Highlights
-
•
TMDs control the intracellular transport of many membrane proteins.
-
•
The length and hydrophobicity of TMDs determine their sorting.
-
•
Some membrane receptors for sorting TMDs have been identified.
-
•
Lipid partitioning may also participate in the sorting of TMDs.
Keywords: transmembrane domains, protein sorting, protein traffic, lipid domains, transmembrane receptors, endomembrane system
Abstract
The transmembrane domains (TMDs) of integral membrane proteins have emerged as major determinants of intracellular localization and transport in the secretory and endocytic pathways. Unlike sorting signals in cytosolic domains, TMD sorting determinants are not conserved amino acid sequences but physical properties such as the length and hydrophilicity of the transmembrane span. The underlying sorting machinery is still poorly characterized, but several mechanisms have been proposed, including TMD recognition by transmembrane sorting receptors and partitioning into membrane lipid domains. Here we review the nature of TMD sorting determinants and how they may dictate transmembrane protein localization and transport.
Transmembrane domains: not just anchors
Transmembrane proteins account for 20–30% of all proteins encoded in the genome of eukaryotic organisms [1]. They include bitopic proteins that span the membrane only once and polytopic proteins that cross the membrane multiple times. To perform their functions, transmembrane proteins must be transported and localized to the correct intracellular compartment [e.g., endoplasmic reticulum (ER), Golgi apparatus, plasma membrane, endosomes] and in many cases traffic in a regulated manner between different compartments (e.g., cycling of the transferrin receptor between the plasma membrane and endosomes, ligand-induced endocytosis of signaling receptors) (Figure 1 ). Intracellular localization and traffic are often determined by information contained within the cytosolic domains of the transmembrane proteins. This information generally comprises linear amino acid motifs or folded domains that interact with components of protein coats, thus leading to selective incorporation of membrane proteins into transport vesicles [2]. However, although cytosolic sorting determinants have received the most attention in the protein-trafficking field, numerous studies over the past 25 years have shown that TMDs also contribute to protein localization and transport.
Figure 1.
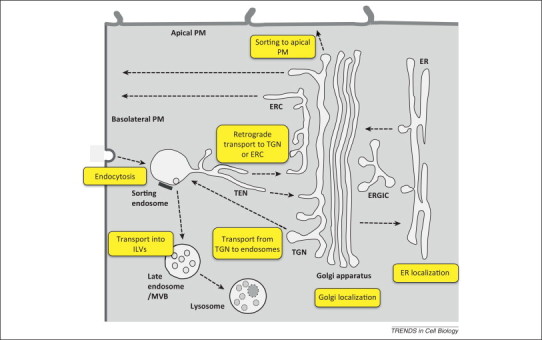
Sorting processes mediated by transmembrane domains (TMDs). Schematic representation of intracellular transport pathways (arrows) and processes in which TMDs participate in protein sorting (yellow boxes). Newly synthesized transmembrane proteins can be transported from the endoplasmic reticulum (ER) to the Golgi apparatus, from where they return to the ER (ER–Golgi recycling) or continue on to the trans-Golgi network (TGN) and the plasma membrane (PM) (secretory pathway). The ER–Golgi intermediate compartment (ERGIC) may play a role in both anterograde and retrograde transport steps. In polarized epithelial cells, the PM is specialized in apical and basolateral domains to which proteins are differentially sorted (polarized sorting). PM proteins can be internalized into endosomes (endocytosis) from where they can return to the PM via the endocytic recycling compartment (ERC) (endocytic recycling) or undergo transport to the TGN via the tubular endosomal network (TEN) (retrograde transport) or to late endosomes/multivesicular bodies (MVBs) and then to lysosomes (lysosomal transport). In MVBs, proteins can either remain in the limiting membrane or be transported into intraluminal vesicles (ILVs) (MVB pathway). Some proteins cycle between the TGN and endosomes (TGN–endosome recycling).
TMDs usually comprise a stretch of 17–25 (average 21) hydrophobic amino acid residues [3] that are structured as an α-helix [4]. A bioinformatics analysis of integral membrane proteins encoded in eukaryotic genomes revealed a strong correlation between the intracellular localization of the proteins and the exact length and amino acid composition of their TMDs [5]. For instance, the TMDs of ER proteins were found to be shorter than those of plasma membrane proteins. Furthermore, the TMDs of plasma membrane proteins exhibit an asymmetric distribution of amino-acids along the α-helix, with more valine and glycine residues toward the exofacial side and more leucine residues toward the endofacial side of the membrane [5]. By contrast, the TMDs of ER proteins do not display such asymmetry. These findings provided a global perspective on previous experimental analyses of the contribution of TMDs to various sorting events, including localization to the ER and the Golgi apparatus, endocytosis from the plasma membrane, transport from endosomes to the trans-Golgi network (TGN), and entry into intraluminal vesicles (ILVs) of multivesicular bodies (MVBs) (Figure 1 and Table 1 ). Here we review the critical roles played by TMDs in intracellular sorting and discuss the mechanisms that have been proposed to explain TMD-mediated sorting.
Table 1.
Examples of TMD sorting determinants
| Protein | Sorting event | TMD determinant | Refs |
|---|---|---|---|
| Unassembled α subunit of the T cell receptor | ERAD targeting | 20-aa TMD containing two critical basic residues | [13] |
| Unassembled mIgM subunit of the B cell receptor | ER localization | 26-aa TMD containing ten polar residues, four of which are critical for ER localization | [12] |
| p24 ER cargo receptor | ER localization | 19-aa TMD containing critical Glu residue | [15] |
| UBC6 ubiquitin-conjugating enzyme | ER localization | 17-aa TMD | [18] |
| Cosmc ER chaperone | ER localization | 18-aa TMD containing critical Cys residue | [19] |
| Coronavirus E1 glycoprotein | Golgi localization | First of three TMDs; 22 aa long, containing critical polar residues | [63] |
| β-1,4-Galactosyl-transferase | Golgi localization | 20-aa TMD containing critical Cys and polar residues | [28] |
| Syntaxin 5 t-SNARE | Golgi localization | 17-aa TMD | [26] |
| Transferrin receptor mutant | Internalization and recycling | Placement of three polar residues within the 25-aa TMD promotes internalization and inhibits recycling | [30] |
| CD1b mutant | Internalization | Shortening of TMD from 21 to 18 aa promotes internalization | [31] |
| TGN38 TGN protein | Endosome-to-TGN transport | 21-aa TMD; lengthening to 24 aa decreases transport | [32] |
| Pep12 t-SNARE | MVB sorting | Placement of acidic residues in the 18-aa TMD diverts protein into ILVs | [35] |
Abbreviation: aa, amino acid.
Sorting events mediated by TMDs
ER retention and degradation
Retention of proteins in the ER was one of the first sorting processes found to depend on TMDs (Table 1). Studies on the assembly and transport of hetero-oligomeric membrane complexes such as the T-cell antigen receptor (TCR) (comprising eight type I transmembrane subunits, αβγδɛ2ζ2) showed that only fully assembled complexes reach the cell surface, whereas unassembled subunits or partial complexes are retained in the ER 6, 7. For some unassembled subunits, ER retention is followed by degradation 6, 7 through a ubiquitin/proteasome-dependent process known as ER-associated degradation (ERAD) [8]. Information leading to ER retention and ERAD targeting was mapped to the TMDs of the proteins [7]. The TMDs of TCR subunits are unusual in that they contain one or two charged residues (basic in the TCR-α and -β subunits; acidic in the CD3-γ, -δ, -ɛ, and -ζ subunits) that contribute to ER retention and ERAD targeting, as demonstrated by the fact that mutating them to hydrophobic residues disrupts both processes [7]. Moreover, simple placement of a charged or strongly polar residue (i.e., asparagine, glutamine) in the TMD of a reporter plasma membrane protein can confer localization to the ER and targeting to ERAD [9]. Whether strongly polar residues cause just ER retention or additional ERAD targeting depends on the nature of the residue and its position within the TMD. These outcomes are also dependent on the length of the TMD, with shorter TMDs enhancing and longer TMDs diminishing the effects of charged residues [10]. Similar to the TCR subunits, the α-chain of the high-affinity IgE receptor contains a charged residue [11] and the membrane IgM (mIgM) subunit of the B-cell antigen receptor (BCR) [12] contains a large number of polar residues (nine serine and threonine residues) that contribute to retention of the unassembled subunits in the ER. The TMDs of these proteins also mediate subunit interactions, such that subunit assembly abrogates ER retention and ERAD targeting [13]. By coupling oligomer assembly with export from the ER, the TMD-dependent sorting machinery participates in quality control, ensuring that only fully assembled complexes reach the plasma membrane.
Similar determinants account for the localization of ER-resident proteins and some viral envelope glycoproteins to the ER (Table 1). For example, charged or hydrophilic residues in the TMDs contribute to the ER localization of the cellular proteins cytochrome P450 2C1 [14] and p24 [15], as well as envelope glycoproteins from hepatitis C virus [16] and Dengue virus [17]. The shorter length of TMDs from UBC6 (17 residues) [18] also determines ER localization. Finally, the TMD of the ER chaperone Cosmc directs ER retention, although in this case it is by virtue of a cysteine residue that participates in disulfide-bonded dimerization [19].
Golgi localization
TMDs are also a major determinant of protein localization to the Golgi apparatus, albeit often in cooperation with other topologic domains (Table 1). This role of TMDs was first demonstrated for the coronavirus E1 glycoprotein [20] and later found to apply to other viral envelope glycoproteins 17, 21, 22. Additionally, TMDs contribute to Golgi localization of a large number of glycosylation enzymes that process the carbohydrate chains of newly synthesized glycoproteins and glycolipids as they traverse the Golgi apparatus 23, 24. Likewise, other Golgi proteins such as GOLPH2 [25] and Syntaxin 5 [26] depend on their TMDs for Golgi localization. Dissection of Golgi localization determinants in these proteins revealed a requirement for hydrophilic, sulfhydryl, and/or aromatic residues in the TMDs as well as shorter-than-average TMDs 26, 27, 28, 29.
Sorting at the plasma membrane and endosomes
The role of TMDs in protein sorting is not limited to early compartments of the secretory pathway but also extends to the plasma membrane and endosomes (Table 1). Rapid endocytosis of plasma membrane proteins is primarily mediated by endocytic signals encoded within the cytosolic domain [2]. However, alterations in the TMD can affect the rate of endocytosis. For example, placement of polar residues (i.e., threonine, glutamine) in the TMD of the transferrin receptor increases receptor internalization and reduces its recycling to the plasma membrane, contributing to its downregulation from the cell surface [30]. Similarly, shortening the TMD of a CD1b reporter protein from 21 to 18 residues promotes internalization through increased capture into clathrin-coated pits [31]. Endosomes are a major site of protein sorting from where transmembrane proteins can be routed for recycling to the plasma membrane or the TGN or for delivery to lysosomes (or the yeast vacuole) via the MVB pathway. TMDs also play roles in these endosomal routes 32, 33, 34, 35. For instance, the 21-residue TMD of TGN38 promotes ‘retrograde’ transport of this protein from early endosomes to the TGN; lengthening of the TGN38 TMD to 24 residues impairs this process [32]. In yeast, polar residues in TMDs promote delivery of proteins from the TGN to endosomes and then to ILVs, whereas more hydrophobic TMDs restrict proteins to the limiting membrane of MVBs 34, 35. Finally, sorting of the influenza virus hemagglutinin from the TGN or endosomes to the apical plasma membrane domain of polarized epithelial cells is also determined by the protein's TMD [36].
Mechanisms of TMD-mediated sorting
Similarities between TMD sorting determinants
Remarkably, TMD determinants that control sorting in different intracellular compartments share common properties. Shorter and more hydrophilic TMDs tend to localize proteins to early compartments of the secretory pathway (ER–Golgi), whereas longer and more hydrophobic TMDs favor localization to the plasma membrane. Similar but subtler differences influence sorting at the plasma membrane and in endosomes. Thus, proteins with short or hydrophilic TMDs are excluded from the cell surface first because they are retained in the ER or Golgi apparatus and second because the fraction of these proteins that escapes these early compartments is subsequently targeted to endosomal/lysosomal compartments. These experimentally determined roles of TMDs in protein sorting are broadly in line with bioinformatic analyses, which concluded that ER and Golgi proteins exhibit shorter TMDs than plasma membrane proteins and that endosomal proteins may have TMDs of intermediate length [5].
Overall, these observations indicate that TMD sorting determinants are not conserved sequences or motifs but global properties such as the length and hydrophilicity of the transmembrane spans. TMD length and hydrophilicity may be related properties, because the presence of polar residues in a TMD shortens the hydrophobic span and, reciprocally, shortening a TMD could force flanking polar residues into the membrane. Currently, it is unclear how the presence of other residues such as cysteine or the asymmetric distribution of residues along the TMD α-helix influence sorting.
Two distinct mechanisms have been proposed for TMD-dependent sorting: recognition by transmembrane proteins that function as receptors or adaptors and spontaneous partitioning into distinct membrane lipid domains. Evidence in favor of these two mechanisms at specific transport steps is discussed in the next sections.
Sorting at the ER–Golgi interface: Erv14 and Rer1 transmembrane receptors
A study of fluorescently tagged, C-terminally anchored proteins expressed in mammalian cells provided clues about how TMDs might control sorting at ER exit sites [37]. This study showed that a protein with a short TMD localizes to ER cisternae but not to sites of ER exit for transport to the Golgi apparatus. By contrast, the same protein with a longer TMD is recruited to ER exit sites. The protein that does not enter ER exit sites has a high diffusion coefficient, excluding the possibility that this protein is immobilized by interaction with a putative ER matrix. Genetic analyses in yeast revealed that the transmembrane protein Erv14 acts as a sorting receptor for sorting proteins with long TMDs to ER exit sites. Indeed, Erv14 was shown to interact specifically with proteins having long TMDs and to target them to COPII-coated vesicles that transport cargo from the ER to the Golgi apparatus (Figure 2 ). Loss of Erv14 abrogates efficient transport of proteins with long TMDs to the cell surface. Another study uncovered receptor-mediated retrieval from the Golgi apparatus as another mechanism contributing to TMD-dependent ER localization. Analysis of the glycosylation patterns of proteins with short/hydrophilic TMDs in yeast showed that these proteins can gain access to early Golgi compartments, from where they are continuously and specifically retrieved back to the ER. Retrieval was found to be dependent on COPI, another protein coat involved in retrograde transport from the Golgi apparatus to the ER [38]. Screening for yeast mutants defective for ER localization of the transmembrane protein Sec12, which localizes to the ER by virtue of its TMD, identified a requirement for Rer1, a transmembrane protein that cycles between the Golgi apparatus and the ER [39]. These studies indicated that Sec12 is exported from the ER, but once in the Golgi apparatus its TMD is recognized by Rer1. The Rer1 cytosolic domain in turn interacts with COPI, leading to retrieval of the Sec12–Rer1 complex to the ER 38, 40. Interaction with Rer1 mediates the ER localization of several other ER-resident proteins, such as Mns1 [41] and Sec71 [42], as well as the unassembled Fet3 subunit of a reductive iron transporter [43]. Of note, these proteins have multiple TMDs, and it seems likely that Rer1 recognizes the folding status of the TMDs. In summary, Rer1 functions as a receptor that recognizes the TMDs of a subset of ER proteins, enabling their continuous retrieval to the ER by the COPI transport machinery (Figure 3A). The coordinate action of Erv14 and Rer1 ensures the efficient export of proteins with long TMDs from the ER and the retrieval of proteins with short TMDs to the ER and is probably capable of recognizing more complex determinants such as the folding state of polytopic membrane proteins (Figure 2). Other receptors such as Gsf2 and Dip5 have also been proposed to mediate export of specific subsets of membrane proteins from the ER [44].
Figure 2.
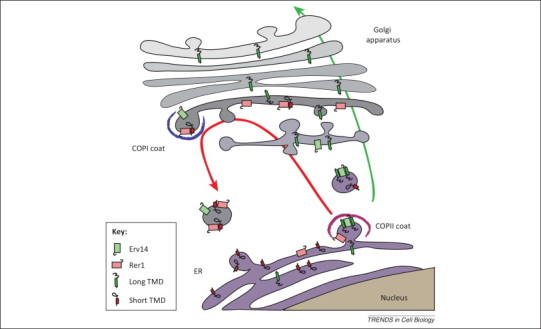
TMD recognition in the early secretory pathway. To be expressed at the cell surface, membrane proteins must be transported along the secretory pathway from the endoplasmic reticulum (ER) to the Golgi apparatus and then to the plasma membrane (green arrow). In the early steps of the secretory pathway, transmembrane domains (TMDs) are recognized at least at two distinct sites. During the formation of COPII-coated vesicles, ER proteins with long TMDs are concentrated in budding vesicles due to their interaction with Erv14 and are thus efficiently transported to the cis-Golgi apparatus and beyond. By contrast, proteins with short TMDs are bound by Rer1 in the cis-Golgi and concentrated in retrograde COPI-coated vesicles destined for the ER (red arrow). These two systems, probably in concert with partitioning into different lipid domains, ensure efficient transport of proteins with long TMDs along the secretory pathway and localization of proteins with short TMDs to the ER. This model assumes that both Erv14 and Rer1 bind their targets in a regulated manner, to capture them in one compartment (e.g., the ER for Erv14) and release them in another (e.g., the Golgi for Erv14) before returning to their original location, but the mechanisms involved are unknown. This simplified scheme also does not depict other putative sorting receptors involved in the export of distinct subsets of membrane proteins from the ER.
Figure 3.
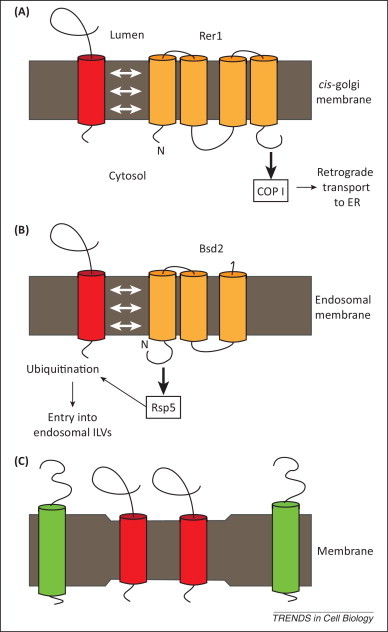
Molecular mechanisms of transmembrane domain (TMD)-mediated sorting. (A) Interaction with sorting receptors. In cis-Golgi cisternae, Rer1 interacts with the TMD of a subset of transmembrane proteins such as Sec12, Sec71, and Mns1. The Rer1 cytosolic domain then recruits the COPI coat, which returns bound proteins to the endoplasmic reticulum (ER). Similarly, Erv14 binds proteins with long TMDs in the ER and concentrates them in COPII-coated ER exit vesicles. (B) Interaction with transmembrane ubiquitination adaptors. Recognition of polar residues in the TMD of Cps1 by the adaptor protein Bsd2 allows recruitment of the cytosolic ubiquitin ligase Rsp5. This causes ubiquitination of the cytosolic domains of Cps1 and its targeting to intraluminal vesicles (ILVs). (C) Lipid partitioning. In reconstituted lipid bilayers, short TMDs segregate into thinner membrane domains whereas long TMDs are found in thicker membranes. In living cells, a similar mechanism coupled to the formation of transport vesicles may ensure differential transport of transmembrane proteins.
Targeting to endosomal ILVs: role of the ubiquitination machinery
Studies in yeast have shown that the TMDs of some proteins can be recognized by transmembrane components of the ubiquitination machinery, leading to ubiquitin-dependent sorting of the proteins. In the ER, the presence of hydrophilic residues in the TMDs of misfolded or unassembled proteins can be detected by transmembrane ubiquitin ligases such as Hrd1 [45], resulting in targeting to the ERAD pathway. A similar TMD recognition mechanism has been shown to mediate transport from the late Golgi apparatus to endosomes and then into ILVs. One study showed that targeting of the transmembrane vacuolar hydrolases carboxypeptidase S (Cps1) and polyphosphate phosphatase (Phm5) into ILVs depends on recognition of polar TMD residues by the transmembrane ubiquitin ligase Tul1 [46]. A later study revealed an alternative sorting mechanism involving recognition of polar TMD residues in Cps1 and Phm5a by the membrane protein Bsd2, which functions as an adaptor for the cytosolic ubiquitin ligase Rsp5 [47] (Figure 3B). In both cases, recognition of polar residues in the TMD of a protein ultimately leads to ubiquitination of its cytosolic domain, a signal for targeting to ILVs by the MVB pathway [48].
TMD-mediated sorting through partitioning into lipid domains
The physical properties of TMDs may also determine spontaneous partition of transmembrane proteins into different lipid domains. Minimizing exposure of hydrophobic groups to the aqueous environment is a driving force for the assembly of cellular membranes and consequently one might expect lipid bilayers and proteins with similar hydrophobic spans to segregate together. In principle, the length and hydrophobicity of TMDs could simply determine partitioning of the corresponding proteins into different membrane environments, with short/hydrophilic and long/hydrophobic TMDs promoting protein segregation to thinner and thicker membranes, respectively (Figure 3C). Indeed, electron microscopy of fixed cells has revealed a gradient of membrane thickness from the ER (thinner) to the plasma membrane (thicker) [49]. A study of synthetic transmembrane peptides embedded in reconstituted lipid bilayers provided experimental support for lipid-dependent sorting: when short transmembrane peptides were inserted into reconstituted membranes, they segregated together with shorter lipids into discrete microdomains excluding long transmembrane peptides. Segregation was maximal when the mismatch between transmembrane peptide length and membrane thickness was highest. This lateral sorting occurred only in the presence of cholesterol, which presumably straightened lipid acyl chains and increased the energetic cost of mixing peptides and lipids of different hydrophobic length [50].
An affinity of some TMDs for specific lipids (e.g., cholesterol [51]) could also drive partitioning into specific membrane microdomains. Indeed, membrane cholesterol content has also been shown to increase from the ER to the plasma membrane 52, 53. More generally, membrane microdomains of specific lipid and protein composition (e.g., lipid rafts, caveolae) have been studied extensively [54] and segregation to such domains may also be linked to intracellular sorting and transport. A recent study revealed the existence of multiple microdomains at the plasma membrane of yeast cells [55], pointing to a much greater diversity of membrane domains than has been envisioned so far.
Although the general principle of lipid partitioning provides a sound basis for explaining TMD-dependent protein localization, it remains to be established how this principle operates in living cells at specific transport steps. The relative contributions of protein-based and lipid-based mechanisms of TMD-dependent sorting are also unclear, although cooperation of both mechanisms is likely.
Concluding remarks
The studies reviewed here make it abundantly clear that TMDs play key roles in transmembrane protein sorting in the endomembrane system. Although understanding of TMD sorting determinants has lagged behind that of cytosolic sorting signals, there is growing awareness that information in both topologic domains contributes to defining the precise location and traffic of transmembrane proteins within cells. The interplay between TMD and cytosolic determinants could take several forms. TMD and cytosolic determinants within the same protein could mediate different sorting steps (e.g., retention and retrieval). Alternatively, one determinant could influence the function of the other in the same sorting step. For example, TMD-dependent segregation into certain lipid domains or TMD–receptor interactions could change the oligomeric state of transmembrane proteins, thus modulating the avidity of cytosolic domains for protein coats.
Compelling genetic evidence indicates that sorting by TMDs relies at least partly on protein–protein interactions within membranes. To date, the argument in favor of lipid partitioning of TMDs is based on less direct evidence and its relative importance in intracellular sorting of TMDs remains to be established. Our understanding of protein-based and lipid-based mechanisms of TMD-mediated sorting is far from complete and does not exclude less conventional explanations. For example, proteins like Rer1 may not function strictly as sorting receptors but rather organize lipid domains into which proteins partition for recycling to the ER. Likewise, loose networks of TMD interactions could mediate lateral segregation of transmembrane proteins leading to partitioning of a whole group of proteins into a lipid domain.
The role of TMDs in specific sorting events is but one of many processes that are known to involve molecular recognition within lipid bilayers. The function of intramembrane chaperones such as BAP31 [56], proteases such as presenilins [57], and viral accessory proteins such as HIV-1 Vpu [58] also rely on substrate recognition via TMDs. A detailed understanding of all of these processes will require more extensive genetic and proteomic analysis, as well as the development of more powerful methods for examining the behavior of proteins in lipid environments. For example, X-ray crystallography and NMR [59] are beginning to provide insights into how TMDs interact among themselves and with structural lipids (i.e., those that are tightly associated with proteins). Mass spectrometry [60] as well as fluorescence spectroscopy and imaging techniques 61, 62 are also shedding new light into protein–protein and protein–lipid interactions in the membrane. Application of these methods to the problem of TMD-mediated sorting should contribute to answering several key questions. How do various TMDs interact among themselves and with lipids? Does binding of specific structural lipids (e.g., cholesterol, sphingomyelin) to TMDs confer partitioning into specific lipid microdomains? How do interactions between luminal and cytosolic domains influence TMD–TMD and TMD–lipid interactions? How do post-translational modifications such as palmitoylation modulate TMD-mediated sorting? Addressing these questions will expand our understanding of molecular recognition events that occur in the hydrophobic milieu of membranes, bringing it up to par with the detailed knowledge of the mechanisms of signal recognition in the aqueous environment of the cytosol.
References
- 1.Wallin E., von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Traub L.M. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat. Rev. Mol. Cell Biol. 2009;10:583–596. doi: 10.1038/nrm2751. [DOI] [PubMed] [Google Scholar]
- 3.Pasquier C. A novel method for predicting transmembrane segments in proteins based on a statistical analysis of the SwissProt database: the PRED-TMR algorithm. Protein Eng. 1999;12:381–385. doi: 10.1093/protein/12.5.381. [DOI] [PubMed] [Google Scholar]
- 4.Engelman D.M. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 5.Sharpe H.J. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 2010;142:158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lippincott-Schwartz J. Degradation from the endoplasmic reticulum: disposing of newly synthesized proteins. Cell. 1988;54:209–220. doi: 10.1016/0092-8674(88)90553-3. [DOI] [PubMed] [Google Scholar]
- 7.Bonifacino J.S. A peptide sequence confers retention and rapid degradation in the endoplasmic reticulum. Science. 1990;247:79–82. doi: 10.1126/science.2294595. [DOI] [PubMed] [Google Scholar]
- 8.McCracken A.A., Brodsky J.L. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J. Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonifacino J.S. Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J. 1991;10:2783–2793. doi: 10.1002/j.1460-2075.1991.tb07827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lankford S.P. Transmembrane domain length affects charge-mediated retention and degradation of proteins within the endoplasmic reticulum. J. Biol. Chem. 1993;268:4814–4820. [PubMed] [Google Scholar]
- 11.Cauvi D.M. Transport of the IgE receptor alpha-chain is controlled by a multicomponent intracellular retention signal. J. Biol. Chem. 2006;281:10448–10460. doi: 10.1074/jbc.M510751200. [DOI] [PubMed] [Google Scholar]
- 12.Williams G.T. The sequence of the mu transmembrane segment determines the tissue specificity of the transport of immunoglobulin M to the cell surface. J. Exp. Med. 1990;171:947–952. doi: 10.1084/jem.171.3.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonifacino J.S. Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell. 1990;63:503–513. doi: 10.1016/0092-8674(90)90447-m. [DOI] [PubMed] [Google Scholar]
- 14.Szczesna-Skorupa E., Kemper B. Endoplasmic reticulum retention determinants in the transmembrane and linker domains of cytochrome P450 2C1. J. Biol. Chem. 2000;275:19409–19415. doi: 10.1074/jbc.M002394200. [DOI] [PubMed] [Google Scholar]
- 15.Fiedler K., Rothman J.E. Sorting determinants in the transmembrane domain of p24 proteins. J. Biol. Chem. 1997;272:24739–24742. doi: 10.1074/jbc.272.40.24739. [DOI] [PubMed] [Google Scholar]
- 16.Ciczora Y. Contribution of the charged residues of hepatitis C virus glycoprotein E2 transmembrane domain to the functions of the E1E2 heterodimer. J. Gen. Virol. 2005;86:2793–2798. doi: 10.1099/vir.0.81140-0. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh S.C. The length of and nonhydrophobic residues in the transmembrane domain of dengue virus envelope protein are critical for its retention and assembly in the endoplasmic reticulum. J. Virol. 2010;84:4782–4797. doi: 10.1128/JVI.01963-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang M. The transmembrane domain of a carboxyl-terminal anchored protein determines localization to the endoplasmic reticulum. J. Biol. Chem. 1997;272:1970–1975. doi: 10.1074/jbc.272.3.1970. [DOI] [PubMed] [Google Scholar]
- 19.Sun Q. The transmembrane domain of the molecular chaperone Cosmc directs its localization to the endoplasmic reticulum. J. Biol. Chem. 2011;286:11529–11542. doi: 10.1074/jbc.M110.173591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Machamer C.E., Rose J.K. A specific transmembrane domain of a coronavirus E1 glycoprotein is required for its retention in the Golgi region. J. Cell Biol. 1987;105:1205–1214. doi: 10.1083/jcb.105.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerrard S.R., Nichol S.T. Characterization of the Golgi retention motif of Rift Valley fever virus G(N) glycoprotein. J. Virol. 2002;76:12200–12210. doi: 10.1128/JVI.76.23.12200-12210.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaecher S.R. The transmembrane domain of the severe acute respiratory syndrome coronavirus ORF7b protein is necessary and sufficient for its retention in the Golgi complex. J. Virol. 2008;82:9477–9491. doi: 10.1128/JVI.00784-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munro S. Sequences within and adjacent to the transmembrane segment of alpha-2,6-sialyltransferase specify Golgi retention. EMBO J. 1991;10:3577–3588. doi: 10.1002/j.1460-2075.1991.tb04924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teasdale R.D. The signal for Golgi retention of bovine beta 1,4-galactosyltransferase is in the transmembrane domain. J. Biol. Chem. 1992;267:13113. [PubMed] [Google Scholar]
- 25.Hu L. The Golgi localization of GOLPH2 (GP73/GOLM1) is determined by the transmembrane and cytoplamic sequences. PLoS ONE. 2011;6:e28207. doi: 10.1371/journal.pone.0028207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watson R.T., Pessin J.E. Transmembrane domain length determines intracellular membrane compartment localization of syntaxins 3, 4, and 5. Am. J. Physiol. Cell Physiol. 2001;281:C215–C223. doi: 10.1152/ajpcell.2001.281.1.C215. [DOI] [PubMed] [Google Scholar]
- 27.Munro S. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 1995;14:4695–4704. doi: 10.1002/j.1460-2075.1995.tb00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masibay A.S. Mutational analysis of the Golgi retention signal of bovine beta-1,4-galactosyltransferase. J. Biol. Chem. 1993;268:9908–9916. [PubMed] [Google Scholar]
- 29.Sousa V.L. Importance of Cys, Gln, and Tyr from the transmembrane domain of human alpha 3/4 fucosyltransferase III for its localization and sorting in the Golgi of baby hamster kidney cells. J. Biol. Chem. 2003;278:7624–7629. doi: 10.1074/jbc.M209325200. [DOI] [PubMed] [Google Scholar]
- 30.Zaliauskiene L. Down-regulation of cell surface receptors is modulated by polar residues within the transmembrane domain. Mol. Biol. Cell. 2000;11:2643–2655. doi: 10.1091/mbc.11.8.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercanti V. Transmembrane domains control exclusion of membrane proteins from clathrin-coated pits. J. Cell Sci. 2010;123:3329–3335. doi: 10.1242/jcs.073031. [DOI] [PubMed] [Google Scholar]
- 32.Chia P.Z. Rab9-dependent retrograde transport and endosomal sorting of the endopeptidase furin. J. Cell Sci. 2011;124:2401–2413. doi: 10.1242/jcs.083782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J. The transmembrane domain of acid trehalase mediates ubiquitin-independent multivesicular body pathway sorting. Mol. Biol. Cell. 2007;18:2511–2524. doi: 10.1091/mbc.E06-11-0995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rayner J.C., Pelham H.R. Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J. 1997;16:1832–1841. doi: 10.1093/emboj/16.8.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reggiori F. Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol. Biol. Cell. 2000;11:3737–3749. doi: 10.1091/mbc.11.11.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin S. Mutations in the middle of the transmembrane domain reverse the polarity of transport of the influenza virus hemagglutinin in MDCK epithelial cells. J. Cell Biol. 1998;142:51–57. doi: 10.1083/jcb.142.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ronchi P. Transmembrane domain-dependent partitioning of membrane proteins within the endoplasmic reticulum. J. Cell Biol. 2008;181:105–118. doi: 10.1083/jcb.200710093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Letourneur F., Cosson P. Targeting to the endoplasmic reticulum in yeast cells by determinants present in transmembrane domains. J. Biol. Chem. 1998;273:33273–33278. doi: 10.1074/jbc.273.50.33273. [DOI] [PubMed] [Google Scholar]
- 39.Nishikawa S., Nakano A. Identification of a gene required for membrane protein retention in the early secretory pathway. Proc. Natl. Acad. Sci. U.S.A. 1993;90:8179–8183. doi: 10.1073/pnas.90.17.8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato K. Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer. J. Cell Biol. 2001;152:935–944. doi: 10.1083/jcb.152.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Massaad M.J., Herscovics A. Interaction of the endoplasmic reticulum alpha 1,2-mannosidase Mns1p with Rer1p using the split-ubiquitin system. J. Cell Sci. 2001;114:4629–4635. doi: 10.1242/jcs.114.24.4629. [DOI] [PubMed] [Google Scholar]
- 42.Sato K. Rer1p, a retrieval receptor for ER membrane proteins, recognizes transmembrane domains in multiple modes. Mol. Biol. Cell. 2003;14:3605–3616. doi: 10.1091/mbc.E02-12-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato M. Endoplasmic reticulum quality control of unassembled iron transporter depends on Rer1p-mediated retrieval from the golgi. Mol. Biol. Cell. 2004;15:1417–1424. doi: 10.1091/mbc.E03-10-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herzig Y. A systematic approach to pair secretory cargo receptors with their cargo suggests a mechanism for cargo selection by Erv14. PLoS Biol. 2012;10:e1001329. doi: 10.1371/journal.pbio.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato B.K. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol. Cell. 2009;34:212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reggiori F., Pelham H.R. A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat. Cell Biol. 2002;4:117–123. doi: 10.1038/ncb743. [DOI] [PubMed] [Google Scholar]
- 47.Hettema E.H. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 2004;23:1279–1288. doi: 10.1038/sj.emboj.7600137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henne W.M. The ESCRT pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 49.Sandelius A.S. Isolation of highly purified fractions of plasma membrane and tonoplast from the same homogenate of soybean hypocotyls by free-flow electrophoresis. Plant Physiol. 1986;81:177–185. doi: 10.1104/pp.81.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaiser H.J. Lateral sorting in model membranes by cholesterol-mediated hydrophobic matching. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16628–16633. doi: 10.1073/pnas.1103742108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanson M.A. A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lange Y. Disposition of intracellular cholesterol in human fibroblasts. J. Lipid Res. 1991;32:329–339. [PubMed] [Google Scholar]
- 53.van Meer G. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inder K.L. Ripples in the pond - using a systems approach to decipher the cellular functions of membrane microdomains. Mol. Biosyst. 2013;9:330–338. doi: 10.1039/c2mb25300c. [DOI] [PubMed] [Google Scholar]
- 55.Spira F. Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat. Cell Biol. 2012;14:640–648. doi: 10.1038/ncb2487. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y., Williams D.B. Assembly of MHC class I molecules within the endoplasmic reticulum. Immunol. Res. 2006;35:151–162. doi: 10.1385/IR:35:1:151. [DOI] [PubMed] [Google Scholar]
- 57.Wolfe M.S. Structure, mechanism and inhibition of gamma-secretase and presenilin-like proteases. Biol. Chem. 2010;391:839–847. doi: 10.1515/BC.2010.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magadan J.G., Bonifacino J.S. Transmembrane domain determinants of CD4 downregulation by HIV-1 Vpu. J. Virol. 2012;86:757–772. doi: 10.1128/JVI.05933-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lacapere J.J. Determining membrane protein structures: still a challenge! Trends Biochem. Sci. 2007;32:259–270. doi: 10.1016/j.tibs.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 60.Barrera N.P., Robinson C.V. Advances in the mass spectrometry of membrane proteins: from individual proteins to intact complexes. Annu. Rev. Biochem. 2011;80:247–271. doi: 10.1146/annurev-biochem-062309-093307. [DOI] [PubMed] [Google Scholar]
- 61.Contreras F.X. Molecular recognition of a single sphingolipid species by a protein's transmembrane domain. Nature. 2012;481:525–529. doi: 10.1038/nature10742. [DOI] [PubMed] [Google Scholar]
- 62.Betaneli V. The role of lipids in VDAC oligomerization. Biophys. J. 2012;102:523–531. doi: 10.1016/j.bpj.2011.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swift A.M., Machamer C.E. A Golgi retention signal in a membrane-spanning domain of coronavirus E1 protein. J. Cell Biol. 1991;115:19–30. doi: 10.1083/jcb.115.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]


