Abstract
TAR DNA-binding protein 43 (TDP-43) is a nuclear protein involved in RNA splicing and a major protein component in ubiquitin-positive, tau-negative inclusions of frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS). Under disease conditions TDP-43 redistributes to the cytoplasm where it can be phosphorylated, ubiquitinated, and proteolytically cleaved. Enzymes responsible for TDP-43 proteolytic processing in brain remain largely unreported. Using a mass spectrometry approach, we identified two truncated TDP-43 peptides, terminating C-terminal to asparagines 291 (N291) and 306 (N306). The only documented mammalian enzyme capable of cleaving C-terminal to asparagine is asparaginyl endopeptidase (AEP). TDP-43-immunoreactive fragments (~35 and 32 kDa) predicted to be generated by AEP cleavage at N291 and N306 were observed by western blot analyses of post-mortem FTLD brain tissue and cultured human cells over-expressing TDP-43. Studies in vitro determined that AEP can directly cleave TDP-43 at seven sites, including N291 and N306. Western blots of brain homogenates isolated from AEP-null mice and wild-type littermate controls revealed that TDP-43 proteolytic fragments were substantially reduced in the absence of AEP in vivo. Taken together, we conclude that TDP-43 is cleaved by AEP in brain. Moreover, these data highlight the utility of combining proteomic strategies in vitro and in vivo to provide insight into TDP-43 biology that will fuel the design of more detailed models of disease pathogenesis.
Keywords: Asparaginyl endopeptidase, degradomics, frontotemporal dementia, neurodegeneration, TAR DNA-binding protein 43
1 Introduction
Frontotemporal lobar degeneration (FTLD) is the second leading cause of dementia in the population under the age of 65 and is clinically characterized by prominent behavioral abnormalities, including personality changes and language dysfunction [1, 2]. The most common underlying pathology in FTLD is ubiquitin-positive inclusions that lack tau or α-synuclein and contain the RNA binding protein, TAR DNA-binding protein 43 (TDP-43) [3–5]. Ubiquitinated TDP-43 inclusions are also present in spinal cord of individuals with amyotrophic lateral sclerosis (ALS), a severe form of motor neuron disease which can co-occur with FTLD. Moreover, more than thirty TDP-43 (TARDBP) mutations have been identified in ALS cases and rare FTLD patients [5]. Under normal physiological conditions, TDP-43 predominantly resides in the nucleus, but in disease TDP-43 redistributes to the cytoplasm where it is phosphorylated, ubiquitinated, and forms inclusions [4]. Immunoblot analyses of detergent-insoluble fractions from postmortem human brain tissue indicated that short TDP-43 C-terminal fragments (CTFs) are enriched in FTLD cases compared to control or Alzheimer’s disease (AD) brains [4, 6]. These studies suggest that proteolytic cleavage of TDP-43 may play a unique role in disease pathogenesis; however, the underlying molecular mechanisms that contribute to FTLD disease progression and neurodegeneration remain to be elucidated.
Abnormal proteolytic processing and aggregation of truncated products is a commonality among neurodegenerative diseases, including AD, Parkinson’s disease, and polyglutamine disorders [7–10]. Studies comparing frontal cortex and spinal cord from FTLD and ALS cases, respectively, indicate that TDP-43 CTFs are enriched selectively in brain [6, 11]. It is proposed that TDP-43 CTFs initiate and/or promote TDP-43 aggregation, and purification of TDP-43 CTFs from FTLD brains revealed that Arg 208 is a likely cleavage site [11]. Furthermore, Nonaka et al. suggest that proteolytic processing at Met 218 – Asp 219 or Glu 246 – Asp 247 may generate TDP-43 CTFs in FTLD [12]. Several studies also indicate that caspases may mediate TDP-43 CTF production in brain [13–15]; however, other TDP-43-cleaving enzymes remain unreported.
In this study we employed a mass spectrometry approach to catalog TDP-43 proteolytic fragments from post-mortem human brain samples. Comparison of the detergent-insoluble proteome from FTLD and control cases revealed an enrichment of TDP-43 C-terminal peptides in disease tissue, which was consistent with previous findings [4, 16–19]. A semi-tryptic TDP-43 peptide terminating in asparagine 291 (N291) was identified in FTLD tissue and parallel analysis of a cultured human cell line revealed an additional prematurely truncated TDP-43 peptide, terminating C-terminal to N306. Notably, asparaginyl endopeptidase (AEP), a late endosomal/lysosomal cysteine protease that is expressed in brain, is the only known mammalian enzyme that cleaves C-terminal to asparagine. In vitro assays utilizing recombinant AEP and TDP-43 revealed that AEP can directly cleave TDP-43 at seven sites, including N291 and N306. Furthermore, western blot analyses of murine frontal cortex homogenates indicated that TDP-43-immunoreactive fragments (~33–37 kDa), predicted to be generated by AEP cleavage, were noticeably absent in AEP null mice compared to littermate controls. Taken together, we propose that TDP-43 is a substrate of AEP in human and mouse brain.
2 Materials and methods
2.1 Protein extraction and digestion from brain tissue
FTLD cases included in this study underwent extensive neuropathologic characterization required for diagnosis based on consensus criteria in the Alzheimer’s Disease Research Center Neuropathology Core [20, 21]. The cases characterized as FTLD exhibited ubiquitin/TDP-43-positive, tau- and α-synuclein-negative cytoplasmic inclusions in neurons of both the superficial frontal cortex and dentate gyrus of the hippocampus. Furthermore, these cases did not meet criteria for the neuropathological diagnosis of AD (CERAD [22] or NIA-Reagan [23] or Lewy Body disease [24], nor did they display tau pathology consistent with a tauopathy [20]. Frontal cortex from four FTLD cases (confirmed TDP-43 positive) and five control cases, matched as closely as possible by age and post-mortem interval (PMI) (Supplemental Table S1), were weighed individually (~ 1 g), homogenized (Dounce homogenizer), and detergent-insoluble protein extraction was performed by differential ultracentrifugation as described previously [25].
2.2 Western blotting and antibodies
Immunoblotting was performed according to standard procedures as reported previously [26, 27]. Briefly, samples were resolved by SDS-PAGE and stained with Coomassie Blue G-250 or transferred to Immobilon-P membranes (Millipore, Bedford, MA). Blots were blocked with TBS plus blocking buffer (USB Corporation, Cleveland, OH) at room temperature for 30 min and probed with primary antibodies in TBS plus 0.1 % Tween-20 plus blocking buffer overnight at 4°C. The following day, blots were rinsed and incubated with secondary antibodies conjugated to fluorophores (Molecular Probes/Invitrogen) for one hour at room temperature. Images were captured using an Odyssey Image Station (LiCor, Lincoln, NE). Antibodies used: rabbit anti-TDP-43 (ProteinTech, 10782-1-AP), mouse monoclonal anti-HA (Covance, Princeton NJ), mouse monoclonal β-tubulin (Promega, G712A).
2.3 Peptide analysis by LC-MS/MS
Urea fraction proteins (50 µg per sample) corresponding to pooled control or FTLD samples were reduced with 10 mM dithiothreitol (DTT) and alkylated in the dark with 50 mM iodoacetamide for 30 min. The samples were then separated on a 10% SDS gel (0.75 mm thickness) and stained with Coomassie Blue G-250 to both confirm equal sample loading and visualize proteins for subsequent processing. Each sample lane was then cut into twenty gel bands and subjected to in-gel digestion (12.5 µg/mL trypsin). Purified peptides were analyzed by reverse-phase liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) and all samples were analyzed in technical replicate. Briefly, peptide mixtures were loaded onto a C18 column (75 µm i.d., 10 cm long, 3 µM resin from Michrom Bioresources, Inc., Auburn, CA) and eluted over a 5–30% gradient (Buffer A: 0.4% acetic acid, 0.005% heptafluorobutyric acid, and 5% AcN; Buffer B: 0.4% acetic acid, 0.005% heptafluorobutyric acid, and 95% AcN). Eluates were monitored in a MS survey scan followed by nine data-dependent MS/MS scans on an LTQ-Orbitrap ion trap mass spectrometer (Thermo Finnigan, San Jose, CA). The LTQ acquired MS/MS spectra (2 m/z isolation width, 35% collision energy, 5,000 AGC target, 150 ms maximum ion time). The Orbitrap collected MS scans (300–1600 m/z, 1,000,000 AGC target, 750 ms maximum ion time, resolution 60,000). Acquired MS/MS spectra were searched against the human reference database of the National Center for Biotechnology Information (November 19, 2008) using the SEQUEST Sorcerer algorithm (version 2.0, SAGE-N). Searching parameters included: partially tryptic restriction, parent ion mass tolerance (± 50 ppm), product ion tolerance (± 0.5 m/z), fixed modification on carboxyamidomethylated Cys (+57.0215 Da); dynamic modifications for oxidized Met (+15.9949 Da), 2 maximal modification sites and 2 maximal missed cleavages. Only b and y ions were considered during the database match. To assess false discovery rate (FDR), all original protein sequences were reversed to generate a decoy database that was concatenated to the original database. The FDR was estimated by the number of decoy matches (nd) and total number of assigned matches (nt). FDR = 2*nd/nt, assuming mismatches in the original database were the same as in the decoy database. To remove false positive matches, assigned peptides were grouped by a combination of tryptic state (fully and semi) and precursor ion-charge state (2+, 3+, and 4+). All groups were first filtered by mass accuracy (10 ppm), and then XCorr and ΔCn values to reduce FDR to less than 1%. XCorr is the cross-correlation score between experimental and theoretical spectra, and ΔCn is the normalized difference of XCorr scores between the 1st and 2nd peptide matches. The MS/MS spectra of TDP-43 peptides were manually inspected. Representative MS/MS spectra for TDP-43 peptides are listed in supplemental data (Supplemental Table S3) with amino acid sequence, matching scores (XCorr and ΔCn) and mass accuracy.
2.4 TDP-43 expression in HEK293 cells
Plasmid expressing recombinant TDP-43 harboring an N-terminal hemagglutinin (HA) tag (HA-TDP-43) was generated as previously described [28]. Human embryonic kidney 293 (HEK293) cells were maintained in DMEM with 10% fetal bovine serum (FBS) (Invitrogen) and 1% penicillin/streptomycin (Cellgro/Mediatech Inc., Herndon, VA). Equivalent amounts of cells were plated and transfected with the indicated constructs using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. Cells were lysed and prepared for LC-MS/MS and Western blot.
2.5 In vitro AEP cleavage assay
Plasmid expressing TDP-43 fused to GST (pGS21-TDP-43) was transformed into the E. coli BL21 cells and a monoclonal culture was expanded in 150 mL TB media until reaching an OD600 of 0.6 and induced for 6h with 0.1M IPTG (Sigma, St. Louis MO) at 28°C. Isolation of GST-TDP-43 and binding to glutathione sepharose (Amersham, Piscataway NJ) was carried out as previously reported [29]. Isolated TDP-43 on beads (5 µg) was washed in reaction buffer (50 mM sodium citrate, pH 6.0, containing 1 mM EDTA, 1 mM DTT, 60 mM Na2HPO4 and 0.01% CHAPS). Recombinant human AEP (R&D Systems) was added (20 ng/uL) in reaction buffer or buffer alone as controls. Samples were incubated for 0, 30, 60 or 120 min at 37°C and subsequently analyzed by western blot or LC-MS/MS as described above. Representative MS/MS spectra for AEP cleaved TDP-43 peptides are provided in (Supplemental Table S4).
2.6 Western blot analysis of AEP−/− brain
AEP−/− mice were previously generated and characterized [30, 31]. Brains were excised from individual AEP−/− mice or AEP+/+ littermate controls and whole brain homogenates were lysated in enzyme reaction buffer (25 mM sodium citrate [pH 6.0], 25 mM KCI, 5 mM MgCl2, 1mM 2-mercaptoethanol, 1mM PMSF, plus protein inhibitor cocktail) as previously reported [30]. Protein concentration was determined by the bicinchoninic acid method and samples were subjected to Western blot.
3 Results and discussion
3.1 Identification of TDP-43 proteolytic fragments in FTLD insoluble proteome
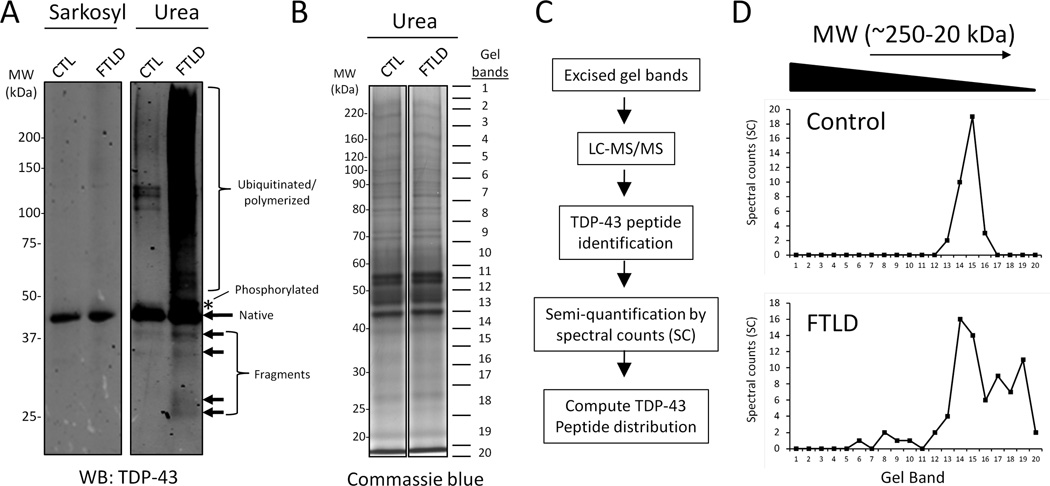
Like other neurodegenerative diseases, FTLD is characterized by intracellular deposition of detergent-insoluble protein aggregates, and the most common underlying pathology is ubiquitinated TDP-43 inclusions. The previously reported observation that TDP-43 C-terminal fragments (CTFs) are enriched in detergent-insoluble fractions from postmortem FTLD cases suggests that proteolytic processing of TDP-43 may be disrupted in FTLD. Therefore, we sought to catalog the TDP-43 proteolytic fragments in human post-mortem control and FTLD brain to facilitate the identification of enzymes responsible for TDP-43 processing. In this strategy, frontal cortex from control or FTLD cases was serially extracted with buffers of increasing stringency, first by sarkosyl detergent containing buffer and subsequently by urea, to enrich for detergent-insoluble TDP-43. An equivalent amount of urea extract from each case was combined for pooled control and FTLD samples. To confirm the disease-specific TDP-43 biochemical signature reported for FTLD, western blot analysis was performed using antibody directed against the N-terminus of TDP-43 (Fig. 1A). TDP-43 immunoreactivity was observed at ~43 kDa in detergent-soluble (sarkosyl) and detergent-insoluble (urea) control and FTLD cases. Notably in the FTLD detergent-insoluble sample, there was a selective enrichment for phosphorylated (~45 kDa) and high molecular weight (MW) TDP-43 immunoreactivity, suggesting protein polymerization and/or ubiquitination [4]. Additionally, TDP-43 fragments (~25–40 kDa) were detected in detergent-insoluble control and FTLD samples as previously described [4, 16–19]. To identify TDP-43 proteolytic cleavage products, pooled control and FTLD samples were subjected to SDS-PAGE and twenty gel bands were excised for LC-MS/MS analyses (Fig. 1B and C). First, the pooled control sample was analyzed by LC-MS/MS, and then following two consecutive washes, the pooled FTLD sample was analyzed. The majority of sequenced TDP-43 peptides in control brain were sharply distributed between 34 and 46 kDa, whereas TDP-43 peptides were more broadly distributed between ~15 and 140 kDa in FTLD (Fig. 1D). These results are consistent with the pattern of TDP-43 immunoreactivity observed in control and FTLD detergent-insoluble fractions (Fig. 1A). Breakdown of individual TDP-43 peptide and spectral count distribution in each MW region indicated that in FTLD, below gel band 17 (<30 kDa), the C-terminal region of TDP-43 is predominantly observed (Supplemental Table S2). This finding is consistent with reported observations documenting accumulation of TDP-43 CTFs in FTLD. Analysis of semi-tryptic TDP-43 peptides in the FTLD sample revealed a prematurely truncated peptide terminating at asparagine 291 (N291) (Fig. 2A and Table 1). The truncated N291 peptide was only sequenced in the FTLD sample, however based on the peptide retention time and the mass accuracy afforded by the Orbitrap, a detectable signal for the N291 peptide was observed in the control sample, suggesting that TDP-43 fragmentation at N291 may not be disease-specific.
Figure 1. Identification of TDP-43 proteolytic fragments from post-mortem control and FTLD brain.
(A) Western blot analysis of sarkosyl (soluble) and insoluble fractions show enrichment of post-translationally modified TDP-43 (i.e. ubiquitination/polymerization, phosphorylation and proteolytic cleavage) in control and FTLD brain. (B and C) Detergent-resistant fractions were resolved by SDS-PAGE and the control and FTLD samples were excised into 20 gel slices, trypsin digested, and analyzed via LC-MS/MS on a high resolution Orbitrap mass spectrometer. (D) Differences in the TDP-43 peptide distribution between control and FTLD. Spectral counts (SCs) (x-axis) refer to the number of unique TDP-43 peptide sequencing events in each gel band (y-axis).
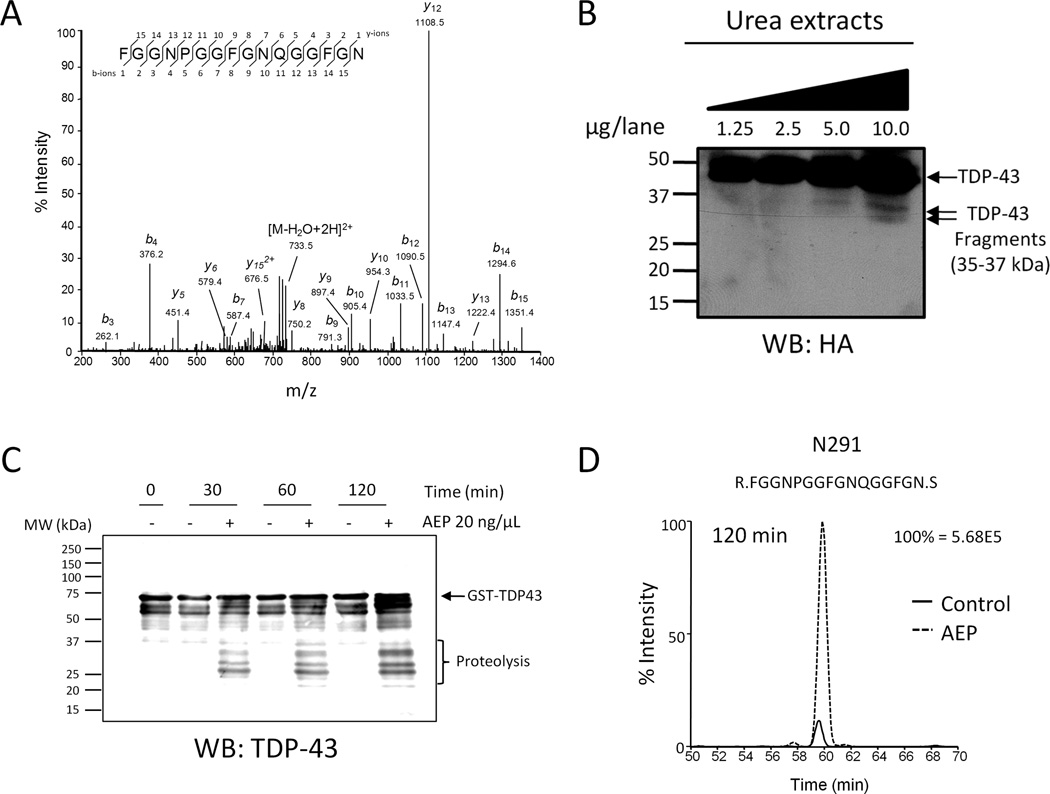
Figure 2. TDP-43 is an AEP substrate.
(A) Representative MS/MS spectra of TDP-43 partial tryptic peptides prematurely ending with C-terminal asparagine 291 (N291), that were identified in FTLD and HEK293 cell samples. (B) Western blot analysis of N-terminal TDP-43 cleavage products from cells expressing HA-TDP-43. HEK293 cells were transfected with plasmid expressing N-terminally tagged HA-TDP-43 and sarkosyl-insoluble fractions were prepared for parallel LC-MS/MS and western blot analyses. HA-immunoreactive TDP-43 cleavage products with predicted molecular weights of approximately 32 and 35 kDa were observed in urea extracts. (C) Asparaginyl endopeptidase (AEP) cleaves TDP-43 in vitro. Recombinant TDP-43 fused to GST was incubated with or without recombinant AEP for the indicated times at 37°C. Western blot revealed increased TDP-43 immunoreactivity ≤37 kDa over time in samples containing AEP. (D) Representative extracted ion chromatogram of N291 peptide from in vitro samples. TDP-43 semi-tryptic peptides prematurely ending with N291 were identified in samples with and without AEP at the 120 min time point, however the presence of AEP increased the abundance of these peptides approximately 10-fold. Data representative of three independent experiments.
Table 1.
TDP-43 peptides identified in the detergent-insoluble proteome in human brain and cells over-expressing recombinant TDP-43
| Sequence | Trypticity | Position | HEK293 | Brain |
|---|---|---|---|---|
| R.LVEGILHAPDAGWGNLVYVVNYPK.D | Fully | 56–79 | Yes | Yes |
| R.KMDETDASSAVK.V | Fully | 84–95 | Yes | Yes |
| K.MDETDASSAVK.V | Fully | 85–95 | Yes | Yes |
| K.TSDLIVLGLPWK.T | Fully | 103–114 | Yes | Yes |
| K.TTEQDLK.E | Fully | 115–121 | Yes | No |
| K.EYFSTFGEVLM*VQVK.K | Fully | 122–136 | Yes | No |
| R.FTEYETQVK.V | Fully | 152–160 | Yes | Yes |
| R.HMIDGR.W | Fully | 166–171 | Yes | No |
| K.QSQDEPLR.S | Fully | 182–189 | No | Yes |
| R.EFFSQYGDVM*DVFIPK.P | Fully | 209–224 | Yes | Yes |
| R.EFFSQYGDVMDVFIPK.P | Fully | 209–224 | Yes | Yes |
| K.GISVHISNAEPK.H | Fully | 252–263 | Yes | Yes |
| I.SVHISNAEPK.H | Semi | 254–263 | No | Yes |
| R.FGGNPGGFGNQGGFGN.S | Semi | 276–291 | Yes | Yes |
| R.FGGNPGGFGNQGGFGNSR.G | Fully | 276–293 | Yes | Yes |
| R.GGGAGLGNNQGSN.M | Semi | 294–306 | Yes | No |
| R.GGGAGLGNNQGSNM*GGGM*NFGAF.S | Semi | 294–316 | Yes | No |
| R.GGGAGLGNNQGSNM*GGGMNFGAF.S | Semi | 294–316 | Yes | No |
| R.GGGAGLGNNQGSNMGGGM*NFGAF.S | Semi | 294–316 | Yes | No |
| R.GGGAGLGNNQGSNMGGGMNFGAF.S | Semi | 294–316 | Yes | No |
| M.LASQQNQSGPSGNNQNQGNMQR.E | Semi | 340–361 | Yes | No |
represents oxidized methionine (M)
3.2 Identification of TDP-43 proteolytic fragments in HEK293 cells
In parallel to proteomic analyses of post-mortem human brain samples, we transiently expressed recombinant TDP-43 harboring an N-terminal hemagglutinin (HA) tag (HA-TDP-43) in human embryonic kidney cells (HEK) 293 cells and profiled the detergent-insoluble proteome by LC-MS/MS. Due to low endogenous expression of TDP-43 in cultured human cells, over-expression of TDP-43 was necessary to facilitate detection of TDP-43 peptides in the detergent-insoluble fraction. Twenty-four hours post-transfection, HEK293 cells were harvested for LC-MS/MS and western blot analyses. Comparison of detergent-insoluble proteomes from human brain samples and cells expressing HA-TDP-43 revealed 10 shared peptides, including peptides truncated at N291 (Table 1). Additionally, a semi-tryptic peptide truncated at N306 was identified in HA-TDP-43 expressing cells. Proteolytic cleavage of TDP--43 at N291 and N306 is expected to generate N-terminal TDP-43 fragments of ~35 and 32 kDa (Fig. 4). Notably, two TDP-43-immunoreactive products, ~35 and 32 kDa, were observed in detergent-insoluble fractions from FTLD cases by western blot using TDP-43 antibody targeted against the N-terminus (Fig. 1A). Similarly, western blot analyses of detergent-insoluble fractions prepared from HEK293 cells expressing HA-TDP-43 revealed HA-immunoreactivity at ~35 and 32 kDa (Fig. 2B). The only reported mammalian enzyme that cleaves C-terminal to asparagine is asparaginyl endopeptidase (AEP), a late endosomal/lysosomal cysteine protease that is expressed in brain [30–32]. AEP, also known as legumain, hydrolyzes the C-terminal side of asparagine residues in the substrate and is highly active under acidic conditions [32]. Notably, AEP and caspases are structurally similar, share similar roles in apoptosis, and hypothesized to be derived from a common protein ancestor [33]. AEP also plays a critical role in processing antigens for class II major histocompatibility complex (MHC) presentation as well as Toll-like receptors and extracellular matrix proteins [34–36]. Importantly, AEP is not restricted to the late endosome/lysosome compartment in neurons; Liu et al. report that under hypoxia conditions AEP proteins levels are substantially increased in the neuronal cell body and neuritic processes where it cleaves the DNase inhibitor SET [30].
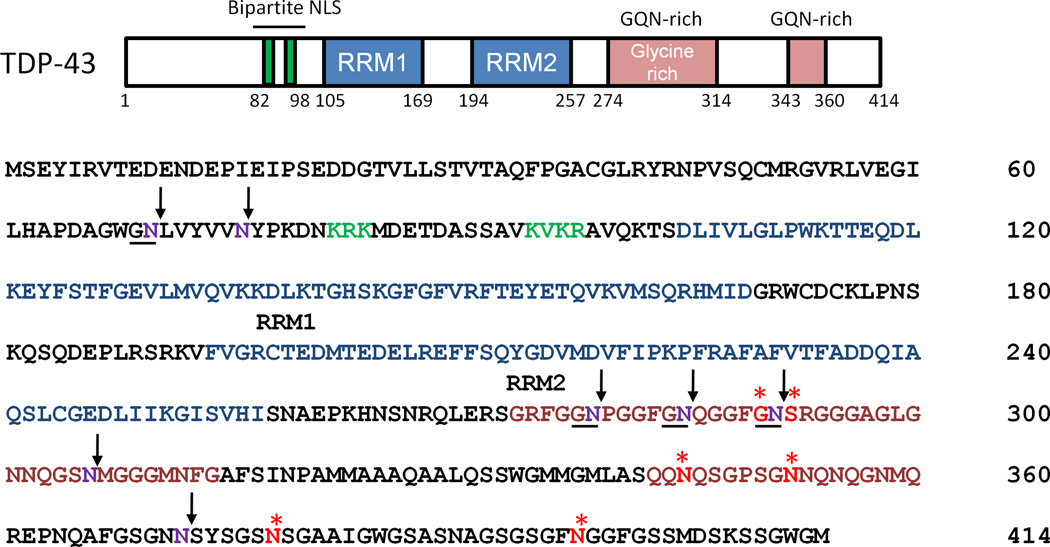
Figure 4. Proposed AEP cleavage sites in TDP-43.
TDP-43 amino acid sequence illustrates functional motifs and putative AEP recognition sites. Arrows designate asparagines predicted to be AEP cleavage sites (residues in purple) based on compiled LC-MS/MS data in this report. Stars indicate residues (highlighted in red) that are known genetic mutations in patients with amyotrophic lateral sclerosis (ALS). RRM, RNA recognition motif (highlighted in blue); NLS, nuclear localization signal (highlighted in green), Glycine, glutamine, asparagine (G/Q/N) rich domain (highlighted in pink).
3.3 TDP-43 is cleaved by AEP in vitro
Based on results from our LC-MS/MS and western blot analyses of post-mortem human cases and human cells expressing HA-TDP-43, we hypothesized that AEP directly cleaves TDP-43 at N291 and N306. To test this hypothesis, recombinant TDP-43 fused to GST was produced in bacteria and isolated with glutathione sepharose. Purified TDP-43 was incubated with or without recombinant human AEP for 0, 30, 60 or 120 min at 37°C and samples were subjected to SDS-PAGE. Western blot analysis with N-terminal specific TDP-43 antibody revealed a relative absence of TDP-43 immunoreactivity below the 37 kDa molecular weight marker in samples lacking AEP (Fig. 2C). However in the presence of AEP, the intensity of TDP-43-immunoreactive fragments below 37 kDa increased over time. Parallel LC-MS/MS analysis indicated seven putative AEP cleavage sites in TDP-43, including N291 and N306 (Table 2 and Fig. 4). At the 120 min time point TDP-43 peptides truncated at N291 were also observed in samples absent for AEP, suggesting that non-specific fragmentation at N291 can occur in vitro. However, comparison of extracted ion chromatograms reveal that TDP-43 peptides terminating at N291 were ~10-fold more abundant in samples containing AEP (Fig. 2D). Based on these in vitro results, we conclude that AEP can directly cleave TDP-43.
Table 2.
AEP specific TDP-43 cleavage sites determined in vitro
| Sequence | Trypticity | Position | Site |
|---|---|---|---|
| N.LVYVVNYPK.D | Semi | 71–79 | N70 |
| R.LVEGILHAPDAGWGNLVYVVN.Y | Semi | 56–76 | N76 |
| N.PGGFGNQGGFGNSR.G | Semi | 280–293 | N279 |
| R.FGGNPGGFGNQGGFGN.S | Semi | 276–291 | N291 |
| R.FGGNPGGFGN.Q | Semi | 276–285 | N285 |
| R.GGGAGLGNNQGSN.M | Semi | 294–306 | N306 |
| R.EPNQAFGSGNN.S | Semi | 362–372 | N372 |
3.4 TDP-43 fragments are reduced in AEP−/− mice
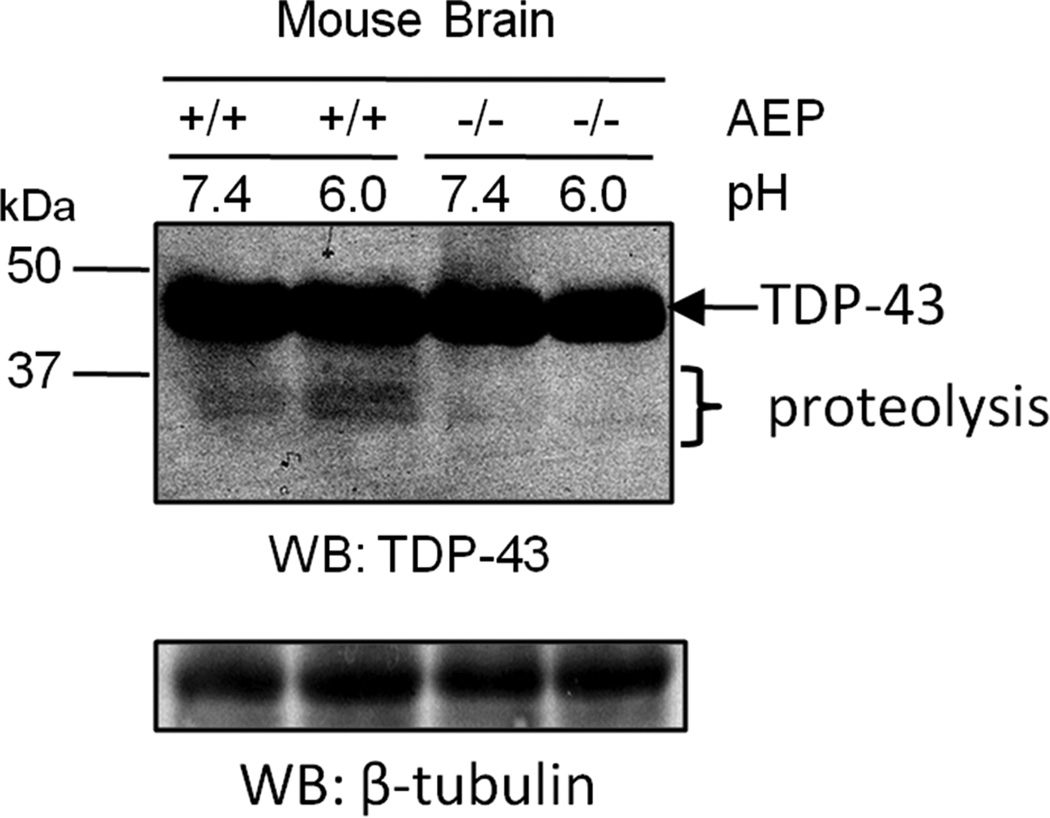
Our in vitro studies highlight the possibility that TDP-43 fragmentation at N291 can occur in the absence of AEP. Therefore, we sought to determine if AEP plays a role in TDP-43 cleavage in vivo. Western blot of post-mortem human brain tissue and human cells expressing HA-TDP-43 displayed N-terminal TDP-43-immunoreactive fragments of ~35 and 32 kDa (Fig. 1A and 2B). Based on our LC-MS/MS analyses and in vitro studies, we predicted that these TDP-43 isoforms are generated by AEP cleavage at N291 and N306 (Fig. 4). To assess if TDP-43 fragmentation is observed in the absence of AEP in vivo, frontal cortex homogenates from adult AEP-null (AEP−/−) and wild-type littermate control (AEP+/+) mice were lysed in buffer at pH 6.0 or pH 7.4 [30] and samples were subjected to Western blot analysis using TDP-43 antibody targeted against the N-terminus. No change in the level of full-length TDP-43 protein was observed, however TDP-43-immunoreactive fragments (~35 and 32 kDa) were noticeably absent in AEP−/− brain homogenates compared to AEP+/+ littermates (Fig. 3). Moreover, the intensity of TDP-43-immunoreactive bands below 37 kDa were increased in AEP+/+ samples lysed at pH 6.0. These findings are consistent with previous reports documenting that AEP substrate cleavage is maximal around pH 6.0 [32]. Based on these results and the in vitro and the post-mortem human case studies described above, we conclude that TDP-43 is directly cleaved by AEP in brain.
Figure 3. TDP-43 fragments are reduced in AEP−/− brain.
Comparison of whole brain homogenates from AEP-null (−/−) and wild-type (+/+) littermate controls by western blot revealed that N-terminal TDP-43-immunoreactive fragments (~35 and 32 kDa) were substantially reduced in the absence of AEP. β-tubulin was used as a western blot loading control. Data representative of two independent experiments.
4 Concluding remarks
TDP-43 is a nuclear protein that harbors two highly conserved RNA recognition motifs (RRM) and an intrinsically disordered "prion-like" C-terminal domain rich with glycine, glutamine, and asparagine (G/Q/N) (Fig. 4). Under physiological conditions this domain mediates protein – protein interactions and is critical for TDP-43’s role in alternative splicing as well as transcriptional regulation [37]. Many prion-like proteins self-aggregate when over-expressed [38, 39], and TDP-43’s C-terminal domain is hypothesized to enhance TDP-43 aggregation in FTLD and ALS. Importantly, the TDP-43 C-terminus harbors nearly all the genetic mutations associated with familial ALS and FTLD, suggesting that alterations in the function of this protein region may contribute to disease pathogenesis [40]. In this report, the majority of putative AEP cleavage sites identified in TDP-43 in vivo and in vitro reside in the C-terminus (Fig. 4). The biological implications of TDP-43 processing by AEP remain to be determined; however the sites of cleavage, especially N291, raises questions regarding the possibility that TDP-43 genetic mutations associated with familial ALS patients and FTLD patients may impact TDP-43 proteolysis by AEP. Specifically, mutations substituting glycine 290 and serine 292 to alanine and asparagine, respectively, may structurally alter TDP-43 and abrogate AEP cleavage at N291 or introduce a novel cleavage site at N292 (Fig. 4). Other ALS mutations map directly to asparagines at the extreme C-terminus of TDP-43, including N345, N352, N378, and N398 [40]. Although these asparagines were not identified in our LC-MS/MS analyses as terminal residues in prematurely truncated peptides, it is possible that the use of trypsin limited our coverage of peptides originating from this region of TDP-43. Alternative methods of protein digestion, such as using chymotrypsin, may be necessary to catalog other AEP cleavage sites. In conclusion, we provide strong evidence that TDP-43 is an AEP substrate and that N291 is a candidate AEP recognition site in human brain. These insights may prove an exciting avenue for future studies investigating the impact of familial ALS and FTLD genetic mutations on this aspect of TDP-43 biology.
Supplementary Material
Acknowledgements
We thank members of the Lah/Levey and Seyfried labs for constructive discussion regarding this manuscript. This work was supported by the National Institutes of Health through the Emory Alzheimer’s Disease Center grant (P50AG025688), the Emory Neuroscience NINDS Core Facilities (P30NS055077), and NIH training grant (F32AG032848-02 to N.T.S.). Acknowledgement is made to the donors of ADR, a program of the American Health Assistance Foundation, for support of this research.
Abbreviations
- AEP
Asparaginyl endopeptidase
- TDP-43
TAR DNA-binding protein 43
- FTLD
Frontotemporal lobar degeneration
- ALS
Amyotrophic lateral sclerosis
- CTFs
C-terminal fragments
References
- 1.Josephs KA. Frontotemporal dementia and related disorders: Deciphering the enigma. Annals of Neurology. 2008;64:4–14. doi: 10.1002/ana.21426. [DOI] [PubMed] [Google Scholar]
- 2.Forman MS, Farmer J, Johnson JK, Clark CM, et al. Frontotemporal dementia: clinicopathological correlations. Ann Neurol. 2006;59:952–962. doi: 10.1002/ana.20873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackenzie IR, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- 4.Neumann M, Sampathu DM, Kwong LK, Truax AC, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 5.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Igaz LM, Kwong LK, Xu Y, Truax AC, et al. Enrichment of C-terminal fragments in TAR DNA-binding protein-43 cytoplasmic inclusions in brain but not in spinal cord of frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Am J Pathol. 2008;173:182–194. doi: 10.2353/ajpath.2008.080003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardy J, Allsop D. Amyloid deposition as the central event in the aetiology of Alzheimer's disease. Trends Pharmacol Sci. 1991;12:383–388. doi: 10.1016/0165-6147(91)90609-v. [DOI] [PubMed] [Google Scholar]
- 8.Li W, West N, Colla E, Pletnikova O, et al. Aggregation promoting C-terminal truncation of α-synuclein is a normal cellular process and is enhanced by the familial Parkinson's disease-linked mutations. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2162–2167. doi: 10.1073/pnas.0406976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarlac V, Storey E. Role of proteolysis in polyglutamine disorders. Journal of Neuroscience Research. 2003;74:406–416. doi: 10.1002/jnr.10746. [DOI] [PubMed] [Google Scholar]
- 10.Wang YP, Biernat J, Pickhardt M, Mandelkow E, Mandelkow E-M. Stepwise proteolysis liberates tau fragments that nucleate the Alzheimer-like aggregation of full-length tau in a neuronal cell model. Proceedings of the National Academy of Sciences. 2007;104:10252–10257. doi: 10.1073/pnas.0703676104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Igaz LM, Kwong LK, Chen-Plotkin A, Winton MJ, et al. Expression of TDP-43 C-terminal Fragments in Vitro Recapitulates Pathological Features of TDP-43 Proteinopathies. J. Biol. Chem. 2009;284:8516–8524. doi: 10.1074/jbc.M809462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nonaka T, Kametani F, Arai T, Akiyama H, Hasegawa M. Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Human Molecular Genetics. 2009;18:3353–3364. doi: 10.1093/hmg/ddp275. [DOI] [PubMed] [Google Scholar]
- 13.Zhang YJ, Xu YF, Dickey CA, Buratti E, et al. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J Neurosci. 2007;27:10530–10534. doi: 10.1523/JNEUROSCI.3421-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y-J, Xu Y-F, Cook C, Gendron TF, et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proceedings of the National Academy of Sciences. 2009;106:7607–7612. doi: 10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dormann D, Capell A, Carlson AM, Shankaran SS, et al. Proteolytic processing of TAR DNA binding protein-43 by caspases produces C-terminal fragments with disease defining properties independent of progranulin. Journal of Neurochemistry. 2009;110:1082–1094. doi: 10.1111/j.1471-4159.2009.06211.x. [DOI] [PubMed] [Google Scholar]
- 16.Cairns NJ, Neumann M, Bigio EH, Holm IE, et al. TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am J Pathol. 2007;171:227–240. doi: 10.2353/ajpath.2007.070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackenzie IR, Bigio EH, Ince PG, Geser F, et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol. 2007;61:427–434. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- 18.Neumann M, Kwong LK, Truax AC, Vanmassenhove B, et al. TDP-43-positive white matter pathology in frontotemporal lobar degeneration with ubiquitin-positive inclusions. J Neuropathol Exp Neurol. 2007;66:177–183. doi: 10.1097/01.jnen.0000248554.45456.58. [DOI] [PubMed] [Google Scholar]
- 19.Mackenzie IR, Rademakers R. The molecular genetics and neuropathology of frontotemporal lobar degeneration: recent developments. Neurogenetics. 2007 doi: 10.1007/s10048-007-0102-4. [DOI] [PubMed] [Google Scholar]
- 20.Cairns N, Bigio E, Mackenzie I, Neumann M, et al. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathologica. 2007;114:5–22. doi: 10.1007/s00401-007-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackenzie I, Neumann M, Bigio E, Cairns N, et al. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathologica. 2009;117:15–18. doi: 10.1007/s00401-008-0460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirra SS, Heyman A, McKeel D, Sumi SM, et al. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–486. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- 23.Consensus recommendations for the postmortem diagnosis of Alzheimer's disease. The National Institute on Aging, and Reagan Institute Working Group on Diagnostic Criteria for the Neuropathological Assessment of Alzheimer's Disease. Neurobiol Aging. 1997;18:S1–S2. [PubMed] [Google Scholar]
- 24.McKeith IG, Dickson DW, Lowe J, Emre M, et al. Diagnosis and management of dementia with Lewy bodies. Neurology. 2005;65:1863–1872. doi: 10.1212/01.wnl.0000187889.17253.b1. [DOI] [PubMed] [Google Scholar]
- 25.Gozal YM, Duong DM, Gearing M, Cheng D, et al. Proteomics analysis reveals novel components in the detergent-insoluble subproteome in Alzheimers disease. Journal of Proteome Research. 2009 doi: 10.1021/pr900474t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herskowitz JH, Seyfried NT, Duong DM, Xia Q, et al. Phosphoproteomic analysis reveals site-specific changes in GFAP and NDRG2 phosphorylation in frontotemporal lobar degeneration. J Proteome Res. 2010;9:6368–6379. doi: 10.1021/pr100666c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herskowitz JH, Seyfried NT, Gearing M, Kahn RA, et al. Rho kinase II phosphorylation of the lipoprotein receptor LR11/SORLA alters amyloid-beta production. J Biol Chem. 2011;286:6117–6127. doi: 10.1074/jbc.M110.167239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seyfried NT, Gozal YM, Dammer EB, Xia Q, et al. Multiplex SILAC analysis of a cellular TDP-43 proteinopathy model reveals protein inclusions associated with SUMOylation and diverse polyUb chains. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M800390-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frangioni JV, Neel BG. Solubilization and Purification of Enzymatically Active Glutathione S-Transferase (pGEX) Fusion Proteins. Analytical Biochemistry. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z, Jang S-W, Liu X, Cheng D, et al. Neuroprotective Actions of PIKE-L by Inhibition of SET Proteolytic Degradation by Asparagine Endopeptidase. Molecular Cell. 2008;29:665–678. doi: 10.1016/j.molcel.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirahama-Noda K, Yamamoto A, Sugihara K, Hashimoto N, et al. Biosynthetic Processing of Cathepsins and Lysosomal Degradation Are Abolished in Asparaginyl Endopeptidase-deficient Mice. Journal of Biological Chemistry. 2003;278:33194–33199. doi: 10.1074/jbc.M302742200. [DOI] [PubMed] [Google Scholar]
- 32.Chen J-M, Dando PM, Rawlings ND, Brown MA, et al. Cloning, Isolation, and Characterization of Mammalian Legumain, an Asparaginyl Endopeptidase. Journal of Biological Chemistry. 1997;272:8090–8098. doi: 10.1074/jbc.272.12.8090. [DOI] [PubMed] [Google Scholar]
- 33.Shutov AD, Blattner FR, Kakhovskaya IA, Müntz K. New aspects of the molecular evolution of legumains, Asn-specific cysteine proteinases. Journal of Plant Physiology. doi: 10.1016/j.jplph.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Manoury B, Hewitt EW, Morrice N, Dando PM, et al. An asparaginyl endopeptidase processes a microbial antigen for class II MHC presentation. Nature. 1998;396:695–699. doi: 10.1038/25379. [DOI] [PubMed] [Google Scholar]
- 35.Watts C, Matthews SP, Mazzeo D, Manoury B, Moss CX. Asparaginyl endopeptidase: case history of a class II MHC compartment protease. Immunological Reviews. 2005;207:218–228. doi: 10.1111/j.0105-2896.2005.00312.x. [DOI] [PubMed] [Google Scholar]
- 36.Sepulveda FE, Maschalidi S, Colisson R, Heslop L, et al. Critical Role for Asparagine Endopeptidase in Endocytic Toll-like Receptor Signaling in Dendritic Cells. Immunity. 2009;31:737–748. doi: 10.1016/j.immuni.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 37.Baloh RH. TDP-43: the relationship between protein aggregation and neurodegeneration in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. FEBS Journal. 278:3539–3549. doi: 10.1111/j.1742-4658.2011.08256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Udan M, Baloh RH. Implications of the prion-related Q/N domains in TDP-43 and FUS. Prion. 5:1–5. doi: 10.4161/pri.5.1.14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polymenidou M, Cleveland, Don W. The Seeds of Neurodegeneration: Prion-like Spreading in ALS. Cell. 147:498–508. doi: 10.1016/j.cell.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Human Molecular Genetics. 19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.