Abstract
Conventional anti-cancer drug screening is typically performed in the absence of accessory cells of the tumor microenvironment, which can profoundly alter anti-tumor drug activity. To address this major limitation, we developed the tumor cell-specific in vitro bioluminescence imaging (CS-BLI) assay. Tumor cells (e.g. myeloma, leukemia and solid tumors) stably expressing luciferase are co-cultured with non-malignant accessory cells (e.g. stromal cells) for selective quantification of tumor cell viability, in presence vs. absence of stromal cells or drug treatment. CS-BLI is high-throughput scalable and identifies stroma-induced chemoresistance in diverse malignancies, including imatinib-resistance in leukemic cells. A stromal-induced signature in tumor cells correlates with adverse clinical prognosis and includes signatures for activated Akt, Ras, NF-κB, HIF-1α, myc, hTERT, and IRF4; signatures for biological aggressiveness and for self-renewal. Unlike conventional screening, CS-BLI can also identify agents with increased activity against tumor cells interacting with stroma. One such compound, reversine, exhibits more potent activity in an orthotopic model of diffuse myeloma bone lesions than in conventional subcutaneous xenografts. Use of CS-BLI, therefore, enables refined screening of candidate anti-cancer agents to enrich preclinical pipelines with potential therapeutics that overcome stroma-mediated drug resistance and can act in a synthetic lethal manner in the context of tumor-stromal interactions.
INTRODUCTION
Tumor-stromal interactions are increasingly recognized as critical components of tumor biology 1, including invasion and metastatic potential 2. The interactions of bone marrow stromal cells (BMSCs) with tumor cells play an important role in tumor drug resistance 3, 4 through complex cytokine and adhesion-mediated mechanisms 3,5. Such microenvironment-mediated drug resistance accounts, at least in part, for the inability of conventional chemotherapies to cure such diseases as multiple myeloma (MM), the second most commonly diagnosed hematologic malignancy, as well as other disseminated neoplasias. Recently, FDA-approved novel agents including thalidomide, lenalidomide and bortezomib (PS-341) have been shown to overcome drug resistance mediated by BMSCs in preclinical models and to be clinically active even in cases of resistance to conventional chemotherapy6–11.
Conventional in vitro assays used for high-throughput drug screening have limitations that prevent their use in co-culture settings. For example, conventional viability (e.g. MTT, Alamar Blue) or cytotoxicity (e.g. LDH release) assays do not distinguish normal from neoplastic cells, and the high background signal from stromal cells in co-cultures confounds quantification of tumor specific viability. In addition, assays that can distinguish tumor cells from accessory cells measure cell proliferation rather than viability (3H-thymidine incorporation), require radioactivity (e.g. 3H-thymidine incorporation, Cr release assay) or involve laborious steps of limited high-throughput scalability (e.g. flow cytometry).
To address these limitations, we developed an in vitro tumor cell-specific bioluminescence imaging (CS-BLI) assay for the study of tumor-stromal co-cultures. In these assays, malignant cells stably expressing luciferase (luc) are co-cultured with luc negative stromal cells, and bioluminescent signal is proportional to the number of viable luc-expressing tumor cells. This allows for specific quantification of tumor cell viability in co-cultures with any luc negative accessory cells of the tumor microenvironment. Using CS-BLI, we validated prior studies of stromal-induced drug resistance in MM, extended these studies to leukemia and solid tumors, and performed open-ended screening to identify novel anti-cancer agents active in the context of tumor-stromal interactions. Specifically, we observed that dexamethasone (Dex) and doxorubicin (Doxo) activity was decreased when MM cells were cultured in the presence of stromal cells. In addition, imatinib and nilotinib activity was decreased in certain leukemia cell lines when they interact with stroma. Importantly, CS-BLI is scalable to high-throughput application, which allows the study of tumor cell heterogeneity in terms of their response to stroma and various drugs. Using CS-BLI, we identified reversine, an agent with increased activity in the presence vs. absence of stromal cells in vitro. Further testing of reversine in xenograft murine models with subcutaneous vs. diffuse tumors in bone provided results concordant with our in vitro observations. CS-BLI, therefore, is a unique drug screening platform which takes into account tumor-microenvironmental interactions, thus providing a level of information that cannot be gained from conventional drug screening assays.
RESULTS
Detection of stromal-induced proliferation and drug resistance of tumor cells
Luc-expressing MM.1S cells were plated at increasing cell concentrations and increasing doses of luciferin substrate. The bioluminescent signal had a statistically significant linear correlation with the number of viable cells in each well for all luciferin concentrations tested, both in the presence and absence of HS-5 stromal cells (P<0.001, R2 ≥ 0.99, Supplementary Fig. 1). In the absence of stromal cells, the CS-BLI technique yielded results concordant with conventional techniques for evaluation of tumor cell viability (Supplementary Fig. 1), without requiring cell lysis. Co-culture of MM cells with BMSCs for 48 h at various stroma-tumor ratios distinguished cell lines with increased number of viable tumor cells (e.g. MM.1S, MM.1R, KMS-18), from lines (e.g. KMS-34, OPM-1) which did not exhibit such a response (Fig. 1a). Interestingly, OPM-1 cell proliferation is reduced in the presence of stroma (Fig. 1a), illustrating that stroma can have growth inhibitory effects in some settings. We then compared the response of MM cells to various anti-cancer agents in the presence vs. absence of stromal cells. Co-culture with BMSCs attenuated the responses of luc+ MM.1S and MM.1R cells to Dex and Doxo treatment (Fig. 1b, full dose-response curve shown in Supplementary Fig. 2a–d); but not to PS-341 (Fig. 2; full dose-response curve shown in Supplementary Fig. 2e, f). Evaluation of drug-induced death of tumor cells in the context of tumor-stromal co-culture using CS-BLI assay was consistent with flow cytometric analysis (Supplementary Fig. 1e).
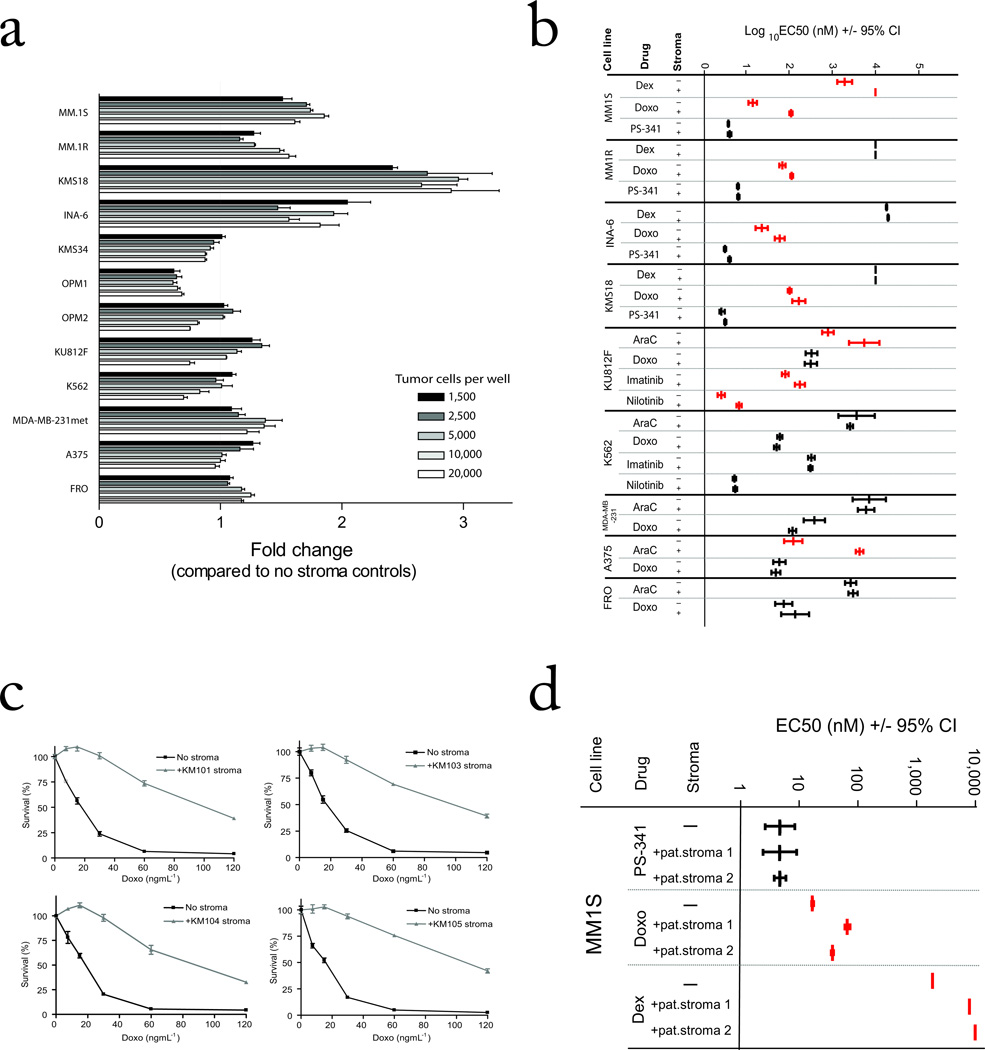
FIGURE 1. Stromal cells modify the response of diverse tumor cell types to various agents.
(a) CS-BLI based viability measurement was performed on Luc+ tumor cell lines plated in the presence and absence of HS-5 BMSCs (10,000 per well) at increasing numbers of tumor cells. (b) Tumor cells were plated in the presence or absence of BMSCs and treated with increasing doses of drug. The log10 EC50 (+/− 95% Confidence Intervals of log10 of EC50) of each drug and cell line in the presence or absence of BMSCs is displayed. Experiments with increased EC50 value in the presence of stromal cells are shown in red (stroma-induced drug resistance) and those with similar EC50 value in the presence vs. absence of stromal cells are shown in black (full dose-response curves are shown in Supplemental Figs 2–4), (c). Viability of MM.1S-GFP/luc cells treated with Doxo in the presence or absence of different BMSC lines was measured by CS-BLI. (d) Viability of MM.1S-GFP/luc cells treated with various anti-MM agents in the presences or absence of primary BMSCs from individuals with MM were analyzed (full response curves shown in Supplementary Fig 5).
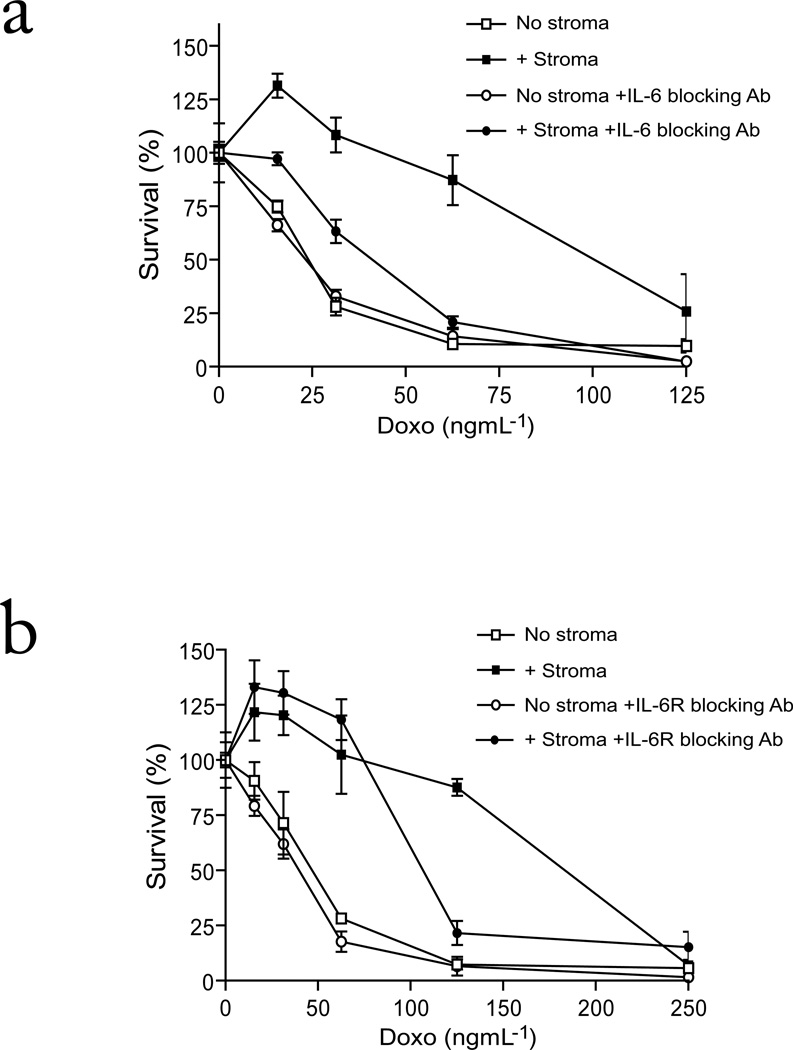
FIGURE 2. Effect of blocking IL-6 and IL-6 Receptor on MM cell co-cultures with BMSCs.
Viability of MM.1S-GFP/luc cells treated with Doxo for 48 h, in the presence or absence of HS-5 stromal cells, in the presence or absence of (a) IL-6 blocking antibody or (b) IL-6 Receptor blocking Ab was measured by CS-BLI. Values are normalized to each respective Doxo-free control. There is a significant difference in MM cell survival in the presence of stroma which is attenuated by IL-6 blocking antibody (2-way ANOVA; Drug P<0.0001; Ab P<0.0001; Interaction P<0.0001) and by the IL-6R blocking antibody (2-way ANOVA; Drug P<0.0001; Ab P<0.0001; Interaction P<0.0001).
We next expanded the application of CS-BLI co-culture testing to other classes of tumors and drugs. We tested anti-leukemia agents, including AraC, Doxo, imatinib, and nilotinib, against luc-expressing Bcr-Abl+ K562 and KU812F leukemic cells in the presence vs. absence of HS-5 stromal cells. The interaction with stromal cells attenuated the response of KU812F cells to AraC or the bcr/abl inhibitors imatinib and nilotinib, but not to Doxo (Fig. 1b; full dose-response curve shown in Supplementary Fig. 3). In contrast, stromal cells protected K562 cells against low AraC doses, but not against imatinib, nilotinib or Doxo at all doses (Fig. 1b; full dose-response curve shown in Supplementary Fig. 3). Co-culture with stromal cells modestly decreased response of the breast cancer cell line MDA-MB-231met to AraC; drastically decreased the sensitivity of A375 melanoma cells to AraC; and decreased the response of the anaplastic thyroid carcinoma cell line FRO to Doxo, but not AraC (Fig. 1b; full dose-response curves shown in Supplementary Fig. 4). These data indicate that the micro-environmental effects of stromal cells can protect both hematological and solid tumors from various classes of anti-cancer drugs.
The protection conferred to MM cells against various drugs was next assessed in co-cultures with different types of stromal cells, including stromal cell lines and primary stromal cells from individuals with MM. Doxo resistance was observed in MM.1S cells co-cultured with multiple BMSC lines (e.g.; KM101, KM103, KM104 and KM105 stromal lines; Fig. 1c). Importantly, in the presence vs. absence of primary BMSCs from individuals with MM, the sensitivity of MM.1S and MM.1R cells to Dex, Doxo and PS-341 (Fig. 1c; full dose-response curve shown in Supplementary Fig. 5) were consistent with results obtained with BMSC lines.
Mechanistic evaluation of stromal-mediated drug resistance
To further evaluate the mechanism of stromal-mediated drug resistance, we next probed the role of interleukin-6 (IL-6) signaling in the CS-BLI system. We focused on IL-6, a major cytokine secreted by stromal cells and a key regulator of MM growth and survival 3, 4. MM-BMSC co-cultures were exposed to anti-IL-6 (Fig. 2a) or anti-IL-6 receptor (Fig. 2b) neutralizing antibodies, both of which suppressed, but did not completely abrogate, stroma-induced resistance of MM.1S cells to Doxo. This observation suggested that other mechanisms in addition to IL-6 signaling contribute to stromal-mediated drug resistance.
To evaluate other potential mechanisms in an open-ended manner, we compared molecular profiles of GFP+ MM cells in the absence vs. presence of GFP− BMSCs to identify genes modulated by tumor-stromal interaction. We observed that this interaction triggers in at least one of these cell lines, upregulation of transcripts (Fig. 3 and Supplementary Table 1) for: (i) cytokine and growth factors implicated in proliferation, survival, and resistance of tumor cells to apoptosis, including IL-6; members of insulin-like growth factor (IGF) signaling cascade12; and members of the Notch pathway 13; (ii) oncogenic kinases, (e.g. PIM1, VAV1; VRK2; SGK1), as well as Ras pathway members (e.g. KRAS); (iii) other molecular targets functionally required for survival of oncogenically addicted MM and other tumor cells, including heat shock proteins (e.g. different isoforms of hsp90, hsp70 and other heat shock proteins) and ubiquitin/proteasome pathway members (e.g. several 26S proteasome subunits, ubiquitin B, and ubiquitin conjugating enzymes); (iv) transcription factors critical for tumor cells in general (e.g. MYC, RELA) 14,15 or MM cells in particular (e.g. IRF4 or BCL6) 5,16,17; (v) regulators of chromatin remodeling, including histone deacetylases (e.g. HDAC1 and HDAC8) which have emerged as novel therapeutic targets in MM and other neoplasias 18,19; (vi) cell cycle regulators (e.g. D-type cyclins; CDK1, CDK5, CDK6; BUB1, BUB3), as well as AURKB 20, and DNA repair enzymes (e.g. TOP2A, TOP2B, or ERCC1); (vii) caspase inhibitors (e.g. CFLAR, BIRC3, BIRC5)15,18,21; anti-apoptotic Bcl-2 family members (e.g. MCL1) 22; and the mitochondrial regulators of cell viability VDAC1, VDAC2 or VDAC3, which have emerged as promising targets for MM therapy 23; (viii) extracellular matrix proteins and adhesion molecules; (ix) targets involved in remodeling of the microenvironment (e.g. matrix metalloproteinases) or tumor cell responses to hypoxia (e.g. HIF-1α-responsive genes); (x) regulators of angiogenesis (e.g. IL8) 24; (xi) positive regulators of bone resorption in MM (e.g. IL1B) 25; as well as (xii) chemokines (e.g. CXCL1, CXCL2, CXCL3, and CXCL5) and chemokine receptors (e.g. CXCR4) proposed to be involved in inflammatory responses, angiogenesis, or chemotaxis of tumor cells to the bone milieu26. Immunoblotting analyses and transcription factor activity assays confirmed at a protein and functional level the upregulation of select molecules and pathways identified in our transcriptional profiling studies (data not shown).
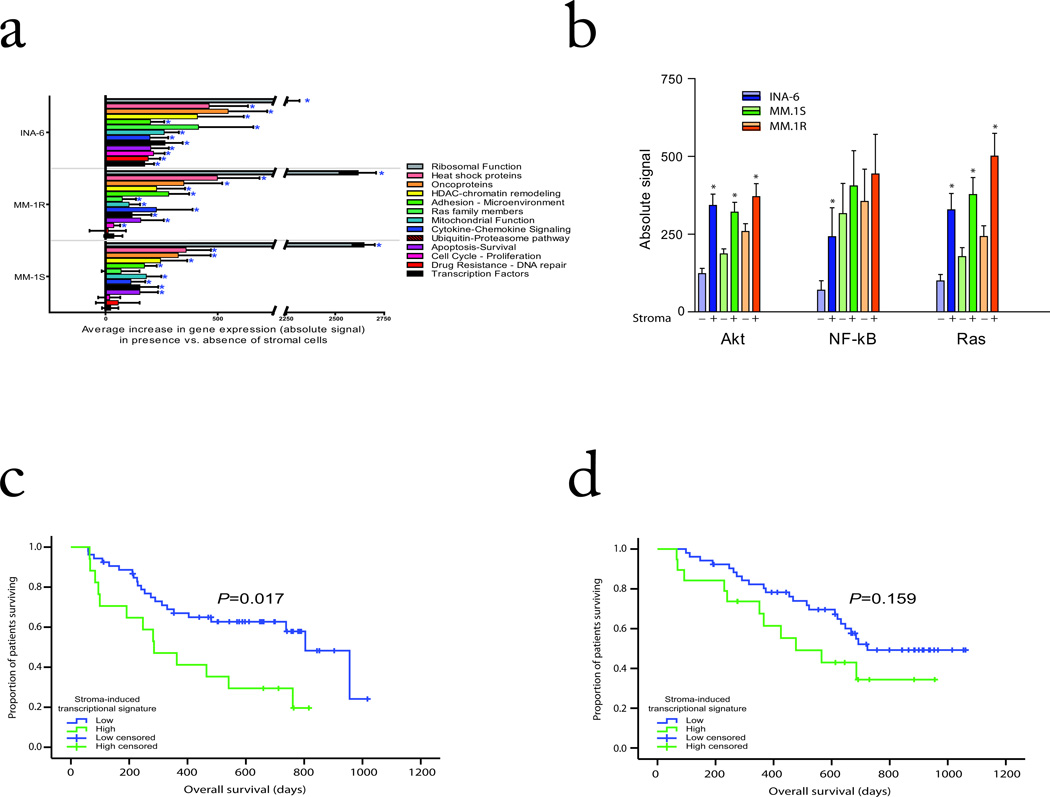
FIGURE 3. Mechanisms of drug sensitivity modulation in the context of tumor-stromal interactions.
GFP+ tumor cell lines were FACS-sorted following culture in the presence and absence of GFP− HS-5 stromal cells for 24 h. RNA was isolated and cDNA generated for analysis on U133 2.0 Plus Affymetrix chips. (a) We compared select gene signatures in stroma-responsive MM cells in the presence vs. absence of stromal cells. (b) Comparison of average absolute signal (+/− SEM) in the stromal responsive cells lines in the presence vs. absence of stromal cells for NF-κB, Ras, and Akt transcriptional signatures (P<0.05). (c) A transcriptional signature of genes induced in MM cells by their interaction with stromal cells was used to classify Bortezomib- or Dex-treated patients with relapsed and relapsed/refractory MM in the randomized phase III APEX trial as having high vs. low expression of stroma-responsive genes. Patients with high expression of stroma-responsive genes had significantly shorter overall survival compared to those with low expression levels in the Dex arm (P=0.017, log-rank test). (e) In contrast, no significant difference was observed in the Bortezomib treatment arm between patients with high vs low transcriptional signature of stroma responsive genes (P=0.159, log-rank test).
To further evaluate how interaction with stromal cells modulates the activation status of different signaling pathways in MM cells, we studied the expression patterns of downstream target genes of these pathways. We observed significant increases in both ubiquitin/proteasome and ribosomal signatures; transcriptional signatures of Akt, NF-kB, and Ras signaling activation (Fig. 3b; P<0.05); transcriptional signatures of self renewal implicated in normal and cancer stem cell biology, such as myc 27,28, IRF4 17, hTERT 29, Hedgehog 30, Notch 31, as well as transcriptional signatures of genes upregulated in undifferentiated human embryonic stem cells 32; genes downregulated by p53 function 33 and a transcriptional signature previously reported to correlate with unfavorable clinical prognosis of MM patients 34 (Supplementary Fig. 6). These observations suggested that the transcriptional signature of MM cell response to stroma is enriched in genes which may correlate with clinical outcome. We therefore identified a transcriptional index of MM cell response to stroma (described in Supplement) and applied this index to gene expression data of relapsed/refractory MM patients enrolled in a randomized clinical trial 11 comparing bortezomib vs. Dex. High expression of stroma-responsive genes was associated with significantly shorter overall survival in Dex- treated patients (P=0.017, log-rank test, Fig. 3c), but did not reach statistical significance in bortezomib-treated patients (P=0.159, log-rank test, Fig. 3d).
High-throughput CS-BLI screening
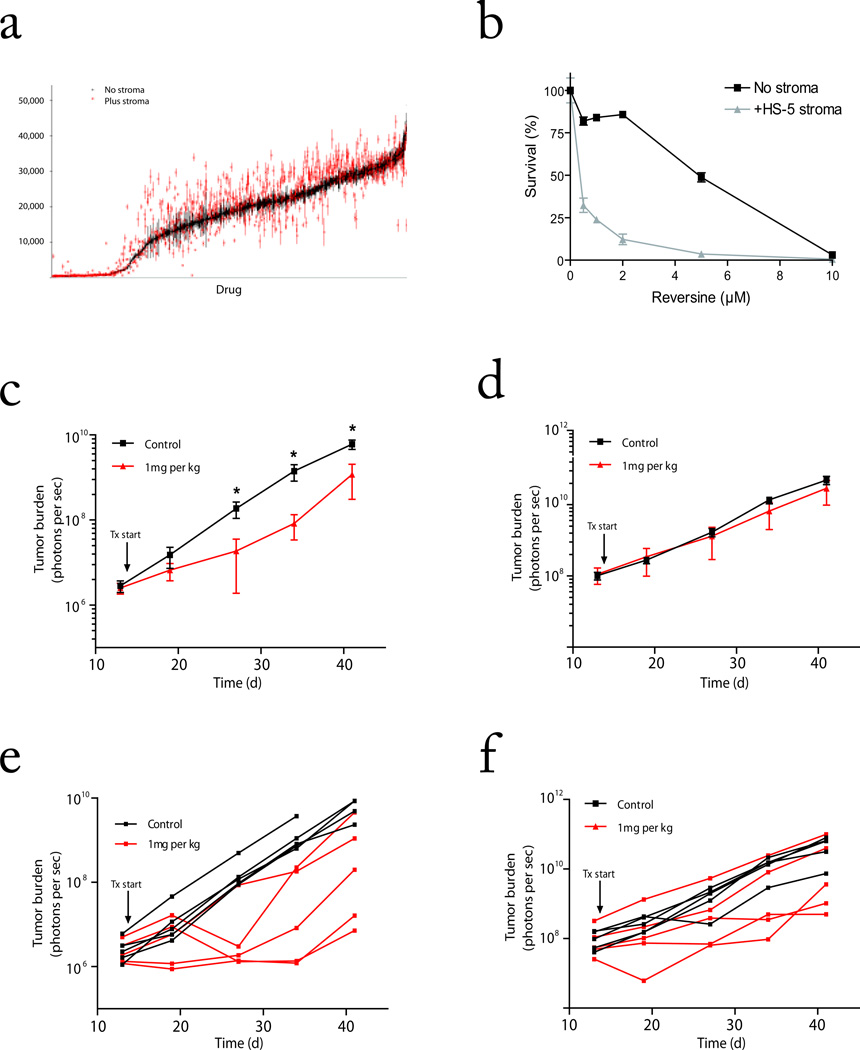
Our mechanistic observation that stromal-induced molecular changes in tumor cells correlates with clinical drug resistance led us to hypothesize that targeting those molecular pathways may sensitize tumors to drug treatment during interaction with stroma. To evaluate this concept and address the high-throughput scalability of CS-BLI, we screened anti-tumor activity of over 3,000 compounds, including a library of bioactive compounds (Fig. 4a) and a library of kinase inhibitors (Table 1). Specifically, we highlight results from three luc+ cell lines (MM.1S, MM.1R, and KU812F) screened in the presence and absence of stromal cells against a panel of documented kinase inhibitors (Table 1). We observed consistent results between MM.1S and MM.1R cells (e.q. BAY 11-7082); showed qualitative differences in KU812F response compared to MM cells (e.g. Piceatannol); and confirmed that the majority of compounds were less active in the presence vs. absence of stroma (including Ro 31-8220, Sphingosine).
Figure 4. Enhanced activity of reversine in the presence of stromal cells.
(a) A library of ~1,000 small molecule inhibitors was screened for activity against MM.1S-GFP/Luc cells in the presence vs. absence of BMSCs. MM viability is shown for each compound (average ± SEM) in absence (black) and presence of stroma (red) and ranked along the X-axis in descending order of anti-MM activity in the absence of stroma. (b) MM.1S-GFP/luc viability was evaluated following reversine treatment in the presence and absence of HS-5 BMSCs by CS-BLI. (c–e) We compared the in vivo activity of reversine against MM.1S-GFP/luc cells in a diffuse bone lesion model vs. subcutaneous model in which tumor cells do not interact with BMSCs. Mice were inoculated with MM.1S-GFP/luc cells i.v. (c, e) or subcutaneously (d, f). Tumor burden was assessed weekly (IVIS imaging) and following engraftment mice were treated with reversine (1 mg kg−1) or vehicle control. Average tumor burden is plotted on logarithmic scale (log10 of average ± log10 of SEM) for the i.v. (c) or subcutaneous (d) model. Mouse tumor burden was significantly reduced in the i.v. model in treated mice at day 27, 33 and 41 (Mann-Whitney 2-tailed test; Day 27 P=0.008, Day 33 P=0.008, Day 41 P=0.032), but was not significantly altered in the subcutaneous model at any time point. (e, f). Tumor burden for each mouse is also plotted for the i.v. (e) and s.c. (f) model. All mice eventually died of tumor burden, indicating that tumor engraftment occurred in all mice.
Table 1. Activity of select compounds in the presence and absence of stromal cells.
Luc+ MM.1S, MM.1R, and KU812F cells were screened against a panel of kinase inhibitors and phosphatase inhibitors at 0.1, 1 and 10 µM, both in the presence and absence of stromal cells. Cells were cultured in the presence of drug for 48 h, and the tumor cell viability was assessed using CS-BLI. Survival of tumor cells was normalized to DMSO controls in the absence of stromal cells. Samples were run in duplicate per plate, and the average of 3 plates is shown for select active agents.
| Percent Survival +/− SEM (relative to control) | ||||||||
|---|---|---|---|---|---|---|---|---|
| MM.1S | MM.1R | KU812F | ||||||
| Compound Name | Known Targeta | Conc. (uM) | No stroma | Plus stroma | No stroma | Plus stroma | No stroma | Plus stroma |
| Staurosporine | Pan-specific | 10 | 1.43 +/− 0.24 | 3.32 +/− 0.24 | 0.76 +/− 0.14 | 0.76 +/− 0.14 | 6.72 +/− 1.33 | 2.79 +/− 0.3 |
| Staurosporine | Pan-specific | 1 | 1.45 +/− 0.08 | 2.59 +/− 0.19 | 0.8 +/− 0.08 | 1.38 +/− 0.11 | 18.32 +/− 2.43 | 13.09 +/− 1.75 |
| Ro 31-8220 | PKC | 10 | 2.49 +/− 0.33 | 5.2 +/− 0.66 | 2.84 +/− 0.63 | 3.98 +/− 0.71 | 13.26 +/− 5.24 | 14.22 +/− 4.51 |
| Sphingosine | PKC | 10 | 4.05 +/− 0.49 | 103.34 +/− 19.93.93 | 5.38 +/− 0.72 | 36.35 +/− 9.3 | 4.24 +/− 0.9 | 32.99 +/− 16.45 |
| HBDDE | PKC alpha, PKC gamma | 10 | 5.83 +/− 0.4 | 42.29 +/− 5.53 | 19.56 +/− 2.28 | 76.74 +/− 8.05 | 68.81 +/− 3.96 | 67.99 +/− 11.24 |
| Staurosporine | Pan-specific | 0.1 | 6.75 +/− 0.31 | 10.25 +/− 0.95 | 6.28 +/− 0.55 | 6.03 +/− 0.49 | 11.7 +/− 1.66 | 10.01 +/− 2.74 |
| 5-Iodotubercidin | ERK2, CK1, CK2 | 10 | 8.13 +/− 0.39 | 11.81 +/− 0.29 | 15.44 +/− 1.7 | 14.98 +/− 1.78 | 43.09 +/− 3.59 | 40.68 +/− 2.17 |
| Lavendustin A | EGFRK | 10 | 9.05 +/− 1.3 | 99.34 +/− 7.33 | 12.28 +/− 1.61 | 52.96 +/− 5.96 | 76.62 +/− 8.71 | 68 +/− 9.79 |
| Rottlerin | PKC delta | 10 | 11.67 +/− 0.41 | 16.91 +/− 0.62 | 13.87 +/− 1.65 | 19.01 +/− 1.24 | 5.48 +/− 0.31 | 10.64 +/− 1.06 |
| Tyrphostin 9 | PDGFRK | 10 | 12.1 +/− 0.69 | 8.82 +/− 0.74 | 15.94 +/− 1 | 14.16 +/− 1.73 | 3.16 +/− 0.73 | 6.62 +/− 1.29 |
| SU4312 | Flk1 | 10 | 12.94 +/− 0.93 | 28.94 +/− 2.73 | 24.38 +/− 4 | 31.18 +/− 4.27 | 5.58 +/− 0.56 | 7.56 +/− 0.56 |
| Hypericin | PKC | 10 | 15.76 +/− 0.67 | 22.03 +/− 0.64 | 18.67 +/− 1.42 | 21.03 +/− 1.43 | 13.77 +/− 3.75 | 38.9 +/− 10.93 |
| Piceatannol | Syk | 10 | 21.11 +/− 17.79 | 56.83 +/− 4.97 | 11.37 +/− 1.91 | 49.79 +/− 4.79 | 86.95 +/− 13.51 | 67.1 +/− 13.68 |
| GF 109203X | PKC | 10 | 21.82 +/− 10.43 | 25.75 +/− 1.11 | 27 +/− 3.66 | 26.67 +/− 1.37 | 57.45 +/− 10.63 | 50.58 +/− 13.34 |
| Ro 31-8220 | PKC | 1 | 25.78 +/− 2.45 | 68.39 +/− 5.32 | 49.54 +/− 9.21 | 69.84 +/− 7.33 | 64.21 +/− 4.55 | 82.87 +/− 6.73 |
| AG-879 | NGFRK | 10 | 26.42 +/− 0.91 | 26.84 +/− 1.57 | 30.13 +/− 3.57 | 23.72 +/− 2.1 | 30.06 +/− 3.98 | 50.33 +/− 9.02 |
| HDBA | EGFRK, CaMKII | 10 | 30.71 +/− 7.25 | 153.95 +/− 10.02 | 15.92 +/− 1.3 | 85.63 +/− 4.65 | 93.27 +/− 6.7 | 92.94 +/− 4.06 |
| Wortmannin | PI3-K | 10 | 42.57 +/− 1.98 | 39.15 +/− 1.56 | 48.24 +/− 4.01 | 29.96 +/− 2.67 | 45.26 +/− 9.17 | 47.64 +/− 4.52 |
| Tyrphostin 9 | PDGFRK | 1 | 44.23 +/− 4.75 | 36.33 +/− 0.6 | 53.72 +/− 4.56 | 51.45 +/− 4.28 | 48.03 +/− 12.5 | 35.38 +/− 9.23 |
| Kenpaullone | GSK-3beta | 10 | 45.81 +/− 4.67 | 34.48 +/− 4.49 | 96.31 +/− 6.57 | 4.48 +/− 4.49 | 16.42 +/− 2.47 | 18.54 +/− 6.81 |
| PP1 | Src family | 10 | 52.05 +/− 3.36 | 71.11 +/− 7.49 | 50.5 +/− 4.02 | 67.13 +/− 7.12 | 4.82 +/− 0.74 | 33.09 +/− 12.39 |
| 5-Iodotubercidin | ERK2, CK1, CK2 | 1 | 53.2 +/− 2.1 | 101.21 +/− 4.27 | 81.51 +/− 8.18 | 126.92 +/− 10.59 | 44.81 +/− 5.35 | 68.47 +/− 11.5 |
| Tyrphostin 25 | EGFRK | 10 | 57.41 +/− 7.78 | 95.81 +/− 22.44 | 5.75 +/− 0.57 | 35.03 +/− 2.58 | 79.12 +/− 8.86 | 104.46 +/− 9.51 |
| PD-98059 | MEK | 10 | 59.02 +/− 6.01 | 105.34 +/− 15.78 | 88.65 +/− 8.96 | 65.73 +/− 21.61 | 16.12 +/− 2.65 | 43.69 +/− 9.45 |
| GF 109203X | PKC | 1 | 62.91 +/− 3.87 | 67.77 +/− 18.4 | 77.73 +/− 4.09 | 64.42 +/− 6.45 | 90.81 +/− 10.86 | 87.69 +/− 17.15 |
| Tyrphostin 51 | EGFRK | 10 | 63.83 +/− 11.59 | 152.16 +/− 5.85 | 29.27 +/− 3.03 | 87.82 +/− 7.98 | 91.75 +/− 8.45 | 89.9 +/− 11.62 |
| Erbstatin analog | EGFRK | 10 | 66.99 +/− 3.5 | 142.35 +/− 3.37 | 82.16 +/− 5.67 | 123.9 +/− 4.96 | 17.49 +/− 2.09 | 21.08 +/− 2.31 |
| U-0126 | MEK | 10 | 74.6 +/− 3.73 | 111.69 +/− 5.03 | 48.2 +/− 4.11 | 51.98 +/− 5.56 | 13.73 +/− 3.22 | 19.93 +/− 2.85 |
| Indirubin-3'-monoxime | GSK-3beta | 10 | 79.24 +/− 7.91 | 74.92 +/− 8.1 | 120.21 +/− 3.86 | 134.51 +/− 12.02 | 43.49 +/− 4.71 | 33.06 +/− 5.42 |
| Kenpaullone | GSK-3beta | 1 | 79.46 +/− 11.35 | 154.02 +/− 18.31 | 147.06 +/− 12.71 | 168.97 +/− 11.22 | 36.5 +/− 4.15 | 26.5 +/− 3.87 |
| PP2 | Src family | 10 | 89.89 +/− 6.56 | 108.38 +/− 8.38 | 95.93 +/− 9.3 | 132.25 +/− 10.05 | 35.05 +/− 13.05 | 35.79 +/− 9.02 |
| SB-203580 | p38 MAPK | 10 | 103.16 +/− 6.72 | 144.5 +/− 5.74 | 64.97 +/− 4.23 | 85.87 +/− 5.1 | 125.82 +/− 11.49 | 159.29 +/− 24.13 |
| SU4312 | Flk1 | 1 | 105.95 +/− 8.23 | 71.07 +/− 7.7 | 87.69 +/− 15.19 | 80.57 +/− 23.07 | 21.14 +/− 4.51 | 38.21 +/− 7.08 |
According to BIOMOL library classification
Interestingly, a small fraction of compounds were more active in the presence of stromal cells (Table 1). This pattern was also observed in the screening of a larger library of compounds (~1,000 different chemical entities) against luc+ MM.1S cells (Fig. 4a). Heterogeneity of tumor response in the context of the microenvironment can therefore be screened using the CS-BLI technique.
In vitro and in vivo anti-tumor activity of reversine
We extended the use of CS-BLI to testing of other compounds beyond those of Fig. 4a. In the process of these tumor-stromal interaction studies, we identified 2-(4-morpholinoanilino)-6-cyclohexylaminopurine (reversine) to be more active against tumor cells in the presence vs. absence of stromal cells (Fig. 4b), and observed in an in vitro cell-free assay that this compound does not spuriously inhibit luc enzymatic activity (Supplementary Fig. 7a). We then evaluated the in vivo activity of reversine (1 mg kg−1 twice weekly) in a subcutaneous tumor model, where tumor cells do not interact with BMSCs, compared to a model of diffuse tumor lesions in which MM cells home to bone. Importantly, we observed significant decrease in tumor burden in the diffuse lesion model with MM-BM interaction (Fig 4c and Supplementary Fig. 8a; 2-way ANOVA, P=0.0003 for Drug; P<0.0001 for Time; P<0.0001 for the interaction), but not in the subcutaneous model (Fig. 4d and Supplementary Fig. 8b; 2-way ANOVA, P=0.2486 for Drug; P<0.0001 for Time; P=0.5974 for the interaction).
Mechanistically, the chemical similarity of reversine to ATP suggested that this agent may function as a kinase inhibitor. Indeed, in vitro kinase activity assays showed a distinct pattern of activity against kinases such as Auroras, JAK2, and SRC, with no activity against other important kinases for MM survival, including AKT1, 2, or 3, FGFR3, or GSK3 (Supplementary Fig. 7b).
DISCUSSION
Conventional preclinical models in cancer drug discovery, including in vitro drug screens and in vivo subcutaneous xenograft models, do not take into account the tumor-stromal interactions which influence cancer pathophysiology. Consequently, the first exposure of a new drug to microenvironment-mediated resistance is in patients during clinical trails. To address this problem, which may contribute to the sub-optimal translation of pre-clinical studies to clinical results, analysis of tumor-microenvironment interactions early in the drug development process is imperative. We therefore developed in vitro tumor cell-specific bioluminescence imaging (CS-BLI), a high throughput-scalable strategy which quantifies the effect of anti-cancer agents on tumor cell viability, both in the presence and absence of non-neoplastic cells interacting with the tumor.
A major strength of CS-BLI is its ability to identify how accessory cells modulate drug activity across different classes of tumors, accessory cell types, and drugs. In their microenvironment, tumor cells behave heterogeneously, with each precise response depending on the particular tumor, accessory cell, and drug combination evaluated. For instance, tumor cell lines with similar patterns of growth and drug response in the absence of stromal cells can have drastically divergent responses in the presence of stroma. The evaluation of all these variables requires the ability to screen their large number of permutations in a high-throughput manner. CS-BLI addresses this need and can conceivably be used to evaluate multiple cell types of the tumor milieu, modeling complex interactions between multiple cells, such as stromal cells, osteoclasts, osteoblasts, and tumor cells of the bone or other relevant cell interactions in diverse neoplasias.
The ability of stromal cells to modify drug-responsiveness of tumor cells is explained mechanistically, at least in part, by our molecular profiling studies. MM cells interacting with stroma exhibit increased activation signatures for pathways associated with tumor cell proliferation, survival, drug-resistance, and self renewal of stem cells, including Akt, Ras, NF-κB, HIF-1α, myc, hTERT, IRF4, and Notch. The biological relevance of these pleiotropic molecular events is further supported by the correlation of a distinct stromal-response signature with adverse clinical prognosis in MM. This further supports the biological significance of stroma-mediated drug resistance as highlighted in several large randomized clinical trials in MM 11,35–37 which have shown that drugs with in vitro stromal-mediated drug resistance (i.e. Dex) have inferior clinical anti-tumor activity compared to drugs whose activity is not blocked by stromal cells (i.e. PS-341).
A distinctive advantage of CS-BLI is the identification of compounds more active in the presence than the absence of stromal cells. Such drugs, which may target various signaling pathways triggered during tumor-stromal interaction, may have been missed using traditional screening methods which test only tumor cells in isolation. For example, the anti-MM activity of reversine is modest in the absence of stromal cells, but increased in their presence. Consistent with in vitro observations, reversine remains more active in a model of diffuse MM bone lesions, which simulates the interaction of tumors with their microenvironment in patients, compared to a subcutaneous tumor model lacking bone marrow stroma. This observation further supports the notion that preclinical models for testing of anti-cancer drugs should take into account the microenvironment with which tumor cells interact.
Our observation of stroma-induced sensitization to certain agents has many parallels to the traditional definition of synthetic lethality. Historically, the concept of synthetic lethality has focused on how tumor cells harboring specific constitutive oncogenetic lesions can be responsive to certain agents, while activity is not observed in the absence of these genetic events. Our study introduces the notion that a synthetic lethal phenotype, rather than being exclusively genotype-dependent, can also be driven by the extrinsic influences of the tumor microenvironment. Importantly, CS-BLI can probe both genetically- and microenvironment-determined synthetic lethality in a high-throughput scalable manner. This allows the testing of a large number of permutations, including multiple candidate therapeutics, cell lines, and non-malignant accessory cells, thus enabling the previously intractable large-scale evaluation of how genetics and microenvironment interact to modulate cancer cell response to treatment.
Anti-cancer drug discovery is a serial process with high attrition at each step. Target identification, target validation, cell-based activity screens, toxicity screens, and animal efficacy studies all eliminate drugs from the discovery pipeline. In contrast, CS-BLI can add to the pipeline those agents with enhanced anti-tumor activity in the context of tumor-microenvironment interactions. These agents are currently eliminated from the process, because conventional preclinical in vitro drug screens and in vivo subcutaneous xenograft studies do not take into account the importance of tumor-microenvironment interactions modulating in vivo response to therapy. By introducing the element of tumor-microenvironment interactions at earlier in vitro stages of drug development, CS-BLI allows the reprioritization of those drugs which are active in the tumor microenvironment and bridges the gap between the over-simplicity of conventional anti-cancer drug screening in monolayer cultures with the intractable complexity of in vivo studies where tumor cells interact with their microenvironment.
MATERIAL AND METHODS
Tumor cell-specific bioluminescence imaging in co-cultures with stromal cells
We plated luciferase+ myeloma (MM.1S, MM.1R, KMS-18, INA-6, KMS-34, OPM-1, OPM-2), leukemia (KU812F, K562), and solid tumor (MDA-MB-231met, A375, FRO) cell lines in 96-well optical plates (Corning 3903) in the presence or absence of pre-plated luciferase− stromal cells (HS-5, KM101, KM103, KM104, KM105, primary stromal cells), with or without drug treatment, as indicated in each experiment. We measured tumor-specific viability using a Luminoskan luminometer (Labsystems) following addition of luciferin substrate (Xenogen Corp).
Molecular profiling of MM cells co-cultured in vitro with BMSCs
We co-cultured GFP+ tumor cell lines (MM.1S, MM.1R, INA-6) with GFP− HS-5 cells for 24 hours. Using fluorescent activated cell sorting (FACS), we isolated GFP+ MM cells (>95% purity in CD38+ CD138+ MM cells) from GFP− BMSCs. We then analyzed the gene expression profiling of the MM cell compartment using U133 2.0 Plus oligonucleotide microarrays (Affymetrix). We compared molecular profiles of MM cells cultured alone vs. cultured in the presence of BMSCs, as described in Supplement.
High-throughput screening of compound libraries in tumor-stroma co-cultures using CS-BLI
We plated stromal cells in 384-well flat bottom optical plates (Corning 3704) and allowed them to adhere overnight. We added luc+ MM.1S, MM.1R (Dex resistant MM.1S) or KU812F cells to cultures and treated them with small-molecule inhibitors from commercially available compound libraries (Biomol 2832 Kinase Inhibitor Library, Biomol International L.P); known bioactives libraries (Institute of Chemistry and Cell Biology (ICCB), Harvard Medical School); and natural products libraries (ICCB, Harvard Medical School). We incorporated DMSO controls and untreated controls on each plate and tested each condition in duplicate per plate, with 3 replicate plates before incubating cells at 37 °C for 48 h. We then added luciferin substrate and read the plates on a Luminoskan luminometer or Envision plate reader (PerkinElmer).
In vivo anti-tumor activity of reversine
We evaluated the in vivo anti-MM activity of reversine in an established model of diffuse MM lesions in SCID/Beige mice 12,38. Briefly, we housed male (6 to 8-week old) SCID/Beige mice (Jackson Laboratories), at the Animal Research Facility of the Dana-Farber Cancer Institute. We irradiated (150 rads) mice using a Cs137 γ-irradiator source and performed (24 hours post-irradiation) tail intravenous (i.v.) or subcutaneous (s.c.) injections of 106 MM.1S-GFP/luc cells per mouse in phosphate buffered saline (PBS). We monitored daily for changes in body weight, signs of infection or paralysis. In accordance with institutional guidelines, we sacrificed mice by CO2 inhalation in the event of paralysis or moribund state. All experimental procedures and protocols were approved by the Animal Care and Use Committee of the Dana-Farber Cancer Institute. We measured tumor burden weekly using the IVIS imaging system (Xenogen Corp) and Living Image software (Xenogen Corp) for i.v. injected mice or calipers for s.c. injected mice. We performed 2-way ANOVA statistical analysis using Prism software (Prism Software Corp).
Cell lines, reagents, cell survival assays, flow cytometric analyses, in vitro kinase and luciferase activity assays
Detailed information on cell lines from MM, leukemia, or solid tumors; primary MM tumor cells; stromal cells; reagents; conventional cell survival assays; flow cytometric analyses; as well as in vitro kinase and luciferase activity assays are included in the Supplement.
Supplementary Material
Acknowledgments
Supported by “Dunkin Donuts Rising Stars” Program at the Dana-Farber Cancer Institute (C.S.M), the Chambers Medical Foundation (C.S.M. and P.G.R), the Steven Cobb Foundation (D.W.M, C.S.M), and US National Institutes of Health grant R01CA050947 (C.S.M and K.C.A.). We wish to thank Towia Libermann and Marie Joseph-Bruno (Harvard Institutes of Medicine Genomics Core, Boston, MA) for generation of gene expression data and Leutz Buon (Dana-Farber Cancer Institute) for help with bioinformatic analyses.
Footnotes
Author Contributions
D.W.M. conducted experiments, performed analysis and wrote the manuscript: J.D. conducted experiments, performed analysis; E.W. conducted experiments, performed analysis; J.M.N. conducted experiments, performed analysis; D.C.G. conducted experiments, performed analysis; S.K. generated cell lines; N.M. performed analysis; R.L.S. provided primary tissue samples; N.C.M. provided primary tissue samples; A.L.K. performed analysis and participated in writing of the manuscript; J.D.G. provided cell lines; P.G.R. provided primary tissue samples. K.C.A. participated in writing of the manuscript; C.S.M. performed analysis, wrote manuscript and supervised the project.
REFERENCES
- 1.Mueller MM, Fusenig NE. Friends or foes - bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 2.Karnoub AE, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 3.Mitsiades CS, Mitsiades N, Munshi NC, Anderson KC. Focus on multiple myeloma. Cancer Cell. 2004;6:439–444. doi: 10.1016/j.ccr.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 4.Grigorieva I, Thomas X, Epstein J. The bone marrow stromal environment is a major factor in myeloma cell resistance to dexamethasone. Exp Hematol. 1998;26:597–603. [PubMed] [Google Scholar]
- 5.Hurt EM, et al. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell. 2004;5:191–199. doi: 10.1016/s1535-6108(04)00019-4. [DOI] [PubMed] [Google Scholar]
- 6.Hideshima T, et al. Thalidomide and its analogs overcome drug resistance of human multiple myeloma cells to conventional therapy. Blood. 2000;96:2943–2950. [PubMed] [Google Scholar]
- 7.Hideshima T, et al. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 2001;61:3071–3076. [PubMed] [Google Scholar]
- 8.Richardson PG, et al. Immunomodulatory drug CC-5013 overcomes drug resistance and is well tolerated in patients with relapsed multiple myeloma. Blood. 2002;100:3063–3067. doi: 10.1182/blood-2002-03-0996. [DOI] [PubMed] [Google Scholar]
- 9.Richardson PG, et al. A phase 2 study of bortezomib in relapsed, refractory myeloma. N Engl J Med. 2003;348:2609–2617. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- 10.Richardson PG, et al. A randomized phase 2 study of lenalidomide therapy for patients with relapsed or relapsed and refractory multiple myeloma. Blood. 2006;108:3458–3464. doi: 10.1182/blood-2006-04-015909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson PG, et al. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med. 2005;352:2487–2498. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- 12.Mitsiades CS, et al. Inhibition of the insulin-like growth factor receptor-1 tyrosine kinase activity as a therapeutic strategy for multiple myeloma, other hematologic malignancies, and solid tumors. Cancer Cell. 2004;5:221–230. doi: 10.1016/s1535-6108(04)00050-9. [DOI] [PubMed] [Google Scholar]
- 13.Nefedova Y, Cheng P, Alsina M, Dalton WS, Gabrilovich DI. Involvement of Notch-1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell lines. Blood. 2004;103:3503–3510. doi: 10.1182/blood-2003-07-2340. [DOI] [PubMed] [Google Scholar]
- 14.Shou Y, et al. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc Natl Acad Sci U S A. 2000;97:228–233. doi: 10.1073/pnas.97.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitsiades CS, et al. Activation of NF-kappaB and upregulation of intracellular anti-apoptotic proteins via the IGF-1/Akt signaling in human multiple myeloma cells: therapeutic implications. Oncogene. 2002;21:5673–5683. doi: 10.1038/sj.onc.1205664. [DOI] [PubMed] [Google Scholar]
- 16.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaffer AL, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitsiades CS, et al. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc Natl Acad Sci U S A. 2004;101:540–545. doi: 10.1073/pnas.2536759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitsiades N, et al. Molecular sequelae of histone deacetylase inhibition in human malignant B cells. Blood. 2003;101:4055–4062. doi: 10.1182/blood-2002-11-3514. [DOI] [PubMed] [Google Scholar]
- 20.Keen N, Taylor S. Aurora-kinase inhibitors as anticancer agents. Nat Rev Cancer. 2004;4:927–936. doi: 10.1038/nrc1502. [DOI] [PubMed] [Google Scholar]
- 21.Mitsiades N, et al. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc Natl Acad Sci U S A. 2002;99:14374–14379. doi: 10.1073/pnas.202445099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Opferman JT, et al. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 23.Yagoda N, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith DR, et al. Inhibition of interleukin 8 attenuates angiogenesis in bronchogenic carcinoma. J Exp Med. 1994;179:1409–1415. doi: 10.1084/jem.179.5.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lust JA, Donovan KA. The role of interleukin-1 beta in the pathogenesis of multiple myeloma. Hematol Oncol Clin North Am. 1999;13:1117–1125. doi: 10.1016/s0889-8588(05)70115-5. [DOI] [PubMed] [Google Scholar]
- 26.Roodman GD. Role of the bone marrow microenvironment in multiple myeloma. J Bone Miner Res. 2002;17:1921–1925. doi: 10.1359/jbmr.2002.17.11.1921. [DOI] [PubMed] [Google Scholar]
- 27.Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu D, Cozma D, Park A, Thomas-Tikhonenko A. Functional validation of genes implicated in lymphomagenesis: an in vivo selection assay using a Myc-induced B-cell tumor. Ann N Y Acad Sci. 2005;1059:145–159. doi: 10.1196/annals.1339.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindvall C, et al. Molecular characterization of human telomerase reverse transcriptase-immortalized human fibroblasts by gene expression profiling: activation of the epiregulin gene. Cancer Res. 2003;63:1743–1747. [PubMed] [Google Scholar]
- 30.Ingram WJ, Wicking CA, Grimmond SM, Forrest AR, Wainwright BJ. Novel genes regulated by Sonic Hedgehog in pluripotent mesenchymal cells. Oncogene. 2002;21:8196–8205. doi: 10.1038/sj.onc.1205975. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen BC, et al. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006;20:1028–1042. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhattacharya B, et al. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood. 2004;103:2956–2964. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- 33.Kannan K, et al. DNA microarrays identification of primary and secondary target genes regulated by p53. Oncogene. 2001;20:2225–2234. doi: 10.1038/sj.onc.1204319. [DOI] [PubMed] [Google Scholar]
- 34.Shaughnessy JD, Jr, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109:2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 35.Dimopoulos M, et al. Lenalidomide plus dexamethasone for relapsed or refractory multiple myeloma. N Engl J Med. 2007;357:2123–2132. doi: 10.1056/NEJMoa070594. [DOI] [PubMed] [Google Scholar]
- 36.Weber DM, et al. Lenalidomide plus dexamethasone for relapsed multiple myeloma in North America. N Engl J Med. 2007;357:2133–2142. doi: 10.1056/NEJMoa070596. [DOI] [PubMed] [Google Scholar]
- 37.San Miguel JF, et al. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N Engl J Med. 2008;359:906–917. doi: 10.1056/NEJMoa0801479. [DOI] [PubMed] [Google Scholar]
- 38.Mitsiades CS, et al. Fluorescence imaging of multiple myeloma cells in a clinically relevant SCID/NOD in vivo model: biologic and clinical implications. Cancer Res. 2003;63:6689–6696. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.