Although mRNAs encoding secreted and membrane proteins are believed to associate with the ER only upon translation, they access the membrane independently of both translational control and the signal recognition particle. Thus, alternate paths exist for RNA delivery to and retention at the ER.
Abstract
mRNAs encoding secreted/membrane proteins (mSMPs) are believed to reach the endoplasmic reticulum (ER) in a translation-dependent manner to confer protein translocation. Evidence exists, however, for translation- and signal recognition particle (SRP)–independent mRNA localization to the ER, suggesting that there are alternate paths for RNA delivery. We localized endogenously expressed mSMPs in yeast using an aptamer-based RNA-tagging procedure and fluorescence microscopy. Unlike mRNAs encoding polarity and secretion factors that colocalize with cortical ER at the bud tip, mSMPs and mRNAs encoding soluble, nonsecreted, nonpolarized proteins localized mainly to ER peripheral to the nucleus (nER). Synthetic nontranslatable uracil-rich mRNAs were also demonstrated to colocalize with nER in yeast. This mRNA–ER association was verified by subcellular fractionation and reverse transcription-PCR, single-molecule fluorescence in situ hybridization, and was not inhibited upon SRP inactivation. To better understand mSMP targeting, we examined aptamer-tagged USE1, which encodes a tail-anchored membrane protein, and SUC2, which encodes a soluble secreted enzyme. USE1 and SUC2 mRNA targeting was not abolished by the inhibition of translation or removal of elements involved in translational control. Overall we show that mSMP targeting to the ER is both translation- and SRP-independent, and regulated by cis elements contained within the message and trans-acting RNA-binding proteins (e.g., She2, Puf2).
INTRODUCTION
Targeted mRNA delivery to specific subcellular compartments and localized protein synthesis are crucial for various cellular and physiological functions. One major and well-studied mRNA targeting pathway is the translation-dependent delivery of secreted/membrane proteins (mSMPs) into the endoplasmic reticulum (ER), as initially described by Blobel and Dobberstein (1975). According to this model, ribosomes engaged in mSMP translation are recognized by the signal recognition particle (SRP) via the signal peptide present at the amino terminus of the newly synthesized polypeptide. The SRP binds the ribosome–mRNA–nascent polypeptide chain complex via the signal peptide and traffics it from the cytoplasm to the ER. mRNA anchoring is then achieved through binding of the SRP to its receptor on the ER membrane, and protein elongation across the membrane is carried out via a protein-conducting channel called the translocon (Schwartz, 2007). According to this model, the cis-acting element directing mRNA localization is not a sequence contained within the mRNA but instead the amino acid sequence of the newly translated signal peptide, which serves as a positive selection signal to recruit the SRP. Although the SRP model has been widely accepted, it is unclear whether all mRNAs target the ER in a translation-dependent manner or whether other pathways for RNA delivery exist. Based mainly on studies using prokaryotes (Bibi, 2012), an alternative model suggests that mRNA and ribosomes can undergo independent targeting to membranes by which ribosomes access membranes via an association with the SRP receptor, whereas mRNAs are targeted independently of translation by cis-acting sequences contained within the transcript.
Evidence for SRP-independent mRNA localization to the ER has emerged (reviewed in Kraut-Cohen and Gerst, 2010). Several groups have examined the distribution of mRNAs coding for cytosolic and membrane-bound proteins between the cytosol and ER-bound polyribosome fractions (Diehn et al., 2000; Lerner et al., 2003; Pyhtila et al., 2008) and found that a large number of mRNA species (>9000 in mammalian cells) are shared between the two compartments. More recently, Nicchitta and colleagues examined the partitioning of mRNAs encoding three classes of proteins —cytosolic, endomembrane, and secretory cargo—between the ER and cytosol. All three classes were found spread between the two compartments, with mRNAs encoding secretory cargo showing a broad spectrum of distribution and mRNAs encoding proteins that lack a signal sequence fractionating with the ER (Chen et al., 2011). Thus lack of a signal sequence does not prevent the localization of mRNAs encoding cytosolic proteins to ER membranes (Nicchitta et al. 2005; Pyhtila et al., 2008; Chen et al., 2011). This pattern of localization was not altered in cells either lacking a component of the SRP complex or treated with protein synthesis inhibitors (Lerner et al., 2003; Pyhtila et al., 2008), whereas the disruption of polysome aggregation (using EDTA) or removal of the signal sequence also did not alter the distribution of mRNAs from the ER (Pyhtila et al., 2008). Similarly, certain mRNAs in Escherichia coli were also found to translocate to the inner membrane independently of translation (Nevo-Dinur et al., 2011). Thus these different studies provide substantial evidence that alternative paths may direct mRNAs to the ER even when the SRP pathway and translation are interrupted. Of importance, this finding may explain why loss of the SRP pathway in yeast and in higher eukaryotes does not result in lethality (Mutka and Walter, 2001; Lakkaraju et al., 2007).
According to the classic SRP model, an mRNA must be translated in order for the signal peptide to serve as an ER-targeting device. Sequence analysis of mSMPs from different organisms, however, reveals the existence of unique features (Palazzo et al., 2007; Prilusky and Bibi, 2009) that suggest that mRNAs might contain SRP-independent targeting information. For example, a recent nonbiased analysis of mRNA nucleotide composition (Prilusky and Bibi, 2009) showed that mRNAs encoding membrane proteins are strongly biased toward containing uracil (U-biased; Prilusky and Bibi, 2009) due to the use of uracil in codons for hydrophobic amino acids. Of probable relevance, a uracil-rich sequence contained within the extensive 3′ untranslated region (UTR; which likely results from a premature transcription termination site) of the yeast PMP1 gene is sufficient to mediate mRNA targeting in a translation-independent manner (Loya et al., 2008). Moreover, genomic analyses demonstrate that nucleotide stretches encoding signal sequence–coding regions (SSCRs) in eukaryotes have decreased use of adenines (A) and create “no-A” stretches in these tracts (Palazzo et al., 2007). Together the data suggest that mSMPs include sequence information that distinguishes them from the general mRNA population. Finally, although ER-targeting motifs for mRNAs are not well defined, several RNA-binding proteins (RBPs) in yeast (e.g., She2, Scp160, Puf1, Puf2, and Whi3) preferentially bind mRNAs associated with the secretory pathway and may serve in SRP-independent mRNA-delivery pathways (Li et al., 2003; Gerber et al., 2004; Jambhekar et al., 2005; Schmid et al., 2006; Aronov et al., 2007; Colomina et al., 2008; Gelin-Licht et al., 2012).
In this study, we describe the localization of endogenously expressed mSMPs that encode ER-resident proteins and proteins that either localize later in the secretory pathway or are secreted from the cell. By using three different and complementary procedures—in vivo fluorescence microscopy, in situ hybridization, and subcellular fractionation and reverse transcription (RT)-PCR—we show that mSMPs reside on ER membranes and, in particular, on ER that is peripheral to the nucleus (nER). mRNA association with the ER was not abolished by the inhibition of translation or inactivation of SRP, the latter accomplished using a temperature-sensitive allele of SRP65, and even synthetic nontranslatable uracil-rich mRNAs were found to be ER localized. In contrast, non-mSMPs (e.g., FSH3) were either distributed equally between the ER and cytosol or enriched in the cytosolic fraction. Together the results indicate that multiple mRNAs are targeted to the ER in a manner that is independent of translation and SRP but may involve cis-acting sequence elements and trans-acting RBPs.
RESULTS
Endogenously expressed mSMPs colocalize with ER, as shown by fluorescence microscopy
To visualize endogenously expressed mSMPs in living yeast, we used m-TAG, a chromosomal tagging procedure that inserts 12 MS2 aptamer loop (MS2L) sequences between the open reading frame (ORF) and 3′UTR for any gene of interest via homologous recombination (Haim et al., 2007; Haim-Vilmovsky and Gerst, 2009). Subsequently the MS2 aptamer-tagged mRNA can be visualized upon the induced expression of the bacteriophage MS2 coat protein (MS2-CP) fused to several copies of green fluorescent protein (GFP; e.g., MS2-CP-GFP(x3) or (x4)). This technique has been used to successfully localize in vivo endogenously expressed mRNAs to the ER, mitochondria, peroxisome, and actin patches (Haim et al., 2007; Zipor et al., 2009; Gadir et al., 2011; Casolari et al., 2012).
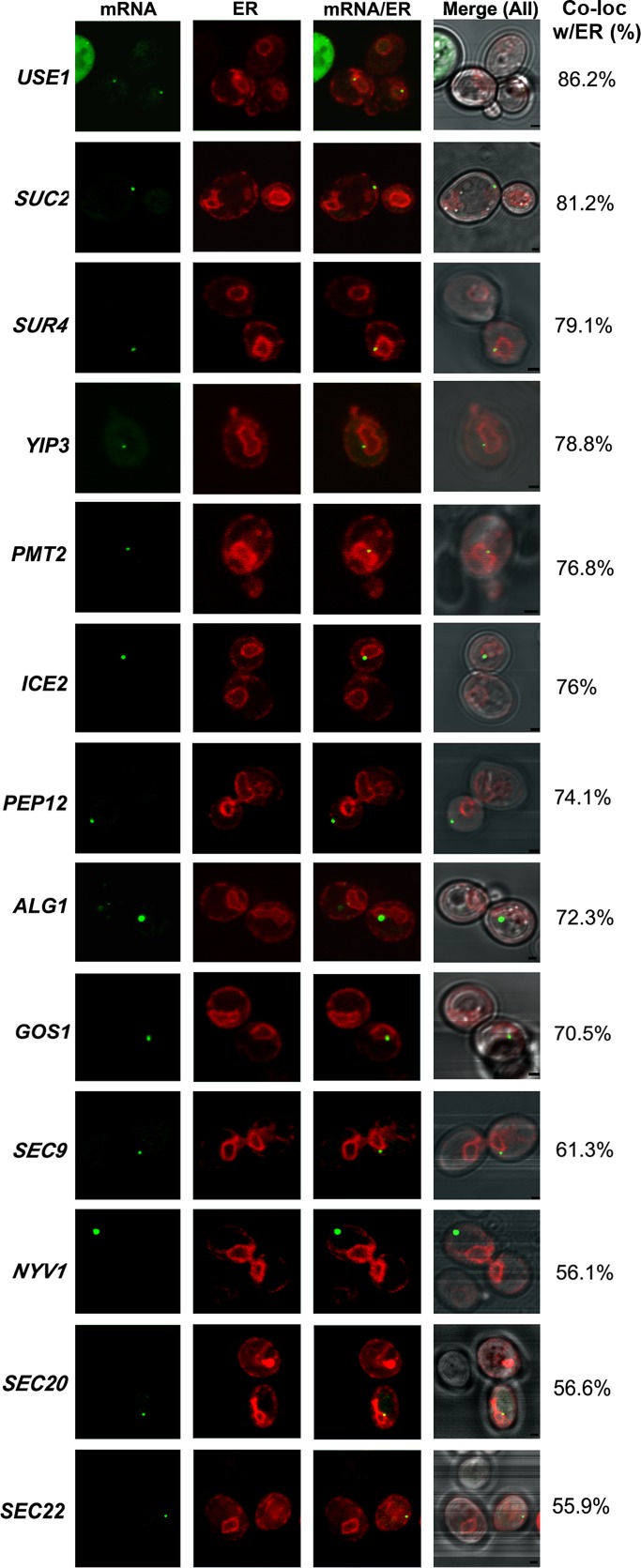
We determined mSMP localization by examining colocalization of the GFP-labeled mRNA granules with red fluorescent protein (RFP)–tagged Sec63, an ER marker. ER localization was divided into four categories: nuclear ER (nER), cortical ER (cER), both nuclear and cortical ER (both), and non-ER localization (Supplemental Figure S1). We tagged 11 genes coding mSMPs (ALG1, GOS1, ICE2, NYV1, PEP12, PMT1, SEC22, SUC2, SUR4, USE1, and YIP3) and two non-SMP controls that encode non–membrane-anchored, soluble N-ethylmaleimide–sensitive factor attachment protein receptors (SNAREs; SEC9, SEC20) of the secretory pathway that localize to the plasma membrane and ER, respectively. Representative images of the different tagged strains are shown in Figure 1, and statistics for the scoring of mRNA localization to the ER (∼100 cells scored for each experiment; n = 2 or 3 experiments) are given in Table 1. In the cases in which fluorescent granules were hard to detect using MS2-CP-GFP(x3), probably due to low levels of gene expression, we used MS2-CP-GFP(x4) (i.e., fused to four GFP molecules; Zipor et al., 2009). Because we found that MS2-CP-GFP(x4) can induce the accumulation of fluorescent aggregates in a low percentage (8%) of wild-type cells (Zipor et al., 2009; and data not shown), we took care not to count cells containing large aggregates and minimized MS2-CP-GFP(x4) induction by growing cells in medium containing methionine (i.e., without direct induction), which results in residual gene expression from the somewhat leaky MET25 promoter. Under these conditions typically only one RNA granule per cell was observed. In addition, we found no significant difference in the localization of mSMPs using MS2-CP-GFP(x3) or MS2-CP-GFP (x4). For example, fluorescent aptamer-tagged USE1 granules showed 86% colocalization with ER using MS2-CP-GFP(x3) for detection and 83% colocalization using MS2-CP-GFP(x4) (Table 1).
FIGURE 1:
Visualization of endogenous mSMPs and their colocalization with ER. Cells bearing the MS2L aptamer sequence integrated into genes encoding various mSMPs and secretory pathway components were transformed with plasmids expressing MS2-CP-GFP(x3) or MS2-CP-GFP(x4) (see Table 1 for details) and Sec63-RFP (an ER marker). Cells expressing MS2-CP-GFP(x3) were grown on liquid synthetic medium and induced with the same medium lacking methionine for 1 h before visualization. Cells expressing MS2-CP-GFP(x4) were grown on liquid synthetic medium without induction. For SUC2 mRNAs, cells were grown on low glucose (0.1%)–containing medium to induce SUC2 expression. Percentage of fluorescent RNA granules colocalized with ER (% Local.). mRNA, localization of GFP-labeled RNA granules; ER, localization of ER labeled with Sec63-RFP; mRNA/ER, merger of ER and mRNA windows; Merge (All), merger of light (differential interference contrast), ER, and mRNA windows. Scale bar, 1 μm.
TABLE 1:
Localization to the ER of endogenous mRNAs encoding secretory pathway proteins.
| Percentage of RNA granules colocalized with ER | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene accession number | Gene name (strain background) | INTa | Granules detected by CP-GFP(x3) | Granules detected by CP-GFP(x4) | Cortical ER | Nuclear ER | Both | Non-ER | Total ER localization |
| YGL098W | USE1 (WT) | + | + | 26.0 | 52.1 | 8.2 | 13.7 | 86.2 | |
| YGL098W | USE1 (WT) | + | + | 23.5 | 55.6 | 4.3 | 16.6 | 83.4 | |
| YIL162W | SUC2 (WT) | + | + | 31.2 | 42.5 | 7.5 | 18.8 | 81.2 | |
| YLR372W | SUR4 (WT) | + | + | 20.7 | 52.8 | 5.6 | 20.9 | 79.1 | |
| YNL044W | YIP3 (WT) | + | + | 29.1 | 39.5 | 10.2 | 20.9 | 78.8 | |
| YAL023C | PMT2 (WT) | + | + | 28.8 | 44.2 | 3.8 | 23.2 | 76.8 | |
| YIL090W | ICE2 (WT) | + | + | 34.0 | 35.0 | 7.0 | 24.0 | 76.0 | |
| YOR036W | PEP12 (WT) | + | + | 46.0 | 22.1 | 6.0 | 25.9 | 74.0 | |
| YBR110W | ALG1 (WT) | + | + | 33.7 | 31.0 | 7.6 | 26.9 | 72.3 | |
| YHL031C | GOS1 (WT) | + | + | 44.0 | 17.7 | 8.8 | 29.5 | 70.5 | |
| YGR009C | SEC9 (WT) | + | + | 34.2 | 21.4 | 5.7 | 38.7 | 61.3 | |
| YLR093C | NYV1 (WT) | + | + | 34.3 | 18.8 | 3.1 | 43.8 | 56.2 | |
| YDR498C | SEC20 (WT) | + | 16.6 | 35.0 | 5.0 | 43.4 | 56.6 | ||
| YLR268W | SEC22 (WT) | + | + | 32.8 | 20 | 3.10 | 44.1 | 55.9 | |
| USE1 (she2Δ) | + | + | 34.2 | 40.3 | 5.6 | 19.9 | 80.1 ± 7.3 | ||
| USE1 (whi3Δ) | + | + | 36.1 | 44.6 | 3.1 | 16.2 | 83.8 ± 2.6 | ||
| USE1 (puf1Δ) | + | + | 34.7 | 40.9 | 7.6 | 16.8 | 83.2 ± 5.1 | ||
| USE1 (puf2Δ) | + | + | 23.2 | 42.2 | 4.8 | 29.8 | 70.2 ± 6.6 | ||
| SUC2 (she2Δ) | + | + | 32.6 | 28.8 | 1.0 | 37.6 | 62.4 ± 6.4 | ||
| SUC2 (whi3Δ) | + | + | 38.7 | 35.5 | 6.5 | 19.3 | 80.7 ± 0.0 | ||
| SUC2 (puf1Δ) | + | + | 31.8 | 39.9 | 3.9 | 24.4 | 75.6 ± 2.9 | ||
| SUC2 (puf2Δ) | + | + | 31.7 | 27.7 | 0.9 | 39.7 | 60.3 ± 1.8 | ||
| SUC2 (puf2Δ she2Δ) | + | + | 21.5 | 30.8 | 1.4 | 46.3 | 53.7 ± 1.9 | ||
| USE1 10’CHX | + | + | 33.7 | 46.2 | 5 | 15.1 | 84.9 | ||
| USE1 30’CHX | + | + | 22.5 | 61.9 | 1.4 | 14.2 | 85.8 | ||
| SUC2 10’CHX | + | + | 34.6 | 36.6 | 5.9 | 22.8 | 77.1 | ||
| SUC2 30’CHX | + | + | 47.4 | 26.2 | 2.0 | 24.4 | 75.6 | ||
We examined mRNA localization by confocal microscopy and found that 9 of 11 mSMPs tagged (ALG1, GOS1, ICE2, PEP12, PMT2, SUC2, SUR4, USE1, and YIP3) showed high levels of granule colocalization with ER (>70%; Figure 1 and Table 1), and 2 (NYV1 and SEC22) showed lower levels of ER localization (56%; Figure 1 and Table 1). A similar low level of ER localization was observed with two mRNAs that encode soluble SNAREs (SEC9 and SEC20) known to associate with the plasma and ER membranes (Figure 1 and Table 1). Of interest, some preference toward nER localization was observed with mSMPs such as PMT2, SUC2, SUR4, USE1, and YIP3, whereas in contrast, most of the mRNAs encoding SNAREs (e.g., GOS1, NYV1, PEP12, SEC9, and SEC22) demonstrated a preference for cER (Table 1). It is important to note that unlike mRNAs encoding polarity and secretion factors (mPOLs; e.g., SRO7, SEC3, SEC4, CDC42, etc.), which localize with cER at the incipient bud site or bud tip (Aronov et al., 2007), mSMP localization was restricted to the mother in small-budded cells (i.e., early in the cell cycle). Thus mSMPs are not polarized, unlike ASH1 mRNA (Takizawa et al., 1997; Long et al., 1997), mPOLs (Aronov et al., 2007), and other mRNAs that use the She2 RBP for localization (Shepard et al., 2003).
The highest levels of mSMP granule colocalization with ER were observed with USE1 (86% colocalization), which encodes an essential tail-anchored SNARE involved in Golgi–ER retrograde transport (Belgareh-Touze et al., 2003; Dilcher et al., 2003), and SUC2 (81% colocalization), which encodes the secreted enzyme invertase (Carlson et al., 1983). Both mRNAs exhibit ER localization throughout the cell cycle (Supplemental Movies S1 and S2, respectively). SUC2 mRNA localization was performed in cells shifted to 0.1% glucose for 1.5 h to induce expression of the mRNA encoding the secreted form of invertase instead of the constitutively expressed short form of SUC2, which lacks the signal sequence (Carlson and Botstein, 1982; Ozcan et al., 1997).
Endogenously expressed mSMPs colocalize with ER, as shown by subcellular fractionation
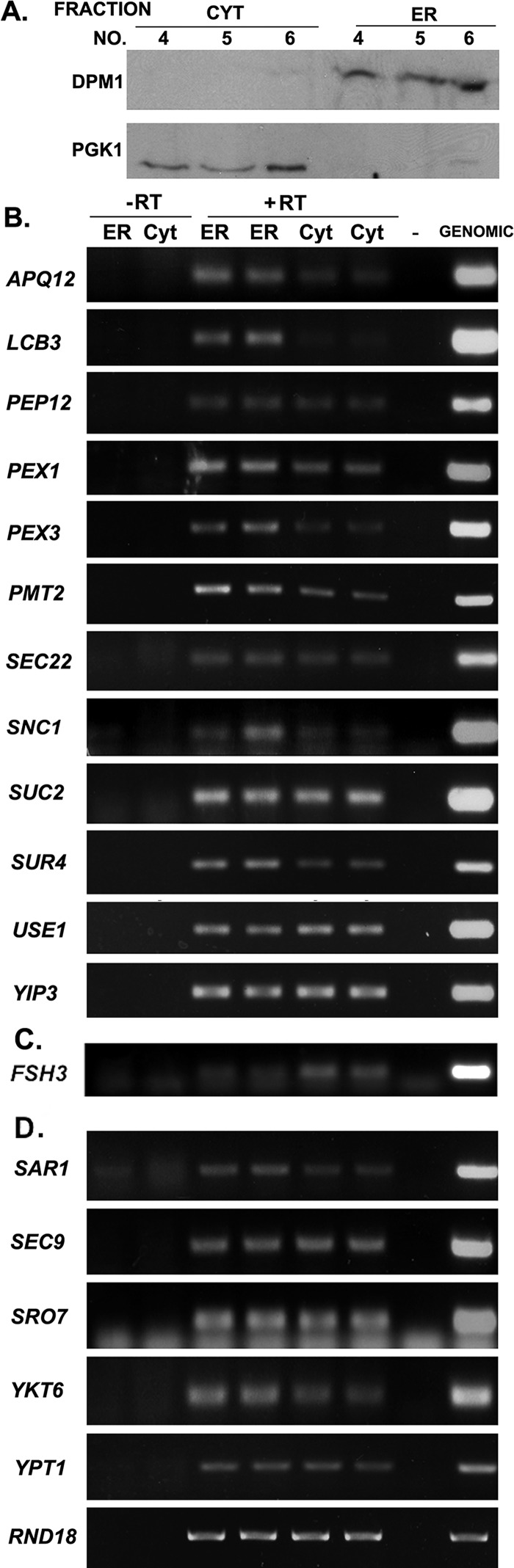
To verify that mSMPs physically associate with ER and that the MS2 aptamer sequence is not solely responsible for ER targeting, we lysed wild-type cells pretreated for 15 min with cycloheximide (to prevent the dissociation of translating RNAs) under conditions that preserve RNA and separated the postnuclear supernatant into cytosolic and microsomal fractions using nonlinear density gradient centrifugation. Separation of the cytosolic and ER-enriched microsome fractions was verified using antibodies to phosphoglycerate kinase (Pgk1) and dolichol phosphate mannose synthase (Dpm1), respectively (Figure 2A). We also verified that the ER microsome fraction was not enriched with mitochondrial membranes, using an antibody to Tom70, a mitochondrial membrane protein, and that the level of mitochondrial contamination was minor (Supplemental Figure S2). After RNA extraction from the different fractions, both mSMPs and control mRNAs were detected in the fractions by semiquantitative RT-PCR (performed in duplicate). To avoid false positives that can arise from overamplification, we increased the number of cycles of amplification gradually for each gene examined. We found that mSMPs (e.g., APQ12, LCB3, PEP12, PEX3, PMT2, SEC22, SNC1, SRO7, SUC2, SUR4, USE1, and YIP3) were detected in both the ER and cytosolic fractions, with some, like APQ12, LCB3, PEX3, and SNC1, showing enrichment in the ER fraction (Figure 2B). In contrast to these mSMPs, an mRNA that encodes a cytosolic serine hydrolase, FSH3, was found enriched in the cytosolic fraction (Figure 2C). Thus not all RNAs are enriched with ER membranes. Similarly, non-mSMP RNAs that encode extralumenal membrane-associated proteins of the secretory pathway (e.g.. SAR1, SEC9, YKT6, and YPT1), as well as a control rRNA (RDN18), were found to be detected in both the ER and cytosolic fractions (Figure 2D).
FIGURE 2:
mSMPs cofractionate with ER in WT cells. (A) Fractionation of lysates into ER (microsomal) and cytosolic fractions. WT yeast were lysed and subjected to sucrose density gradient centrifugation as described in Materials and Methods. Aliquots from the different fractions obtained through centrifugation were subjected to SDS–PAGE and detected in blots with antibodies against an ER marker, dolichol phosphate mannose (anti-Dpm1) synthase, or a cytosolic marker, phosphoglycerate kinase (anti-PGK1). Note lack of overlap between markers in the different fractions. (B) Distribution of mSMPs. Total mRNA isolated from the fractions (ER or cytosol [Cyt]) obtained using density gradient centrifugation was subjected to DNase I treatment and RT-PCR with oligonucleotide pairs specific to each mSMP. Genomic DNA served as a positive control for the PCR (Genomic), and RT without RNA template served as a negative control (–). PCR amplification of RNA from the ER and cytosolic fractions without reverse transcription (–RT) served as controls to detect DNA contamination. PCR amplification of RNA from the ER and cytosolic fractions with reverse transcription (+RT) is shown in duplicate. Samples were electrophoresed on agarose gels and documented by ethidium bromide labeling. (C) Distribution of an mRNA encoding a cytosolic protein. Same as in B except that an oligonucleotide pair specific to FSH3 was used. (D) Distribution of mRNAs not encoding mSMPs. Same as in B except that oligonucleotide pairs specific to mRNAs encoding proteins not translocated into the ER, but instead associated either with the cytosolic leaflet of the secretory pathway (SAR1, SEC9, SRO7, YKT6, YPT1) or ribosome (RDN18) were used.
In contrast to our imaging data, we did not see a significant enrichment of SUC2 mRNA in the ER fraction derived from cells grown on high glucose (i.e., glucose-repressed cells). We noted, however, that glucose derepression is typically required for the induction of the secreted (signal peptide–containing) form of invertase. Thus we probably detected the RNA encoding only the nonsecreted isoform in this experiment (Figure 2A). We therefore repeated the experiment using derepressed cells grown on 0.1% glucose and examined for the presence of SUC2 mRNA. Under these conditions, we found that nearly all SUC2 mRNA was associated with the ER fraction (Supplemental Figure S3). We also note that USE1 and several other strongly ER-localized mRNAs (Figure 1) were not enriched in the ER fraction (Figure 2), which might indicate that the preparative procedure can have differential effects on RNA binding to the ER. Overall, however, both the microscopy and fractionation results shown here parallel earlier microarray studies demonstrating RNA association with the ER (Diehn et al., 2000; Lerner et al., 2003; Pyhtila et al., 2008; Chen et al., 2011) and suggest that mSMPs target ER membranes (reviewed in Kraut-Cohen and Gerst, 2010).
Endogenously expressed SUC2 mRNA colocalizes with ER, as shown by single-molecule fluorescence in situ hybridization
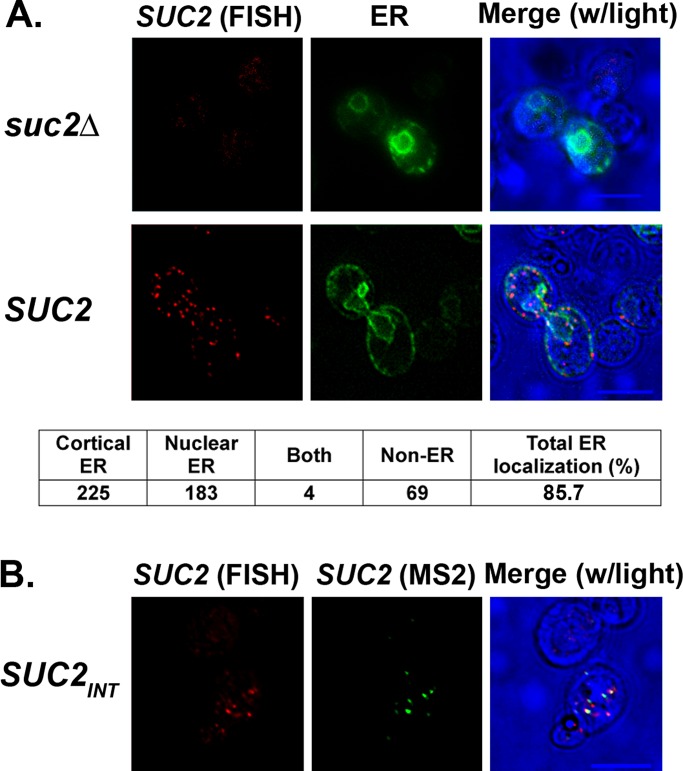
To further demonstrate that the presence of the MS2 aptamer does not affect (i.e., drive) RNA localization to the ER, we used single-molecule fluorescence in situ hybridization (smFISH; Zenklusen et al., 2008) to examine the distribution of SUC2 mRNA between the ER and cytosol. We used WT cells that express native SUC2 mRNA (i.e., without the MS2 aptamer) from the genome or, as a negative control, cells lacking SUC2 (suc2Δ), using low-glucose-containing medium to induce expression. On fixation and hybridization with multiple (20) nonoverlapping tetramethylrhodamine (TAMRA)-labeled oligodeoxynucleotide probes (22-mers) complementary to SUC2, we observed multiple small fluorescent dots that appeared similar to RNA granules, but only in the wild-type (WT) cells and not in the suc2Δ control. Scoring of the WT cells revealed that 85.7% of these labeled SUC2 mRNA granules were associated with the ER (Figure 3A), similar to the values obtained using the m-TAG MS2 system (Figure 1 and Table 1). Thus both methods of detection indicate that SUC2 transcripts localize preferentially to the ER, which, together with our subcellular fractionation studies (Figure 2B and Supplemental Figure S3), confirms that presence of the MS2 aptamer does not confer transcript localization to the ER per se.
FIGURE 3:
SUC2 mRNA colocalizes with ER using smFISH labeling. (A) Endogenous SUC2 mRNA localizes to the ER using smFISH. WT and suc2Δ yeast expressing SEC63-GFP from a single-copy plasmid were grown to mid log phase and shifted to low glucose–containing medium for 1.5 h to induce SUC2 expression. Cells were processed for smFISH labeling according to Materials and Methods, using nonoverlapping, TAMRA-labeled FISH probes complementary to SUC2. Labeling of SUC2 mRNA (red fluorescence; SUC2 [FISH]) was observed only in WT cells and colocalized mainly with Sec63-GFP fluorescence (ER). Merge (w/light), merger of the FISH, ER, and transmitted light windows. Bottom, scoring of SUC2 mRNA localization, in terms of granules that localize to cortical ER, nuclear ER, both, or neither. (B) smFISH detects MS2-CP-GFP(x3)–labeled RNA granules. WT yeast bearing the MS2L aptamer sequence integrated into the SUC2 locus (SUC2INT) were transformed with a plasmid expressing MS2-CP-GFP(x3), grown to mid log phase, induced on selective medium lacking methionine, and processed for smFISH. Cells were examined for smFISH labeling (red fluorescence; SUC2 [FISH]) or MS2-CP-GFP(x3) labeling of the tagged transcript (green; SUC2 [MS2]). Merge (w/light), merger of the smFISH, MS2, and transmitted light windows. Scale bar, 5 μm.
Of interest, we observed more mRNA granules per cell using smFISH than typically seen using m-TAG. To compare the two techniques for mRNA visualization, we examined suc2Δ cells expressing MS2 aptamer–tagged SUC2 from a single-copy plasmid, along with MS2-CP-GFP(x3), using both methods of detection. Under these expression conditions, we found that the smFISH probes detected all of the SUC2 mRNA granules that could be detected with m-TAG (Figure 3B). Additional smFISH-labeled granules were apparent, however, that were not labeled by the MS2 system. This is likely to indicate that smFISH is more sensitive than m-TAG (which we showed to label granules containing two or more tagged mRNAs; Haim et al., 2007), as expected. Alternatively, whereas smFISH should label mRNAs that are in the process of translation, it is likely that bound MS2-CP-GFP(x3) reporter proteins necessitate removal to allow for ribosome readthrough of tagged transcripts. Thus smFISH is likely to label transcripts that the MS2 system is unable to, and this could be yet another reason for the lower degree of granule detection.
Determination of the cis elements that control SUC2 and USE1 mRNA localization
To investigate the elements that target mSMPs to the ER, we used the USE1 and SUC2 mRNAs as models for mSMPs encoding integral membrane proteins or soluble secreted enzymes, respectively. To do this, we expressed MS2 aptamer–tagged, full-length SUC2 and USE1 mRNAs (including their respective 3′UTRs) under the control of a methionine-starvation-inducible promoter from single-copy plasmids in wild-type cells. In addition, both MS2-CP-GFP(x3) and RFP-tagged Sec63 were coexpressed to visualize RNA and ER, respectively.
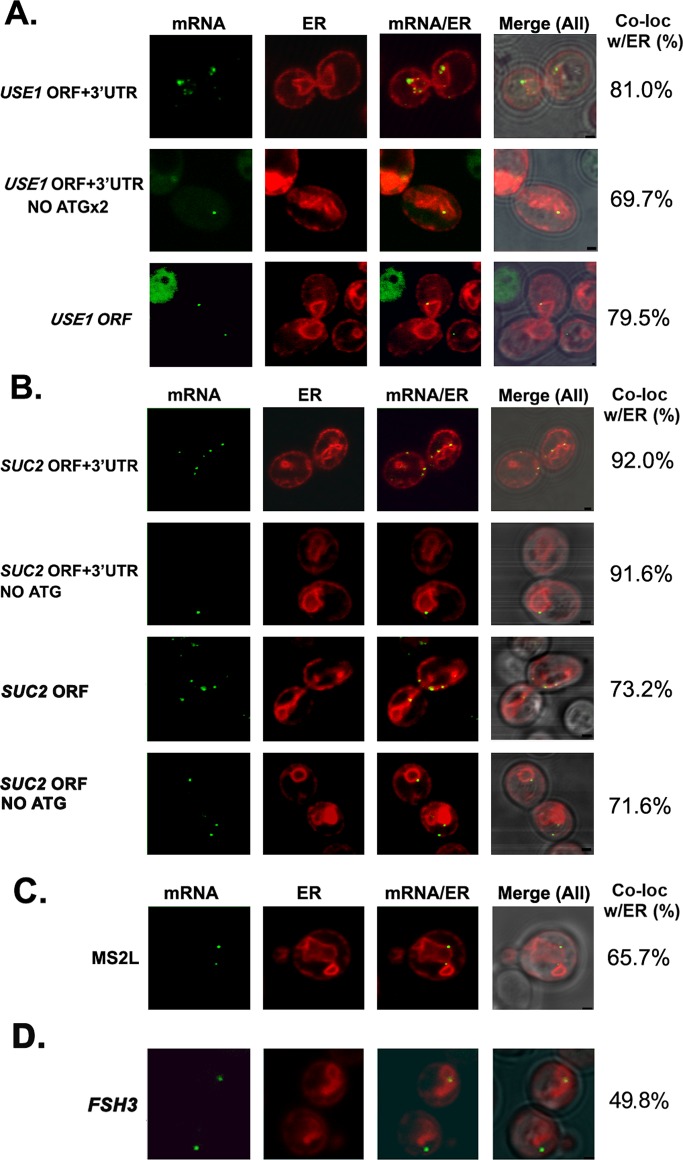
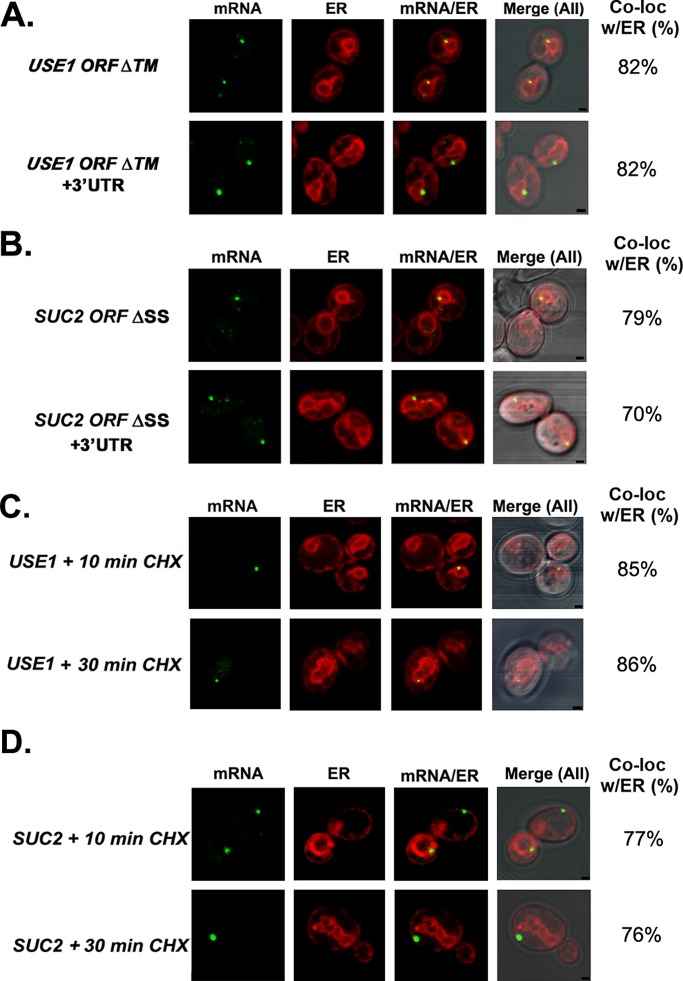
Because multiple studies show that the 3′UTR includes regulatory cis elements (Jambhekar and Derisi, 2007; Andreassi and Riccio, 2009), we compared the level of ER localization of full-length USE1 and SUC2 (i.e., open reading frames [ORFs] + 3′UTRs) mRNAs versus mRNAs lacking their 3′UTRs (i.e., ORFs alone). We found that 81% of full-length USE1 mRNA granules colocalized with RFP-Sec63–labeled ER (Figure 4A and Table 2), which was similar to what was observed with endogenously expressed USE1 mRNA (86% colocalization; Figure 1 and Table 1), whereas removal of the 3′UTR essentially did not change localization (79% colocalization; Figure 4A and Table 2). On the other hand, the ER localization of full-length SUC2 mRNA colocalization was somewhat higher than with the endogenous message (92 vs. 81%) but was reduced (73% localization) upon removal of the 3′UTR (Figure 4B and Table 2). Overall, however, it appears that the 3′UTR is not essential for the association of these mSMPs with ER.
FIGURE 4:
Role of sequence elements in SUC2 and USE1 colocalization with ER. (A, B) Four MS2L-tagged USE1 or SUC2 mRNAs (native full-length [ORF + 3′UTR]; lacking the 3′UTR [ORF, no 3′UTR]; bearing a mutated start codon [ORF + 3′UTR, no ATG]; lacking the 3′UTR and bearing a mutated start codon [ORF, no 3′UTR, no ATG]) were expressed from single-copy plasmids under the control of a methionine starvation–inducible promoter and examined for localization in WT cells by confocal microscopy. (A) Neither the 3′UTR nor translation initiation is required for the association of USE1 mRNA with the ER. Representative confocal microscopy images of cells transformed with the different MS2L-tagged USE1 plasmids (as labeled) and expressing MS2-CP-GFP(x3) and Sec63-RFP. Percentage of RNA granules that colocalize with ER (% Local.). mRNA, localization of GFP-labeled RNA granules; ER, localization of ER labeled with Sec63-RFP; mRNA/ER, merger of the ER and mRNA windows; Merge (All), merger between light, ER, and mRNA windows. Scale bar, 1 μm. (B) SUC2 mRNA localization to the ER may be partially 3′UTR dependent but does not depend on translation initiation. Same as in A except that representative images of cells transformed with plasmids expressing the different MS2L-tagged SUC2 plasmids (as labeled), as well as MS2-CP-GFP(x3) and Sec63-RFP, are shown. (C) The MS2 aptamer sequence also associates with the ER. Representative images of cells expressing the MS2L sequence without additional gene sequences, along with both MS2-CP-GFP(x3) and Sec63-RFP. Labeled as in A. (D) FSH3 mRNA localizes to both the cytoplasm and ER. Representative images of cells expressing MS2L-tagged full-length FSH3 (ORF + 3′UTR) from a single-copy plasmid under a methionine starvation–inducible promoter, along with MS2-CP-GFP(x3) and Sec63-RFP. Labeled as in A.
TABLE 2:
USE1 and SUC2 mRNA localization to ER.
| Percentage of RNA granules colocalized with ER | |||||
|---|---|---|---|---|---|
| Gene expressed | Cortical ER | Nuclear ER | Both | Non-ER | Total ER localization |
| USE1 ORF + 3′UTR (full length) | 14.0 | 61.0 | 6.0 | 19.0 | 81.0 |
| USE1 ORF (no 3′UTR) | 27.1 | 49.5 | 2.9 | 20.5 | 79.5 |
| USE1 ORF + 3′UTR, no ATG | 15.8 | 68.0 | — | 16.2 | 83.8 |
| USE1 ORF + 3′UTR, no ATGx2 | 50.1 | 18.6 | 1 | 30.3 | 69.7 |
| USE1 ORF + 3′UTR, ∆TMD | 21.6 | 53 | 7.2 | 18.2 | 81.8 |
| USE1 ORF, ∆TMD | 32 | 43 | 7 | 18 | 82.0 |
| SUC2 ORF + 3′ UTR (full length) | 28.7 | 56.4 | 6.9 | 8 | 92.0 |
| SUC2 ORF (no 3′ UTR) | 21.9 | 48.5 | 2.8 | 26.8 | 73.2 |
| SUC2 ORF + 3′UTR, no ATG | 25 | 61.6 | 5 | 8.4 | 91.6 |
| SUC2 ORF (no 3′ UTR), no ATG | 25.5 | 42 | 4.1 | 28.2 | 71.6 |
| SUC2 ORF + 3′UTR, ∆SS | 22.8 | 53 | 3.3 | 20.9 | 79.3 |
| SUC2 ORF (no 3′UTR), ∆SS | 11.9 | 52.4 | 5.9 | 29.8 | 70.2 |
| FSH3 ORF + 3′UTR (full length) | 14.9 | 33.6 | 1.3 | 50.2 | 49.8 |
| MS2L | 7.3 | 54.8 | 3.6 | 34.3 | 65.7 |
Of interest, we noted that the polymeric MS2 aptamer sequence (MS2L; ∼750 bases; 12 loops) alone could colocalize to some degree with ER when detected with either MS2-CP-GFP(x1) (53% after 1 h of induction) or with MS2-CP-GFP(x3) (49% for uninduced cells vs. 66% after 1 h of induction; Figure 4C and Table 2). Thus the MS2 aptamer sequence associates with the ER, although not with mitochondria (∼0% colocalization; data not shown). Because our smFISH and subcellular fractionation results clearly demonstrate that untagged mRNAs associate with ER microsomes (Figures 2B and 3A), however, we do not believe that the presence of the aptamer sequence is the means by which tagged transcripts localize with ER membranes. This does suggest, however, that naive RNAs target the ER by default, and this could indicate that ER membranes act as a trap for RNA molecules. Because FSH3 mRNA was enriched in the cytosolic fraction (Figure 2C), we examined its localization by aptamer tagging and fluorescence microscopy. We found that ∼50% of the RNA granules showed colocalization with ER (Figure 4D). Therefore all large coding and noncoding (e.g., RDN18 and MS2L) RNAs tested, as well as an RNA encoding a cytosolic protein (FSH3), appear to have some propensity to associate with ER membranes. As a consequence, we were unable to identify a negative control for the association of large RNAs to the ER, using either microscopy or cell fractionation.
Removal of the translation initiation codon does not alter SUC2 and USE1 mRNA localization to the ER
According to the classic model, mSMPs target the ER in a translation-dependent manner via the SRP. Because all mSMPs examined colocalize with ER, we tested whether this association is translation dependent. First, we examined localization of the USE1 and SUC2 mRNAs lacking their start codon (in order to prevent translation). Because USE1 also possesses an internal translation initiation site (nucleotides [nts] 211–213), we used a double mutant that lacks both the first and second ATG codons and cannot give rise to a functional Use1 protein (Supplemental Figure S4). We found that RNA granules of full-length USE1 mRNA with both initiation sites (USE1 ORF + 3′UTR, no ATGx2) still colocalized with the ER (70% colocalization; Figure 4A and Table 2). Similarly, full-length SUC2 mRNA lacking its start codon (SUC2 ORF + 3′UTR, no ATG) showed 92% colocalization with the ER, whereas SUC2 mRNA lacking both the start codon and 3′UTR (SUC2 ORF, no 3′UTR, no ATG) gave 72% colocalization (Figure 4B and Table 2). Thus removal of the translation initiation signal did not prevent the association of either USE1 or SUC2 message with the ER.
Of interest, Western analysis revealed production of a truncated, presumably intracellular, nonglycosylated form of Suc2 protein that lacks the signal peptide (encoded by the SSCR located between the first and second start codons; nts 4–57) from either the full-length SUC2 mRNA construct lacking the first start codon or from SUC2 in which the SSCR was removed (Supplemental Figure S5A). Thus either a block in translation of the encoded SSCR or its removal likely prevents interaction of the nascent peptide with SRP and leads to the accumulation of truncated Suc2 in the cytosol. To verify this further, we determined the enzymatic activities of plasmid-expressed, full-length Suc2, Suc2 lacking its first start codon, and Suc2 bearing a deletion in its SSCR in both lysed and nonlysed cells, which represent the intracellular versus secreted forms of invertase, respectively. By using a standard invertase assay (Supplemental Figure S5B), we found that essentially all invertase activity of SUC2 lacking the first start codon or the SSCR was intracellular. In contrast, most of activity expressed from full-length SUC2 was secreted. Therefore, although SUC2 mRNA lacking the start codon localizes to the ER, it gives rise to a protein that cannot be secreted. This finding alone indicates that SUC2 mRNA localization is likely to be SRP-independent.
Removal of the signal sequence–coding region of SUC2 or transmembrane domain–coding region of USE1 does not inhibit mRNA colocalization with ER
The SRP recognizes the newly translated signal peptide or signal anchor sequence of type II membrane proteins emerging from ribosomes engaged in mSMP translation (Martoglio and Dobberstein, 1998). To determine the necessity of the signal sequence and transmembrane domain (TMD)–coding regions for SUC2 and USE1 mRNA delivery to the ER, respectively, we expressed SUC2 mRNA lacking its SSCR (SUC2 ORF + 3′UTR ∆SS) and USE1 mRNA lacking the encoded TMD (USE1 ORF + 3′UTR ∆TMD) in wild-type cells. We could not, however, detect a reduction in ER localization for USE1 mRNA either lacking the encoded TMD (82% ER localization) or both the TMD and the 3′UTR (USE1 ORF [no 3′UTR] ∆TMD; 82% ER localization; Figure 5A and Table 2). Similarly, removal of the SSCR had little effect on SUC2 mRNA delivery to the ER. SUC2 mRNA lacking the SSCR (SUC2 ORF + 3′UTR ∆SS) showed 79% ER localization, whereas SUC2 mRNA lacking both the SSCR and 3′UTR (SUC2 ORF [no 3′UTR] ∆SS) showed 70% ER localization (Figure 5B and Table 2). Thus neither the SUC2 SSCR nor encoded USE1 TMD appears essential for ER localization. More important, mSMP localization to the ER occurs despite removal of these elements and implies that mRNAs can be delivered in an SRP-independent manner.
FIGURE 5:
Localization of SUC2 and USE1 mRNAs to the ER is not dependent on the SSCR or TMD and is not sensitive to CHX treatment. (A) USE1 colocalization with the ER does not depend on its TMD. Representative confocal microscopy images of WT cells expressing MS2-CP-GFP(x3), Sec63-RFP, and either MS2L-tagged USE1 ORF + 3′UTR or ORF alone in which the TMD (∆TM) has been removed. Cells were grown overnight on liquid selective medium containing glucose and then shifted to the same medium lacking methionine for 1 h. Percentage of fluorescent RNA granules that colocalize with ER (% Local.). mRNA, localization of GFP-labeled RNA granules; ER, localization of ER labeled with Sec63-RFP; mRNA/ER, merger of the ER and mRNA windows; Merge (All), merger of the light, ER, and mRNA windows. Scale bar, 1 μm. (B) SUC2 mRNA localizes to the ER after deletion of the SSCR. Representative WT cells expressing MS2-CP-GFP(x3), Sec63-RFP, and the MS2L-tagged ORF + 3′UTR or ORF alone in which the SSCR (∆SS) has been removed. Labeled as in A. (C) Endogenous USE1 mRNA associates with the ER even on CHX treatment. Representative confocal microscopy images of cells expressing aptamer-tagged USE1 from its genomic locus (USE1int), as well as MS2-CP-GFP(x3) and Sec63-RFP from plasmids. Cells were induced with medium lacking methionine 1 h before visualization, treated with 100 μg/ml CHX for 10 or 30 min, and immediately fixed. Labeled as in A. (D) Endogenous SUC2 mRNA associates with the ER even on CHX treatment. Cells expressing aptamer-tagged SUC2 from its genomic locus (SUC2int), as well as MS2-CP-GFP(x4) and Sec63-RFP from plasmids. Cells were grown and treated as in C, except that the medium contained 0.1% glucose (to induce SUC2).
Cycloheximide treatment does not alter SUC2 and USE1 mRNA localization to the ER
To verify translation-independent delivery of the USE1 and SUC2 mRNAs to the ER, we examined the localization of endogenously expressed, aptamer-tagged USE1 and SUC2 mRNAs in cells treated with the translation elongation inhibitor, cycloheximide (CHX). Aptamer-integrated cells grown to mid log phase were induced in medium lacking methionine (1 h) to express MS2-CP-GFP(x3) before CHX treatment. Because CHX induces cell cycle arrest (Dieci et al., 1995), we initially examined its effect on wild-type cells. Within 1 h of treatment we detected an arrest in cell proliferation (data not shown). Because many of the cells were distorted morphologically by this time, however, we examined mRNA–ER colocalization at earlier time points (10 and 30 min after CHX treatment) using the aptamer-integrated cells. We found that USE1 mRNA localization was not altered under these conditions, because treatment with CHX for either 10 or 30 min did not change ER localization (85% colocalization; Figure 5C and Table 1). Similarly, SUC2 mRNA colocalization with the ER was only very slightly effected by the CHX treatment, with 77% of RNA granules colocalized with the ER after 10 min and 75% colocalized after 30 min of CHX treatment (Figure 5D and Table 1). This result implies that either SUC2 and USE1 mRNAs are targeted to the ER in an essentially translation-independent manner or CHX treatment anchors the SUC2/USE1 mRNA pool to the ER during the course of the experiment. Of interest, a shift in the distribution of SUC2 RNA granules on the ER was observed, as the majority of granules (47%) were found on cER after CHX treatment, unlike in untreated cells, in which the majority of granules (42%) were found on nER. This change in granule distribution may represent a stress response to translation inhibition, since mRNAs have been shown to sequester on ER during stress instead of aggregating in stress granules (Kilchert et al. 2010). Finally, to verify that CHX-mediated translation inhibition occurred within the time frame of the RNA localization measurements, we incubated WT cells with CHX for 5 or 15 min, followed by a 15-min pulse with [35S]methionine to label newly synthesized proteins. CHX treatment led to the inhibition of protein synthesis, as almost no [35S]methionine label was incorporated into protein after as little as 5 min of treatment (Supplemental Figure S6).
Overall our results show that mSMPs like SUC2 and USE1 are retained on the ER even in the absence of elements crucial for translation initiation, SRP-dependent ER targeting, or integration into the membrane (TMD) and are resistant to the effects of translation inhibition by CHX. Thus mSMPs are likely to use translation- and SRP-independent routes to reach the ER.
Synthetic mRNAs target the ER without translation
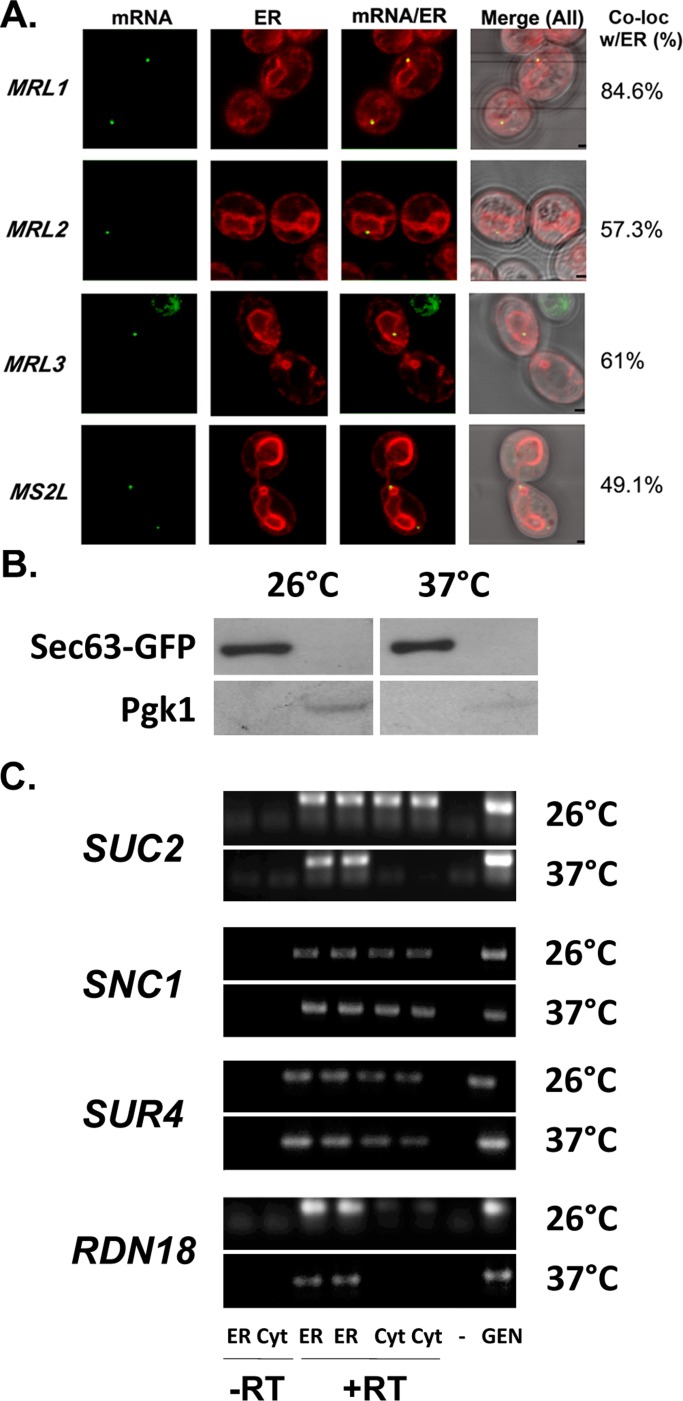
Prilusky and Bibi (2009) demonstrated that mRNAs encoding membrane proteins have a higher uracil content (U-rich) than mRNAs encoding cytosolic proteins and that ∼60-nt coding stretches that contain ∼40% uracil residues invariably encode TMDs. This is because the codons for hydrophobic amino acids are U-rich, and this phenomenon is conserved from prokaryotes to eukaryotes. To investigate whether U-richness alone can promote RNA delivery to the ER in yeast, we expressed four MS2 aptamer–tagged synthetic nontranslatable RNA sequences and examined their localization in vivo. All RNA sequences expressed were ∼400 nt long, lacking both their ATGs and ribosome-binding sequences, and included stop codons before the U-rich regions (Supplemental Figure S7). Therefore these sequences are nontranslatable. We analyzed the following constructs: 1) MRL1, an RNA sequence based on bacterial lacY that acts as a representative U-rich mRNA coding for a bacterial membrane protein; 2) MRL2, an RNA sequence based on bacterial lacZ that acts as a representative mRNA coding for a cytosolic protein; 3) MRL3, a U-rich random RNA sequence resembling a membrane protein (Prilusky and Bibi, 2009); and 4) MRL4, a random RNA sequence that is U-poor (Supplemental Figure S7).
On induction in medium lacking methionine, these constructs typically yielded >10 granules/cell (data not shown). Therefore, to avoid artifacts arising due to overexpression, we performed RNA localization without direct induction of MS2-CP-GFP(x3) (via expression due to leakiness of the MET25 promoter), which led to ≤5 granules per cell. We observed that MRL1–3 could form detectable granules, whereas MRL4 was undetectable under these conditions. MRL1 granules showed a high level of colocalization with ER (85%), whereas MRL2 and 3 showed more moderate levels of colocalization (57 and 61% respectively; Figure 6A and Table 3). Although ER localization of the MS2 aptamer (MS2L) alone was not insignificant and showed 49% colocalization under these conditions (Figure 6A and Table 3), the experiment does reveal that nontranslatable mRNAs that do not bind ribosomes still reach the ER. In addition, U-rich MRL1 sequence localized preferentially to ER in comparison to its parallel U-poor cohort (MRL2). Because MRL4 granules could not be quantified, we could not compare MRL3 and MRL4. Nonetheless, the ability of MRL3, as well as the MS2 aptamer, to arrive at the ER implies that the ER may serve as a default destination for cellular mRNAs.
FIGURE 6:
Synthetic nontranslatable mRNAs localize to the ER, and SRP inactivation does not block RNA association with ER membranes. (A) Synthetic nontranslatable RNAs can target the ER. Representative confocal microscopy images of cells expressing MS2L-tagged synthetic nontranslatable mRNA sequences, MRL1-3 (Supplemental Figure S5), or the MS2 aptamer (MS2L) alone, in cells coexpressing MS2-CP-GFP(x3) and RFP-Sec63. Cells were grown on liquid selective medium to mid log phase without induction in medium lacking methionine. Percentage of fluorescent RNA granules that colocalize with ER (% Local.). mRNA, localization of GFP-labeled RNA granules; ER, localization of ER labeled with Sec63-RFP; mRNA/ER, merger of the ER and mRNA windows; Merge (All), merger of the light, ER, and mRNA windows. Scale bar,1 μm. (B, C) SRP inactivation does not block RNA association with ER membranes. (B) Fractionation of sec65-1 cell lysates into ER (microsomal) and cytosolic fractions. Temperature-sensitive sec65-1 cells expressing Sec63-GFP were grown to mid log phase on selective medium and maintained at 26°C or shifted for 6 h to 37°C. Cells were lysed and subjected to differential centrifugation to obtain protein and RNA as described in Materials and Methods. Protein aliquots from the different fractions obtained through centrifugation (ER-enriched membrane fraction [ER] and cytosolic fraction [Cyt]) were subjected to SDS–PAGE and detected in blots with antibodies against an ER marker, Sec63-GFP (Sec63-GFP; using anti-GFP antibodies), or a cytosolic marker, phosphoglycerate kinase (PGK1). (C) Distribution of mRNAs to ER microsomes and cytosol in sec65-1 cells. After subcellular fractionation, RNA isolated from the different fractions (ER or Cyt) was subjected to DNase I treatment and RT-PCR with oligonucleotide pairs specific to each RNA (as indicated). Genomic DNA (Gen) served as a positive control for the PCR, and reverse transcription without an RNA template served as a negative control (–). PCR amplification of RNA from the ER and cytosolic fractions without reverse transcription (–RT) served as controls to detect DNA contamination. PCR amplification of RNA from the ER and cytosolic fractions with reverse transcription (+RT) is shown in duplicate. Samples were electrophoresed on agarose gels and documented by ethidium bromide labeling.
TABLE 3:
Localization of synthetic nontranslatable RNAs to the ER.
| Percentage of RNA granules colocalized with ER | |||||
|---|---|---|---|---|---|
| Gene expressed | Cortical ER | Nuclear ER | Both | Non-ER | Total ER localization |
| MRL1 | 13.6 | 62.0 | 9.0 | 15.4 | 84.6 |
| MRL2 | 11.4 | 42.6 | 3.3 | 42.7 | 57.3 |
| MRL3 | 21.0 | 40 | — | 39 | 61.0 |
| MS2L | 13.8 | 33.8 | 1.5 | 50.9 | 49.1 |
Expression was not induced by medium lacking methionine
SRP inactivation does not lead to a decrease in mRNA localization to the ER
To obtain additional proof of SRP-independent mRNA delivery to the ER, we examined the distribution of endogenously expressed SUC2, SNC1, and SUR4 mRNAs in sec65-1 temperature-sensitive SRP mutant cells (RSY457 strain; Supplemental Table S1) under conditions in which SRP is inactivated (incubation at 37°C). First, to both monitor and verify SRP inactivation, we analyzed sec65-1 cells expressing DsRed bearing a Kar2 signal sequence at the N-terminus and an HDEL Golgi–ER retrieval sequence (Kar2SS-DsRed-HDEL) that allows for the fusion protein to enter into and remain within the ER. At 26°C we observed Kar2SS-DsRed-HDEL to reside within the ER. On sec65-1 inactivation at 37°C, we observed its time-dependent localization in the cytosol, indicating blocked entry into the ER (Supplemental Figure S8). Next we fractionated both temperature-shifted and nonshifted sec65-1 cells into cytosolic and ER fractions (Figure 6B) and examined them for the presence of RNAs using RT-PCR (Figure 6C). We found that SEC65 inactivation did not alter the distribution of SNC1 and SUR4 mRNAs, whereas the level of SUC2 mRNA localization to the cytosol greatly decreases. We observed a similar result for the RDN18 rRNA. Thus SRP inactivation does not appear to be involved in the localization of these RNAs to the ER.
Involvement of RNA-binding proteins in targeting of SUC2 and USE1 mRNA
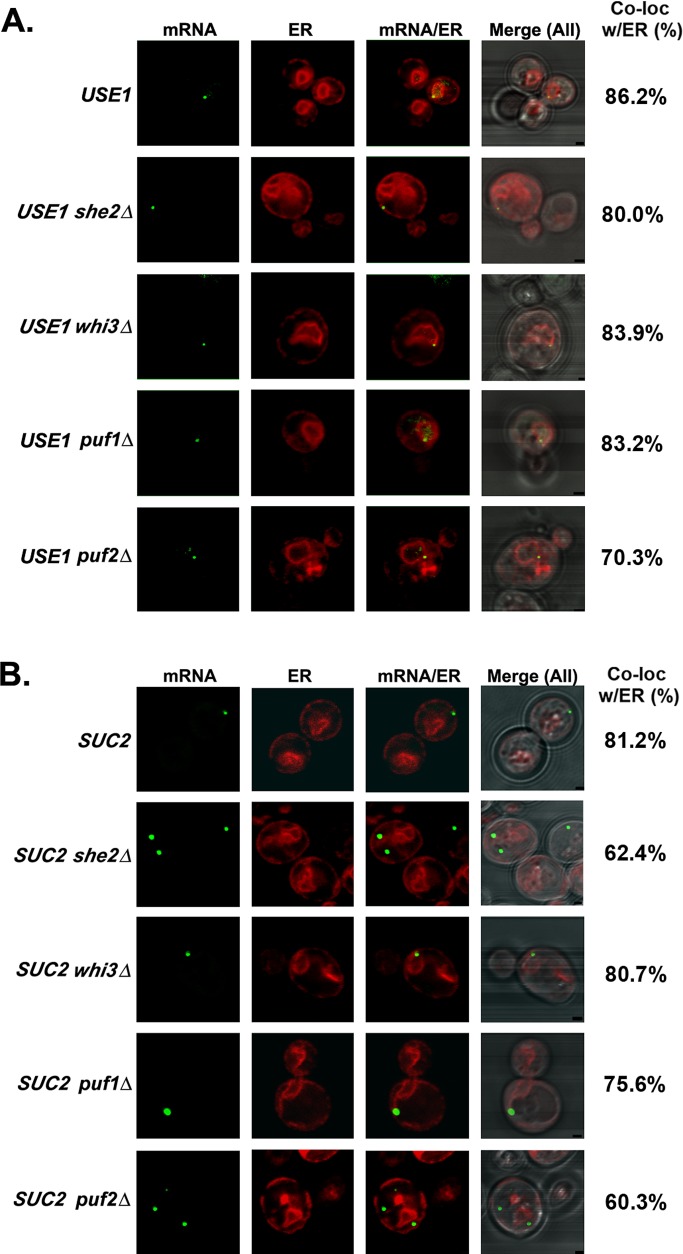
Because several RBPs affect RNA localization to yeast ER (Gerber et al., 2004; Aronov et al., 2007; Colomina et al., 2008), we determined their contribution to delivery of the SUC2 and USE1 mRNAs to the ER. We deleted genes encoding RBPs that were reported to either bind mSMPs (e.g., Puf1/2, Whi3) or be involved in ER targeting (e.g., She2) in the aptamer-integrated USE1 and SUC2 strains. We found that USE1 mRNA remained localized to ER in all of the deletion mutants, although we observed a slight decrease in puf2Δ cells (Figure 7A and Table 1). Deletion of either SHE2 or PUF2, however, significantly reduced the localization of SUC2 mRNA with ER (60 and 62%, respectively, vs. 81% in wild-type cells; Figure 7B and Table 1). To assess whether the deletion of both SHE2 and PUF2 could have a cumulative effect, we scored the localization of SUC2 mRNA in puf2Δ she2Δ cells. We observed decreased SUC2 mRNA localization to the ER in these cells (54%; Table 1), but in comparison to the individual deletions, this effect did not appear to be additive. In contrast, the deletion of PUF1 or WHI3 did not significantly change SUC2 mRNA colocalization with ER (75.6 and 80.7% respectively; Figure 7B and Table 1).
FIGURE 7:
Localization of USE1 and SUC2 mRNAs in cells lacking ER-associated RBPs. (A) Localization of USE1 mRNA in cells lacking ER-associated RBPs. Representative images of endogenously expressed MS2L-tagged, full-length USE1 mRNA in WT, she2Δ, puf1Δ, puf2Δ, or whi3Δ cells coexpressing MS2-CP-GFP(x3) and Sec63-RFP. Cells were grown on selective medium to mid log phase without induction in medium lacking methionine. Percentage of fluorescent RNA granules that colocalize with ER (% Local.). mRNA, localization of GFP-labeled RNA granules; ER, localization of ER labeled with Sec63-RFP; mRNA/ER, merger of the ER and mRNA windows; Merge (All), merger of the light, ER, and mRNA windows. Scale bar, 1 μm. (B) Localization of SUC2 mRNA in cells lacking ER-associated RBPs. Representative images of endogenously expressed, MS2L-tagged SUC2 mRNA in WT, she2Δ, puf1Δ, puf2Δ, or whi3Δ cells coexpressing MS2-CP-GFP(x3) and Sec63-RFP. Cells were grown on synthetic selective medium and then shifted to low glucose (0.1%)–containing medium lacking methionine for 1 h. Labeled as in A.
Although SUC2 mRNA is not polarized to the daughter cells, like ASH1 and POL mRNAs, its localization to the ER was affected in she2Δ cells. By comparison to the known consensus sequence for She2 binding (Olivier et al., 2005), we found a potential She2-binding site in the SUC2 coding region (base pairs 280–345) that might serve as a cis-acting element. We then verified whether SUC2 mRNA is indeed bound to She2, by expressing Myc-tagged She2 in wild-type cells that were shifted to low (0.1%) glucose for 2 h, followed by immunoprecipitation with anti-Myc antibodies (Supplemental Figure S9). Along with ASH1 mRNA, SUC2 mRNA was also detected by RT-PCR specifically in cells expressing Myc-She2 but not the control vector alone (Supplemental Figure S9A). Similarly, Myc-She2 could be detected only in immunoprecipitates derived from cells expressing MYC-SHE2 and not from the control cells (Supplemental Figure S9B). Thus, upon its induction, endogenous SUC2 mRNA is likely to be transported with ASH1 mRNA in the same RNA granule.
In summary, our results suggest the existence of translation-independent delivery of mRNAs to the ER, which is regulated by both cis-acting sequence elements and trans-acting protein partners.
DISCUSSION
Multiple studies have demonstrated SRP-dependent delivery of mSMPs to ER membranes in vitro; however, this path necessitates translation as the preliminary step that promotes SRP binding and cotransport of the translating ribosome and SRP complex to the ER (Schwartz, 2007). Studies that use biochemical fractionation followed by microarray analysis already demonstrated the existence of numerous mRNAs, both mSMPs and mRNAs encoding cytosolic proteins, on the ER (Lerner et al., 2003; Pyhtila et al., 2008; Chen et al., 2011). Thus the question remains open of whether translation and SRP involvement are absolutely required for mRNA association with ER.
In this study, we localized in vivo 11 endogenously expressed yeast mSMPs (ALG1, GOS1, ICE2, NYV1, PEP12, PMT2, SEC22, SUC2, SUR4, USE1, and YIP3) and two non-mSMP RNAs that encode soluble proteins that act from the cytosol upon the secretory pathway (SEC9, SEC20) using m-TAG (Figure 1). All of these mRNAs colocalized to a large degree with the ER, whereas some preference was demonstrated for either nER (PMT2, SUC2, SUR4, USE1, and YIP3 mRNAs) or cER (GOS1, NYV1, PEP12, SEC9 and SEC22 mRNAs; Figure 1 and Table 1). Because none these mRNAs or their encoded proteins are polarized (i.e., to the bud tip, where polarized mRNAs, POL proteins, and cER colocalize; Aronov et al., 2007; Gelin-Licht et al., 2012), it is unclear how or why such a preference might exist. That said, however, we note that mSMPs encoding tail-anchored SNARE proteins (GOS1, NYV1, PEP12, SEC22), which likely use the GET pathway for insertion into the ER membrane (Schuldiner et al., 2008), appear to show a preference for the cER. This might suggest that some areas of the ER have different properties that could potentiate the import and localization of tail-anchored proteins. We also note that aptamer-tagging and RNA visualization revealed that several mRNAs, including NYV1, SEC22, SEC20, a control mRNA encoding a cytosolic protein, FSH3, and even the MS2L aptamer, showed a roughly equal distribution between the ER and cytoplasm (Figures 1 and 4, C and D, and Table 1). Moreover, the MS2 imaging data were verified by subcellular fractionation, in which the different (nontagged) mRNAs examined all fractionated to some extent between both the ER and cytosolic fractions (Figure 2B and Table 1). Similarly, we demonstrated, using smFISH, that a nontagged transcript (SUC2) colocalized to the same degree with the ER as an MS2 aptamer–tagged SUC2 transcript using m-TAG (Figure 3A). Thus the presence of the MS2 aptamer sequence neither is necessary nor appears to enhance mRNA localization to the ER. Moreover, the results shown here validate previous cell fractionation and microarray studies showing that numerous mRNAs are associated with ER membranes (Diehn et al., 2000; Lerner et al., 2003; Pyhtila et al., 2008; Chen et al., 2011) and suggest that all RNAs, either coding or noncoding (e.g., RDN18, MS2L), have some inherent propensity to interact with ER membranes.
By using the SUC2 and USE1 mRNAs as a representative mSMPs encoding for secreted (Ozcan et al., 1997) and TMD-containing proteins (Dilcher et al., 2003), respectively, we further investigated mRNA targeting to the ER. Although the targeting of USE1 mRNA was unaffected by removal of its 3′UTR, that of SUC2 mRNA showed a modest reduction (∼20 percentage point change) in localization that effectively resulted in a threefold increase in the level of RNA mislocalization to the cytoplasm (Figure 4 and Table 2). This suggests that the 3′UTR of SUC2 contains cis elements that may contribute to ER localization. No other mutation (e.g., removal of the translation initiation codon or SSCR) alone was found to affect SUC2 mRNA localization significantly, and combinations of these mutations (along with removal of the 3′UTR) were not additive (Table 2). Similarly, USE1 colocalization with the ER was robust, and no mutation—not even removal of the TMD or start codons—could abolish this (Figure 4A and Table 2). Of interest, the mammalian orthologue of Use1, p31, is required for the maintenance of tubular ER structures (cER) and cell survival (Uemura et al., 2009). ER morphology is abnormal in p31-knockout mouse embryo fibroblasts, resulting in impaired protein transport and disorganization of the ER, which leads to ER stress and apoptosis. Thus, besides being an essential SNARE, Use1 could serve as a regulator of ER tubules and their integrity. This suggests that the ER localization of mRNAs encoding Use1 and other tail-anchored proteins (e.g., Gos1, Nyv1, Pep12, and Sec22) may enhance the likelihood of their translation products encountering GET-complex components and undergoing insertion into the membrane. Moreover, any secondary role these proteins have in maintaining tubular ER structures would also necessitate correct placement into the cER, which seems be a preferred site for the localization of their mRNAs (Figure 1 and Table 1). Future studies are necessary to decipher the cis elements responsible for localization of mRNAs encoding tail-anchored proteins.
In addition to using the SUC2 and USE1 mRNAs to dissect the sequence elements involved in ER localization, we examined whether their delivery is translation-dependent. Several lines of evidence suggest that translation is not obligatory for mRNA targeting to the ER. First, endogenous SUC2 and USE1 mRNA could localize within the proximity of the ER after treatment with the translation inhibitor CHX (Figure 5, C and D, and Table 1). Similarly, treatment with puromycin or pactamycin, two other translation inhibitors, did not disrupt the association of mRNAs encoding either cytosolic proteins or ER-targeted proteins in mammalian cells (Lerner et al., 2003; Pyhtila et al., 2008). Second, both USE1 and SUC2 mRNAs bearing mutations in their translation initiation sites still localized to the ER (Figure 4, A and B). In the case of SUC2 mRNA, this led to the appearance of a truncated cytosolic form of invertase (Supplemental Figure S5), which indicates that not only translation, but even the association with SRP, is not requisite for RNA localization. This idea is supported by the fact that deletion of the SSCR of SUC2 (which prevents SUC2 mRNA association with the SRP-ribosome complex and thereby abolishes cotranslational delivery) had only a small effect on ER targeting (Figure 5B). Moreover, cell fractionation studies using membranes derived from a temperature-sensitive mutant of the Sec65 SRP subunit demonstrated that SUC2 mRNA remains ER associated even on SRP inactivation (Figure 6C). Thus our work supports the idea of translation- and SRP-independent mRNA localization to ER membranes. Consistent with these findings, others have shown that deletion of the encoded SSCR of GRP94, an mRNA enriched at the ER, inhibits only delivery of the protein and not the message to the ER in Cos-7 cells (Pyhtila et al., 2008). Correspondingly, knockdown of the essential 54-kDa subunit of the SRP in HeLa cells did not alter mRNA delivery to the ER (Pyhtila et al., 2008). Finally, a recent study demonstrated translation- and ribosome-independent localization of a subset of RNAs and identified an ER membrane protein, p180, as a putative mRNA receptor (Cui et al., 2012). Thus results from a wide variety of systems and sources all indicate the existence of alternative translation- and SRP-independent paths for mRNA delivery to the ER.
Overall, numerous studies by Nicchitta and colleagues (Diehn et al., 2000; Lerner et al., 2003; Pyhtila et al., 2008; Chen et al., 2011; Reid and Nicchitta, 2012) demonstrated that mRNAs coding for cytosolic and secretory proteins can partition to the ER in both a ribosome-dependent and -independent manner. Of interest, biochemically distinct modes of mRNA association with the ER can be ascribed (Chen et al., 2011). For example, mRNAs encoding secreted proteins dissociate from the membrane faction under conditions that release ribosomes (high salt/EDTA treatment), whereas mRNAs coding for resident ER proteins have a tighter level of membrane binding and remain attached under the same conditions (Chen et al., 2011). Although we modeled the localization of mRNAs encoding secreted (SUC2) and ER-resident (USE1) proteins, we did not examine the strength of their association with membranes. Nevertheless, our results imply that USE1 mRNA delivery to the ER is translation-independent, whereas that of SUC2 mRNA is translation-, SSCR-, and SRP-independent. Therefore RNA localization to the ER is likely to be ribosome independent in both cases.
One additional measure we used to verify that translation is not required for mRNA delivery to the ER was to examine the localization of mRNAs that cannot engage and be translated by ribosomes (e.g., MRL1-3; Figure 6A). Of importance, these mRNAs also colocalized strongly with ER, which indicates both that their delivery is translation-independent and even prokaryote-based sequences undergo targeting within eukaryotic cells. According to the work of Prilusky and Bibi (2009), mRNAs that encode integral membrane proteins are highly U-rich (in the TMD-coding region). By comparing a U-rich sequence (MRL1) and a U-poor one (MRL2), we found that the former showed a greater preference for the ER. This suggests that U-rich sequences could serve as cis-acting elements for ER targeting.
We found that all mSMPs, along with mRNAs encoding soluble SNAREs (which lack a signal sequence or TMD), which localize to the cytosolic aspect of the secretory pathway, showed a robust ability to associate with ER (Figures 1 and 2 and Table 1). Because the noncoding MS2 aptamer MS2L and nontranslatable synthetic message MRL3 also localized to ER, this suggests that this organelle is both a general anchoring site for RNA and, perhaps, a default sorting center for RNA after nuclear export, although it is important to stress that not all RNAs target the ER directly (Zipor et al., 2009; Gadir et al., 2011). In support of this idea, our lab recently demonstrated that a mutant form of OXA1 mRNA, which encodes a mitochondria-localized protein, that lacked both its 3′UTR and translation initiation codon was mislocalized to nER (Gadir et al., 2011). Thus OXA1 mRNA mislocalizes to the ER in the absence of normal mitochondrial targeting information. Another reason for the high abundance of mRNA on the ER is that ER anchoring enables transcripts to be cotransported efficiently to the bud (Gerst, 2008). Therefore RNA–ER associations likely allow not only for the efficient import of protein into the ER, but also for localized translation/translocation at distal regions of the cell.
In addition to the cis elements involved in RNA targeting, trans-acting RBPs involved in the binding of mSMPs have been identified in various organisms (reviewed in Kraut-Cohen and Gerst, 2011). In Saccharomyces cerevisiae, several RBPs (Puf1, Puf2, Whi3, She2, Scp160) bind mSMPs, although in this work we could not find one that regulates USE1 mRNA targeting to the ER. We were able to see, however, a modest effect of She2 and Puf2 on the localization of SUC2 mRNA and verified that endogenous SUC2 mRNA immunoprecipitates with She2 (Figure 6C, Table 1, and Supplemental Figure S9). Although a double deletion of PUF2 and SHE2 did not lead to an additive decrease in SUC2 mRNA localization (Table 1), it may be that functional redundancy between these different RBPs (Puf1/2, Whi3, and She2) results in the minimal effects observed on mSMP targeting and cell viability upon their deletion. Further studies involving the affinity purification of these mRNAs are needed to determine what trans factors specifically contribute to SRP-independent mRNA targeting to ER.
According to the classic model, mRNAs are targeted to the ER in an SRP- and translation-dependent manner (Schwartz, 2007), and, although other pathways have recently been suggested to regulate the import of signal sequence-containing proteins (Ast et al., 2013), no translation-independent mechanism for RNA delivery to the ER has been put forth. Although our results do not preclude translation-dependent mRNA delivery, it is highly apparent that RNAs can use alternative means (as proposed in Bibi, 2011, 2012; Chen et al., 2011) to associate with membranes in a translation-independent manner. Our work strongly supports the idea of translation-independent mRNA targeting to the ER in eukaryotes, and, although the mechanism is unclear, it probably involves cis-element recognition by trans-acting RBPs.
MATERIALS AND METHODS
Yeast strains, growth conditions, and mRNA imaging
Supplemental Table S1 lists yeast strains used in this study. Yeast were grown to mid log phase in standard rich growth medium containing either 2% glucose (YPD) or synthetic medium containing glucose (synthetic complete [SC] and selective SC drop-out medium lacking an amino acid or nucleotide base). For fluorescence microscopy, the induction of MS2-CP fused to three GFPs (MS2-CP-GFP(x3)) was performed by growing yeast strains in synthetic selective medium to mid log phase, followed by a shift to the same medium lacking methionine for 1 h at 26°C before visualization by confocal microscopy. For the induction of MS2-CP-GFP(x4), cells were either maintained on synthetic selective medium (in which some MS2-CP-GFP expression occurs due to leakiness of the MET25 promoter) to minimize reporter expression and aggregation or induced for 1 h in the same medium lacking methionine. Aptamer-integrated SUC2 cells were grown to mid log phase on selective synthetic medium containing 0.1% glucose and shifted to the same medium lacking methionine for 1 h. Cells were then collected and fixed in a solution containing 4% (wt/vol) paraformaldehyde and 4% (wt/vol) sucrose and taken directly for imaging. Representative images were acquired using either a Zeiss LSM510 Meta confocal microscope (Carl Zeiss, Jena, Germany) and a Plan Apo 100×/oil objective or a LSM710 Meta and a Plan Apo 63×/oil objective. The following wavelengths were used: for GFP, excitation at 480 nm and emission at 530 nm; for monomeric RFP, excitation at 545 nm and emission at 560–580 nm. Colocalization of MS2 aptamer–tagged mRNAs with ER was performed using cells that coexpressed Sec63-RFP. mRNA granule localization was scored in 50–100 cells for each tagged RNA. At least two individual transformants for any given mRNA were used for statistical analysis. Table 1 shows statistics for the percentage of granules that colocalized with ER.
Plasmids and DNA manipulation
Plasmid pSL1180, which bears 12 loops of the viral MS2-CP binding site, was provided by R. Singer (Albert Einstein College of Medicine, Bronx, NY). Plasmid pSM1960, which expresses Sec63-RFP, was provided by S. Michaelis (Johns Hopkins University, Baltimore, MD). Plasmid pSH47, which expresses Cre recombinase from a galactose-inducible promoter, was obtained from the European Saccharomyces cerevisiae Archive for Functional Analysis (Frankfurt, Germany). Plasmid pLOXHIS5MS2L was used to generate integration constructs for genomic insertion of the MS2 aptamer by PCR, as described in the m-TAG procedure (Haim et al., 2007; Haim-Vilmovsky and Gerst, 2009). Plasmid pMS2CPGFP(x3), which expresses MS2-CP-GFP(x3), and pMS2CPGFP(x4), which expresses MS2-CP-GFP(x4), were previously described (Haim et al., 2007; Zipor et al., 2009). Plasmids expressing MS2L-tagged SUC2, USE1, or FSH3 were created using a modified pUG36 plasmid, pUG36w/oGFP, lacking the GFP gene (removed using XbaI and religated). Next a BamHI-BglII fragment containing the MS2 loop sequence was excised from pSL1180 and ligated to the BamHI site of pUG36w/oGFP, followed by blunt-ending and ligation to the SmaI site to yield pUG36MS2L. Next the SUC2 and USE1 ORFs were amplified by PCR using primers bearing the SpeI restriction site and inserted into the SpeI site of the polylinker of pUG36MS2L to yield plasmids pSUC2MS2L and pUSE1MS2L, respectively. The FSH3 ORF was amplified by PCR using primers bearing the BglII restriction site and inserted into the BamHI restriction site of pUG36MS2L to yield pFSH3MS2L. The SUC2 and USE1 3′UTRs (500 base pairs) were amplified by PCR using primers bearing XhoI restriction sites and cloned into the XhoI site of pSUC2MS2L and pUSE1MS2L to yield plasmids pSUC2MS2L+3′UTR and pUSE1MS2L+3′UTR, respectively. The FSH3 3′UTR (600 base pairs) was amplified by PCR using primers bearing XhoI and HindIII restriction sites and cloned into the XhoI and HindIII restriction sites of pFSH3MS2L to yield plasmid pFSH32MS2L+3′UTR. Plasmids pSUC2MS2L+3′UTR-ATGmut and pUSE1MS2L+3′UTR-ATGmut, which express MS2L-tagged full-length SUC2 or USE1 mRNAs bearing mutations in the first ATG of SUC2 and USE1, respectively, were created using Pfu DNA polymerase, specific mutagenic primers, and plasmids pSUC2MS2L+3′UTR or pUSE1MS2L+3′UTR as templates, respectively. Plasmid pUSE12MS2L+3′UTR-ATGx2mut, which expresses MS2L-tagged full-length USE1 mRNA bearing mutations in both the first and second ATGs, was created using Pfu DNA polymerase, specific mutagenic primers, and plasmid pUSE1MS2L+3′UTR-ATGmut as template. Plasmid pMS2LSUC2-ATGmut, which expresses MS2L-tagged SUC2 mRNA lacking its 3′UTR and bearing a mutation in the initial ATG, was created using Pfu DNA polymerase, specific mutagenic primers, and plasmid pSUC2MS2L as a template. Plasmids pSUC2MS2L+3′UTR-SS∆ and pSUC2MS2L-SS∆, which lack the SUC2 SSCR (nucleotides 4–57), were created using Pfu DNA polymerase, specific deletion primers, and plasmids pSUC2MS2L+3′UTR and pSUC2MS2L as templates, respectively. Plasmids pUSE1MS2L+3′UTR-TM∆ and pUSE1MS2L-TM∆ lacking the USE1 TMD-coding region (nucleotides 655–717) were created using Pfu DNA polymerase, specific deletion primers, and plasmids pUSE1MS2L+3′UTR and pUSE1MS2L as templates, respectively. Plasmids pMRL1-MS2L, pMRL2-MS2L, pMRL3-MS2L, and pMRL4-MS2L were created by amplifying sequences MRL1–4 (Supplemental Figure S6) from a bacterial vector (pUC57) by PCR. Amplification products were digested with BamHI and ligated into pUG36w/oGFP to yield the foregoing plasmids. All constructs were verified by DNA sequencing. Supplemental Table S2 lists the primers.
To evaluate the role of Whi3, Puf1, Puf2, and She2 in USE1 and SUC2 mRNA localization, the genes were deleted in the corresponding MS2 aptamer–integrated SUC2 and USE1 strains using deletion constructs as described in Gadir et al. (2011).
Genomic integration of the MS2 aptamer sequence
For each gene to be tagged with the MS2 aptamer, a forward oligonucleotide complementary to the 3′-end of the coding region (overlapping by ∼40 base pairs including stop codon) and 5′-end of the loxP::Sphis5+::loxP::MS2L cassette was used, along with a reverse oligonucleotide complementary to the 3′UTR (by ∼40 base pairs) and 3′-end of the loxP::Sphis5+::loxP::MS2L cassette, in the PCR with pLOXHIS5MS2L as the template (Haim et al., 2007; Haim-Vilmovsky and Gerst, 2009, 2011). PCR products of the correct size were transformed into yeast and grown on plates containing SC medium lacking histidine for 3–5 d at 26°C. To confirm integration, genomic DNA was extracted from single colonies, and PCR amplification, using a forward primer complementary to the coding region and reverse primer complementary to the loxP::Sphis5+::loxP::MS2L cassette and 3′UTR, was performed. PCR products were sized on agarose gels and sequenced for verification. Yeast bearing correct loxP::Sphis5+::loxP::MS2L integrations were transformed with plasmid pSH47 and grown on SC medium lacking histidine and uracil. Cre recombinase expression was induced by growing transformed cells in SC medium containing 3% galactose and lacking uracil for 16 h at 26°C. Cells were then diluted, plated, and grown on SC medium lacking uracil and replica plated to determine the presence or absence of the Sphis5+ auxotrophic marker. Yeast bearing the loxP::MS2L integration (∼790 base pairs including loxP and MS2L) were verified by PCR amplification (using oligonucleotides complementary to the coding region and 3′UTR, respectively) and DNA sequencing. MS2-CP binding sites (MS2L) were integrated into wild-type yeast between the coding region and 3′UTR of the genes.
Subcellular fractionation and protein/RNA detection
Separation of the ER and cytosol was performed using two methods. Microsome fractionation using the first method was performed as described in Aronov et al. (2007). In brief, wild-type BY4743Hα yeast were grown at 26°C in selective medium (400 ml) to OD600 of 0.6. Cycloheximide was added to a final concentration of 100 μg/ml, and cultures were grown for another 15 min at 26°C. Cells were harvested and washed with ice-cold SK buffer (1.2 M sorbitol, 0.1 M KPO4, pH 7.5, and 100 μg/ml cycloheximide). After 5 min of incubation on ice, cells were centrifuged for 3 min at 500 × g, mechanically disrupted in liquid nitrogen using a mortar and pestle, and resuspended in 1 ml of ice-cold lysis buffer. Lysates were further homogenized using 30 slow strokes with an ice-cold Dounce homogenizer. Crude total cell lysates (TCLs) were obtained by centrifugation for 5 min at 500 × g. Lysates (900 μl total volume each) were then equilibrated with a solution of 5.37 ml of 2.5 M sucrose, 190 μl of 3 M KCl, and 40 μl of 1 M Tris, pH 7.5, to yield a final concentration of 2.1 M sucrose. Next a discontinuous sucrose gradient was formed by adding consecutive layers of 2.5 M (volume, 2 ml), 2.1 M (volume, 6.25 ml; the sucrose-equilibrated cell lysate), 2 M (volume, 6.25 ml), and 1.3 M (volume, 2 ml) sucrose, respectively, in a 17-ml ultracentrifuge tube. Gradients were centrifuged at 25,000 rpm (90,000 × g) in an SW32 rotor (Beckman Coulter, Indianapolis, IN) at 4°C for 12 h. Next 400-μl samples were collected, and total RNA from the membrane interface (interface between 1.3 and 2.05 M sucrose layers) and cytosolic fraction (2.1 M sucrose layer) was isolated using a MasterPure Yeast RNA Purification Kit (including DNase I treatment) according to manufacturer's recommendations (Epicentre Biotechnologies, Madison, WI).
The second method is based on ultracentrifugation of total cell lysates for 1 h at 48,000 × g, with subsequent analysis of RNA extracted (as described) from supernatant and pellet, corresponding to cytosol and ER, respectively. This method was used to analyze the distribution of SUC2 mRNA in both repressed and derepressed cells, as well as to analyze RNA distribution in the sec65-1 temperature-sensitive strain at both permissive and restrictive temperatures. In brief, WT BY4743Hα cells expressing SEC63-GFP from a single-copy plasmid (as an ER marker) were grown at 26°C in selective medium (400 ml) to OD600 of 0.6. Cycloheximide was added to a final concentration of 100 μg/ml, and cultures were grown for another 15 min at 26°C. Cells were harvested and washed with ice-cold SK buffer. After 5 min of incubation on ice, cells were centrifuged for 3 min at 500 × g, resuspended in a buffer solution containing 50 mM Tris (pH 7.6), 150 mM NaCl, 200 U of RNasin Ribonuclease Inhibitor (Promega, Madison, WI), Complete Protease Inhibitor Cocktail (Roche Diagnostics, Basel, Switzerland), and cycloheximide 100 μg/ml, and disrupted using glass beads and vortexing for 10 min at 4°C. Crude lysates were centrifuged for 10 min at 1000 × g to remove cell debris, and then 1 ml of each lysate was subjected to ultracentrifugation for 1 h at 48,000 × g. The resulting pellet was then resuspended in 500 μl of buffer containing 50 mM Tris (pH 7.6), 150 mM NaCl, 80 U/ml RNasin Ribonuclease Inhibitor, Complete Protease Inhibitor Cocktail, and 100 μg/ml cycloheximide. Next total RNA from both the membrane fraction (resuspended pellets) and the cytosolic fraction (supernatants) was isolated using the MasterPure Yeast RNA Purification Kit (including DNase I treatment) according to the manufacturer's recommendations.
For each method of cell fractionation performed, reverse transcription (RT) was performed using Moloney murine leukemia virus RT RNase H(–) (Promega) under conditions suggested by the manufacturer. For each mRNA to be detected, the RT reaction was performed for the same quantity of total RNA (2.5 μg). cDNA used for PCR amplification was tested by serial dilution in order to avoid reaching a plateau phase (saturation) during PCR. Specific oligonucleotide pairs were then used to amplify genes of interest. For each mRNA to be tested, the same quantity of cDNA from the RT reaction was taken from both the ER and cytosolic fractions for PCR. Negative controls (PCR amplification of RNA from ER and cytosolic fractions without RT or PCR without template) were performed in parallel and yielded no products after PCR, with PCR on genomic DNA serving as positive control. Samples of the PCR products were electrophoresed on 1% agarose gels and documented. To detect proteins by immunoblotting, 25 μg of protein from the ER and cytosolic fractions was subjected to SDS–PAGE on 7 or 10% acrylamide gels. Immunoblots were blocked in 5% nonfat milk in phosphate-buffered saline and incubated with a polyclonal anti-DPM1 antibody (1:1000; Molecular Probes, Eugene, OR) to detect the ER and either anti–phosphoglycerate kinase antibody (diluted 1:1000; gift of Z. Elazar, Weizmann Institute of Science, Rehovot, Israel) or monoclonal anti-GFP antibodies (Roche Diagnostics) to detect cytosolic Pgk1 or GFP-Sec63, respectively. Rabbit polyclonal antibodies against Kar2 (diluted 1:5000) and Tom70 (diluted 1:10,000), used to detect ER and mitochondria, respectively, were gifts from M. Schuldiner (Weizmann Institute of Science) and Y. Arava (Technion, Haifa, Israel).
smFISH
Yeast cells were grown to OD600 of 0.1–0.2 at 26°C. Induction of endogenous SUC2 expression was performed by shifting the cells to rich (YPD) medium containing 0.1% glucose for 1.5 h. Plasmid-based expression of SUC2 under MET25 promoter was induced by shifting the cells on selective synthetic medium without methionine for 1.5 h. After SUC2 induction, cells were fixed in the same medium with added formaldehyde (3.7% final concentration) for 45 min. Cells were gently washed twice with 0.1 M potassium phosphate buffer, pH 7.4, containing 1.2 M sorbitol, after which cells were spheroplasted in 1 ml of a freshly prepared spheroplast buffer (0.1 M potassium phosphate buffer, pH 7.4, 1.2 M sorbitol, 20 mM ribonucleoside vanadyl complexes [Sigma-Aldrich, St. Louis, MO], 1× Complete Protease Inhibitor Cocktail, 28 mM β-mercaptoethanol, 120 U/ml RNasin Ribonuclease Inhibitor, Zymolase [10 kU/ml]) for 20 min at 30°C. The spheroplasts were centrifuged for 4 min at 1300 × g at 4°C and washed twice in 0.1 M potassium phosphate buffer, pH 7.4, with 1.2 M sorbitol. Spheroplasts were then resuspended in 70% ethanol and incubated overnight at 4°C. Afterward, cells were centrifuged at 1300 × g at 4°C for 4 min, washed with WASH buffer (0.3 M sodium chloride, 30 mM sodium citrate, and 10% formamide), and incubated in the hybridization solution, containing 0.3 M sodium chloride, 30 mM sodium citrate, 10% dextran sulfate, 10% formamide, 2 mM ribonucleoside vanadyl complexes, and the TAMRA-labeled Stellaris probe mix for SUC2 (Biosearch Technologies, Novato, CA) overnight at 30°C in a dark chamber. After hybridization, labeled spheroplasts were centrifuged at 1300 × g, the hybridization solution aspirated, and the spheroplasts incubated for 30 min at 30°C in WASH buffer. Next cells were centrifuged and resuspended in a solution containing 0.3 M sodium chloride and 30 mM sodium citrate. SUC2 mRNA colocalization with the ER was visualized using a DeltaVision imaging system (Applied Precision, Issaquah, WA). Images were processed by deconvolution. The localization of a total of 481 granules was scored in 58 WT cells using smFISH in Figure 3A.
Invertase assay
Invertase secretion was measured as described previously (Goldstein and Lampen, 1975).
Cell preparation for the invertase assay was performed as described in Novick and Schekman (1979). Secreted and nonsecreted activities were expressed in units based on absorption at 540 nm (1 U = 1 μmol glucose released/min per 100 mg dry cells).
Movies
Cells endogenously expressing MS2 aptamer–tagged mRNAs were transformed with plasmids expressing MS2-CP-GFP(x3 or x4) to label the mRNA and Sec63-RFP to label ER. Cells were grown to mid log phase at 26°C on liquid synthetic medium and then shifted to medium lacking methionine for 1 h. For SUC2 mRNA, the cells were grown on low glucose (0.1%)–containing medium to induce SUC2 expression. Cells were placed on a thin agar block containing synthetic medium lacking methionine and visualized for up to 0.5 h using a DeltaVision imaging system. Images were processed by deconvolution.
Cycloheximide treatment
MS2 aptamer–integrated SUC2 or USE1 cells were grown to mid log phase at 26°C in selective medium and then shifted to medium lacking methionine for 1 h. Cycloheximide was then added to a final concentration of 100 μg/ml and incubated for the indicated times (10 or 30 min) at 26°C. The cells were immediately fixed, as described in Haim-Vilmovsky and Gerst (2009), before RNA visualization.
[35S]Methionine protein labeling
WT yeast were grown to mid log phase at 26°C on medium lacking methionine. Aliquots of cells (3.75 OD600 units) were treated with CHX (final concentration, 100 μg/ml) for 5 or 15 min and then pulse labeled with [35S]methionine (125 μCi/ml; PerkinElmer-Cetus, Waltham, MA) for 15 min to label newly synthesized proteins. In parallel, control cells that were not treated with CHX were pulse labeled with [35S]methionine for 15 min. Cells were collected and washed in a buffer solution containing 50 mM Tris (pH 7.6) and 150 mM NaCl. Cells were lysed as described in Kushnirov (2000) and separated by SDS–PAGE on 10% polyacrylamide gels.
Immunoprecipitation of protein and RNA
Immunoprecipitation (IP) of Myc-She2 was done as described in Gelin-Licht et al. (2012). Briefly, WT yeast (BY4743Hα) expressing either Myc-tagged She2 or the Myc tag empty vector were grown on 200 ml of synthetic selective medium to OD600 of 1, after which they were grown for an additional 2 h in selective medium containing 0.1% glucose to induce SUC2 expression. Cells were centrifuged at 3000 × g for 5 min at 4°C and resuspended in 1 ml of lysis buffer (20 mM Tris-Cl, pH 7.5, 150 mM KCl, 5 mM MgCl2, 100 U/ml RNasin, 0.1% NP-40, 1 mM dithiothreitol, 2 μg/ml aprotinin, 1 μg/ml pepstatin, 0.5 μg/ml leupeptin, and 0.01 μg/ml benzamidine). Cells (200 OD600 units) were broken by vortexing with glass beads (0.5 mm) for 1 h at 4°C and centrifuged at 4000 × g for 10 min at 4°C to yield the TCL. For IP and Western analysis, 1 mg of TCL was diluted in lysis buffer (final volume, 1 ml) and subjected to IP with 2 μl of monoclonal anti-Myc antibody (9E10; Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C with rotation. Then 20 μl of a protein G agarose slurry (Santa Cruz Biotechnology) was added and the samples incubated for an additional 1 h at 4°C with rotation. The precipitates were washed (×3) with lysis buffer, electrophoresed on 14% SDS–PAGE gels, and detected in immunoblots with the anti-Myc antibodies. In parallel, 50-μg samples of the TCL were also electrophoresed and detected in blots with anti-Suc2 antibodies (gift from C. Barlowe, Dartmouth University, Hanover, NH), anti-actin antibodies (MP Biomedicals, Solon, OH), and the anti-Myc antibodies. For IP and RNA detection analysis, 3 mg of TCL was diluted in lysis buffer (final volume, 1 ml) and incubated with 10 μl of anti-Myc antibody overnight at 4°C with rotation. Then 30 μl of a protein G agarose slurry (Santa Cruz Biotechnology) was added and the samples incubated for an additional 1 h at 4°C. The agarose beads were washed (×3) with lysis buffer, and protein–RNA complexes were eluted by incubation in 300 μl of elution buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10 mM EDTA, 0.1% SDS), in diethyl pyrocarbonate–treated water with 100 μg/ml proteinase K (Promega) at 65°C for 30 min.
RNA extraction from immunoprecipitated protein–RNA complexes and RT-PCR
RNA was isolated from the IP eluate by adding 175 μl of MPC Protein Precipitation Reagent (MasterPure RNA Isolation Kit; Epicentre Biotechnologies) per 300 μl volume, as described by the manufacturer, followed by centrifugation at 10,000 × g for 10 min at 4°C. The supernatant was transferred into a fresh microcentrifuge tube, and NaOAc (pH 5.4; 0.3 M final concentration) and glycogen (8 μg/ml final concentration; Fermentas/Thermo Scientific, Waltham, MA) were added, followed by thorough mixing. An equal volume of EtOH was then added to the tube, followed by vortexing and incubation of 20 min at −70°C. After centrifugation at 10,000 × g for 10 min at 4°C, the RNA pellet was rinsed once with 70% ethanol, air dried, and dissolved in 25 μl of Ultra Pure Water (Biological Industries, Kibbutz Beit-Haemek, Israel). The recovered RNA was subjected to treatment with RQ1 RNase-free DNase (Promega), as detailed by the manufacturer, and 15 μl from this reaction was taken for RT using M-MLV Reverse Transcriptase RNase, H Minus, Point Mutant (Promega), and a random hexamer mixture (Fermentas, Burlington, Canada) in a final volume of 25 μl. For the detection of specific transcripts, 3 μl of reverse-transcribed RNA was amplified by PCR (28 cycles, 20-μl total reaction volume) using Taq Purple Mix (Lambda Biotech, St. Louis, MO) and oligonucleotide primer pairs specific for ASH1 or SUC2. Control reactions (PCR amplification of genomic DNA, PCR mix without template, and PCR amplification of pulled-down RNA without RT) were performed in parallel. PCR products were electrophoresed on 1% agarose gels and photodocumented.
Supplementary Material
Acknowledgments
We thank Yoav Arava, Charles Barlowe, Noga Gadir, Kerry Bloom, Zvulun Elazar, Susan Michaelis, Maya Schuldiner, and Robert Singer for reagents. This study was supported by grants to J.E.G. from the Minerva Foundation, Germany, and the Y. Leon Benoziyo Institute for Molecular Medicine and the Center for Scientific Excellence, Weizmann Institute, Israel. J.E.G. holds the Besen-Brender Chair in Microbiology and Parasitology, Weizmann Institute. E.B. is supported by an Israel Science Foundation First Award and holds the Jerome A., Freda, and Edward M. Siegel Chair, Weizmann Institute.
Abbreviations used:
- cER
cortical endoplasmic reticulum
- CHX
cycloheximide
- ER
endoplasmic reticulum
- INT
integrated
- GFP
green fluorescent protein
- mPOL
mRNA encoding polarity and secretion factor
- MS2-CP
MS2 bacteriophage coat protein
- MS2L
MS2 aptamer loop
- mSMP
mRNA encoding secreted or membrane proteins
- m-TAG
MS2 aptamer genome-tagging procedure
- nER
nuclear ER (ER peripheral to the nucleus)
- ORF
open reading frame
- RBP
RNA-binding protein
- RFP
red fluorescent protein
- RT-PCT
reverse transcription-PCR
- smFISH
single-molecule fluorescence in situ hybridization
- SNARE
soluble N-ethylmaleimide–sensitive factor attachment protein receptors
- SRP
signal recognition particle
- SSCR
signal-sequence coding region
- TAMRA
tetramethylrhodamine
- TMD
transmembrane domain
- UTR
untranslated region
- YPD
yeast extract–bactopeptone–dextrose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-01-0038) on July 31, 2013.
*These authors contributed equally.
REFERENCES
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3’UTR ends. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Aronov S, Gelin-Licht R, Zipor G, Haim L, Safran E, Gerst JE. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:3441–3455. doi: 10.1128/MCB.01643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast T, Cohen G, Schuldiner M. A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell. 2013;152:1134–1145. doi: 10.1016/j.cell.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Belgareh-Touze N, Corral-Debrinski M, Launhardt H, Galan JM, Munder T, Le Panse S, Haguenauer-Tsapis R. Yeast functional analysis: identification of two essential genes involved in ER to Golgi trafficking. Traffic. 2003;4:607–617. doi: 10.1034/j.1600-0854.2003.00116.x. [DOI] [PubMed] [Google Scholar]
- Bibi E. Early targeting events during membrane protein biogenesis in Escherichia coli. Biochim Biophys Acta. 2011;1808:841–850. doi: 10.1016/j.bbamem.2010.07.025. [DOI] [PubMed] [Google Scholar]
- Bibi E. Is there a twist in the E. coli signal recognition particle pathway. Trends Biochem Sci. 2012;37:1–6. doi: 10.1016/j.tibs.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B. Transfer of proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J Cell Biol. 1975;67:852–862. doi: 10.1083/jcb.67.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5’ ends encode secreted with intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Carlson M, Taussig R, Kustu S, Botstein D. The secreted form of invertase in Saccharomyces cerevisiae is synthesized from mRNA encoding a signal sequence. Mol Cell Biol. 1983;3:439–447. doi: 10.1128/mcb.3.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Thompson MA, Salzman J, Champion LM, Moerner WE, Brown PO. Widespread mRNA association with cytoskeletal motor proteins and identification and dynamics of myosin-associated mRNAs in S. cerevisiae. PLoS One. 2012;7:e31912. doi: 10.1371/journal.pone.0031912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Jagannathan S, Reid D, Zheng T, Nicchitta C. Hierarchical regulation of mRNA partitioning between the cytoplasm and the endoplasmic reticulum of mammalian cells. Mol Biol Cell. 2011;22:2646–2658. doi: 10.1091/mbc.E11-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomina N, Ferrezuelo F, Wang H, Aldea M, Gari E. Whi3, a developmental regulator of budding yeast, binds a large set of mRNAs functionally related to the endoplasmic reticulum. J Biol Chem. 2008;283:28670–28679. doi: 10.1074/jbc.M804604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui XA, Zhang H, Palazzo AF. p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. PLoS Biol. 2012;10:e1001336. doi: 10.1371/journal.pbio.1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Duimio L, Peracchia G, Ottonello S. Selective inactivation of two components of the multiprotein transcription factor TFIIIB in cycloheximide growth-arrested yeast cells. J Biol Chem. 1995;270:13476–13482. doi: 10.1074/jbc.270.22.13476. [DOI] [PubMed] [Google Scholar]
- Diehn M, Eisen MB, Botstein D, Brown PO. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- Dilcher M, Veith B, Chidambaram S, Hartmann E, Schmitt HD, Fischer von Mollard G. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 2003;22:3664–3674. doi: 10.1093/emboj/cdg339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadir N, Haim-Vilmovsky L, Kraut-Cohen J, Gerst JE. Localization of mRNAs coding for mitochondrial proteins in the yeast, Saccharomyces cerevisiae. RNA. 2011;17:1551–1565. doi: 10.1261/rna.2621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelin-Licht R, Paliwal S, Conlon P, Levchenko A, Gerst JE. Scp160-dependent mRNA trafficking mediates pheromone gradient sensing and chemotropism in yeast. Cell Rep. 2012;1:483–494. doi: 10.1016/j.celrep.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerst JE. Message on the web: mRNA and ER co-trafficking. Trends Cell Biol. 2008;18:68–76. doi: 10.1016/j.tcb.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Goldstein A, Lampen JO. Beta-D-fructofuranoside fructohydrolase from yeast. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- Haim-Vilmovsky L, Gerst JE. m-TAG: a PCR-based genomic integration method to visualize the localization of specific endogenous mRNAs in vivo in yeast. Nat Protocols. 2009;4:1274–1284. doi: 10.1038/nprot.2009.115. [DOI] [PubMed] [Google Scholar]
- Haim-Vilmovsky L, Gerst JE. Visualizing endogenous mRNAs in living yeast using m-TAG, a PCR-based genomic integration method, and fluorescence microscopy. Methods Mol Biol. 2011;714:237–247. doi: 10.1007/978-1-61779-005-8_15. [DOI] [PubMed] [Google Scholar]
- Haim L, Zipor G, Aronov S, Gerst JE. A genomic integration method to visualize localization of endogenous mRNAs in living yeast. Nat Methods. 2007;4:409–412. doi: 10.1038/nmeth1040. [DOI] [PubMed] [Google Scholar]
- Jambhekar A, Derisi JL. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA. 2007;13:625–642. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A, McDermott K, Sorber K, Shepard KA, Vale RD, Takizawa PA, DeRisi JL. Unbiased selection of localization elements reveals cis-acting determinants of mRNA bud localization in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2005;102:18005–18010. doi: 10.1073/pnas.0509229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchert C, Weidner J, Prescianotto-Baschong C, Spang A. Defects in the secretory pathway and high Ca2+ induce multiple P-bodies. Mol Biol Cell. 2010;21:2624–2638. doi: 10.1091/mbc.E10-02-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut-Cohen J, Gerst JE. Addressing mRNAs to the ER: cis sequences act up. Trends Biochem Sci. 2010;35:459–469. doi: 10.1016/j.tibs.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;30:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Lakkaraju AK, Luyet PP, Parone P, Falguieres T, Strub K. Inefficient targeting to the endoplasmic reticulum by the signal recognition particle elicits selective defects in post-ER membrane trafficking. Exp Cell Res. 2007;313:834–847. doi: 10.1016/j.yexcr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Lerner RS, Seiser RM, Zheng T, Lager PJ, Reedy MC, Keene JD, Nicchitta CV. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AM, Watson A, Fridovich-Keil JL. Scp160p associates with specific mRNAs in yeast. Nucleic Acids Res. 2003;31:1830–1837. doi: 10.1093/nar/gkg284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- Loya A, Pnueli L, Yosefzon Y, Wexler Y, Ziv-Ukelson M, Arava Y. The 3’-UTR mediates the cellular localization of an mRNA encoding a short plasma membrane protein. RNA. 2008;14:1352–1365. doi: 10.1261/rna.867208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martoglio B, Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- Mutka SC, Walter P. Multifaceted physiological response allows yeast to adapt to the loss of the signal recognition particle-dependent protein-targeting pathway. Mol Biol Cell. 2001;12:577–588. doi: 10.1091/mbc.12.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. Translation-independent localization of mRNA in E. coli. Science. 2011;331:1081–1084. doi: 10.1126/science.1195691. [DOI] [PubMed] [Google Scholar]
- Nicchitta CV, Lerner RS, Stephens SB, Dodd RD, Pyhtila B. Pathways for compartmentalizing protein synthesis in eukaryotic cells: the template-partitioning model. Biochem Cell Biol. 2005;83:687–695. doi: 10.1139/o05-147. [DOI] [PubMed] [Google Scholar]
- Novick P, Scheckman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier C, Poirier G, Gendron P, Boisgontier A, Major F, Chartrand P. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol Cell Biol. 2005;25:4752–4766. doi: 10.1128/MCB.25.11.4752-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Vallier LG, Flick JS, Carlson M, Johnston M. Expression of the SUC2 gene of Saccharomyces cerevisiae is induced by low levels of glucose. Yeast. 1997;13:127–137. doi: 10.1002/(SICI)1097-0061(199702)13:2<127::AID-YEA68>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Springer M, Shibata Y, Lee CS, Dias AP, Rapoport TA. The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol. 2007;5:e322. doi: 10.1371/journal.pbio.0050322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prilusky J, Bibi E. Studying membrane proteins through the eyes of the genetic code revealed a strong uracil bias in their coding mRNAs. Proc Natl Acad Sci USA. 2009;106:6662–6666. doi: 10.1073/pnas.0902029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyhtila B, Zheng T, Lager PJ, Keene JD, Reedy MC, Nicchitta CV. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA. 2008;14:445–453. doi: 10.1261/rna.721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D, Nicchitta C. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem. 2012;287:5518–5527. doi: 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jaedicke A, Du TG, Jansen RP. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr Biol. 2006;16:1538–1543. doi: 10.1016/j.cub.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz TU. Origins and evolution of cotranslational transport to the ER. Adv Exp Med Biol. 2007;607:52–60. doi: 10.1007/978-0-387-74021-8_4. [DOI] [PubMed] [Google Scholar]
- Shepard KA, Gerber AP, Jambhekhar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci USA. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- Uemura T, Sato T, Aoki T, Yamamoto A, Okada T, Hirai R, Harada R, Mori K, Tagaya M, Harada A. p31 deficiency influences endoplasmic reticulum tubular morphology and cell survival. Mol Cell Biol. 2009;29:1869–1881. doi: 10.1128/MCB.01089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipor G, Haim-Vilmovsky L, Gelin-Licht R, Gadir N, Brocard C, Gerst JE. Localization of mRNAs coding for peroxisomal proteins in the yeast, Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2009;106:19848–19853. doi: 10.1073/pnas.0910754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.