Abstract
We report a high-quality draft sequence of the genome of the horse (Equus caballus). The genome is relatively repetitive, but has little segmental duplication. Chromosomes appear to have undergone few historical rearrangements – 48% of equine chromosomes show conserved synteny to a single human chromosome. Equine chromosome 11 is shown to have an evolutionary novel centromere devoid of centromeric satellite DNA, suggesting that centromeric function may arise prior to satellite repeat accumulation. Linkage disequilibrium, showing the influences of early domestication of large herds of female horses, is intermediate in length between dog and human, and there is long-range haplotype sharing among breeds.
As one of the earliest domesticated species, the horse Equus caballus, has played an important role in human exploration of novel territories. Belonging to the order perissodactyla; i.e. odd-toed animals with hooves, the genus Equus radiated into 8-9 species around three million years ago (1). Members of the family equidae exhibit diverged karyotypes (2) and variable centromeric positioning (1). With over 90 hereditary conditions, which may serve as models for human disorders (3, 4) (e.g. infertility, inflammatory diseases, and muscle disorders), the horse has much to offer as a model species.
DNA from a single mare of the Thoroughbred breed was sequenced to 6.8× coverage (see SOM text), resulting in a high-quality draft assembly (designated EquCab2.0) with a 112 kb N50 contig size and a 46 Mb N50 scaffold size (Tables S1, S2), and >95% of the sequence anchored to the 64 (2N) equine chromosomes. The 2.5-2.7 Gb genome size is somewhat larger than dog (2.5Gb) and smaller than the human and bovine (2.9 Gb) genomes (5-7). Segmental duplications (8) comprise < 1% of the equine genome, and most are intra-chromosomal duplications; such as are seen in many other mammalian genomes (SOM). Repetitive sequences, many equine specific, comprise 46% of the genome assembly (SOM). The predominant repeat classes include LINEs dominated by L1 and L2 types (Tables S3, S4) (19% of bases) and SINEs including the recent ERE1/2 and the ancestral MIRs (7% of bases). Comparison of horse and human chromosomes reveals strong conserved synteny between these species (Fig. S1). Indeed, seventeen horse chromosomes (53%) comprise material from a single human chromosome (dog, 29%).
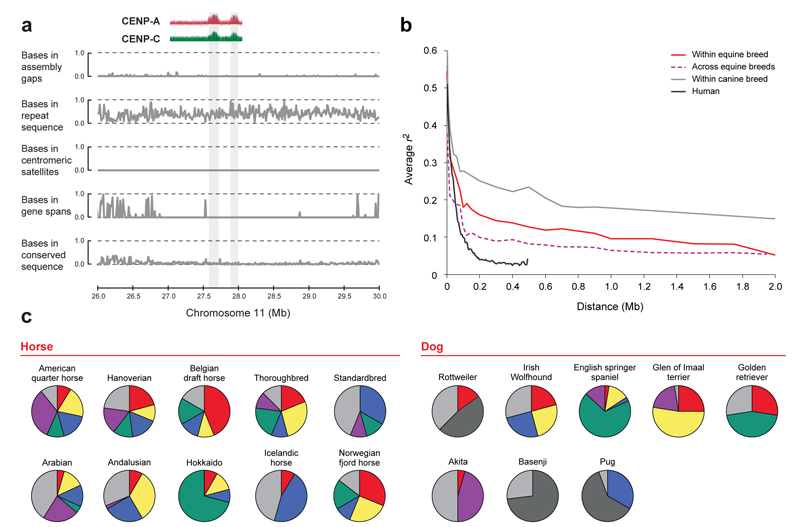
One unexpected feature of the horse genome landscape was the identification of an evolutionary new centromere (ENC) on chromosome 11 (ECA11) captured in an immature state. Several ENCs have been generated in the genus Equus by centromere repositioning (shift of centromeric position without chromosome rearrangement)(1). Mammalian centromeres are typically complex structures characterized by the presence of satellite tandem repeats. ENCs are believed to form initially by unknown mechanisms in repeat free regions and then progressively acquire extended arrays of satellite tandem repeats that may contribute to functional stability (9). The centromere of ECA11 resides in a large region of conserved synteny with many mammals, where horse is the only species with a centromere present, strongly suggesting that this centromere is evolutionarily new. The ECA11 centromere is the only horse centromere lacking any hybridization signal in FISH experiments probing with the two major horse satellite sequences (Fig. S2a; Table S8; SOM) - as if it had not had enough time to acquire satellite DNA. We cytogenetically localized the primary constriction (Fig. S2b), then precisely mapped, at the sequence level, the centromeric function using ChIP-on-chip experiments (Fig. S5). In this region, we found only five sequence gaps (none > 200 bp), no protein coding sequences, normal levels of non-coding conserved elements and typical levels of interspersed repetitive sequences, but no satellite tandem repeated sequences (Fig 1a). We also found no evidence of accumulation of L1 transposons (10) or KERV-1 elements (11) previously hypothesized to influence ENC formation. In conclusion, we propose that the ECA11 centromere was formed very recently during the evolution of the horse lineage and, in spite of being functional and stable in all horses, has not yet acquired the marks typical of mammalian centromeres.
Figure 1. Major findings of the genome analysis.
(a) Analysis of the primary centromeric constriction of ECA11: 26,000,000–30,000,000; (i) ChIP-on-chip analysis with antibodies against centromeric proteins (CENP-A and CENP-C) shows two regions (136 and 99kb) bound by kinetochore proteins; (ii) there are no un-captured and few captured gaps; (iii) a normal fraction of bases in repeat sequences; (iv) no satellite tandem repeats (v) no protein coding sequences are present nearby (vi) normal levels of non-coding conserved elements (29 eutherians). (b) Horse LD is intermediate between human and dog. (c) Horses exhibit more long-range across-breed haplotype sharing than dogs. Haplotypes have the same color across breeds, haplotypes in < 5% of all individuals (light gray), haplotypes in > 5% of all individuals but a single breed (dark gray). Data show LD regions of ECA18 (first 100kb) and dog chromosome 12 (first 100kb) which are representative. Full data are in Table S11.
The equine gene set is similar to other eutherian mammals and has a predicted 20,322 protein-coding genes (Ensembl build 52.2b) of which 16,617, 17,106 and 17,106 have evidenced orthology to human, mouse and dog, respectively. The remainder comprises projected protein-coding genes, novel protein-coding genes, and pseudogenes. One-to-one orthologs with human account for 15,027 horse gene predictions (SOM). Transcriptome analysis of eight equine samples confirms expression of 87% of the 18,039 non-overlapping genes predicted by ENSEMBL and 88% of the 169,073 predicted exons. Gene family analysis shows paralogous expansion in horses compared to both human and bovine (SOM) for several interesting families; keratin genes related to the condition of pachyonychia (nail bed thickening) in humans (12) - perhaps affecting hoof formation, and opsin genes for photoreception - possibly advantageous for visual perception of predators (Table S9).
The history of horse domestication, which has important implications for trait mapping strategies, differs in important ways from that of the domestic dog, but is perhaps similar to that of the cow. Horses do not appear to have undergone a tight domestication bottleneck and the presence of many matrilines in domestic horse history has been postulated (13). Screening the horse Y chromosome revealed a limited number of patrilines, consistent with a strong sex-bias in the domestication process (14).
We first generated a single nucleotide polymorphism (SNP) map of more than one million markers at an average density of one SNP per 2kb by lightly sequencing seven horses from different breeds and by mining the assembly for SNPs (Table S10).
We characterized the haplotype structure within and across breeds by genotyping 1,007 SNPs from ten regions of the genome (SOM) in twelve populations, including eleven breed sets (each with 24 representatives), and one set of individual representatives from 24 other breeds and equids. 98% of SNPs were validated with an average of 69% being polymorphic in alternate breeds (SOM). Like the bovine (15), within-breed linkage disequilibrium (LD) is moderate, dropping to twofold the background levels (r2) at 100-150kb (Fig. 1b). The majority of breeds showed similar LD, (SOM, Fig. S7) and major haplotypes were frequently shared among diverse populations (Fig. 1c). Based on the length of LD in the horse, the number of haplotypes within haplotype blocks, and the polymorphism rate, power calculations suggest that ~100,000 SNPs are sufficient for association mapping within all breeds as well as across breeds (SOM, Fig. S8).
Phylogenetic relationships among breeds were inconsistent across re-sequenced regions (Fig. S9)-most likely a consequence of the close relationships of horse breeds world-wide. We were unable to phylogenetically separate E. przewalskii from the domesticated horses despite its different karyotype (2N=66 versus 2N=64 for horse), in agreement with recent findings (16), whereas donkey (E. africanus) is clearly a distinct taxon (Fig. S9, Table S14, SOM). This suggests that either inter-mixing of E. przewalskii and E. caballus has occurred after subspecies separation or that E. przewalskii is recently derived from E. caballus.
We demonstrated the utility of the equine genome sequence and a SNP map by applying these resources to mutation detection for the Leopard Complex (LP) spotting locus (SOM). LP (Appaloosa spotting) is defined by patterns of white occurring with or without pigmented spots (Fig. S10). Homozygosity confers a phenotype associated with Congenital Stationary Night Blindness in the Appaloosa breed (17). Fine mapping of a 2Mb region followed by regional sequence capture and sequencing (300kb) found no indications of associated copy number variants or insertion-deletions but found 42 associated SNPs. Of these 21 reside within an associated haplotype near a candidate gene melastatin 1 (TRPM1), which is expressed in eye and melanocytes (18). Two conserved SNPs may be good candidates for the causal mutation.
Our analysis of the first high-quality draft sequence of a horse (Equus caballus) distinguishes Equus caballus from earlier eutherian genomes by its large synteny with humans and the identification of a centromere repositioning event which may provide an effective model to study epigenetic factors responsible for centromere function. Our results demonstrate that horse population history has led to across breed haplotype sharing, increasing the feasibility of across breed mapping. Mapping projects in the horse are likely to accelerate in the coming years and will identify mutations in genes related to morphology, immunology, and metabolism, and may benefit human health.
Supplementary Material
Acknowledgments
We thank the Kentucky Horse Park and L. Chemnick for samples, L. Gaffney for graphics and M. Daly for useful discussions. Supported by the National Human Genome Research Institute, the Dorothy Russell Havemeyer Foundation, the Volkswagen Foundation, the Morris Animal Foundation and Programmi di Ricerca Scientifica di Rilevante Interesse Nazionale (PRIN-2006). KLT is a EURYI funded by ESF. Sequences have Genbank accessions AAWR02000001-AAWR02055316. SNP in dbSNP
References
- 1.Carbone L, et al. Genomics. 2006 Jun;87:777. doi: 10.1016/j.ygeno.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Trifonov VA, et al. Chromosome Res. 2008;16:89. doi: 10.1007/s10577-007-1201-7. [DOI] [PubMed] [Google Scholar]
- 3.Online Mendelian Inheritance in Animals. University of Sydney; 2009. [Google Scholar]
- 4.Chowdhary BP, Paria N, Raudsepp T. Anim Reprod Sci. 2008 Sep;107:208. doi: 10.1016/j.anireprosci.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Lindblad-Toh K, et al. Nature. 2005 Dec 8;438:803. [Google Scholar]
- 6.Lander ES, et al. Nature. 2001 Feb 15;409:860. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Elsik CG, et al. Science. 2009 Apr 24;324:522. [Google Scholar]
- 8.Mikkelsen TS, et al. Nature. 2007 May 10;447:167. [Google Scholar]
- 9.Ventura M, et al. Science. 2007 Apr 13;316:243. doi: 10.1126/science.1140615. [DOI] [PubMed] [Google Scholar]
- 10.Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KH, Wong LH. PLoS Genet. Jan 2009;5:e1000354. doi: 10.1371/journal.pgen.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carone DM, et al. Chromosoma. 2009 Feb;118:113. doi: 10.1007/s00412-008-0181-5. [DOI] [PubMed] [Google Scholar]
- 12.Wu JW, et al. J Eur Acad Dermatol Venereol. Feb 2009;23:174. doi: 10.1111/j.1468-3083.2008.02752.x. [DOI] [PubMed] [Google Scholar]
- 13.Vila C, et al. Science. 2001 Jan 19;291:474. doi: 10.1126/science.291.5503.474. [DOI] [PubMed] [Google Scholar]
- 14.Lindgren G, et al. Nat Genet. 2004 Apr;36:335. doi: 10.1038/ng1326. [DOI] [PubMed] [Google Scholar]
- 15.Gibbs RA, et al. Science. 2009 Apr 24;324:528. [Google Scholar]
- 16.Lau AN, et al. Mol Biol Evol. 2009 Jan;26:199. doi: 10.1093/molbev/msn239. [DOI] [PubMed] [Google Scholar]
- 17.Sandmeyer LS, Breaux CB, Archer S, Grahn BH. Vet Ophthalmol. 2007 Nov-Dec;10:368. doi: 10.1111/j.1463-5224.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- 18.Bellone RR, et al. Genetics. 2008 Aug;179:1861. doi: 10.1534/genetics.108.088807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.