Abstract
The transcriptional control of primary cilium formation and ciliary motility are beginning to be understood, but little is known about the transcriptional programs that control cilium number and other structural and functional specializations. One of the most intriguing ciliary specializations occurs in multiciliated cells (MCCs), which amplify their centrioles to nucleate hundreds of cilia per cell, instead of the usual monocilium. Here we report that the transcription factor MYB, which promotes S phase and drives cycling of a variety of progenitor cells, is expressed in postmitotic epithelial cells of the mouse airways and ependyma destined to become MCCs. MYB is expressed early in multiciliogenesis, as progenitors exit the cell cycle and amplify their centrioles, then switches off as MCCs mature. Conditional inactivation of Myb in the developing airways blocks or delays centriole amplification and expression of FOXJ1, a transcription factor that controls centriole docking and ciliary motility, and airways fail to become fully ciliated. We provide evidence that MYB acts in a conserved pathway downstream of Notch signaling and multicilin, a protein related to the S-phase regulator geminin, and upstream of FOXJ1. MYB can activate endogenous Foxj1 expression and stimulate a cotransfected Foxj1 reporter in heterologous cells, and it can drive the complete multiciliogenesis program in Xenopus embryonic epidermis. We conclude that MYB has an early, crucial and conserved role in multiciliogenesis, and propose that it promotes a novel S-like phase in which centriole amplification occurs uncoupled from DNA synthesis, and then drives later steps of multiciliogenesis through induction of Foxj1.
Keywords: Myb, Multiciliogenesis, Centriole amplification
INTRODUCTION
Cilia are microtubule-based organelles that project from almost every vertebrate cell (Gerdes et al., 2009). A primary cilium is nucleated from the mother centriole of the centrosome during interphase of the cell cycle. It contains nine peripheral and no central microtubule doublets (9+0) and functions in intercellular signaling and mechanosensing (Gerdes et al., 2009). Many cells modify their cilium, generating a wide diversity of cilium types. Cells of the embryonic node form a 9+0 motile cilium that generates fluid flow to determine left-right asymmetry (Nonaka et al., 1998), whereas hair cells of the inner ear form a non-motile cilium with two central microtubules (9+2) that transduces sound (Nayak et al., 2007), and sperm have a 9+2 motile cilium that propels them through the reproductive tract (Fisch and Dupuis Williams, 2011). This diversity of cilium types is reflected in the wide range of human diseases caused by ciliary defects (Davis and Katsanis, 2012).
One of the most striking ciliary specializations is the formation of multicilia, in which cells generate hundreds of 9+2 motile cilia, the coordinated beating of which generates fluid flow over the tissue. Multiciliated cells (MCCs) that line the airways are part of an essential defense mechanism that moves inhaled pathogens and debris out of the lungs (Knowles and Boucher, 2002). MCCs are also found in other tissues including the brain ependyma where they promote cerebrospinal fluid flow (Del Bigio, 2010), the fallopian tubes where they propel the egg outward (Hagiwara et al., 2004), and in the skin of Xenopus laevis embryos (Deblandre et al., 1999).
Each motile cilium is nucleated by a specialized centriole called the basal body (Sorokin, 1968; Hagiwara et al., 2004). Centrioles along with pericentriolar material form the centrosome, which organizes the interphase microtubule skeleton and mitotic spindle. Centriole duplication is tightly regulated and coordinated with DNA synthesis in the S phase of cycling cells (Hinchcliffe and Sluder, 2001; Tsou and Stearns, 2006). Extraneous centrioles can lead to genomic instability and aberrant cell signaling (Mahjoub and Stearns, 2012). MCCs somehow escape this regulation to generate hundreds of centrioles and basal bodies per cell (Sorokin, 1968), which dock at the apical cell surface and initiate a ciliary axoneme (Fig. 1A).
Fig. 1.
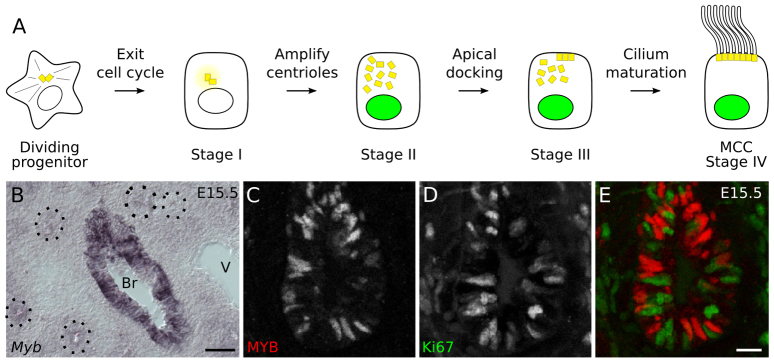
Myb expression in the developing bronchial epithelium. (A) Schematic of multiciliogenesis in developing airway epithelial cells. FOXJ1 expression is indicated by green nuclei. Stage I: the presumptive multiciliated cell (MCC) has exited the cell cycle and pericentriolar material components begin to accumulate near the centrosome. Stage II: new centrioles begin to appear. Stage III: centrioles migrate and dock at the apical cell surface. Stage IV: each docked centriole (now called a basal body) nucleates a motile 9+2 ciliary axoneme in a mature MCC (Vladar and Stearns, 2007). (B) In situ hybridization of Myb on a section of E15.5 mouse lung. Myb mRNA is detected only in the large proximal airways (bronchus, Br), in a ‘salt and pepper’ pattern, and not in smaller, more distal airways (outlined) that have not yet begun to differentiate or in surrounding mesenchyme or blood vessels (V). (C-E) Immunostaining of E15.5 lung section for MYB and Ki67, a marker of cycling cells. MYB is not expressed in cells that express Ki67. Scale bars: 50 μm in B; 10 μm in C-E.
Although the transcriptional program that controls primary cilium formation has begun to be elucidated (Chu et al., 2010; Piasecki et al., 2010; Thomas et al., 2010), much less is known about the transcriptional programs that control multiciliogenesis and other ciliary specializations. The best understood ciliary transcription factor is forkhead box J1 (FOXJ1), which is expressed and required in all cells with motile cilia (Brody et al., 2000; Gomperts et al., 2004; Pan et al., 2007; Yu et al., 2008). Recently, a pathway that controls MCC development was identified in mouse airway epithelium and X. laevis epidermis. Downregulation of Notch signaling in MCC precursor cells (Tsao et al., 2009; Morimoto et al., 2010) leads to induction of multicilin (Mcin; Mcidas - Mouse Genome Informatics), which functions upstream of centriole amplification and induction of Foxj1 (Stubbs et al., 2012). Although MCIN expression is necessary and sufficient for MCC formation (Stubbs et al., 2012), how this novel nuclear protein controls centriole amplification and later steps in the multiciliogenesis program is not known.
Here we identify the product of the myeloblastosis proto-oncogene Myb (c-Myb) as a crucial early transcription factor in the multiciliogenesis program. MYB belongs to a family of transcription factors involved in cell cycle regulation and progenitor cell proliferation (Ramsay and Gonda, 2008; Lipsick, 2010). Mybl2 (b-Myb) is ubiquitously expressed and is required for the G1/S transition (Bessa et al., 2001), whereas Mybl1 (a-Myb) (Toscani et al., 1997) and Myb show restricted expression. Myb is required for the self-renewal of hematopoietic stem cells (Lieu and Reddy, 2009) and promotes the proliferation of progenitor/stem cells in the colon (Malaterre et al., 2007) and brain (Malaterre et al., 2008; Ramsay and Gonda, 2008). Misregulation of Myb has been implicated in cancers of the blood, gut, lung and many other tissues (Ramsay and Gonda, 2008).
In contrast to its role in proliferative progenitor cells, we found that Myb is expressed in developing MCCs of the mouse lung just after they exit the cell cycle and initiate multiciliogenesis. Conditional inactivation of Myb delays or blocks the initiation of centriole amplification and FOXJ1 expression, and airways do not become fully ciliated. We provide evidence that MYB functions as a transcription factor in a conserved multiciliogenesis program downstream of Notch signaling and MCIN and upstream of FOXJ1, and we propose a model that reconciles its postmitotic function in this program with its well-established role in proliferating progenitor cells.
MATERIALS AND METHODS
Mice
Timed pregnant CD1 female mice were obtained from Charles Rivers Laboratories. Noon of the day that a vaginal plug was observed was considered day (E) 0.5 of gestation. Mybfl/fl mice, in which exon II is flanked by loxP sites, were crossed to an HPRT-Cre strain (Tang et al., 2002) to obtain a null allele (Myb-) that produces no detectable protein (Bender et al., 2004). We did not detect any differences between Mybfl/fl and Mybfl/- mice (either alone or when crossed to Cre driver alleles) and so used them interchangeably. The Shh driver line (ShhCre) (Harfe et al., 2004), which has a Cre-GFP knock-in at the Shh locus that is active in the lung epithelium throughout development (Harris et al., 2006), and the Nkx2.1 driver line (Nkx2.1-CreTg) (Xu et al., 2008), which has an Nkx2.1 BAC transgene that expresses Cre in the lung epithelium during development (Tiozzo et al., 2009), have been described. The Cre reporter line R26-mTmG (Muzumdar et al., 2007) labels cells with membrane-bound GFP following Cre recombination. Foxj1 null mice (Foxj1-/-), in which exon I is deleted, produce no detectable mRNA or protein (Brody et al., 2000). All control animals were littermates of experimental animals. All procedures involving animals were approved by the Institutional Animal Care and Use Committee in accordance with established guidelines for animal care.
Lung fixation and sectioning
Embryonic lungs were dissected, fixed in 4% paraformaldehyde (PFA) pH 7 for 1 hour at 4°C, equilibrated in 30% sucrose/PBS for 30 minutes at 4°C, embedded in OCT compound (Sakura Finetek) and stored at -80°C. Postnatal lungs were inflated intratracheally with cold 4% PFA, removed from the body cavity, submerged in 4% PFA for 1 hour at 4°C, washed in PBS overnight at 4°C and equilibrated in 30% sucrose/PBS overnight at 4°C. Individual lobes were embedded and stored at -80°C. Tissue blocks were sectioned on a Leica CM3050 S cryostat.
Lung in situ hybridization of Myb and other transcription factors
A literature search using PubMed and Google Scholar was conducted of 1971 known and predicted mouse transcription factor genes for expression data linking any of the genes to the lung (F.E.T., PhD Thesis, Stanford University, CA, USA, 2011). The lung developmental expression patterns of selected genes were examined by in situ hybridization using probes prepared from sequence-verified I.M.A.G.E. clones (Open Biosystems, Research Genetics); I.M.A.G.E. clone 695394 was used for Myb. Genes not in the I.M.A.G.E. collection were amplified from E15.5 lung and brain cDNA (RETROscript Kit, Life Technologies), cloned into pCR4-BLUNT (Life Technologies) and sequence verified. Amplicons containing the insert and flanking RNA polymerase promoters were used as templates for in vitro transcription of digoxigenin-labeled antisense probes (Roche) (Sambrook and Russell, 2001).
Cryosections (20 μm) of E13.5, E15.5 and E17.5 lungs were mounted on a single Superfrost Plus glass slide (Thermo Fisher Scientific) and stored at -80°C. Mounted sections were thawed, postfixed in 4% PFA, acetylated for 10 minutes, rinsed in DEPC-treated PBS and then incubated in DEPC-treated 5× SSC. Sections were pretreated in hybridization solution for 2 hours then incubated with probe {400 ng/ml in hybridization solution [50% formamide, DEPC-treated 5× SSC, 4× Denhardt’s solution (50× Denhardt’s solution is 0.1% Ficoll 400, 0.1% polyvinylpyrrolidone, 0.1% BSA, w/v)], 400 ng/ml salmon or herring sperm DNA, 200 ng/ml yeast tRNA in DEPC-treated water} at 58°C for 2 days. Sections were washed three times for 1 hour each at 65°C in 0.2× SSC, then incubated with alkaline phosphatase-conjugated anti-digoxigenin Fab fragments diluted 1:5000 in 1% Blocking Reagent (Roche) at 4°C overnight. After washing, sections were incubated with NBT/BCIP (Roche) diluted 1:50 in AP reaction buffer (0.1 M Tris-Cl pH 9.5, 50 mM MgCl2, 0.1 M NaCl) for up to 6 hours, mounted in Aqua-Poly/Mount (Polysciences) and visualized under brightfield optics on a Zeiss Axiophot microscope equipped with an AxioCam HRC camera.
Lung immunostaining
Immunostaining was performed on 10 μm cryosections of E11.5 through E17.5 lungs. Tissue sections were washed in PBST (PBS with 0.1% Tween 20), blocked for 1 hour with 10% goat serum in PBS, and incubated with primary antibodies diluted in 1.5% goat serum at 4°C overnight. After washing, Alexa Fluor-conjugated secondary antibodies (Life Technologies) diluted 1:500 in 1.5% goat serum were applied for 1 hour at room temperature. Slides were mounted with Vectashield containing DAPI (Vector Laboratories) for nuclear labeling. To quench endogenous reporters, stained tissues from transgenic animals carrying the mTmG reporter were ascended and descended through a graded methanol/PBS series after secondary antibody incubations. Images were captured on a Leica TCS SP2 SE confocal microscope.
Primary antibodies were mouse monoclonal anti-MYB (clone D-7, Santa Cruz Biotechnology; used at 1:500 dilution), mouse monoclonal anti-FOXJ1 (clone 2A5, eBioscience; 1:500), chicken anti-GFP (Abcam, ab13970; 1:500), rabbit anti-RFP (Rockland, Gilbertsville, PA, USA, 600-401-379; 1:250), mouse monoclonal anti-Ki67 (BD Biosciences, 550609; 1:500), rabbit anti-Ki67 (Vector Laboratories, VP-K451; 1:500), mouse monoclonal anti-acetylated tubulin (Sigma, T6793; 1:1000), rat anti-cadherin 1 (Life Technologies, 13-1900; 1:1000) and mouse monoclonal anti-pericentrin (BD Biosciences, 611814; 1:500). Isotype-specific anti-mouse secondary antibodies were used to simultaneously detect multiple mouse monoclonal primary antibodies. Cells were counted with CellProfiler (Carpenter et al., 2006).
Mouse tracheal epithelial cell culture
Mouse tracheal epithelial cell (MTEC) cultures were prepared as described (You et al., 2002; Vladar and Stearns, 2007). Briefly, epithelial cells obtained from adult mouse tracheas by Pronase (Roche) digestion were grown to confluence on Transwell-Clear filters (Corning) at 37°C/5% CO2, then switched to an air-liquid interface (ALI) to induce differentiation of MCCs and other tracheal epithelial cell types. Immunostaining was performed as described (Vladar and Stearns, 2007) and specimens were imaged on a Leica TCS SP2 or SP5 confocal microscope. RNA expression levels of Myb family transcription factors were determined by DNA microarray analysis of FACS-sorted MTEC cultures prepared from FOXJ1-GFP transgenic mice (Hoh et al., 2012). For Notch inhibition, MTECs were treated with 1 μM DAPT (Sigma) from ALI -1 day to ALI +3 days and cell counts were performed with the Nucleus Counter plug-in in ImageJ (NIH).
For lentivirus-mediated gene transfer into MTECs, epitope-tagged recombinant MCIN (Stubbs et al., 2012) and other lentiviruses were prepared and MTECs were infected as described (Vladar and Stearns, 2007). The mouse Myb mRNA coding region from pcRM-Myb (from Dr Timothy Bender, University of Virginia, Charlottesville, VA, USA) was cloned into pLVX-tdTomato-C1 (Life Technologies) to generate an N-terminal tdTomato-MYB fusion construct that was subcloned into pLentiPGK to create pLenti-td-Tomato-MYB for virus production (Vladar and Stearns, 2007). Cell counts were performed on filters from at least two independent infections.
Brain immunostaining
Dissected brain tissues were incubated with primary and secondary antibodies in 10% goat serum in PBS/1% Triton X-100 for 48 hours at 4°C as described (Mirzadeh et al., 2008; Mirzadeh et al., 2010). After staining, ventricular walls were dissected from underlying parenchyma as slivers of tissue 200-300 μm in thickness and mounted. Confocal images were captured on a Leica SP5 microscope.
RNA microinjection of Xenopus embryos
X. laevis embryos were obtained following in vitro fertilization (Sive et al., 1998). Embryos were injected at the two-cell stage with in vitro synthesized capped mRNAs (typically 1-5 ng/embryo) (Turner and Weintraub, 1994) encoding MCIN-HGR, activated Notch (ICD), membrane RFP (mRFP) and Hyls1-GFP to mark centrioles as described (Stubbs et al., 2012). X. laevis Myb was inserted into the CS2 vector to generate a myc-tagged Myb construct (MT-Myb) (supplementary material Table S1).
To analyze the effects of MYB overexpression, embryos were injected at the two-cell stage with MT-Myb RNA (0.5 pg/embryo) along with RNA encoding mRFP (0.1 pg/embryo) and Hysl1-GFP (0.5 pg/embryo). Control embryos were injected with just mRFP and Hyls1-GFP. At stage 28, the embryos were fixed, immunostained with anti-acetylated tubulin antibody and imaged by confocal microscopy as described (Stubbs et al., 2012). Two random fields of the flank of four to seven embryos were imaged and scored for MCCs (multiple basal bodies and acetylated tubulin staining), proton-secreting cells (small apical domain) or outer cells (remaining cells).
For quantitative (q) RT-PCR analysis of gene expression, embryos were injected at the two-cell stage with ICD RNA or with both ICD (0.2 pg/embryo) and MCIN-HGR RNA (0.08 pg/embryo). Animal caps were dissected at stage 10, incubated until stage 11, and then treated with dexamethasone (10 μM) to induce MCIN activity. Total RNA was harvested 6 hours later. cDNA templates were generated from 3 μg RNA using Superscript III reverse transcriptase (Life Technologies). qPCR reactions were performed in triplicate using an ABI Prism 7900HT thermal cycler (Life Technologies) and primers for Foxj1 and Myb, and normalized with ornithine decarboxylase (Odc) as an internal control (supplementary material Table S2). qPCR data were analyzed with Sequence Detection System (SDS, Applied Biosystems) software.
Cotransfection assay
A 1 kb region upstream of the human FOXJ1 translation start site (hg19, chr17:74136502-74137569; nucleotides -876 to +162 with respect to the transcription initiation site) was cloned by PCR upstream of the coding sequence for firefly luciferase in the pGL4.20 vector (Promega) to create pGL4.20-Foxj1. This fragment includes the -872 to +108 region upstream of the transcription start site used to generate the Foxj1-GFP mouse line (Ostrowski et al., 2003). Human MYB (clone ID 6069320) and FOXJ1 (clone ID 5171637) coding sequences were obtained from Open Biosystems (Thermo Fisher Scientific) and cloned into the pCDH-CMV-MCS-EF1-PuroGreen (pCDH) vector (Systems Bioscience) to generate pCDH-Myb and pCDH-Foxj1. The MCIN expression vector was described previously (Stubbs et al., 2012).
HEK293T cells (5×104 cells in 100 μl per well) were transfected using FuGENE6 (Promega) in a 96-well plate with a total of 50 ng DNA (6:1 FuGENE6:DNA ratio) consisting of 15 ng pGL4.20-Foxj1 DNA, a total of 32.5 ng transcription factor DNA (empty pCDH vector, pCDH-MYB, pCDH-MCIN, pCDH-Foxj1) and 2.5 ng plasmid containing a thymidine kinase promoter-driven Renilla luciferase (pRL-TK). Cells were incubated at 37°C/5% CO2 for 3 days, then lyzed and assayed for firefly and Renilla luciferase activity using the Dual-Glo Luciferase Assay System (Promega) in a Synergy H1 hybrid multi-mode microplate reader (Biotek, Winooski, VT, USA). Three or four independent transfections were performed in triplicate, and the ratio of firefly to Renilla luciferase signal was normalized to the values obtained for empty pCDH vector.
RESULTS
Myb is expressed in postmitotic cells of the developing bronchial airway epithelium
In an in situ hybridization screen for transcription factors expressed during Mus musculus (mouse) lung development (see Materials and methods), we found that Myb was expressed in a ‘salt and pepper’ pattern in the large diameter proximal airway epithelium, but not in the small distal epithelial bud tips or surrounding mesenchyme (Fig. 1B). Expression initiated at E13.5 and expanded down the bronchial tree as branching morphogenesis progressed, but never extended into the undifferentiated portion of the distal airway epithelium. The density of cells expressing Myb peaked at ∼E16.5 and began to decline by E17.5 (supplementary material Fig. S1) and was completely absent by early postnatal life (data not shown).
Myb is expressed in cycling progenitor cells of the blood, colon and brain. Such cell cycle phase-specific expression could account for the ‘salt and pepper’ appearance of Myb transcripts in the developing bronchial epithelium. To test this, we labeled E14.5 through E17.5 lungs for MYB and Ki67 (MKI67 - Mouse Genome Informatics), a cell cycle marker that is absent only in quiescent (G0) cells (Scholzen and Gerdes, 2000). Surprisingly, there was almost no colocalization between MYB and Ki67 expression (0-2% colocalization, n>98 cells scored at each stage), indicating that MYB is expressed in cells that have exited the cell cycle (Fig. 1C-E; supplementary material Table S2). Thus, in contrast to other tissues, MYB is expressed in postmitotic cells in the developing bronchial epithelium.
MYB is expressed early and transiently in developing MCCs
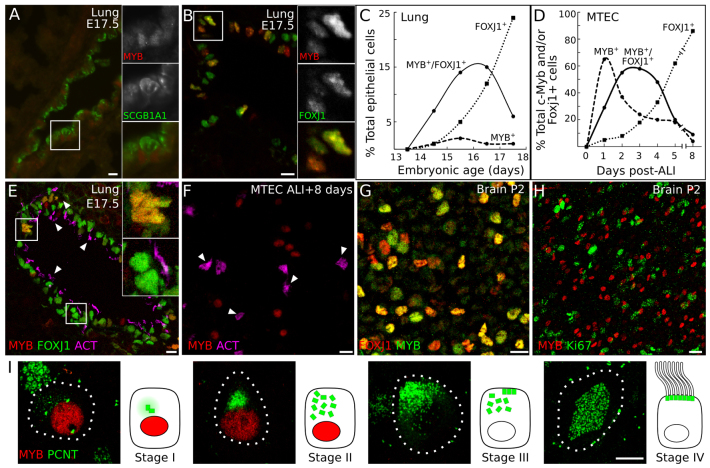
The MYB expression pattern is reminiscent of markers of differentiating cell types such as Clara cells and MCCs, which are interspersed in the bronchial epithelium. These cell types appear during lung development in a proximal-to-distal wave of differentiation, in which cells in the larger proximal airways have begun differentiation before those in the smaller distal airways. To determine whether MYB is expressed in developing secretory Clara cells or MCCs, we co-stained E17.5 airways for MYB and either secretoglobin 1a1 (SCGB1A1), a canonical Clara cell marker, or FOXJ1, which labels MCCs. MYB expression did not overlap with that of SCGB1A1 (Fig. 2A). However, most MYB-expressing cells also expressed FOXJ1 (Fig. 2B), indicating that MYB is expressed in developing MCCs of the lung.
Fig. 2.
MYB is expressed early and transiently during multiciliogenesis in the lung and brain. (A,B) Immunostaining of E17.5 mouse lung sections for MYB and either SCGB1A1 (Clara cells) or FOXJ1 (MCCs). MYB is not detected in SCGB1A1-expressing cells (boxed region, insets in A) but many MYB-positive cells also express FOXJ1 (boxed region, insets in B). (C) Sections of E14.5 through E17.5 lungs were stained for MYB, FOXJ1 and SOX2 and the number of epithelial cells (marked by SOX2) expressing MYB only, FOXJ1 only, or both MYB and FOXJ1 were counted. Total SOX2-positive cells scored was >1000 per age. (D) Mouse tracheal epithelial cell (MTEC) cultures were immunostained for MYB and FOXJ1 at the times indicated after exposure to an air-liquid interface (ALI) to promote differentiation. The number of cells that were MYB+, FOXJ1+ or double-positive were counted. Similar to in vivo, the FOXJ1+ population continually increased during culturing, whereas the percentage of MYB+ and double-positive cells initially increased and then declined. Total cells scored was >100 per time point. (E) Immunostaining of E17.5 lung section for MYB, FOXJ1 and acetylated tubulin (ACT, cilia marker). MYB is seen in developing (upper inset) but not mature (lower inset) MCCs. (F) Immunostaining of ALI +8 days MTEC cultures for MYB and ACT. MYB is not detected in mature MCCs. Arrowheads (E,F), ciliary tufts of mature MCCs. (G,H) Whole-mount ventricles of P2 mouse brain stained for FOXJ1 and MYB (G) or Ki67 and MYB (H). (I) Stage-specific expression of MYB during multiciliogenesis in MTECs. ALI +6 days MTECs were stained for MYB and pericentrin (PCNT), a centriole marker, and cells were staged as described in Fig. 1A. MYB is detected at stages I and II, before and during centriole amplification, but not during stages III or IV. Dotted lines show cell outline. Scale bars: 10 μm in A,B,E-H; 5 μm in I.
We compared the temporal expression patterns of MYB and FOXJ1 in the developing bronchial epithelium from E13.5 through E17.5 (Fig. 2C; supplementary material Fig. S2). Expression of both MYB and FOXJ1 initiated between E13.5 and E14.5, and the number of cells expressing MYB and/or FOXJ1 increased over the next 2 days. However, whereas the number of FOXJ1-expressing cells continued to increase through E17.5, the number of MYB-expressing cells began to decline (Fig. 2C). This suggests that MYB is expressed early and turns off at later stages of the multiciliogenesis program. Indeed, triple immunostaining of E17.5 lungs for MYB, FOXJ1 and acetylated tubulin (ACT), which labels the ciliary tufts of mature MCCs, showed that FOXJ1 expression is maintained in mature MCCs whereas MYB expression is not (Fig. 2E).
We observed a similar pattern of MYB expression in mouse tracheal epithelial cell (MTEC) cultures, an in vitro model of airway epithelial differentiation, in which primary airway epithelial cells proliferate and then, upon establishment of an air-liquid interface (ALI), differentiate into MCCs and other airway epithelial cells (You et al., 2002). Both MYB and FOXJ1 expression initiated after proliferation ceased, ∼1 day after ALI (ALI +1) (supplementary material Fig. S3A-H). Expression of both proteins initially increased during the culture period (Fig. 2D), but whereas FOXJ1 expression was maintained in mature MCCs, MYB expression ceased (Fig. 2F). Expression profiling showed a dramatic transcriptional induction of MYB in multiciliating cells. This induction was specific to Myb, as Mybl1 and Mybl2 were not similarly upregulated (supplementary material Fig. S3I). Careful examination of the progressive stages of multiciliogenesis in MTECs (Vladar and Stearns, 2007) showed that MYB is expressed during centriolar material buildup and centriole amplification, but expression is lost as centrioles dock at the apical membrane (Fig. 2I).
We also examined MYB and FOXJ1 expression during the development of brain ependymal cells, where Myb had previously been implicated in neural progenitor cell development (Malaterre et al., 2008). We found that, similar to the lung epithelium, in the lateral ventricular walls of the brain (Mirzadeh et al., 2010) MYB was co-expressed with FOXJ1 in developing MCCs (Fig. 2G; supplementary material Fig. S4), and expression initiated just after presumptive MCCs had exited the cell cycle and no longer expressed Ki67 (Fig. 2H; supplementary material Fig. S5A-C). As in the lung, MYB was not detected in mature MCCs (supplementary material Fig. S5D-F). These data suggest that developing MCCs first express MYB, followed closely by FOXJ1, and that FOXJ1 expression is maintained in MCCs whereas MYB is downregulated before the MCC matures.
MYB functions upstream of FOXJ1 in multiciliogenesis
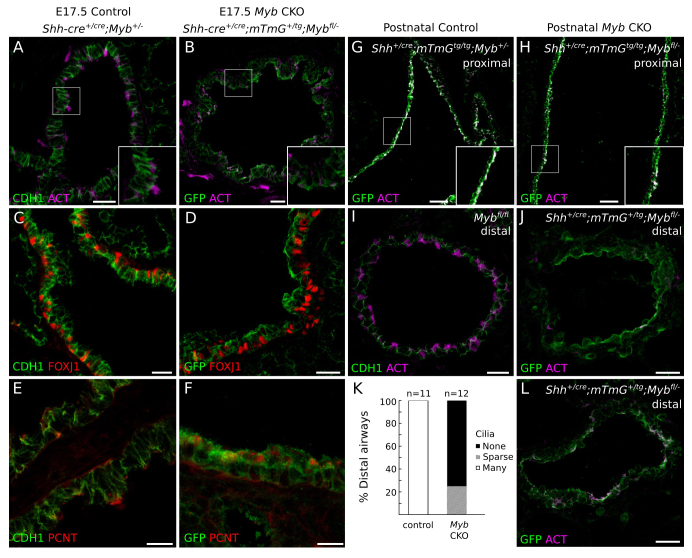
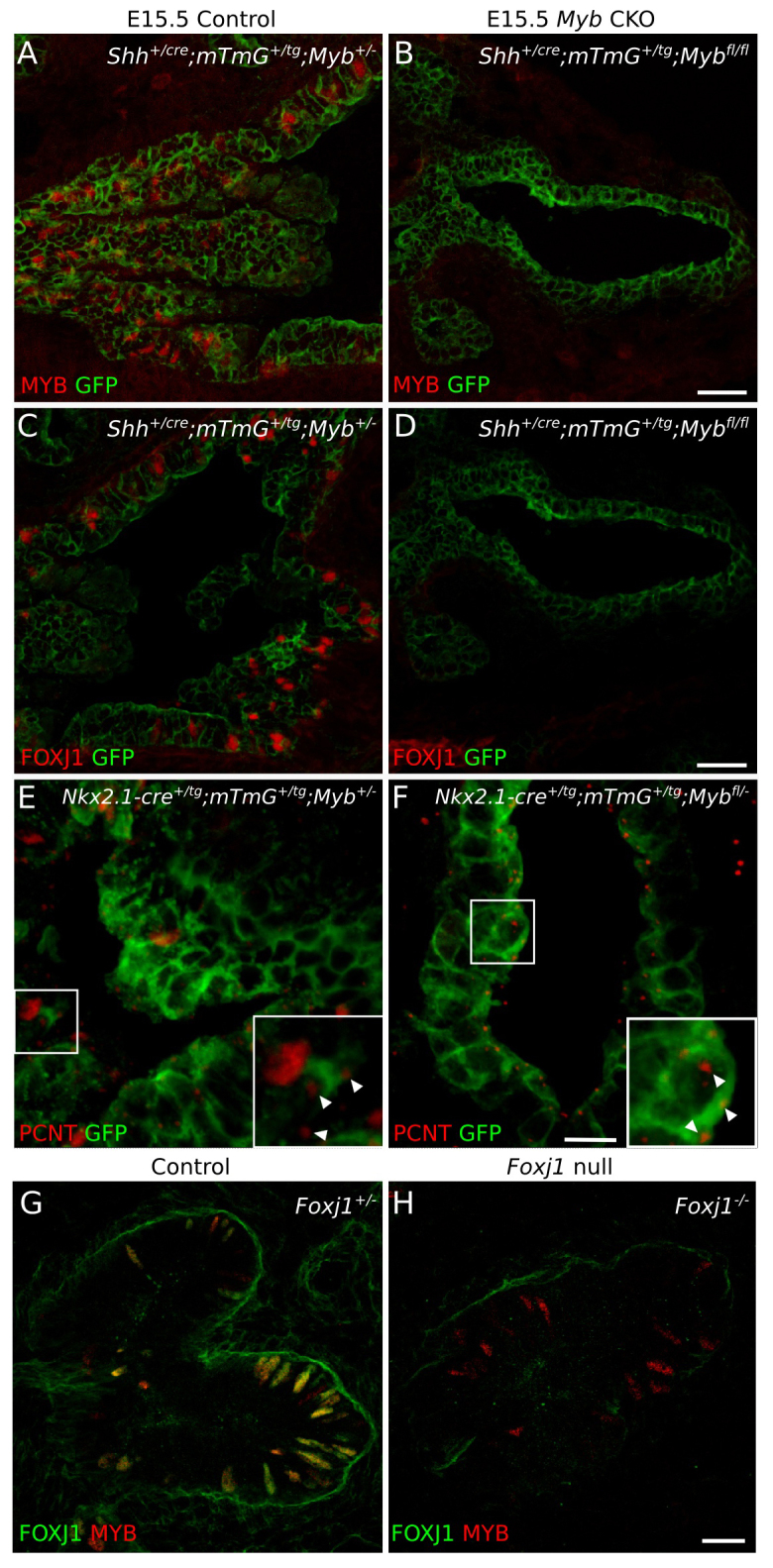
To determine whether MYB is required for FOXJ1 expression, we examined FOXJ1 expression in a Myb mutant. Because Myb null mutants die at ∼E15.5 (Mucenski et al., 1991), we used a conditional Myb allele (Mybfl) (Bender et al., 2004) and inactivated it throughout the developing lung epithelium (Fig. 3A,B) with ShhCre (Harfe et al., 2004; Harris et al., 2006) to generate a Myb conditional knockout (CKO). There was no detectable expression of FOXJ1 at E15.5 in the Myb CKO (Fig. 3C,D). The same result was obtained when Mybfl was inactivated with another lung epithelial driver, Nkx2.1-CreTg (Xu et al., 2008) (data not shown). Conversely, we found that MYB expression was unperturbed in a Foxj1 null mutant (Brody et al., 2000) (Fig. 3G,H). Thus, Myb functions upstream of Foxj1 in the transcriptional regulatory program of MCCs.
Fig. 3.
Effect of Myb deletion on initiation of FOXJ1 expression in the lung. (A-F) Mice carrying a conditional allele of Myb (Mybfl) and a Cre recombination reporter (mTmG) were crossed to mice carrying Shh-Cre or Nkx2.1-Cre transgenes to selectively delete Myb from the developing airway epithelium (Myb CKO), and lungs from E15.5 control (Myb+/fl) and Myb CKO mice were immunostained for the proteins indicated. (A,B) MYB immunostaining confirms loss of MYB expression in Myb CKO tissue. (C,D) FOXJ1 expression is not detected in Myb CKO tissue. (E,F) Large concentrations of PCNT, indicative of ciliating cells with amplified centrioles (inset in E), are not observed in Myb CKO tissue (inset in F). PCNT staining of centrosomes (arrowheads) is seen in both control and Myb CKO non-MCCs. (G,H) Lungs from E15.5 Foxj1 control heterozygous and homozygous knockout (Foxj1-/-) mice immunostained for MYB. MYB is expressed normally in the absence of Foxj1. Scale bars: 10 μm.
We also tested the role of MYB in the regulation of Foxj1 by overexpressing MYB in the MTEC system. When MTECs were infected with a lentivirus that expressed a tdTomato-MYB fusion protein, 44% of infected cells expressed FOXJ1, as compared with 13% of cells infected with a control lentivirus that expressed RFP alone (Fig. 4A-C). Thus, MYB is able to induce FOXJ1 expression in MTECs. MYB also increased the expression of a Foxj1 reporter gene in a cotransfection assay in HEK293T cells (Fig. 4D), although the effect was small (1.5-fold), perhaps because HEK293T cells lack co-factors present in developing MCCs. FOXJ1 and MCIN also increased expression of the Foxj1 reporter, consistent with previous results (Venugopalan et al., 2008; Stubbs et al., 2012), and MCIN together with MYB gave additive effects (Fig. 4D).
Fig. 4.
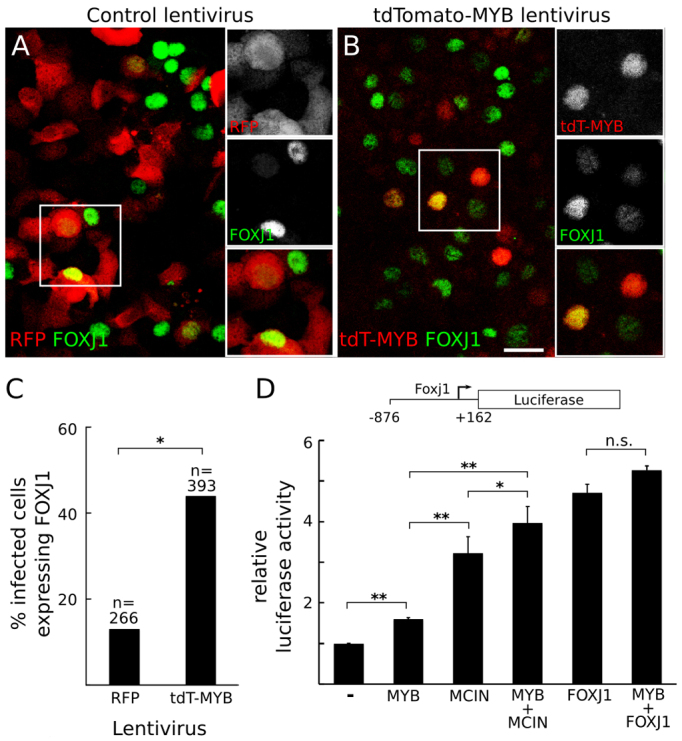
Effect of MYB on FOXJ1 expression in MTECs and a HEK293T cell cotransfection assay. (A,B) MTECs infected with a lentivirus expressing either a tdTomato-MYB fusion protein (tdT-MYB) or RFP alone were harvested at ALI +4 days and stained for RFP (A) or tdTomato (B) and FOXJ1. Scale bar: 20 μm. (C) Quantification of the percentage of infected cells that express FOXJ1. *P<0.0001 by chi-square test. (D) Luciferase activity driven by the Foxj1 reporter illustrated (nucleotides -876 to +162 with respect to the Foxj1 transcription initiation site) cotransfected into human HEK293T cells with pCDH empty vector (-) or pCDH constructs expressing the indicated proteins. Reporter activity is normalized to the vector-only (pCDH) control. *P<0.05, **P<0.003; n.s., not significant (Student’s t-test). Error bars indicate s.e.m.
MYB is required for timely initiation of centriole amplification
We tested whether Myb is required for centriole amplification, a fundamental early step in multiciliogenesis that is not affected by loss of FOXJ1 (You et al., 2004), by assessing the expression of the centriole marker pericentrin (PCNT). In Myb CKO airway epithelial cells at E15.5, no large concentrations of PCNT were observed, in contrast to the amplified centrioles found at this stage in normal MCCs (Fig. 3E,F). Only one or two puncta of PCNT were seen, resembling those in cells with the standard pair of centrioles. Thus, Myb is required for the timely initiation of centriole amplification during multiciliogenesis.
The multiciliogenesis program can partially recover from loss of Myb
At E17.5, the major airways are normally well ciliated. When we examined Myb CKO epithelium at E17.5, however, no mature cilia were detectable by ACT staining (Fig. 5A,B). This indicates a dramatic arrest of the multiciliogenesis program, as observed 2 days earlier with the early MCC markers (Fig. 3E,F). However, FOXJ1 expression (Fig. 5C,D) and multiple centrioles (Fig. 5E,F) were now detectable in the large airways at levels indistinguishable from the control, implying that the multiciliogenesis program had begun to recover. When we examined Myb CKO airways at postnatal day (P) 14 and P21, we found that, at both stages, large proximal airways were ciliated at levels similar to control tissues (Fig. 5G,H). However, the small diameter distal airways were sparsely ciliated, if at all (Fig. 5I-L). Thus, recovery of the multiciliogenesis program in the absence of Myb is incomplete, at least in the distal airways. Likewise, we found that in MTECs generated from Myb CKO mice, MCC differentiation is reduced early but partially recovers at later time points (supplementary material Fig. S8).
Fig. 5.
Effect of Myb conditional knockout on multiciliogenesis in the lung. Lungs from Myb CKO mice (E17.5 in B,D,F and P14 or P21 in H,J,L) or littermate controls (E17.5 in A,C,E and P14 or P21 in G,I) were immunostained for the indicated markers. Cadherin 1 (CDH1) was used to visualize the epithelium in control tissues without mTmG reporter. (A,B) Immunostaining for ACT shows that the large apical tufts of ACT of mature MCCs (inset in A) are not present in Myb CKO tissue (inset in B) at E17.5. Immunostaining for FOXJ1 (C,D) and PCNT (E,F) shows that the multiciliogenesis program has begun to recover (compare with E15.5 in Fig. 3C-F). (G-L) ACT staining in postnatal lungs shows that multiciliogenesis in the large proximal airways recovers by P14 (G,H) (note that white indicates overlap of green and purple). However, small distal airways of Myb CKO lungs lack mature cilia (J) or are only sparsely ciliated (L) even at P21, as compared with control distal airways (I), as quantified in K. n, number of airways scored in three mice for each condition. Samples were stained simultaneously and scanned with the same confocal laser settings. Scale bars: 20 μm in A-F,I,J,L; 50 μm in G,H.
Conservation of MYB in the multiciliogenesis pathway
Mcin controls centriole amplification, FOXJ1 expression and multiciliogenesis (Stubbs et al., 2012). To determine whether Mcin controls MYB expression, we examined the expression of MYB in MTECs expressing myc-tagged MCIN (MCIN-myc) or myc-tagged dominant-negative MCIN (DN MCIN-myc) (Fig. 6A-C). The former causes infected MTECs to become multiciliated, whereas the latter blocks multiciliogenesis (Stubbs et al., 2012). We found that 80% of MCIN-myc-expressing cells expressed MYB, whereas only 5% of control cells infected with the control lentivirus expressed MYB (Fig. 6A,D; supplementary material Fig. S6). In parallel experiments with DN-MCIN-myc, almost no infected cells (<2%) expressed MYB (Fig. 6D). Thus, Myb lies downstream of Mcin in MTECs.
Fig. 6.
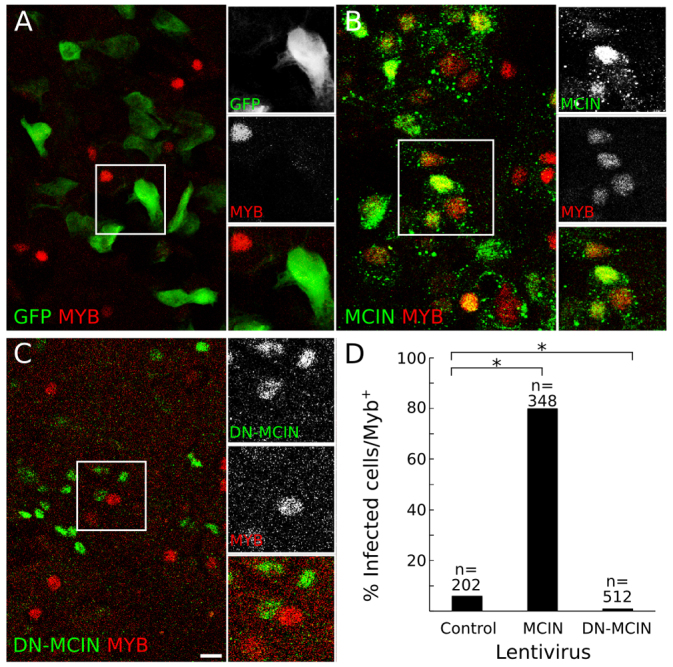
Effect of the multiciliogenesis regulator multicilin (MCIN) on MYB expression in MTECs. (A-C) MTECs were infected with a control lentivirus expressing GFP (A), a myc-tagged wild-type MCIN (B) or dominant-negative MCIN (DN-MCIN) (C), and stained for MYB and GFP or myc at ALI +4 days. (D) Quantification of the percentage of infected cells that express MYB. *P<0.001 by chi-square test. For examples of infected cells, see supplementary material Fig. S6. Scale bar: 10 μm.
Notch signaling controls Mcin expression in MTECs (Stubbs et al., 2012). To determine whether Myb expression is also controlled by Notch signaling, MYB expression was examined in MTECs treated with the Notch inhibitor DAPT. This increased the number of MYB-expressing cells at early time points in culture (ALI +1 and ALI +2) (supplementary material Fig. S9). We conclude that Myb lies downstream of Notch and Mcin but upstream of Foxj1 during MCC differentiation in MTECs (Fig. 8).
Fig. 8.
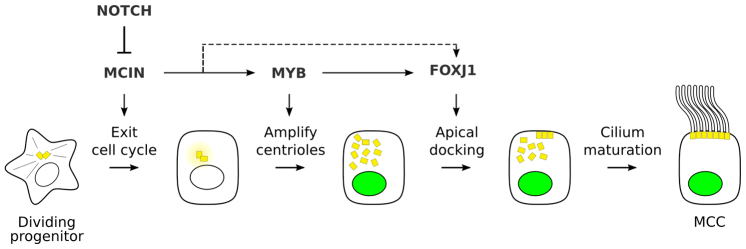
Proposed molecular pathway governing multiciliogenesis. MYB and other transcriptional regulators associated with each step in the program are shown along with the regulatory relationships among them, together with the Notch signaling pathway that controls initiation of the program. The dashed line indicates that there is an auxiliary pathway that can partially bypass the requirement for MYB, at least in some MCCs.
MCCs similar to those of mammalian airways line the embryonic epidermis of X. laevis, interspersed with other epidermal cell types (Fig. 7B) (Dubaissi and Papalopulu, 2011). We tested whether Myb function is conserved in the X. laevis multiciliogenesis program by injecting X. laevis embryos at the two-cell stage with mRNA encoding the Notch intracellular domain (ICD), which inhibits MCC differentiation, or with mRNAs encoding ICD and MCIN-HGR, an inducible form of MCIN that restores MCC differentiation. qPCR demonstrated a 4-fold increase in Myb transcript levels in the ICD+MCIN-HGR-injected animal caps as compared with ICD-injected animal caps (Fig. 7A). Thus, as in mouse airways, Myb is downstream of Mcin in X. laevis multiciliogenesis.
Fig. 7.
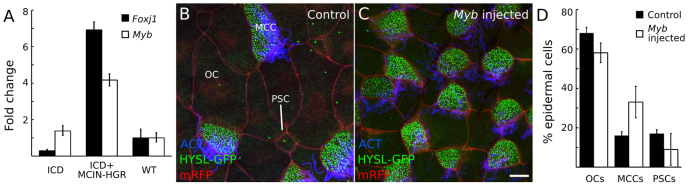
Myb acts downstream of Mcin and can promote multiciliogenesis in Xenopus laevis embryonic epidermis. (A) Two-cell stage X. laevis embryos were injected with Notch intracellular domain (ICD) mRNA alone to repress multiciliogenesis or together with MCIN (ICD/MCIN-HGR) to induce it, and Foxj1 and Myb mRNA levels were measured by qRT-PCR, normalized to Odc mRNA levels as an internal control. Values are mean ± s.d. (B-D) Two-cell stage embryos were injected with control (B) or Myb mRNA (C) and Hysl-GFP mRNA to visualize centrioles and membrane RFP mRNA to visualize cell boundaries and stained for acetylated tubulin (ACT) to label cilia. Scale bar: 10 μm. (D) Quantification of B and C. Note that the percentage of MCCs is increased in Myb mRNA-injected embryos, whereas the percentage of outer cells (OCs) and proton-secreting cells (PSCs) declines slightly. P<0.05 for all three cell types by Student’s t-test. Error bars indicate s.d.
We examined the effects of exogenous MYB in X. laevis by injecting two-cell stage embryos with Myb mRNA and assessing the epidermal cell type distribution at stage 28. The epidermis of embryos injected with Myb mRNA had twice as many MCCs as control, whereas other cell types slightly decreased (Fig. 7B-D). Thus, Myb can drive the complete multiciliogenesis program in X. laevis epidermis.
DISCUSSION
We have shown that Myb is a crucial early transcription factor in the multiciliogenesis program of lung development. Myb is expressed as presumptive MCCs exit the cell cycle and amplify their centrioles, and switches off as centrioles dock and MCCs mature. Deletion of Myb in the developing airway epithelium blocked or delayed centriole amplification and FOXJ1 expression, and the lung failed to become fully ciliated. Overexpression of MYB in cell culture was sufficient to turn on FOXJ1 expression, although it did not on its own induce centriole amplification (supplementary material Fig. S7). Primary culture studies showed that MYB functions downstream of Notch signaling and MCIN, a coiled-coil protein that controls multiciliogenesis in X. laevis skin and MTECs (Stubbs et al., 2012), and upstream of FOXJ1, which controls centriole docking and ciliary motility. This places MYB in a central position in the multiciliogenesis program (Fig. 8). We propose that MCIN promotes cell cycle exit and induction of Myb, and MYB then promotes centriole amplification and induction of Foxj1, which in turn activates centriole docking and the assembly of a motile cilium.
MYB functions similarly in the development of two other multiciliated cell types that we examined. As in the lung, MYB is expressed early and transiently during multiciliogenesis of brain ependymal cells, after the progenitors exit the cell cycle, and deletion of Myb in these cells appears to disrupt the multiciliogenesis process (Malaterre et al., 2008). During multicilia formation in X. laevis epidermal cells, we showed that MYB functions downstream of MCIN and can drive the full multiciliogenesis program when ectopically expressed. A ciliogenesis defect was also identified recently in the multiciliated epithelial cells of zebrafish kidneys following Myb disruption (Wang et al., 2013). Thus, the central role for MYB in the multiciliogenesis program appears to be conserved across diverse tissues and species.
Although MYB plays a key conserved role in the multiciliogenesis program, there must be at least one other factor in the program that can at least partially compensate for loss of Myb because we observed a partial recovery of multiciliogenesis in certain cell types following Myb deletion. Several days after Myb deletion in the lung, early steps in the program can be detected in the proximal airways. Several weeks later these regions appear to have a normal number of cilia, whereas MCCs in the distal airways almost completely lack cilia. Myb deletion in ependymal cells results in hydrocephalus, implying that the multiciliogenesis program in these cells is permanently impaired (Malaterre et al., 2008). One possibility is that MCIN contributes to this compensatory activity because it can induce Foxj1 expression in the apparent absence of MYB, at least in cotransfection experiments (Fig. 4D, Fig. 8). It will be important to identify the auxiliary factor(s) and to determine how MCIN, which lacks a DNA-binding domain (Stubbs et al., 2012), can induce Myb and Foxj1 expression, and to identify direct targets of MYB, in addition to Foxj1, in the multiciliogenesis program.
There is an apparent paradox in the proposed role for MYB in promoting the differentiation of MCCs and its classical function in hematopoietic and other stem and progenitor cells, where MYB promotes S phase and cell cycling; indeed, loss of Myb typically leads to progenitor cell differentiation and depletion (Malaterre et al., 2007; Malaterre et al., 2008; Lieu and Reddy, 2009). These contrasting roles of MYB can be reconciled if its role in multiciliogenesis is actually a variant of its cell cycle role. During a normal S phase, both chromosomes and centrioles are duplicated; these processes typically begin at the onset of S phase (Hinchcliffe and Sluder, 2001). Cells in G0 do not generate new chromosomes or centrosomes. We speculate that during multiciliogenesis MYB promotes an S phase-like state, which we refer to as S*, during which centriole amplification occurs but not the eponymous hallmark of S phase - DNA synthesis. Interestingly, the mouse homologue of MCIN called IDAS (Stubbs et al., 2012) and another related coiled-coil domain protein named Geminin have both been implicated in the regulation of S phase (Pefani et al., 2011). In our model, DNA synthesis must be specifically repressed in S*. An intriguing candidate for this function is cyclin A1, which is a tissue-restricted homolog of the ubiquitous S-phase cyclin, cyclin A2, which is expressed at unusually high levels in MCCs (Hoh et al., 2012). Cyclin A2 prevents replication complexes from binding to origins of replication that have already fired (Coverley et al., 2002). Perhaps cyclin A1 functions in a similar manner to inactivate replication complexes in MCCs.
In the last 15 years, much progress has been made in understanding the transcriptional program that drives canonical primary cilium formation. RFX3, a transcription factor that apparently functions in all cilia, drives expression of crucial intraflagellar transport components and controls the expression of proteins found at the basal body and the transition zone (Chu et al., 2010; Piasecki et al., 2010; Thomas et al., 2010). We propose that cells deploy specific transcriptional modules that modify this basal ciliogenic program to form different types of cilia (supplementary material Fig. S10). Such modules govern cilium number and other structural attributes found across multiple cell and tissue types. For example, ciliary motility is governed by FOXJ1, which is expressed and required in all cells with motile cilia (Brody et al., 2000; Yu et al., 2008). Our work identifies MYB as a crucial regulator of centriole amplification and multiciliogenesis in multiple tissues and species. We presume that there is at least one other, as yet unidentified, transcription factor and associated transcriptional module that determines the 9+2 axoneme structure instead of the standard 9+0. These transcriptional modules may be coupled; for example, deployment of Myb and the multiplicity module also induces Foxj1 and ciliary motility and a 9+2 axonemal structure (supplementary material Fig. S10). Evolution appears to have ‘mixed and matched’ these ciliary diversity modules in different ciliated cells (supplementary material Fig. S10) and then further modified some ciliary programs for tissue- or cell type-specific purposes, such as the sensory cilium in Drosophila neurons (Newton et al., 2012) and the light-sensing cilium of photoreceptors (Gerdes et al., 2009). Elucidating the full transcriptional programs that control cilium number and diversification will also help us to understand and hopefully treat the many distinct ciliopathies that result from alterations in these programs.
Supplementary Material
Acknowledgments
We thank Dr Jennifer Stubbs (Salk Institute) for initiating the Xenopus Myb experiments; Dr Gray Camp (Stanford University) for help with bioinformatics; Drs Timothy Bender (University of Virginia) and Steven Brody (Washington University) for mouse lines; Dr Peter Lee for help with CellProfiler; and members of the M.A.K. lab and Dr Joe Lipsick (Stanford University) for expert advice and comments on the manuscript.
Footnotes
Funding
This work was supported by the National Institutes of Health (NIH) [Progenitor Cell Biology Consortium grant 5U01HL099995 to M.A.K., R01 GM096021 to C.K., and Cellular and Molecular Biology Training program NIH 5T32 GM07276 to F.E.T.] and fellowships from the A. P. Giannini Foundation (E.K.V.), Howard Hughes Medical Institute fellowship of the Helen Hay Whitney Foundation (L.C.F.), and the Damon Runyon Cancer Research Foundation and the Parker B. Francis Foundation (F.H.E.). M.A.K. is an investigator of the Howard Hughes Medical Institute. Deposited in PMC for immediate release.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
F.E.T., E.K.V. and M.A.K. conceived and analyzed the experiments. F.E.T. and E.K.V. designed and executed experiments. L.M. and C.K. performed X. laevis experiments and L.C.F. performed ependymal experiments. R.H. and T.S. performed microarray analysis of MTECs. F.H.E. compiled the annotated list of mouse transcription factors. F.E.T. and M.A.K. wrote the manuscript. All authors discussed results and edited the manuscript. J.D.A. supported the work of E.K.V.; A.A.-B. supported the work of L.C.F.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.094102/-/DC1
References
- Bender T. P., Kremer C. S., Kraus M., Buch T., Rajewsky K. (2004). Critical functions for c-Myb at three checkpoints during thymocyte development. Nat. Immunol. 5, 721–729 [DOI] [PubMed] [Google Scholar]
- Bessa M., Joaquin M., Tavner F., Saville M. K., Watson R. J. (2001). Regulation of the cell cycle by B-Myb. Blood Cells Mol. Dis. 27, 416–421 [DOI] [PubMed] [Google Scholar]
- Brody S. L., Yan X. H., Wuerffel M. K., Song S. K., Shapiro S. D. (2000). Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am. J. Respir. Cell Mol. Biol. 23, 45–51 [DOI] [PubMed] [Google Scholar]
- Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., Friman O., Guertin D. A., Chang J. H., Lindquist R. A., Moffat J., et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J. S. C., Baillie D. L., Chen N. (2010). Convergent evolution of RFX transcription factors and ciliary genes predated the origin of metazoans. BMC Evol. Biol. 10, 130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley D., Laman H., Laskey R. A. (2002). Distinct roles for cyclins E and A during DNA replication complex assembly and activation. Nat. Cell Biol. 4, 523–528 [DOI] [PubMed] [Google Scholar]
- Davis E. E., Katsanis N. (2012). The ciliopathies: a transitional model into systems biology of human genetic disease. Curr. Opin. Genet. Dev. 22, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblandre G. A., Wettstein D. A., Koyano-Nakagawa N., Kintner C. (1999). A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos. Development 126, 4715–4728 [DOI] [PubMed] [Google Scholar]
- Del Bigio M. R. (2010). Ependymal cells: biology and pathology. Acta Neuropathol. 119, 55–73 [DOI] [PubMed] [Google Scholar]
- Dubaissi E., Papalopulu N. (2011). Embryonic frog epidermis: a model for the study of cell-cell interactions in the development of mucociliary disease. Dis. Model. Mech. 4, 179–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch C., Dupuis-Williams P. (2011). Ultrastructure of cilia and flagella - back to the future! Biol. Cell 103, 249–270 [DOI] [PubMed] [Google Scholar]
- Gerdes J. M., Davis E. E., Katsanis N. (2009). The vertebrate primary cilium in development, homeostasis, and disease. Cell 137, 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomperts B. N., Gong-Cooper X., Hackett B. P. (2004). Foxj1 regulates basal body anchoring to the cytoskeleton of ciliated pulmonary epithelial cells. J. Cell Sci. 117, 1329–1337 [DOI] [PubMed] [Google Scholar]
- Hagiwara H., Ohwada N., Takata K. (2004). Cell biology of normal and abnormal ciliogenesis in the ciliated epithelium. Int. Rev. Cytol. 234, 101–141 [DOI] [PubMed] [Google Scholar]
- Harfe B. D., Scherz P. J., Nissim S., Tian H., McMahon A. P., Tabin C. J. (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517–528 [DOI] [PubMed] [Google Scholar]
- Harris K. S., Zhang Z., McManus M. T., Harfe B. D., Sun X. (2006). Dicer function is essential for lung epithelium morphogenesis. Proc. Natl. Acad. Sci. USA 103, 2208–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe E. H., Sluder G. (2001). ‘It takes two to tango’: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 15, 1167–1181 [DOI] [PubMed] [Google Scholar]
- Hoh R. A., Stowe T. R., Turk E., Stearns T. (2012). Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS ONE 7, e52166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles M. R., Boucher R. C. (2002). Mucus clearance as a primary innate defense mechanism for mammalian airways. J. Clin. Invest. 109, 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieu Y. K., Reddy E. P. (2009). Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc. Natl. Acad. Sci. USA 106, 21689–21694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsick J. S. (2010). The C-MYB story - is it definitive? Proc. Natl. Acad. Sci. USA 107, 17067–17068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub M. R., Stearns T. (2012). Supernumerary centrosomes nucleate extra cilia and compromise primary cilium signaling. Curr. Biol. 22, 1628–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaterre J., Carpinelli M., Ernst M., Alexander W., Cooke M., Sutton S., Dworkin S., Heath J. K., Frampton J., McArthur G., et al. (2007). c-Myb is required for progenitor cell homeostasis in colonic crypts. Proc. Natl. Acad. Sci. USA 104, 3829–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaterre J., Mantamadiotis T., Dworkin S., Lightowler S., Yang Q., Ransome M. I., Turnley A. M., Nichols N. R., Emambokus N. R., Frampton J., et al. (2008). c-Myb is required for neural progenitor cell proliferation and maintenance of the neural stem cell niche in adult brain. Stem Cells 26, 173–181 [DOI] [PubMed] [Google Scholar]
- Mirzadeh Z., Merkle F. T., Soriano-Navarro M., Garcia-Verdugo J. M., Alvarez-Buylla A. (2008). Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell 3, 265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z., Doetsch F., Sawamoto K., Wichterle H., Alvarez-Buylla A. (2010). The subventricular zone en-face: wholemount staining and ependymal flow. J. Vis. Exp. 39, e1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto M., Liu Z., Cheng H. T., Winters N., Bader D., Kopan R. (2010). Canonical Notch signaling in the developing lung is required for determination of arterial smooth muscle cells and selection of Clara versus ciliated cell fate. J. Cell Sci. 123, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucenski M. L., McLain K., Kier A. B., Swerdlow S. H., Schreiner C. M., Miller T. A., Pietryga D. W., Scott W. J., Jr, Potter S. S. (1991). A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65, 677–689 [DOI] [PubMed] [Google Scholar]
- Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 [DOI] [PubMed] [Google Scholar]
- Nayak G. D., Ratnayaka H. S. K., Goodyear R. J., Richardson G. P. (2007). Development of the hair bundle and mechanotransduction. Int. J. Dev. Biol. 51, 597–608 [DOI] [PubMed] [Google Scholar]
- Newton F. G., zur Lage P. I., Karak S., Moore D. J., Göpfert M. C., Jarman A. P. (2012). Forkhead transcription factor Fd3F cooperates with Rfx to regulate a gene expression program for mechanosensory cilia specialization. Dev. Cell 22, 1221–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y., Kido M., Hirokawa N. (1998). Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829–837 [DOI] [PubMed] [Google Scholar]
- Ostrowski L. E., Hutchins J. R., Zakel K., O’Neal W. K. (2003). Targeting expression of a transgene to the airway surface epithelium using a ciliated cell-specific promoter. Mol. Ther. 8, 637–645 [DOI] [PubMed] [Google Scholar]
- Pan J., You Y., Huang T., Brody S. L. (2007). RhoA-mediated apical actin enrichment is required for ciliogenesis and promoted by Foxj1. J. Cell Sci. 120, 1868–1876 [DOI] [PubMed] [Google Scholar]
- Pefani D. E., Dimaki M., Spella M., Karantzelis N., Mitsiki E., Kyrousi C., Symeonidou I. E., Perrakis A., Taraviras S., Lygerou Z. (2011). Idas, a novel phylogenetically conserved geminin-related protein, binds to geminin and is required for cell cycle progression. J. Biol. Chem. 286, 23234–23246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecki B. P., Burghoorn J., Swoboda P. (2010). Regulatory Factor X (RFX)-mediated transcriptional rewiring of ciliary genes in animals. Proc. Natl. Acad. Sci. USA 107, 12969–12974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay R. G., Gonda T. J. (2008). MYB function in normal and cancer cells. Nat. Rev. Cancer 8, 523–534 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Scholzen T., Gerdes J. (2000). The Ki-67 protein: from the known and the unknown. J. Cell. Physiol. 182, 311–322 [DOI] [PubMed] [Google Scholar]
- Sive H. L., Grainger R. M., Harland R. M. (1998). Early Development of Xenopus Laevis: A Laboratory Manual. Plainsview, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Sorokin S. P. (1968). Centriole formation and ciliogenesis. Aspen Emphysema Conf. 11, 213–216 [PubMed] [Google Scholar]
- Stubbs J. L., Vladar E. K., Axelrod J. D., Kintner C. (2012). Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat. Cell Biol. 14, 140–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S. H., Silva F. J., Tsark W. M., Mann J. R. (2002). A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis 32, 199–202 [DOI] [PubMed] [Google Scholar]
- Thomas J., Morlé L., Soulavie F., Laurençon A., Sagnol S., Durand B. (2010). Transcriptional control of genes involved in ciliogenesis: a first step in making cilia. Biol. Cell 102, 499–513 [DOI] [PubMed] [Google Scholar]
- Tiozzo C., De Langhe S., Yu M., Londhe V. A., Carraro G., Li M., Li C., Xing Y., Anderson S., Borok Z., et al. (2009). Deletion of Pten expands lung epithelial progenitor pools and confers resistance to airway injury. Am. J. Respir. Crit. Care Med. 180, 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toscani A., Mettus R. V., Coupland R., Simpkins H., Litvin J., Orth J., Hatton K. S., Reddy E. P. (1997). Arrest of spermatogenesis and defective breast development in mice lacking A-myb. Nature 386, 713–717 [DOI] [PubMed] [Google Scholar]
- Tsao P. N., Vasconcelos M., Izvolsky K. I., Qian J., Lu J., Cardoso W. V. (2009). Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development 136, 2297–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou M. F., Stearns T. (2006). Mechanism limiting centrosome duplication to once per cell cycle. Nature 442, 947–951 [DOI] [PubMed] [Google Scholar]
- Turner D. L., Weintraub H. (1994). Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 8, 1434–1447 [DOI] [PubMed] [Google Scholar]
- Venugopalan S. R., Amen M. A., Wang J., Wong L., Cavender A. C., D’Souza R. N., Akerlund M., Brody S. L., Hjalt T. A., Amendt B. A. (2008). Novel expression and transcriptional regulation of FoxJ1 during oro-facial morphogenesis. Hum. Mol. Genet. 17, 3643–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar E. K., Stearns T. (2007). Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 178, 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Fu C., Fan H., Du T., Dong M., Chen Y., Jin Y., Zhou Y., Deng M., Gu A., et al. (2013). miR-34b regulates multiciliogenesis during organ formation in zebrafish. Development 140, 2755–2764 [DOI] [PubMed] [Google Scholar]
- Xu Q., Tam M., Anderson S. A. (2008). Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol. 506, 16–29 [DOI] [PubMed] [Google Scholar]
- You Y., Richer E. J., Huang T., Brody S. L. (2002). Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am. J. Physiol. 283, L1315–L1321 [DOI] [PubMed] [Google Scholar]
- You Y., Huang T., Richer E. J., Schmidt J. E., Zabner J., Borok Z., Brody S. L. (2004). Role of f-box factor foxj1 in differentiation of ciliated airway epithelial cells. Am. J. Physiol. 286, L650–L657 [DOI] [PubMed] [Google Scholar]
- Yu X., Ng C. P., Habacher H., Roy S. (2008). Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat. Genet. 40, 1445–1453 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.