Background: An interaction between arrestin-3 and MKK7 has never been elucidated.
Results: Arrestin-3 directly binds MKK7 and promotes JNK3α2 phosphorylation by MKK7 in vitro and in intact cells.
Conclusion: Arrestin-3 recruits JNK3α2 and both upstream MKKs.
Significance: Arrestin-3 promotes full JNK3α2 activation; MKK binding is regulated by JNK3α2.
Keywords: Arrestin, Cell Signaling, Jun N-terminal Kinase (JNK), MAP Kinases (MAPKs), Protein Kinases, Protein Phosphorylation
Abstract
Arrestin-3 was previously shown to bind JNK3α2, MKK4, and ASK1. However, full JNK3α2 activation requires phosphorylation by both MKK4 and MKK7. Using purified proteins we show that arrestin-3 directly interacts with MKK7 and promotes JNK3α2 phosphorylation by both MKK4 and MKK7 in vitro as well as in intact cells. The binding of JNK3α2 promotes an arrestin-3 interaction with MKK4 while reducing its binding to MKK7. Interestingly, the arrestin-3 concentration optimal for scaffolding the MKK7-JNK3α2 module is ∼10-fold higher than for the MKK4-JNK3α2 module. The data provide a mechanistic basis for arrestin-3-dependent activation of JNK3α2. The opposite effects of JNK3α2 on arrestin-3 interactions with MKK4 and MKK7 is the first demonstration that the kinase components in mammalian MAPK cascades regulate each other's interactions with a scaffold protein. The results show how signaling outcomes can be affected by the relative expression of scaffolding proteins and components of signaling cascades that they assemble.
Introduction
Arrestins specifically bind active phosphorylated G-protein-coupled receptors, blocking further G protein activation, targeting receptors for internalization, and initiating alternative signaling (for review, see Refs. 1 and 2). Among four mammalian subtypes (3), two ubiquitously expressed non-visual arrestins, arrestin-22 and -3, act as multifunctional adaptors, interacting with dozens of non-receptors partners, including mitogen-activated protein (MAP)3 kinases (2, 4). MAP kinases form highly conserved signaling cascades stimulated by a wide variety of extracellular signals, such as growth factors, cytokines, and environmental stress (5). Canonical MAP kinase cascades consist of three kinases that successively phosphorylate and activate the downstream components (5, 6). In many cases signal propagation through these kinase cascades is regulated by scaffolding proteins that assemble the kinases into multiprotein complexes (7–9). Arrestins were reported to serve as scaffolds promoting the activation of all three main subfamilies of MAP kinases: JNK (10), ERK (11), and p38 (12).
The JNK signaling pathway is involved in the regulation of many cellular events, including growth control, transformation, and apoptosis (13). The JNK family consists of several ubiquitously expressed JNK1 and JNK2 isoforms and neuron-specific JNK3, which is also expressed in the heart and testes (14). Differential splicing and exon usage yield 10 different JNK isoforms (15). JNKs are activated by concomitant phosphorylation of a threonine and a tyrosine residue within a conserved Thr-Pro-Tyr (TPY) motif in the activation loop of the kinase domain. Interestingly, two upstream MAP kinase kinases, MKK4 and MKK7, preferentially phosphorylate distinct JNK activation sites: MKK4 phosphorylates tyrosine, whereas MKK7 phosphorylates threonine. The monophosphorylation of JNKs on either residue is reportedly sufficient to partially activate the kinase and induce phosphorylation of downstream substrates; however, full activation of JNKs requires both MKK4 and MKK7 (14, 16). Previous studies showed that biological functions of MKK4 and MKK7 are non-redundant despite the fact that they share several upstream activators and scaffold proteins (17). The actual mechanism is determined by different stimuli, physiological and pathological processes, and differential tissue distribution of MKKs (18). The biological significance and the underlying molecular mechanisms of differential activation of JNK kinases by distinct MKKs remain largely unknown.
Arrestin-3 was shown to bind ASK1, MKK4, and JNK3 and promote JNK3 activation (10, 19–22). Arrestin-3 scaffolding of the ASK1-MKK4-JNK3 cascade has been extensively studied (10, 19–23). However, whether arrestin-3 is able to promote JNK3 phosphorylation by MKK7 remains unknown. Here, we provide the first evidence that arrestin-3 scaffolds the MKK7-JNK3α2 signaling complex. We found that the phosphorylation of the threonine residue within the TPY motif was promoted by arrestin-3 in COS-7 cells. We showed that arrestin-3 directly binds to MKK7, comparably to MKK4. Moreover, we found that JNK3α2 enhanced the association between arrestin-3 with MKK4 while reducing arrestin-3 binding to MKK7. We also compared the binding of arrestin-3 to active and inactive MKK4 and MKK7. In both cases active MKKs bind arrestin-3 as well as inactive ones, and JNK3α2 differentially regulates the binding of the two upstream MKKs to arrestin-3. This differential regulation of the binding of upstream kinases by JNK3α2 may underlie the higher optimal arrestin-3 concentration for JNK3α2 phosphorylation by MKK7 than by MKK4. We found that the two MKKs compete for arrestin-3 binding. Finally, we found that arrestin-3 facilitates JNK3α2 activation by either MKK in intact cells. These findings represent the first evidence that arrestin-3 is not only capable of scaffolding MKK4-JNK3α2 but also the MKK7-JNK3α2 signaling complex. This is also the first demonstration that the kinase components in mammalian mitogen-activated protein kinase (MAPK) cascades regulate each other's interactions with scaffold proteins.
EXPERIMENTAL PROCEDURES
Materials
[γ-32P]ATP was from Perkin-Elmer (Waltham, MA). All restriction enzymes were from New England Biolabs (Ipswich, MA). Cell culture reagents and media were from Mediatech (Manassas, VA) or Invitrogen. All other chemicals were from sources previously described (22).
Protein Purification
Arrestin-3, JNK3α2, GST-MKK4, and GST-tagged proteins were purified as previously described (22, 24).
Purification of Tag-less MKK4
GST-MKK4 was bound to glutathione-Sepharose high performance beads, which were re-suspended in 10 ml of a suspension buffer containing 140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, 0.1% 2-mercaptoethanol, and 20% glycerol, pH 7.3). 20 units of thrombin (Novagen) were added to the suspension and incubated at room temperature overnight. The column was washed with ∼3 column volumes of the same buffer and analyzed by SDS-PAGE. Tag-less MKK4 fractions were pooled and further purified by a Mono Q HR 10/10 anion exchange column that was pre-equilibrated with Mono Q buffer (20 mm Tris, pH 8.0, 0.03% Brij-30 (v/v), 0.1% β-mercaptoethanol (v/v)). The column was eluted with 15–17 column volumes of Mono Q buffer with a linear gradient of 0–0.5 m NaCl. Fractions of pure tag-less MKK4 were collected, dialyzed, and stored in buffer S (25 mm HEPES, pH 7.5, 50 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 2 mm DTT) containing 20% glycerol at −80 °C until further use.
Expression and Purification of (WT) GST-MKK7-His6
The pGEX 4T1 vector containing DNA encoding full-length Mus musculus mitogen-activated protein kinase kinase 7 (GenBankTM accession number NM_011944) with N-terminal cleavable GST tag, and C-terminal cleavable His tag was prepared as described previously (22) and transformed into BL21 (DE3) electro-competent cells. Cells from a single colony were used to inoculate 40 ml of Terrific Broth medium containing 100 μg/ml ampicillin and grown overnight at 37 °C. The culture was diluted 100-fold into the same media containing 50 μg/ml ampicillin and grown at 37 °C with shaking at 250 rpm to an optical density of 0.6 at 600 nm. The cells were then induced with 25 μm isopropylthio-β-d-galactoside and cultured for an additional 20 h at 25 °C with shaking at 250 rpm. The cells were then harvested, immediately flash-frozen in liquid nitrogen, and stored at −80 °C. Cells were then lysed in 200 ml of buffer A (20 mm Tris, pH 8.0, 0.03% Brij-30, 0.1% (v/v) β-mercaptoethanol, 5 mm imidazole, 1 mm benzamidine, 0.1 mm PMSF, and 0.1 mm tosylphenylalanyl chloromethyl ketone) containing 0.5 m NaCl, 0.2 mg/ml lysozyme, 1 mm MgCl2, and 20% glycerol. The mixture was incubated at 4 °C for 30 min, then Triton-100 was added to a final concentration of 1% (v/v), and the incubation at 4 °C was extended for another 30 min. The cell lysate was sonicated (at 5 s pulses with 5-s intervals) at 4 °C for 5 min and then cleared by centrifugation at 18,000 × g for 30 min. The supernatant was then agitated with nickel-nitrilotriacetic acid beads for 1 h at 4 °C. After washing the beads with 200 ml of buffer A containing 10 mm imidazole and 20% glycerol, the GST-MKK7-His6 protein was eluted with 40 ml buffer A, pH 8.0, containing 200 mm imidazole and 20% glycerol. The eluted protein was collected and dialyzed into buffer S (25 mm HEPES pH 7.5, 50 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 2 mm DTT) containing 20% glycerol. Protein concentration was measured using the Bradford reagent.
Expression and Purification of Maltose-binding Protein (MBP)-tagged Arrestin-3
To make MBP-arrestin-3, arrestin-3 cDNA was subcloned into pMal-p2T (25) (a generous gift from Dr. Noriyuki Matsuda, Tokyo Institute of Medical Science) between EcoRI and NotI sites. Escherichia coli strain BL21-codon plus (DE3)-RIL was used for expression. The cells were grown in LB at 30 °C overnight to A600 = 1.5–2.0 and then induced with 1 mm isopropylthio-β-d-galactoside at 30 °C for 5–6 h. The cells from 1 liter were pelleted by centrifugation, resuspended in buffer containing 150 mm NaCl, and 20 mm Tris-HCl, pH 7.5 (buffer A), and lysed by freezing and thawing in the presence of lysozyme (3 mg/liter) followed by sonication (3 × 15 s; 80% amplitude; Sonic Dismembrator Model 500, Fisher). After centrifugation (9000 rpm for 90 min, Sorvall SLA-3000 rotor), the supernatant was incubated with 1 ml of amylose beads (50% slurry, New England Biolabs) at 4 °C for 2 h. Supernatant was removed by centrifugation (3000 rpm for 10 min), and the beads were washed by 20 ml of buffer A 3 times. 5 ml of elution buffer containing 50 mm maltose was added to elute the MBP-arrestin-3 proteins. The eluate was dialyzed 3 times against buffer A to remove maltose (>3 h each time). The protein samples were collected, concentrated to ∼1 mg/ml, and stored at −80 °C. MBP was expressed in empty pMal-p2T vector and purified using the same procedure.
Active MKK4 and MKK7
GST-MKK4 (full-length wild-type human mitogen-activated protein kinase kinase 4 (MAP2K4) (GenBankTM accession number NM_003010) with an N-terminal cleavable GST tag) was expressed, purified, and activated essentially as described previously (21).
GST-MKK7-His6
4 μm GST-MKK7-His6 was activated by incubation with 2 μm GST-MEKK1c (C-terminal 320 amino acids corresponding to the catalytic domain) (21) and 4 mm ATP for 60 min at 30 °C in 10 ml of activation buffer B (25 mm Hepes, pH 7.5, 2 mm dithiothreitol, 20 mm MgCl2, 0.1 mm EDTA, 0.1 mm EGTA) and re-purified using a nickel affinity column that separates the active GST-MKK7-His6 from GST-MEKK1c. The activated GST-MKK7-His6 was stored in buffer S containing 20% glycerol at −80 °C until further use.
MBP Pulldown
Binding of MKK4 and MKK7 to arrestin-3 was assayed by MBP pulldown with MBP-arrestin-3 immobilized on amylose resin (New England Biolabs) according to the manufacturer's instructions. Briefly, 25 μl of purified MBP-arrestin-3 (5 μg) were incubated with 25 μl of amylose resin (50% slurry) in binding buffer (50 mm HEPES-Na, pH 7.3, 150 mm NaCl) at 4 °C with gentle rotation for 2 h. Subsequently, 100 μl of protein solutions containing active (phosphorylated by GST-MEKK1c) or inactive MKK4 or MKK7 (5 μg) along with different amounts of JNK3α2 (0, 2, 5, 10 μg) were added, and the suspensions were incubated at 4 °C for 2 h. The suspension was transferred to centrifuge filters (Ultrafree, Millipore) and washed three times with binding buffer. The proteins were eluted from resin by the addition of 100 μl of elution buffer (50 mm maltose, 50 mm HEPES-Na, pH 7.3, and 150 mm NaCl). Eluates were analyzed by SDS-PAGE and Western blotting. Samples obtained with MBP bound to the resin served as controls for nonspecific binding. To evaluate the effects of MKK7 on the arrestin3-MKK4 binding, the interaction between arrestin-3 and MKK4 was assayed using immobilized MBP-arrestin-3 (5 μg) to pull down tag-less MKK4 (5 μg) in the presence of various amounts of GST-MKK7 (0, 5, 10 μg). Eluates were analyzed by SDS-PAGE and Western blotting.
GST Pulldown
To test how many MKK molecules can bind arrestin-3 simultaneously, we performed GST pulldown using GST-MKK7 to pull down tag-less MKK4 in the presence of different amounts of arrestin-3. Briefly, 25 μl of GST-MKK7 (3 μg) was incubated with 25 μl of glutathione-agarose beads (50% slurry, Sigma) at 4 °C for 2 h. Subsequently, 50 μl of protein solutions containing MKK4 (3 μg) and different amounts of arrestin-3 (0, 5, 10 μg) were added, and the suspensions were incubated at 4 °C for 2 h. The suspension was transferred to centrifuge filters (Ultrafree, Millipore) and washed 3 times with binding buffer. The proteins were eluted from resin by the addition of 100 μl of elution buffer (50 mm reduced glutathione, 4 mm DTT, 50 mm HEPES-Na, pH 7.3, 150 mm NaCl). Eluates were analyzed by SDS-PAGE and Western blotting.
Cell Culture and Transient Transfection
African green monkey cells (COS-7) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen) plus penicillin and streptomycin at 37 °C in a humidified incubator with 5% CO2. The cells were plated at 80–90% confluence and transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were serum-starved overnight before all experiments and used 48 h post-transfection.
Immunoprecipitation
Cells (60-mm plates) were lysed in 0.5 ml of lysis buffer (50 mm Tris, pH 7.4, 2 mm EDTA, 250 mm NaCl, 10% glycerol, 0.5% Nonidet P-40, 20 mm NaF, 1 mm sodium orthovanadate, 10 mm N-ethylmaleimide, 2 mm benzamidine, and 1 mm phenylmethylsulfonyl fluoride) for 60 min at 4 °C. After centrifugation (10 min at 9000 rpm, 4 °C), supernatants containing around 500 μg of total protein were precleared by incubation with 35 μl of protein G-agarose (50% slurry, Millipore). Supernatant was incubated with primary antibodies for 2 h or overnight followed by the addition of 35 μl of protein G-agarose beads (50% slurry, Millipore) for 2 h. The beads were washed 3 times with 1 ml of lysis buffer, and the proteins were eluted with 50 μl of SDS sample buffer, boiled for 5 min, and analyzed by Western blot.
In Vitro JNK3a2 Phosphorylation
The effect of arrestins on the phosphorylation of JNK3α2 by MKK7 or MKK4 was analyzed by an in vitro kinase activity assay, as previously described (22). Briefly, the assays were conducted in 10 μl containing the following final concentrations: 50 nm active MKK7 or MKK4, 0.5 μm JNK3α2, and 0–30 μm arrestin-3. The reactions were initiated by the addition of 0.2 mm ATP (4 μCi of γ-32P), and the mixtures were incubated individually at 30 °C for the indicated times. The reactions were stopped by the addition of 15 μl of Laemmli SDS sample buffer (Sigma) and subjected to SDS-PAGE (10%). The gels were stained with Coomassie Blue and dried. Phosphorylated JNK3α2 was visualized by autoradiography. The bands were cut out, and the radioactivity was measured in a Tri-Carb liquid scintillation counter (PerkinElmer Life Sciences).
Mathematical Modeling
A simple interaction model that only requires concentrations of components and equilibrium affinity constants as an input (KinTek Explorer Professional Version 3.0.2394) from KinTek Corp. was used. Based on these values, the model calculates the concentrations of all possible complexes.
RESULTS
Arrestin-3 Promotes the Phosphorylation of Both Tyrosine and Threonine on JNK3α2
Activation of JNK relies on two upstream MAP kinase kinases, MKK4 and MKK7, each of which preferentially phosphorylates a distinct site, tyrosine (MKK4) and threonine (MKK7) (Fig. 1B) (16, 17). Although to reach the maximal activity of JNK both tyrosine and threonine must be phosphorylated, JNK monophosphorylated at either site was reported to be capable of phosphorylating downstream substrates (16, 26). How JNK makes the appropriate choice of its upstream kinases is a challenging question, as the activation of JNK depends on a wide variety of factors, including the nature of a stimulus and relevant physiological and pathological processes as well as differential tissue distribution of MKKs and scaffolding proteins. Previous studies showed that scaffold proteins play a significant role in determining the differential requirements of upstream kinases for JNK activation (27). Although arrestin-3 was shown to scaffold the JNK3 signaling cascade, previous studies focused on the ASK1-MKK4-JNK3 module (10, 19–22). It was never established whether arrestin-3 facilitates only the monophosphorylation of JNK3 on tyrosine by MKK4 or the dual phosphorylation of JNK3.
FIGURE 1.
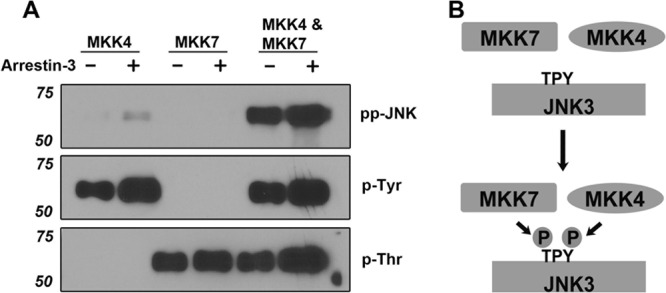
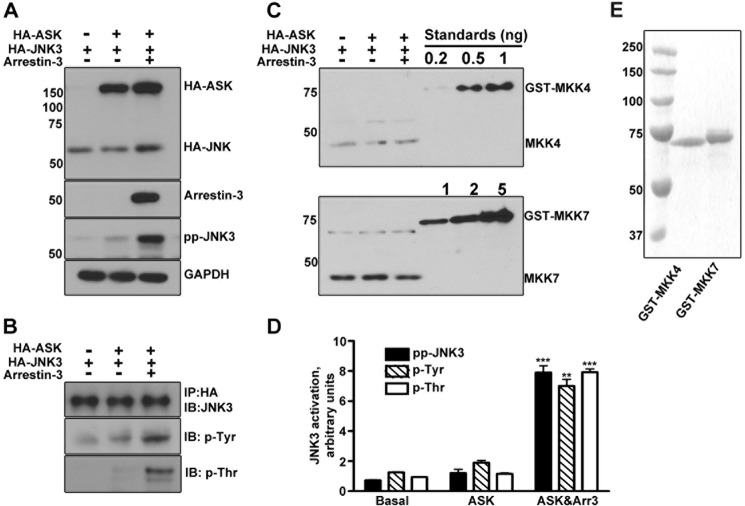
The specificity of MKK4 and MKK7 toward JNK3α2 is not affected by arrestin-3. A, Western blot of MKK-phosphorylated JNK3α2. In the presence (+) or absence (−) of arrestin-3 (1 μm), 1 μm JNK3α2 was incubated with active MKK4, MKK7, or MKK4+MKK7 combination for 3 min. The reactions were stopped by SDS sample buffer and analyzed by Western blot with phospho-JNK3α2 (ppJNK), phosphotyrosine (p-Tyr), and phosphothreonine (p-Thr) antibodies, as indicated. B, a schematic showing that full activation of JNK3α2 requires both MKK4 and MKK7 that phosphorylate Tyr-223 and Thr-221, respectively.
To answer this question, we first examined the specificity of each MKK toward JNK3α2 in the absence and presence of arrestin-3 by using purified components in vitro. Fig. 1A shows Western blots of JNK3α2 samples phosphorylated by MKK4, MKK7, or the combination of MKK4 and MKK7, visualized by three different antibodies recognizing phosphotyrosine (Tyr(P)), phosphothreonine (Thr(P)), and double-phosphorylated JNK (ppJNK). Phospho-amino acid-specific antibodies only detect JNK3α2 phosphorylated by MKK4 (Tyr(P)) or MKK7 (Thr(P)). The phospho-JNK antibody has a striking preference for double-phosphorylated ppJNK3α2 (by MKK4 + MKK7) over either of the mono-phosphorylated species. It was reported that MKK4 could not phosphorylate JNK3α1 alone, because the threonine residue had to be phosphorylated first by MKK7 (26). Our data show that either MKK4 or MKK7 alone can effectively phosphorylate its substrate JNK3α2 (Fig. 1). In agreement with previous reports (16, 17, 26), the tyrosine and threonine located in the TPY motif of JNK3α2 can only be phosphorylated by MKK4 and MKK7, respectively. The presence of arrestin-3 does not alter the specificity of either MKK for its target (Fig. 1A).
To determine whether the phosphorylation of either site in JNK3α2 is enhanced by arrestin-3 in the cellular environment, we expressed ASK1, JNK3α2, and arrestin-3 individually and in different combinations in COS-7 cells. Co-expression of arrestin-3 with ASK1+JNK3α2 increased the level of phospho-JNK3α2 ∼10-fold (Fig. 2A), in agreement with previous reports (19–21). ASK1-initiated JNK3α2 activation apparently relied on the endogenous MKK4 and MKK7. To test whether the exogenous expression of JNK3α2, ASK1, or arrestin-3 influences endogenous MKK4 and MKK7 expression in cells, we quantified MKK4 and MKK7 using purified GST-MKK4 and GST-MKK7 proteins as standards (Fig. 2C). The results showed that the levels of endogenous MKK4 and MKK7 were not affected by the overexpression of ASK1, JNK3α2, and arrestin-3 individually or in different combinations, further supporting the notion that the expression of arrestin-3 underlies the observed increase in phospho-JNK3α2. We found that in COS-7 cells the amounts of endogenous MKK7 (31.2 ± 10.1 fmol/10 μg of cell lysate protein) were ∼8-fold higher than MKK4 (3.9 ± 0.3 fmol/10 μg of cell lysate protein) (Fig. 2C).
FIGURE 2.
The phosphorylation of both tyrosine and threonine on JNK3α2 is enhanced by arrestin-3. A, COS-7 cells were transfected with JNK3α2 alone or in combination with HA-ASK1 with or without arrestin-3. Cell lysates were analyzed by Western blot using indicated antibodies. GADPH served as the loading control. B, HA-JNK3α2 in cell lysates was immunoprecipitated (IP) by an anti-HA antibody. The immunoprecipitate was analyzed by Western blot (IB) using anti-HA, Tyr(P) (p-Tyr), or Thr(P) (p-Thr) antibodies. C, cell lysates containing 10 or 5 μg of total protein were used to quantify endogenous MKK4 and MKK7, respectively, by Western blot with antibodies recognizing MKK4 (upper panel) or MKK7 (lower panel). Purified GST-MKK4 (0.2, 0.5, 1 ng) and GST-MKK7 (1, 2, 5 ng) were used as standards. D, quantitative analysis of arrestin-3 effect on JNK3α2 phosphorylation (representative blots shown in A (ppJNK) and B (Tyr(P) and Thr(P)). Means ± S.D. (n = 3) of relative intensity of phospho-JNK3 (***, p < 0.001, as compared with the basal level in cells co-expressing ASK1 and JNK3 without arrestin-3), Tyr(P) (**, p < 0.01), and Thr(P) (***, p < 0.001) bands are shown. E, Coomassie Blue-stained SDS-PAGE gel showing the purity of GST-MKK4 and GST-MKK7 used as standards to generate calibration curves (1 μg of each protein was loaded).
We used phospho-Tyr and phospho-Thr antibodies to determine whether arrestin-3 predominantly promotes monophosphorylation of the tyrosine site or full activation of JNK3α2. To avoid interference from other kinases, HA-JNK3α2 was immunoprecipitated with anti-HA high affinity antibody (rat, Roche Applied Science) (Fig. 2B). We found that co-expression of arrestin-3 increased not only phospho-Tyr JNK3α2 (∼5-fold) but also phospho-Thr JNK3α2 (∼8-fold) (Fig. 2, B and D). The facilitation by arrestin-3 of threonine phosphorylation of JNK3α2 was a surprising finding, because MKK7 is the only MAP kinase kinase known to phosphorylate threonine residues on JNK3. To date no previous study using co-immunoprecipitation and mass spectrometry-based proteomics has reported an association between arrestin-3 and MKK7 (4, 10). These results demonstrate that arrestin-3 is able to promote JNK3α2 phosphorylation by MKK7, suggesting that the molecular mechanism of this phenomenon must be elucidated.
JNK3α2 Binding Differentially Affects Direct Association of Arrestin-3 with MKK4 and MKK7
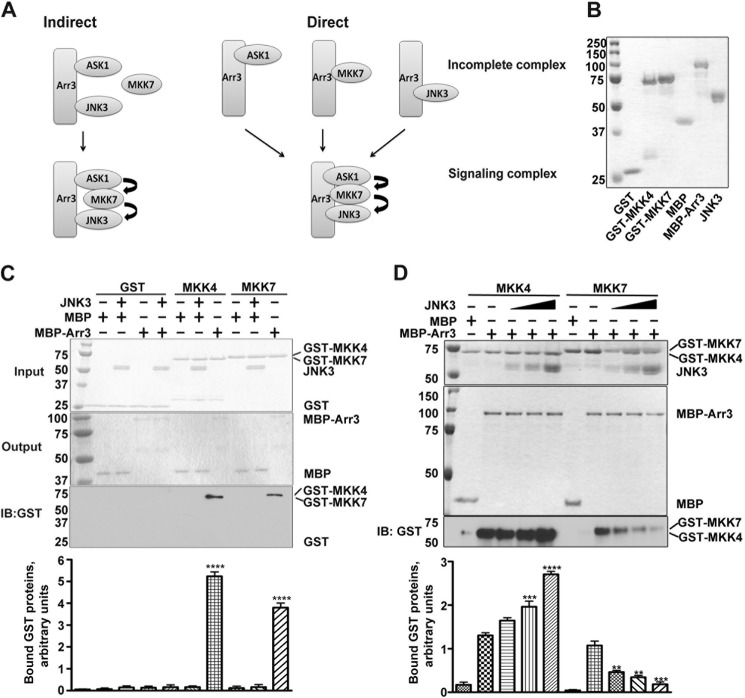
Two distinct models can explain how arrestin-3 facilitates JNK3α2 activation by MKK7. It is conceivable that arrestin-3 directly associates with ASK1 (MAPKK kinase) and JNK3α2 (MAPK), whereas MKK7 (MAPKK) is recruited by ASK1 and JNK3 without directly associating with arrestin-3. It is also possible that arrestin-3 binds all three kinases directly to form a complete signaling module (Fig. 3A). Because co-immunoprecipitation data are inherently ambiguous, earlier studies interpreted essentially the same findings as evidence of indirect (10) or direct (19–21) arrestin-3 interaction with MKK4. Recently we definitively demonstrated direct interaction between MKK4 and arrestin-3 using two purified proteins (22). Therefore, we investigated the mechanism of arrestin-3-dependent JNK3α2 phosphorylation by MKK7 using the least ambiguous approach (Fig. 3). To this end, we used purified proteins (Fig. 3B) in the in vitro pulldown assays to compare the interactions of MKK7, MKK4, and MBP-arrestin-3, with equal amounts of MBP as negative controls (Fig. 3, C and D). Because the same bait protein, MBP-arrestin-3, was used to pull down target proteins carrying the same tag (GST), this MBP pulldown assay allowed us to quantitatively compare the binding of two different MKKs (Fig. 3, C and D). We found that arrestin-3 directly binds both MKK4 and MKK7 with somewhat higher affinity for MKK4 than MKK7. No nonspecific binding to MBP was detected (Fig. 3C). These data provide the first experimental evidence that arrestin-3 directly binds MKK7.
FIGURE 3.
JNK3α2 has an opposite effect on arrestin-3 binding to MKK4 and MKK7. A, two proposed models of arrestin-3-mediated scaffolding of ASK1-MKK7-JNK3 cascade. Indirect model (left panel), MKK7 is recruited to the complex through ASK1 and JNK3 without direct binding to arrestin-3. Direct model (right panel), arrestin-3 directly binds all three kinases to form a complete signaling complex. B, Coomassie-stained SDS-PAGE gel showing the purity of proteins used in the assays shown in C and D. C, an MBP pulldown assay showed that both MKK4 and MKK7 were retained by MBP-arrestin-3 but not by MBP (control). No nonspecific binding of GST to MBP-arrestin-3 or MBP was detected. Top panel, Coomassie Blue-stained SDS-PAGE gel showing the input of GST fusion proteins and JNK3α2. Middle panel, Coomassie Blue staining of MBP and MBP-Arr3 eluted from the amylose column. Bottom panels, Western blot (IB) of the eluates from the indicated columns and bar graph showing the quantification of the Western blot; means ± S.D. (n = 3) of the relative intensity of bands are shown (****, p < 0.0001, as compared with MBP control). Note that both GST-tagged MKK4 and MKK7 are retained by MBP-Arr3, but not by MBP, and no nonspecific effects of JNK3α2 on the pulldown were observed. D, JNK3α2 enhanced the binding of MKK4 but decreased the binding of MKK7 to arrestin-3. Top panel, Coomassie staining of input of the indicated proteins (10%). Middle panel, Coomassie staining of eluted proteins (20%). Bottom panel, Western blots of ⅕ of the eluate to detect the indicated GST-tagged proteins. The amounts of JNK3α2 in the assay were (left to right): 2, 5, and 10 μg. Representative gels are shown. Bar graph, quantification of the Western blot data. Means ± S.D. (n = 3) of the relative intensity of bands are shown (**, p < 0.01: ***, p < 0.001; ****, p < 0.0001, as compared with corresponding control binding in the absence of JNK3α2).
It was reported previously that MKK4 binding to arrestin-3 was significantly enhanced in the presence of JNK3 (10, 19). These studies used co-immunoprecipitation from cell lysates, where the interaction could have been mediated and/or regulated by any of the hundreds of other cellular proteins. Therefore, it remained unclear whether the observed increases of arrestin-3 association with MKK4 were caused by JNK3α2 or by other proteins. Direct binding of arrestin-3 to MKK7 (Fig. 3) also raises the question as to whether JNK3α2 has any effect on the recruitment of MKK7 to arrestin-3. Therefore, we performed MBP pulldown of MKK4 and MKK7 in the absence or presence of varying amounts of purified JNK3α2 (0, 2, 5, 10 μg). We did not detect any nonspecific effects of JNK3α2 on MBP pulldown of GST-MKK7, GST-MKK4, or control GST (Fig. 3C). We found that JNK3α2 enhanced arrestin-3 binding to MKK4 (Fig. 3D), in agreement with previous reports (10, 19–21). Unexpectedly, the presence of increasing amounts of JNK3α2 progressively decreased arrestin-3 association with MKK7 (Fig. 3D). These data suggest that JNK3α2 binding differentially regulates the recruitment of MKK4 and MKK7 to arrestin-3.
All earlier measurements of the binding of arrestins with the kinases from the JNK3 cascade were performed with either inactive kinases or kinases expressed in intact cells. Thus, how the phosphorylation of these kinases affects their association with arrestin-3 remained unexplored. This question is particularly important in the case of MKK4 and MKK7 because the inactive forms serve as the substrates for ASK1, whereas the active forms are upstream kinases for JNK3. Therefore, we performed MBP pulldown to compare the binding of active and inactive MKK4 or MKK7 to arrestin-3. We found that both inactive and active MKKs bind arrestin-3 with comparable affinities (Fig. 4). Furthermore, we observed the same differential regulation by JNK3α2 of the interactions of arresitn-3 with active MKK4 and MKK7; JNK3α2 increased the binding of MKK4 but reduced the binding of MKK7 (Fig. 4). Thus, the phosphorylation of MKK4 and MKK7 does not significantly change their recruitment to arrestin-3.
FIGURE 4.
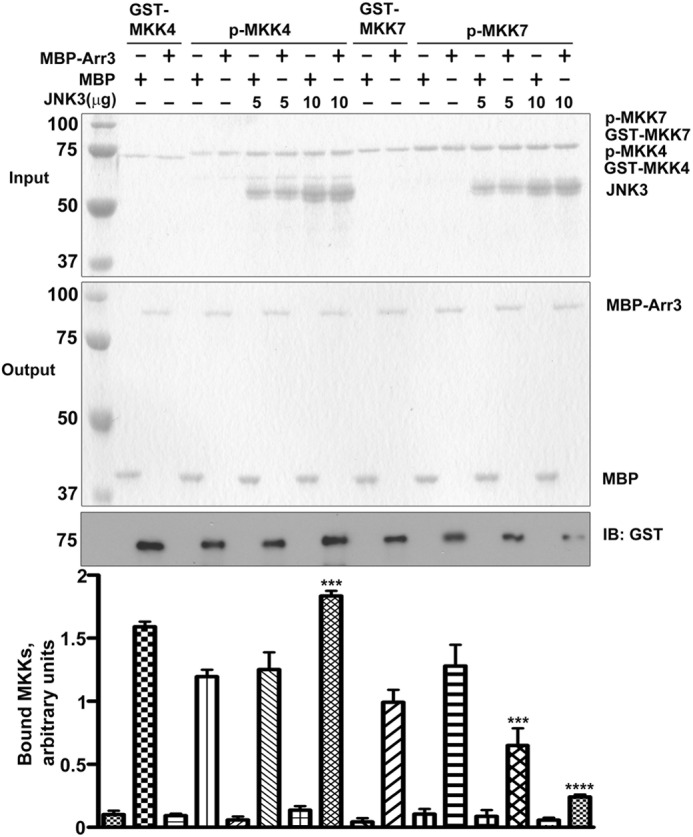
Arrestin-3 binding to active MKK4 and active MKK7 is regulated by JNK3α2. The binding of active MKKs to arrestin-3 was measured and compared with their inactive forms by MBP pulldown assay. The effects of JNK3α2 on the interaction of arrestin-3 with active MKK4 and 7 were also analyzed. Top panel, Coomassie Blue-stained gels of input of prey proteins (JNK3, GST-MKK4, active GST-MKK4 (p-MKK4), GST-MKK7, and active GST-MKK7 (p-MKK7). Middle panel, Coomassie Blue-stained gel of the eluted bait proteins (MBP and MBP-arrestin-3). Bottom panel, the amount of GST-tagged MKK4 or MKK7 retained was analyzed by Western blot. Representative gels are shown. Bar graph, quantification of the Western blot data. Means ± S.D. (n = 3) of the relative intensity of bands are shown (***, p < 0.001; ****, p < 0.0001, as compared with corresponding control binding in the absence of JNK3α2). IB, immunoblot.
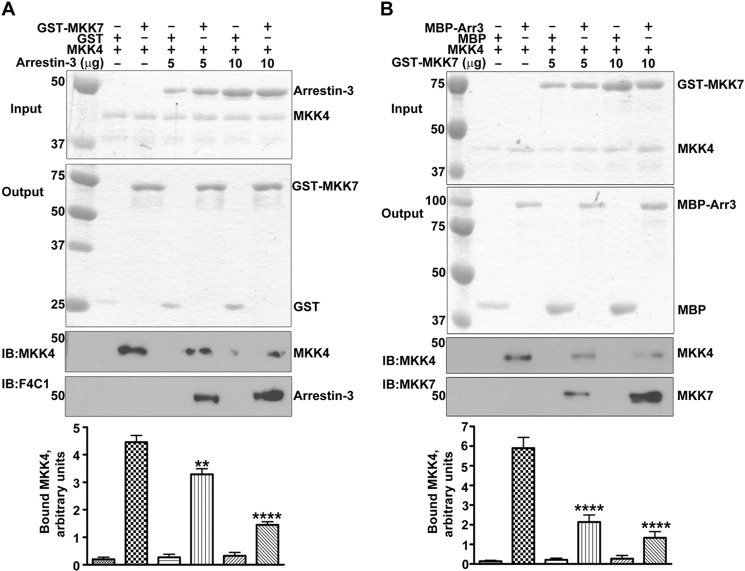
Direct association of arrestin-3 with both upstream kinases raises a new question: whether MKK4 and MKK7 can bind arrestin-3 simultaneously or compete for the same or overlapping binding site(s)? To address this issue, we performed in vitro GST pulldown assays using immobilized GST-MKK7 (5 μg) as bait along with tag-free MKK4 (2 μg) in the presence of varying amounts of arrestin-3 (0, 5, 10 μg), with equal amounts of GST as negative control (Fig. 5A). If arrestin-3 associates with both MKK4 and MKK7 simultaneously, the presence of arrestin-3 could have increased the amounts of MKK4 retained by GST-MKK7 via the formation of an MKK4-arrestin-3-MKK7 complex. The results show that MKK4 was retained by GST-MKK7, which indicates that MKK4 and MKK7 can form hetero-oligomers. The addition of arrestin-3 decreased the amount of MKK4 retained by MKK7 (Fig. 5A). Thus, the MKK4-arrestin-3-MKK7 complex did not form under the conditions tested. These data suggest that arrestin-3 binding prevents the hetero-dimerization between MKK4 and MKK7.
FIGURE 5.
MKK4 and MKK7 compete for arrestin-3 binding. A, pulldown of MKK4 by GST-MKK7 but not by GST (control). MKK4/MKK7 interaction was decreased by arrestin-3. Top panel, Coomassie Blue stained gel of input of prey proteins (arrestin-3 and MKK4). Middle panel, Coomassie Blue-stained gel of the output of bait proteins (GST and GST-MKK7). Lower panels, the amounts of retained MKK4 and arrestin-3 were analyzed by Western blot with indicated antibodies. Representative gels are shown. Bar graph, quantification of the MKK4 retained by GST-MKK7. Means ± S.D. (n = 3) of the relative intensity of bands (**, p < 0.01; ****, p < 0.0001, as compared with control without arrestin-3). IB, immunoblot. B, MKK4 and MKK7 compete for arrestin-3 binding. The amount of MKK4 retained by immobilized MBP-arrestin-3 progressively decreased in the presence of increasing concentrations of GST-MKK7. Coomassie Blue-stained gels of input of prey proteins (MKK4 and GST-MKK7) and output of bait proteins (MBP and MBP-Arr3) are shown in top two panels, respectively; the amount of MKK4 or GST-MKK7 retained was analyzed by Western blot using indicated antibodies. Representative gels are shown. Bar graph, quantification of the MKK4 retained by MBP-arrestin-3. Means ± S.D. (n = 3) of the relative intensity of bands (****, p < 0.0001, as compared with control without GST-MKK7).
Therefore, we tested whether MKK4 and MKK7 compete for arrestin-3 (Fig. 5B). We tested the binding of immobilized MBP-arrestin-3 (5 μg) and tag-free MKK4 (5 μg) in the presence of different amounts of GST-MKK7 (0, 5, 10 μg). The amount of MKK4 bound to MBP-arrestin-3 significantly decreased with the addition of MKK7 (Fig. 5B). These data indicate that MKK4 and MKK7 compete for arrestin-3, implying that the binding sites for the two MKKs likely overlap and suggesting that arrestin-3 is unlikely to bind both MKKs simultaneously.
Arrestins can assume several distinct conformations (28–33). The original hypothesis that different binding partners preferentially interact with arrestins in a particular conformation (34) was supported by the subsequent demonstration that ubiquitin ligases Mdm2 (35) and parkin (36) prefer “inactive” free arrestins, whereas ERK1/2 prefers a receptor-associated state (37). Interestingly, JNK3α2 was found to bind both equally well (35). The model where the binding of JNK3α2 induces a conformational change in the arrestin-3 molecule, which differentially affects the binding of the two MKKs, provides the simplest explanation of our data (Fig. 3). The ability of JNK3α2 to induce a conformational rearrangement in arrestin is consistent with the finding that it interacts with both arrestin domains (19). Characterization of the conformational changes in arrestin-3 induced by JNK3α2 is necessary to understand the structural basis of the differential regulation by JNK3α2 of the binding of MKK4 and MKK7. Regardless of the exact mechanism, these data are the first demonstration that kinases of the JNK3 cascade regulate each other's interactions with a scaffolding protein.
Biphasic Effect of Arrestin Concentration on JNK3α2 Phosphorylation by MKK7
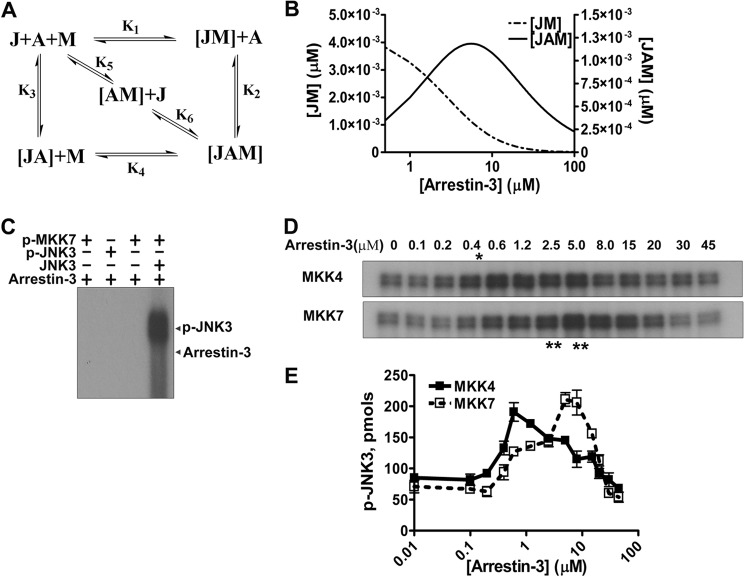
Using purified proteins, we recently showed that arrestin-3 acts as a simple scaffold, facilitating JNK3 phosphorylation by MKK4 by bringing the two kinases together (22). To directly test how arrestin-3 promotes JNK3α2 phosphorylation by MKK7, we reconstituted arrestin-3-MKK7-JNK3α2 signaling module from purified proteins (Fig. 6). We first tested whether arrestin-3 can be phosphorylated by either MKK7 or JNK3α2 and found that neither active MKK7 nor active JNK3α2 phosphorylated arrestin-3 under our assay conditions (Fig. 6C). To evaluate the functional role of arrestin-3, we kept the concentrations of active MKK7 (50 nm) and its substrate unphosphorylated JNK3α2 (0.5 μm) constant and measured the extent of JNK3α2 phosphorylation in the presence of various arrestin-3 concentrations (0–45 μm). JNK3α2 phosphorylation by MKK7 demonstrated a biphasic dependence on arrestin-3 concentration, being enhanced by low and inhibited by higher concentrations (Fig. 6, D and E). Under these conditions, the optimal arrestin-3 concentration for the facilitation of JNK3α2 phosphorylation by MKK7 was ∼5–8 μm, which is about 10 times higher than the optimal arrestin-3 concentration that promotes JNK3α2 phosphorylation by MKK4 (0.6 μm) (Fig. 6D, E; see also Ref. 22).
FIGURE 6.
Biphasic effects of arrestin-3 on JNK3α2 activation by MKK7 and MKK4. A, a model showing the scaffolding mechanism of the two-kinase signaling module. A, J, and M stand for arrestin-3, JNK3α2, and upstream kinase MKK4 or MKK7, respectively. Kinases can exist in three different complexes: (a) directly interacting JM; (b) bound to the scaffold to form incomplete complexes containing a single kinase (JA or AM); (c) simultaneously tethered by a scaffold to form a complete two-kinase signaling complex JAM. Six affinity constants (K1--K6) describe the indicated binding equilibria. B, calculated concentrations of JM (dotted line, left y axis) and JAM (solid line, right y axis) complexes (KinTek Explorer 3.0; all six Kd values were set at 5 μm). C, active MKK7 and JNK3α2 do not phosphorylate arrestin-3. Purified arrestin-3 (1 μm) was incubated with active MKK7 (50 nm), active JNK3α2 (50 nm), or inactive JNK3α2 (0.5 μm) for 5 min. The reactions were stopped with SDS sample buffer, and the proteins were separated on 10% SDS-PAGE. The gel was dried and exposed to x-ray film. A representative autoradiogram from one of four experiments is shown. D, representative autoradiograms showing JNK3α2 phosphorylated by MKK4 (upper panel) and MKK7 (lower panel) at the indicated concentration of arrestin-3 (10-s incubation). The optimal arrestin-3 concentrations are indicated (*, for MKK4; **, for MKK7). E, the effect of arrestin-3 concentration on JNK3α2 phosphorylation by both MKK4 and MKK7 is biphasic. The bands from the autoratiogram gels were cut out, and the radioactivity was measured in a Tri-Carb liquid scintillation counter to quantify the incorporation of [32P]phosphate from [γ-32P]ATP into JNK3α2. Means ± S.D. (n = 3) are shown.
These results are consistent with our MBP pulldown data, which show direct interaction between arrestin-3 and MKK7 (Fig. 3). The higher optimal concentration of arrestin-3 necessary to promote JNK3α2 phosphorylation by MKK7 compared with MKK4 is consistent with the differential regulation of MKK4 and MKK7 binding by JNK3α2 (Fig. 3). Optimal concentrations of scaffold proteins are mainly determined by the binding affinities between a scaffold protein and its partners (38–40). Positive or negative cooperativity of the simultaneous binding of the two components to the scaffold protein may also contribute to affect the optimal concentration (39).
This biphasic dependence on scaffold concentration would be observed when the scaffold acts by directly binding both kinases and bringing them together without activating either MKK or its substrate (38–42). In the presence of scaffold, the activation of the downstream kinase (JNK3) is determined by the concentrations of the following complexes (where A stands for arrestin-3, J for JNK3, and M for MKK7): the complete scaffolded complex JAM, incomplete complexes (JA and AM), and scaffold-free MKK-JNK complex formed via free diffusion of these kinases (JM) (Fig. 6A). For the scaffold to facilitate JNK3 activation (Fig. 6, D and E), the rate of JNK3 phosphorylation in the scaffolded complex (JAM) must be higher than in the JM complex, whereas the incomplete complexes (JA or AM) actually prevent kinase activation by separating the two kinases (Fig. 6A). Without scaffold and at low scaffold concentration, JNK3 activation is dominated by the output of the JM complex. An increase in scaffold concentration in the lower range enhances the formation of an active (JAM) complex containing both kinases, thereby facilitating the activation of the downstream kinase. A further increase in scaffold concentration enhances the probability of downstream and upstream kinases associating with different molecules of the scaffold protein, forming incomplete complexes (JA and AM) that prevent the phosphorylation of JNK3. Therefore, a scaffold has a biphasic effect on the activation of a downstream kinase.
The experimentally measured arrestin-3 concentration optimal for facilitation of JNK3 phosphorylation by MKK7 (Fig. 6, D and E) is ∼5 μm. This value is higher than the concentration of either JNK3α2 or MKK7 in these experiments, which appears counterintuitive. Therefore, we used the model shown in Fig. 6A (along with thermodynamic limitations inherent in this system of equilibria: K1 × K2 = K3 × K4 = K5 × K6) to test whether this is possible. To model the effect of arrestin-3 on JNK3 activation by MKK4 or MKK7, we calculated the predicted formation of two “meaningful” complexes that include JNK3 and its upstream kinase, JM and JAM, as a function of arrestin-3 concentration (Fig. 6B). The model requires the values of six equilibrium dissociation constants, K1 through K6 (Fig. 6A). Only the binding affinity between JNK3 and arrestin-3 was previously measured experimentally and found to be ∼4–5 μm (22). To simplify the modeling, we set all constants at 5 μm. As could be expected, the model showed that the concentration of JM progressively decreased with an increasing concentration of arrestin-3 (Fig. 6B). The concentration of JAM demonstrated a biphasic dependence on the concentration of arrestin-3 and peaked at ∼5.5 μm arrestin-3 (Fig. 6B). Thus, the modeling shows that the highest level of JAM can be achieved at a scaffold concentration exceeding that of either kinase.
The rates of enzymatic reactions are governed by factors that influence the steady-state levels of catalytic species; thus the effects of scaffold proteins on MAPK activation may be reflective of cooperativity in MAPKK-MAPK binding, which results in an increase in the steady-state concentration of an active scaffold complex. Thus, even though arrestin-3 binds MKK4 more tightly than it binds MKK7 (Fig. 3, C and D), the difference in the concentration of arrestin-3 that optimally promotes MKK4- and MKK7-dependent JNK3α2 phosphorylation may also reflect the differential effects of JNK3 on the binding of arrestin-3 to MKK4 and MKK7 (38, 40, 41). Our data suggest that JNK3α2 phosphorylation is characterized by a faster rate of the scaffolded complex JAM compared with the binary complex JM. Presumably, the formation of JAM occurs through JA or AM, and its steady-state level may be sensitive to the cooperativity of upstream and downstream kinase binding. Although an increase of JA or AM is expected to result in a higher steady-state level of the scaffolded complex JAM, which facilitates JNK3 activation, a further increase in scaffold concentration results in a higher ratio of JA and AM, which is predicted to inhibit JNK3 activation. Thus, the ratio of the concentrations of JA, AM, JAM, and JM is predicted to determine the output of phosphorylated JNK3. With positive cooperativity, a second component (e.g. MKK4) may prefer to bind the JA complex, inducing the formation of JAM at lower scaffold concentration. In contrast, with negative cooperativity (e.g. MKK7) a higher scaffold concentration may be required to reach the optimal steady-state level of JAM for highest output of p-JNK3 (Fig. 6, D and E). In this case a positive effect of JNK3α2 on MKK4 association results in lower optimal arrestin-3 concentration, whereas a negative effect increases the optimal arrestin-3 concentration in the case of MKK7 (Figs. 3 and 6). Previously, a theoretical model was proposed that suggests that positive cooperativity lowers the optimal concentration of scaffold protein, whereas negative cooperativity moves the optimal concentration in the opposite direction (39). To the best of our knowledge, our data represent the first experimental support for this model. However, further work is required to critically evaluate the mechanism in detail.
Arrestin-3 Comparably Enhanced the Phosphorylation of JNK3α2 by MKK4 and MKK7 in Intact Cells
Next we tested whether arrestin-3 facilitates MKK7-mediated JNK3α2 phosphorylation in living cells. To this end, we overexpressed JNK3α2 individually or in combination with MKK4, MKK7, and arrestin-3 in COS-7 cells and monitored JNK3α2 phosphorylation (Fig. 7). Arrestin-3 significantly increased JNK3α2 phosphorylation by either MKK4 (∼10-fold) or MKK7 (∼7-fold). The level of double-phosphorylated JNK3α2 selectively recognized by ppJNK antibodies (Fig. 1) is higher when MKK7 is overexpressed than with MKK4 in the absence of arrestin-3 (Fig. 7). These data suggest that endogenous scaffolding proteins in COS-7 cells promote the assembly of MKK7-JNK3α2 complexes more efficiently than MKK4-JNK3α2. This explains the greater detectable effect of exogenous arrestin-3 on JNK3α2 phosphorylation by MKK4 than by MKK7 (Fig. 7). The data show that arrestin-3 can recruit both upstream MKKs to phosphorylate two distinct residues in JNK3α2 catalytic loop. Although JNK3α2 binding dramatically increased the binding of MKK4 and decreased that of MKK7 (Fig. 3D), arrestin-3 effectively promotes JNK3α2 phosphorylation by MKK7 in the cellular environment (Fig. 7). Collectively, our data suggest that arrestin-3-mediated JNK3α2 activation is strictly regulated by the complex interplay of expression levels of MKK4, MKK7, arrestin-3, and other proteins scaffolding JNK3 activation cascade.
FIGURE 7.
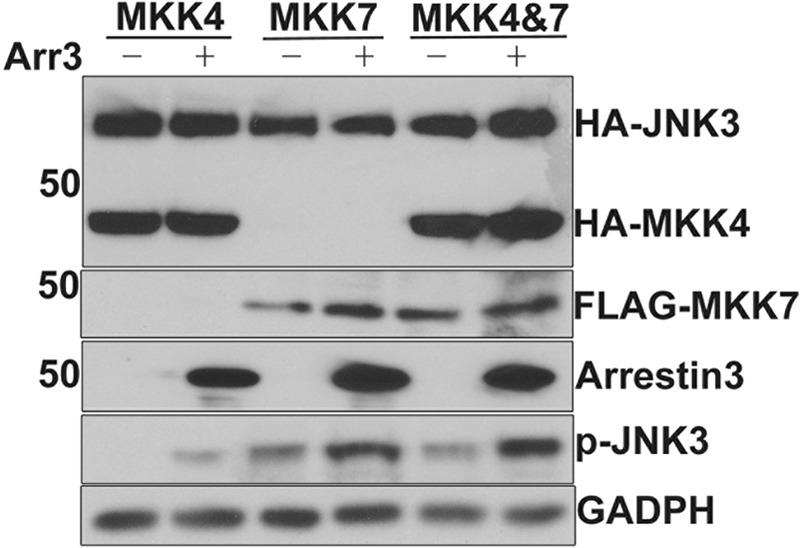
Arrestin-3 enhances the phosphorylation of JNK3α2 by MKK4 or MKK7 in intact cells. COS-7 cells were transfected with HA-JNK3α2 in combination with HA-MKK4, FLAG-MKK7, or both with (+) or without (−) arrestin-3. Cell lysates were analyzed by Western blot using the indicated antibodies. Note that co-expression of arrestin-3 enhances JNK3α2 phosphorylation by either MKK. GADPH served as the loading control.
DISCUSSION
Scaffolding proteins have emerged as critical elements in determining the specificity of intracellular signaling pathways (9). All evolutionarily conserved MAPK signaling pathways are organized by scaffold proteins (9, 43). MAPK scaffolds are extremely diverse, including kinase suppressor of Ras (KSR), JNK-interacting protein (JIP), paxillin involved in cell adhesion, MAPKK kinase (MEKKK1), etc. Non-visual arrestins were also reported to scaffold MAPK pathways leading to the activation of JNK3 (10, 19–21, 23), ERK1/2 (11, 37, 44), and p38 (12, 45). Most scaffolds are believed to use a simple tethering mechanism to facilitate the efficiency of signal transduction (38, 39, 42), although this model is rarely proved experimentally. Some scaffolds were shown to be directly involved in complex allosteric control of their binding partners. One well characterized example of these is yeast Ste5, which does not simply bring the kinases together, but also catalytically unlocks the Fus3 MAP kinase for activation (46, 47).
MAP kinases are fully activated when they are phosphorylated on both threonine and tyrosine in the activation loop (16). MKK4 and MKK7 are the upstream kinases for JNKs, and each preferentially phosphorylates a distinct site, tyrosine (MKK4) or threonine (MKK7). Importantly, the requirements for MKK4 and MKK7 for JNK activation are different, depending on the nature of the stimulus (18, 48). Targeted gene disruption in mice showed that simultaneous disruption of MKK4 and MKK7 genes was required to block JNK activation caused by environmental stress, whereas the disruption of the MKK7 gene alone was sufficient to prevent JNK activation caused by proinflammatory cytokines (49). The differential role of MKK4 and MKK7 in JNK activation can also be determined by distinct scaffold proteins (27). Previous studies performed in mouse embryonic fibroblasts showed that axin-mediated JNK activation depends mainly on MKK7 and Dvl-induced JNK activation almost equally depends on MKK4 and MKK7, whereas Epstein-Barr virus latent membrane protein-1-mediated JNK activation is primarily dependent on MKK4 (27). There can be various reasons for differential requirement of MKK4 and MKK7 for JNK activation, such as different expression levels of MKK4 and MKK7, scaffolds, and other regulators in particular cell types. The mechanisms responsible for distinct roles of these two MKKs in JNK activation by different stimuli remain largely unknown.
Arrestin-3 dependent JNK3 activation was discovered more than a decade ago (10), and subsequent studies revealed some mechanistic details of this process. Arrestin-3 scaffolding of the JNK3 signaling cascade was first reported to depend on arrestin-3 binding to G-protein-coupled receptors (10), whereas later the same group (23) and others (19–22) showed that free arrestin-3 is capable of performing this function. Moreover, structure-function analysis of arrestin-3 revealed that binding of all three kinases, ASK1, MKK4, and JNK3α2, is a function that can be separated from JNK3α2 activation (20) and that an arrestin-3 mutant that efficiently binds all three kinases but does not promote JNK3α2 phosphorylation can act as dominant-negative silent scaffold, suppressing JNK3α2 activation in the cell by recruiting it away from productive scaffolds (21). Our recent demonstration that arrestin-3-dependent facilitation of JNK3 phosphorylation by MKK4 can be reproduced with purified proteins in the absence of G-protein-coupled receptors (22) confirmed the idea that this is a receptor-independent process. This model is also supported by the finding that an arrestin-3 mutant with the deletion in the interdomain hinge, which greatly impairs its ability to bind receptors (50, 51), facilitated JNK3 activation as effectively as WT arrestin-3 (19, 21). Although all four mammalian arrestin subtypes bind the kinases in the ASK1-MKK4-JNK3 cascade (35, 52), only arrestin-3 facilitates JNK3 activation (10, 19, 20, 23). Our recent finding that arrestin-2 has much lower affinity for MKK4 than arrestin-3 (22) likely explains why arrestin-2 does not promote JNK3α2 phosphorylation even though it is expressed at 10–20-fold higher levels in most cell types (53, 54).
All previous studies focused on the ASK1-MKK4-JNK3 cascade. Full JNK activation requires the phosphorylation of both Tyr (by MKK4) and Thr (by MKK7) residues (16), but it remained unclear whether arrestin-3 can promote the phosphorylation of JNK3 by MKK7. Therefore, we used purified proteins to investigate the role of arrestin-3 in JNK3α2 phosphorylation by MKK4, MKK7, and the combination of MKK4 + MKK7. We found that arrestin-3 did not alter the sites of JNK3α2 phosphorylated by either MKK (Fig. 1). Using specific phospho-Tyr and phospho-Thr antibodies, we demonstrated that arrestin-3 facilitated the phosphorylation of both Tyr and Thr residues within the TPY motif of JNK3α2 in vitro (Fig. 1) and in COS-7 cells (Fig. 2). The enhancement of Thr phosphorylation by arrestin-3 demonstrates that it facilitates JNK3α2 activation by MKK7.
The binding of MKK4 to arrestin-3 is harder to detect than that of ASK1 or JNK3α2 (10, 19). The authors of the initial study found that although it was hard to detect the binding of MKK4 or MKK7 to arrestin-3 in cell lysates, MKK4 binding to arrestin-3 was significantly enhanced upon co-expression of ASK1 and JNK3 individually or together, whereas the interaction of MKK7 with arrestin-3 was still undetectable under the same conditions (10). Recently we demonstrated direct interaction of MKK4 with arrestin-3 using purified proteins (22). Here we extended our findings by comparing the binding of purified MKK4 and MKK7 to arrestin-3 and found that arrestin-3 binds both MKK7 and MKK4 directly (Fig. 3). We also evaluated the binding of active MKK4 and MKK7 to arrestin-3 and showed that the phosphorylation of MKK4 or MKK7 does not significantly affect their association with arrestin-3 (Fig. 4). Unexpectedly, we found that the binding of the downstream kinase JNK3α2 enhanced the association of arrestin-3 with MKK4 while reducing the binding of MKK7 (Fig. 3). JNK3 phosphorylation by either MKK4 or MKK7 demonstrated a biphasic dependence on arrestin-3 concentration (Fig. 6), which indicates that arrestin-3 functions as a scaffold bringing the two kinases together in both cases (38, 39). We also investigated whether both upstream MKKs bind arrestin-3 simultaneously or compete with each other. The data showed that MKK4 and MKK7 compete for arrestin-3 binding (Fig. 5), suggesting that these two MKKs engage the same or a significantly overlapping site(s) on arrestin-3. Demonstrated positive cooperativity of JNK3α2 and MKK4 binding to arrestin-3 (Fig. 3) may contribute to the relatively low optimal concentration of arrestin-3 in facilitating JNK3α2 phosphorylation by MKK4 (Fig. 6). In contrast, negative cooperativity of JNK3α2 and MKK7 binding to arrestin-3 (Fig. 3) may contribute to the ∼10-fold higher optimal concentration of arrestin-3 to promote JNK3α2 phosphorylation by MKK7 (Fig. 6). Our findings are in agreement with theoretical models (38, 39). It was predicted that the concentration of a simple scaffold that directly binds two components and brings them together should biphasically affect signal transduction (38, 39), as our data demonstrate (Fig. 6). It was predicted that the scaffold concentration necessary for efficient signaling is determined by the binding affinities of the scaffold for its partners (38, 39). Our data (Fig. 6) suggest that positive or negative cooperativity between the proteins recruited by a scaffold may also determine the optimal scaffold concentrations (38, 39).
Scaffold proteins regulate the output by bringing signaling proteins into close proximity to each other, holding them in a particular orientation as well as by allosteric regulation of the enzyme and/or its substrate (9). Most scaffold proteins facilitate signal transduction simply by simultaneous recruitment of several components of the pathway. This tethering mechanism can significantly increase the effective local concentrations of enzymes and their substrates. However, there are functional limitations of this simple scaffolding mechanism, such as the inhibition of signaling by high concentrations of scaffold proteins (Fig. 6, D and E). To improve the efficiency, some scaffold proteins mediate the enzyme-substrate reactions through orientation and allosteric regulation. For example, to stimulate substrate ubiquitination effectively, a scaffold cullin-RING-F-box complex has to orient the substrate properly for the upstream E2 ubiquitin-conjugating enzyme (56, 57). Allosteric regulation was observed in the yeast mating system, where Ste5 scaffold allosterically “unlocks” its binding partner MAPK Fus3 to make it a good substrate for the MKK Ste7 (47). The cooperativity of the binding of JNK3α2 and both MKKs to arrestin-3 observed here (Fig. 3) appears to represent a more sophisticated regulatory mechanism (Fig. 6, D and E). Considering the lower levels of endogenous MKK4 (Fig. 2C), a positive effect of JNK3α2 on its binding could ensure that arrestin-3 recruits MKK4 more effectively than other binding partners, such as MKK7 expressed at much higher level (Fig. 2C). We observed that relatively high arrestin-3 concentrations are optimal for JNK3 phosphorylation by this kinase (∼5 μm, Fig. 6, D and E), potentially reflecting the negative cooperativity of binding. Modeling describing the formation of various complexes (Fig. 6A) predicts that at the arrestin-3 concentration optimal for the formation of the JAM complex, the concentration of the JM complex is reduced ∼4-fold (Fig. 6B). Thus, the observed enhancement of JNK3α2 phosphorylation by arrestin-3 (Fig. 6, D and E) indicates that in the arrestin-3-MKK7-JNK3α2 complex the phosphorylation of JNK3α2 is significantly more efficient than in the MKK7-JNK3α2 complex. These results suggest that arrestin-3 regulates JNK3α2 phosphorylation by MKKs not just by tethering the two kinases together but potentially also by ensuring their optimal orientation relative to each other, which can account for a significant increase in the efficiency of the reaction between two proteins (58).
Qualitatively our data fit existing mathematical models remarkably well, but some quantitative aspects remain unexplained. Existing models (38–42, 59) do not take into account additional effects of scaffolds, particularly the fact that scaffolds impose a particular relative orientation of the two proteins that can increase reaction rates by orders of magnitude (58). In addition, scaffolds can “concentrate” reaction components; even low affinity binding to a scaffold would increase local concentration of both kinases in its vicinity, thereby making the formation of JAM complexes more likely than when the kinases are evenly distributed in the solution. Our data suggest that more sophisticated mathematical models of scaffolded signaling cascades need to be developed. The positive effect of JNK3α2 binding on MKK4 recruitment is biologically relevant; it likely helps MKK4 expressed at low levels (Fig. 2) to compete with other arrestin-3 binding partners, including higher expressed MKK7.
Arrestin-3 was shown to bind dozens of different partners (4), whereas the size of arrestin-3 suggests that it can accommodate no more than 3–4 proteins simultaneously (1). To perform their functions, non-visual arrestins are often required to assemble particular combinations of proteins, such as three kinases belonging to the same MAP pathway, to form a productive signaling complex. How exactly arrestins manage to recruit proper combinations of partners remains largely unknown. Mechanistically, the change in arrestin conformation upon binding of certain partners could regulate the association of others. The best characterized example of this type of regulation is arrestin association with the internalization machinery; G-protein-coupled receptor binding induces the release of the arrestin C-tail (29–32), exposing clathrin (60) and AP2 (61) binding sites localized on this element. Arrestins exist in several distinct conformations: free, revealed by most crystal structures (22, 62–66), receptor-bound (29, 30), and microtubule-bound (28, 51). Recent structures of truncated “preactivated” (67) arrestin-2 in complex with a multiphosphorylated C-terminal peptide of vasopressin V2 receptor and a nanobody (68) and of a short splice variant of arrestin-1 (69) likely represent an intermediate state between free and receptor-bound conformations. Several arrestin binding partners show distinct preference for particular conformations; ERK1/2 prefers receptor-bound arrestins (37), whereas ubiquitin ligases Mdm2 and parkin preferentially interact with arrestins in the basal conformation (35, 36). Although we showed unambiguous evidence that free arrestin-3 binds MKK4, MKK7, and JNK3α2 and facilitates JNK3α2 phosphorylation, the role of the receptor in arrestin3-mediated JNK3α2 activation remains unclear. The most straightforward approach to elucidation of the role of G-protein-coupled receptors in this process is the use of direct biochemical and biophysical methods to study arrestin-3-MAPK complexes reconstructed with purified proteins in the presence and absence of receptors under carefully controlled conditions.
Human cells express >20 different MAPKK kinases (MAPKKK), 7 MAPKK, and 11 MAPKs (70), forming an amazing variety of signaling modules with distinct cellular functions (9, 59, 71). Scaffolding proteins play critical roles in keeping signals accurately and specifically transmitted through these pathways. Differential regulation of the binding of arrestin-3 to MKK4 and MKK7 by JNK3α2 (Fig. 3) demonstrates one possible mechanism of how a scaffold selects proper binding partners upon recruiting another component in the same pathway. Our data (Fig. 6) support theoretical models (38, 39) suggesting that this process is regulated by relative concentrations of scaffolds and kinases involved. Although the JNK3α2-arrestin-3 complex prefers MKK4 (Fig. 3), under appropriate conditions arrestin-3 facilitates JNK3α2 activation by MKK7 as well (Figs. 6 and 7). These findings support the idea that differential requirements for MKK4 and MKK7 for JNK3α2 activation depend not only on the type of stimulus (59) but also on the expression levels of relevant scaffolding proteins. These data are the first direct experimental evidence that components of mammalian MAPK cascades can regulate the binding of each other to scaffold proteins. Our data also suggest that in rod photoreceptors, where arrestin-1 is expressed at very high levels, reaching up to 2 mm concentration in the cell body in the dark (72–74), its reported interactions with JNK3α2 (35) and upstream kinases (55) is likely to suppress the signaling in this pathway rather than enhance it.
In conclusion, we demonstrated that arrestin-3 promotes JNK3 phosphorylation by MKK7. The data suggest that although arrestin-3 binds MKK7 with only somewhat lower affinity than MKK4, JNK3 binding has the opposite effect on arrestin-3 interactions with MKK4 and MKK7, enhancing the former and strongly suppressing the latter. This differential regulation suggests that arrestin-3 might undergo a significant conformational change upon the binding of JNK3α2. Further experimentation with pure proteins to explore this conformational change is necessary. To better understand the scaffolding function of arrestin-3, the effects of other components (ASK1, receptor) need to be carefully explored as well. It will be important to test whether other scaffold proteins in MAPK pathways are regulated in a similar way to maintain the accuracy and specificity of signal transduction.
Acknowledgment
We are grateful to Dr. Noriyuki Matsuda, Tokyo Institute of Medical Science, for pMal-p2T plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grants EY011500, GM077561, and GM081756 (to V. V. G.) and GM059802 and CA167505 (to K. N. D.). This work was also supported by the Welch Foundation (F-1390; to K. N. D.).
We use the systematic names of arrestin proteins: arrestin-1 (historic names S-antigen, 48-kDa protein, visual or rod arrestin), arrestin-2 (β-arrestin or β-arrestin1), arrestin-3 (β-arrestin2 or hTHY-ARRX), and arrestin-4 (cone or X-arrestin; for unclear reasons its gene is called “arrestin 3” in the HUGO database).
- MAP
- mitogen-activated protein
- MBP
- maltose-binding protein
- MKK
- mitogen-activated protein kinase kinase.
REFERENCES
- 1. Gurevich V. V., Gurevich E. V. (2006) The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol. Ther. 110, 465–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DeWire S. M., Ahn S., Lefkowitz R. J., Shenoy S. K. (2007) β-Arrestins and cell signaling. Annu. Rev. Physiol. 69, 483–510 [DOI] [PubMed] [Google Scholar]
- 3. Gurevich E. V., Gurevich V. V. (2006) Arrestins. Ubiquitous regulators of cellular signaling pathways. Genome Biol. 7, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xiao K., McClatchy D. B., Shukla A. K., Zhao Y., Chen M., Shenoy S. K., Yates J. R., 3rd, Lefkowitz R. J. (2007) Functional specialization of β-arrestin interactions revealed by proteomic analysis. Proc. Natl. Acad. Sci. U.S.A. 104, 12011–12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pearson G., Robinson F., Beers Gibson T., Xu B. E., Karandikar M., Berman K., Cobb M. H. (2001) Mitogen-activated protein (MAP) kinase pathways. Regulation and physiological functions. Endocr. Rev. 22, 153–183 [DOI] [PubMed] [Google Scholar]
- 6. Morrison D. K., Davis R. J. (2003) Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 19, 91–118 [DOI] [PubMed] [Google Scholar]
- 7. Dard N., Peter M. (2006) Scaffold proteins in MAP kinase signaling. More than simple passive activating platforms. Bioessays 28, 146–156 [DOI] [PubMed] [Google Scholar]
- 8. Zeke A., Lukács M., Lim W. A., Reményi A. (2009) Scaffolds. Interaction platforms for cellular signalling circuits. Trends Cell Biol. 19, 364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Good M. C., Zalatan J. G., Lim W. A. (2011) Scaffold proteins. Hubs for controlling the flow of cellular information. Science 332, 680–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McDonald P. H., Chow C. W., Miller W. E., Laporte S. A., Field M. E., Lin F. T., Davis R. J., Lefkowitz R. J. (2000) β-Arrestin 2. A receptor-regulated MAPK scaffold for the activation of JNK3. Science 290, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 11. Luttrell L. M., Roudabush F. L., Choy E. W., Miller W. E., Field M. E., Pierce K. L., Lefkowitz R. J. (2001) Activation and targeting of extracellular signal-regulated kinases by β-arrestin scaffolds. Proc. Natl. Acad. Sci. U.S.A. 98, 2449–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bruchas M. R., Macey T. A., Lowe J. D., Chavkin C. (2006) Kappa opioid receptor activation of p38 MAPK is GRK3- and arrestin-dependent in neurons and astrocytes. J. Biol. Chem. 281, 18081–18089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Davis R. J. (2000) Signal transduction by the JNK group of MAP kinases. Cell 103, 239–252 [DOI] [PubMed] [Google Scholar]
- 14. Gupta S., Barrett T., Whitmarsh A. J., Cavanagh J., Sluss H. K., Dérijard B., Davis R. J. (1996) Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 15, 2760–2770 [PMC free article] [PubMed] [Google Scholar]
- 15. Waetzig V., Herdegen T. (2005) Context-specific inhibition of JNKs. Overcoming the dilemma of protection and damage. Trends Pharmacol. Sci. 26, 455–461 [DOI] [PubMed] [Google Scholar]
- 16. Lawler S., Fleming Y., Goedert M., Cohen P. (1998) Synergistic activation of SAPK1/JNK1 by two MAP kinase kinases in vitro. Curr. Biol. 8, 1387–1390 [DOI] [PubMed] [Google Scholar]
- 17. Haeusgen W., Herdegen T., Waetzig V. (2011) The bottleneck of JNK signaling. Molecular and functional characteristics of MKK4 and MKK7. Eur. J. Cell Biol. 90, 536–544 [DOI] [PubMed] [Google Scholar]
- 18. Wang X., Destrument A., Tournier C. (2007) Physiological roles of MKK4 and MKK7. Insights from animal models. Biochim Biophys. Acta 1773, 1349–1357 [DOI] [PubMed] [Google Scholar]
- 19. Song X., Coffa S., Fu H., Gurevich V. V. (2009) How does arrestin assemble MAPKs into a signaling complex? J. Biol. Chem. 284, 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seo J., Tsakem E. L., Breitman M., Gurevich V. V. (2011) Identification of arrestin-3-specific residues necessary for JNK3 kinase activation. J. Biol. Chem. 286, 27894–27901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Breitman M., Kook S., Gimenez L. E., Lizama B. N., Palazzo M. C., Gurevich E. V., Gurevich V. V. (2012) Silent scaffolds. Inhibition OF c-Jun N-terminal kinase 3 activity in cell by dominant-negative arrestin-3 mutant. J. Biol. Chem. 287, 19653–19664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhan X., Gimenez L. E., Gurevich V. V., Spiller B. W. (2011) Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual subtypes. J. Mol. Biol. 406, 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller W. E., McDonald P. H., Cai S. F., Field M. E., Davis R. J., Lefkowitz R. J. (2001) Identification of a motif in the carboxyl terminus of β-arrestin2 responsible for activation of JNK3. J. Biol. Chem. 276, 27770–27777 [DOI] [PubMed] [Google Scholar]
- 24. Gurevich V. V., Benovic J. L. (2000) Arrestin. Mutagenesis, expression, purification, and functional characterization. Methods Enzymol. 315, 422–437 [DOI] [PubMed] [Google Scholar]
- 25. Matsuda N., Kitami T., Suzuki T., Mizuno Y., Hattori N., Tanaka K. (2006) Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J. Biol. Chem. 281, 3204–3209 [DOI] [PubMed] [Google Scholar]
- 26. Lisnock J., Griffin P., Calaycay J., Frantz B., Parsons J., O'Keefe S. J., LoGrasso P. (2000) Activation of JNK3 α1 requires both MKK4 and MKK7. Kinetic characterization of in vitro phosphorylated JNK3 α1. Biochemistry 39, 3141–3148 [DOI] [PubMed] [Google Scholar]
- 27. Zou H., Li Q., Lin S. C., Wu Z., Han J., Ye Z. (2007) Differential requirement of MKK4 and MKK7 in JNK activation by distinct scaffold proteins. FEBS Lett. 581, 196–202 [DOI] [PubMed] [Google Scholar]
- 28. Wu N., Hanson S. M., Francis D. J., Vishnivetskiy S. A., Thibonnier M., Klug C. S., Shoham M., Gurevich V. V. (2006) Arrestin binding to calmodulin. A direct interaction between two ubiquitous signaling proteins. J. Mol. Biol. 364, 955–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim M., Vishnivetskiy S. A., Van Eps N., Alexander N. S., Cleghorn W. M., Zhan X., Hanson S. M., Morizumi T., Ernst O. P., Meiler J., Gurevich V. V., Hubbell W. L. (2012) Conformation of receptor-bound visual arrestin. Proc. Natl. Acad. Sci. U.S.A. 109, 18407–18412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhuang T., Chen Q., Cho M.-K., Vishnivetskiy S. A., Iverson T. M., Gurevich V. V., Sanders C. R. (2013) Involvement of distinct arrestin-1 elements in binding to different functional forms of rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 110, 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vishnivetskiy S. A., Francis D., Van Eps N., Kim M., Hanson S. M., Klug C. S., Hubbell W. L., Gurevich V. V. (2010) The role of arrestin α-helix I in receptor binding. J. Mol. Biol. 395, 42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hanson S. M., Francis D. J., Vishnivetskiy S. A., Kolobova E. A., Hubbell W. L., Klug C. S., Gurevich V. V. (2006) Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc. Natl. Acad. Sci. U.S.A. 103, 4900–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carter J. M., Gurevich V. V., Prossnitz E. R., Engen J. R. (2005) Conformational differences between arrestin2 and pre-activated mutants as revealed by hydrogen exchange mass spectrometry. J. Mol. Biol. 351, 865–878 [DOI] [PubMed] [Google Scholar]
- 34. Gurevich V. V., Gurevich E. V. (2003) The new face of active receptor bound arrestin attracts new partners. Structure 11, 1037–1042 [DOI] [PubMed] [Google Scholar]
- 35. Song X., Raman D., Gurevich E. V., Vishnivetskiy S. A., Gurevich V. V. (2006) Visual and both non-visual arrestins in their “inactive” conformation bind JNK3 and Mdm2 and relocalize them from the nucleus to the cytoplasm. J. Biol. Chem. 281, 21491–21499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ahmed M. R., Zhan X., Song X., Kook S., Gurevich V. V., Gurevich E. V. (2011) Ubiquitin ligase parkin promotes Mdm2-arrestin interaction but inhibits arrestin ubiquitination. Biochemistry 50, 3749–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Coffa S., Breitman M., Hanson S. M., Callaway K., Kook S., Dalby K. N., Gurevich V. V. (2011) The effect of arrestin conformation on the recruitment of c-Raf1, MEK1, and ERK1/2 activation. PLoS ONE 6, e28723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Levchenko A., Bruck J., Sternberg P. W. (2004) Regulatory modules that generate biphasic signal response in biological systems. Syst. Biol. (Stevenage) 1, 139–148 [DOI] [PubMed] [Google Scholar]
- 39. Levchenko A., Bruck J., Sternberg P. W. (2000) Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc. Natl. Acad. Sci. U.S.A. 97, 5818–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ferrell J. E., Jr. (2000) What do scaffold proteins really do? Sci. STKE 2000, pe1. [DOI] [PubMed] [Google Scholar]
- 41. Bray D., Lay S. (1997) Computer-based analysis of the binding steps in protein complex formation. Proc. Natl. Acad. Sci. U.S.A. 94, 13493–13498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burack W. R., Shaw A. S. (2000) Signal transduction. hanging on a scaffold. Curr. Opin. Cell Biol. 12, 211–216 [DOI] [PubMed] [Google Scholar]
- 43. Kyriakis J. M., Avruch J. (2001) Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81, 807–869 [DOI] [PubMed] [Google Scholar]
- 44. Coffa S., Breitman M., Spiller B. W., Gurevich V. V. (2011) A single mutation in arrestin-2 prevents ERK1/2 activation by reducing c-Raf1 binding. Biochemistry 50, 6951–6958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schattauer S. S., Miyatake M., Shankar H., Zietz C., Levin J. R., Liu-Chen L. Y., Gurevich V. V., Rieder M. J., Chavkin C. (2012) Ligand directed signaling differences between rodent and human κ-opioid receptors. J. Biol. Chem. 287, 41595–41607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Good M., Tang G., Singleton J., Reményi A., Lim W. A. (2009) The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell 136, 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bhattacharyya R. P., Reményi A., Good M. C., Bashor C. J., Falick A. M., Lim W. A. (2006) The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science 311, 822–826 [DOI] [PubMed] [Google Scholar]
- 48. Yamasaki T., Kawasaki H., Nishina H. (2012) Diverse Roles of JNK and MKK pathways in the brain. J. Signal Transduct. 2012, 459265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tournier C., Dong C., Turner T. K., Jones S. N., Flavell R. A., Davis R. J. (2001) MKK7 is an essential component of the JNK signal transduction pathway activated by proinflammatory cytokines. Genes Dev. 15, 1419–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vishnivetskiy S. A., Hirsch J. A., Velez M.-G., Gurevich Y. V., Gurevich V. V. (2002) Transition of arrestin into the active receptor-binding state requires an extended interdomain hinge. J. Biol. Chem. 277, 43961–43968 [DOI] [PubMed] [Google Scholar]
- 51. Hanson S. M., Cleghorn W. M., Francis D. J., Vishnivetskiy S. A., Raman D., Song X., Nair K. S., Slepak V. Z., Klug C. S., Gurevich V. V. (2007) Arrestin mobilizes signaling proteins to the cytoskeleton and redirects their activity. J. Mol. Biol. 368, 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Song X., Gurevich E. V., Gurevich V. V. (2007) Cone arrestin binding to JNK3 and Mdm2. Conformational preference and localization of interaction sites. J. Neurochem. 103, 1053–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gurevich E. V., Benovic J. L., Gurevich V. V. (2004) Arrestin2 expression selectively increases during neural differentiation. J. Neurochem. 91, 1404–1416 [DOI] [PubMed] [Google Scholar]
- 54. Gurevich E. V., Benovic J. L., Gurevich V. V. (2002) Arrestin2 and arrestin3 are differentially expressed in the rat brain during postnatal development. Neuroscience 109, 421–436 [DOI] [PubMed] [Google Scholar]
- 55. Gurevich V. V., Hanson S. M., Song X., Vishnivetskiy S. A., Gurevich E. V. (2011) The functional cycle of visual arrestins in photoreceptor cells. Prog. Retin. Eye Res. 30, 405–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Saha A., Deshaies R. J. (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zheng N., Schulman B. A., Song L., Miller J. J., Jeffrey P. D., Wang P., Chu C., Koepp D. M., Elledge S. J., Pagano M., Conaway R. C., Conaway J. W., Harper J. W., Pavletich N. P. (2002) Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 [DOI] [PubMed] [Google Scholar]
- 58. Schlosshauer M., Baker D. (2004) Realistic protein-protein association rates from a simple diffusional model neglecting long-range interactions, free energy barriers, and landscape ruggedness. Protein Sci. 13, 1660–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Locasale J. W., Chakraborty A. K. (2008) Regulation of signal duration and the statistical dynamics of kinase activation by scaffold proteins. PLoS Comput. Biol. 4, e1000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Goodman O. B., Jr., Krupnick J. G., Santini F., Gurevich V. V., Penn R. B., Gagnon A. W., Keen J. H., Benovic J. L. (1996) β-Arrestin acts as a clathrin adaptor in endocytosis of the β2-adrenergic receptor. Nature 383, 447–450 [DOI] [PubMed] [Google Scholar]
- 61. Laporte S. A., Oakley R. H., Zhang J., Holt J. A., Ferguson S. S., Caron M. G., Barak L. S. (1999) The β2-adrenergic receptor/β-arrestin complex recruits the clathrin adaptor AP-2 during endocytosis. Proc. Natl. Acad. Sci. U.S.A. 96, 3712–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Granzin J., Wilden U., Choe H. W., Labahn J., Krafft B., Büldt G. (1998) X-ray crystal structure of arrestin from bovine rod outer segments. Nature 391, 918–921 [DOI] [PubMed] [Google Scholar]
- 63. Schubert C., Hirsch J. A., Gurevich V. V., Engelman D. M., Sigler P. B., Fleming K. G. (1999) Visual arrestin activity may be regulated by self-association. J. Biol. Chem. 274, 21186–21190 [DOI] [PubMed] [Google Scholar]
- 64. Han M., Gurevich V. V., Vishnivetskiy S. A., Sigler P. B., Schubert C. (2001) Crystal structure of β-arrestin at 1.9 Å. Possible mechanism of receptor binding and membrane translocation. Structure 9, 869–880 [DOI] [PubMed] [Google Scholar]
- 65. Milano S. K., Pace H. C., Kim Y. M., Brenner C., Benovic J. L. (2002) Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry 41, 3321–3328 [DOI] [PubMed] [Google Scholar]
- 66. Sutton R. B., Vishnivetskiy S. A., Robert J., Hanson S. M., Raman D., Knox B. E., Kono M., Navarro J., Gurevich V. V. (2005) Crystal structure of cone arrestin at 2.3 Å. Evolution of receptor specificity. J. Mol. Biol. 354, 1069–1080 [DOI] [PubMed] [Google Scholar]
- 67. Kovoor A., Celver J., Abdryashitov R. I., Chavkin C., Gurevich V. V. (1999) Targeted construction of phosphorylation-independent β-arrestin mutants with constitutive activity in cells. J. Biol. Chem. 274, 6831–6834 [DOI] [PubMed] [Google Scholar]
- 68. Shukla A. K., Manglik A., Kruse A. C., Xiao K., Reis R. I., Tseng W. C., Staus D. P., Hilger D., Uysal S., Huang L. Y., Paduch M., Tripathi-Shukla P., Koide A., Koide S., Weis W. I., Kossiakoff A. A., Kobilka B. K., Lefkowitz R. J. (2013) Structure of active β-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature 497, 137–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kim Y. J., Hofmann K. P., Ernst O. P., Scheerer P., Choe H. W., Sommer M. E. (2013) Crystal structure of pre-activated arrestin p44. Nature 497, 142–146 [DOI] [PubMed] [Google Scholar]
- 70. Johnson G. L. (2011) Defining MAPK interactomes. ACS Chem. Biol 6, 18–20 [DOI] [PubMed] [Google Scholar]
- 71. Dhanasekaran D. N., Kashef K., Lee C. M., Xu H., Reddy E. P. (2007) Scaffold proteins of MAP-kinase modules. Oncogene 26, 3185–3202 [DOI] [PubMed] [Google Scholar]
- 72. Strissel K. J., Sokolov M., Trieu L. H., Arshavsky V. Y. (2006) Arrestin translocation is induced at a critical threshold of visual signaling and is superstoichiometric to bleached rhodopsin. J. Neurosci. 26, 1146–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Song X., Vishnivetskiy S. A., Seo J., Chen J., Gurevich E. V., Gurevich V. V. (2011) Arrestin-1 expression level in rods. Balancing functional performance and photoreceptor health. Neuroscience 174, 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hanson S. M., Gurevich E. V., Vishnivetskiy S. A., Ahmed M. R., Song X., Gurevich V. V. (2007) Each rhodopsin molecule binds its own arrestin. Proc. Natl. Acad. Sci. U.S.A. 104, 3125–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]