Abstract
Living cells are continually exposed to DNA-damaging agents that threaten their genomic integrity. Although DNA repair processes rapidly target the damaged DNA for repair, some lesions nevertheless persist and block genome duplication by the cell’s replicase. To avoid the deleterious consequence of a stalled replication fork, cells use specialized polymerases to traverse the damage. This process, termed “translesion DNA synthesis” (TLS), affords the cell additional time to repair the damage before the replicase returns to complete genome duplication. In many cases, this damage-tolerance mechanism is error-prone, and cell survival is often associated with an increased risk of mutagenesis and carcinogenesis. Despite being tightly regulated by a variety of transcriptional and posttranslational controls, the low-fidelity TLS polymerases also gain access to undamaged DNA where their inaccurate synthesis may actually be beneficial for genetic diversity and evolutionary fitness.
Cells use specialized polymerases to traverse DNA lesions. However, these polymerases are error-prone, and despite their tight regulation, they sometimes gain access to undamaged DNA.
The Watson–Crick (WC) DNA structure was published in 1953 (Watson and Crick 1953). Coincidentally, it was in 1953 when Weigle discovered that λ bacteriophage that had been killed by exposure to UV radiation could, in essence, be brought back to life by irradiation of its Escherichia coli host (Weigle 1953). The reactivation of λ, however, was accompanied by a sizable increase in phage mutagenesis (Weigle 1953). With the benefit of nearly five decades of hindsight, the increase in phage survival and mutagenesis observed by Weigle turns out to have been caused by the action of translesion DNA synthesis (TLS) polymerases. Currently, there are three TLS polymerases (Pols) in E. coli, and many of the 15 known polymerases in eukaryotes have the capacity to promote some degree of TLS (Table 1) (Goodman 2002; Bebenek and Kunkel 2004; Waters et al. 2009; Lange et al. 2011; Sale et al. 2012).
Table 1.
Properties of key TLS polymerases across all domains of life
| Organism | Gene name | Protein name | Characteristicsa |
|---|---|---|---|
| E. coli | polB | Pol II | B-family polymerase Regulated by LexA; damage inducible Creates −2 frameshift mutations during TLS of N2-dG-AAF adducts |
| E. coli | dinB | Pol IV | Regulated by LexA; damage inducible Prone to making −1 frameshifts when overexpressed in vivo Bypasses N2-dG adducts efficiently and accurately Involved in TLS of alkylation damage in vivo Major polymerase involved in stress-induced mutagenesis |
| E. coli | umuDC | Pol V | Regulated by LexA; damage inducible Major TLS polymerase in E. coli Composed of a heterotrimer of UmuD′2 (∼24 kDa) and UmuC (∼48 kDa) to form an ∼72-kDa UmuD′2C complex Interacts with RecA and ATP to form Pol V Mut |
| Sulfolobus solfataricus | dpo4 | Dpo4 | Archaeal ortholog of E. coli Pol IV Numerous crystal structures of Dpo4 in the process of TLS have been solved |
| Sulfolobus acidocaldarius | dbh | Dbh | First archaeal DinB homology identified Less processive than Dpo4 |
| Saccharomyces cerevisiae | REV3 | Rev3p | B-family polymerase Together with Rev7, Pol31 and Pol32 forms Pol ζ4 Can bypass lesions unassisted Works with other TLS polymerases to facilitate the extension step of TLS Stimulated by an interaction with Rev1 |
| S. cerevisiae | REV1 | Rev1p | Specifically incorporates dCMP opposite abasic sites and undamaged template dG Interacts with Pol ζ to stimulate Pol ζ–dependent TLS in vivo |
| S. cerevisiae | RAD30 | Pol η | Bypasses a thymine–thymine CPD relatively accurately and efficiently |
| Homo sapiens | REV3L | REV3 | B-family polymerase Catalytic subunit of human Pol ζ Very large protein consisting of 3130 amino acids Murine homozygous knockout is embryonic lethal |
| H. sapiens | REV1 | REV1 | Similar to S. cerevisiae Rev1 protein, it specifically incorporates dCMP opposite dG and abasic sites Acts as scaffold protein that interacts with TLS polymerases ζ, η, ι, and κ Generates mutations at G-C base pairs during immunoglobulin gene somatic hypermutation |
| H. sapiens | POLH/ XPV/ RAD30A | Pol η | Bypasses a thymine–thymine CPD relatively efficiently and accurately Defects lead to the sunlight-sensitive and cancer-prone xeroderma pigmentosum variant (XP-V) phenotype Regulated by ubiquitination and phosphorylation Generates mutations at A-T base pairs during immunoglobulin gene somatic hypermutation |
| H. sapiens | POLI/RAD30B | Pol ι | Has unique replication fidelity; incorporates opposite template dA reasonably accurately, but opposite template dT in a highly error-prone manner In vivo functions remain unknown |
| H. sapiens | POLK/DINB1 | Pol κ | Prone to making −1 frameshift mutations, but can accurately and efficiently bypass a number of N2-dG lesions Plays additional roles in repair synthesis steps of nucleotide excision repair |
aUnless specified, the TLS polymerase belongs to the Y-family of DNA polymerases (Ohmori et al. 2001).
In this review, we revisit the concepts and experiments in E. coli centered on the induction and regulation of mutagenesis that began in the mid 1970s, continuing throughout the 1980s, and culminating with the identification of a new family (Y-family) of error-prone DNA polymerases (Ohmori et al. 2001). We review early TLS models based principally on genetic data from E. coli that were generated in the 1980s and discuss recent biochemical data leading to current models of TLS in E. coli and humans. The typically poor fidelity of TLS polymerases can now be understood thanks to high-resolution X-ray structures that indicate a more spacious active cleft that can accommodate a variety of non-WC base pairs. Owing to their potential to cause excessive numbers of deleterious mutations compared with replication polymerases, we describe how TLS Pol expression is tightly regulated, at both the transcriptional and posttranslational levels. However, owing to the need for generating beneficial mutations, we also review recent data showing that TLS Pols are involved in providing evolutionary fitness in bacteria and in generating immunological diversity in higher vertebrates. Recognizing that the cellular function(s) of many of the human TLS Pols remain unknown, it may turn out that lesion bypass is but one important property of TLS Pols, and perhaps not even their most salient one.
DNA DAMAGE-INDUCED REGULATION OF GENE EXPRESSION IN E. coli
Weigle’s 1953 phage reactivation experiment suggested the possibility of an induced cellular DNA repair process. The presence of a bacterial repressor system that could be inactivated by excessive DNA damage was proposed by Witkin in 1967 (Witkin 1967) as a molecular model to explain UV-induced phage reactivation. The pivotal connection to mutagenesis was made by Radman, who proposed the SOS model for an inducible error-prone mechanism in E. coli for the repair of DNA (Radman 1974). The conceptual framework provided by Witkin and Radman paved the way for several decades of genetic and biochemical studies aimed at elucidating the molecular basis of “SOS error-prone repair” (Witkin 1976; Walker 1984; Friedberg et al. 2006; Schlacher and Goodman 2007).
The damage-inducible SOS regulon requires interplay of the LexA and RecA proteins. The lexA gene encodes the LexA transcriptional repressor, which binds to sequences in the operator region of the >40 genes under its control (Fernández de Henestrosa et al. 2000; Courcelle et al. 2001). There is a minimal 6-bp operator consensus sequence, but it is the nonconsensus surrounding bases that determine repressor-operator binding constants, so that the earliest expressed genes are those whose operators are bound weakly by LexA (Lewis et al. 1994; Fernández de Henestrosa et al. 2000). It is estimated that there are roughly 8000 RecA molecules in an undamaged cell (Boudsocq et al. 1997). When the cellular DNA is damaged, RecA assembles as a nucleoprotein filament on single-stranded DNA (ssDNA), requiring the presence of ATP (Kuzminov 1999). This nucleoprotein filament, commonly referred to as RecA*, acts as a coprotease to mediate self-cleavage of LexA molecules in solution (Little et al. 1980). As the overall cellular concentration of intact LexA diminishes, only those genes that bind LexA tightly remain repressed.
The earliest genes to be induced after DNA damage are those involved in error-free repair, such as nucleotide excision repair (uvrA, uvrB, uvrD, and cho), as well as recombinational repair (recA, recN). E. coli’s three TLS polymerases are also regulated by LexA. The operators for Pol II (polB) and Pol IV (dinB) bind LexA weakly (Fernández de Henestrosa et al. 2000) and are induced early in the SOS response. In contrast, the umuDC operon (encoding Pol V) has one of the tightest LexA-binding sites and is induced late in the response, 30–40 min after DNA damage (Sommer et al. 1993). Given their early induction and relatively high basal expression levels in the absence of DNA damage, it is thought that Pol II and Pol IV are likely to participate in mostly error-free TLS of specific DNA lesions. In contrast, Pol V can traverse a wide range of DNA lesions and does so in a much more error-prone manner. Its induction late in the SOS response therefore suggests that E. coli only uses Pol V as a last resort, once all other error- free repair pathways have been exhausted (Sommer et al. 1993).
E. coli AS A MODEL SYSTEM FOR TLS AND MUTAGENESIS
By the mid-1970s, early 1980s, genetic experiments had led to the identification of the principal players involved in damage-induced mutagenesis, lexA, recA, along with umuD and umuC. The same mechanism used by RecA* to mediate LexA cleavage (Little et al. 1980) is also used to cleave UmuD to form UmuD′ (Burckhardt et al. 1988; Shinagawa et al. 1988) and is an absolute requirement for SOS mutagenesis (Nohmi et al. 1988). By isolating RecA mutants that were UV non-mutable, despite retaining the ability to cleave LexA and UmuD, Devoret and colleagues showed that RecA* also had a separate direct role in SOS mutagenesis (Dutreix et al. 1989). This mutagenic role for RecA* remained a mystery until 2009 (Jiang et al. 2009); its biochemical mechanism is discussed below.
Early TLS Models
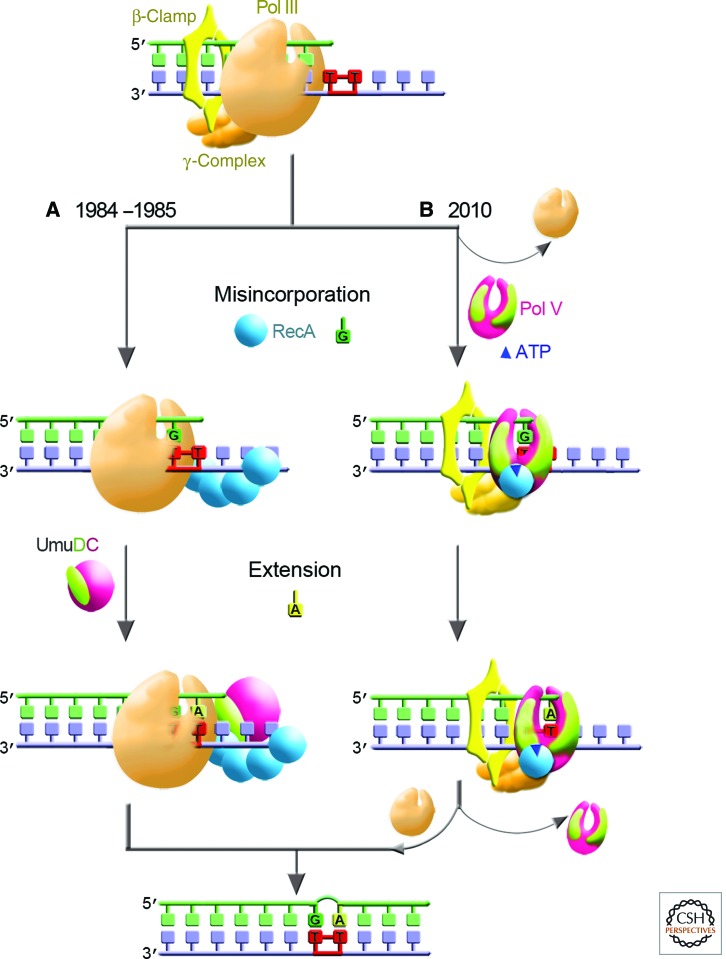
The first model to address the function of the Umu proteins during UV-induced TLS was proposed by Bridges and Woodgate in 1985 (Bridges and Woodgate 1985a,b). According to this model, TLS occurred in a two-step process in which Pol III incorporates a nucleotide opposite the first (3′) T of a T-T CPD, requiring the presence of RecA protein bound to the template proximal to the lesion. Pol III would subsequently interact with the UmuDC proteins to incorporate another nucleotide at the second (5′) T of the CPD (Fig. 1A). At least one of the two incorporations would be non-WC, thereby causing a mutation targeted at the site of the CPD.
Figure 1.
Two-step model for UV mutagenesis in E. coli. (A) The two-step model for UV mutagenesis proposed in 1984/1985 assumed that TLS in vivo is performed by the replicative Pol III. The first step, nucleotide misincorporation opposite a 3′ T of a CPD, was hypothesized to be mediated by the RecA protein (represented as blue sphere). The misincorporated base was subsequently fixed as a mutation in a second extension/bypass step that was dependent on UmuC (purple) and UmuD (lime) (Bridges and Woodgate 1984, 1985a,b). (B) Subsequent studies revealed that rather than being accessory factors of Pol III, the products of umuDC genes encode a bona fide DNA polymerase, Pol V (shown as purple UmuC and two lime UmuD′ subunits assembled in the shape of a right hand) (Reuven et al. 1999; Tang et al. 1999), which interacts with RecA* in trans (not shown) from which molecules of RecA (blue sphere) and ATP (dark blue triangle) are transferred from the 3′-filament tip to generate Pol V Mut (Jiang et al. 2009). Pol V Mut can perform both the (mis)insertion and extension steps of TLS. After traversing the damaged DNA, Pol V Mut is replaced by Pol III holenzyme, which resumes high-fidelity chromosomal duplication.
A model proposed by Echols and Goodman in 1990 (Echols and Goodman 1990) envisioned complete blockage of the Pol III holoenzyme (Pol III core, β-sliding clamp, γ-clamp-loading complex) when encountering a template lesion, followed by the assembly of a damage-localized nucleoprotein complex involving RecA, UmuC, UmuD′, SSB, and Pol III holoenzyme—a “mutasome”—to copy past a template lesion (Woodgate et al. 1989; Echols and Goodman 1991). This model took into account the finding that RecA* facilitates cleavage of UmuD to a mutagenically active form, UmuD′ (Burckhardt et al. 1988; Nohmi et al. 1988; Shinagawa et al. 1988). Subsequently, it was shown that it was actually a dimeric UmuD2 that is cleaved to UmuD′2 and that subsequently interacts with UmuC to form a stable complex of UmuD′2C (Woodgate et al. 1989; Bruck et al. 1996), identified in 1999 as Pol V (Tang et al. 1999). Whereas a Pol III replisome performs rapid and processive “error-free” genome replication on undamaged DNA (Johnson and O’Donnell 2005; McHenry 2011), a mutasome would perform slow, poorly processive, “error-prone” TLS.
Biochemical Characterization of the E. coli Umu Proteins and Evolution of TLS Models
At the time of the mutasome model, UmuD′ could be obtained in high purity and yield; not so for UmuC, which formed insoluble inclusion bodies in E. coli cell lysates. Woodgate, working with Echols, denatured and then renatured UmuC to obtain soluble UmuC that was included in a biochemical reconstitution assay along with UmuD′, SSB, β-sliding clamp, γ-clamp loader, and Pol III core. Using this combination of proteins to copy a template DNA containing an abasic moiety, a small amount of primer was extended past the lesion (Rajagopalan et al. 1992). The difficulties with obtaining sufficient quantities of UmuC for detailed biochemical characterization were circumvented in 1996, by the isolation and purification of the UmuD′2C complex (from 200 L of E. coli!) (Bruck et al. 1996). When UmuD′2C was used in replication assays, robust TLS was observed. Remarkably, TLS also occurred in the absence of Pol III core (Tang et al. 1998). It appeared, therefore, that UmuD′2C by itself had the ability to copy undamaged and damaged DNA templates, and, about a year later, UmuD′2C was conclusively shown to be a DNA polymerase, Pol V (Reuven et al. 1999; Tang et al. 1999).
A key feature of the mutasome model is the assembly of RecA* on ssDNA proximal to the lesion (Fig. 1A). When a replication fork encounters a lesion, an uncoupling of leading- and lagging-strand synthesis may ensue. Then, one of the TLS Pols can replace Pol III on the β-clamp and copy the damaged DNA. If the lesion were present on the leading strand, then DNA unwinding by the DnaB helicase downstream from the lesion would leave a region of ssDNA on which a RecA* filament could form. In the case of a lagging-strand lesion, ssDNA is likely to be present as a consequence of Okazaki fragment synthesis and processing. Thus, for both leading- and lagging-strand lesions, one can easily envision how RecA* could assemble in cis, that is, on the template strand being copied, proximal to the lesion. However, the presence of RecA bound in cis to the template blocks DNA synthesis and would assuredly have to be removed to permit synthesis to occur beyond the lesion (Pham et al. 2001). Because both RecA* assembly and disassembly normally occur in a 5′→3′ direction (Kuzminov 1999), removal of a blocking RecA* ahead of an advancing Pol V would entail filament disassembly in an “abnormal” 3′→5′ direction (Pham et al. 2001).
The problematic issue of RecA* formation and dissolution on the template strand would be irrelevant if RecA* were absent from the DNA template, but rather acted in trans by forming on a non-template ssDNA strand (Schlacher and Goodman 2007; Patel et al. 2010). And that appears to be what is happening. Transactivation of Pol V by RecA* to perform TLS has been shown to take place in vitro (Schlacher et al. 2006; Jiang et al. 2009), giving rise to a Pol V mutasome model for TLS (Fig. 1B). The unanticipated role for RecA* in the transactivation of Pol V provided the key to establishing its biochemical function. By incubating Pol V with RecA* bound to resin and then spinning down the resin-bound RecA*, what remained in solution along with “unreacted” Pol V was a new form of Pol V, named Pol V Mut=UmuD′2C-RecA-ATP (Jiang et al. 2009). Once formed, Pol V Mut copies undamaged and damaged DNA (i.e., performs TLS) in the absence of RecA*.
Thus, the direct role of RecA* in SOS-mediated TLS, described by Devoret and colleagues in 1989 (Dutreix et al. 1989), is to transfer a molecule of RecA from its 3′-filament tip along with a molecule of ATP to convert UmuD′2C into Pol V Mut, which can traverse a variety of DNA lesions unassisted (Fig. 1B). Pol V Mut has the unique property that it exists in two conformational states, either activated (able to copy DNA) or deactivated (unable to copy DNA), perhaps depending on where RecA-ATP is bound in relation to the polymerase subunits UmuD′2 and UmuC (Jiang et al. 2009). A deactivated Pol V Mut is completely reactivated when re-exposed to RecA*, which is used to replace the old RecA-ATP with new RecA-ATP from the 3′-filament tip (Jiang et al. 2009). This type of repeating on–off switching has not been observed for any other DNA polymerase, and perhaps provides a way for E. coli to protect undamaged DNA from gratuitous mutation, enabling the cell to activate Pol V Mut whenever replication forks have stalled at DNA template damage sites.
POL V ORTHOLOGS
E. coli Pol V has been the subject of intense study, but many orthologs are now known to exist in a wide range of prokaryotes and self-transmissible plasmids that they harbor (Vaisman et al. 2012). Of particular interest are Pol V orthologs MucA′B and RumA′B found on the IncN R-plasmid R46/pKM101 (Perry and Walker 1982) and integrating conjugative element (ICE) R391 (Kulaeva et al. 1995), respectively. The ability of MucA′B to increase mutagenesis in strains of Salmonella typhimurium exposed to a variety of DNA-damaging agents plays an important role in increasing the efficacy of the “Ames test” that was proposed as a possible way to identify human carcinogens (McCann et al. 1975). Although the MucA′B proteins appear to be significantly more mutagenic than their E. coli counterparts, the RumA′B proteins are the most potent at promoting cellular mutagenesis than any other Pol V ortholog identified so far (Mead et al. 2007). This is especially worrisome, because the rum genes are found on natural isolates of the STX/R391 ICE family of mobile elements that are involved in antibiotic resistance spread among γ-proteobacteria, including virulent strains of Vibrio cholera and Proteus mirabilis, and their ability to provide genetic diversification to the host bacterium may pose a significant risk to human health (Beaber et al. 2004).
E. coli TLS POL II AND POL IV
Special mention should be made regarding Pol II in relation to SOS and TLS. The discovery in 1970 of Pol II (Knippers 1970) in the De Lucia and Cairns polA mutant (De Lucia and Cairns 1969) that had a severely compromised Pol I protein was followed shortly thereafter by the discovery of Pol III (Gefter et al. 1971). The isolation of a conditionally lethal temperature-sensitive Pol III mutant showed that Pol III was absolutely required for E. coli survival (Gefter et al. 1971), acting as the principal replicative polymerase responsible for copying almost the entire bacterial genome (Johnson and O’Donnell 2005; McHenry 2011).
Mutations in Pol II, however, proved to be non-informative, at least not until relatively recently (Foster et al. 1995; Rangarajan et al. 1997; Frisch et al. 2010). Pol II was “rediscovered” in 1988, as a polymerase activity isolated from UV-irradiated cells that could replicate past abasic template lesions (Bonner et al. 1988). This activity was induced sevenfold in response to UV radiation that depended on expression of the lexA gene. It became evident upon purification that the induced lesion-copying protein was Pol II (Bonner et al. 1988) and that the DNA damage-inducible dinA gene identified in 1980 by Kenyon and Walker (Kenyon and Walker 1980) encodes Pol II (Bonner et al. 1990; Iwasaki et al. 1990) and is allelic with the polB gene that had been identified in 1970 (Qiu and Goodman 1997). In addition to bypassing abasic sites, Pol II appears adept at bypassing N2-deoxyguanosine-acetyl aminofluorene (AAF) adducts in an error-prone manner that produces −2 frameshift mutations (Table 1) (Fuchs and Fujii 2007).
The dinB gene was identified by Kenyon and Walker in 1980 as being induced as part of the cellular SOS response to DNA damage (Kenyon and Walker 1980). However, its function remained largely unknown for many years aside from a paper in 1986 implicating a role in producing “untargeted” mutagenesis of bacteriophage λ (Brotcorne-Lannoye and Maenhaut-Michel 1986). Some 15 years after the dinB locus was first identified, Ohmori et al. (1995) discovered a damage-inducible gene, dinP, in E. coli that mapped to the same general region of the E. coli chromosome as dinB. A short time thereafter, the dinB/P genes were also shown to be allelic (Kim et al. 1997) and encode the Y-family DNA Pol IV (Wagner et al. 1999). DinB orthologs are the most ubiquitous of Y-family polymerases, because they are found in all domains of life (Ohmori et al. 2001). However, like many other TLS polymerases, they are not absolutely essential for life, and some organisms (such as Saccharomyces cerevisiae) lack any obvious dinB ortholog. DinB’s role in TLS was largely overlooked because it is not highly mutagenic (unlike Pol V). Indeed, Pol IV appears to bypass certain N2-dG adducts (such as N2-furfuryl-dG) in an error-free manner (Table 1) (Jarosz et al. 2006) and can even copy past N2-N2-guanine interstrand cross-links with high fidelity (Kumari et al. 2008).
TLS POLYMERASES IN ARCHAEA AND EUKARYOTES
Ever since the discovery of “reversionless” (REV) strains of S. cerevisiae (Lemontt 1971) and non-mutable (umu/uvm) strains of E. coli (Kato and Shinoura 1977; Steinborn 1978) in the 1970s, it was known that cellular mutagenesis is not a passive process but instead required the active participation of certain so-called mutagenesis proteins, which we now know are bona fide DNA polymerases. Over the years, the number of mutagenesis proteins discovered increased steadily, and with the advent of whole-genome sequencing projects, orthologs were identified at an ever-increasing pace. By the mid 1990s, orthologs of the Rev1, Rev3, UmuC, and DinB proteins had been identified in bacteria, archaea, and eukaryotes.
The first archaeal ortholog was reported in the literature in 1996, as a DinB homolog (Dbh) from what was originally believed at the time to be Sulfolobus solfataricus (Table 1) (Kulaeva et al. 1996). However, analysis of the sequenced S. solfataricus genome (She et al. 2001) identified a different DinB ortholog (Dpo4), which is only 53% identical to the Dbh protein (Boudsocq et al. 2001). Subsequent studies suggest that the original dbh gene appears to be from the closely related organism Sulfolobus acidocaldarius instead of S. solfataricus (Chen et al. 2005; McDonald et al. 2006). Both archaeal Dbh and Dpo4 proteins have proven extremely useful for subsequent structure–function studies into TLS (Table 1) (Ling et al. 2001, 2003, 2004a,b; Silvian et al. 2001; Zhou et al. 2001; Boudsocq et al. 2004; Yang and Woodgate 2007; Wilson and Pata 2008; Pata 2010) (see below).
S. cerevisiae RAD30 was identified in 1997, as an ortholog of E. coli UmuC and DinB proteins that is involved in error-free repair of UV damage (McDonald et al. 1997; Roush et al. 1998). In 1999, the RAD30 gene was subsequently shown to encode Pol η, which can bypass a T-T CPD efficiently and with the same accuracy as undamaged DNA (Johnson et al. 1999b). A human ortholog of S. cerevisiae Pol η was also identified in 1999 and shown to possess similar properties to the yeast enzyme (Masutani et al. 1999a; Johnson et al. 2000b; Matsuda et al. 2001), but perhaps more importantly, was found to be defective in humans with the sunlight-sensitive and cancer-prone xeroderma pigmentosum variant (XP-V) syndrome (Johnson et al. 1999a; Masutani et al. 1999a,b). A second Rad30 ortholog (McDonald et al. 1999) and E. coli DinB ortholog (Gerlach et al. 1999; Ogi et al. 1999) were identified in humans and mice and were subsequently shown to encode DNA polymerases ι and κ, respectively (Johnson et al. 2000a; Ohashi et al. 2000; Tissier et al. 2000; Gerlach et al. 2001; Zhang et al. 2001). Thus, within a period of a roughly 18 mo, a significant number of the phylogenetically related “mutagenesis proteins,” which were once thought to be merely accessory factors to replicative polymerases, evolved into the “Y-family” of DNA polymerases (Ohmori et al. 2001) that can perform unassisted TLS.
To date, only defects in Pol η have been associated with a human disease (XP-V), and this phenotype has been recapitulated in mouse models (Dumstorf et al. 2006; Ohkumo et al. 2006). A loss of Pol η function is also responsible for the absence of mutations at A-T sites in immunoglobulin variable regions during somatic hypermutation (Rogozin et al. 2001; Zeng et al. 2001). Thus, Pol η is responsible for copying damaged DNA (i.e., pyrimidine dimers) accurately, in the avoidance of skin cancer, and for copying undamaged DNA (i.e., Ig V-gene repair gaps) inaccurately, to generate mutations that contribute to antibody diversity (Peled et al. 2008). Mice with defects in Pol η, Pol ι, or Rev1 are viable but do not show any gross phenotypic changes (Ogi et al. 2002; Schenten et al. 2002; McDonald et al. 2003; Jansen et al. 2006). In contrast, homozygous inactivation of murine Rev3 (catalytic subunit of Pol ζ) is embryonic lethal, suggesting that Pol ζ plays additional cellular roles besides TLS (Bemark et al. 2000; Esposito et al. 2000; Wittschieben et al. 2000, 2006). Distinct roles for Pol ζ is supported by the recent discovery of a four-subunit complex, Pol ζ4 (Rev3-Rev7-Pol31-Pol32) (Baranovskiy et al. 2012; Johnson et al. 2012; Makarova et al. 2012), where the binding of the Pol31 and Pol32 accessory subunits of Pol δ are required for damaged-induced mutagenesis in yeast (Johnson et al. 2012; Makarova et al. 2012).
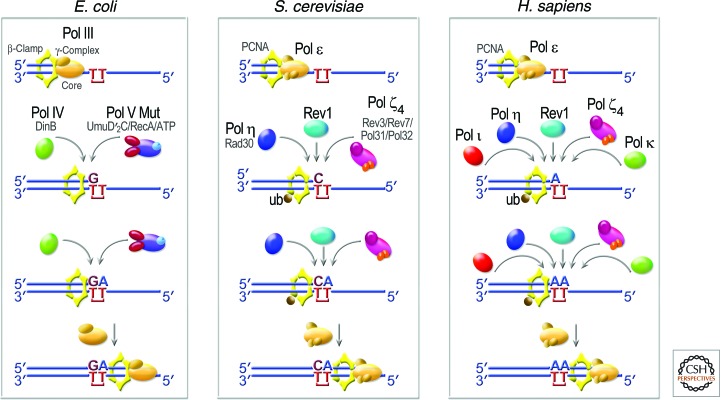
The molecular mechanism of TLS appears to be conserved from E. coli to humans (Fig. 2). Similar to the original TLS model proposed by Bridges and Woodgate in 1985, it is generally thought to occur in two kinetically separable steps: misincorporation followed by extension. In principle, a single TLS enzyme (such as E. coli Pol V or human Pol η) can facilitate both steps. However, it is also evident that some TLS polymerases are more proficient at extension (such as Pol η and Pol ζ) than incorporation; thus, TLS not only occurs in two steps but may also require two polymerases for complete lesion bypass to occur (Fig. 2). Support for the proposed polymerase switching model comes from biochemical studies that reveal a dynamic exchange process in which E. coli Pol IV (Indiani et al. 2005, 2009; Furukohri et al. 2008) and Pol II (Indiani et al. 2009) can replace Pol III on the β-sliding clamp at a stalled replication fork, with a subsequent resumption of rapid DNA synthesis following the displacement of Pol IV and Pol II by Pol III.
Figure 2.
Evolutionarily conserved roles of TLS polymerases. Replicative polymerases such as E. coli Pol III or eukaryotic Pol ε stall at the site of a DNA lesion. TLS polymerases are recruited to the site via interactions with the replicative sliding processivity clamp (β-subunit in E. coli and PCNA in eukaryotes). E. coli has a choice of three TLS polymerases: Pol II, Pol IV, and Pol V Mut (composed of UmuD′2C-RecA-ATP) (Jiang et al. 2009). S. cerevisiae also has a choice of three TLS polymerases: Pol ζ4, Rev1, and Pol η. In humans, at least five TLS polymerases can be recruited to sites of arrested replication, including Pol η, Pol ι, Pol κ, Rev1, and Pol ζ4. Additional human DNA polymerases, such as Pol β, Pol λ, Pol θ, and/or Pol ν (Maga et al. 2007; Seki and Wood 2008; Shtygasheva et al. 2008; Yamanaka et al. 2010; Hogg et al. 2011; Villani et al. 2011), may also facilitate TLS under certain conditions, but these have been omitted for clarity. The likelihood that this insertion step is error-prone, as shown for E. coli and S. cerevisiae, or error-free, as shown for humans, will depend on the DNA lesion encountered and the polymerase used for TLS. The extension step may be facilitated by the same enzyme that performed the (mis)insertion or by a different polymerase. Once the nascent DNA chain has been extended beyond the lesion, the TLS polymerase is replaced by the cell’s replicative DNA polymerase so as to complete genome duplication.
STRUCTURAL INSIGHTS INTO TLS AND MUTAGENIC SPECIFICITY
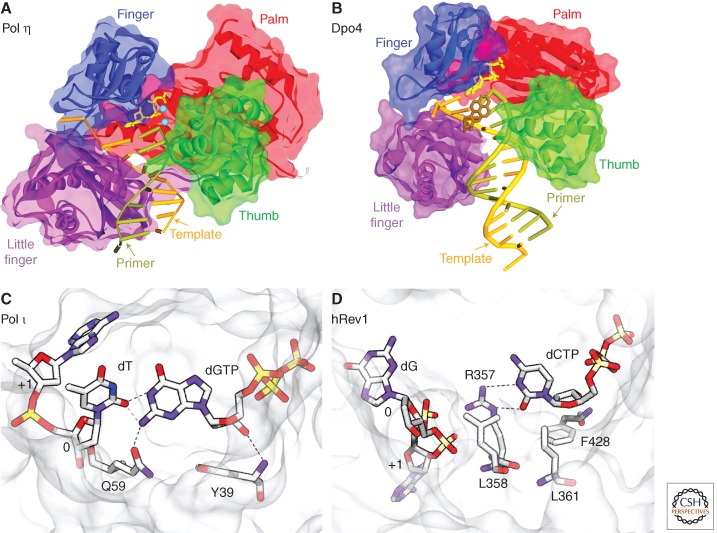
Probably the biggest surprise to researchers studying TLS was the fact that the mutagenesis proteins turned out to be bona fide (Y-family) DNA polymerases, because the primary amino acid sequence of the proteins show virtually no sequence homology to DNA polymerases from other families. However, quite remarkably, crystallographical studies of archaeal Dbh and Dpo4 revealed a topology similar to replicases, including domains likened to a right-hand with palm, thumb, and fingers subdomains (Ling et al. 2001; Silvian et al. 2001; Zhou et al. 2001). The TLS polymerases also have an additional domain that has been called the little finger (to coincide with the analogy to a hand) (Ling et al. 2001) or polymerase-associated domain (Fig. 3A,B) (Trincao et al. 2001). As with high-fidelity replicases, residues required for catalysis are located in the palm domain of the Y-family polymerases. In contrast to the replicases, the remaining domains are much smaller and are stubby.
Figure 3.
Structural insights into TLS polymerases and their mutagenic specificity. In panels A and B, the main domains are color-coded: (red) palm; (green) thumb; (blue) fingers; (purple) little finger. (A) Crystal structure of human Pol η in a ternary complex with a CPD. In this view, the 3′T of the CPD is in the active site and is correctly paired with incoming dATP (PDB: 3MR3) (Biertümpfel et al. 2010). (Rust) The template strand; (olive green) the primer; (yellow) the incoming dNTP. (Burgundy stick) The position of the CPD; (small blue spheres) the metal ions. The protein backbone is represented by the ribbon surrounded by the semitransparent solvent-accessible surface. (B) Crystal structure of the S. solfataricus Dpo4 in a ternary complex with DNA containing a benzo[a]pyrene lesion (brown) and incoming nucleotide (yellow) (PDB: 1S0M_BP-2) (Ling et al. 2004b). As can be seen, the benzo[a]pyrene lesion is flipped into the major groove, so as to accommodate base pairing. (C) Human Pol ι making a G:T mispair (PDB:3GV8); note that the template dT and incoming dG are both in an anti conformation (Kirouac and Ling 2009) and the mispair is stabilized through hydrogen bonds with Gln59. (D) Arg357-directed dC incorporation by human Rev1 (PDB: 3GQC) (Swan et al. 2009). (C,D) (White/gray) The protein surface; (dotted lines) hydrogen bonds.
As a consequence, the active site of the Y-family TLS polymerases is much more spacious and solvent exposed (Ling et al. 2001), and it can often accommodate large bulky DNA lesions that are unable to fit into the tighter constraints of the high-fidelity replicases. Based on a thermodynamic model, it has been proposed that high-fidelity Pols enhance WC base-pairing specificities by partially excluding bulk water from the Pol active cleft (Petruska et al. 1986; Petruska and Goodman 1995). By having a more open architecture that enables water to compete with nucleobases in forming H-bonds, the error-prone Pols generally show less ability to discriminate between forming WC and non-WC base pairs. Having said that, “not all Y-family TLS polymerases are created equal.” Pol η appears to have evolved to bypass CPDs efficiently and accurately. It does so by acting as a “molecular splint” straightening the normally kinked damaged DNA backbone, so that the covalently linked pyrimidines of the CPD can be “read” and accurately copied (Fig. 3A) (Biertümpfel et al. 2010).
Structural studies have also revealed that key residues within the active sites of Pol ι and Rev1 contribute to their unique mutagenic “signature.” For example, Pol ι is best characterized in vitro for its ability to misincorporate dG opposite template T threefold to 10-fold better than the normal base dA (Table 1) (Johnson et al. 2000a; Tissier et al. 2000; Zhang et al. 2000). This is in contrast to efficient and accurate (for a Y-family polymerase) incorporation of T opposite dA (Johnson et al. 2000a; Tissier et al. 2000; Zhang et al. 2000).
Such diverse properties can be explained by the presence of amino acids (most notably Gln59) of the finger domain that protrude into the active-site cavity and direct the orientation of the templating base. In the case of the G:T mispair, the template base, dT, is always held in an anti conformation irrespective of the incoming dNTP. The mispaired dG also adopts an anti conformation and is further stabilized by hydrogen bonds with Gln59 (Fig. 3C) (Kirouac and Ling 2009). In contrast to a dT, a template dA is driven into a syn conformation by the incoming dTTP, which remains in an anti conformation. As a consequence, base pairing is limited so that dA makes a Hoogsteen, rather than WC base pair with the incoming dTTP (Nair et al. 2004, 2006). Similarly, Rev1 is characterized as a dCMP transferase (Nelson et al. 1996a), but this specificity is not dependent on bonding of the incoming dCMP with a template base, but rather is determined by a particular arginine residue (K324 in S. cerevisiae and K357 in humans) of the little finger domain of the Rev1 protein (Fig. 3D) (Nair et al. 2005; Swan et al. 2009).
Structural studies with archaeal Dpo4 have been particularly enlightening because the protein readily crystallizes in a ternary complex with an incoming nucleotide and lesion-containing template DNA. There are more than 60 different structures in the PDB database with individual “snapshots” of Dpo4 in the process of facilitating TLS (Vaisman et al. 2012). What is evident is that the enzyme can perform a wide range of molecular gymnastics with either the template base or incoming nucleotide so as to allow TLS to occur. An example is shown in Figure 3B, in which a large bulky benzo[a]pyrene adduct linked to dA is flipped into the major groove of the template DNA, so as to allow base pairing of the adducted dA with the incoming T.
Last, we mention E. coli Pol II. The apo structure of Pol II was first reported in 1994 (Anderson et al. 1994), but only recently has the ternary complex with template DNA and incoming nucleotide been solved. It appears that Pol II contains many molecular “nooks and crannies” near the active site of the polymerase that allow for manipulation of the lesion-containing template DNA (Wang and Yang 2009). In the case of an N2-dG-AAF adduct, the template loops out in such a manner that two bases are skipped, leading to the −2 frameshift mutagenesis that is a characteristic of Pol II bypass of AAF adducts (Fuchs and Fujii 2007).
REGULATION OF TLS POLYMERASES
The ability to traverse a wide array of DNA lesions often comes at the price of reduced fidelity when replicating undamaged DNA. With the exception of E. coli Pol II, which has intrinsic 3′–5′ exonucleolytic proofreading, none of the other TLS polymerases has the capacity to excise errors made when duplicating an undamaged DNA template. It is therefore not surprising that cells use several mechanisms to keep the error-prone TLS polymerases in check, so that they are only used as, and when required. Perhaps the best-studied example is in E. coli, where the three TLS polymerases are first negatively regulated at the transcriptional level by the LexA repressor. Although no additional regulation of Pol II is required because of its high fidelity ensured by intrinsic proofreading, the Y-family polymerases, Pol IV and Pol V, are exonuclease deficient and are subject to a variety of posttranslational controls.
Most notably, the UmuD′ subunit of Pol V has to be activated via damage-inducible cleavage of UmuD (Nohmi et al. 1988), and the UmuD, UmuD′, and UmuC proteins are all subject to rapid proteolytic degradation by the Lon and ClpXP proteases (Frank et al. 1996; Gonzalez et al. 1998, 2000; Neher et al. 2003). Any UmuD′2C complex that escapes degradation is essentially catalytically inactive until activated through protein interactions with RecA* to form Pol V Mut, whose activity is enhanced by binding to the β-clamp (Schlacher et al. 2006; Karata et al. 2012). Pol IV activity is enhanced through protein interactions with UmuD, RecA, and the β-clamp (Wagner et al. 2001; Becherel et al. 2002; Godoy et al. 2007).
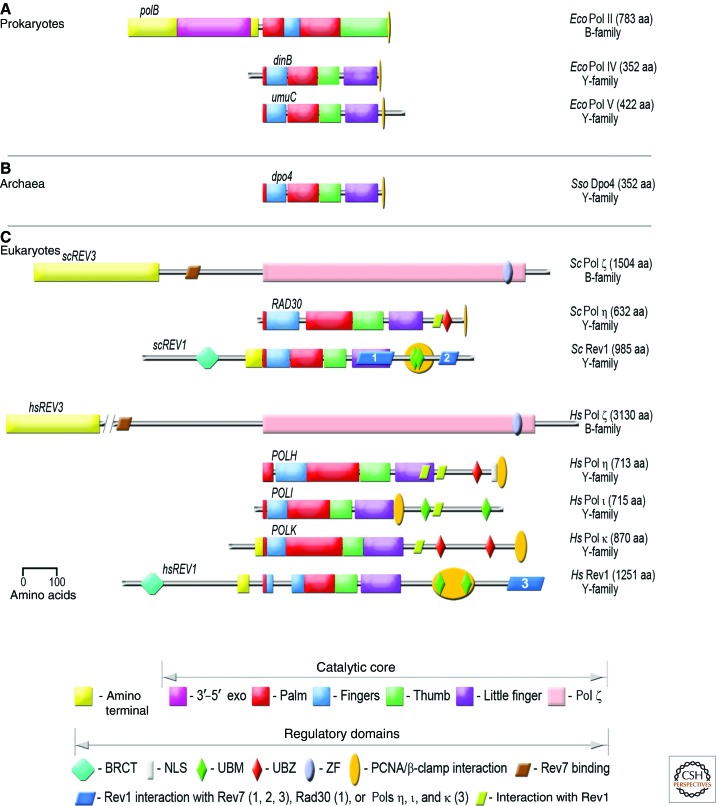
Although the S. cerevisiae RAD30 (Pol η) transcript appears to be induced roughly threefold after DNA damage (McDonald et al. 1997), much of the regulation of TLS polymerases in eukaryotes appears to rely on their posttranslational modification and/or specific protein–protein interactions that target them to the site of DNA damage. A key player in this process is ubiquitin. In S. cerevisiae, ubiquitination of Rad30 (Pol η) leads to its rapid degradation by the proteasome (Skoneczna et al. 2007). Similarly, ubiquitination of human Pol η by the E3 ubiquitin ligase Pirh2 results in its degraded by the 20S proteasome (Jung et al. 2010). In addition to ubiquitination via conventional E3 ligases, the human Y-family polymerases can undergo self-ubiquitination that is mediated by their UBMs (found in Pol ι or Rev1) or UBZ motifs (found in Pol η and Pol κ) (Fig. 4) (Bienko et al. 2005, 2010; Guo et al. 2006; Plosky et al. 2006; Jung et al. 2011). These motifs bind ubiquitin in a noncovalent manner and induce a conformational change in the TLS polymerase that prevents any subsequent interaction with other ubiquitinated proteins, such as PCNA. As a consequence, the enzymes are no longer correctly targeted to sites of DNA damage, which precludes their ability to facilitate TLS (Bienko et al. 2005, 2010; Plosky et al. 2006).
Figure 4.
Domain organization of selected TLS polymerases. (A) Prokaryotes; (B) archaea; (C) eukaryotes. The name and phylogenetic family relationship of each of the TLS polymerases along with number of amino acid residues in each polymerase are indicated on the right-hand side of the figure. The structural catalytic domains present in all of the polymerases are color-coded as follows: (red) palm; (green) thumb; (blue) finger; (purple) little finger; (violet) 3′–5′ exonuclease of Pol II; (yellow) amino-terminal domain of Pol II and Rev3; (pink) uncharacterized catalytic domain of Rev3. Additional domains involved in localization and regulation of the TLS polymerases are as follows: (teal octagon) breast cancer-associated protein-1 carboxy-terminal domain (BRCT); (gray rectangle) nuclear localization signal (NLS); (green diamond) ubiquitin binding motif (UBM); (red diamond) ubiquitin binding zinc-finger motif (UBZ); (grayish oval) zinc finger (ZF); (gold oval) PCNA/β-clamp motif; (brown or blue rhomboid) Rev7 binding region; (olive rhomboid) Rev1-interacting region. Species: Ec, E. coli; Ss, S. solfataricus; Sc, S. cerevisiae; Hs, H. sapiens.
The eukaryotic Y-family TLS polymerases are also regulated through key protein–protein interactions. One such interaction is with the replicative clamp, PCNA. The Y-family polymerases possess specific PCNA-binding motifs (PIP-box) that allow for a direct interaction with the clamp in a manner similar to β-clamp binding by the bacterial polymerases (Fig. 4) (Warbrick 1998, 2000; Dalrymple et al. 2001). Upon DNA damage, PCNA is monoubquitinated (Hoege et al. 2002), and this further strengthens the interaction with the TLS polymerases because they bind to the monoubiquitinated PCNA via the PIP box and the UBM/UBZ motifs (Bienko et al. 2005, 2010; Guo et al. 2006; Plosky et al. 2006). Another important protein interaction regulating the eukaryotic TLS polymerases is with the Rev1 protein. In addition to its catalytic dCMP transferase activity, Rev1 plays a crucial structural function by interacting with Pol ζ (Lawrence and Hinkle 1996; Nelson et al. 1996b) and with Pol η, Pol ι, and Pol κ (Fig. 4) (Guo et al. 2003; Ohashi et al. 2004; Tissier et al. 2004) and targeting them to sites of DNA damage.
CONCLUDING REMARKS: IS TLS PERHAPS THE TIP OF THE ICEBERG?
Although the TLS polymerases are best characterized by their ability to traverse lesions that would otherwise block replicases, it is becoming increasingly obvious that they also participate in other cellular pathways. In some cases, such as Pol IV–dependent stress-induced mutagenesis in E. coli (Bull et al. 2001; Tompkins et al. 2003) or Pol η/Rev1-dependent somatic hypermutation of immunoglobulin genes in humans (Rogozin et al. 2001; Zeng et al. 2001), this occurs during error-prone replication of undamaged DNA and generates various mutations, the ultimate source of genetic diversity (Peled et al. 2008). However, there is also evidence that human Pol κ participates in nucleotide excision repair (Ogi et al. 2010) and human Pol η in recombinational repair (Kawamoto et al. 2005; McIlwraith et al. 2005), both of which are thought to be essentially error-free processes.
The heavy emphasis that has been accorded as principal roles for SOS E. coli Pols, and more generally for eukaryotic TLS Pols, namely, copying damaged DNA templates, has to a large extent ignored what could well turn out to be an even more important biological function in long-term survival and evolutionary fitness in the absence of exogenous DNA damage. In E. coli, eliminating any one of the E. coli SOS Pols (Pol II, IV, or V) causes a severe loss of cell fitness, so that when wild-type cells are grown in the presence of cells lacking either Pol II, Pol IV, or Pol V, the wild-type cells take over as 100% of the population of surviving cells, thus driving each of the strains lacking one of the SOS Pols to extinction within ∼6–7 days of continuous growth in rich medium (Yeiser et al. 2002). For this relative fitness determination, wild-type and mutant cells compete for nutrient resources when both are inoculated at low but equal population densities (∼102–103 cells/mL) and undergo co-outgrowth while growing exponentially. Thus, when competing for nutrient resources that are diminishing with time, the elimination of any one of the SOS Pols is sufficient to ensure the mutant’s eventual demise when competing against wild-type E. coli.
Our understanding of the molecular mechanisms of TLS has increased dramatically in the past few years, but there is still much to be learned. Although we now know of several regulatory pathways that target the TLS polymerase sites of DNA damage, how the physical switch between the replicative and TLS polymerases occurs in a living cell is still largely unknown, and insights into this process should be forthcoming with the advent of single-molecule visualization of DNA polymerases at replication forks in vivo (Reyes-Lamothe et al. 2010; Lia et al. 2012). Crystal structures of the catalytic domains of the eukaryotic TLS polymerase have been reported along with isolated PIP-box and UBM domains (Bomar et al. 2007, 2010; Hishiki et al. 2009), but to date, there have been no reports of a full-length eukaryotic TLS polymerase. Indeed, such a structure would provide a major advance by allowing us to visualize how the various regulatory domains interact with the catalytic core of the TLS polymerase. It is evident that the TLS polymerases are regulated through a multitude of direct protein–protein interactions and structures of such multiprotein complexes as Pol V Mut (UmuD′2C-RecA-ATP) (Jiang et al. 2009) and Pol ζ4 (Rev3-Rev7-Pol31-Pol32) (Baranovskiy et al. 2012; Nelson et al. 1996b; Makarova et al. 2012) are critical to further our understanding of TLS.
ACKNOWLEDGMENTS
We thank Alexandra Vaisman (National Institutes of Health/National Institute of Child Health and Human Development) for the graphic artwork depicted in this review. R.W. is supported by funds from the NIH/NICHD Intramural Research Program and M.F.G. by NIH grants GM21422 and ES012259.
Footnotes
Editors: Stephen D. Bell, Marcel Méchali, and Melvin L. DePamphilis
Additional Perspectives on DNA Replication available at www.cshperspectives.org
REFERENCES
- Anderson WF, Prince DB, Yu H, McEntee K, Goodman MF 1994. Crystallization of DNA polymerase II from Escherichia coli. J Mol Biol 238: 120–122 [DOI] [PubMed] [Google Scholar]
- Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, Tahirov TH 2012. DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ. J Biol Chem 287: 17281–17287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaber JW, Hochhut B, Waldor MK 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427: 72–74 [DOI] [PubMed] [Google Scholar]
- Bebenek K, Kunkel TA 2004. Functions of DNA polymerases. Adv Protein Chem 69: 137–165 [DOI] [PubMed] [Google Scholar]
- Becherel OJ, Fuchs RP, Wagner J 2002. Pivotal role of the β-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair 1: 703–708 [DOI] [PubMed] [Google Scholar]
- Bemark M, Khamlichi AA, Davies SL, Neuberger MS 2000. Disruption of mouse polymerase ζ (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr Biol 10: 1213–1216 [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. 2005. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310: 1821–1824 [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Sabbioneda S, Crosetto N, Matic I, Hibbert RG, Begovic T, Niimi A, Mann M, Lehmann AR, et al. 2010. Regulation of translesion synthesis DNA polymerase η by monoubiquitination. Mol Cell 37: 396–407 [DOI] [PubMed] [Google Scholar]
- Biertümpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W 2010. Structure and mechanism of human DNA polymerase η. Nature 465: 1044–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomar MG, Pai MT, Tzeng SR, Li SS, Zhou P 2007. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase η. EMBO Rep 8: 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomar MG, D’Souza S, Bienko M, Dikic I, Walker GC, Zhou P 2010. Unconventional ubiquitin recognition by the ubiquitin-binding motif within the Y family DNA polymerases ι and Rev1. Mol Cell 37: 408–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner CA, Randall SK, Rayssiguier C, Radman M, Eritja R, Kaplan BE, McEntee K, Goodman MF 1988. Purification and characterization of an inducible Escherichia coli DNA polymerase capable of insertion and bypass at abasic lesions in DNA. J Biol Chem 263: 18946–18952 [PubMed] [Google Scholar]
- Bonner CA, Hays S, McEntee K, Goodman MF 1990. DNA polymerase II is encoded by the DNA damage-inducible dinA gene of Escherichia coli. Proc Natl Acad Sci 87: 7663–7667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq F, Campbell M, Devoret R, Bailone A 1997. Quantitation of the inhibition of Hfr × F− recombination by the mutagenesis complex UmuD′C. J Mol Biol 270: 201–211 [DOI] [PubMed] [Google Scholar]
- Boudsocq F, Iwai S, Hanaoka F, Woodgate R 2001. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): An archaeal DNA polymerase with lesion-bypass properties akin to eukaryotic polη. Nucleic Acids Res 29: 4607–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq F, Kokoska RJ, Plosky BS, Vaisman A, Ling H, Kunkel TA, Yang W, Woodgate R 2004. Investigating the role of the little finger domain of Y-family DNA polymerases in low-fidelity synthesis and translesion replication. J Biol Chem 279: 32932–32940 [DOI] [PubMed] [Google Scholar]
- Bridges BA, Woodgate R 1984. Mutagenic repair in Escherichia coli, X. The umuC gene product may be required for replication past pyrimidine dimers but not for the coding error in UV mutagenesis. Mol Gen Genet 196: 364–366 [DOI] [PubMed] [Google Scholar]
- Bridges BA, Woodgate R 1985a. Mutagenic repair in Escherichia coli: Products of the recA gene and of the umuD and umuC genes act at different steps in UV-induced mutagenesis. Proc Natl Acad Sci 82: 4193–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges BA, Woodgate R 1985b. The two-step model of bacterial UV mutagenesis. Mutat Res 150: 133–139 [DOI] [PubMed] [Google Scholar]
- Brotcorne-Lannoye A, Maenhaut-Michel G 1986. Role of RecA protein in untargeted UV mutagenesis of bacteriophage λ: Evidence for the requirement for the dinB gene. Proc Natl Acad Sci 83: 3904–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruck I, Woodgate R, McEntee K, Goodman MF 1996. Purification of a soluble UmuD′C complex from Escherichia coli: Cooperative binding of UmuD′C to single-stranded DNA. J Biol Chem 271: 10767–10774 [DOI] [PubMed] [Google Scholar]
- Bull HJ, Lombardo MJ, Rosenberg SM 2001. Stationary-phase mutation in the bacterial chromosome: Recombination protein and DNA polymerase IV dependence. Proc Natl Acad Sci 98: 8334–8341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckhardt SE, Woodgate R, Scheuermann RH, Echols H 1988. UmuD mutagenesis protein of Escherichia coli: Overproduction, purification and cleavage by RecA. Proc Natl Acad Sci 85: 1811–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Brugger K, Skovgaard M, Redder P, She Q, Torarinsson E, Greve B, Awayez M, Zibat A, Klenk HP, et al. 2005. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J Bacteriol 187: 4992–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158: 41–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA 2001. A universal protein–protein interaction motif in the eubacterial DNA replication and repair systems. Proc Natl Acad Sci 98: 11627–11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia P, Cairns J 1969. Isolation of an E. coli strain with a mutation affecting DNA polymerase. Nature 224: 1164–1166 [DOI] [PubMed] [Google Scholar]
- Dumstorf CA, Clark AB, Lin Q, Kissling GE, Yuan T, Kucherlapati R, McGregor WG, Kunkel TA 2006. Participation of mouse DNA polymerase ι in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc Natl Acad Sci 103: 18083–18088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutreix M, Moreau PL, Bailone A, Galibert F, Battista JR, Walker GC, Devoret R 1989. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol 171: 2415–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols H, Goodman MF 1990. Mutation induced by DNA damage: A many protein affair. Mutat Res 236: 301–311 [DOI] [PubMed] [Google Scholar]
- Echols H, Goodman MF 1991. Fidelity mechanisms in DNA replication. Annu Rev Biochem 60: 477–511 [DOI] [PubMed] [Google Scholar]
- Esposito G, Godindagger I, Klein U, Yaspo M, Cumano A, Rajewsky K 2000. Disruption of the Rev3l-encoded catalytic subunit of polymerase ζ in mice results in early embryonic lethality. Curr Biol 10: 1221–1224 [DOI] [PubMed] [Google Scholar]
- Fernández de Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R 2000. Identification of additional genes belonging to the LexA-regulon in Escherichia coli. Mol Microbiol 35: 1560–1572 [DOI] [PubMed] [Google Scholar]
- Foster PL, Gudmundsson G, Trimarchi JM, Cai H, Goodman MF 1995. Proofreading-defective DNA polymerase II increases adaptive mutation in Escherichia coli. Proc Natl Acad Sci 92: 7951–7955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank EG, Ennis DG, Gonzalez M, Levine AS, Woodgate R 1996. Regulation of SOS mutagenesis by proteolysis. Proc Natl Acad Sci 93: 10291–10296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood R, Schultz RA, Ellenberger T 2006. DNA repair and mutagenesis. ASM Press, Washington, DC [Google Scholar]
- Frisch RL, Su Y, Thornton PC, Gibson JL, Rosenberg SM, Hastings PJ 2010. Separate DNA Pol II– and Pol IV–dependent pathways of stress-induced mutation during double-strand-break repair in Escherichia coli are controlled by RpoS. J Bacteriol 192: 4694–4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs RP, Fujii S 2007. Translesion synthesis in Escherichia coli: Lessons from the NarI mutation hot spot. DNA Repair 6: 1032–1041 [DOI] [PubMed] [Google Scholar]
- Furukohri A, Goodman MF, Maki H 2008. A dynamic polymerase exchange with Escherichia coli DNA polymerase IV replacing DNA polymerase III on the sliding clamp. J Biol Chem 283: 11260–11269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefter ML, Hirota Y, Kornberg T, Wechsler JA, Barnoux C 1971. Analysis of DNA polymerases II and III in mutants of Escherichia coli thermosensitive for DNA synthesis. Proc Natl Acad Sci 68: 3150–3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach VL, Aravind L, Gotway G, Schultz RA, Koonin EV, Friedberg EC 1999. Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc Natl Acad Sci 96: 11922–11927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach VL, Feaver WJ, Fischhaber PL, Friedberg EC 2001. Purification and characterization of polκ, a DNA polymerase encoded by the human DINB1 gene. J Biol Chem 276: 92–98 [DOI] [PubMed] [Google Scholar]
- Godoy VG, Jarosz DF, Simon SM, Abyzov A, Ilyin V, Walker GC 2007. UmuD and RecA directly modulate the mutagenic potential of the Y family DNA polymerase DinB. Mol Cell 28: 1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M, Frank EG, Levine AS, Woodgate R 1998. Lon-mediated proteolysis of the Escherichia coli UmuD mutagenesis protein: In vitro degradation and identification of residues required for proteolysis. Genes Dev 12: 3889–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M, Rasulova F, Maurizi MR, Woodgate R 2000. Subunit-specific degradation of the UmuD/D′ heterodimer by the ClpXP protease: The role of trans recognition in UmuD′ stability. EMBO J 19: 5251–5258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem 71: 17–50 [DOI] [PubMed] [Google Scholar]
- Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC 2003. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J 22: 6621–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC 2006. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol 26: 8892–8900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishiki A, Hashimoto H, Hanafusa T, Kamei K, Ohashi E, Shimizu T, Ohmori H, Sato M 2009. Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J Biol Chem 284: 10552–10560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Hogg M, Seki M, Wood RD, Doublie S, Wallace SS 2011. Lesion bypass activity of DNA polymerase θ (POLQ) is an intrinsic property of the Pol domain and depends on unique sequence inserts. J Mol Biol 405: 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiani C, McInerney P, Georgescu R, Goodman MF, O’Donnell M 2005. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell 19: 805–815 [DOI] [PubMed] [Google Scholar]
- Indiani C, Langston LD, Yurieva O, Goodman MF, O’Donnell M 2009. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc Natl Acad Sci 106: 6031–6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Nakata A, Walker GC, Shinagawa H 1990. The Escherichia coli polB gene, which encodes DNA polymerase II, is regulated by the SOS system. J Bacteriol 172: 6268–6273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N 2006. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med 203: 319–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC 2006. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 439: 225–228 [DOI] [PubMed] [Google Scholar]
- Jiang Q, Karata K, Woodgate R, Cox MM, Goodman MF 2009. The active form of DNA polymerase V is UmuD′2C-RecA-ATP. Nature 460: 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, O’Donnell M 2005. Cellular DNA replicases: Components and dynamics at the replication fork. Annu Rev Biochem 74: 283–315 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, Prakash L 1999a. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285: 263–265 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L 1999b. Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, polη. Science 283: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L 2000a. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406: 1015–1019 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Prakash S, Prakash L 2000b. Fidelity of human DNA polymerase η. J Biol Chem 275: 7447–7450 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash L, Prakash S 2012. Pol 31 and Pol 32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc Natl Acad Sci 109: 12455–12460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YS, Liu G, Chen X 2010. Pirh2 E3 ubiquitin ligase targets DNA polymerase η for 20S proteasomal degradation. Mol Cell Biol 30: 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YS, Hakem A, Hakem R, Chen X 2011. Pirh2 E3 ubiquitin ligase monoubiquitinates DNA polymerase η to suppress translesion DNA synthesis. Mol Cell Biol 19: 3997–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karata K, Vaisman A, Goodman MF, Woodgate R 2012. Simple and efficient purification of E. coli DNA polymerase V: Cofactor requirements for optimal activity and processivity in vitro. DNA Repair 11: 431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Shinoura Y 1977. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet 156: 121–131 [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, et al. 2005. Dual roles for DNA polymerase η in homologous DNA recombination and translesion DNA synthesis. Mol Cell 20: 793–799 [DOI] [PubMed] [Google Scholar]
- Kenyon CJ, Walker GC 1980. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci 77: 2819–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: An overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc Natl Acad Sci 94: 13792–13797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirouac KN, Ling H 2009. Structural basis of error-prone replication and stalling at a thymine base by human DNA polymerase ι. EMBO J 28: 1644–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knippers R 1970. DNA polymerase II. Nature 228: 1050–1053 [DOI] [PubMed] [Google Scholar]
- Kulaeva OI, Wootton JC, Levine AS, Woodgate R 1995. Characterization of the umu-complementing operon from R391. J Bacteriol 177: 2737–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva OI, Koonin EV, McDonald JP, Randall SK, Rabinovich N, Connaughton JF, Levine AS, Woodgate R 1996. Identification of a DinB/UmuC homolog in the archeon Sulfolobus solfataricus. Mutat Res 357: 245–253 [DOI] [PubMed] [Google Scholar]
- Kumari A, Minko IG, Harbut MB, Finkel SE, Goodman MF, Lloyd RS 2008. Replication bypass of interstrand cross-link intermediates by Escherichia coli DNA polymerase IV. J Biol Chem 283: 27433–27437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol Mol Biol Rev 63: 751–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange SS, Takata K, Wood RD 2011. DNA polymerases and cancer. Nat Rev Cancer 11: 96–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CW, Hinkle DC 1996. DNA polymerase ζ and the control of DNA damage induced mutagenesis in eukaryotes. In Cancer surveys: Genetic instability in cancer (ed. T Lindahl), pp. 21–31 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- Lemontt JF 1971. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics 68: 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LK, Harlow GR, Gregg-Jolly LA, Mount DW 1994. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J Mol Biol 241: 507–523 [DOI] [PubMed] [Google Scholar]
- Lia G, Michel B, Allemand JF 2012. Polymerase exchange during Okazaki fragment synthesis observed in living cells. Science 335: 328–331 [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Woodgate R, Yang W 2001. Crystal structure of a Y-family DNA polymerase in action: A mechanism for error-prone and lesion-bypass replication. Cell 107: 91–102 [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W 2003. Replication of a cis–syn thymine dimer at atomic resolution. Nature 424: 1083–1087 [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Woodgate R, Yang W 2004a. Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol Cell 13: 751–762 [DOI] [PubMed] [Google Scholar]
- Ling H, Sayer JM, Plosky BS, Yagi H, Boudsocq F, Woodgate R, Jerina DM, Yang W 2004b. Crystal structure of a benzo[a]pyrene diol epoxide adduct in a ternary complex with a DNA polymerase. Proc Natl Acad Sci 101: 2265–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JW, Edmiston SH, Pacelli LZ, Mount DW 1980. Cleavage of the Escherichia coli lexA protein by the recA protease. Proc Natl Acad Sci 77: 3225–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U 2007. 8-Oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature 447: 606–608 [DOI] [PubMed] [Google Scholar]
- Makarova AV, Stodola JL, Burgers PM 2012. A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res 10.1093/nar/gks948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F 1999a. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J 18: 3491–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F 1999b. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399: 700–704 [DOI] [PubMed] [Google Scholar]
- Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA 2001. Error rate and specificity of human and murine DNA polymerase η. J Mol Biol 312: 335–346 [DOI] [PubMed] [Google Scholar]
- McCann JC, Springarn NE, Kobari J, Ames BN 1975. Detection of carcinogens as mutagens: Bacterial tester strains with R factor plasmids. Proc Natl Acad Sci 72: 979–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JP, Levine AS, Woodgate R 1997. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147: 1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JP, Rapic-Otrin V, Epstein JA, Broughton BC, Wang X, Lehmann AR, Wolgemuth DJ, Woodgate R 1999. Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase η. Genomics 60: 20–30 [DOI] [PubMed] [Google Scholar]
- McDonald JP, Frank EG, Plosky BS, Rogozin IB, Masutani C, Hanaoka F, Woodgate R, Gearhart PJ 2003. Identification of a nonsense mutation in DNA polymerase ι from 129-derived strains of mice and its effect on somatic hypermutation. J Exp Med 198: 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JP, Hall A, Gasparutto D, Cadet J, Ballantyne J, Woodgate R 2006. Novel thermostable Y-family polymerases: Applications for the PCR amplification of damaged or ancient DNAs. Nucleic Acids Res 34: 1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry CS 2011. DNA replicases from a bacterial perspective. Annu Rev Biochem 80: 403–436 [DOI] [PubMed] [Google Scholar]
- McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC 2005. Human DNA polymerase η promotes DNA synthesis from strand invasion intermediates (D-loops) of homologous recombination. Mol Cell 20: 783–792 [DOI] [PubMed] [Google Scholar]
- Mead S, Vaisman A, Valjavec-Gratian M, Karata K, Vandewiele D, Woodgate R 2007. Characterization of polVR391: A Y-family polymerase encoded by rumA′B from the IncJ conjugative transposon, R391. Mol Microbiol 63: 797–810 [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK 2004. Replication by human DNA polymerase ι occurs by Hoogsteen base-pairing. Nature 430: 377–380 [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK 2005. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science 309: 2219–2222 [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK 2006. An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-ι active site. Structure 14: 749–755 [DOI] [PubMed] [Google Scholar]
- Neher SB, Sauer RT, Baker TA 2003. Distinct peptide signals in the UmuD and UmuD′ subunits of UmuD/D′ mediate tethering and substrate processing by the ClpXP protease. Proc Natl Acad Sci 100: 13219–13224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC 1996a. Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382: 729–731 [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC 1996b. Thymine–thymine dimer bypass by yeast DNA polymerase ζ. Science 272: 1646–1649 [DOI] [PubMed] [Google Scholar]
- Nohmi T, Battista JR, Dodson LA, Walker GC 1988. RecA-mediated cleavage activates UmuD for mutagenesis: Mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci 85: 1816–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T, Kato TJ, Kato T, Ohmori H 1999. Mutation enhancement by DINB1, a mammalian homologue of the Escherichia coli mutagenesis protein DinB. Genes Cells 4: 607–618 [DOI] [PubMed] [Google Scholar]
- Ogi T, Shinkai Y, Tanaka K, Ohmori H 2002. Polk protects mammalian cells against the lethal and mutagenic effects of polycyclic hydrocarbons. Proc Natl Acad Sci 99: 15548–15553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, et al. 2010. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell 37: 714–727 [DOI] [PubMed] [Google Scholar]
- Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H 2000. Error-prone bypass of certain DNA lesions by the human DNA polymerase κ. Genes Dev 14: 1589–1594 [PMC free article] [PubMed] [Google Scholar]
- Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H 2004. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells 9: 523–531 [DOI] [PubMed] [Google Scholar]
- Ohkumo T, Kondo Y, Yokoi M, Tsukamoto T, Yamada A, Sugimoto T, Kanao R, Higashi Y, Kondoh H, Tatematsu M, et al. 2006. UV-B radiation induces epithelial tumors in mice lacking DNA polymerase η and mesenchymal tumors in mice deficient for DNA polymerase ι. Mol Cell Biol 26: 7696–7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H, Hatada E, Qiao Y, Tsuji M, Fukuda R 1995. dinP, a new gene in Escherichia coli, whose product shows similarities to UmuC and its homologues. Mutat Res 347: 1–7 [DOI] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RPP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. 2001. The Y-family of DNA polymerases. Mol Cell 8: 7–8 [DOI] [PubMed] [Google Scholar]
- Pata JD 2010. Structural diversity of the Y-family DNA polymerases. Biochim Biophys Acta 1804: 1124–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M, Jiang Q, Woodgate R, Cox MM, Goodman MF 2010. A new model for SOS-induced mutagenesis: How RecA protein activates DNA polymerase V. Crit Rev Biochem Mol Biol 45: 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD 2008. The biochemistry of somatic hypermutation. Annu Rev Immunol 26: 481–511 [DOI] [PubMed] [Google Scholar]
- Perry KL, Walker GC 1982. Identification of plasmid (pKM101) coded proteins involved in mutagenesis and UV resistance. Nature 300: 278–281 [DOI] [PubMed] [Google Scholar]
- Petruska J, Goodman MF 1995. Enthalpy–entropy compensation in DNA melting thermodynamics. J Biol Chem 270: 746–750 [DOI] [PubMed] [Google Scholar]
- Petruska J, Sowers LC, Goodman MF 1986. Comparison of nucleotide interactions in water, proteins, and vacuum: Model for DNA polymerase fidelity. Proc Natl Acad Sci 83: 1559–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham P, Bertram JG, O’Donnell M, Woodgate R, Goodman MF 2001. A model for SOS-lesion targeted mutations in E. coli involving pol V, RecA, SSB and β sliding clamp. Nature 409: 366–370 [DOI] [PubMed] [Google Scholar]
- Plosky BS, Vidal A, Fernández de Henestrosa AR, McLenigan MP, McDonald JP, Mead S, Woodgate R 2006. Controlling the subcellular localization of DNA polymerases ι and η via interactions with ubiquitin. EMBO J 25: 2847–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Goodman MF 1997. The Escherichia coli polB locus is identical to dinA, the structural gene for DNA polymerase II. Characterization of Pol II purified from a polB mutant. J Biol Chem 272: 8611–8617 [DOI] [PubMed] [Google Scholar]
- Radman M 1974. Phenomenology of an inducible mutagenic DNA repair pathway in Escherichia coli: SOS repair hypothesis. In Molecular and environmental aspects of mutagenesis (ed. Prakash L, et al. ), pp. 128–142 Charles C. Thomas, Springfield, IL [Google Scholar]
- Rajagopalan M, Lu C, Woodgate R, O’Donnell M, Goodman MF, Echols H 1992. Activity of the purified mutagenesis proteins UmuC, UmuD′ and RecA in replicative bypass of an abasic DNA lesion by DNA polymerase III. Proc Natl Acad Sci 89: 10777–10781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan S, Gudmundsson G, Qiu Z, Foster PL, Goodman MF 1997. Escherichia coli DNA polymerase II catalyzes chromosomal and episomal DNA synthesis in vivo. Proc Natl Acad Sci 94: 946–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuven NB, Arad G, Maor-Shoshani A, Livneh Z 1999. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J Biol Chem 274: 31763–31766 [DOI] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Sherratt DJ, Leake MC 2010. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 328: 498–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA 2001. Somatic mutation hotspots correlate with DNA polymerase η error spectrum. Nat Immunol 2: 530–536 [DOI] [PubMed] [Google Scholar]
- Roush AA, Suarez M, Friedberg EC, Radman M, Siede W 1998. Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol Gen Genet 257: 686–692 [DOI] [PubMed] [Google Scholar]
- Sale JE, Lehmann AR, Woodgate R 2012. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol 13: 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenten D, Gerlach VL, Guo C, Velasco-Miguel S, Hladik CL, White CL, Friedberg EC, Rajewsky K, Esposito G 2002. DNA polymerase κ deficiency does not affect somatic hypermutation in mice. Eur J Immunol 32: 3152–3160 [DOI] [PubMed] [Google Scholar]
- Schlacher K, Goodman MF 2007. Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat Rev Mol Cell Biol 8: 587–594 [DOI] [PubMed] [Google Scholar]
- Schlacher K, Cox MM, Woodgate R, Goodman MF 2006. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature 442: 883–887 [DOI] [PubMed] [Google Scholar]
- Seki M, Wood RD 2008. DNA polymerase θ (POLQ) can extend from mismatches and from bases opposite a (6–4) photoproduct. DNA Repair 7: 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, et al. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci 98: 7835–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa H, Iwasaki H, Kato T, Nakata A 1988. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci 85: 1806–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtygasheva AA, Belousova EA, Rechkunova NI, Lebedeva NA, Lavrik OI 2008. DNA polymerases β and λ as potential participants of TLS during genomic DNA replication on the lagging strand. Biochemistry (Mosc) 73: 1207–1213 [DOI] [PubMed] [Google Scholar]
- Silvian LF, Toth EA, Pham P, Goodman MF, Ellenberger T 2001. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nat Struct Biol 8: 984–989 [DOI] [PubMed] [Google Scholar]
- Skoneczna A, McIntyre J, Skoneczny M, Policinska Z, Sledziewska-Gojska E 2007. Polymerase η is a short-lived, proteasomally degraded protein that is temporarily stabilized following UV irradiation in Saccharomyces cerevisiae. J Mol Biol 366: 1074–1086 [DOI] [PubMed] [Google Scholar]
- Sommer S, Bailone A, Devoret R 1993. The appearance of the UmuD′C protein complex in Escherichia coli switches repair from homologous recombination to SOS mutagenesis. Mol Microbiol 10: 963–971 [DOI] [PubMed] [Google Scholar]
- Steinborn G 1978. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol Gen Genet 165: 87–93 [DOI] [PubMed] [Google Scholar]
- Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK 2009. Structure of the human Rev1–DNA–dNTP ternary complex. J Mol Biol 390: 699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Bruck I, Eritja R, Turner J, Frank EG, Woodgate R, O’Donnell M, Goodman MF 1998. Biochemical basis of SOS-induced mutagenesis in Escherichia coli: Reconstitution of in vitro lesion bypass dependent on the UmuD′2C mutagenic complex and RecA. Proc Natl Acad Sci 95: 9755–9760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Shen X, Frank EG, O’Donnell M, Woodgate R, Goodman MF 1999. UmuD′2C is an error-prone DNA polymerase, Escherichia coli, DNA pol V. Proc Natl Acad Sci 96: 8919–8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A, McDonald JP, Frank EG, Woodgate R 2000. polι, a remarkably error-prone human DNA polymerase. Genes Dev 14: 1642–1650 [PMC free article] [PubMed] [Google Scholar]
- Tissier A, Kannouche P, Reck MP, Lehmann AR, Fuchs RP, Cordonnier A 2004. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase polη and REVl protein. DNA Repair 3: 1503–1514 [DOI] [PubMed] [Google Scholar]
- Tompkins JD, Nelson JL, Hazel JC, Leugers SL, Stumpf JD, Foster PL 2003. Error-prone polymerase, DNA polymerase IV, is responsible for transient hypermutation during adaptive mutation in Escherichia coli. J Bacteriol 185: 3469–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK 2001. Structure of the catalytic core of S. cerevisiae DNA polymerase η. Implications for translesion DNA synthesis. Mol Cell 8: 417–426 [DOI] [PubMed] [Google Scholar]
- Vaisman A, McDonald JP, Woodgate R 2012. Translesion DNA synthesis. In EcoSal—Escherichia coli and Salmonella: Cellular and molecular biology (ed. Böck A, Curtiss R III, Kaper JB, Karp PD, Neidhardt FC, Slauch JM, Squires CL). ASM Press, Washington, DC [Google Scholar]
- Villani G, Hubscher U, Gironis N, Parkkinen S, Pospiech H, Shevelev I, di Cicco G, Markkanen E, Syvaoja JE, Tanguy Le Gac N 2011. In vitro gap-directed translesion DNA synthesis of an abasic site involving human DNA polymerases ε, λ, and β. J Biol Chem 286: 32094–32104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, Fuchs RPP, Nohmi T 1999. The dinB gene encodes a novel Escherichia coli DNA polymerase, DNA Pol IV, involved in mutagenesis. Mol Cell 4: 281–286 [DOI] [PubMed] [Google Scholar]
- Wagner J, Fujii S, Gruz P, Nohmi T, Fuchs RP 2001. The β clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep 1: 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GC 1984. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev 48: 60–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Yang W 2009. Structural insight into translesion synthesis by DNA Pol II. Cell 139: 1279–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warbrick E 1998. PCNA binding through a conserved motif. Bioessays 20: 195–199 [DOI] [PubMed] [Google Scholar]
- Warbrick E 2000. The puzzle of PCNA's many partners. Bioessays 22: 997–1006 [DOI] [PubMed] [Google Scholar]
- Waters LS, Minesinger BK, Wiltrout ME, D’Souza S, Woodruff RV, Walker GC 2009. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Molec Biol Rev 73: 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FH 1953. Molecular structure of nucleic acids; A structure for deoxyribose nucleic acid. Nature 171: 737–738 [DOI] [PubMed] [Google Scholar]
- Weigle JJ 1953. Induction of mutation in a bacterial virus. Proc Natl Acad Sci 39: 628–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RC, Pata JD 2008. Structural insights into the generation of single-base deletions by the Y family DNA polymerase Dbh. Mol Cell 29: 767–779 [DOI] [PubMed] [Google Scholar]
- Witkin EM 1967. The radiation sensitivity of Escherichia coli B: A hypothesis relating filament formation and prophage induction. Proc Natl Acad Sci 57: 1275–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin EM 1976. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev 40: 869–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben J, Shivji MK, Lalani E, Jacobs MA, Marini F, Gearhart PJ, Rosewell I, Stamp G, Wood RD 2000. Disruption of the developmentally regulated REV3L gene causes embryonic lethality. Curr Biol 10: 1217–1220 [DOI] [PubMed] [Google Scholar]
- Wittschieben JP, Reshmi SC, Gollin SM, Wood RD 2006. Loss of DNA polymerase ζ causes chromosomal instability in mammalian cells. Cancer Res 66: 134–142 [DOI] [PubMed] [Google Scholar]
- Woodgate R, Rajagopalan M, Lu C, Echols H 1989. UmuC mutagenesis protein of Escherichia coli: Purification and interaction with UmuD and UmuD′. Proc Natl Acad Sci 86: 7301–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Minko IG, Takata K, Kolbanovskiy A, Kozekov ID, Wood RD, Rizzo CJ, Lloyd RS 2010. Novel enzymatic function of DNA polymerase ν in translesion DNA synthesis past major groove DNA–peptide and DNA–DNA cross-links. Chem Res Toxicol 23: 689–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Woodgate R 2007. What a difference a decade makes: Insights into translesion DNA synthesis. Proc Natl Acad Sci 104: 15591–15598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeiser B, Pepper ED, Goodman MF, Finkel SE 2002. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc Natl Acad Sci 99: 8737–8741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ 2001. DNA polymerase η is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol 2: 537–541 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Wu X, Wang Z 2000. Preferential incorporation of G opposite template T by the low-fidelity human DNA polymerase ι. Mol Cell Biol 20: 7099–7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Wu X, Taylor J-S, Wang Z 2001. Response of human DNA polymerase ι to DNA lesions. Nucleic Acids Res 29: 928–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Pata JD, Steitz TA 2001. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol Cell 8: 427–437 [DOI] [PubMed] [Google Scholar]