Abstract
It has been difficult to examine the role of TGF-ß in post-natal tooth development due to perinatal lethality in many of the signaling deficient mouse models. To address the role of Tgfbr2 in postnatal tooth development, we generated a mouse in which Tgfbr2 was deleted in odontoblast-and bone-producing mesenchyme. Osx-Cre;Tgfbr2fl/fl mice were generated (Tgfbr2cko) and postnatal tooth development was compared in Tgfbr2cko and control littermates. X-ray and μCT analysis showed that in Tgfbr2cko mice radicular dentin matrix density was reduced in the molars. Molar shape was abnormal and molar eruption was delayed in the mutant mice. Most significantly, defects in root formation, including failure of the root to elongate, were observed by postnatal day 10. Immunostaining for Keratin-14 (K14) was used to delineate Hertwig's epithelial root sheath (HERS). The results showed a delay in elongation and disorganization of the HERS in Tgfbr2cko mice. In addition, the HERS was maintained and the break up into epithelial rests was attenuated suggesting that Tgfbr2 acts on dental mesenchyme to indirectly regulate the formation and maintenance of the HERS. Altered odontoblast organization and reduced Dspp expression indicated that odontoblast differentiation was disrupted in the mutant mice likely contributing to the defect in root formation. Nevertheless, expression of Nfic, a key mesenchymal regulator of root development, was similar in Tgfbr2cko mice and controls. The number of osteoclasts in the bone surrounding the tooth was reduced and osteoblast differentiation was disrupted likely contributing to both root and eruption defects. We conclude that Tgfbr2 in dental mesenchyme and bone is required for tooth development particularly root formation.
Keywords: odontoblast, osteoclast, HERS, eruption
Introduction
Mammalian teeth develop from the reciprocal interactions between oral epithelium and underlying neural crest derived mesenchyme (Thesleff and Nieminen, 1996). The formation of tooth crown is characterized by a series of well-defined stages, epithelial thickening, bud, cap and bell (Caton and Tucker, 2009). The development of tooth root is initiated postnatally after completion of crown formation (Lungova et al., 2011). Root formation is initiated at postnatal day 5 with the formation of a bilayered epithelial structure called the Hertwig's epithelial root sheath (HERS). After the bell stage, inner and outer enamel epithelium continues to grow apically and fuse below the level of the crown cervical enamel to form the HERS (Luan et al., 2006). Morphologically, the HERS bends inwards towards the pulp chamber at the early stage of root development to guide root elongation (Luan et al., 2006). The root continues to elongate for approximately 3 weeks. Reciprocal interactions between the HERS and dental pulp mesenchyme play an important role in the regulation of odontoblast differentiation in the root promoting radicular (root) dentinogenesis, development of the periodontium and cementogenesis (Thomas, 1995). The HERS is a transient structure and fragments into epithelial rests of Malassez (ERM) later during tooth development (Luan et al., 2006).
Epithelial-mesenchymal interactions during tooth development have been studied for decades. Conserved signal pathways mediating epithelial-mesenchymal interactions during tooth development include the TGFβ, Wnt, FGF, Hedgehog, and a TNF signal called ectodysplasin (Eda) [reviewed in (Bei, 2009)]. The TGFβ family of signaling peptides includes TGFβs, BMPs, activins and related proteins, which regulate many developmental processes including tooth development [reviewed in (Massague, 2000)]. TGFβ signals through a dual receptor system of type I and type II transmembrane serine/threonine kinases (Janssens et al., 2005). Upon TGFβ binding to type II receptor (Tgfbr2), which is a constitutively active kinase, type I receptor (Tgfbr1) can be recruited to the complex and becomes phosphorylated and activated. The canonical Tgfbr2 signaling pathway is mediated by a group of cytoplasmic signal transducers called Smads. Smads regulate organ development and disease by directly regulating the transcription of target genes (Janssens et al., 2005).
The role of members of the TGFβ super family in specific aspects of tooth development and pathology is most clearly illustrated in mice with mutations or targeted deletions in their respective genes (Ferguson et al., 1998; Gao et al., 2009; Ko et al., 2007). Many studies have focused on the action of TGFβ and Smad signalling on tooth development. The TGFβ type 2 receptor (Tgfbr2) expresses in both epithelium and neural crest derived mesenchyme. Deletion of Tgfbr2 in Wnt1 expressing mesenchyme results in defects in odontoblast differentiation and dentin formation in the crown (Ito et al., 2003; Oka et al., 2007). The ablation of Tgfbr2 signaling in Wnt1 expressing populations leads to cleft palate and perinatal death so the role of Tgfbr2 in postnatal root development was not addressed. Smad4 is a central intracellular effector of both TGFβ and BMP signaling. Mice with a conditional deletion of Smad4 in neural-crest derived mesenchymal cells do not survive through mid-gestation; however, loss of Smad4 resulted in arrested tooth development at the lamina stage in a transplant model (Ko et al., 2007). In contrast, mice with a knockout of Smad4 in Osteocalcin-Cre expressing odontoblasts survive and demonstrate disruption of root development (Gao et al., 2009). Conditional deletion of Smad4 in dental epithelium also results in a failure in the elongation of the HERS, indirectly disrupting development of root dentin (Huang et al., 2010). Since Smad4 affects both TGF-ß and BMP signaling it has been difficult to sort out which signaling pathways are involved in root development. Furthermore, many Smad4-independent TGF-ß and BMP signaling pathways exist (Xu et al., 2008).
Nuclear factor I transcription protein C (Nfic) has been shown to be a key regulator of postnatal root formation (Lee et al., 2009a; Lee et al., 2009b; Park et al., 2007; Steele-Perkins et al., 2003). Mice lacking Nfic had short and abnormal roots due to a disturbance in odontoblast proliferation and differentiation, and subsequent apoptosis of aberrant odontoblasts. A recent study suggested that root formation is mediated through a Smad4-Shh-Nfic signaling cascade (Huang et al., 2010). In this model, Smad4 in the HERS regulates expression of Shh, which is secreted and acts on the dental mesenchyme through Nfic to regulate radicular dentin formation in the root. In contrast, others have shown that Nfic acts upstream of TGF-ß in the dental mesenchyme to down-regulate signaling via dephosphorylation of Smad proteins (Lee et al., 2009a). Most recently, it was shown that TGF-ß and Nfic regulate each other's activity in cultured dental mesenchyme. Nfic down-regulates TGF-ß signals, while TGF-ß promotes the degradation of Nfic (Lee et al., 2011).
Osteoclasts are required to remodel bone and are required for tooth eruption and root elongation (Aioub et al., 2007; Helfrich, 2005). Osteoclast formation and activity are regulated by a cascade of signaling molecules including, macrophage colony-stimulating factor-1 (CSF-1), receptor activator of nuclear factor-kappa B ligand (RANKL), and osteoprotegerin (OPG). CSF-1 is required to recruit the osteoclast precursors to the dental follicle, and RANKL is both necessary and sufficient for the complete differentiation of the precursor cells into mature osteoclasts [reviewed in (Khosla, 2001)]. RANKL is typically expressed on the surface of pre-osteoblasts, and binds to RANK receptor on osteoclast precursors after its release into the bone microenvironment. OPG is a soluble decoy of RANKL and it is secreted by osteoblasts to inhibit osteoclast formation (Khosla, 2001). TGF-ß signaling in osteoblasts has been shown to control osteoclast numbers in long bone and calvaria (Filvaroff et al., 1999; Qiu et al., 2010). Mice expressing a dominant-negative form of Tgfbr2 in mature osteoblasts demonstrate osteopetrosis and fewer osteoclasts (Filvaroff et al., 1999). Deletion of Tgfbr2 in osteoblasts via Osteocalcin-Cre results in a similar phenotype (Qiu et al., 2010).
To investigate the role of TGFβ signalling in postnatal tooth development, our laboratory established a mouse model in which Tgfbr2 was conditionally deleted in odontoblast-producing mesenchyme using an Osterix (Osx/ Sp7) promoter driven Cre recombinase (Tgfbr2cko) (Rodda and McMahon, 2006). We show that Tgfbr2 in the dental mesenchyme is required for normal tooth development including elongation of the root. Radicular odontoblast differentiation was disrupted and a significant decrease in osteoclast number was detected in the alveolar bone surrounding the tooth.
Materials and Methods
Mice
All experiments were carried out with the approval of the UAB institutional animal care committee. The generation of Tgfbr2fl/fl mice was previously described (Chytil et al., 2002). Generation of the Osx-Cre mice has been previously described (Rodda and McMahon, 2006). Osx-Cre mice were crossed to Tgfbr2fl/fl mice to obtain Osx-Cre+;Tgfbr2fl/wt mice, which were subsequently back crossed to Tgfbr2fl/fl mice to get Osx-Cre+;Tgfbr2fl/fl (CKO) as the experimental group and Osx-Cre− as controls. The Cre reporter strain Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP) Luo/J (ROSA26mTmG) was obtained from Jackson Laboratories. ROSA26mTmG mice were crossed to Osx-Cre mice to generate Osx-Cre; ROSA26mTmG mice to trace Cre activity.
X-Ray analysis and micro-computed tomography
Four 3-week old mice from experimental and control groups were sacrificed and the mandible from each mouse was dissected and fixed in 4% paraformaldehyde overnight. Mandibles were radiographed using a Faxitron radiograph inspection unit. The mandible, incisors, and the first molars were dissected and radiographed. A microCT-40 computed tomography system (Scanco Medical, Bassersdorf, Switzerland) with analysis software was used to analyze the volume, density and root length of the first molar from the CKO and controls. A student's t-test was performed and the significant level was set as P < 0.05. Four control and four mutant mice were measured.
Histology, Immunostaining, and In situ Hybridization
At least three mice from each group were sacrificed at postnatal 0, 5, 7, 10 and 14 days. The mandibles were fixed in 4% paraformaldehyde overnight, followed by de-calcification. The specimens were then embedded in Optimal Cutting Temperature (OCT) compound, and sectioned at 10 μm. For histological analysis, sections were stained with hematoxylin and eosin. For immunofluorescence and immunohistochemical staining, sections were post-fixed in cold acetone for 5 min. The sections were then immersed in 3% H2O2/PBS for 10 min to quench the endogenous peroxidase activity. After blocking in 5% BSA for 1 hr, the sections were incubated 2 hrs at room temperature with a certain dilution of primary antibodies. Antibodies used for this experiment include keratin 14 polyclonal antibody (1:1000, Covance), polyclonal Amelogenin (1:85;Sigma) Ki67 (1:500, Abcam), and Nfic polyclonal antibody (1:200) generated against the synthesized peptide LRPTRPLQTVPLWD in Dr. MacDougall's laboratory. Biotinylated goat-anti-rabbit IgG (1:200, Vector Laboratories) was added as the secondary antibody and incubated for 10 min at room temperature. Sections were then reacted with avidin-biotin-peroxidase complex (Vector Laboratories) for 30 min, followed by the color development using 3,3'-Diaminobenzidine (DAB) substrate (Vector Laboratories). The sections were counterstained with methyl green (Sigma). For immunofluorescence, fluorescent signal was detected with avidin-conjugated Cy3 (Invitrogen).
In situ hybridization was performed using a DIG labeled riboprobes to Dspp described in (Wang et al., 2009; Wang et al., 2004). The procedure was done as previously described (Sohn et al., 2010).
TRAP staining
To identify osteoclasts, consecutive sections were stained for tartrate resistant acid phosphatase (TRAP). Briefly, mandibles from control and CKO were fixed in 4% paraformaldehyde, decalcified in EDTA and then sectioned after OCT embedding. TRAP staining was carried out per manufacturer's protocol (387A-1KT; Sigma-Aldrich). Images of the first molar were obtained, and the number of TRAP-positive cells lining on the alveolar bone surface was calculated for every section using Bioquant software. Briefly, the perimeter of the alveolar bone surface is measured at 20x. Next, the number of TRAP stained osteoclasts is counted (manual object counting) in the same field.
Quantitative real-time RT-PCR
Alveolar bone samples were isolated from 14 (bone) day old Tgfbr2CKO and control littermates. Cells from inside the tooth pulp chamber, including pulp and odontoblasts, were flushed from 10 day-old mouse molars and labeled dental pulp. RNA was isolated using the standard Trizol method (Paaske et al., 1987). RNA concentration was determined by Nanodrop Spectrophotometer. QuantiFast SYBR Green RT-PCR Kit (Qiagen) was used for quantitative real-time RT-PCR analysis with specific primers (Table 1). Data was normalized to three housekeeping genes: ß-2 microglobulin (ß2m), Glyceralderhyde 3 phosphate dehydrogenase (Gapdh), and Hypoxanthine-guanine phosphoribosyltransferase (Hprt) (Bustin et al., 2009). Primers for Hprt1 and Gapdh were obtained from the RTPrimerDB database. Pth1r primers were designed using qPrimerDepot (Cui et al., 2007). Rankl primers were designed using QuantPrime (Arvidsson et al., 2008). For dental pulp, we used three biological samples for each control and Tgfbr2cko. For alveolar bone we had three control and two Tgfbr2cko samples. Data analysis was performed with REST software (www.qiagen.com/Products/REST2009Software.aspx?r=8042; (Pfaffl et al., 2002).
Table 1.
Primers used in real-time PCR
| Gene Name | Forward Primer (5'–3') | Reverse Primer (5'–3') |
|---|---|---|
| β2-microglobulin | GCCGTGTGAACCATGTGACTTT | CCAAATGCGGCATCTTCAAA |
| Bglap | ATAGCTCGTCACAAGCAGGG | TGACAAAGCCTTCATGTCCA |
| Csf-1 | TACTGTGCCATCTCTAGCCAAGCA | TGGGAAACAGAGGGCATCAGAACA |
| Dmp1 | CATTCTCCTTGTGTTCCTTTGGG | TGTGGTCACTATTTGCCTGTG |
| Dspp | AGTTCGATGACGAGTCC | GTCTTCTCCCGCATGT |
| Gapdh | TGTGTCCGTCGTGGATCTGA | TTGCTGTTGAAGTCGCAGGAG |
| Hprt | TCAGTCAACGGGGGACATAAA | GGGGCTGTACTGCTTAACCAG |
| Nfi-c | GGAACCGGACCCAACTTCTC | CGTCCTCTTCCATCGAGCC |
| Opg | GGAACCCCAGAGCGAAATACA | CCTGAAGAATGCCTCCTCACA |
| Pth1r | TCC TTC TCT GCT GCC CAG T | GGT GCA GCA GGA AAA TCT GT |
| RankL | TCC TGT ACT TTC GAG CGC AGA TG | AGA GTC GAG TCC TGC AAA CCT G |
Results
Tgfbr2 is required for molar tooth eruption and root formation
Osx is a transcription factor that has been best characterized as a regulator of osteogenesis (Long, 2011). The expression pattern of Osx in rodent molars has been described. In post-natal stages, Osx mRNA is localized to odontoblasts, dental pulp cells, cementoblasts, and alveolar bone (Chen et al., 2009). Likewise, Osx protein was observed by immunoflourescent staining in odontoblasts, osteoblasts, and cementoblasts. Osx was not detected in the HERS (Hirata et al., 2009). The Osx-Cre mice used in this study express a GFP-Cre fusion protein under the control of the Osx promoter (Rodda and McMahon, 2006). We used the GFP tag in these mice to determine the Osx-Cre expression pattern in teeth (Figure 1). At post-natal day 5, GFP was detected sporadically in dental pulp cells, with more cells expressing GFP near the single layer of odontoblasts (Figure 1B, D). The highest levels of GFP were detected in mature odontoblasts (Figure 1B, D) and osteoblasts (Figure 1C) in the alveolar bone. GFP was excluded from epithelial derivatives including ameloblasts (Figure 1B) and the HERS (Figure 1D). We saw a similar GFP expression pattern in newborn mice (data not shown). Since the GFP-Cre fusion only detects Cre when it is actively expressed, we also used a ROSA26 reporter strain (ROSA26mTmG) that expresses a membrane bound GFP tag in cells that have expressed Cre at any point in their lifetime to determine which cell types in the tooth are derived from Osx-Cre expressing cells Figure 1 (E–H). The reporter used, ROSA26mTmG, also expresses a membrane bound Tomato red fluorescent protein in cells that have not seen Cre. In the first molar from P7 day Osx-Cre; ROSA26mTmG mice, cells derived from dental mesenchyme, including odonotoblasts and pulp, demonstrated green staining indicating these cells were derived from Osx-Cre expressing cells. More importantly, cells derived from the epithelium, including the HERS, were stained red, indicating that Cre had never been expressed in these cells (Figure 1 F, G). In the crown, odontoblasts stained very strongly for the membrane bound GFP protein. Odontoblastic cell processes were outlined by GFP staining in the molar dentin whereas, ameloblasts were stained red, indicating no Cre activity in this cells type (Figure 1H). Other groups have used similar methods to trace Osx-Cre activity in the tooth (Ono et al., 2013; Rakian et al., 2012). Similar to our results, Cre activity resulted in reporter activity in odontoblasts, dental pulp, cementoblasts, and osteoblasts (Ono et al., 2013). Reporter activity was not detected in tooth derived epithelial cells (Rakian et al., 2012).
Figure 1. Cre-GFP expression and activity in tooth.
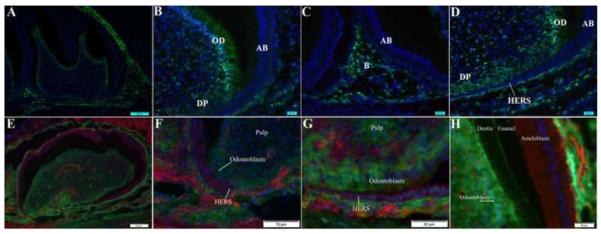
Cre expression was detected as green fluorescence in cryosections from P5 Cre+;Tgfbr2loxP/wt mice (A–D). Nuclei in the sections were counterstained with DAPI (blue). Cre-GFP was detected in dental mesenchyme including mature odontoblasts (B, D; OD), and some of the cells in the dental pulp (B, D; DP). Cre was also seen in the alveolar bone (C; B). GFP staining was not detected in cells of epithelial origin including ameloblasts (A–D, AB), or the HERS (D; small arrow). Low magnification image is shown in (A). Lineage tracing of Osx-Cre activity up to P7 days was determined using Osx-Cre+; ROSA25mTmG reporter mice (E–H). Lineage tracing of Cre activity is shown as green membrane staining. Cells in which Cre was never active are shown with red membrane staining. Nuclei are blue. Cre activity was present in cells derived from dental mesenchyme including odontoblasts (F, G). Epithelial structures including the HERS stained red, indicating no Cre activity during the lifetime of those cells (F, G). In the crown, Cre activity was detected in odontoblasts whereas ameloblasts demonstrated no Cre activity and were stained red (H). Odontoblastic cell processes in the dentin were outlined with GFP fluorescence (H). A low magnification image of the first molar from a P7 Osx-Cre+; ROSA25mTmG mouse is shown in (E).
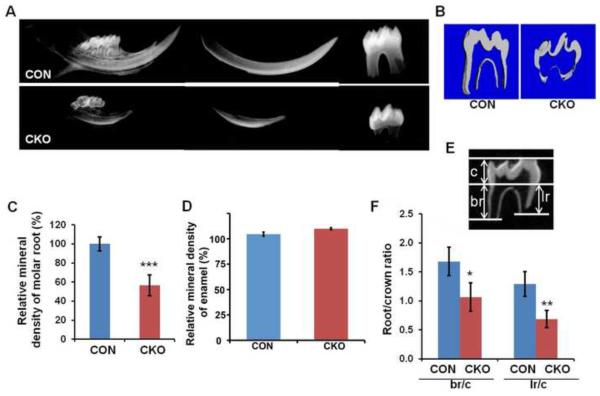
Tgfbr2cko mice survived post-natal, although they were significantly smaller than the Cre-negative and Cre-positive, Tgfbr2fl/wt controls (data not shown). By three weeks of age, we noticed that the Tgfbr2cko mice showed significant defects in their teeth upon x-ray analysis (Figure 2A). Most notably, the mineral density of the root dentin was reduced as measured by microCT (Figure 2B, C), whereas, the enamel density was not affected (Figure 2B, D). The roots of the first molar were significantly shorter on both the lingual and buccal aspects of the tooth compared to the controls (Figure 2 E, F). In addition, molars in mutant mice had not erupted although molars in control mice erupted at approximately two weeks of age (Figure 2A and data not shown). The phenotype indicated that Tgfbr2 in Osx-Cre expressing cells is required for normal molar root development and eruption.
Figure 2. Root formation is disrupted in Tgfbr2CKO mice.
(A) Representative X-ray of mandibular incisors and the first molars from 3-week old control (CON) and Tgfbr2cko (CKO) mice. (B) Representative μCT images of the first mandibular molar from 3 weeks old CON and CKO mice. The parameters measured were (C) the mineral density in the root and (D) the mineral density of the enamel (E) Parameters used to evaluate tooth: crown (c), buccal root (br), and lingual root (lr) length. (F) The ratio of root length to crown length was calculated. Loss of Tgfbr2 in Osx-Cre expressing cells results in defects in root length, volume, and mineral density. Enamel density was not affected. N = 4; *: P < 0.05; **: P < 0.01; ***: P < 0.001.
HERS is disrupted in Tgfbr2 conditional knockout mice
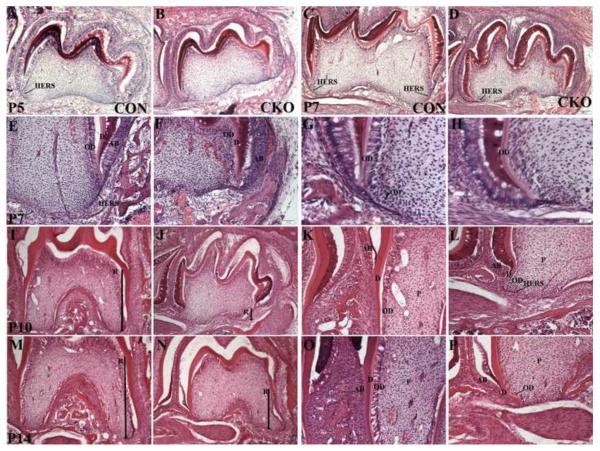
To begin to identify the basis of the root phenotype, we compared H & E stained sections of the first molar from Tgfbr2cko and control mice at varying ages, from newborn to 14 days old. At birth, the molars in control and Tgfbr2cko mice were comparable (data not shown). At P5 days, the crown in control and mutant mice were similar. In control mice, the epithelial HERS was detected on the distal side of the tooth near the second molar (Figure 3A arrow). In contrast, extension of the HERS was not detected in Tgfbr2cko molars at this stage (Figure 3B). At P7 days, some disorganization of the odontoblasts in the crown was observed in Tgfbr2cko mice (Figure 3 C–H). In control mice, the HERS on both the distal and proximal side of the first molar could be seen elongating downward into the mesenchyme (Figure 3 C, E arrow). The shape of the newly forming roots was evident. In contrast, at P7 days in the mutant teeth, the HERS were either missing or severely truncated and there was no evidence of root elongation (Figure 3 F, H). Furthermore, crown odontoblasts were disorganized in the Tgfbr2 mutants. In control teeth, the nuclei of odontoblasts in the crown to the crown root junction were polarized and presumptive odontoblasts were starting to undergo cytodifferentiation along the HERS (Figure 3G). In contrast, in the Tgfbr2 mutants the odontoblast nuclei in the crown root junction were not uniformly polarized as seen in the controls. In addition, pulp cells were not becoming aligned on the truncated HERS in the mutant teeth (Figure 3H). At P10 and P14 days in the first molar of control mice, the formation of the root was well established (Figure 3H, M). Radicular dentin was deposited and odontoblasts in the root were well organized (Figure 3K, O). In contrast, the first molar roots in the mutant mice were not well developed (Figure 3 J, N). The HERS were missing or truncated and the odontoblast in the root were not well organized (Figure 3 L, P). The histology of the mutant teeth suggested that the function of the HERS and/or odontoblasts may be altered in the absence of Tgfbr2 leading to defects in root development.
Figure 3. Comparison of tooth histology.
Representative images of H&E stained mandibular first molar from postnatal day (P) 5 (A, B), 7 (C–H), 10 (I–L) and 14 (M–P) Tgfbr2cko (CKO; A, C, E, G, I, K, M, O) and control (CON; B, D, F, H, J, L, N, P) mice. 100× (A–F, I, J, N, N) and 200× (K, L, O, P) and 400× (G, H) images from sagittal sections are shown. The second molar is to the left in all images. R- root length designated by thick arrow; OD- odontoblast; D- Dentin; AB- Ameloblasts; P- Dental pulp. Tgfbr2cko mice demonstrate alterations in the histology of odontoblasts and HERS. Representative images from three separate control and mutant mice are shown.
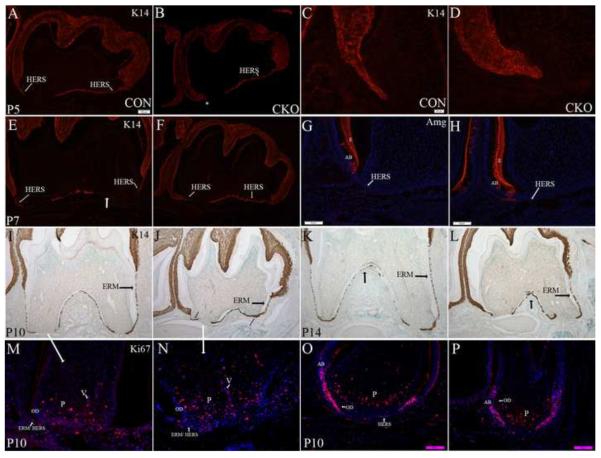
To better see the structure of the HERS, sections from the first molar at each stage were immunostained with K14 (Figure 4). K14 expression has been used as an epithelial cell marker during normal tooth development (Luan et al., 2006). At P5 days, epithelium from the proximal side of the tooth was observed underneath and parallel to the tooth in both control and Tgfbr2cko mice (Figure 4 A, B). In the P5 control mice, the HERS could be seen starting to extend down from the crown on the distal side of the tooth (Figure 4A); however, the HERS was not detected on the distal side of P5 mutant molars (Figure 4B). In control mice, the HERS was composed of a bi- layer of cells extending down from the epithelium of the crown (Figure 4C). In contrast, in Tgfbr2cko mice, the area where the HERS would normally form consisted of a short, thick, and multilayered epithelium (Figure 4D). By P7 days, the HERS continued to extend down the distal side of the control tooth but was still truncated in the mutant molars (Figure 4 E, F). The epithelium that ran under and parallel to the tooth had broken apart in control mice so that the HERS could now be seen extending down from the proximal side of the tooth (Figure 4E, G). In contrast, this epithelium was maintained in the mutant molar. High magnification images showed that this layer of cells was disorganized and multilayered (Figure 4 H). At P10 and P14 days, the distal HERS and root were severely truncated in Tgfbrcko mice as compared to the controls (Figure 4 I–L). The proximal root was not as severely affected. In control teeth, the HERS had started to break down into ERM by P10 days (Figure 4 I, K). Formation of ERM appeared attenuated in the mutant teeth (Figure 4 J, L). We also looked at cell proliferation using antibodies to the Ki67 antigen, which is present in proliferating cells. In the first molar of control mice, proliferating cells were concentrated in the dental pulp at the bottom of the tooth (Figure 4M). Staining could also be seen in the invading vasculature (Figure 4M). Some cells in the remnants of the HERS at the base of the root were also stained with Ki67 (Figure 4M). Odontoblasts at this stage had withdrawn from the cell cycle and were not proliferating (Figure 4M). The pattern of Ki67 staining was similar in the Tgfbr2 mutants (Figure 4N). Most of the proliferating cells were concentrated in the bottom of the tooth within the pulp root apex with staining in the invading vasculature also seen (Figure 4N). The odontoblast layer and truncated HERS we severely disorganized so it was difficult to identify the cells and determine if they stained for Ki67 (Figure 4N). We also looked at Ki67 staining in the third molar of P10 mice since the third molar represents an earlier stage of development (Figure 4O, P). In the control third molar, most of the staining was localized to pulp apex cells at the extending end of the tooth root as well as the epithelial cells at this same region of the tooth. Staining could also be seen in some of the cells in the HERS (Figure 4O). The HERS was not detectable in the mutant mice but similar to controls a high level of Ki67 staining was observed in the epithelial cells at the bottom of the tooth that would be expected to form the HERS (Figure 4P). Since Osx-Cre targets dental mesenchyme and does not target the epithelial-derived HERS directly, the results suggest that TGF-ß signaling in the dental mesenchyme affects the development and maintenance of the HERS indirectly, perhaps through paracrine interactions with the odontoblasts.
Figure 4. HERS is disrupted in Tgfbr2CKO mice.
Representative images from K14 stained sagittal sections (red fluorescence A–F; DAB brown I–L) of the first mandibular molar from postnatal P5 (A–D), P7 (E,F), P10 (I, J) and P14 (K, L) Tgfbr2cko (CKO; B, D, F, J, L) and control (CON; A, C, E, I, K) mice. The second molar is to the left. Magnification: 100× (A, B, E, F, I–L) and 400× (C, D, G, H). HERS is designated with the small white arrows. Discontinuous parts of the HERS and of ERM are designated with thick arrows. Missing HERS at P7 designated with an *. There is a delay in the elongation of the HERS on the side adjacent to the second molar (C, D). The opposite HERS elongates but is not a single layer and does not break up or extend down to form the root (E, F, I, J). Formation of ERM is delayed (I–L). Amelogenin marks the enamel and ameloblasts (G, H). Amelogenin expression is excluded from root epithelium in control (G) and mutant (H) P7 teeth (Magnification: 200×). Ki67 staining to localize proliferating cells is shown (M –P). Sections from the first mandibular molar in control (M) and Tgfbr2 mutant (N) mice demonstrate areas of proliferation (pink; Magnification 400×). Sections from the 3rd mandibular molar in control (O) and mutant (P) mice are also shown (Magnification 200×). OD- odontoblast, P- pulp, V- vasculature, AB- ameloblasts.
Odontoblast differentiation is disrupted in Tgfbr2cko root
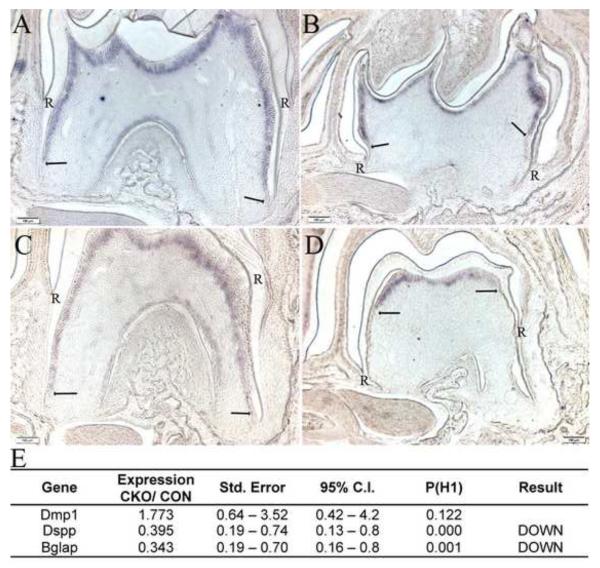
Tgfbr2 has been shown to regulate odontoblast differentiation in the crown and deletion of Smad4 can disrupt differentiation in the root (Ito et al., 2003; Oka et al., 2007). Furthermore, proper odontoblast differentiation is required for formation and elongation of the HERS (Park et al., 2007). We used in situ hybridization and real time quantitative PCR to measure localization and expression of dentin sialophosphoprotein (Dspp) a marker of odontoblast differentiation in control and Tgfbr2cko molars (Figure 5). In control teeth at P10 and P14 days, clear homogenous expression of Dspp mRNA was detected in odontoblasts in both the crown and the root. The level of Dspp mRNA expression extended to the tip of the root (Figure 5A, C; arrows). In contrast, in mutant teeth, Dspp mRNA expression was more limited in the crown and did not extend into the root. Staining within the crown odontoblasts layer was absent in the crown root junction region well before the transition to root odontoblasts (Figure 5 B, D; arrows). This same pattern of Dspp expression was observed in Nfic-deficient mice (Park et al., 2007). We then isolated mRNA from the molar root pulp of P10 control and Tgfr2cko mice and performed real time RT-PCR to determine the relative levels of Dspp and other odontoblast markers including Dmp1 and Osteocalcin (Bglap) (Figure 5E). Results showed a significant decrease in Dspp and Bglap mRNA expression in mutant versus control teeth suggesting that loss of Tgfb signaling in the tooth mesenchyme disrupts differentiation of odontoblasts in the root. Dmp1 mRNA levels were not altered. It has been shown that Dmp1 regulates expression of later markers of odontoblast differentiation like Bglap and Dspp (Narayanan et al., 2001). Therefore it is possible that Tgfbr2 acts at a point in odontoblast differentiation driving the cells to be more mature odontoblasts after expression of Dmp1 but before expression of more terminal markers of odontoblast differentiation like Bglap and Dspp.
Figure 5. Odontoblast differentiation is disrupted in Tgfbr2CKO mice.
Representative images showing in situ hybridization for Dspp mRNA in the first mandibular molar form P10 (A, B) and P14 (C, D) day control (A, C) and Tgfbr2cko (B, D) mice. Hybridization to the Dspp probe is seen as purple staining. The termination of the expression domain is marked by arrows. Dspp expression observed in the lateral sides and bifurcation in control teeth is absent in the Tgfbr2cko teeth as is staining in the crown to root transition. R denotes where the root starts. (E) Expression of odontoblast markers in dental pulp of P10 day Tgfbr2cko mice (CKO) vs. control (CON) mice as determined by quantitative real time RT-PCR. Both beta-2-microglobulin (B2m) and glyceraldehyde 3 phospho dehydrogenase (Gapdh) were used as normalization controls.
Nfic levels and localization are not affected by loss of Tgfbr2
It has been shown that the transcription factor Nfic plays a critical role in root formation (Lee et al., 2009a; Lee et al., 2009b; Park et al., 2007; Steele-Perkins et al., 2003). Nfic is widely expressed in both dental epithelium and mesenchyme as well as alveolar bone and the molar roots of Nfic−/− mice fail to develop. More recently it was shown that TGF-ß and Nfic have a complex relationship in the root where TGF-ß promotes the degradation of Nfic while Nfic down-regulates TGF-ß/ Smad signals (Lee et al., 2011). To determine if deletion of Tgfbr2 in dental mesenchyme and alveolar bones affected the expression or localization of Nfic, we used real time RT-PCR and immunohisotochemistry to compare Nfic in control and Tgfbr2cko mice. To localize Nfic at the protein level, we compared immunostaining on sections of the first molar from Tgfbr2cko mice and controls. Alterations in localization of Nfic were not detected at any stage examined (Figure 6A–H). Nfic was strongly detected in the nuclei of pulp cells as well as in osteoblasts of the alveolar bone in both control and mutant teeth. Nfic was strongly detected in the nuclei of odontoblast cells in the crown and root of control teeth, highlighting the organized nature of this cell layer. As described above, the odontoblast layer in the crown was disorganized in mutant teeth and Nfic staining appeared to be reduced but this may be a consequence of the disorganization of the cells (arrows; Figure 6 B, D). Nevertheless, in the truncated root in mutants, Nfic localization was comparable to controls (Figure 6, E–H). Next, RNA was isolated from dental pulp and alveolar bone of P10 day Tgfbr2cko and control mice. Quantitative real-time RTPCR was used to determine the relative expression levels of Nfic in these cell types. No statistically significant differences in Nfic mRNA expression were detected (Figure 6I). Together, these data suggest that Tgfbr2 regulates root formation independent of detectable changes in Nfic localization or expression levels.
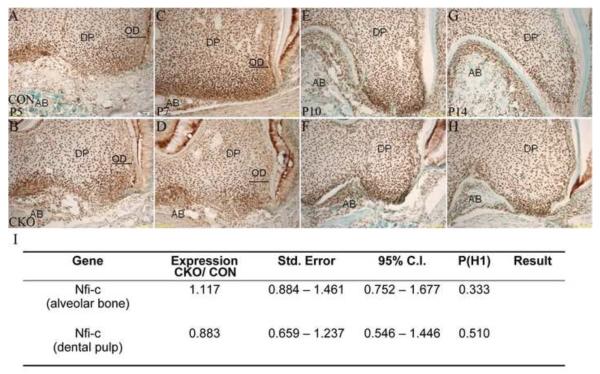
Figure 6. Nfic expression in Tgfbr2CKO mice.
Representative images of Nfic immunostaining in the first mandibular molar from P5 (A, B), 7 (C, D), 10 (E, F) and 14 (G, H) Tgfbrcko (CKO; B, D, F, H) and control (CON; A, C, E, G) mice. DP: dental pulp, AB: alveolar bone, OD (arrow): crown odontoblasts. Magnification: 400×. No differences in the broad Nfic dental tissue localization were observed; however, the odonotoblast layer was disorganized in the mutant mice (OD, arrows). (I) Expression of Nfic mRNA in alveolar bone and dental pulp of P10 day Tgfbr2cko (CKO) vs. control (CON) mice as determined by real time quantitative RT-PCR. Quantitative alterations in the level of Nfic mRNA were not detected in cells isolated from dental pulp or alveolar bone inTgfbr2cko or control mice.
Osteoblast differentiation was disrupted and osteoclast numbers were reduced in alveolar bone surrounding the tooth in Tgfbr2cko mice
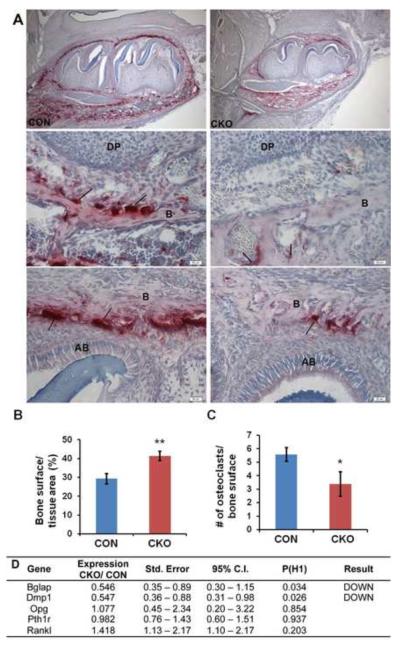
In addition to defects in root formation, the molars in Tgfbr2cko mice did not erupt by 3 weeks after birth although control teeth erupted around 2 weeks of age. Osteoclasts play an important role in regulating both root morphogenesis and tooth eruption (Wise, 2009) (Heinrich et al., 2005; Rani and MacDougall, 2000). During eruption, there is extensive remodeling of the surrounding bone. To look at osteoclasts in the bone surrounding the tooth, we analyzed osteoclast activity using tartrate-resistant acid phosphatase (TRAP) staining and histomorphometry of the bone surrounding the first mandibular molar from P7 days old mice (Figure 7). TRAP staining was reduced in the alveolar bone at the base of the tooth (Figure 7A top and middle) and on the upper occlusal region (Figure 7A top an bottom). Histomorphometry of the occlusal region confirmed that there was a significant reduction in the number of ostoclasts per unit of bone surface in the mutant mice (Figure 7B, C). We did not detect alterations in osteoclast numbers in long bones from 4-week old mice (OC # / bone surface: Control 2.23; CKO 1.98; n=4 each). The results suggest that alterations in bone remodeling in Tgfbr2cko mice could play a role in mediating the root elongation and tooth eruption phenotypes observed in the mice. Since Osx-Cre is expressed in osteoblasts and not in the osteoclasts directly, we propose Tgfbr2 would indirectly regulate osteoclast function in the bone surrounding the tooth through effects on osteoblast function. Dmp1 and Bglap, which are normally expressed in differentiated osteoblasts were down regulated in mutant alveolar bone confirming disruption of osteoblast differentiation (Figure 7 D).
Figure 7. Osteoclast number is reduced in bone from Tgfbr2cko mice.
(A) Representative TRAP-stained images of mandibular sections from P7 control (CON; left) and Tgfbr2cko (CKO; right) mice. The top panels show the whole tooth and surrounding bone. The middle panels show alveolar bone near the root apex. The bottom panels are images of bone at the tooth crown. Magnification: 100× (top) and 400× (middle and bottom). Histomorphometry was used to evaluate (B) bone surface per bone area at the crown and (C) osteoclast number per unit bone surface at the crown. N = 3; *: P < 0.05; **: P < 0.01. Fewer TRAP-stained cells were seen in Tgfbr2cko mice. (D) Expression of osteoblast markers and factors that regulate osteoclast formation in bone. RNA was isolated from alveolar bone of P14 day control (CON) and Tgfbr2cko (CKO) mice. Quantitative real time RT-PCR was used to determine the relative levels of osteoblast differentiation markers and osteoclast regulators in the samples.
To investigate potential osteoblast derived signaling molecules involved in reduced osteoclast numbers, we isolated alveolar bones from P14 day Tgfbr2cko mice. RNA was extracted and quantitative real-time PCR was performed to examine mRNA expression levels of molecules that are known to regulate osteoclast number and behavior and the tooth eruption cycle including RANKL, OPG, CSF-1, and PTH1R (Garlet et al., 2006) (Heinrich et al., 2005; Rani and MacDougall, 2000; Werner et al., 2007). CSF-1 expression was below the level of detection in the samples tested. RANKL, OPG, and PTH1R mRNAs were not significantly different in control and Tgfbr2cko bone.
Discussion
The interaction between the epithelium and the mesenchyme regulates tooth morphogenesis in mammals. Several conserved signaling pathways are involved in this process, including TGF-β signals. The TGF-β type 2 receptor (Tgfbr2) is expressed in both epithelium and neural crest derived mesenchyme, and studies have been performed to conditionally knockout Tgfbr2 independently in both cell types; however, the role of Tgfbr2 in postnatal tooth formation remains unclear since these animals die at birth (Ito et al., 2003; Oka et al., 2007). Our laboratory has established a mouse model in which Tgfbr2 is conditionally deleted in odontoblast-producing mesenchyme using an Osterix promoter driven Cre recombinase (Tgfbr2cko). These mice survive postnatally but with significant defects in the development of bone and teeth evident as early as 7 days postnatal. Tooth root formation was disrupted by postnatal day 10, mimicking radicular dentin dysplasia type 1 (DTDP1; OMIM 125400), a rare human genetic disease associated with short or absent roots. Delayed dental eruption can also be observed in this condition but is atypicial (Rocha et al., 2011). There are few mouse models available to study root development. Here we use a conditional knockout mouse model to study the role of Tgfbr2 in postnatal molar root development and tooth eruption. In the Tgfbr2cko mice, the HERS was disorganized and did not extend deeply into the under-laying mesenchyme to form the root. Delayed dissociation of the truncated HERS to form the epithelial rests of Malassez (ERM) was also observed. Odontoblast crown and tooth cytodifferentiation was disrupted; however, changes in Nfic expression or localization were not observed in the Tgfbr2cko mice. Molar eruption was delayed in the Tgfbr2 mutants, and reduced osteoclast numbers, which would be expected to affect eruption and root elongation, were detected in the surrounding alveolar bone. Our results suggest that Tgfbr2 is required for root formation likely though regulation of odontoblast differentiation in the root to crown transition and osteoclast activity in the surrounding alveolar bone.
Root development requires cytodifferentiation and maintainence of odontoblasts. Interactions between the HERS and dental pulp play an important role in the regulation of odontoblast differentiation. It is believed that factors from the HERS induce the differentiation of mesenchymal cells in the dental pulp apex region to form the odontoblasts that are then responsible for radicular dentin formation. Odontoblasts signal in turn to regulate growth and morphogenesis of the HERS (Luan et al., 2006; Zeichner-David et al., 2003). Osx expression is primarily localized to the dental mesenchyme (Chen et al., 2009; Hirata et al., 2009; Ono et al., 2013; Rakian et al., 2012). Although there is a report of low, transient Osx expression in tooth epithelium in the embryonic stages (Chen et al., 2009), we do not detect Osx-Cre activity in epithelial derivatives of the tooth using a lineage tracing Cre reporter. In addition, Osx-Cre;Tgfbr2cko mice have normal enamel density in contrast to K14-Cre; Smad4cko mice (Huang et al., 2010) suggesting that TGF-ß signaling is intact in the tooth epithelium. The HERS in Osx-Cre;Tgfbrcko mice was disrupted but since Osx-Cre does not target the epithelium, we propose that failure of the HERS to elongate was likely secondary to alterations of TGF-ß signaling in the dental mesenchyme. We found using in situ hybridization that the expression domain of Dspp, a marker of odontoblast differentiation, was more highly restricted in Tgfbr2cko molars relative to controls. In addition, RNA isolated from Tgfbr2cko dental pulp had significantly less Dspp mRNA than the controls. We propose that this is a direct effect of Tgfbr2 on dental mesenchyme and that disruption of the HERS and altered root formation in Tgfbr2cko mice is a consequence of impaired odontoblast differentiation. Previously, disruption of the Smad4 gene in dental mesenchyme using Osteocalcin-Cre, which is active later than Osx-Cre, was also shown to result in shortened molar roots and delayed odontoblast differentiation although less severe than the phenotype we report in Tgfbr2cko mice (Gao et al., 2009).
Nfic is a transcription factor that is required for proper root formation (Park et al., 2007). Nfic is required in the dental mesenchyme to promote radicular odontoblast differentiation independent from crown odontoblasts. In the absence of Nfic, root odontoblast differentiation is disrupted and roots are absent. Since Tgfbr2cko mice had abnormal short roots as well as alterations in root odontoblast differentiation, we tested the hypothesis that Tgfbr2 regulates Nfic. We did not detect any differences in Nfic localization by immunohistochemistry or the levels of Nfic expression as determined by real time RT-PCR in isolated root dental pulp or alveolar bone in Tgfbr2cko versus control mice. This suggests Nfic is not a downstream target of Tgfbr2 signaling in root dental pulp cells or alveolar bone. Previously, it was shown that disruption of Smad4 in dental epithelium results in reduced Nfic expression in the dental pulp; however, this effect was indirect and mediated by Shh secreted from the epithelium (Huang et al., 2010). More recently, it was shown that Nfic and TGF-ß can regulate each other's activity in the dental pulp (Lee et al., 2011). TGF-ß induced degradation of Nfic protein in dental pulp cell cultures during early stages of odontoblast differentiation. At later stages of differentiation, Nfic inhibited TGF-ß signaling through dephosphorylation of Smad3.
Osteoclasts are required to remodel bone and defects in osteoclast numbers or activity have been shown to affect root development as well as tooth eruption (Berdal et al., 2011; Castaneda et al.; Wang and McCauley; Wise, 2009) (Heinrich et al., 2005). Msx2 is a transcription factor that is expressed in osteoclasts at active sites of bone remodeling associated with eruption and root elongation (Aioub et al., 2007). Deletion of Msx2 in osteoclasts resulted in reduced activity and defects in tooth eruption and root morphology. RANK signaling, which is required for osteoclast activation, was modulated by Msx2. Furthermore, over-expression of RANK in the alveolar osteoclasts resulted in early tooth eruption and accelerated root elongation (Castaneda et al.). Tgfbr2cko mice demonstrated a significant decrease in TRAP positive cells in the bone surrounding the tooth. These mice also demonstrated defects in tooth eruption and molar root formation a likely consequence of defects in osteoclast function. Since Osx-Cre is not active in osteoclasts, we propose that disruption of Tgfbr2 in alveolar osteoblasts results in down-regulation of osteoclast function. Disruption of osteoblast differentiation was confirmed by reduced expression of Dmp1 and Bglap, which are normally expressed in differentitated osteoblasts. Cross-talk between osteoblasts and osteoclasts to regulate bone homeostasis is well documented (Phan et al., 2004). It was previously shown that inhibition of TGF-ß receptor signaling in the osteoblasts results in decreased bone remodeling by affecting the number and activity of osteoclasts in the long bones (Filvaroff et al., 1999; Qiu et al., 2010). Osteoblasts express a number of osteoclast regulatory molecules including RANKL and PTH1R. It has been shown that reduced PTHrP signaling results in failure of tooth eruption as well as alterations in the early morphogenesis of the tooth (Kitahara et al., 2002; Philbrick et al., 1996; Wysolmerski et al., 2001). Likewise, constitutive activation of PTH1R also demonstrated delayed tooth eruption and short roots indicating that precise levels of PTHrP signaling are required for normal development of the tooth (Tsutsui et al., 2008). RANKL is also required for tooth eruption (Kong et al., 1999). We were not able to detect alterations in the level of mRNA for several known regulatory factors suggesting that either additional factors are involved or Tgfbr2 does not regulate activity of these molecules at the level of the mRNA.
Highlights
-
-
Tgfbr2 in dental mesenchyme is required for formation of the tooth root.
-
-
Odontoblast differentiation in the root was disrupted in Tgfbr2cko mice.
-
-
The HERS was disorganized and formation of epithelial rests was attenuated.
-
-
Eruption of the molars was disrupted.
-
-
Reduced numbers of osteoclasts were found in the bone surrounding the tooth.
Acknowledgements
This study was supported by grants from the National Institutes of Health, R01- AR053860 to RS and the Institute of Oral Health Research UAB School of Dentistry (UAB SOD). YW and MKC are supported by the UAB SOD NIDCR Dental Academic Research Training Grant (DART) T32-DE017607/ T90DE022736. YW was previously supported by an NIH Institutional Training Grant T32 - AR047512 in bone biology and disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aioub M, Lezot F, Molla M, Castaneda B, Robert B, Goubin G, Nefussi JR, Berdal A. Msx2 −/− transgenic mice develop compound amelogenesis imperfecta, dentinogenesis imperfecta and periodental osteopetrosis. Bone. 2007;41:851–859. doi: 10.1016/j.bone.2007.07.023. [DOI] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riano-Pachon DM, Mueller-Roeber B. QuantPrime--a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics. 2008;9:465. doi: 10.1186/1471-2105-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bei M. Molecular genetics of tooth development. Curr Opin Genet Dev. 2009;19:504–510. doi: 10.1016/j.gde.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdal A, Castaneda B, Aioub M, Nefussi JR, Mueller C, Descroix V, Lezot F. Osteoclasts in the dental microenvironment: a delicate balance controls dental histogenesis. Cells Tissues Organs. 2011;194:238–243. doi: 10.1159/000324787. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Castaneda B, Simon Y, Jacques J, Hess E, Choi YW, Blin-Wakkach C, Mueller C, Berdal A, Lezot F. Bone resorption control of tooth eruption and root morphogenesis: Involvement of the receptor activator of NF-kappaB (RANK) J Cell Physiol. 226:74–85. doi: 10.1002/jcp.22305. [DOI] [PubMed] [Google Scholar]
- Caton J, Tucker AS. Current knowledge of tooth development: patterning and mineralization of the murine dentition. J Anat. 2009;214:502–515. doi: 10.1111/j.1469-7580.2008.01014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Gluhak-Heinrich J, Wang YH, Wu YM, Chuang HH, Chen L, Yuan GH, Dong J, Gay I, MacDougall M. Runx2, osx, and dspp in tooth development. J Dent Res. 2009;88:904–909. doi: 10.1177/0022034509342873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chytil A, Magnuson MA, Wright CV, Moses HL. Conditional inactivation of the TGF-beta type II receptor using Cre:Lox. Genesis. 2002;32:73–75. doi: 10.1002/gene.10046. [DOI] [PubMed] [Google Scholar]
- Cui W, Taub DD, Gardner K. qPrimerDepot: a primer database for quantitative real time PCR. Nucleic Acids Res. 2007;35:D805–809. doi: 10.1093/nar/gkl767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson CA, Tucker AS, Christensen L, Lau AL, Matzuk MM, Sharpe PT. Activin is an essential early mesenchymal signal in tooth development that is required for patterning of the murine dentition. Genes Dev. 1998;12:2636–2649. doi: 10.1101/gad.12.16.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filvaroff E, Erlebacher A, Ye J, Gitelman SE, Lotz J, Heillman M, Derynck R. Inhibition of TGF-beta receptor signaling in osteoblasts leads to decreased bone remodeling and increased trabecular bone mass. Development. 1999;126:4267–4279. doi: 10.1242/dev.126.19.4267. [DOI] [PubMed] [Google Scholar]
- Gao Y, Yang G, Weng T, Du J, Wang X, Zhou J, Wang S, Yang X. Disruption of Smad4 in odontoblasts causes multiple keratocystic odontogenic tumors and tooth malformation in mice. Mol Cell Biol. 2009;29:5941–5951. doi: 10.1128/MCB.00706-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlet GP, Cardoso CR, Silva TA, Ferreira BR, Avila-Campos MJ, Cunha FQ, Silva JS. Cytokine pattern determines the progression of experimental periodontal disease induced by Actinobacillus actinomycetemcomitans through the modulation of MMPs, RANKL, and their physiological inhibitors. Oral Microbiol Immunol. 2006;21:12–20. doi: 10.1111/j.1399-302X.2005.00245.x. [DOI] [PubMed] [Google Scholar]
- Heinrich J, Bsoul S, Barnes J, Woodruff K, Abboud S. CSF-1, RANKL and OPG regulate osteoclastogenesis during murine tooth eruption. Arch Oral Biol. 2005;50:897–908. doi: 10.1016/j.archoralbio.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Helfrich MH. Osteoclast diseases and dental abnormalities. Arch Oral Biol. 2005;50:115–122. doi: 10.1016/j.archoralbio.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Hirata A, Sugahara T, Nakamura H. Localization of runx2, osterix, and osteopontin in tooth root formation in rat molars. J Histochem Cytochem. 2009;57:397–403. doi: 10.1369/jhc.2008.952192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Xu X, Bringas P, Jr., Hung YP, Chai Y. Smad4-Shh-Nfic signaling cascade-mediated epithelial-mesenchymal interaction is crucial in regulating tooth root development. J Bone Miner Res. 2010;25:1167–1178. doi: 10.1359/jbmr.091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Yeo JY, Chytil A, Han J, Bringas P, Jr., Nakajima A, Shuler CF, Moses HL, Chai Y. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 2003;130:5269–5280. doi: 10.1242/dev.00708. [DOI] [PubMed] [Google Scholar]
- Janssens K, ten Dijke P, Janssens S, Van Hul W. Transforming growth factor-beta1 to the bone. Endocr Rev. 2005;26:743–774. doi: 10.1210/er.2004-0001. [DOI] [PubMed] [Google Scholar]
- Khosla S. Minireview: the OPG/RANKL/RANK system. Endocrinology. 2001;142:5050–5055. doi: 10.1210/endo.142.12.8536. [DOI] [PubMed] [Google Scholar]
- Kitahara Y, Suda N, Kuroda T, Beck F, Hammond VE, Takano Y. Disturbed tooth development in parathyroid hormone-related protein (PTHrP)-gene knockout mice. Bone. 2002;30:48–56. doi: 10.1016/s8756-3282(01)00669-x. [DOI] [PubMed] [Google Scholar]
- Ko SO, Chung IH, Xu X, Oka S, Zhao H, Cho ES, Deng C, Chai Y. Smad4 is required to regulate the fate of cranial neural crest cells. Dev Biol. 2007;312:435–447. doi: 10.1016/j.ydbio.2007.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong YY, Yoshida H, Sarosi I, Tan HL, Timms E, Capparelli C, Morony S, Oliveira-dos-Santos AJ, Van G, Itie A, Khoo W, Wakeham A, Dunstan CR, Lacey DL, Mak TW, Boyle WJ, Penninger JM. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- Lee DS, Park JT, Kim HM, Ko JS, Son HH, Gronostajski RM, Cho MI, Choung PH, Park JC. Nuclear factor I-C is essential for odontogenic cell proliferation and odontoblast differentiation during tooth root development. J Biol Chem. 2009a;284:17293–17303. doi: 10.1074/jbc.M109.009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DS, Yoon WJ, Cho ES, Kim HJ, Gronostajski RM, Cho MI, Park JC. Crosstalk between nuclear factor I-C and transforming growth factor-beta1 signaling regulates odontoblast differentiation and homeostasis. PLoS One. 2011;6:e29160. doi: 10.1371/journal.pone.0029160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TY, Lee DS, Kim HM, Ko JS, Gronostajski RM, Cho MI, Son HH, Park JC. Disruption of Nfic causes dissociation of odontoblasts by interfering with the formation of intercellular junctions and aberrant odontoblast differentiation. J Histochem Cytochem. 2009b;57:469–476. doi: 10.1369/jhc.2009.952622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2011;13:27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- Luan X, Ito Y, Diekwisch TG. Evolution and development of Hertwig's epithelial root sheath. Dev Dyn. 2006;235:1167–1180. doi: 10.1002/dvdy.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lungova V, Radlanski RJ, Tucker AS, Renz H, Misek I, Matalova E. Tooth-bone morphogenesis during postnatal stages of mouse first molar development. J Anat. 2011;218:699–716. doi: 10.1111/j.1469-7580.2011.01367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- Narayanan K, Srinivas R, Ramachandran A, Hao J, Quinn B, George A. Differentiation of embryonic mesenchymal cells to odontoblast-like cells by overexpression of dentin matrix protein 1. Proc Natl Acad Sci U S A. 2001;98:4516–4521. doi: 10.1073/pnas.081075198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka S, Oka K, Xu X, Sasaki T, Bringas P, Jr., Chai Y. Cell autonomous requirement for TGF-beta signaling during odontoblast differentiation and dentin matrix formation. Mech Dev. 2007;124:409–415. doi: 10.1016/j.mod.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono W, Ono N, Kroneneberg H. PTH/PTHrP receptor in Osterix-expressing prgenitors is essential for root formation. J. Dental Res. 2013;92:1023. [Google Scholar]
- Paaske PB, Witten J, Schwer S, Hansen HS. Results in treatment of carcinoma of the external auditory canal and middle ear. Cancer. 1987;59:156–160. doi: 10.1002/1097-0142(19870101)59:1<156::aid-cncr2820590130>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Park JC, Herr Y, Kim HJ, Gronostajski RM, Cho MI. Nfic gene disruption inhibits differentiation of odontoblasts responsible for root formation and results in formation of short and abnormal roots in mice. J Periodontol. 2007;78:1795–1802. doi: 10.1902/jop.2007.060363. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TC, Xu J, Zheng MH. Interaction between osteoblast and osteoclast: impact in bone disease. Histol Histopathol. 2004;19:1325–1344. doi: 10.14670/HH-19.1325. [DOI] [PubMed] [Google Scholar]
- Philbrick WM, Wysolmerski JJ, Galbraith S, Holt E, Orloff JJ, Yang KH, Vasavada RC, Weir EC, Broadus AE, Stewart AF. Defining the roles of parathyroid hormone-related protein in normal physiology. Physiol Rev. 1996;76:127–173. doi: 10.1152/physrev.1996.76.1.127. [DOI] [PubMed] [Google Scholar]
- Qiu T, Wu X, Zhang F, Clemens TL, Wan M, Cao X. TGF-beta type II receptor phosphorylates PTH receptor to integrate bone remodelling signalling. Nat Cell Biol. 2010;12:224–234. doi: 10.1038/ncb2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakian A, Gluhak-Heinrich J, Yang W, Harris MA, Chun YP, Cui Y, Villarreal D, Harris S. BMP-2 controls links between odontoblasts, stem cells, and angiogenesis. J. Dental Res. 2012;91:166. [Google Scholar]
- Rani CS, MacDougall M. Dental cells express factors that regulate bone resorption. Mol Cell Biol Res Commun. 2000;3:145–152. doi: 10.1006/mcbr.2000.0205. [DOI] [PubMed] [Google Scholar]
- Rocha CT, Nelson-Filho P, Silva LA, Assed S, Queiroz AM. Variation of dentin dysplasia type I: report of atypical findings in the permanent dentition. Braz Dent J. 2011;22:74–78. doi: 10.1590/s0103-64402011000100013. [DOI] [PubMed] [Google Scholar]
- Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- Sohn P, Cox M, Chen D, Serra R. Molecular profiling of the developing mouse axial skeleton: a role for Tgfbr2 in the development of the intervertebral disc. BMC Dev Biol. 2010;10:29. doi: 10.1186/1471-213X-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Perkins G, Butz KG, Lyons GE, Zeichner-David M, Kim HJ, Cho MI, Gronostajski RM. Essential role for NFI-C/CTF transcription-replication factor in tooth root development. Mol Cell Biol. 2003;23:1075–1084. doi: 10.1128/MCB.23.3.1075-1084.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thesleff I, Nieminen P. Tooth morphogenesis and cell differentiation. Curr Opin Cell Biol. 1996;8:844–850. doi: 10.1016/s0955-0674(96)80086-x. [DOI] [PubMed] [Google Scholar]
- Thomas HF. Root formation. Int J Dev Biol. 1995;39:231–237. [PubMed] [Google Scholar]
- Tsutsui TW, Riminucci M, Holmbeck K, Bianco P, Robey PG. Development of craniofacial structures in transgenic mice with constitutively active PTH/PTHrP receptor. Bone. 2008;42:321–331. doi: 10.1016/j.bone.2007.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XP, O'Connell DJ, Lund JJ, Saadi I, Kuraguchi M, Turbe-Doan A, Cavallesco R, Kim H, Park PJ, Harada H, Kucherlapati R, Maas RL. Apc inhibition of Wnt signaling regulates supernumerary tooth formation during embryogenesis and throughout adulthood. Development. 2009;136:1939–1949. doi: 10.1242/dev.033803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XP, Suomalainen M, Jorgez CJ, Matzuk MM, Werner S, Thesleff I. Follistatin regulates enamel patterning in mouse incisors by asymmetrically inhibiting BMP signaling and ameloblast differentiation. Dev Cell. 2004;7:719–730. doi: 10.1016/j.devcel.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Wang Z, McCauley LK. Osteoclasts and odontoclasts: signaling pathways to development and disease. Oral Dis. 17:129–142. doi: 10.1111/j.1601-0825.2010.01718.x. [DOI] [PubMed] [Google Scholar]
- Werner SA, Gluhak-Heinrich J, Woodruff K, Wittrant Y, Cardenas L, Roudier M, MacDougall M. Targeted expression of csCSF-1 in op/op mice ameliorates tooth defects. Arch Oral Biol. 2007;52:432–443. doi: 10.1016/j.archoralbio.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise GE. Cellular and molecular basis of tooth eruption. Orthod Craniofac Res. 2009;12:67–73. doi: 10.1111/j.1601-6343.2009.01439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysolmerski JJ, Cormier S, Philbrick WM, Dann P, Zhang JP, Roume J, Delezoide AL, Silve C. Absence of functional type 1 parathyroid hormone (PTH)/PTH-related protein receptors in humans is associated with abnormal breast development and tooth impaction. J Clin Endocrinol Metab. 2001;86:1788–1794. doi: 10.1210/jcem.86.4.7404. [DOI] [PubMed] [Google Scholar]
- Xu X, Han J, Ito Y, Bringas P, Jr., Deng C, Chai Y. Ectodermal Smad4 and p38 MAPK are functionally redundant in mediating TGF-beta/BMP signaling during tooth and palate development. Dev Cell. 2008;15:322–329. doi: 10.1016/j.devcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeichner-David M, Oishi K, Su Z, Zakartchenko V, Chen LS, Arzate H, Bringas P., Jr. Role of Hertwig's epithelial root sheath cells in tooth root development. Dev Dyn. 2003;228:651–663. doi: 10.1002/dvdy.10404. [DOI] [PubMed] [Google Scholar]