Abstract
Background. The pathogenesis of intra-abdominal candidiasis is poorly understood.
Methods. Mice were intraperitoneally infected with Candida albicans (1 × 106 colony-forming units) and sterile stool. nanoString assays were used to quantitate messenger RNA for 145 C. albicans genes within the peritoneal cavity at 48 hours.
Results. Within 6 hours after infection, mice developed peritonitis, characterized by high yeast burdens, neutrophil influx, and a pH of 7.9 within peritoneal fluid. Organ invasion by hyphae and early abscess formation were evident 6 and 24 hours after infection, respectively; abscesses resolved by day 14. nanoString assays revealed adhesion and responses to alkaline pH, osmolarity, and stress as biologic processes activated in the peritoneal cavity. Disruption of the highly-expressed gene RIM101, which encodes an alkaline-regulated transcription factor, did not impact cellular morphology but reduced both C. albicans burden during early peritonitis and C. albicans persistence within abscesses. RIM101 influenced expression of 49 genes during intra-abdominal candidiasis, including previously unidentified Rim101 targets. Overexpression of the RIM101-dependent gene SAP5, which encodes a secreted protease, restored the ability of a rim101 mutant to persist within abscesses.
Conclusions. A mouse model of intra-abdominal candidiasis is valuable for studying pathogenesis and C. albicans gene expression. RIM101 contributes to persistence within intra-abdominal abscesses, at least in part through activation of SAP5.
Keywords: Candida albicans, intra-abdominal candidiasis, in vivo gene expression, nanoString, RIM101, SAP5
Invasive candidiasis is the most common fungal disease in US hospitals [1]. Candidemia and intra-abdominal candidiasis are the 2 most common types of invasive candidiasis [2, 3]. Intra-abdominal candidiasis is associated with mortality rates as high as 65% [2–5]. The disease stems from introduction of Candida into the normally sterile peritoneal cavity through direct inoculation, such as with infected peritoneal dialysis catheters, or, more commonly, as a result of leakage or perforation of the gastrointestinal tract. Indeed, intra-abdominal candidiasis is diagnosed in up to 39% of patients who experience gastrointestinal leaks or perforations [6–8]. Intra-abdominal candidiasis manifests as 2 conditions: peritonitis and abscess formation. Peritonitis is inflammation resulting from the host response to Candida within the peritoneal cavity. Abscess formation occurs over subsequent days as Candida cells that invade abdominal organs from the peritoneal cavity are contained by the inflammatory response, walled off, and finally eliminated. Epidemiologic data on the prevalence of these entities are conflicting, as various studies have reported that Candida peritonitis and intra-abdominal abscesses account for 62%–63% [9, 10] and 67% of intra-abdominal candidiasis cases [11], respectively.
Despite the clinical importance of intra-abdominal candidiasis, its pathogenesis is not well studied. Mouse models of C. albicans peritonitis and intra-abdominal abscesses have been developed, but they have not been widely used to study the full spectrum of intra-abdominal candidiasis. In the peritonitis model, C. albicans strains inoculated intra-peritoneally persist for several days in peritoneal fluid and directly invade abdominal organs such as the liver and spleen [12–14]. The model has been used to compare the relative virulence of mutant strains for secreted aspartic proteinases and several other proteins and to evaluate in vivo expression of the corresponding genes [12]. However, abscess formation is not prominent in this model, transcriptional profiling of more than a handful of genes has not been undertaken, and host responses within the peritoneal cavity have not been assessed. In the intra-abdominal abscess model, sterilized mouse stool is included in the inoculum as a potentiating agent [15, 16]. The infection mimics fecal soilage that follows gastrointestinal tract disruption and results in C. albicans–containing abscesses that peak in number over the course of a week. The model has been used to study antifungal regimens and host factors relevant to abscess formation but not to investigate peritonitis or C. albicans virulence mechanisms.
In this study, we adapted previously described mouse models to recapitulate the progression of C. albicans intra-abdominal candidiasis from peritonitis to abscesses [15–18]. We used our model to study the C. albicans–host interaction during the course of disease and to perform large-scale transcriptional profiling of C. albicans. For the latter, we used the nanoString nCounter System, a new technology that digitally measures >100 target messenger RNAs (mRNAs) with a sensitivity that is greater than that of microarray and comparable to that of quantitative real-time polymerase chain reaction [19–21]. In a recent study of mouse oropharyngeal candidiasis, nanoString afforded highly reproducible measurements of C. albicans gene expression that agreed well with previously published data [19]. Finally, we validated the mouse model and expression data by demonstrating that the highly expressed gene RIM101 and its major target, SAP5, facilitated the persistence of C. albicans within abscesses.
METHODS
C. albicans Strains and Growth Conditions
C. albicans strains are listed in Table 1. Strains were routinely grown in yeast extract-peptone-dextrose medium (1% yeast extract, 2% Bacto peptone, and 2% α-d-glucose) at 30°C unless otherwise noted.
Table 1.
Candida albicans Strains Used in This Study
| Strain | Genotypes | Reference |
|---|---|---|
| SC5314 | Clinical isolate | … |
| DAY25 | ura3Δ::λimm434 HIS1::his1::hisG arg4::hisG rim101::ARG4 ura3Δ::λimm434 his1::hisG arg4::hisG rim101::URA3 | [29] |
| DAY44 | ura3Δ::λimm434 his1::hisG arg4::hisG rim101::ARG4::pRIM101::HIS1 ura3Δ::λimm434 his1::hisG arg4::hisG rim101::URA3 | [23] |
| CJN793 | ura3Δ::λimm434/ura3Δ::λimm434 arg4::hisG/arg4::hisG his1::hisG::pHIS1-NRG1/his1::hisG rim101::dpl200/rim101::dpl200 nrg1::ARG4/nrg1::URA3 | [35] |
| CJN1111 | ura3Δ::λimm434/ura3Δ::λimm434 arg4::hisG/arg4::hisG his1::hisG::pHIS1-NRG1/his1::hisG rim101::dpl200/rim101::dpl200 nrg1::ARG4/nrg1::URA3 TEF1-SAP5::NAT1/SAP5 | [35] |
Mouse Model of Intra-abdominal Candidiasis
Murine experiments were performed according to the University of Pittsburgh Institutional Animal Care and Use Committee guidelines. C. albicans strains were grown overnight in Sabouraud dextrose broth at 30°C. Mice were infected intraperitoneally with 100 µL of suspension containing 1 × 106 colony-forming units (CFU) of C. albicans without or with mouse feces. Feces from uninfected mice were ground in a tissue grinder, suspended in normal saline solution to form a 5% weight/volume mixture, and sterilized in a steam autoclave (15 minutes, 200 kPa, 120°C) [15]. Abdominal cavities were explored at given time points, and the number of abscesses >1 mm in diameter were counted. Abscesses were removed and homogenized for colony enumeration. To assess for peritonitis, the peritoneum was washed with 1 mL of phosphate-buffered saline, and cell counts, C. albicans levels, and pH of the lavage fluid were determined [22]. To assess for tissue invasion, liver and spleen were homogenized for C. albicans enumeration. We did not evaluate burdens within kidneys, which are retroperitoneal and did not yield consistent results in previous studies [12]. For histopathologic analysis, tissues and abscesses were fixed with formalin and embedded in paraffin, and thin sections were stained with hematoxylin-eosin and Gomori methenamine silver. Tissue burdens for mice infected with each strain are presented as mean (±SD) log10 CFU/gram of tissue. The difference in tissue burdens between mice infected with mutant versus comparator strains was determined by the Wilcoxon test. A P value of <.05 was considered statistically significant.
nanoString Profiling
The peritoneal cavity was washed with normal saline solution, and fluid was immediately placed in RNAlater buffer (Ambion), stored at 4°C overnight, and then stored at −20°C. RNA was isolated using a Qiagen kit and a bead beater and was mixed with a custom-designed nanoString probe set as previously described [19, 20]. Probe sets included 145 genes that were chosen on the basis of published microarray data and our nanoString in vitro data (Supplementary data). Specifically, we selected representative genes that are involved in processes such as hyphal growth, in various stress responses, and in adhesion; control genes for high, moderate, and low expression; and internal normalization genes [19, 20]; we previously showed that control gene expression varies little in microarray and nanoString studies [19, 20]. For each experiment, data were obtained in triplicate from 3 independently infected mice. The raw counts were adjusted for technical variability, using positive and negative controls of irrelevant RNA sequences included in the code set, and then normalized for total input C. albicans RNA, using the geometric mean of robustly expressed genes. Data of normalized counts were presented as mean values (±SD) of 3 independent experiments. To identify RIM101-dependent genes in vivo, we infected mice with strains DAY44 and DAY25 and harvested peritoneal fluid for nanoString determination. The mean values of normalized data for each gene were log transformed, and the differences in gene expression in vivo between strains DAY25 and DAY44 were compared using the Student t test; a P value of <.05 was considered statistically significant.
RESULTS
Mouse Model of Intra-abdominal Candidiasis
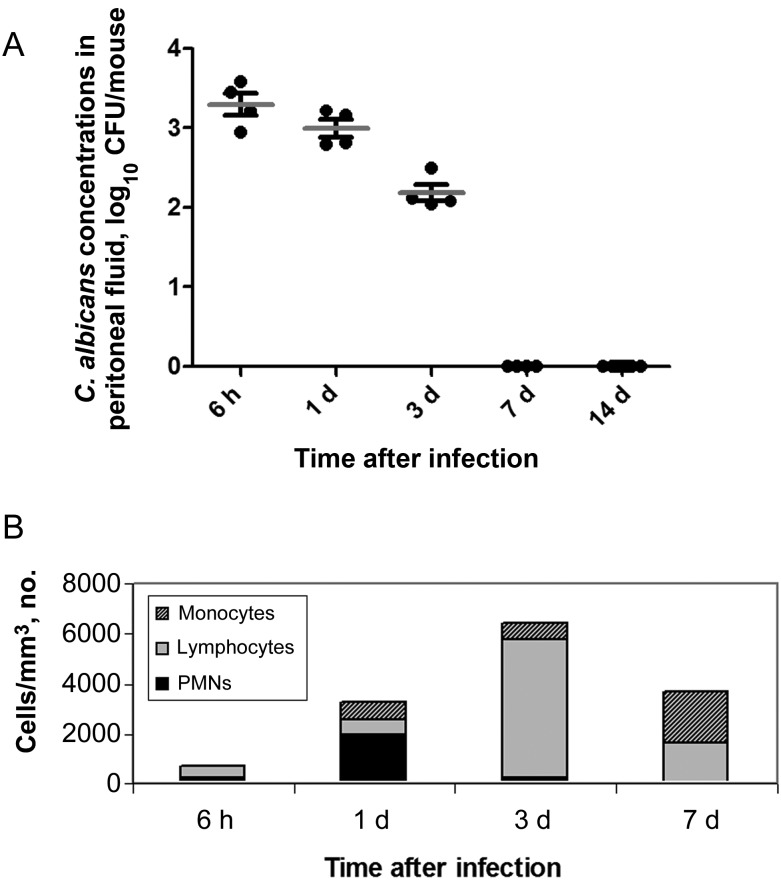
Mice infected intraperitoneally with C. albicans SC5314 (1 × 106 CFU) and sterile stool survived for 30 days. The mean C. albicans burden (±SD) in the peritoneal cavity at 6 hours was 3.30 ± 0.28 log10 CFU/mouse (Figure 1A). Mean burdens (±SD) progressively decreased at 1 and 3 days (3.00 ± 0.22 and 2.18 ± 0.21 log10 CFU/mouse, respectively), and C. albicans was fully eradicated at 7 days. C. albicans cells were evident as yeasts, rather than hyphae, at each time point. White blood cell (WBC) counts within peritoneal fluid progressively increased from 6 hours to 1 and 3 days and then decreased at 7 days (Figure 1B). WBCs were predominantly neutrophils at 6 hours and 1 day, with a lymphocyte predominance on day 3 and a lymphocyte and monocyte predominance on day 7. The mean pH (±SD) of peritoneal fluid on day 1 was 7.9 ± 0.1.
Figure 1.
Tissue burdens (A) and white blood cell (WBC) counts (B) in peritoneal fluid of mice infected with C. albicans intraperitoneally. Abbreviations: CFU, colony-forming units; PMN, polymorphonuclear cell.
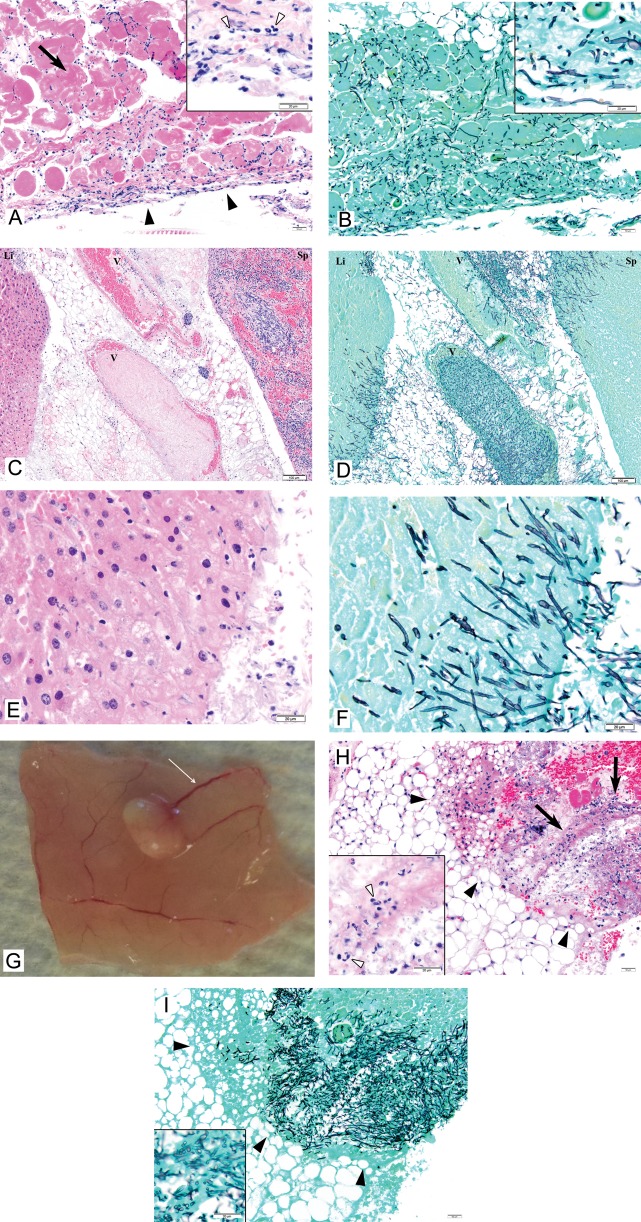
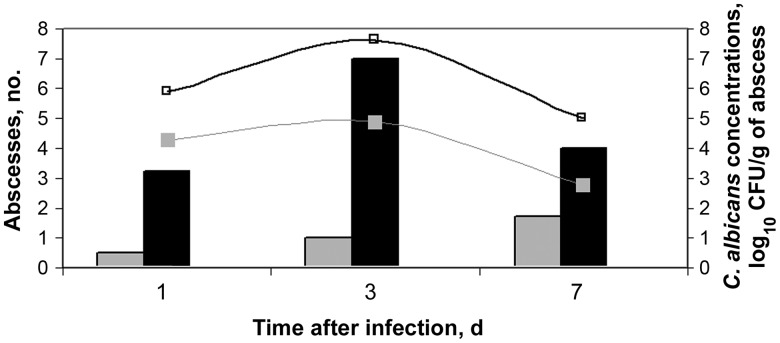
C. albicans hyphae were shown to directly invade the peritoneum and intra-abdominal organs at 6 hours (Figure 2A–F). Tissue invasion elicited acute inflammatory responses characterized by neutrophil predominance. Small vesicles resembling abscesses by gross pathologic analysis were first evident at 1 day, affixed to the peritoneal surface, bowel, and intra-abdominal organs (Figure 2G). Abscess numbers and intra-abscess C. albicans burdens increased from days 1–3 and then decreased at day 7 and resolved at day 14 (Figure 3). Histopathologic analysis of dissected abscesses revealed C. albicans yeasts and hyphae surrounded by a mixed inflammatory infiltrate comprised predominantly of neutrophils with a few admixed lymphocytes (Figure 2H and 2I). The mean intra-abscess pH (±SD) was 6.9 ± 0.3.
Figure 2.
Necropsy findings and histopathology of tissues of mice infected with C. albicans intraperitoneally. 2A and 2B. Peritoneum on day 1 of infection. 2A: Acute inflammation of the peritoneum is apparent (solid arrowheads). The inflammatory response is strongest on the surface, consistent with an inflammatory stimulus arising in the peritoneal cavity. The inflammatory response penetrates the serosa, resulting in necrosis of underlying diaphragmatic muscle (arrow). The inflammatory response comprises predominantly neutrophils (inset, empty arrowheads) with a smaller number of admixed lymphocytes and macrophages. 2B: C. albicans within the peritoneum are a mixture of hyphae and yeasts (inset). The density of organisms is greatest in the serosa, a finding in keeping with the gradient of the inflammatory response (Hematoxylin Eosin (A) and Grocott Methenamine Silver (B), original x 100, insets original x 600). Bars at low magnification: 50 μm. Bars at high magnification: 20 μm. 2C-2F. Liver and spleen on day 1 of infection. 2C, 2D: C. albicans infiltrating liver (Li, left), veins (V, center), and spleen (Sp, right). 2E, 2F: Higher magnification of liver showing inflammation (predominantly neutrophils) and mats of hyphae. Hematoxylin Eosin (2C, 2E) and Grocott Methenamine Silver (2D, 2F), original x 100 (2F, 2H), original x 600 (2G, 2I). Bars at low magnification: 50 μm. Bars at high magnification: 20 μm. 2G. Necropsy on day 3 of infection shows an abscess on the peritoneal membrane. Note the engorged blood vessels (thin arrow) consistent with tissue inflammation. 2H and 2I. Mesenteric abscess on day 3 of infection. 2H: Mesenteric fat showing a large area of fat necrosis (solid arrowheads) with fibrin exudates and acute inflammation (arrows). The mixed inflammatory infiltrate comprises predominantly neutrophils (inset, empty arrowheads) and few admixed lymphocytes. 2I: Grocott stain of the same area highlights numerous hyphae and yeasts in the area of necrosis and inflammation (Hematoxylin Eosin (2H) and Grocott Methenamine Silver (2I), original x 100, insets original x 600). Bars at low magnification: 50 μm. Bars at high magnification: 20 μm.
Figure 3.
Impact of addition of sterile stool on number of abscesses and burdens within abscess in mice infected with C. albicans intraperitoneally. Mice were infected with 1x106 CFU. Bar graphs represent the mean number of abscesses for 8 mice infected with C. albicans either alone (gray bar) or with sterile stool (black bar). Curved lines represent the mean number of log10 CFU for mice infected with C. albicans alone (gray line) or with sterile stool (gray bar). All abscesses resolved by day 14 (data not shown). Abbreviation: CFU, colony-forming units.
Exclusion of stool from inocula did not influence C. albicans burdens within peritoneal fluid, cellular morphology, WBC counts, peritoneal fluid pH, or the degree of invasion into the liver or spleen (as assessed by tissue burden and histopathologic analysis; data not shown). In contrast, at each time point the number of abscesses and the C. albicans burden within abscesses were greater in the presence of stool than in the absence of stool (Figure 3). Intraperitoneal inoculations of sterile stool without C. albicans elicited minimal formation of peritoneal fluid and induced sterile (ie, culture-negative) abscesses that were small and resolved by day 3. Sterile stool was included in studies of gene expression and virulence, since this condition mimics infections in humans following gastrointestinal perforations.
C. albicans Transcriptional Profiling During Intra-abdominal Candidiasis
Mice were infected intraperitoneally, and mRNA levels for 145 C. albicans genes were quantitated within peritoneal fluid by nanoString at 48 hours [21]. There was excellent correlation between mRNA levels for each gene in samples collected from independently infected mice (R2 = 0.97; Supplementary Figure 1). The genes with the greatest and least expression (and the biologic processes to which they contribute) are presented in Table 2, Supplementary Figure 1, and Supplementary Table 1.
Table 2.
Genes Most Upregulated During Peritonitis
| Biological Process | Genes |
|---|---|
| Response to pH | PHR1,RIM101, TEC1, ENA2, CAT1, GNP1, PHO89, RBT5, ARG1, FRP2 |
| Response to polymorphonuclear cells | DDR48, CAP1, FRP3, GCN4, TRR1, TRX1 |
| Response to stress | CAT1, ENA21, GPD2, HOG1, HSP70, SSB1, TRR1, TRX1, orf19.5517 |
| Adhesion | ALS1, ALS3, ALS4, SAP9, TDH3, TEC1 |
| Transporter | ENA21, ENA2, GNP1, SIT1, HGT7 |
| Cellular process | GCN4, GPD2 |
To validate the relevance of the transcriptional profiling data to pathogenesis, rim101 null mutant and RIM101 complemented strains were tested in the model. RIM101 was selected because it was among the most highly expressed genes in the nanoString probe set. Mice infected with the rim101 mutant had significantly lower organism burdens in peritoneal fluid at 6 hours than mice infected with the complemented strain (Table 3). Moreover, the null mutant elicited greater neutrophil responses at both 1 hour (absolute mean neutrophil counts [±SD], 35.2 ± 13.0 vs 8.9 ± 4.0/mm3; P = .06) and 6 hours (absolute mean neutrophil counts [±SD], 700.6 ± 31.8 vs 299.6 ± 142.7/mm3; P = .03). Organism burdens and neutrophil counts within peritoneal fluid did not differ between the strains at later time points.
Table 3.
Peritoneal Fluid, Organ Invasion, and Intra-abdominal Abscesses of Mice Infected With Various Candida albicans Strains
| Specimen, Sampling Time |
C. albicans Concentrations by Strain, Log10 CFU, Mean ± SD |
P | |
|---|---|---|---|
| DAY44a | DAY25b | ||
| Peritoneal fluid | |||
| 1 h | 5.61 ± 0.17 | 5.66 ± 0.21 | .66 |
| 6 h | 4.33 ± 0.12 | 3.75 ± 0.66 | .04 |
| 1 d | 3.49 ± 0.72 | 3.71 ± 0.14 | .4 |
| 3 d | 3.10 ± 0.58 | 3.15 ± 0.88 | .89 |
| 7 d | 0.76 ± 0.86 | 0.32 ± 0.60 | .25 |
| 10 d | No growth | No growth | |
| Organs | |||
| 6 h | |||
| Liver | 4.02 ± 0.51 | 2.55 ± 0.38 | .006 |
| Spleen | 4.64 ± 0.38 | 3.27 ± 1.26 | .03 |
| 1 d | |||
| Liver | 2.77 ± 1.03 | 2.48 ± 0.60 | .50 |
| Spleen | 3.60 ± 0.97 | 3.74 ± 0.67 | .73 |
| 3 d | |||
| Liver | 2.68 ± 1.14 | 2.41 ± 0.79 | .60 |
| Spleen | 2.81 ± 0.39 | 2.99 ± 0.78 | .57 |
| 7 d | |||
| Liver | 1.83 ± 1.04 | 1.94 ± 0.95 | .87 |
| Spleen | No growth | No growth | |
| 10 d | |||
| Liver | 0.21 ± 0.36 | 0.32 ± 0.41 | .70 |
| Spleen | No growth | No growth | |
| Abscesses | |||
| 1 d | 2.59 ± 3.58 | 0.83 ± 2.34 | .26 |
| 3 d | 6.65 ± 0.50 | 4.03 ± 3.02 | .03 |
| 7 d | 4.74 ± 0.30 | 4.03 ± 0.30 | .01 |
| 10 d | 3.17 ± 2.28 | 0.36 ± 0.36 | .02 |
A P value of <.05 was considered statistically significant.
Abbreviation: CFU, colony-forming units.
a rim101 mutant with reinserted RIM101.
b rim101 mutant.
To study the invasion of intra-abdominal organs from the peritoneal cavity, livers and spleens were assessed for colony counts. The rim101 mutant caused significantly lower tissue burdens within livers and spleens at 6 hours but not at later time points (Table 3). Histopathologic analysis revealed that both strains invaded the liver and spleen as mats of hyphae. There was a trend toward lower intra-abscess burdens of the mutant strain than the complemented strain from the earliest stages of abscess formation at 1 day; differences in burdens were statistically significant at 3, 7 and 10 days (Table 3). Indeed, 67% of mice (6/9) infected with the mutant had no abscesses at 10 days, compared with 11% of mice (1/9) infected with the complemented strain (P = .049, by the Fisher exact test). C. albicans morphology and the host cell response within abscesses were comparable in mice infected with either strain.
Identification of Rim101 Targets In Vivo
nanoString experiments were performed on peritoneal fluid recovered at 48 hours from mice infected with the rim101 null mutant or RIM101 complemented strains. The expression of 49 genes in the probe set was significantly different between the mutant and complemented strain, and expression of 38 genes varied by >2-fold (Supplementary Table 2). Among the 33 genes that were significantly downregulated in the null mutant, 61% (20/33) were previously shown to be Rim101 dependent in vitro [23, 24]. Among the 16 genes significantly upregulated in the mutant, 19% (3/16; NRG1, RBR1, and RBR2) were previously identified in vitro.
SAP5 was among the genes most strongly regulated by RIM101 during intra-abdominal candidiasis. To validate the relevance of the nanoString data, mice were infected with a rim101 mutant or a rim101 mutant that overexpressed SAP5. SAP5 overexpression restored the ability to persist within abscesses, as burdens of the overexpression strain were significantly greater than burdens of the rim101 mutant at 3 and 7 days (Table 4). SAP5 overexpression had no impact on burdens within peritoneal fluid, livers, or spleens (Table 4 and data not shown).
Table 4.
Investigation of the Role of SAP5 in the Virulence of Intra-abdominal Candidiasis
| Specimen, Sampling Time |
C. albicans Concentrations by Strain, Log10 CFU, Mean ± SD |
P | |
|---|---|---|---|
| CJN1111a | CJN793b | ||
| Peritoneal fluid | |||
| 1 d | 3.57 ± 0.26 | 2.85 ± 1.39 | .20 |
| 3 d | 2.84 ± 0.25 | 2.70 ± 0.51 | .48 |
| Abscesses | |||
| 3 d | 6.17 ± 0.30 | 5.71 ± 1.39 | .20 |
| 7 d | 5.21 ± 0.32 | 4.07 ± 1.07 | .01 |
| 10 d | 4.21 ± 0.49 | 1.93 ± 1.88 | .004 |
A P value of <.05 was considered statistically significant.
Abbreviation: CFU, colony-forming units.
a rim101 mutant with PTEF1-SAP5.
b rim101 mutant.
DISCUSSION
To our knowledge, this is the first study to use an animal model to investigate the pathogenesis of intra-abdominal candidiasis as it progresses from peritonitis to abscesses. Our mouse model is simple, reproducible, and, most importantly, suitable for studying C. albicans gene expression in vivo and measuring the relative virulence of infecting strains. There are 4 major findings from the study. First, using nanoString assays for 145 carefully chosen C. albicans genes, we identified a transcriptional signature for C. albicans within the peritoneal cavity. The data revealed biologic processes involved in adaptation to the peritoneal environment. Second, we validated the expression data by demonstrating that the alkaline pH–regulated transcription factor Rim101, which is encoded by a gene that was among the most highly expressed in vivo, plays a significant role in mediating early peritonitis and persistence of C. albicans within intra-abdominal abscesses. Third, we showed that RIM101 significantly influenced the expression of known and previously unidentified Rim101 targets during intra-abdominal candidiasis. SAP5, which encodes a secreted aspartic proteinase, was among the genes most strongly dependent on Rim101 in vivo, consistent with the relationship in vitro [23, 24]. Finally, we demonstrated that the contribution of RIM101 to the persistence of intra-abdominal abscesses was dependent, at least in part, on SAP5 expression. Taken together, the data afford insights into the pathogenesis of intra-abdominal candidiasis and highlight the power of in vivo transcriptional profiling for understanding gene function during infection.
The study of C. albicans gene expression in infected hosts has been limited by technical difficulties, most notably challenges in detecting relatively small concentrations of pathogen RNA amid abundant host RNA [25]. This issue is less relevant in the intra-abdominal candidiasis model than in hematogenously disseminated or oropharyngeal candidiasis because significant burdens of C. albicans are easily accessible in the peritoneal cavity, which has relatively few host cells. Moreover, the nanoString platform can detect C. albicans transcripts within in vivo samples when the C. albicans RNA level is as low as approximately 100 ng and constitutes <0.1% of total RNA [19, 20]. Indeed, we generated expression data that were highly reproducible and organized around several common themes.
In addition to RIM101, the most highly expressed genes on our probes included ENA2 and ENA21 (sodium efflux pumps), ALS1 and ALS4 (adhesins), GPD2 (glycerol biosynthesis enzyme), SSB1 (heat shock protein), CAT1 (catalase), and TRR1 and TRX1 (thioredoxins). The expression of RIM101 was in keeping with the alkaline pH within the peritoneal cavity. RIM101 is also a positive regulator of ENA2, so it makes sense that expression of the genes may be coordinated. A second theme was reflected by ENA2, ENA21, and GPD2, which are homologs of Saccharomyces cerevisiae high-osmolarity adaptation genes. A third theme was the upregulation of genes associated with stress response, such as SSB1, TRR1, TRX1, and CAT1. Expression of SSB1 is induced by high temperature, oxidative stress, and nutritional deficiencies in vitro, [26] conditions relevant to the peritoneal cavity. Likewise, TRR1 and TRX1 are induced by exposure to neutrophils [27], and CAT1 expression renders cells more resistant to neutrophils, oxidative stress, and peroxide [28]. Fourth, hyphal genes like HWP1, HYR1, and ECE1 were among the most weakly expressed genes in the probe set, consistent with the growth of C. albicans as yeasts within peritoneal fluid. Taken with our finding that many highly expressed genes are involved in adherence (SAP9, TDH3, TEC1, and ALS), the data suggest that yeast cells in the peritoneal cavity are attempting to bind to the surfaces of contiguous organs.
RIM101 significantly influenced the expression of 49 genes during intra-abdominal candidiasis. Sixty-one percent of downregulated genes (20/33) and 19% of upregulated genes (3/16) in our rim101 mutant were previously identified Rim101 targets in vitro [23, 24, 29]. More interesting was the identification of novel Rim101-suppressed genes, the majority of which are involved in stress responses (HSP70, HSP104, TYE7, HGT7, and CAP1). Our results are consistent with accumulating evidence that unique environmental factors at sites of C. albicans infection influence and reshape the spectrum of transcription factor–responsive genes [13, 14, 30, 31]. Rewiring of transcriptional pathways is a feature that distinguishes C. albicans from the avirulent baker's yeast S. cerevisiae and may account for the former's remarkable versatility as a human pathogen [19, 20]. The transcriptional profiling data highlight the importance of studying and validating C. albicans gene expression in diverse models, as results cannot be extrapolated from other in vivo systems or in vitro conditions [13, 14, 19].
RIM101 was previously implicated in virulence in mouse models of hematogenous disseminated candidiasis, oropharyngeal candidiasis, and keratitis [24, 29, 32]. The role of RIM101 during intra-abdominal candidiasis, however, differed in important ways from its role during other diseases. Most notably, RIM101's contribution to pathogenesis during intra-abdominal candidiasis was not related to hyphal formation. Previous studies demonstrated that Rim101 promotes hyphal growth under alkaline conditions in vitro [23, 33, 34], and the attenuated virulence of rim101 mutants during disseminated candidiasis and keratitis was associated with impaired hyphal formation [24, 32]. As mentioned, C. albicans grew exclusively as yeasts during peritonitis, despite alkaline pH and high levels of RIM101 expression. Moreover, the rim101 mutant formed dense hyphae during invasion of the spleen and liver from the peritoneal cavity and within intra-abdominal abscesses. Nevertheless, the disruption of RIM101 resulted in reduced C. albicans burdens within abscesses. Clearly, therefore, the roles of RIM101 in regulating the pathogenesis of intra-abdominal candidiasis and hyphal formation are independent.
RIM101 plays a more restricted role in the pathogenesis of disseminated candidiasis than in the pathogenesis of intra-abdominal candidiasis. Following intravenous inoculation, a rim101 mutant colonized mouse kidneys as well as wild-type but did not proliferate and sustain the infection [29]. During intra-abdominal candidiasis, RIM101 was important for both maintenance of intra-abdominal abscesses and survival of C. albicans in the first hours of peritonitis and tissue invasion. Interestingly, the temporal contributions of RIM101 to peritonitis and tissue invasion were the opposite of the contributions to disseminated candidiasis, as the rim101 mutant was attenuated at late but not early time points in the latter model. Along these lines, the mutant induced significantly greater neutrophil influx into the peritoneal cavity in the first 6 hours, which may account for reduced infectious burdens. Consistent with this hypothesis, RIM101 was important for C. albicans immune evasion during keratitis [32]. As wild-type C. albicans induces greater neutrophil influx into the peritoneal cavity over subsequent hours, it is plausible that early differences in virulence are lost. The reduced tissue invasion of the rim101 mutant at 6 hours may also stem from diminished adherence. Since adherence is a highly redundant process dependent on many genes, it is feasible that early differences are not sustained as tissue invasion progresses. Taken together, the data demonstrate that RIM101 makes distinct temporal-spatial contributions to the pathogenesis of intra-abdominal candidiasis, which could not be predicted from disseminated candidiasis or other models.
Similarly, our finding that Rim101 exerts its effects within abscesses at least in part through activation of SAP5 is noteworthy. In previous studies, overexpression of SAP5 in rim101 mutants rescued the ability to invade reconstituted human epithelium but failed to restore damage of FaDu epithelial cells [24, 35]. In fact, recent studies have cast doubt on whether SAP5 and other SAP genes play roles in virulence, as previously ascribed contributions to disseminated candidiasis and reconstituted human epithelium infections were shown to be artifacts of the ura-blaster gene disruption technique [36–38]. Our data clearly implicate SAP5 in the maintenance of intra-abdominal abscesses but not at earlier stages of peritonitis or tissue invasion. These results are consistent with previous studies of mouse peritonitis, which also found that SAP5 did not facilitate direct organ invasion after intraperitoneal inoculation [12]. The mechanisms by which SAP5 mediates persistence within abscesses are not clear but may include effects on nitrogen supply and evasion of host defenses [14, 39, 40]. On the basis of these data, we conclude that different Rim101-targeting genes contribute to various stages of intra-abdominal candidiasis.
Finally, C. albicans–host interactions during intra-abdominal candidiasis afford additional insights into pathogenesis. Following the introduction of C. albicans and stool to the peritoneal cavity, the rapid influx of neutrophils established control of the infectious burden within 24 hours. As C. albicans burdens were diminished at later time points, there was a switch to lymphocytic predominance. Invasion of intra-abdominal organs was apparent within 6 hours, and host control in the form of a brisk neutrophil response, sequestration and early abscess formation was observed at 24 hours. The number of abscesses and intra-abscess tissue burdens peaked on day 3 and fully resolved by day 14. Interestingly, C. albicans hyphae were abundant within livers and spleens, which was in sharp contrast to the paucity of hyphae and the lack of abscesses within these organs following lateral tail vein injection [41]. The reasons for the differences in the models are unclear and merit investigation in future studies. At present, we may conclude that the host limited C. albicans spread and replication during intra-abdominal candidiasis through the action of neutrophils, which was aided by structural containment within the peritoneal cavity or abscesses. Of note, C. albicans in the absence of sterile stool induced fewer abscesses and lower intra-abscess burdens. In contrast, C. albicans burdens and the host inflammatory response during peritonitis were comparable in the presence or absence of stool. In humans, C. albicans peritonitis resulting from infected peritoneal dialysis catheters is rarely associated with abscess formation, unlike peritonitis following gastrointestinal leaks or ruptures. Our data suggest that stool, as a potentiating agent that depletes complement-derived opsonins and impairs phagocytic killing [42, 43], may account for the more aggressive course in the latter cases.
In conclusion, the mouse model of intra-abdominal candidiasis is a powerful tool for studying pathogenesis and C. albicans gene expression. It should be easily adaptable to study other aspects of intra-abdominal candidiasis, including C. albicans–bacteria dual infections and detailed analyses of host responses. The model will be useful for understanding how C. albicans adapts to diverse host environments and in identifying new approaches for diagnosing, preventing, and treating intra-abdominal candidiasis.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support. This work was supported by the Department of Medicine, University of Pittsburgh (to C. J. C. and M. H. N.) and the National Institutes of Health (grant R01 AI070272 to A. P. M.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Pappas PG. Invasive candidiasis. Infect Dis Clin North Am. 2006;20:485–506. doi: 10.1016/j.idc.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Blot SI, Vandewoude KH, De Waele JJ. Candida peritonitis. Curr Opin Crit Care. 2007;13:195–9. doi: 10.1097/MCC.0b013e328028fd92. [DOI] [PubMed] [Google Scholar]
- 3.Carneiro HA, Mavrakis A, Mylonakis E. Candida peritonitis: an update on the latest research and treatments. World J Surg. 2011;35:2650–9. doi: 10.1007/s00268-011-1305-2. [DOI] [PubMed] [Google Scholar]
- 4.Party BSfACW. Management of deep Candida infection in surgical and intensive care unit patients. Intensive Care Med. 1994;20:522–8. [PubMed] [Google Scholar]
- 5.Blot S, De Waele JJ. Critical issues in the clinical management of complicated intra-abdominal infections. Drugs. 2005;65:1611–20. doi: 10.2165/00003495-200565120-00002. [DOI] [PubMed] [Google Scholar]
- 6.Sandven P, Qvist H, Skovlund E, Giercksky KE. Significance of Candida recovered from intraoperative specimens in patients with intra-abdominal perforations. Crit Care Med. 2002;30:541–7. doi: 10.1097/00003246-200203000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Sotto A, Lefrant JY, Fabbro-Peray P, et al. Evaluation of antimicrobial therapy management of 120 consecutive patients with secondary peritonitis. J Antimicrob Chemother. 2002;50:569–76. doi: 10.1093/jac/dkf167. [DOI] [PubMed] [Google Scholar]
- 8.Charles PE. Multifocal Candida species colonization as a trigger for early antifungal therapy in critically ill patients: what about other risk factors for fungal infection? Crit Care Med. 2006;34:913–4. doi: 10.1097/01.CCM.0000202435.98240.ED. [DOI] [PubMed] [Google Scholar]
- 9.Calandra T, Bille J, Schneider R, Mosimann F, Francioli P. Clinical significance of Candida isolated from peritoneum in surgical patients. Lancet. 1989;2:1437–40. doi: 10.1016/s0140-6736(89)92043-6. [DOI] [PubMed] [Google Scholar]
- 10.Rutledge R, Mandel SR, Wild RE. Candida species. Insignificant contaminant or pathogenic species. Am Surg. 1986;52:299–302. [PubMed] [Google Scholar]
- 11.Marsh PK, Tally FP, Kellum J, Callow A, Gorbach SL. Candida infections in surgical patients. Ann Surg. 1983;198:42–7. doi: 10.1097/00000658-198307000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felk A, Kretschmar M, Albrecht A, et al. Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs. Infect Immun. 2002;70:3689–700. doi: 10.1128/IAI.70.7.3689-3700.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staib P, Binder A, Kretschmar M, Nichterlein T, Schroppel K, Morschhauser J. Tec1p-independent activation of a hypha-associated Candida albicans virulence gene during infection. Infect Immun. 2004;72:2386–9. doi: 10.1128/IAI.72.4.2386-2389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staib P, Kretschmar M, Nichterlein T, Hof H, Morschhauser J. Transcriptional regulators Cph1p and Efg1p mediate activation of the Candida albicans virulence gene SAP5 during infection. Infect Immun. 2002;70:921–7. doi: 10.1128/IAI.70.2.921-927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vonk AG, Netea MG, van Krieken JH, van der Meer JW, Kullberg BJ. Delayed clearance of intraabdominal abscesses caused by Candida albicans in tumor necrosis factor-alpha- and lymphotoxin-alpha-deficient mice. J Infect Dis. 2002;186:1815–22. doi: 10.1086/345818. [DOI] [PubMed] [Google Scholar]
- 16.Vonk AG, Netea MG, van Krieken JH, Verweij PE, van der Meer JW, Kullberg BJ. Treatment of intra-abdominal abscesses caused by Candida albicans with antifungal agents and recombinant murine granulocyte colony-stimulating factor. Antimicrob Agents Chemother. 2003;47:3688–93. doi: 10.1128/AAC.47.12.3688-3693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawyer RG, Adams RB, Rosenlof LK, May AK, Pruett TL. Effectiveness of fluconazole in murine Candida albicans and bacterial C. albicans peritonitis and abscess formation. J Med Vet Mycol. 1995;33:131–6. [PubMed] [Google Scholar]
- 18.Sawyer RG, Adams RB, Rosenlof LK, May AK, Pruett TL. The role of Candida albicans in the pathogenesis of experimental fungal/bacterial peritonitis and abscess formation. Am Surg. 1995;61:726–31. [PubMed] [Google Scholar]
- 19.Fanning S, Xu W, Solis N, Woolford CA, Filler SG, Mitchell AP. Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryot Cell. 2012;11:896–904. doi: 10.1128/EC.00103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finkel J, Xu W, Huang D, et al. Portrait of Candida albicans adherence regulators. PLoS Pathog. 2012;8(2):e1002525. doi: 10.1371/journal.ppat.1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 22.Ford CW, Hamel JC, Stapert D, Yancey RJ. Establishment of an experimental model of a Staphylococcus aureus abscess in mice by use of dextran and gelatin microcarriers. J Med Microbiol. 1989;28:259–66. doi: 10.1099/00222615-28-4-259. [DOI] [PubMed] [Google Scholar]
- 23.Davis D, Wilson RB, Mitchell AP. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol Cell Biol. 2000;20:971–8. doi: 10.1128/mcb.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nobile CJ, Solis N, Myers CL, et al. Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell Microbiol. 2008;10:2180–96. doi: 10.1111/j.1462-5822.2008.01198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown AJ, Odds FC, Gow NA. Infection-related gene expression in Candida albicans. Curr Opin Microbiol. 2007;10:307–13. doi: 10.1016/j.mib.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Maneu V, Roig P, Gozalbo D. Complementation of Saccharomyces cerevisiae mutations in genes involved in translation and protein folding (EFB1 and SSB1) with Candida albicans cloned genes. Res Microbiol. 2000;151:739–46. doi: 10.1016/s0923-2508(00)01139-6. [DOI] [PubMed] [Google Scholar]
- 27.Enjalbert B, Moran GP, Vaughan C, et al. Genome-wide gene expression profiling and a forward genetic screen show that differential expression of the sodium ion transporter Ena21 contributes to the differential tolerance of Candida albicans and Candida dubliniensis to osmotic stress. Mol Microbiol. 2009;72:216–28. doi: 10.1111/j.1365-2958.2009.06640.x. [DOI] [PubMed] [Google Scholar]
- 28.Nakagawa Y, Kanbe T, Mizuguchi I. Disruption of the human pathogenic yeast Candida albicans catalase gene decreases survival in mouse-model infection and elevates susceptibility to higher temperature and to detergents. Microbiol Immunol. 2003;47:395–403. doi: 10.1111/j.1348-0421.2003.tb03376.x. [DOI] [PubMed] [Google Scholar]
- 29.Davis D, Edwards JE, Jr, Mitchell AP, Ibrahim AS. Candida albicans RIM101pH response pathway is required for host-pathogen interactions. Infect Immun. 2000;68:5953–9. doi: 10.1128/iai.68.10.5953-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White SJ, Rosenbach A, Lephart P, et al. Self-regulation of Candida albicans population size during GI colonization. PLoS Pathog. 2007;3:e184. doi: 10.1371/journal.ppat.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenbach A, Dignard D, Pierce JV, Whiteway M, Kumamoto CA. Adaptations of Candida albicans for growth in the mammalian intestinal tract. Eukaryot Cell. 2010;9:1075–86. doi: 10.1128/EC.00034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan X, Mitchell BM, Hua X, Davis DA, Wilhelmus KR. The RIM101 signal transduction pathway regulates Candida albicans virulence during experimental keratomycosis. Invest Ophthalmol Vis Sci. 2010;51:4668–76. doi: 10.1167/iovs.09-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bensen ES, Martin SJ, Li M, Berman J, Davis DA. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol Microbiol. 2004;54:1335–51. doi: 10.1111/j.1365-2958.2004.04350.x. [DOI] [PubMed] [Google Scholar]
- 34.Ramon AM, Porta A, Fonzi WA. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J Bacteriol. 1999;181:7524–30. doi: 10.1128/jb.181.24.7524-7530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villar CC, Kashleva H, Nobile CJ, Mitchell AP, Dongari-Bagtzoglou A. Mucosal tissue invasion by Candida albicans is associated with E-cadherin degradation, mediated by transcription factor Rim101p and protease Sap5p. Infect Immun. 2007;75:2126–35. doi: 10.1128/IAI.00054-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Correia A, Lermann U, Teixeira L, et al. Limited role of secreted aspartyl proteinases Sap1 to Sap6 in Candida albicans virulence and host immune response in murine hematogenously disseminated candidiasis. Infect Immun. 2010;78:4839–49. doi: 10.1128/IAI.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lermann U, Morschhauser J. Secreted aspartic proteases are not required for invasion of reconstituted human epithelia by Candida albicans. Microbiology. 2008;154(Pt 11):3281–95. doi: 10.1099/mic.0.2008/022525-0. [DOI] [PubMed] [Google Scholar]
- 38.Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67:400–28. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaminishi H, Miyaguchi H, Tamaki T, et al. Degradation of humoral host defense by Candida albicans proteinase. Infect Immun. 1995;63:984–8. doi: 10.1128/iai.63.3.984-988.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruchel R. Cleavage of immunoglobulins by pathogenic yeasts of the genus Candida. Microbiol Sci. 1986;3:316–9. [PubMed] [Google Scholar]
- 41.Lionakis MS, Lim JK, Lee CC, Murphy PM. Organ-specific innate immune responses in a mouse model of invasive candidiasis. J Innate Immun. 2011;3:180–99. doi: 10.1159/000321157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finlay-Jones JJ, Davies KV, Sturm LP, Kenny PA, Hart PH. Inflammatory processes in a murine model of intra-abdominal abscess formation. J Leukoc Biol. 1999;66:583–7. doi: 10.1002/jlb.66.4.583. [DOI] [PubMed] [Google Scholar]
- 43.Finlay-Jones JJ, Kenny PA, Nulsen MF, Spencer LK, Hill NL, McDonald PJ. Pathogenesis of intraabdominal abscess formation: abscess-potentiating agents and inhibition of complement-dependent opsonization of abscess-inducing bacteria. J Infect Dis. 1991;164:1173–9. doi: 10.1093/infdis/164.6.1173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.