SUMMARY
Vital to bacterial survival is the faithful propagation of cellular signals, and in Caulobacter crescentus ChpT is an essential mediator within the cell cycle circuit. ChpT functions as a histidine-containing phosphotransfer protein (HPt) that shuttles a phosphoryl group from the receiver domain of CckA, the upstream hybrid histidine kinase (HK), to one of two downstream response regulators (RRs)—CtrA or CpdR—that controls cell cycle progression. To understand how ChpT interacts with multiple signaling partners, we solved the crystal structure of ChpT at 2.3 Å resolution. ChpT adopts a pseudo-HK architecture but does not bind ATP. We identified two point mutation classes affecting phosphotransfer and cell morphology: one that globally impairs ChpT phosphotransfer, and a second that mediates partner selection. Importantly, a small set of conserved ChpT residues promotes signaling cross-talk and contributes to the branched signaling that activates the master regulator CtrA while inactivating the CtrA degradation signal, CpdR.
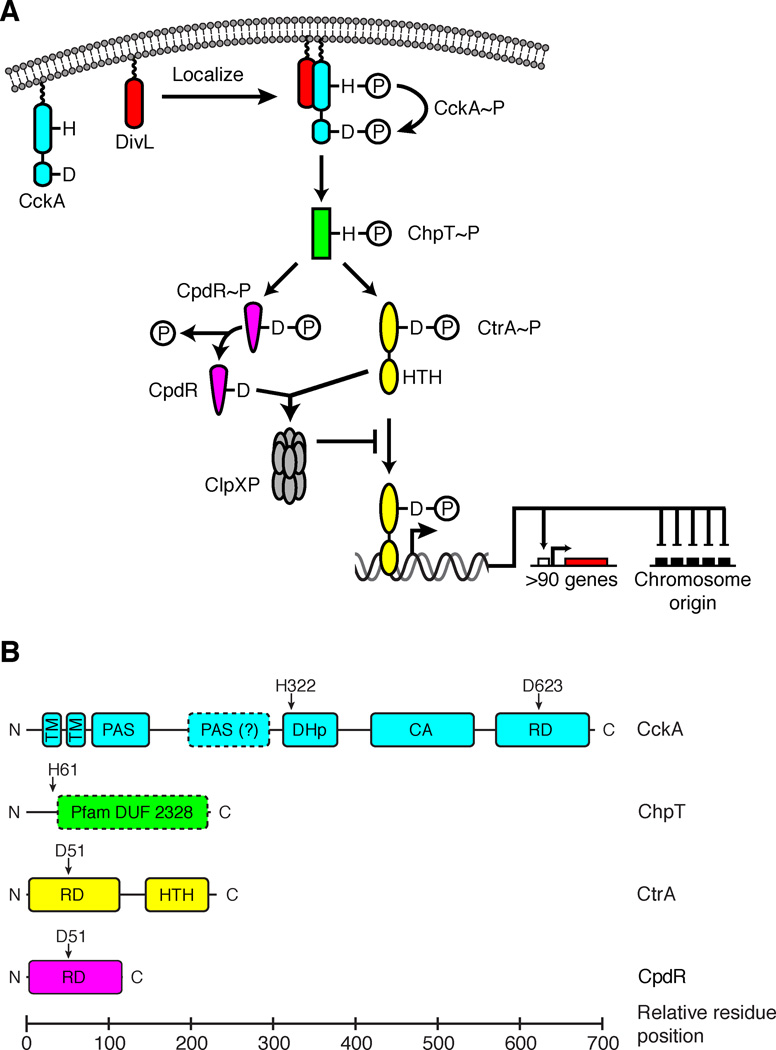
To thrive in dilute freshwater, its environmental niche, Caulobacter crescentus has evolved sophisticated two-component signaling (TCS) systems that integrate environmental sensing and asymmetric cellular proliferation (Curtis and Brun, 2010; Skerker et al., 2005). The CckA-ChpT-CtrA-CpdR pathway is the core signaling circuit controlling the cell cycle (Biondi et al., 2006). In canonical TCS pathways a histidine kinase (HK) senses a signal and undergoes autophosphorylation: the catalytic, ATP-binding (CA) domain binds ATP and catalyzes transfer of the γ-phosphate of ATP to a conserved histidine residue in the dimerization and histidine phosphotransfer (DHp) domain (Gao and Stock, 2009). Subsequent transfer of the phosphoryl group to an aspartic acid residue in the receiver domain (RD) of a response regulator (RR) activates the RR, which induces an appropriate cellular response (e.g. activation of transcription) (Gao and Stock, 2009). The CckA-ChpT-CtrA-CpdR pathway has an additional step, using a bifurcated His-Asp-His-Asp phosphorelay (Fig. 1A). CckA is a hybrid HK with a C-terminal RD in addition to its CA and DHp domains (Angelastro et al., 2010; Chen et al., 2009; Jacobs et al., 2003; Jacobs et al., 1999) (Fig. 1B). Upon localization to the new cell pole, mediated by the DivL pseudo-histidine kinase (Iniesta et al., 2010), CckA autophosphorylates His322 of its DHp domain, followed by an intramolecular phosphotransfer reaction that passes the phosphoryl group from His322 to Asp623 in the RD (Chen et al., 2009) (Fig. 1A). ChpT then accepts the phosphoryl group from CckA-RD and donates it either to CtrA or CpdR, two downstream RRs that coordinate the Caulobacter cell cycle (Biondi et al., 2006; Chen et al., 2009).
FIGURE 1.
The CckA-ChpT-CtrA-CpdR signaling pathway controls cell cycle progression in Caulobacter. A. Schematic of the CckA-ChpT-CtrA-CpdR signaling pathway. B. Domain organization of the TCS proteins in the CckA-ChpT-CtrA-CpdR pathway (transmembrane helix, TM; Per-Arnt-Sim sensor domain, PAS; dimerization and histidine phosphotransfer domain, DHp; catalytic and ATP-binding domain, CA; receiver domain, RD; helix-turn-helix DNA binding domain, HTH; domain of unknown function, DUF). Vertical arrows point to residues involved in phosphotransfer.
CckA and ChpT are essential to cell-cycle progression, as they regulate both activation by phosphorylation of CtrA and proteolytic degradation of CtrA, a master transcriptional regulator controlling the cell cycle and the initiation of DNA replication (Angelastro et al., 2010; Biondi et al., 2006; Iniesta et al., 2010; Iniesta and Shapiro, 2008; Jacobs et al., 2003; Jacobs et al., 1999). The phosphorylation state of CtrA is a vital checkpoint in the Caulobacter cell cycle: active phospho-CtrA (CtrA~P) binds to the origin of replication (Quon et al., 1996; Quon et al., 1998), thereby silencing replication initiation in swarmer cells and ensuring that Caulobacter replicates its DNA only once per cell cycle. To relieve this inhibition, a feedback loop—mediated by the single domain RR CpdR—regulates CtrA~P proteolysis (Iniesta et al., 2006; Iniesta and Shapiro, 2008). ChpT~P transfers phosphate to Asp51 on CpdR (Biondi et al., 2006; Chen et al., 2009) to keep CpdR in an inactive state in swarmer cells. After the swarmer-to-stalk transition, unphosphorylated CpdR localizes to the stalked pole, recruiting the ClpXP protease that degrades CtrA (Iniesta et al., 2006; Iniesta and Shapiro, 2008).
ChpT is interesting because it sits at a TCS pathway branch point, where it is a conduit for shuttling biochemical information among three independent RD domains (CckA, CtrA and CpdR, Fig. 1A). ChpT has no autokinase activity, and its only activation partner is CckA (Biondi et al., 2006). ChpT is also conserved among alphaproteobacteria, and the Pfam database (Fig. 1B) annotates part of ChpT as a domain of unknown function (DUF2328) related to HKs (Punta et al., 2012). A search for remote structural homologs of ChpT using HHpred (Soding et al., 2005) suggested that ChpT is structurally similar to the DHp-CA architectures of HKs. However, with no close structural homolog, our molecular level understanding of how this key signaling molecule functions was incomplete. Therefore, we solved the crystal structure of ChpT, demonstrating that ChpT adopts a fold remarkably similar to DHp-CA domains found in HKs—albeit with a pseudo-CA domain—and we showed that ChpT neither binds nor hydrolyzes ATP. The structure suggests ChpT-RR interactions are analogous to that of RR-HK interactions. We tested this hypothesis by mutating specific amino acids in the putative binding surfaces, measuring wild-type and mutant ChpT phosphotransfer activity to its multiple signaling partners, and evaluating cellular morphology.
RESULTS
Structure determination and the overall structure
In the deposited genomes for Caulobacter crescentus, chpT is misannotated (Chen et al., 2009; Christen et al., 2011). We therefore chose to clone residues 29–253, representing the correct ORF for chpT, as annotated in GenBank (GI:13425192). We cloned chpT as an N-terminal His6 fusion with a thrombin cleavage site, expressed this construct in Escherichia coli, and purified it to greater than 90% purity. We use 1–225 amino acid numbering for ChpT: Met-1 is annotated as Met-29 in GenBank.
Purified ChpT crystallized from several conditions using commercial screening kits, but all crystals diffracted poorly. Removal of the His6-tag and subsequent optimization of crystallization conditions improved the diffraction quality significantly. Native data (up to 2.3 Å resolution) and heavy atom derivative data (up to 3.0 Å) were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) on beamline BL12-2. We determined the ChpT crystal structure by multiple-wavelength anomalous diffraction (MAD) phasing using a gold derivative.
We refined the final model to an Rcryst of 18.4% and an Rfree of 21.4%. The ChpT model displays good geometry with an all-atom clash score of 5.7, and the Ramachandran plot calculated by MolProbity (Chen et al., 2010) shows all residues lie in allowed regions with 98.9% in favored regions. The model contains four ChpT protein chains (A/17–225 and B/C/D/18–225) in the asymmetric unit (asu, Fig. S1A) along with four sodium ions, four glycerol molecules, and 311 water molecules. The residual purification tag and the first 17 residues were disordered and were not included in the final model. Additionally, 17 side-chains on the protein surface were not modeled due to poor density. We summarize the data processing and refinement statistics in Table 1.
TABLE 1.
Data collection and refinement statistics (PDB ID 4fmt)
| Data collection | Native | Au derivative | |
|---|---|---|---|
| Wavelength (Å) | 0.9795 | 1.0395 | 0.8731 |
| Resolution range (Å) | 65–2.3 | 48–2.95 | 48.0–3.07 |
| No. observations | 181765 | 92087 | 83223 |
| No. unique reflections | 52631 | 24702 | 21925 |
| Completeness (%)a | 97.3 (97.5) | 97.7 (98.0) | 97.6 (98.5) |
| Mean I/σ (I)a | 10.7 (1.8) | 11.2 (2.3) | 10.8 (2.9) |
| Rmerge on I (%)a | 8.7 (83.5) | 9.3 (49.8) | 9.9 (41.5) |
| Rmeas on I (%)a | 10.3 (99.7) | 12.5 (67.8) | 13.4 (56.5) |
| Rpim on I (%)a | 5.5 (53.5) | 8.3 (45.7) | 8.9 (38.1) |
| High resolution shell | 2.42–2.3 | 3.11–2.95 | 3.24–3.07 |
| Model and refinement statistics | |||
| No. reflections (total) | 52619 | ||
| No. reflections (test) | 2682 | ||
| Cutoff criteria | |F|>0 | ||
| Rcryst | 18.4 | ||
| Rfree | 21.4 | ||
| Restraints (RMS observed) | |||
| Bond angle (°) | 1.02 | ||
| Bond length (Å) | 0.010 | ||
| MolProbity Scores | |||
| All-atom clash score | 5.7 | ||
| Ramachandran plot Favored (allowed, %) | 98.9 (100) | ||
| Rotamer outliers (%) | 0.9 | ||
| Average isotropic B-value (Å2)b | 54.2 (51.4) | ||
| ESU based on Rfree (Å) | 0.17 | ||
| Protein chains/residues/atoms | 4/833/6118 | ||
Highest resolution shell in parentheses.
This value represents the total B that includes TLS and residual B components. Wilson B-value in parenthesis.
Rmerge= ∑hkl ∑i|Ii(hkl)−<I(hkl)>|/∑hkl ∑ iIi(hkl), Rmeas(redundancy-independent Rmerge) = ∑hkl[Nhkl/(Nhkl−1)]½ ∑i|Ii(hkl) −<I(hkl)>|/ ∑hkl ∑iIi(hkl), and Rpim(precision-indicating Rmerge)= ∑hkl[1/(Nhkl−1)]½ ∑i|Ii(hkl)−<I(hkl)>|/ ∑hkl ∑iIi(hkl).
Rcryst = ∑| |Fobs|−|Fcalc| | / ∑|Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively. Rfree = as for Rcryst, but for 5.0% of the total reflections chosen at random and omitted from refinement.
ESU = Estimated overall coordinate error.
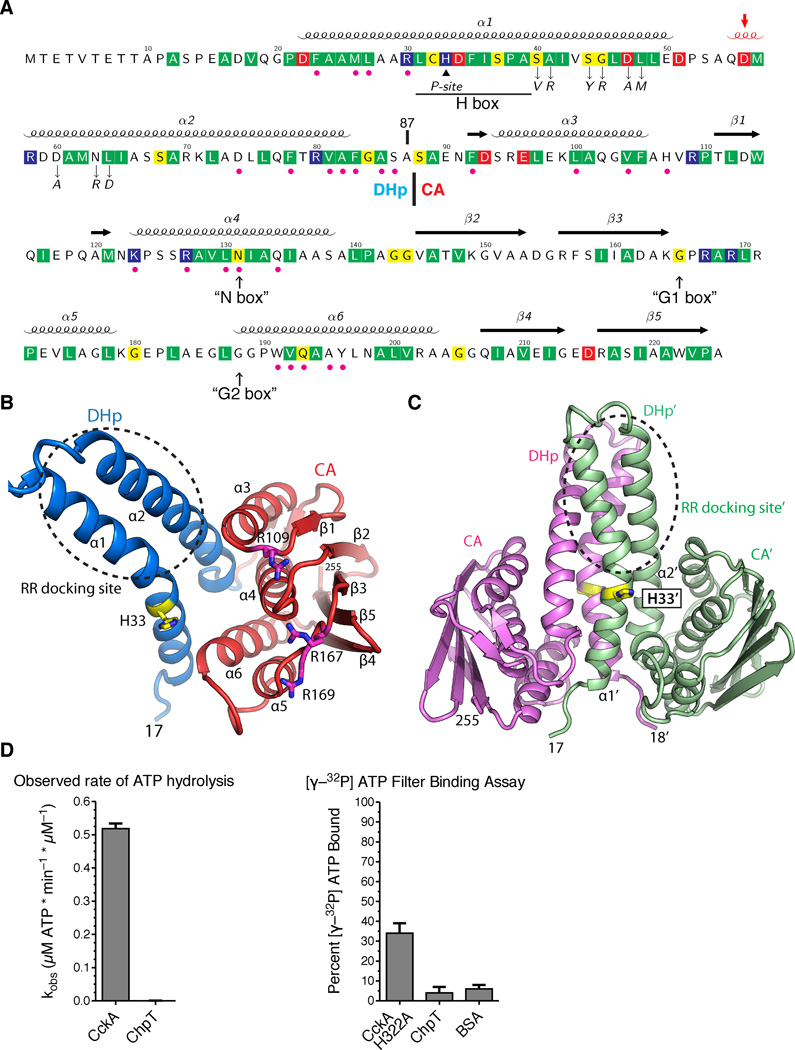
The four ChpT monomers in the asu are almost identical with an average RMSD of only 0.28 Å (208 Cα). The domain organization of ChpT is similar to HKs: each ChpT monomer (Fig. 2A–B) consists of a DHp domain (α1–α2, residue range 17–87) and a CA domain (residue range 88–225). The CA domain (α3–α6 and β1–β5) is related to other members of the GHKL superfamily (Dutta and Inouye, 2000). Analysis of protein interfaces in the crystal lattice suggested these four monomers form two ChpT dimers (Fig. S1A), which is in agreement with size exclusion chromatography that suggested a ChpT dimer in solution with an observed molecular weight of 60.7 ± 14 kDa; the calculated dimer molecular weight is 51.1 kDa (Table S1). Hydrophobic interactions between DHp domains mediate the dimer interface (Fig. 2C), burying a total surface area of 3650 Å2. Interestingly, one of the dimer-dimer interfaces involves two DHp domains packing perpendicular to each other (Fig. S1A), resembling the binding of the HK KinB by its inhibitor protein Sda (Bick et al., 2009).
FIGURE 2.
The ChpT crystal structure shows a pseudo-HK fold. A. The annotated ChpT primary sequence with the secondary structure elements shown above (the 310 helix in red and indicated by an arrow). Conserved residues among ChpT orthologs are highlighted in colors (red, acidic; blue, basic; yellow, polar uncharged; and green, hydrophobic nonpolar). Magenta dots denote the residues at the DHp-CA interfaces, and the black triangle denotes the site of phosphorylation (His33). Residues that are subjected to point mutations are denoted by down arrows. The expected locations for degenerate HK sequence motifs are shown at the bottom (in quotes and up arrows). The domain boundary between the DHp and CA domains is located at residue 87. B. The domain organization of a ChpT monomer colored by domain (CA - red, DHp - blue). His33 and conserved arginines on the surface of CA domain are shown as sticks. C. A ChpT homodimer contains a four-helix bundle formed by two DHp domains. One ChpT molecule is colored magenta; the other is colored sea-green. His33, the site of phosphorylation, is shown in sticks. A circle highlights the site for interaction with RRs. D. A coupled-enzyme assay confirms ChpT cannot catalyze ATP hydrolysis. E. An ATP filter binding assay confirms ChpT cannot bind ATP (see also Fig. S1).
ChpT is structurally related to HKs
The ChpT structure is similar to several structurally characterized HKs, including HK853 (Casino et al., 2009; Marina et al., 2005) and ThkA (Yamada et al., 2009) from Thermotoga maritima, and KinB from Bacillus subtilis (Bick et al., 2009), as well as the B. subtilis HPt Spo0B (Varughese et al., 1998). ChpT more closely resembles HKs than Spo0B, in both its sequence and structure. ChpT shares ~16% sequence identity [HHpred (Soding et al., 2005), E-value ~1.0e-29] with HKs HK853 (PDB ID 2c2a) (Marina et al., 2005) or KinB (PDB ID 3d36) (Bick et al., 2009), and <10% sequence identity (E-value 8.1e-07) with Spo0B (PDB ID 1ixm) (Varughese et al., 1998). The ChpT-CA domain can be superposed to HK853-CA with an RMSD of 3.0 Å (127 Cα, Dali Z=12.5), compared to an RMSD of 2.9 Å (109 Cα, Z=8.3) between the ChpT-CA and the Spo0B-CA. The ChpT-CA domain contains all the core secondary structural elements present in the CA domain of HKs, while Spo0B is missing one helix that is critical for ATP binding (α6, Fig. S2).
The CA domain of ChpT contains residual sequence signatures from the ATP binding motifs (N, G1, and G2 boxes) of the canonical HKs (Parkinson and Kofoid, 1992) (Fig. 2A). In contrast to typical HKs that harbor G1, F and G2 boxes in the β3-α6 region, ChpT is missing the hFxxF motif that is characteristic of the F-box (Fig 2A, Fig. S2B). Thus, the ChpT ATP binding site is degenerate. Using in vitro assays, we determined that ChpT neither binds ATP nor catalyzes ATP hydrolysis (Fig. 2D–E), consistent with this structure.
Phosphorylation and RR-binding sites
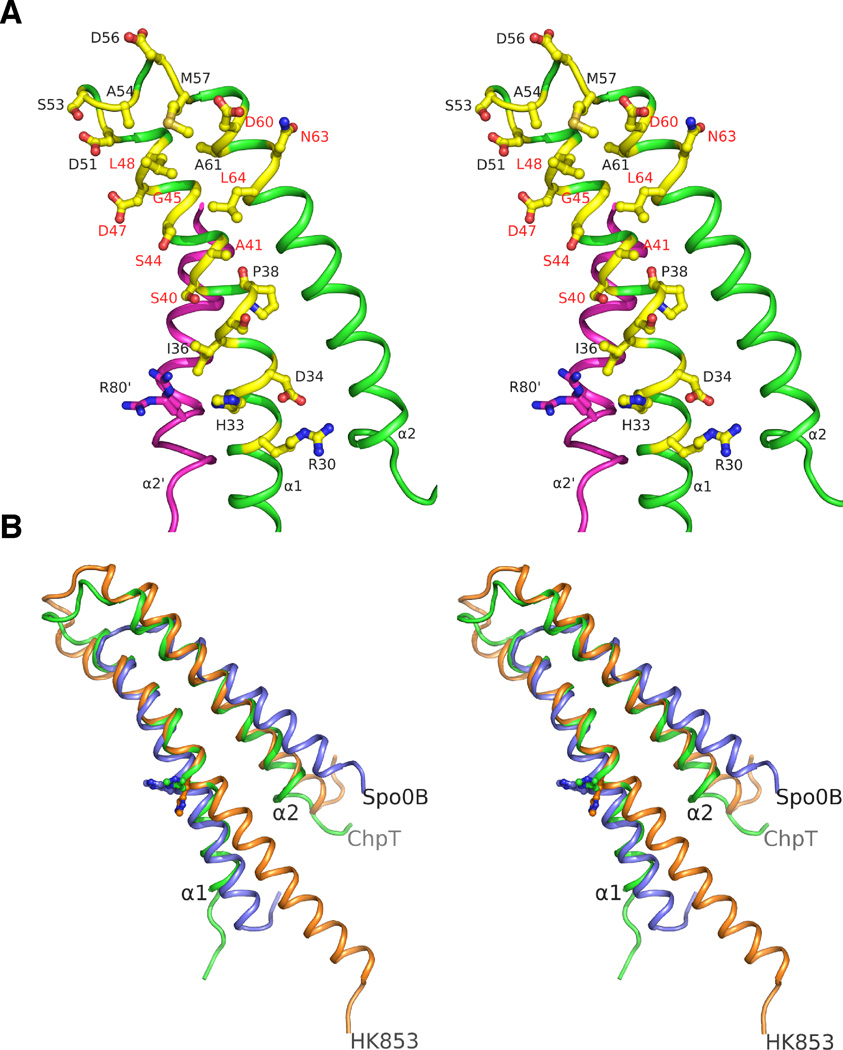
The phosphorylation site of ChpT (His33) (Biondi et al., 2006) resides on the third turn of helix α1 (Fig. 2C). The side-chain conformation exposes the Nε phosphorylation site to solvent, similar to the yeast phosphotransfer protein YPD1 (Xu and West, 1999) and E. coli ArcB (Kato et al., 1997). His33 is part of a sequence motif (hCHDhhsPs, h=hydrophobic, s=residues with small side-chains) that bears significant resemblance to the H-box of HKs. A proline (Pro38) induces a small kink in helix α1 (Fig. 3A). His33 is located near two conserved arginines (Arg30 and Arg80’). Arg30 is hydrogen bonded with Asp34 and residues from the CA domain. Arg80’, from the adjacent protomer in the dimer, is conformationally flexible (Fig. 3A). The side-chain of Arg80’ is located in a position similar to that of Lys67 of YPD1, which functions to stabilize the phosphorylated histidine (Janiak-Spens et al., 2005; Janiak-Spens and West, 2000).
FIGURE 3.
Stereoview of the ChpT recognition surface for RRs. A. Residues (shown in yellow/blue/red ball-and-sticks) in the ChpT DHp region (green) hypothesized to be involved in binding RRs or phosphotransfer. The residues mutated in this study are labeled in red colored texts. B. Structural alignments of the DHp regions of ChpT (green), HK853 (orange) and Spo0B (blue). ChpT has a shorter α1 helix like Spo0B compared to HK853. Histidine residues involved in phosphotransfer are shown as balls-and-sticks (see also Fig. S2).
The ChpT DHp domain is similar to that of HK853 (Fig. 3B). However, the first DHp helix α1 is shorter on the N-terminal end for both the HK-like HPts ChpT and Spo0B. This portion of α1 in HKs is usually connected to upstream sensor domains but is not needed for HPt function. The most structurally conserved region between ChpT and HK853 maps to a helical hairpin formed by the C-terminal portion of α1 and the N-terminal portion of α2 (RMSD ~1.4 Å for 46 equivalent Cα atoms defined by ChpT residues 31–50 and 57–82). Previous studies of Spo0B and HK853 indicated that this region is involved in RR recognition (Casino et al., 2009; Zapf et al., 2000). In this conserved mechanism, Spo0B and HK853 mainly recognize the first helix of RRs through hydrophobic interactions. The protein surface for this region of ChpT shares similar properties with related HKs, where exposed residues on α1 following His33 generally have small side-chains (Ser37, Ser40, Ala41, Ser44, and Gly45). A small hydrophobic surface patch (residues Pro38, Ala41, Gly45, Leu48, Ala54, Met57, and Leu64) with hydrophilic residues at the perimeter forms the expected RR-binding surface of ChpT (Fig. 3A). The helical hairpin of ChpT or HK853 is longer than that of Spo0B (Fig. 3B), which may extend its RR binding surface at the perimeter.
Phosphotransfer activity of ChpT variants
To test whether ChpT uses a similar surface as HKs to phosphotransfer to RRs, we measured the ability of ChpT variants to transfer 32P-phosphate to its three RD partners and to receive phosphate from CckA~P (Fig. 4, Fig. S3). To determine the position of relevant amino acid substitutions in ChpT, we aligned the ChpT DHp domain sequence with DHp domain sequences of HKs from E. coli EnvZ, RstB and CpxA, and from T. maritima HK853 (Fig. 2A, Fig. S4A). This alignment identified a set of nine ChpT DHp residues that are crucial for HK-RR binding specificity (Capra et al., 2012a; Capra et al., 2012b; Skerker et al., 2008), and we selected the substituting residue by considering which amino acid was most chemically distinct from the native residue. This set was: S40V, A41R, S44Y, G45R, D47A, L48M, D60A, N63R, and L64D (Fig. 2A). All nine variants, as well as wild-type ChpT, were purified as N-terminal His6 fusions using Ni-NTA affinity purification. Wavelength scan circular dichroism spectra of ChpT-S40V, ChpT-A41R, ChpT-S44Y, ChpT-G45R, and ChpT-L64D were similar to those exhibited by ChpT wild-type (Fig. S4C). Furthermore, analytical size exclusion chromatography revealed that each ChpT variant purified as a homodimer at the same retention volume as ChpT wild-type (Table S1). Taken together, these data suggest that the point mutation variants of ChpT fold similarly to wild-type. In addition, we purified CpdR as described (Abel et al., 2011), CckA without its N-terminal transmembrane helices (amino acids 70–691) as an N-terminal His6 fusion, and CtrA as an N-terminal His6-SUMO domain fusion.
FIGURE 4.
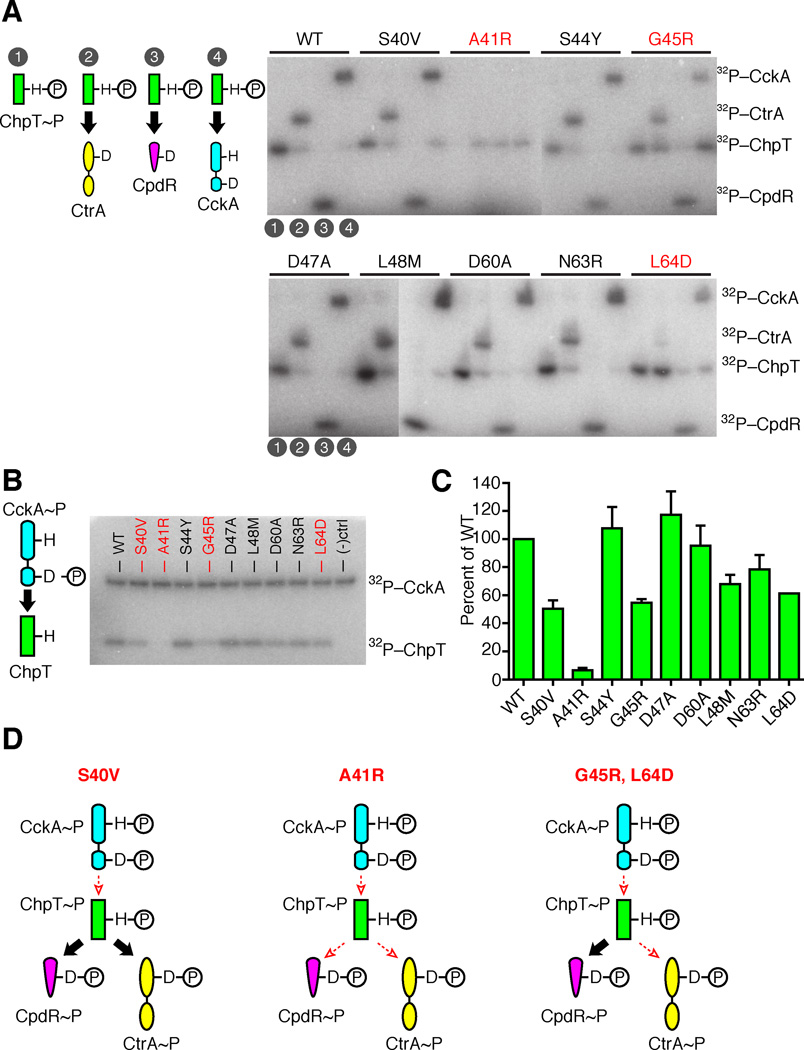
An essential surface on ChpT governs its RR interactions. A. For each ChpT mutant, ChpT~P was generated by incubation with FLAG-CckA, then subsequently purified to remove FLAG-CckA and ATP. Purified ChpT~P mutants were incubated for 10 s with either (1) ChpT~P only, (2) ChpT~P + SUMO-CtrA, (3) ChpT~P + CpdR, (4) ChpT~P + FLAG-CckA. B. A phosphotransfer assay between CckA and ChpT variants with point mutations in the putative RR binding region. CckA~P autophosphorylated in [γ-32P] ATP was mixed with each ChpT variant and allowed to react for 10s before quenching. The phosphoproteins were separated by SDS-PAGE and imaged by phosphor storage. Negative control lane was a reaction mixture lacking ChpT. C. Quantitation of three replicate phosphotransfer assays between CckA~P and ChpT variants with mean intensity and standard deviation of each ChpT~P band shown. D. Classification of ChpT mutants as deficient in CckA-ChpT phosphotransfers, deficient in all phosphotransfers, or deficient in phosphotransfers between CckA-ChpT and ChpT-CtrA. Large black arrows indicate efficient phosphotransfer between signaling partners, whereas small, dashed, red arrows indicate diminished phosphotransfer (see also Fig. S3).
To examine how ChpT mutations impacted each phosphotransfer step, we characterized single-turnover phosphotransfer from purified ChpT~P to each RD: SUMO-CtrA, ChpT, and CckA. ChpT~P was generated by incubation with FLAG-CckA and [g-32P] ATP for 45 minutes, and then FLAG-CckA was removed using anti-FLAG M2 magnetic beads. Purified ChpT~P was incubated with hexokinase and glucose for 10 minutes to turnover the remaining ATP into ADP. Individual ChpT reactions were set up in a 10:1 RD:ChpT molar ratio, allowed to incubate for 10 seconds before quenching, and separated by PAGE for each ChpT mutant: (1) ChpT~P only, (2) ChpT~P to CtrA transfer, (3) ChpT~P to CpdR transfer, and (4) ChpT~P to CckA transfer (Fig. 4A). All ChpT phosphotransfers occurred in less than 10 seconds for wild-type ChpT, and S40V, S44Y, D47A, L48M, D60A, and N63R. While no phosphotransfers were observed for ChpT A41R, mutations at G45R and L64D only impacted the phosphotransfers to CtrA and CckA, while preserving phosphotransfers to CpdR. Equilibrium phosphorylation of the CckA-ChpT-CtrA or CckA-ChpT-CpdR phosphorelays for 30 minutes confirmed these results (Fig. S3A): ChpT A41R and G45R accumulated phosphate on all RDs poorly, and ChpT S40V and L64D showed decreased accumulation only for SUMO-CtrA~P. CtrA~P and CpdR~P half lives were longer than 60 minutes in vitro (Chen et al., 2009), therefore spontaneous RR~P dephosphorylation likely did not play a major role in phosphorylation equilibrium experiments.
To test for impact on the kinetics of the forward reaction from CckA to ChpT phosphotransfer, we compared the nine ChpT variants for their ability to accept 32P-phosphate from autophosphorylated CckA under a short time frame. We incubated CckA with [g-32P] ATP for one hour at room temperature to induce autophosphorylation; the reaction products were then incubated with each ChpT variant at room temperature for 10 seconds. An aliquot from each reaction was analyzed similar to the above phosphotransfer reactions. ChpT variants S40V, A41R, G45R, and L64D displayed severely reduced capacity to receive phosphate from autophosphorylated CckA relative to wild-type (Fig. 4B–C). Additionally, ChpT variants L48M and N63R displayed a mild, but detectable, decrease in phosphotransfer relative to wild-type. These disruptions in phosphotransfer suggest that ChpT residues Ser40, Ala41, Gly45, Leu48, Asn63 and Leu64 contribute to the ChpT phosphotransfer interaction surface. Interestingly, the ChpT S40V mutation selectively and severely impaired CckA to ChpT phosphotransfer (Fig. 4C) while having no effect on other ChpT phosphotransfers (Fig. 4A).
Surface plasmon resonance (SPR) between CckA and ChpT
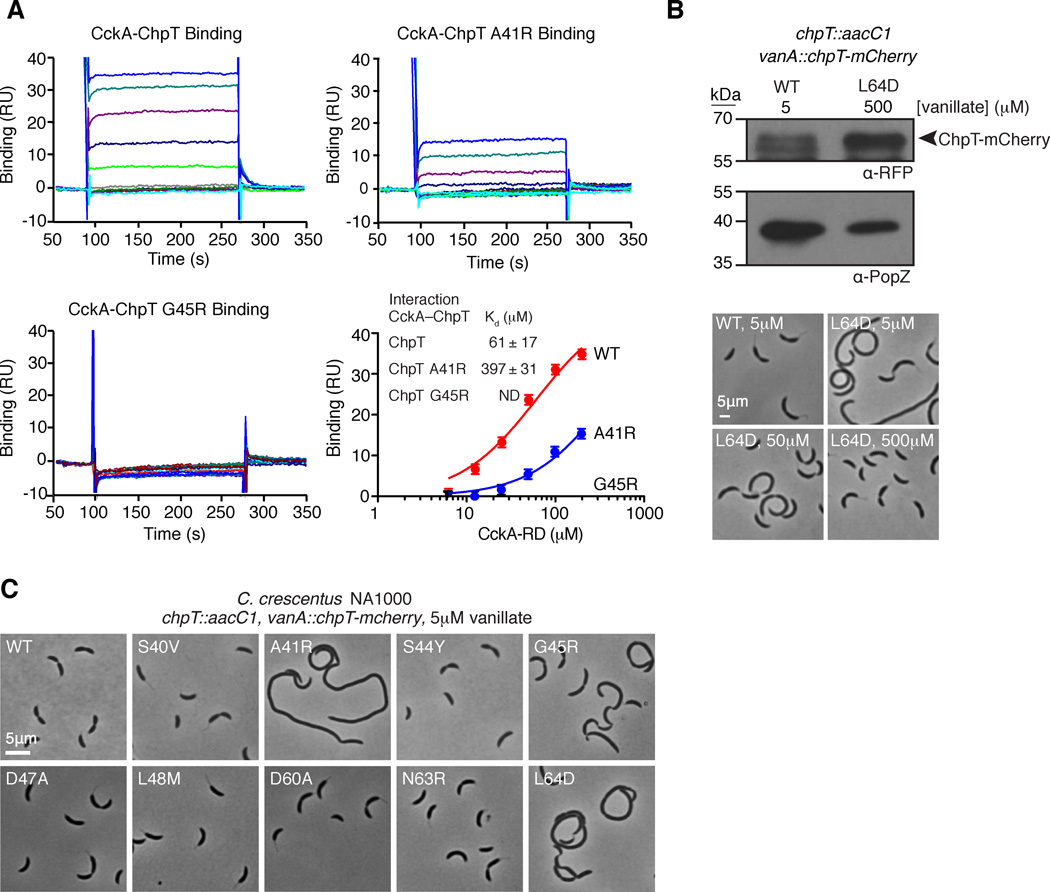
Point mutations along the ChpT DHp domain affect phosphotransfer, possibly by ablating binding interactions with CckA. We designed an SPR assay to measure direct binding of the CckA receiver domain (CckA-RD, amino acids 571–691) to immobilized His6-ChpT, His6-ChpT-A41R, or His6-ChpT-G45R. Our data demonstrated that CckA-RD binds to wild-type ChpT with a Kd of 61 µM (Fig. 5A). In contrast, CckA-RD bound to His6-ChpT-A41R with a Kd of 397 µM, and we were unable to detect binding of CckA-RD to His6-ChpT-G45R (Fig. 5A). Therefore, the weak binding affinity is a contributing factor to the phosphotransfer redcution between CckA and ChpT mutants A41R and G45R.
FIGURE 5.
A. Surface plasmon resonance demonstrates that His6-ChpT and His6-ChpT-A41R interact with CckA70–691, but His6-ChpT-G45R does not. Each SPR trace represents an increasing concentration of CckA-RD (1.6, 3.2, 6.3, 12.5, 25, 50, 100 and 200 µM). The mean response units (RU) from the CckA-ChpT interaction are plotted as a function of the CckA-RD concentration and fit to a one site specific binding model. Error bars represent the maximum deviation from the mean. B. ChpT point mutations along the putative RR binding interface affect Caulobacter crescentus morphology. Phase contrast micrographs of Caulobacter NA1000 strains whose sole chpT copy is either wild-type chpT-mcherry or a chpT-mcherry variant harboring a chpT RR binding mutation. C. Overexpression of chpT-mcherry-L64D overcomes cell morphology defects. Western blot analysis of ChpT-mCherry levels using an anti-RFP antibody confirms ChpT-mCherry L64D overexpression at 0.5mM vanillate relative to ChpT-mCherry expression levels at 5 µM vanillate. Probing the same samples with anti-PopZ sera provides a loading control. Phase contrast micrographs of chpT-mcherry-L64D cells reveal normal morphologies at 0.5 mM vanillate (see also Fig. S5).
Point mutations in the ChpT DHp domain disrupt the Caulobacter cell cycle
We asked if point mutations in the ChpT DHp domain that impact ChpT phosphotransfers also affect cell division and cell morphology. We constructed strains whose only copy of chpT harbored the mutations described above and included a C-terminal mcherry. The chpT-mcherry construct complemented the loss of the native allele. Also, each strain with chpT-mcherry showed mostly delocalized ChpT-mCherry throughout the cell (Fig. S5). The vanA::chpT-mcherry, chpT::aacC1 strains exhibited morphology identical to NA1000 wild-type cells (Fig. 5C), consistent with published reports (Biondi et al., 2006). Likewise, chpT::aacC1, vanA::chpT-mcherry-S40V, -S44Y, -D47A, -L48M, -D60A, and -N63R mutants exhibited no discernible phenotype (Fig. 5C). Alleles that impaired phosphotransfers in vitro also exhibited morphological defects in vivo; for example, ChpT-A41R exhibited diminished phosphotransfer between all ChpT binding partners (Fig. 4) and cells harboring these alleles replicated as long, filamentous cells (Fig. 5C). Likewise, ChpT-L64D and ChpT-G45R neither efficiently transferred phosphate to SUMO-CtrA nor received phosphate from CckA, and chpT::aacC1, vanA::chpT-mcherry-L64D/G45R showed morphological defects (Fig. 5C). In contrast, chpT::aacC1, vanA::chpT-mcherry-S40V exhibited normal morphologies (Fig. 5C) despite inefficient phosphotransfers from CckA to ChpT-S40V (Fig. 4B).
Because ChpT L64D transferred phosphate in vitro at a reduced rate, we asked if the morphology of ChpT L64D mutant cells could be complemented by overexpression of the mutant gene upon vanillate induction. Cells expressing the wild-type chpT-mcherry displayed normal shapes when cultured with 5 µM vanillate. As we supplemented the growth medium with higher vanillate concentrations, we found that those cells expressing the L64D variant began to resemble the wild-type (Fig. 5B). Immunoblotting confirmed that higher concentrations of vanillate increased the abundance of ChpT-mCherry.
Interactions between ChpT and the CckA, CtrA, and CpdR receiver domains
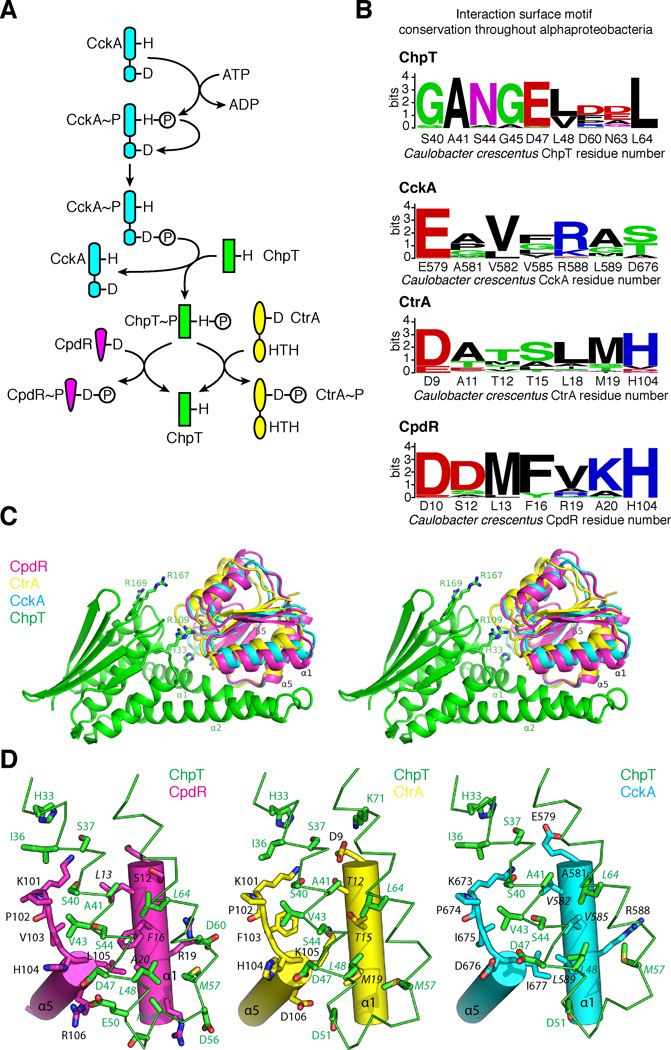
Structure and mutational analysis indicates ChpT interacts with a RR domain using a conserved TCS mechanism (Fig. 6A) (Casino et al., 2009; Xu et al., 2003; Zapf et al., 2000). Alignment of ChpT, CtrA, CpdR, and CckA sequences from a set of alphaproteobacteria (Brilli et al., 2010) allowed us to construct sequence logos (Crooks et al., 2004; Schneider and Stephens, 1990) representing each interaction surface (Fig. 6B). Notably, the CpdR surface is highly conserved, while the CtrA and CckA-RD interaction surfaces display more variability. Among the RR residues that play a role in kinase-RR binding, only CckA(Glu579), CtrA(Asp9), and CpdR(Asp10) are highly conserved, while the remaining residues on this proposed interaction surface are much less conserved.
FIGURE 6.
Computational models of ChpT bound to its cognate RR domains. A. Schematic of the CckA-ChpT-CtrA-CpdR signaling pathway. B. Interaction site sequence conservation logos throughout alphaproteobacteria for ChpT, CckA-RD, CtrA, and CpdR. C. Stereoview of homology models of CckA (blue), CtrA (yellow), or CpdR (magenta) docked onto ChpT (green). D. Close-up views of the ChpT DHp-RR binding interface. Potential residues likely involved in binding are drawn as sticks and labeled.
To understand how a single molecule interacts with three different RRs, we generated homology models for each ChpT-RR pair. CtrA, CpdR, and CckA RDs were docked onto ChpT based on the signaling HK-RD complex HK853/RR489 (Casino et al., 2009). The resulting models (Fig. 6C–D) suggest that it is possible for the different RDs to bind ChpT without significant conformational changes. The CckA-RD and CtrA-RD adopt a similar conformation, which deviates from the CpdR conformation. The structural elements involved in the binding include helix α1 and the β5-α5 loop. The CA domain helps define a concave binding surface. Most notably, three conserved arginines (Arg109, Arg167, and Arg169) on the CA domain could make contacts with RDs (Fig. 6C).
The predicted binding interfaces between ChpT and RDs reveal differences in binding between the RDs. The hydrophobic patch on ChpT is complemented by mostly hydrophobic residues on one of the solvent-exposed surface of the helix α1 of RDs (Fig. 6D, Fig. S4B). Most of these residues have small side-chains, except for Phe16 in CpdR (Fig. 6B, Fig. 6D, Fig. S4B). This bulkier Phe side-chain of CpdR is mainly responsible for the larger differences in the docking orientation of CpdR with regard to ChpT compared to CtrA-RD or CckA-RD. The adjacent contact area formed by the β5-α5 loop region of RD is more conserved (Fig. 6D), containing a Lys-Pro motif ubiquitous in all RDs. Our model also suggests that the linker region between α1 and α2 of ChpT may also be involved in the interactions in an RR-dependent manner and thus may contribute to specificity. Structural elements similar to the linker between these helices are observed in other monomeric HPts such as YPD1 (Porter and West, 2005) and ShpA (Xu et al., 2009).
DISCUSSION
ChpT is the crux of the core signaling circuit that controls Caulobacter cell cycle progression, and our data show it coordinates biochemical signal transductions by using a conserved protein-recognition mode shared between HKs and RRs. How has ChpT evolved to adopt this pivotal role in an essential network? Two distinct classes of HPts have been characterized to date, and each folds into a four-helix bundle. One class involves a monomeric four-helix bundle, while the other class adopts a dimer of two-helix monomers to form its four-helix bundle. The dimeric HK-like HPts represented by Spo0B (Varughese et al., 1998) and ChpT are distinct from monomeric HPts, such as yeast YPD1 (Xu et al., 2003) and ShpA from Caulobacter (Xu et al., 2009). Whereas the monomeric HPts harbor only one phosphorylated histidine residue, the two phosphorylation sites of the ChpT (or Spo0B) dimer are equivalent and may interact with two RRs simultaneously. Both monomeric and dimeric HPts, as well as HKs, recognize RRs using a similar mechanism, involving mostly the central helical bundles (Casino et al., 2009; Xu et al., 2003; Zapf et al., 2000). It remains an open question why there are two families of HPts, which presumably convergently evolved to perform the same function.
The vast majority of two-component systems have evolved to promote orthogonal HK-RR pairs that minimize cross-talk (Biondi et al., 2006; Skerker et al., 2005). In contrast to HKs, HPt proteins are commonly found at the center of highly branched signaling networks. In the case of ChpT, it is the hub of cell-cycle signaling in which it receives input from CckA and transfers outputs to CtrA and CpdR (Fig. 6A). This branch point allows ChpT to simultaneously activate CtrA and prevent CtrA degradation. We discovered three mutations within ChpT's RR docking interface that directly alter cell morphology. One mutation (A41R) severely weakened phosphotransfers between all three partners, and a second set (G45R, L64D) selectively impaired phosphotransfers between CckA-ChpT and ChpT-CtrA but not CpdR (Fig. 4D). A mutation at S40V also caused diminished CckA~P to ChpT phosphotransfer (Fig. 4B–C) but displayed normal cell morphologies.
In our structural model, residue Ala41 is strictly conserved throughout alphaproteobacteria ChpTs, and it also interacts with the most highly conserved residue among ChpT's signaling partners: CckA(E579), CtrA(D9), and CpdR(D10). Introduction of an arginine residue likely disrupts binding or alters the binding configuration. Meanwhile, two ChpT mutations, G45R and L64D, were found to selectively decrease phosphotransfer between CckA-ChpT and ChpT-CtrA with no impact on ChpT-CpdR. Indeed, in our computational models, residue Leu64 of ChpT interacts with similar surfaces composed of smaller residues in CckA (Val582, Val585) and CtrA (Thr15, Thr12), while interacting with bulkier residues on CpdR (Leu12, Phe16). Similarly, ChpT G45R interacts with small residues in CckA (Val582) and CtrA (Thr12), and interacts with bulkier Leu13 residue in CpdR. Therefore, the amino acids at position 45 and 64 are crucial for the maintenance of branched signaling to distinct RR binding surfaces (Fig. 6). In bacteria, orthogonal TCS signal wiring of protein interaction surfaces between HKs and RRs allows accurate processing of signaling information while avoiding destructive cross -talk. In the context of phosphotransfer proteins, bacteria have exploited constructive cross-talk using this highly tunable interaction surface to enable increasingly complex decision making.
Postscript
During manuscript preparation, Fioravanti and co-workers reported the crystal structure of ChpT from Caulobacter with no biochemical analysis (Fioravanti et al., 2012). Their structural model (PDB ID 4fpp, deposit date 2012-06-22) is essentially identical to ours (PDB ID 4fmt, deposit date 2012-06-18) with an RMSD of 0.35 Å for backbone atoms of 205 residues. The crystals belong to different space groups, but the molecular packing interactions within the crystals are very similar.
EXPERIMENTAL PROCEDURES
Materials
General methods, materials and detailed cloning procedures are described in the Supplementary Information. The plasmids, strains, and oligonucleotides used in this study are listed in Tables S2, S3, and S4, respectively.
Protein Purification
His6-ChpT and His6-ChpT harboring point mutations were expressed in E. coli, then affinity purified using Ni-NTA agarose (QIAGEN) as previously (Skerker et al., 2005). The His6-tag was removed using solid-supported thrombin (CleanCleave thrombin, Sigma) overnight at 4°C, then incubated with Ni-NTA agarose for 1 h to remove undigested His6-ChpT. The ChpT eluate was further purified by size exclusion chromatography equilibrated in kinase storage buffer (50 mM HEPES·KOH pH 8.0 at 20°C, 200 mM KCl, 10% (v/v) glycerol, 0.1 mM EDTA, 1 mM DTT), yielding 33 mg of ChpT, then flash frozen in liquid nitrogen and stored at –80°C. Expression constructs were generated for His6-CckA70–691 (the full-length cytoplasmic region of CckA), His6-CckA70–691-H322A (a variant that cannot autophosphorylate), and CckA-RD. Full-length CckA constructs were purified by Ni-NTA chromatography, anion exchange chromatography, and gel filtration chromatography. CckA-RD was purified as described for ChpT, including His6-tag removal. CpdR was expressed and purified as described (Abel et al., 2011). His6-SUMO-CtrA was expressed in E. coli, purified by Ni-affinity chromatography and used without SUMO-domain cleavage. Detailed methods are described in the Supplementary Information.
Crystallization
Initial crystallization conditions for ChpT were obtained using the sparse matrix screening method (Emerald Biosystems). The conditions were manually optimized to improve crystal quality. The crystals used for structure solution were obtained using hanging-drop vapor diffusion at 22.5°C. The reservoir well contained 500 µL 0.2 M NaCl, 0.1 M MES pH 6.0 and 18% PEG 8000, while the drop contained 1.2 µL of ChpT (concentration 8 mg/mL) mixed with 1.2 µL of the reservoir solution. The crystals were cryoprotected in reservoir solution containing 30% PEG 8000 then flash frozen in liquid nitrogen. The data were indexed and processed in the monoclinic space group P21 with unit cell dimensions of a=65.2 Å, b=94.3 Å, c=100.6 Å and β =92.26. To obtain heavy atom derivatives, crystals were soaked in cryo solution containing 10 mM KAu(CN)2 for 190 mins.
Data collection, structure determination, and refinement
Native data, and multiwavelength anomalous diffraction (MAD) data for the gold derivative were collected at the SSRL Beamline 12-2 at 100 K using a Pilatus 6M pixel array detector (Dectris). Each data set was processed using XDS (Kabsch, 2010). Gold sites were located with SHELXD (Sheldrick, 2008). Phase refinement based on the MAD data (FOM=0.25 for 6 Au sites) and automatic model building were performed using autoSHARP (Vonrhein et al., 2007) and BUCCANEER (Cowtan, 2006). Model completion and refinement were performed with COOT (Emsley and Cowtan, 2004) and BUSTER (Bricogne G., 2011). Each protein monomer was refined as one TLS group.
Sequence Logos and Molecular modeling
We extracted HK-RR co-varying interaction residues (Capra et al., 2012a; Capra et al., 2012b; Skerker et al., 2008) from aligned ChpT, CckA, and CtrA alphaproteobacteria homologs (Brilli et al., 2010) and created sequence logos using Weblogo (Crooks et al., 2004; Schneider and Stephens, 1990). We built CckA, CtrA, and CpdR RD homology models from an OmpR/PhoB RD template (Buckler et al., 2002) (PDB ID 1kgs) using WHAT IF (Vriend, 1990) and compared to models generated using the I-TASSER server (Zhang, 2008). Complexes of ChpT and an RD were based on the HK853/RR468 complex (Casino et al., 2009) (PDB ID 3dge) by superposing the RD to RR468 and ChpT onto HK853-DHp and were optimized in ROSETTA software (Das and Baker, 2008). Detailed methods are described in the Supplementary Information.
Phosphotransfer assays
We performed phosphotransfer assays as described (Biondi et al., 2006; Skerker et al., 2005). Briefly, phosphotransfer from CckA to ChpT was measured in triplicate by incubating 30 µL of 5 µM 32P-phosphorylated His6-CckA70–691 with 30 µL of 5 µM His6-ChpT or His6-ChpT variants. After 10s, 8 µL of the reaction mixture was removed and quenched into 2 µL of 5× Laemmli Sample Buffer. Each reaction was loaded onto 12% Tris-HCl gels for PAGE, the proteins separated by electrophoresis, the radioactivity in the wet gels recorded on a phosphor storage plate for 3 hrs, and the images were recorded on a Typhoon fluorescence imager (Molecular Dynamics). Band intensities were analyzed using ImageJ (Schneider et al., 2012). The data were analyzed using Origin software (OriginLab) and plotted as the mean percent of wild-type with standard deviation. Detailed methods of these procedures are described in the Supplementary Information. Prior to biochemical analysis, any inactive protein aggregates were spun down in a TL-100 ultracentrifuge at 80,000 rpm (278,000 g) for 30 min and supernatant protein concentrations were measured. ChpT~P was generated by incubating 5 µM ChpT, 1 µM FLAG-CckA, [g-32P] ATP and anti-FLAG M2 magnetic beads (Sigma) for 45 minutes. ChpT was separated from FLAG-CckA and incubated with 1.5 U hexokinase (VWR) and 5 mM D-glucose (VWR) to convert residual ATP into ADP. ChpT~P was incubated with 5 µM SUMO-CtrA, CpdR, or CckA for 10s at a 1:10 molar ratio, then 10 µL reaction mixture samples were quenched into 10 µL of 2× Laemmli Sample Buffer. Reaction products were separated via PAGE on 4–20% Tris-HCl gradient gels and analyzed as described above.
Coupled-enzyme activity assay
ATPase activity was measured using a coupled-enzyme assay (Kiianitsa et al., 2003; Lindsley, 2001). ChpT constructs were mixed in kinase buffer supplemented with 1 mM ATP, 10 mM MgCl2, 1 mM phosphoenolpyruvate, 0.2 mM NADH, 2 units of pyruvate kinase and 6.6 units of lactate dehydrogenase (P0294, Sigma). Reactions were performed in triplicate in a 200 µL volume and loaded into a clear polystyrene 96 well-plate. Each reaction was initiated by the addition of protein, and 340 nm absorbance was recorded every 10s for 30 minutes on a SpectraMax M5 microplate reader (Molecular Devices). The slope of a stable, linear absorbance decay was measured to calculate ATP hydrolysis rates, using a NADH Kpath value of 3248 OD M–1 (Kiianitsa et al., 2003). Background rates of ATP hydrolysis and NADH oxidation were measured and subtracted from observed ChpT construct ATP hydrolysis rates. The mean observed rate and standard deviation were determined and analyzed using Prism 5 (GraphPad).
Filter binding assays
Filter binding assays were designed to capture transiently bound [g-32P] ATP (Ertel et al., 1968; Miller and Weissbach, 1974). Solutions of CckA70–691-H322A, ChpT, and BSA at 10 µM protein concentration was mixed with 27.5 fmol of [g-32P] ATP in kinase buffer at room temperature for 30 min. Solutions were then passed over a nitrocellulose membrane filter, and washed 3 times with 1 mL of kinase buffer. Bradford assays indicated >95% of protein remained bound to the nitrocellulose. Subsequently, nitrocellulose filters were scintillation counted to quantify bound [g-32P] ATP. [g-32P] ATP only solutions were also passed through nitrocellulose membranes to quantify non-specific binding. The data were analyzed using Origin software (OriginLab) and plotted as the mean percent binding of [g-32P] ATP and the standard deviation.
Construction of chpT::aacC1, chpT-mcherry substitution variants
The construction of each ChpT variant plasmid is described in the Supplemental Information. To isolate strains whose sole copy of chpT harbored point mutations, we isolated mereodiploid strains containing native chpT and each vanA::chpT-mcherry with a chpT point mutation as before and then performed generalized transduction to generate strains chpT::aacC1, vanA::chpT-mcherry S40V, A41R, S44Y, G45R, D47A, L48M, D60A, N63R, and L64D strains (JWK1437–JWK1441, JWK1450–1453).
Microscopy
Strains JWK1436–JWK1441, JWK1450–1453 were cultured overnight in PYE with 5 µM vanillate (or 0.5 mM vanillate in overexpression studies) and sub-cultured into fresh media for six hours or until they reached an OD600 of 0.4. Cell suspensions were then dried onto agarose pads (1.5% agarose in PYE) containing 5 µM vanillate (or 0.5 mM vanillate in overexpression studies) and imaged in phase-contrast on a Leica DM 6000 B microscope with a HCX PL APO 100×/1.40 Oil PH3 CS objective, Hamamatsu EM-CCD C9100 camera and Metamorph (Molecular Devices).
Surface plasmon resonance (SPR) of CckA-ChpT interactions
SPR experiments were designed to measure CckA-RD binding to immobilized His6-ChpT similar to methods described by Bell et al. (Bell et al., 2010). Purified His6-ChpT, His6-ChpT-A41R, and His6-ChpT-G45R were immobilized on the Biacore CM5 sensor chips via an anti-His6 antibody, and CckA-RD was used as the analyte. SPR experiments were performed on a Biacore 3000 system at 25°C using a flow rate of 30 µL min−1 in running buffer (50 mM HEPES·KOH pH 8.0, 200 mM KCl, 1 mM DTT). For each data set the signal in a control flow cell that lacked immobilized ligand was subtracted to account for non-specific binding and analyzed using the BIAevaluation software (Biacore). In saturation binding experiments, data was fit to a one site-specific binding model where y=Bmax*X/(Kd + X). Error for each SPR experiment is the maximum deviation from the SPR trace mean at binding equilibrium and was found to be ~1.2 RU.
Supplementary Material
HIGHLIGHTS.
∙ChpT serves as a signaling branch point between three receiver domains.
∙The ChpT crystal structure reveals a pseudo-histidine kinase (HK) fold.
∙Phosphotransfer occurs through a conserved HK-RR interaction surface.
∙Specific ChpT point mutants mediate HK-RR partner selection.
ACKNOWLEDGEMENTS
We thank Ruth Sommese for assistance with CD and Peter Chien (UMass Amherst) for sharing strains BPC223 and BPC250. Also, we thank the Shapiro and Harley McAdams lab members for insights, discussions, and coffee. Portions of this research were performed at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The DOE Office of Biological and Environmental Research, the NIH NIGMS (including P41GM103393), and the NCRR (P41RR001209) support the SSRL Structural Molecular Biology Program. The following grants supported this work: NIH Grants R01 GM51426 and R01 GM32506 to LS, NIH Postdoctoral Fellowship F32 A1082915 to JAB and F32 GM099173 to JWK, and the Jane Coffin Childs Memorial Fund to WSC. NIH, NIGMS, Protein Structure Initiative grants U54 GM094586 and GM074898 support the JCSG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
We deposited the atomic coordinates and experimental structure factors for ChpT at 2.3 Å resolution in the Protein Data Bank under accession code 4fmt.
REFERENCES
- Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, Baker TA, Jenal U. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol. Cell. 2011;43:550–560. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelastro PS, Sliusarenko O, Jacobs-Wagner C. Polar localization of the CckA histidine kinase and cell cycle periodicity of the essential master regulator CtrA in Caulobacter crescentus. J. Bacteriol. 2010;192:539–552. doi: 10.1128/JB.00985-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CH, Porter SL, Strawson A, Stuart DI, Armitage JP. Using structural information to change the phosphotransfer specificity of a two-component chemotaxis signalling complex. PLoS Biol. 2010;8:e1000306. doi: 10.1371/journal.pbio.1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick MJ, Lamour V, Rajashankar KR, Gordiyenko Y, Robinson CV, Darst SA. How to switch off a histidine kinase: crystal structure of Geobacillus stearothermophilus KinB with the inhibitor Sda. J. Mol. Biol. 2009;386:163–177. doi: 10.1016/j.jmb.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- Bricogne G, B E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart OS, Vonrhein C, Womack TO. BUSTER version 2.10.0. Cambridge, United Kingdom: Global Phasing Ltd.; 2011. [Google Scholar]
- Brilli M, Fondi M, Fani R, Mengoni A, Ferri L, Bazzicalupo M, Biondi EG. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst. Biol. 2010;4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler DR, Zhou Y, Stock AM. Evidence of intradomain and interdomain flexibility in an OmpR/PhoB homolog from Thermotoga maritima. Structure. 2002;10:153–164. doi: 10.1016/s0969-2126(01)00706-7. [DOI] [PubMed] [Google Scholar]
- Capra EJ, Perchuk BS, Ashenberg O, Seid CA, Snow HR, Skerker JM, Laub MT. Spatial tethering of kinases to their substrates relaxes evolutionary constraints on specificity. Mol. Microbiol. 2012a;86:1393–1403. doi: 10.1111/mmi.12064. [DOI] [PubMed] [Google Scholar]
- Capra EJ, Perchuk BS, Skerker JM, Laub MT. Adaptive mutations that prevent crosstalk enable the expansion of paralogous signaling protein families. Cell. 2012b;150:222–232. doi: 10.1016/j.cell.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YE, Tsokos CG, Biondi EG, Perchuk BS, Laub MT. Dynamics of two Phosphorelays controlling cell cycle progression in Caulobacter crescentus. J. Bacteriol. 2009;191:7417–7429. doi: 10.1128/JB.00992-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B, Abeliuk E, Collier JM, Kalogeraki VS, Passarelli B, Coller JA, Fero MJ, McAdams HH, Shapiro L. The essential genome of a bacterium. Mol. Syst. Biol. 2011;7:528. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis PD, Brun YV. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol. Mol. Biol. Rev. 2010;74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with Rosetta. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ertel R, Brot N, Redfield B, Allende JE, Weissbach H. Binding of guanosine 5'-triphosphate by soluble factors required for polypeptide synthesis. Proc. Natl. Acad. Sci. U.S.A. 1968;59:861–868. doi: 10.1073/pnas.59.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioravanti A, Clantin B, Dewitte F, Lens Z, Verger A, Biondi EG, Villeret V. Structural insights into ChpT, an essential dimeric histidine phosphotransferase regulating the cell cycle in Caulobacter crescentus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012;68:1025–1029. doi: 10.1107/S1744309112033064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA, Hillson NJ, Shapiro L. Cell pole-specific activation of a critical bacterial cell cycle kinase. Proc. Natl. Acad. Sci. U.S.A. 2010;107:7012–7017. doi: 10.1073/pnas.1001767107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA, Shapiro L. A bacterial control circuit integrates polar localization and proteolysis of key regulatory proteins with a phospho-signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16602–16607. doi: 10.1073/pnas.0808807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C, Ausmees N, Cordwell SJ, Shapiro L, Laub MT. Functions of the CckA histidine kinase in Caulobacter cell cycle control. Mol. Microbiol. 2003;47:1279–1290. doi: 10.1046/j.1365-2958.2003.03379.x. [DOI] [PubMed] [Google Scholar]
- Jacobs C, Domian IJ, Maddock JR, Shapiro L. Cell cycle–dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- Janiak-Spens F, Cook PF, West AH. Kinetic analysis of YPD1-dependent phosphotransfer reactions in the yeast osmoregulatory phosphorelay system. Biochemistry. 2005;44:377–386. doi: 10.1021/bi048433s. [DOI] [PubMed] [Google Scholar]
- Janiak-Spens F, West AH. Functional roles of conserved amino acid residues surrounding the phosphorylatable histidine of the yeast phosphorelay protein YPD1. Mol. Microbiol. 2000;37:136–144. doi: 10.1046/j.1365-2958.2000.01973.x. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Mizuno T, Shimizu T, Hakoshima T. Insights into multistep phosphorelay from the crystal structure of the C-terminal HPt domain of ArcB. Cell. 1997;88:717–723. doi: 10.1016/s0092-8674(00)81914-5. [DOI] [PubMed] [Google Scholar]
- Kiianitsa K, Solinger JA, Heyer WD. NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Anal. Biochem. 2003;321:266–271. doi: 10.1016/s0003-2697(03)00461-5. [DOI] [PubMed] [Google Scholar]
- Lindsley J. Use of a real-time, coupled assay to measure the ATPase activity of DNA topoisomerase II. Methods Mol. Biol. 2001;95:57–64. doi: 10.1385/1-59259-057-8:57. [DOI] [PubMed] [Google Scholar]
- Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 2005;24:4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DL, Weissbach H. Elongation factor Tu and the aminoacyl-tRNA-EFTu-GTP complex. Methods Enzymol. 1974;30:219–232. doi: 10.1016/0076-6879(74)30024-9. [DOI] [PubMed] [Google Scholar]
- Parkinson JS, Kofoid EC. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- Porter SW, West AH. A common docking site for response regulators on the yeast phosphorelay protein YPD1. Biochim. Biophys. Acta. 2005;1748:138–145. doi: 10.1016/j.bbapap.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon KC, Marczynski GT, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. U.S.A. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallogr. A, Found. Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varughese KI, Madhusudan, Zhou XZ, Whiteley JM, Hoch JA. Formation of a novel four-helix bundle and molecular recognition sites by dimerization of a response regulator phosphotransferase. Mol. Cell. 1998;2:485–493. doi: 10.1016/s1097-2765(00)80148-3. [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. 29. [DOI] [PubMed] [Google Scholar]
- Xu Q, Carlton D, Miller MD, Elsliger MA, Krishna SS, Abdubek P, Astakhova T, Burra P, Chiu HJ, Clayton T, et al. Crystal structure of histidine phosphotransfer protein ShpA, an essential regulator of stalk biogenesis in Caulobacter crescentus. J. Mol. Biol. 2009;390:686–698. doi: 10.1016/j.jmb.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Porter SW, West AH. The yeast YPD1/SLN1 complex: insights into molecular recognition in two-component signaling systems. Structure. 2003;11:1569–1581. doi: 10.1016/j.str.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Xu Q, West AH. Conservation of structure and function among histidine-containing phosphotransfer (HPt) domains as revealed by the crystal structure of YPD1. J. Mol. Biol. 1999;292:1039–1050. doi: 10.1006/jmbi.1999.3143. [DOI] [PubMed] [Google Scholar]
- Yamada S, Sugimoto H, Kobayashi M, Ohno A, Nakamura H, Shiro Y. Structure of PAS-linked histidine kinase and the response regulator complex. Structure. 2009;17:1333–1344. doi: 10.1016/j.str.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Zapf J, Sen U, Madhusudan, Hoch JA, Varughese KI. A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Structure. 2000;8:851–862. doi: 10.1016/s0969-2126(00)00174-x. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.