CXCL1, and CXCL10) known to operate in any sterile tissues colonized by invasive microbes, whether they are nonparasitic or parasitic (Fig. 8 C). Interestingly, the GRA24-dependent changes are exacerbated in cells infected with the type II clonal lineage, possibly as a consequence of allelic polymorphism between the strain types (Fig. 1 B), which leads to a concomitant up-regulation of 13 genes linked to M1-MØ activation and down-regulation of 17 genes associated with the alternative M2–MØ pathway (Fig. 8 B). We have also noticed that both IFN-γ– and iNOS-coding transcript populations were also up-regulated in a GRA24-dependent manner by 12- and 3-fold, respectively (Fig. 8, A and B). Of note, at least within the time frame (24 h) examined in these experiments, the BMDM supernatants display no difference in IFN-γ levels as assessed by ELISA (unpublished data).
Figure 8.
GRA24 alters the host-cell transcriptome. C57BL/6 BMDMs were infected with either type I RHku80 Δgra24 or type II Pruku80 Δgra24 mutant parasites and compared with infection with their respective parental Toxoplasma strains. Cells were infected at an MOI of 1:3 and total RNAs were extracted 18–20 h after infection. cDNA synthesis, labeling, and differential gene expression analysis are detailed in Materials and methods. The differentially regulated genes were identified after normalization of the raw data and filtering genes with a fold change cutoff of twofold or more and p-value ≤0.05. (A) Heat map of the GRA24-regulated cytokine-related genes identified by KEGG analysis. Mean log2 gene expression values were median-centered and genes were clustered according to the biological pathways identified. The complete set of genes is listed in GEO dataset accession no. GSE38782. (B) Heat map representation of the GRA24-regulated genes associated with classical (M1) or alternative (M2) MØ phenotypes after infection of BMDM with type II strains. Gene symbols and the fold change (FC, at least twofold mean difference) in gene expression are listed for GRA24II-dependent genes for comparison with those reported by Jensen et al. (2011) as ROP16I- and GRA15II-regulated. (C) KEGG analysis of the differentially expressed genes when comparing WT versus Δgra24-infected cells. Genes listed in GEO dataset accession no. GSE38782 (fold change cutoff twofold or more) were analyzed to determine pathways that were statistically overrepresented (p-value <0.05).
GRA24-dependent IL-12p40 and MCP-1 production by mouse macrophages hosting tachyzoites
As stated above, in laboratory mice, the T. gondii– and mouse-driven generation of immune type 1 effectors that prevent both uncontrolled tachyzoite population expansion and subsequent host death is characterized by the IL-12p40 production by tachyzoites-hosting macrophages (Scharton-Kersten et al., 1996; Sher et al., 2003; Yap et al., 2006). GRA24 is clearly playing a major role in regulating IL-12p40 synthesis, as its loss severely compromises both IL-12 mRNA and protein levels in mouse BMDM (Fig. 8 B and Fig. 9 A). Similarly, on day 4 after infection, mice infected with a Pruku80 Δgra24 strain had significantly less IL-12p40 in their i.p. cavities and serum than mice infected with the parental strain (Fig. 9 B). However, despite an efficient GRA24-mediated p38α activation (Fig. 4), type I parasites fail to trigger IL-12 secretion in BMDM (Fig. 8 A and not depicted), a phenotype which might reflect a dominant IL-12 suppressive activity driven by the ROP16 type I allele (Saeij et al., 2007; Yamamoto et al., 2009; Butcher et al., 2011). In addition, other effectors, such as GRA15, are involved in the control of IL-12 synthesis through the activation of the NF-κB pathway (Rosowski et al., 2011). Although GRA15 does not trigger p38α phosphorylation (Fig. 9 C), GRA24 does not affect NF-κB (not depicted), highlighting that several T. gondii strain-specific or -shared effectors display synergistic or antagonist effects on the regulation of IL-12p40–coding gene expression. In support of GRA24 key contribution to the IL-12 response, GRA24 is found to critically account for the typical increase in MCP-1, CXCL1, CCL5, and CXCL10 chemokine synthesis (Fig. 9 D) triggered by a type II tachyzoite lineage. Remarkably, MCP-1 was shown to play a substantial role in the recruitment, to T. gondii–loaded tissues, of Gr-1+ inflammatory monocytes that, in turn, produce nitric oxide and are essential for the local control of the tachyzoite population size (Robben et al., 2005; Dunay et al., 2008). The above results argue that the GRA24-dependent MCP-1 secretion might be involved in the recruitment of Gr-1+ monocytes in tissues transiently remodeled as cell-cycling tachyzoite-protective niches. To examine the functional consequences in vivo of GRA24-dependent MCP-1 secretion, we challenged C57BL/6 mice with 103 tachyzoites of Pruku80 WT or Pruku80 Δgra24 strains and assessed parasite numbers in the peritoneal exudate as well as in the spleen. A larger number of parasites are detected beyond day 7 in the peritoneal contents of mice infected with Pruku80 Δgra24 than in mice infected with the parental strain (Fig. 9 E). Parasites are also found in the spleen, indicating that the infection becomes quickly systemic, but in this tissue no significant deviation in parasite loads was detected between the WT and the Δgra24 strains at any of the time points analyzed (Fig. 9 E). These results suggest that the deletion of GRA24, while suppressing MCP-1 production, most likely prevents Gr-1+ recruitment and contributes to impair the ability of the mice to suppress parasite numbers at the site of infection in the gut. We then assess if GRA24 is altering the overall host survival of C57BL/6 mice. A dose of 5 × 102 tachyzoites required to achieve a near LD100 when using the Pruku80 strain was i.p. inoculated to C57BL/6 mice whose survival was monitored over the acute phase of infection. Mice infected by either the WT Pruku80 or Pruku80 Δgra24 parasites succumb to infection within 10 to 20 d (unpublished data; n = 6 mice per strain). Therefore, GRA24-deficient parasites did not fully recapitulate the phenotype of MCP-1−/− and CCR2−/− mice that were shown to be profoundly susceptible to acute toxoplasmosis (Robben et al., 2005; Dunay et al., 2008). This difference might be a result of the genetic background of the parasite strains, the type II Pruku80 strain (Fox et al., 2011) being more virulent and less cystogenic than the type II ME49 strain used in most Toxoplasma infection including those using MCP-1−/− and CCR2−/− mice (Robben et al., 2005).
Figure 9.
GRA24 promotes chemokine secretion and contributes to the control of early parasite replication in vivo at the site of infection. (A) IL-12p40 cytokine production by uninfected (u.i.) or 24-h-infected (Pruku80 Δgra24 or parental strains) BMDM measured using ELISA. Means of three independent experiments ± SD are shown (*, P < 0.005, two sample Student’s t tests). (B) Peritoneal lavage fluid and serum were collected on days 2 and 4 after infection of C57BL/6 mice that had received an i.p. dose of 103 Pruku80 WT or Pruku80 Δgra24 tachyzoites. Concentrations of IL-12p40 were determined by ELISA. Data shown are means ± SD with n = 3 individual mice per parasite genotype at each time point. Error bars represent SD from one experiment. *: significantly different from WT (P < 0.05, Student’s t test). (C) Subcellular detection in J774 infected with Δgra24 and Δgra15 mutants demonstrated that *P-p38α and Egr-1 induction is dependent of GRA24 and independent of GRA15. Data are representative of two experiments. (D) Comparison of GRA24-dependent and -independent chemokine expression profiles after BMDM infection by Toxoplasma. Quantification is given in digital light units (DLU) drawn from the whole chemokine array. Error bars represent SD (two sample Student’s t tests; *, P < 0.006). (E) WT C57BL/6 mice were i.p. infected with a dose of 103 Pruku80 WT or Pruku80 Δgra24 tachyzoites. Parasites were enumerated using recovered i.p. contents or from homogenates of spleen at the indicated times after infection. Data shown are means ± SD (from three mice per parasite genotype) and representative of two independent experiments. *: significantly different from WT (P < 0.05, Student’s t test).
GRA24–p38a complex regulates the expression of Egr-1 and c-Fos transcription factors
Finally, as a link between the cytokine response and the direct GRA24-dependent p38α activation, we identified several transcription factors up-regulated in a GRA24-dependent fashion in MØ, among which those that either control the immediate early gene response or are immediate early prototype gene products like Egr-1 and c-Fos (Fig. 10 A). Of note, first, Egr-1 and c-Fos belong to a restricted set of 30 core genes whose expression is equally regulated by major p38α MAPK-activating stimuli (i.e., osmo-stress, the cytokine TNF, and the protein synthesis inhibitor anisomycin; Ferreiro et al., 2010), and second, the Egr-1 promoter activity is blocked by a pharmacologically inactivated p38α kinase (Tur et al., 2010). These observations strongly argue for a direct function of GRA24 on classical p38α MAPK-regulated genes and confirm previous studies on the induction of Egr-1 and its paralog Egr-2 during Toxoplasma infection (Phelps et al., 2008; Wiley et al., 2011). By Western blotting and immunofluorescence, we indeed confirmed at the protein level that Egr-1 is induced in the nuclei of human stromal cells and mouse macrophages hosting WT T. gondii, a phenotype which is not induced in cells hosting Δgra24 Toxoplasma or Neospora (Fig. 10, B and C; and not depicted). Reintroduction of the GRA24 gene restored the Toxoplasma ability to up-regulate Egr-1 (Fig. 10 B). Egr-1 induction was also validated in 293 cells conditionally expressing GRA24 (Fig. 10 D), and the level of expression was reduced in a dose-dependent fashion by the inhibitor SB203580, thereby establishing the link between Egr-1 and the autophosphorylation of p38α by GRA24 (Fig. 5 B). Finally, we identified the GRA24-KIM as the basis of p38α binding and autophosphorylation that further entails Egr-1 activation (Fig. 6 B). Besides Egr-1, c-fos is also regulated at the transcriptional level by GRA24 (Fig. 10 A). In particular, this transcription factor is phosphorylated on serine 32 in a GRA24-dependent fashion in both murine MØ and human fibroblasts (Fig. 10 E and not depicted). Unexpectedly, Toxoplasma elicits a prolonged induction of Egr-1 and c-Fos, which correlates with the nuclear persistence of the GRA24–p38α complex during infection but differs from the transient usual canonical activation of these genes. The induction of these transcription factors by GRA24 is most likely followed by a second wave of host cell transcriptome alteration because both Egr-1 and c-Fos then operate in a hierarchical manner by inducing the expression of their downstream genes (e.g., Egr-1-regulated genes, see GEO accession no. GSE38782).
Figure 10.
GRA24 triggers up-regulated expression of the host transcription factors Egr-1 and c-FOS. (A) Heatmap representation of host transcription factors up-regulated in a GRA24-dependent fashion in BMDM infected by the aforementioned strains. Egr-1 and c-FOS are indicated in red. (B) IFA of GRA24-dependent induction of Egr-1 (red) in uninfected (u.i.) or infected (24 h) HFF by the indicated strains. Images are representative of at least four experiments. Bar, 20 µm. (C) Cellular fractionation of J774 cells left uninfected (u.i.) or infected (24 h) by the indicated strains. Cytoplasmic (C) and nuclear (N) fractions were analyzed by Western blotting using indicated antibodies. TBP (host-specific) and toxofilin (parasite-specific) are shown as loading controls. Data are representative of three experiments. (D) Western blot analysis of nuclear fractions of 293-TRex cell line expressing GRA24-HF using indicated antibodies. Cells were either left untreated (− Tet) or treated (+ Tet) with 1 µg/ml tetracycline for 20 h. AcH4 was used to assess host nuclear protein enrichment. Data are representative of three experiments. (E) IFA showing the serine 32 phosphorylation status of c-FOS (in red) in serum-starved confluent HFF left uninfected (u.i.) or after 24 h of infection by RHku80, RHku80 Δgra24, and RHku80 Δgra24, GRA24+ parasites. Images are representative of two experiments. Bar, 20 µm.
DISCUSSION
Host cell invasion is a critical step by which Toxoplasma zoites rapidly establish themselves and further complete their developmental program as bradyzoite-containing cysts known to be located in the tissue of prey that are ingested by predators. Recent research has highlighted that early during the invasion process, the parasite secretes several rhoptry bulb resident proteins with a majority of kinases and phosphatases that co-opt host cells by interfacing with their signaling pathways (Melo et al., 2011). Once internalized, a Toxoplasma tachyzoite resides within a dynamic PV that is constantly reshaped through molecular exchanges with the host cell cytoplasm and that support parasite growth. Although nonenzymatic molecules secreted by DG were first thought of as parasite effectors restricted to the PV biogenesis and maturation, a recent study (Bougdour et al., 2013) and the present study highlight that DG-resident nonenzymatic proteins are secreted into the vacuole space, cross the PVM, and reach the host cell nucleus where they modulate host gene expression. Thus, GRA16 and GRA24 emerge as a subfamily of DG-scaffolding proteins exported beyond the tachyzoite-hosting vacuole to operate as effector molecules in infected cells.
In the present study, we have identified GRA24 as a strong interactor of host p38α, with the resulting complex acting as a regulator of gene expression in the host cell nucleus. Indeed, we establish that GRA24 interacts with the p38α enzyme, triggers its autophosphorylation, and consequently its activation. GRA24 bypasses the typical MKK-dependent MAPK phosphorylation cascade upstream of p38 and is the first parasitic microbe agonist so far described to promote p38α autophosphorylation through a dimeric stable complex. Interestingly, GRA24 specifically promotes Thr180 autophosphorylation of p38α because it neither activates ERK1,2 (Thr202/Tyr204 and Thr185/Tyr187, respectively) nor alters the JNK (Thr183/Tyr185) phosphorylation (unpublished data). In agreement with these results, none of the aforementioned kinases copurify with GRA24, reinforcing the notion of a high specificity of GRA24 function on p38α. Another unique feature of GRA24 function lies in its property to promote a sustained phosphorylation of p38α that correlates with the long-lasting secretion of GRA24. This kinetics of activation is remarkably distinct from the transient activation mediated by the prototypic MAPK pathway and has important consequences because it likely avoids conventional negative feedback by phosphatases. Consistent with this idea, we provide evidence that the carboxy-terminal repeat of GRA24 is essential for binding to p38α and its subsequent autophosphorylation (Fig. 6 B). Furthermore, molecular modeling reveals that GRA24 repeat contains a p38α docking KIM motif that is phylogenetically related to those of the PTP tyrosine phosphatase family (Fig. 6 D). Collectively, these data suggest that GRA24 has evolved a structurally optimized KIM domain that might be able to compete accurately with the phosphatases for binding to MAPK.
Toxoplasma-induced p38α activation was initially reported as TAK1- and Myd88-independent and proposed to occur through an association with the scaffold protein TAB1 (Kim et al., 2005). But—of note—first, TAB1 does not activate p38α after TNF exposure of fibroblasts derived from the MKK3-MKK6 double knockout mice (Brancho et al., 2003), and second, once bound to p38α, TAB1 prevents the p38α nuclear localization (Lu et al., 2006) disproving the validity of this proposal. In contrast, our many complementary data allow us to highlight a major role for GRA24 as the shuttling and scaffolding protein that allows not only the p38α nuclear localization but also its autophosphorylation, at least in two distinct cell lineages—fibroblasts and macrophages—that T. gondii tachyzoites do subvert as host cells. Moreover, a contribution of parasite-derived tyrosine kinase is also very unlikely because bypassing the recourse to tachyzoites, through the ectopic expression of GRA24 in mammalian cells, is clearly sufficient to promote p38α autophosphorylation. It is noteworthy that GRA24 has multiple alternative splice variants. Specifically, the presence of the exon E3b in the mRNA isoform 2 introduces a premature stop codon, leading to a protein variant devoid of the terminal repeats R1 and R2 (Fig. 1 A). In this fashion, the parasite might use alternative splicing to turn off GRA24-dependent p38α autophosphorylation under certain growth or developmental conditions, which remain to be discovered.
The ability of GRA24 to directly promote host p38α autophosphorylation places it as a potential regulator of the synthesis of growth factors and/or cytokines/chemokines and associated receptors, at least in macrophages once they are subverted as tachyzoite-hosting cells. Using microarray analysis, we showed that GRA24 controls, at the transcriptional level, an impressive network of genes encoding for cytokines/chemokines (Fig. 8), well beyond those associated previously with the secretion of GRA15 (Jensen et al., 2011; Rosowski et al., 2011). Chief among the cytokines is the proinflammatory cytokine IL-12p40, with a 20-fold transcript excess that typifies a robust M1 profile in MØ hosting type II tachyzoites. As reported previously, IL-12 synthesis induction by Toxoplasma-infected MØ occurs at lower levels and with distinct kinetics relative to endotoxin stimulation; in brief, although IL-12 levels peaks within 6 h of LPS stimulation (Kim et al., 2005), parasite-induced IL-12 is detected within a 24–40-h window after macrophage invasion (Fig. 9 A and Kim et al., 2005). This timing is consistent with the kinetics of GRA24 secretion and the requisite for de novo synthesis of p38-activated transcription factors driving IL-12 synthesis. Of note, once macrophages are hosting tachyzoites, both IL-1β and IL-18 signaling use MyD88 as an adaptor, leading to NF-κB and MAPK activation and subsequent IL-12 production (Adachi et al., 1998). It is also noteworthy that IL-18 is approximately sixfold induced by GRA24 in MØ infected with type II strain (Fig. 8 A). It is therefore possible that autocrine production of IL-18 results in GRA24-mediated IL-12 release. Anyhow, the massive production of IL-12 induced by type II strains when tachyzoites start to proliferate is most likely controlled by the synergistic action of GRA15 and GRA24, which keep parasite numbers low, thus facilitating long-term parasitism. This is consistent with the observation that GRA24 is one among the few polymorphic highly expressed genes that are under positive selection in Toxoplasma population (Minot et al., 2012). We can also speculate that GRA24 loss in Neospora caninum correlates with the inability of Neospora to properly modulate the host immune response, an argument which has been put forward earlier to explain why Neospora has a more limited host range than Toxoplasma (Reid et al., 2012).
Besides triggering a strong Th1 response, GRA24 also significantly contributes to suppress M2 markers at the transcriptional level in MØ hosting type II tachyzoites, a situation which powers the M1 response. The repertoire of M2 genes that are down-regulated in a GRA24-dependent manner again significantly exceeds that of GRA15, with Arg1 being distinctively ∼60-fold repressed by GRA24 (Jensen et al., 2011; Fig. 8 B). However, neither arginase-1 nor the mannose receptor type C (Mrc1/CD206) are induced at the protein level in Δgra24-infected BMDMs (unpublished data), suggesting that the suppression of the GRA24-dependent transcriptional repression of M2 genes in type II strain is not sufficient to induce the production of the corresponding protein product, consistent with previous studies (Butcher et al., 2011; Jensen et al., 2011). Additionally, GRA24 is found to critically account for the typical increase at the protein level of four chemokines including MCP-1/CCL2 (Fig. 8 A and Fig. 9 D). Remarkably, in mice lacking MCP-1 or its receptor CCR2, no recruitment, in T. gondii–hosting tissues, of inflammatory Gr-1+ monocytes, is detectable, a situation which likely accounts for the sustained parasite burden and early host mortality (Robben et al., 2005; Dunay et al., 2008). Consistent with these data, we show that mice infected with Δgra24 tachyzoites fail to control parasite numbers in the gut (Fig. 9 E), revealing a contributory role for GRA24 in resistance to acute toxoplasmosis, most likely through the regulation of cytokine expression and secretion including MCP-1.
GRA24 also significantly induces the secretion of CXCL10/IP-10 (Fig. 8 A and Fig. 9 D). This chemokine is required for preventing uncontrolled T. gondii tachyzoite burden (Khan et al., 2000), and its depletion prevents T cell recruitment and effector function in mouse tissues where the tachyzoite to bradyzoite developmental transition is known to occur. In this regard, it was recently shown that the CXCL10/IP-10 chemokine enhances the ability of CD8+ T cells to control tachyzoite reexpansion in cyst-hosting mouse brains (Harris et al., 2012). Collectively, these studies underscore the importance of GRA24 in orchestrating a cell response during Toxoplasma infections by promoting the induction of a selective set of chemokines, which contributes to the recruitment of inflammatory monocytes previously described as a first line of defense controlling intestinal pathogens, including T. gondii (Dunay et al., 2008).
Given that GRA24 was involved in nonprototypical direct p38α activation, we next sought to trace the factors that act downstream of this unusual signaling pathway. This led us to show that several transcription factors, including Egr-1 and c-Fos (Fig. 10 A), inhabit key downstream positions within this coordinated acute inflammatory process driven by tachyzoites nested within macrophage PV. Several factors are needed for full, regulated transactivation of the Egr1 (Tur et al., 2010) or c-Fos (O’Donnell et al., 2012) genes. As such, the mitogen- and stress-induced phosphorylation of CREB (CRE binding protein) at Ser133, especially by p38α, has been linked to the transcription of several immediate early genes, including c-fos, junB, and Egr-1 (Tur et al., 2010; O’Donnell et al., 2012). In this regard, the Toxoplasma tachyzoite, in a GRA24-dependent fashion, positively affects the transcription of two CREB orthologs, i.e., CREB2/ATF2 and CREBL2 (Fig. 10 A), as well as CREB-dependent downstream genes (GEO accession no. GSE38782). We first thought that GRA24 might activate p38α, which in turn directly phosphorylates CREB-related factors in macrophages hosting tachyzoites. But this is apparently more complicated because Toxoplasma induces the phosphorylation of both CREB-Ser133 and ATF2-Thr-71 independently of GRA24 (Fig. 4 D). Therefore, GRA24 might act upstream in the pathway. It was previously shown that the phosphorylation of Ser 133 facilitates the ability of CREB to interact with CBP (CREB-binding protein) and its paralog p300, therefore leading to the transcription of CREB-responsive genes (Johannessen et al., 2004). CBP and p300 possess an intrinsic acetylation activity and are thought to acetylate specific histone tails, thereby assuring relaxation of the chromatin environment (Johannessen et al., 2004). As such, the sustained presence of GRA24-p38α in the nucleus of the tachyzoites-hosting macrophages might reflect the need for the complex to be embedded in the chromatin to operate in the vicinity of p38α MAPK-responsive genes. A GRA24-dependent response might alternatively be mediated through Egr-1 that physically and functionally interacts with CBP/p300 to modulate gene transcription (Silverman et al., 1998). Although interesting, this hypothesis deserves further investigation.
In conclusion, GRA24 extends the scope of the function of T. gondii DG proteins beyond their initial and dedicated role in PV formation and maturation. Moreover, the specific attributes of the GRA24-dependent p38α activation unveil a novel regulatory path for p38α-controlled transcriptional changes. This path not only shortcuts the canonical MAPK signaling cascade turned on in response to various stress-related stimuli and intracellular pathogens (Ferreiro et al., 2010; Krachler et al., 2011) but it also differs from the alternate p38 activation pathways described so far involving the scaffolding protein TAB1 or the T cell–restricted kinase ZAP-70 (Brancho et al., 2003; Salvador et al., 2005).
MATERIALS AND METHODS
Parasites and host cells.
HFF primary cells, 293-TRex, and J774 cell lines were cultured in DMEM (Invitrogen) supplemented with 10% heat inactivated FBS (Invitrogen), 10 mM Hepes buffer, pH 7.2, 2 mM l-glutamine, and 50 µg/ml penicillin and streptomycin (Invitrogen). Cells were incubated at 37°C in 5% CO2. BMDMs were obtained from female C57BL/6 mice. Bone marrow was isolated by flushing hind tibias and femurs using a 25-gauge needle followed by passages through an 18-gauge needle to disperse cell clamps. Cells were suspended in DMEM supplemented with 10% heat-inactivated FBS and 20 ng/ml recombinant M-CSF (Invitrogen), and incubated in a tissue culture-treated flask for 8–12 h. Then, nonadherent cells were harvested and transferred in 55 cm2 non–tissue culture–treated plates (Corning) at 4–6 × 106 cells per plate and further incubated at 37°C, with 5% CO2 in humidified air. After 6 d, cells were washed with PBS to remove nonadherent cells, harvested by dislodging with a cell scraper in ice-cold PBS, and replated for the assay. This method yielded a highly pure population of F4/80+ macrophages by IFA. The Toxoplasma strains used in this study are listed in Table S1. All T. gondii strains were maintained in vitro by serial passage on monolayers of HFFs.
Reagents.
Antibodies against HA (3F10; Roche), MIC2 (provided by D. Sibley, Washington University School of Medicine, St. Louis, MO), Toxofilin (provided by I. Tardieux, INSERM, U1016, Institut Cochin, Paris, France), GRA1 and GRA7 (provided by J.-F. Dubremetz, UMR 5235 Centre National de la Recherche Scientifique, Montpellier, France), p38 MAPK (Cell Signaling Technology), Phospho-p38 MAPK (Cell Signaling Technology), Phospho-c-Fos (Ser32; Cell Signaling Technology), H4 Acetylated (EMD Millipore), TgHDAC3 (Bougdour et al., 2009), EGR1 (Cell Signaling Technology), and anti-TBP (Abcam) were used in the immunofluorescence assay and/or in Western blotting. Immunofluorescence secondary antibodies were coupled with Alexa Fluor 488 or Alexa Fluor 594 (Invitrogen). Secondary antibodies used in Western blotting were conjugated to alkaline phosphatase (Promega). The inhibitors SB203580, SP600125, and PD98059 were purchased from InvivoGen.
Immunofluorescence microscopy.
Immunofluorescence assays were performed as described in Bougdour et al. (2009). Cells grown on coverslips were washed in PBS and fixed for 20 min at room temperature with PBS containing 3% (vol/vol) formaldehyde. Fixed cells were permeabilized with PBS-0.1% Triton X-100 (vol/vol) for 10 min and blocked in PBS-3% BSA for 1 h at room temperature. Samples were incubated in PBS containing 3% BSA with the primary antibodies indicated in the figures, followed by the secondary antibodies coupled with Alexa Fluor 488 or Alexa Fluor 568 (Invitrogen) at a 1:1,000 dilution each in PBS–3% BSA. Nuclei of both host cells and parasites were stained for 10 min at room temperature with Hoechst 33258 at 2 µg/ml in PBS. After four washes in PBS, coverslips were mounted on a glass slide with Mowiol mounting medium. Images were acquired with a fluorescence microscope (Axioplan 2; Carl Zeiss).
Plasmid constructs.
To construct the vector pLIC-GRA24-HF, the coding sequence of GRA24 was amplified using primers LICF-117710_F and LICR-117710_R using RH genomic DNA as template. The resulting PCR product was cloned into the pLIC-HF-dhfr vector (Bougdour et al., 2013) using the LIC cloning method previously reported (Huynh and Carruthers, 2009). The C-terminal truncated versions of GRA24 were amplified and cloned in the aforementioned pLIC-HF-dhfr vector to generate pLIC-GRA24ΔR2-HF and pLIC-GRA24ΔR1/R2-HF. The pLIC-PGRA24-GRA24-HF was obtained by amplifying the promoter and coding region of GRA24 (strain GT1, Chromosome VIII, position 664,928–671,201) with LICF–5′ region–117710_F2 and LICR-117710_R2 primers. The resulting PCR product was cloned in the aforementioned pLIC-HF-dhfr vector. To construct the mammalian expression vector pcDNA-GRA24-HF, the coding sequence of GRA24 (residues 25–542) was amplified from a poly(A)+ enriched RH cDNA library using pcDNA4-LIC-117710-HAFlag_F and pcDNA4-LIC-117710-HAFlag_R primers. The resulting PCR product was cloned by LIC into the pcDNA-LIC-HF plasmid (Bougdour et al., 2013). Likewise, pcDNA4-FlagHA-LIC-117710_F and pcDNA4-FlagHA-LIC-117710_R primers were used to generate the plasmid pcDNA-HF-GRA24 harboring an N-terminal HAFlag tag. The vector pDEST14 KOGRA24 was generated to construct the deletion/insertion mutation of GRA24 in Toxoplasma strains RHku80 (type I, Huynh and Carruthers, 2009) and Pruku80 (type II, Fox et al., 2011). The Multisite Gateway Pro 3-fragment Recombination system was used to clone the dhfr cassette flanked by the 5′ and 3′ surrounding regions of GRA24 coding sequence of RH genomic DNA. The 5′ flanking region of GRA24 of RH strain was amplified using primers attB1-117710_F and attB4-117710_R, and was cloned into the plasmid pDONR221 P1-P4 (Invitrogen). The 3′ flanking region of GRA24 was amplified using primers attB3-117710_F and attB2-117710_R, and was cloned into the plasmid pDONR221 P3-P2 (Invitrogen). The resulting vectors, pDONR221/5′GRA24 and pDONR221/3′GRA24, respectively, were then recombined with pDONR221/DHFR (Bougdour et al., 2013) into the destination vector pDEST14, yielding the pDEST14 KOGRA24. The vector pDEST14 KOGRA15 was generated to construct the deletion/insertion mutation of gra15 in T. gondii Pruku80 strain. The 5′ flanking region of GRA15 of type II Pru strain was amplified using primers attB1-275470_F and attB4-275470_R, and was cloned into the plasmid pDONR221 P1-P4 (Invitrogen). The 3′ flanking region of gra15 was amplified using primers attB3-275470_F and attB2-275470_R, and was cloned into the plasmid pDONR221 P3-P2 (Invitrogen). The resulting vectors, pDONR221/5′gra15 and pDONR221/3′gra15, respectively, were then recombined with the pDONR221/DHFR (Bougdour et al., 2013) into the destination vector pDEST14, yielding the pDEST14 KOGRA15.
Toxoplasma transfection.
Vectors were transfected into RHku80 and Pruku80 parasites by electroporation. Electroporation was done in a 2-mm cuvette in a BTX ECM 630 (Harvard Apparatus) at 1,100 V, 25 Ω, and 25 μF. Stable integrants were selected in media with 1 µM pyrimethamine and cloned by limiting dilution.
Endogenous gene tagging in Toxoplasma.
We used the RHku80 strain (Huynh and Carruthers, 2009) to endogenously tag GRA24 in Toxoplasma. In brief, ∼20 µg of each DNA construct was linearized within the region of homology. Linearized vectors pLIC-GRA24-HF (BsiWI), pLIC-GRA24ΔR2-HF (BlpI), and pLIC-GRA24ΔR1/R2-HF (BlpI) were phenol-chloroform extracted and ethanol precipitated. Constructions were resuspended in TlowE buffer (10 mM Tris-HCl, pH 8.0, and 0.1 mM EDTA) for transfection. Stable recombinants were selected with pyrimethamine, single-cloned by limiting dilution, and verified by IFA. The resulting strains are RHku80 GRA24-HF, RHku80 GRA24ΔR2-HF, and RHku80 GRA24ΔR1/R2-HF, respectively (Table S1).
Generation of knockout and complementation.
To construct the deletion/insertion mutation of GRA24 in type I (RHku80) and type II (Pruku80) strains of Toxoplasma, the plasmid pDEST14 KOGRA24 was amplified by PCR using primers attB1-117710_F and attB2-117710_R. After sodium acetate/ethanol precipitation, DNA was resuspended in TlowE buffer (10 mM Tris-HCl, pH 8.0, and 0.1 mM EDTA) and ∼20 µg of PCR product was used for transfection. Recombinants were single-cell cloned by limiting dilution and verified by PCR analysis as follows. PCR with either a forward primer (5′ region–117710_F2) upstream of the 5′ flanking region used to generate the knockout construction and a reverse primer within the dhfr cassette (dhfr-KO-ol1_R) or a forward primer within the dhfr cassette (dhfr-KO-ol1_F) and a reverse primer (3′ region–117710_R1) downstream of the 3′ flanking region of GRA24 confirmed a disruption in the GRA24 locus. Further PCRs were performed to confirm the inability to amplify GRA24 coding sequence using primers (pcDNA4-FlagHA-LIC-117710_F and pcDNA4-FlagHA-LIC-117710_R) in both type I and type II strains. The deletion/insertion mutation of GRA15 in Pruku80 was constructed as described above with the plasmid pDEST14 KOGRA15 amplified by PCR using primers attB1-275470_F and attB2-275470_R. Disruption in the GRA15 locus was confirmed by PCR analysis with either a forward primer (5′ region–275470_F) upstream of the 5′ flanking region used to generate the knockout construction and a reverse primer within the dhfr cassette (dhfr-KO-ol1_R) or a forward primer within the dhfr cassette (dhfr-KO-ol1_F) and a reverse primer (3′ region–275470_R) downstream of the 3′ flanking region of gra15. To generate a complemented RHku80Δgra24 strain, vector pLIC-PGRA24-GRA24-HF was co-electroporated with the pMiniHX at 1:10 ratio, followed by selection with mycophenolic acid and xanthine, and cloned by limiting dilution. The resulting strain is named RHku80 Δgra24, GRA24+ (Table S1).
293-TRex transfection.
24 h before transfection, the cells were plated (80% confluency) in 6-well tissue culture dishes. 1 µg Flag–fusion protein–expressing plasmids and 0.1 µg puromycin selection plasmid were cotransfected into 293-TRex cells with Lipofectamine reagent (Invitrogen) according to the manufacturer’s instructions. 72 h later, cells were diluted in the presence of 5 µg/ml puromycin (Sigma-Aldrich) for selection. Individual drug-resistant clones were expanded and tested for tetracycline-inducible gene expression.
Chromatographic purification of GRA24-containing complexes.
Nuclear extracts from 293-TRex cells stably expressing Flag-tagged protein were incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich) for 1 h at 4°C. Beads were washed with 10-column volumes of BC500 buffer (20 mM Tris-HCl, pH 8.0, 500 mM KCl, 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.1% NP-40, 0.5 mM PMSF, and 1 µg ml−1 each of aprotinin, leupeptin, and pepstatin). Bound peptides were eluted stepwise with 250 µg ml−1 FLAG peptide (Sigma-Aldrich) diluted in BC100 buffer. For size-exclusion chromatography, protein eluates were loaded onto a Superose 6 HR 10/30 column equilibrated with BC500. Flow rate was fixed at 0.35 ml/min, and 0.5-ml fractions were collected.
Mass spectrometry and peptide sequencing.
Protein bands were excised from colloidal blue–stained gels (Invitrogen), treated with DTT and iodoacetamide to alkylate the cysteines, and then immediately subjected to in-gel tryptic digestion. Peptides were extracted with 5% vol/vol formic acid solution and acetonitrile, and injected into an Ultimate 3000 (Dionex) nanoLC system that was directly coupled to a LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific). MS and MS/MS data were acquired using Xcalibur (Thermo Fischer Scientific) and processed automatically using Mascot Daemon software (Matrix Science). Tandem mass spectra were searched against a compiled T. gondii database using the MASCOT program (Matrix Sciences).
Cell fractionation.
For cytosolic and nuclear fractionation analysis, cells were washed in ice-cold PBS and harvested using a cell scraper. After centrifugation, cells were resuspended in buffer D (10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M Sucrose, 10% Glycerol, 1 mM DTT, and 1× Roche protease inhibitor cocktail) and lysed for 8 min with Triton X-100 at a final concentration of 0.1%. Cell lysates were centrifuged at 1,300 g for 5 min and the supernatant containing the cytosolic fraction was collected. The pellet, containing the nuclear fraction, was washed once in buffer D and resuspended in cell extraction buffer (Invitrogen) for protein extraction. The supernatant and pellet fractions were clarified by centrifugation at 20,000 g for 10 min at 4°C. Supernatants were mixed with protein sample buffer (Invitrogen) for Western blot analysis.
Western blot.
Proteins were separated by SDS-PAGE, transferred to a polyvinylidene fluoride membrane (Immobilon-P; Millipore) by liquid transfer and Western blots were probed using appropriate primary antibodies followed by phosphatase-conjugated goat secondary antibodies (Promega). Signals were detected using NBT-BCIP (Amresco).
In vitro cytokine ELISA.
C57BL/6 BMDMs were seeded (4 × 105 per well) in 96-well plates and left to adhere overnight at 37°C in 5% CO2. Cells were infected with freshly lysed T. gondii tachyzoites at MOI = 3, and supernatants (200 µl) were collected 24 h after infection and stored at −20°C if necessary. IL-12p40 levels were determined using a commercially available ELISA kit (Mouse IL-12 p40 ELISA kit; Thermo Fisher Scientific) according to the manufacturer’s instructions.
Chemokine array.
For qualitative determination of the kinds of chemokines secreted by infected BMDM, chemokine Ab arrays were performed with culture supernatant. Macrophages were left uninfected or infected with Pruku80 or Pruku80 Δgra24 strain parasites at an MOI of 3. Culture supernatant was harvested after 24 h and used in mouse chemokine Ab arrays according to the manufacturer’s instructions (R&D Systems). In brief, supernatant was incubated with an array membrane on which 25 different mouse chemokine Abs were bound. The membrane was then incubated with a mixture of biotinylated anti-chemokine Abs followed by HRP-conjugated streptavidin. Detection was by chemiluminescence reagents provided by the manufacturer.
Quantitative real-time RT-PCR.
Total RNA was isolated from HFFs or BMDM infected with Pru or RH strains using TRIzol reagent (Invitrogen). cDNA was synthesized with random hexamers by using the High Capacity RNA-to-cDNA kit (Applied Biosystems). Samples were analyzed by real-time quantitative PCR for IL-12b, IFN-γ, CD70, and CD200 using TaqMan Gene Expression Master Mix (Applied Biosystems) according to the manufacturer’s instructions. β2-microglobulin was used as an internal control gene.
Structural studies.
Based on sequence similarities, we have generated a GRA24-KIM 16-aa (GLLERRGVSELPPLYI) model using the HePTP phosphatase KIM domain (RLQERRGSNVALMLDV) from structure 2GPH (Zhou et al., 2006) as a template, with appropriate residue mutations. The conformation of the side-chains and orientation of the main-chain for N- and C-terminal regions of peptides were modeled based on the bound peptide conformations in crystal structures of ERK2-pepHePTPm (2GPH) and p38-pepMEF2 (1LEW) peptide complexes, respectively. Similar work has been done with GRA24-KIM2(519–534), i.e., with an R instead of Y, with no significant differences. A p38–GRA24-KIM binary complex was then prepared using Coot modeling software (Emsley et al., 2010). The structure of p38, isolated from the p38–pepMEF2 complex (PDB accession no. 1LEW; Chang et al., 2002) was used as template for p38–GRA24-KIM docking and energy minimization calculations. The p38–GRA24-KIM complex was then subjected to energy minimization cycles under the MacroModel software (Schrödinger, LLC) using the OPLS-2005 (Optimized Potential for Liquid Simulations) force field and water as a solvent with constant dielectric value of 1. All the atoms in protein as well as peptide models were allowed to move freely. Energy minimization of structures were completed via the PRCG (Polak-Ribiere Conjugate Gradient) energy minimization procedure at 10,000 iterations and performed to a convergence threshold of 0.02 kJ mol−1-Å).
Microarray hybridization and data analysis.
BMDMs were plated at 8–10 × 106 cells per 55 cm2 plate, infected (MOI = 3) for 18 h with RHku80, RHku80 Δgra24, Pruku80, and Pruku80 Δgra24. RNA was isolated using TRIzol reagent (Invitrogen), followed by phenol/chloroform/isoamyl extraction. Transcripts were obtained from three biological replicates each. RNA Labeling and Array Hybridization of BMDM/RHku80 and BMDM/Pruku80 experiments were done by ArrayStar as follows: sample labeling and array hybridization were performed according to the One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology). In brief, total RNA from each sample was linearly amplified and labeled with Cy3-UTP. The labeled cRNAs were purified by the RNeasy Mini kit (QIAGEN). The concentration and specific activity of the labeled cRNAs (pmol Cy3/µg cRNA) were measured by NanoDrop ND-1000. 1 µg of each labeled cRNA was fragmented by adding 11 µl of 10× blocking agent and 2.2 µl of 25× fragmentation buffer, the mixture was heated at 60°C for 30 min, and finally, 55 µl of 2× GE Hybridization buffer was added to dilute the labeled cRNA. 100 µl of hybridization solution was dispensed into the gasket slide and assembled to the gene expression microarray slide. The Mouse 4× 44K Gene Expression Array was manufactured by Agilent Technologies. The slides were incubated for 17 h at 65°C in a hybridization oven (Agilent Technologies). The hybridized arrays were washed, fixed, and scanned using the DNA Microarray Scanner (part number G2505B; Agilent Technologies). Microarray data analysis of acquired array images was performed using the Feature Extraction software (version 10.7.3.1; Agilent Technologies). Quantile normalization and subsequent data processing were performed using the GeneSpring GX software package (version 11.5.1; Agilent Technologies). After quantile normalization of the raw data, genes in which at least six out of nine samples have flags in Detected or Not Detected (All Targets Value) were chosen for further data analysis. Differentially expressed genes with statistical significance were identified through Volcano Plot filtering between two groups (WT versus GRA24 mutant) of compared arrays. The thresholds are fold change ≥ 2.0, p-value ≤ 0.05. GO analysis and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis were performed using the DAVID Bioinformatics Resources. Microarray data has been uploaded to GEO Datasets under accession no. GSE38782.
Online supplemental material.
Fig. S1 shows a schematic representation of the strategy and analysis of GRA24 knockout. Table S1 provides a list of the vectors and the oligonucleotides used in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20130103/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Vern B. Carruthers (University of Michigan Medical School) and Dr. David J. Bzik (Dartmouth Medical School) for sharing the RHku80 and Pruku80 strains, respectively. We thank Dr. Geneviève Milon for her review and fruitful discussion on the manuscript.
This work was supported by the ANR grants ANR Blanc 2010 APImiR (ANR 2010 BLAN 1315 01), ANR Jeune Chercheur 2012 ToxoEffect (ANR-12-JSV3-0004-01), and the Laboratoire d’Excellence (LabEx) ParaFrap (ANR-11-LABX-0024), as well as by the FINOVI foundation.
The authors have no competing financial interests.
Footnotes
Abbreviations used:
- BMDM
- BM-derived macrophage
- DG
- dense granule
- HFF
- human foreskin fibroblast
- KIM
- kinase interacting motif
- MOI
- multiplicity of infection
- PV
- parasitophorous vacuole
- PVM
- PV membrane
References
- Adachi O., Kawai T., Takeda K., Matsumoto M., Tsutsui H., Sakagami M., Nakanishi K., Akira S. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 9:143–150 10.1016/S1074-7613(00)80596-8 [DOI] [PubMed] [Google Scholar]
- Ashwell J.D. 2006. The many paths to p38 mitogen-activated protein kinase activation in the immune system. Nat. Rev. Immunol. 6:532–540 10.1038/nri1865 [DOI] [PubMed] [Google Scholar]
- Blader I.J., Manger I.D., Boothroyd J.C. 2001. Microarray analysis reveals previously unknown changes in Toxoplasma gondii-infected human cells. J. Biol. Chem. 276:24223–24231 10.1074/jbc.M100951200 [DOI] [PubMed] [Google Scholar]
- Bougdour A., Maubon D., Baldacci P., Ortet P., Bastien O., Bouillon A., Barale J.C., Pelloux H., Ménard R., Hakimi M.A. 2009. Drug inhibition of HDAC3 and epigenetic control of differentiation in Apicomplexa parasites. J. Exp. Med. 206:953–966 10.1084/jem.20082826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougdour A., Durandau E., Brenier-Pinchart M.P., Ortet P., Barakat M., Kieffer S., Curt-Varesano A., Curt-Bertini R.L., Bastien O., Coute Y., et al. 2013. Host cell subversion by Toxoplasma GRA16, an exported dense granule protein that targets the host cell nucleus and alters gene expression. Cell Host Microbe. 13:489–500 10.1016/j.chom.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Brancho D., Tanaka N., Jaeschke A., Ventura J.J., Kelkar N., Tanaka Y., Kyuuma M., Takeshita T., Flavell R.A., Davis R.J. 2003. Mechanism of p38 MAP kinase activation in vivo. Genes Dev. 17:1969–1978 10.1101/gad.1107303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher B.A., Fox B.A., Rommereim L.M., Kim S.G., Maurer K.J., Yarovinsky F., Herbert D.R., Bzik D.J., Denkers E.Y. 2011. Toxoplasma gondii rhoptry kinase ROP16 activates STAT3 and STAT6 resulting in cytokine inhibition and arginase-1-dependent growth control. PLoS Pathog. 7:e1002236 10.1371/journal.ppat.1002236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.I., Xu B.E., Akella R., Cobb M.H., Goldsmith E.J. 2002. Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol. Cell. 9:1241–1249 10.1016/S1097-2765(02)00525-7 [DOI] [PubMed] [Google Scholar]
- Coulthard L.R., White D.E., Jones D.L., McDermott M.F., Burchill S.A. 2009. p38(MAPK): stress responses from molecular mechanisms to therapeutics. Trends Mol. Med. 15:369–379 10.1016/j.molmed.2009.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A., Nebreda A.R. 2010. Mechanisms and functions of p38 MAPK signalling. Biochem. J. 429:403–417 10.1042/BJ20100323 [DOI] [PubMed] [Google Scholar]
- Diskin R., Lebendiker M., Engelberg D., Livnah O. 2007. Structures of p38alpha active mutants reveal conformational changes in L16 loop that induce autophosphorylation and activation. J. Mol. Biol. 365:66–76 10.1016/j.jmb.2006.08.043 [DOI] [PubMed] [Google Scholar]
- Dunay I.R., Damatta R.A., Fux B., Presti R., Greco S., Colonna M., Sibley L.D. 2008. Gr1(+) inflammatory monocytes are required for mucosal resistance to the pathogen Toxoplasma gondii. Immunity. 29:306–317 10.1016/j.immuni.2008.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreiro I., Joaquin M., Islam A., Gomez-Lopez G., Barragan M., Lombardía L., Domínguez O., Pisano D.G., Lopez-Bigas N., Nebreda A.R., Posas F. 2010. Whole genome analysis of p38 SAPK-mediated gene expression upon stress. BMC Genomics. 11:144 10.1186/1471-2164-11-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox B.A., Falla A., Rommereim L.M., Tomita T., Gigley J.P., Mercier C., Cesbron-Delauw M.F., Weiss L.M., Bzik D.J. 2011. Type II Toxoplasma gondii KU80 knockout strains enable functional analysis of genes required for cyst development and latent infection. Eukaryot. Cell. 10:1193–1206 10.1128/EC.00297-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D.M., Różycki B., Koveal D., Hummer G., Page R., Peti W. 2011. Structural basis of p38α regulation by hematopoietic tyrosine phosphatase. Nat. Chem. Biol. 7:916–924 10.1038/nchembio.707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garai Á., Zeke A., Gógl G., Törő I., Fördős F., Blankenburg H., Bárkai T., Varga J., Alexa A., Emig D., et al. 2012. Specificity of linear motifs that bind to a common mitogen-activated protein kinase docking groove. Sci. Signal. 5:ra74 10.1126/scisignal.2003004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzinelli R.T., Wysocka M., Hayashi S., Denkers E.Y., Hieny S., Caspar P., Trinchieri G., Sher A. 1994. Parasite-induced IL-12 stimulates early IFN-gamma synthesis and resistance during acute infection with Toxoplasma gondii. J. Immunol. 153:2533–2543 [PubMed] [Google Scholar]
- Ge B., Gram H., Di Padova F., Huang B., New L., Ulevitch R.J., Luo Y., Han J. 2002. MAPKK-independent activation of p38alpha mediated by TAB1-dependent autophosphorylation of p38alpha. Science. 295:1291–1294 10.1126/science.1067289 [DOI] [PubMed] [Google Scholar]
- Goldszmid R.S., Caspar P., Rivollier A., White S., Dzutsev A., Hieny S., Kelsall B., Trinchieri G., Sher A. 2012. NK cell-derived interferon-γ orchestrates cellular dynamics and the differentiation of monocytes into dendritic cells at the site of infection. Immunity. 36:1047–1059 10.1016/j.immuni.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gum R.J., McLaughlin M.M., Kumar S., Wang Z., Bower M.J., Lee J.C., Adams J.L., Livi G.P., Goldsmith E.J., Young P.R. 1998. Acquisition of sensitivity of stress-activated protein kinases to the p38 inhibitor, SB 203580, by alteration of one or more amino acids within the ATP binding pocket. J. Biol. Chem. 273:15605–15610 10.1074/jbc.273.25.15605 [DOI] [PubMed] [Google Scholar]
- Harris T.H., Banigan E.J., Christian D.A., Konradt C., Tait Wojno E.D., Norose K., Wilson E.H., John B., Weninger W., Luster A.D., et al. 2012. Generalized Lévy walks and the role of chemokines in migration of effector CD8+ T cells. Nature. 486:545–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J.C., Hunn J.P., Steinfeldt T. 2011. The IRG protein-based resistance mechanism in mice and its relation to virulence in Toxoplasma gondii. Curr. Opin. Microbiol. 14:414–421 10.1016/j.mib.2011.07.002 [DOI] [PubMed] [Google Scholar]
- Hunter C.A., Sibley L.D. 2012. Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nat. Rev. Microbiol. 10:766–778 10.1038/nrmicro2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh M.H., Carruthers V.B. 2009. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot. Cell. 8:530–539 10.1128/EC.00358-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K.D., Wang Y., Wojno E.D., Shastri A.J., Hu K., Cornel L., Boedec E., Ong Y.C., Chien Y.H., Hunter C.A., et al. 2011. Toxoplasma polymorphic effectors determine macrophage polarization and intestinal inflammation. Cell Host Microbe. 9:472–483 10.1016/j.chom.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen M., Delghandi M.P., Moens U. 2004. What turns CREB on? Cell. Signal. 16:1211–1227 10.1016/j.cellsig.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Khan I.A., MacLean J.A., Lee F.S., Casciotti L., DeHaan E., Schwartzman J.D., Luster A.D. 2000. IP-10 is critical for effector T cell trafficking and host survival in Toxoplasma gondii infection. Immunity. 12:483–494 10.1016/S1074-7613(00)80200-9 [DOI] [PubMed] [Google Scholar]
- Kim L., Del Rio L., Butcher B.A., Mogensen T.H., Paludan S.R., Flavell R.A., Denkers E.Y. 2005. p38 MAPK autophosphorylation drives macrophage IL-12 production during intracellular infection. J. Immunol. 174:4178–4184 [DOI] [PubMed] [Google Scholar]
- Krachler A.M., Woolery A.R., Orth K. 2011. Manipulation of kinase signaling by bacterial pathogens. J. Cell Biol. 195:1083–1092 10.1083/jcb.201107132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna M., Narang H. 2008. The complexity of mitogen-activated protein kinases (MAPKs) made simple. Cell. Mol. Life Sci. 65:3525–3544 10.1007/s00018-008-8170-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Kang Y.J., Han J., Herschman H.R., Stefani E., Wang Y. 2006. TAB-1 modulates intracellular localization of p38 MAP kinase and downstream signaling. J. Biol. Chem. 281:6087–6095 10.1074/jbc.M507610200 [DOI] [PubMed] [Google Scholar]
- Martin A.M., Liu T., Lynn B.C., Sinai A.P. 2007. The Toxoplasma gondii parasitophorous vacuole membrane: transactions across the border. J. Eukaryot. Microbiol. 54:25–28 10.1111/j.1550-7408.2006.00230.x [DOI] [PubMed] [Google Scholar]
- Masek K.S., Fiore J., Leitges M., Yan S.F., Freedman B.D., Hunter C.A. 2006. Host cell Ca2+ and protein kinase C regulate innate recognition of Toxoplasma gondii. J. Cell Sci. 119:4565–4573 10.1242/jcs.03206 [DOI] [PubMed] [Google Scholar]
- Mashayekhi M., Sandau M.M., Dunay I.R., Frickel E.M., Khan A., Goldszmid R.S., Sher A., Ploegh H.L., Murphy T.L., Sibley L.D., Murphy K.M. 2011. CD8α(+) dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity. 35:249–259 10.1016/j.immuni.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo M.B., Jensen K.D., Saeij J.P. 2011. Toxoplasma gondii effectors are master regulators of the inflammatory response. Trends Parasitol. 27:487–495 10.1016/j.pt.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S., Melo M.B., Li F., Lu D., Niedelman W., Levine S.S., Saeij J.P. 2012. Admixture and recombination among Toxoplasma gondii lineages explain global genome diversity. Proc. Natl. Acad. Sci. USA. 109:13458–13463 10.1073/pnas.1117047109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M.M., Jones A.R., Carmen J.C., Sinai A.P., Burchmore R., Wastling J.M. 2008. Modulation of the host cell proteome by the intracellular apicomplexan parasite Toxoplasma gondii. Infect. Immun. 76:828–844 10.1128/IAI.01115-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell A., Odrowaz Z., Sharrocks A.D. 2012. Immediate-early gene activation by the MAPK pathways: what do and don’t we know? Biochem. Soc. Trans. 40:58–66 10.1042/BST20110636 [DOI] [PubMed] [Google Scholar]
- Phelps E.D., Sweeney K.R., Blader I.J. 2008. Toxoplasma gondii rhoptry discharge correlates with activation of the early growth response 2 host cell transcription factor. Infect. Immun. 76:4703–4712 10.1128/IAI.01447-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plattner F., Soldati-Favre D. 2008. Hijacking of host cellular functions by the Apicomplexa. Annu. Rev. Microbiol. 62:471–487 10.1146/annurev.micro.62.081307.162802 [DOI] [PubMed] [Google Scholar]
- Reid A.J., Vermont S.J., Cotton J.A., Harris D., Hill-Cawthorne G.A., Könen-Waisman S., Latham S.M., Mourier T., Norton R., Quail M.A., et al. 2012. Comparative genomics of the apicomplexan parasites Toxoplasma gondii and Neospora caninum: Coccidia differing in host range and transmission strategy. PLoS Pathog. 8:e1002567 10.1371/journal.ppat.1002567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reményi A., Good M.C., Bhattacharyya R.P., Lim W.A. 2005. The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network. Mol. Cell. 20:951–962 10.1016/j.molcel.2005.10.030 [DOI] [PubMed] [Google Scholar]
- Robben P.M., LaRegina M., Kuziel W.A., Sibley L.D. 2005. Recruitment of Gr-1+ monocytes is essential for control of acute toxoplasmosis. J. Exp. Med. 201:1761–1769 10.1084/jem.20050054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosowski E.E., Lu D., Julien L., Rodda L., Gaiser R.A., Jensen K.D., Saeij J.P. 2011. Strain-specific activation of the NF-κB pathway by GRA15, a novel Toxoplasma gondii dense granule protein. J. Exp. Med. 208:195–212 10.1084/jem.20100717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeij J.P., Coller S., Boyle J.P., Jerome M.E., White M.W., Boothroyd J.C. 2007. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature. 445:324–327 10.1038/nature05395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador J.M., Mittelstadt P.R., Guszczynski T., Copeland T.D., Yamaguchi H., Appella E., Fornace A.J., Jr, Ashwell J.D. 2005. Alternative p38 activation pathway mediated by T cell receptor-proximal tyrosine kinases. Nat. Immunol. 6:390–395 10.1038/ni1177 [DOI] [PubMed] [Google Scholar]
- Scharton-Kersten T.M., Wynn T.A., Denkers E.Y., Bala S., Grunvald E., Hieny S., Gazzinelli R.T., Sher A. 1996. In the absence of endogenous IFN-γ, mice develop unimpaired IL-12 responses to Toxoplasma gondii while failing to control acute infection. J. Immunol. 157:4045–4054 [PubMed] [Google Scholar]
- Sher A., Collazzo C., Scanga C., Jankovic D., Yap G., Aliberti J. 2003. Induction and regulation of IL-12-dependent host resistance to Toxoplasma gondii. Immunol. Res. 27:521–528 10.1385/IR:27:2-3:521 [DOI] [PubMed] [Google Scholar]
- Silverman E.S., Du J., Williams A.J., Wadgaonkar R., Drazen J.M., Collins T. 1998. cAMP-response-element-binding-protein-binding protein (CBP) and p300 are transcriptional co-activators of early growth response factor-1 (Egr-1). Biochem. J. 336:183–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tur G., Georgieva E.I., Gagete A., López-Rodas G., Rodríguez J.L., Franco L. 2010. Factor binding and chromatin modification in the promoter of murine Egr1 gene upon induction. Cell. Mol. Life Sci. 67:4065–4077 10.1007/s00018-010-0426-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley M., Teygong C., Phelps E., Radke J., Blader I.J. 2011. Serum response factor regulates immediate early host gene expression in Toxoplasma gondii-infected host cells. PLoS ONE. 6:e18335 10.1371/journal.pone.0018335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Standley D.M., Takashima S., Saiga H., Okuyama M., Kayama H., Kubo E., Ito H., Takaura M., Matsuda T., et al. 2009. A single polymorphic amino acid on Toxoplasma gondii kinase ROP16 determines the direct and strain-specific activation of Stat3. J. Exp. Med. 206:2747–2760 10.1084/jem.20091703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap G.S., Shaw M.H., Ling Y., Sher A. 2006. Genetic analysis of host resistance to intracellular pathogens: lessons from studies of Toxoplasma gondii infection. Microbes Infect. 8:1174–1178 10.1016/j.micinf.2005.10.031 [DOI] [PubMed] [Google Scholar]
- Zhang Y.Y., Wu J.W., Wang Z.X. 2011. A distinct interaction mode revealed by the crystal structure of the kinase p38α with the MAPK binding domain of the phosphatase MKP5. Sci. Signal. 4:ra88 10.1126/scisignal.2002241 [DOI] [PubMed] [Google Scholar]
- Zhou T., Sun L., Humphreys J., Goldsmith E.J. 2006. Docking interactions induce exposure of activation loop in the MAP kinase ERK2. Structure. 14:1011–1019 10.1016/j.str.2006.04.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.