Abstract
Background and Aims
In habitat mosaics, plant populations face environmental heterogeneity over short geographical distances. Such steep environmental gradients can induce ecological divergence. Lowland rainforests of the Guiana Shield are characterized by sharp, short-distance environmental variations related to topography and soil characteristics (from waterlogged bottomlands on hydromorphic soils to well-drained terra firme on ferralitic soils). Continuous plant populations distributed along such gradients are an interesting system to study intrapopulation divergence at highly local scales. This study tested (1) whether conspecific populations growing in different habitats diverge at functional traits, and (2) whether they diverge in the same way as congeneric species having different habitat preferences.
Methods
Phenotypic differentiation was studied within continuous populations occupying different habitats for two congeneric, sympatric, and ecologically divergent tree species (Eperua falcata and E. grandiflora, Fabaceae). Over 3000 seeds collected from three habitats were germinated and grown in a common garden experiment, and 23 morphological, biomass, resource allocation and physiological traits were measured.
Key Results
In both species, seedling populations native of different habitats displayed phenotypic divergence for several traits (including seedling growth, biomass allocation, leaf chemistry, photosynthesis and carbon isotope composition). This may occur through heritable genetic variation or other maternally inherited effects. For a sub-set of traits, the intraspecific divergence associated with environmental variation coincided with interspecific divergence.
Conclusions
The results indicate that mother trees from different habitats transmit divergent trait values to their progeny, and suggest that local environmental variation selects for different trait optima even at a very local spatial scale. Traits for which differentiation within species follows the same pattern as differentiation between species indicate that the same ecological processes underlie intra- and interspecific variation.
Keywords: Eperua falcata, E. grandiflora, habitat mosaics, intrapopulation divergence, maternal family inheritance, common garden experiment, ecological traits
INTRODUCTION
Environmental variation occurring at the local scale creates complex habitat patchiness which has been found to contribute to shaping the great diversity observed in tropical rainforests (Ricklefs, 1977; Wright, 2002; Vincent et al., 2011). A common explanation for these diversity patterns is the divergence of preferences for edaphic conditions among tree species, as repeatedly shown throughout the Neotropics (e.g. ter Steege et al., 1993; Sabatier et al., 1997; Clark et al., 1999; Valencia et al., 2004; Baraloto et al., 2007; John et al., 2007; Kanagaraj et al., 2011). Community-level differences in functional traits have been found to underlie such differences (Kraft et al., 2008): for instance, Lopez et al. (2003) and Engelbrecht et al. (2007) showed that divergence in species distribution between hilltops and bottomlands is determined by variations in tolerance to drought and waterlogging.
It has been shown that bottomland, slope and hilltop habitats actually differ in many ways that may explain their impact on forest community composition. Generally speaking, water availability in lowland tropical forests is strongly associated with topography and soil characteristics (Sabatier et al., 1997). Large variations occur in soil drainage and moisture between hilltops, slopes and bottomlands (ter Steege et al., 1993; Clark et al., 1999; Webb and Peart, 2000). Bottomland soils are subject to frequent periods of flooding and undergo cyclical changes in O2 availability that strongly affect the metabolism of root tissues and thus tree establishment and growth (Ponnamperuma, 1972; Kozlowski, 1997; Perata et al., 2011). In contrast, thin soils on slopes undergo lateral drainage, which increases their susceptibility to water shortage during dry periods (Sabatier et al., 1997). Finally, hilltops are usually characterized by deep soils and display deep vertical drainage, with strong seasonal variations in soil water availability (Sabatier et al., 1997). Beside differences in water availability constraints, these habitats also differ in nutrient content, with lower nitrogen and higher phosphorus content in bottomlands than on plateaus (Luizao et al., 2004; Ferry et al., 2010). Moreover, soil respiration decreases in bottomlands as root biomass and soil carbon content decreases (Epron et al., 2006). These variations in soil characteristics have an additional impact on forest dynamics, with slopes and bottomlands exhibiting more frequent light gaps than hilltops, and therefore higher irradiance reaching the understorey (Ferry et al., 2010).
The widespread links between gradients of soil properties and species-specific habitat preferences suggest that ecological specialization has recurrently arisen through evolutionary processes such as adaptation and species divergence (Endler, 1977; Schluter, 2001; Rundle and Nosil, 2005; Savolainen et al., 2007). Evolutionary dynamics may play a major role in the build-up of lowland rain forest community diversity, and the role of genetic diversity (including sensu lato both allelic and gene expression variability) in ecological processes has been widely acknowledged (Ford, 1964; Randall Hughes et al., 2008). In other words, if ecological sorting of functional traits has occurred across different habitats and has led to the emergence of ecologically different species, it is sensible to expect that such processes are also currently occurring within species. Therefore, in species with continuous stands growing in different, contiguous habitats, we should be able to observe ‘highly local’ intraspecific divergence (sensu Salvaudon et al., 2008) between sub-populations submitted to divergent local environmental conditions; moreover, we expect that divergence between intraspecific sub-populations growing in different habitats should co-occur with divergence between species with different ecological preferences for those habitats. Here, we use the term ‘highly local’ to characterize patterns observed at scales for which environmental turnover occurs at shorter distances than gene flow (i.e. the average distance between patches of different habitat types is shorter than the average gene dispersal distance, implying that gene flow occurs among different habitats).
Tree populations in general are known to harbour large amounts of heritable variation for several putatively adaptive characters (Cornelius, 1994; González-Martínez et al., 2006); Neotropical rain forest trees are no exception (Navarro et al., 2004; Scotti et al., 2010). If adaptation contributes to divergence between sub-populations occupying different habitats, these sub-populations should be differentiated at potentially adaptive traits (sensu Howe and Brunner, 2005). The goal of the present study was therefore to test whether populations of tree species growing as continuous stands across different habitats could be sub-divided into habitat-associated sub-populations displaying phenotypic divergence for such traits [i.e. divergence caused by differentiation in (multi-locus) gene frequencies, by maternal effects or by inheritance of stable gene expression patterns (‘epigenetic inheritance’)]. The test was performed in two congeneric rain forest tree species of the Guiana Shield (Eperua falcata and E. grandiflora), that display partially divergent habitat preferences (Sabatier et al., 1997; Baraloto et al., 2007) but occur, even in low abundance, in multiple habitat types. In Eperua species, gene flow is expected to be restricted – mainly due to heavy seeds – but still intense at the distances considered here (estimate of mean parent–offspring distance for E. grandiflora: 166–343 m; Hardy et al., 2006). In spite of such dispersal distances, a recent study, performed partly on the same populations as those studied in the present paper (Audigeos et al., 2013), has shown that molecular divergence occurs (in E. falcata) at a highly local scale for genes involved in response to soil water content-related stress, against an overall background of no genetic differentiation at other loci.
Two specific questions are asked in this study about phenotypic divergence in these two congeneric species. (1) Do seedlings from different local habitats diverge phenotypically? (2) Are patterns of intraspecific phenotypic divergence similar to those observed at the interspecific level?
MATERIALS AND METHODS
Study species
Eperua falcata and E. grandiflora are abundant in the Guiana Shield, and grow sympatrically in different but partially overlapping habitats. This allowed us to compare intraspecific and interspecific patterns of divergence in the same phylogenetic context and ecological background. Eperua falcata (Aubl.) (Fabaceae) has a preference for seasonally waterlogged bottomlands, whereas E. grandiflora (Aubl.) Benth (Fabaceae) is mostly restricted to hilltops and slopes (Baraloto et al., 2007). The two species differ in several morphological and functional traits, but their seedlings display similar degrees of tolerance to drought or hypoxia under controlled conditions (Baraloto et al., 2007), indicating that they are potential generalists for soil water conditions, at least at the younger life stages. Both species are bat pollinated (Cowan, 1975) and disperse their heavy seeds by explosive dehiscence and gravity at short distances of a few metres (Forget, 1989). Gene dispersal distance is about 150–350 m for E. grandiflora (Hardy et al., 2006) and probably similar for E. falcata (O. Hardy, pers. comm.), well beyond the size of the habitat patches studied here. Data from nuclear genetic markers (Audigeos et al., 2013) suggest that E. falcata is allogamous with no significant selfing.
Study site
The experiment was performed in Plot 6 at the Paracou forest inventory site (5°18′N, 52°53′W) (Gourlet-Fleury et al., 2004) located in an undisturbed forest in coastal French Guiana, South America. The sampling area covers 9 ha and is characterized by a rugged landscape formed by the alternation of 40–50 m high hills, slopes and bottomlands, varying in soil drainage type and water table depth (Gourlet-Fleury et al., 2004). In such a habitat mosaic, variations occur on geographical distances of the same order of magnitude as pollen and seed dispersal but do not occur monotonically (i.e. there is no continuous gradient in a given spatial direction). Three habitat types have been identified in the study area (Supplementary Data, Fig. S1) based on elevation, soil drainage and waterlogging characteristics (Ferry et al., 2010): ‘bottomlands’ (B) with hydromorphic soils and a water table between 0 and 60 cm in depth depending on the season (Supplementary Data, Fig. S1); ‘slopes’ (S) with surface drainage conditions, and a water table consistently below 100 cm; and ‘hilltops’ (H) with deep soils, deep vertical drainage and a water table consistently below 150 cm.
Seed sampling
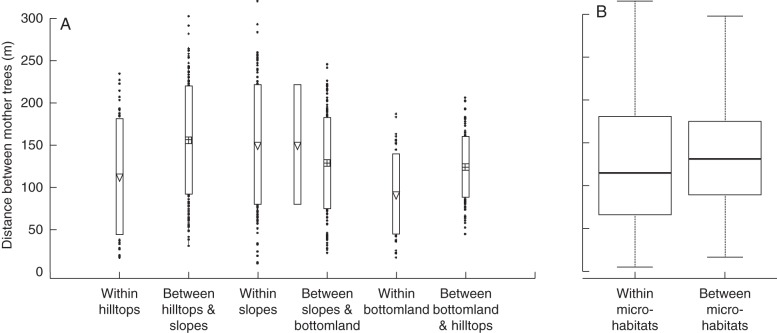
A total of 267 E. falcata trees and 67 E. grandiflora trees were identified in the study area. Operators visited the plot at least three times a week in February–March 2006, 2007 and 2008 to hand-collect seeds on the ground from 44 fruiting trees. The choice of the mother tree set was based on several considerations: (a) tree fertility; (b) balanced sampling from all habitats; and (c) non-overlapping tree crowns. Pairwise distances between same-habitat fruiting trees were not statistically smaller than between trees in different habitats (Fig. 1).
Fig. 1.
(A) Pairwise spatial distances between mother trees within and between microhabitats. Boxes show the standard deviation of each group. (B) Boxplots showing the distribution of pairwise spatial distance between mother trees within and between all microhabitats.
Seeds collected for our experimental study were assigned to the same habitat as their mother tree, thus forming three different native habitat types (‘B’, ‘S’ and ‘H’). When crowns of conspecific trees overlapped, seeds were collected at opposite sides of the crown. Each seed was assigned to a maternal family corresponding to its mother tree. A total of 3122 seeds were collected over the three seed production years.
Glasshouse experiment
The seeds were weighed and laid down in germination boxes that were filled with a substrate made of river sand which was kept damp using an automatic sprinkler system. Germination success rate was about 60 % for both species. Two months after germination, the seedlings were transplanted into individual 12 L pots filled with a mixture of sand and an A-horizon soil (30/70, v/v), then transferred to a glasshouse. The A-horizon had been collected in the same plot as the seeds and contained about 1·4–1·9 g kg−1 of nitrogen (Ferry et al., 2010).
About 4 % of the seedlings died before transfer to the glasshouse. The remaining seedlings were grown in the glasshouse for 24 months, until the study ended; then they were harvested. The 1637 seedlings (Supplementary Data, Table S1) were randomly assigned to each of 103 blocks of 16 plants. Each block contained four seedlings from each of four randomly drawn maternal families, so that each family was combined randomly with a different set of other families in each of the blocks in which it was represented (see Supplementary Data, Method 1 for details). The seedlings were placed under non-limiting conditions, which prevented both drought and hypoxia (expected to occur in the field on hilltops/slopes and in bottomlands, respectively; see above). Moreover, seedlings grown in the glasshouse experienced higher light levels and milder competition than in natural conditions, favouring optimal growth. A layer of neutral shade-cloth was used to reduce irradiance received to about 13 % of full sun [maximum photosynthetic photon flux density (PPFD) approx. 300 µmol m−2 s−1] to simulate solar radiation levels received by seedlings in gap openings. Seedlings were watered 2–3 times per week to maintain the substrate close to field capacity (approx. 0·25 m3 m−3). The pots were fertilized every 6 months (5 g of complete fertilizer per pot, 12/12/17/2 N/P/K/Mg). Pots were distributed in the glasshouse following an incomplete randomized block layout (for the details of the experimental design, see Supplementary Data, Method 1).
A total of 1637 seedlings survived until month 24. For measures taken at 24 months, the sample used in the present study was restricted to 656 seedlings of E. falcata and 170 seedlings of E. grandiflora (Supplementary Data, Tables S1 and S2), since two-thirds of the seedlings grown in this experiment were set aside for a companion experiment involving different soil water content treatments.
Phenotypic traits
We recorded 23 functional traits (Table 1) related to plant growth, biomass allocation, leaf structure and leaf physiology (photosynthetic capacity and carbon isotope composition). These traits are commonly used as proxies of plant fitness in general (Kraft et al., 2008) and their ecological significance as proxies of fitness in seedlings has been established by several studies (Westoby et al., 2002; Cornelissen et al., 2003; Wright et al., 2004).
Table 1.
List of abbreviations and units of phenotypic traits
| Abbreviation | Trait | Units |
|---|---|---|
| Growth and biomass allocation | ||
| (1) Seedling dimensions | ||
| H6 | Height at 6 months | cm |
| H12 | Height at 12 months | cm |
| H18 | Height at 18 months | cm |
| H24 | Height at 24 months | cm |
| H612 | Elongation rate from 6 to 12 months | cm month−1 |
| H1218 | Elongation rate from 12 to 18 months | cm month−1 |
| H1824 | Elongation rate from 18 to 24 months | cm month−1 |
| D18 | Diameter at 18 months | mm |
| D24 | Diameter at 24 months | mm |
| D1824 | Radial growth rate from 18 to 24 months | mm month−1 |
| (2) Biomass and allocation | ||
| TM24 | Total dry mass at 24 months | g |
| RM24 | Root dry mass at 24 months | g |
| LM24 | Total leaf dry mass at 24 months | g |
| LA24 | Total leaf area at 24 months | cm2 |
| LMR24 | Leaf/total mass ratio at 24 months | g g−1 |
| RMR24 | Root/total mass ratio at 24 months | g g−1 |
| LAR24 | Leaf area/total biomass ratio at 24 months | cm2 g−1 |
| Leaf traits | ||
| LMA18 | Leaf mass/area ratio at 18 months | g m−2 |
| LMA24 | Leaf mass/area ratio at 24 months | g m−2 |
| %C18 | Carbon content in leaves at 18 months | % |
| %N18 | Nitrogen content in leaves 18 months | % |
| Asat | Light-saturated carbon assimilation rate at 18 months | μmol m−2 s−1 |
| δ13C | Carbon isotope composition of leaves at 18 months | ‰ |
Plant height and stem diameter at collar were measured every 6 months. Net CO2 assimilation rate under saturating irradiance (Asat, μmol m−2 s−1) was recorded in vivo at 18 months on one leaf per plant with a portable photosynthesis system (CIRAS1, PP-Systems, Hoddesdon, UK) operating in open mode and fitted with a Parkinson leaf cuvette, under the following microclimate: ambient air CO2 concentration = 380 µmol mol−1; PPFD = 600 ± 20 µmol m−2 s−1; vapour pressure deficit = 1·0 ± 0·5 kPa; ambient air temperature = 28·7 ± 2·0 °C. Full stabilization was obtained after about 3–5 min. Measurements were conducted between 0900 h and 1300 h to avoid mid-day depression of photosynthesis. After gas exchange measurements, 6–8 mature and fully expanded leaflets were collected per plant close to the top of the stem. Fresh leaf area (LA) was then measured in the laboratory with an area meter (Li-2100, Licor, Lincoln, NE, USA). The leaves were subsequently dried to constant weight at 60 °C for about 3 d, then finely ground to measure carbon (C) and nitrogen (N) content and carbon isotope composition (δ13C, ‰) as a surrogate for intrinsic water-use efficiency (WUEi; Farquhar et al., 1982). Elemental and isotopic analyses were conducted on a sub-sample of about 1 mg of dry leaf powder with an isotopic ratio spectrometer (Delta-S Finnigan Mat, Bremen, Germany). Leaf mass to area ratio (LMA, g m−2) was calculated as the ratio of dry mass to LA.
At 24 months, all the plants were harvested and the leaves, stems, and roots were separated for biomass measurements. Total leaf area was measured with the same area meter as above. All three compartments were dried at 60 °C to constant weight for about 3–4 d and then weighed. Leaf area to total biomass ratio (LAR, m2 g−1) was obtained by dividing the total LA of a given plant by its total dry weight. Leaf mass ratio (LMR, g g−1) and root mass ratio (RMR, g g−1) were calculated as the ratio of leaf or root dry mass to total plant dry mass (Table 1). Growth rates for height and diameter growth between two dates were calculated as ΔP/Δt = (Pt2 – Pt1)/(t2 – t1), where P indicates the phenotypic value and t1, t2 the times of the two different measurements.
Linear model of character variation
We fitted a classical linear model for the partition of individual phenotypic values, including species, native habitat, maternal family, year of seed collection and seed mass as sources of trait variation in a hierarchical framework. To produce unbiased estimates of progeny and native habitat type effects, interannual variation and seed mass effects were used as cofactors in the model, as they capture, at least partially, environmental effects mediated by maternal allocation to seeds, and thus represent ‘maternal effects’ related to resource availability (Rice et al., 1993; Leiva and Fernández-Alés, 1998; González-Rodríguez et al., 2012). Our hierarchical framework allowed us to estimate the effects of each habitat type for each species, and the effect of each maternal family in each native habitat and each species. The linear model for all traits is as follows:
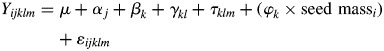 |
(1) |
where Yijklm is the phenotypic value of the i-th individual, μ the global mean, αj the effect of the j-th year of seed sampling and cultivation, βk the effect of the k-th species, γkl the effect of the l-th native habitat type within the k-th species, τklm the effect of the m-th progeny within the l-th native habitat within the k-th species, φk the regression coefficient between trait value and seed mass in the k-th species, seed massi the fresh mass of the i-th seed, and ɛijklm the residual variation of the i-th individual.
Model parameters and effects were estimated in a Bayesian framework (see Supplementary Data, Methods 2 for details) using the WINBUGS® software (Lunn et al., 2000). Bayesian methods can easily accommodate unbalanced/incomplete experimental designs (Browne and Draper, 2006) [erratic seed output (Supplementary Data, Tables S1 and S2) made a balanced design impossible in our study].
Conventional hypothesis testing of the significance of effects can be performed using the 95 % posterior distribution of effects (Qian and Shen, 2007). In this context, credible intervals are treated as the Bayesian analogues of confidence intervals: an estimated parameter has 95 % chance to be within the credible interval (Ellison, 1996): parameters for which zero falls outside the credible interval are considered significantly different from zero. The statistical consequences of multiple testing were evaluated by computing the Bayesian analogue of the false discovery rate (FDR; Benjamini and Hochberg, 1995; Miranda-Moreno et al., 2007).
Bayesian estimation of maternal family variance effects
We computed the ratio of maternal family variance (which includes truly genetic, epigenetic and possibly non-genetic maternal effects, and which we summarize as σ2M) to total phenotypic variance (σ2P). To estimate variances, we used a reduced version of linear model (1) restricted to family variations within each species. Phenotypic values were broken down as follows:
| (2) |
and the ratio of maternal family variance to total variance was estimated as:
Maternal family effects were estimated by fitting a quantitative–genetic hierarchical model by a Bayesian inference method of variance partitioning (Supplementary Data, Methods 3). This simplified model was preferred to the full model to compute variance components because (1) it is designed to estimate variance components directly, thus saving computation time and (2) the maternal family-level component (σ2M) we wished to obtain included all sources of among-family variation, including habitat, but did not include species effects (each species is treated separately).
Phenotypic correlations between traits
We estimated phenotypic correlations both at the individual (seedling) and at the maternal family level, using observed individual phenotypic values of seedlings and Bayesian estimates of maternal family values, respectively. The latter were computed as the sum of all sensu lato ‘genetic’ factors from eqn (1): Y'klm = μ + βk + γkl + τklm.. The sum of these factors conveys the mean phenotypic value of each progeny free from seed mass and year effects (which represent ‘environmental maternal effects’). Phenotypic correlations were calculated using Pearson's coefficient. Significance at two-tailed α = 0·05 was tested by the cor.test function in R (R Development Core Team, 2008). The FDR (Benjamini and Hochberg, 1995) was computed for all correlation matrices.
RESULTS
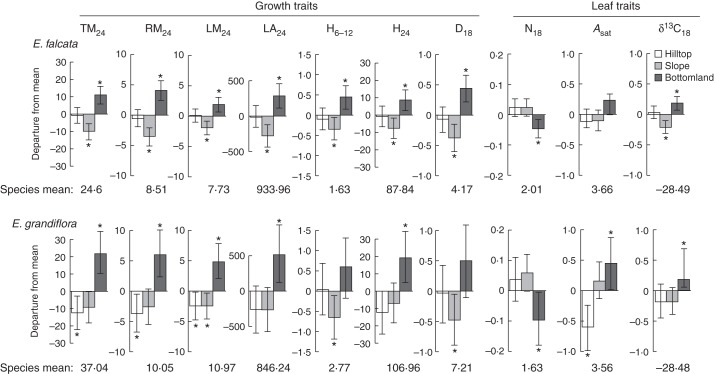
At the intraspecific level, native habitat had a significant effect on 18 out of 23 traits in E. falcata, and 15 out of 23 in E. grandiflora (Fig. 2; Supplementary Data, Tables S3–S6). Both species displayed significant variation among native habitats for growth traits (including height and diameter; height and diameter growth rates; total, root and leaf mass): seedlings from bottomlands grew faster and produced more biomass than those from slopes and hilltops. Growth rate varied significantly among native habitats at early stages in both species, but this effect vanished after 12 and 18 months for E. grandiflora and E. falcata, respectively. In both species, δ13C, leaf area and leaf mass were larger, and N content smaller, in seedlings from bottomlands than those from the other two habitats. Eperua falcata seedlings from bottomlands showed lower LAR, but higher LMA, than those from slopes and hilltops. For E. grandiflora, Asat was higher in bottomland seedlings than in those from hilltops. We did not find any significant variation in RMR among native habitats. We estimated the expected rate of false positives (FDR) as 0·8 % with a single test α = 5 % as used here.
Fig. 2.
Phenotypic differentiation among habitat types for growth, biomass allocation and leaf traits for Eperua falcata (top) and E. grandiflora (below) sampled at Paracou, French Guiana. Bayesian estimates of departures of each group from the global mean are shown as boxes; t-bars show the 95 % Bayesian credible interval of the estimated parameters. Figures above each plot provide the within-species trait means, which correspond to the zero value in the plots. Units for each trait are provided in Table 1. Hilltop, slope and bottomland are as indicated in the key in the top panel. Asterisks indicate a significant effect of habitat type (95 % CI intervals do not overlap 0).
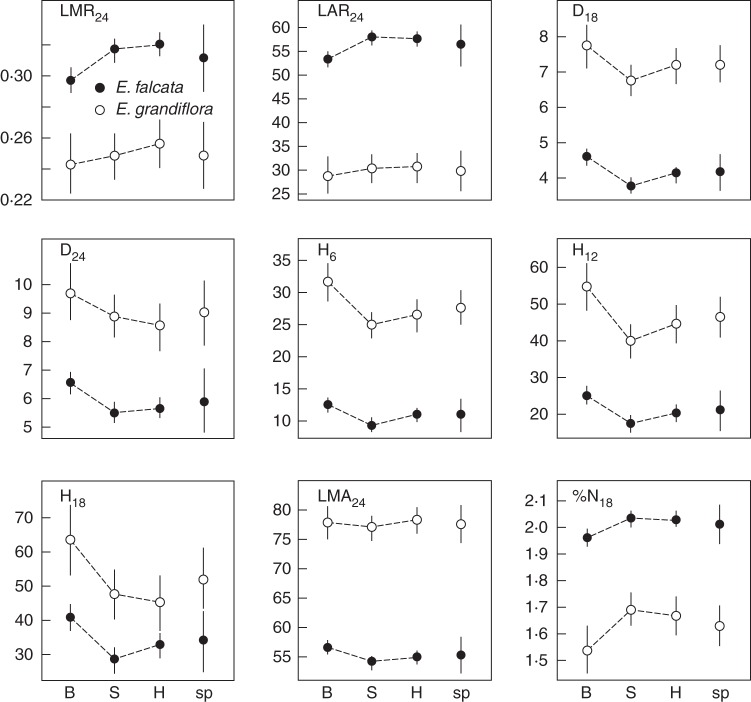
The two species displayed significant differences for a sub-set of the recorded traits (Supplementary Data, Tables S3–S6): E. falcata seedlings had significantly smaller stems, higher LA and LMR, higher %N and lower LMA than E. grandiflora. No difference was detected for growth rate, biomass accumulation, Asat or δ13C. Nine traits (LMR24, LAR24, D18, D24, H6, H12, H18, LMA24 and %N18; Supplementary Data, Tables S3–S6) had significant differences at both the intra- and interspecific level. For these traits, intraspecific trends ran contrary to the interspecific traits (Fig. 3; Supplementary Data, Tables S3–S6), i.e. the overall direction of change between same-species hilltop and bottomland sub-populations was contrary to the change between hilltop-preferring E. grandiflora and bottomland-preferring E. falcata. None of the traits showing significant differences among hilltop and bottomland sub-populations also showed significant differences in the same direction between hilltop-preferring E. grandiflora and bottomland-preferring E. falcata. Four traits (RMR24, D1824, H1218 and Asat; Supplementary Data, Tables S3–S6) showed such a trend, but for none of them were the effects significant both at the species and at the sub-population level. Cofactors representing maternally transmitted environmental effects (year of fruit set and seed mass) also influenced several traits (Supplementary Data, Tables S3–S6).
Fig. 3.
Comparison of the direction of trait value change between habitats (within species) and between species for traits with significant differences both at the intraspecific and at the interspecific level (see Supplementary Data, Tables S3–S6 for raw results). Trait names and units are as described in Table 1. B, bottomlands; S, slopes; H, hilltops; sp, species-level values. Bayesian posterior medians for Eperua falcataand E. grandiflora as indicated in the key in the first graph. Error bars are Bayesian 95 % credible intervals; non-overlapping credible intervals between two values imply significant differences (95 % CI intervals do not overlap 0).
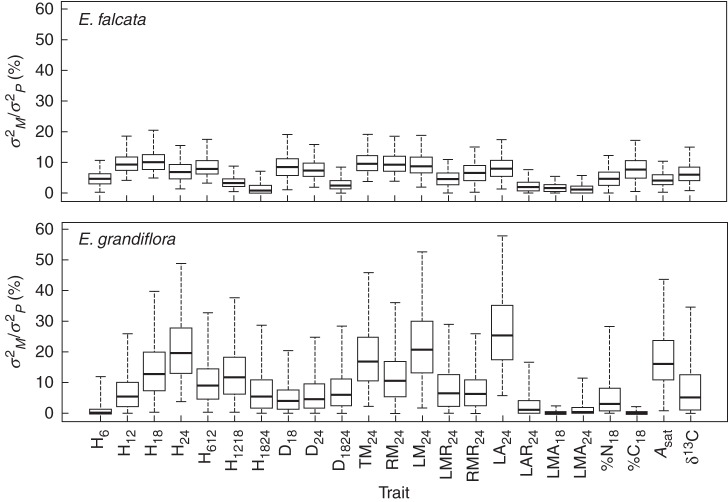
The maternal family effect (which is obtained independently from the native habitat effect described above) was significant for all traits in both species (Supplementary Data, Tables S3–S6). Ratios of maternal family-to-total variance (σ2M/σ2P) ranged between 1·2 % [H1824; 95 % credible interval (c.i.0·95) = 0·007–7·2 %] and 10·1 % (H18; c.i.0·95 = 4·9–20·5 %) in E. falcata, and from 0·02 % (LMA18; c.i.0·95 = 0·00003–2·61 %) to 25·4 % (LA24; c.i.0·95 = 6–58 %) in E. grandiflora (Fig. 4; Supplementary Data, Table S7). Credible intervals were larger in E. grandiflora than in E. falcata (Supplementary Data, Table S7) probably due to differences in sample size (Supplementary Data, Table S1).
Fig. 4.
Boxplots of Bayesian posteriors of σ2M/σ2P for all traits for Eperua falcata and E. grandiflora. Trait names are as described in Table 1.
Correlation matrices were very similar between the two species (Supplementary Data, Figs S3 and S4). Most traits showed significant correlation at the individual seedling level (raw phenotypic data), but not at the maternal family level (capturing maternally inherited effects on traits). Seedling-level and progeny-level trait correlation matrices, if both significant, always had the same sign; we did not observe any significant family-level correlation without matching significant seedling-level correlation. At the seedling level, two main correlation groups emerged: dimensions, biomass and leaf traits (Table 1; Supplementary Data, Figs S3 and S4) were tightly correlated; allocation traits were all negatively correlated with the remaining traits and had a mixed pattern of correlation to each other. Leaf mass per area (LMA) was somewhat intermediate, showing both positive and negative correlations with dimension, leaf and biomass traits and positive correlation with RMR. At the maternal family level, traits such as Asat and δ13C retained their positive correlation with biomass traits (but not with dimension traits) and their negative correlation with allocation traits; the latter globally retained their negative correlation with all other traits and the positive correlation between LAR and LMR (although fewer correlations were significant in E. grandiflora than in E. falcata). The FDR was <2 % for all matrices for both the 5 % and the 1 % significance threshold (Supplementary Data, Figs S3 and S4).
DISCUSSION
Divergence among sub-populations and maternal families was apparent for several traits, indicating the presence of maternally inherited variability in both species, in agreement with existing estimates of quantitative trait diversity in wild tree populations (Cornelius, 1994; Coutand et al., 2010; Scotti et al., 2010).
After removal of environmentally derived maternal effects (as described by seed mass and year of fructification), native habitat explained a significant fraction of phenotypic differentiation for several leaf- or plant-level traits. These effects are relatively small (Supplementary Data, Fig. S2) but significant, which is quite surprising, considering the small spatial scale at which they occur. A sub-set of these traits may show divergence between sub-populations only because they are correlated with traits that are involved in some adaptively meaningful divergence (Lande and Arnold, 1983). The analysis of phenotypic correlations at the progeny level actually reveals that 20 of the 23 traits (61 %) showing some degree of divergence are correlated to at least another divergent trait. Because maternal family-level correlations were estimated on mean maternal family phenotypic values (which do not include seed mass and year of production effects), the correlations between traits is probably driven by several factors [including epigenetic effects, pleiotropy, and physical quantitative trait locus (QTL) linkage], which we cannot break apart with the current data set.
Nine traits (Fig. 3) displayed divergence both between species and between sub-populations within species. For all these traits, the intraspecific patterns ran opposite to the interspecific one. This suggests that intraspecific trait distributions may be unimodal functions of environmental variables with peak positions that differ between species (‘reaction norm shift’: Fig. 3; fig. 5 in Albert et al., 2010; Crispo, 2007). In such conditions, if the span of environmental conditions sampled is limited relative to the extent of such unimodal distributions, one may observe the kind of patterns reported here, with intraspecific trends contrary to interspecific ones (Albert et al., 2010). Four additional traits (RMR24, D1824, H1218 and Amax18; Supplementary Data, Tables S3–S6) had monotonic intraspecific trends that were concordant with interspecific ones, but without significant effects at either the species or the population level, or both. These results show that, at least for a relatively large sub-set of traits (nine out of 23, or 39 %), it is possible to detect intraspecific variation for those traits showing interspecific variation along the same environmental gradients. This is in agreement with the hypothesis that the differentiation processes currently affecting within-population diversity may be the same as those that caused species divergence, although our observations require confirmation by functional–ecological experiments.
The maternally transmitted component of both trait divergence and trait correlations may have multiple origins.
(1) Environmentally driven maternal effects (i.e. variation in resource availability transmitted to seedlings through seed resources) can influence seedling growth (González-Rodríguez et al., 2011, 2012). In our study, these were controlled through modelling of the effect of both seed mass and year of seed set, which are estimated separately from maternal family effect; therefore, we suggest that these effects should be negligible in our estimation of sensu lato genetic factors, although some cases of maternal background × environmental effects have been reported (Rice et al., 1993; González-Rodríguez et al., 2011).
(2) ‘Epigenetic’ maternal effects (mainly due to the transient transmission of gene expression states through the embryo) can contribute to similarity of traits within maternal families, thus inflating maternal family effects. Epigenetic inheritance has been proven to occur in trees (Rix et al., 2012), although its overall impact on trait variance was negligible. It is not possible to estimate the importance of such effects in our study, and they can clearly contribute to trait divergence among maternal families from different native habitats, if mother trees transmit environmentally induced gene expression states to their progeny. These variations in epigenetic state may have an adaptive meaning, if epigenetically inherited trait values confer higher fitness in the maternal habitat.
(3) Truly heritable (additive and non-additive) genetic effects may also contribute to trait divergence, and also have an adaptive meaning, for the same reasons as in (2). Two arguments let us think that ‘truly genetic’ effects may account for at least part of the observed divergence between sub-populations. First, the same E. falcata adult tree population used for the present study displayed molecular–genetic divergence between habitats for genes involved in response to stresses related to soil water content (Audigeos et al., 2013); this supports the possibility that genetic structuring can occur in these populations. Secondly, we have shown that there are significant phenotypic differences between maternal families within habitats. If habitat-driven differentiation were only caused by epigenetic effects related to environmental differences, variation between same-habitat maternal families should be negligible, which is not the case in our results. Traits that had large maternal family variance components (σ2M/σ2P) in our study (e.g. height and biomass traits, and LA; Fig. 4) often also showed high heritability in other tropical or temperate tree species (Vásquez and Dvorak, 1996; Hodge et al., 2002; Carnegie et al., 2004; Navarro et al., 2004; Scotti-Saintagne et al., 2004; Costa e Silva et al., 2005; Sotelo Montes et al., 2007; Callister and Collins, 2008; Ward et al., 2008a, b; Scotti et al., 2010), suggesting that a non-negligible part of the phenotypic divergence among maternal families may be due to true genetic factors; it has to be noted that heritability estimates are generally obtained at the species or at the whole-population level, without considerations for environmental sub-division, and therefore our σ2M/σ2P estimates are properly comparable with previous studies. Finally, it has been proven that plant populations can show genetic divergence at functional traits even if they are potentially connected by migration (Hovenden and Vander Schoor, 2004; Byars et al., 2007) or have been shown to undergo strong gene flow (Gonzalo-Turpin and Hazard, 2009).
Whatever the mechanistic base of phenotypic divergence between sub-populations from different native habitats, how likely is it that these differences have arisen because of neutral processes, e.g. spatial genetic structure (due to local inbreeding)? Our study plot is a 300 m sided square, and the largest possible distance between trees is approx. 425 m, within Eperua gene dispersal distance (Hardy et al., 2006); gene flow is thus possible between the different habitat types. Moreover, seeds were sampled in a habitat mosaic, and mother trees inhabiting the same habitat type are not on average closer than trees inhabiting different habitats (Fig. 1). Thus, neutral divergence induced by neutral spatial genetic structure seems unlikely.
Several studies on plants have shown divergence in adaptive traits along environmental gradients (Kawecki and Ebert, 2004; Carlson et al., 2011), particularly with respect to edaphic factors and waterlogging conditions (Silva et al., 2010). The existence of sensu lato heritable traits showing highly local divergence between sub-populations suggests that local adaptation at short geographical distances may occur (Ehrlich and Raven, 1969; Schemske, 1984; Jump et al., 2006; Turner et al., 2010) in the presence of gene flow, which is precisely the sense given by Kawecki (2004) to the term ‘local adaptation’. Conditions for highly local adaptation are not unlikely in tropical rain forests, based on evidence about local species distribution (ter Steege and Hammond, 2001) and the association between functional traits and habitats (Baraloto et al., 2005) over short spatial scales (<50 m) (Kraft and Ackerly, 2010).
Functional considerations can help the interpretation of the observed differences among seedlings native from different habitats. A higher productivity of seedlings from bottomlands as compared with the other two habitats is consistent with larger leaf area and higher Asat, since these seedlings are therefore able to assimilate more carbon, use it to synthesize more biomass and, eventually, allocate it to growth. This is consistent with the results of previous studies revealing a trend towards increasing growth performances from drier to wetter habitats (Russo et al., 2005; Kariuki et al., 2006; Sanchez-Gomez et al., 2006; Ferry et al., 2010). The LMR and LAR were slightly lower in E. falcata seedlings from bottomlands, suggesting that they invest more biomass in roots and stems than in leaves. This is consistent with frequent waterlogging events that drastically reduce O2 availability in the soil and decrease hydraulic conductivity of roots, with consequences similar to those of drought (Ponnamperuma, 1972). Lower LMR and LAR would also contribute to reducing water loss through a lower LA per unit of plant mass (Poorter and Markesteijn, 2008). Higher LMA in bottomland seedlings also permits a reduction of water loss through the reduction of transpiring LA at the leaf level (Poorter et al., 2009). In parallel, higher investment in root biomass would enhance water capture ability during dry periods as well as root O2 absorption during wet periods. Furthermore, bottomland seedlings of both species display higher WUE (i.e. less negative δ13C) than slope or hilltop seedlings, which means that, during photosynthesis, they use less water for the same amount of CO2 assimilation (Farquhar et al., 1982). This trade-off in water and carbon use at leaf level is an efficient strategy when soil water resources are limiting (e.g. Ehleringer and Cooper, 1988), not only on hilltops but also in the bottomlands (Baraloto et al., 2007). Finally, variations of N content are well identified as a determinant of photosynthetic capacities (Reich et al., 1994), as revealed by the strong correlations between leaf nitrogen and Asat. In natural conditions, leaf nitrogen and foliar N:P ratios are known to be highly dependent upon soil chemical properties (Townsend et al., 2007), and the dependence of Amax on N is expected to be stronger in N-limiting habitats than in P- or Ca-limiting habitats. Bottomlands have higher N content and lower P content than hilltop habitats (Luizao et al., 2004; Ferry et al., 2010), and we observe here lower %N in bottomland than in hilltop seedlings. This suggests that the faster growing bottomland seedlings, which also have higher photosynthetic rates, have lower nitrogen content, contrary to what is expected – at the interspecific level – according to the World Leaf Economic Spectrum (Donovan et al., 2011).
Conclusions
We detected phenotypic divergence for growth and physiological traits occurring over very short spatial distances within a habitat mosaic. This suggests that large reservoirs of within-species adaptive potential are maintained by trait filtering caused by niche partitioning and habitat associations (Russo et al., 2005; Kraft and Ackerly, 2010), and possibly by local adaptive processes. Species displaying such variation may respond more easily to environmental changes through microevolution (by being able to react adaptively to the expected impact of global change), if at least part of the variation is heritable or is caused by adaptive plasticity. It is worth remembering that epigenetic (maternal) effects can be considered as heritable in the broad sense (Bossdorf et al., 2008; Klironomos et al., 2013). The mechanisms underlying such local intraspecific divergence may also turn out to play a major role in the generation of the outstanding diversity in tropical forest ecosystems and, more generally, to be a fundamental mechanism in the maintenance of trait variation in natural populations.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Saintano Dufort, Laetitia Ruelle, Jean Weigel, Jocelyn Cazal, Saint-Omer Cazal, Valérie Troispoux, Caroline Duret, Ann-Magret Amui-Vedel, Vincent Vedel and the CIRAD Kourou forest technical team for seed sampling, plant culture and phenotypic measurements. We thank Jean-Jacques Boreux and Marie-Pierre Etienne for their advice on Bayesian modelling. We thank Erwin Dreyer, Jérôme Chave and Christian Lexer who carefully and critically read this manuscript, and Lynda Delph for insightful discussions. Louise Brousseau was supported by a ‘Contrat Jeune Scientifique’ assistantship from INRA. This work was supported by the French MEDD ministry ‘Ecofor – Ecosystèmes forestiers’ programme and by the EU-funded PO-FEDER 2007–2013 ENERGIRAVI programme. This work has benefited from an “Investissement d'Avenir” grant managed by Agence Nationale de la Recherche (CEBA, ref. ANR-10-LABX-0025).
LITERATURE CITED
- Albert CH, Thuiller W, Yoccoz NG, Soudant A, Boucher F, Saccone P, Lavorel S. Intraspecific functional variability: extent, structure and sources of variation. Journal of Ecology. 2010;98:604–613. [Google Scholar]
- Audigeos D, Brousseau L, Traissac S, Scotti-Saintagne C, Scotti I. Molecular divergence in tropical tree populations occupying environmental mosaics. Journal of Evolutionary Biology. 2013;26:529–544. doi: 10.1111/jeb.12069. [DOI] [PubMed] [Google Scholar]
- Baraloto C, Goldberg D, Bonal D. Performance trade-offs among tropical tree seedlings in contrasting microhabitats. Ecology. 2005;86:2461–2472. [Google Scholar]
- Baraloto C, Morneau F, Bonal D, Blanc L, Ferry B. Seasonal water stress tolerance and habitat associations within four neotropical tree genera. Ecology. 2007;88:478–489. doi: 10.1890/0012-9658(2007)88[478:swstah]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rates: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- Bossdorf O, Richards CL, Pigliucci M. Epigenetics for ecologists. Ecology Letters. 2008;11:106–115. doi: 10.1111/j.1461-0248.2007.01130.x. [DOI] [PubMed] [Google Scholar]
- Browne W, Draper D. A comparison of Bayesian and likelihood-based methods for fitting multilevel models. Bayesian Analysis. 2006;1:473–514. [Google Scholar]
- Byars SG, Papst W, Hoffmann AA. Local adaptation and cogradient selection in the alpine plant, Poa hiemata, along a narrow altitudinal gradient. Evolution. 2007;61:2925–2941. doi: 10.1111/j.1558-5646.2007.00248.x. [DOI] [PubMed] [Google Scholar]
- Callister A, Collins S. Genetic parameter estimates in a clonally replicated progeny test of teak (Tectona grandis Linn. f.) Tree Genetics and Genomes. 2008;4:237–245. [Google Scholar]
- Carlson JE, Holsinger KE, Prunier R. Plant responses to climate in the cape floristic region of South Africa: evidence for adaptive differentiation in the Proteaceae. Evolution. 2011;65:108–124. doi: 10.1111/j.1558-5646.2010.01131.x. [DOI] [PubMed] [Google Scholar]
- Carnegie A, Johnson I, Henson M. Variation among provenances and families of blackbutt (Eucalyptus pilularis) in early growth and susceptibility to damage from leaf spot fungi. Canadian Journal of Forest Research. 2004;34:2314–2326. [Google Scholar]
- Clark DB, Palmer MW, Clark DA. Edaphic fators and the landscape-scale distribution of tropical rain forest trees. Ecology. 1999;80:2662–2675. [Google Scholar]
- Cornelissen JHC, Lavorel S, Garnier E, et al. A handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Australian Journal of Botany. 2003;51:335–380. [Google Scholar]
- Cornelius J. Heritabilities and additive genetic coefficients of variation in forest trees. Canadian Journal of Forest Research. 1994;24:372–379. [Google Scholar]
- Costa e Silva J, Dutkowski G, Borralho N. Across-site heterogeneity of genetic and environmental variances in the genetic evaluation of Eucalyptus globulus trials for height growth. Annals of Forest Science. 2005;62:183–191. [Google Scholar]
- Coutand C, Chevolot M, Lacointe A, Rowe N, Scotti I. Mechanosensing of stem bending and its interspecific variability in five neotropical rainforest species. Annals of Botany. 2010;105:341–347. doi: 10.1093/aob/mcp286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan RS. Washington, DC: Smithsonian Institution Press; 1975. A monograph of the genus Eperua (Leguminosae-Caesalpinioideae) [Google Scholar]
- Crispo E. The Baldwin effect and genetic assimilation: revisiting two mechanisms of evolutionary change mediated by phenotypic plasticity. Evolution. 2007;61:2469–2479. doi: 10.1111/j.1558-5646.2007.00203.x. [DOI] [PubMed] [Google Scholar]
- Donovan LA, Maherali H, Caruso CM, Huber H, de Kroon H. The evolution of the worldwide leaf economics spectrum. Trends in Ecology and Evolution. 2011;26:88–95. doi: 10.1016/j.tree.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Ehleringer JR, Cooper TA. Correlations between carbon isotope ratio and microhabitat in desert plants. Oecologia. 1988;76:562–566. doi: 10.1007/BF00397870. [DOI] [PubMed] [Google Scholar]
- Ehrlich PR, Raven PH. Differentiation of populations. Science. 1969;165:1228–1232. doi: 10.1126/science.165.3899.1228. [DOI] [PubMed] [Google Scholar]
- Ellison AM. An introduction to Bayesian inference for ecological research and environmental decision-making. Ecological Applications. 1996;6:1036–1046. [Google Scholar]
- Endler J. Geographic variation, speciation and clines. Princeton, NJ: Princeton University Press; 1977. [PubMed] [Google Scholar]
- Engelbrecht BMJ, Comita LS, Condit R, et al. Drought sensitivity shapes species distribution. Nature. 2007;447:80–83. doi: 10.1038/nature05747. [DOI] [PubMed] [Google Scholar]
- Epron D, Bosc A, Bonal D, Freycon V. Spatial variation of soil respiration across a topographic gradient in a tropical rain forest in French Guiana. Journal of Tropical Ecology. 2006;22:565–574. [Google Scholar]
- Farquhar GD, Oleary MH, Berry JA. On the relationship between carbon isotope discrimination and the inter-cellular carbon-dioxide concentration in leaves. Australian Journal of Plant Physiology. 1982;9:121–137. [Google Scholar]
- Ferry B, Morneau F, Bontemps J-D, Blanc L, Freycon V. Higher treefall rates on slopes and waterlogged soils result in lower stand biomass and productivity in a tropical rain forest. Journal of Ecology. 2010;98:106–116. [Google Scholar]
- Ford EB. Ecological genetics. London: Chapman & Hall; 1964. [Google Scholar]
- Forget JM. La régénération naturelle d'une espece autochore de la foret guyanaise: Eperua falcata Aublet (Caesalpiniaceae) Biotropica. 1989;21:115–125. [Google Scholar]
- González-Martínez SC, Krutovsky KV, Neale DB. Forest-tree population genomics and adaptive evolution. New Phytologist. 2006;170:227–238. doi: 10.1111/j.1469-8137.2006.01686.x. [DOI] [PubMed] [Google Scholar]
- González-Rodríguez V, Villar R, Navarro-Cerrillo RM. Maternal influences on seed mass effect and initial seedling growth in four Quercus species. Acta Oecologica. 2011;37:1–9. [Google Scholar]
- González-Rodríguez V, Barrio IC, Villar R. Within-population variability influences early seedling establishment in four Mediterranean oaks. Acta Oecologica. 2012;41:82–89. [Google Scholar]
- Gonzalo-Turpin H, Hazard L. Local adaptation occurs along altitudinal gradient despite the existence of gene flow in the alpine plant species Festuca eskia. Journal of Ecology. 2009;97:742–751. [Google Scholar]
- Gourlet-Fleury S, Guelh JM, Laroussine O. Ecology and management of a Neotropical rainforest: lessons drawn from Paracou, a long-term experimental research site in French Guiana. Amsterdam: Elsevier; 2004. [Google Scholar]
- Hardy OJ, Maggia L, Bandou E, et al. Fine-scale genetic structure and gene dispersal inferences in 10 Neotropical tree species. Molecular Ecology. 2006;15:559–571. doi: 10.1111/j.1365-294X.2005.02785.x. [DOI] [PubMed] [Google Scholar]
- Hodge G, Dvorak W, Urueña H, Rosales L. Growth, provenance effects and genetic variation of Bombacopsis quinata in field tests in Venezuela and Colombia. Forest Ecology and Management. 2002;158:273–289. [Google Scholar]
- Hovenden MJ, Vander Schoor JK. Nature vs nurture in the leaf morphology of Southern beech, Nothofagus cunninghamii (Nothofagaceae) New Phytologist. 2004;161:585–594. doi: 10.1046/j.1469-8137.2003.00931.x. [DOI] [PubMed] [Google Scholar]
- Howe GT, Brunner AM. An evolving approach to understanding plant adaptation. New Phytologist. 2005;167:1–5. doi: 10.1111/j.1469-8137.2005.01469.x. [DOI] [PubMed] [Google Scholar]
- John R, Dalling JW, Harms KE, et al. Soil nutrients influence spatial distributions of tropical tree species. Proceedings of the National Academy of Sciences, USA. 2007;104:864–869. doi: 10.1073/pnas.0604666104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jump AS, Hunt JM, Martinez-Izquierdo JA, Penuelas J. Natural selection and climate change: temperature-linked spatial and temporal trends in gene frequency in Fagus sylvatica. Molecular Ecology. 2006;15:3469–3480. doi: 10.1111/j.1365-294X.2006.03027.x. [DOI] [PubMed] [Google Scholar]
- Kanagaraj R, Wiegand T, Comita LS, Huth A. Tropical tree species assemblages in topographical habitats change in time and with life stage. Journal of Ecology. 2011;99:1441–1452. [Google Scholar]
- Kariuki M, Rolfe M, Smith RGB, Vanclay JK, Kooyman RM. Diameter growth performance varies with species functional-group and habitat characteristics in subtropical rainforests. Forest Ecology and Management. 2006;225:1–14. [Google Scholar]
- Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecology Letters. 2004;7:1225–1241. [Google Scholar]
- Klironomos FD, Berg J, Collins S. How epigenetic mutations can affect genetic evolution: model and mechanism. BioEssays. 2013;35:571–578. doi: 10.1002/bies.201200169. [DOI] [PubMed] [Google Scholar]
- Kozlowski TT. Responses of woody plants to flooding and salinity. Tree Physiology Monograph. 1997;17:1–29. [Google Scholar]
- Kraft NJB, Ackerly DD. Functional trait and phylogenetic tests of community assembly across spatial scales in an Amazonian forest. Ecological Monographs. 2010;80:401–422. [Google Scholar]
- Kraft NJB, Valencia R, Ackerly DD. Functional traits and niche-based tree community assembly in an Amazonian forest. Science. 2008;322:580–582. doi: 10.1126/science.1160662. [DOI] [PubMed] [Google Scholar]
- Lande R, Arnold S. The measurement of selection on correlated characters. Evolution. 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. [DOI] [PubMed] [Google Scholar]
- Leiva MJ, Fernández-Alés R. Variability in seedling water status during drought within a Quercus ilex subsp. ballota population, and its relation to seedling morphology. Forest Ecology and Management. 1998;111:147–156. [Google Scholar]
- Lopez OR, Kursar TA. Does flood tolerance explain tree species distribution in tropical seasonally flooded habitats? Oecologia. 2003;136:193–204. doi: 10.1007/s00442-003-1259-7. [DOI] [PubMed] [Google Scholar]
- Luizao RC, Luizao FJ, Paiva RQ, Monteiro TF, Sousa LS, Kruijt B. Variation of carbon and nitrogen cycling processes along a topographic gradient in a central Amazonian forest. Global Change Biology. 2004;10:592–600. [Google Scholar]
- Lunn DJ, Thomas A, Best N, Spiegelhalter D. WinBUGS – a Bayesian modelling framework: concepts, structure, and extensibility. Statistics and Computing. 2000;10:325–337. [Google Scholar]
- Miranda-Moreno LF, Labbe A, Fu L. Bayesian multiple testing procedures for hotspot identification. Accident Analysis and Prevention. 2007;39:1192–1201. doi: 10.1016/j.aap.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Navarro C, Montagnini F, Hernández G. Genetic variability of Cedrela odorata Linnaeus: results of early performance of provenances and families from Mesoamerica grown in association with coffee. Forest Ecology and Management. 2004;192:217–227. [Google Scholar]
- Perata P, Armstrong W, Voesenek LACJ. Plants and flooding stress. New Phytologist. 2011;190:269–273. doi: 10.1111/j.1469-8137.2011.03702.x. [DOI] [PubMed] [Google Scholar]
- Ponnamperuma FN. The chemistry of submerged soils. Advances in Agronomy. 1972;24:29–96. [Google Scholar]
- Poorter H, Niinemets Ü, Poorter L, Wright IJ, Villar R. Causes and consequences of variation in leaf mass per area (LMA): a meta-analysis. New Phytologist. 2009;182:565–588. doi: 10.1111/j.1469-8137.2009.02830.x. [DOI] [PubMed] [Google Scholar]
- Poorter L, Markesteijn L. Seedling traits determine drought tolerance of tropical tree species. Biotropica. 2008;40:321–331. [Google Scholar]
- Qian SS, Shen Z. Ecological applications of multilevel analysis of variance. Ecology. 2007;10:2489–2495. doi: 10.1890/06-2041.1. [DOI] [PubMed] [Google Scholar]
- Randall Hughes A, Inouye BD, Johnson MTJ, Underwood N, Vellend M. Ecological consequences of genetic diversity. Ecology Letters. 2008;11:609–623. doi: 10.1111/j.1461-0248.2008.01179.x. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. www.r-project.org . [Google Scholar]
- Reich PB, Walters MB, Ellsworth DS, Uhl C. Photosynthesis–nitrogen relations in amazonian tree species.1. Patterns among species and communities. Oecologia. 1994;97:62–72. doi: 10.1007/BF00317909. [DOI] [PubMed] [Google Scholar]
- Rice KJ, Gordon DR, Hardison JL, Welker JM. Phenotypic variation in seedlings of a ‘keystone’ tree species (Quercus douglasii): the interactive effects of acorn source and competitive environment. Oecologia. 1993;96:537–547. doi: 10.1007/BF00320511. [DOI] [PubMed] [Google Scholar]
- Ricklefs RE. Environmental heterogeneity and plant species diversity: a hypothesis. American Naturalist. 1977;111:376–381. [Google Scholar]
- Rix K, Gracie A, Potts B, Brown P, Spurr C, Gore P. Paternal and maternal effects on the response of seed germination to high temperatures in Eucalyptus globulus. Annals of Forest Science. 2012;69:673–679. [Google Scholar]
- Rundle HD, Nosil P. Ecological speciation. Ecology Letters. 2005;8:336–352. [Google Scholar]
- Russo SE, Davies SJ, King DA, Tan S. Soil-related performance variation and distributions of tree species in a Bornean rain forest. Journal of Ecology. 2005;93:879–889. [Google Scholar]
- Sabatier D, Grimaldi M, Prévost M-F, et al. The influence of soil cover organization on the floristic and structural heterogeneity of a Guianan rain forest. Plant Ecology. 1997;131:81–108. [Google Scholar]
- Salvaudon L, Giraud T, Shykoff JA. Genetic diversity in natural populations: a fundamental component of plant–microbe interactions. Current Opinion in Plant Biology. 2008;11:135–143. doi: 10.1016/j.pbi.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Sanchez-Gomez D, Valladares F, Zavala MA. Performance of seedlings of Mediterranean woody species under experimental gradients of irradiance and water availability: trade-offs and evidence for niche differentiation. New Phytologist. 2006;170:795–805. doi: 10.1111/j.1469-8137.2006.01711.x. [DOI] [PubMed] [Google Scholar]
- Savolainen O, Pyhäjärvi T, Knürr T. Gene flow and local adaptation in trees. Annual Review of Ecology, Evolution, and Systematics. 2007;38:595–619. [Google Scholar]
- Schemske DW. Population structure and local selection in Impatiens pallida (Balsaminaceae), a selfing annual. Evolution. 1984;38:817–832. doi: 10.1111/j.1558-5646.1984.tb00354.x. [DOI] [PubMed] [Google Scholar]
- Schluter D. Ecology and the origin of species. Trends in Ecology and Evolution. 2001;16:372–380. doi: 10.1016/s0169-5347(01)02198-x. [DOI] [PubMed] [Google Scholar]
- Scotti-Saintagne C, Bodénès C, Barreneche T, Bertocchi E, Plomion C, Kremer A. Detection of quantitative trait loci controlling bud burst and height growth in Quercus robur L. Theoretical and Applied Genetics. 2004;109:1648–1659. doi: 10.1007/s00122-004-1789-3. [DOI] [PubMed] [Google Scholar]
- Scotti I, Calvo-Vialettes L, Scotti-Saintagne C, Citterio M, Degen B, Bonal D. Genetic variation for growth, morphological, and physiological traits in a wild population of the Neotropical shade tolerant rainforest tree Sextonia rubra (Mez) van der Werff (Lauraceae) Tree Genetics and Genomes. 2010;6:319–329. [Google Scholar]
- Silva DCG, Carvalho MCCG, Ruas PM, Ruas CF, Medr ME. Evidence of ecotypic differentiation between populations of the tree species Parapiptadenia rigida due to flooding. Genetics and Molecular Research. 2010;2:797–810. doi: 10.4238/vol9-2gmr736. [DOI] [PubMed] [Google Scholar]
- Sotelo Montes C, Beaulieu J, Hernández RE. Genetic variation in wood shrinkage and its correlations with tree growth and wood density of Calycophyllum spruceanum at an early age in the Peruvian Amazon. Canadian Journal of Forest Research. 2007;37:966–976. [Google Scholar]
- ter Steege H, Hammond DS. Character convergence, diversity, and disturbance in tropical rain forest in Guyana. Ecology. 2001;82:3197–3212. [Google Scholar]
- ter Steege H, Jetten VG, Polak AM, Werger MJA. Tropical rain forest types and soil factors in a watershed area in Guyana. Journal of Vegetation Science. 1993;4:705–716. [Google Scholar]
- Townsend AR, Cleveland CC, Asner GP, Bustamante MMC. Controls over foliar N:P ratios in tropical rain forests. Ecology. 2007;88:107–118. doi: 10.1890/0012-9658(2007)88[107:cofnri]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nature Genetics. 2010;42:260–263. doi: 10.1038/ng.515. [DOI] [PubMed] [Google Scholar]
- Valencia R, Foster RB, Villa G, et al. Tree species distributions and local habitat variation in the Amazon: large forest plot in eastern Ecuador. Journal of Ecology. 2004;92:214–229. [Google Scholar]
- Vásquez J, Dvorak WS. Trends in variances and heritabilities with stand development of tropical pines. Canadian Journal of Forest Research. 1996;26:1473–1480. [Google Scholar]
- Vincent G, Molino J-F, Marescot L, et al. The relative importance of dispersal limitation and habitat preference in shaping spatial distribution of saplings in a tropical moist forest: a case study along a combination of hydromorphic and canopy disturbance gradients. Annals of Forest Science. 2011;68:357–370. [Google Scholar]
- Ward S, Wightman K, Rodriguez Santiago B. Early results from genetic trials on the growth of Spanish cedar and its susceptibility to the shoot borer moth in the Yucatan Peninsula, Mexico. Forest Ecology and Management. 2008a;255:356–364. [Google Scholar]
- Ward SE, Wightman KE, Santiago BR. Early results from genetic trials on the growth of Spanish cedar and its susceptibility to the shoot borer moth in the Yucatan Peninsula, Mexico. Forest Ecology and Management. 2008b;255:356–364. [Google Scholar]
- Webb CO, Peart DR. Habitat associations of trees and seedlings in a Bornean rain forest. Journal of Ecology. 2000;88:464–478. [Google Scholar]
- Westoby M, Falster D, Moles A, Vesk P, Wright IJ. Plant ecological strategies: some leading dimensions of variation between species. Annual Review of Ecology and Systematics. 2002;33:125–159. [Google Scholar]
- Wright IJ, Reich PB, Westoby M, et al. The worldwide leaf economics spectrum. Nature. 2004;428:821–827. doi: 10.1038/nature02403. [DOI] [PubMed] [Google Scholar]
- Wright SJ. Plant diversity in tropical forests: a review of mechanisms of species coexistence. Oecologia. 2002;130:1–14. doi: 10.1007/s004420100809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.