Background: Traffic of the amyloid precursor protein (APP) determines its processing to neurotoxic Aβ peptides.
Results: Mint3 is recruited to the Golgi by APP and is required for export to a LAMP1+ compartment.
Conclusion: Mint3 and a LAMP1+ compartment are important components in APP traffic.
Significance: Mint3 is an essential component in normal APP traffic with the potential to shift the balance in Aβ generation.
Keywords: Adaptor Proteins, Amyloid Precursor Protein, Golgi, Lysosomes, Membrane Trafficking, ADP-ribosylation Factor, Mint3, Recycling Endosomes
Abstract
The amyloid precursor protein (APP) is a ubiquitously expressed single-pass transmembrane protein that undergoes proteolytic processing by secretases to generate the pathogenic amyloid-β peptide, the major component in Alzheimer plaques. The traffic of APP through the cell determines its exposure to secretases and consequently the cleavages that generate the pathogenic or nonpathogenic peptide fragments. Despite the likely importance of APP traffic to Alzheimer disease, we still lack clear models for the routing and regulation of APP in cells. Like the traffic of most transmembrane proteins, the binding of adaptors to its cytoplasmic tail, which is 47 residues long and contains at least four distinct sorting motifs, regulates that of APP. We tested each of these for effects on the traffic of APP from the Golgi by mutating key residues within them and examining adaptor recruitment at the Golgi and traffic to post-Golgi site(s). We demonstrate strict specificity for recruitment of the Mint3 adaptor by APP at the Golgi, a critical role for Tyr-682 (within the YENPTY motif) in Mint3 recruitment and export of APP from the Golgi, and we identify LAMP1+ structures as the proximal destination of APP after leaving the Golgi. Together, these data provide a detailed view of the first sorting step in its route to the cell surface and processing by secretases and further highlight the critical role played by Mint3.
Introduction
The amyloid precursor protein (APP)2 is a ubiquitously expressed single-pass transmembrane protein of unknown function in mammals. APP is important in the brain as its proteolytic processing can generate the Aβ peptide, which contributes to the pathogenesis of Alzheimer disease (1, 2). APP is synthesized in the rough endoplasmic reticulum (ER), travels to the Golgi where it is glycosylated into its mature form, and then exits the Golgi before arriving at the plasma membrane. From the plasma membrane it can be internalized through the endocytic pathway (1, 3) and traffic to endosomes. Throughout this journey, APP may be cleaved by α- and γ-secretases to generate nontoxic protein fragments or by β- and γ-secretases to generate the pathogenic fragment Aβ. These peptide fragments oligomerize and later can be deposited into extracellular amyloid plaques, the pathogenic hallmark of Alzheimer disease (4–6). The different secretases responsible for the cleavage of APP have distinct distributions throughout the cell. Nonamyloidogenic processing of APP occurs predominantly at the plasma membrane where APP is exposed to α-secretase (7). Although still controversial, the amyloidogenic processing is thought to occur predominantly in the endomembrane system (for a review of APP processing see Ref. 8). Both the routing and the kinetics of APP traffic through the cell are thought to determine the extent of exposure to the different secretases and thereby the rate and extent of processing into amyloidogenic or nonamyloidogenic species.
The sorting of APP at each membrane and its traffic through the cell are influenced by its binding to adaptor proteins that are recruited from cytosol to initiate carrier biogenesis (9–12). Adaptors interact with cargo via sorting motifs contained in the cytoplasmic tail. Adaptors can bind directly to the cargo and facilitate both the concentration of the cargo into nascent carriers as well as recruit other proteins that together form a coated carrier (11–16). The mature carrier requires machinery to pinch it off from the donor membrane, as well as information ensuring its movement to a specific membrane destination and machinery involved in uncoating and fusion at that site. Thus, the recruitment of specific protein adaptors to APP is directly linked to its routing throughout the endomembrane system.
The cytoplasmic tail of APP is 47 residues long and contains at least four distinct sorting motifs (see Fig. 3A) (17–19), each with the potential to regulate APP traffic at one or more sites (20). APP is expressed in mammals in several splice variants of different lengths. The most prominent variant in human brain is 695 residues long, and this variant was used in all of our studies. The numbering of residues is based on this variant. The membrane-proximal YXXφ (where X is any amino acid and φ is a hydrophobic one) adaptin-binding motif (653YTSI656) functions in the basolateral sorting of APP in at least some cell types (21, 22), as well as in internalization at the plasma membrane (23). The more membrane-distal portion of the tail contains a YXNPXY motif (682YENPTY687) that binds to the Munc18-interacting proteins (Mint1–3 (24)). We have previously shown that the monomeric adaptor Mint3 is important for export from the Golgi, as depletion of Mint3 or truncation of the tail of APP to remove the YENPTY motif alter APP traffic (25). The shorter NPXY motif is found within the YENPTY motif and binds to Fe65 and Dab2 (19), although reports vary as to the effect of Fe65 on the processing of APP (26, 27). Snx17 also binds to APP through the YENPTY motif at early endosomes to affect Aβ production (28). Finally, the membrane-distal YKFFE motif (686YKFFE690) mediates the binding of the tetrameric adaptin, AP-4, and can affect the processing of APP to pathogenic species (29, 30). AP-4 also has been implicated in regulating traffic of APP from the Golgi. Indeed, it was reports that AP-4 is the mediator of export of APP from the Golgi (29) that prompted this study to assess the contribution of Mint3 to this process.
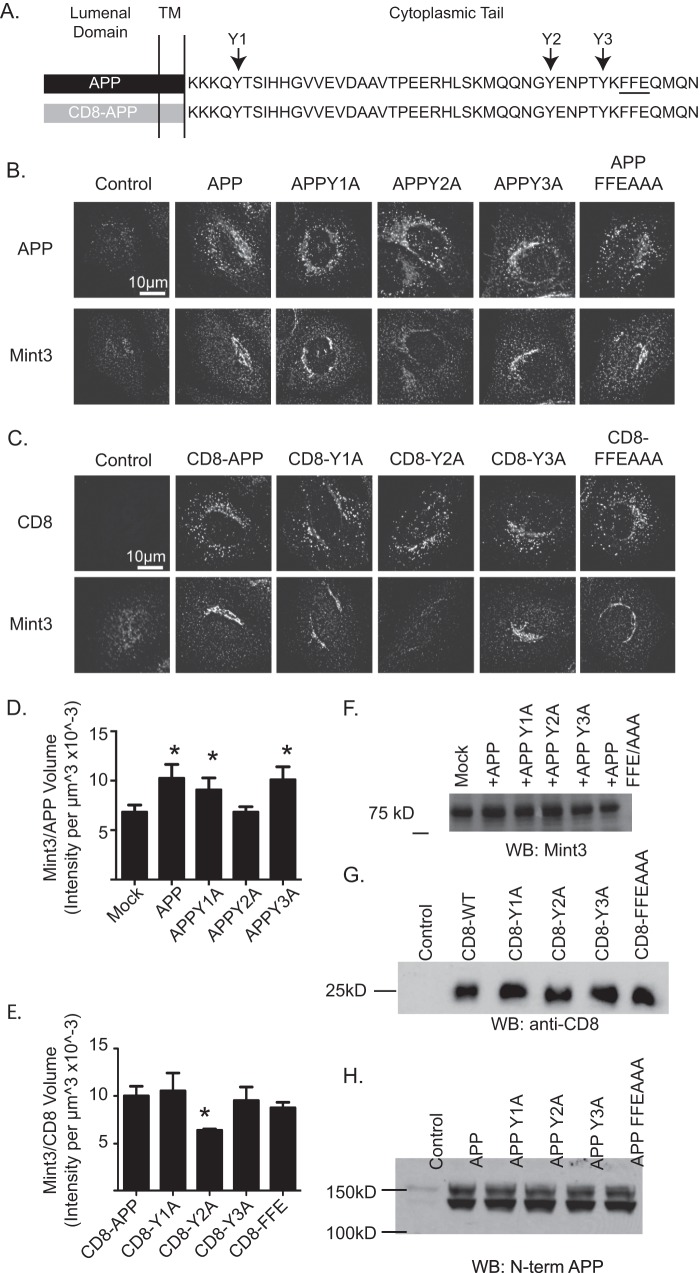
FIGURE 3.
Mutation of Y2 to alanine results in the inability to recruit Mint3 in CD8-APP or APP. A, schematic representation of APP, consisting of the lumenal, transmembrane (TM), and cytosolic (tail) domains showing the primary sequence of only the cytoplasmic tail. The three tyrosines mutated (to alanines) are shown above as Y1, Y2, or Y3 and the three-residue FFE motif, mutated to AAA, is underlined. The lower construct indicates that the lumenal and transmembrane domains of APP were replaced with those of CD8. HeLaM cells were transfected with the cargos indicated and 18 h later were fixed and stained with antibodies against APP and Mint3 (B) or CD8 and Mint3 (C). Confocal images are shown. Wild type and all other mutants recruit Mint3 comparably, but the Y2A mutation uniquely loses Mint3 recruitment. D and E, Mint3 recruitment was quantified in cells treated as described above. Stacks of wide field images were collected and deconvolved, and isosurfaces were generated based on APP staining (D) or CD8 staining (E). The total pixel intensity within each isosurface was calculated and expressed as a ratio of Mint3 intensity/cargo volume. n >7 cells were used per condition. ANOVA was use to compare groups to mock-transfected cells (D) or CD8-APP (E). Asterisks indicate statistical significance compared with mock-transfected (D) or CD8-APP (E) (p < 0.05). F, total cellular Mint3 is unaffected by expression of APP mutants. HeLaM cells were transfected with the CD8-APP (G) or full-length APP (H) constructs indicated and treated as described in F. Immunoblots were probed with an antibody against CD8 (G) or the N terminus of APP (H), as described under “Experimental Procedures.” Overall, none of the mutations cause a change in the level of expression in total cell homogenates. WB, Western blot.
Because of the importance of membrane traffic to the localization and processing of APP, this protein has been the subject of a large number of studies (8). Importantly, although there appears to be strong evidence that the generation of the pathogenic Aβ occurs largely in endosomes, there are several reports claiming that the Golgi is also an important site of processing (29–32). The Golgi is the first site of sorting divergence for membrane proteins, allowing their export to distinct destinations. Although the mechanisms by which this sorting decision is made and carried out are not completely understood, it is likely that the adaptors have a large influence on the destination of each transmembrane cargo, i.e. if APP has more than a single means for exit from the Golgi the nature of the adaptor(s) it recruits will likely result in different routes to the plasma membrane. This has the potential for delivering APP to different sites for processing, resulting in differences in the location and extent of Aβ generation. Clearly, a better understanding of the regulation of export of APP from the Golgi will provide not only a better understanding of the regulation of this process but may also provide targets for clinically relevant intervention.
Previous work from our laboratory demonstrated that APP recruits Mint3 for export from the Golgi, and a truncation of the cytoplasmic tail of APP to eliminate the YENPTY motif eliminated Mint3 recruitment and altered APP export (25). However, this truncation also eliminated other known adaptor-binding motifs, most importantly to our interpretation is the motif that binds to AP-4. Thus, we wanted to refine these studies using site-directed mutagenesis to determine the impact on binding and functionality of a much more discrete number of binding partners. To evaluate the effect of the sorting motifs on the export of APP from the Golgi and proximal destination, we mutated key residues within the cytosolic tail of APP and evaluated effects on adaptor recruitment at the Golgi and traffic to post-Golgi sites. We also took advantage of previously described protocols that can arrest protein export from the Golgi and accumulate a bolus of cargo that is more easily tracked (20 °C temperature block) or strip the Golgi of Arf-dependent adaptors (short term exposure to the drug brefeldin A (BFA)) to ask specific questions about the impact of specific residues in the cytoplasmic tail of APP on adaptor recruitment and Golgi export and proximal destination.
EXPERIMENTAL PROCEDURES
Cell Culture
HeLaM (generous gift from Dr. Margaret Robinson) cells were maintained in 10% fetal bovine serum (Atlanta Biologicals, catalog no. S11150) in DMEM (Invitrogen catalog no. 11965, v/v) in a water-jacketed incubator and maintained at 5% CO2 and 37 °C.
Plasmids and Transfections
Generation of the CD8-tail constructs is described by Caster et al. (33). Each of these constructs expresses the ectodomain and transmembrane domain of CD8 fused to the cytoplasmic tail of APP with the indicated mutations. Mutations were introduced by amplifying the region encoding the cytoplasmic tail of APP using primers that incorporated the desired changes. All constructs were sequenced to confirm the mutation desired and ensure against additional changes. pFUW-APP was described by Shrivastava-Ranjan et al. (25) and directs expression of the 695-residue variant of human APP, under control by the ubiquitin C promoter.
Plasmids were transfected using FuGENE 6 transfection reagent (Roche Applied Science catalog no. 11814443001). Cells were plated onto 6-well dishes 1 day prior to transfection at a density resulting in 80% confluence at the time of transfection. Each well of a 6-well plate received 1 μg of DNA in 100 μl of warmed Opti-MEM medium (Invitrogen catalog no. 11058). After a 5-min incubation, 6 μl of FuGENE 6 was added to the DNA/Opti-MEM mixture and incubated at room temperature for 20 min. During this incubation, cells were rinsed once with 1 ml of pre-warmed Opti-MEM and then placed in 1 ml of pre-warmed Opti-MEM. At the conclusion of the 20 min-incubation, the DNA/FuGENE/Opti-MEM solution was added dropwise to the well. Cells were then returned to 37 °C for 4 h. Cells were then split by taking up cells with 0.5 ml of 0.05% trypsin/EDTA (Invitrogen catalog no. 25300), adding 3 ml of medium, and plating onto 6-cm dishes containing Matrigel-coated coverslips (BD Biosciences, catalog no. 356234). Cells were allowed to attach overnight.
Antibodies
Primary antibodies reported in this study are as follows: C-terminal APP (Synaptic Systems catalog no. 127002) at 1:1000 (WB) and 1:500 (ICC), N-terminal APP (Chemicon, catalog no. MAB348) at 1:1000 for ICC; CD8 (Santa Cruz Biotechnology, catalog no. SC7188) was used at 1:500 for immunoblotting; CD8 (Ancell Corp., catalog no. 153020) was used for ICC at 1:1000; Mint3 (BD Transduction Laboratories, catalog no. 611380) was used at 1:500 for WB and 1:200 for ICC; Mint3 (Santa Cruz Biotechnology, catalog no. SC-28970) was used at 1:500 for ICC; ϵ4 (AP-4, BD Biosciences, catalog no. 612018) was used at 1:100 for ICC; pre-immune serum and μ4 (AP-4) antibody were generous gifts from Dr. Juan Bonifacino (NICHD, National Institutes of Health) used at 1:200 (ICC); Fe65 (Upstate EMD Millipore, catalog no. 05-758) at 1:1000 (ICC); FAPP2 (gift from Antonella DeMatteis) was used at 1:1000; giantin (Covance, catalog no. PRB-114C) was used at 1:1000; GM130 (BD Transduction Laboratories, catalog no. 610823) was used at 1:1000; EEA1 (Abcam, catalog no. ab2900) used at 1:1000; transferrin receptor (Invitrogen, catalog no. 13-680) was used at 1:1000; LAMP1 (catalog no. H4A3), LAMP2 (catalog no. H4B4), and AP-3 (ΔSA4) were purchased from the Developmental Studies Hybridoma Bank, University of Iowa and used at 1:1000 (ICC); TGN46 (AbD Serotec (Bio-Rad.), catalog no. AHP500) was used at 1:1000; AP-1 (γ-adaptin, BD Transduction Laboratories, catalog no. 610502) was used at 1:100. Secondary antibodies reported in this manuscript are: Alexa488 and Alexa594 (both from Invitrogen) were used at 1:500 for ICC.
Immunoblotting
Cells were lysed in RIPA buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40) containing a mixture of protease inhibitors (Sigma catalog no. P2714-1PTL). Protein concentrations were determined using the Bradford protein assay (Bio-Rad catalog no. 500-0113). Proteins (20 μg) were resolved in SDS-polyacrylamide gels prior to transfer to nitrocellulose, which were later placed in blocking solution consisting of phosphate-buffered saline (PBS; 137 mm NaCl, 2.7 mm KCl, 10 mm sodium phosphate dibasic, 2 mm potassium phosphate monobasic, pH 7.4) with 5% powdered milk and 0.02% sodium azide for 1 h. Primary antibodies were diluted in blocking solution and applied to the blot overnight at 4 °C. Blots were then washed three times in PBS containing 0.01% Tween 20. Secondary antibodies were diluted in blocking solution and applied to the blot for 1 h at room temperature. Blots were developed using SuperSignal (Thermo Scientific, catalog no. 34075), and films were exposed for the times indicated in the figures.
BFA Treatment
Cells maintained and transfected as described above were plated onto 6-cm dishes containing Matrigel-coated coverslips. BFA was diluted in methanol to a 10× working stock and applied to cells so the final concentration was 7.5 μg/ml. Two minutes after application of drug, cells were rinsed once with pre-warmed medium, and 4 ml of medium was applied to the cells for the remainder of the recovery period, and cells were fixed as described below. Cells treated with methanol (final concentration 1% v/v) vehicle control were morphologically indistinguishable from untreated cells (data not shown).
Temperature Block and Immunocytochemistry
Medium was aspirated from cells and replaced with 4 ml of 20 mm HEPES, 10% fetal bovine serum in DMEM (Invitrogen catalog no. 11965). Cells were then placed in the 19.5 °C water bath for 4 h. Following this temperature block, cells were either immediately fixed or returned to 37 °C for varying times of “release,” prior to fixation. Release was performed by replacing medium with fresh, pre-warmed (37 °C) medium without HEPES, and dishes were placed in a gassed incubator. Cells were fixed in 2% paraformaldehyde in PBS for 20 min at room temperature. Following fixation, cells were rinsed with PBS for 5 min, a total of four times. Individual coverslips were then placed on a parafilm-coated 24-well dish and ∼200 μl of blocking solution, 1% bovine serum albumin, and 0.05% saponin in PBS for 20 min at room temperature. Primary antibodies were diluted in blocking solution, and ∼100 μl was added to a coverslip, covered, and placed at 4 °C overnight. Coverslips were then washed four times with washing solution (0.05% saponin in PBS) for 5 min each wash. Secondary antibodies were diluted in blocking solution at a 1:500 dilution and applied to cells for 1 h at room temperature. Coverslips were then washed twice with washing solution for 5 min. Hoechst 33342 (Invitrogen) was diluted 1:5000 in blocking solution and applied to cells for 5 min. Cells were washed twice for 5 min with washing solution and then placed in PBS for 5 min. Coverslips were mounted using Mowiol (Calbiochem, catalog no. 475904), prepared as described in Valnes and Brandtzaeg (34).
Image Acquisition and Quantification
Confocal images were acquired on an Olympus IX81 microscope using a ×100 objective, NA 1.45. Each channel was manually thresholded using red/blue intensity indicators. Channels were excited and imaged sequentially. Wide field images were collected on a Nikon TE300 using a ×63 objective lens with a numerical aperture of 1.4 and a Quantix cooled CCD camera. Stacks of images were collected with a 1316 × 1035 pixel aspect ratio, a 16-bit file depth, and with a step size of 0.2 μm.
Structured illumination microscopy images were collected on a Nikon Eclipse Ti microscope using a ×100 objective with NA 1.49 and an Andor DU-897 camera. The 594- and 488-nm channels were collected with a 100- and 300-ms exposure time, respectively, each with a conversion gain of ×1. Images were reconstructed using Nikon Elements software with the following parameter settings: structured illumination contrast of 1.5, apodization filter parameter of 1.0, and width of 3D-SIM filter of 0.15.
Quantitative Image Analysis
Stacks of wide field images were deconvolved using Huygens SVI deconvolution software. Each channel was deconvolved using the theoretical point spread function appropriate for the fluorophore indicated. Each stack of images was iteratively deconvolved until a threshold of 0.1 total image change was achieved. Deconvolved images were imported into Imaris where isosurfaces were defined based on cell marker staining. Note that once an isosurface is defined, Imaris generates many parameters that describe the structure, including but not limited to volume and surface area. We chose to use sum pixel intensity within an isosurface volume, rather than surface area, due to concerns that invaginations or extensions of membrane surfaces or adjacent planar membranes may contribute more to errors in the latter. This method and surrounding issues are described in more detail in Caster and Kahn (35).
RESULTS
Mint3 Is Specifically Recruited in Response to APP Expression
The cytoplasmic tail of APP is 47 residues in length and contains four previously described sorting motifs that bind different proteins and potentially contribute to APP traffic from the Golgi: YXXφ, YENPXY, NPXY (contained within the larger YENPTY motif), and KFFE. We mutated key residues in these motifs to evaluate their effect on the following: (i) the recruitment of adaptors, (ii) the rate of export of APP from the Golgi, and (iii) the proximal destination of APP after leaving the Golgi.
The APP gene is expressed in humans as a number of alternatively spliced transcripts, resulting in different lengths of proteins. We based our constructs and numbering of residues on the transcript that encodes a protein of 695 residues, as it is the major isoform expressed in the brain and is most commonly studied by Alzheimer disease researchers. The lumenal domain of APP has been shown to interact with other proteins, notably the transmembrane protein LR11/SORLA (36), and this has been predicted to influence the retention, sorting, and kinetics of its export from the Golgi. To allow us to discriminate between the cytoplasmic tail and combined lumenal plus cytoplasmic tail interactions, we generated a fusion protein consisting of the lumenal and transmembrane domain of CD8 with only the cytoplasmic tail of APP. Homologous CD8 fusion proteins have been used in several laboratories and found to retain key sorting information for protein cargos, including the mannose 6-phosphate receptor and furin (37, 38). The CD8-APP proteins expressed are each 257 amino acids in length, with an apparent molecular mass of ∼28 kDa. Thus, our studies were performed with both full-length APP and the CD8 fusion constructs, and the results were quite consistent, indicating only minimal, if any, effects of lumenal interactions on APP in our assays.
APP or CD8-APP proteins were transiently expressed in HeLaM cells to allow quantification and localization using immunofluorescence. This cell line was chosen due to its flattened morphology when grown on coverslips, providing advantages for imaging and low background of adaptor staining on endomembranes, facilitating quantification. The use of fusion proteins and protein overexpression required that attention be given to the potential for artifactual results. To minimize these concerns, the following steps were taken: (i) results from CD8 fusions were always compared with those from use of the full-length (untagged) APP; (ii) we selected for analysis only those cells that displayed clear but minimal protein expression; (iii) we confirmed by visual inspection that cells expressing higher levels of exogenous protein than those used in quantification yielded the same qualitative results; and (iv) DNA concentrations in transfections were optimized such that each mutant in the series was expressed to the same extent as the nonmutated proteins (we found that this could vary with the DNA preparation but was not consistently different among the mutants). Additional controls included untransfected cells, empty vector transfections, and CD8 lumenal and transmembrane domain with no cytoplasmic tail. This last control displayed diffuse staining throughout the cell, which confirmed the functional importance of the cytoplasmic tail, but was not used in quantification due to its completely diffuse nature (data not shown).
We evaluated each of the well characterized Golgi adaptors for membrane recruitment in response to expression of our APP constructs and found that only Mint3 was differentially affected. HeLaM cells were transfected with the indicated cargo, fixed ∼20 h later, and stained with antibodies against either APP or CD8 and COPI, GGA1, AP-1, AP-3, AP-4, or Mint3. Endogenous APP staining in HeLaM cells is punctate and widely distributed throughout the cytoplasm, with a higher density of staining in the perinuclear region that overlaps with Golgi markers (Fig. 1A). With expression of APP or CD8-APP, staining with the antibody directed against the C terminus of APP or to CD8, respectively, resulted in brighter staining that is otherwise the same as that of endogenous APP (Fig. 1A). Each of the adaptors tested exist in a cytosolic pool from which they are recruited to membranes as a result of direct interactions with some combination of a transmembrane cargo, activated Arfs, and phospholipids (14, 39–42). The levels of adaptors in the cells did not change in response to cargo expression (e.g. see Fig. 3F or Caster et al. (33)). Thus, increased adaptor staining at a membrane is interpreted as evidence of recruitment to that organelle from the soluble pool. We observed no changes in the staining of COPI, GGAs, AP-3 (data not shown), AP-1 (Fig. 1A), or AP-4 (Fig. 1A) in response to increased expression of APP or CD8-APP. In marked contrast, Mint3 staining was seen to specifically increase (Fig. 1A), and the intensity of Mint3 staining correlated, by visual inspection, with the level of APP or CD8-APP staining in the perinuclear region (data not shown).
FIGURE 1.
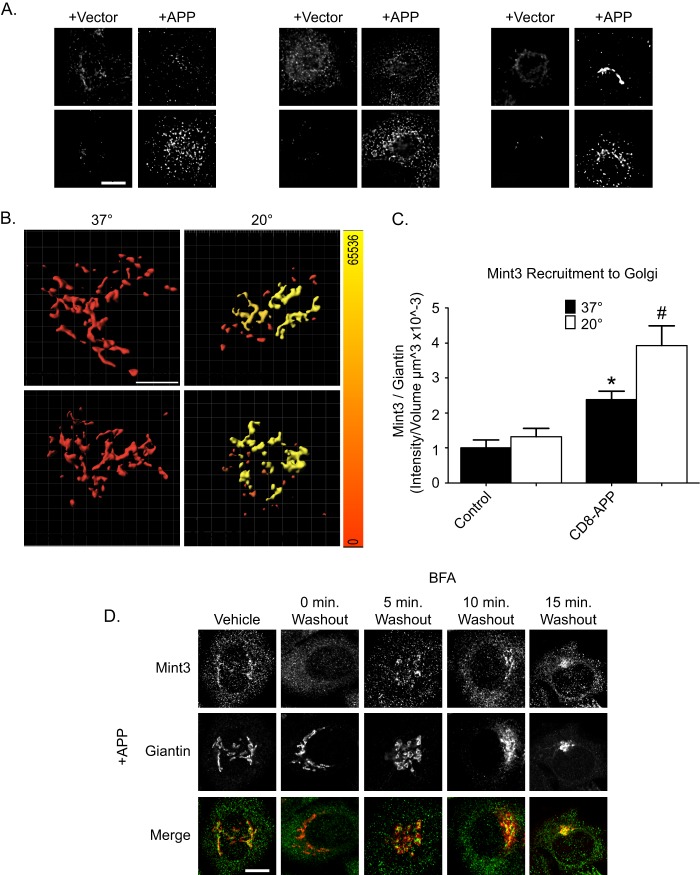
APP specifically recruits Mint3 to the Golgi, in an Arf-dependent fashion. A, cells were transfected with empty vector (pFUGW) or pFUGW-APP as indicated. Cells were fixed 1 day later and stained with antibodies against the C terminus of APP and γ-adaptin (AP-1), ϵ4 (AP-4), or Mint3. Maximum intensity projections of wide field images are shown. B, cells were transfected with APP or CD8-APP, as indicated, and stained with antibodies to APP and GM130 (top panels) or CD8 and giantin (bottom panels). Isosurfaces were built based on GM130 (top panels) or giantin (bottom panels) staining, and the total intensity of APP (top panels) or CD8 (bottom panels) within the isosurface was calculated. A heat map of intensities ranging from 0 to 65,536 was generated (color bar on right). C, mock-transfected cells or cells expressing CD8-APP were treated as described in B and stained with antibodies against Mint3 and giantin. Wide field stacks of images were collected, deconvolved, and imported into Imaris where isosurfaces were built based on giantin staining; the total amount of Mint3 within the isosurface was recorded and expressed as a ratio of intensity/giantin volume (intensity/μm3 × 10−3). Values were normalized to control cells maintained at 37 °C. Statistical significance is indicated with #, p < 0.01, and *, p < 0.05, when analyzed using ANOVA. D, APP-dependent Mint3 recruitment is BFA-sensitive and rapidly reversible. HeLaM cells expressing APP were treated with either vehicle or BFA for 2 min (see “Experimental Procedures”). Fresh, pre-warmed medium was then used to wash out the drug, and cells were allowed to recover for the times indicated, before fixation and staining with antibodies against Mint3 and giantin. Confocal images are shown.
The cytoplasmic tail of APP also binds a number of other proteins, most of which have no clear role in membrane traffic. The NPXY motif is directly involved in binding to Fe65 and Dab2 (19, 28), and each of these has been found capable of changing the amount of Aβ secreted by cells, although through unknown mechanisms or sites of action within cells. Like the adaptors assessed in the previous section, each of these proteins reside in the cytosol from which they can be recruited to binding sites on different membranes. We also tested FAPP2, which localizes at the late Golgi in an Arf- and phosphatidylinositol 4-phosphate-dependent fashion and serves as a control for nonspecific Golgi recruitment as there is no published evidence linking FAPP2 to APP or other transmembrane cargos transiting the Golgi. We expressed APP or CD8-APP in HeLaM cells, fixed cells ∼20 h later, and then stained with antibodies against APP and Fe65, Dab2, or FAPP2. As FAPP2 served as our negative control, it was not surprising to find that FAPP2 staining was not changed in cells expressing even the highest levels of APP or CD8-APP. Similarly, Dab2 and Fe65 staining is localized to small puncta throughout the periphery of the cell, and each was unaltered by APP or CD8-APP expression (data not shown). Thus, of the known adaptors acting at the Golgi and previously described proteins that bind the cytoplasmic tail of APP, only Mint3 was recruited to the Golgi in an APP-dependent fashion in HeLaM cells.
Both APP and Mint3 Are Further Enriched at the Golgi in Response to Temperature Block
APP traffics the secretory and endocytic pathway at different rates in different cells, and its steady state distributions are quite different among cell types (3). Our goal was to focus on APP traffic at the Golgi, and we developed a method termed three-dimensional isosurface image intensity (3D3I) for quantification of immunofluorescence imaging data, described in detail under “Experimental Procedures” and previous publications (33, 35). Briefly, we use wide field fluorescence imaging to collect a z-stack that is deconvolved using Huygen's SVI software. Deconvolved stacks are imported into Imaris, and an isosurface is generated based upon the staining of a previously characterized marker of an organelle. The fluorescence intensity of the APP or CD8-APP staining within that isosurface is then calculated. Alternatively, we can quantify any changes in adaptor localization within a cargo-defined isosurface. We also took advantage of a temperature block (20 °C, 4 h) that inhibits export from the Golgi (although ER to Golgi traffic continues) and results in an increase in the amount of cargo at the Golgi. The temperature block is rapidly reversible and allows us to monitor the abundance of adaptors recruited in response to specific cargos in a temporally and spatially highly controlled fashion.
APP or CD8-APP displayed punctate staining throughout the cytoplasm with little evidence of localization to the Golgi in HeLaM cells (Fig. 1A) without imposition of the temperature block. This low steady state localization at the Golgi contributed to a large fold-increase in the staining of APP or CD8-APP in response to the 20 °C block (Fig. 1B). In this experiment, 1 day after transfection, fixed cells were stained with antibodies against the C terminus of APP and GM130 or CD8-APP and giantin. Isosurfaces were generated based on GM130 or giantin staining (Fig. 1B), and the fluorescence intensity of APP (Fig. 1B, top panels) or CD8-APP (Fig. 1B, bottom panels) within the GM130 or giantin isosurfaces, respectively, was calculated. This switch in Golgi marker was necessary to facilitate double labeling of our fixed cells as the APP and CD8 antibodies were raised in different species. Each of the Golgi markers was color-coded as a heat map in Fig. 1B, to indicate the amount of APP or CD8-APP within the isosurface at 37 or 20 °C. The scale of sum intensities per volume (μm3) is shown on the right of Fig. 1B. Incubation at 20 °C significantly increased the amount of APP or CD8-APP in GM130 or giantin isosurfaces. We also calculated the percent of total APP present within giantin-defined isosurfaces. To do this, two isosurfaces were defined: one was generated based on the total APP fluorescence within the cell, then an isosurface based on giantin staining was identified, and the amount of APP within it was calculated. At steady state, 10.5 ± 2.8% of total APP fluorescence was found within giantin-defined isosurfaces. Following the 4-h incubation at 20 °C, APP fluorescence within giantin-defined isosurfaces increased to 75.2 ± 4.1% (results typical of three independent experiments, >5 cells per condition). Thus, as expected and shown previously for Man-6-P receptor or CD8-Man-6-P receptor (33), the amount of APP and CD8-APP at the Golgi were significantly increased by temperature block in HeLaM cells.
As shown above in Fig. 1A, expression of APP or CD8-APP resulted in increased recruitment of Mint3 to the perinuclear region. To establish the site of Mint3 recruitment, we performed double labeling with markers of several different endomembranes (including ER, recycling endosomes, lysosomes, and early, medial, and late Golgi), and we found the strongest overlap with those of the Golgi. We next quantified the amount of Mint3 recruited to giantin-positive isosurfaces in response to CD8-APP expression using 3D3I (Fig. 1C). Temperature block has little or no effect on the amount of Mint3 within a giantin-defined isosurface in control (mock-transfected) cells (Fig. 1C, left panel). Expression of CD8-APP significantly increased the amount of Mint3 within giantin-defined isosurfaces at 37 °C, and this was further increased by the 4-h incubation at 20 °C (Fig. 1C, right panel). Similar results were obtained using APP (data not shown). We also examined the recruitment of other adaptors to APP accumulated at the Golgi by the temperature block. We observed no changes in the recruitment of other proteins or adaptors such as AP-1, GGAs, FAPP2, Dab2, or Fe65 in APP-expressing cells following temperature blockade, despite the increased presence of APP or CD8-APP (see below). We conclude that increasing the amount of cellular APP or CD8-APP results in a higher concentration of either cargo at the Golgi, with parallel increases in Mint3, and that imposition of a temperature block further increased the levels of these cargos and Mint3 at the Golgi, but not those of the other adaptors or binding partners examined.
Increasing APP at the Golgi Leads to Greater Recruitment of Mint3 That Retains BFA Sensitivity
The use of BFA as a rapid, reversible inhibitor of Arf-dependent recruitment of adaptins (e.g. AP-1) and GGAs to cargos carrying tyrosine or acidic dileucine motifs is well established but has only rarely been used to assess recruitment of other adaptors to cargos carrying different sorting motifs, e.g. the YENPTY motif of APP. Mint3 has been shown to bind directly to active (GTP-bound) Arf GTPases through the C-terminal portion of their phosphotyrosine binding domains (PTB) and the C terminus of two PDZ domains (43). Mint1 and Mint2 are neuron-specific adaptors (44), whereas Mint3 is ubiquitously expressed (43, 45), localizes to the TGN in multiple cells lines, and is the only Mint paralog expressed in HeLa cells (43, 46). Like many other protein adaptors at the Golgi, Mint3 is recruited to membranes in an Arf-dependent fashion (43). To ensure that the pool of Mint3 recruited to the Golgi is predominantly recruited in an Arf-dependent fashion, we treated cells with the membrane-permeable inhibitor BFA that strips endomembranes of activated Arf, and subsequently Arf-dependent adaptors, within minutes of addition to live cells (38, 47–49). These responses to short term exposure to BFA are in marked contrast to the much more dramatic and well known loss of Golgi integrity and retrograde movement of many Golgi components back to the ER, seen with longer exposures or higher concentrations of the drug (50, 51). APP-expressing HeLaM cells were treated with vehicle or 7.5 μg/ml BFA for 2 min and then were either fixed or the drug was washed out, and cells were allowed to recover for the times indicated (Fig. 1D). APP localization was unaltered by addition of BFA compared with vehicle-treated cells (data not shown), but Mint3 staining at the Golgi was undetectable after 2 min of BFA treatment. Recovery of Mint3 staining at the Golgi was evident by 5 min after washout of the drug and continued to increase back toward steady state levels over the first 15 min. Thus, the recruitment of Mint3 to Golgi in response to increased levels of APP (or furin, see below) parallels in all respects that of other more extensively studied Arf-dependent adaptors to other cargos.
APP Traffics to a LAMP1+ Compartment from the Golgi
As noted above, temperature block led to a significant accumulation of APP cargos and Mint3 at the Golgi, and this afforded us the opportunity to follow a bolus of APP as it leaves the Golgi and to assess the proximal destination. We used temperature block and release of APP overexpressing cells, fixing at 5-min intervals after return to 37 °C. Cells were double-labeled with antibodies against the C terminus of APP and organelle markers (including EEA1, transferrin receptor, LAMP1, LAMP2, Rab5, Rab11, and cathepsin D) and evaluated for overlap in staining. Early endosomes (EEA1 and Rab5), recycling endosomes (transferrin receptor and Rab11), and a subset of lysosomes (cathepsin D (data not shown) and LAMP2 (Fig. 2C)) showed very little overlap with APP at any of the early times after release from temperature block. In contrast, LAMP1 staining displayed a high level of overlap with that of APP at 5 and 10 min after release (Fig. 2, A, B, and D). Note that at 0-min release, APP and LAMP1 co-localization is quite low, although LAMP1 staining is more centralized compared with cells maintained at 37 °C (Fig. 2A, compare 37 °C to 0′). The co-localization of APP and LAMP1 was transient as it was clearly reduced by 15 min and not evident in cells that had never been exposed to the block. We were surprised that APP staining co-localized with one lysosomal membrane marker, LAMP1 (Fig. 2, A, B, and D), but not another, LAMP2 (Fig. 2C), under the same conditions. Although these two lysosomal membrane proteins are often used interchangeably and are described as staining lysosomes, there is little or no evidence we could find that they necessarily are found in the same lysosomes. Because we do see some changes in LAMP1 staining profiles before and after temperature block, it is possible that the changes observed result from alterations in either the traffic of APP, LAMP1, or both following release from temperature block. However, note that LAMP1 staining did not increase in overlap with Golgi markers at any time or with APP prior to release from the blockade. Thus, we conclude that there exists two distinct compartments, with different lysosomal markers, and APP moves only through the LAMP1-positive structures at early times after exit from the Golgi. Similar results were obtained using CD8-APP (see below).
FIGURE 2.
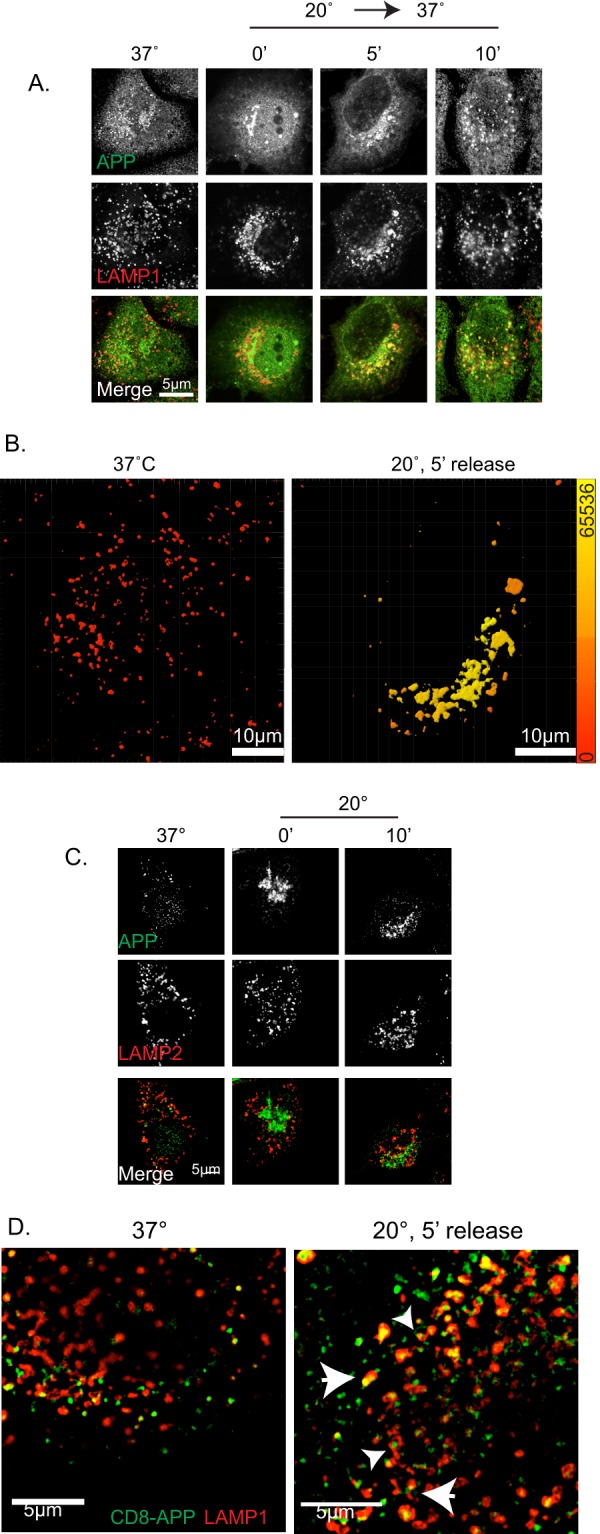
APP exits from the Golgi and arrives at LAMP1+ structures. HeLaM cells were transfected with APP (A–C) or CD8-APP (D). The next day, cells were maintained at 37 or 20 °C for 4 h and then returned to 37 °C for the times indicated. A, cells were fixed and stained with antibodies against the C terminus of APP and LAMP1. Maximum intensity projections of wide field images are shown. B, isosurfaces were generated based on the LAMP1 staining shown in A, as described under “Experimental Procedures.” Isosurfaces were then color-coded using a heat map, indicating the sum intensity of APP within the LAMP1+ isosurfaces. Color range is indicated on the vertical bar on the right. C, cells expressing APP were treated as described in A, fixed, and stained with antibodies against APP and LAMP2. APP does not overlap with LAMP2 at steady state or at any point after release from the temperature blockade. Maximum intensity projections of wide field images are shown. D, cells expressing CD8-APP, and treated as described above, were stained and fixed with antibodies against CD8 and LAMP1 at the times indicated. Cells were imaged on a Nikon structured illumination microscope (see “Experimental Procedures”), and images were reconstructed. Large arrowheads indicate examples of overlapping CD8-APP and LAMP1 compartment, and smaller arrowheads indicate examples of CD8-APP that is closely apposed to LAMP1 staining. 5 min after release from blockade, CD8-APP overlap with LAMP1 is increased.
We were surprised to observe APP and CD8-APP traffic from the Golgi to a potentially degradative compartment as we do not observe a decrease in total cellular APP levels post-release from temperature block. In efforts to better characterize the relationship of APP to LAMP1, we used the higher resolution, structured illumination microscopy to visualize this compartment. As seen in Fig. 2D, there is little overlap of the two markers at steady state, but this is quite different at early times after release from temperature blockade. These images reveal close apposition of LAMP1 and APP or CD8 staining that appears adjacent to one another, at times appearing to represent two parts of the same structure, with different extents of overlap (yellow staining) among the closely related structures (Fig. 2D, small arrowheads). Other structures even appear to have staining of one marker wrapping around the other (Fig. 2D, large arrowheads). We currently interpret these data as evidence of a transient mixing and quick resolution of two compartments.
Y2A Mutant of APP Specifically Fails to Recruit Mint3 to the Golgi
To assess the importance of each of the four previously described sorting signals in the cytoplasmic tail of APP to its localization and export from the Golgi, we generated a series of mutants in APP and CD8-APP. Each of the constructs that were used in our studies are depicted in Fig. 3A. Because of the importance of tyrosines in sorting motifs and the presence of three of these in the cytoplasmic tail of APP, we refer to Tyr-653 within the YTSI motif as Y1, Tyr-682 within the larger YENPXY motif as Y2, and Tyr-687 within the NPXY motif as Y3 (Fig. 3A). Mutation of 688FFE690 to AAA is referred to as FFE/AAA.
We tested each of the Tyr → Ala and FFE/AAA mutations for their effect on the recruitment of Mint3 to the Golgi (Fig. 3). Expression of APP (Fig. 3B) or CD8-APP (Fig. 3C) with the mutants Y1A, Y3A, or FFE/AAA resulted in Mint3 staining that was indistinguishable as far as intensity of Mint3 staining or its location in cells. In contrast, the Y2A mutants failed to promote Mint3 recruitment and instead showed staining comparable with that of untransfected cells or those transfected with the vector that lacked insert (Fig. 3, B and C). Recruitment of Mint3 to each cargo was quantified using 3D3I. Wide field images of at least seven cells in each case were analyzed. Computational isosurfaces were generated based on APP (Fig. 3D) or CD8-APP (Fig. 3E) staining, and the sum intensity of Mint3 within the isosurfaces was determined. The Mint3 staining within the APP-Y2A-defined isosurface was not statistically different from control cells that expressed no exogenous proteins (Fig. 3D). Similarly, CD8-Y2A displayed significantly less (p < 0.05) Mint3 staining within cargo-defined isosurfaces, compared with CD8-APP or the other three mutants (Fig. 3E). Note that in this case we could not compare with mock-transfected or untransfected controls, as they display no CD8 staining. The Y3A mutant fully supported Mint3 recruitment that was indistinguishable from the nonmutated tail, despite being found within the larger YENPXY motif that included Y2.
The cytoplasmic tail of APP is expected to be largely or wholly unstructured (52), so a point mutation is unlikely to cause changes in overall protein folding and consequent stability. In contrast, even a point mutation can alter traffic, post-translational modifications, protease sensitivity, or other changes that would be expected to alter the protein half-life, readily seen as a change in the level of expression versus controls. To ensure we were comparing adaptor recruitment under comparable conditions of protein expression, transfected cells were collected 18 h later, and cell homogenates were analyzed for Mint3 (Fig. 3F) or CD8-APP (Fig. 3G) expression by immunoblots from denaturing SDS gels. When the cell lysates were immunoblotted using a commercially available (Chemicon) antibody to the N terminus of APP, all the proteins were found to be expressed to comparable levels (Fig. 3H). The same pattern was found for the CD8-APP constructs (Fig. 3G).
Failure of APP-Y2A to Recruit Mint3 Is Correlated with a Delay in Its Export from the Golgi
With the near complete lack of Mint3 recruitment to the Y2A mutant of APP or CD8-APP, we asked if the lack of Mint3 binding caused a defect in export from the Golgi. This was deemed risky because it is common for transmembrane protein cargos to have alternative routes of traffic and so the lack of one route was typically compensated by an alternative route. Nevertheless, we compared the rate of exit from the Golgi and the subsequent time it took to arrive at LAMP1+ structures after release from temperature block, for the CD8-APP and CD8-Y2A constructs. Isosurfaces were generated using TGN46 staining (Fig. 4, A and B), and the amount of CD8 fluorescence within those structures was determined. CD8-APP accumulated in the TGN following temperature block (Fig. 4A) and then returned to levels comparable with those seen at 37 °C by 10 min post-release. CD8-Y2A accumulated in the TGN to essentially the same levels as CD8-APP during temperature block (Fig. 4B). However, CD8-Y2A failed to exit the TGN during the 15-min recovery period, as seen by the unchanging overlap with TGN46 (Fig. 4B), in contrast to the result with CD8-APP (Fig. 4A).
FIGURE 4.
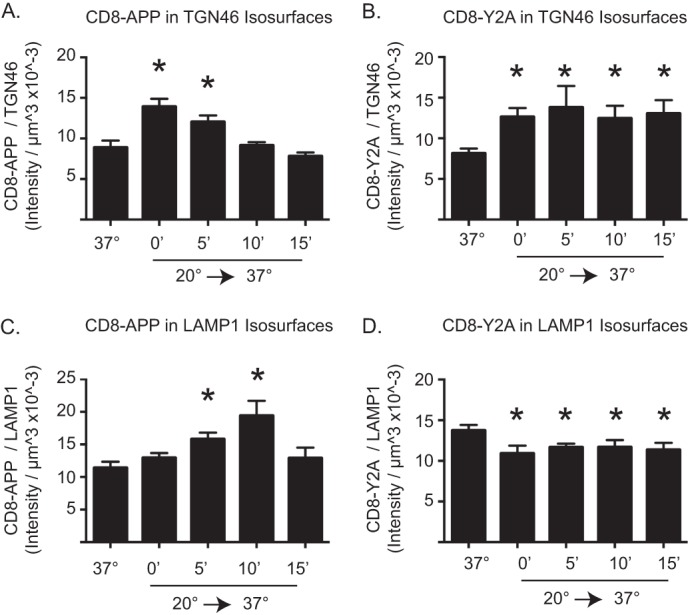
Mutation of Y2A alters APP exit from the Golgi and arrival at LAMP1+ structures. The amount of CD8-APP (A and C) or CD8-Y2A (B and D) in TGN46 isosurfaces (A and B) or LAMP1 isosurfaces (C and D) was quantified using 3D3I analysis. Cells treated as described above were stained with antibodies against CD8 and TGN46 (A and B) or CD8 and LAMP1 (C and D). Isosurfaces were generated based on TGN46 (A and B) or LAMP1 staining (C and D). The intensity of CD8 staining within each isosurface was totaled and expressed as a ratio of CD8 intensity per isosurface volume. Asterisks indicate statistical significance (p < 0.05) using ANOVA, comparing each group to cells maintained at 37 °C.
The kinetics of CD8-APP or CD8-Y2A arrival at LAMP1+ structures were also evaluated. Isosurfaces were generated based on LAMP1 staining, and the amount of CD8 fluorescence within those isosurfaces was determined. The amount of CD8-APP within LAMP1+ structures was unchanged by temperature block (Fig. 4C, compare 37 °C to 0′ release), as it was quite low under each condition. The amount of CD8-APP in LAMP1+ isosurfaces was significantly increased 5 min post-release from blockade. This value was increased even further at 10 min post-release and returned to levels indistinguishable from control (37 °C) by 15 min, consistent with our observations using APP described above. In contrast, CD8-Y2A displayed greater overlap in staining within LAMP1+ isosurfaces at 37 °C (Fig. 4D), prior to temperature block. In this case, the block reduced the amount of CD8-Y2A in LAMP1+ isosurfaces and did not return during the 15-min recovery. These results are consistent with Y2A having a defect in the rate of export from the Golgi.
We followed up on the observation that APP-Y2A displayed a greater extent of overlap with LAMP1+ structures than did APP in cells maintained at 37 °C (Fig. 4, C and D) by including each of our mutants in these analyses of steady state distributions. We found no differences between APP and mutants in their overlap with EEA1 or transferrin receptor (data not shown). However, the APP-Y2A and to an even greater extent APP-Y3A staining displayed enhanced overlap with that of LAMP1 (Fig. 5A). Selected mutants shown in Fig. 5A were analyzed using 3D3I to quantify the amount of overlap of cargo with LAMP1 at steady state (Fig. 5B). APP-Y2A and APP-Y3A are increased at LAMP1+ endosomes at steady state, compared with APP and APP-FFE/AAA. These data are consistent with both Y2 and Y3 playing roles in traffic of APP in HeLaM cells, potentially through interactions with other adaptors, although not necessarily at the Golgi based on this result alone. Together, the data support the conclusion that Y2 and Mint3 recruitment are critical to the export of APP from the Golgi and that the greater YENPTY motif, or at least Y3 at its C terminus, has at least one additional role in traffic to or from the LAMP1+ compartment. Of course, the traffic to that site is not limited to late Golgi/TGN, but it could include other (e.g. endosomal) sources of APP. The fact that we also observe overlap in LAMP1 and APP or CD8 staining at steady state with the Y2A and Y3A mutants suggests that this co-localization at the LAMP1+ compartment is not an artifact of temperature block or release.
FIGURE 5.
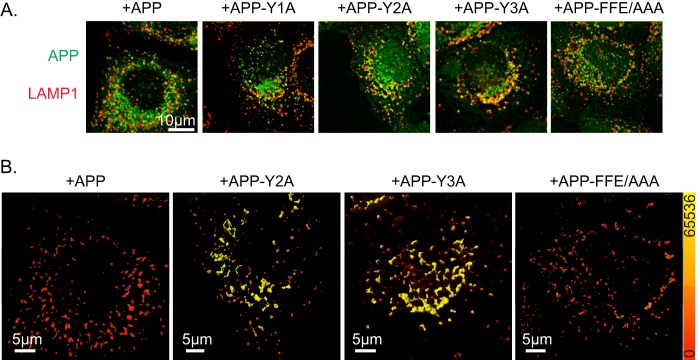
APP-Y3A or APP-Y2A display increased overlap with LAMP1 at steady state. HeLaM cells were transfected with the APP mutants indicated. The next day, cells were fixed and stained with antibodies against APP and LAMP1. A, maximum intensity projections of wide field images are shown. B, images shown in A were imported into Imaris, and isosurfaces were generated based on LAMP1 staining. Isosurfaces were then falsely colored with a heat map (shown on right, ranging from 0 to 65,536) indicating the sum intensity of APP staining within the isosurfaces.
APP and Furin Each Recruit Mint3 to the Golgi but with Opposing Outcomes
Arf-dependent adaptors regulate membrane traffic specifically in the initiation of nascent vesicular carriers for export from a donor compartment. In contrast to the critical role played by Mint3 in export of APP from the Golgi, Mint3 has been shown previously to play an equally important role in the retention of another cargo at the Golgi, furin. To compare the characteristics of the recruitment of Mint3 to these two different cargos, which result in opposing outcomes as far as export, we used FLAG-furin or CD8-furin expression in HeLaM cells. Furin is an endoprotease that acts at the Golgi, being retained there via a mechanism that includes direct binding of the cytoplasmic tail to Mint3 (53). The residues involved in furin binding to Mint3 are less well characterized than those in the APP tail but are thought to include a stretch of acidic residues 773SDSEEDE779 (numbered using the human furin sequence), based upon deletion analysis (53). We generated a plasmid that directs expression of a CD8 fusion protein containing the entire 56-residue cytoplasmic tail of human furin, homologous to the CD8-APP construct. Expression of CD8-furin in HeLaM cells led to a steady state distribution that was highly enriched in Golgi structures (33), clearly more so than was seen for CD8-APP. This is despite the fact that immunoblotting with anti-CD8 revealed very similar levels of CD8-APP and CD8-furin were expressed in HeLaM cells (Fig. 6A). CD8-furin expression resulted in enhanced Mint3 recruitment to the Golgi at steady state, and this recruitment is only slightly enhanced following temperature block (33).When we quantified total cellular staining for CD8-APP and CD8-furin and then determined the fraction that was within isosurfaces identified by TGN46 staining, we found 12.8% of CD8-APP and 64.3% of CD8-furin in this region of the Golgi. Imposition of the temperature block only slightly enhanced CD8-furin (76.0% (33)) at that site but dramatically increased the fraction of cellular CD8-APP (68.1%) there. We attribute the less dramatic changes in CD8-furin localization in response to temperature block to the already high percentage of CD8-furin there prior to the block, but it also suggests that the rate of export may be substantially lower than that of APP.
FIGURE 6.
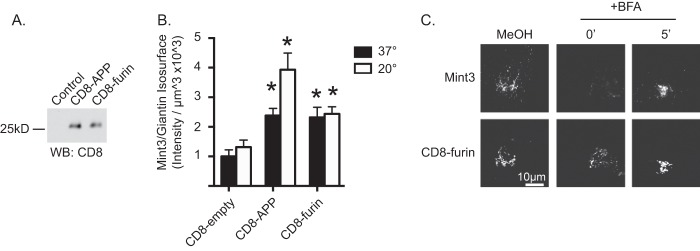
CD8-furin recruits Mint3 to the Golgi in a BFA-sensitive fashion. A, HeLaM cells or those expressing CD8-APP or CD8-furin, as indicated, were collected 1 day after transfection, and total cell homogenates (20 μg) were analyzed by immunoblotting with anti-CD8. WB, Western blot. B, HeLaM cells expressing CD8-empty (no cytoplasmic tail), CD8-APP, or CD8-furin were maintained at 37 °C or subjected to a 20 °C block, before fixation and staining for Mint3 and giantin. Isosurfaces based on giantin staining were generated, and the sum intensity of Mint3 staining within those isosurfaces was determined. Graphs represent an average (n ≥7 cells per condition) and are representative of at least three independent experiments. Error bars indicate S.E., and bars are normalized to Mint3 recruitment to giantin isosurfaces in cells not overexpressing CD8-empty at 37 °C. Asterisks indicate a p < 0.01 when analyzed using ANOVA, each compared with control staining. C, HeLaM cells were transfected with a plasmid directing the expression of CD8-furin. The next day cells were treated with methanol vehicle (methanol) or BFA for 2 min. Drug was then washed from cells with pre-warmed medium and incubated for the times indicated. Cells were then fixed and stained with antibodies against CD8 and Mint3. Confocal images are shown.
Like APP and CD8-APP, expression of furin or CD8-furin resulted in increased staining of Mint3 at the Golgi, as defined here by giantin isosurfaces (Fig. 6B). No changes were seen in total cellular Mint3 (data not shown), indicating that the adaptor is recruited from cytosol. One day (∼18 h) after transfections, we compared the relative abilities of these two cargos to recruit Mint3 to the Golgi (giantin isosurfaces) at steady state and after temperature block (Fig. 6B). Transfected cells were maintained at 37 °C (Fig. 6B, black bars) or switched to 20 °C (white bars) for 4 h and then fixed and stained for Mint3 and giantin. CD8-APP and CD8-furin resulted in similar levels of Mint3 recruitment to giantin-defined isosurfaces at 37 °C, each significantly higher than control cells (CD8-empty). CD8-APP-dependent recruitment of Mint3 was further and significantly increased during temperature block, but CD8-furin-mediated recruitment was not (Fig. 6B).
Finally, we evaluated the BFA sensitivity of Mint3 recruited to the Golgi by CD8-furin expression. HeLaM cells were transfected with plasmids directing expression of CD8-furin (Fig. 6C) and after ∼20 h were treated with vehicle (methanol) or 7.5 μg/ml BFA for 2 min. The drug was then washed out with pre-warmed medium, and cells were allowed to recover for 0, 5 (Fig. 6C), 10, or 15 min (data not shown). Treatment with BFA caused the essentially complete loss of Mint3 staining that returned to the Golgi, evident already at 5 min and increasing back to steady state levels by 15 min. We conclude that the cytoplasmic tails of APP and furin each recruit Mint3 to the Golgi in an Arf-dependent manner despite the fact that the consequences differ, with one cargo (APP) being dependent on it for export and the other (furin) for its retention.
DISCUSSION
In this study, we investigated the role of specific residues within sorting motifs in the cytoplasmic tail of APP to assess the specificity for and roles of adaptors involved in export from the Golgi. Parallel studies using full-length untagged APP and CD8-APP ensured that the results were both dependent upon the cytoplasmic tail and representative of the native protein. The combined use of minimal exogenous protein expression, point mutations in the tail, temperature block and release, and BFA treatment and recovery afforded us the opportunity to assess the role of Mint3 in APP export specifically from the Golgi and also allowed us to propose LAMP1+ structures as the proximal destination of those carriers. We conclude the following: (i) Mint3 is specifically recruited to the Golgi in response to increases in the levels of APP/CD8-APP; (ii) Y2, the first tyrosine within the YENPXY motif, is critical for APP-dependent Mint3 recruitment to the Golgi; (iii) mutation of Y2 to alanine results in a strong delay in export of APP from the Golgi; (iv) APP increases at LAMP1+ structures soon after leaving the Golgi; and (v) Mint3 can be recruited to the Golgi by different cargos, using different sorting motifs and resulting in different outcomes as far as cargo export or retention.
We conclude that Mint3 recruitment and Y2 are critical for export of APP from the Golgi in HeLaM cells based upon the following observations: (i) APP or CD8-APP carrying the Y2A mutation failed to recruit Mint3 to the Golgi; (ii) the Y2A mutants showed a delay in export after the block; and (iii) the Y2A mutants failed to demonstrate the transient wave of localization to LAMP1+ structures after release from block, compared with wild type. These results are consistent with and extend our previous observations that APP and Mint3 were found together on post-Golgi vesicles (25) and that truncation of the 10 amino acids near the cytoplasmic tail of APP (includes both YENPXY and FFE motifs) failed to recruit Mint3 (25, 53).
The truncation mutant used in our previous study exited the Golgi by 15 min after temperature block, but we did not identify its destination in that study. The fact that the truncated APP still exited the Golgi is consistent with alternative routes for egress. Our earlier study used the 15-min post-release as the earliest time point evaluated and found that the truncated form of APP resulted in enlarged carriers. Additionally, depletion of Mint3 was evaluated under steady state conditions, and we found increased overlap with syntaxin6 (a SNARE protein that localizes to the Golgi and endosomes) and EEA1. This study enhances the previous findings in that we looked at earlier time points after release from temperature block and identified an earlier structure (LAMP1) to which APP localizes very soon (within 5 min) after release. We also add context to the earlier work in that the overall effects of Mint3 depletion were likely due to traffic events occurring at sites other than the Golgi. Alternatively, it is possible that residue(s) within the C-terminal 10-amino acid deletion are involved in retention of APP at the Golgi. Although there is currently scant experimental evidence for such speculation, such a mechanism could have important ramifications to APP traffic and human disease.
Note that a strength of this study is the specific examination of directional traffic from a single site, as induced by imposition and release of the temperature block or BFA treatment. By generating a bolus of cargo in the Golgi as it is released, we can monitor subsequent changes in localization of cargo and associated traffic components. However, as the period of release increases, it is increasingly difficult to assign changes in traffic as resulting solely from the cargo leaving the Golgi.
AP-4 has also been reported to affect APP traffic at the Golgi in HeLa cells (29). In their studies, a change in the steady state distribution of APP-CFP was observed either when the APP-CFP construct contained the FFE/AAA mutations or when the μ4 subunit of AP-4 was knocked down by siRNA. The APP-CFP changed from dispersed puncta (presumably EEA1+ endosomes) to perinuclear staining displaying increased overlap with TGN46. They also show that AP-4 is concentrated in the perinuclear region of HeLa cells and partially overlaps with TGN46 staining. The authors interpreted these data as evidence that AP-4 is important for APP export from the Golgi in HeLa cells. In contrast, we were unable to find any APP- or CD8-APP-dependent changes in the staining of AP-4 in HeLaM cells. Neither did we find evidence of perinuclear clustering of AP-4, using the commercially available (BD Biosciences) antibody directed against the ϵ4 subunit (29) of AP-4 (Fig. 1A). We obtained an aliquot of the antibody directed against the μ4 subunit of AP-4, kindly provided by Dr. Juan Bonifacino (29, 54), but again we did not detect any APP-dependent changes in the staining of AP-4 in HeLaM cells nor staining that overlapped to any appreciable extent with Golgi markers (data not shown). As a positive control, the same antibody was used to stain primary cultures of mouse cortical neurons, and we observed very clear perinuclear staining that overlapped extensively with Golgi markers. Thus, AP-4 expression appears to be quite low in HeLaM cells and possibly lower than in HeLa cells. We also treated APP-expressing HeLaM cells with BFA and found no changes to AP-4 staining patterns with either antibody. This suggests either that the staining observed with AP-4 antibodies is not Arf-dependent or that the AP-4 recruited to endomembranes in HeLaM cells is mediated by an Arf GEF that is not sensitive to BFA. We interpret these data as evidence that AP-4 is not recruited to the Golgi in response to increased levels of APP in HeLaM cells, and as a consequence, it is unlikely to serve as the protein adaptor for carrier biogenesis or export. Thus, of the Arf-dependent adaptors functional in carrier biogenesis at the Golgi, only Mint3 was found to concentrate there in response to increased APP expression.
A trivial explanation for these differences could be that HeLa and HeLaM differ in some important way to affect APP traffic, e.g. levels of AP-4 expressed, although this would be surprising for cell lines that have common origins. Although our studies address fundamental aspects of membrane traffic relevant to all eukaryotic cells, the traffic of APP is particularly important in the brain because changes in traffic can alter exposure to secretases, increase Aβ generation, and promote neurodegeneration (8). Thus, it will be important to expand our observations into other cell types and specifically test the extent to which levels of expression of different binding partners or post-translational modifications to APP or partners may influence outcomes. Burgos et al. (29) did not report on AP-4 distribution with and without APP overexpression, neither did they look at time-resolved traffic of APP constructs in their cells. Thus, detailed comparisons of datasets are not possible. Nevertheless, based upon our results, we believe that Mint3 is the adaptor involved in APP export from the Golgi, although we cannot conclude that AP-4 plays a role in its export in other cells.
These data are consistent with the model that a BFA-sensitive Arf GEF at the Golgi is responsible for activation of the Arf(s) that recruit the adaptor Mint3 to that site for binding to the cytoplasmic tail of APP. This is in notable contrast to results obtained with CD8-Man-6-P receptor that lost its Arf-dependent adaptors, GGAs and AP-1, during exposure to BFA, but recovery was delayed by almost 15 min and occurred initially at recycling endosomes (33). When combined with results from this closely related study (33), we find that at least three different cargos (the cation-independent mannose 6-phosphate receptor, furin, and APP) each display a high level of specificity in Arf-dependent adaptor recruitment to the Golgi or endosomes. As the number of cargos that are discovered to share these properties rises, and the number of Arf GEFs there is limited, we speculate that the cargo itself contains within it the information that dictates specificity in adaptor recruitment. Our simplest model to explain this would be if cargo activates an Arf GEF, leading to activation and recruitment of one or more Arfs at the Golgi, which leads to recruitment of adaptor(s) that bind both activated Arf and motifs in the cytoplasmic tail of the cargo. This idea of dual binding sites is a form of coincidence detection that can provide the level of specificity found in cells. This speculative model suggests the formation of a multisubunit complex of cargo-GEF-Arf-adaptor that is likely to be quite transient and difficult to document in cells. The later role of an Arf GAP and consequent dissociation of the Arf GEF and Arfs are also predicted as carriers mature, but these were not addressed at all in this study.
Our data also lend support to the conclusion that at least one other APP binding partner plays a significant role in its traffic, other than Mint3 or AP-4. This comes from the observation that the Y3A and Y2A mutants each display increased staining at LAMP1+ structures at steady state (no temperature block). Both Y3 and FFE have been implicated in AP-4 binding, but the fact that the FFE/AAA mutant did not increase at the LAMP1+ compartment indicates that this effect of Y3A is likely not mediated by AP-4. The observation that these two mutants each cause increased steady state accumulation at LAMP1+ structures also seems to suggest that this compartment is perhaps more than a transient stop for APP exiting the Golgi. The finding that APP-Y3A is concentrated there, relative to the wild type protein, should also allow the means to identify the binding partner responsible for what we tentatively interpret as a delay in exit from this organelle.
What is the nature of the LAMP1+ compartment where APP accumulates and APP arrival correlates temporally with its export from the Golgi? We do not believe that it represents a degradative compartment as the association with LAMP1 is transient and the levels of APP fluorescence and protein do not decrease in synchrony with its traffic to/through that compartment, despite the fact that more than 50% of cellular APP was predicted to do so. It is more likely to represent a sorting compartment that happens to also contain LAMP1 but not LAMP2. These are often used interchangeably as markers of lysosomes, although they are known to traffic to endosomes and to the TGN (55). Note, however, that we did not observe increased staining of either LAMP with markers of the Golgi or TGN during temperature block. Rather, the overlap in APP staining was decidedly after release. The higher resolution images provided by structured illumination microscopy (e.g. Fig. 2D) indicate a close apposition of markers with at least some mixing and are encouraging that further studies may provide more details and potentially functions of this novel compartment. The fact that the Y2A and Y3A mutants each displayed increased overlap in staining with LAMP1 at steady state is evidence that the presence of APP at this compartment cannot be dismissed as an unexplained artifact resulting from the temperature block. Future studies of this organelle, and the binding partners involved in APP's import and export, are planned to further expand our detailed understanding of the routes and mechanisms of APP traffic.
Finally, we were interested in comparing results in our assays for two different cargos, each of which recruits Mint3 to the Golgi, although by using distinct sorting motifs and with opposing consequences to cargo traffic. We focused on the CD8 fusion proteins to allow more direct and comparable quantitative data, but the same (qualitative) results were obtained with the full-length proteins. In comparison with CD8-APP, a much higher fraction of cellular CD8-furin is present at the Golgi in steady state cells, and this fraction does not change with temperature block. Indeed, because the two cargos are expressed to comparable levels and recruit very similar percentages of cellular Mint3 to the Golgi, yet severalfold more CD8-furin is at the Golgi compared with CD8-APP, it appears that CD8-APP has a higher “specific activity” as far as Mint3 recruitment to the Golgi. This is consistent with the fact that furin also binds AP-1 and PACS-1 at the Golgi and thus may have a lower percentage of Mint3 sites accessible for binding. The commonalities between cargo-dependent Mint3 recruitment for export and retention suggest the possibility that other cargos may be recruiting Arf-dependent adaptors for retention that are currently interpreted solely as a role in export. This highlights a point that will require much more detailed follow-up studies than can be included here but is one that we believe will be important in the evolution of models of the regulation of membrane traffic.
Acknowledgments
We thank Victor Faundez (Emory University) and Elizabeth Sztul (University of Alabama at Birmingham) for reading versions of the manuscript and providing critical feedback. We gratefully acknowledge Gary Thomas (Oregon Health and Science University) and Margaret Robinson and Matthew Seaman (University of Cambridge) for sharing their plasmids and/or cell lines and Antonella De Matteis (Telethon Institute of Genetics and Medicine) and Juan Bonifacino (National Institutes of Health, NICHD) for their generous gifts of the FAPP2 and AP-4 antibodies, respectively.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM067226 from NIGMS and Grant P30 NS055077 (Imaging Core of the Emory Neuroscience NINDS Core Facilities).
- APP
- amyloid precursor protein
- 3D3I
- three-dimensional isosurface image intensity
- BFA
- brefeldin A
- ER
- endoplasmic reticulum
- LAMP
- lysosome-associated membrane protein
- Man-6-P
- mannose 6-phosphate
- Mint3
- Munc18 interacting protein 3
- Aβ
- amyloid-β peptide
- TGN
- trans-Golgi network
- ANOVA
- analysis of variance
- CFP
- cyan fluorescent protein
- GEF
- guanine nucleotide exchange factor
- WB
- Western blot
- ICC
- immunocytochemistry.
REFERENCES
- 1. Selkoe D. J. (1998) The cell biology of β-amyloid precursor protein and presenilin in Alzheimer's disease. Trends Cell Biol. 8, 447–453 [DOI] [PubMed] [Google Scholar]
- 2. Rajendran L., Annaert W. (2012) Membrane trafficking pathways in Alzheimer's disease. Traffic 13, 759–770 [DOI] [PubMed] [Google Scholar]
- 3. Selkoe D. J. (1994) Cell biology of the amyloid β-protein precursor and the mechanism of Alzheimer's disease. Annu. Rev. Cell Biol. 10, 373–403 [DOI] [PubMed] [Google Scholar]
- 4. Selkoe D. J., Yamazaki T., Citron M., Podlisny M. B., Koo E. H., Teplow D. B., Haass C. (1996) The role of APP processing and trafficking pathways in the formation of amyloid β-protein. Ann. N.Y. Acad. Sci. 777, 57–64 [DOI] [PubMed] [Google Scholar]
- 5. Lah J. J., Levey A. I. (2000) Endogenous presenilin-1 targets to endocytic rather than biosynthetic compartments. Mol. Cell. Neurosci 16, 111–126 [DOI] [PubMed] [Google Scholar]
- 6. Greenfield J. P., Tsai J., Gouras G. K., Hai B., Thinakaran G., Checler F., Sisodia S. S., Greengard P., Xu H. (1999) Endoplasmic reticulum and trans-Golgi network generate distinct populations of Alzheimer β-amyloid peptides. Proc. Natl. Acad. Sci. U.S.A. 96, 742–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sisodia S. S. (1992) β-Amyloid precursor protein cleavage by a membrane-bound protease. Proc. Natl. Acad. Sci. U.S.A. 89, 6075–6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thinakaran G., Koo E. H. (2008) Amyloid precursor protein trafficking, processing, and function. J. Biol. Chem. 283, 29615–29619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bonifacino J. S., Glick B. S. (2004) The mechanisms of vesicle budding and fusion. Cell 116, 153–166 [DOI] [PubMed] [Google Scholar]
- 10. Bonifacino J. S., Lippincott-Schwartz J. (2003) Coat proteins: shaping membrane transport. Nat. Rev. Mol. Cell Biol. 4, 409–414 [DOI] [PubMed] [Google Scholar]
- 11. Rothman J. E., Wieland F. T. (1996) Protein sorting by transport vesicles. Science 272, 227–234 [DOI] [PubMed] [Google Scholar]
- 12. Robinson M. S., Bonifacino J. S. (2001) Adaptor-related proteins. Curr. Opin. Cell Biol. 13, 444–453 [DOI] [PubMed] [Google Scholar]
- 13. Donaldson J. G., Jackson C. L. (2011) ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12, 362–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bonifacino J. S., Traub L. M. (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447 [DOI] [PubMed] [Google Scholar]
- 15. Lee M. C., Miller E. A., Goldberg J., Orci L., Schekman R. (2004) Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 20, 87–123 [DOI] [PubMed] [Google Scholar]
- 16. Rothman J. E., Orci L. (1992) Molecular dissection of the secretory pathway. Nature 355, 409–415 [DOI] [PubMed] [Google Scholar]
- 17. Marks M. S., Woodruff L., Ohno H., Bonifacino J. S. (1996) Protein targeting by tyrosine- and di-leucine-based signals: evidence for distinct saturable components. J. Cell Biol. 135, 341–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nordeng T. W., Bakke O. (1999) Overexpression of proteins containing tyrosine- or leucine-based sorting signals affects transferrin receptor trafficking. J. Biol. Chem. 274, 21139–21148 [DOI] [PubMed] [Google Scholar]
- 19. Borg J. P., Ooi J., Levy E., Margolis B. (1996) The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol. Cell. Biol. 16, 6229–6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee M. S., Kao S. C., Lemere C. A., Xia W., Tseng H. C., Zhou Y., Neve R., Ahlijanian M. K., Tsai L. H. (2003) APP processing is regulated by cytoplasmic phosphorylation. J. Cell Biol. 163, 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Icking A., Amaddii M., Ruonala M., Höning S., Tikkanen R. (2007) Polarized transport of Alzheimer amyloid precursor protein is mediated by adaptor protein complex AP1–1B. Traffic 8, 285–296 [DOI] [PubMed] [Google Scholar]
- 22. Haass C., Koo E. H., Capell A., Teplow D. B., Selkoe D. J. (1995) Polarized sorting of β-amyloid precursor protein and its proteolytic products in MDCK cells is regulated by two independent signals. J. Cell Biol. 128, 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perez R. G., Soriano S., Hayes J. D., Ostaszewski B., Xia W., Selkoe D. J., Chen X., Stokin G. B., Koo E. H. (1999) Mutagenesis identifies new signals for β-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Aβ42. J. Biol. Chem. 274, 18851–18856 [DOI] [PubMed] [Google Scholar]
- 24. Borg J. P., Yang Y., De Taddéo-Borg M., Margolis B., Turner R. S. (1998) The X11α protein slows cellular amyloid precursor protein processing and reduces Aβ40 and Aβ42 secretion. J. Biol. Chem. 273, 14761–14766 [DOI] [PubMed] [Google Scholar]
- 25. Shrivastava-Ranjan P., Faundez V., Fang G., Rees H., Lah J. J., Levey A. I., Kahn R. A. (2008) Mint3/X11γ is an ADP-ribosylation factor-dependent adaptor that regulates the traffic of the Alzheimer's precursor protein from the trans-Golgi network. Mol. Biol. Cell 19, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sabo S. L., Lanier L. M., Ikin A. F., Khorkova O., Sahasrabudhe S., Greengard P., Buxbaum J. D. (1999) Regulation of β-amyloid secretion by FE65, an amyloid protein precursor-binding protein. J. Biol. Chem. 274, 7952–7957 [DOI] [PubMed] [Google Scholar]
- 27. Saito Y., Akiyama M., Araki Y., Sumioka A., Shiono M., Taru H., Nakaya T., Yamamoto T., Suzuki T. (2011) Intracellular trafficking of the amyloid β-protein precursor (APP) regulated by novel function of X11-like. PLoS One 6, e22108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee J., Retamal C., Cuitiño L., Caruano-Yzermans A., Shin J. E., van Kerkhof P., Marzolo M. P., Bu G. (2008) Adaptor protein sorting nexin 17 regulates amyloid precursor protein trafficking and processing in the early endosomes. J. Biol. Chem. 283, 11501–11508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burgos P. V., Mardones G. A., Rojas A. L., daSilva L. L., Prabhu Y., Hurley J. H., Bonifacino J. S. (2010) Sorting of the Alzheimer's disease amyloid precursor protein mediated by the AP-4 complex. Dev. Cell 18, 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choy R. W., Cheng Z., Schekman R. (2012) Amyloid precursor protein (APP) traffics from the cell surface via endosomes for amyloid β (Aβ) production in the trans-Golgi network. Proc. Natl. Acad. Sci. U.S.A. 109, E2077–E2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Siman R., Velji J. (2003) Localization of presenilin-nicastrin complexes and γ-secretase activity to the trans-Golgi network. J. Neurochem. 84, 1143–1153 [DOI] [PubMed] [Google Scholar]
- 32. Xu H., Sweeney D., Wang R., Thinakaran G., Lo A. C., Sisodia S. S., Greengard P., Gandy S. (1997) Generation of Alzheimer β-amyloid protein in the trans-Golgi network in the apparent absence of vesicle formation. Proc. Natl. Acad. Sci. U.S.A. 94, 3748–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Caster A. H., Sztul E., Kahn R. A. (2013) A role for cargo in the activation of ADP-ribosylation factors (Arf) and adaptor recruitment. J. Biol. Chem. 288, 14788–14804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Valnes K., Brandtzaeg P. (1985) Retardation of immunofluorescence fading during microscopy. J. Histochem. Cytochem. 33, 755–761 [DOI] [PubMed] [Google Scholar]
- 35. Caster A. H., Kahn R. A. (2012) Computational method for calculating fluorescence intensities within three-dimensional structures in cells. Cell. Logist. 2, 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Andersen O. M., Reiche J., Schmidt V., Gotthardt M., Spoelgen R., Behlke J., von Arnim C. A., Breiderhoff T., Jansen P., Wu X., Bales K. R., Cappai R., Masters C. L., Gliemann J., Mufson E. J., Hyman B. T., Paul S. M., Nykjaer A., Willnow T. E. (2005) Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. U.S.A. 102, 13461–13466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seaman M. N. (2004) Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 165, 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hirst J., Seaman M. N., Buschow S. I., Robinson M. S. (2007) The role of cargo proteins in GGA recruitment. Traffic 8, 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aridor M., Traub L. M. (2002) Cargo selection in vesicular transport: the making and breaking of a coat. Traffic 3, 537–546 [DOI] [PubMed] [Google Scholar]
- 40. Cockcroft S., Thomas G. M., Fensome A., Geny B., Cunningham E., Gout I., Hiles I., Totty N. F., Truong O., Hsuan J. J. (1994) Phospholipase D: a downstream effector of ARF in granulocytes. Science 263, 523–526 [DOI] [PubMed] [Google Scholar]
- 41. Godi A., Pertile P., Meyers R., Marra P., Di Tullio G., Iurisci C., Luini A., Corda D., De Matteis M. A. (1999) ARF mediates recruitment of PtdIns-4-OH kinase-β and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1, 280–287 [DOI] [PubMed] [Google Scholar]
- 42. Jones D. H., Morris J. B., Morgan C. P., Kondo H., Irvine R. F., Cockcroft S. (2000) Type I phosphatidylinositol 4-phosphate 5-kinase directly interacts with ADP-ribosylation factor 1 and is responsible for phosphatidylinositol 4,5-bisphosphate synthesis in the Golgi compartment. J. Biol. Chem. 275, 13962–13966 [DOI] [PubMed] [Google Scholar]
- 43. Hill K., Li Y., Bennett M., McKay M., Zhu X., Shern J., Torre E., Lah J. J., Levey A. I., Kahn R. A. (2003) Munc18 interacting proteins: ADP-ribosylation factor-dependent coat proteins that regulate the traffic of β-Alzheimer's precursor protein. J. Biol. Chem. 278, 36032–36040 [DOI] [PubMed] [Google Scholar]
- 44. Duclos F., Boschert U., Sirugo G., Mandel J. L., Hen R., Koenig M. (1993) Gene in the region of the Friedreich ataxia locus encodes a putative transmembrane protein expressed in the nervous system. Proc. Natl. Acad. Sci. U.S.A. 90, 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Okamoto M., Nakajima Y., Matsuyama T., Sugita M. (2001) Amyloid precursor protein associates independently and collaboratively with PTB and PDZ domains of Mint on vesicles and at cell membrane. Neuroscience 104, 653–665 [DOI] [PubMed] [Google Scholar]
- 46. Okamoto M., Südhof T. C. (1998) Mint3: a ubiquitous Mint isoform that does not bind to Munc18-1 or -2. Eur. J. Cell Biol. 77, 161–165 [DOI] [PubMed] [Google Scholar]
- 47. Szul T., Garcia-Mata R., Brandon E., Shestopal S., Alvarez C., Sztul E. (2005) Dissection of membrane dynamics of the ARF-guanine nucleotide exchange factor GBF1. Traffic 6, 374–385 [DOI] [PubMed] [Google Scholar]
- 48. Robinson M. S., Kreis T. E. (1992) Recruitment of coat proteins onto Golgi membranes in intact and permeabilized cells: effects of brefeldin A and G protein activators. Cell 69, 129–138 [DOI] [PubMed] [Google Scholar]
- 49. Donaldson J. G., Lippincott-Schwartz J., Bloom G. S., Kreis T. E., Klausner R. D. (1990) Dissociation of a 110-kD peripheral membrane protein from the Golgi apparatus is an early event in brefeldin A action. J. Cell Biol. 111, 2295–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lippincott-Schwartz J., Yuan L. C., Bonifacino J. S., Klausner R. D. (1989) Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56, 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Strous G. J., Berger E. G., van Kerkhof P., Bosshart H., Berger B., Geuze H. J. (1991) Brefeldin A induces a microtubule-dependent fusion of galactosyltransferase-containing vesicles with the rough endoplasmic reticulum. Biol. Cell 71, 25–31 [DOI] [PubMed] [Google Scholar]
- 52. Ramelot T. A., Gentile L. N., Nicholson L. K. (2000) Transient structure of the amyloid precursor protein cytoplasmic tail indicates preordering of structure for binding to cytosolic factors. Biochemistry 39, 2714–2725 [DOI] [PubMed] [Google Scholar]
- 53. Han J., Wang Y., Wang S., Chi C. (2008) Interaction of Mint3 with Furin regulates the localization of Furin in the trans-Golgi network. J. Cell Sci. 121, 2217–2223 [DOI] [PubMed] [Google Scholar]
- 54. Hirst J., Bright N. A., Rous B., Robinson M. S. (1999) Characterization of a fourth adaptor-related protein complex. Mol. Biol. Cell 10, 2787–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pols M. S., van Meel E., Oorschot V., ten Brink C., Fukuda M., Swetha M. G., Mayor S., Klumperman J. (2013) hVps41 and VAMP7 function in direct TGN to late endosome transport of lysosomal membrane proteins. Nat. Commun. 4, 1361. [DOI] [PubMed] [Google Scholar]