Abstract
Bone tissue regeneration is orchestrated by the surrounding supporting tissues and involves the build-up of osteogenic cells, which orchestrate remodeling/healing through the expression of numerous mediators and signaling molecules. Periodontal regeneration models have proven useful for studying the interaction and communication between alveolar bone and supporting soft tissue. We applied a quantitative proteomic approach to analyze and compare proteins with altered expression in gingival soft tissue and alveolar bone following tooth extraction. For target identification and validation, hard and soft tissue were extracted from mini-pigs at the indicated times after tooth extraction. From triplicate experiments, 56 proteins in soft tissue and 27 proteins in alveolar bone were found to be differentially expressed before and after tooth extraction. The expression of 21 of those proteins was altered in both soft tissue and bone. Comparison of the activated networks in soft tissue and alveolar bone highlighted their distinct responsibilities in bone and tissue healing. Moreover, we found that there is crosstalk between identified proteins in soft tissue and alveolar bone with respect to cellular assembly, organization, and communication. Among these proteins, we examined in detail the expression patterns and associated networks of ATP5B and fibronectin 1. ATP5B is involved in nucleic acid metabolism, small molecule biochemistry, and neurological disease, and fibronectin 1 is involved in cellular assembly, organization, and maintenance. Collectively, our findings indicate that bone regeneration is accompanied by a profound interaction among networks regulating cellular resources, and they provide novel insight into the molecular mechanisms involved in the healing of periodontal tissue after tooth extraction.
Healthy dental gingival tissue and the alveolar bone that surrounds the teeth are essential for the proper function of teeth, as well as for a good appearance and good general health. Socket healing after tooth extraction is a useful experimental model for investigating the communication between gingival tissue and alveolar bone after tooth extraction. Preservation of the alveolar socket after tooth extraction requires the formation of a biological connection between the living and osseous tissue, which has to be created during the healing process. The success of such dental remodeling is dependent on the establishment of a soft tissue barrier that is able to shelter the underlying osseous structures and the osseo-integration of the soft tissue surrounding the alveolar bone. Understanding the processes governing soft and hard tissue healing and maintenance around the alveolar socket is paramount for oral health.
Several studies have reported significant structural changes and bone reabsorption in fresh sockets following tooth extraction, with important dimensional changes in the surrounding alveolar bone (1–3). A reduction of alveolar bone may present problems after tooth extraction, especially in aged individuals in whom bone volume is important for both physiological and medical reasons. Although it has been shown that reduction defects in alveolar bone can be completely repaired using surgical techniques such as guided bone regeneration (4, 5), bone autograft, bone allograft, and xenograft (6, 7), these techniques are not broadly applicable (8). However, the introduction of biomimetic agents such as enamel matrix derivatives (9), platelet-rich plasma (10), platelet-derived growth factor (11, 12), and bone morphogenic proteins (BMPs)1 (9) promises potentially better outcomes with bone regeneration treatments, although their efficacy remains controversial.
The proteins present in bone are essential for all of the life processes ongoing in bone, and they are the most important final products of the homeostatic signaling pathways. Profiling those proteins is vital for a thorough understanding of bone biology. To date, proteome research on bone has been focused mainly on in vitro analysis of bone-forming cells (osteoblasts and osteoclasts) to determine which proteins are expressed under a given set of experimental conditions (13–16). Although important, such studies cannot identify the actual protein profile in oral alveolar bone. Recently, the extraction of proteins directly from skull bone for proteome analysis was reported (17, 18). The extracted proteins were first separated using two-dimensional gel electrophoresis, after which spots of interest were excised and the proteins were identified via mass spectrometry (MS). However, using two-dimensional gel electrophoresis to analyze extreme proteins (e.g. extremely basic or acidic, extremely small or large, extremely hydrophobic) is challenging. Shotgun proteomics, which is a method of high-throughput proteome analysis (19–21), avoids the intrinsic limitations of two-dimensional gel electrophoresis. Despite an interesting need for large-scale characterization of the bone proteome, one study has been reported to apply shotgun proteomics for proteome analysis of rat femur bone (22). However, they identified only 133 proteins, because they analyzed bone proteins using a one-step method without a demineralization stage. The other report showed only that bone proteins extracted from the skull bone of an adult beagle are carried using a demineralization step (23). There are no reports regarding the interaction between alveolar bone and soft tissue yet.
The efficient extraction of bone proteins is a critical issue for proteome analysis (24). Because bone is largely mineralized, and therefore nearly solid, classical protein extraction methods used for soft tissues and cells may not be appropriate for bone. It is therefore necessary to develop methods to efficiently extract protein from bone. In earlier bone proteome analyses (17, 18, 22), the bones were first ground to powder, after which the proteins were extracted by means of incubating the powder in lysis buffer. However, mechanically breaking bones down into powder is laborious, especially for large animal bones. More important, large amounts of collagen and proteoglycans also are extracted, and this can impair the detection of low-abundance proteins and strongly affect isoelectric focusing (25). For the present study, we adopted an alternative method of demineralizing bone tissue and then investigated the efficiency of protein extraction from the demineralized bone tissue. This method was based on a recently reported sequential protein extraction protocol that was used to extract proteins from skull for comprehensive analysis of its proteome. Two-dimensional high-performance liquid chromatography–tandem mass spectrometry (LC-MS/MS) was then applied to analyze the protein extracts, enabling the identification of 2479 proteins (23). We employed a similar method to extract and identify proteins in tooth alveolar bone.
Given that a large number of proteins are likely involved in the healing of bone, as well as of soft tissues, another goal of the present study was to examine protein expression and putative signaling during bone healing after tooth extraction. Here, we used nano-UPLC-MSE-based label-free quantitative proteomics to analyze alveolar bone and the adjacent soft tissue. The environment surrounding healing bone would be expected to affect the specific signaling networks involved in bone regeneration. We suggest that determining the protein networks in alveolar bone and gingival tissue will enable improvement of the soft tissue interface, aspects of the hard tissue, and dental appearance during and after therapy.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
All chemicals were purchased from Sigma-Aldrich, Merck, or USB (Affymetrix, Inc., Santa Clara, CA).
Animal Experimental Groups
Mini-pigs (miniature pig; PWG Genetics Korea, Ltd) were maintained under specific-pathogen-free conditions. All animal-related procedures were reviewed and approved under the Animal Care Regulations of Chonnam National University (accession number: CNU IACUC YB-2011-3). For proteomic analysis, nine pigs were subdivided into three groups (n = 3 in each group) and were sacrificed without tooth extraction, 1 week after tooth extraction, or 2 weeks after tooth extraction, respectively. After a mini-pig was sacrificed, bone fragments were harvested from the surgical area (the first to the fourth premolars on the right and left sides of the maxillary and mandibular regions). After tooth extraction, the fragments were trimmed to separate the gingival soft tissue from the alveolar bone. Both soft and hard tissues were stored at −70 °C until analyzed.
Preparation of Soft Tissue Proteins
For protein extraction, gingival soft tissue was homogenized in buffer containing 1% Triton X-100, 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 2.5 mm sodium pyrophosphate, 1 mm β-glycerolphosphate, 1 mm sodium orthovanadate, 25 mm sodium fluoride, 1 μg/ml leupeptin, and 1 mm PMSF. The proteins were extracted over a 4-h period at 4 °C, after which the extract was sonicated and cleared via centrifugation at 13,000 rpm for 30 min at 4 °C. The protein concentration in the cleared extract was measured using a BCA protein assay (Thermo Scientific, Rockford, IL), and samples containing 200 μg of protein were subjected to 10% SDS-PAGE. The separated proteins were then visualized by staining with Coomassie Blue R-250, and the gels were washed three times in distilled water prior to tryptic digestion. For efficient trypsinization, the separated proteins were collected from the gels by excising 10 slices of gel for each patient sample, as described previously (26). The gel slices were then chopped into 1-mm3 pieces and destained overnight via incubation in 50% acetonitrile and 50 mm ammonium bicarbonate. Each gel piece was then reduced using 10 mm dithiothreitol and alkylated using 55 mm N-ethylmaleimide in 100 mm ammonium bicarbonate. Following tryptic digestion (2 μg/sliced gel; Promega, Madison, WI) for 16 h at 37 °C, the peptides were recovered and extracted from the sliced gels using 5% formic acid and 50% acetonitrile. After extraction, the peptides were dried in a vacuum centrifuge and combined into a single tube. The combined samples were desalted using a solid-phase Oasis HLB C18 microelution plate (Waters, Inc., Milford, MA) and then stored at −80 °C until subjected to nano-LC-MS/MS for comparative proteomics.
Preparation of Alveolar Bone Proteins
Protein was extracted from alveolar bone using the method described by Jiang et al., with some modifications (Fig. 1A) (23). Alveolar bone fragments (about 10 mm long × 10 mm wide × 10−20 mm thick) were trimmed of soft tissue in liquid nitrogen and then incubated overnight in PBS (pH 7.4) containing a protease inhibitor mixture at 4 °C to remove contaminants. Thereafter, about 150 mg of bone fragments were incubated in 1.2 m HCl overnight at 4 °C to demineralize the bone (200 mg of bone slice per milliliter of solution). After centrifugation, the supernatant was collected as Extract 1. The pellet was washed with water and incubated in buffer containing 100 mm Tris, 6 m guanidine-HCl (pH 7.4), and a protease inhibitor mixture for 72 h at 4 °C. After centrifugation, this supernatant was collected as Extract 2. The pellet was extracted further in the extraction solution (100 mm Tris, 6 m guanidine-HCl, pH 7.4) containing 0.5 m tetrasodium EDTA for 72 h at 4 °C, and the supernatant was collected as Extract 3 after centrifugation. Finally, the remaining pellet was incubated in 6 m HCl at 4 °C, and the supernatant was collected as Extract 4 after centrifugation. All centrifugations carried out during the extraction protocol were at 12,000 rpm for 20 min at 4 °C, and all crude protein extracts were precipitated with acetone overnight at −20 °C. The four samples of precipitated protein (Extracts 1–4) were then separately dissolved in 200 μl of buffer containing 100 mm Tris and 6 m guanidine-HCl (pH 8.1). Once in solution, the extracts were combined (total volume: 800 μl) and the protein concentration was determined using Bradford assays (Bio-Rad Laboratories), after which 200-μg aliquots of protein were subjected to 10% SDS-PAGE. Thereafter, tryptic digestion was carried out as described for soft tissue.
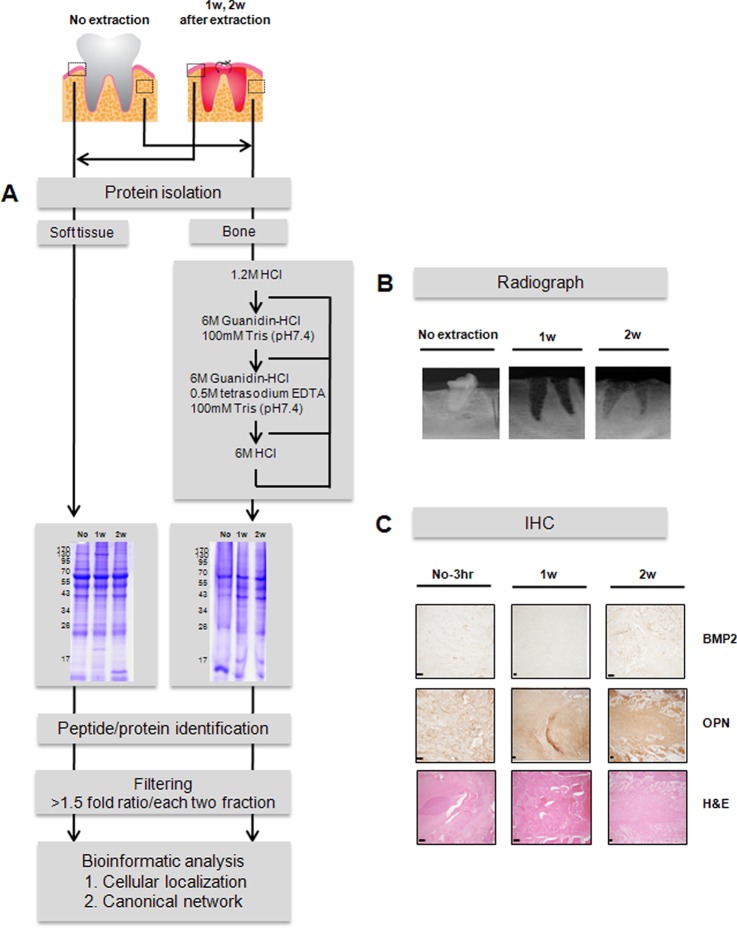
Fig. 1.
Quantitative proteomic analysis of soft tissue and bone following tooth extraction. A, strategy for identifying proteins with altered expression in gingival soft tissue and alveolar bone following tooth extraction in a porcine model. After tooth extraction, the proteins isolated from soft tissue and bone were subjected to SDS-PAGE and tryptic digestion. The digested peptides were subjected to LC-MS/MS. The regions outlined by dotted lines in the upper left indicate the locations of the extracted samples from soft tissue and bone. “No” indicates no tooth extraction; 1w, 1 week after tooth extraction; 2w, 2 weeks after tooth extraction. B, intraoral x-ray images. C, immunohistochemical images of osteoblastic markers. N-3 h, adjacent non-tooth extraction region at 3 h post-extraction; BMP2, bone morphogenetic protein 2; OPN, osteopontin; H&E, hematoxylin and eosin stain. Magnification, 100×; scale bars, 100 μm.
Protein Identification and Quantitative Analysis Using Nano-UPLC-MSE Tandem Mass Spectrometry
Separations were performed on a nano-UPLC C18 RP column (75 μm × 250 mm; particle size, 1.7 μm) and an enrichment Symmetry C18 RP column (180 μm × 20 mm; particle size, 5 μm) using a nano-ACQUITY Ultra PerformanceTM chromatography system (Waters Corporation, Milford, MA). The LC gradient program and operation mode were as reported previously (27). Trypsin-digested peptides (5 μl) were loaded onto the enrichment column with mobile phase A (3% acetonitrile in water with 0.1% formic acid), and a step gradient was used at a flow rate of 300 nl/min. This included a 3%–40% mobile phase B (97% acetonitrile in water with 0.1% formic acid) over 95 min and a 40%–70% mobile phase B over 20 min, followed by a sharp increase to 80% B within 10 min. Sodium formate (1 μm) dissolving of 10 mm NaOH in isopropanol and 0.2% HCOOH (1:1, v/v) was used to externally calibrate the TOF analyzer in the range of m/z 50–2000 with series of singly or doubly charged ion clusters prior to LC-MS/MS experiments (28).
The mass accuracy of the raw data was corrected against the monoisotope ion of [Glu1]-fibrinopeptide (m/z 785.8426 Da [M + 2H]2+). The [Glu1]-fibrinopeptide (200 fmol/μl, 600 nl/min flow rate) was infused every 1 min into the mass spectrometer for lock mass correction during sample analysis. During data acquisition, the collision energy in the low-energy MS mode and in the elevated-energy mode (MSE) were set at 4 and 15–40 eV energy ramping mode, respectively. One cycle of MS and MSE was performed every 3.2 s. In each cycle, MS spectra were acquired for 1.5 s using a 0.1-s interscan delay (m/z 300–1990), and the ions exceeding 50 counts were selected for MSE fragmentation in the collision cell (m/z 50–2000).
LC-MSE data were processed and searched using ProteinLynx GlobalServer version 2.3.3 (Waters Corporation) to reconstruct MS/MS spectra by combining all masses with identical retention times. A database search against the Sus scrofa database, which was retrieved from the NCBI website and included 27,041 entries, was performed using the LC-MS/MS data. For the protein identification, the parent ion tolerance was set at 100 ppm, and the fragment ion tolerance was set at 0.2 Da. Carbamidomethylation (+57 Da) of cysteine and methionine oxidation (+16 Da) were chosen as the fixed and variable modifications, respectively. Protein identification was also performed with at least two peptides and allowed only if the confidence was greater than 95% on the basis of the IDENTITYE algorithm (21). The false-positive rate for protein identification was set at 5% in the data bank search query option based on the automatically generated reversed database in ProteinLynx GlobalServer 2.3.3. ExpressionTM software generated results as an exact mass retention time table with quantitative protein and peptide values (supplemental Fig. S2). Analysis of quantitative changes in protein abundance, based on measurements of peptide ion peak intensities observed in the low-collision-energy mode (MS) in a triplicate set, was carried out using ExpressionTM software (version 2.3.3., Waters Corporation, Milford, MA). Peptides and their fragmented ions clustered chemically identical components together from each triplicate set per sample. Precursors were clustered to align identical components on the basis of a retention time deviation threshold (<0.25 min) and mass precision (<15 ppm) in ProteinLynx GlobalServer 2.3.3. For normalization of the dataset, each sample was spiked with a tryptic digested standard protein mixture including yeast alcohol dehydrogenase (1 pmole), rabbit glycogen phosphorylase B (0.5 pmole), yeast enolase (4 pmole), and bovine serum albumin (8 pmole) (Mass PREP Digestion Standard Mixture, Part No. 186002866, Waters) as an internal standard. Yeast alcohol dehydrogenase was mainly used for normalization between datasets.
Bioinformatic Analysis
Ingenuity Pathway Analysis (IPA version 8.0, Ingenuity Systems Inc., Redmond City, CA) was used for knowledge-based network and canonical pathway analysis of the nano-LC-MSE comparative proteomics data with the built-in function. STRING 9.0 was queried using human protein symbols from supplemental Fig. S1. The selected parameters were set to medium confidence (0.400) and prediction options “Co-expression,” “Experiments,” and “Databases.”
Quantitative Real-time PCR
Total RNA was isolated from porcine gingival tissue (3 h, 1 week, or 2 weeks after tooth extraction) using QIAzol® RNA Lysis reagent (Qiagen Sciences, Valencia, CA), after which cDNAs were synthesized using a PrimeScript™ RT Reagent Kit for real-time PCR (Takara Biotechnology, Japan) according to the manufacturer's instructions. Quantitative PCR was performed using an ABI 7300 Prism SDS real-time PCR detection system (Applied Biosystems, Foster City, CA) with a SYBR® Premix Ex Tag kit (Takara Biotechnology) and a standard temperature protocol. The results obtained using a cycle threshold are expressed as relative quantities and were calculated using the 2-ΔΔCT method (expressed as the relative fold ratio). Hypoxanthine phosphoribosyltransferase 1 was used as a control gene for normalization, and three separate experiments were performed. Supplemental Table S1 lists the porcine primers (Sus Scrofa) used for the quantitative real-time PCR.
Western Blotting and Immunohistochemistry
Samples (30 μg) of protein extracted from porcine gingiva (soft tissues) were separated on 12% polyacrylamide gels. The separated proteins were transferred onto polyvinylidene fluoride membranes (Bio-Rad Laboratories), after which the membranes were incubated first with anti-ATPB (NBP1–54700, Novus Biologicals, Littleton, CO) or anti-β-actin (A5441, Sigma-Aldrich) as the primary antibody and then with HRP-conjugated secondary antibody (Cell Signaling Technology, Danvers, MA). Immunoreactive proteins were detected using an ECL system (iNtRON, South Korea).
Maxillae, including the teeth, were removed and immersed for 24 h in 10% neutralized buffered formalin solution containing 0.1 mol/l phosphate buffer (pH 7.4). After decalcification in 10% EDTA solution for 4 weeks at 4 °C, the specimens were embedded in paraffin, and sagittal sections of the teeth were cut at a thickness of 4 μm. The sections were then processed for H&E staining and immunohistochemistry. All unstained slides were stained with anti-BMP2 (1:50, ab82511, Abcam), anti-osteopontin (1:50, ab8448, Abcam, Cambridge, UK), anti-ATP5B (1:50, NBP1–54700, Novus Biologicals), and anti-fibronectin (1:200, ab23751, Abcam) antibodies using a Ventana Ultraview DAB detection kit in a Ventana BenchMark XT processor (Ventana, Tucson, AZ). Antigen retrieval was not performed for any antibodies. Applied antibodies were incubated for 60 min at 37 °C. As negative controls, serial sections of the same specimens were processed, substituting commercially available mouse non-immune immunoglobulin G serum (DAKO, Carpinteria, CA) for the primary antibody.
RESULTS
Experimental Strategy for Comparative Proteomics in Gingival Tissues and Alveolar Bone After Tooth Extraction
We used a porcine model with standard radiography and immunohistochemical analysis of osteogenic markers to characterize the quantitative proteomic differences in soft tissue and bone before and after tooth extraction. We found that sockets filled with particles of regenerated bone within 1 week after tooth extraction (Fig. 1B). By 2 weeks post-extraction, BMP2 immunoreactivity was present in the soft tissue, mainly in fibroblasts and adipose tissue, and osteopontin was seen in the matrix (Fig. 1C). Notably, bone regeneration actively arose after tooth extraction in this pig model, confirming the model's suitability for use in identifying the proteins involved in active bone healing.
The strategy we used is illustrated in Fig. 1A. In three replicates, proteins extracted from gingival soft tissue and alveolar bone were digested with trypsin, after which the digested peptides were desalted using C18 and analyzed using a label-free quantitative proteomics approach. This entailed separation of the digested materials using nano-UPLC followed by analysis online using a Q-TOF tandem mass spectrometer. For efficient separation, we used a 25-cm reversed-phase column packed with small particles (1.7 μm) combined with ultrahigh pressure (∼7000 psi), which provided adequate separation in a single chromatography run. We also used MSE technology, which entailed continuous switching between low and high collision energies to gather precursor ion masses (low collision) and fragment ions (high collision) at 1-s intervals and yielded more than five times more information from a single LC run. Using this approach to measure the peak intensities of individual peptide masses obtained in separate chromatography runs without the incorporation of stable isotope tags confirmed the reproducibility of the results obtained using nano-UPLC.
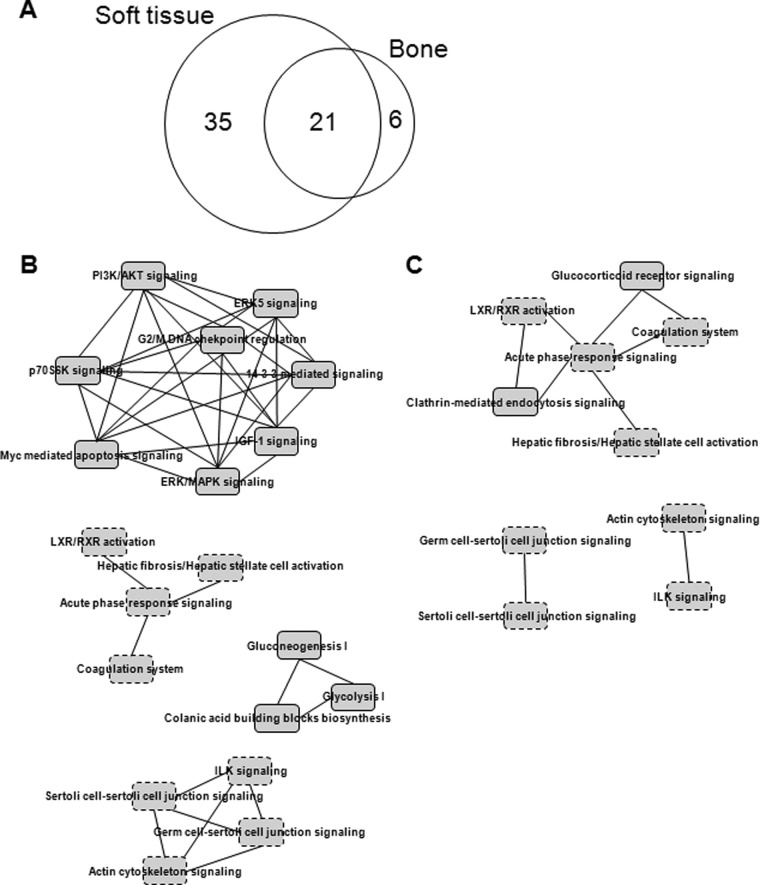
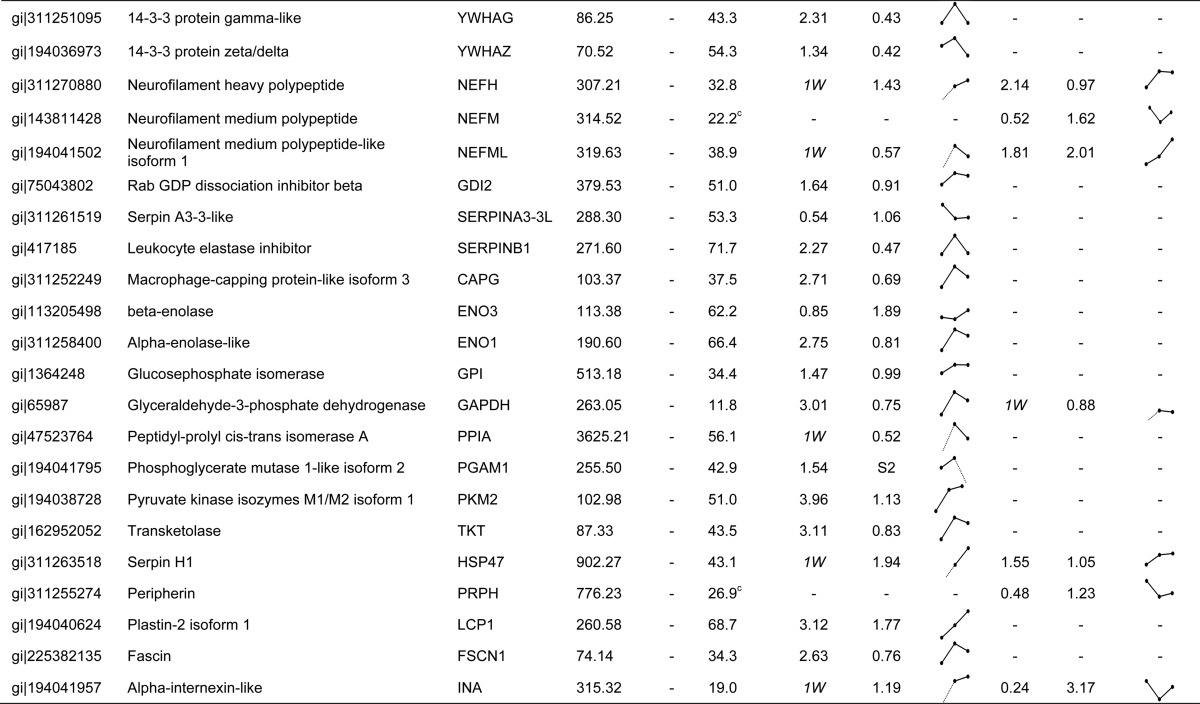
To further ensure the reliability of the quantitative profiling results, three samples (no extraction, 1 week after tooth extraction, and 2 weeks after tooth extraction) were prepared and analyzed in triplicate, which allowed the rates of expressional change above 1.1-fold change and below 0.9-fold change to be determined with high significance (p < 0.01) (supplemental Fig. S2). A total of 16,961 MS/MS spectra in soft tissue and 17,163 in bone were designated as peptides and were detected more than once, leading to the confident identification of a total of 946 proteins in soft tissue and 940 in bone with an average of at least two unique peptides per protein (supplemental Table S2). From among the total identified protein sets, 340 proteins for which we had precise quantitative values in more than one of the three samples were selected from soft tissue and bone (supplemental Table S3). Table I lists 62 proteins found to be differentially expressed with fold changes ranging between more than 1.5 (ratio ≥ 1.5) and less than 0.6 (ratio ≤ 0.6) through the comparison of sample pairs (supplemental Table S4). Of these, 56 proteins were differentially expressed in gingival soft tissue and 27 proteins were differentially expressed in alveolar bone, comparing among the three samples (Fig. 2A). These proteins belong to extracellular matrix (15), plasma membrane (3), mitochondria (4), endoplasmic reticulum (3), and cytoplasm and nucleus (14). In summary, 62 identified proteins may be targets affected by bone regeneration, and vice versa.
Table I. List of proteins differentially expressed in soft tissue and bone in a porcine model after tooth extraction.
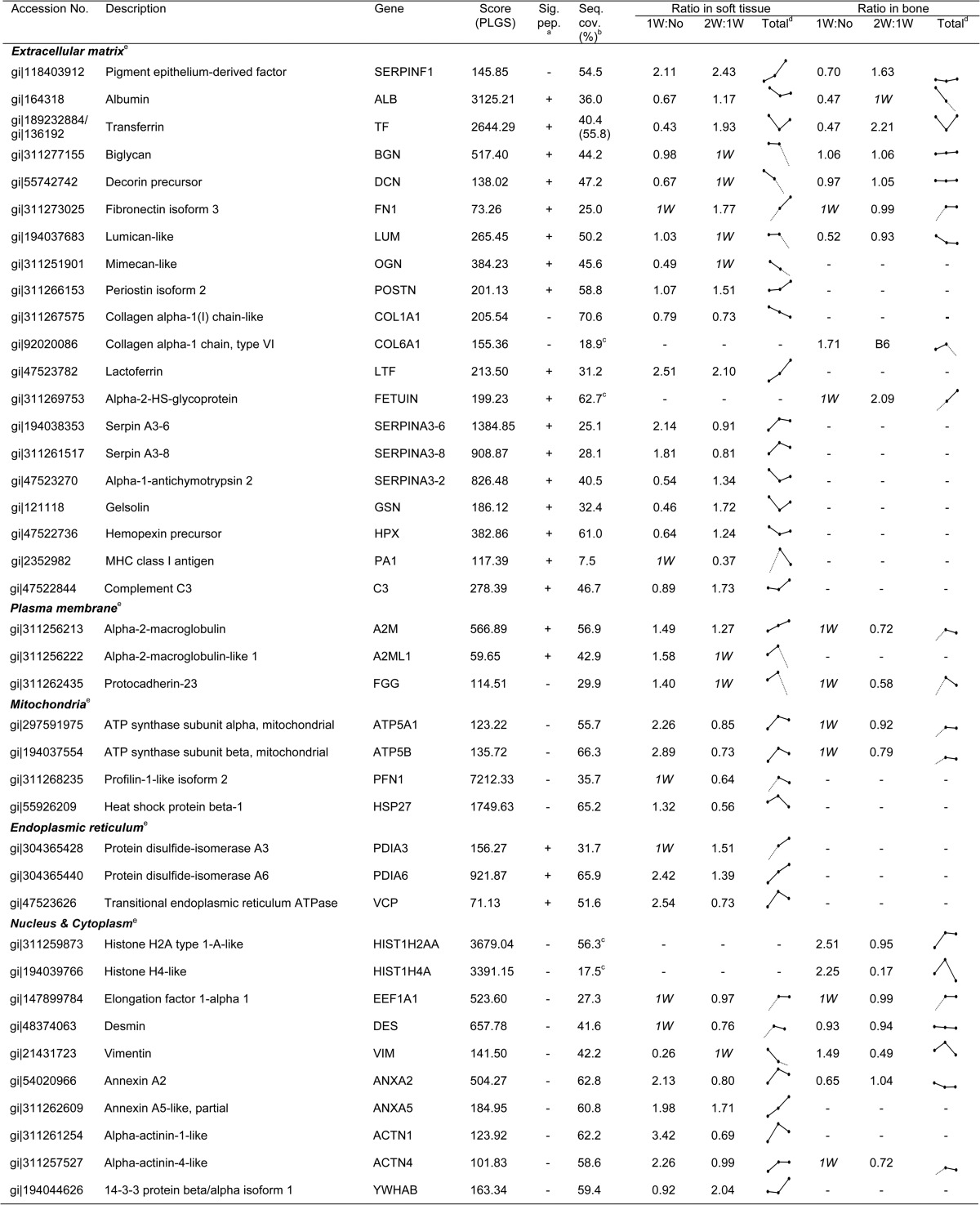
a Predicted signal peptide in the protein sequence determined by Phobius.
b Sequence coverage (%) calculated in soft tissue.
c Ratio comparisons made to obtain the quantitative values 1W:No and 2W:1W.
d Predicted cellular localization analyzed by Cello (v 2.5) program.
e Sequence coverage (%) calculated in bone.
Fig. 2.
Molecular network candidates in soft tissue and bone. A, Venn diagram illustrating the overlap between differentially expressed proteins (Table I) identified in soft tissue and bone after tooth extraction. B, canonical overlapping pathways of proteins identified in soft tissue. C, canonical overlapping pathways of proteins identified in bone. Each network comprised arrangements of at least two molecules. The rectangles outlined by solid lines indicate unique networks in soft tissue or bone; those outlined by dotted lines indicate common networks in soft tissue and bone. ERK5, mitogen-activated protein kinase 7 (MAPK7); p70S6K, p70 ribosomal S6 kinase; IGF-1, insulin-like growth factor 1; LXR/RXR, liver X receptor/retinoid X receptor.
Classification of Expression Differences between Soft Tissue and Bone
In subsequent bioinformatics analyses, we focused on the 62 proteins differentially expressed in soft tissue and bone. In order to provide an overview of the functional roles of the differentially expressed proteins, the 62 proteins were annotated based on Ingenuity functional analysis assignments (Figs. 2B and 2C). Their most prominent and relevant actions in soft tissue included MAPK/ERK-related signaling (ERK5, 14-3-3-mediated, PI3K/AKT, IGF-1, and p70S6K signaling), p53-related signaling (Myc-mediated apoptosis, 14-3-3-mediated signaling, and G2/M checkpoint regulation), and glucose metabolic processes (rectangles outlined by solid lines in Fig. 2B), whereas receptor-mediated signaling (glucocorticoid receptor and clathrin-mediated endocytosis signaling) was clearly overrepresented in bone (rectangles outlined by solid lines in Fig. 2C). Networks of the altered proteins in both soft tissue and bone after tooth extraction were related to cell–cell communication (junction signaling and integrin-linked kinase signaling), cellular maintenance (liver X receptor/retinoid X receptor activation and actin cytoskeleton signaling), and injury-inducible responses (acute phase response signaling, hepatic fibrosis activation, and coagulation system). Protein interactions between and within the affected correlations are presented as an interaction network, which also reveals that various proteins involved in gene regulation are differentially expressed in soft tissue and bone after tooth extraction. These evident effects on several fundamental and essential cellular processes demonstrate that there are profound differences between the functional networks active in soft tissue and in bone responding to bone healing. To obtain further insight into key proteomic differences, we arranged functional profiles of the proteins involved in these processes, as detailed below.
Distinct Protein Networks of Targets in Soft Tissue and Bone
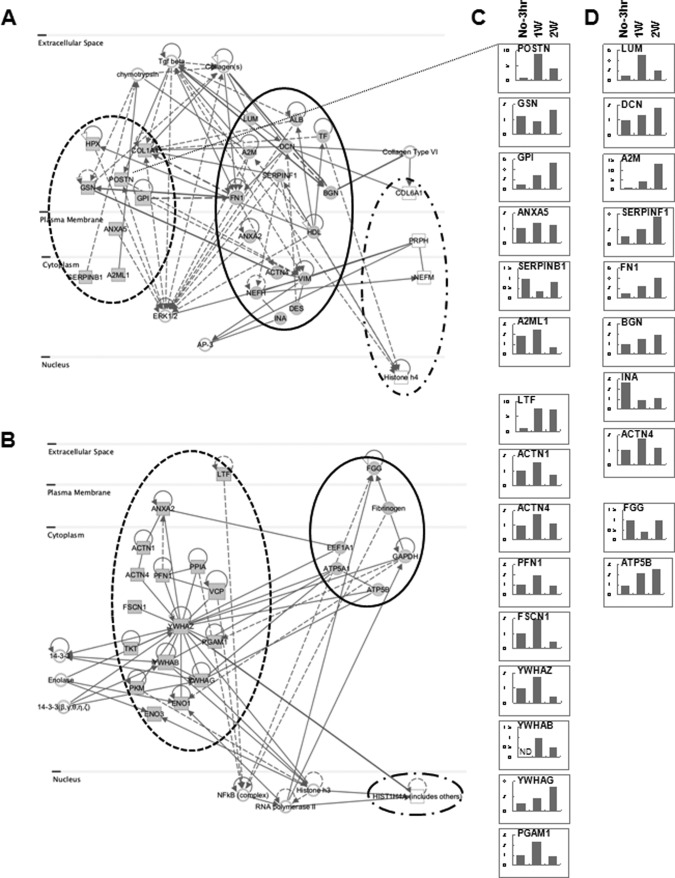
To further validate the correlation among proteins showing expressional alteration, we evaluated the connection between associated networks of targets in soft tissue and bone (supplemental Table S5). The majority of the proteins involved in these networks were found to have disease- and disorder-related functions in soft tissue (21 proteins) and cellular organization and maintenance-related functions in bone (17 proteins). Of the 21 quantified proteins related to a disease condition, 12 (ACTN1, ACTN4, ANXA2, ATP5A1, ATP5B, EEF1A1, ENO1, LTF, PFN1, PKM, TKT, and VCP) showed a fold increase 1 week post-extraction, and 7 (FGG, FSCN1, GAPDH, PGAM1, PPIA, YWHAB, and YWHAG) showed significantly higher levels 1 week post-extraction, relative to no extraction. At the transcriptional level, several candidates (LTF, ACTN1, ACTN4, PFN1, FSCN1, YWHAZ, YWHAB, YWHAG, and PGAM1) were significantly increased in soft tissue 1 week post-extraction (Fig. 3C). The expression levels of both ATP5B (ATP synthase subunit beta, mitochondrial) mRNA and protein showed the same pattern of increase in soft tissue after tooth extraction (Table I and Fig. 3D). In addition, YWHAB and ENO3 were dramatically increased in soft tissue 2 weeks post-extraction (Table I). This network (Fig. 3B) was centered on 14-3-3 protein zeta/delta (YWHAZ), which is known to be a key mediator in several signal transduction pathways. For example, YWHAZ has been implicated in the initiation and progression of cancer and is overexpressed in various cancer types, including oral carcinoma (29, 30). Overexpression of YWHAZ is associated with anchorage-independent growth and survival advantage under stress conditions (31). The cellular functions of these networks were ultimately associated with neurological disease, skeletal and muscular disorders, and dermatological diseases and conditions (supplemental Table S5). In addition, all six genes (ATP5A1, ATP5B, EEF1A1, FGG, FN1, and GAPDH) in bone tissue that have connections to targets in soft tissue were increased after tooth extraction (Fig. 3B). Interestingly, at the protein level these targets were not detected in unstimulated bone (no tooth extraction) (Table I). Moreover, HIST1H4A, which was detected only in bone, was dramatically increased 1 week post-extraction (ratios: 1W:No = 2.25; 2W:1W = 0.17 in Table I). All proteins involved in these networks were significantly increased in both soft tissue and bone after tooth extraction. We therefore suggest that these proteins might be required for bone regeneration and healing.
Fig. 3.
Complementary protein interaction network of expression-altered proteins. An interaction network was built based on the 62 significantly differentially expressed proteins. A, reciprocal network of proteins involved in cellular assembly and organization. B, reciprocal network of proteins relevant to disease. Gray rectangles represent targets differentially expressed only in soft tissue; white rectangles represent targets differentially expressed only in bone; gray circles represent targets differentially expressed in both soft tissue and bone. C, D, qPCR analysis of gene expression in soft tissue in A and B. C, targets detected only in soft tissue in A and B. D, targets detected in both soft tissue and bone in A and B. Y-axis is the relative fold ratio based on data from the No-3 h samples (= 1). No-3 h, adjacent non-tooth extraction region at 3 h post-extraction; 1w, 1 week after tooth extraction; 2w, 2 weeks after tooth extraction. ND, not detected.
Our data analysis also revealed a highly interconnected group of 27 proteins involved in the dynamics of cellular organization and maintenance that were differentially expressed in soft tissue and bone (Fig. 3A). This network was mainly centered on targets and stimuli within extracellular matrix (ECM), which is a composite of collagens and elastic fibers embedded in a viscoelastic gel of proteoglycans, hyaluronan, and assorted glycoproteins (32). These molecules interact through entanglement, cross-linking, and charge-dependent interactions to form bioactive polymers that, in part, regulate the biomechanical properties of tissues and their cellular phenotypes (33). The relative contributions of different ECM molecules can vary with tissue type and exhibit mechanical and chemical properties appropriate to each environment. The actions of several targets in this group that were affected by growth factors such as transforming growth factor β (TGF-β) are thought to activate the ERK pathway. In soft tissue, biglycan (BGN), decorin (DCN), lumican, and mimecan were reduced or absent 2 weeks post-extraction (Table I). In contrast, in bone tissue following tooth extraction, the expression of BGN, DCN, and mimecan was maintained at the normal level (Table I), though lumican levels were somewhat reduced, suggesting the importance of glycosylation for protein functionality. BGN, DCN, lumican, and mimecan are all members of a small leucine-rich proteoglycan family and are found in connective tissues such as bone, muscle, and blood vessels, as well as in the keratinocyte layer. They reportedly organize ECM assembly, bone formation and absorption, and osteogenesis (34, 35). Levels of α-2-macroglobulin protein were increased in soft tissue (ratios: 1W:No = 1.49; 2W:1W = 1.27; Table I) and bone (ratios: 1W:No, detected only in 1W; 2W:1W = 0.72; Table I) after tooth extraction, and increased levels of transcript were also detected in soft tissue (Fig. 3D). It was previously reported that α-2-macroglobulin exerts effects on collagen fiber organization at the healing tendon–bone interface (36, 37) and that α-2-macroglobulin mRNA, which is known to share vascular, osteogenic, and cartilage functions relevant for avascular necrosis of the femoral head, is overexpressed in rats with steroid-induced avascular necrosis of the femoral head (37). Interestingly, serpin peptidase inhibitor, clade F, member 1 (SERPINF1) was continuously increased in soft tissue after tooth extraction at both protein and mRNA levels (Table I and Fig. 3D). SERPINF1, also known as pigment epithelium-derived factor, was originally identified as a potent inhibitor of angiogenesis, the proliferation and migration of endothelial cells, retinal vascular permeability, and tumor activity (38). SERPINF1 expression changes with the progression of various tumor types (39), which suggests that SERPINF1 may direct cellular fate. It was also recently reported that a frameshift mutation in exon 4 of the SERPINF1 gene that reduced expression of the transcription/translation product and causes progressively deforming osteogenesis imperfecta is likely a key factor in bone deposition and remodeling (40).
The expression of fibronectin 1 (FN1) protein and mRNA was enhanced in both soft tissue and bone after tooth extraction. Cellular fibronectin has numerous functions involved in cell adhesion, growth, migration, and differentiation (41). In particular, fibronectin profoundly affects wound healing, including the formation of a proper substratum for cell migration and growth during the development and organization of granulation tissue, as well as synthesis and remodeling of the connective tissue matrix (42, 43). In addition, RUNX2 appears to up-regulate FN1 in osteoblasts in vitro (44) and to be required for osteoblast differentiation and mineralization (45, 46). These proteins have been shown to modulate multiple processes related to ECM function, including the interaction of cytoskeletal and ECM constituents. Our network analysis highlighted the modification and reconstruction of ECM and communication between gingival soft tissue and alveolar bone for bone healing.
Interaction of ATP5B and FN1 with Cellular Functionality
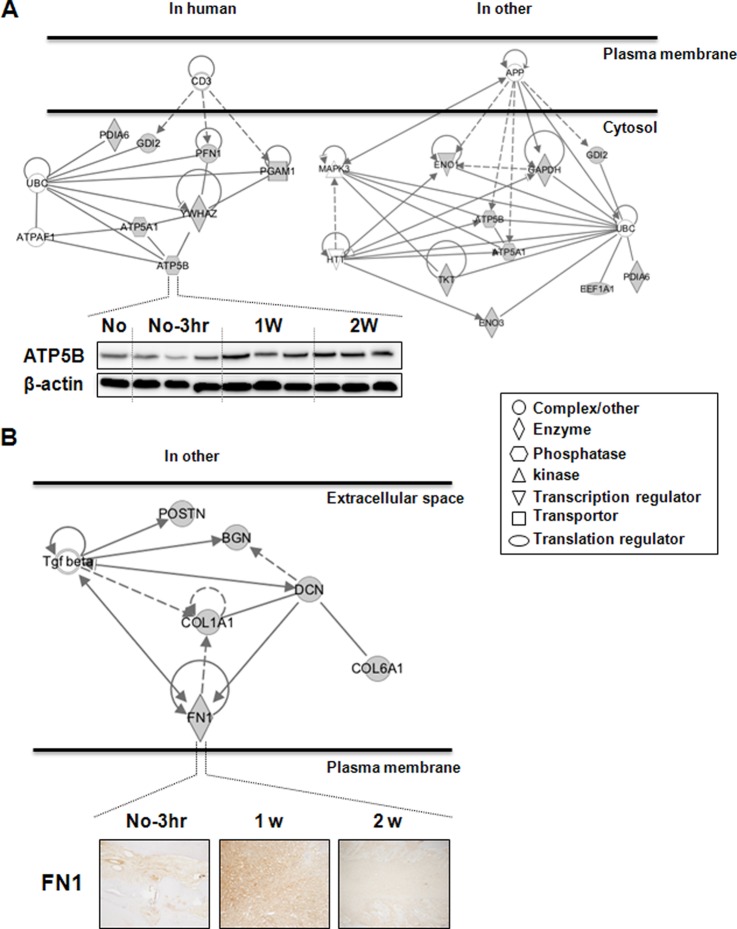
Among the 62 identified proteins, we examined the protein interactions involving ATP5B and FN1 in cellular functionality in greater detail. We observed the predicted association between various proteins and ATP5B or FN1, based on their observed patterns of simultaneous expression (supplemental Fig. S1 and supplemental Table S6). Consistent with the MS result (ratios: 1W:No = 2.89; 2W:1W = 0.73 in Table I), ATP5B expression was increased in soft tissue after tooth extraction (Fig. 4A). In humans, ATP5B is involved with molecular transport, nucleic acid metabolism, and small molecule biochemistry, along with ATP5A1, PFN1, PDIA6, YWHAZ, PGAM1, and GDI2 (left-hand side of Fig. 4A). This network is central to the interaction between ATP5B and YWHAZ. Previously, MS was used to show that the levels of these two proteins are lower in osteosarcoma than in benign bone tumors, including osteoblastoma, chondroblastoma, and giant cell tumor of bone (47). In addition, levels of ATP5B and YWHAZ mRNA are unstable during cutaneous wound healing (48). In other species, including M. musculus, S. cerevisiae, and C. elegans, ATP5B has been connected to nucleic acid metabolism, small molecule biochemistry, and neurological disease, along with GAPDH, ENO1, ENO3, PDIA6, ATP5A1, GDI2, TKT, and EEF1A1 (right-hand side of Fig. 4B). More specifically, the primary stimulus for this connection was focused on neuronal disorders caused by aberrant posttranslational modifications, including ubiquitination and proteolytic cleavage (49). These proteins are thought to play a collateral role in bone regeneration via a network linking protein stability and mitochondrial signaling.
Fig. 4.
Cellular network of proteins co-expressed with ATP5B or FN1. A, ATP5B-related subnetwork in humans and other species. Panels show Western blot data for ATP5B in soft tissue. β-actin was used as a loading control, and three repeated samples were loaded onto the same acrylamide gel. B, FN1-related subnetwork in other species. Panels show immunohistochemical data for FN1 in soft tissue. These connections were based on the co-expression patterns shown in supplemental Fig. S1. Gray figures indicate targets identified in this study; white figures indicate stimulatory complexes/molecules. “No” indicates no tooth extraction. No-3 h, adjacent non-tooth extraction region at 3 h post-extraction; 1w and 2w, 1 week and 2 weeks after tooth extraction, respectively; H&E, hematoxylin and eosin stain; CD3, T cell receptor (TCR); UBC, polyubiquitin C; ATPAF1, ATP synthase mitochondrial F1 complex assembly factor 1; APP, amyloid precursor protein; MAPK3, mitogen-activated protein kinase 3; HTT, huntingtin; Tgf beta, transforming growth factor beta.
IHC showed that FN1 levels were increased 1 week after tooth extraction and had declined by 2 weeks post-extraction (Fig. 4B). In vitro, FN1 up-regulates mineralized osteoblasts relative to nonmineralized osteoblasts (46). As expected, within the extracellular space, FN1 was coordinated with cellular assembly and organization and cellular maintenance, along with DCN, COL1A1, BGN, COL6A1, and POSTN. The possible link between these proteins is TGF-β, which is crucial for connective tissue regeneration and bone remodeling (50, 51). TGF-β affects osteoblast differentiation and bone formation (52, 53) and increases transcription of osteoblast differentiation markers and alkaline phosphatase activity in murine bone marrow stromal cells (54). FN1, which is a TGF-β target gene, might be a trigger factor induced during bone healing and regeneration, as it has interactions involving cellular adhesion and migration processes, including wound healing, blood coagulation, host defenses, and metastasis. Taken together, our data show that ATP5B and FN1 are differentially expressed in soft tissue and bone after tooth extraction, which suggests their participation in the transduction of bone regeneration signals.
DISCUSSION
Our aims in this study were to compare the patterns of protein expression in gingival soft tissue and alveolar bone following tooth extraction and to identify specific targets for bone regeneration (hard tissue) within soft tissue. Quantitative proteomics data represent an important source that can be used to expand our understanding of the molecular basis of the communication between soft tissue and hard tissue during bone healing.
Bone is a dynamic and heterogeneous tissue and is continually being remodeled in physiological circumstances through the bone-forming activities of osteoblasts and the bone-resorption activities of osteoclasts. Bone mass and turnover are maintained through the coordinated activities of these two cell types under the regulation of numerous systemic and local factors (55, 56). Osteoblasts are bone-forming cells that synthesize an ECM and then participate in the mineralization of that matrix (46). Similar repair and regeneration after bone damage is the main goal of bone therapy; however, the potential of bone regeneration appears to be limited by our incomplete understanding of the cellular and molecular events ongoing within the supporting and surrounding stromal tissue. Regarding that issue, the periodontal regeneration model is a useful tool with which to verify the interaction and communication between target (bone) tissue and supporting stromal (soft) tissue. With the exception of teeth, periodontal tissue is surrounded by periodontal supporting tissue, including cementum, periodontal ligament, and alveolar bone. Several studies have reported that tooth extraction leads to alteration of the residual alveolar bone (57, 58). Regeneration of the periodontal structure involves complex processes regulated by the actions and interactions of various cells and stimulation factors (59). These factors affect cell proliferation, chemotaxis, and differentiation among epithelium, bone, and connective tissue. They act by binding to specific cell-surface receptors present on target cells such as osteoblasts, cementoblasts, and periodontal ligament fibroblasts. The resultant changes in cellular morphology and the induction of locomotion in response to extracellular stimuli are mediated mainly through the concerted reorganization of the target cells' cytoskeleton, which forms a dynamic structural framework around which cell shape, polarity, motility, and mitosis are defined. Given that bone formation is the desired outcome, the observation that several factors participate in all related cell communications suggests that the exogenous application of these proteins during bone regeneration might represent an alternative approach to bone therapy. The earliest altered process in repair and healing contained a sequence of controlled events including the formation of ECM, which is primarily composed of fibrin, fibronectin, and vitronectin (60–63). In this study, a variety of altered proteins in proteomics data (Table I) are connected with the compartment of the ECM. As a consequence, cell–cell communication and information are transferred from the matrix to the cell interior through binding to ECM proteins, giving rise to alterations in cell activity and features.
Our data strongly suggest that the gene networks activated by tooth extraction differ between soft tissue and bone. Whereas the interactions of the affected proteins in soft tissue were focused mainly on cellular function and the activation of signaling and metabolic pathways, interactions in bone were involved in accepter signaling—for example, glucocorticoid receptor signaling and signaling related to clathrin-mediated endocytosis (Fig. 2B). In particular, IGF-1 is secreted by mature osteoblasts and can be stored locally in the bone matrix until its release during bone resorption. IGF-1 acts via specific membrane receptors to stimulate osteoblast proliferation and differentiation, which contribute to the induction of bone matrix proteins (64) and promote longitudinal bone growth by augmenting chondrocyte hypertrophy (65). IGF-1 also interacts with AKT and the p38 MAPK pathway leading to TGF-β (52, 66). In addition, p70S6K signaling is involved in osteoblast activity through BMP-4-stimulated VEGF synthesis mediated by p38 MAPK (67). Recently, ERK5 activation was shown to promote proliferation (68) and cytoskeletal reorganization (69) in osteoblastic cells subjected to fluid shear stress. Proteomic analysis revealed that several members of the 14-3-3 family are involved in the mediation of matrix vesicle secretion by osteoblasts into the ECM to initiate mineralization (70). Soft tissue also showed up-regulated expression of genes related to metabolic pathways such as glycolysis, gluconeogenesis, and colanic acid building block biosynthesis. Although the exact connections between metabolic pathways and bone regeneration and remodeling remain unknown, bone, acting through osteoblasts, has recently emerged as an endocrine organ regulating glucose metabolism (71).
Within bone, the regulation of glucocorticoid receptor signaling and clathrin-mediated endocytosis is very important for maintaining bone mass. Glucocorticoid administration is widely used to treat arthritic and inflammatory disorders; however, prolonged use of glucocorticoids accelerates bone loss and skeletal deterioration and increases the risk of osteoporotic fracture and osteonecrosis (72, 73). Impairments of osteoblast survival and differentiation are prominent reactions that contribute to glucocorticoid-induced bone loss (74, 75). Similarly, the bone-resorption activity of osteoclasts is regulated at many levels, including via clathrin-mediated endocytosis (76), which showed significant colocalization with RANKL-RANK complexes, indicating that their internalization is clathrin dependent (77). Ultimately, these signaling networks must mesh with mechanisms underlying overall bone turnover, as bone is continuously being repaired and remodeled through the coordinated activities of mediators and signaling molecules in the surrounding environment.
We confirmed the expression pattern and partner networks of ATP5B and FN1 in soft tissue (Fig. 4). Proteomic analysis previously revealed that ATP5B expression is altered by disease-related stress (78–81). ATP synthase, the enzyme catalyzing the synthesis of ATP, projects from the inner mitochondrial membrane to the mitochondrial matrix, where ATP5B catalyzes the rate-limiting step in ATP synthesis. For a long time, F1F0 ATP synthase expression was believed to occur only in mitochondria, where most cellular ATP synthesis takes place. However, recent studies suggest that components of ATP synthase are also present on the outer surface of the plasma membrane, where they function as receptors for various ligands and are known to be involved in lipid formation, the regulation of endothelial cell proliferation and differentiation, and immune responses in tumor cells (82–85). It is also known that intracellular ATP levels regulate the inverse correlation between osteoclast survival and bone resorption through an unknown mechanism, and that extracellular ATP likely exerts inhibitory effects on osteoclast survival and the bone remodeling and resorption induced by mature osteoclasts through the alteration of cytoskeletal structures (86). The inverse correlation between osteoclast survival and bone resorption regulated by ATP5B provides new insight into the molecular mechanisms regulating bone homeostasis.
The dynamic assembly of the FN1 matrix is cell specific and a key event regulating cell adhesion, migration, and differentiation (87). Cell adhesion to ECM is essential for the development, maintenance, and remodeling of osseous tissues (88). Adhesive interactions with ECM components, including fibronectin and type I collagen, play critical roles in osteoblast survival, proliferation, and differentiation and in matrix mineralization and bone formation (89). For that reason, we will next focus on analyzing these ECM regulatory proteins in more detail using more refined bone-healing models and bone grafting. We anticipate that our findings will shed new light on the interaction between gingival soft tissue and alveolar bone and on the cellular signaling underlying bone remodeling.
Supplementary Material
Footnotes
* This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (No. 2012-0009425) and by a grant from the Korean Health Technology R&D Project, Ministry of Health & Welfare (A111455), Republic of Korea.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ATP5B
- ATP synthase subunit beta, mitochondrial
- BGN
- biglycan
- BMP
- bone morphogenetic protein
- DCN
- decorin
- ECM
- extracellular matrix
- FN1
- fibronectin 1
- MSE
- high/low-collision-energy mass spectrometry
- SERPINF1
- serpin peptidase inhibitor, clade F, member 1
- TGF-β
- transforming growth factor β
- UPLC
- ultraperformance liquid chromatography.
REFERENCES
- 1. Araujo M. G., Lindhe J. (2005) Dimensional ridge alterations following tooth extraction. An experimental study in the dog. J. Clin. Periodontol. 32, 212–218 [DOI] [PubMed] [Google Scholar]
- 2. Cardaropoli G., Araujo M., Lindhe J. (2003) Dynamics of bone tissue formation in tooth extraction sites. An experimental study in dogs. J. Clin. Periodontol. 30, 809–818 [DOI] [PubMed] [Google Scholar]
- 3. Zubillaga G., Von Hagen S., Simon B. I., Deasy M. J. (2003) Changes in alveolar bone height and width following post-extraction ridge augmentation using a fixed bioabsorbable membrane and demineralized freeze-dried bone osteoinductive graft. J. Periodontol. 74, 965–975 [DOI] [PubMed] [Google Scholar]
- 4. Sclar A. G. (2004) Strategies for management of single-tooth extraction sites in aesthetic implant therapy. J. Oral Maxillofac. Surg. 62, 90–105 [DOI] [PubMed] [Google Scholar]
- 5. Becker W., Dahlin C., Becker B. E., Lekholm U., van Steenberghe D., Higuchi K., Kultje C. (1994) The use of e-PTFE barrier membranes for bone promotion around titanium implants placed into extraction sockets: a prospective multicenter study. Int. J. Oral Maxillofac. Implants 9, 31–40 [PubMed] [Google Scholar]
- 6. Artzi Z., Tal H., Dayan D. (2000) Porous bovine bone mineral in healing of human extraction sockets. Part 1: histomorphometric evaluations at 9 months. J. Periodontol. 71, 1015–1023 [DOI] [PubMed] [Google Scholar]
- 7. Cammack G. V., 2nd, Nevins M., Clem D. S., 3rd, Hatch J. P., Mellonig J. T. (2005) Histologic evaluation of mineralized and demineralized freeze-dried bone allograft for ridge and sinus augmentations. Int. J. Periodontics Restorative Dent. 25, 231–237 [PubMed] [Google Scholar]
- 8. Klein C., de Groot K., Chen W., Li Y., Zhang X. (1994) Osseous substance formation induced in porous calcium phosphate ceramics in soft tissues. Biomaterials 15, 31–34 [DOI] [PubMed] [Google Scholar]
- 9. Hoffmann T., Richter S., Meyle J., Gonzales J. R., Heinz B., Arjomand M., Sculean A., Reich E., Jepsen K., Jepsen S., Boedeker R. H. (2006) A randomized clinical multicentre trial comparing enamel matrix derivative and membrane treatment of buccal class II furcation involvement in mandibular molars. Part III: patient factors and treatment outcome. J. Clin. Periodontol. 33, 575–583 [DOI] [PubMed] [Google Scholar]
- 10. Pradeep A. R., Pai S., Garg G., Devi P., Shetty S. K. (2009) A randomized clinical trial of autologous platelet-rich plasma in the treatment of mandibular degree II furcation defects. J. Clin. Periodontol. 36, 581–588 [DOI] [PubMed] [Google Scholar]
- 11. Nevins M., Camelo M., Nevins M. L., Schenk R. K., Lynch S. E. (2003) Periodontal regeneration in humans using recombinant human platelet-derived growth factor-BB (rhPDGF-BB) and allogenic bone. J. Periodontol. 74, 1282–1292 [DOI] [PubMed] [Google Scholar]
- 12. Camelo M., Nevins M. L., Schenk R. K., Lynch S. E., Nevins M. (2003) Periodontal regeneration in human Class II furcations using purified recombinant human platelet-derived growth factor-BB (rhPDGF-BB) with bone allograft. Int. J. Periodontics Restorative Dent. 23, 213–225 [PubMed] [Google Scholar]
- 13. Kubota K., Wakabayashi K., Matsuoka T. (2003) Proteome analysis of secreted proteins during osteoclast differentiation using two different methods: two-dimensional electrophoresis and isotope-coded affinity tags analysis with two-dimensional chromatography. Proteomics 3, 616–626 [DOI] [PubMed] [Google Scholar]
- 14. Wang D., Park J. S., Chu J. S., Krakowski A., Luo K., Chen D. J., Li S. (2004) Proteomic profiling of bone marrow mesenchymal stem cells upon transforming growth factor beta1 stimulation. J. Biol. Chem. 279, 43725–43734 [DOI] [PubMed] [Google Scholar]
- 15. Chang E. J., Kwak H. B., Kim H., Park J. C., Lee Z. H., Kim H. H. (2004) Elucidation of CPX-1 involvement in RANKL-induced osteoclastogenesis by a proteomics approach. FEBS Lett. 564, 166–170 [DOI] [PubMed] [Google Scholar]
- 16. Salasznyk R. M., Westcott A. M., Klees R. F., Ward D. F., Xiang Z., Vandenberg S., Bennett K., Plopper G. E. (2005) Comparing the protein expression profiles of human mesenchymal stem cells and human osteoblasts using gene ontologies. Stem Cells Dev. 14, 354–366 [DOI] [PubMed] [Google Scholar]
- 17. Fan Y., Liu J., Wang S., Wang H., Shi F., Xiong L., He W., Peng X. (2005) Functional proteome of bones in rats with osteoporosis following ovariectomy. Life Sci. 76, 2893–2901 [DOI] [PubMed] [Google Scholar]
- 18. Pastorelli R., Carpi D., Airoldi L., Chiabrando C., Bagnati R., Fanelli R., Moverare S., Ohlsson C. (2005) Proteome analysis for the identification of in vivo estrogen-regulated proteins in bone. Proteomics 5, 4936–4945 [DOI] [PubMed] [Google Scholar]
- 19. Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 20. Xie C., Ye M., Jiang X., Jin W., Zou H. (2006) Octadecylated silica monolith capillary column with integrated nanoelectrospray ionization emitter for highly efficient proteome analysis. Mol. Cell. Proteomics 5, 454–461 [DOI] [PubMed] [Google Scholar]
- 21. Peng J., Elias J. E., Thoreen C. C., Licklider L. J., Gygi S. P. (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43–50 [DOI] [PubMed] [Google Scholar]
- 22. Schreiweis M. A., Butler J. P., Kulkarni N. H., Knierman M. D., Higgs R. E., Halladay D. L., Onyia J. E., Hale J. E. (2007) A proteomic analysis of adult rat bone reveals the presence of cartilage/chondrocyte markers. J. Cell. Biochem. 101, 466–476 [DOI] [PubMed] [Google Scholar]
- 23. Jiang X., Ye M., Liu G., Feng S., Cui L., Zou H. (2007) Method development of efficient protein extraction in bone tissue for proteome analysis. J. Proteome Res. 6, 2287–2294 [DOI] [PubMed] [Google Scholar]
- 24. Saravanan R. S., Rose J. K. (2004) A critical evaluation of sample extraction techniques for enhanced proteomic analysis of recalcitrant plant tissues. Proteomics 4, 2522–2532 [DOI] [PubMed] [Google Scholar]
- 25. Hermansson M., Sawaji Y., Bolton M., Alexander S., Wallace A., Begum S., Wait R., Saklatvala J. (2004) Proteomic analysis of articular cartilage shows increased type II collagen synthesis in osteoarthritis and expression of inhibin betaA (activin A), a regulatory molecule for chondrocytes. J. Biol. Chem. 279, 43514–43521 [DOI] [PubMed] [Google Scholar]
- 26. Yang H. Y., Kwon J., Cho E. J., Choi H. I., Park C., Park H. R., Park S. H., Chung K. J., Ryoo Z. Y., Cho K. O., Lee T. H. (2010) Proteomic analysis of protein expression affected by peroxiredoxin V knock-down in hypoxic kidney. J. Proteome Res. 9, 4003–4015 [DOI] [PubMed] [Google Scholar]
- 27. Xu D., Suenaga N., Edelmann M. J., Fridman R., Muschel R. J., Kessler B. M. (2008) Novel MMP-9 substrates in cancer cells revealed by a label-free quantitative proteomics approach. Mol. Cell. Proteomics 7, 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moon Y. J., Kwon J., Yun S. H., Lim H. L., Kim M. S., Kang S. G., Lee J. H., Choi J. S., Kim S. I., Chung Y. H. (2012) Proteome analyses of hydrogen-producing hyperthermophilic archaeon Thermococcus onnurineus NA1 in different one-carbon substrate culture conditions. Mol. Cell. Proteomics 11, M111.015420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arora S., Matta A., Shukla N. K., Deo S. V., Ralhan R. (2005) Identification of differentially expressed genes in oral squamous cell carcinoma. Mol. Carcinog. 42, 97–108 [DOI] [PubMed] [Google Scholar]
- 30. Zhang J., Chen F., Li W., Xiong Q., Yang M., Zheng P., Li C., Pei J., Ge F. (2012) 14-3-3zeta interacts with stat3 and regulates its constitutive activation in multiple myeloma cells. PLoS One 7, e29554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Neal C. L., Yu D. (2010) 14-3-3zeta as a prognostic marker and therapeutic target for cancer. Expert Opin. Ther. Targets 14, 1343–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hay E. D. (1991) Cell Biology of Extracellular Matrix, Springer, New York [Google Scholar]
- 33. Wight T. N., Potter-Perigo S. (2011) The extracellular matrix: an active or passive player in fibrosis? Am. J. Physiol. Gastrointest. Liver Physiol. 301, G950–G955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Igwe J. C., Gao Q., Kizivat T., Kao W. W., Kalajzic I. (2011) Keratocan is expressed by osteoblasts and can modulate osteogenic differentiation. Connect. Tissue Res. 52, 401–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang X., Harimoto K., Xie S., Cheng H., Liu J., Wang Z. (2010) Matrix protein biglycan induces osteoblast differentiation through extracellular signal-regulated kinase and Smad pathways. Biol. Pharm. Bull. 33, 1891–1897 [DOI] [PubMed] [Google Scholar]
- 36. Bedi A., Kovacevic D., Hettrich C., Gulotta L. V., Ehteshami J. R., Warren R. F., Rodeo S. A. (2010) The effect of matrix metalloproteinase inhibition on tendon-to-bone healing in a rotator cuff repair model. J. Shoulder Elbow Surg. 19, 384–391 [DOI] [PubMed] [Google Scholar]
- 37. Kerachian M. A., Cournoyer D., Harvey E. J., Chow T. Y., Begin L. R., Nahal A., Seguin C. (2010) New insights into the pathogenesis of glucocorticoid-induced avascular necrosis: microarray analysis of gene expression in a rat model. Arthritis Res. Ther. 12, R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chandolu V., Dass C. R. (2012) Cell and molecular biology underpinning the effects of PEDF on cancers in general and osteosarcoma in particular. J. Biomed. Biotechnol. 2012, 740295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Broadhead M. L., Dass C. R., Choong P. F. (2009) In vitro and in vivo biological activity of PEDF against a range of tumors. Expert Opin. Ther. Targets 13, 1429–1438 [DOI] [PubMed] [Google Scholar]
- 40. Venturi G., Gandini A., Monti E., Dalle Carbonare L., Corradi M., Vincenzi M., Valenti M. T., Valli M., Pelilli E., Boner A., Mottes M., Antoniazzi F. (2012) Lack of expression of SERPINF1, the gene coding for pigment epithelium-derived factor, causes progressively deforming osteogenesis imperfecta with normal type I collagen. J. Bone Miner. Res. 27, 723–728 [DOI] [PubMed] [Google Scholar]
- 41. Pankov R., Yamada K. M. (2002) Fibronectin at a glance. J. Cell Sci. 115, 3861–3863 [DOI] [PubMed] [Google Scholar]
- 42. Grinnell F. (1984) Fibronectin and wound healing. J. Cell. Biochem. 26, 107–116 [DOI] [PubMed] [Google Scholar]
- 43. Valenick L. V., Hsia H. C., Schwarzbauer J. E. (2005) Fibronectin fragmentation promotes alpha4beta1 integrin-mediated contraction of a fibrin-fibronectin provisional matrix. Exp. Cell Res. 309, 48–55 [DOI] [PubMed] [Google Scholar]
- 44. Komori T. (2010) Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 339, 189–195 [DOI] [PubMed] [Google Scholar]
- 45. Tavella S., Bellese G., Castagnola P., Martin I., Piccini D., Doliana R., Colombatti A., Cancedda R., Tacchetti C. (1997) Regulated expression of fibronectin, laminin and related integrin receptors during the early chondrocyte differentiation. J. Cell Sci. 110 (Pt 18), 2261–2270 [DOI] [PubMed] [Google Scholar]
- 46. Alves R. D., Eijken M., Swagemakers S., Chiba H., Titulaer M. K., Burgers P. C., Luider T. M., van Leeuwen J. P. (2010) Proteomic analysis of human osteoblastic cells: relevant proteins and functional categories for differentiation. J. Proteome Res. 9, 4688–4700 [DOI] [PubMed] [Google Scholar]
- 47. Li Y., Liang Q., Wen Y. Q., Chen L. L., Wang L. T., Liu Y. L., Luo C. Q., Liang H. Z., Li M. T., Li Z. (2010) Comparative proteomics analysis of human osteosarcomas and benign tumor of bone. Cancer Genet. Cytogenet. 198, 97–106 [DOI] [PubMed] [Google Scholar]
- 48. Turabelidze A., Guo S., DiPietro L. A. (2010) Importance of housekeeping gene selection for accurate reverse transcription-quantitative polymerase chain reaction in a wound healing model. Wound Repair Regen. 18, 460–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ehrnhoefer D. E., Sutton L., Hayden M. R. (2011) Small changes, big impact: posttranslational modifications and function of huntingtin in Huntington disease. Neuroscientist 17, 475–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Patil A. S., Sable R. B., Kothari R. M. (2011) An update on transforming growth factor-beta (TGF-beta): sources, types, functions and clinical applicability for cartilage/bone healing. J. Cell. Physiol. 226, 3094–3103 [DOI] [PubMed] [Google Scholar]
- 51. Janssens K., ten Dijke P., Janssens S., Van Hul W. (2005) Transforming growth factor-beta1 to the bone. Endocr. Rev. 26, 743–774 [DOI] [PubMed] [Google Scholar]
- 52. Lee K. S., Hong S. H., Bae S. C. (2002) Both the Smad and p38 MAPK pathways play a crucial role in Runx2 expression following induction by transforming growth factor-beta and bone morphogenetic protein. Oncogene 21, 7156–7163 [DOI] [PubMed] [Google Scholar]
- 53. Ripamonti U., Ferretti C., Teare J., Blann L. (2009) Transforming growth factor-beta isoforms and the induction of bone formation: implications for reconstructive craniofacial surgery. J. Craniofac. Surg. 20, 1544–1555 [DOI] [PubMed] [Google Scholar]
- 54. Zhao L., Jiang S., Hantash B. M. (2010) Transforming growth factor beta1 induces osteogenic differentiation of murine bone marrow stromal cells. Tissue Eng. Part A 16, 725–733 [DOI] [PubMed] [Google Scholar]
- 55. Kawamura N., Kugimiya F., Oshima Y., Ohba S., Ikeda T., Saito T., Shinoda Y., Kawasaki Y., Ogata N., Hoshi K., Akiyama T., Chen W. S., Hay N., Tobe K., Kadowaki T., Azuma Y., Tanaka S., Nakamura K., Chung U. I., Kawaguchi H. (2007) Akt1 in osteoblasts and osteoclasts controls bone remodeling. PLoS One 2, e1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Harada S., Rodan G. A. (2003) Control of osteoblast function and regulation of bone mass. Nature 423, 349–355 [DOI] [PubMed] [Google Scholar]
- 57. Schropp L., Wenzel A., Kostopoulos L., Karring T. (2003) Bone healing and soft tissue contour changes following single-tooth extraction: a clinical and radiographic 12-month prospective study. Int. J. Periodontics Restorative Dent. 23, 313–323 [PubMed] [Google Scholar]
- 58. Cardaropoli G., Wennstrom J. L., Lekholm U. (2003) Peri-implant bone alterations in relation to inter-unit distances. A 3-year retrospective study. Clin. Oral Implants Res. 14, 430–436 [DOI] [PubMed] [Google Scholar]
- 59. Dereka X. E., Markopoulou C. E., Vrotsos I. A. (2006) Role of growth factors on periodontal repair. Growth Factors 24, 260–267 [DOI] [PubMed] [Google Scholar]
- 60. Juhasz I., Murphy G. F., Yan H. C., Herlyn M., Albelda S. M. (1993) Regulation of extracellular matrix proteins and integrin cell substratum adhesion receptors on epithelium during cutaneous human wound healing in vivo. Am. J. Pathol. 143, 1458–1469 [PMC free article] [PubMed] [Google Scholar]
- 61. Velvart P., Peters C. I., Peters O. A. (2005) Soft tissue management: suturing and wound closure. Endodontic Topics 11, 179–195 [Google Scholar]
- 62. Green R. J., Usui M. L., Hart C. E., Ammons W. F., Narayanan A. S. (1997) Immunolocalization of platelet-derived growth factor A and B chains and PDGF-alpha and beta receptors in human gingival wounds. J. Periodontal Res. 32, 209–214 [DOI] [PubMed] [Google Scholar]
- 63. Clark R. (1996) Wound repair: overview and general considerations, in The Molecular and Cellular Biology of Wound Repair (Clark R., ed.), pp. 3–50, Plenum Press, New York [Google Scholar]
- 64. Power R. A., Iwaniec U. T., Wronski T. J. (2002) Changes in gene expression associated with the bone anabolic effects of basic fibroblast growth factor in aged ovariectomized rats. Bone 31, 143–148 [DOI] [PubMed] [Google Scholar]
- 65. Wang J., Zhou J., Bondy C. A. (1999) Igf1 promotes longitudinal bone growth by insulin-like actions augmenting chondrocyte hypertrophy. FASEB J. 13, 1985–1990 [DOI] [PubMed] [Google Scholar]
- 66. Ochiai H., Okada S., Saito A., Hoshi K., Yamashita H., Takato T., Azuma T. (2012) Inhibition of insulin-like growth factor-1 (IGF-1) expression by prolonged transforming growth factor-beta1 (TGF-beta1) administration suppresses osteoblast differentiation. J. Biol. Chem. 287, 22654–22661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tokuda H., Hatakeyama D., Shibata T., Akamatsu S., Oiso Y., Kozawa O. (2003) p38 MAP kinase regulates BMP-4-stimulated VEGF synthesis via p70 S6 kinase in osteoblasts. Am. J. Physiol. Endocrinol. Metab. 284, E1202–E1209 [DOI] [PubMed] [Google Scholar]
- 68. Li P., Ma Y. C., Sheng X. Y., Dong H. T., Han H., Wang J., Xia Y. Y. (2012) Cyclic fluid shear stress promotes osteoblastic cells proliferation through ERK5 signaling pathway. Mol. Cell. Biochem. 364, 321–327 [DOI] [PubMed] [Google Scholar]
- 69. Li P., Ma Y. C., Shen H. L., Han H., Wang J., Cheng H. J., Wang C. F., Xia Y. Y. (2012) Cytoskeletal reorganization mediates fluid shear stress-induced ERK5 activation in osteoblastic cells. Cell. Biol. Int. 36, 229–236 [DOI] [PubMed] [Google Scholar]
- 70. Xiao Z., Camalier C. E., Nagashima K., Chan K. C., Lucas D. A., de la Cruz M. J., Gignac M., Lockett S., Issaq H. J., Veenstra T. D., Conrads T. P., Beck G. R., Jr. (2007) Analysis of the extracellular matrix vesicle proteome in mineralizing osteoblasts. J. Cell. Physiol. 210, 325–335 [DOI] [PubMed] [Google Scholar]
- 71. Fukumoto S., Martin T. J. (2009) Bone as an endocrine organ. Trends Endocrinol. Metab. 20, 230–236 [DOI] [PubMed] [Google Scholar]
- 72. de Nijs R. N., Jacobs J. W., Lems W. F., Laan R. F., Algra A., Huisman A. M., Buskens E., de Laet C. E., Oostveen A. C., Geusens P. P., Bruyn G. A., Dijkmans B. A., Bijlsma J. W. (2006) Alendronate or alfacalcidol in glucocorticoid-induced osteoporosis. N. Engl. J. Med. 355, 675–684 [DOI] [PubMed] [Google Scholar]
- 73. Zaidi M., Sun L., Robinson L. J., Tourkova I. L., Liu L., Wang Y., Zhu L. L., Liu X., Li J., Peng Y., Yang G., Shi X., Levine A., Iqbal J., Yaroslavskiy B. B., Isales C., Blair H. C. (2010) ACTH protects against glucocorticoid-induced osteonecrosis of bone. Proc. Natl. Acad. Sci. U.S.A. 107, 8782–8787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Weinstein R. S., Jilka R. L., Almeida M., Roberson P. K., Manolagas S. C. (2010) Intermittent parathyroid hormone administration counteracts the adverse effects of glucocorticoids on osteoblast and osteocyte viability, bone formation, and strength in mice. Endocrinology 151, 2641–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rauch A., Seitz S., Baschant U., Schilling A. F., Illing A., Stride B., Kirilov M., Mandic V., Takacz A., Schmidt-Ullrich R., Ostermay S., Schinke T., Spanbroek R., Zaiss M. M., Angel P. E., Lerner U. H., David J. P., Reichardt H. M., Amling M., Schutz G., Tuckermann J. P. (2010) Glucocorticoids suppress bone formation by attenuating osteoblast differentiation via the monomeric glucocorticoid receptor. Cell. Metab. 11, 517–531 [DOI] [PubMed] [Google Scholar]
- 76. Tat S. K., Padrines M., Theoleyre S., Couillaud-Battaglia S., Heymann D., Redini F., Fortun Y. (2006) OPG/membranous–RANKL complex is internalized via the clathrin pathway before a lysosomal and a proteasomal degradation. Bone 39, 706–715 [DOI] [PubMed] [Google Scholar]
- 77. Narducci P., Bortul R., Bareggi R., Nicolin V. (2010) Clathrin-dependent endocytosis of membrane-bound RANKL in differentiated osteoclasts. Eur. J. Histochem. 54, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Xu C., Zhang X., Yu C., Lu G., Chen S., Xu L., Ding W., Shi Q., Li Y. (2009) Proteomic analysis of hepatic ischemia/reperfusion injury and ischemic preconditioning in mice revealed the protective role of ATP5beta. Proteomics 9, 409–419 [DOI] [PubMed] [Google Scholar]
- 79. Han D., Moon S., Kim H., Choi S. E., Lee S. J., Park K. S., Jun H., Kang Y., Kim Y. (2011) Detection of differential proteomes associated with the development of type 2 diabetes in the Zucker rat model using the iTRAQ technique. J. Proteome Res. 10, 564–577 [DOI] [PubMed] [Google Scholar]
- 80. Severino V., Locker J., Ledda-Columbano G. M., Columbano A., Parente A., Chambery A. (2011) Proteomic characterization of early changes induced by triiodothyronine in rat liver. J. Proteome Res. 10, 3212–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Datta A., Jingru Q., Khor T. H., Teo M. T., Heese K., Sze S. K. (2011) Quantitative neuroproteomics of an in vivo rodent model of focal cerebral ischemia/reperfusion injury reveals a temporal regulation of novel pathophysiological molecular markers. J. Proteome Res. 10, 5199–5213 [DOI] [PubMed] [Google Scholar]
- 82. Arakaki N., Nagao T., Niki R., Toyofuku A., Tanaka H., Kuramoto Y., Emoto Y., Shibata H., Magota K., Higuti T. (2003) Possible role of cell surface H+ -ATP synthase in the extracellular ATP synthesis and proliferation of human umbilical vein endothelial cells. Mol. Cancer Res. 1, 931–939 [PubMed] [Google Scholar]
- 83. Martinez L. O., Jacquet S., Esteve J. P., Rolland C., Cabezon E., Champagne E., Pineau T., Georgeaud V., Walker J. E., Terce F., Collet X., Perret B., Barbaras R. (2003) Ectopic beta-chain of ATP synthase is an apolipoprotein A-I receptor in hepatic HDL endocytosis. Nature 421, 75–79 [DOI] [PubMed] [Google Scholar]
- 84. Scotet E., Martinez L. O., Grant E., Barbaras R., Jeno P., Guiraud M., Monsarrat B., Saulquin X., Maillet S., Esteve J. P., Lopez F., Perret B., Collet X., Bonneville M., Champagne E. (2005) Tumor recognition following Vgamma9Vdelta2 T cell receptor interactions with a surface F1-ATPase-related structure and apolipoprotein A-I. Immunity 22, 71–80 [DOI] [PubMed] [Google Scholar]
- 85. Burwick N. R., Wahl M. L., Fang J., Zhong Z., Moser T. L., Li B., Capaldi R. A., Kenan D. J., Pizzo S. V. (2005) An inhibitor of the F1 subunit of ATP synthase (IF1) modulates the activity of angiostatin on the endothelial cell surface. J. Biol. Chem. 280, 1740–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Miyazaki T., Iwasawa M., Nakashima T., Mori S., Shigemoto K., Nakamura H., Katagiri H., Takayanagi H., Tanaka S. (2012) Intracellular and extracellular ATP coordinately regulate the inverse correlation between osteoclast survival and bone resorption. J. Biol. Chem. 287, 37808–37823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lin W., Wang S. M., Huang T. F., Fu W. M. (2002) Differential regulation of fibronectin fibrillogenesis by protein kinases A and C. Connect. Tissue Res. 43, 22–31 [PubMed] [Google Scholar]
- 88. Damsky C. H. (1999) Extracellular matrix-integrin interactions in osteoblast function and tissue remodeling. Bone 25, 95–96 [DOI] [PubMed] [Google Scholar]
- 89. Garcia A. J., Reyes C. D. (2005) Bio-adhesive surfaces to promote osteoblast differentiation and bone formation. J. Dent. Res. 84, 407–413 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.