Abstract
The advent of next generation sequencing technologies is providing new insight into HIV-1 diversity and evolution, which has created the need for bioinformatics tools that could be applied to the characterization of viral quasispecies. Here we present Nautilus, a bioinformatics package for the analysis of HIV-1 targeted deep sequencing data. The DeepHaplo module determines the nucleotide base frequency and read depth at each position and computes the haplotype frequencies based on the linkage among polymorphisms in the same next generation sequence read. The Motifs module computes the frequency of the variants in the setting of their sequence context and mapping orientation, which allows for the validation of polymorphisms and haplotypes when strand bias is suspected. Both modules are accessed through a user-friendly GUI, which runs on Mac OS X (version 10.7.4 or later), and are based on Python, JAVA, and R scripts. Nautilus is available from www.hivresearch.org/research.php?ServiceID=5&SubServiceID=6.
Within an infected individual, HIV-1 viral populations can exhibit an enormous level of genetic diversity, which presents major obstacles for the sustained control of viral replication by host immune responses and antiretroviral treatments.1 Until recently, the molecular tools for the characterization of viral quasispecies were extremely arduous and costly.2 The advent of next generation sequencing (NGS) technologies, with their expanded sampling depth and capacity for automation,3 is providing new insight into viral diversity and evolution.4 The main experimental approaches of NGS have been whole genome sequencing,5 whole gene sequencing,6 and targeted deep sequencing (TDS).7–9 The latter examines a defined subgenomic region of interest at great sampling and sequencing depth to determine the frequency of the different variants. As the capacity to obtain longer reads has increased over the past years, it is now possible to accurately determine, rather than just infer, the linkage among measured polymorphisms. The quantity and the quality of the data generated in TDS experiments present major challenges for traditional analysis tools. Unfortunately, most of the existing NGS bioinformatics tools10,11 have been developed for the analysis of haploid or diploid organisms, preventing their seamless application to HIV-1 populations.
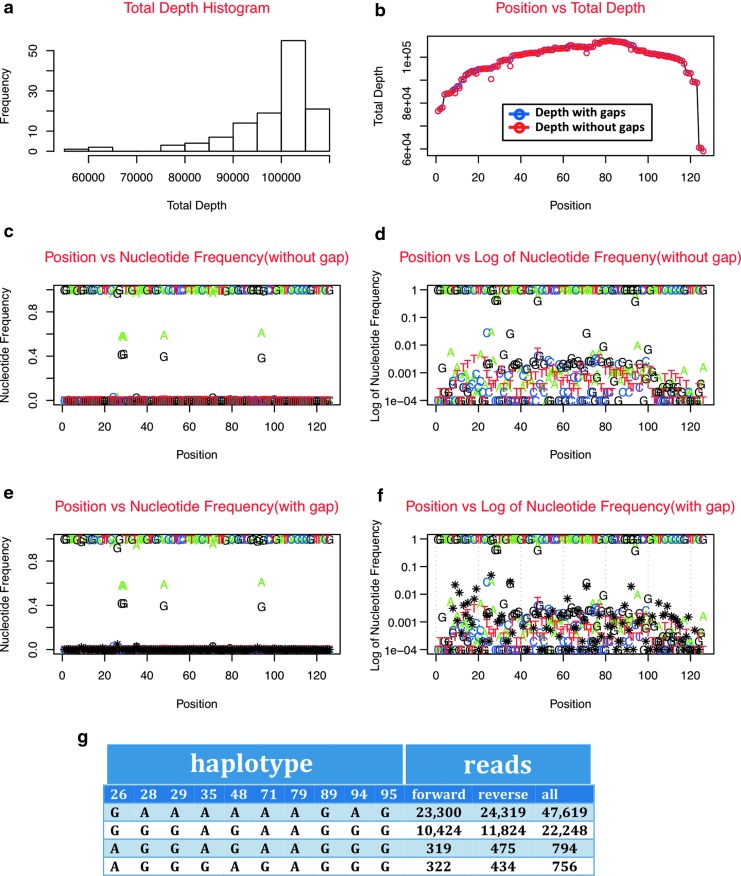
Here we present Nautilus, a bioinformatics package for the analysis of HIV-1 TDS data. The program consists of a graphical user interface (GUI) with two modules: DeepHaplo and Motifs. Using as an input an alignment file in the SAM format,10 DeepHaplo computes the nucleotide base frequency and read depth at each position, and presents the results in tabular and graphic formats (Fig. 1a–f). To facilitate the visualization of the different facets of the data, results are represented including or omitting alignment gaps, and in linear or logarithmic scales. A novel feature of DeepHaplo is the implementation of a hash algorithm (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/aid) to efficiently compute the frequency of haplotypes (i.e., polymorphisms that are present in the same NGS read). Positions of interest are either entered by the user or are identified by the software based on a user-defined threshold for minor-allele frequency (MAF) (Fig. 1g).
FIG. 1.
Read depth and frequencies of single nucleotide variants and haplotypes can be computed by the DeepHaplo module. (a) Histogram of the distribution of sequencing depth at each position. (b) Scatterplot of the sequencing depth at each position acknowledging or ignoring alignment gaps (blue and red symbols, respectively). The frequency of each variant at each position can be visualized either acknowledging (d, f) or ignoring alignment gaps (c, e) in linear (c, e) or logarithmic scales (d, f). (g) The frequency of haplotypes that involve linkage among polymorphisms in positions of interest is computed using a hash algorithm. The number of reads in each mapping orientation that support each haplotype can be used to discern true signals from sequencing artifacts.
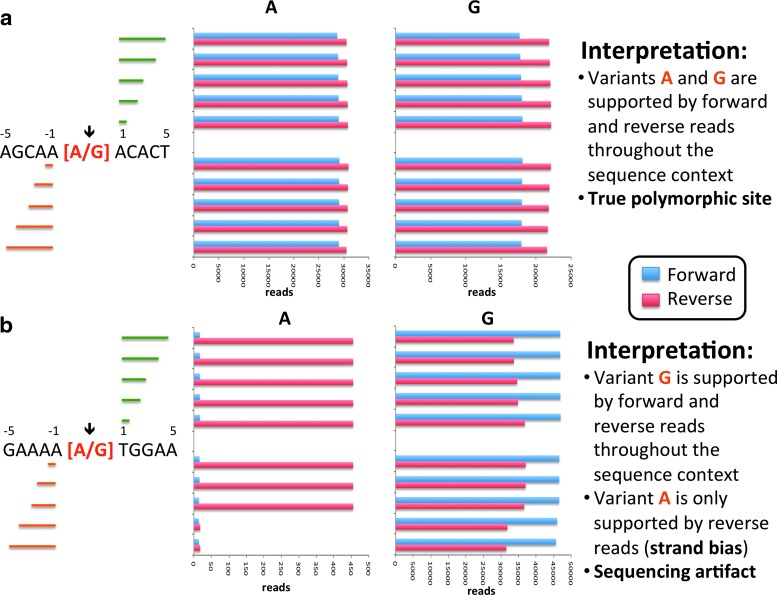
DeepHaplo uses the mapping orientation information provided in the bitwise FLAG value in the SAM file10 to compute the frequencies of nucleotide bases at each position and the haplotypes in each orientation. This feature, combined with the analysis of the Motifs module, allows the validation of polymorphisms and haplotypes when strand bias is suspected. In Motifs, interrogated positions are identified through a user-defined threshold for MAF, and the frequency of variants at each position is computed for the forward and reverse orientations. Motifs also calculates the number of forward and reverse reads supporting a given variant in the setting of the sequence context surrounding the candidate variant, as this has been shown to strongly influence strand bias (e.g., homopolymers).12 Figure 2a shows a real case of a polymorphic position where the variants are equally supported by reads in both orientations (compare the blue and red bars), whereas Fig. 2b shows that the A variant is observed only in reads in the reverse orientation, likely reflecting a sequencing artifact.
FIG. 2.
The Motifs module provides information about the frequency of single nucleotide variants based on mapping orientation and the sequence context surrounding the putatively polymorphic position. (a) Profile of a true polymorphic site. The detected variants along with the sequence context of the position are shown. Each cluster of bars in the chart represents the count of reads supporting the variant in question in each orientation (color coded) and context. For example, the top cluster depicts the number of forward and reverse reads supporting AACACT and GACACT, while the bottom cluster depicts the number of forward and reverse reads supporting AGCAAA and AGCAAG. Intermediate clusters indicate shorter sequence contexts. (b) Profile of a sequencing artifact due to strand bias. In this case, the G variant is supported by forward and reverse reads in various sequence contexts, whereas the A variant is supported only by reads from the reverse mapping orientation.
In summary, Nautilus represents a new suite of bioinformatics tools to support the analysis of TDS data in order to facilitate the application of NGS to the characterization of HIV-1 populations and evolution. Nautilus runs on Mac OS X (version 10.7.4 or later), and is based on Python, JAVA, and R scripts (required packages are stated in the accompanying user manual), and is freely available from www.hivresearch.org/research.php?ServiceID=5&SubServiceID=6.
Supplementary Material
Acknowledgments
This work was supported in part by an Interagency Agreement (Y1-AI-2642-12) between the U.S. Army Medical Research and Materiel Command and the National Institute of Allergy and Infectious Diseases. This work was also supported by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine and the U.S. Department of Defense.
The opinions expressed in this article are those of the authors and do not represent the official views of the U.S. Department of Health and Human Services, the National Institute of Allergy and Infectious Diseases, the U.S. Department of Defense, or the Department of the Army.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Coffin J. Swanstrom R. HIV pathogenesis: Dynamics and genetics of viral populations and infected cells. Cold Spring Harbor Perspect Med. 2013;3:a012526. doi: 10.1101/cshperspect.a012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shankarappa R. Margolick JB. Gange SJ, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glenn TC. Field guide to next-generation DNA sequencers. Mol Ecol Resour. 2011;11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 4.Beerenwinkel N. Gunthard HF. Roth V. Metzner KJ. Challenges and opportunities in estimating viral genetic diversity from next-generation sequencing data. Front Microbiol. 2012;3:329. doi: 10.3389/fmicb.2012.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henn MR. Boutwell CL. Charlebois P, et al. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 2012;8:e1002529. doi: 10.1371/journal.ppat.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischer W. Ganusov VV. Giorgi EE, et al. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS One. 2010;5:e12303. doi: 10.1371/journal.pone.0012303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsibris AM. Korber B. Arnaout R, et al. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One. 2009;4:e5683. doi: 10.1371/journal.pone.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kijak G. Sanders-Buell E. Rolland M, et al. AIDS Vaccine. P05.06. Boston, MA: 2012. 2012. Incident cases characterization, deep sequencing provide new insight into multiplicity of infection, HIV evolution in very early acute infection. [Google Scholar]
- 9.Shao W. Boltz VF. Spindler JE, et al. Analysis of 454 sequencing error rate, error sources, and artifact recombination for detection of low-frequency drug resistance mutations in HIV-1 DNA. Retrovirology. 2013;10:18. doi: 10.1186/1742-4690-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H. Handsaker B. Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;5:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Fernandez H. Glez-Pena D. Reboiro-Jato M, et al. PileLineGUI: A desktop environment for handling genome position files in next-generation sequencing studies. Nucleic Acids Res. 2011;39:W562–566. doi: 10.1093/nar/gkr439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balzer S. Malde K. Jonassen I. Systematic exploration of error sources in pyrosequencing flowgram data. Bioinformatics. 2011;27:i304–309. doi: 10.1093/bioinformatics/btr251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.