Abstract
The defining characteristic of recessive diseases is the absence of a phenotype in the heterozygous carriers. Nonetheless, subtle manifestations may be detectable by new methods, such as expression profiling. Ataxia telangiectasia (AT) is a typical recessive disease, and individual carriers cannot be reliably identified. As a group, however, carriers of an AT disease allele have been reported to have a phenotype that distinguishes them from normal control individuals: increased radiosensitivity and risk of cancer. We show here that the phenotype is also detectable, in lymphoblastoid cells from AT carriers, as changes in expression level of many genes. The differences are manifested both in baseline expression levels and in response to ionizing radiation. Our findings show that carriers of a recessive disease may have an “expression phenotype.” In the particular case of AT, this suggests a new approach to the identification of carriers and enhances understanding of their increased cancer risk. More generally, we demonstrate that genomic technologies offer the opportunity to identify and study unaffected carriers, who are hundreds of times more common than affected patients.
Ataxia telangiectasia (AT [MIM 208900]) is an autosomal recessive disease caused by mutations in the gene ataxia telangiectasia mutated (ATM) on human chromosome 11q22-23 (Savitsky et al. 1995). The ATM product is a protein kinase that plays a role in DNA-damage repair through cell cycle regulation. AT is a rare disease, with a frequency of ∼1/40,000. However, heterozygous carriers are not rare; their frequency is ∼1/100. Although the disease AT is recessive, epidemiological studies have suggested that AT carriers, as a group, have a shortened lifespan and an elevated risk of cancer, especially breast cancer; cellular studies have shown increased sensitivity to ionizing radiation (IR) (Swift et al. 1986, 1991; Athma et al. 1996; Broeks et al. 2000). Some studies have estimated that AT carriers have a fivefold increased risk of breast cancer compared with control individuals and that they may account for 8%–18% of all patients with breast cancer in the United States (Swift et al. 1976). These findings are controversial; other studies have shown minimal-to-absent contribution of heterozygous ATM mutations to risk of breast cancer (FitzGerald et al. 1997; Chen et al. 1998).
The lack of a reliable diagnostic assay for the identification of individual AT carriers has hampered studies in this area. ATM is a large gene (∼150 kb), and there are no common mutations causing AT (Wright et al. 1996), so sequence-based diagnostic methods are difficult (Concannon and Gatti 1997). Existing protein and cell-based assays are inaccurate for the identification of carriers and are time and labor intensive (Telatar et al. 1996). ATM mRNA levels are normal in almost all patients with AT and carriers of AT. Although ATM protein levels are significantly decreased in ∼85% of the patients, they are decreased only in some AT carriers (Becker-Catania et al. 2000). Thus, testing for ATM transcript or protein levels will not allow detection of AT carriers. The main goal of our study was to determine whether AT carriers have a phenotype at the gene expression level that differs from that of control individuals. This finding would be of biological significance and might lead to methods for identifying carriers individually.
Material and Methods
Study Subjects
Baseline expression levels
We used Epstein-Barr virus–transformed lymphoblastoid cell lines (Coriell Cell Repositories) from 10 obligate AT carriers (GM08931, GM03334, GM03382, GM03188, GM09588, GM00736, GM02781, GM09585, GM09583, and GM09579) and 10 control individuals (GM06995, GM06997, GM07014, GM10832, GM10835, GM10848, GM10849, GM10860, GM06987, and GM07038) for our analysis. None of the subjects were known to be blood relatives. Reference samples used in all hybridizations were made with RNA from six CEPH individuals (GM06987, GM07038, GM06995, GM06997, GM07014, and GM07042).
Expression response to IR
In the first experiment, lymphoblastoid cell lines from seven obligate AT carriers (GM09583, GM09579, GM08930, GM08931, GM03334, GM03382, and GM03188) and six control individuals (GM10832, GM10835, GM10860, GM10848, GM10849, and GM07057) were studied. In the replication experiment, we studied the lymphoblastoid cells lines from five new obligate AT carriers (GM00736, GM02781, GM03187, GM09585, and GM09588) and six new control individuals (GM06987, GM07038, GM06995, GM06997, GM07014, and GM07042). None of the study participants were known to be blood relatives. Reference samples used in all hybridizations were made with RNA from six CEPH individuals, as described above.
cDNA Microarrays
We randomly selected 2,880 cDNA clones from a sequence-verified cDNA clone set (Research Genetics). The clones were grown in Luria broth–ampicillin as overnight cultures. The cultures were then diluted (1:10 with Tris-EDTA), were boiled at 95°C for 3 min, and were used as DNA templates for PCR amplifications. DNA was amplified with 0.4 μM vector-specific primers (T3/T7 or M13 forward and reverse), 200 μM dNTP, 2.5 mM MgCl2, 2.5 U Taq DNA polymerase (Perkin Elmer or Promega), and 1× PCR buffer. Amplifications were performed in 96-well plates by an initial denaturation at 96°C for 5 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 5 min. Of the amplicons in each 96-well plate, 10% were checked by gel electrophoresis. If the success rate of amplifications was ⩾90%, the amplicons were precipitated with ethanol and dried; otherwise, the amplifications were repeated. The amplicons were reconstituted in 2× sodium chloride–sodium citrate (SSC), 0.01% sarkosyl for arraying onto aminoalkylsilane-coated microscope slides (Sigma), using a pin-and-ring arrayer model 417 (Affymetrix). The DNA samples on the array were moistened over gentle steam and were then UV-crosslinked for attachment onto the glass surface. The glass arrays were denatured at 95°C for 3 min and were immediately placed into ice-cold ethanol. The arrays were then dried by centrifugation at 1,268 g for 2 min. The arrays were prehybridized with 5× SSC, 0.2% SDS, and 1% BSA at 42°C for 1 h.
Probe Preparation
Lymphoblastoid cells of the subjects were grown in Roswell Park Memorial Institute (RPMI) 1640 medium with 15% fetal bovine serum, 1% penicillin-streptomycin, and 1% l-glutamine at 37°C in a humidified 5% CO2 chamber. Cells were grown to a density of ∼1 × 106/ml. They were irradiated with 3 grays (Gy) IR in a 137Cs gamma irradiator. To minimize variations caused by culture conditions, all cells were irradiated 24 h after the addition of fresh medium. The cells were harvested before IR and at 2, 6, 12, and 24 h after IR. Total RNA was extracted from cell pellets, using the RNeasy midi kit (Qiagen).
In each reaction, total RNA was reverse transcribed into fluorescently (Cy3 or Cy5) labeled cDNA, using the Genisphere 3DNA expression array detection kit (Genisphere). In brief, for each reaction, 10 μg total RNA was reverse transcribed using 50 nM oligo dT–Genisphere capture primer, 0.5 mM dNTP, 200 U Superscript II (Gibco BRL) in 1× first-strand Superscript II buffer at 42°C for 2 h. The RNA from the DNA/RNA hybrids was denatured with 0.07 M NaOH. The reaction was then neutralized to pH 7.5, using Tris-HCl. Then, for each array hybridization, 10% of the cDNA mixture was incubated with 2.5 μl Cy3 or Cy5 dendrimer in Expresshyb (Clontech) at 55°C for 30 min; 2 μg denatured Cot1DNA (Gibco BRL) was added, and the entire mixture was added to the prehybridized array for hybridization at 62°C for at least 12 h. After hybridization, the arrays were washed with 2× SSC and 0.2% SDS at 55°C for 7 min, followed by a wash with 2× SSC and another with 0.2× SSC at room temperature for 7 min each. The hybridization signals were read using a dual-laser fluorescent scanner model 428 (Affymetrix).
Analysis of Replicated Microarrays
The scanned images were analyzed using Spotfinder (TIGR). For each data point (observation), the hybridization signals from the image analysis yield values for Cy3 (experimental) and Cy5 (reference). Signal intensities for the Cy3 and the Cy5 channels were “normalized”; the Cy3 measurements were multiplied by a scaling factor to make the mean Cy3:Cy5 ratio for all the spots on the slide = 1.0. (We assume that, on average, the genes have the same expression level in the experimental and the reference samples.) Spotfinder assigns a signal intensity of zero to an observation when the signal is less than the background. If the Cy5 value was zero, that observation was not included in the analysis. (If there is no signal in the pooled (reference) RNA, we cannot use that observation). The Cy3 value was divided by the Cy5 value, to give the expression ratio (R) for each replicate of each gene.
We were particularly concerned to deal appropriately with data for genes that are expressed in some individuals but not in others. The Cy3 value (and R) for these genes will be zero for some arrays. To allow us to take the logarithm of the R (see below), we replaced the R values that are zero with R′, the smallest nonzero R found for any gene on that slide. We completed the transformation of the observations by calculating log2 of the R and R′ values.
Replicates
A number of studies emphasize the variability of individual results from microarray studies (Lee et al. 2000; Newton et al. 2001). In this project, there were four replicates for each array hybridization. Our present procedure for analysis can be used with any experiment that includes three or more replicate observations. The goal is to eliminate single observations that are aberrant for technical reasons and to retain only observations that are valid, while avoiding the usual arbitrary or subjective definition of outliers.
For some genes, individual replicate observations were discarded because the Cy5 value for that replicate was zero. If fewer than three replicate observations remained, we discarded that gene from the analysis. If all four values remained, we found the replicate value that was most deviant from the mean of the replicates and discarded it. This trimming was not done if there were only three valid observations. The trimming procedure tended to discard the highest value somewhat more often (∼57% of time) than the lowest one but was applied equally to the data from carriers and from control individuals. Since our significance levels are based only on the nonparametric permutation test described below, this reduction in variance does not bias the resulting P values.
The rest of the analysis was performed with only the remaining three observations. Thus, the final set of data consists of logarithms of the original R values, and these have been trimmed by discarding the most extreme replicate value. For each gene, we calculated the mean of the remaining replicate observations and used these for further analysis. The software for performing this analysis of replicated microarray data is available on the authors' Web site, Gene Expression Profiling of Carriers of the ATM Mutation.
Baseline Experiment
In view of the problems of multiple testing and correlations among observations, we did not use any parametric tests of significance. We chose the t statistic, because of its familiar properties, and used it solely as a measure of the difference between carriers and control individuals in gene expression levels, not for a t test (Callow et al. 2000; Tusher et al. 2001). To determine whether there were more differences between carriers and control individuals than would be expected by chance, we used a permutation test (Manly 1997) as follows: We randomly assigned each individual to one of two groups of 10 (corresponding to the real sample sizes); we then calculated the t score (absolute value) for each gene. After repeating this procedure 3,000 times, we had a distribution of t scores for each gene, which was based on the 20 values actually observed for expression level but with group membership randomly assigned.
For each gene, we determined whether the t score (absolute value) from the real data fell among the largest 1% of values resulting from permutation. When several t scores among the 3,000 had the same value, we assigned the percentile of the real value (conservatively) as the least extreme in the set. Among the 2,880 genes/ESTs on the array, there were 71 whose t scores fell in the largest 1% obtained from permutation. Similarly, we used the 3,000 sets of permutations to provide 3,000 estimates for the number of extreme t scores expected under the null hypothesis that no genes differ in expression level between carriers and control individuals. The frequency of permutations in which this number was ⩾71 (14/3,000 in our data) is our estimate of the significance level for rejecting the null hypothesis. The procedure used is similar to that of Storey and Tibshirani (see “Electronic Database Information” section).
Conventional stepwise linear discriminant analysis (SPSS) was performed with data from the 12 genes with observations on all 20 individuals, and it identified the four genes named in the “Classification of AT Carriers and Control Individuals” subsection below. Adding genes beyond the four selected did not yield discrimination that was significantly better (P<.05). Assignment to the two groups was performed with the “leave-one-out” cross-validation procedure.
IR Response (Time-Course) Experiment
Standardization over experiments was performed as follows: For each time point, the set of observations for all genes was considered separately for carriers (pooled RNA) and control individuals (pooled RNA). Most of the cell lines for the IR response experiment were from individuals used previously in the baseline experiment. The mean and standard deviation were determined (for that time point), and each observation was expressed as units of standard deviation from the mean for all genes.
Discriminant analysis was performed, using the statistical package SPSS, on the standardized expression ratio(s) for only those genes that had valid observations at all the time points. The variables are the expression levels at five time points after IR. These were measured on the 2,880 genes (“members” of the carrier group) in one RNA sample pooled from carriers. The same five variables were measured on 2,880 genes (the “members” of the control group) in pooled RNA from control individuals. Thus, the carrier and control groups consist of gene expression observations on the same set of genes but in different pools of RNA. The discriminant function was estimated using the pooled covariance matrix for carriers and control individuals. The discriminant score (linear combination of five observations) for each gene in each group was used to assign a gene expression pattern to one of the two groups (with cross-validation). For each gene, two expression patterns were assigned, one with the observations from carriers, one with those from control individuals. This approach detects the genes with the most different levels of expression after IR but does not directly address differences in time trends.
In the first experiment, this procedure resulted in 442 genes correctly assigned for both groups; in the replication experiment, this resulted in 183 (of the 442) genes correctly assigned for both groups. To identify particular genes with the most distinctive expression patterns, we compared the discriminant scores for assigning each gene to the carrier or the control group. Genes were ranked by the difference in these two scores. This was done for the first and the replication experiments, and the mean of the two values was found. The genes with largest mean difference are taken to be those with most distinctive expression patterns.
Real-Time Quantitative RT-PCR
Five of the 183 genes that were correctly assigned in the time-course experiments were assayed using real-time quantitative RT-PCR. Sequences of these genes were obtained from UCSC Genome Browser. Primers were designed using Primer Express software (Applied Biosystems). The reactions were performed with 1× SYBR-Green PCR master mix buffer (Applied Biosystems), 300-nM forward and reverse primers, and cDNA. cDNA from lymphoblastoid cell lines from two AT carriers (GM09585 and GM09588) and two control individuals (GM06987 and GM06997) was analyzed. Assays were performed in triplicate, using an ABI 7000 instrument. The fold change was calculated using a standard curve analysis and was normalized to the level of β-actin. For each gene, the data from the two AT carriers were averaged, as were those from the two control individuals. These results from RT-PCR were compared with the corresponding microarray results by calculating the correlation coefficient of the expression ratio over the five time points.
Results
Baseline Expression Differences
We used cDNA microarrays to compare the expression levels of genes in lymphoblastoid cells from 10 AT carriers and 10 control individuals. The Cy3-labeled cDNA from each individual was cohybridized with the Cy5 reference cDNA onto microarrays containing ∼3,000 human known genes/ESTs. All hybridizations were done with four replicates. We removed the most deviant observation from every set of four replicates and represented the expression level of each gene by the average of the three remaining measurements (see “Analysis of Replicated Microarrays” in the “Material and Methods” section).
In this “baseline” experiment, we used the t statistic to compare the expression level of each gene in cells from AT carriers and control individuals. Our goal was to show that there are statistically significant differences between carriers and control individuals in the number of genes that differ in expression level, and to identify the genes that are most likely to show consistent differences. The large number of genes/ESTs resulted in a severe multiple-testing problem, and the possible correlations between genes posed an additional problem, so we did not perform standard t tests. Instead, we assessed the significance of the t scores for each gene empirically by a permutation test. Among the 2,880 cDNA clones on the arrays, we found 71 clones whose t scores ranked in the top 1% of the 3,000 random permutations we performed with the data from the 10 AT carriers and 10 control individuals; the absolute values of these t scores were 1.6–5.1. Because of the large number of tests, the corresponding P values ⩽.01 are “nominal”; nevertheless, the true significance of the number of genes (71 clones) can be assessed, as described below.
To determine the statistical significance of our overall result, we used the permutations described above. These provided 3,000 estimates of the number of genes that have nominal P⩽.01 by chance—that is, under the null hypothesis that no gene among the 2,880 differs significantly between carriers and control individuals. Among the 3,000 permutations, we found only 14 (0.47%), with ⩾71 clones, that met this criterion. The mean number of genes from the permutations was 27.4. We conclude that there are significantly (P⩽.005) more genes that differ in baseline expression level than would be expected under the null hypothesis.
Among the 71 clones, 29 are known genes (WEE1 is represented by two clones), and 41 are ESTs. We list the 29 known genes (represented by 30 cDNA clones) in table 1. Among them, 15 genes (HDAC1, MAPKAP3, WEE1, LIM, CDKN2D, THBS1, SSI2, TSSC6, CCNE1, CHN2, G6PD, TXN2, RPA1, GCP3, and DIO1) regulate cell growth and maintenance through various pathways, including cell cycle control and regulation of apoptosis (Heald et al. 1993; Liu and Weaver 1993; McLaughlin et al. 1996; Roberts 1996; Minamoto et al. 1997; Ueno et al. 1999; Ashburner 2000; Juan et al. 2000; Tuttle et al. 2000). Table 1 shows the t score and the P value for each known gene. The complete list of 71 cDNA clones, including known genes and ESTs is available at the Gene Expression Profiling of Carriers of the ATM Mutation Web site, under “Baseline Experiment.” The most marked excess of small permutation P values occurs for the 22 clones with P<.0017 (see list of 71 cDNA clones at the authors' Web site, under “Baseline Experiment”). This observation suggests that these genes and ESTs are the most likely to be “true positives.”
Table 1.
The 30 cDNA Clones (Representing 29 Known Genes) with the Largest Difference (Nominal P<.01, by Permutation Test) in Baseline Expression Level between AT Carriers and Control Individuals[Note]
| GeneSymbol | Gene Name | df | t Score(Carrier vs. Control) | P |
| CSF2RA | Colony-stimulating factor 2 receptor alpha | 9 | 2.85 | <.0003 |
| HDAC1 | Histone deacetylase 1 | 12 | −3.30 | <.0003 |
| MAPKAPK3 | Mitogen-activated protein kinase–activated protein kinase 3 | 14 | −3.04 | <.0003 |
| SLC25A6 | Solute carrier family 25, member 6 | 18 | 3.61 | <.0003 |
| WEE1 | WEE1 | 18 | −3.37 | .0003 |
| TFRC | Transferrin receptor (p90) | 18 | −3.15 | .0007 |
| LIM | LIM protein | 18 | 3.81 | .0010 |
| OGT | O-linked N-acetylglucosamine transferase | 15 | 2.18 | .0010 |
| WEE1 | WEE1 | 16 | −3.20 | .0010 |
| CDKN2D | Cyclin-dependent kinase inhibitor 2D (p19) | 18 | −3.31 | .0013 |
| THBS1 | Thrombospondin 1 | 17 | 3.57 | .0013 |
| SSI2 | STAT induced STAT inhibitor | 16 | 3.77 | .0023 |
| TSSC6 | Pan-hematopoietic expression | 14 | 3.76 | .0023 |
| CCNE1 | Cyclin E1 | 13 | −2.57 | .0027 |
| CHN2 | Chimerin | 18 | −3.14 | .0030 |
| G6PD | Glucose-6-phosphate dehydrogenase | 17 | 3.80 | .0033 |
| PLOD3 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 | 17 | −3.27 | .0033 |
| TXN2 | Thioredoxin | 16 | 3.27 | .0037 |
| ARF6 | ADP-ribosylation factor 6 | 18 | −2.98 | .0047 |
| CSF3R | Colony stimulating factor 3 receptor | 16 | −2.14 | .0047 |
| NDST1 | N-deacetylase 1 | 15 | 3.22 | .0047 |
| GOSR2 | Golgi SNAP receptor complex member 2 | 13 | −3.00 | .0053 |
| KIAA0204 | Ste20-related serine/threonine kinase | 11 | −2.35 | .0057 |
| RPA1 | Replication protein A1 | 17 | −2.19 | .0060 |
| GCP3 | Spindle pole body protein 3 | 16 | 1.98 | .0067 |
| SULT1C1 | Sulfotransferase family, 1C, member 1 | 13 | 3.04 | .0073 |
| HNRPD | Heterogeneous nuclear ribonucleoprotein D | 15 | −3.27 | .0080 |
| DIO1 | Death inducer-obliterator 1 | 12 | −2.86 | .0083 |
| SLC7A6 | Solute carrier family 7, member 6 | 7 | 3.01 | .0083 |
| PPP1R2 | Protein phosphatase 1, regulatory subunit 2 | 17 | −2.97 | .0097 |
Note.— Genes are ranked by P value.
Classification of AT Carriers and Control Individuals
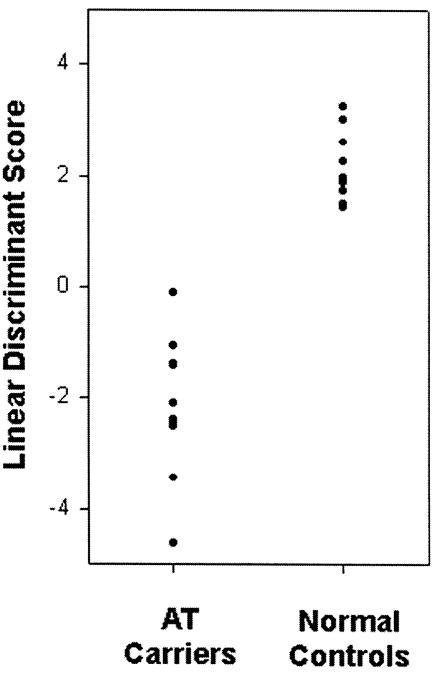
To explore the biological differences between AT carriers and control individuals, we wanted to identify the largest set of genes that differ in expression between the two groups. However, for classification purposes, we want to have the smallest set of genes whose expression levels, when combined in a discriminant procedure, would yield highly accurate classification. To find that set, we performed discriminant analysis with a subset of the 71 clones with a nominal P⩽.01. In the analysis, we used the 12 genes/ESTs for which we have expression values in all 20 individuals. By stepwise discriminant analysis, we selected four genes (LIM, CDKN2D, TFRC, and ARF6) and determined the best linear discriminant function. The discriminant scores for the 10 AT carriers and 10 control individuals are shown in figure 1. The apparent greater variance in carriers is not surprising, in view of the known heterogeneity of AT mutations. However, the two distributions do not overlap, so the discriminant function provides highly accurate assignment of these 20 individuals to the two groups. We also assessed how accurately we would classify individuals who were not part of the “training” sample. For this purpose, we performed “cross-validation”; the individual to be assigned was left out of both the selection of genes and the calculation of the discriminant function and then was assigned on the basis of the data from the other 19 individuals. With this more stringent requirement, we were able to classify 95% of the 20 individuals correctly. Of the 20 cross-validation “trials,” 9 resulted in exactly the same set of four genes as were reported in the complete data. Of the other 11, LIM was included in 5, CDKN2D in 9, TFRC in 10, and ARF6 in 6 trials. These findings indicate the tendency for the four genes to be selected even in cross-validation.
Figure 1.
Discriminant scores for gene expression levels in cells from 10 AT carriers and 10 control individuals. Four genes (LIM, CDKN2, TFRC, and ARF6) were selected by stepwise linear discriminant analysis. (Some points overlap with others in the plot and cannot be distinguished.)
Since the 12 genes/ESTs in the starting set were selected from those with the largest t scores, the high level of accurate classification for these 20 individuals is not surprising. Our purpose was not to show that classification is possible but to confirm that it can be achieved with a small number of genes and ESTs.
Differences in Transcriptional Response to IR
In view of the increased sensitivity to IR among patients with AT, we also studied the changes in expression profiles of AT carriers in response to IR. Lymphoblastoid cells from AT carriers and control individuals were exposed to low-dose IR (3 Gy). In the “baseline” experiment above, we tested expression profiles of individuals. To reduce the number of hybridizations in this time-course experiment, we used pooled RNA samples instead of samples from individuals. This allowed us to obtain expression profiles at five time points (instead of at one time point, as in the previous experiment) and nevertheless to continue our rigorous quality control by performing four replications of all array hybridizations. Lymphoblastoid cells from seven AT carriers and six control individuals were studied. Cells were harvested immediately before IR, and at 2, 6, 12, and 24 h after IR (3 Gy). At each time, total RNA was extracted from the cells and assembled into two pools, one from AT carriers and the other from control individuals. Each pooled RNA sample was reverse-transcribed, with Cy3 fluorescent tags, into labeled cDNA. The Cy3-labeled AT carrier cDNA and the Cy3-labeled control cDNA were separately hybridized with a common Cy5-labeled reference cDNA onto microarrays containing ∼3,000 human genes/ESTs. Thus, there were two kinds of hybridization (AT carriers and control individuals) for each of the five time points. All the hybridizations were carried out with four replicates. The replicate measurements were treated, as described in the “Baseline Expression Differences” subsection, by removing the most deviant observations and performing the analysis on the remaining three measurements.
Our goal was to identify the genes that differ most between AT carriers and control individuals in expression levels at several time points after IR. This is done to reveal the biological differences between AT carriers and control individuals, not for classification purposes. The discriminant-analysis procedure allows us to replace the set of five observations on each gene by a single discriminant score; this score is used to assign the expression pattern of a gene to the carrier or the control group. Under our null hypothesis, we expect only random differences between the expression patterns of a gene tested in the two cell types. Thus, when tested in AT carriers, the expression pattern of a gene would have a 50% chance of being correctly assigned as carrier; when the same gene is tested in cells, the expression pattern again would have a 50% chance of being correctly assigned as normal. The random chance of correct assignment of the expression pattern in both cases together is 25%.
We analyzed only genes that gave three or four “valid” replicate observations at all five time points in both cell types. In the first experiment, 1,382 cDNA clones met this criterion. Among these, the discriminant score (with cross-validation) correctly assigned 442 clones (32%) in both the AT carriers and control individuals; this is significantly higher than the 346 clones (25%) expected (χ2=36; P≪.001).
To follow up these results, we performed a replication experiment with only the 442 clones identified in the first experiment; we studied pools of RNA from five new AT carriers and six new control individuals. Among the 442 clones, there were 377 with valid observations (at least three replicates) at five time points in both cell types. Among these 377 clones, 183 (48.5%) were assigned correctly in both groups, significantly more than the 94 clones (25%) expected under the null hypothesis (χ2=111; P≪.001). Thus, the expression differences between carriers and control cells resulted in correct classification of approximately twice as many genes as expected by chance.
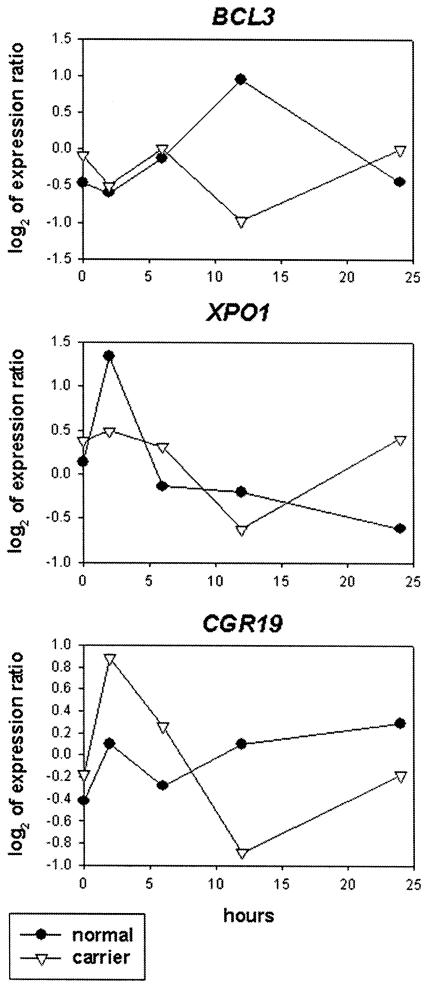
Thus, by two successive rounds of classifications, we identified 183 clones whose expression levels in AT carriers over five time points after IR are different from the levels in control individuals. Among these 183 cDNA clones, there were 101 uncharacterized ESTs and 82 known genes. When the 82 known genes were grouped into Gene Ontology categories (Ashburner et al. 2000), the two largest categories were cell growth and maintenance (25 genes; e.g., B-cell lymphoma 3 [BCL3], exportin 1 [XPO1], and cell growth regulator 19 [CGR19]) and signal transduction (15 genes; e.g., interferon receptor α2, jagged 2, IκB kinase complex–associated protein). Compared with the proportions of genes in these categories present on the array (11% in cell growth and maintenance, 13% in signal transduction), the proportions observed are higher (P<.001) for cell growth and maintenance. For illustration, we show the expression profiles of three genes (BCL3, XPO1, and CGR19) from the cell growth and maintenance category (fig. 2).
Figure 2.
Time course of gene expression after exposure to IR (3 Gy). Expression levels were determined in pooled RNA from lymphoblastoid cells from seven carriers and from six control individuals.
We expected some correct classification by chance (false positives). We ranked the 183 genes by the differences in discriminant scores between AT carriers and control individuals in the first and the replication studies. The top-ranked genes are most likely to be true positives. Among the 183 genes, the 10 known genes with the largest differences in discriminant scores between AT carriers and control individuals are listed in order in table 2. The complete ranked list of 183 genes/ESTs is available at the authors' Web site under “IR responses.”
Table 2.
Known Genes with the Largest Expression Differences between AT Carriers and Control Individuals in Post-IR Time Course[Note]
| Gene | Gene Name | Functiona |
| KLK5 | Kallikrein 5 | Protease |
| COL15A1 | Collagen, type XV, alpha 1 | Connective tissue development and maintenance |
| BCL3 | B-cell CLL/lymphoma 3 | Cell cycle control |
| NRP2 | Neuropilin 2 | Receptor for semaphorins |
| KIAA0993 | Unknown | |
| XPO1 | Exportin 1 | Cell cycle–regulated nuclear export |
| IFNAR2 | Interferon receptor 2 | Signal transduction |
| CGR19 | Cell growth regulatory with ring finger domain | Negative control of cell proliferation |
| MLF2 | Myeloid leukemia factor 2 | Cell growth |
| PLCB2 | Phospholipase C, β2 | Hydrolyzes phosphatidylinositol 4,5-bisphosphate |
Note.— Genes are ranked by difference in discriminant scores.
Determined using LocusLink of the National Center for Biotechnology Information.
Independent confirmation of the microarray results was obtained by quantitative RT-PCR for 5 of the 183 genes: BCL3, CCN1, CHEK1, COL15A1, and DAPK. For each gene, we calculated the correlation coefficient for the corresponding observations from microarrays and RT-PCR on cDNA from carriers and from control individuals. There was good agreement between the data from microarray and RT-PCR analysis. The average of the correlation coefficient was 0.76, with a range of 0.63–0.95.
Discussion
Autosomal recessive diseases that result from single mutations can have many, apparently unrelated, manifestations (pleiotropy). However, there is usually no marked heterozygote phenotype that permits identification of individual heterozygous carriers of a recessive disease. Patients with AT, who have mutations in both copies of the ATM gene, have a wide variety of manifestations, from neurodegeneration and immune deficiency to malignancies. AT carriers as a group, who constitute ∼1% of the population, have been found in some epidemiological studies to share a specific phenotype: increased cellular radiosensitivity and risk of cancer. However, these differences do not reliably distinguish individuals as carriers. In the present study, we provide evidence that the phenotype extends to differences in expression of many genes, both at baseline and in response to low-dose IR. DNA microarrays have been used successfully by others (Golub et al. 1999; Alizadeh et al. 2000; Bittner et al. 2000; Perou et al. 2000) to classify somatic mutations in cancers; here, we demonstrate their power in classifying individual carriers of recessive germline mutations. Our results for baseline expression levels suggest that carriers of mutations for other autosomal recessive diseases might be identified by the same approach.
The causes and pathogenesis of malignancies in AT carriers are poorly understood. A major obstacle has been the lack of methods for reliably identifying carriers in the population. In our analysis of baseline gene expression, we found significantly more genes than expected by chance (71 vs. 27) that differed between lymphoblastoid cell lines from AT carriers and from control individuals. The expression levels of these genes individually are not very different between AT carriers and control individuals. This is not surprising, since the differences between AT carriers and control individuals are subtle. However, when the genes are considered in aggregate, their expression levels are significantly different (P⩽.005) between AT carriers and control individuals. The existing evidence that AT carriers have elevated risk for breast cancer comes only from epidemiological studies, since AT carriers cannot be identified individually by available diagnostic tests or physical examinations. Here, we showed that most of the discrimination between our present samples of AT carriers and control individuals could be achieved using just four (LIM, CDKN2D, TFRC, and ARF6) of the 71 genes.
For future studies, we do not propose that expression levels of these four genes alone will allow reliable classification of all AT carriers, since our results are based on small samples. However, to the extent that our results are representative, the baseline expression levels of a small number of the genes will allow the identification of the majority of carriers of the ATM mutations. The present results are based on relatively small samples of carriers and control individuals. For this reason, and because of the known heterogeneity of AT mutations, the present study will need to be replicated to identify the set of genes that consistently provide discrimination. It will also be necessary to determine the accuracy of our expression assay in identifying AT carriers in the general population, where the frequency of AT carriers is much lower and where there are carriers of syndromes similar to AT. As a practical matter, it will be necessary to extend results to peripheral blood lymphocytes so that the expression assays can be performed using blood samples, thereby eliminating the need to prepare cell lines.
We designed our study to identify genes that account for the differences between AT carriers and control individuals in cellular response to IR. We identified 183 cDNA clones (101 ESTs and 82 known genes) that differ between AT carriers and control individuals in expression patterns over five time points after IR. A complete biological explanation will require functional analysis of these genes, which will help to explain why AT carriers are at increased risk of malignancies.
Cellular assays have shown that cells from AT carriers, unlike control cells, have incomplete cell cycle arrest in response to IR (West et al. 1995; Xu and Baltimore 1996; Barlow et al. 1999). Our data agree with this finding. Among the genes that have different expression patterns between the two cell types at baseline and in response to IR are several that play a role in proliferation, apoptosis, and cell cycle regulation. In the baseline study, we found that cyclin-dependent kinase inhibitor (CDKN2D), which regulates G1-to-S transition by controlling the phosphorylation of RB1, and WEE1, which regulates G2-to-M transition, were both decreased in AT carriers relative to control individuals. This finding suggests that AT carriers may be less effective in regulation of the cell cycle at various checkpoints when compared with control individuals. In the IR study, the expression level of exportin 1, which mediates nuclear export of cyclin B, is up-regulated in control cells in response to IR, compared with cells from AT carriers. In addition, several genes involved in DNA repair, such as BRCA2, are up-regulated in control cells, compared with AT carriers. This also supports the observation that cells from AT carriers are less efficient when compared with cells from control individuals in response to IR-induced DNA damage. ATM protein is a protein tyrosine kinase involved in early steps of response to DNA damage. Thus it is perhaps not surprising to find that mutations in one copy of the ATM gene can cause a plethora of changes in the genes downstream of ATM—with many of them being involved in the regulation of cell proliferation and cell death.
Since carriers of recessive diseases are usually several hundred times more common than affected patients, there is considerable practical significance in understanding how they differ from control individuals. This point is illustrated with AT. Clinically, it is important to develop methods for identifying AT carriers and to understand their radiosensitivity. Previous studies have indicated that IR used in diagnosis and treatment may trigger the development of cancer in AT carriers (Swift et al. 1986, 1991; Athma et al. 1996; Broeks et al. 2000). The dose of IR (3 Gy) used in the present study is relatively low. As a point for comparison, the maximum permissible exposure for IR workers is 5 Gy per year. Thus the differences detected in expression of relevant genes are not the result of unusually high levels of IR. This observation reinforces the need to take a more active role in identifying AT carriers, to minimize their risk of developing radiation-induced cancers. The use of expression analysis, to identify carriers of other recessive diseases and to improve understanding of their phenotype, is likely to have correspondingly extensive implications.
Acknowledgments
We thank Melissa Arcaro for sequencing the cDNA clones, and we thank Robert L. Nussbaum, Haig H. Kazazian, and an anonymous reviewer for advice and comments. This work is supported in part by the W. W. Smith Endowed Chair in Pediatric Genomics, Foerderer Funds from The Children’s Hospital of Philadelphia (to V.G.C.) and by National Institutes of Health grants DC00154 (to V.G.C.) and DK47481 and HG02386 (both to R.S.S.).
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- Gene Expression Profiling of Carriers of the ATM Mutation, http://genomics.med.upenn.edu/athet (for analysis of replicated microarrays)
- LocusLink, http://www.ncbi.nlm.nih.gov/LocusLink/ [Google Scholar]
- Method for multiple testing correction by Storey and Tibshirani, http://www-stat.stanford.edu/~jstorey/papers/dep.pdf
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AT [MIM 208900]) [PubMed]
- TIGR, http://www.tigr.org
- UCSC Genome Bioinformatics, http://genome.cse.ucsc.edu/
References
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, et al (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403:503–511 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athma P, Rappaport R, Swift M (1996) Molecular genotyping shows that ataxia telangiectasia heterozygotes are predisposed to breast cancer. Cancer Genet Cytogenet 92:130–134 [DOI] [PubMed] [Google Scholar]
- Barlow C, Eckhaus MA, Schaffer AA, Wynshaw-Boris A (1999) Atm haploinsufficiency results in increased sensitivity to sublethal doses of ionizing radiation in mice. Nat Genet 21:359–360 [DOI] [PubMed] [Google Scholar]
- Becker-Catania SG, Chen G, Hwang MJ, Wang Z, Sun X, Sanal O, Bernatowska-Matuszkiewicz E, Chessa L, Lee EY, Gatti RA (2000) Ataxia-telangiectasia: phenotype/genotype studies of ATM protein expression, mutations and radiosensitivity. Mol Genet Metab 70:122–133 [DOI] [PubMed] [Google Scholar]
- Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, et al (2000) Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature 406:536–540 [DOI] [PubMed] [Google Scholar]
- Broeks A, Urbanus JH, Floore AN, Dahler EC, Klijn JG, Rutgers EJ, Devilee P, Russell NS, van Leeuwen FE, van’t Veer LJ (2000) ATM-heterozygous germline mutations contribute to breast cancer-susceptibility. Am J Hum Genet 66:494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow MJ, Dudoit S, Gong EL, Speed TP, Rubin EM (2000) Microarray expression profiling identifies genes with altered expression in HDL-deficient mice. Genome Res 10:2022–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Birkholtz GG, Lindblom P, Rubio C, Lindblom A (1998) The role of ataxia-telangiectasia heterozygotes in familial breast cancer. Cancer Res 58:1376–1379 [PubMed] [Google Scholar]
- Concannon P, Gatti RA (1997) Diversity of ATM gene mutations detected in patients with ataxia-telangiectasia. Hum Mutat 10:100–107 [DOI] [PubMed] [Google Scholar]
- FitzGerald MG, Bean JM, Hegde SR, Unsal H, MacDonald DJ, Harkin DP, Finkelstein DM, Isselbacher KJ, Haber DA (1997) Heterozygous ATM mutations do not contribute to early onset of breast cancer. Nat Genet 15:307–310 [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286:531–537 [DOI] [PubMed] [Google Scholar]
- Heald R, McLoughlin M, McKeon F (1993) Human WEE1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated Cdc2 Kinase. Cell 74:463–474 [DOI] [PubMed] [Google Scholar]
- Juan LJ, Shia WJ, Chen MH, Yang WM, Seto E, Lin YS, Wu CW (2000) Histone deacetylases specifically down-regulate p53-dependent gene activation. J Biol Chem 275:20436–20443 [DOI] [PubMed] [Google Scholar]
- Lee ML, Kuo FC, Whitmore GA, Sklar J (2000) Importance of replication in microarray gene expression studies: statistical methods and evidence from repetitive cDNA hybridization. Proc Natl Acad Sci USA 97:9834–9839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu VF, Weaver DT (1993) The ionizing radiation-induced replication protein A phosphorylation response differs between ataxia telangiectasia and normal human cells. Mol Cell Biol 13:7222–7231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly BJF (1997) Randomization, bootstrap and Monte Carlo methods in biology. 2nd ed. Chapman & Hall, London [Google Scholar]
- McLaughlin MM, Kumar S, McDonnell PC, Van Horn S, Lee JC, Livi GP, Young PR (1996) Identification of mitogen-activated protein (MAP) kinase-activated protein kinase-3, a novel substrate of CSBP p38 MAP kinase. J Biol Chem 271:8488–8492 [DOI] [PubMed] [Google Scholar]
- Minamoto S, Ikegame K, Ueno K, Narazaki M, Naka T, Yamamoto H, Matsumoto T, Saito H, Hosoe S, Kishimoto T (1997) Cloning and functional analysis of new members of STAT induced STAT inhibitor (SSI) family: SSI-2 and SSI-3. Biochem Biophys Res Comm 237:79–83 [DOI] [PubMed] [Google Scholar]
- Newton MA, Kendziorski CM, Richmond CS, Blattner FR, Tsui KW (2001) On differential variability of expression ratios: improving statistical inference about gene expression changes from microarray data. J Comput Biol 8:37–52 [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumours. Nature 406:747–752 [DOI] [PubMed] [Google Scholar]
- Roberts DD (1996) Regulation of tumor growth and metastasis by thrombospondin-1. FASEB J 10:1183–1191 [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S (1995) A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 268:1749–1753 [DOI] [PubMed] [Google Scholar]
- Swift M, Morrell D, Cromartie E, Chamberlin AR, Skolnick MH, Bishop DT (1986) The incidence and gene frequency of ataxia-telangiectasia in the United States. Am J Hum Genet 39:573–583 [PMC free article] [PubMed] [Google Scholar]
- Swift M, Morrell D, Massey RB, Chase CL (1991) Incidence of cancer in 161 families affected by ataxia-telangiectasia. N Engl J Med 325:1831–1836 [DOI] [PubMed] [Google Scholar]
- Swift M, Sholman L, Perry M, Chase C (1976) Malignant neoplasms in the families of patients with ataxia telangiectasia. Cancer Res 36:209–215 [PubMed] [Google Scholar]
- Telatar M, Wang Z, Udar N, Liang JT, Bernatowska-Matuszkiewicz E, Lavin M, Shiloh Y, Concannon P, Good RA, Gatti RA (1996) Ataxia-telangiectasia: mutations in ATM cDNA detected by protein truncation screening. Am J Hum Genet 59:40–44 [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle S, Stamato T, Perez ML, Biaglow J (2000) Glucose-6-phosphate dehydrogenase and the oxidative pentose phosphate cycle protect cells against apoptosis induced by low doses of ionizing radiation. Radiat Res 153:781–787 [DOI] [PubMed] [Google Scholar]
- Ueno M, Masutani H, Arai RJ, Yamauchi A, Hirota K, Sakai T, Inamoto T, Yamaoka Y, Yodoi J, Nikaido T (1999) Thioredoxin-dependent redox regulation of p53-mediated p21 activation. J Biol Chem 274:35809–35815 [DOI] [PubMed] [Google Scholar]
- West CM, Elyan SA, Berry P, Cowan R, Scott D (1995) A comparison of the radiosensitivity of lymphocytes from normal donors, cancer patients, individuals with ataxia telangiectasia and A-T heterozygotes. Int J Radiat Biol 68:197–203 [DOI] [PubMed] [Google Scholar]
- Wright J, Teraoka S, Onengut S, Tolun A, Gatti RA, Ochs HD, Concannon P (1996) A high frequency of distinct ATM gene mutations in ataxia-telangiectasia. Am J Hum Genet 59:839–846 [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Baltimore D (1996) Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev 10:2401–2410 [DOI] [PubMed] [Google Scholar]