Abstract
Medulloblastoma (MB) is one of the most common pediatric cancers, likely originating from abnormal development of cerebellar progenitor neurons. MicroRNA (miRNA) has been shown to play an important role in the development of the central nervous system. Microarray analysis was used to investigate miRNA expression in desmoplastic MB from patients diagnosed at a young age (1 or 2 years old). Normal fetal or newborn cerebellum was used as control. A total of 84 differentially expressed miRNAs (64 downregulated and 20 upregulated) were found. Most downregulated miRNAs (32/64) were found to belong to the cluster of miRNAs at the 14q32 locus, suggesting that this miRNA locus is regulated as a module in MB. Possible mechanisms of 14q32 miRNAs downregulation were investigated by the analysis of publicly available gene expression data sets. First, expression of estrogen-related receptor-γ (ESRRG), a reported positive transcriptional regulator of some 14q32 miRNAs, was found downregulated in desmoplastic MB. Second, expression of the parentally imprinted gene MEG3 was lower in MB in comparison to normal cerebellum, suggesting a possible epigenetic silencing of the 14q32 locus. miR-129-5p (11p11.2/7q32.1), miR-206 (6p12.2), and miR-323-3p (14q32.2), were chosen for functional studies in DAOY cells. Overexpression of miR-129-5p using mimics decreased DAOY proliferation. No effect was found with miR-206 or miR-323 mimics.
Keywords: 14q32 miRNA cluster, desmoplastic medulloblastoma, ESRRG, miR-129-5p, miRNA profile
Introduction
Medulloblastoma (MB) is an embryonic tumor of the cerebellum and the most common malignant brain tumor in childhood, likely originating from abnormal development of cerebellar progenitor neurons (1, 2). Transcriptional profiling of large number of MB samples unraveled the existence of at least four distinct molecular subgroups: (i) Wingless (Wnt) group, (ii) Sonic Hedgehog (SHH) group, (iii) Group 3 and (iv) Group 4 (3, 4). These various subtypes of MB are suggested to arise from different populations of precursor or stem cells which form the cerebellum (5–8). This transcriptome-based classification has opened new avenues for the understanding of the molecular mechanism contributing to MB.
MicroRNAs (miRNAs) are suggested to play an important role in controlling the development of the central nervous system (CNS) by regulating cell proliferation and differentiation, as well as apoptosis (9). miRNAs are small non-coding RNA molecules of ∼22–25 nucleotides that post-transcriptionally downregulate gene expression by binding the 3′-untranslated region (UTR) of protein coding transcripts, resulting in either mRNA cleavage or translational repression (10, 11). miRNA expression profiling of both mouse and human MB has led to the identification of signatures associated with the molecular subgroups of MB, tumor diagnosis, and response to treatment, as well as novel targets of potential clinical relevance (12–15). Previous studies, however, interrogated limited number of miRNAs and included adult cerebellum in the normal control group. We investigated the expression profile of 847 miRNA in primary human desmoplastic MB of younger children in comparison to normal fetuses or newborn cerebellum. Eighty-four miRNAs were found to be differential expressed in MB, most of them belonging to the cluster 14q32. Possible mechanisms of 14q32 locus downregulation were investigated by the analysis of publicly available gene expression data set. Functional studies using mimic miR-129-5p (11p11.2/7q32.1), miR-206 (6p12.2), and miR-323-3p (14q32.2) and the DAOY cell line, suggested a suppressive role for miR-129-5p in MB proliferation.
Materials and Methods
Primary medulloblastoma tissue samples
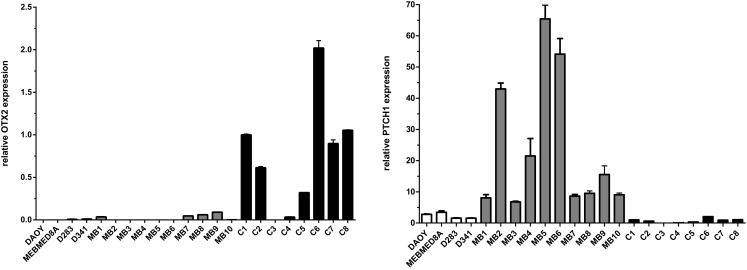
Surgical specimens were obtained from 1 to 5 years old children (n = 10), with desmoplastic MB (Table 1). Of note, microarray analyses were performed with MB samples from children with 1–2 years old. Desmoplastic MBs belong, with rare exceptions, to the SHH molecular subgroup (13–16). All MB samples used in the present study had high mRNA levels of PTCH1 and low levels of OTX2 (Figure A1 in Appendix), in comparison to normal cerebellum, which is in keeping with the differential transcriptional profile of SHH tumors (3). Normal cerebellum tissues were obtained from 22 to 39 weeks old fetal and newborn (NW) autopsy (n = 8) (Table 2). Ethical approvals were obtained from the Ethical Research Committee of the Faculdade de Ciências Médicas (n°656/2009), CAISM (n°064/2010), the Ethical Research Committee of Centro Infantil Boldrini (n°1.90-030710), and National Committee of Ethics in Research (CONEP) n°0005.0.144.146-09. Subtyping of MB was obtained by histological analysis.
Table 1.
Summary of the medulloblastoma samples included in the study.
| Medulloblastoma | Age at diagnosis | Gender | Histology |
|---|---|---|---|
| MB 1a | 1 | M | N/D |
| MB 2a | 1 | F | N/D |
| MB 3a | 2 | M | D |
| MB 4a | 2 | M | N/D |
| MB 5a | 1 | M | N/D |
| MB 6a | 2 | M | N/D |
| MB 7 | 5 | M | N/D |
| MB 8 | 5 | M | D |
| MB 9 | 4 | M | D |
| MB 10 | 5 | M | N/D |
aSamples used in Affymetrix miRNA microarray analysis; F = female; M = male; D = desmoplastic; N/D = nodular/desmoplastic.
Table 2.
Summary of the normal cerebellum tissues.
| Normal cerebellum | Gestational age | Gender | Diagnosis |
|---|---|---|---|
| C1 | 37 | M | Bilateral renal agenesis |
| C2a | 39 | M | Hydropsy |
| C3a | 22 | – | NM |
| C4a | 31 | M | NM |
| C5a | 36 | – | NM |
| C6a | 24 | M | NM |
| C7 | 30 | – | Cardiopathy |
| C8a | 26 | M | NM |
aSamples used in Affymetrix miRNA microarray analysis; F = female; M = male; NM = no malformation and no aneuploidy.
Total RNA isolation and analysis of global miRNA expression
Total RNA was extracted by Trizol™ (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions, with an additional overnight precipitation step at −20°C with isopropanol (Merck). RNA quantification was carried out in a Qubit Quantitation Platform (Invitrogen) and RNA quality was analyzed via gel electrophoresis. Five hundred nanograms of RNA from 12 samples (6 MB and 6 fetal cerebellum) were labeled with the 3′-DNA FlashTag Biotin HSR kit (Genisphere, Hatfield, PA, USA) and hybridized to GeneChip miRNA Array 1.0 (Affymetrix Inc., Santa Clara, CA, USA), which comprises 847 human miRNAs. Data was acquired using a GeneChip Scanner 3000 7G (Affymetrix).
Validation of miRNA deregulation by quantitative real-time PCR
Reverse transcription (RT) and quantitative real-time RT-PCR (RT-qPCR) analysis were carried out using commercially available TaqMan microRNA assays (Applied Biosystems, Foster City, CA, USA) and a 7500 Real-time PCR System (Applied Biosystems). RT reactions (50 ng of total RNA) were performed in a 15 μl final volume containing specific stem-loop primers for each miRNA (129-5p, 206, 323-3p, 495, and internal control small RNA, RNU6B), 10× RT Buffer, dNTPs, reverse transcriptase, RNase inhibitor, and water in 96-well plates. Thermal cycling included 30 min at 16°C, 30 min at 42°C, and a final step of RT inactivation for 5 min at 85°C. PCR reactions were performed in a 10 μl final volume containing 5 μl TaqMan Universal Master Mix II, without UNG (Applied Biosystems), 3.5 μl water, 0.5 μl TaqMan microRNA assay, and 1 μl cDNA. Thermal cycling included an initial step of 10 min at 95°C for Taq activation followed by 40 cycles of 15 s denaturation at 95°C and 1 min of annealing/extension at 60°C. Each reaction was performed in triplicate and the miRNAs expression levels were normalized against RNU6B. The threshold cycle numbers (Ct) were calculated by relative quantification using the 2−ΔΔCt method, as described by Livak and Schmittgen (17). One of the control samples was chosen as calibrator.
Cell lines
Four human MB cell lines were utilized: DAOY (HTB 186), D283 Med (HTB185), and D431 Med (HTB-187) were obtained from American Type Culture Collection (ATCC). The MB cell line, MEB-Med-8A, was kindly provided by Prof. T. Pietsch (18). The MB cell lines DAOY, D283 Med, and MEB-Med-8A were maintained in High Glucose Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 1 mM sodium pyruvate (PAA), l-glutamine, 1% penicillin/streptomycin (Invitrogen, Karlsruhe, Germany), and 10% fetal bovine serum (FBS, Invitrogen). The MB cell line D341 Med was maintained in DMEM with l-glutamine supplemented with 1 mM sodium pyruvate, 1% penicillin/streptomycin, and 10% Human Serum (HS, PAA, UK).
Transient transfection of miRNAs
DAOY cells (1.5 × 105) were seeded in six-well plates in 2 ml of RPMI-1640 medium (Cultilab, Campinas, Brazil) supplemented with 10% FBS (Sigma-Aldrich, St. Louis, MO, USA) and penicillin/streptavidin (Cultilab). Transfection of miRVana miRNA mimics (Invitrogen Ambion, Austin, TX, USA) of miR-206, miR-129-5p, miR-323-3p, or miRVana miRNA mimic negative control #1 (referred to as scrambled) was carried out 24 h after seeding, in a final concentration of 3 nM, using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer’s recommendation. Efficiency of transfection was evaluated 24 post-transfection by RT-qPCR using total RNA.
Cell viability: MTS assay
Cell survival/proliferation after the transfection with mimic-miRNAs was evaluated by using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Wallisellen, Switzerland), a colorimetric [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)]-2H-tetrazolium inner salt (MTS) assay. Briefly, mimic-miR-206, mimic-miR-129-5p, and mimic-miR-323-3p or mimic-negative control #1 transfected cells were harvested 20 h after transfection and seeded in triplicate in 96-well plate (1,500 cells/well) in serum-free RPMI-1640 (Cultilab). At 24, 48, or 72 h post-transfection (i.e., 4, 28, or 52 h after passage to the 96-well plate) cells were incubated for 1 h with MTS reagent and absorbance read at 492 nm (reference wavelength 620 nm) using an ASYS Expert Plus Microplate Reader (Biochrom, Holliston, MA, USA). Three independent experiments were performed.
Apoptosis assay
DAOY cells transfected with miR-206, 129-5p, 323-3p, or scramble mimics were cultured for 24 h in serum-free RPMI-1640 (Cultilab), harvested and part of it was resuspended in the appropriate binding buffer, stained with FITC-conjugated Annexin V (BD Biosciences, San Jose, CA, USA) and propidium iodide at room temperature for 15 min, and subsequently analyzed by flow cytometry in a FACS Canto II (Becton Dickinson). The remaining cells were replated in six-well plates for another 24 h culture period in serum-free RPMI-1640 (Cultilab) and harvested 48 h post-transfection for Annexin V labeling.
Statistical analysis and bioinformatics methods to signaling pathway prediction
MicroRNA expression was analyzed in R environment1 using the packages Affy and RankProd from Bioconductor (19– 21). The MB miRNA profile was compared to the cerebellum profile. Differentially expressed miRNAs were selected according to the fold change ≥2.00 and p-value ≤0.05. Heat maps were created using tools of the MetaboAnalyst 2.02. Signaling pathways were prospected by DIANA-miRPath (microT-v4.0, beta version)3. The input dataset enrichment analysis was performed by Pearson’s chi-squared test and each pathway was represented by the negative natural logarithm of the P-value (−ln P). The Ingenuity Pathway Analysis (IPA) software4 was used to identify possible pathways associated to differentially expressed miRNAs.
Comparisons of RT-qPCR values from MB versus normal cerebellum were performed by the Mann–Whitney test. Cell proliferation results, at each time point, from mimic miRNA transfections versus mimic-negative control #1 were analyzed by the two-tailed unpaired t-test. Alpha error of P = 0.05 was tolerated. The GraphPad Prism 5 software was used throughout.
Results
Identification of differentially expressed miRNAs in desmoplastic MBs of 1–2 years old children
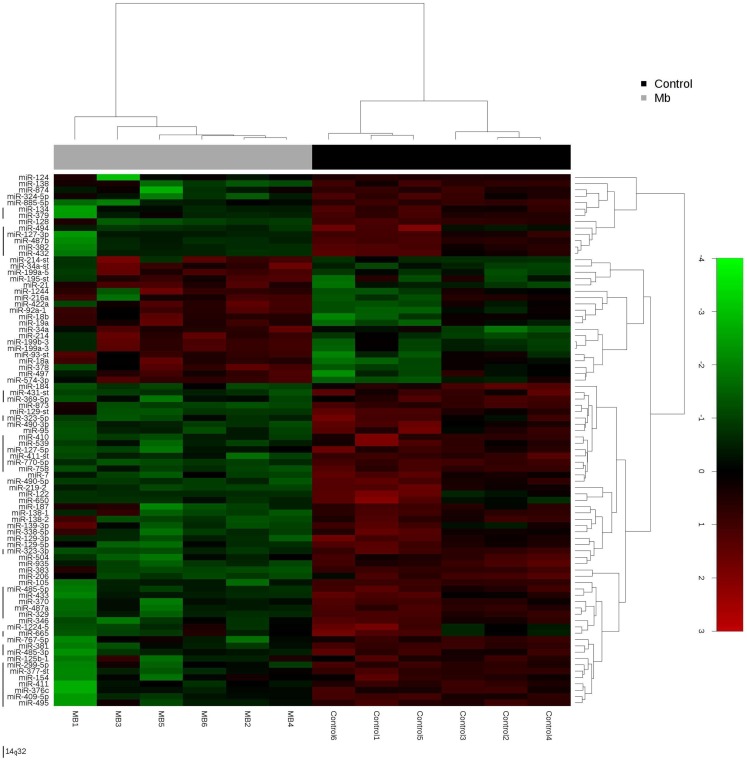
Global miRNA profiles were generated for primary MB of the desmoplastic subtype and most likely SHH molecular subgroup (n = 6), and normal fetal/NW cerebellum (n = 6). Eighty-four miRNAs (64 miRNAs downregulated and 20 miRNAs upregulated) were considered to be differentially expressed (fold change ≥2.0, p ≤ 0.05) in MB in comparison to normal fetal/NW cerebellum (Table 3; Figure 1). Among these 84 miRNAs, 46 had been previously described as deregulated in human primary MB (Table 3), and only 8 were previously validated by functional assays (Table 4). Upregulation of miRNAs from the miR-17 ∼ 92 cluster (in this work miR-18a, 19a, and 92a-1) and downregulation of miR-324-5p were previously described in human MB of the SHH subgroup (12, 13). Of especial note, 32 of the 64 downregulated miRNAs belong to a large cluster on human chromosome 14q32 (Figure 1; Table 3).
Table 3.
Deregulated miRNA in desmoplastic medulloblastoma compared to normal cerebellum.
| Our miRNA profile (84) | Chromosomal localization | Fold change | Reference |
|---|---|---|---|
| DOWNREGULATED | |||
| hsa-miR-206 | 6p12.2 | −7.53 | (29) |
| hsa-miR-219-2-3p | 9q33.3 | −6.64 | (52) |
| hsa-miR-383 | 8p22 | −6.56 | (12, 55, 56) |
| hsa-miR-138 | 16q13.3/3p21.32 | −5.16 | (12, 14) |
| hsa-miR-323-3p | 14q32.2 | −4.96 | (12, 52) |
| hsa-miR-122 | 18q21.31 | −4.82 | |
| hsa-miR-105 | Xq28 | −4.66 | |
| hsa-miR-129-5p | 11p11.2/7q32.1 | −4.56 | (23) |
| hsa-miR-935 | 19q13.43 | −4.53 | (52) |
| hsa-miR-329 | 14q32.2 | −4.48 | |
| hsa-miR-129-3p | 11p11.2/7q32.1 | −4.43 | |
| hsa-miR-650 | 22q11.21 | −4.19 | |
| hsa-miR-184 | 15q24.3 | −4.14 | |
| hsa-miR-370 | 14q32.2 | −3.99 | (12) |
| hsa-miR-433 | 14q32.2 | −3.96 | (29) |
| hsa-miR-138-2* | 16q13.3/3p21.32 | −3.91 | |
| hsa-miR-487b | 14q32.2 | −3.82 | (29) |
| hsa-miR-487a | 14q32.2 | −3.78 | |
| hsa-miR-758 | 14q32.2 | −3.65 | |
| hsa-miR-485-5p | 14q32.2 | −3.60 | |
| hsa-miR-138-1* | 16q13.3/3p21.32 | −3.55 | |
| hsa-miR-382 | 14q32.2 | −3.53 | (12, 29) |
| hsa-miR-504 | Xq26.3 | −3.45 | (52) |
| hsa-miR-128 | 2q21.3/3p22.3 | −3.43 | (12, 14, 51, 59) |
| hsa-miR-490-5p | 7q33 | −3.42 | |
| hsa-miR-770-5p | 14q32.2 | −3.35 | |
| hsa-miR-410 | 14q32.2 | −3.30 | (29) |
| hsa-miR-432 | 14q32.2 | −3.29 | |
| hsa-miR-485-3p | 14q32.2 | −3.02 | |
| hsa-miR-490-3p | 7q33 | −2.88 | |
| hsa-miR-381 | 14q32.2 | −2.73 | (12) |
| hsa-miR-377* | 14q32.2 | −2.72 | |
| hsa-miR-7 | 15q25.3/19p13.3/9q21.32 | −2.72 | (12, 14) |
| hsa-miR-124 | 20p23.1/8q12.3/8p23.1 | −2.71 | (12, 14, 29, 48, 49) |
| hsa-miR-323-5p | 14q32.31 | −2.69 | (12) |
| hsa-miR-873 | 9p21.1 | −2.65 | |
| hsa-miR-129* | 11p11.2/7q32.1 | −2.63 | |
| hsa-miR-338-5p | 17q25.3 | −2.61 | (14) |
| hsa-miR-409-5p | 14q32.2 | −2.61 | |
| hsa-miR-874 | 5q31.2 | −2.46 | |
| hsa-miR-495 | 14q32.2 | −2.46 | (52) |
| hsa-miR-885-5p | 3p25.3 | −2.45 | |
| hsa-miR-376c | 14q32.2 | −2.43 | (52) |
| hsa-miR-299-5p | 14q32.2 | −2.41 | |
| hsa-miR-539 | 14q32.2 | −2.40 | (52) |
| hsa-miR-127-5p | 14q32.2 | −2.35 | (12, 29) |
| hsa-miR-127-3p | 14q32.2 | −2.35 | (52, 59) |
| hsa-miR-411* | 14q32.2 | −2.30 | |
| hsa-miR-125b-1* | 11q24.1/21q21.1 | −2.27 | |
| hsa-miR-411 | 14q32.2 | −2.23 | (29) |
| hsa-miR-379 | 14q32.2 | −2.22 | (29, 52) |
| hsa-miR-431* | 14q32.2 | −2.22 | |
| hsa-miR-767-5p | Xq28 | −2.20 | |
| hsa-miR-139-3p | 11q13.4 | −2.17 | |
| hsa-miR-154 | 14q32.2 | −2.16 | (12) |
| hsa-miR-1224-5p | 3q27.2 | −2.15 | |
| hsa-miR-187 | 18q12.1 | −2.14 | (12) |
| hsa-miR-95 | 4p16.1 | −2.10 | (14) |
| hsa-miR-369-5p | 14q32.2 | −2.05 | |
| hsa-miR-665 | 14q32.2 | −2.05 | |
| hsa-miR-494 | 14q32.2 | −2.03 | (52) |
| hsa-miR-134 | 14q32.2 | −2.03 | (12, 29) |
| hsa-miR-346 | 10q23.2 | −2.01 | (12, 13) |
| hsa-miR-324-5p | 17p13.1 | −2.00 | (12, 50) |
| UPREGULATED | |||
| hsa-miR-199b-3p | 9q33.3 | 4.56 | (12) |
| hsa-miR-199a-3p | 19p13.2/1q24.1 | 4.49 | |
| hsa-miR-199a-5p | 19p13.2/1q24.1 | 4.14 | (28) |
| hsa-miR-21 | 17q22 | 3.70 | (12–14, 29, 31) |
| hsa-miR-214 | 1q24.2 | 3.59 | |
| hsa-miR-19a | 13q31.3 | 3.11 | (12–14) |
| hsa-miR-92a-1* | 13q31.3/Xq26.2 | 3.06 | |
| hsa-miR-214* | 1q24.2 | 2.93 | |
| hsa-miR-34a | 1p36.23 | 2.78 | (13, 30, 53) |
| hsa-miR-18b | Xq26.2 | 2.74 | |
| hsa-miR-422a | 15q22.2 | 2.72 | (14) |
| hsa-miR-34a* | 1p36.23 | 2.58 | (14) |
| hsa-miR-574-3p | 4p14 | 2.49 | (14) |
| hsa-miR-378 | 5q32 | 2.39 | (14) |
| hsa-miR-1244 | 12p13.2/12p13.31/2q37.1/5q23.1 | 2.39 | |
| hsa-miR-18a | 13q31.3 | 2.39 | (12–14) |
| hsa-miR-93* | 7q22.1 | 2.26 | |
| hsa-miR-497 | 17p13.1 | 2.17 | (13) |
| hsa-miR-195* | 17p13.1 | 2.14 | |
| hsa-miR-216a | 2p16.1 | 2.07 | (14) |
Figure 1.
Hierarchical clustering analysis of medulloblastoma and normal cerebellum. Unsupervised hierarchical cluster analysis representing the 84 miRNAs expressed in medulloblastoma samples versus normal cerebellum. 14q32 miRNAs are depicted by a left vertical line. -st, star.
Table 4.
miRNAs validated by functional assay in human and mouse medulloblastoma.
| miRNA | Deregulation | Cells | Functional assaya | Target genes | Reference |
|---|---|---|---|---|---|
| 124 | Down | Primary human MB, cell lines | ↑Cell cycle progression at G1 | CDK6 | (48, 49) |
| ↓Cell proliferation | SLC16A1 | ||||
| 324-5p | Down | Primary human and mouse MB, cell lines | ↓Cell proliferation | SMO | (50) |
| 326 | GLI1 | ||||
| 125p | |||||
| 9 | Down | Primary human MB, cell lines | ↑Apoptosis | Trkc | (12) |
| 125a | ↓Cell proliferation | ||||
| 199b-5p | Up | Primary human and mouse MB, cell lines | ↑Cell cycle progression at G1 | HES1 | (28) |
| ↓Cell proliferation | |||||
| 128 | Down | Primary human and mouse MB | ↓Cell proliferation | BMI-1 | (51) |
| ↑Cell senescence | |||||
| 21 | Up | Primary human MB, cell lines | ↓Cell migration | PDCD4 | (31) |
| 935 | Down | Primary human MB, cell lines | – | KIAA0232 | (52 ) |
| SLC5A3 | |||||
| TBC1D9 | |||||
| ZFAND6 | |||||
| 34a | Down | MB cell lines | ↑Apoptosis | MAGE-A | (30, 53) |
| ↑Cell cycle progression at S/phase and G2/M | Dll1 | ||||
| ↓Cell proliferation | Notch1 | ||||
| ↑Cell senescence | Notch2 | ||||
| 512-5p | Down | Primary human MB, cell lines | – | MYCC | (54 ) |
| 383 | Down | Primary human MB, cell lines | ↑Apoptosis | PRDX3 | (55, 56) |
| ↑Cell cycle progression at G1 | |||||
| ↓Cell proliferation | |||||
| 183 ∼96 ∼ | Down | MB cell lines | ↑Cell cycle progression at | AKT | (57) |
| 182 | G0/G1 and G2 | ||||
| ↓Cell migration | |||||
| ↓Cell proliferation | |||||
| 218 | Down | MB cell lines | ↓Cell migration | CDK6 | (58) |
| ↓Cell proliferation | REST |
Underlined miRNAs are miRNAs also found deregulated in the present study;
aresults of ectopic expression or knockdown assays; MB = medulloblastoma.
Signaling pathways analysis by DIANA
Signaling pathways putatively altered by MB deregulated miRNA were depicted by DIANA-miRPath. The list of the top 20 pathways is shown in Table 5. The Ribosome pathway was only pointed by the list of downregulated miRNAs. Adherens junction, oxidative phosphorylation, and TGF-beta signaling pathways showed higher enrichment when the list of downregulated miRNAs was used in the analysis. On the other hand, the MAPK pathway and genes associated to cancer showed higher enrichment when upregulated miRNAs were used in the analysis.
Table 5.
Top 20 pathways predicted by DIANA-miRPath analysis.
| Pathway signaling | All deregulated miRNAs | Downregulated miRNAs | Upregulated miRNAs | 14q32 miRNAs |
|---|---|---|---|---|
| P-valuea | ||||
| Ribosome | 30.03 | 25.34 | – | 17.78 |
| Axon guidance | 24.96 | 19.98 | 17.95 | 17.7 |
| Wnt signaling pathway | 18.6 | 19.34 | 17.71 | 16.99 |
| Focal adhesion | 16.65 | 17.02 | 15.13 | 14.73 |
| Adherens junction | 16.23 | 20.37 | 12.18 | 16.6 |
| Oxidative phosphorylation | 14.95 | 14.76 | 7.34 | 14.45 |
| ErbB signaling pathway | 14.8 | 11.03 | 9.14 | 9.02 |
| Metabolism of xenobiotics by cytochrome P450 | 14.06 | 15.33 | 3.15 | 10.01 |
| Renal cell carcinoma | 13.48 | 13.21 | 9.35 | 11.64 |
| TGF-beta signaling pathway | 12.4 | 14.11 | 5.64 | 17.56 |
| Regulation of actin cytoskeleton | 12.25 | 11.52 | 7.86 | 10.63 |
| Chronic myeloid leukemia | 12.06 | 11.02 | 8.14 | 10.54 |
| MAPK signaling pathway | 11.86 | 9.81 | 17.06 | 12.4 |
| Colorectal cancer | 11.72 | 13.24 | 9.79 | 16.92 |
| Glioma | 10 | 8.47 | 16.21 | 7.23 |
| Pancreatic cancer | 9.69 | 9.61 | 13.18 | 5.83 |
| Melanogenesis | 9.4 | 9.32 | 9.07 | 10.59 |
| Ubiquitin mediated proteolysis | 9.28 | 10.09 | 6.63 | 11.11 |
| Prostate cancer | 9.1 | 7 | 18.2 | 6.06 |
| Insulin signaling pathway | 9 | 8.54 | 5.86 | 4.65 |
aThe negative natural logarithm of the enrichment P-value calculated for the specific pathway. Underlined shows higher enrichment in downregulated, upregulated, or 14q32 miRNAs lists.
Interestingly, oxidative phosphorylation, TGF-beta signaling pathway, and ubiquitin mediated proteolysis were enriched in the list of 14q32 miRNAs.
Ingenuity pathway analysis
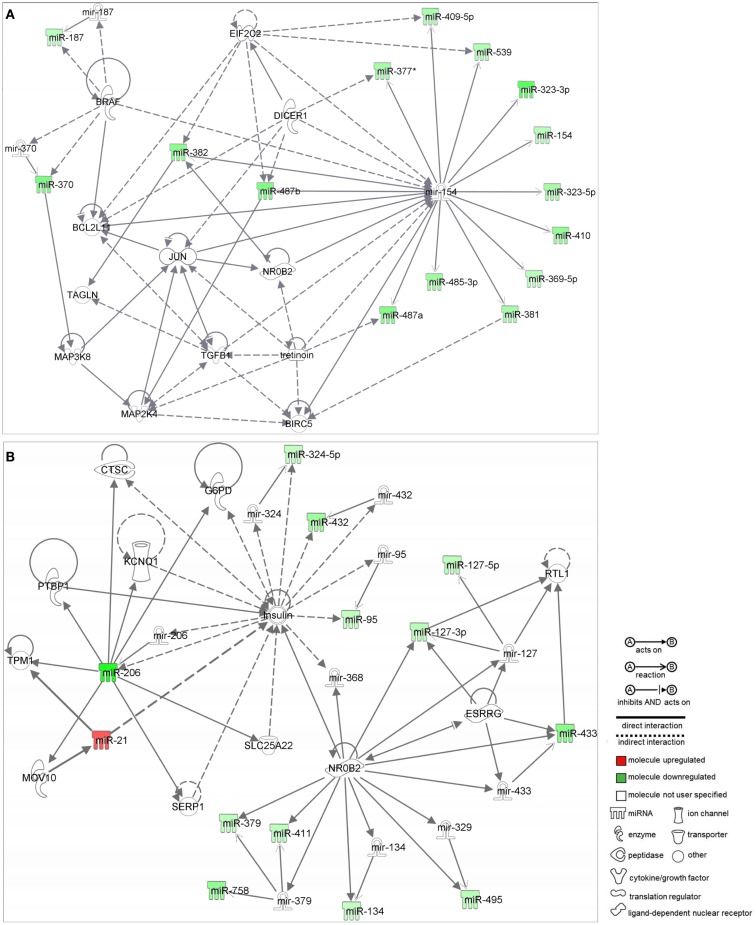
Network analysis by IPA identified two networks as putative targets for 73 out of the 84 MB miRNAs. Networks were prospected considering only relationships that were experimentally observed. Interestingly, both networks were enriched with miRNAs belonging to the 14q32 cluster.
Network 1 (Figure 2A) included 13 miRNAs of the 14q32 cluster (also known as miR-154 cluster), which were all downregulated in MB samples (miR-154, 323-3p, 323-5p, 369-5p, 377*, 381, 382, 409-5p, 410, 485-3p, 487a, 487b, 539) and were depicted by IPA as having direct interactions with BCL2L11, JUN, BIRC5, MAP2K4, and NR0B2. BIRC5 and BCL-2 have anti-apoptotic roles, and are expected to be at increasing levels in MB as all miRNAs connecting to these genes were found downregulated (Figure 2A). NR0B2 and JUN were suggested in this network as candidate genes controlling the expression of the 14q32 miRNA cluster.
Figure 2.
Ingenuity pathway analysis networks (IPA). (A) Network 1; (B) Network 2; both were constructed with deregulated miRNAs in medulloblastoma.
Again, most miRNAs shown in Network 2 (Figure 2B) belong to the 14q32 cluster (miR-127-3p, 127-5p, 134, 379, 411, 432, 433, 495, and 758). In this case, NR0B2 and estrogen-related receptor-γ (ESRRG) were suggested as candidate genes controlling the expression of the 14q32 miRNA cluster. Insulin appeared in Network 2 as indirectly controlling the expression of miR-206, 324-5p, 432, and 95.
RT-qPCR validation of some deregulated miRNAs
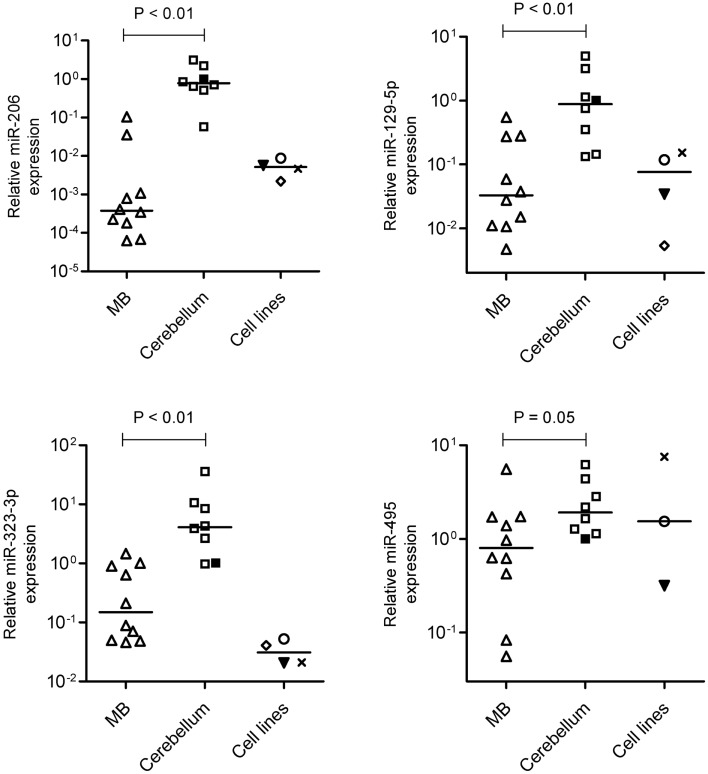
miR-323-3p and 495, both belonging to cluster 14q32, were chosen for validation by RT-qPCR. In addition, miR-206 and miR-129-5p were chosen for analysis because of their high fold change (see Table 3), lack of previous functional studies and possible oncogenic role. miR-206 expression was reported to inhibit cell proliferation in breast cancer cells (22). miR-129 is reported to be significantly downregulated in pediatric brain tumors compared to normal tissues (23). Most importantly, miR-129 downregulation is associated to SOX4 overexpression in endometrial and gastric cancers (24, 25). SOX4 is upregulated and has prognostic impact in MB (26, 27).
Real-time RT-qPCR analysis were performed with all samples used in the microarray analysis (n = 6 MB and n = 6 cerebellum) plus four other samples of MB (Table 1) and two new fetal/NW cerebellum controls (Table 2). As expected, miR-206 (p = 0.0001; Mann–Whitney test), miR-129-5p (p = 0.002), miR-323-3p (p = 0.014), and miR-495 (p = 0,054), had lower expression in MB in comparison to normal cerebellum (Figure 3), thus confirming our microarray findings. Expression of miR-206, 129-5p, 323-3p, and 495 were also investigated in a representative panel of MB cell lines. Compared with normal human cerebellum, miR-206, 129-5p, and 323-3p expression were found to be downregulated in all MB cell line tested (Figure 3).
Figure 3.
Validation of miR-206, 129-5p, 323-3p, and 495 downregulation by reverse transcriptase-quantitative PCR. Expression levels of miR-206, 129-5p, 323-3p, and 495 were performed in MB, normal fetal/newborn cerebellum and four different MB cell lines. Expression of miR-495 was not performed in D341 cell line. ○, DAOY; ▾, Meb-Med-8A; x, D283Med; ♢, D341. Comparisons of MB versus normal cerebellum were done by the Mann–Whitney test.
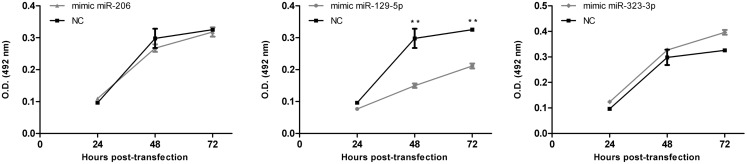
Upregulation of miR-206, 129-5p, and 323-3p in DAOY cells
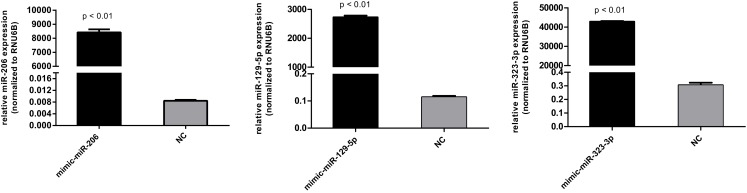
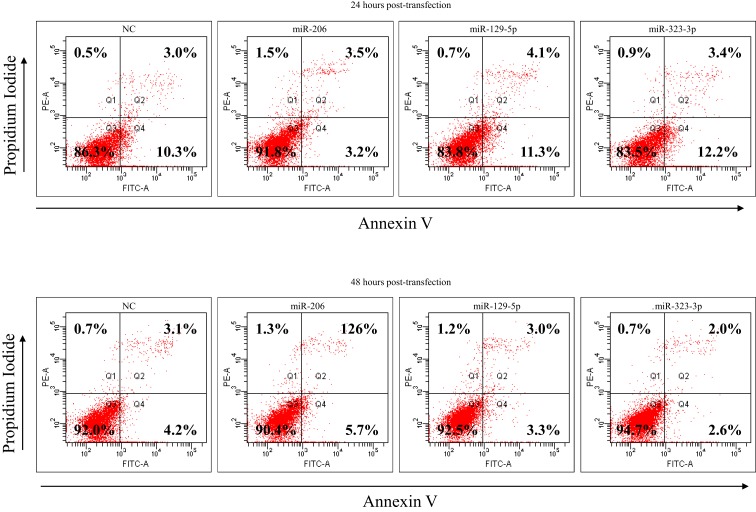
As a first approach to investigate the functional significance of miRNAs downregulation in MB, DAOY cells were transiently transfected with mimics of miR-206, 129-5p, 323-3p, or negative control #1. Transfection efficiency was confirmed by RT-qPCR (Figure A2 in Appendix). Twenty hours post-transfection, cells were collected and seeded in 96-well plates in serum-free medium. DAOY cells survive and even proliferate in serum-free medium for a short period of time. As shown in Figure 4, no consistent differences in proliferation were found in transfections with miR-206 and miR-323-3p. On the contrary, transfections with miR-129-5p resulted in a significant decrease in DAOY cell proliferation, as evaluated by the MTS assay (Figure 4). Similar experiments were conducted to evaluate cell survival and apoptosis by the Annexin V and propidium iodide staining methodology. No significant differences were found on cell viability or apoptosis after miR-206, miR-129-5p, and miR-323-3p transfections in comparison to control (Figure 4; Figure A3 in Appendix, respectively).
Figure 4.
Effect of miR-206, 129-5p, and 323-3p overexpression in DAOY cells proliferation. DAOY cells were transiently transfected with miRNAs mimic and replated after 20 h at a 1,500 cells/well density. Cell proliferation was evaluated by the MTS assay at 4, 28, and 52 h after re-plating, thus after 24, 48, and 72 h post-transfection. Results are representative of three independent experiments. Mean ± SE are shown; **P < 0.01 according to two-tailed t-test; NC, negative control miRNA mimic (scrambled).
Discussion
We investigated the expression profile of miRNAs in primary human MB of desmoplastic histology and SHH molecular subgroup, in comparison to normal fetal/newborn cerebellum. Eighty-four miRNAs were found to be differentially expressed in MB. The majority of these differentially expressed miRNAs were downregulated in comparison to normal cerebellum, corroborating previous studies. Most upregulated miRNAs identified in our study (12 out of 20) had been previously described in MB (12–14, 23, 28–32). On the contrary, 31 out of the 64 downregulated miRNAs are here described for the first time in association to MB (Table 3). Differences may be explained by the fact that a more comprehensive version of Affymetrix miRNA microarray was used in the present study. Moreover, previous studies included different subtypes of MB and a mix of children and adults cerebellum samples in their analysis (12, 13, 29). We believe that analysis on more uniform groups of both cancer and control samples may have helped us in detecting some smaller but consistent differences between groups.
A computational analysis was performed to predict the network and signaling pathways collectively targeted by the 64 downregulated and 20 upregulated miRNAs. Downregulated miRNAs in MB were predicted to target genes related to the ribosome, adherens junction, oxidative phosphorylation, metabolism of xenobiotics by cytochrome P450, and transforming growth factor-beta (TGF-β) signaling pathways. Axon guidance, TGF-β, WNT, insulin signaling pathways are known to play an important role in neurulation, CNS developmental, and/or MB pathogenesis (3, 33, 34). Since miRNA act as negative regulators of gene expression, a simpler interpretation of these findings is that MB has increased activation of these pathways in comparison to normal cerebellum.
Most importantly, half (32/64) of downregulated miRNAs reported in our study were found to belong to the cluster at 14q32 locus (also known as miR-154 cluster). This is in keeping with a previous study in a mouse model of MB, reporting that activation of SHH signaling leads to downregulation of the miR-154 cluster (35). Moreover, previous publications with primary MB found downregulation of some 14q32 miRNAs in MBs of the molecular subgroups WNT, SHH, and C as compared to normal cerebellum and MBs of subgroup D (13, 29). However, this is the first time that so many 14q32 miRNAs are shown to be downregulated in MB, thus suggesting a co-regulatory control of this cluster’s expression.
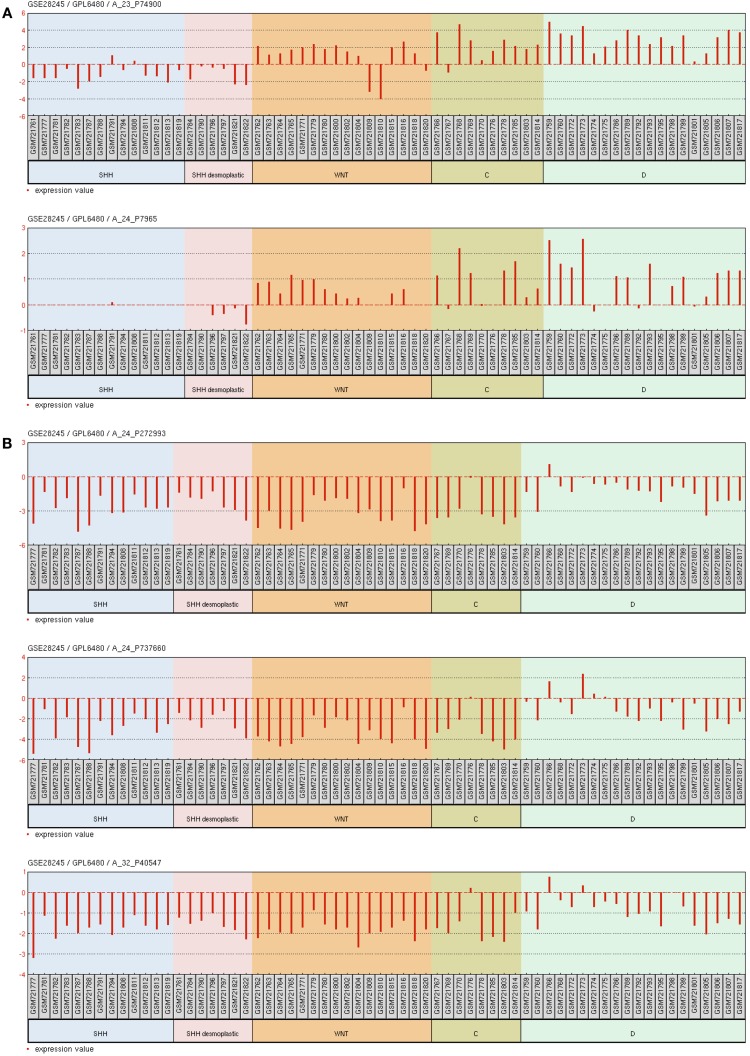
Deletions at locus 14q32 would be one possible explanation to the decreased 14q32 miRNAs expression. The recent analysis of somatic copy number aberrations in 1,087 MB samples report significant losses of chromosome arm 14q in the SHH subgroup of MBs, though not restricted to 14q32 (36). Alternative explanations are discussed below. Our Ingenuity pathways analysis pointed to nuclear orphan receptor NR0B2 (also known as Small Heterodimer Partner, SHP) and (ESRRG) as possible controllers of 14q32 miRNA cluster expression. There is indeed experimental evidence in mouse showing that NR0B2 is a repressor while ESRRG is and activator of a miRNA cluster in chromosome 12, which is ortholog to the 14q32 cluster in humans (37). Our analysis of microarray mRNA expression data for 64 primary human MB samples, accessible through GEO Series accession number GSE28245 (38) in NCBI’s Gene Expression Omnibus (39) revealed that NR0B2 is not expressed in MB. Interestingly, ESRRG expression was found to be relatively high in MBs of the molecular subgroup D, intermediate in MBs of the WNT and C subgroups, and very low or absent in MBs of the SHH subgroup (Figure A4A in Appendix), thus reflecting 14q32 miRNAs abundance in each of the MB subgroup. These findings were confirmed by the analysis of gene expression data of an independent cohort of 90 primary MB samples (accession number GSE21166) deposited by Northcott et al. (13) (data not shown). ESRRG suppress cell proliferation in prostate cancer cells (40) and the estrogen receptor beta agonist diarylpropionitrile (DPN) exhibit a pro-apoptotic and anti-proliferative effect on MB (41). Experiments are warranted to investigate a possible causal connection between ESRRG and 14q32 miRNA cluster expression in MB.
The miRNA cluster at 14q32 lies within a parentally imprinted chromosomal area spanning genes Dlk1, Meg3, Rtl1, Meg8, and Dio3 (42). Dlk1, Rtl1, and Dio3 are paternally-, whereas Meg3 and Meg8 are maternally expressed transcripts (43). Imprinting of 14q32 is regulated, to some extent, by two intergenic differentially methylated regions known as IG-DMR and MEG3-DMR (44, 45). Deletions of the regulatory regions and/or epigenetic modifications may in theory cause aberrant 14q32 silencing in cancer. The recent 1,000 genome study of somatic copy number aberrations shows no recurrent focal deletions at locus 14q32 in MB (36). However, our analysis of public mRNA microarray expression data GSE28245 (38) revealed that MEG3 is downregulated in MB in comparison to normal cerebellum. MBs of the molecular group C and WNT have the lowest expression, SHH has intermediate levels while group D have MEG3 levels closer to normal cerebellum (Figure A4B in Appendix). Thus MEG3 expression seems to correlate with the expression of 14q32 miRNAs among the different MB molecular groups, suggesting that the 14q32 miRNA locus may be under epigenetic regulation in MB. However, a genome wide analysis of promoter methylation on four primary MB samples showed no consistent methylation of 14q32 gene promoters (46). Although higher number of samples should be analyzed, this result corroborates findings in osteosarcoma, a tumor also presenting with downregulated 14q32 miRNAs expression and with no consistent changes in the methylation patterns at 14q32. Instead, silencing of 14q32 miRNA in osteosarcoma seems to be mediated by histone modification(s) (47).
Preliminary functional studies were performed in DAOY cells by ectopic expression of miR-129-5p, 206, and 323-3p mimics. Mimics for miR-206 and 323-3p had no significant effect on DAOY cells. miR-129-5p overexpression resulted in decreased cell proliferation, which may suggest a tumor suppressor role in MB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Carlos A Scrideli for helpful discussion and for the cell line DAOY and Dr. Priscila P. Zenatti and Dr. Angelo B. A. Laranjeira for technical assistance with FACS analysis. We also acknowledge the Microarray Laboratory at Brazilian Biosciences National Laboratory, CNPEM-ABTLuS, Campinas, Brazil for their support with the use of Fluidics station and Scanner GeneChip equipments. This work was supported by grant from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; 2010/07020-9). DRL have Coordenação de Aperfeiçoamento de Pessoal de Nível Superior PhD scholarship (2012/890912-1).
Appendix
Figure A1.
Expression of OTX2 and PTCH1 in the different primary medulloblastomas (MB), medulloblastoma cell lines (named), and normal cerebellum (C). Total RNA (100 ng) was reverse transcribed and gene expression was measured by qPCR, in triplicates, using the SYBR method. Primers are available upon request to the authors. Values were normalized to the HPRT endogenous control gene. Relative expression values were calculated using the 2−ΔΔCt method.
Figure A2.
Transfection efficiency of miR-206, miR-129-5p, and miR-323-3p in DAOY cells as evaluated by RT-qPCR. Cells transfected with the scrambled mimic were included as negative control (NC). Values were normalized to the RUN6B endogenous control RNA. Relative expression values were calculated using the 2−ΔΔCt method. Bars represent mean ± SD. Comparisons were performed by the Mann–Whitney test.
Figure A3.
Apoptosis analysis of DAOY cells at 24 and 48 h post-transfection with miR-206, 129-5p, 323-3p, or negative control mimics, as evaluated by Annexin V and propidium iodide staining and FACS analysis. DAOY cells grown in serum-free RPMI-1640 medium. The percentage of necrotic (Q1), late apoptotic (Q2), viable (Q3), and early apoptotic (Q4) cells are shown in the corresponding quadrants. NC, negative control (scrambled mimic).
Figure A4.
Gene expression graph for different microarray probes of interest. Data from 64 primary human MB samples accessible through GEO Series accession number GSE28245 (38) in NCBI’s Gene Expression Omnibus. (A) probe sets for ESRRG. (B) Probe sets for MEG3. Samples were grouped according to the molecular MB subgroups into WNT, SHH, C, and D. SHH desmoplastic was separated from SHH classic.
Footnotes
References
- 1.Ellison D. Classifying the medulloblastoma: insights from morphology and molecular genetics. Neuropathol Appl Neurobiol (2002) 28:257–82 10.1046/j.1365-2990.2002.00419.x [DOI] [PubMed] [Google Scholar]
- 2.Gulino A, Di Marcotullio L, Ferretti E, De Smaele E, Screpanti I. Hedgehog signaling pathway in neural development and disease. Psychoneuroendocrinology (2007) 32:S25–56 10.1016/j.psyneuen.2007.03.017 [DOI] [PubMed] [Google Scholar]
- 3.Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van SluisP, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS ONE (2008) 3:e3088. 10.1371/journal.pone.0003088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol (2010) 29:1408–14 10.1200/JCO.2009.27.4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbertson RJ, Ellison DW. The origins of meduloblastoma subtypes. Annu Rev Pathol (2008) 3:341–65 10.1146/annurev.pathmechdis.3.121806.151518 [DOI] [PubMed] [Google Scholar]
- 6.Schüller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form SHH-induced medulloblastoma. Cancer Cell (2008) 14:123–34 10.1016/j.ccr.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell (2008) 14(2):135–45 10.1016/j.ccr.2008.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson P, Tong Y, Robinson G, Thomson MC, Currie DS, Eden C, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature (2010) 468:1095–9 10.1038/nature09587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen SM. MiRNAs in CNS Development and Neurodegeneration: Insights from Drosophila Genetics. Springer; (2010). Available from: http://www.springer.com/978-3-642-04297-3 [Google Scholar]
- 10.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A (2006) 103(8):2746–51 10.1073/pnas.0511045103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature (2010) 466(7308):835–41 10.1038/nature09267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferretti E, De Smaele E, Po A, Di Marcotullio L, Tosi E, Espínola MSB, et al. MiRNA profiling in human meduloblastoma. Int J Cancer (2009) 124:568–77 10.1002/ijc.23948 [DOI] [PubMed] [Google Scholar]
- 13.Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, et al. The miR-17/92 polycistron is upregulated in Sonic Hedgehog driven medulloblastomas and induced by N-myc in Sonic Hedgehog-treated cerebellar neural precursors. Cancer Res (2009) 69:3249–55 10.1158/0008-5472.CAN-08-4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol (2011) 29(11):1424–30 10.1200/JCO.2010.28.5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez LA, Northcott PA, Taylor MD, Kenney AM. Normal and oncogenic roles for microRNAs in the developing brain. Cell Cycle (2009) 8(24):4049–54 10.4161/cc.8.24.10243 [DOI] [PubMed] [Google Scholar]
- 16.Pfister SM, Korshunov A, Kool M, Hasselblatt M, Eberhart C, Taylor MD. Molecular diagnostics of CNS embryonal tumors. Acta Neuropathol (2010) 120:553–66 10.1007/s00401-010-0751-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods (2001) 25:402–8 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 18.Pietsch T, Scharmann T, Fonatsch C, Schmidt D, Ockler R, Freihoff D, et al. Characterization of five new cell lines derived from human primitive neuroectodermal tumors of the central nervous system. Cancer Res (1994) 54:3278–87 [PubMed] [Google Scholar]
- 19.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett (2004) 573:83–92 10.1016/j.febslet.2004.07.055 [DOI] [PubMed] [Google Scholar]
- 20.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy – analysis of Affymetrix GeneChip data at the probe level. Bioinformatics (2004) 20:307–15 10.1093/bioinformatics/btg405 [DOI] [PubMed] [Google Scholar]
- 21.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol (2004) 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J, Tian Y, Li J, Lu B, Sun M, Zou Y, et al. miR-206 is down-regulated in breast cancer and inhibits cell proliferation through the up-regulation of cyclinD2. Biochem Biophys Res Commun (2013) 433(2):207–12 10.1016/j.bbrc.2013.02.084 [DOI] [PubMed] [Google Scholar]
- 23.Birks DK, Barton VN, Donson AM, Handler MH, Vibhakar R, Foreman NK. Survey of MicroRNA expression in pediatric brain tumors. Pediatr Blood Cancer (2011) 56(2):211–6 10.1002/pbc.22723 [DOI] [PubMed] [Google Scholar]
- 24.Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, et al. Epigenetic repression of microRNA-129-2 leads to upregulation of SOX4 oncogene in endometrial cancer. Cancer Res (2009) 69(23):9038–46 10.1158/0008-5472.CAN-09-1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen R, Pan S, Qi S, Lin X, Cheng S. Epigenetic repression of microRNA-129-2 leads to upregulation of SOX4 in gastric cancer. Biochem Biophys Res Commun (2010) 394(4):1047–52 10.1016/j.bbrc.2010.03.121 [DOI] [PubMed] [Google Scholar]
- 26.Neben K, Korshunov A, Benner A, Wrobel G, Hahn M, Kokocinski F, et al. Microarray-based screening for molecular markers in medulloblastoma revealed STK15 as independent predictor for survival. Cancer Res (2004) 64(9):3103–11 10.1158/0008-5472.CAN-03-3968 [DOI] [PubMed] [Google Scholar]
- 27.de Bont JM, Kros JM, Passier MM, Reddingius RE, Sillevis Smitt PA, Luider TM, et al. Differential expression and prognostic significance of SOX genes in pediatric medulloblastoma and ependymoma identified by microarray analysis. Neuro Oncol (2008) 10(5):648–60 10.1215/15228517-2008-032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garzia L, Andolfo I, Cusanelli E, Marino N, Petrosino G, De Martino D, et al. MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS ONE (2009) 4:4998. 10.1371/journal.pone.0004998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gokhale A, Kunder R, Goel A, Sarin R, Moiyadi A, Shenoy A, et al. Distinctive microRNA signature of medulloblastomas associated with the WNT signaling pathway. J Cancer Res Ther (2010) 6:521–9 10.4103/0973-1482.77072 [DOI] [PubMed] [Google Scholar]
- 30.de Antonellis P, Medaglia C, Cusanelli E, Andolfo I, Liguori L, De VitaG, et al. MiR-34a targeting of notch ligand delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma. PLoS ONE (2011) 6(9):e24584. 10.1371/journal.pone.0024584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grunder E, D’Ambrosio R, Fiaschetti G, Abela L, Arcaro A, Zuzak T, et al. MicroRNA-21 suppression impedes medulloblastoma cell migration. Eur J Cancer (2011) 47:2479–90 10.1016/j.ejca.2011.06.041 [DOI] [PubMed] [Google Scholar]
- 32.Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, et al. The miR-17 92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci USA (2009) 106:2812–7 10.1073/pnas.0809579106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chédotal A, Kerjan G, Moreau-Fauvarque C. The brain within the tumor: new roles for axon guidance molecules in cancers. Cell Death Differ (2005) 12(8):1044–56 10.1038/sj.cdd.4401707 [DOI] [PubMed] [Google Scholar]
- 34.Aref D, Moffatt CJ, Agnihotri S, Ramaswamy V, Dubuc AM, Northcott PA, et al. Canonical TGF-b pathway activity is a predictor of SHH-driven medulloblastoma survival and delineates putative precursors in cerebellar development. Brain Pathol (2013) 23:178–91 10.1111/j.1750-3639.2012.00631.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo X, Liu J, Cheng SY. The role of microRNAs during the genesis of medulloblastoma induced by the hedgehog pathway. Biomed Res (2011) 25(1):42–8 10.1016/S1674-8301(11)60005-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature (2012) 488(7409):49–56 10.1038/nature11327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song G, Wang L. miR-433 and miR-127 arise from independent overlapping primary transcripts encoded by the miR-433-127 locus. PLoS ONE (2008) 3:e3574. 10.1371/journal.pone.0003574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Remke M, Hielscher T, Korshunov A, Northcott PA, Bender S, Kool M, et al. FSTL5 is a marker of poor prognosis in non-WNT/non-SHH medulloblastoma. J Clin Oncol (2011) 29(29):3852–61 10.1200/JCO.2011.36.2798 [DOI] [PubMed] [Google Scholar]
- 39.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res (2002) 30:207–10 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu S, Wang X, Ng CF, Chen S, Chan FL. ERRgamma suppresses cell proliferation and tumor growth of androgen-sensitive and androgen-insensitive prostate cancer cells and its implication as a therapeutic target for prostate cancer. Cancer Res (2007) 67(10):4904–14 10.1158/0008-5472.CAN-06-3855 [DOI] [PubMed] [Google Scholar]
- 41.Mancuso M, Leonardi S, Giardullo P, Pasquali E, Borra F, Stefano ID, et al. The estrogen receptor beta agonist diarylpropionitrile (DPN) inhibits medulloblastoma development via anti-proliferative and pro-apototic pathways. Cancer Lett (2011) 308(2):197–202 10.1016/j.canlet.2011.05.004 [DOI] [PubMed] [Google Scholar]
- 42.da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet (2008) 24:306–16 10.1016/j.tig.2008.03.011 [DOI] [PubMed] [Google Scholar]
- 43.Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, et al. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet (2003) 35:97–102 10.1038/ng1233 [DOI] [PubMed] [Google Scholar]
- 44.Takada S, Paulsen M, Tevendale M, Tsai CE, Kelsey G, Cattanach BM, et al. Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: implications for imprinting control from comparison with Igf2-H19. Hum Mol Genet (2002) 11:77–86 10.1093/hmg/11.1.77 [DOI] [PubMed] [Google Scholar]
- 45.Kagami M, O’Sullivan MJ, Green AJ, Watabe Y, Arisaka O, Masawa N, et al. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet (2010) 6:e1000992. 10.1371/journal.pgen.1000992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diede SJ, Guenthoer J, Geng LN, Mahoney SE, Marotta M, Olson JM, et al. DNA methylation of developmental genes in pediatric medulloblastomas identified by denaturation analysis of methylation differences. Proc Natl Acad Sci U S A (2010) 107(1):234–9 10.1073/pnas.0907606106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thayanithy V, Park C, Sarver AL, Kartha RV, Korpela DM, Graef AJ, et al. Combinatorial treatment of DNA and chromatin-modifying drugs cause cell death in human and canine osteosarcoma cell lines. PLoS ONE (2012) 7(9):e43720. 10.1371/journal.pone.0043720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierson J, Hostager B, Fan R, Vibhakar R. Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol (2008) 90:1–7 10.1007/s11060-008-9624-3 [DOI] [PubMed] [Google Scholar]
- 49.Li KK, Pang JC, Ching AK, Wong CK, Kong X, Wang Y, et al. miR-124 is frequently down-regulated in meduloblastoma and is a negative regulator of SLC16A1. Hum Pathol (2009) 40(9):1234–43 10.1016/j.humpath.2009.02.003 [DOI] [PubMed] [Google Scholar]
- 50.Ferretti E, De Smaele E, Miele E, Laneve P, Pó A, Pelloni M, et al. Concerted miRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J (2008) 27:2616–27 10.1038/emboj.2008.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venkataraman S, Alimova I, Fan R, Harris P, Foreman N, Vibhakar R. MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS ONE (2010) 5(6):e10748. 10.1371/journal.pone.0010748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Genovesi LA, Carter KW, Gottardo NG, Giles KM, Dallas PB. Integrated analysis of miRNA and mRNA expression in childhood medulloblastoma compared with neural stem cells. PLoS ONE (2011) 6(9):e23935. 10.1371/journal.pone.0023935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weeraratne SD, Amani V, Neiss A, Teider N, Scott DK, Pomeroy SL, et al. miR-34a confers chemosensitivity through modulation of MAGE-A and p53 in medulloblastoma. Neuro Oncol (2011) 13(2):165–75 10.1093/neuonc/noq179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lv SQ, Kim YH, Giulio F, Shalaby T, Nobusawa S, Yang H, et al. Genetic alterations in MicroRNAs in medulloblastomas. Brain Pathol (2012) 22:230–9 10.1111/j.1750-3639.2011.00523.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang XM, Zhang SF, Cheng ZQ, Peng QZ, Hu JT, Gao LK, et al. MicroRNA383 regulates expression of PRDX3 in human medulloblastomas. Zhonghua Bing Li Xue Za Zhi (2012) 41(8):547–52 10.3760/cma.j.issn.0529-5807.2012.08.009 [DOI] [PubMed] [Google Scholar]
- 56.Li KKW, Pang JCS, Lau KM, Zhou L, Mao Y, Wang Y, et al. MiR-383 is downregulated in medulloblastoma and targets peroxiredoxin 3 (PRDX3). Brain Pathol (2013) 23(4):413–25 10.1111/bpa.12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ, et al. Pleiotropic effects of miR-183 96 182 converge to regulate cell survival, proliferation and migration in meduloblastoma. Acta Neuropathol (2012) 123:539–52 10.1007/s00401-012-0969-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Venkataraman S, Birks DK, Balakrishnan I, Alimova I, Harris PS, Patel PR, et al. MicroRNA 218 acts as a tumor suppressor by targeting multiple cancer phenotype-associated genes in medulloblastoma. J Biol Chem (2013) 228(3):1918–28 10.1074/jbc.M112.396762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei L, Yan-hua G, Teng-fei C, Xiao-zhong P, Jian-gang Y, Zhen-yu M, et al. Identification of differentially expressed microRNAs by microarray: a possible role for microRNAs gene in medulloblastomas. Chin Med J (2009) 122(20):2405–11 [PubMed] [Google Scholar]