Abstract
Intracellular proteins with long lifespans have recently been linked to age-dependent defects, ranging from decreased fertility to the functional decline of neurons. Why long-lived proteins exist in metabolically active cellular environments and how they are maintained over time remains poorly understood. Here we provide a system-wide identification of proteins with exceptional lifespans in the rat brain. These proteins are inefficiently replenished despite being translated robustly throughout adulthood. Using nucleoporins as a paradigm for long-term protein persistence, we found that nuclear pore complexes (NPCs) are maintained over a cell’s life through slow but finite exchange of even its most stable subcomplexes. This maintenance is limited, however, as some nucleoporin levels decrease during aging, providing a rationale for the previously observed age-dependent deterioration of NPC function. Our identification of a long-lived proteome reveals cellular components that are at increased risk for damage accumulation, linking long-term protein persistence to the cellular aging process.
Introduction
The majority of cellular proteins are rapidly degraded and replaced with newly synthesized copies, minimizing accumulation of potentially toxic damage and ensuring a functional proteome throughout a cell’s lifetime. Several studies have measured global protein turnover rates in yeast and mammals and reported an average protein half-life of ~1.5 hours to 1-2 days, respectively (Belle et al., 2006; Cambridge et al., 2011; Price et al., 2010). In certain post-mitotic tissues, however, a handful of proteins exhibit limited turnover and have been shown to persist for months or even years (D’Angelo et al., 2009; Masters et al., 1977; Piha et al., 1966; Rodrijguez de Lores et al., 1971; Savas et al., 2012; Verzijl et al., 2000). The persistence of these proteins is remarkable as, in contrast to DNA, another long-lived molecule (Spalding et al., 2005), dedicated repair mechanisms have not been established for proteins with long lifespans.
Based on localization within different tissues, long-lived proteins fall into at least two categories. The first group, best exemplified by crystallin and collagen, have half-lives on the order of years in humans (Masters et al., 1977; Verzijl et al., 2000) and are confined to organelle-free lens fiber cells or the extracellular matrix. The lack of turnover of these proteins can be explained by their existence in metabolically inactive environments that lack protein synthesis and degradation machineries (Bassnett, 2002). In contrast, a recently identified second class of long-lived proteins is present in metabolically active cells with intact protein synthesis, degradation, and quality control mechanisms. For example, the protein cohesin has been shown to mediate sister chromatid cohesion during female meiosis from birth until ovulation, which can span decades in humans, without being efficiently replenished (Tachibana-Konwalski et al., 2010). While cohesin’s lifespan might be confined to oocytes, the recent report of long-lived nuclear pore proteins in post-mitotic cells of C. elegans and rat (D’Angelo et al., 2009; Savas et al., 2012) suggests that exceptional protein lifespans might be more common than previously thought. How proteins like cohesin and nucleoporins (Nups) are maintained over long periods of time and whether additional proteins exist that exhibit no or limited turnover remains unclear.
The identification and characterization of long-lived proteins has potential implications for age-related defects in tissue homeostasis. For both crystallin and collagen, it was shown that their lifespans resulted in accumulation of damage that manifested as cataract eye lenses and cartilage stiffening, respectively (Bloemendal et al., 2004; Haus et al., 2007; Kragstrup et al., 2011; Roy and Spector, 1976; Sell and Monnier, 2011; Sharma and Santhoshkumar, 2009; Wilmarth et al., 2006). In the case of sister chromatid cohesion, the inability of oocytes to replenish cohesin may contribute to age-related meiosis I errors and decreased fertility during aging (Tachibana-Konwalski et al., 2010). Analogous to cohesin’s role in chromatin organization, protein persistence of nucleoporins, which form the nuclear transport channels that mediate all nuclear/cytoplasmic transport in mammalian cells, might be linked to the long-term maintenance of the nuclear compartment and genomic integrity. For long-lived scaffold Nups, oxidative damage during aging is associated with defects in nuclear trafficking and the breakdown of the nuclear permeability barrier (D’Angelo et al., 2009). It is also possible that long-lived histones (Commerford et al., 1982; Duerre and Lee, 1974; Piha et al., 1966) might be linked to loss of youthful gene expression observed in adult tissues (Lee et al., 2000). In summary, proteins with exceptional lifespans, particularly those that are not shielded from harmful metabolites, might be sources of vulnerability in the mammalian proteome, and their identification is thus critical for our understanding of the aging process in post-mitotic tissues.
Results
Identification of long-lived proteome in the brain
Previous studies that identified long-lived proteins relied on a priori hypotheses and were thus targeted approaches or limited in scope (D’Angelo et al., 2009; Masters et al., 1977; Savas et al., 2012; Shapiro et al., 1991; Verzijl et al., 2000). We sought to conduct a systematic and unbiased analysis of the long-lived proteome for an entire organism during aging. To achieve this, we employed a unique pulse-chase labeling time course of a rat (Figure 1A) and used mass spectrometry (MS) to comprehensively identify long-lived proteins in the brain and to probe for evidence of their maintenance during aging (McClatchy et al., 2007). In brief, female rats were fed a diet containing exclusively the 15N isotope just after weaning. Progeny born from these females were efficiently labeled with the 15N isotope (McClatchy et al., 2007) and were continued on this diet for 6 weeks after birth (pulse). After this period, the pulse-labeled animals were switched to a normal 14N diet (chase). Rats were sacrificed at 0, 4, 6, 9, and 12 months post-chase, and all tissues were harvested and stored for analysis. Tissue samples were fractionated into nuclear, endoplasmic reticulum (ER), mitochondrial, and cytoplasmic fractions, digested with trypsin, and analyzed by MudPIT (multi-dimensional protein identification technology) LC/LC-MS/MS (multi-dimensional liquid chromatography-tandem mass spectrometry) to identify peptides and their 15N/14N abundance ratios (Washburn et al., 2001). Persistence of 15N peptides in pulse-chased animals indicates a lack of degradation of their corresponding proteins, and we define these proteins as long-lived.
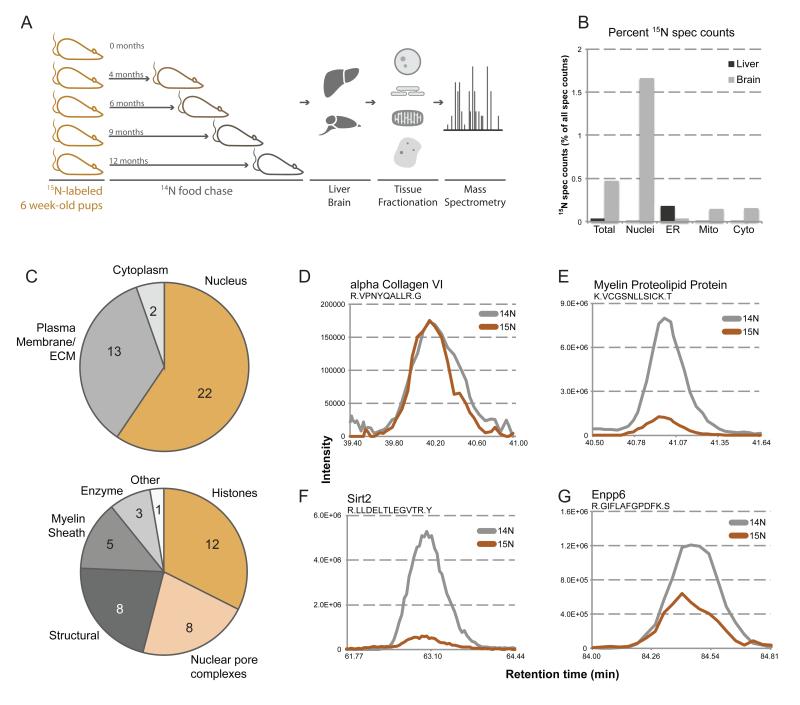
Figure 1.
Discovery of new members of the long-lived proteome. (A) Pulse-chase labeling of whole rats. Depicted is a schematic of the pulse-chase labeling procedure. Litters of rats were fully 15N-labeled through feeding a 15N diet starting from a previous generation. Fully labeled rats were then switched to a normal 14N diet (chase) at 6 weeks post-natal, and sacrificed a 0, 4, 6, 9, and 12 months post-chase. Tissues were harvested, fractionated, and analyzed by MS (B) Tissue localization of long-lived proteins. 15N spectral counts were calculated and plotted as a percentage of total spectral counts in each fraction of liver (grey) and brain (black) tissues from an animal 6-months post-chase. (C) Cellular localization and processes of long-lived proteins. Identified long-lived proteins were sorted by subcellular localization (upper) and cellular process (lower), and plotted as a pie chart, inset numbers representing the number of proteins per localization. (D-G) MS1 traces of representative peptides. Aligned elution profile MS1 traces are plotted for representative peptides for alpha collagen VI (D), myelin proteolipid protein (E), Sirt2 (F), and Enpp6 (G), with 15N signal in orange and 14N signal in grey. See also Figure S1-3.
To establish the validity of this method, we first analyzed the eye lens, which harbors the known long-lived crystallin proteins (Masters et al., 1977). Lenses were isolated and homogenized from rat eyes at time points corresponding to 0, 6, and 12 months post-chase. The soluble pool of proteins, composed mostly of crystallin, was then analyzed by LC/LC-MS/MS (Figure S1A,B). As expected, many proteins were found to be present with average 15N fractional peptide abundances (the 15N fraction of a particular peptide calculated by: 15N abundance / (15N + 14N abundance)) of at least 20%, even after 6 months chase (Figure S1C, D). The presence of 14N peptides in chase lenses is likely due to the proliferation of new fiber cells at the outer layers of the lens, which continues to grow through adulthood (Bassnett et al., 2011).
As mentioned above, since differentiated lens fiber cells lack organelles such as mitochondria and have a highly specialized proteome, crystallin longevity can be explained by the absence of protein synthesis and degradation machineries. To test if long-lived proteins exist in metabolically active cells, we analyzed the brain, which is composed of a vast array of different cell types each harboring robust protein turnover mechanisms. In addition, we compared the proteome of the adult brain, an organ with limited capacity for regeneration and with very long-lived cells, to that of the liver, which in rodents has been shown to renew its constituent cells within <6 months (Arber et al., 1988). Our subcellular fractionation strategy allowed the identification of over 11,500 distinct proteins in each tissue, as well as many of their 15N versus 14N abundances. Analysis of 15N spectral counts across the different fractions revealed a 10-fold greater 15N content in the brain over the liver (Figure 1B). Using the program CENSUS, we defined proteins as long-lived when multiple peptides with greater than 5% 15N fractional abundances at the 6-month chase time point were identified per protein (see full methods) (Table 1) (Park et al., 2008). This analysis was also performed on chromatin and histone fractions in addition to those listed above, as a many long-lived proteins were found to reside in the cell nucleus (Figure 1C). We frequently found long-lived proteins to be components of large protein assemblies, as is the case with histones, nuclear pore complex (NPC) proteins, lamins and myelin proteins (Fig. 1C-E, S2, S3A, B). These results suggest that limited protein turnover might be a characteristic of specific subcellular structures that, like the nucleus, have to be maintained for long periods of time. We were therefore surprised to find several long-lived soluble proteins, including enzymes such as the plasma membrane-localized lysophospholipase Enpp6, the cytoplasmic deacetylase Sirt2, and the phosphodiesterase Cnp1 (Tsukada, 1992) (Figure 1F-G, S3C), that are not thought to be structural components. In summary, our pulse-chase labeling strategy suggests that, at least in the brain, post-mitotic tissue function appears to rely on the long-term persistence of key regulatory proteins found in cellular structures with exceptional lifespans.
Table 1.
Identified long-lived proteins. Listed are the names (column 1), average 15N fractional abundance at 6-months post-chase (column 2), number of peptides used to determine 15N fractional abundance (column 3), subcellular localization as plotted in Figure 1C (column 4), and cellular process as plotted in Figure 1C (column 5).
| Protein | % | Peptides | Localization | Process |
|---|---|---|---|---|
| Histone H1.0 | 5.36 | 6 | Nucleus | Histones |
| Histone H1.1 | 7.87 | 3 | Nucleus | Histones |
| Histone H1.2 isol | 10.64 | 2 | Nucleus | Histones |
| Histone H1.2 iso2 | 2.13 | 1 | Nucleus | Histones |
| Histone H1.2 iso3 | 62.50 | 1 | Nucleus | Histones |
| Histone H1.5 | 35.23 | 15 | Nucleus | Histones |
| H2A | 15.63 | 10 | Nucleus | Histones |
| Macro H2A | 9.10 | 3 | Nucleus | Histones |
| H2A.x | 17.42 | 6 | Nucleus | Histones |
| Histone H2B | 21.70 | 19 | Nucleus | Histones |
| Histone H3.1 | 90.39 | 3 | Nucleus | Histones |
| Histone H4 | 29.54 | 22 | Nucleus | Histones |
| Nup205 | 47.18 | 14 | Nucleus | NPC |
| Nup155 | 39.43 | 10 | Nucleus | NPC |
| Nup93 | 50.25 | 9 | Nucleus | NPC |
| Nup107 | 13.82 | 5 | Nucleus | NPC |
| Nup160 | 20.12 | 8 | Nucleus | NPC |
| Nup96 | 19.48 | 9 | Nucleus | NPC |
| Nup188 | 8.33 | 2 | Nucleus | NPC |
| Nup85 | 12.24 | 6 | Nucleus | NPC |
| MBP | 20.28 | 30 | PM/ECM | Myelin Sheath |
| PLP | 18.53 | 11 | PM/ECM | Myelin Sheath |
| MOG | 14.33 | 4 | PM/ECM | Myelin Sheath |
| Nef3 | 5.91 | 7 | PM/ECM | Myelin Sheath |
| Cnp1 | 4.82 | 13 | PM/ECM | Myelin/Enzyme |
| Enpp6 | 34.39 | 6 | PM/ECM | Enzyme |
| Sirt2 | 11.49 | 6 | Cyto/Nucleus | Enzyme |
| Asrgl1 | 78.70 | 2 | Cyto | Enzyme |
| igsf8 | 11.99 | 4 | PM/ECM | other |
| Lamin-B1 | 13.18 | 32 | Nucleus | Structural |
| Lamin-B2 | 6.63 | 9 | Nucleus | Structural |
| Cspg2 | 17.21 | 4 | PM/ECM | Structural |
| Lamc1 | 10.62 | 19 | ECM | Structural |
| Col6a1 | 35.61 | 9 | ECM | Structural |
| Col6a3 | 36.72 | 9 | ECM | Structural |
| Col4a2 | 18.51 | 3 | ECM | Structural |
| Col1a1 | 9.10 | 4 | ECM | Structural |
Protein lifespans vary within individual complexes
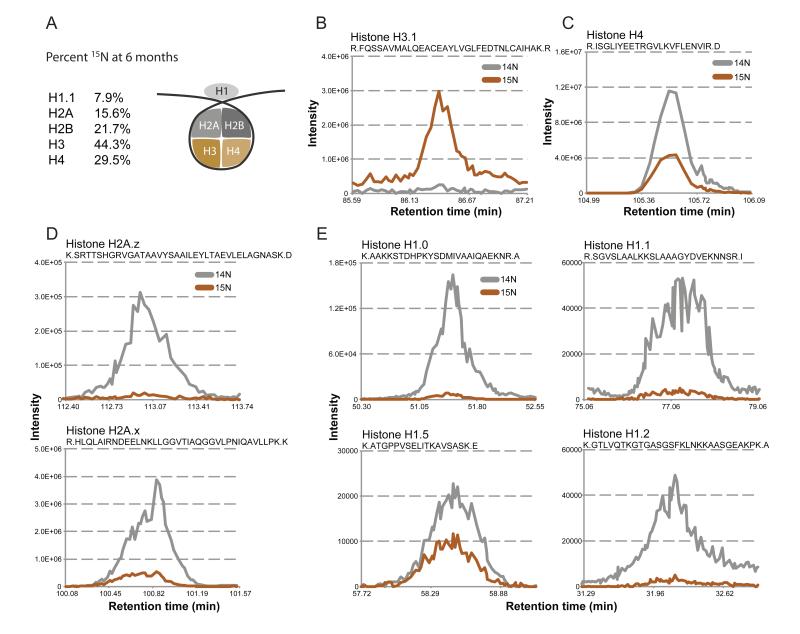
Our analysis of the long-term persistence of proteins in the brain showed that 15N content varied greatly among long-lived proteins, ranging from greater than 90% (Histone H3.1) to ~2.5% (Histone H1.2 isomer) 15N fractional abundances (Table 1). Although most long-lived proteins were part of larger assemblies, not all components or variants of a given structure had identical 15N enrichment. For example, two components of the nuclear lamina, LaminB1 and LaminB2, had 13.2% and 6.6% 15N-fractional abundances at 6 months, respectively (Table 1, S3A). Furthermore, two members of the core histone octamer, H3 and H4, were more 15N-enriched than canonical H2A and H2B (Figure 2A-C). These disparate stabilities are consistent with reports that H2A/H2B dimer exchange precedes H3/H4 exchange from the histone octamer (Kimura and Cook, 2001; Kireeva et al., 2002). Variants within the same histone family also displayed different 15N-fractional abundances, such as histone H2A.x (17.4%) versus H2A.z (N/A) (Table 1, Figure 2D) and Histone H1 variants H1.0 (5.4%), H1.1 (7.9%), H1.5 (35.2%) and H1.2 isoforms (10.6%, 2.1%, 62.5%) (Table 1, Figure 2E).
Figure 2.
Long-lived histones at 6 months. (A) Long-lived histone octamer. Schematic of the histone octamer, and 15N fractional abundance at 6-months post-chase for indicated histones. For H2A and H2B, the 15N fractional abundance was calculated from peptides that map to representative core (not variant) histones, and H3 fractional abundance determined from peptides common to all three major H3 variants (H3.1, H3.2, H3.3). (B-E) Example histone MS1 traces. MS1 elution profiles are plotted as described earlier from 6-months post-chase brain tissue for, (B) the single unique peptide for histone H3.1, (C) histone H4, (D) histone H2A variants H2A.z and H2A.x, and (E) H1 variants H1.0, H1.1, H1.5, and H1.2.
The heterogeneity of protein lifespans within one complex, however, was particularly striking for the NPC, a structure that is composed of multiple copies of over 30 different nucleoporins (Nups) (Cronshaw et al., 2002; D’Angelo and Hetzer, 2008). MS data were acquired on all NPC proteins; however, the only long-lived Nups were found to be part of two scaffold components, the Nup107/160 and Nup205 subcomplexes (Figure S2). Importantly, similar 15N abundance ratios were obtained when the analysis was performed on purified NPCs, confirming that these long-lived Nups reside in the nuclear membrane and do not represent nuclear aggregates (Figure S2B). Together with previously measured residence times of some Nups (Rabut et al., 2004), our data suggest that, at any given time during adulthood, the NPC is a mosaic structure of individual constituents of varying age, with the oldest components forming the scaffold structure of the nuclear pore.
Robust translation of long-lived proteins
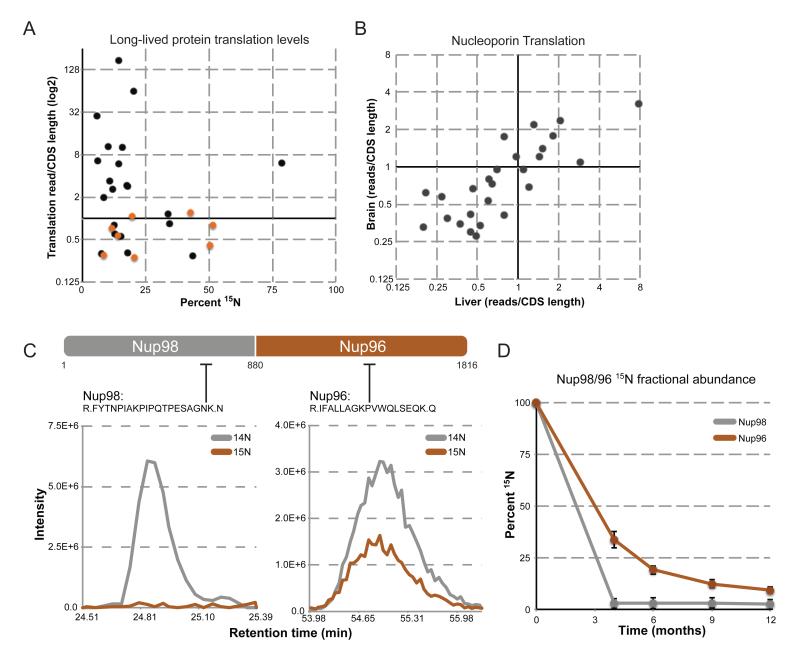
In the case of eye lens crystallin, the lack of protein translation (and degradation) in lens fiber cells provides a rationale for their exceptional lifespan. To determine if lack of synthesis might explain the exceptional lifespans of the cellular proteins identified above, we determined the level of translation for all proteins expressed in liver and brain tissue through deep sequencing of ribosome-protected mRNA footprints (Ingolia et al., 2009). Translation levels (i.e. density of ribosome footprints) were determined for over 11,000 proteins in 6-month old liver and brain, and, unlike crystallin, we found evidence for translation of almost every long-lived protein. To test if there was a correlation between a protein’s synthesis rate and its lifespan, we plotted translation levels versus 15N fractional abundance at 6-months post-chase (Figure 3A). Intriguingly, no strong correlation could be determined, with long-lived proteins possessing translation levels that span 3 orders of magnitude. Prevalent translation was also seen for all Nups regardless of their protein lifespan or tissue type (Figure 3A, B) and is particularly noteworthy for the NPC proteins Nup98 and Nup96. These nucleoporins are translated as a single precursor polypeptide that is auto-catalytically cleaved into Nup98 and Nup96 (Fontoura et al., 1999). MS data on these two proteins 6 months post-chase reveal that, despite their identical translation rates, Nup96 retains 15N signal while Nup98 has been completely replaced with newly synthesized copies (Figure 3C, D). This difference is consistent with the structural and functional properties of these two Nups: while Nup98 is a peripheral and highly mobile nucleoporin with very low residence time at the NPC, Nup96 is a member of the Nup107/160 subcomplex and is therefore critical for the assembly and structural integrity of the nuclear pore (Hoelz et al., 2011; Rabut et al., 2004). It thus appears that protein longevity is the result of protein deposition into a stable complex rather than a lack of expression. Since the overall levels of Nup96 and NPC numbers do not increase with age (see below), these results also suggest that newly synthesized copies of Nup96, which are not incorporated into the NPC, are rapidly degraded.
Figure 3.
Protein translation does not correlate with protein lifespan. (A) Protein translation levels of long-lived proteins. Translation levels (reads/CDS length) of long-lived proteins are plotted (log2) against their corresponding 15N fractional abundance at 6 months post-chase. Translation levels of long-lived nucleoporins are plotted in orange. (B) Translation levels of NPC proteins in liver and brain tissue. Translation levels of all NPC proteins were determined in liver (horizontal axis) and brain (vertical axis) tissue, and plotted against each other (log2). (C) Translation and stability of Nup98/96. Top: Schematic of the Nup98/96 translated peptide, as well as the cleavage site (a.a. 880) that produces the separate Nup98 and Nup96 proteins. Lower: Elution profile MS1 traces of the indicated peptides from the Nup98 and Nup96 region, plotted as describe for Figure 1 D-G. (D) Stability of Nup98/96 over 12 months. Average 15N fractional abundance for Nup98 (grey) and Nup96 (orange) was determined from multiple peptides for each indicated time point and plotted over time.
Long-lived proteins in neurons and glia
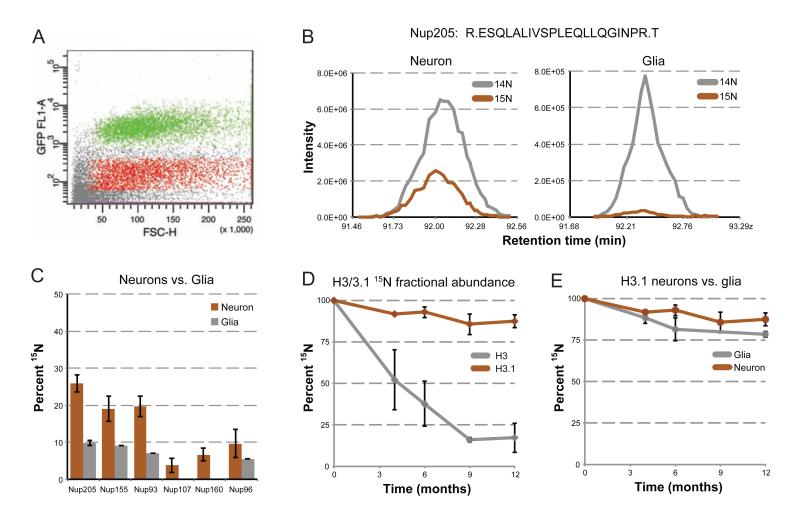
When analyzing total brain homogenates, we observed that most long-lived proteins nonetheless exhibit a gradual in their 15N fractional abundances. In principle, this observed reduction in 15N signal can be attributed to either (i) the renewal of cells and consequently the dilution of old protein through cell division, or (ii) turnover of individual proteins in non-dividing cells. A critical aspect of our study is the ability to discriminate between these two possibilities. Though no proteins were found to be 15N labeled in the liver, this is most likely due to cell division and proliferation. To determine the contribution of cell turnover to the observed decrease of 15N fractional abundance, we analyzed two different populations of brain cells: glial cells, which are much more abundant and have some ability to self-renew, versus neurons, which have been shown to exhibit minimal cell division in most regions of the adult brain (Kuhn et al., 1996). To distinguish these two cell populations, we fluorescently labeled purified nuclei from pulse-chase brains with the NeuN antibody, which is specific for neuronal nuclei, and used fluorescence-activated cell sorting (FACS) to purify NeuN-positive (neuronal-enriched) and NeuN-negative (glial-enriched) populations of nuclei (Figure 4A), as described previously (Spalding et al., 2005). Fluorescence microscopy confirmed the efficient enrichment of the fluorescent marker (Figure S4A), and sorted samples were subjected to analysis by MS. At 1 year post-chase, we observed a greater than 2-fold difference in 15N fractional abundance of three components of the Nup205 complex and three components of the Nup107/160 complex between neuronal and glial cells (Figure 4B,C). In all cases, Nup 15N fractional abundances were lower in the glia-enriched population, although the extent of this difference was variable between different animals (Figure S4B,C). Interestingly, the 15N fractional abundance in neuronal nuclei was similar across all animals tested, and most of the variability was seen for the glial-enriched populations, suggesting a contribution of cell proliferation to 15N content decay in glia cells (Figure S4B,C).
Figure 4.
15N fractional abundance decay rates in neuronal versus glial nuclei. (A) FACS sorting of brain nuclei. Brain nuclei were purified and labeled with a fluorescent marker for neuronal nuclei (NeuN), and sorted for NeuN positive and negative populations. Plotted is a scatter plot of representative sorted events, with positive (green) and negative (red) sorted populations highlighted. (B) 15N fractional abundance decay of Nup205 in neurons versus glia. Elution profile MS1 traces, as described earlier, are plotted for the same peptide from Nup205, originating from neuron-enriched (left) and glial-enriched (right) sorted nuclei from a 1-year post-chase rat. (C) 15N fractional abundance decay of Nups in neurons versus glia. Average 15N fractional abundance for each indicated Nup was determined from multiple peptides from the same neuron-enriched (orange) and glial-enriched (grey) nuclei described in B. (D) Histone H3.1 stability. 15N fractional abundance was determined for the single unique H3.1 peptide at 0, 4, 6, 9, and 12 months post-chase from multiple animals (orange), and plotted against peptides from the same time points which were common to all histone H3 variants (grey). (E) Histone H3.1 in neurons versus glia. 15N fractional abundance was determined for the single unique H3.1 peptide at 0, 4, 6, 9, and 12 months post-chase from neuron-enriched (orange) and glial-enriched (grey) sorted nuclei, and plotted over time. Note: we did not observe any peptides for glia 9-months post-chase. All error bars are plotted as a standard deviation. See also Figure S4.
To specifically address the contribution of cell division to the decrease in 15N fractional abundance over time, we analyzed neuronal and glial data for the specific histone H3.1. Histone H3.1 is a replication-dependent histone, and upon exit from the cell cycle, is no longer deposited into nucleosomes (Wu et al., 1982). When displaced in non-dividing cells, H3.1 is replaced with the variant, H3.3 (Ahmad and Henikoff, 2002; Schwartz and Ahmad, 2005). Thus, the exchange of 15N H3.1 for 14N H3.1 only occurs through cell division. Although the sequence similarity between H3.1 and H3.3 is high, with only a single peptide capable of distinguishing the two by MS (Figure S4D), we were able to identify (Figure S4D) and extract quantitative data for the H3.1 polypeptide. Over the course of a 12-month chase, H3.1 had a lower 15N fractional abundance decrease in neuronal nuclei than any other protein, remaining as high as 85% 15N (Figure 4D). This is in contrast to H3 peptides that were common to all H3 variants, which decreased to below 25% 15N fractional abundance. H3.1 was the most stable long-lived protein we observed in all our studies, confirming the replication-dependence of this H3 variant and also providing proteomic evidence that there is very little renewal of neuronal cells over the course of 12 months. Comparison of H3.1 peptides in neuronal versus glial-enriched nuclei showed more 15N fractional abundance decay in the glial population, also consistent with greater cell turnover among glia than neurons (Figure 4E).
Long-term persistence of nucleoporins despite robust translation
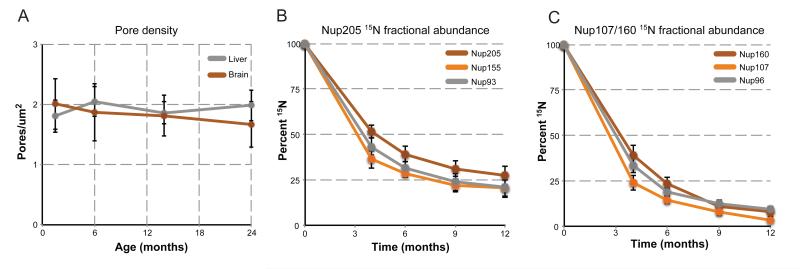
The persistence of histone H3.1 strongly supports the idea that the vast majority of neurons do not turn over in the adult brain. This further implies that the reduction to less than 25% 15N fractional abundance observed in Nups in the neuronal fraction is a result of protein replenishment that occurs in post-mitotic cells slowly over several months. We first tested the possibility that this 15N content decrease is due to the insertion of new NPCs that dilute out existing 15N NPCs, and thereby lower 15N fractional abundances. In this scenario, a decrease in 15N fractional abundance ~25% by 6-months post-chase would require ã300% increase in pore number between 6 weeks and 6 months of age. To test for such an increase, we quantified the number of NPCs per nucleus using structured illumination microscopy (SIM) to visualize immunostained for nucleoporins in rats from 6 weeks of age to 2 years. NPC density remained unchanged over this time period in both liver and brain nuclei (Figure 5A). The corresponding nuclei surface area and total nuclear pore numbers also did not change more than one standard deviation (Figure S5), suggesting that new NPC insertion cannot explain the observed decrease of NPC 15N fractional abundance. However, it remained possible that new NPC insertion could be coupled to whole NPC removal. Alternatively, individual Nups within an NPC might be exchanged at very slow rates in the absence of any NPC turnover. The primary point of distinction between these models is that all stable Nups would turnover at the same rate in the first model, while stable Nups could turnover at different rates in the latter. To this end, we compared 15N fractional abundance data for 3 components of the Nup205 complex (Nup205, Nup155, and Nup93) and 3 components of the Nup107/160 complex (Nup160, Nup107, Nup96) over the entire 12-month time course from neuronal-enriched nuclei. Both subcomplexes displayed a reduction of 15N fractional abundance over time, with very similar rates observed for proteins within the same subcomplex (Figure 5B,C). Comparison between the Nup205 and Nup107/160 complexes, however, revealed differences in the kinetics of 15N-signal decay between the two complexes, with the Nup107/160 complex being completely replaced by 14N at 12-months post-chase, while the Nup205 complex retaining ~25% 15N fractional abundance. The disparate decay kinetics between these subcomplexes within the same NPC suggests that NPCs do not turnover as an entire unit. Rather, NPCs themselves appear to be stable over the lifespan of the organism with their constituent subcomplexes being able to exchange at extremely low rates. This is consistent with the observed slow, but finite, exchange of Nup107/160 complex components seen in cultured cells (Rabut et al., 2004), as well as the continued translation of all Nups we observe in the brain.
Figure 5.
Long-term maintenance of the NPC. (A) NPC pore density through aging. Brain and liver nuclei were purified from 6 week, 6, 13, and 24 month-old rats and fixed. NPCs were stained and visualized by super-resolution microscopy, and individual pores counted for each nucleus (n > 30). Plotted are the average pore densities (pores per nuclei surface area [μM]) for brain (orange) and liver (grey) nuclei across the entire time span. See also Figure S5. (B) 15N fractional abundance decay of the Nup205 complex. 15N fractional abundance was determined from multiple peptides for the indicated members of the Nup205 complex, from neuronal-enriched nuclei from all time points of the pulse-chase and plotted over time. (C) 15N fractional abundance decay of the Nup107/160 complex. 15N fractional abundance was determined from multiple peptides for the indicated members of the Nup107/160 complex, from neuronal-enriched nuclei from all time points of the pulse-chase and plotted over time. All error bars represent standard deviations.
Long-lived proteins in aging
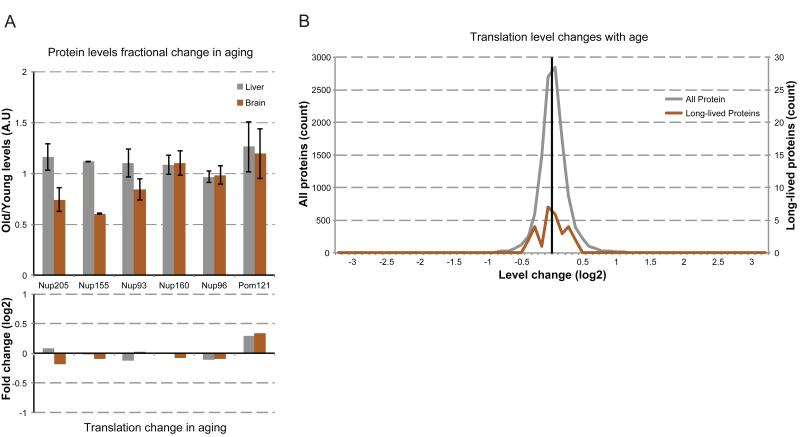
The finding that the Nup205 and Nup107/160 NPC subcomplexes display different kinetics of 15N fractional abundance decay suggests that distinct NPC subcomplexes are individually replaced, with the Nup205 complex having the longest residence time. As a decrease in NPC function is seen in aged nuclei (D’Angelo et al., 2009), we asked if this decay may be due to selective loss of scaffold NPC subcomplexes (Figure S2). To address this possibility, we quantified Nup levels from purified NPCs from young (6-month) and old (24-month) liver and brain nuclei. Previous data suggested that protein levels of the Nup107/160 complex do not change with age (D’Angelo et al., 2009). Consistent with this finding, we observed no changes in Nup160 or Nup96 protein levels with age in either liver or brain nuclei, nor did we detect changes in the transmembrane Nup Pom121 (Figure 6A). In striking contrast, all three long-lived members of the Nup205 complex were reduced to less than 85% in old brain nuclei but unchanged in liver.
Figure 6.
NPC composition changes with age. (A) NPC levels with age. Upper: NPCs were purified from brain (orange) and liver (grey) nuclei from rats at 6 (young) and 24 (old) months of age. Indicated Nup levels were determined by western blot, normalized to Nup107 levels. Plotted are the old levels normalized to the young levels. Lower: Translation levels of the indicated Nups from 6-month-old rat brains were compared to translation levels of 24-month-old rat brains, and plotted as old/young translation levels (log2 scale). (B) Changes in translation levels (old rate/young rate, log2 scale) were binned by log2 fold change (0.1 unit bins) and plotted as a histogram for all proteins (grey, >11,000 proteins, left vertical scale) and the identified long-lived proteins (orange, right vertical scale). See also Figure S5.
The partial loss of the Nup205 complex from the NPC might be caused by a change in gene expression of these NPC proteins, as alterations in transcript levels of some genes have been reported to occur during aging (Lee et al., 2000). To address this possibility, we determined translation levels of all expressed genes in 24-month old liver and brain tissue, and compared them to our data from 6-month old tissues. Over the more than 11,000 genes analyzed, more than 98% experienced an age-dependent change in translation level of less than 2-fold (Figure 6B). This robust stability of protein translation levels through age is consistent with the previously reported stability of the majority of the aging proteome (Walther and Mann, 2011). None of the long-lived proteins experienced a higher than 1.5-fold change in translation level, and similar to the rest of the proteome, the distribution of their changes centered on zero change (Figure 6B). More specifically, translation levels of long-lived Nups changed less than 0.15-fold with age (Figure 6A), suggesting that the decrease in levels of Nup205 complex members with age might be due to inefficient replacement at the NPC. This deficiency of Nup205 subcomplex maintenance in aged nuclei, as well as the previously observed breakdown of the nuclear permeability barrier with age (D’Angelo et al., 2009), provide an exciting new link between long-term protein maintenance and cellular aging.
Discussion
Proteins with exceptional lifespans have been thought to be a rare phenomenon restricted to highly specialized and protected environments with limited metabolism such as eye lens crystallin and collagens. Here we provide a comprehensive, system-wide identification and characterization of proteins with limited turnover in the adult brain and show that long-lived proteins are more common than previously appreciated. Importantly, the long-lived proteome is composed of functionally diverse proteins that regulate a myriad of cellular functions and includes Nups, components of the myelin sheath, specific canonical histones and histone variants, as well as enzymes. Importantly, the long lifespan of these proteins does not seem to be rooted in a cell’s inability to express these proteins, as we found evidence for robust translation of almost all long-lived proteins identified. Using the NPC as a paradigm for long-lived protein maintenance, we found that this stable structure experiences slow but finite turnover of its individual subcomplexes, which may help to clear potentially damaged components.
Although the existence of a long-lived proteome is now evident, the prevailing question still remains: Why would the cell employ proteins with limited turnover if their persistence places them at increased risk for damage accumulation? A common, but not exclusive, characteristic of the long-lived proteins we identified here is their involvement in large and stable cellular structures. Histones, for example, play a critical role in chromatin organization and compaction. Some of the most stable histones we observed, H3.1 and H1.5, are often associated with heterochromatin, with H1.5 also specifically associated with gene repression, SIRT1 binding, and H3K9me2 enrichment (Hake et al., 2006; Li et al., 2012). These roles in transcriptional repression, chromatin compaction, and maintenance of epigenetic memory may require exceptional protein lifespan and stability, as frequent exchange of these histones could transiently compromise chromatin structure, resulting in aberrant expression at their resident loci. Whether chemical or structural defects in old histones are responsible for age-dependent changes in gene expression in post-mitotic tissues remains to be tested.
Nucleoporins are also engaged in a large stable structure, the nuclear envelope-embedded NPC. The lifelong persistence of Nups might be born out of a necessity to maintain structural and functional integrity of the NPC and the nuclear membrane. This is evidenced by the fact that the only time NPCs are known to disassemble is during nuclear envelope (NE) breakdown in mitosis (Guttinger et al., 2009). In post-mitotic cells, the disassembly of entire NPCs might not be possible because dismantling the nuclear pore might place the nucleus at risk for partial NE breakdown or transient loss of nuclear/cytoplasmic compartmentalization. With these limitations in mind, we provide new insights into the long-term maintenance of the NPC. This large protein complex is composed both of members that experience fast turnover as well as members that are extremely stable. Even among the stable components, two subcomplexes that each manifest distinct 15N decay kinetics can be identified. This finding suggests that, unlike other large protein complexes such as ribosomes or proteasomes (Cambridge et al., 2011; Price et al., 2010), the NPC does not turnover as an entire complex; rather, individual subcomplexes are exchanged at specific rates with newly synthesized copies. A remarkable observation of our studies is that ~25% of Nup205 complexes have not been replaced even after one year, making these nucleoporins together with histone H3.1, among the most persistent and stable cellular proteins known to exist in mammals. Such protein lifespan is remarkable since scaffold nucleoporins may not be protected from potentially harmful metabolites, which, in combination with inefficient replacement, may eventually result in the age-dependent loss of the stable Nup205 subcomplex and concomitant deterioration of nuclear integrity (D’Angelo et al., 2009).
Besides histones and Nups, several other long-lived proteins are engaged in clear stable structures, such as collagens in the extracellular matrix, lamins in the nuclear lamina, and proteins of the myelin sheath. Some long-lived proteins, however, lacked association with any apparent stable structures, and include the enzymes Sirt2, Enpp6, and Cnp1. How or why seemingly soluble proteins persist through aging is puzzling, particularly for proteins with enzymatic activity. How these unique long-lived proteins evade turnover, whether they are part of large protein assemblies that have to be maintained over time, and if they exhibit any age-dependent loss of function will be of great interest for future studies.
Another surprising result from our study is the continued translation of almost all long-lived proteins. This phenomenon is most clearly illustrated by the Nup98/Nup96 proteins, which are translated at identical rates from the same mRNA but exhibit divergent lifespans. While Nup98 turns over rapidly, Nup96 is incorporated into the NPC scaffold, where it persists for months. This observation implies that the majority of newly synthesized Nup96 is immediately degraded in post-mitotic cells. Why the coupling of translation of these two proteins is evolutionarily conserved when their cellular lifespans are divergent remains a mystery. Also unclear is the continuous production of long-lived proteins despite the fact that they are stably embedded in a cellular structure. One possibility is that long-lived proteins may exist in multiple functional populations within the cell, whereby one population is stable and long-lived, while another is dynamic and short-lived. Thus, translation would be needed to maintain constant turnover of the short-lived population. Alternatively, most of the newly synthesized copies may be unincorporated and subject to degradation. This possibility is consistent with protein turnover serving as a mechanism to buffer intracellular amino acid levels and with the reported immediate degradation of ~30% of all translated proteins (Schubert et al., 2000; Vabulas and Hartl, 2005).
The long-lived proteome may certainly not be limited to the proteins described here. First, these results are only from two tissues, the liver and brain, and other long-lived proteins may be discovered in other tissues such as the heart. Further, the isotopic pulse-chase method cannot distinguish between true “turnover” of a protein from simple dilution of stable proteins with newly synthesized copies. Indeed, we observed robust translation of most of the long-lived proteins, proving that, despite their long lifespan, they are continuously expressed. Thus, a number of proteins may have long-lived populations that are diluted over time, evading their detection by our methods. Identification of the long-lived proteome is critical, as there is a well-established relationship between protein lifespan and damage, as illustrated by eye lens crystallin. Our study used the NPC as a paradigm for long-lived protein maintenance, finding that despite the NPC’s stability, it can be maintained through slow but finite turnover of its constituent subcomplexes. As many of the long-lived proteins identified in this study are components of essential and fundamental biological processes, maintaining their function through adulthood is crucial. It remains to be seen, however, if they reside in long-lived protein complexes that have the ability to maintain or even repair inevitable chemical modifications and damage.
Experimental Procedures
Detailed experimental procedures can be found in the extended experimental procedures
Pulse-chase labeling of rats
Rats were pulse labeled as described earlier (McClatchy et al., 2007; Savas et al., 2012). Rats from multiple litters were then sacrificed at 4, 6, 9, and 12 months post-chase, tissues harvested and flash frozen.
Tissue fractionations and purifications
Liver and brain nuclei were purified according to protocol (Blobel and Potter, 1966; Lovtrup-Rein and McEwen, 1966). Supernatants of the nuclei purifications were diluted 5x with corresponding nuclei purification buffer with no sucrose, and spun at 13,000rcf for 15 minutes. The pellet was resuspended in nuclei purification buffer, representing the mitochondria-enriched fraction. The supernatant was then spun at 100,000rcf for 20 minutes, and the pellet resuspended in nuclei purification buffer representing the ER-enriched fraction. The supernatant of this fraction represented the cytosol-enriched fraction.
NeuN nuclei labeling, sorting, and analysis
Nuclei were labeled with Alexa Fluor488-conjugated NeuN antibody (Millipore) and sorted as described using a Vantage SE DiVa (Spalding et al., 2005).
Mass spectrometry and analysis
All MS samples were digested and analyzed as described previously (Savas et al., 2012). Protein identification and quantification and analysis were done with Integrated Proteomics Pipeline - IP2 (Integrated Proteomics Applications, Inc., San Diego, CA. http://www.integratedproteomics.com/) using ProLuCID, DTASelect2, Census, and QuantCompare. Spectrum raw files were extracted into ms1 and ms2 files from raw files using RawExtract 1.9.9 (http://fields.scripps.edu/downloads.php) (McDonald et al., 2004), and the tandem mass spectra were searched against EBI IPI mouse protein database (EBI-IPI_rat_3.30_con_06-28-2007).
Determination of translation rates
Ribosome footprinting and deep sequencing was performed as previously described (Ingolia et al., 2009), adapted for use on tissue samples. See extended experimental procedures for details.
NPC counting
Nuclei were purified from liver and brain tissue as described earlier from at least 2 different rats of 6 weeks, 6, 13, or 24 months of age. NPCs were stained with the antibody mab414 and then imaged with a Zeiss Elyra structured illumination super resolution microscope. After 3D reconstructions, pore numbers were quantitated using the spot finder tool in Imaris (Bitplane) and surfaces area using the surfaces tool. At least 30 nuclei were quantitated from at least 2 rats for each time point. See extended experimental procedures for details.
NPC protein levels with age
Liver and brain nuclei were purified as described previously from 6 and 24 month-old rat tissues, NPCs subsequently purified according the protocol (Cronshaw et al., 2002), and quantified by western blot. See extended experimental procedures for details.
Supplementary Material
Highlights.
Metabolic pulse-chase labeling of rats identified long-lived proteins in rats
Long-lived proteins include nucleoporins, histone variants and enzymes
Long-lived proteins cannot be replaced despite their robust translation
Nuclear pores are maintained over the lifespan of the organism
Acknowledgments
We thank members of the Hetzer lab and E.Q. Toyama for critical reading of the manuscript. B.H.T is supported by the Hewitt Foundation; M.W.H by the NIH grant R01GM098749, the Glenn Aging Foundation, the American Cancer Society Award Number P30CA014195 and the Ellison Medical Foundation; J.N.S. by NIH fellowship F32AG039127; JRY and JNS by NIH P41 RR011823, P01 AG031097, and R01 MH067880; N.T.I. by the Searle Scholars Program. The RAW files and complete parameter files will be publically available at http://fields.scripps.edu/published/ELLP2/ upon publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
References
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Molecular cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Arber N, Zajicek G, Ariel I. The streaming liver. II. Hepatocyte life history. Liver. 1988;8:80–87. doi: 10.1111/j.1600-0676.1988.tb00972.x. [DOI] [PubMed] [Google Scholar]
- Bassnett S. Lens organelle degradation. Experimental eye research. 2002;74:1–6. doi: 10.1006/exer.2001.1111. [DOI] [PubMed] [Google Scholar]
- Bassnett S, Shi Y, Vrensen GF. Biological glass: structural determinants of eye lens transparency. Philosophical transactions of the Royal Society of London Series B. Biological sciences. 2011;366:1250–1264. doi: 10.1098/rstb.2010.0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle A, Tanay A, Bitincka L, Shamir R, O’Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Potter VR. Nuclei from rat liver: isolation method that combines purity with high yield. Science. 1966;154:1662–1665. doi: 10.1126/science.154.3757.1662. [DOI] [PubMed] [Google Scholar]
- Bloemendal H, de Jong W, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A. Ageing and vision: structure, stability and function of lens crystallins. Progress in biophysics and molecular biology. 2004;86:407–485. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Cambridge SB, Gnad F, Nguyen C, Bermejo JL, Kruger M, Mann M. Systems-wide proteomic analysis in mammalian cells reveals conserved, functional protein turnover. Journal of proteome research. 2011;10:5275–5284. doi: 10.1021/pr101183k. [DOI] [PubMed] [Google Scholar]
- Commerford SL, Carsten AL, Cronkite EP. Histone turnover within nonproliferating cells. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:1163–1165. doi: 10.1073/pnas.79.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. The Journal of cell biology. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends in cell biology. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerre JA, Lee CT. In vivo methylation and turnover of rat brain histones. Journal of neurochemistry. 1974;23:541–547. doi: 10.1111/j.1471-4159.1974.tb06057.x. [DOI] [PubMed] [Google Scholar]
- Fontoura BM, Blobel G, Matunis MJ. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. The Journal of cell biology. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nature reviews Molecular cell biology. 2009;10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. The Journal of biological chemistry. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- Haus JM, Carrithers JA, Trappe SW, Trappe TA. Collagen, cross-linking, and advanced glycation end products in aging human skeletal muscle. J Appl Physiol. 2007;103:2068–2076. doi: 10.1152/japplphysiol.00670.2007. [DOI] [PubMed] [Google Scholar]
- Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annual review of biochemistry. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. The Journal of cell biology. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Molecular cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- Kragstrup TW, Kjaer M, Mackey AL. Structural, biochemical, cellular, and functional changes in skeletal muscle extracellular matrix with aging. Scandinavian journal of medicine & science in sports. 2011;21:749–757. doi: 10.1111/j.1600-0838.2011.01377.x. [DOI] [PubMed] [Google Scholar]
- Kuhn HG, Dickinson-Anson H, Gage FH. Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1996;16:2027–2033. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nature genetics. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- Li JY, Patterson M, Mikkola HK, Lowry WE, Kurdistani SK. Dynamic distribution of linker histone H1.5 in cellular differentiation. PLoS genetics. 2012;8:e1002879. doi: 10.1371/journal.pgen.1002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovtrup-Rein H, McEwen BS. Isolation and fractionation of rat brain nuclei. The Journal of cell biology. 1966;30:405–415. doi: 10.1083/jcb.30.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters PM, Bada JL, Zigler JS., Jr. Aspartic acid racemisation in the human lens during ageing and in cataract formation. Nature. 1977;268:71–73. doi: 10.1038/268071a0. [DOI] [PubMed] [Google Scholar]
- McClatchy DB, Dong MQ, Wu CC, Venable JD, Yates JR., 3rd 15N metabolic labeling of mammalian tissue with slow protein turnover. Journal of proteome research. 2007;6:2005–2010. doi: 10.1021/pr060599n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, Yates JR., 3rd MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid communications in mass spectrometry: RCM. 2004;18:2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- Park SK, Venable JD, Xu T, Yates JR., 3rd A quantitative analysis software tool for mass spectrometry-based proteomics. Nature methods. 2008;5:319–322. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piha RS, Cuenod M, Waelsch H. Metabolism of histones of brain and liver. The Journal of biological chemistry. 1966;241:2397–2404. [PubMed] [Google Scholar]
- Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. Analysis of proteome dynamics in the mouse brain. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14508–14513. doi: 10.1073/pnas.1006551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nature cell biology. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Rodrijguez de Lores A, Alberici de Canal M, De Robertis E. Turnover of proteins in subcellular fractions of rat cerebral cortex. Brain research. 1971;31:179–184. doi: 10.1016/0006-8993(71)90642-1. [DOI] [PubMed] [Google Scholar]
- Roy D, Spector A. Absence of low-molecular-weight alpha crystallin in nuclear region of old human lenses. Proceedings of the National Academy of Sciences of the United States of America. 1976;73:3484–3487. doi: 10.1073/pnas.73.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas JN, Toyama BH, Xu T, Yates JR, 3rd, Hetzer MW. Extremely long-lived nuclear pore proteins in the rat brain. Science. 2012;335:942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes & development. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sell D, Monnier V. Comprehensive Physiology (American Physiological Society) 2011. Aging of Long-Lived Proteins: Extracellular Matrix (Collagens, Elastins, Proteoglycans) and Lens Crystallins; pp. 235–305. [Google Scholar]
- Shapiro SD, Endicott SK, Province MA, Pierce JA, Campbell EJ. Marked longevity of human lung parenchymal elastic fibers deduced from prevalence of D-aspartate and nuclear weapons-related radiocarbon. The Journal of clinical investigation. 1991;87:1828–1834. doi: 10.1172/JCI115204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma KK, Santhoshkumar P. Lens aging: effects of crystallins. Biochimica et biophysica acta. 2009;1790:1095–1108. doi: 10.1016/j.bbagen.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisen J. Retrospective birth dating of cells in humans. Cell. 2005;122:133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- Tachibana-Konwalski K, Godwin J, van der Weyden L, Champion L, Kudo NR, Adams DJ, Nasmyth K. Rec8-containing cohesin maintains bivalents without turnover during the growing phase of mouse oocytes. Genes & development. 2010;24:2505–2516. doi: 10.1101/gad.605910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Kurihara T. 2′,3′-Cyclic nucleotide 3′-phosphodiesterase: Molecular characterization and possible functional significance. In: Martenson RE, editor. Myelin: Biology and Chemistry. CRC Press; Boca Raton, FL: 1992. pp. 449–480. [Google Scholar]
- Vabulas RM, Hartl FU. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- Verzijl N, DeGroot J, Thorpe SR, Bank RA, Shaw JN, Lyons TJ, Bijlsma JW, Lafeber FP, Baynes JW, TeKoppele JM. Effect of collagen turnover on the accumulation of advanced glycation end products. The Journal of biological chemistry. 2000;275:39027–39031. doi: 10.1074/jbc.M006700200. [DOI] [PubMed] [Google Scholar]
- Walther DM, Mann M. Accurate quantification of more than 4000 mouse tissue proteins reveals minimal proteome changes during aging. Molecular & cellular proteomics: MCP. 2011;10:M110–004523. doi: 10.1074/mcp.M110.004523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature biotechnology. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wilmarth PA, Tanner S, Dasari S, Nagalla SR, Riviere MA, Bafna V, Pevzner PA, David LL. Age-related changes in human crystallins determined from comparative analysis of post-translational modifications in young and aged lens: does deamidation contribute to crystallin insolubility? Journal of proteome research. 2006;5:2554–2566. doi: 10.1021/pr050473a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RS, Tsai S, Bonner WM. Patterns of histone variant synthesis can distinguish G0 from G1 cells. Cell. 1982;31:367–374. doi: 10.1016/0092-8674(82)90130-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.