Abstract
Pulsed electron paramagnetic resonance at the microwave Ka band (~30 GHz) was used to study the coordination of adenosine nucleotides to Mn2+ at the active site of myosin ATPase and in solution. We have found that the electron spin echo (ESE) field sweep, electron–nuclear double resonance (ENDOR) and ESE envelope modulation (ESEEM) techniques are not sufficiently specific for reliable differentiation between the solvated and myosin-bound Mn·nucleotide complexes. Therefore, to directly detect binding of the Mn·nucleotide to myosin, we used nonhydrolizable nucleotide analogs, site-directed spin labeling, and pulsed electron–electron double resonance to detect spin probe–manganese dipolar interaction. We found that under substoichiometric conditions, both Mn·AMPPNP and Mn·ADP·AlF4 form a complex with myosin, and Mn·ADP does not form such a complex. This correlates well with the biological dissociation of Mg·ADP from myosin after the hydrolysis of ATP. The analysis of 31P ENDOR spectra reveals that in Mn·AMPPNP, Mn·ATP, and Mn·ADP at myosin or in solution, the nucleotide is coordinated to Mn2+ by two phosphate groups, whereas in Mn·ADP·AlF4, only one phosphate group is coordinated. The observation of two phosphates and one nitrogen in the coordination sphere of Mn·ADP in solution by ESEEM spectroscopy suggests that a significant population of Mn ions is coordinated by two ADP molecules, one of which is coordinated by phosphates, and the other one, by a nitrogen atom. The developed approach will be generally useful for monitoring the metal–protein binding when such binding does not provide reliable spectroscopic signatures.
INTRODUCTION
Myosin is a molecular motor producing force in muscle. It works cyclically, interacting with two ligands, actin and ATP, and changing conformation two times per cycle. When ATP binds to actomyosin (myosin is in the post power stroke structural state), actomyosin dissociates, and myosin performs the recovery stroke and is ready for subsequent actin binding. It was proposed that ATP hydrolysis fuels the myosin conformational change, but recent kinetics studies1,2 place ATP hydrolysis after the recovery stroke, which means that ATP hydrolysis does not fuel the recovery stroke in myosin. Actin binds to myosin after ATP hydrolysis,3 and its binding activates the power stroke. Thus, ATP hydrolysis must occur before the actin binding and the power stroke, and it does not fuel the power stroke, either. What, then, is the role of ATP hydrolysis in myosin and other ATPases? Does ATP hydrolysis fuel the molecular motor, or is it only needed to remove the nucleotide from the active site? How is ATP hydrolysis coupled to the mechanical cycle of a myosin motor?
To study the details of ATP hydrolysis at the myosin active site, a reporting probe should be incorporated in ATP or nearby. Unfortunately, fluorescent and spin-labeled ATP analogs are sensitive only to the binding to the active site,4,5 not to the hydrolysis. Tryptophan residues introduced in the vicinity of the myosin ATP binding site do not reflect the hydrolysis event.6 An alternative approach is based on using paramagnetic Mn2+ instead of diamagnetic Mg2+ that is naturally complexed with ATP and applying electron paramagnetic resonance (EPR) spectroscopy to examine the coordination environment of the Mn2+ ion. Previously, EPR has been used to study Mn2+ coordination in various biological environments, and a substitution of Mg2+ by Mn2+ has been employed to study the structure of the metal binding sites in myosin,7,8 creatine kinase,9 F1 ATPase,10,11 etc. These studies have demonstrated that Mn2+ EPR can provide the structural details required for monitoring ATP hydrolysis; in particular, it can detect the interactions with the coordinated phosphates of the nucleotide.
At lower-frequency microwave (mw) bands (below W-band, ~ 90 GHz), the continuous wave (CW) EPR spectrum of Mn2+ is significantly broadened by the crystal field interactions (cfi) and, in most cases, does not reveal the hyperfine interactions (hfi) with the ligand nuclei. Therefore, the most informative EPR studies were done using high-resolution pulsed EPR techniques of electron–nuclear double resonance (ENDOR) and electron spin echo (ESE) envelope modulation (ESEEM).10–16 To our knowledge, however, such high resolution studies have never been performed for myosin.
In this work, we performed a pulsed EPR investigation of the myosin·Mn·nucleotide complexes with adenosine nucleotides, ADP, ADP·AlF4, and AMPPNP. The experiments were performed in the mw Ka band (~30 GHz) to reduce the detrimental effects of cfi on the ESEEM and ENDOR spectra.17–22 The main purpose of this investigation was to develop strategies for (a) determination of the number of phosphate groups, coordinated by manganese in the myosin·Mn·nucleotide complex, and (b) monitoring the myosin·Mn·nucleotide complex formation. The first objective was achieved by applying the pulsed ENDOR technique and referencing the normalized 31P ENDOR amplitudes to the known atomic structures obtained by the X-ray crystallography.23,24 The second aim was addressed by using a spin-labeled myosin construct and applying the pulsed electron–electron double resonance (ELDOR) technique. It was shown that the monitoring of the nucleotide binding to myosin ATPase is important for meaningful data analysis and interpretation. For example, Mn·ADP exhibits a significant 14N ESEEM and does not form a stable complex with myosin under substoichiometric conditions, which are essential in our experiments for unambiguous interpretation of the results. Knowing that the myosin·Mn·ADP complex is not formed, one can rule out the interpretation of the 14N ESEEM in terms of unusual myosin·nucleotide interaction and conclude that in solution, Mn coordinates two ADP molecules, one of them via the adenosine nitrogen.
MATERIALS AND METHODS
Chemicals
ATP, ADP, AMPPNP, maleimide spin label (MSL), AlCl3, and NaF were obtained from Sigma-Aldrich (Milwaukee, WI). All chemicals were of reagent grade. The chemical structures of the nucleotides are given in the Supporting Information.
Protein Preparation and Labeling
A639C:K498C double Cys mutant of myosin motor domain was expressed in Dictyostelium discoudeum and purified as described previously.25 The protein was labeled with MSL by incubating myosin with a 4-fold excess of MSL for 12 h on ice. Labeling buffer contained 20 mM MOPS and 50 mM KCl, pH 7.5. Unreacted excess MSL was washed out with size-exclusion spin columns (Pierce, Rockford IL) in the buffer containing 20 mM 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid, pH 8.0 (experimental buffer). The labeled myosin was then centrifuged for 20 min at 100 000g, T = 4 °C, to remove aggregated myosin, and concentrated to 300–500 μM with Microcon concentrators (Millipore, Billerica MA) for preparation of myosin·Mn·nucleotide complexes. The concentration of unlabeled myosin was determined spectrophotometrically at 280 nm using an extinction coefficient of 0.69 (mg/mL)−1 cm−1. The concentration of the labeled myosin was determined by Bradford assay. The final myosin concentration was 140–412 μM in EPR experiments, 2.8 μM in Mn2+–ATPase assay, and 8.4 μM in Mg2+–ATPase assay.
Preparation of Mn·Nucleotide Complexes
Mn2+–nucleotide (ADP, ATP, or AMPPNP) complexes were obtained by mixing MnCl2 with the corresponding nucleotide at a 1:10 Mn/nucleotide ratio in the experimental buffer at 20 °C, then 50% glycerol was added, and the solution was loaded into the EPR tube and flash-frozen in liquid nitrogen. The final concentration of Mn was 0.5–1.25 mM.
Preparation of Myosin·Mn·Nucleotide Complexes
Complexes of myosin with nucleotides were obtained as described previously26 by incubation of myosin with premixed Mn·nucleotide (1:0.8, myosin/Mn ratio; 1:10 Mn/nucleotide ratio) for 5 min at 20 °C. A substoichiometric Mn/myosin ratio was chosen to maximize the signal from myosin·Mn·nucleotide complex and minimize the signal of the Mn·nucleotide in solution. Glycerol (40%) was added to the sample, and the solution was loaded into the EPR tube and flash-frozen in liquid nitrogen. The complex of myosin with Mn·ADP·AlF4 (myosin·Mn·ADP·AlF4) was obtained by incubation of the myosin·Mn·ADP solution with 20 mM NaF for 5 min at 20 °C and subsequent incubation with 5 mM AlCl3 for 25 min at 20 °C. Then 40% glycerol was added, and the solution was loaded into the EPR tube and flash-frozen in liquid nitrogen.
ATPase Assays
Myosin ATPase activity was monitored by measuring the release of orthophosphate at 25 °C in 10 mM Tris and 5 mM ATP, pH 7.5, in the presence of 5 mM MgCl2 or 5 mM MnCl2 using a malachite green-enhanced phosphate assay.27 Myosin MnATPase activity was 3.6 times as high as that of MgATPase (0.86 s−1 vs 0.24 s−1).
Pulsed EPR Experiments
The pulsed EPR experiments were performed on the Ka-band pulsed EPR spectrometer of the University of Arizona28 at a mw frequency of ~30 GHz. The measurement temperature was 15 K. The detailed experimental conditions are given in the figure captions.
RESULTS AND DISCUSSION
Number of Coordinated Phosphates in Mn·Nucleotide and Myosin·Mn·Nucleotide Complexes
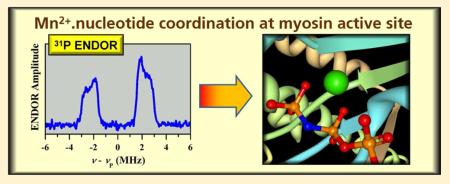
All of the Mn·nucleotide and myosin·Mn·nucleotide complexes studied in this work were characterized with two-pulse ESE field sweeps, ESEEM, and pulsed ENDOR, with the main purpose of detecting the coordinated nucleotide phosphates. As an example, Figure 1a shows the field sweep spectra of myosin·Mn·AMPPNP (trace 1) and Mn·AMPPNP (trace 2). Trace 1 in Figure 1b shows the expanded central part of the spectrum of myosin·Mn·AMPPNP. Similar spectra were obtained for other studied complexes. The spectra show a characteristic pattern of six relatively narrow lines of |−1/2⟩ ↔ | 1/2⟩ transitions of the Mn2+ spin S = 5/2. The splitting between these lines (~9.5 mT) is caused by the isotropic hfi of the Mn nucleus (I = 5/2), and their broadening results from second-order effects of the cfi. Other transitions of the Mn electronic spin (| ± 3/2⟩ ↔ | ± 1/2⟩, | ± 5/2⟩ ↔ | ± 3/2⟩) are broadened by the cfi to first order and form a broad, featureless bell-shaped line that has a characteristic width of ~150 mT and is located underneath the six narrow lines of |−1/2⟩ ↔ |1/2⟩ transitions (Figure 1a). Spectral simulations, accounting for only the quadrupolar terms of the cfi (D and E), result in D/gβ ~ 25 mT.
Figure 1.
Panel a, two-pulse ESE field sweep spectra of myosin·Mn·AMPPNP (trace 1) and Mn·AMPPNP (trace 2). Panel b, central part of two-pulse ESE field sweep spectra of myosin·Mn·AMPPNP (trace 1) and MSL·myosin·Mn·AMPPNP (trace 2). Experimental conditions: νmw = 30.108 GHz; mw pulses, 10 and 15 ns; time interval between the mw pulses, τ = 200 ns; boxcar integration gate, 15 ns; temperature, 15 K. The resonance positions of the pumping (νpmp) and observation (νobs) mw frequencies as used in pulsed ELDOR experiments are indicated.
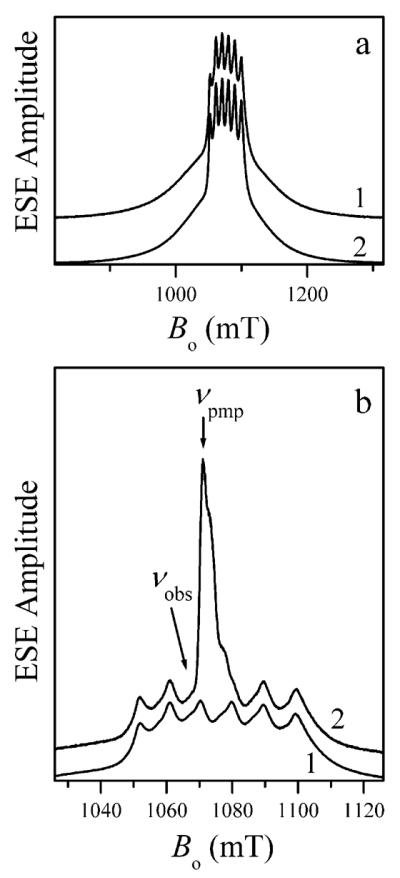
In most cases, the field sweep spectra did not reveal any interactions with the ligand nuclei, with the notable exception of the myosin·Mn·ADP·AlF4 sample, in which the lines of the |−1/2⟩ ↔ |1/2⟩ transitions exhibited a ~2 mT doublet splitting due to the hfi with the 19F ligand nucleus (see Figure 2).
Figure 2.
The central part of the two-pulse ESE field sweep spectrum of myosin·Mn·ADP·AlF4 showing the 19F hfi splittings of the Mn transition lines. Experimental conditions: νmw = 30.136 GHz; mw pulses, 80 and 160 ns; time interval between the mw pulses, τ = 400 ns; boxcar integration gate, 150 ns; temperature, 15 K.
The Ka-band, two-pulse ESEEM experiments (discussed below) have shown mostly weak 1H and 14N ESEEM. The 31P ESEEM was very weak (mostly the sum combination line at the frequency of ~36 MHz was observed), and we have therefore concluded that the Ka band ESEEM is not sufficiently sensitive to coordinated phosphates. Decreasing the mw frequency makes the 31P ESEEM more observable,10,11 but this can also generate interpretational problems resulting from the increase in the relative significance of the Mn2+ cfi.17,19–22
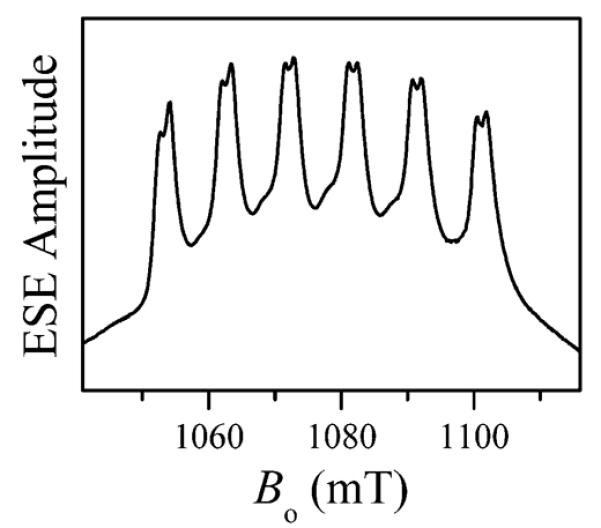
As an alternative, we have used the refocused Mims ENDOR technique,29–32 which proved successful in detecting the nearby 31P nuclei. The 31P ENDOR spectra are shown in Figure 3. The spectra consist of two lines located symmetrically with respect to the 31P Zeeman frequency, νP (νP ~ 18.5 MHz at Bo ~ 1070 mT). The average splitting between these lines, Δν ~ 4–5 MHz, represents an effective hfi constant, aPef, which corresponds to the true one-electron hfi constant aP = 2S·Δν ~ 20–25 MHz (the scaling factor of 2S represents the ratio of the total and one-electron SZ; see the Supporting Information). The multiplication by 2S implies that only one of the unpaired electrons residing in the Mn2+ d orbitals and forming the S = 5/2 multiplet is responsible for this hfi constant. The hfi constant on the order of tens of megahertz indicates that the corresponding phosphate group is coordinated to the Mn2+ ion. Similar 31P hfi constants were earlier observed for the phosphate groups coordinated to the Mo(V) center of sulfite oxidase33,34 and in the complexes with oxovanadium.16,35,36 The 31P hfi constants in Mn2+-coordinated nucleotides obtained earlier for different systems at X band10,11 are also similar, although the reported values were not corrected for the spin projection factor.
Figure 3.
Solid traces, refocused Mims ENDOR spectra of myosin·Mn·AMPPNP (trace 1), Mn·AMPPNP (trace 2), Mn·ATP (trace 3), Mn·ADP (trace 4), and myosin·Mn·ADP·AlF4 (trace 5). The spectra were normalized by the ESE signal amplitudes without RF. Experimental conditions: νmw = 30.108 GHz (traces 1–4) and 30.136 GHz (trace 5); Bo = 1070.3 mT (traces 1–4) and 1072.8 mT (trace 5); mw pulses, 8, 8, 8, and 14 ns; time interval between the first and second mw pulses, τ = 80 ns; time interval between the second and third mw pulses, T = 25 μs; time interval between the third and fourth (refocusing) pulses, t = 380 ns; RF pulse, 22 μs; temperature, 15 K. Dashed traces superimposed on the solid traces are simulated with parameters shown in Table 1.
The structure of the 31P ENDOR lines (the peaks at smaller splittings and the shoulders at larger splittings) observed in the spectra of myosin·Mn·AMPPNP and myosin·Mn·ADP·AlF4 (traces 1 and 5 in Figure 3, respectively) is most likely produced by the anisotropic hfi. A numerical simulation of the ENDOR spectrum of myosin·Mn·AMPPNP (assuming an axial hfi tensor) results in the effective isotropic hfi constant aPef = 4.3 MHz (corresponds to the true aP = 2S·aPef = 21.5 MHz) and the anisotropic hfi constant T⊥ = −0.9 MHz. In the point dipole approximation, this anisotropic hfi constant corresponds to the Mn–P distance of ~3.3 Å, in good agreement with the distances of 3.2–3.3 Å determined from the corresponding crystal structures of myosin.23,24 This agreement, however, is most likely coincidental, because it does not take into account the spin density delocalization on the phosphate ligand (see the Supporting Information). The simulated ENDOR spectrum of myosin·Mn·AMPPNP as well as the simulated spectra of other samples are shown by the dashed traces in Figure 3. These simulations did not account for the effects of the cfi17,18 because at the Ka band, the cfi is much weaker than the electronic Zeeman interaction (D/gβBo ~ 1/40). The results are summarized in Table 1.
Table 1.
Hyperfine Interaction Parameters of 31P Nuclei in Phosphate Ligands Obtained by Numerical Simulations of the Pulsed ENDOR Spectra Recorded for the Samples of Mn2+ with Various Nucleotides, with or without Myosina
| sample | aP (MHz) | T⊥ (MHz) | ΔaP (MHz) |
|---|---|---|---|
| myosin·Mn·AMPPNP | 21.5 | −0.9 | 3.5 |
| Mn·AMPPNP | 20.5 | −0.9 | 8 |
| Mn·ATP | 22 | −0.9 | 8 |
| myosin·Mn·ADP and Mn·ADP | 26 | −0.9 | 14 |
| myosin·Mn·ADP·AlF4 | 26.5 | −0.9 | 4 |
See Figure 3. ΔaP is the width of the Gaussian distribution of aP between the maximum slope points. The true values of aP and ΔaP are obtained from the respective effective values (that would directly correspond to the ENDOR splittings) by multiplication by 2S = 5, as explained in the text.
The 31P lines in the spectra of Mn·AMPPNP, Mn·ATP, Mn·ADP (traces 2–4 in Figure 3, respectively), unlike those detected for myosin·Mn·AMPPNP and myosin·Mn·ADP·AlF4, are essentially featureless and resemble simple Gaussian lines. This loss of detail is explained by the greater degree of structural disorder in the phosphate coordination in solution, which leads to a more broad statistical distribution of the hfi constants, aP. Interestingly, the spectra of Mn·ADP (trace 4) and myosin·Mn·ADP (not shown) are identical, which raises a question about the Mn·ADP's binding to myosin when Mn·ADP is in substoichiometric amounts.
The spectra shown in Figure 3 are normalized by the ESE signal amplitudes without RF. The purpose of such a normalization is to enable the comparison of the 31P line amplitudes in different samples to estimate the relative number of coordinated phosphates. This comparison is not straightforward, however, because the linewidths in the ENDOR spectra of different samples are different. The hfi constants are also somewhat different (see Table 1), which results in the necessity to take into account the amplitude scaling factor of the Mims ENDOR, (1-cos(2πAτ))/2,29,30 where A = aPef + TZZ, and TZZ is the diagonal part of the anisotropic hfi. Counterbalancing these minor complications, there is a simplifying consideration that in the weak hfi limit (νP ⪢ aPef/2) the nuclear transition probabilities under the RF excitation are to first order independent of the hfi constant or the orientation of the hfi tensor with respect to the magnetic field.
Therefore, to obtain the relative number of coordinated phosphate groups, the normalized ENDOR spectra were corrected for the Mims amplitude scaling factor and then integrated. A comparison of the integrals has revealed that the number of coordinated phosphates for all of the samples except myosin·Mn·ADP·AlF4 is the same. In myosin·Mn·ADP·AlF4, the number of coordinated phosphates is one-half that obtained for other nucleotides.
A similar approach to determine the number of nearby magnetic nuclei (1H, 13C, 17O) was previously used for Gd3+ complexes.12,37,38 The dramatic differences in the cfi of different Gd3+ complexes in those works have necessitated a somewhat more elaborate procedure based on obtaining the separate ENDOR spectra for the |−1/2⟩ ↔ |1/2⟩ transition of the Gd3+ spin S = 7/2 and then normalizing these spectra by the ESE amplitude associated with this transition. In this work, a simpler normalization by the total ESE signal (contributed to by all of the Mn spin transitions) is used because the cfi in the studied complexes are of the same order of magnitude, and near the center of the EPR spectrum, the variations of the relative contributions of the narrow and broad lines are only ~10%.
We found that the number of coordinated phosphates in myosin·Mn·AMPPNP is two times larger than in myosin·Mn·ADP·AlF4. This finding is in agreement with the X-ray crystallographic data for myosin complexed with nonhydrolyzable nucleotide analogs,23,24 which show that a divalent cation coordinates one phosphate in myosin·ADP·AlF4 and two phosphates in myosin·AMPPNP. Thus, the comparison with the X-ray data not only confirms the relative number of coordinated phosphates but also actually provides a reference for the absolute quantification. We can conclude, therefore, that in the myosin-bound Mn·ADP·AlF4, only one phosphate is coordinated to Mn2+, whereas in all of the other samples, two phosphates are coordinated.
One has to mention that the number of coordinated phosphates was estimated previously by W-band pulsed ENDOR for Mn·ADP and Mn·ATPγS.12 It was found that at [Mn2+]/[ADP] ~ 1 two phosphates are coordinated, whereas for [Mn2+]/[ADP] ⪡ 1, an additional phosphate coordination was inferred from the observed changes in the ENDOR lineshapes. These results are in apparent contradiction with our results, which indicate that only two phosphates are coordinated at [Mn2+]/[ADP] = 0.1. Although this discrepancy requires additional investigation, we have to note that the W-band ENDOR quantification of coordinated phosphates12 was done indirectly, from the estimated number of coordinated water molecules, and is therefore less accurate than the quantification based on referencing the ENDOR intensities to the available X-ray crystallographic data.
Monitoring Myosin·Mn·Nucleotide Complex Formation Using Pulsed ELDOR
Mn·nucleotide complexes form regardless of the myosin presence, which is readily established by 31P pulsed ENDOR. A formation of the myosin·Mn·nucleotide complexes in the studied three-component systems, however, is less obvious, especially taking into account the similarity of the 31P ENDOR spectra of Mn·ADP and myosin·Mn·ADP. According to the X-ray structures of myosin, the divalent cation is coordinated to the oxygens of Ser and Thr (other ligands being two water molecules and two phosphates of the nucleotide).23,24,39,40 These protein-derived ligands are EPR-silent, and therefore, ESEEM and ENDOR techniques cannot be used to monitor the Mn binding to myosin (this would not be the case if the ligands were nitrogen atoms11,15,41–43). The shape and width of the EPR spectrum are also not very sensitive to the protein binding. For example, trace 2 in Figure 1 shows the ESE field sweep spectrum of Mn·AMPPNP. This spectrum is slightly narrower than that of myosin·Mn·AMPPNP (trace 1), but it is not immediately clear if such a minor change can be used as unequivocal evidence of the myosin's binding.
An alternative way to monitor the Mn–myosin binding is based on using a spin-labeled myosin and applying the pulsed ELDOR technique to detect the dipole interaction between the Mn2+ ion and the nitroxide spin label. For this purpose, we used a double-Cys myosin construct A639C:K498C44 labeled with maleimide spin probe (MSL·myosin). The conformation and structural kinetics of the force generation region of that construct were characterized before,1,44 and for consistency of the results, we used the same double-Cys construct in this work, as well. The expected Mn–nitroxide distances are ~22 and 45 Å (the distance between metal cation and Cβ of A639 and K498, respectively39).
The ESE field sweep spectrum of MSL·myosin·Mn·AMPPNP is shown by trace 2 in Figure 1b. The narrow, intense signal in the center of the Mn2+ spectrum is that of the nitroxide spin label, MSL. In the pulsed ELDOR experiment, the pumping mw frequency, νpmp, was in resonance with the sharp low-field component of the nitroxide EPR spectrum, and the observation frequency, νobs, was in resonance with Mn2+ (as indicated in Figure 1b). The four-pulse ELDOR trace obtained for MSL·myosin·Mn·AMPPNP is shown by trace 1 in Figure 4. This trace shows the presence of a fast-damping oscillation with the first half-period traceable to about 230 ns, which corresponds to a characteristic frequency of ~2.2 MHz. Trace 2 in the same Figure is obtained with both νpmp and νobs being in resonance with Mn2+, which was achieved by changing the magnetic field, Bo. This trace does not show any oscillations. These observations indicate that the ~2.2 MHz oscillation in trace 1 is caused by the dipole interaction between Mn2+ and the nitroxide spin label(s).
Figure 4.
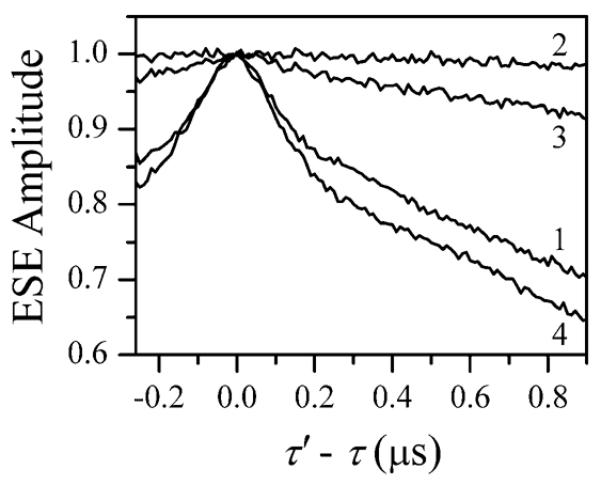
Pulsed ELDOR traces of the samples with spin-labeled myosin obtained using the four-pulse ELDOR sequence. Traces 1, 3, and 4 are obtained for MSL·myosin·Mn·AMPPNP, MSL·myosin·M-n·ADP, and myosin·Mn·ADP·AlF4, respectively, with νpmp in resonance with MSL (as indicated in Figure 1b). Trace 2, the ELDOR trace obtained for MSL·myosin·Mn·AMPPNP with νpmp off-resonance with MSL (i.e., at a different magnetic field). Experimental conditions: νpmp = 30.108 GHz; νobs = 30.236 GHz; Bo = 1071.2 mT (traces 1, 3, and 4) and 1089.5 mT (trace 2); observation mw pulses, 40, 50, 50 ns (traces 1–3) and 20, 30, 30 ns (trace 4); pumping pulse, 40 ns (traces 1–3) and 31 ns (trace 4); time interval between the first and second observation pulses, τ = 300 ns; time interval between the second and third observation pulses, T = 1.6 μs; temperature, 15 K. τ′ is the interval between the second observation pulse and the pumping pulse.
To assign the observed ELDOR oscillation to a specific spin label, at C498 or C639, we have to note that with the pumping mw pulse durations of >30 ns (see Figure 4), the spin inversion probabilities for the spin label are below 21%. With such small probabilities, the ELDOR effects from the two spin labels are to good accuracy additive and can be considered separately. The distance corresponding to the dipole interaction constant, D = 2.2 MHz, is ~31 Å and has to be assigned to the spin label at C639. The 9 Å difference between this distance and the X-ray distance from Mn to C639 (22 Å39) is within the range of uncertainty (±10 Å) associated with the size of the spin probe. The distance to the other cysteine, C498, is 45 Å,39 which could result in Mn–MSL distances from 35 Å (D ≈ 1.26 MHz) to 55 Å (D ≈ 0.32 MHz). Since no clear oscillation with the frequency near 1 MHz was observed, we conclude that the distance between Mn and MSL at C498 is probably closer to the 55 Å limit, and the frequency of the corresponding dipolar oscillation is too low to be observed within the time window of our ELDOR experiments (~1 μs, see Figure 4).
The pulsed ELDOR experiments were also performed for MSL·myosin·Mn·ADP and MSL·myosin·Mn·ADP·AlF4. In the case of ADP, the binding of Mn to myosin was not observed under the same conditions as those used for AMPPNP (see ELDOR trace 3 in Figure 4), but for MSL·myosin·Mn·ADP·AlF4, the binding of Mn to myosin was confirmed by pulsed ELDOR (trace 4 in Figure 4).
Nitrogen Coordination in Mn·Nucleotide Complexes
Figure 5 shows the low-frequency part of the cosine Fourier transform (FT) spectra of two-pulse ESEEM recorded for various Mn·nucleotide and myosin·Mn·nucleotide complexes. The 1H fundamental matrix line (at ~45 MHz) and the 31P sum combination line (at ~36 MHz) are beyond the shown frequency range. The assignment of various spectral features is indicated in the Figure. The 14N ESEEM observed for the Mn·ADP complex (trace 4, Figure 5) is significantly stronger than that detected for other studied complexes. This spectrum is characteristic of the weak hfi condition: νN > aNef/2, where νN is the Zeeman frequency of 14N and aNef is the effective (i.e., uncorrected for the electronic spin) hfi constant. The line with negative amplitude at ~14.2 MHz is a sum combination of the frequencies of ΔmI = 2 transitions corresponding to the electron spin manifolds involved in the formation of the ESE signal (mostly, with mS = ±1/2). The fundamental features (the lines with positive amplitude) are not sufficiently resolved to provide an accurate estimate of the hfi. The hyperfine sublevel correlation (HYSCORE) spectrum (Figure 6), however, shows a pair of correlation peaks at the frequencies of MHz and MHz (where the superscripts “α” and “β” denote the electron spin manifolds involved in the transition). The difference between these frequencies provides an estimate of aNef ≈ 2.5 MHz, which corresponds to the true hfi constant aN ≈ 2SaNef ≈ 12.5 MHz.
Figure 5.
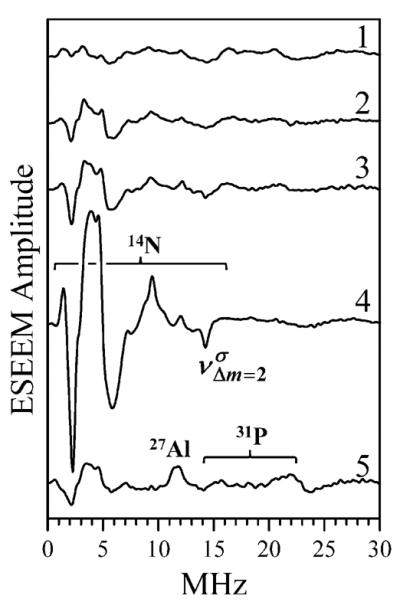
Two-pulse ESEEM spectra (cosine FT) of myosin·M-n·AMPPNP (trace 1), Mn·AMPPNP (trace 2), Mn·ATP (trace 3), Mn·ADP (trace 4), and myosin·Mn·ADP·AlF4 (trace 5). Experimental conditions: νmw = 30.108 GHz (traces 1–4) and 30.136 GHz (trace 5); Bo = 1070.3 mT (traces 1 – 4) and 1072.8 mT (trace 5); mw pulses, 8 and 14 ns; temperature, 15 K. Assignments of various features to different magnetic nuclei are indicated.
Figure 6.
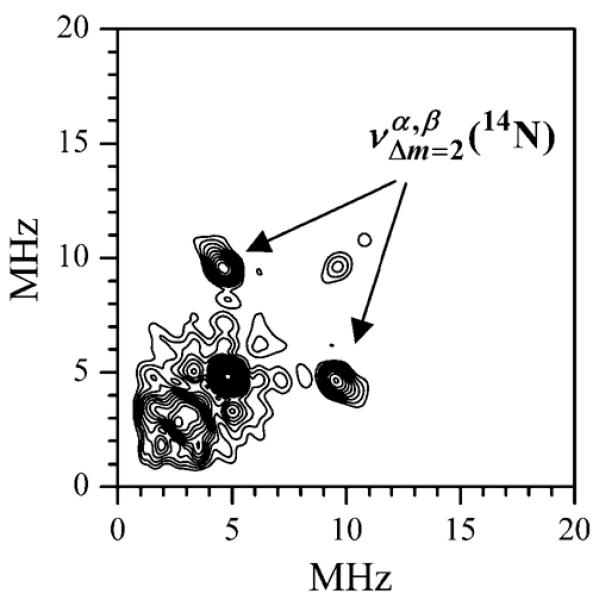
HYSCORE spectrum of Mn·ADP. Experimental conditions: νmw = 30.108 GHz; Bo = 1070.3 mT; mw pulses: 8, 8, 15, and 8 ns; temperature, 15 K. The spectrum represents the sum of the spectra obtained at time intervals between the first and second mw pulses τ = 120, 150, and 180 ns. The correlation features is indicated.
The shift of the sum combination frequency MHz observed in the primary ESEEM spectrum from 4νN ≈ 13.2 MHz is mostly explained by the second-order effect of the nuclear quadrupole interaction (nqi):
| (1) |
where k = e2Qq/4h is the reduced quadrupole coupling constant and η is the asymmetry parameter of the nqi tensor. This equation is readily obtained from the well-known expression for the frequencies of the ΔmI = 2 transitions:45 . It neglects the second order shift due to the anisotropic hfi, (where T⊥ is the perpendicular component of the anisotropic hfi), because with |T⊥| ~ 0.6 MHz14 and νN ~ 3.3 MHz, the estimated hfi-related shift is only ~0.12 MHz (about 12% of the experimental one). The cfi-related shifts17,18 can also be neglected: MHz, and with the orientation of the cfi tensor typically being disordered with respect to the molecular frame, it leads to a broadening rather than to a well-defined shift. Using eq 1 and taking into account that η ∈ [0, 1], one can estimate the quadrupole coupling constant, e2Qq/h, to be between 2.6 and 3.1 MHz. Both the hfi and nqi estimates are in a good agreement with those made elsewhere for the Mn2+–adenosine nucleotide using X-band ESEEM spectroscopy.11,15,41–43
The observed 14N hfi constant, aN, is too large for a second-sphere nitrogen atom, and it is similar to the constants obtained for the nitrogen atoms directly coordinated to Mn2+.11,15,41–43 Considering simultaneous observation of two coordinated phosphates and a coordinated nitrogen in the Mn·ADP complex, a tridentate coordination of ADP could formally be proposed. A more likely explanation, however, invokes a bidentate phosphate coordination of one nucleotide and a simultaneous nitrogen coordination of the second nucleotide. This explanation is in agreement with the W-band 31P ENDOR study of Mn·ADP12 that had detected the presence of a weakly coupled 31P in the vicinity of the Mn2+ ion in the samples with [Mn2+]/[ADP] ⪡ 1. Similar 31P lines (corresponding to T⊥ ≈ −0.27 MHz) were also observed by us in the Mims ENDOR spectrum of Mn·ADP recorded at the time interval τ between the first two mw pulses of 500 ns. The possibility of Mn2+ coordination by two nucleotides would also explain the observation of minor 14N ESEEM for the samples containing the nucleotides other than ADP.
Stability of Myosin·Mn·Nucleotide Complexes
The lack of the Mn·ADP binding to myosin reflected by pulsed ELDOR explains the similarity of the field sweep, ESEEM, and pulsed ENDOR spectra obtained for Mn·ADP and myosin·Mn·ADP samples. According to the literature, all myosin complexes studied in this work have different stabilities. The myosin·Mg·ADP·AlF4 complex is the most stable, with a dissociation rate of ~1.14 × 10−5 s−1.46 Myosin complexed with Mg·AMPPNP is stable, as well, with a dissociation rate of ~2 × 10−2 s−1.47 The myosin·Mg·ADP complex is the least stable, and therefore, in the X-ray crystallographic studies in which myosin·Mg·ADP complex was observed, the crystals were grown from solutions containing a large (~20-fold) excess of Mg and ADP.24 The reported dissociation rate for myosin·Mg·ADP is ~1.4 s−1 and KD ~ 4 μM for rabbit skeletal S1·Mn·ADP.7,47D. discoideum myosin·Mg·ADP has a KD of 5.8 μM6 to 40 μM.48 Replacing magnesium by manganese could destabilize the D. discoideum myosin·metal·ADP complex even further. In this work, we did not observe the formation of the myosin·Mn·ADP complex. The maximum concentration of Mn2+ in our myosin·Mn·ADP experiments was 330 μM, with [myosin] = 410 μM and [ADP] = 3.3 mM. If each Mn cation formed a complex with ADP, the concentration of Mn·ADP in the sample was 330 μM. The experimental threshold for the observation of the MSL·myosin·Mn·ADP complex in our ELDOR experiments was ≤30 μM. We can therefore estimate the dissociation constant of the myosin·Mn·ADP complex as KD = [Mn·ADP]·[MSL·myosin]/[MSL·myosin·Mn·ADP] ≥ (330 μM·410 μM)/30 μM = 4.5 mM at T = 20 °C.
The above estimate assumes that ADP does not bind to myosin without a divalent metal cation.49 If this assumption is not valid, the competitive inhibition of the myosin·Mn·ADP complex formation should be considered. In this case, our experimental data provide an apparent dissociation constant, Kapp = KD (1 + [ADP]/KADP), in which KADP is the dissociation constant of the myosin·ADP complex without metal cation. To ensure the timely release of ADP from the active site in the myosin–nucleotide cycle, KADP should be greater than KD. Then, KD for the D. discoideum myosin·Mn·ADP complex is estimated at 1.2–4.5 mM.
CONCLUSIONS
In this work, we report on the first high-resolution pulsed EPR study of Mn·nucleotide binding to the myosin active site. We used pulsed EPR at the Ka-band to study the Mn2+–nucleotide coordination and Mn·nucleotide binding to myosin for diphosphate and triphosphate adenosine nucleotides. Since the solvent- and protein-derived ligands are both oxygens, the ESE field sweeps, pulsed ENDOR, and ESEEM spectra were not sufficiently specific to reliably distinguish between the solvated and myosin-bound Mn·nucleotide complexes. Therefore, to directly detect the binding of the Mn·nucleotide complexes to the active site of the myosin motor domain, a novel approach based on the pulsed ELDOR of the spin-labeled A639C:K498C mutant of myosin was used. Myosin was found to form a complex with Mn·AMPPNP and Mn·ADP·AlF4, whereas in the preparations with Mn·ADP, no myosin·Mn·nucleotide complex formation was detected, and the complex of Mn·ADP in solution was observed instead. This observation is in agreement with the general mechanism of myosin ATPase, which implies the liberation of the nucleotide (ADP) after ATP hydrolysis.
The number of coordinated phosphate groups was determined by referencing the integrated intensities of the normalized 31P Mims ENDOR spectra to the known X-ray crystallographic structures of myosin·nucleotide complexes. The coordination of Mn2+ by two phosphate groups was established for all of the nucleotides used except ADP·AlF4, for which the coordination by one phosphate and one AlF4 group was determined. The solution complex of Mn·ADP reveals a relatively strong hfi with one of the nitrogens of the nucleotide indicative of the nitrogen coordination (in addition to two phosphates). This nitrogen is tentatively assigned to the second coordinated ADP molecule. The results of this work prepare the stage for time-resolved studies of ATP hydrolysis reaction on myosin using a rapid freeze-pulsed EPR technique.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant AR59621, Wachovia fellowship, and by funds provided by the University of North Carolina at Charlotte (Y.E.N). We thank Richard Tran for assistance with myosin motor domain preparation. A.V.A. acknowledges the NIH/NCRR funding for acquisition of the new EPR spectrometer for the EPR Facility of the University of Arizona (Grant no. 1S10RR26416-01).
ABBREVIATIONS
- ADP
adenosine diphosphate
- AMPPNP
adenylyl-imidodiphosphate
- ATP
adenosine triphosphate
- ATPγS
adenosine 5′-O-(3-thiotriphosphate)
- cfi
crystal field interaction
- ELDOR
electron–electron double resonance
- ENDOR
electron–nuclear double resonance
- EPR
electron paramagnetic resonance
- ESEEM
electron spin echo envelope modulation
- hfi
hyperfine interaction
- HYSCORE
hyperfine sublevel correlation spectroscopy
- mw
microwave
- MSL
maleimide spin label
- nqi
nuclear quadrupole interaction
- RF
radiofrequency
Footnotes
Supporting Information Structures of the nucleotides studied in this work; Interpretation of the ligand hfi parameters observed in the complexes of high-spin transition metal ions. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- (1).Nesmelov YE, Agafonov RV, Negrashov IV, Blakely SE, Titus MA, Thomas DD. Structural kinetics of myosin by transient time-resolved FRET. Proc. Natl. Acad. Sci. U.S.A. 2011;108(5):1891–6. doi: 10.1073/pnas.1012320108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Gyimesi M, Kintses B, Bodor A, Perczel A, Fischer S, Bagshaw CR, Malnasi-Csizmadia A. The mechanism of the reverse recovery step, phosphate release, and actin activation of Dictyostelium myosin II. J. Biol. Chem. 2008;283(13):8153–63. doi: 10.1074/jbc.M708863200. [DOI] [PubMed] [Google Scholar]
- (3).Lymn RW, Taylor EW. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry. 1971;10(25):4617–24. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- (4).Jahn W. The association of actin and myosin in the presence of gamma-amido-ATP proceeds mainly via a complex with myosin in the closed conformation. Biochemistry. 2007;46(33):9654–64. doi: 10.1021/bi700318t. [DOI] [PubMed] [Google Scholar]
- (5).Naber N, Purcell TJ, Pate E, Cooke R. Dynamics of the nucleotide pocket of myosin measured by spin-labeled nucleotides. Biophys. J. 2007;92(1):172–84. doi: 10.1529/biophysj.106.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kovacs M, Malnasi-Csizmadia A, Woolley RJ, Bagshaw CR. Analysis of nucleotide binding to Dictyostelium myosin II motor domains containing a single tryptophan near the active site. J. Biol. Chem. 2002;277(32):28459–67. doi: 10.1074/jbc.M202180200. [DOI] [PubMed] [Google Scholar]
- (7).Bagshaw CR, Reed GH. Investigations of equilibrium complexes of myosin subfragment 1 with the manganous ion and adenosine diphosphate using magnetic resonance techniques. J. Biol. Chem. 1976;251(7):1975–83. [PubMed] [Google Scholar]
- (8).Webb MR, Ash DE, Leyh TS, Trentham DR, Reed GH. Electron paramagnetic resonance studies of Mn(II) complexes with myosin subfragment 1 and oxygen 17-labeled ligands. J. Biol. Chem. 1982;257(6):3068–72. [PubMed] [Google Scholar]
- (9).Leyh TS, Goodhart PJ, Nguyen AC, Kenyon GL, Reed GH. Structures of manganese(II) complexes with ATP, ADP, and phosphocreatine in the reactive central complexes with creatine kinase: electron paramagnetic resonance studies with oxygen-17-labeled ligands. Biochemistry. 1985;24(2):308–16. doi: 10.1021/bi00323a011. [DOI] [PubMed] [Google Scholar]
- (10).Schneider B, Sigalat C, Amano T, Zimmermann JL. Evidence for changes in the nucleotide conformation in the active site of H(+)-ATPase as determined by pulsed EPR spectroscopy. Biochemistry. 2000;39(50):15500–12. doi: 10.1021/bi0014599. [DOI] [PubMed] [Google Scholar]
- (11).Zoleo A, Lippe G, Contessi S, Brustolon M, Dabbeni-Sala F, Maniero AL. Conformational role of the divalent metal in bovine heart mitochondrial F1-ATPase: an electron spin echo envelope modulation study. Biochemistry. 2007;46(46):13443–50. doi: 10.1021/bi700949e. [DOI] [PubMed] [Google Scholar]
- (12).Potapov A, Goldfarb D. Quantitative Characterization of the Mn2+ Complexes of ADP and ATPgS by W-band ENDOR. Appl. Magn. Reson. 2006;30:461–72. [Google Scholar]
- (13).Petersen J, Gessner C, Fisher K, Mitchell CJ, Lowe DJ, Lubitz W. Mn2+-adenosine nucleotide complexes in the presence of the nitrogenase iron-protein: detection of conformational rearrangements directly at the nucleotide binding site by EPR and 2D-ESEEM (two-dimensional electron spin-echo envelope modulation spectroscopy) Biochem. J. 2005;391(Pt 3):527–39. doi: 10.1042/BJ20050226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Hoogstraten CG, Grant CV, Horton TE, DeRose VJ, Britt RD. Structural Analysis of Metal Ion Ligation to Nucleotides and Nucleic Acids Using Pulsed EPR Spectroscopy. J. Am. Chem. Soc. 2002;124:834–42. doi: 10.1021/ja0112238. [DOI] [PubMed] [Google Scholar]
- (15).Morrissey SR, Horton TE, Grant CV, Hoogstraten CG, Britt RD, DeRose VJ. Mn2+–nitrogen interactions in RNA probed by electron spin-echo envelope modulation spectroscopy: Application to the hammerhead ribozyme. J. Am. Chem. Soc. 1999;121:9215–18. [Google Scholar]
- (16).Buy C, Matsui T, Andrianambinintsoa S, Sigalat C, Girault G, Zimmermann JL. Binding sites for Mg(II) in H(+)-ATPase from Bacillus PS3 and in the α-3 β-3 γ subcomplex studied by one-dimensional ESEEM and two-dimensional HYSCORE spectroscopy of oxovanadium(IV) complexes: a possible role for β-His-324. Biochemistry. 1996;35(45):14281–93. doi: 10.1021/bi961811b. [DOI] [PubMed] [Google Scholar]
- (17).Astashkin AV, Raitsimring AM. Electron spin echo envelope modulation theory for high electron spin systems in weak crystal field. J. Chem. Phys. 2002;117:6121–32. [Google Scholar]
- (18).Astashkin AV, Raitsimring AM, Caravan P. Pulsed ENDOR study of water coordination to Gd3+ complexes in orientationally disordered systems. J. Phys. Chem. A. 2004;108:1990–2001. [Google Scholar]
- (19).Coffino AR, Peisach J. Nuclear modulation effects in high-spin electron systems with small zero-field splittings. J. Chem. Phys. 1992;97:3072–91. [Google Scholar]
- (20).Larsen RJ, Halkides C, Singel DJ. Geometrical representation of nuclear modulation effects: the effects of high spin multiplicity on the electron spin envelope modulation spectra of Mn2+ complexes of N-ras p21. J. Chem. Phys. 1993;98:6704–21. [Google Scholar]
- (21).Tan X, Bernardo M, Thomann H, Scholes CP. 17O hyperfine and quadrupole interactions for water ligands in frozen solutions of high spin Mn2+ J. Chem. Phys. 1995;102:2675–90. [Google Scholar]
- (22).Jeschke G, Schweiger A. Hyperfine decoupling in electron spin resonance. J. Chem. Phys. 1997;106:9979–91. [Google Scholar]
- (23).Fisher AJ, Smith CA, Thoden JB, Smith R, Sutoh K, Holden HM, Rayment I. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP·BeFx and MgADP·AlF4. Biochemistry. 1995;34(28):8960–72. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- (24).Gulick AM, Bauer CB, Thoden JB, Rayment I. X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain. Biochemistry. 1997;36(39):11619–28. doi: 10.1021/bi9712596. [DOI] [PubMed] [Google Scholar]
- (25).Korman VL, Anderson SE, Prochniewicz E, Titus MA, Thomas DD. Structural dynamics of the actin–myosin interface by site-directed spectroscopy. J. Mol. Biol. 2006;356(5):1107–17. doi: 10.1016/j.jmb.2005.10.024. [DOI] [PubMed] [Google Scholar]
- (26).Agafonov RV, Nesmelov YE, Titus MA, Thomas DD. Muscle and nonmuscle myosins probed by a spin label at equivalent sites in the force-generating domain. Proc. Natl. Acad. Sci. U.S.A. 2008;105(36):13397–402. doi: 10.1073/pnas.0801342105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 1979;100(1):95–7. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- (28).Astashkin AV, Enemark JH, Raitsimring AM. 26.5–40 GHz Kα-band pulsed EPR spectrometer. Concepts in Magnetic Resonance Part B: Magnetic Resonance Engineering. 2006;29B:125–36. [Google Scholar]
- (29).Mims WB. Pulsed Endor Experiments. Proc. R. Soc. London A. 1965;283:452–57. [Google Scholar]
- (30).Liao PF, Hartmann SR. Determination of Cr-Al hyperfine and electric quadrupole interaction parameters in ruby using spin-echo electron-nuclear double resonance. Phys. Rev. B. 1973;8:69–80. [Google Scholar]
- (31).Astashkin AV, Kawamori A, Kodera Y, Kuroiwa S, Akabori K. An electron spin echo envelope modulation study of the primary acceptor quinone in Zn-substituted plant photosystem II. J. Chem. Phys. 1995;102:5583–88. [Google Scholar]
- (32).Doan PE, Hoffman BM. Making hyperfine selection in Mims ENDOR independent of deadtime. Chem. Phys. Lett. 1997;269:208–14. [Google Scholar]
- (33).Klein EL, Raitsimring AM, Astashkin AV, Rajapakshe A, Johnson-Winters K, Arnold AR, Potapov A, Goldfarb D, Enemark JH. Identity of the Exchangeable Sulfur-Containing Ligand at the Mo(V) Center of R160Q Human Sulfite Oxidase. Inorg. Chem. 2012;51:1408–18. doi: 10.1021/ic201643t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Pacheco A, Basu P, Borbat P, Raitsimring AM, Enemark JH. Multifrequency ESEEM Spectroscopy of Sulfite Oxidase in Phosphate Buffer: Direct Evidence for Coordinated Phosphate. Inorg. Chem. 1996;35(24):7001–08. doi: 10.1021/ic9607806. [DOI] [PubMed] [Google Scholar]
- (35).Dikanov SA, Liboiron BD, Thompson KH, Vera E, Yuen VG, McNeill JH, Orvig C. In Vivo Electron Spin-Echo Envelope Modulation (ESEEM) Spectroscopy: First Observation of Vanadyl Coordination to Phosphate in Bone. J. Am. Chem. Soc. 1999;121:11004–5. [Google Scholar]
- (36).Mustafi D, Telser J, Makinen MW. Molecular Geometry of Vanadyl-Adenine Nucleotide Complexes Determined by EPR, ENDOR, and Molecular Modeling. J. Am. Chem. Soc. 1992;114:6219–26. [Google Scholar]
- (37).Zech SG, Sun WC, Jacques V, Caravan P, Astashkin AV, Raitsimring AM. Probing the water coordination of protein-targeted MRI contrast agents by pulsed ENDOR spectroscopy. ChemPhysChem. 2005;6(12):2570–7. doi: 10.1002/cphc.200500250. [DOI] [PubMed] [Google Scholar]
- (38).Raitsimring AM, Astashkin AV, Baute D, Goldfarb D, Poluektov OG, Lowe MP, Zech SG, Caravan P. Determination of the hydration number of gadolinium(III) complexes by high-field pulsed 17O ENDOR spectroscopy. ChemPhysChem. 2006;7(7):1590–7. doi: 10.1002/cphc.200600138. [DOI] [PubMed] [Google Scholar]
- (39).Bauer CB, Holden HM, Thoden JB, Smith R, Rayment I. X-ray structures of the apo and MgATP-bound states of Dictyostelium discoideum myosin motor domain. J. Biol. Chem. 2000;275(49):38494–9. doi: 10.1074/jbc.M005585200. [DOI] [PubMed] [Google Scholar]
- (40).Smith CA, Rayment I. X-ray structure of the magnesium-(II)·ADP·vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 A resolution. Biochemistry. 1996;35(17):5404–17. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- (41).Lee HC, Goroncy AK, Peisach J, Cavada BS, Grangeiro TB, Ramos MV, Sampaio AH, Dam TK, Brewer CF. Demonstration of a conserved histidine and two water ligands at the Mn2+ site in Diocleinae lectins by pulsed EPR spectroscopy. Biochemistry. 2000;39(9):2340–6. doi: 10.1021/bi992102b. [DOI] [PubMed] [Google Scholar]
- (42).McCracken J, Peisach J, Bhattacharyya L, Brewer F. Electron spin echo envelope modulation studies of lectins: evidence for a conserved Mn(2+)-binding site. Biochemistry. 1991;30(18):4486–91. doi: 10.1021/bi00232a016. [DOI] [PubMed] [Google Scholar]
- (43).Tipton PA, Peisach J. Pulsed EPR analysis of tartrate dehydrogenase active-site complexes. Biochemistry. 1991;30(3):739–44. doi: 10.1021/bi00217a024. [DOI] [PubMed] [Google Scholar]
- (44).Agafonov RV, Negrashov IV, Tkachev YV, Blakely SE, Titus MA, Thomas DD, Nesmelov YE. Structural dynamics of the myosin relay helix by time-resolved EPR and FRET. Proc. Natl. Acad. Sci. U.S.A. 2009;106(51):21625–30. doi: 10.1073/pnas.0909757106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Dikanov SA, Tsvetkov YD, Bowman MK, Astashkin AV. Parameters of quadrupole coupling of 14N nuclei in chlorophyll a cations determined by the electron spin echo method. Chem. Phys. Lett. 1982;90:149–53. [Google Scholar]
- (46).Phan BC, Cheung P, Stafford WF, Reisler E. Complexes of myosin subfragment-1 with adenosine diphosphate and phosphate analogs: probes of active site and protein conformation. Biophys. Chem. 1996;59(3):341–9. doi: 10.1016/0301-4622(95)00127-1. [DOI] [PubMed] [Google Scholar]
- (47).Trybus KM, Taylor EW. Transient kinetics of adenosine 5′-diphosphate and adenosine 5′-(β,γ-imidotriphosphate) binding to subfragment 1 and actosubfragment 1. Biochemistry. 1982;21(6):1284–94. doi: 10.1021/bi00535a028. [DOI] [PubMed] [Google Scholar]
- (48).Ritchie MD, Geeves MA, Woodward SK, Manstein DJ. Kinetic characterization of a cytoplasmic myosin motor domain expressed in Dictyostelium discoideum. Proc. Natl. Acad. Sci. U.S.A. 1993;90(18):8619–23. doi: 10.1073/pnas.90.18.8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Lowey S, Luck SM. Equilibrium binding of adenosine diphosphate to myosin. Biochemistry. 1969;8(8):3195–9. doi: 10.1021/bi00836a010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


