PKC activation enhances myosin II contractility in the central growth cone domain while decreasing actin density and increasing actin network flow rates in the peripheral domain. This dual mode of action has mechanistic implications for interpreting reported effects of PKC on growth cone guidance and neuronal regeneration.
Abstract
Protein kinase C (PKC) can dramatically alter cell structure and motility via effects on actin filament networks. In neurons, PKC activation has been implicated in repulsive guidance responses and inhibition of axon regeneration; however, the cytoskeletal mechanisms underlying these effects are not well understood. Here we investigate the acute effects of PKC activation on actin network structure and dynamics in large Aplysia neuronal growth cones. We provide evidence of a novel two-tiered mechanism of PKC action: 1) PKC activity enhances myosin II regulatory light chain phosphorylation and C-kinase–potentiated protein phosphatase inhibitor phosphorylation. These effects are correlated with increased contractility in the central cytoplasmic domain. 2) PKC activation results in significant reduction of P-domain actin network density accompanied by Arp2/3 complex delocalization from the leading edge and increased rates of retrograde actin network flow. Our results show that PKC activation strongly affects both actin polymerization and myosin II contractility. This synergistic mode of action is relevant to understanding the pleiotropic reported effects of PKC on neuronal growth and regeneration.
INTRODUCTION
The protein kinase C (PKC) family of serine/threonine kinases plays a central role in regulation of many cellular processes (Nishizuka, 1986; Larsson, 2006; Rosse et al., 2010). Original PKC family members include calcium-dependent conventional isoforms (cPKC) and calcium-independent novel isoforms (nPKC). Both cPKC and nPKC are activated by the second messenger diacylglycerol downstream of phospholipase C (Liu and Heckman, 1998; Nishizuka, 1988). Numerous studies demonstrate that PKC contributes to changes in morphology and migration in a variety of cell types, including smooth muscle cells (Hai et al., 2002), neutrophils (Downey et al., 1992), fibroblasts (Dugina et al., 1987), and epithelial cells (Xiao et al., 2009).
Recent studies also place PKC downstream of repulsive neuronal guidance signals, such as myelin-associated glycoproteins (Hasegawa et al., 2004), Nogo (Domeniconi et al., 2005), chondroitin sulfate proteoglycans (Sivasankaran et al., 2004), and ephrin (Wong et al., 2004). In agreement with the repulsive trend, PKC activation has been associated with growth cone collapse in cervical ganglion explants (Ozsarac et al., 2003), retraction responses in dorsal root ganglia (DRG; Bonsall and Rehder, 1999) and hippocampal neurons (Mattson et al., 1988), and decreased neurite sprouting in neuroblastoma cells (Cressman and Shea, 1995). PKC activation also induced axon loss at the neuromuscular junction of neonatal rats (Lanuza et al., 2002). In complementary studies, PKC inhibition increased neurite outgrowth (Hasegawa et al., 2004) and prevented growth cone collapse (Wong et al., 2004; Conrad et al., 2007) in vitro, improved axon regeneration after injury (Sivasankaran et al., 2004), and prevented dendritic spine loss due to stress (Hains et al., 2009) in vivo.
Cell and growth cone motility ultimately depend on the coordinated behavior of cytoskeletal proteins, including actin filaments and microtubules (MTs; Rodriguez et al., 2003). Many studies describe strong morphological effects of PKC; however, how PKC activation affects the cytoskeletal protein dynamics that mediates these morphological changes is not well understood. Quantitative analysis of actin filament dynamics as a function of PKC activity has not been reported and could further our understanding of relevant cell biological mechanisms.
In the marine mollusk Aplysia, there are two phorbol ester–responsive PKC isoforms: Apl I, a cPKC homologue, and Apl II, an nPKC homologue (Kruger et al., 1991). Both are abundantly expressed in the nervous system and exhibit activity-dependent association with actin filaments and MTs (Sossin et al., 1993; Nakhost et al., 1998, 2002). Previously, we reported that PKC regulates MT dynamic instability in Aplysia bag cell neuronal growth cones (Kabir et al., 2001). Specifically, PKC activation decreased MT catastrophes and increased rescue frequencies, resulting in longer MT lifetimes and MT advance into the growth cone peripheral (P) domain. We also noted that PKC activation markedly affected actin filament structure, but we did not pursue this further. We return to the question of how PKC activity affects actin dynamics and structure in the present study. Addressing these issues could have translational impact, given that PKC activity has been implicated as a negative factor in nerve regeneration (Sivasankaran et al., 2004).
Using a combination of immunocytochemistry, electron microscopy (EM), and quantitative fluorescent speckle microscopy, we provide evidence that PKC regulates actin filament dynamics and structure in growth cones via two synergistic mechanisms. In the central (C) domain, PKC activation enhances myosin II regulatory light chain phosphorylation through activation of C-kinase–potentiated protein phosphatase inhibitor (CPI-17) independent of Rho kinase activity, resulting in increased contractility that pulls actin networks inward and alters growth cone geometry. In the P domain, PKC activation depletes Arp2/3 complex and actin polymerization sites from the leading edge, dramatically reducing the density of actin veil networks. This combination of decreased actin network density and increased myosin II activity is associated with faster rates of retrograde actin flow. We consider mechanisms by which these parallel PKC actions could modulate neurite outgrowth and regeneration.
RESULTS
PKC activation induces C-domain contraction and depletes P-domain actin veil networks
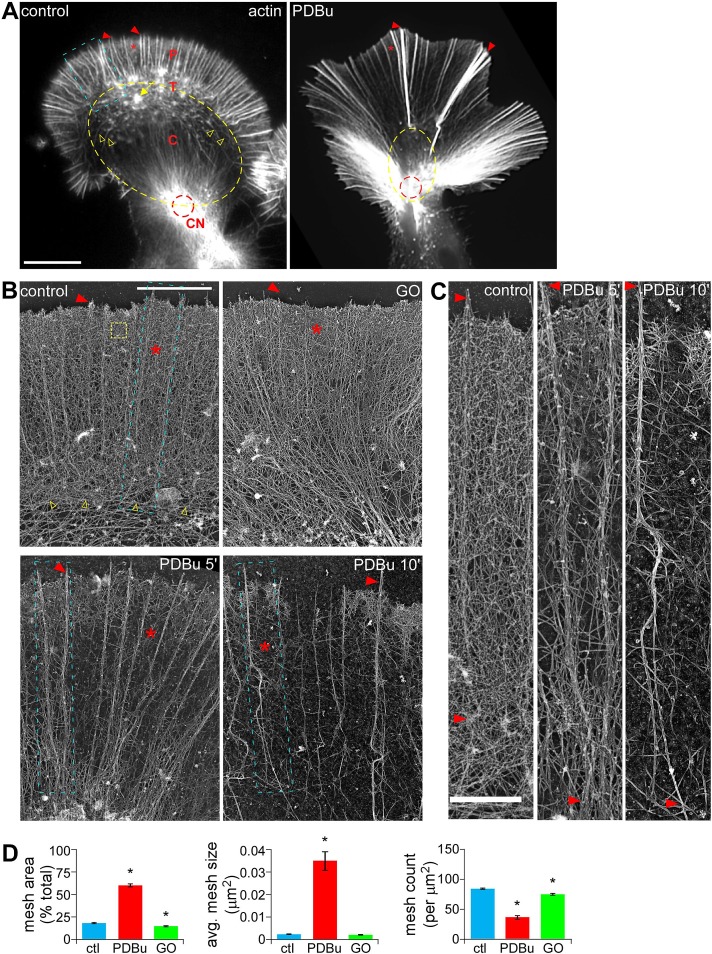
Aplysia bag cell growth cones display two distinct cytoplasm domains (Figure 1A, left; Bridgman and Dailey, 1989; Lowery and Van Vactor, 2009): a P domain containing radial filopodial actin bundles (arrowhead) and intervening actin veils (asterisk), and a C domain circumscribed by contractile actin arc structures (yellow open arrowheads; Schaefer et al., 2002) that often converge at the contractile node (CN; red dashed circle; Burnette et al., 2008), where the growth cone meets the axon shaft. The transition (T) zone between the P and C domains is populated with actin arcs (Schaefer et al., 2002) and occasional intrapodia actin structures (yellow arrow; Rochlin et al., 1999).
FIGURE 1:
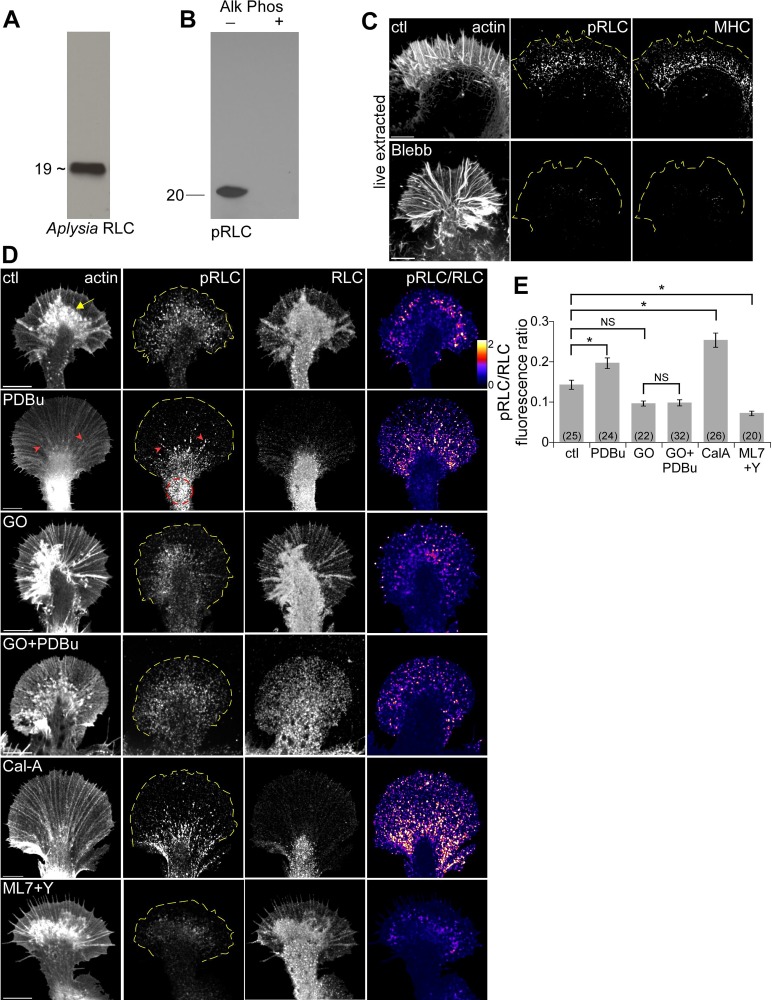
PKC activation induced retraction response and depleted P-domain actin veil networks in Aplysia growth cones (GCs). (A) Representative phalloidin labeling of control and PDBu-treated (100 nM, 10 min) GCs after regular fixation. (B) Representative rotary shadowed electron micrographs of the P domain and T zone (similar to blue boxed region in A) of GCs treated with DMSO (control), Go6976 (10 μM, 10 min), or PDBu (100 nM, 5 and 10 min). (C) High magnification of areas marked by blue boxes in B, showing filopodial bundles and intervening veil regions. (D) Quantification of actin veil meshwork parameters from 1 × 1–μm2 P-domain regions (similar to yellow dotted box in B). n = 102 regions from nine GCs for control; 73 regions from five GCs for PDBu (100 nM, 10 min); and 73 regions from four GCs for Go6976. C, central domain; CN, contractile node; P, peripheral domain; T, transition zone; red arrowhead, filopodium; asterisk, actin veil, yellow open arrowhead, actin arcs; yellow arrow, intrapodia; red dotted circle, contractile node. p < 0.01 two-tailed unpaired t test. Scale bars, 10 μm (A), 5 μm (B), 1 μm (C).
Treatment with the PKC activator phorbol 12,13-dibutyrate (PDBu; 100 nM, 10 min) resulted in a dramatic reorganization of actin filaments characterized by widening of the P domain and elongation of peripheral structures, disappearance of intrapodia, constriction of the C domain (yellow dotted lines), and buildup of actin in the proximal contractile node region (Figure 1A, right; Burnette et al., 2008). Differential interference contrast (DIC) imaging showed that PDBu application induced contraction of the C domain and proximal extension of the P domain within minutes (Supplemental Figure S1A, left and middle). This effect was reversed when a PKC inhibitor, bisindolylmaleimide-1 (BIS; 10 μM), was added in the continued presence of PDBu (Supplemental Figure S1A, right). Either BIS or a structurally distinct PKC inhibitor, Go6976 (10 μM, 10-min pretreatment), completely blocked the actin reorganization associated with PDBu treatment (Supplemental Figure S1B). Bisindolylmaleimides and indolocarbazoles (such as Go6976) are two of the most commonly used PKC-specific inhibitors (Martiny-Baron et al., 1993; Wu-Zhang and Newton, 2013). These agents inhibit both classical and novel isoforms of Aplysia PKC by competing with ATP binding (Manseau et al., 2001) and are well characterized in bag cell neurons (Kabir et al., 2001). Because BIS and GO6976 had essentially identical inhibitory effects, we chose to use GO6976 for the remaining studies.
To test whether the foregoing effects depended on MT dynamics, we pretreated cells with Taxol (1 μM, 30-min pretreatment), which inhibits MT dynamic instability and results in rapid clearance of MTs from the P domain without significantly altering actin-based motility (Kabir et al., 2001; Suter et al., 2004). Taxol pretreatment completely blocked PDBu-induced MT advance, as previously reported, but did not affect actin network reorganization (Supplemental Figure S2), indicating that these PKC effects on actin filament structure do not depend on MT dynamics or peripheral MT–actin interactions.
At the ultrastructural level, the P domain of control or Go6976-treated growth cones (Figure 1B, top) contained bundles of unipolar actin filaments comprising filopodia (arrowheads) and isotropic networks of short filaments in the veil regions (asterisks), similar to that previously described (Lewis and Bridgman, 1992; Schaefer et al., 2002). High magnifications (Figure 1C, left) show that control veils contain a dense network of actin filaments. To quantify actin network density, we measured the amount of filament-free space in 1-μm2 regions of interest (ROIs) near the leading edge and thereby defined network mesh size and mesh count parameters (see Materials and Methods). Mesh space accounted for only ∼17% of the total area under control conditions (ROI similar to yellow box in Figure 1B), indicating a densely packed actin filament network (Figure 1D). In contrast, actin veil network density progressively decreased and mesh area increased to ∼60% in PDBu-treated growth cones (Figure 1, B–D). This was accompanied by a significant increase in average mesh element size and a significant decrease in mesh count (Figure 1D) after a 10-min treatment with 100 nM PDBu. Filopodial bundles also elongated (Figure 1, B, bottom, and C) with PKC activation. In contrast, treatment with the PKC inhibitor Go6976 (10 μM, 10 min) slightly but significantly decreased total mesh area and mesh count (Figure 1D).
Collectively our results suggest two main effects of PKC activation on the structure and distribution of actin filaments in growth cones: 1) C-domain compression and 2) depletion of actin veil networks in the P domain.
PKC activation increases C-domain contractility and actin network translocation throughout the growth cone
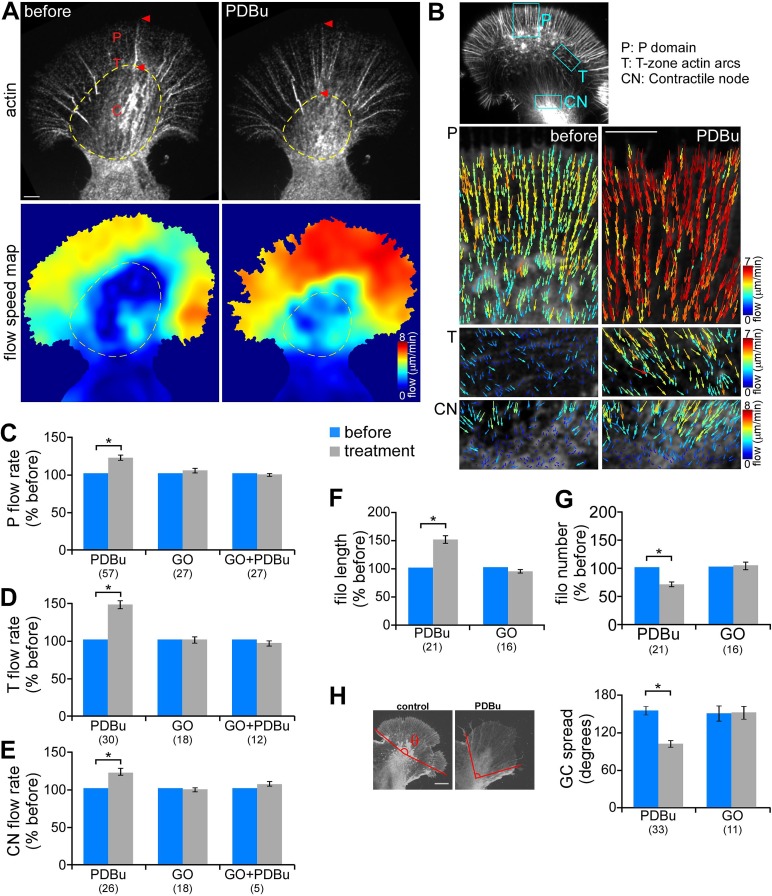
Compression of the C domain and widening of the P domain could be driven by a contractile response. To investigate this further, we quantified actin dynamics before and after PDBu treatment (100 nM, 10 min) using fluorescent speckle microscopy (FSM; Schaefer et al., 2002; Figure 2A). Briefly, trace amount of fluorescently labeled actin monomers (G-actin) or phalloidin were injected into the neurons and allowed to incorporate into the polymer lattice. Actin speckle movement was quantified using a cross-correlation–based tracking algorithm (Ji and Danuser, 2005). FSM time-lapse imaging showed that after PKC activation filopodial bundles elongated proximally (Figure 2A, red arrowhead) as C-domain structures contracted (yellow dotted circle; also see Supplemental Figure S4B and Supplemental Movies S1 and S2). Flow speed maps, generated from spatial and temporal averaging of the magnitude of speckle displacements, indicated that PDBu treatment led to increased rates of actin network translocation throughout the growth cone (Figure 2A, bottom). Flow vectors overlaid on FSM images revealed a significant increase of actin flow rates in P domain by 20.1 ± 3.6% compared with the case before PDBu addition. Retrograde movement of actin arcs in the T zone and contractile node also significantly increased after PDBu treatment by 46.3 ± 5.1 and 21.0 ± 5.4%, respectively (Figure 2, B–E, and Supplemental Figure S4, A and B). The presence of the PKC inhibitor Go6976, which by itself did not significantly affect actin network translocation, prevented PDBu-induced flow rate changes (Figure 2, C–E). Increased flow rates throughout the growth cone persisted for up to 1.5 h and were reversible after PDBu washout (30–60 min) or addition of Go6976 in the continued presence of PDBu (data not shown).
FIGURE 2:
PKC activation increased retrograde actin filament flow rate and contraction of central actin networks. (A) Representative actin FSM images (top) and corresponding spatially averaged flow speed maps (bottom) from a growth cone injected with Alexa 594–phalloidin before and after PDBu treatment (100 nM, 10 min). Yellow dotted circle marks the C-domain boundary. (B) Representative flow vector fields in the P domain (top), T zone enriched with actin arcs (middle), and contractile node (bottom) before and after PDBu treatment. Each field was obtained from an area similar to the boxed region in the inset, respectively. On both flow speed and vector maps, colors encode flow magnitude (see color bar). Vectors also indicate the flow direction. (C–E) Summary of relative changes in P-domain (C), T-zone (D), and contractile node (E) actin network translocation rates in response to PDBu (100 nM, 10–30 min), Go6976 (10 μM, 10–30 min), or pretreatment with Go6976 (10 min) followed by cotreatment with Go6976 and PDBu (10–30 min). Translocation rates were calculated by averaging the vector magnitude within regions similar to those shown in B or by kymograph analysis. (F, G) Average filopodium length (F) and number (G) measured in growth cones before and after treatment with PDBu or Go6976. Data are normalized to the values before drug addition for each growth cone. (H) Growth cone spread before and after treatment with PDBu or Go6976 measured as the angle formed by outermost filopodial actin bundles (θ in schematic). Numbers in parentheses, growth cones measured. *p < 0.01, two-tailed paired t test. Scale bars, 5 μm.
In addition to actin flow rate increases, PDBu treatment resulted in ∼50% increase in filopodial actin bundle length (Figure 2F). The average number of filopodia per growth cone decreased by ∼30% (Figure 2G). This appeared to occur by merging of existing filopodia as they elongated proximally (Supplemental Figure S3 and Supplemental Movie S3). PDBu treatment also markedly decreased growth cone spread, as measured by the angle formed between the outermost radial filopodial bundles (Figure 2H; average spread: control, 151.8°; after PDBu, 99.5°). Go6976 did not significantly affect filopodial bundle length or number or growth cone geometry. Taken together, these findings suggest PKC activation results in a contractile response associated with accelerated actin network translocation into the contractile node region.
PKC activation enhances myosin II colocalization with contractile structures
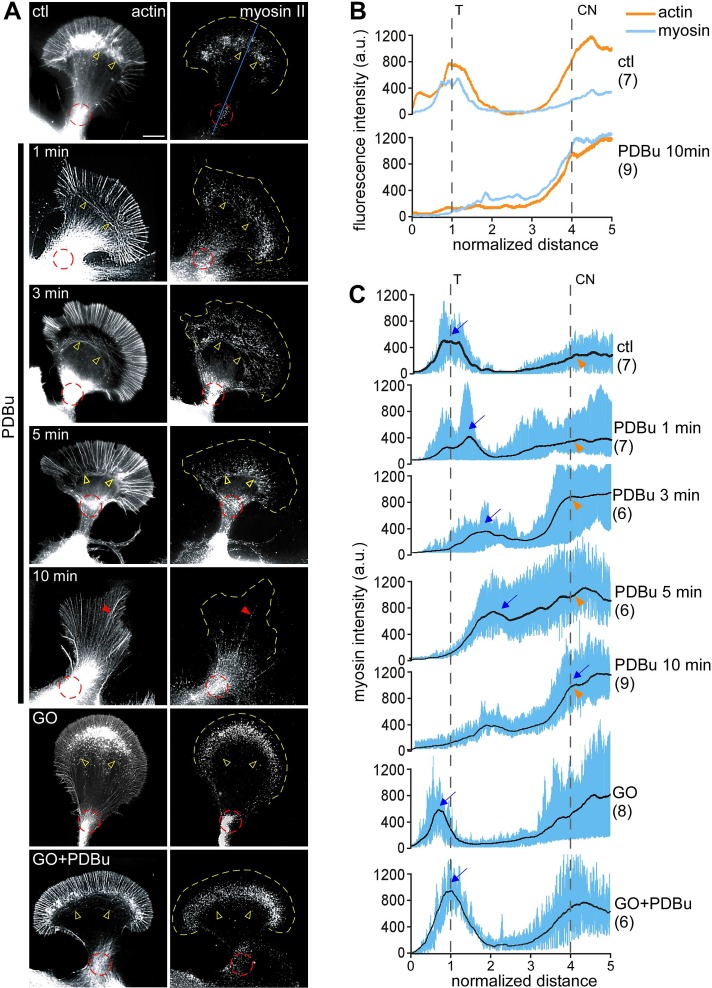
PKC activation was correlated with significant increases in rate of retrograde movement of actin arcs in the T zone and C-domain actin bundles into the contractile node. Both of these actin structures support myosin II–based contractility (Medeiros et al., 2006; Burnette et al., 2008). Rho- and myosin II–dependent increases in actin arc contractility have also been implicated in neurite retraction in response to LPA (Zhang et al., 2003). To investigate a potential role for myosin II in PKC effects here, we visualized cytoskeleton-associated myosin II at various time points after PKC activation (Figure 3). After live extraction (Figure 3A, top), in control cells, myosin II was concentrated in a band corresponding to the T zone that overlapped with the roots of filopodia and actin arcs (yellow open arrowheads) and in a cluster at the neck of the growth cone where the contractile node was situated (Figure 3A, red dotted circle), as previously observed (Medeiros et al., 2006; Burnette et al., 2008; Van Goor et al., 2012). After PDBu treatment (100 nM), the band of myosin II at the T zone shifted progressively rearward into the C domain, accompanied by ring-like constriction of the actin arcs (Figure 3A, rows 2–4, yellow open arrowheads). Line scan population analysis clearly shows the proximal shift in peak myosin II fluorescence (Figure 3, B and C, blue arrows) over time with PKC activation. The intensity of myosin II labeling at the contractile node increased concurrently (Figure 3, A, red dotted circles, and C, orange arrowheads). After 10 min in PDBu, myosin II labeling was concentrated near the contractile node (Figure 3, A and C, row 5), with some punctuate labeling present on elongated filopodial bundles (Figure 3A, red arrowhead). Actin filament intensities in the contractile node area were comparable between control and PDBu-treated growth cones (Figure 3B, top vs. bottom, orange traces), suggesting that PKC activation increased myosin II colocalization with actin filaments in this region (Figure 3B). Go6976 (10 μM, 10-min pretreatment) completely blocked the PDBu-induced constriction of actin arcs and redistribution of myosin II (Figure 3, A and C, bottom two rows). Taken together, these results indicate that PKC activation promotes myosin II colocalization with actomyosin structures, which may contribute to the observed increases in contractility and actin network flow in the C domain.
FIGURE 3:
PKC activation promoted a ring-like contractile response mediated by redistribution of myosin II to actin contractile node. (A) Fluorescence labeling of growth cones with myosin II heavy chain tail antibody (right) and tetramethylrhodamine isothiocyanate (TRITC)–phalloidin (left) after live-cell extraction. Growth cones were treated with DMSO, PDBu (100 μM, 1–10 min), or Go6976 (10 μM, 10 min) or pretreated with Go6976 (10 min), followed by Go6976 and PDBu (10 min). Yellow open arrowhead, actin arc; red dotted circle, contractile node; red arrowhead, filopodia; yellow dotted line traces the leading edge. (B) Line scan analysis of actin (orange) and myosin II (blue) fluorescence intensities under control conditions or after 10-min treatment with PDBu. Line scans (50 pixels in width, five times the P domain in length) were sampled along the growth axis, as represented by the blue line on the top in A. Traces in this graph are the average values of multiple growth cones. (C) Line scan analysis of myosin II localization in growth cones under the conditions in A. Samples were measured in a manner similar to B. Blue lines represent data from individual growth cones. Black lines represent the population average. CN, contractile node; T, transition zone; blue arrow, peak of myosin II fluorescence in T zone; orange arrowhead, myosin II fluorescence at contractile node. Number in parentheses, growth cones measured. Scale bar, 10 μm.
Myosin II inhibition prevents PKC-induced increases in retrograde network flow
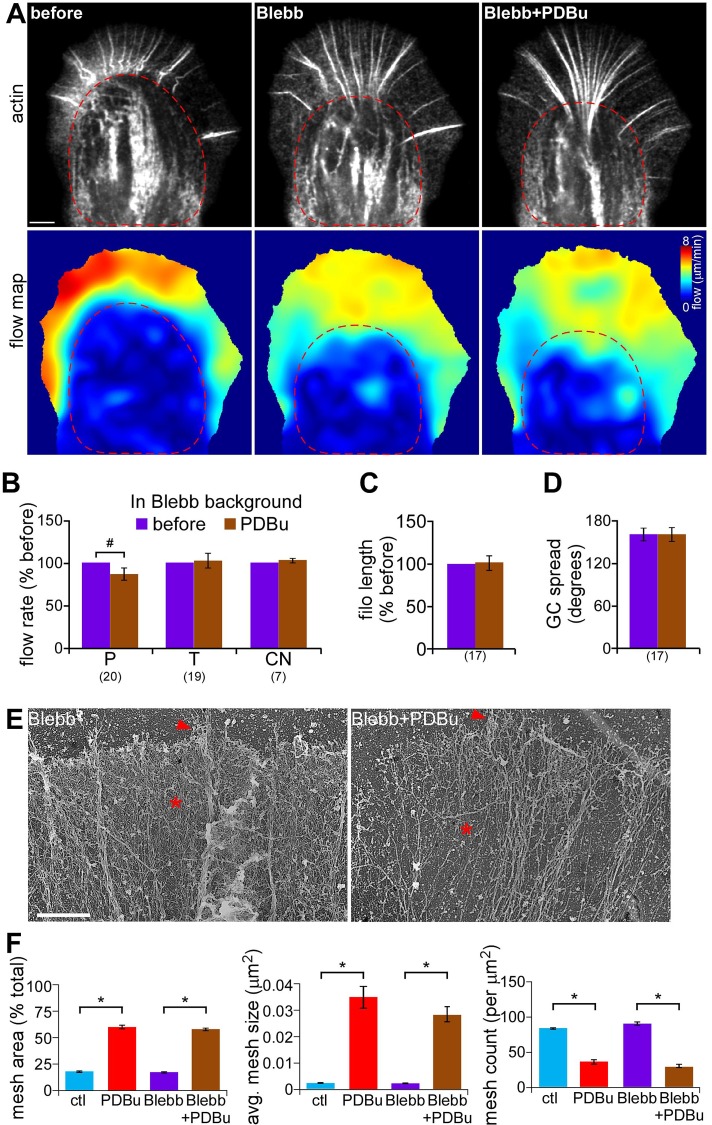
To further investigate myosin II's role in PKC effects on actin networks, we used blebbistatin, a specific nonmuscle myosin II ATPase inhibitor (Straight et al., 2003; Allingham et al., 2005; Medeiros et al., 2006). Pretreatment with blebbistatin alone (60 μM, 10 min) decreased rates of actin network translocation in both P and C domains (Figure 4A, flow speed maps; Supplemental Figure S4, C and D), as previously observed (Medeiros et al., 2006; Burnette et al., 2008; Van Goor et al., 2012; Yang et al., 2012). In blebbistatin backgrounds, PDBu treatment (100 nM, 10–30 min) no longer resulted in C-domain contraction (Figure 4A, red dashed circles) or significant changes in T-zone arc or contractile node actin flow rates (Figure 4B). Instead, PDBu treatment now resulted in decreased rates of actin network flow in the P domain (−12.6 ± 6.8%; Figure 4B). In addition, myosin II inhibition suppressed PDBu-induced changes in filopodium length and growth cone geometry (Figure 4, C and D). Note that blebbistatin pretreatment alone increased filopodial bundle length (Supplemental Figure S4E) due to attenuated filopodia recycling, as previously reported (Medeiros et al., 2006), without affecting growth cone spread (Supplemental Figure S4F). Together these results indicate that myosin II activity is crucial for the contractile response and increased network translocation associated with PKC activation. In contrast, PKC-induced decreases in actin veil network density were independent of myosin II activity (Figure 4E, asterisk), and changes in meshwork parameters were comparable to those observed with PDBu treatment alone (Figure 4F). The apparently independent PKC effects on myosin contractility and actin network structure are addressed sequentially in what follows.
FIGURE 4:
Myosin II inhibition prevented PKC-induced actin flow increase and contractile response but not the depletion of P-domain actin networks. (A) Representative actin FSM images (top) and corresponding spatially averaged flow speed maps (bottom) from a growth cone injected with Alexa 594–phalloidin under control conditions (left), in blebbistatin (60 μM, 10 min), and after PDBu (100 nM, 10 min) addition in blebbistatin background. Red dotted line marks C-domain boundary. (B) Summary of relative changes in P-domain, T-zone, and contractile node retrograde flow rates in response to PDBu (100 nM, 10–30 min) in blebbistatin background. (C, D) Average filopodium length (C) and growth cone spread (D) in growth cones treated as in B. Number in parentheses, growth cones measured. (E) EM of growth cone P domain after treatment with blebbistatin (60 μM, 10 min) or pretreatment with blebbistatin followed by cotreatment with blebbistatin and 100 nM PDBu for 10 min. Arrowhead, filopodium; asterisk, actin veil. (F) Quantification of actin veil meshwork parameters for growth cones treated as in E. Data for control and PDBu from Figure 1D are shown for comparison. N = 68 regions from five GCs for blebbistatin and 62 regions from four GCs for blebbistatin and PDBu. *p < 0.01 and #p < 0.05 with two-tailed paired (B–D) and unpaired (F) t tests. Scale bar, 5 μm (A), 2 μm (E).
PKC-dependent myosin II activation is correlated with CPI-17 phosphorylation
The foregoing results suggested that PKC activation was increasing myosin II activity. Myosin II is regulated by phosphorylation of its regulatory light chain (RLC) at a conserved serine residue (Ser-16 of Aplysia RLC) by myosin light chain kinase (MLCK) or Rho kinase. RLC phosphorylation promotes myosin minifilament formation, actin binding, and subsequent ATP-dependent force generation (Fukata et al., 2001; Vicente-Manzanares et al., 2009). RLC dephosphorylation by myosin phosphatase inactivates myosin II. In agreement, we noted that calyculin A (Ishihara et al., 1989), an inhibitor of myosin phosphatase, induced contractile responses and significantly accelerated actomyosin II arc flow in growth cones (Zhang et al., 2003), which were very similar to observations with PDBu treatment.
To investigate PKC-dependent changes in myosin II activity, we cloned myosin II regulatory light chain (Supplemental Figure S8) and produced a polyclonal antibody that labeled a single ∼19-kDa band in Western blots of Aplysia CNS tissue (Figure 5A). We used a phospho-specific antibody (Totsukawa et al., 2000) that recognized a similar–molecular weight band in control but not alkaline phosphatase–treated Aplysia CNS tissue samples (Figure 5B) to assess RLC phosphorylation (pRLC) levels. In immunolabeled growth cones, pRLC colocalized with Aplysia myosin II heavy chain after live-cell extraction that solubilized and removed the weakly bound myosin II fraction (Medeiros et al., 2006; Figure 5C, top; compare with Figure 3A, control labeling). pRLC and myosin II heavy chain labeling was strongly attenuated after blebbistatin treatment (Figure 5C, bottom), consistent with the pRLC antibody labeling active nonmuscle myosin II.
FIGURE 5:
PKC activation increased myosin II regulatory light chain phosphorylation. (A) Western blots of Aplysia CNS proteins probed with rabbit anti–Aplysia RLC sera recognizing ∼19-kDa band. (B) Western blot of control and alkaline phosphatase–treated Aplysia CNS homogenate with a mouse monoclonal antibody against conserved pSer19 on human myosin II regulatory light chain (pRLC). (C) Growth cones were live cell extracted and labeled with TRITC–phalloidin (left), antibody against conserved pSer19 on human myosin II regulatory light chain (pRLC; middle), and antibody against Aplysia myosin II heavy chain (MHC; right) after treatment with DMSO or blebbistatin (60 μM, 10 min). (D) Immunolabeling of growth cones with pRLC antibody and total Aplysia RLC antibody after normal fixation. F-actin was visualized with phalloidin. Right, ratio of pRLC and Aplysia RLC after background subtraction, encoded in a linear pseudocolor lookup table (see color bar). Growth cones were treated with DMSO, PDBu (100 nM, 10 min), Go6976 (10 μM, 10 min), Go6976 and PDBu (10 min) after Go6976 pretreatment, calyculin A (50 nM, 20 min), or combination of Y27632 and ML7 (10 μM each, 20 min). Yellow arrow, intrapodia; red arrowhead, filopodium; red dotted circle, contractile node; yellow dotted line traces the leading edge. (E) Quantification of average pRLC/RLC fluorescence ratio in entire growth cones for each of the conditions in D. Numbers in parentheses, growth cones measured. p < 0.0001 with single-factor ANOVA. Asterisk indicates significant difference using Tukey's HSD post hoc analysis. NS, not significant. Scale bars, 10 μm.
To quantify myosin II activity, we colabeled growth cones with pRLC and total RLC using a normal fixation procedure and used ratiometric imaging of pRLC to total RLC labeling to estimate the relative fraction of activated myosin II under different conditions (Figure 5, D and E). Aplysia total RLC labeling was punctuate in the P and C domains and appeared to distribute with cytoplasmic volume with the C-domain and intrapodia (yellow arrow) exhibiting highest intensities (Figure 5D, column 3). In contrast, pRLC labeling was highest in the T zone, where active myosin II is normally localized (Medeiros et al., 2006), and lower in the C domain (Figure 5D, column 2). After PDBu treatment (100 nM, 10 min), pRLC labeling was concentrated in the contractile node region (Figure 5D, red dotted circles) and also colocalized with filopodial bundles (red arrowheads), reminiscent of the myosin II redistribution observed in live-extracted cells (Figure 3A). Under control conditions myosin II activity was localized to the T zone (Figure 5D, column 4). PDBu treatment resulted in a proximal shift of highest myosin II activity (Figure 5D, column 4) and a corresponding ∼37% increase in growth cone myosin II activity relative to controls (Figure 5E). In contrast, Go6976 treatment (10 μM, 10 min) modestly decreased myosin II activity and completely blocked PKC effects (Figure 5, D and E). As positive and negative controls, growth cones were treated with calyculin A (50 nM, 10 min) or a combination of ML-7 plus Y27632 (10 μM each, 10 min), which inhibit MLCK and Rho kinase, respectively (Figure 5D, bottom two rows). As expected, calyculin A alone increased the fraction of active myosin II by ∼77% over controls; in contrast, concurrent treatment with ML-7 and Y27632 decreased myosin II activity by ∼50% (Figure 5E). These results suggest that PKC activation enhances myosin II activity through increased RLC phosphorylation.
We next investigated possible mechanisms for PKC-dependent myosin II activation. Phosphorylation of myosin RLC is Ca2+/calmodulin dependent. PKC has been reported to modulate trafficking of Ca2+ channels with subsequent effects on intracellular Ca2+ levels in both Aplysia and vertebrate neurons (Knox et al., 1992; Herlitze et al., 2001). To test whether intracellular Ca2+ changes could be involved in these effects, we did ratiometric Ca2+ imaging in tandem with assessment of actin dynamics during PKC activation (Supplemental Figure S5). Whereas actin FSM indicated typical acceleration of peripheral retrograde actin flow and C-domain contraction during PKC activation, intracellular Ca2+ levels did not significantly change (Supplemental Figure S5). Note that we cannot formally rule out involvement of rapid Ca2+ fluctuations such as those resulting from voltage-dependent Ca2+-channel activity from these experiments, due to the recording bandwidth (0.1 Hz). If rapid Ca2+ fluctuations were involved, however, they are not of sufficient average strength to significantly alter baseline Ca2+ levels. In summary, our results suggest that intracellular Ca2+ changes do not play a significant role in the observed actin network reorganization.
The small GTPase Rho and its effector Rho kinase constitute another major pathway for regulation of myosin II–dependent contractility. Rho kinase can directly phosphorylate RLC and sensitize it to MLCK action via inhibition of myosin phosphatase (Fukata et al., 2001). We previously showed that Rho–Rho kinase activation increased actin arc stability and translocation rates, as well as C-domain contractility (Zhang et al., 2003). PKC has been reported to associate with and activate Rho (Slater et al., 2001; Pan et al., 2005). In some cell types, inhibition of Rho–Rho kinase signaling prevented PKC-induced morphological changes (Lai et al., 2003). To test whether this was the case in our system, we treated growth cones with PDBu in the presence of Rho kinase inhibitor Y27632 (10 μM, 20-min pretreatment; Supplemental Figure S6). Rho kinase inhibition alone attenuated actin arc movement in the T zone and contractile node region without affecting retrograde flow in the P domain (Supplemental Figure S6, B and C; Zhang et al., 2003); however, Rho kinase inhibition did not block the changes in actin filament reorganization (Supplemental Figure S6A; cf. Figure 1A), increased C-domain contractility (Supplemental Figure S6B, yellow dotted circles), and increased actin flow rates typically observed after PKC activation (Supplemental Figure S6, B, flow maps, and D). Despite slower baseline rates of actin network translocation in the C domain in Y27632 (Supplemental Figure S6C), PDBu treatment still increased contractility in this region (i.e., in both the T zone and node; Supplemental Figure S6D). These results are inconsistent with Rho kinase being a downstream PKC effector here and suggested an alternative mechanism.
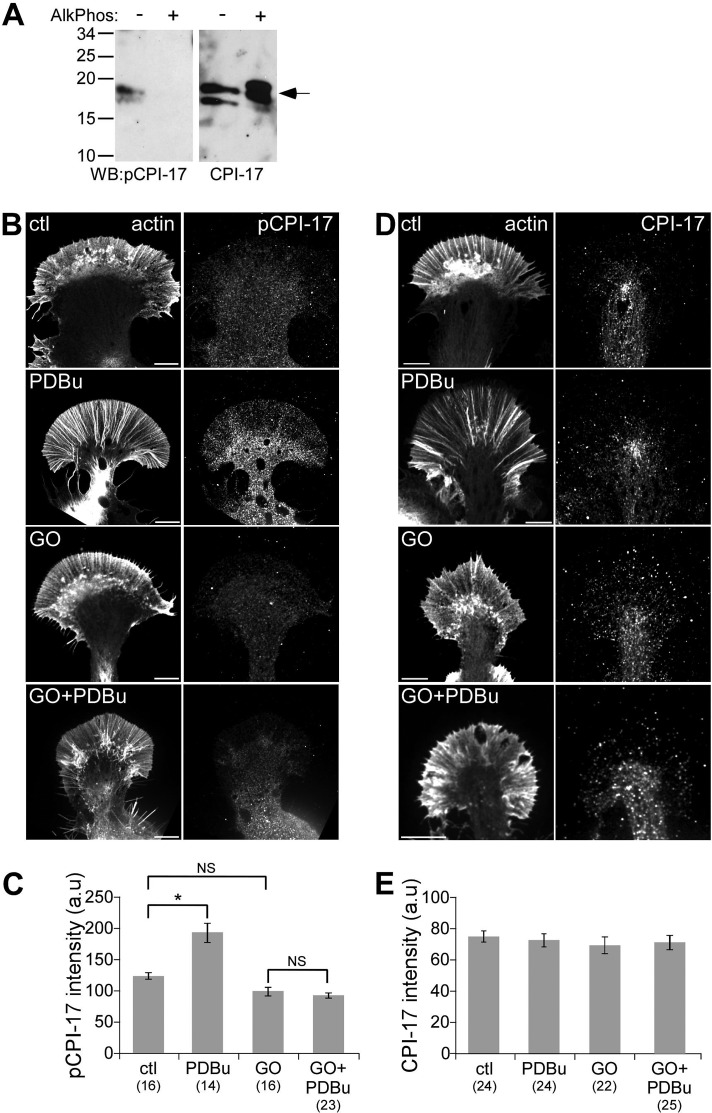
Possible alternative candidates are C kinase–potentiated protein phosphatase-1 inhibitor (CPI-17; Kitazawa et al., 1999; Eto et al., 2000; Ohki et al., 2001) and its homologues kinase-enhanced protein phosphatase 1 inhibitor (Liu et al., 2002; Gong et al., 2005) and gastrointestinal and brain-specific PP1-inhibitory protein (Liu et al., 2004). These proteins possess potent inhibitory activity toward myosin phosphatase when phosphorylated by PKC at a conserved threonine residue (Thr-38 of human CPI-17). Moreover, CPI-17 phosphorylation has been correlated with contractile responses and cytoskeletal changes in neurons and other motile cells (Eto et al., 2000, 2002; Watanabe et al., 2001). To test a possible role for CPI-17 activity, we used a phospho-specific CPI-17 antibody that recognized two bands at ∼17 kDa on Western blots of control but not alkaline phosphatase–treated Aplysia CNS homogenates (Figure 6A, left). As a control, an antibody against nonphosphorylated CPI-17 recognized bands of the same molecular weight in control and phosphatase-treated CNS homogenates (Figure 6A, right). The doublet band may represent multiple CPI-17 isoforms in Aplysia, as the antibodies were made against a conserved peptide sequence. Under control conditions, immunolabeling revealed a diffuse pattern of phosphorylated or total CPI-17 in the growth cone with higher labeling densities in the C domain, where the path length is longer (Figure 6, B and D). PDBU treatment (100 nM, 10 min) increased p-CPI-17 labeling in the growth cone by ∼54% relative to controls (Figure 6C; note that ratiometric imaging could not be used for quantification in this case since both antibodies were from the same host species). Go6976 treatment alone decreased baseline p-CPI-17 labeling (down ∼21%), although this did not meet statistical significance. Cotreatment with PDBu blocked increases in p-CPI-17 labeling (Figure 6, B and C). Note that neither PDBu nor Go6976 significantly changed total CPI-17 labeling levels (Figure 6, D and E). These results are consistent with PKC activation of CPI-17 as a mechanism for the observed increased myosin II activity and related contractile effects on actin dynamics.
FIGURE 6:
PKC activation increased CPI-17 phosphorylation. (A) Western blot of control and alkaline phosphatase–treated Aplysia CNS homogenate with antibodies against total and pThr38 CPI-17. (B, D) Immunolabeling of growth cones with antibodies against p-CPI-17 (B) or total CPI-17 (D) after normal fixation. F-actin was visualized with phalloidin. Growth cones were treated with DMSO, PDBu (100 nM, 10 min), Go6976 (10 μM, 10 min), or Go6976 and PDBu (10 min) after Go6976 pretreatment. (C, E) Quantification of average p-CPI-17 (C) and total CPI-17 (E) fluorescence intensity in entire growth cones for each of the conditions in B and D. Numbers in parentheses, growth cones measured. For C, p < 0.0001 with-single factor ANOVA. Asterisk indicates significant difference using Tukey's HSD post hoc analysis. NS, not significant. For E, p = 0.89 with single-factor ANOVA. Scale bars, 10 μm.
PKC activation delocalizes Arp2/3 complex from the leading edge independently of myosin II activity
PKC effects on contractility and actin network translocation depended on nonmuscle myosin II activity; however, the observed changes in actin veil density persisted after nonmuscle myosin II inhibition (Figure 4, E and F). These results suggested the presence of a PKC pathway affecting actin filament dynamics functioning in parallel with the contractile effects of nonmuscle myosin II as described earlier.
The Arp2/3 complex is a conserved seven-protein nucleator of actin filament assembly responsible for formation of dense branched networks in the lamellipodia of motile cells (Pollard, 2007) and growth cone veils (Mongiu et al., 2007; Korobova and Svitkina, 2008; Yang et al., 2012). We recently reported that acute suppression of Arp2/3 complex activity with small-molecule inhibitors (Yang et al., 2012) drastically reduced actin veil network density (Korobova and Svitkina, 2008) and accelerated P-domain retrograde flow rates (Yang et al., 2012) in a manner strikingly similar to what we observed after PKC activation.
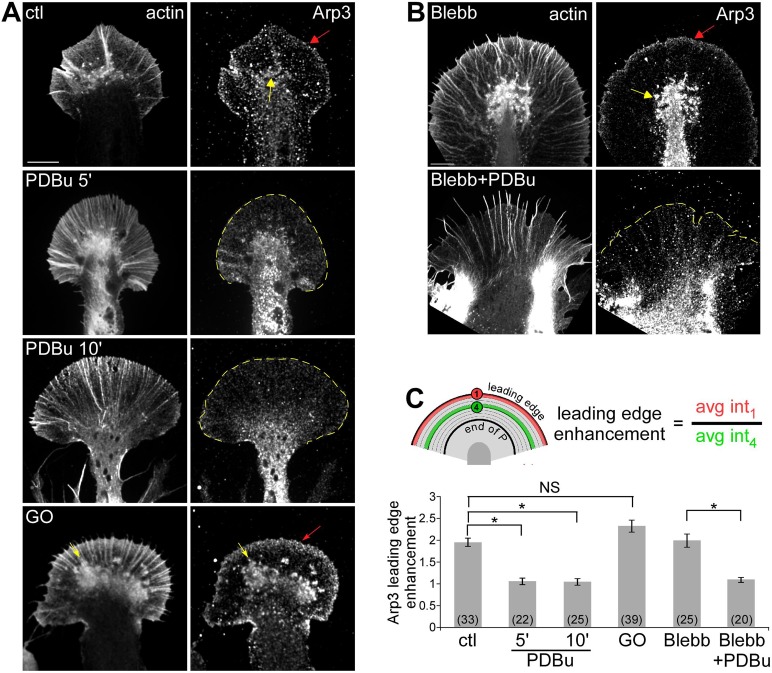
To investigate whether the Arp2/3 complex might play a role in these PKC effects, we first compared Arp2/3 complex localization in growth cones under control conditions and after PKC activation. Under control conditions, Arp2/3 complex density was enhanced by ∼1.8-fold near the leading edge when compared with adjacent regions (Figure 7A, red arrow; Mongiu et al., 2007; Korobova and Svitkina, 2008; Yang et al., 2012). Intrapodia actin structures near the T zone (Rochlin et al., 1999; Figure 7A, yellow arrows) also exhibited relatively high Arp2/3 complex densities. Leading edge enhancement of Arp2/3 complex was significantly reduced after treatment with PDBu (100 nM) for either 5 or 10 min (Figure 7, A and C). Furthermore, blebbistatin pretreatment (60 μM, 10 min) had no effect on control Arp2/3 complex distribution (Yang et al., 2012). PDBu treatment in the presence of blebbistatin, however, continued to attenuate Arp2/3 complex leading edge enhancement to about the same degree (Figure 7, B and C). These observations suggest that PKC activation results in delocalization of Arp2/3 complex from the leading edge independent of myosin II activity. Conversely, to some extent PKC inhibition with Go6976 (10 μM, 10 min) increased Arp2/3 complex localization to the leading edge (Figure 7A, bottom, and C) and the presence of Arp2/3 complex–positive intrapodia.
FIGURE 7:
PKC activation disrupted Arp2/3 complex localization at the leading edge independent of myosin II activity. (A, B) Fluorescence labeling of growth cones with phalloidin and Arp3 antibody after normal fixation. (A) Growth cones were treated with DMSO, PDBu (100 nM, 5 and 10 min), or Go6976 (10 μM, 10 min). (B) Growth cones treated with blebbistatin (60 μM, 10 min) or pretreatment with blebbistatin followed by cotreatment with blebbistatin and 100 nM PDBu for 10 min. Red arrow, band of concentrated Arp2/3 complex localization at the leading edge; yellow arrow, intrapodia. Yellow dotted line traces the leading edge. Scale bar, 10 μm. (C) Quantification of Arp2/3 complex enrichment at the leading edge for each condition in A and B. The enrichment was calculated as the ratio of Arp3 fluorescence along the leading edge to a parallel region in the adjacent P domain (schematic). p < 0.0001 with single-factor ANOVA, Asterisk indicates significant difference using Tukey's HSD post hoc analysis. NS, not significant.
We wanted to further probe the mechanism by which PKC may delocalize Arp2/3 complex from the leading edge. Protein interacting with C-kinase 1 (PICK1) can bind to and interfere with Arp2/3 complex function upon association with active PKC (Rocca et al., 2008). If PICK1 recruitment constitutes a pathway for PKC-mediated Arp2/3 complex inhibition, PICK1 would be expected to colocalize with Arp2/3 complex early during PKC activation. To test this hypothesis, we obtained Aplysia-specific PICK1 antibodies and visualized PICK1 localization before and after PKC activation. PICK1 exhibited a punctate labeling pattern in growth cones under control conditions, with slightly higher densities in the C domain, consistent with increased cellular volume relative to the P domain (Supplemental Figure S7). Treatment with PDBu at various time points resulted in progressive changes in actin structure but no obvious recruitment of PICK1 to the leading edge. PICK1 labeling generally decreased over time in the P domain, consistent with observed decreases in actin filament density (Figure 1).
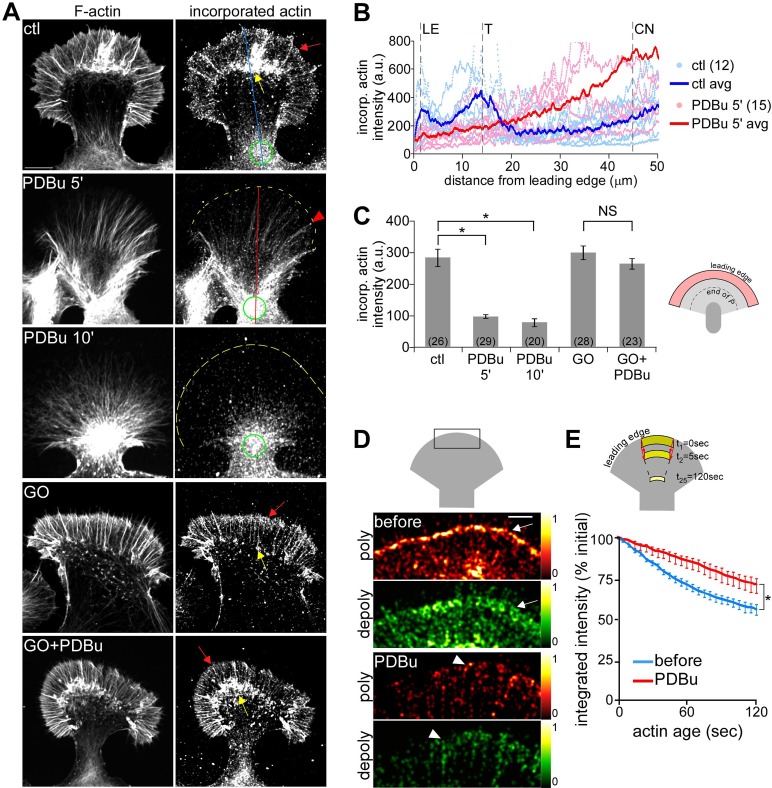
Arp2/3 complex activity generates barbed-end actin nucleation sites in motile cells (Bailly et al., 1999) and growth cones (Yang et al., 2012). To investigate possible PKC effects on nucleation site generation, we assessed the density and distribution of polymerization-competent barbed ends using an in vitro actin incorporation assay (Symons and Mitchison, 1991; Chan et al., 1998). Under control conditions, barbed-end density was high near the leading edge (Figure 8A, red arrow) and in the T zone, where there is abundant intrapodia activity (Figure 8A, yellow arrow; cf. line scans in Figure 8B, dark blue lines; Van Goor et al., 2012; Yang et al., 2012). After 5–10 min in PDBu (100 nM) barbed-end density was markedly attenuated at the leading edge and T zone (Figure 8, A and B, dark red line). Barbed ends were still present at the tips of filopodia, where Arp2/3 complex–independent actin nucleation likely persists (Figure 8A, red arrowhead), and near the contractile node (green circles), where significant actin recycling is likely to occur (Van Goor et al., 2012). On average, barbed-end density in the distal half of P domain decreased by ∼70% with PDBu treatment (Figure 8C). These effects were completely suppressed in the presence of Go6976 (10 μM, 10-min pretreatment), confirming that they indeed depend on PKC activity (Figure 8, A, bottom, and C).
FIGURE 8:
PKC activation reduced barbed-end density and actin polymerization event density along the leading edge. (A) Growth cones were live extracted and labeled with TRITC–phalloidin to show total F-actin (left) and Alexa 488 G-actin, which incorporated at filament barbed ends (right). Growth cones were treated with DMSO (top), PDBu (100 nM, 5 and 10 min), or Go6976 (10 μM, 10 min) or pretreated with Go6976 followed by cotreatment with Go6976 and PDBu for 10 min. Red arrow, band of concentrated barbed ends near the leading edge; yellow arrow, intrapodia; red arrowheads, filopodia; yellow dotted line traces the leading edge; green circles, contractile node. (B) Line scan analysis of barbed-end localization in control and PDBu-treated (100 nM, 5 min) growth cones. Line scans (50 pixels in width) were sampled along the growth axis of growth cones (similar to blue and red lines in the respective panels of A). Scattered dots represent data set from individual growth cones. Solid lines represent the population average. CN, contractile node; LE, leading edge; T, transition zone. (C) Average barbed-end intensities in the distal half of the P domain for different treatment conditions in A. p < 0.0001 with single-factor ANOVA. Asterisk indicates significant difference using Tukey's HSD post hoc analysis. NS, not significant. Numbers in parentheses, growth cones measured. (D) Steady-state map of actin assembly (red) and disassembly (green) events near the leading edge of a growth cone (schematic) before and after PDBu (100 nM, 10 min) treatment. Maps were generated from FSM images of a growth cone injected with Alexa 568 G-actin. Colors indicate relative assembly or disassembly event densities (see color bars). Red and green arrows, respectively, mark the band of strong assembly near the leading edge and the region of strong disassembly proximally juxtaposed to the assembly band. Arrowheads mark filopodia tips. (E) Population average of changes in integrated actin network fluorescence intensity within the flow-displaced regions before and after PDBu (100 nM, 10–30 min) measured by the ROI-based turnover algorithm (schematic). n = 9 growth cones, 3–5 ROIs per growth cone. *p < 0.01, two-tailed paired t test. Scale bars, 10 μm (A), 5 μm (D).
The decreased actin nucleation site density at the leading edge after PKC activation is consistent with the reduction in actin veil network density and delocalization of Arp2/3 complex described in Figures 1 and 7, respectively. Because the Arp2/3 complex is important for maintaining steady-state actin polymerization rates at the leading edge of growth cones (Yang et al., 2012), we investigated PKC effects on actin dynamics using FSM. Polymerization and depolymerization in the distal P domain (Figure 8D, schematic) were calculated from single-speckle tracking (Ponti et al., 2004) over 2-min sampling periods. Under control conditions, strong polymerization, indicated by a high density of speckle “birth” events, occurred in a narrow band along the leading edge (Figure 8D, top, arrow). Treatment with PDBu (100 nM, 10 min) markedly decreased the number of speckle births recorded during the same sampling interval and eliminated the band of strong leading edge polymerization observed under control conditions (Figure 8D, bottom). A punctate pattern of actin polymerization remained, some of which appeared to be associated with linear filopodial bundles (Figure 8D, arrowheads).
Of interest, PKC activation also affected actin depolymerization. Under control conditions, depolymerization, estimated from the integrated density of speckle “death” events, was distributed throughout the P domain with a strong band juxtaposed to the leading edge polymerization zone (Figure 8D, top, arrow). After PDBu treatment, the overall depolymerization event density was reduced in the P domain, and the band of strong depolymerization observed near the leading edge under control conditions was absent. Depolymerization appeared to be associated with residual filopodial actin bundles, which persist during PDBu treatment (Figure 8D, bottom, arrowhead; cf. Figure 1B). Similar results were obtained using a recently developed algorithm (Van Goor et al., 2012) that estimates net actin turnover rates by tracking integrated actin speckle intensity within an ROI as it moves progressively across the P domain with retrograde flow (Figure 8E, schematic). Over the 2-min tracking period, the integrated intensity in the control ROI decreased to ∼50% of its initial value as it moved through the P domain under control conditions (Figure 8F, blue line). After PDBu treatment (100 nM, 10–30 min), the integrated intensity within the same ROI decreased at a significantly slower rate, retaining ∼70% its initial value at 2 min (Figure 8F, red line), indicating a slower net rate of actin turnover. The observed reduction in actin polymerization frequency and network turnover after PKC activation is consistent with a P domain sparsely populated (Figure 1, B and C) with more-stable actin filaments relative to control conditions. In summary, the remaining actin filaments undergoing retrograde flow after PKC activation tend to move faster (Figure 2) and be more stable (Figure 8) than those comprising flow under control conditions.
DISCUSSION
We investigated effects of phorbol ester activation of PKC on the actin cytoskeleton of neuronal growth cones and provided evidence that PKC simultaneously modulates actin polymerization and myosin II contractility to affect growth cone motility. Figure 9 summarizes our results.
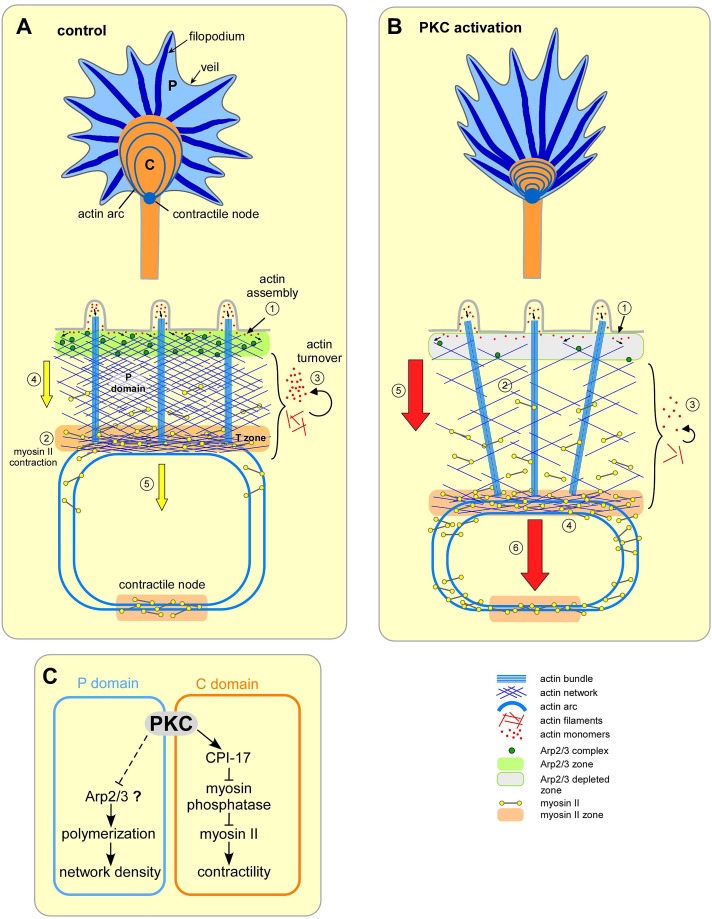
FIGURE 9:
Summary of key results. (A) Control conditions. Top, relevant structures in the P and C growth cone domains. Bottom: 1) Arp2/3 complex–dependent actin veil polymerization at the leading edge. 2) Myosin II–dependent network contraction at the T zone and contractile node. 3) Network recycling throughout the P domain. 4) P-domain retrograde flow. 5) T-zone flow. (B) PKC activation. Top, actin veil retracts and C-domain compresses. Bottom, 1) Delocalization of Arp2/3 complex and reduced assembly sites at the leading edge. 2) Decreased actin veil network density. 3) Reduced network turnover in the P domain. 4) Activation and enhanced localization of myosin II on actin arcs and at the contractile node. 5) Increased P-domain retrograde flow. 6) Increased central actin flow. (C) Proposed signaling pathways for dual cytoplasmic domain-specific effects of PKC activation.
Actin veils of neuronal growth cones contains Arp2/3 complex–dependent branched actin networks resembling the lamellipodium of motile cells (Ponti et al., 2004; Korobova and Svitkina, 2008; Yang et al., 2012). In addition, growth cones contain long linear actin bundles comprising filopodia and display myosin motor–assisted actin array treadmilling (Lin et al., 1996; Medeiros et al., 2006; Craig et al., 2012; Figures 1 and 4). The steady-state balance between actin polymerization, myosin II driven contractility, and actin filament turnover determines actin network behavior and the rate of classic P-domain retrograde flow (Medeiros et al., 2006; Van Goor et al., 2012; Yang et al., 2012). Polarized actin bundles comprising filopodia appear to be preferentially recycled in the T zone (Medeiros et al., 2006). Less-polarized actin veil networks undergo local network contraction in the T zone, forming actin arcs, which continue to exhibit myosin II–dependent movement rearward through the C domain and into the contractile node (Figure 9A, steps 1–5; Zhang et al., 2003; Medeiros et al., 2006).
Decreased actin network density is a hallmark of PKC activation in many cell types, but the underlying mechanisms are not well characterized. Some of the earliest studies on phorbol esters noted actin filament loss and general network disorganization in epithelial (Schliwa et al., 1984) and smooth muscle cells (Dugina et al., 1987), effects later identified as being mediated by PKC (Hai et al., 2002). Similar changes were observed with PKC activation in trigeminal ganglion cell (Ozsarac et al., 2003) and DRG growth cones (Bonsall and Rehder, 1999) and in hippocampal dendritic spines (Calabrese and Halpain, 2005). In agreement with previous studies, we observed significant decreases in P-domain actin density with PKC activation (Figure 1, Supplemental Figures S2 and S4B, and Supplemental Movies S1 and S2). The observed decreases in peripheral actin veil network density (Figure 1) occurred by a mechanism that involved 1) delocalization of Arp2/3 complex from the leading edge (Figure 7) and 2) rapid decrease in the density of barbed-end actin nucleation sites in the same region (Figure 8, A and B). The latter was corroborated by quantification of actin polymer dynamics (Figure 8, D and E). Conversely, treatment with the PKC inhibitor GO6976 alone increased actin veil network density, indicating that basal PKC activity levels affect peripheral actin network structures under control conditions (Figures 1 and 7).
The decreases in actin veil density and increases in P-domain retrograde flow after PKC activation (Figures 1 and 2, Supplemental Movies S1 and S2, and Supplementary Figure S4) were similar to what we previously observed after acute Arp2/3 complex inhibition (Yang et al., 2012). Moreover, when myosin II activity was inhibited by blebbistatin pretreatment, retrograde flow slowed in response to PKC activation (Figure 4, A and B)—also as observed after acute Arp2/3 complex inhibition in low–myosin II activity backgrounds (Yang et al., 2012). Attenuation of retrograde flow would be expected if PKC activation decreases leading edge assembly–dependent pushing forces in a treadmilling actin filament array system (Mogilner and Oster, 2003; Medeiros et al., 2006).
Are PKC effects on the Arp2/3 complex direct or indirect? Although phosphorylation is an important regulatory mechanism for Arp2/3 complex function (Vadlamudi et al., 2004; LeClaire et al., 2008) and activated PKC translocates to the membrane (Sossin et al., 1993; Liu and Heckman, 1998) close to where the Arp2/3 complex is localized, no PKC phosphorylation sites have been reported for any of the Arp2/3 complex subunits. We therefore suggest that action of PKC on Arp2/3 complex function involves other molecular players. One candidate is PICK1, a scaffold protein that associates with activated PKC that can bind to and block actin nucleation by Arp2/3 complex (Rocca et al., 2008). PICK1 knockdown increased actin content and induced aberrant branching in hippocampal neurons (Rocca et al., 2008), consistent with elevated Arp2/3 complex activity. Recently Aplysia PICK1 was cloned and shown to affect sensory neuron habituation through interaction with PKC (Wan et al., 2012). Our results (Supplemental Figure S7), however, cast doubt on PICK1 involvement in the observed effects on actin assembly and Arp2/3 complex localization. Further studies will be needed to address the mechanism of PKC effects on actin assembly.
Evidence suggests that actin arcs in growth cones are functionally similar to actomyosin II bundles involved in cytokinesis and cellular wound healing, which increase their rates of contractile movement with PKC activation (Driedger and Blumberg, 1977; Bement and Capco, 1991). In neuronal cells, PKC activation can result in neurite retraction (Cressman and Shea, 1995; Lanuza et al., 2002; Conrad et al., 2007). Consistent with the foregoing, PKC-dependent increases in retrograde flow rates were highest in the T zone, where contractile actin arcs normally form (Figures 2D and 3). PKC activation also resulted in more-stable actin filament structures with longer turnover times (Figure 8, D and E). The presence of generally longer, less branched, and more-stable actin filaments (Figure 1C) may facilitate more efficient contractile force production, leading to the increased rates of retrograde flow observed in the presence of myosin II activity.
PKC has also been suggested to up-regulate myosin II activity by increasing Rho and Rho kinase activities (Brandt et al., 2002; Lai et al., 2003; Domeniconi et al., 2005; Pan et al., 2005; Conrad et al., 2007). In growth cones, actin arc contractility is regulated by Rho kinase (Zhang et al., 2003); we found, however, that PKC acts by an independent mechanism, since essentially the same effects on actin dynamics were present in a background of Rho kinase inhibition (Supplemental Figure S6). A more compelling mechanism for PKC potentiation of myosin II activity here is activation of the myosin phosphatase inhibitor CPI-17 (Kitazawa et al., 1999; Eto et al., 2000; Ohki et al., 2001; Watanabe et al., 2001). This hypothesis is supported by our observation of PKC-dependent increases in phosphorylated CPI-17 in the same regions where myosin II activity levels increased (Figures 5 and 6).
Note that PKC has also been reported to down-regulate myosin II activity via phosphorylation of multiple serine and threonine residues at the N-terminus of heavy meromyosin (Nishikawa et al., 1984). This scenario is unlikely here, since these amino acids are not conserved in the Aplysia myosin II heavy chain (Medeiros et al., 2006). Parenthetically, inhibitory phosphorylation by PKC does not appear to have a significant effect on vertebrate myosin II activity in vivo, and so the role of PKC in down-regulation of myosin II function is not clear (Turbedsky et al., 1997; Beach et al., 2011). In summary, we suggest that lower peripheral network density facilitates higher rates of myosin II–dependent network contraction in the P domain, and CPI-17 activation promotes higher levels of myosin II contractility in the T zone and C domain. The net result of these actions is the acceleration of peripheral retrograde flow and rearward compression of the C domain observed after PKC activation.
PKC and Rho activity have both been implicated in signaling processes that inhibit neurite outgrowth (Tang, 2003; Teng and Tang, 2006). Conversely, inhibition of either Rho kinase (Fournier et al., 2003) or PKC (Sivasankaran et al., 2004) has been reported to promote neurite extension on normally inhibitory myelin-associated glycoprotein substrates and improve axon regeneration in animal models. In the present study, we describe synergistic domain-specific mechanisms by which PKC activity regulates growth cone motility: 1) inhibition of actin nucleation and consequent attenuation of actin veil network density in the P domain and 2) increasing myosin II contractility in the C domain. These effects were independent of Rho kinase. Our findings suggest that Rho and PKC signal via functionally independent mechanisms to control growth cone motility. This parallel signaling structure might be exploited in combinatorial intervention schemes that target Rho and PKC simultaneously to inhibit axon degeneration and/or promote regrowth. In addition, this two-tiered effect of PKC activation could prompt development of novel therapeutic approaches to overcome repulsive responses encountered after CNS injury and/or lend insight into mechanisms of growth cone guidance.
MATERIALS AND METHODS
Cell culture and chemicals
Primary culture of Aplysia bag cell neurons was as previously described (Forscher and Smith, 1988), with the following modifications. Coverslips were treated with 20 μg/ml poly-l-lysine for 15 min and washed with water before plating. A 1:10,000 dilution of fetal bovine serum was added to the culture medium 2 h after plating the neurons. PDBu, Go6976, blebbistatin, Y27632, Taxol, ML-7, and calyculin A were purchased from Calbiochem (La Jolla, CA). Actin monomers purified from rabbit muscle and tagged with Alexa 568 or Alexa 488, Alexa 594–phalloidin, conjugated secondary antibodies, including goat anti-rabbit immunoglobulin G (IgG) and goat anti-mouse IgG, calcium green-1 (CG-1), and conjugated dextrans, molecular weight 10 kDa, were from Invitrogen (Carlsbad, CA). Anti-Arp3 and anti-pThr38 of CPI-17 rabbit polyclonal antibodies were from Upstate (Millipore, Billerica, MA). Anti-pSer19 of myosin II regulatory light chain mouse monoclonal antibody was from Cell Signaling Technology (Beverly, MA). Anti–CPI-17 rabbit polyclonal antibody was from Signalway (College Park, MD). Anti-tubulin mouse monoclonal antibody was from Sigma-Aldrich. Aplysia PICK1 antibodies were kindly provided by Thomas Abrams (Department of Pharmacology, University of Maryland School of Medicine, Baltimore, MD; Wan et al., 2012). Dimethyl sulfoxide (DMSO) and other chemicals were from Sigma-Aldrich unless otherwise stated.
Solutions
Artificial seawater (ASW) contained 400 mM NaCl, 10 mM KCl, 15 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 10 mM CaCl2, and 55 mM MgCl2 at pH 7.8. ASW was supplemented with 3 mg/ml bovine serum albumin (BSA), 0.5 mM vitamin E, and 1 mg/ml carnosine before experiments. Ca2+ injection buffer consisted of 100 mM potassium aspartate and 10 mM HEPES at pH 7.4. Lysis buffer for homogenizing Aplysia CNS tissue contained 50 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM ethylene glycol tetraacetic acid (EGTA), 5 mM dithiothreitol with 0.01 mg/ml aprotinin, 0.01 mg/ml leupeptin, 5 μg/ml pepstatin, 0.4 μg/ml Pefabloc, and 1 mM Na3VO4. Tris-buffered saline (TBS) contained 25 mM Tris base, 137 mM NaCl, and 2 mM KCl. Live-cell extraction buffer (LE) contained 100 mM 1,4-piperazinediethanesulfonic acid (PIPES), pH 6.9, 10 mM KCl, 100 mM NaCl, 5 mM EGTA, 5 mM MgCl2, 4% polyethylene glycol, average molecular weight 35,000, and 20% sucrose at ∼1000 mOsm/l and was supplemented with 1% Triton X-100, 10 μM phalloidin, 10 μM Taxol, and 10 mg/ml 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid immediately before use. Washing buffer for live-cell extraction (WLE) contained 80 mM PIPES, pH 6.9, 5 mM EDTA, and 5 mM MgCl2. Block solution for antibody labeling was 5% BSA in phosphate-buffered saline (PBS) containing 0.1% Triton X-100 (PBS-T). MOWIOL (Calbiochem) for cell mounting in immunocytochemistry was prepared following manufacturer's protocol with 10% MOWIOL, 25% glycerol, 0.2 M Tris, pH 8.5, and 20 mM n-propylgalate.
Aplysia neuronal myosin II RLC cloning and antibody production
The full-length primary sequence of Aplysia neuronal myosin II RLC (167 amino acids; Supplemental Figure S6B) was obtained with degenerate PCR and rapid amplification of cDNA ends. The full-length gene was cloned into a bacterial expression vector (Novagen, Gibbstown, NJ) and recombinant proteins expressed with an N-terminal hexahistidine affinity tag for purification. Purified RLC was dialyzed into PBS overnight at 4°C, concentrated to 1 mg/ml using a centrifugal filter device (Millipore), and used as antigen for polyclonal antibody production in rabbit host (Proteintech Group, Chicago, IL).
Western blotting
CNS tissue from each Adult Aplysia was homogenized in 1 ml of ice-cold lysis buffer with a rotor stator, and SDS was added to a final concentration of 1%. Lysates were cleared by centrifugation, boiled for 3 min, resolved on 12% SDS–PAGE, and transferred to nitrocellulose membranes (GE Healthcare, Piscataway, NJ) by semidry transfer (Bio-Rad TransBlot SD; Bio-Rad, Hercules, CA). After probing with primary and secondary antibodies, the membrane was developed by electrochemiluminescence (Thermo Scientific, Waltham, MA) and exposed to x-ray films (GE Healthcare).
Microinjection
Neurons were microinjected with Alexa 568 actin monomers (needle concentration, 0.4 mg/ml) and Alexa 594–phalloidin (needle concentration, 20 μM; Lin et al., 1996; Schaefer et al., 2002). Alexa 488 or Alexa 647 dextran (10 kDa; needle concentration, 2 mg/ml) was often coinjected as a volume marker. Reagent solution injections were typically ∼10% of cell volume. After microinjection, cells were incubated in culture medium 1 h before imaging.
Calcium imaging with actin dynamics
Neurons were injected with Ca2+ dye CG-1, Alexa 647 dextran as a volume tracer, and Alexa 594–phalloidin (needle concentration 11.5 mg/ml, 0.9 mg/ml, and 38 μM, respectively). DIC, Ca2+ level, volume tracer, and actin FSM images were recorded in tandem at 10-s sampling intervals using a confocal microscope. Paired images with comparable intensities of CG-1 and Alexa 647 were subjected to Gaussian convolution to reduce noise levels, and a binary mask was used to eliminate noise amplification outside the cell. The ratio images (CG-1/volume) were created by dividing background-corrected intensity values of CG-1 fluorescence by volume fluorescence and converted into time-lapse montages for data analysis as reported previously (Zhang and Forscher, 2009). Average pixel intensity values were obtained from the entire growth cone. The Ca2+ changes over time were expressed as ΔF/Fo, where ΔF = Ft − Fo and Fo is the average Ca2+ level sampled during the 3- to 5-min baseline period before drug treatment. ΔF/Fo > 10% is considered significant.
Immunocytochemistry
Cells were fixed with 4% formaldehyde and 400 mM sucrose in ASW or low-ionic-strength ASW (100 mM NaCl, 10 mM KCl, 15 mM HEPES, 5 mM CaCl2, 5 mM MgCl2, and 628 mM betaine at pH 7.8) and permeabilized with 1% Triton X-100 (Forscher and Smith, 1988). Actin filaments were labeled with 0.66 μM Alexa 594–phalloidin stock in PBS-T. For antibody labeling, cells were blocked for 20 min with the blocking solution, incubated with 5 μg/ml affinity-purified primary antibody or 1:1000 of unpurified antisera for 30 min, washed three times with blocking solution, and incubated for 15 min with 10 μg/ml secondary antibody. To label actin monomers, 5 μg/ml Oregon green DNaseI was used after blocking. Cells were then washed three times in PBS-T and mounted in MOWIOL.
Live-cell extractions were performed as previously described (Schaefer et al., 2002; Medeiros et al., 2006), with modifications. Extraction with LE for 3–5 min was followed by washing with WLE for 2 min. Free barbed ends were labeled by incubating cells with 350 nM Alexa 488 actin monomers in WLE for 1 min and washed again (Symons and Mitchison, 1991). Cells were fixed for 20 min in 4% formaldehyde in WLE for fluorescence or 6.5% glutaraldehyde in WLE for electron microscopy. Extracted cells were labeled with antibodies as described.
Platinum/palladium replica electron microscopy
Cells were live extracted and fixed as described, followed by treatment with 6.5% glutaraldehyde in water, 0.2% tannic acid, and 0.2% uranyl acetate for 20 min each with water washes in between (Svitkina et al., 1995). Samples were dehydrated with a graded series of ethanol concentrations before critical point drying. Specimens were rotary shadowed with platinum/palladium at a 45° angle to a thickness of 2 nm, followed by carbon coating. Replicas were mounted on carbon/Formvar-coated EM grids and observed by transmission EM at 80 kV.
To quantify actin veil network density, electron micrographs at 20,000× magnification were used to assess 1 × 1–μm2 ROIs near the growth cone leading edge. ROIs were subjected to an intensity threshold process until all the negative spaces bounded by actin filaments were selected, thereby defining the properties of the actin mesh. Thus mesh size is related to the amount of free space bounded by actin filaments and is a measure of the network density. Mesh images were then quantified using the “analyze particle” function in ImageJ (National Institutes of Health, Bethesda, MD). For each condition, 10–15 nonoverlapping regions from three to eight growth cones were analyzed. Regions containing filopodial bundles or intrapodia were excluded.
Confocal microscopy
Confocal fluorescence images were acquired using an Andor Revolution XD spinning disk confocal system (Andor, Belfast, United Kingdom) with a CSU-X1 confocal head (Yokogawa, Tokyo, Japan) mounted on a Nikon TE 2000E inverted microscope with Perfect Focus (Nikon, Melville, NY), using an Andor iXonEM+888 electron-multiplying charge-coupled device camera. Transillumination was provided by a halogen lamp and controlled by a SmartShutter (Sutter Instrument, Novato, CA). Confocal excitation was provided by an Andor Laser Combiner with laser lines at 488, 561, and 647 nm. Emission wavelength was controlled using a Sutter LB10W-2800 filter wheel outfitted with bandpass filters from Chroma Technology (Brattleboro, VT). A Nikon CFI Plan Apo 100×/1.4 numerical aperture objective was used. Image acquisition and all other peripherals were controlled by Micro-Manager (http://valelab.ucsf.edu/∼MM/MMwiki/index.php/Micro-Manager) with a Matlab (MathWorks, Natick, MA) interface.
Epifluorescence microscopy
Epifluorescence images were acquired using a Nikon TE300 inverted microscope equipped with a CoolSNAP HQ camera (Photometrics, Tucson, AZ) and a variable-zoom lens (0.9–2.25×). Epifluorescence illumination was provided by a 100-W mercury arc lamp; two programmable filter wheels with shutters (Ludl Electronic Products, Hawthorne, NY) were outfitted with bandpass filters (Chroma) to select excitation and emission wavelengths. DIC/brightfield illumination was provided by a halogen lamp and controlled by a Uniblitz Shutter (Vincent Associates, Rochester, NY). Image acquisition and peripheral control were provided by MetaMorph software (Universal Imaging, West Chester, PA).
Quantification of actin dynamics
Fluorescent speckle microscopy images of cells injected with Alexa 488 actin monomers incorporated into filaments or low levels of Alexa 594–phalloidin, which specifically binds actin filaments, were acquired using 300- to 700-ms integration. Kymography (Zhang et al., 2003) and automated speckle tracking with the fsmCenter suite of custom Matlab algorithms produced by the Danuser lab were used to determine rates of actin filament movement. Kymographs were generated along the direction of actin network translocation in the P domain, arcs, and the contractile node region of growth cones from actin FMS images using the ImageJ multiple kymograph plug-in. Speckle movement over time was traced using the ImageJ line tool, and the slope of the line was converted to flow rates based on imaging interval and pixel scale. In fsmCenter, an adaptive multiframe correlation algorithm (Ji and Danuser, 2005) was run on every five frames to determine the average flow over five frames. This information was used to initialize a single-particle tracking algorithm (Ponti et al., 2003, 2004; Thomann et al., 2003), which was run on every frame in the movie. Results of the single-particle tracking were used to generate time-averaged actin assembly–disassembly kinetic maps (Ponti et al., 2003, 2004, 2005) and analyze net population turnover with an ROI-based algorithm developed by D. Van Goor (Van Goor et al., 2012).
The ROI-based turnover algorithm is based on conservation of mass principles. Briefly, spatially averaged single-particle tracking vectors are used to track a user-defined ROI as it is displaced by retrograde flow from the leading edge to the peripheral–central interface. The flow-displaced ROI mimics a closed system, assuming that all assembled actin initially identified at the leading edge moves with retrograde flow. Changes in the integrated intensity within the ROI reflect the shifting balance between assembly and disassembly over time. Three to five regions in each growth cone were tracked for 2 min under both control and drug-treated conditions. Intensity changes were normalized to the first frame.
Image processing
Fluorescence images were subjected to background subtraction before quantitative intensity analysis. The only exception was automated speckle tracking, which was performed on raw image data. For ratio calculations, background-subtracted images were divided pixel by pixel and multiplied by a constant to prevent the loss of numerical precision, then subjected to Gaussian blur with a radius of 2 pixels to obtain a smooth contour. For display only, fluorescence images were convolved with a Gaussian kernel, processed with an unsharp mask, and scaled according to a linear lookup table. For display of barbed-end and ratio images, areas outside of the growth cones were cleared with a mask. EM images were inverted and processed with pseudo flat field to eliminate artifacts of uneven illumination, followed by an unsharp mask. Image processing was performed in ImageJ. All images of the same fluorescence label from each experiment were acquired and scaled using the same parameters (excitation intensity, exposure time, gain) and a linear lookup table.
Line scan analysis
Line scans were used to analyze the spatial intensity distribution of fluorescent probes. A 50-pixel-wide line was drawn from the leading edge to a distance specified in the figures or five times the P-domain width along the presumed growth axis of growth cones. The average intensity was measured with the plot profile function in ImageJ and the data exported to Excel (Microsoft, Redmond, WA). Intensity was plotted against either actual distance from the leading edge for single–growth cone line scans or distance normalized to growth cone size for population line scans. When distance is normalized, the beginning (left end) of the line scan is marked as the leading edge, and the 1/5 (given total length = 5× P-domain width) position is marked as the T zone.
Quantification of Arp2/3 complex enrichment
To quantify Arp2/3 complex enrichment along the leading edge, the peripheral domain was divided into eight concentric slices parallel to the leading edge, and the ratio of the average Arp3 intensity in slice 1 versus that in slice 4 was calculated as the enrichment factor, as described previously (Yang et al., 2012).
Statistical analysis
Statistical analysis with pair or unpaired t test, analysis of variance (ANOVA), and post hoc analysis was performed in Excel, with significance established at p < 0.01. Data are presented as mean ± SE of measurement, unless otherwise specified.
Supplementary Material
Acknowledgments
We thank Gaudenz Danuser (Harvard Medical School, Boston, MA) for providing the actin flow tracking software and Barry Piekos for assisting in using EM facilities. We thank Thomas Abrams for the gift of Aplysia PICK1 antibodies and Wayne Sossin and Forscher lab members for critical comments and insightful discussion of this work. This work was supported by National Institutes of Health Grants RO1-NS28695 and RO1-NS051786 to P.F., Yale Medical Scientists Training Program TG-T32GM07205, and the Nikon Partners-in-Research Program.
Abbreviations used:
- C-domain
central domain
- CPI-17
C-kinase–potentiated protein phosphatase inhibitor
- cPKC
conventional or classic protein kinase C
- EM
electron microscopy
- MLCK
myosin light chain kinase
- nPKC
novel protein kinase C
- P-domain
peripheral domain
- PDBu
phorbol 12,13-dibutyrate
- PICK1
protein interacting with C-kinase 1
- PKC
protein kinase C
- RLC
regulatory light chain
- ROI
region of interest
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0289) on August 21, 2013.
*Present address: Jefferson College of Health Sciences, 101 Elm Ave., SE, Roanoke, VA 24013.
Q.Y. designed and performed the majority of experiments, analyzed data, and wrote the article; X.F.Z. performed experiments for Figure 4E, Ca2+ imaging studies in Supplemental Figure S3, and PICK1 staining in Supplemental Figure S5 and edited the manuscript; D.V.G. performed experiments for Figure 1, B and C, contributed to Figures 2, C–E, and 4B and Supplemental Figure S2D, helped with the actin incorporation assay, and wrote the ROI-based actin turnover algorithm; A.P.D. did pilot studies for the project and contributed to Supplemental Figures S1A and 2, C–E; C.H. contributed to Supplemental Figure S5E; N.M. cloned Aplysia myosin II regulatory light chain and characterized RLC antibody in Figure 5A and Supplemental Figure S6; P.F. contributed to experimental design and edited the manuscript.
The authors have no commercial affiliations that would pose a conflict of interest with respect to this research.
REFERENCES
- Allingham JS, Smith R, Rayment I. The structural basis of blebbistatin inhibition and specificity for myosin II. Nat Struct Mol Biol. 2005;12:378–379. doi: 10.1038/nsmb908. [DOI] [PubMed] [Google Scholar]
- Bailly M, Macaluso F, Cammer M, Chan A, Segall JE, Condeelis JS. Relationship between Arp2/3 complex and the barbed ends of actin filaments at the leading edge of carcinoma cells after epidermal growth factor stimulation. J Cell Biol. 1999;145:331–345. doi: 10.1083/jcb.145.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach JR, Licate LS, Crish JF, Egelhoff TT. Analysis of the role of Ser1/Ser2/Thr9 phosphorylation on myosin II assembly and function in live cells. BMC Cell Biol. 2011;12:52. doi: 10.1186/1471-2121-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement WM, Capco DG. Analysis of inducible contractile rings suggests a role for protein kinase C in embryonic cytokinesis and wound healing. Cell Motil Cytoskeleton. 1991;20:145–157. doi: 10.1002/cm.970200207. [DOI] [PubMed] [Google Scholar]
- Bonsall J, Rehder V. Regulation of chick dorsal root ganglion growth cone filopodia by protein kinase C. Brain Res. 1999;839:120–132. doi: 10.1016/s0006-8993(99)01725-4. [DOI] [PubMed] [Google Scholar]
- Brandt D, Gimona M, Hillmann M, Haller H, Mischak H. Protein kinase C induces actin reorganization via a Src- and Rho-dependent pathway. J Biol Chem. 2002;277:20903–20910. doi: 10.1074/jbc.M200946200. [DOI] [PubMed] [Google Scholar]
- Bridgman PC, Dailey ME. The organization of myosin and actin in rapid frozen nerve growth cones. J Cell Biol. 1989;108:95–109. doi: 10.1083/jcb.108.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette DT, Ji L, Schaefer AW, Medeiros NA, Danuser G, Forscher P. Myosin II activity facilitates microtubule bundling in the neuronal growth cone neck. Dev Cell. 2008;15:163–169. doi: 10.1016/j.devcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese B, Halpain S. Essential role for the PKC target MARCKS in maintaining dendritic spine morphology. Neuron. 2005;48:77–90. doi: 10.1016/j.neuron.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Chan AY, Raft S, Bailly M, Wyckoff JB, Segall JE, Condeelis JS. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J Cell Sci. 1998;111:199–211. doi: 10.1242/jcs.111.2.199. [DOI] [PubMed] [Google Scholar]
- Conrad S, Genth H, Hofmann F, Just I, Skutella T. Neogenin-RGMa signaling at the growth cone is bone morphogenetic protein-independent and involves RhoA, ROCK, and PKC. J Biol Chem. 2007;282:16423–16433. doi: 10.1074/jbc.M610901200. [DOI] [PubMed] [Google Scholar]
- Craig EM, Van Goor D, Forscher P, Mogilner A. Membrane tension, myosin force, and actin turnover maintain actin treadmill in the nerve growth cone. Biophys J. 2012;102:1503–1513. doi: 10.1016/j.bpj.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressman CM, Shea TB. Hyperphosphorylation of Tau and filopodial retraction following microinjection of protein kinase C catalytic subunits. J Neurosci Res. 1995;42:648–656. doi: 10.1002/jnr.490420507. [DOI] [PubMed] [Google Scholar]
- Domeniconi M, Zampieri N, Spencer T, Hilaire M, Mellado W, Chao MV, Filbin MT. MAG induces regulated intramembrane proteolysis of the p75 neurotrophin receptor to inhibit neurite outgrowth. Neuron. 2005;46:849–855. doi: 10.1016/j.neuron.2005.05.029. [DOI] [PubMed] [Google Scholar]
- Downey GP, Chan CK, Lea P, Takai A, Grinstein S. Phorbol ester-induced actin assembly in neutrophils: role of protein kinase C. J Cell Biol. 1992;116:695–706. doi: 10.1083/jcb.116.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driedger PE, Blumberg PM. The effect of phorbol diesters on chicken embryo fibroblasts. Cancer Res. 1977;37:3257–3265. [PubMed] [Google Scholar]
- Dugina VB, Svitkina TM, Vasiliev JM, Gelfand IM. Special type of morphological reorganization induced by phorbol ester: reversible partition of cell into motile and stable domains. Proc Natl Acad Sci USA. 1987;84:4122–4125. doi: 10.1073/pnas.84.12.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto M, Bock R, Brautigan DL, Linden DJ. Cerebellar long-term synaptic depression requires PKC-mediated activation of CPI-17, a myosin/moesin phosphatase inhibitor. Neuron. 2002;36:1145–1158. doi: 10.1016/s0896-6273(02)01107-8. [DOI] [PubMed] [Google Scholar]
- Eto M, Wong L, Yazawa M, Brautigan DL. Inhibition of myosin/moesin phosphatase by expression of the phosphoinhibitor protein CPI-17 alters microfilament organization and retards cell spreading. Cell Motil Cytoskeleton. 2000;46:222–234. doi: 10.1002/1097-0169(200007)46:3<222::AID-CM6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Forscher P, Smith SJ. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J Cell Biol. 1988;107:1505–1516. doi: 10.1083/jcb.107.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier AE, Takizawa BT, Strittmatter SM. Rho kinase inhibition enhances axonal regeneration in the injured CNS. J Neurosci. 2003;23:1416–1423. doi: 10.1523/JNEUROSCI.23-04-01416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata Y, Amano M, Kaibuchi K. Rho-Rho-kinase pathway in smooth muscle contraction and cytoskeletal reorganization of non-muscle cells. Trends Pharmacol Sci. 2001;22:32–39. doi: 10.1016/s0165-6147(00)01596-0. [DOI] [PubMed] [Google Scholar]
- Gong JP, Liu QR, Zhang PW, Wang Y, Uhl GR. Mouse brain localization of the protein kinase C-enhanced phosphatase 1 inhibitor KEPI (kinase C-enhanced PP1 inhibitor) Neuroscience. 2005;132:713–727. doi: 10.1016/j.neuroscience.2004.11.052. [DOI] [PubMed] [Google Scholar]
- Hai CM, Hahne P, Harrington EO, Gimona M. Conventional protein kinase C mediates phorbol-dibutyrate-induced cytoskeletal remodeling in a7r5 smooth muscle cells. Exp Cell Res. 2002;280:64–74. doi: 10.1006/excr.2002.5592. [DOI] [PubMed] [Google Scholar]
- Hains AB, Vu MA, Maciejewski PK, van Dyck CH, Gottron M, Arnsten AF. Inhibition of protein kinase C signaling protects prefrontal cortex dendritic spines and cognition from the effects of chronic stress. Proc Natl Acad Sci USA. 2009;106:17957–17962. doi: 10.1073/pnas.0908563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Fujitani M, Hata K, Tohyama M, Yamagishi S, Yamashita T. Promotion of axon regeneration by myelin-associated glycoprotein and Nogo through divergent signals downstream of Gi/G. J Neurosci. 2004;24:6826–6832. doi: 10.1523/JNEUROSCI.1856-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlitze S, Zhong H, Scheuer T, Catterall WA. Allosteric modulation of Ca2+ channels by G proteins, voltage-dependent facilitation, protein kinase C, and Ca(v)beta subunits. Proc Natl Acad Sci USA. 2001;98:4699–4704. doi: 10.1073/pnas.051628998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H, et al. Calyculin A and okadaic acid: inhibitors of protein phosphatase activity. Biochem Biophys Res Commun. 1989;159:871–877. doi: 10.1016/0006-291x(89)92189-x. [DOI] [PubMed] [Google Scholar]
- Ji L, Danuser G. Tracking quasi-stationary flow of weak fluorescent signals by adaptive multi-frame correlation. J Microsc. 2005;220:150–167. doi: 10.1111/j.1365-2818.2005.01522.x. [DOI] [PubMed] [Google Scholar]
- Kabir N, Schaefer AW, Nakhost A, Sossin WS, Forscher P. Protein kinase C activation promotes microtubule advance in neuronal growth cones by increasing average microtubule growth lifetimes. J Cell Biol. 2001;152:1033–1044. doi: 10.1083/jcb.152.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa T, Takizawa N, Ikebe M, Eto M. Reconstitution of protein kinase C-induced contractile Ca2+ sensitization in Triton X-100-demembranated rabbit arterial smooth muscle. J Physiol. 1999;520:139–152. doi: 10.1111/j.1469-7793.1999.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox RJ, Quattrocki EA, Connor JA, Kaczmarek LK. Recruitment of Ca2+ channels by protein kinase C during rapid formation of putative neuropeptide release sites in isolated Aplysia neurons. Neuron. 1992;8:883–889. doi: 10.1016/0896-6273(92)90202-o. [DOI] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol Biol Cell. 2008;19:1561–1574. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger KE, Sossin WS, Sacktor TC, Bergold PJ, Beushausen S, Schwartz JH. Cloning and characterization of Ca(2+)-dependent and Ca(2+)-independent PKCs expressed in Aplysia sensory cells. J Neurosci. 1991;11:2303–2313. doi: 10.1523/JNEUROSCI.11-08-02303.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JM, Hsieh CL, Chang ZF. Caspase activation during phorbol ester-induced apoptosis requires ROCK-dependent myosin-mediated contraction. J Cell Sci. 2003;116:3491–3501. doi: 10.1242/jcs.00660. [DOI] [PubMed] [Google Scholar]
- Lanuza MA, Garcia N, Santafe M, Gonzalez CM, Alonso I, Nelson PG, Tomas J. Pre- and postsynaptic maturation of the neuromuscular junction during neonatal synapse elimination depends on protein kinase C. J Neurosci Res. 2002;67:607–617. doi: 10.1002/jnr.10122. [DOI] [PubMed] [Google Scholar]
- Larsson C. Protein kinase C and the regulation of the actin cytoskeleton. Cell Signal. 2006;18:276–284. doi: 10.1016/j.cellsig.2005.07.010. [DOI] [PubMed] [Google Scholar]
- LeClaire LL, 3rd, Baumgartner M, Iwasa JH, Mullins RD, Barber DL. Phosphorylation of the Arp2/3 complex is necessary to nucleate actin filaments. J Cell Biol. 2008;182:647–654. doi: 10.1083/jcb.200802145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AK, Bridgman PC. Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. J Cell Biol. 1992;119:1219–1243. doi: 10.1083/jcb.119.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Espreafico EM, Mooseker MS, Forscher P. Myosin drives retrograde F-actin flow in neuronal growth cones. Neuron. 1996;16:769–782. doi: 10.1016/s0896-6273(00)80097-5. [DOI] [PubMed] [Google Scholar]
- Liu QR, Zhang PW, Lin Z, Li QF, Woods AS, Troncoso J, Uhl GR. GBPI, a novel gastrointestinal- and brain-specific PP1-inhibitory protein, is activated by PKC and inactivated by PKA. Biochem J. 2004;377:171–181. doi: 10.1042/BJ20030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QR, Zhang PW, Zhen Q, Walther D, Wang XB, Uhl GR. KEPI, a PKC-dependent protein phosphatase 1 inhibitor regulated by morphine. J Biol Chem. 2002;277:13312–13320. doi: 10.1074/jbc.M107558200. [DOI] [PubMed] [Google Scholar]
- Liu WS, Heckman CA. The sevenfold way of PKC regulation. Cell Signal. 1998;10:529–542. doi: 10.1016/s0898-6568(98)00012-6. [DOI] [PubMed] [Google Scholar]
- Lowery LA, Van Vactor D. The trip of the tip: understanding the growth cone machinery. Nat Rev Mol Cell Biol. 2009;10:332–343. doi: 10.1038/nrm2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manseau F, Fan X, Hueftlein T, Sossin W, Castellucci VF. Ca2+-independent protein kinase C Apl II mediates the serotonin-induced facilitation at depressed Aplysia sensorimotor synapses. J Neurosci. 2001;21:1247–1256. doi: 10.1523/JNEUROSCI.21-04-01247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny-Baron G, Kazanietz MG, Mischak H, Blumberg PM, Kochs G, Hug H, Marme D, Schachtele C. Selective inhibition of protein kinase C isozymes by the indolocarbazole Go 6976. J Biol Chem. 1993;268:9194–9197. [PubMed] [Google Scholar]
- Mattson MP, Guthrie PB, Kater SB. Intracellular messengers in the generation and degeneration of hippocampal neuroarchitecture. J Neurosci Res. 1988;21:447–464. doi: 10.1002/jnr.490210236. [DOI] [PubMed] [Google Scholar]
- Medeiros NA, Burnette DT, Forscher P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat Cell Biol. 2006;8:215–226. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- Mogilner A, Oster G. Force generation by actin polymerization II: the elastic ratchet and tethered filaments. Biophys J. 2003;84:1591–1605. doi: 10.1016/S0006-3495(03)74969-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongiu AK, Weitzke EL, Chaga OY, Borisy GG. Kinetic-structural analysis of neuronal growth cone veil motility. J Cell Sci. 2007;120:1113–1125. doi: 10.1242/jcs.03384. [DOI] [PubMed] [Google Scholar]
- Nakhost A, Forscher P, Sossin WS. Binding of protein kinase C isoforms to actin in Aplysia. J Neurochem. 1998;71:1221–1231. doi: 10.1046/j.1471-4159.1998.71031221.x. [DOI] [PubMed] [Google Scholar]
- Nakhost A, Kabir N, Forscher P, Sossin WS. Protein kinase C isoforms are translocated to microtubules in neurons. J Biol Chem. 2002;277:40633–40639. doi: 10.1074/jbc.M205099200. [DOI] [PubMed] [Google Scholar]
- Nishikawa M, Sellers JR, Adelstein RS, Hidaka H. Protein kinase C modulates in vitro phosphorylation of the smooth muscle heavy meromyosin by myosin light chain kinase. J Biol Chem. 1984;259:8808–8814. [PubMed] [Google Scholar]
- Nishizuka Y. Studies and perspectives of protein kinase C. Science. 1986;233:305–312. doi: 10.1126/science.3014651. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. The molecular heterogeneity of protein kinase C and its implications for cellular regulation. Nature. 1988;334:661–665. doi: 10.1038/334661a0. [DOI] [PubMed] [Google Scholar]
- Ohki S, Eto M, Kariya E, Hayano T, Hayashi Y, Yazawa M, Brautigan D, Kainosho M. Solution NMR structure of the myosin phosphatase inhibitor protein CPI-17 shows phosphorylation-induced conformational changes responsible for activation. J Mol Biol. 2001;314:839–849. doi: 10.1006/jmbi.2001.5200. [DOI] [PubMed] [Google Scholar]
- Ozsarac N, Weible M, 2nd, Reynolds AJ, Hendry IA. Activation of protein kinase C inhibits retrograde transport of neurotrophins in mice. J Neurosci Res. 2003;72:203–210. doi: 10.1002/jnr.10568. [DOI] [PubMed] [Google Scholar]
- Pan J, Singh US, Takahashi T, Oka Y, Palm-Leis A, Herbelin BS, Baker KM. PKC mediates cyclic stretch-induced cardiac hypertrophy through Rho family GTPases and mitogen-activated protein kinases in cardiomyocytes. J Cell Physiol. 2005;202:536–553. doi: 10.1002/jcp.20151. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- Ponti A, Matov A, Adams M, Gupton S, Waterman-Storer CM, Danuser G. Periodic patterns of actin turnover in lamellipodia and lamellae of migrating epithelial cells analyzed by quantitative fluorescent speckle microscopy. Biophys J. 2005;89:3456–3469. doi: 10.1529/biophysj.104.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti A, Vallotton P, Salmon WC, Waterman-Storer CM, Danuser G. Computational analysis of F-actin turnover in cortical actin meshworks using fluorescent speckle microscopy. Biophys J. 2003;84:3336–3352. doi: 10.1016/S0006-3495(03)70058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca DL, Martin S, Jenkins EL, Hanley JG. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat Cell Biol. 2008;10:259–271. doi: 10.1038/ncb1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin MW, Dailey ME, Bridgman PC. Polymerizing microtubules activate site-directed F-actin assembly in nerve growth cones. Mol Biol Cell. 1999;10:2309–2327. doi: 10.1091/mbc.10.7.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat Cell Biol. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- Rosse C, Linch M, Kermorgant S, Cameron AJ, Boeckeler K, Parker PJ. PKC and the control of localized signal dynamics. Nat Rev Mol Cell Biol. 2010;11:103–112. doi: 10.1038/nrm2847. [DOI] [PubMed] [Google Scholar]
- Schaefer AW, Kabir N, Forscher P. Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J Cell Biol. 2002;158:139–152. doi: 10.1083/jcb.200203038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliwa M, Nakamura T, Porter KR, Euteneuer U. A tumor promoter induces rapid and coordinated reorganization of actin and vinculin in cultured cells. J Cell Biol. 1984;99:1045–1059. doi: 10.1083/jcb.99.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankaran R, Pei J, Wang KC, Zhang YP, Shields CB, Xu XM, He Z. PKC mediates inhibitory effects of myelin and chondroitin sulfate proteoglycans on axonal regeneration. Nat Neurosci. 2004;7:261–268. doi: 10.1038/nn1193. [DOI] [PubMed] [Google Scholar]
- Slater SJ, Seiz JL, Stagliano BA, Stubbs CD. Interaction of protein kinase C isozymes with Rho GTPases. Biochemistry. 2001;40:4437–4445. doi: 10.1021/bi001654n. [DOI] [PubMed] [Google Scholar]
- Sossin WS, Diaz-Arrastia R, Schwartz JH. Characterization of two isoforms of protein kinase C in the nervous system of Aplysia californica. J Biol Chem. 1993;268:5763–5768. [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- Suter DM, Schaefer AW, Forscher P. Microtubule dynamics are necessary for SRC family kinase-dependent growth cone steering. Curr Biol. 2004;14:1194–1199. doi: 10.1016/j.cub.2004.06.049. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, Borisy GG. Improved procedures for electron microscopic visualization of the cytoskeleton of cultured cells. J Struct Biol. 1995;115:290–303. doi: 10.1006/jsbi.1995.1054. [DOI] [PubMed] [Google Scholar]
- Symons MH, Mitchison TJ. Control of actin polymerization in live and permeabilized fibroblasts. J Cell Biol. 1991;114:503–513. doi: 10.1083/jcb.114.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL. Inhibitors of neuronal regeneration: mediators and signaling mechanisms. Neurochem Int. 2003;42:189–203. doi: 10.1016/s0197-0186(02)00094-3. [DOI] [PubMed] [Google Scholar]
- Teng FY, Tang BL. Axonal regeneration in adult CNS neurons—signaling molecules and pathways. J Neurochem. 2006;96:1501–1508. doi: 10.1111/j.1471-4159.2006.03663.x. [DOI] [PubMed] [Google Scholar]
- Thomann D, Dorn J, Sorger PK, Danuser G. Automatic fluorescent tag localization II: improvement in super-resolution by relative tracking. J Microsc. 2003;211:230–248. doi: 10.1046/j.1365-2818.2003.01223.x. [DOI] [PubMed] [Google Scholar]
- Totsukawa G, Yamakita Y, Yamashiro S, Hartshorne DJ, Sasaki Y, Matsumura F. Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J Cell Biol. 2000;150:797–806. doi: 10.1083/jcb.150.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turbedsky K, Pollard TD, Bresnick AR. A subset of protein kinase C phosphorylation sites on the myosin II regulatory light chain inhibits phosphorylation by myosin light chain kinase. Biochemistry. 1997;36:2063–2067. doi: 10.1021/bi9624651. [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, Li F, Barnes CJ, Bagheri-Yarmand R, Kumar R. p41-Arc subunit of human Arp2/3 complex is a p21-activated kinase-1-interacting substrate. EMBO Rep. 2004;5:154–160. doi: 10.1038/sj.embor.7400079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goor D, Hyland C, Schaefer AW, Forscher P. The role of actin turnover in retrograde actin network flow in neuronal growth cones. PLoS One. 2012;7:e30959. doi: 10.1371/journal.pone.0030959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q, Jiang XY, Negroiu AM, Lu SG, McKay KS, Abrams TW. Protein kinase C acts as a molecular detector of firing patterns to mediate sensory gating in Aplysia. Nat Neurosci. 2012;15:1144–1152. doi: 10.1038/nn.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Ito M, Kataoka Y, Wada H, Koyama M, Feng J, Shiku H, Nishikawa M. Protein kinase C-catalyzed phosphorylation of an inhibitory phosphoprotein of myosin phosphatase is involved in human platelet secretion. Blood. 2001;97:3798–3805. doi: 10.1182/blood.v97.12.3798. [DOI] [PubMed] [Google Scholar]
- Wong EV, Kerner JA, Jay DG. Convergent and divergent signaling mechanisms of growth cone collapse by ephrinA5 and slit2. J Neurobiol. 2004;59:66–81. doi: 10.1002/neu.10342. [DOI] [PubMed] [Google Scholar]
- Wu-Zhang AX, Newton AC. Protein kinase C pharmacology: refining the toolbox. Biochem J. 2013;452:195–209. doi: 10.1042/BJ20130220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Eves R, Yeh C, Kan W, Xu F, Mak AS, Liu M. Phorbol ester-induced podosomes in normal human bronchial epithelial cells. J Cell Physiol. 2009;218:366–375. doi: 10.1002/jcp.21609. [DOI] [PubMed] [Google Scholar]
- Yang Q, Zhang XF, Pollard TD, Forscher P. Arp2/3 complex-dependent actin networks constrain myosin II function in driving retrograde actin flow. J Cell Biol. 2012;197:939–956. doi: 10.1083/jcb.201111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XF, Forscher P. Rac1 modulates stimulus-evoked Ca(2+) release in neuronal growth cones via parallel effects on microtubule/endoplasmic reticulum dynamics and reactive oxygen species production. Mol Biol Cell. 2009;20:3700–3712. doi: 10.1091/mbc.E08-07-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XF, Schaefer AW, Burnette DT, Schoonderwoert VT, Forscher P. Rho-dependent contractile responses in the neuronal growth cone are independent of classical peripheral retrograde actin flow. Neuron. 2003;40:931–944. doi: 10.1016/s0896-6273(03)00754-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.