Abstract
The ING family of tumor suppressors acts as readers and writers of the histone epigenetic code, affecting DNA repair, chromatin remodeling, cellular senescence, cell cycle regulation and apoptosis. The best characterized member of the ING family, ING1, interacts with the proliferating cell nuclear antigen (PCNA) in a UV-inducible manner. ING1 also interacts with members of the 14-3-3 family leading to its cytoplasmic relocalization. Overexpression of ING1 enhances expression of the Bax gene and was reported to alter mitochondrial membrane potential in a p53-dependent manner. Here we show that ING1 translocates to the mitochondria of primary fibroblasts and established epithelial cell lines in response to apoptosis inducing stimuli, independent of the cellular p53 status. The ability of ING1 to induce apoptosis in various breast cancer cell lines correlates well with its degree of translocation to the mitochondria after UV treatment. Endogenous ING1 protein specifically interacts with the pro-apoptotic BCL2 family member BAX, and colocalizes with BAX in a UV-inducible manner. Ectopic expression of a mitochondria-targeted ING1 construct is more proficient in inducing apoptosis than the wild type ING1 protein. Bioinformatic analysis of the yeast interactome indicates that yeast ING proteins interact with 64 mitochondrial proteins. Also, sequence analysis of ING1 reveals the presence of a BH3-like domain. These data suggest a model in which stress-induced cytoplasmic relocalization of ING1 by 14-3-3 induces ING1-BAX interaction to promote mitochondrial membrane permeability and represent a paradigm shift in our understanding of ING1 function in the cytoplasm and its contribution to apoptosis.
Keywords: apoptosis, ING1, mitochondria, BAX
The Inhibitor of Growth1 (ING1) tumor suppressor1 is the founding member of a family of five genes (ING1–5) with conserved plant homeodomains (PHDs).2 ING proteins are implicated in the regulation of cellular senescence,3, 4, 5, 6 chromatin remodeling,7, 8, 9 differentiation,10, 11 DNA damage response,12, 13, 14, 15 cell cycle regulation16, 17, 18 and apoptosis.19, 20, 21, 22, 23, 24 Although mutations in the ING1 gene are rare, diverse human cancers show reduced levels of ING1 protein and in some cases, localization in the cytoplasm, thereby supporting its classification as a type II tumor suppressor.25, 26, 27, 28, 29, 30, 31 Ectopic expression of ING1 also induces apoptosis,19 and although initial reports suggested this role may be p53-dependent,20, 32, 33 more recent evidence suggest that the ING family also has effects on apoptosis that are independent of p53.34, 35, 36, 37 ING1 interacts with proliferating cell nuclear antigen (PCNA) via the PCNA-interacting protein (PIP) domain in a stress-induced manner.13 The PIP domain is necessary for the ability of ING1 to maximally induce apoptosis upon overexpression and in response to DNA damage.13 ING1 also interacts with a PCNA-interacting protein, p15(PAF).38 The endogenous p15(PAF) protein localizes both to the nucleus and the mitochondria.38 The nuclear localization signal (NLS) of ING1 contains three tandemly repeated nucleolar-targeting sequences (NTSs), two of which target ING1 to the nucleoli in response to stresses.12 The NTSs are also required for ING1 apoptotic function. Recently, the cellular senescence-inhibited gene CSIG protein was identified as a binding partner for ING1 in the nucleolus.24
ING proteins bind to and regulate the activities of histone acetyl transferase (HAT) and histone deacetylase (HDAC) chromatin remodeling complexes4, 7, 39, 40, 41 that are responsible for modulating gene expression patterns in response to a variety of stresses, suggesting that ING proteins may have a general role in mediating the cellular response to genotoxic stress.42 This occurs, in part, by the PHD of ING proteins reading the ‘histone code' in a methylation-dependent manner.9, 43, 44 Binding of ING PHDs to trimethylated histones recruits the HDAC complexes to the promoters of proliferation-promoting genes, thus leading to gene repression in response to damage-inducing stimuli. This interaction has also been shown to be important for the DNA repair and apoptotic functions of ING1.9 ING1 interacts with members of the 14-3-3 family, and phosphorylation of Ser199 of ING1 is necessary for this interaction.45 However, although the ING1-14-3-3 interaction has been shown to be necessary for the cytoplasmic localization of ING1, the significance of this nuclear–cytoplasmic relocalization is not fully understood.
The mammalian apoptosis machinery consists of two partially distinct pathways: the intrinsic and the extrinsic pathways. The extrinsic pathway is heavily influenced by the FAS death receptor, a member of the tumor necrosis factor (TNF) receptor superfamily,46 whereas the intrinsic pathway involves the mitochondria as a central factor. When stimulated, the intrinsic pathway leads to the release of cytochrome c from the mitochondria and formation of the apoptosome consisting of cytochrome c, APAF-1 and pro-caspase 9 (reviewed in Letai47 and Brenner and Mak48) Both the extrinsic and intrinsic pathways converge with the cleavage and activation of pro-caspase-3 and imminent cell death.
Death-inducing stimuli, including DNA damage, growth-factor deprivation, oncogene activation and cell signaling pathway perturbations, are communicated through the intrinsic apoptotic pathway. The B-cell CLL/lymphoma 2 (BCL-2) family of proteins are important regulators of this apoptotic pathway.47 BCL-2 family proteins regulate the permeabilization of the outer mitochondrial membrane in response to apoptosis-inducing signals and share homology in 4 α-helical BCL-2-homology (BH) regions (BH1–4), through which they also interact physically. BCL-2-associated X protein (BAX)49 and BCL-2-antagonist/killer (BAK)50 share only the BH1–3 domains and are pro-apoptotic members of the BCL-2 family. Another class of pro-apoptotic BCL-2 family members contains the BAX Homology domain 3 (BH3) domain alone. Prominent members of this class include BH3-interacting domain death agonist (BID), BCL-2 agonist of cell death (BAD), BIM (Bcl-2 Interacting Mediator of cell death), NOXA and PUMA (P53-Upregulated Modulator of Apoptosis).47 The BH3-only proteins are activated through incompletely understood signaling mechanisms; however, once activated, BH3-only proteins induce the activation of BAX and BAK.51, 52 BAX and BAK are absolutely essential for death induced by BH3 proteins, as the deletion of BAX and BAK leads to a profound inhibition of apoptosis in most tissues.53
p53 and ING1 have been shown to functionally cooperate in the activation of apoptosis and physical interactions between the two proteins have been reported.32, 37 Ectopic expression of ING1 can activate p53 target genes such as p21WAF1 and BAX in a number of transformed cell lines;21, 34 however, gene array studies performed with normal human diploid fibroblasts did not reveal significant activation of these genes in response to ING1 overexpression.23 As ING1 is a primarily nuclear protein that has been reported to relocalize to the cytoplasm, and p53 has been reported to be effective in inducing apoptosis when targeted to the mitochondria,54, 55 we investigated whether ING1 localizes to the mitochondria in response to apoptosis-inducing stimuli. Our data suggest a mechanism through which ING1 induces apoptosis by translocation from the nucleus to the mitochondria where interaction between a BAX BH3 motif and a putative ING1 BH3 domain stabilizes BAX, promoting its pro-apoptotic function.
Results
ING1 translocates to a mitochondrial locale after different forms of stress
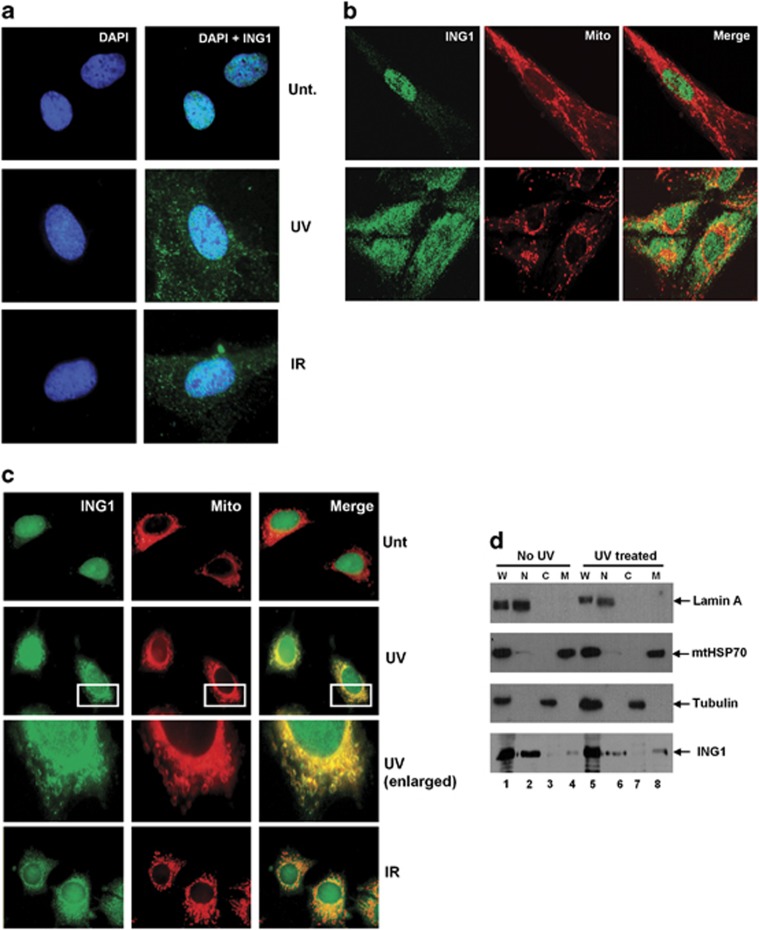
Immunofluorescence analysis of ING1 localization showed that a pool of ING1 translocated to the cytoplasm after UV and IR treatments in primary HS68 fibroblast cells (Figure 1a). As the distribution of ING1 resembled mitochondrial localization, we tested whether ING1 colocalized with the mitochondria. Initial time course experiments showed that by 6 h after UV or IR treatment, significant colocalization occurred (Figure 1b). As conditions for isolating mitochondria had been optimized in the MCF7 epithelial breast cancer cell line that harbors wild-type p53, translocation was examined in these cells. Translocation to the cytoplasm began 3 h after UV and IR treatments; however, maximum colocalization of ING1 and mitochondria was observed 6 h post treatment (Figure 1c). To examine subcellular localization by an independent method, nuclear, cytosolic and mitochondrial fractions with and without UV treatment were prepared from MCF7 cells and immunoblotted with anti-ING1, anti-mtHSP70 (mitochondrial marker), tubulin (cytoplasmic marker) and lamin A (nuclear marker) antibodies. Purity of the fractions was confirmed by the markers and a significant amount of ING1 appeared in the mitochondrial fraction after UV treatment (Figure 1d, lane 8 versus lane 4).
Figure 1.
UV and IR treatments induce ING1 translocation to the mitochondria of HS68 cells. (a) HS68 fibroblasts were either left untreated or irradiated with 60 J/m2 of UVC or 5 Gy of IR and fixed after 6 h. Cells were stained for ING1 and DNA. (b) HS68 fibroblasts were either left untreated or irradiated with 60 J/m2 of UVC and stained with MitoTracker before fixation. Cells were then stained for ING1 and DNA. (c) MCF7 cells were treated with 60 J/m2 UV and 5 Gy of IR. Cells were stained with MitoTracker 30 min before fixation. Cells were stained for ING1 following fixation. (d) MCF7 cells were fractionated into nuclear, cytosolic and mitochondrial fractions, and western blotting was performed with anti-lamin, anti-tubulin, anti-mtHSP70 and anti-ING1 antibodies
ING1 translocation to the mitochondria is independent of p53
Initial studies indicated that ING1 effectively blocked the growth of cells with functional p53,3 and a physical interaction between ING1 and p53 and functional interdependence was later reported.32 ING1 also enhanced levels of p53 acetylation on lysine 382 affecting p53 stability after stress, possibly by inhibiting hSIR2 that can deacetylate p53.34 However, in contrast to reports suggesting an obligatory link between ING1 and p53, a more recent study in knockout mice has indicated that the two proteins can function independently.35
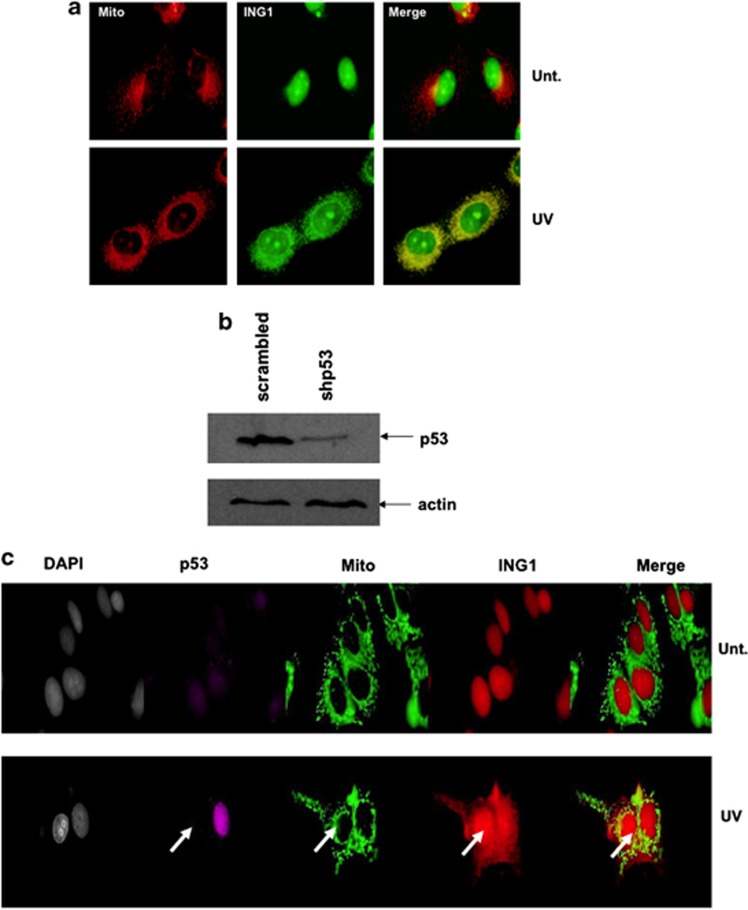
As contrasting reports exist regarding the possible interdependence of ING1 and p53, we asked whether p53 was required for ING1 translocation to the mitochondria in response to UV using H1299 p53 null cells. Figure 2a shows that ING1 translocation to the mitochondria after UV treatment can occur independently of p53. To further confirm that ING1 translocation to the mitochondria was independent of p53, we knocked down p53 in MCF7 cells using short hairpin RNA (shRNA) (Figure 2b). As shown by the arrows in Figure 2c, p53-depleted MCF7 cells showed no difference in ING1 translocation to the mitochondria after UV treatment when compared with cells having significant levels of p53, suggesting that the lack of dependence on p53 was not cell-type-specific.
Figure 2.
ING1 translocates to the mitochondria independent of p53. (a) H1299 cells were either left untreated or were treated with 60 J/m2 of UVC and co-stained for mitochondria and ING1. (b) shRNA knockdown of p53 in MCF7 cells. Cells were transfected either with control shRNA or shRNA against p53. Actin was used as loading control. (c) MCF7 cells were transfected with shRNA against p53 and were either treated with 60 J/m2 of UVC 48 h post transfection or left untreated. Cells were stained for mitochondria, p53, ING1 and DNA 6 h after UV treatment. Arrows indicate the position of a cell with depleted p53 levels as a result of shRNA transfection
The ability of ING1 to induce apoptosis correlates with the degree of mitochondrial translocation
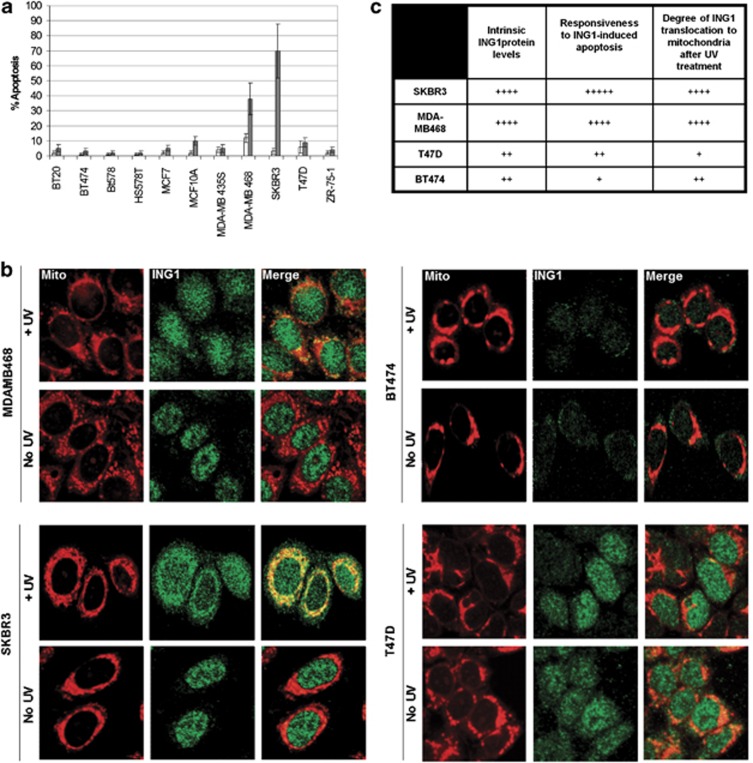
Examination of the ability of an adenoviral ING1 expression construct to induce apoptosis in a panel of 11 breast cancer cell lines showed that sensitivity varied widely as estimated by sub G1 DNA content, 48 h after infection (Figure 3a). To ask whether sensitivity to ING1-induced apoptosis might correlate with the degree of mitochondrial localization after stress, we selected the SKBR3 and MDA-MB468 cell lines that showed high sensitivity to ING1-induced apoptosis, and the BT474 and T47D lines that were significantly less sensitive to ING1-induced apoptosis and examined the degree of ING1 translocation after UV treatment. Figure 3b shows typical fields of the four cell lines before and after UV treatment, and Figure 3c summarizes the relationship between the degree of localization and sensitivity to ING1-induced apoptosis. As seen in Figures 3b and c, SKBR3 and MDA-MB468 cells showed high degrees of ING1 translocation to the mitochondria, whereas T47D and BT474 cells showed significantly less colocalization of ING1 with mitochondria following UV treatment. Thus, the degree of ING1 translocation to the mitochondria correlated very well with its ability to induce apoptosis in the tested cell lines.
Figure 3.
Translocation of ING1 to the mitochondria correlates well with its ability to induce apoptosis. (a) Differential sensitivity of breast cancer cell lines to ING1-induced apoptosis. Eleven breast cancer cell lines were tested for their ability to undergo ING1-induced apoptosis. BT20, BT474, Bt578, HS578T, MCF7, MCF10A, MDA-MB435S, MDA-MB468, SKBR3, T47D and ZR-75-1 breast cancer cells were infected with Ad-GFP or Ad-GFP-ING1 constructs. Apoptosis was estimated with propidium iodide staining for sub-G1 DNA content. SKBR3 and MDA-MB468 cells that responded well to ING1-induced apoptosis and T47D and BT474 cells that were significantly less responsive to ING1-induced apoptosis were chosen for further analysis. Results were obtained using an MOI of 10, 48 h after infection. Data are presented from three independent experiments. (b) Immunofluorescence analysis of ING1 translocation to the mitochondria of SKBR3, MDA-MB468, BT474 and T47D cell lines. All four cell lines were either left untreated or treated with 60 J/m2 of UVC and fixed after 6 h. MitoTracker was added to the media 0.5 h before fixation and cells were stained for ING1 and DNA. (c) Quantification of ING1 translocation to the mitochondria in relation with responsiveness to ING1-induced apoptosis. Translocation of ING1 to the mitochondria was quantified based on immunofluorescence analysis of SKBR3, MDA-MB-468, T47D and BT474 cells. A total of 100 cells were analyzed for each cell line in a blind experimental protocol in three independent experiments by two individuals. Data are presented in relation to the SKBR3 cell line, in which the maximum ING1 mitochondrial translocation (represented as ++++ based on visual estimation of immunofluorescence images) and the maximum relative responsiveness to ING1-induced apoptosis (represented as +++++ based on visual estimation of Figure 3a) were observed. ING1 protein levels were quantified using western blotting and scanning densitometry (data not shown)
ING1 interacts with BAX
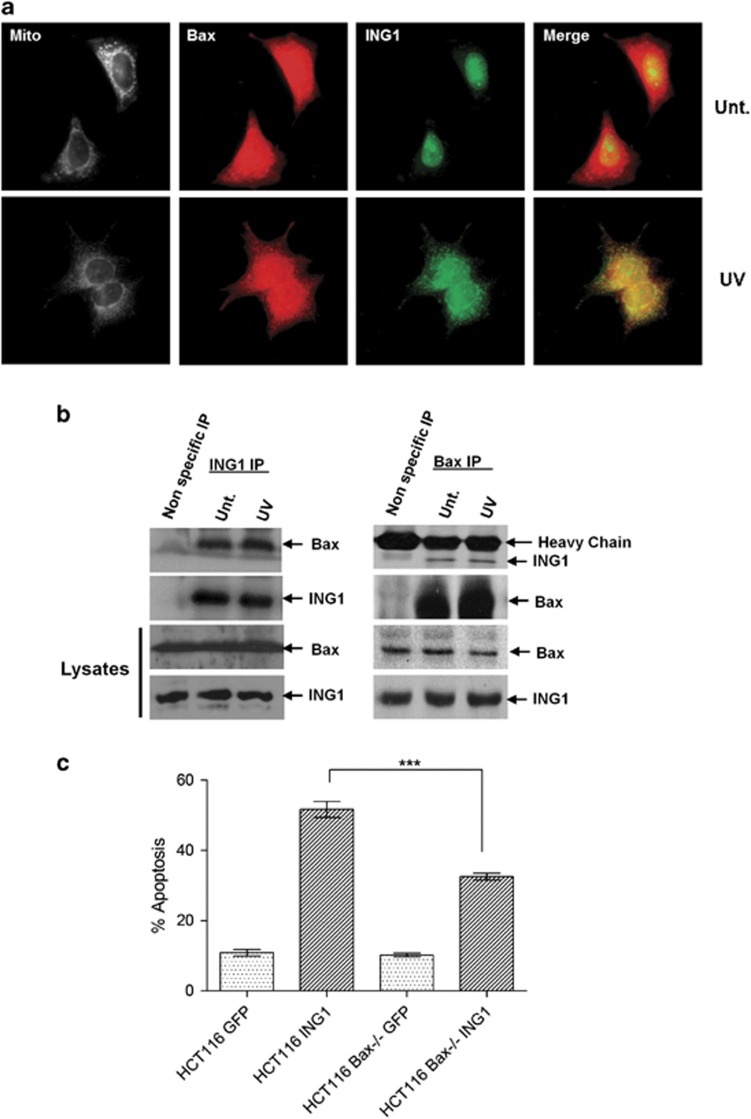
As ING1 has been reported to increase BAX levels21 and BAX is known to act at the mitochondrial membrane to induce apoptosis,56 we next asked whether ING1 colocalized with BAX. Immunofluorescence analysis showed that ING1 and BAX inducibly colocalize with mitochondria after UV treatment (Figure 4a). To further ask whether a physical interaction might occur between ING1 and BAX, we performed reciprocal co-immunoprecipitation–western assays in the absence and presence of UV treatment. We found that BAX is indeed present in ING1 but not in control immunoprecipitates (Figure 4b); however, contrary to expectations, the interaction appeared to be constitutive under the conditions used in IP-western analysis. As a pool of ING1 appears to relocalize to the mitochondria and bind to BAX, this suggested that ING1 binds to and possibly activates BAX for induction of apoptosis. As our results indicated that ING1-induced apoptosis might be BAX-dependent, we tested this by investigating the ability of ING1 to induce apoptosis in the absence of BAX. A significant decrease in ING1-induced apoptosis was observed in HCT116 BAX−/− cells relative to wild-type HCT116 cells after UV treatment (Figure 4c).
Figure 4.
BAX is required for ING1-induced apoptosis. (a) Immunofluorescence analysis of BAX-ING1 colocalization in MCF7 cells. Cells were either left untreated or were treated with 60 J/m2 of UVC and co-stained for mitochondria, ING1 and BAX. (b) Association between endogenous ING1 and BAX in MCF7 cells. MCF7 cell lysates were immunoprecipitated either with anti-GST antibody (non-specific IP) or with anti-ING1 antibody in the presence or absence of UV treatment. Reciprocal IPs were performed by immunoprecipitating MCF7 cell lysates either with rabbit IgG (nonspecific IP) or with anti-BAX antibody in the presence or absence of UV treatment. (c) Flowcytometry analysis of the dependence of ING1-induced apoptosis on cellular BAX levels. HCT116 wild-type and HCT116 BAX−/− cells were transfected with the above mentioned plasmids and treated with UV 48 h after transfection. Cells were stained with Annexin V and 7-AAD 6 h after UV treatment and were analyzed using flowcytometry within 30 min. Data are presented from four independent experiments (***P=0.0002)
Mitochondrially targeted ING1 is more effective in inducing apoptosis than wild-type ING1
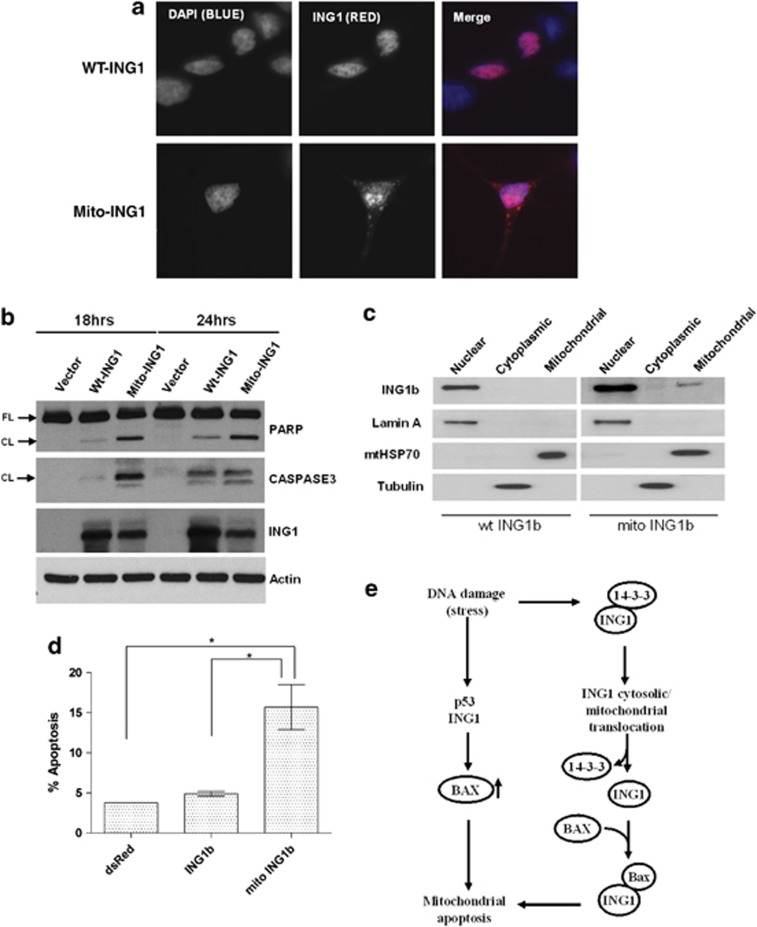
To directly test the possibility that mitochondrial localization of ING1 is necessary for its ability to optimally induce apoptosis, we investigated whether targeting ING1 to the mitochondria was more effective in inducing apoptosis than wild-type (nuclear) ING1. Figure 5a shows representative micrographs of wild-type and mitochondrially targeted ING1 in HEK293 cells. Wild-type ING1 localized exclusively to the nucleus on overexpression. Although nuclear localization persisted for mitochondrially targeted ING1, there was a remarkable increase in cytoplasmic localization with the targeted construct. As previously reported for p53,54 ING1 targeted to the mitochondria induced apoptosis at earlier time points compared with wild-type ING1 as assayed by levels of cleaved Caspase3 and PARP (Figure 5b). Similar results were obtained in MDA-MB468 breast cancer cells where fractionation experiments confirmed effective mitochondrial targeting (Figure 5c) and an increased ability to induce apoptosis compared with wild-type ING1 (Figure 5d). Thus, affinity for mitochondria directly affects the ability of ING1 to induce apoptosis.
Figure 5.
Mitochondrially targeted ING1 is more efficient at inducing apoptosis than wild-type ING1. (a) Representative immunofluorescence images of HEK293 cells transfected with wild-type and mitochondrially targeted ING1. (b) Western blot analysis of Caspase 3 and PARP cleavage in response to the overexpression of wild-type and mitochondrially targeted ING1. HEK293 cells were transfected with empty vector, wild-type ING1 or mitochondrially targeted ING1 constructs. Cell lysates were blotted for ING1, cleaved-Caspase 3 and PARP. Actin was used as a loading control. (c) Western blots analysis comparing the localization of wild-type and mitochondrially targeted ING1 in MDA-MB468 cells. Cells were transfected with wild-type and mitochondrially targeted ING1 constructs and harvested after 12 h. Cells were fractionated into nuclear, cytoplasmic and mitochondrial fractions, and western blotting was performed with anti-lamin, anti-tubulin, anti-mtHSP70 and anti-ING1 antibodies. (d) Flowcytometry analysis of the ability of mitochondrially targeted ING to induce apoptosis. MDA-MB468 cells were transfected with control vector or wild-type or mitochondrially targeted ING1 constructs. Cells were harvested after 12 h and stained with Annexin V and 7-AAD. Flowcytometry was performed within 30 min (*P=0.02). (e) A model for the role of ING1 in the induction of apoptosis. DNA damage and other apoptosis-inducing stimuli trigger the cooperation between ING1 and p53 in the nucleus, leading to the transcriptional activation of pro-apoptotic BAX followed by the induction of apoptosis. Moreover, in response to apoptosis-inducing stimuli, ING1 translocates to the cytosol/mitochondria, possibly by binding to 14-3-3 proteins,45 where it induces apoptosis through BAX binding and activation
ING proteins interact with several mitochondrial proteins
Homologs of human ING proteins exist throughout the plant and animal kingdoms.2 The yeast ING proteins, YNG1, YNG2 and YNG3 (Pho23) exhibit sequence homology with the human ING proteins at their C-terminal regions that affect histone acetylation.14 As members of the ING family of tumor suppressors show significant sequence conservation from yeast to humans, we previously used available yeast interactome data57 to predict mammalian counterparts of yeast ING-interacting partners.58 In silico analysis of ING ‘interologs' indeed showed that ING proteins in yeast interact with 64 mitochondrial proteins with human counterparts (Supplementary Table 1). This list does not include BAX because it is absent in the yeast proteome. Functional classification of the ING-interacting mitochondrial proteins mined from our previous studies is shown in Table 1.
Table 1. Functional classes of mitochondrial proteins interacting with yeast ING proteins. All proteins were previously determined to interact with ING proteins were using TAP-tagging and LC-MS-MS in an interactome analysis. A total of 57 were categorized based upon their localization and function within the mitochondria.
| Function | Number of mitochondrial proteins |
|---|---|
| Metabolism | 21 |
| Mitochondrial transport | 14 |
| Mitochondrial translation | 10 |
| DNA repair and metabolism | 5 |
| Protein modification | 3 |
| RNA metabolism | 2 |
| Uncharacterized | 9 |
ING1 also contains a region showing high homology with BH3 domains. Visual inspection of the ING1 sequence revealed a short sequence adjacent to the Ser199 residue responsible for 14-3-3 binding that shows a high degree of homology to previously characterized BH3 domains. Supplementary Figure 1 indicates that this sequence has overlapping binding sites for 14-3-3 and BH3 domains, suggesting that nuclear to cytoplasmic translocation of ING1 may be coupled through a common sequence element.
Discussion
Previous studies regarding the function of ING1 in apoptosis have focused on nuclear functions.12, 13, 21, 31 As ING1 has been previously implicated in the mitochondrial or intrinsic apoptotic pathway21 and an ING1-interacting protein has been shown to localize to the mitochondria,38 we asked whether it has a function in apoptosis directly through the mitochondria. Immunofluorescence analysis and biochemical fractionation studies showed that ING1 translocates to the mitochondria after treatment of cells with various apoptosis-inducing stimuli. Mitochondrial translocation of ING1 was observed in several different cell types including normal human diploid fibroblasts and epithelial breast cancer cell lines, and the phenomenon was also found to be independent of the p53 status of the cell. This is the first report of ING1 translocating to the mitochondria in response to apoptosis-inducing stimuli. To further understand the significance of this translocation, we asked whether the ability of ING1 to induce apoptosis in several breast cancer cell lines correlated with its ability to colocalize with the mitochondria after treatment with apoptosis-inducing stimuli. The degree of translocation of ING1 to the mitochondria correlated well with susceptibility to ING1-induced apoptosis. Further investigation of the mechanism by which ING1 might act via the mitochondria led us to ask whether ING1 interacts with pro-apoptotic proteins in the mitochondria. A clear UV-inducible colocalization of ING1 with BAX at the mitochondria was seen by using indirect immunfluorescence. Reciprocal co-immunoprecipitation confirmed that ING1 and BAX physically interacted, although the interaction was not UV-inducible under the conditions we used. This may be because of many factors, including UV-induced exposure of epitopes or the association of ING1 with BAX after the disruption of cells for immunoprecipitation; differential localization of ING1 and BAX would not be maintained in cell lysates. Consistent with ING1 interacting directly with BAX, we identified a BH3 domain in ING1 through which it could theoretically bind BAX and other BH3 domain containing proteins (Supplementary Figure 1). As this region is adjacent to the Ser199 residue that defines the ING1-14-3-3-binding site,45 it suggests a mechanism of ING1 relocalization from the nucleus and subsequent targeting to BAX and the mitochondria (Figure 5e). Although ING1 association with and activation of BAX are the simplest interpretations of our results, we have not shown that BAX interacts with ING1 only at the mitochondria or that ING1 is released from 14-3-3 when it translocates to the mitochondria, therefore other possibilities remain; however, a dependence upon p53 has been ruled out.
Our in silico analysis indicates that yeast ING proteins interact with several mitochondrial proteins in yeast and a majority of the interacting partners are conserved in humans. This conservation of interactors across species suggests that ING1 serves evolutionarilly conserved and biologically significant functions at the mitochondria, one of which is in the induction of apoptosis. Although we did not identify mitochondrial translocation sequences in ING1, functional classification of mitochondrial ING-interacting proteins showed that ING interacts with a variety of mitochondrial transport proteins, which might provide a potential mechanism for its transport to the mitochondria. Mitochondrial interaction data coupled with our observation that protein levels of ING1 remain unchanged in MCF7 cells when exposed to apoptosis-inducing stimuli (Supplementary Figure 2) also support the idea that change in subcellular localization is an important mechanism by which ING1 function in apoptosis is regulated. These data parallel closely the effects of p53 on the process of apoptosis and may help explain why in some studies ING1 appears to affect apoptosis independent of p53, although other reports suggest an interdependence of these two tumor suppressors.
Materials and Methods
Cell lines and Plasmids
Normal diploid Hs68 fibroblasts were cultured in low-glucose Dulbecco's Modified Eagles Medium (DMEM) (Thermo Scientific Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum. MCF7, MDA-MB468, BT474 and T47D epithelial breast cancer cells were grown in high-glucose DMEM supplemented with 10% fetal bovine serum. SKBR3, HCT116 wild-type and HCT116 BAX−/− (gift from Dr. Bert Vogelstein)59 cells were grown in McCoy's 5A medium (Gibco, Burlington, ON, Canada) supplemented with 10% fetal bovine serum. All cells were grown at 37 °C with 5% CO2. Except for MCF7 cells, which have wild-type p53, all the other cells lines possess mutations in the p53 gene.
The mitochondria-targeted ING1 plasmid was generated by cloning the ING1b cDNA into the pDsRed2-Mito Vector. The cDNA was amplified with the help of PCR using pCI-ING1b as a template (forward primer: 5′-CGCGGATCCGTTGAGTCCTGCCAACGGG-3′, reverse primer: 5′-CGCGGATCCCTGTTGTAAGCCCTCTCTTTTTTG-3′). The insert was ligated in frame into the BamHI restriction site on the vector such that the fusion product contained ING1b with N-terminal mitochondrial-targeting sequence and C-terminal dsRed. The construct was verified by sequencing. Western blotting with lysates from HEK293 cells transfected with the constructs showed an increase in the relative molecular mass of the fusion product. Mitochondrial localization was verified using immunofluorescence after transfection of HEK293 cells. mCherry-ING1b was constructed by cloning full-length ING1b cDNA cut using XhoI and NotI restriction enzymes and mCherry cDNA at the NotI site of pCI vector. pCI-ING1b and pLEX-mCherry were used as templates for PCR for amplifying the inserts. Adenoviral constructs (Ad-GFP and Ad-GFP-ING1) used in this study have been previously described, and the conditions for infections have been optimized.23, 60
Indirect immunofluorescence
Cells were grown in 6-cm dishes on coverslips. Twenty-four hour after plating, cells were treated with 60 J/m2 of UVC in or with 2 Gy of ionizing radiation. Cells were then labeled with MitoTracker Deep Red (Life Technologies, Burlington, ON, Canada) at 5.5 h and fixed at 6 h post UV treatment with cold 95% methanol at room temperature (RT). Cells were washed with phosphate-buffered saline (PBS) and incubated with primary antibodies for 1 h. Cells were again washed in PBS and incubated with secondary antibody for 0.5 h. Cells were washed in PBS, stained with 4′6-Diamidino-2-phenylindole·(DAPI) for 2 min and mounted on slides for visualization. Microscopy was performed on a LSM 510 META confocal microscope (Zeiss, Toronto, ON, Canada).
Subcellular fractionation and western blotting
The subcellular fractionation protocol has been previously described.61 Briefly, cells were lysed using RIPA buffer (20 mM Tris–HCl (pH 7.4), 150 mM NaCl, 0.5% (v/v) NP-40, protease inhibitor cocktail (Roche, Indianapolis, IN, USA). Nuclear, cytoplasmic and mitochondrial fractions were obtained using a Qproteome mitochondria extraction kit and a Qproteome nucleus extraction kit (Qiagen, Toronto, ON, Canada) according to the manufacturer's protocol.
Cells were washed three times in PBS and collected by scraping in PBS. Pellets were collected by centrifugation at 2500 × g for 5 min and the appropriate amount of Laemmli's sample buffer was added to the cell pellets. Aliquots corresponding to 2 × 105 cells were loaded on 12.5% polyacrylamide gels, and electrophoresis was performed at a constant 100 V. Separated proteins were transferred to nitrocellulose membranes and immunoblotting was performed by incubation with either rabbit anti-BAX (1 : 1000 dilution) or mouse monoclonal anti-ING1 (hybridoma supernatant) after blocking in 5% non-fat milk. After several washes in PBS with 0.5% Tween-20, membranes were incubated with 1 : 5000 dilution of the corresponding secondary antibody conjugated with horseradish peroxidise for 30 min. Membranes were washed with PBS-Tween and exposed to an X-ray film.
Co-immunoprecipitation
Cells were lysed in buffer (10 mmol HEPES PH 7.4, 150 mmol/l NaCl, 1% CHAPS, 1% protease inhibitors) containing a protease inhibitor mixture (Complete Mini, Roche) for 30 min on ice. Cell lysates were sonicated three times for 10 s each on ice. Insoluble material was removed by centrifugation at 14 000 × g for 15 min. Proteins were quantitated using a Bradford assay (Bio-Rad, Mississauga, ON, Canada). Protein G-sepharose (Roche)-precleared lysates were incubated with rabbit anti-Bax (Abcam, Toronto, ON, Canada) or mouse monoclonal anti-ING1 antibody62 conjugated with protein G-sepharose beads for 4 h at 4 °C. Immunocomplexes were collected by centrifugation at 5000 × g and washed three times in ice-cold PBS. After the final wash, the beads were mixed with 30 μl of 2 × Laemmli sample buffer, heated at 100 °C for 5 min and analyzed using western blotting.
Apoptosis assays
For propidium iodide (PI) staining, cells were collected by trypsinization and washed in PBS+5 mM EDTA and fixed in 70% ethanol at 4 °C for 30 min. Cells were washed twice with PBS+5 mM EDTA followed by staining in PI solution (50 mg/ml PI, 20 mg/ml RNase) (Sigma-Aldrich, Oakville, ON, Canada) in PBS in the dark at RT for 30 min. For Annexin V staining, an Annexin V-PE/7AAD kit (BD Biosciences, Mississauga, ON, Canada) was used following the manufacturer's protocol. Flowcytometry was performed on a FACScan flowcytometer (Becton-Dickinson, Mississauga, ON, Canada) at the University of Calgary Flow Cytometry Core Facility.
Identification of orthologs
Yeast ING-interacting partners were identified from data in reference57 and previously partially analyzed in reference.13 BLAST searches were performed to identify human counterparts of the yeast ING-interacting mitochondrial proteins. The results from BLAST search were confirmed using homologene (NCBI) and genecard databases, and biological functions were assigned.
Glossary
- 14-3-3
members of the Tetratrico Peptide Repeat (TPR) superfamily
- APAF-1
Apoptotic Protease Activating Factor 1
- BAD
BCL-2 agonist of cell death
- BAK
BCL-2-antagonist/killer
- BAX
BCL2-Associated X protein
- BCL2
B-cell lymphoma 2 protein
- BH3
BAX Homology domain 3
- BID
BH3-interacting domain death agonist
- BIM
Bcl-2-Interacting Mediator of cell death
- DAPI
the 4′6-Diamidino-2-phenylindole·2HCl DNA stain
- DMEM
Dulbecco's Modified Eagles Medium
- FAS
Fas cell surface death receptor
- HAT
Histone Acetyl Transferase
- HDAC
histone deacetylase
- ING
Inhibitor of Growth
- hSIR2
human sirtuin protein 2; IR, ionizing radiation
- mtHSP70
mitochondrial heat shock protein 70
- NOXA
latin for damagea pro-apoptotic BCL2 family protein
- NLS
Nuclear Localization Signal
- NTS
Nucleolar-Targeting Sequence
- p21
21 kilodalton cyclin-dependent kinase inhibitor
- p53
53 kilodalton tumor suppressor protein
- PARP
Poly ADP Ribose Polymerase
- PBS
phosphate-buffered saline
- PCNA
Proliferating Cell Nuclear Antigen
- PHDPIP
PCNA-interacting protein; plant homeo domain
- PUMA
P53-Upregulated Modulator of Apoptosis
- RIPA
Radio Immunoprecipitation Assay buffer
- shRNA
short-hairpin RiboNucleic Acid
- TNF
tumor necrosis factor
- UV
ultraviolet light
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by M Agostini
Supplementary Material
References
- Garkavtsev I, Kazarov A, Gudkov A, Riabowol K. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat Genet. 1996;14:415–420. doi: 10.1038/ng1296-415. [DOI] [PubMed] [Google Scholar]
- He GH, Helbing CC, Wagner MJ, Sensen CW, Riabowol K. Phylogenetic analysis of the ING family of PHD finger proteins. Mol Biol Evol. 2005;22:104–116. doi: 10.1093/molbev/msh256. [DOI] [PubMed] [Google Scholar]
- Garkavtsev I, Riabowol K. Extension of the replicative life span of human diploid fibroblasts by inhibition of the p33ING1 candidate tumor suppressor. Mol Cell Biol. 1997;17:2014–2019. doi: 10.1128/mcb.17.4.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedeux R, Sengupta S, Shen JC, Demidov ON, Saito S, Onogi H, et al. ING2 regulates the onset of replicative senescence by induction of p300-dependent p53 acetylation. Mol Cell Biol. 2005;25:6639–6648. doi: 10.1128/MCB.25.15.6639-6648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman MA, Berardi P, Pastyryeva S, Bonnefin P, Feng X, Colina A, et al. ING1a expression increases during replicative senescence and induces a senescent phenotype. Aging Cell. 2008;7:783–794. doi: 10.1111/j.1474-9726.2008.00427.x. [DOI] [PubMed] [Google Scholar]
- Abad M, Moreno A, Palacios A, Narita M, Blanco F, Moreno-Bueno G, et al. The tumor suppressor ING1 contributes to epigenetic control of cellular senescence. Aging Cell. 2011;10:158–171. doi: 10.1111/j.1474-9726.2010.00651.x. [DOI] [PubMed] [Google Scholar]
- Vieyra D, Loewith R, Scott M, Bonnefin P, Boisvert FM, Cheema P, et al. Human ING1 proteins differentially regulate histone acetylation. J Biol Chem. 2002;277:29832–29839. doi: 10.1074/jbc.M200197200. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Zhang Y, Erdjument-Bromage H, Tempst P, Reinberg D. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1) Mol Cell Biol. 2002;22:835–848. doi: 10.1128/MCB.22.3.835-848.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena PV, Hom RA, Hung T, Lin H, Kuo AJ, Wong RP, et al. Histone H3K4me3 binding is required for the DNA repair and apoptotic activities of ING1 tumor suppressor. J Mol Biol. 2008;380:303–312. doi: 10.1016/j.jmb.2008.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder KW, Wang X, Escriu C, Ito Y, Schwarz RF, Gillis J, et al. Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol. 2012;14:753–763. doi: 10.1038/ncb2520. [DOI] [PubMed] [Google Scholar]
- Eapen SA, Netherton SJ, Sarker KP, Deng L, Chan A, Riabowol K, et al. Identification of a novel function for the chromatin remodeling protein ING2 in muscle differentiation. PLoS One. 2012;7:e40684. doi: 10.1371/journal.pone.0040684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M, Boisvert FM, Vieyra D, Johnston RN, Bazett-Jones DP, Riabowol K. UV induces nucleolar translocation of ING1 through two distinct nucleolar targeting sequences. Nucleic Acids Res. 2001;29:2052–2058. doi: 10.1093/nar/29.10.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M, Bonnefin P, Vieyra D, Boisvert FM, Young D, Bazett-Jones DP, et al. UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J Cell Sci. 2001;114:3455–3462. doi: 10.1242/jcs.114.19.3455. [DOI] [PubMed] [Google Scholar]
- Cheung KJ, Jr, Mitchell D, Lin P, Li G. The tumor suppressor candidate p33(ING1) mediates repair of UV-damaged DNA. Cancer Res. 2001;61:4974–4977. [PubMed] [Google Scholar]
- Wong RP, Lin H, Khosravi S, Piche B, Jafarnejad SM, Chen DW, et al. Tumor suppressor ING1b maintains genomic stability upon replication stress. Nucleic Acids Res. 2011;39:3632–3642. doi: 10.1093/nar/gkq1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrieu D, Ythier D, Brambilla C, Pedeux R. ING2 controls the G1 to S-phase transition by regulating p21 expression. Cell Cycle. 2010;9:3984–3990. doi: 10.4161/cc.9.19.13208. [DOI] [PubMed] [Google Scholar]
- Zhang X, Xu LS, Wang ZQ, Wang KS, Li N, Cheng ZH, et al. ING4 induces G2/M cell cycle arrest and enhances the chemosensitivity to DNA-damage agents in HepG2 cells. FEBS Lett. 2004;570:7–12. doi: 10.1016/j.febslet.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Wei Q, He W, Lu Y, Yao J, Cao X. Effect of the tumor suppressor gene ING4 on the proliferation of MCF-7 human breast cancer cells. Oncol Lett. 2012;4:438–442. doi: 10.3892/ol.2012.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbing CC, Veillette C, Riabowol K, Johnston RN, Garkavtsev I. A novel candidate tumor suppressor, ING1, is involved in the regulation of apoptosis. Cancer Res. 1997;57:1255–1258. [PubMed] [Google Scholar]
- Shinoura N, Muramatsu Y, Nishimura M, Yoshida Y, Saito A, Yokoyama T, et al. Adenovirus-mediated transfer of p33ING1 with p53 drastically augments apoptosis in gliomas. Cancer Res. 1999;59:5521–5528. [PubMed] [Google Scholar]
- Cheung KJ, Jr, Li G. p33(ING1) enhances UVB-induced apoptosis in melanoma cells. Exp Cell Res. 2002;279:291–298. doi: 10.1006/excr.2002.5610. [DOI] [PubMed] [Google Scholar]
- Vieyra D, Toyama T, Hara Y, Boland D, Johnston R, Riabowol K. ING1 isoforms differentially affect apoptosis in a cell age-dependent manner. Cancer Res. 2002;62:4445–4452. [PubMed] [Google Scholar]
- Feng X, Bonni S, Riabowol K. HSP70 induction by ING proteins sensitizes cells to tumor necrosis factor alpha receptor-mediated apoptosis. Mol Cell Biol. 2006;26:9244–9255. doi: 10.1128/MCB.01538-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhao G, Chen T, Xue L, Ma L, Niu J, et al. Nucleolar protein CSIG is required for p33ING1 function in UV-induced apoptosis. Cell Death Dis. 2012;3:e283. doi: 10.1038/cddis.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama T, Iwase H, Watson P, Muzik H, Saettler E, Magliocco A, et al. Suppression of ING1 expression in sporadic breast cancer. Oncogene. 1999;18:5187–5193. doi: 10.1038/sj.onc.1202905. [DOI] [PubMed] [Google Scholar]
- Ohmori M, Nagai M, Tasaka T, Koeffler HP, Toyama T, Riabowol K, et al. Decreased expression of p33ING1 mRNA in lymphoid malignancies. Am J Hematol. 1999;62:118–119. doi: 10.1002/(sici)1096-8652(199910)62:2<118::aid-ajh11>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Kameyama K, Huang CL, Liu D, Masuya D, Nakashima T, Sumitomo S, et al. Reduced ING1b gene expression plays an important role in carcinogenesis of non-small cell lung cancer patients. Clin Cancer Res. 2003;9:4926–4934. [PubMed] [Google Scholar]
- Tallen G, Kaiser I, Krabbe S, Lass U, Hartmann C, Henze G, et al. No ING1 mutations in human brain tumours but reduced expression in high malignancy grades of astrocytoma. Int J Cancer. 2004;109:476–479. doi: 10.1002/ijc.11715. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Ozaki T, Todo S, Nakagawara A. Decreased expression of the candidate tumor suppressor gene ING1 is associated with poor prognosis in advanced neuroblastomas. Oncol Rep. 2004;12:811–816. [PubMed] [Google Scholar]
- Oki E, Maehara Y, Tokunaga E, Kakeji Y, Sugimachi K. Reduced expression of p33(ING1) and the relationship with p53 expression in human gastric cancer. Cancer Lett. 1999;147:157–162. doi: 10.1016/s0304-3835(99)00288-8. [DOI] [PubMed] [Google Scholar]
- Li XH, Noguchi A, Nishida T, Takahashi H, Zheng Y, Yang XH, et al. Cytoplasmic expression of p33ING1b is correlated with tumorigenesis and progression of head and neck squamous cell carcinoma. Histol Histopathol. 2011;26:597–607. doi: 10.14670/HH-26.597. [DOI] [PubMed] [Google Scholar]
- Garkavtsev I, Grigorian IA, Ossovskaya VS, Chernov MV, Chumakov PM, Gudkov AV. The candidate tumor suppressor p33ING1 cooperates with p53 in cell growth control. Nature. 1998;391:295–298. doi: 10.1038/34675. [DOI] [PubMed] [Google Scholar]
- Shimada H, Liu TL, Ochiai T, Shimizu T, Haupt Y, Hamada H, et al. Facilitation of adenoviral wild-type p53-induced apoptotic cell death by overexpression of p33(ING1) in T.Tn human esophageal carcinoma cells. Oncogene. 2002;21:1208–1216. doi: 10.1038/sj.onc.1205176. [DOI] [PubMed] [Google Scholar]
- Kataoka H, Bonnefin P, Vieyra D, Feng X, Hara Y, Miura Y, et al. ING1 represses transcription by direct DNA binding and through effects on p53. Cancer Res. 2003;63:5785–5792. [PubMed] [Google Scholar]
- Coles AH, Liang H, Zhu Z, Marfella CG, Kang J, Imbalzano AN, et al. Deletion of p37Ing1 in mice reveals a p53-independent role for Ing1 in the suppression of cell proliferation, apoptosis, and tumorigenesis. Cancer Res. 2007;67:2054–2061. doi: 10.1158/0008-5472.CAN-06-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamannai M, Farhangi S, Truss M, Sinn B, Wurm R, Bose P, et al. The inhibitor of growth 1 (ING1) is involved in trichostatin A-induced apoptosis and caspase 3 signaling in p53-deficient glioblastoma cells. Oncol Res. 2010;18:469–480. doi: 10.3727/096504010x12704916124828. [DOI] [PubMed] [Google Scholar]
- Thalappilly S, Feng X, Pastyryeva S, Suzuki K, Muruve D, Larocque D, et al. The p53 tumor suppressor is stabilized by inhibitor of growth 1 (ING1) by blocking polyubiquitination. PLoS One. 2011;6:e21065. doi: 10.1371/journal.pone.0021065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson F, Lammerts van Bueren K, Butterfield N, Bennetts JS, Bowles J, Adolphe C, et al. The PCNA-associated factor KIAA0101/p15(PAF) binds the potential tumor suppressor product p33ING1b. Exp Cell Res. 2006;312:73–85. doi: 10.1016/j.yexcr.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Loewith R, Meijer M, Lees-Miller SP, Riabowol K, Young D. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol Cell Biol. 2000;20:3807–3816. doi: 10.1128/mcb.20.11.3807-3816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Hara Y, Riabowol K. Different HATS of the ING1 gene family. Trends Cell Biol. 2002;12:532–538. doi: 10.1016/s0962-8924(02)02391-7. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Soliman MA, Riabowol K. After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem Sci. 2007;32:509–519. doi: 10.1016/j.tibs.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Martin DG, Baetz K, Shi X, Walter KL, MacDonald VE, Wlodarski MJ, et al. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol. 2006;26:7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W, Russell M, Suzuki K, Riabowol K. Subcellular targeting of p33ING1b by phosphorylation-dependent 14-3-3 binding regulates p21WAF1 expression. Mol Cell Biol. 2006;26:2947–2954. doi: 10.1128/MCB.26.8.2947-2954.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata JM, Pawlowski K, Haas E, Ware CF, Godzik A, Reed JC. A diverse family of proteins containing tumor necrosis factor receptor-associated factor domains. J Biol Chem. 2001;276:24242–24252. doi: 10.1074/jbc.M100354200. [DOI] [PubMed] [Google Scholar]
- Letai AG. Diagnosing and exploiting cancer's addiction to blocks in apoptosis. Nat Rev Cancer. 2008;8:121–132. doi: 10.1038/nrc2297. [DOI] [PubMed] [Google Scholar]
- Brenner D, Mak TW. Mitochondrial cell death effectors. Curr Opin Cell Biol. 2009;21:871–877. doi: 10.1016/j.ceb.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- Chittenden T, Flemington C, Houghton AB, Ebb RG, Gallo GJ, Elangovan B, et al. A conserved domain in Bak, distinct from BH1 and BH2, mediates cell death and protein binding functions. EMBO J. 1995;14:5589–5596. doi: 10.1002/j.1460-2075.1995.tb00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartron PF, Gallenne T, Bougras G, Gautier F, Manero F, Vusio P, et al. The first alpha helix of Bax plays a necessary role in its ligand-induced activation by the BH3-only proteins Bid and PUMA. Mol Cell. 2004;16:807–818. doi: 10.1016/j.molcel.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Green DR, Kroemer G. Cytoplasmic functions of the tumor suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter KG, Hsu YT, Smith CL, Nechushtan A, Xi XG, Youle RJ. Movement of Bax from the cytosol to mitochondria during apoptosis. J Cell Biol. 1997;139:1281–1292. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Gordon PM, Soliman MA, Bose P, Trinh Q, Sensen CW, Riabowol K. Interspecies data mining to predict novel ING-protein interactions in human. BMC Genomics. 2008;9:426. doi: 10.1186/1471-2164-9-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yu J, Park BH, Kinzler KW, Vogelstein B. Role of BAX in the apoptotic response to anticancer agents. Science. 2000;290:989–992. doi: 10.1126/science.290.5493.989. [DOI] [PubMed] [Google Scholar]
- Thakur S, Feng X, Qiao Shi Z, Ganapathy A, Kumar Mishra M, Atadja P, et al. ING1 and 5-azacytidine act synergistically to block breast cancer cell growth. PLoS One. 2012;7:e43671. doi: 10.1371/journal.pone.0043671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn BY, Trinh DL, Zajchowski LD, Lee B, Elwi AN, Kim SW. Tid1 is a new regulator of p53 mitochondrial translocation and apoptosis in cancer. Oncogene. 2010;29:1155–1166. doi: 10.1038/onc.2009.413. [DOI] [PubMed] [Google Scholar]
- Boland D, Olineck V, Bonnefin P, Vieyra D, Parr E, Riabowol K. A panel of CAb antibodies recognize endogenous and ectopically expressed ING1 protein. Hybridoma. 2000;19:161–165. doi: 10.1089/02724570050031202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.