Abstract
8-Hydroxy-2′-deoxyguanosine (8-OHdG) is one of the major forms of oxidative deoxyribonucleic acid (DNA) damage, and is commonly analyzed as an excellent marker of DNA lesions. The purpose of this study was to develop a sensitive method to accurately and rapidly quantify the 8-OHdG by using capillary electrophoresis with laser-induced fluorescence detection (CE-LIF). The method involved the use of specific antibody to detect DNA lesions (8-OHdG) and consecutive fluorescence labeling. Next, the urine sample with 8-OHdG fluorescently labeled along with other constituents was resolved by capillary electrophoretic system and the lesion of interest was detected using fluorescence detector. The limit of detection was 0.18 fmol, which is sufficient sensitivity for detection and quantification of 8-OHdG in untreated urine samples. The relative standard deviation (RSD) was found to be 11.32 % for migration time, and 5.52 % for peak area. To demonstrate the utility of this method, the urinary concentration of 8-OHdG in an Alzheimer’s transgenic mouse model was determined. Collectively, our results indicate that this methodology offers great advantages such as high separation efficiency, good selectivity, low limit of detection (LOD), simplicity and low cost of analysis.
Keywords: Oxidative deoxyribonucleic acid (DNA) damage, 8-Hydroxy-2′-deoxyguanosine (8-OHdG), Alzheimer’s disease (AD), Capillary electrophoresis with laser induced fluorescence detection (CE-LIF), Urine
Introduction
Both nuclear and mitochondrial deoxyribonucleic acid (DNA) from various tissues and white blood cells are subjected to oxidative damage [1]. They interact with reactive oxygen species (ROS), such as superoxide anion radical (O2−), hydrogen peroxide (H2O2), hydroxyl radicals (•OH), and further produce more than 20 oxidized DNA adducts. Among them, 8-hydroxy-2′-deoxyguanosine (8-OHdG) is considered the major type of DNA damage (Fig. 1) and is the most commonly used biomarker for evaluation of cellular oxidative stress. 8-OHdG is implicated in aging and the development of neurodegenerative disease, such as Alzheimer’s disease (AD) [2]. 8-OHdG is probably the best studied biomarker of oxidative stress due to its implication in nucleobase mutations (e.g. CG to AT transversions) [3]. Because guanine has the lowest oxidation potential compared with the other DNA bases, it is easily oxidized and forms one of the most abundant marker of oxidative stress. The mechanism of formation of 8-OHdG involves the attack of carbon 8 (C8) in guanine by hydroxyl radicals, followed by the formation of 8-hydroxyguanine (C8-OH-Gua) adduct radical that may undergo two separate pathways: (1) 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyGua) formation or (2) subsequent oxidation and formation of 8-oxoguanine [4].
Figure 1.

Formation of 8-OHdG by oxidative stress.
Oxidative DNA damage accumulates in the brain, which has been hypothesized to be a major component of aging and a contributing factor for development of AD. Age-dependent increases of 8-OHdG have been reported in various tissues, such as human pituitary gland [5], the liver and kidney of rats [6, 7], human cerebral cortex and cerebellum brain [8]. Because the brain is associated with a high percentage of oxygen consumption and relatively insufficient antioxidant capacity, AD brains are most likely vulnerable and susceptible to oxidative damage [9, 10]. The evidence indicates that oxidative DNA damage is the feature of AD in the brain and ventricular cerebrospinal fluid (CSF) [11, 12]. Using gas chromatography-mass spectrometry (GC-MS), studies of temporal and parietal lobes from the brains of AD patients revealed statistical significant increases of 8-OHdG in AD patients when compared to non-demented control subjects [12]. Studies have also reported that levels of 8-OHdG are increased in the mitochondrial DNA of patients with late AD compared to nuclear DNA [12–14]. Elevated 8-OHdG levels have been detected in mitochondrial DNA in the cortex of AD patients compared with non-demented controls [15]. Another study found higher levels of 8-OHdG in the nuclear DNA of lymphocytes of AD patients [16]. A recent study by Lovell’s group found a significant increase of 8-OHdG in multiple brain regions, including frontal, temporal and parietal lobes in subjects with mild cognitive impairment (MCI) [12]. These findings suggest oxidative DNA damage can be detected from the subjects with MCI, which may serve as a prescreening tool for AD and play a critical role in investigating the AD pathogenesis.
Evidence of oxidative DNA damage can be detected in the urine. After 8-hydroxy-2′-deoxyguanosine 5′-triphosphate (8-OH-dGTP) is formed in the nucleotide pool, the pyrophosphate can be removed by the enzyme MutT Homolog-1 (MTH1) to yield 8-hydroxy-2′-deoxyguanosine 5′-monophosphate (8-OH-dGMP). Afterwards, it is digested by 5′ (3′)-nucleotidase to produce 8-OHdG. Without further metabolism, 8-OHdG is excreted in the urine [17, 18]. The amount of 8-OHdG excreted into the urine can be used as an index of the average rate of oxidative damage in the whole body and as a result, it can be potentially used as a clinical test in the early screening for different diseases. It is notable that measurement of excreted repair products in urine indicates the average rate of the damage in the total body, while the abundance of oxidized bases in nuclear or mitochondrial DNA represents the concentration in the specific local tissues/cells [2]. Compared to DNA samples, urine is a better matrix for analyzing 8-OHdG, since DNA isolation and digestion techniques can be a source of the artificial formation of 8-OHdG [19]. In addition, because 8-OHdG is excreted in the urine, it can be used as a non-invasive assay for determining the levels of in vivo oxidative lesions in the early progression of various diseases, such as cancer, diabetes, atherosclerosis and neurological disorders [20]. Here, we focus on the development of a sensitive assay to monitor 8-OHdG levels in AD urine samples. This method has a great potential to serve as a platform to identify patients at risk for AD and/or the effect of therapies.
To fully understand the extent to which 8-OHdG are involved in disease, different analytical techniques have been used to study 8-OHdG lesions in different matrices most common in the tissue and urine. Enzyme-linked immunosorbent assay (ELISA) is widely used to detect 8-OHdG in urine or other biological samples with limit of detection (LOD) of one 8-OHdG/per 105 deoxyguanosine (dG). Urinary 8-OHdG measurement using ELISA rather than others, such as high performance liquid chromatography (HPLC), saves analytical time, running costs, and sample volume. The disadvantage of this method is that the antibodies may cross react with guanine and other compounds. Cook’s group speculates that high molecular weight compounds, such as proteins and carbohydrates, tend to interfere in the competitive format of the ELISA, which resulted in high 8-OHdG reading [21]. To overcome this problem, immunoaffinity columns have been used to isolate 8-OHdG from DNA hydrolysis followed by ELISA quantification [22].
Aside from immunoassays, chromatographic techniques, such as GC-MS, were widely developed to study oxidative DNA damage. However, it was reported that the background level of 8-OHdG was overestimated. This was suggested to be caused by artificial oxidization of guanine to 8-OH-Gua during the derivatization step [23]. Hence, a purification step to remove intact bases from acid-hydrolysate of DNA is mandated using HPLC prior to the analysis [24]. Another methodology for 8-OHdG detection is to use HPLC with electrochemical column switching system (HPLC-EC) that utilizes the selectivity of different carbon columns. This system can be used to analyze this oxidized DNA adduct in variety of matrices. The reported LOD of this technology was in the range of 25–74 pM [25]. Nevertheless, a significant disadvantage lies in the requirement for column switching to purify the urine sufficiently [26].
Recently, MS-based detection techniques have been increasingly applied to study 8-OHdG. HPLC-tandem mass spectroscopy achieved LOD of 0.5 nM, which requires a minimal amount of sample (12.5 fmol) [27] and does not require pre-treatment and purification steps. 8-OHdG can also be detected using solid phase extraction and HPLC with triple stage quadruple mass detection. The reported LOD for this method is 0.7 nM [28], and allows for inclusion and highly specific determination for relatively large sample volume. With simultaneous measurement of several products and providing structure evidence for analytes, these techniques provide accurate quantification and avoid misleading readings which might be resulted from a single product. Although HPLC-MS and GC-MS are sensitive and accurate, these are not convenient procedures for 8-OHdG detection in clinical settings, because of its elaborate extraction and separation steps, high cost, technical involvement and low throughput.
8-OHdG was also detected using capillary electrophoresis with various detectors, such as UV, electrochemical or amperometric detectors. Capillary electrophoresis method with UV detection was used to quantify 8-OHdG in untreated urine samples. The LOD was reported as 17 uM, where this technology suffers from the low sensitivity [29]. To improve the sensitivity, the pre-concentrated urine analyzed by capillary electrophoresis with electrochemical detection or end-column amperometric detection reached LOD of 20–50 nM [30–32]. Although the detection sensitivity has been improved, these methods require solid phase extraction (SPE) to concentrate samples before the analysis [30, 31]. Therefore, the urine pretreatment could become complicated and time-consuming. Moreover, it has limited application to analyze samples with enhanced levels of 8-OHdG.
A common component of all the above mentioned methods, whether reliant on chromatography techniques (primarily, HPLC or GC or CE coupled ECD or MS or UV detector) or immunoassay (ELISA), few current methods possess sufficient specificity, high throughput, or exclusion of urine pretreatment, without any need for highly trained specialist or expensive equipment. The aim of this study is to develop a simple and sensitive method using capillary electrophoresis with laser induced fluorescence (CE-LIF) detection for detecting 8-OHdG using a primary antibody (Ab) specific for 8-OHdG, followed by binding of a fluorescently labeled secondary antibody to the antibody-8-OHdG complex. The routine measurement of 8-OHdG in urine samples using ELISA is problematic due to the low amount of the analyte as well as interference from other compounds. To address this concern, separation of the analytes based on the charge to size ratio offers great separation efficiency. Furthermore, the LIF detection offers great detection sensitivity by combining both chromatographic separation and immunoassay labeling. This method achieves high specificity and doesn’t require any pre-treatment and pre-concentration of urine samples. In addition, it allows rapid throughput and better accuracy in the detection of this analyte, so this is a simple, fast and sensitive method for detection of 8-OHdG. Our results demonstrate a unique approach of detecting 8-OHdG levels in urine to monitor oxidative stress levels in an AD transgenic (Tg) mouse model.
Materials and Methods
Chemicals and reagents
Aqueous solution of the reagents was prepared using 18 megaohm water from Milli-Q water purification system (Millipore, Bedford, MA). All chemicals were analytical grade quality. 8-OHdG was purchased from Sigma-Aldrich (St. Louis, MO). N45.1 anti 8-OHdG monoclonal antibody was obtained from Genox Corporation (Baltimore, MD). IgG Alexa fluor 488 goat secondary antibody was obtained from Invitrogen (Ontario, Canada). Sodium tetraborate, dimethyl sulfoxide (DMSO), methanol, sodium hydroxide, and hydrochloric acid were obtained from VWR International (Suwanee, GA). Commercially flexible fused silica capillary was obtained from Polymicro Technologies (Phoenix, AZ). The separation buffer consisted of 20 mM sodium tetraborate, pH 9.5.
CE instrument
Beckman Coulter P/ACE MDQ system (Fullerton, CA), a commercial capillary electrophoresis instrument was used for the detection of 8-OHdG. Argon-ion laser was used for excitation (488 nm line, 3 mW). After excitation, the emitted light was filtered with 530DF20 bandpass filter (510–530 nm) before reaching photomultiplier tube (PMT). The PMT output signal was sampled at 4 Hz. Separation were performed on an uncoated 50 μm i.d. fused silica capillary with a 42 cm in total length and 30 cm in effective length.
Labeling of test model: 8-OHdG
A DNA oxidative stress model, 8-OHdG was used to test the labeling scheme and validate the LIF detector response. 8-OHdG (1 mg) was suspended in 50 μL DMSO and 9950 μL of water to reach a final concentration of 100 ng·μL−1 (or 353 μM). Anti-8-OHdG and the secondary Alexa 488 antibody were diluted in water. To generate the calibration curve, five 8-OHdG standards with final concentrations of 10, 8, 6, 4, and 2 ng·μL−1 were prepared. 20 μL 8-OHdG standard, 20 μL primary antibody (40 ng·μL−1), and 10 μL secondary antibody (40 ng·μL−1) complex were mixed. 8-OHdG was incubated on orbital shaker with anti-8-OHdG antibody at room temperature at 500 rpm for 1h. The secondary antibody was added to the mix and the sample was incubated for another hour on orbital shaker at 500 rpm. Five 8-OHdG standard complexes were injected in the CE-LIF instrument to generate the standard curve and further estimate the LOD.
Transgenic mice and genotyping
AD-Tg mice [B6.Cg-Tg, stock no. 005864, Bar Harbor, ME] were purchased from Jackson Laboratory. This model has both amyloid precursor protein (APP) and presenilin (PSEN) mutations (cite original paper describing model). Real time polymerase chain reaction (qPCR) and melting curve analysis (HRM) were used for the genotyping of the AD-Tg mice (data not shown) [33]. The primer sets for APP transgenic gene are (forward 5′-GAC TGA CCA CTC GAC CAG GTT CTG-3′, reverse 5′-CTT GTA AGT TGG ATT CTC ATA TCC G-3′) [34]. DNA was extracted by immersing mouse tail snip in 25 mM NaOH/0.2 mM EDTA at 98 °C for 1 hour [35]. The sequence of APP gene was amplified by quantitative PCR and then analyzed by melting curve analysis using LC 32 Scanner (Idaho Technology, Utah). The melting temperature of App gene is 86 ± 0.6 °C [36].
Urine collection and labeling
Urinary samples were collected from male, 11-months-old, AD-Tg mice. After collection, the samples were centrifuged for 10 min at 4000 rpm and the supernatant was collected and stored at −20°C for further analysis. In order to detect 8-OHdG in the AD mice urine samples, 20 μL urine sample was incubated with 20 μL mix of the primary (40 ng·μL−1) and 10 μL secondary antibody (40 ng·μL−1) for 1h on orbital shaker, 500 rpm, at room temperature.
Separation Condition
Sodium tetraborate buffer (pH 9.5) was used as the carrier electrolyte for all determinations. Samples were injected into the capillary containing the separation buffer using hydrodynamic injection mode at pressure of 3.4 kPa with 5 sec injection time. The separations were performed at 17 kV (400 V/cm) [37]. Measurements were carried out at constant temperature 25°C. The capillaries were preconditioned using methanol for 5 min, followed by 1M NaOH for 10 min, 1M HCl for 10 min, water for 10 min, and 20 mM sodium tetraborate buffer for 10 min. After each run the capillaries were washed with 1M NaOH for 5 min, followed by water for 2 min and sodium tetraborate buffer for 3 min.
Data analysis
Igor Pro Software (Lakemetrics, Lake Oswego, OR), was used to analyze the electropherograms. This program provides intensity values at each migration time point. The fluorescence intensity values detected in the Alexa 488 channel were used to calculate the area associated with the 8-OHdG concentration using the in house written Igor procedure, wide peak analysis [38].
Results and Discussion
We used a procedure for the analysis of 8-OHdG comprising immunoaffinity and indirect fluorescence labeling of 8-OHdG, their separation by CE, and on-line detection by LIF. To monitor the damage to the DNA base, guanine, in transgenic AD mice, we developed an assay that utilizes the very low sensitivity of the capillary electrophoresis coupled with laser induced fluorescence detection. In addition, this assay takes advantage of the specificity provided by the monoclonal antibody N45.1 to 8-OHdG [39]. Preparation of urine was limited to centrifugation, incubation with the antibody, and capillary electrophoresis separation.
Calibration curve using 8-OHdG
To test the feasibility of the assay, we used N45.1 mouse monoclonal antibody that is specific to 8-OHdG [39, 40]. First of all, capillary electrophoresis validated no presence of impurities in primary antibody (Fig. 2 trace 5). Next, it separated the unbound secondary antibody at 135 s and 278 s (Fig. 2 trace 4), and a complex of the secondary antibody along with primary antibody (Fig. 2 trace 3). In addition, a complex of 35 μM 8-OHdG with primary and the secondary antibodies was analyzed for generating a calibration curve (Fig. 2 trace 2). Furthermore, the 3530 μM 8-OHdG complex peak was determined in this study by comparison of the migration time and spiking at 162 s. (Fig. 2 trace 1).
Figure 2.
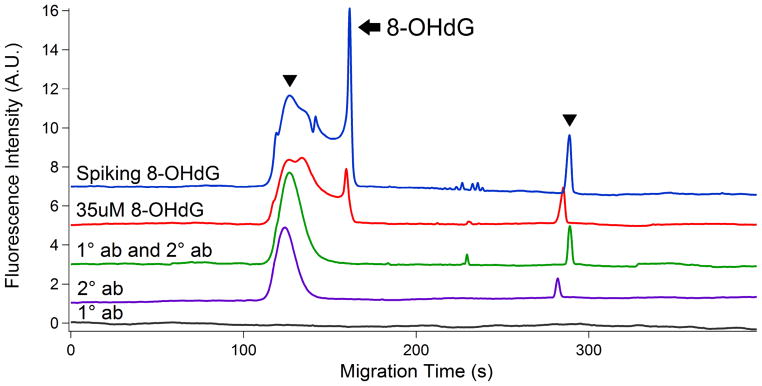
Electropherograms of validation tests in positive and negative controls. (1) positive control #1-- a complex with 3530 uM 8-OHdG standard; (2) Positive control #2 -- a complex with 35 uM 8-OHdG standard; (3) Negative control #1-- a complex with primary and secondary antibody; (4) Negative control #2 -- secondary antibody alone; (5) Negative control #3 -- primary antibody alone. Hydrodynamic injection at pH 9.5 and 3.4 kPa for 5 s; separation in 20 mM sodium tetraborate buffer at 17 kV. Arrowhead: secondary antibody; Arrow: 8-OHdG. Top trace is offset in the y-axis for the clarity.
To estimate the LOD, a calibration curve was constructed by plotting the measurement of the average peak area (y) versus the amount of 8-OHdG (x) in Figure 3. Five standards of the complex of 8-OHdG, primary antibody and secondary antibody were used to generate the standard curve. The five-point calibration from 7 μM to 35 μM showed the linear detection of 8-OHdG with correlation coefficient of 0.96. The injection volume was calculated based on the Hagen-Poiseuille equation, where the values used in this method were the pressure difference (3.4 kPa), the capillary radius (25 μm), the solution viscosity (0.00089 Pa·s), the capillary length (42 cm) and the injection time (5 s), which gives 7.07 nL of the volume of injection. In addition, the LOD and limit of quantitation (LOQ) were estimated as the concentration that gives the signal to noise ratio of 3 and 10, which gives the LOD and LOQ of 8-OHdG are 0.18 and 0.6 fmol. The equation for the regression line is:
| (1) |
Figure 3.
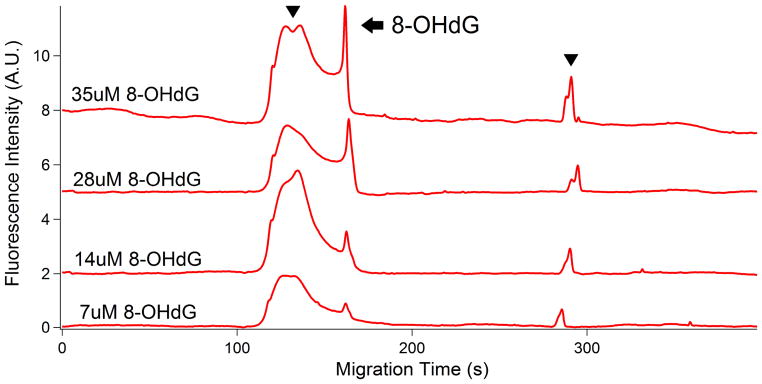
Electropherograms of calibration curve of 8-OHdG standard with different concentrations. Arrowhead: secondary antibody; Arrow: 8-OHdG. All the experimental conditions are the same as in Figure 2. Top trace is offset in the y-axis for the clarity.
3.2 Analysis of 8-OHdG in AD mice urine sample
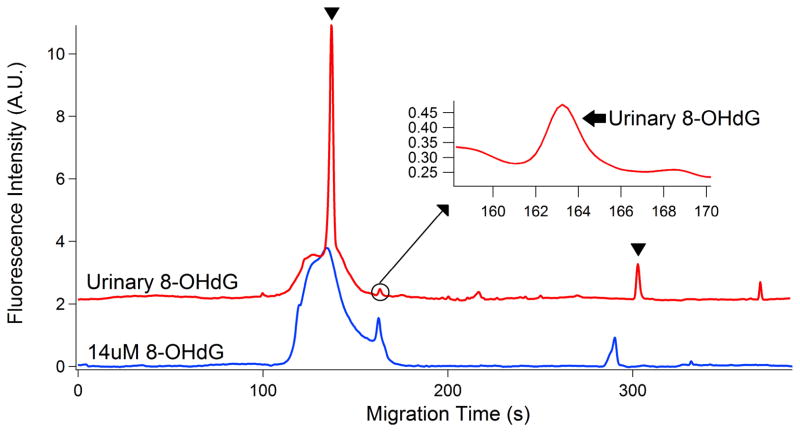
To illustrate the potential of this novel analytical method, we analyzed levels of 8-OHdG in AD-Tg mice urine. We performed experiments using urinary samples from 11-month-old, male, transgenic mice. The electropherogarms of the 8-OHdG in urine and standard is shown in Figure 4. This is a very fast and sensitive method with overall analytical run time for 10 min. The peaks at 164 s indicated the identification of 8-OHdG in urine sample. A good separation was obtained for 8-OHdG complex from secondary antibody and complex of primary and secondary antibody. Based on the Eq. (1), and the measured peak area for the 8-OHdG peak, the total 8-OHdG content in the injection volume for AD-Tg mice urine was calculated as (12.8 ± 0.18) fmole. For the CE analysis, only 7.07 nL volumes of samples were hydrodynamically injected, which leads to urinary concentration of 8-OHdG of (1.82 ± 0.02) μM.
Figure 4.
Electropherograms of urine sample and 8-OHdG standard. Arrowhead: secondary antibody; Arrow: 8-OHdG. All the experimental conditions are the same as in Figure 2. Top trace is offset in the y-axis for the clarity.
The critical issue in detecting 8-OHdG in urinary sample is to avoid interference from other compounds. This is achieved by using antibody that has high selectivity for 8-OHdG compound. Among 500 papers published on urinary 8-OHdG detection, ELISA method has been used primarily. There is ongoing debate whether the kit is specific to recognize other urinary components than 8-OHdG. Toyokuni’s group stated that the antibody N45.1 is highly specific to 8-OHdG, showing weak or no cross reactivity with urine component such as uric acid, urea, creatine and creatinine [39]. Although it can recognize both the modified base (8-oxoguanine) as well as the deoxyribose structure, the modified base has to be at least two orders of magnitude higher in concentration to be a competitor for the monoclonal antibody [39].
The urinary excretion of damaged nucleotides, 8-OHdG from both mtDNA and nDNA serves as an essential biomarker of oxidative stress reflecting the rate of damage in steady state. We have demonstrated this sensitive method combining both CE-LIF separation and immuno-affinity characteristics is capable to study low abundant 8-OHdG in small amount of urine sample. Because 8-OHdG is considered as a biomarker of oxidative DNA damage, we cannot exclusively conclude that the results are only related to AD. However, it allows us to determine oxidative status. Moreover, our results clearly show the screening capability of 8-OHdG using this new developed method from extracellular matrix (urine). The potential of using this method as a diagnostic biomarker in AD progression in a larger set of subjects deserves further study. Additionally, this method can serve as a tool to monitor the oxidative stress response of AD subjects as a function of antioxidant and other treatments.
3.3. Assay repeatability and accuracy
The reproducibility of this method was evaluated with both injection and analytical reproducibility. To assess injection consistency, five replicate injections of labeled 8-OHdG were examined, while five different labeled 8-OHdG samples prepared by the same procedure were studied for analytical repeatability. In the measurement of analysis repeatability, the relative standard deviation (RSD) values for migration time and peak area were 11.32 % and 5.52 %, respectively. In addition, day-to-day assays by replicate injection of the 8-OHdG solution during five consecutive days, were examined to study precision of the method. The RSD values of migration time and peak area were 19.3 % and 24.3 %, respectively.
The accuracy was determined by performing a standard spiking method, where known amounts of labeled 8-OHdG standard was mixed with previous analyzed 8-OHdG and further detected by CE-LIF method in triplicate. The 8-OHdG LOD of 0.18 fmol was subsequently determined by the calibration curve. The percentage of the spiked 8-OHdG recovered by the assay indicates the accuracy of the method was 93.14 %.
3.4 Optimization of separation conditions
For our developed method, even if similar compounds in the urine cross reacted with the antibody, CE with high resolution and high sensitivity offers great promise to separate these interferences. Borate buffer has been widely used to analyze 8-OHdG due to the pH sensitivity and flexibility so that 8-OHdG ionization can be affected in basic or acidic solution. Particularly, mobility of 8-OHdG has been improved in basic separation condition [29]. Weiss et al. demonstrated that 20 mM borate buffer had better 8-OHdG resolution with pH levels of 5.5–9.5, compared to Zwitterionic buffers [41]. Because optimizing pH values affects analyte ionization and enhances sample peak shape and area, the resolution can be improved by a stable pH level of running buffer [42]. It was reported that the resolution of 8-OHdG was poor when pH value was below 8.4. However, the resolution was enhanced while pH was above 9.0 [43]. For the capillary zone electrophoresis, the borate buffer (a mixture of sodium tetraborate and boric acid) at pH 9.5 was employed. Because at this pH level, the boric acid proton dissociation occurs, which further leads to an increase in coupling sugar moiety of 8-OHdG with boric acid [29]. However, both urea and creatinine are uncharged species, the transport of which is controlled by the electroosmotic flow [29]. Therefore, the 8-OHdG and other possible interference can be separated by charge to size ratio. The buffer pH can adjust separation by changing the charge of the analytes. In addition, the concentration of the running buffer has an impact on the electo-osmotic flow (EOF), for instance, decreasing buffer concentration can increase the EOF, which leads to faster migration time, lower electrophoretic current and Joule heat. It was found that the 20 mM borate buffer minimize the effect of Joule heat [44].
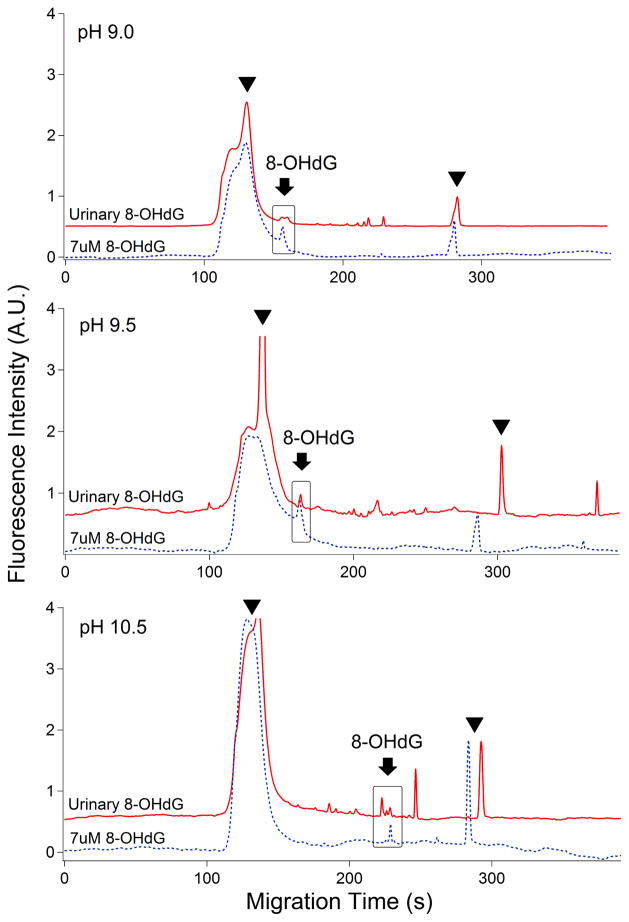
Therefore, to achieve the optimal separation efficiency, 20 mM sodium tetraborate was tested by detecting the 8-OHdG standard complex (7 uM 8-OHdG) and urinary 8-OHdG complex in a range of pH 9.0, 9.5, and 10.5. The separation was improved when pH increased to 9.5. However, 8-OHdG could not be separated from the urinary complex when pH reached to 10.5 shown in Figure 5. We found that 20 mM sodium tetraborate of pH 9.5 was the optimal separation condition under optimal separation voltage (17 kV).
Figure 5.
Electropherograms of the separation efficiency of 8-OHdG standard and urinary 8-OHdG complexes with 20 mM sodium tetraborate buffer at pH 9.0 (top panel); pH 9.5 (middle panel); and pH 10.5 (bottom panel). Arrowhead: secondary antibody; Arrow: 8-OHdG. All the experimental conditions are the same as in Figure 2. Top trace is offset in the y-axis for the clarity.
Conclusions
We developed a CE-LIF system for detection of urinary 8-OHdG. This method demonstrates the detection and screening of 8-OHdG using immuno-affinity labeling coupled with CE-LIF. Our method is unique in that, (1) it reaches high specificity with using CE separation method; also (2) it keeps the ease of use provides by immunoaffinity concept. The sample preparation can be completely excluded, because it requires no solid phase extraction for urine. The ultrasensitive assay described here is not limited to detection of 8-OHdG and can be expanded to other oxidative lesion when appropriate affinity probes are available. Likewise, it has an enormous potential for high throughput clinical applications as well as direct monitoring for the early diagnosis and monitoring DNA oxidative stress progression.
Acknowledgments
This study was supported by the National Institutes of Health and the National Center for Research Resources Grant P20RR016456. Special thanks to Dr. Edgar Arriaga’s group for providing their house written Wide Peak analysis software in our study. We also thank Dr. Spaulding for his great assistance on animal care.
Footnotes
Conflict of Interest The authors state that there are no actual or potential conflicts of interest.
References
- 1.Mohammad Arif YK, Hassan Faizule, Zaved Waise TM, Ehsanul Hoque Mazumder Md, Rahman Shafiqur. Increased DNA damage in blood cells of rat treated with lead as assessed by comet assay. Bangladesh J Pharmacological. 2008;3:97–101. [Google Scholar]
- 2.Lovell MA, Markesbery WR. Oxidative DNA damage in mild cognitive impairment and late-stage Alzheimer’s disease. Nucleic Acids Res. 2007;35:7497–7504. doi: 10.1093/nar/gkm821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G----T and A----C substitutions. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 4.Cadet J, Berger M, Douki T, Ravanat JL. Oxidative damage to DNA: formation, measurement, and biological significance. Rev Physiol Biochem Pharmacol. 1997;131:1–87. doi: 10.1007/3-540-61992-5_5. [DOI] [PubMed] [Google Scholar]
- 5.Kondo T, Ohshima T, Ishida Y. Age-dependent expression of 8-hydroxy-2′-deoxyguanosine in human pituitary gland. Histochem J. 2001;33:647–651. doi: 10.1023/a:1016354417834. [DOI] [PubMed] [Google Scholar]
- 6.Fraga CG, Shigenaga MK, Park JW, Degan P, Ames BN. Oxidative damage to DNA during aging: 8-hydroxy-2′-deoxyguanosine in rat organ DNA and urine. Proc Natl Acad Sci U S A. 1990;87:4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci U S A. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mecocci P, MacGarvey U, Kaufman AE, Koontz D, Shoffner JM, Wallace DC, Beal MF. Oxidative damage to mitochondrial DNA shows marked age-dependent increases in human brain. Ann Neurol. 1993;34:609–616. doi: 10.1002/ana.410340416. [DOI] [PubMed] [Google Scholar]
- 9.Lovell MA, Markesbery WR. Oxidative damage in mild cognitive impairment and early Alzheimer’s disease. J Neurosci Res. 2007;85:3036–3040. doi: 10.1002/jnr.21346. [DOI] [PubMed] [Google Scholar]
- 10.Noseworthy MD, Bray TM. Effect of oxidative stress on brain damage detected by MRI and in vivo 31P-NMR. Free Radic Biol Med. 1998;24:942–951. doi: 10.1016/s0891-5849(97)00383-3. [DOI] [PubMed] [Google Scholar]
- 11.Lovell MA, Gabbita SP, Markesbery WR. Increased DNA oxidation and decreased levels of repair products in Alzheimer’s disease ventricular CSF. J Neurochem. 1999;72:771–776. doi: 10.1046/j.1471-4159.1999.0720771.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Markesbery WR, Lovell MA. Increased oxidative damage in nuclear and mitochondrial DNA in mild cognitive impairment. J Neurochem. 2006;96:825–832. doi: 10.1111/j.1471-4159.2005.03615.x. [DOI] [PubMed] [Google Scholar]
- 13.Markesbery WR, Lovell MA. DNA oxidation in Alzheimer’s disease. Antioxid Redox Signal. 2006;8:2039–2045. doi: 10.1089/ars.2006.8.2039. [DOI] [PubMed] [Google Scholar]
- 14.Lyras L, Cairns NJ, Jenner A, Jenner P, Halliwell B. An assessment of oxidative damage to proteins, lipids, and DNA in brain from patients with Alzheimer’s disease. J Neurochem. 1997;68:2061–2069. doi: 10.1046/j.1471-4159.1997.68052061.x. [DOI] [PubMed] [Google Scholar]
- 15.Mecocci P, MacGarvey U, Beal MF. Oxidative damage to mitochondrial DNA is increased in Alzheimer’s disease. Ann Neurol. 1994;36:747–751. doi: 10.1002/ana.410360510. [DOI] [PubMed] [Google Scholar]
- 16.Mecocci P, Polidori MC, Cherubini A, Ingegni T, Mattioli P, Catani M, Rinaldi P, Cecchetti R, Stahl W, Senin U, Beal MF. Lymphocyte oxidative DNA damage and plasma antioxidants in Alzheimer disease. Arch Neurol. 2002;59:794–798. doi: 10.1001/archneur.59.5.794. [DOI] [PubMed] [Google Scholar]
- 17.Sakumi K, Furuichi M, Tsuzuki T, Kakuma T, Kawabata S, Maki H, Sekiguchi M. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J Biol Chem. 1993;268:23524–23530. [PubMed] [Google Scholar]
- 18.Wiseman H, Halliwell B. Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochem J. 1996;313 (Pt 1):17–29. doi: 10.1042/bj3130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasai H, Nishimura S. Hydroxylation of deoxyguanosine at the C-8 position by ascorbic acid and other reducing agents. Nucleic Acids Res. 1984;12:2137–2145. doi: 10.1093/nar/12.4.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michael C, Peoples HTK. Recent developments in analytical methodology for8-hydroxy-2-deoxyguanosine and related compounds. Journal of Chromatography B. 2005;827:5–15. doi: 10.1016/j.jchromb.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Cooke MS, Olinski R, Loft S. Measurement and meaning of oxidatively modified DNA lesions in urine. Cancer Epidemiol Biomarkers Prev. 2008;17:3–14. doi: 10.1158/1055-9965.EPI-07-0751. [DOI] [PubMed] [Google Scholar]
- 22.Yin B, Whyatt RM, Perera FP, Randall MC, Cooper TB, Santella RM. Determination of 8-hydroxydeoxyguanosine by an immunoaffinity chromatography-monoclonal antibody-based ELISA. Free Radic Biol Med. 1995;18:1023–1032. doi: 10.1016/0891-5849(95)00003-g. [DOI] [PubMed] [Google Scholar]
- 23.Cadet J, Douki T, Ravanat JL. Artifacts associated with the measurement of oxidized DNA bases. Environ Health Perspect. 1997;105:1034–1039. doi: 10.1289/ehp.105-1470384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravanat JL, Duretz B, Guiller A, Douki T, Cadet J. Isotope dilution high-performance liquid chromatography-electrospray tandem mass spectrometry assay for the measurement of 8-oxo-7,8-dihydro-2′-deoxyguanosine in biological samples. J Chromatogr B Biomed Sci Appl. 1998;715:349–356. doi: 10.1016/s0378-4347(98)00259-x. [DOI] [PubMed] [Google Scholar]
- 25.Floyd RA, Watson JJ, Wong PK, Altmiller DH, Rickard RC. Hydroxyl free radical adduct of deoxyguanosine: sensitive detection and mechanisms of formation. Free Radic Res Commun. 1986;1:163–172. doi: 10.3109/10715768609083148. [DOI] [PubMed] [Google Scholar]
- 26.Bogdanov MB, Beal MF, McCabe DR, Griffin RM, Matson WR. A carbon column-based liquid chromatography electrochemical approach to routine 8-hydroxy-2′-deoxyguanosine measurements in urine and other biologic matrices: a one-year evaluation of methods. Free Radic Biol Med. 1999;27:647–666. doi: 10.1016/s0891-5849(99)00113-6. [DOI] [PubMed] [Google Scholar]
- 27.Weimann A, Belling D, Poulsen HE. Quantification of 8-oxo-guanine and guanine as the nucleobase, nucleoside and deoxynucleoside forms in human urine by high-performance liquid chromatography-electrospray tandem mass spectrometry. Nucleic Acids Res. 2002;30:E7. doi: 10.1093/nar/30.2.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renner T, Fechner T, Scherer G. Fast quantification of the urinary marker of oxidative stress 8-hydroxy-2′-deoxyguanosine using solid-phase extraction and high-performance liquid chromatography with triple-stage quadrupole mass detection. J Chromatogr B Biomed Sci Appl. 2000;738:311–317. doi: 10.1016/s0378-4347(99)00542-3. [DOI] [PubMed] [Google Scholar]
- 29.Kvasnicova V, Samcova E, Jursova A, Jelinek I. Determination of 8-hydroxy-2′-deoxyguanosine in untreated urine by capillary electrophoresis with UV detection. J Chromatogr A. 2003;985:513–517. doi: 10.1016/s0021-9673(02)01527-3. [DOI] [PubMed] [Google Scholar]
- 30.Osbourn DM, Weiss DJ, Lunte CE. On-line preconcentration methods for capillary electrophoresis. Electrophoresis. 2000;21:2768–2779. doi: 10.1002/1522-2683(20000801)21:14<2768::AID-ELPS2768>3.0.CO;2-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss DJ, Lunte CE. Detection of a urinary biomaker for oxidative DNA damage 8-hydroxydeoxyguanosine by capillary electrophoresis with electrochemical detection. Electrophoresis. 2000;21:2080–2085. doi: 10.1002/1522-2683(20000601)21:10<2080::AID-ELPS2080>3.0.CO;2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mei SR, Yao QH, Cai LS, Xing J, Xu GW, Wu CY. Capillary electrophoresis with end-column amperometric detection of urinary 8-hydroxy-2′-deoxyguanosine. Electrophoresis. 2003;24:1411–1415. doi: 10.1002/elps.200390181. [DOI] [PubMed] [Google Scholar]
- 33.Zhang C, Rodriguez C, Spaulding J, Aw TY, Feng J. Age-dependent and tissue-related glutathione redox status in a mouse model of Alzheimer’s disease. J Alzheimers Dis. 2012;28:655–666. doi: 10.3233/JAD-2011-111244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Q, Zhang X, Chen J, Miao Y, Sun A. Role of caspase-3 in tau truncation at D421 is restrixted in transgenic mouse models for taupathies. Journal of Neurochemistry. 2009;109:476–484. doi: 10.1111/j.1471-4159.2009.05959.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang C, Kuo CC, Chiu AW, Feng J. Prediction of S-glutathionylated proteins progression in Alzheimer’s transgenic mouse model using principle component analysis. J Alzheimers Dis. 2012;30:919–934. doi: 10.3233/JAD-2012-120028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang C, Rodriguez C, Circu ML, Aw TY, Feng J. S-Glutathionyl quantification in the attomole range using glutaredoxin-3-catalyzed cysteine derivatization and capillary gel electrophoresis with laser-induced fluorescence detection. Anal Bioanal Chem. 2011;401:2165–2175. doi: 10.1007/s00216-011-5311-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muth J, Williams PM, Williams SJ, Brown MD, Wallace DC, Karger BL. Fast capillary electrophoresis-laser induced fluorescence analysis of ligase chain reaction products: human mitochondrial DNA point mutations causing Leber’s hereditary optic neuropathy. Electrophoresis. 1996;17:1875–1883. doi: 10.1002/elps.1150171212. [DOI] [PubMed] [Google Scholar]
- 38.Feng J, Navratil M, Thompson LV, Arriaga EA. Principal component analysis reveals age-related and muscle-type-related differences in protein carbonyl profiles of muscle mitochondria. J Gerontol A Biol Sci Med Sci. 2008;63:1277–1288. doi: 10.1093/gerona/63.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toyokuni S, Tanaka T, Hattori Y, Nishiyama Y, Yoshida A, Uchida K, Hiai H, Ochi H, Osawa T. Quantitative immunohistochemical determination of 8-hydroxy-2′-deoxyguanosine by a monoclonal antibody N45.1: its application to ferric nitrilotriacetate-induced renal carcinogenesis model. Lab Invest. 1997;76:365–374. [PubMed] [Google Scholar]
- 40.de Carvalho LF, Abrao MS, Biscotti C, Sharma R, Agarwal A, Falcone T. Mapping histological levels of 8-hydroxy-2′-deoxyguanosine in female reproductive organs. J Mol Histol. 2012;44:111–116. doi: 10.1007/s10735-012-9454-7. [DOI] [PubMed] [Google Scholar]
- 41.Weiss DJ, Lunte CE. Detection of a urinary biomaker for oxidative DNA damage 8-hydroxydeoxyguanosine by capillary electrophoresis with electrochemical detection. Electrophoresis. 2000;21:2080. doi: 10.1002/1522-2683(20000601)21:10<2080::AID-ELPS2080>3.0.CO;2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao Q-H, Mei S-R, Weng Q-F, Zhang P-d, Yang Q, Wu C-y, Xu G-W. Determination of urinary oxidative DNA damage marker 8-hydroxy-2′-deoxyguanosine and the association with cigarette smoking. Talanta. 2004;63:617–623. doi: 10.1016/j.talanta.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 43.Li MJ, Zhang JB, Li WL, Chu QC, Ye JN. Capillary electrophoretic determination of DNA damage markers: content of 8-hydroxy-2′-deoxyguanosine and 8-nitroguanine in urine. J Chromatogr B Analyt Technol Biomed Life Sci. 879:3818–3822. doi: 10.1016/j.jchromb.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 44.Ullrich O, Grune T. Detection of 8-hydroxydeoxyguanosine in K562 human hematopoietic cells by high-performance capillary electrophoresis. J Chromatogr B Biomed Sci Appl. 1997;697:243–249. doi: 10.1016/s0378-4347(97)00151-5. [DOI] [PubMed] [Google Scholar]