Abstract
The kingdom Stramenopile includes diatoms, brown algae, and oomycetes. Plant pathogenic oomycetes, including Phytophthora, Pythium and downy mildew species, cause devastating diseases on a wide range of host species and have a significant impact on agriculture. Here, we report comparative analyses on the genomes of thirteen straminipilous species, including eleven plant pathogenic oomycetes, to explore common features linked to their pathogenic lifestyle. We report the sequencing, assembly, and annotation of six Pythium genomes and comparison with other stramenopiles including photosynthetic diatoms, and other plant pathogenic oomycetes such as Phytophthora species, Hyaloperonospora arabidopsidis, and Pythium ultimum var. ultimum. Novel features of the oomycete genomes include an expansion of genes encoding secreted effectors and plant cell wall degrading enzymes in Phytophthora species and an over-representation of genes involved in proteolytic degradation and signal transduction in Pythium species. A complete lack of classical RxLR effectors was observed in the seven surveyed Pythium genomes along with an overall reduction of pathogenesis-related gene families in H. arabidopsidis. Comparative analyses revealed fewer genes encoding enzymes involved in carbohydrate metabolism in Pythium species and H. arabidopsidis as compared to Phytophthora species, suggesting variation in virulence mechanisms within plant pathogenic oomycete species. Shared features between the oomycetes and diatoms revealed common mechanisms of intracellular signaling and transportation. Our analyses demonstrate the value of comparative genome analyses for exploring the evolution of pathogenesis and survival mechanisms in the oomycetes. The comparative analyses of seven Pythium species with the closely related oomycetes, Phytophthora species and H. arabidopsidis, and distantly related diatoms provide insight into genes that underlie virulence.
Introduction
Oomycetes are a diverse group of organisms that morphologically resemble Fungi, yet are members of the Straminipila ( = stramenopile) and are more closely related to organisms in aquatic environments such as brown algae and diatoms. There is continued discussion on the higher level nomenclature of Straminipila within the kingdom Chromista [1], [2], [3] which when united with the alveolates, comprise the chromalveolate superkingdom [4], [5]. The algal stramenopiles are secondarily photosynthetic, however, non-photosynthetic stramenopiles, such as the oomycetes, share numerous genes of putative phototrophic origin [6], [7] lending support to the hypothesis that the straminipilous ancestor was photosynthetic [5]. Nevertheless, there has been continuous debate on the existence of chromalveolate hypothesis and the photosynthetic origin of the stramenopiles [8]. The oomycetes include a diverse range of free-living water molds and pathogens of plants, mammals, insects, fish, crustaceans, algae, and various microbes, including fungi [9], [10], [11], [12], [13]. Plant pathogenic oomycetes cause devastating diseases of crop, ornamental, and native species and are thought to not only be the most important group of pathogens of dicotyledonous plants [14] but also often the source of yield reduction in cereal crop species [15], [16], [17]. Some of the most damaging oomycete genera are Aphanomyces [18], Peronospora [19], Phytophthora [20], Plasmopara [21], Pseudoperonospora [22], and Pythium [23] species; the wide host range of these genera, coupled with the diversity of diseases they cause, pose a challenge to the development of durable disease control strategies in plants.
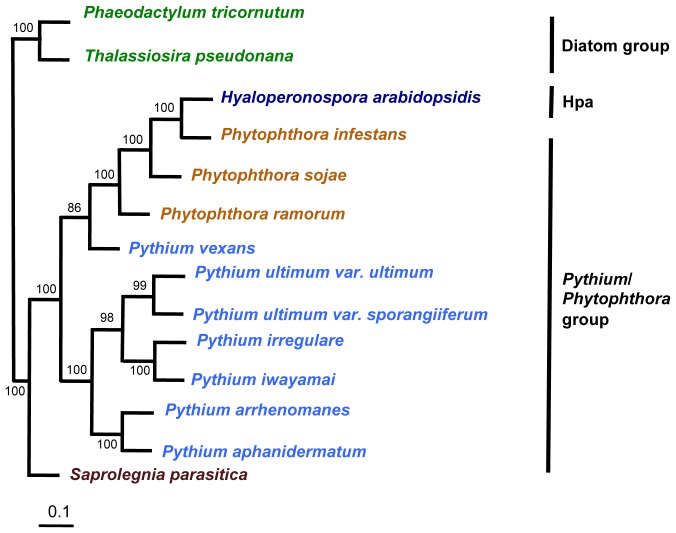
Within the oomycetes, Pythium species belongs to the peronosporalean lineage that includes hemibiotrophic Phytophthora species and the obligate biotrophic Hyaloperonospora species (Figure 1). The genus Pythium comprises more than 250 described species with 50% of these accepted by the community and currently classified into 11 phylogenetic clades [23]. Recently, one of these clades was shown to be closer to Phytophthora and the new genus Phytopythium has been described but official renaming of all Pythium species in clade K has not yet occurred [24], [25]. Most Pythium species are saprobes or facultative plant pathogens causing a wide variety of diseases, including seed rots and damping-off, root, stem and fruit rots, foliar blights, and postharvest decay [26], [27], [28], [29]. Some Pythium species have been reported to be parasitic to fungi and a few have been evaluated for biological control against other oomycete plant pathogens [30], [31], [32]. Some Pythium species are parasites of insects [12], fish [13], algae [33] and at least one species, Pythium insidiosum, infects mammals including humans [11]. Members of the genus Pythium differ from other oomycetes, including Phytophthora species, in morphology, genetic features [13], [14], and lifestyle. Pythium species are primarily necrotrophs and their sporangia produce a vesicle prior to the differentiation and release of zoospores whereas Phytophthora species are hemibiotrophs with zoospore differentiation occurring directly within the sporangia [34].
Figure 1. Phylogeny of oomycetes.
Phylogeny of the large rDNA subunit of select oomycetes as inferred by Bayesian analysis. The phylogenetic tree was constructed using rDNA sequences from 14 stramenopiles. Numbers on the branches are Bayesian posterior probability values calculated using MrBayes [110]. Hpa, Hyaloperonospora arabidopsidis.
Pythium species are genetically diverse [35] and exhibit significant variation in terms of virulence, host range, and distribution [10], [13]. Despite being members of the Pythium lineage that produces filamentous sporangia, Pythium aphanidermatum and Pythium arrhenomanes have distinct temperature optima and levels of virulence [36]. Similar to Pythium ultimum var. ultimum, Pythium aphanidermatum has a broad host range and is frequently found in greenhouses and high temperature conditions [10], [13], [37], [38]. This contrasts with Pythium arrhenomanes which is more restricted in host range with a preference for monocotyledonous plants [10]. Similar to Py. ultimum var. ultimum, Pythium irregulare is a species with globose sporangia, highly virulent at cooler temperatures [10], occurs in a broad eco-geographic range, and exhibits high genetic and morphological diversity [23], [39], [40], [41]. Unlike other species, Pythium vexans, which belongs to clade K and should be renamed as Phytopythium, causes root rot disease in many economically important tropical trees such as durian and rubber plants [42], [43]. Py. vexans also belongs to a species that has a wide range of genetic variation [44]. Py. ultimum var. sporangiiferum is in the P. ultimum species complex which has a wide genetic variation. In this study, we treat Py. ultimum var. sporangiiferum as a separate species because there is no evidence of gene flow between the two Py. ultimum varieties using a large collection of geographically overlapping strains from each group [45]. Pythium iwayamai is pathogenic to monocot grasses, grows at temperatures as low as 10°C, and causes snow rot disease in economically important crops such as turfgrass, barley, and winter wheat [13], [46], [47], [48]. The diversity in host range and optimal environmental conditions for infection makes the genus Pythium a good model to study plant-necrotroph interactions and to identify genes involved in interspecific variation in pathogenicity.
The development of second generation sequencing platforms [49], [50] offers an opportunity to sequence and perform comparative analyses of a large number of genomes, including phytopathogens [51]. A number of genome sequences of plant pathogenic oomycete are now available, including the downy mildew pathogen Hyaloperonospora arabidopsidis [52], three Phytophthora species (Ph. infestans [6], Ph. ramorum, Ph. sojae [7]), and Py. ultimum var. ultimum [53]. To date, comparative analyses of oomycete pathogens have shown variation in genome size, genome content, and evolution of host-pathogen interactions [7], [52], [53], [54], [55], [56]. For example, several gene families that facilitate the infection process are expanded [57] in Phytophthora species and significantly reduced in Py. ultimum var. ultimum and H. arabidopsidis [52], [53]. The availability of genome sequences of two species of diatoms, Phaeodactylum tricornutum [57], and Thalassiosira pseudonana [58], permits comparative analyses within the stramenopiles with respect to evolution of pathogenicity. Here, we describe the genome sequence assemblies and annotation for six additional Pythium species and identify genes involved in pathogenicity and necrotrophic lifestyle. Comparative analyses of seven Pythium species with closely related oomycetes, three Phytophthora species, H. arabidopsidis, and distantly related autotrophic diatoms provided insight into genes that underlie pathogenicity.
Results and Discussion
Genome Sequencing, Assembly, and Annotation
The sequences of six Pythium genomes (Py. aphanidermatum (DAOM BR444 = CBS 132490), Py. arrhenomanes (ATCC 12531 = CBS 324.62), Py. irregulare (DAOM BR486 = CBS 250.28), Py. iwayamai (DAOM 242034 = CBS 132417), Py. ultimum var. sporangiiferum (DAOM BR650 = CBS 219.65), and Py. vexans (DAOM BR484 = CBS 119.80); Table 1) that provide a broad representation of the genus Pythium (Figure 1) were generated using pyrosequencing with the Roche 454 or the Illumina Genome Analyzer (GA) II sequencing-by-synthesis platform. For the five Pythium species (Py. arrhenomanes, Py. irregulare, Py. iwayamai, Py. ultimum var. sporangiiferum, and Py. vexans), 6.6 to 14.4 Gb of purity-filtered (PF) reads were generated by the Illumina GAII/IIx sequencer (Table S1) while for Py. aphanidermatum, 507 Mb of single-end and 137.5 Mb of paired-end reads were generated using pyrosequencing (see Methods). Assembled genomes of six Pythium species yielded 3,685 to 11,542 contigs with an N50 contig length ranging from 9.8 to 37.4 Kb (Table 2, Table S2, Table S3). The total contig length/genome size ranged from 33.9 to 44.7 Mb in the six Pythium species, comparable to 42.8 Mb in Py. ultimum var. ultimum that was sequenced previously using Sanger-based methods [53]. The maximum contig length of the six Pythium species was comparable to Py. ultimum var. ultimum [53], ranging from 96.3 to 222.5 Kb (Table S3). In general, our study shows that Pythium species have smaller genomes than the three Phytophthora species or H. arabidopsidis.
Table 1. Species name, accession numbers, host/substrate and geographical origin of the Pythium strains sequenced in this study.
| Species | CBS accession¶ | Other accession identifiers | Host/substrate | Origin |
| Py. aphanidermatum | 132490 | DAOM BR444 | Cucumis sativus | BC, Canada |
| Py. arrhenomanes | 324.62 | ATCC 12531 | Zea mays | WI, USA |
| Py. irregulare | 250.28 | DAOM BR486 | Phaseolus vulgaris | Netherlands |
| Py. iwayamai | 132417 | DAOM 242034 | Poa annua | CO, USA |
| Py. ultimum var. sporangiiferum | 219.65 | DAOM BR650 | Chenopodium album | USA |
| Py. vexans | 119.80 | DAOM BR484 | Soil | Iran |
CBS: CBS-KNAW fungal biodiversity center, Utrecht, Netherlands (http://www.cbs.knaw.nl); DAOM: Culture collection of Agriculture and Agri-Food Canada, Ottawa, Canada; ATCC: American Type Culture Collection, Manassas, VA, USA.
Table 2. Assembly and annotation statistics† of thirteen stramenopiles.
| Species | Number ofcontigs | N50 contiglength (Kb) | Total contig length/genome size (Mb) ¶ | Number ofgenes | Number oftranscripts |
| This study | |||||
| Py. aphanidermatum | 5,667 | 37.39 | 35.9 | 12,305 | 12,312 |
| Py. arrhenomanes | 10,978 | 9.77 | 44.7 | 13,805 | 13,805 |
| Py. irregulare | 5,887 | 23.22 | 42.9 | 13,804 | 13,805 |
| Py. iwayamai | 11,542 | 11.01 | 43.3 | 14,875 | 14,875 |
| Py. ultimum var. sporangiiferum | 5,482 | 19.11 | 37.7 | 14,086 | 14,096 |
| Py. vexans | 3,685 | 29.24 | 33.9 | 11,957 | 11,958 |
| Published genomes | |||||
| H. arabidopsidis † | 3,138* | NA | 100 | 14,543 | NA |
| Ph. infestans † | 4,921 | 44.5 | 240 | 17,797 | NA |
| Ph. ramorum † | 7,588 | 47.5 | 65 | 15,743 | NA |
| Ph. sojae † | 5,577 | 105.7 | 95 | 19,027 | NA |
| P. tricornutum † | NA | NA | 27.4 | 10,402 | NA |
| Py. ultimum var. ultimum † | 1,747* | 124 | 42.8 | 15,291 | 15,323 |
| T. pseudonana † | NA | NA | 32.4 | 11,776 | NA |
Annotation of the six Pythium species revealed 11,957 to 14,875 predicted genes, comparable with the 15,291 genes annotated in Py. ultimum var ultimum [53]. Overall, the number of genes in any Pythium species is less than in Phytophthora species (15,743 to 19,027 genes) [6], [7] yet similar to H. arabidopsidis (14,543 genes) [52]. Average gene length was similar among all Pythium species, ranging from 1,495 to 1,767 bp (Table S3). Analysis of the intron/exon structure showed that the majority of Pythium species have 1.5 to 1.7 introns per gene, consistent with that of Py. ultimum var. ultimum (1.6 introns per gene) and Ph. infestans (1.7 introns per gene). Average exon length in the six newly sequenced Pythium genomes ranged from 312 to 630 bp, consistent with Py. ultimum var. ultimum (498 bp). Similar to Py. ultimum var. ultimum [53], 58 to 65% of all predicted genes from six Pythium species contain an InterPro protein domain, comparable to that observed with Phytophthora species (55 to 66%) [6], [7].
To aid in our genome annotation, we performed whole transcriptome sequencing (RNA-sequencing (RNA-seq)) of Py. arrhenomanes, Py. irregulare, Py. iwayamai, and Py. vexans. For these four species, a single pooled cDNA library was constructed for each species using RNA isolated from mycelium grown under five different growth conditions (nutrient-rich medium, nutrient-starved condition, fungicide treatment, as well as heat and cold temperature stress) and sequenced using the Illumina GAII. The total number of purity filtered reads ranged from 21.7 to 31.7 million reads per library with 82–88% of reads mapping to the cognate genome (Table S4) indicating a similar performance of library construction and sequencing across the samples. The minimum fragments per kilobase of exon model per million mapped fragments (FPKM) value for all growth conditions was 0 while the maximum FPKM ranged from 10,890 for sylvaticin, an elicitin-like protein in Py. arrhenomanes, to 30,906 for the INF1 elicitin in Py. vexans. The percentage of genes with transcript support ranged from 71% in Py. iwayamai to 81% in Py. vexans (Table S4). A gene was considered expressed if the FPKM value and FPKM 95% confidence interval lower boundary was greater than 0.001 and zero, respectively.
Core and Species-specific Genes and Gene Families in Pythium
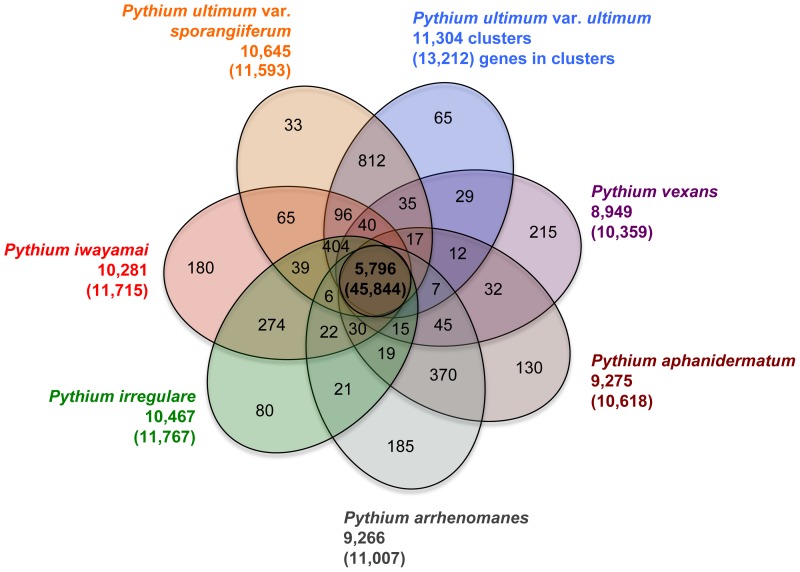
To identify the core Pythium proteome, we clustered orthologs and close paralogs in seven predicted Pythium proteomes (the newly sequenced six Pythium species and the previously sequenced Py. ultimum var. ultimum [53]) using OrthoMCL [59]. Of the 95,668 protein-coding genes, 80,271 genes clustered into 13,803 gene families with 15,397 genes as singletons. A total of 45,844 genes, clustered into 5,796 gene families, were common to all Pythium species, hereafter referred to as the core Pythium proteome (Figure 2). A total of 888 gene families containing 2,233 genes were unique to each species, ranging from 33 gene families in Py. ultimum var. sporangiiferum to 215 gene families in Py. vexans.
Figure 2. Gene families shared by Pythium species.
The predicted proteomes of the seven Pythium species were clustered using OrthoMCL [59] to identify orthologs and close paralogs. The number of gene families shared between the species and total number of clustered genes (numbers in parentheses) are indicated. The numbers outside the Venn diagram show the total number of orthologous clusters and number of genes (in parentheses) within those clusters for each species. Pap, Pythium aphanidermatum; Par, Pythium arrhenomanes; Pir, Pythium irregulare; Piw, Pythium iwayamai; Puls, Pythium ultimum var. sporangiiferum; Pult, Pythium ultimum var. ultimum; Pve, Pythium vexans.
To gain insight into the unique features of the core Pythium genes, we compared the frequency of occurrence of protein family domains in the core Pythium gene family set and the species-specific genes (clustered genes and singletons). First, comparisons were made between core Pythium genes and the rest of the genes from each species. The core Pythium genes were enriched in genes involved in pathogenesis and signaling processes including elicitin (IPR002200), necrosis-inducing (IPR008701), peptidase C1A (IPR000668), protease inhibitor (Kazal-type (IPR011497)), and HECT ubiquitin ligase (IPR000569) whereas genes involved in transport activities (IPR001140, IPR018108) are underrepresented (P<0.05) (Table S5).
Second, the species-specific genes were compared to the rest of the genes from those particular species. Several transporter- (ABC (IPR001140) and nucleotide-sugar transporter-related (IPR007271)) domains were highly over-represented in Py. vexans-specific genes (Chi-square test with Bonferroni correction, P<0.001). The Py. aphanidermatum-specific genes were highly enriched (P<0.01) for aspartic peptidase (IPR021109), endoglucanase (IPR009009), cutinase (IPR000675), and pectate lyase (IPR002022) domains. The highly represented domains in Py. arrhenomanes were protease inhibitor (Kazal-type (IPR011497)), cutinase (IPR000675), necrosis inducing (IPR008701), and pectate lyase (IPR002022). Similarly, pathogenesis related domains such as peptidase (IPR006026) and proteinase inhibitor I25 (cystatin (IPR018073)) were highly represented (P<0.05) in Py. irregulare-specific genes. The leucine-rich repeat (LRR) containing domain (IPR001611), carbonic anhydrase (IPR018338), and chitinase II (IPR011583) were over-represented (P<0.05) in Py. iwayamai-specific genes (Table S5). A number of these protein domains have been shown to be implicated in plant-pathogen interaction in different oomycete pathogens [55], [60]. In general, we observed higher representation of protein domains potentially involved in degradation of host tissues (e.g., glycoside hydrolase) and establishment of infection structure (e.g., elicitin, necrosis inducing proteins, protease inhibitors) in the core Pythium gene set leading to necrotrophic life style.
Metabolism of Complex Carbohydrates
Carbohydrate-active enzymes (CAZymes) are involved in the biosynthesis and degradation of diverse glycoconjugates, oligosaccharides, and polysaccharides [61] and have a central role in the breakdown of the plant cell wall by plant pathogens thereby serving as pathogenicity factors. These enzymes can also be involved in the biosynthesis, breakdown, and modification of the oomycete cell wall and structural polysaccharides as part of growth and development. Thus, comparison of the CAZyme content would provide insights into metabolic and enzymatic diversity in oomycete pathogens. Putative CAZymes in Pythium species were identified using the CAZymes Analysis Toolkit (CAT) [62] and correspondence between CAZyme families and protein family domains was analyzed. The comparison of the glycoside hydrolase (GH), glycosyltransferases (GTs), polysaccharide lyase (PL), and carbohydrate esterase (CE) in the Pythium genomes revealed that these organisms exhibit substantial variation in number of CAZymes (Table S6). The CE and carbohydrate-binding module (CBM) classes were poorly represented in all Pythium genomes.
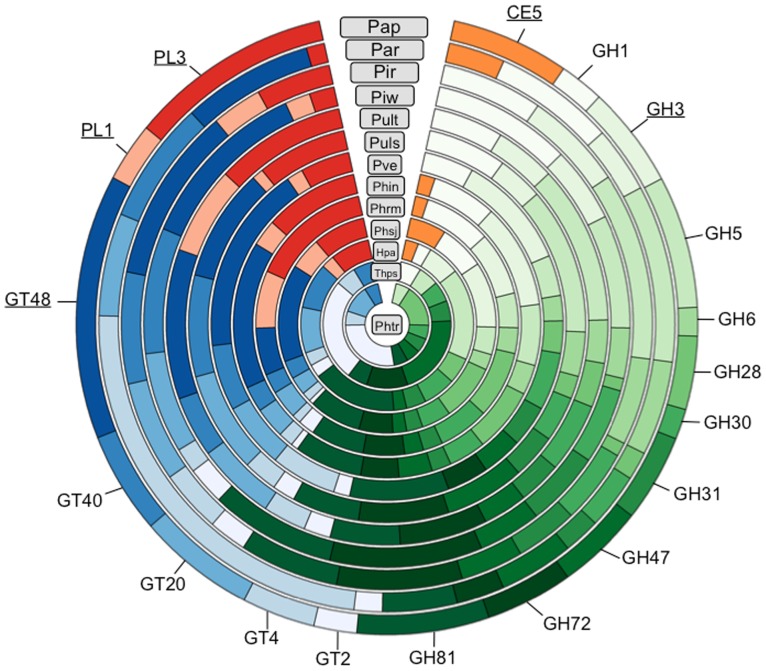
Interestingly, we identified eight and six cutinase-encoding genes (CE5 family) in Py. aphanidermatum and Py. arrhenomanes, respectively, but not in the other Pythium genomes (Figure 3, Table S6, Table S7) suggesting that the evolution of these phytopathogens led to different degrees of reduction in their cutin degrading capabilities. Pythium species have a relatively smaller set of GH-encoding genes (77 to 114 members) compared to all Phytophthora species (166–216 members) yet strikingly larger than the repertoire of the biotroph H. arabidopsidis (72 members) and the diatoms (31–32 members) (Table S6) in agreement with previous findings [52], [53], [56]. The GH superfamily was the most highly represented CAZyme superfamily in all Pythium genomes with PL the least represented (3 families). We observed that in general Pythium species have a highly reduced set of secreted CAZymes when compared to Phytophthora species, which underwent gene expansion [63]. The differential ability of oomycete pathogens to produce different hydrolytic enzymes acting on different complex carbohydrate molecules could determine their infection strategy, host range, and most likely contribute to the different virulence mechanisms between oomycete pathogens. An in-depth study of the Pythium-CAZymes is reported in a companion paper (Zerillo et al. PLoS One, this issue).
Figure 3. Distribution of various carbohydrate-active enzymes (CAZymes) in stramenopile genomes.
The CAZymes coding genes were annotated using the CAZymes Analysis Toolkit- CAT [62] according to the CAZy database [61] in combination with protein family domain analyses. Gene families absent in at least 2 species are underlined. Comparison of total CAZymes from different classes is listed in Table S6. CE, carbohydrate esterase; GH, glycoside hydrolase; GT, glycosyl transferase; PL, polysaccharide lyase; Pap, Pythium aphanidermatum; Par, Pythium arrhenomanes; Pir, Pythium irregulare; Piw, Pythium iwayamai; Pult, Pythium ultimum var. ultimum; Puls, Pythium ultimum var. sporangiiferum; Pve, Pythium vexans; Phin, Phytophthora infestans; Phrm, Phytophthora ramorum; Phsj, Phytophthora sojae; Hpa, Hyaloperonospora arabidopsidis; Thps, Thalassiosira pseudonana; Phtr, Phaeodactylum tricornutum.
The Pythium Secretome
Pythium species, like many oomycete pathogens, secrete effector proteins as well as degradative enzymes that alter host physiology and facilitate colonization. Indeed, the genomes of Ph. infestans, Ph. ramorum, Ph. sojae, Py. ultimum var. ultimum, and H. arabidopsidis contain large complex families of effector genes that encode secreted proteins which have been implicated in pathogenesis [6], [7], [52], [53]. Secreted proteins in the seven Pythium proteomes were predicted using SignalP v3.0 [64] and transmembrane domains predicted with TMHMM [65]. In total, 834 to 1,008 proteins were predicted to be secreted (using the criteria described in Materials and Methods) in the seven Pythium species (Table S8). Genes encoding the predicted secreted proteins were then clustered using OrthoMCL revealing 1,086 clusters containing 4,921 secreted proteins while 1,592 were singletons. A total of 76 clusters containing 782 secreted proteins were common to all Pythium species, hereafter referred to as core Pythium secretome (Figure S1). Of the total, 745 clusters have secreted proteins from at least three different species. The largest secretome gene family contains 25 members from all Pythium species, and encodes a polysaccharide lyase involved in host cell wall degradation. Other families of secreted core proteins in Pythium include elicitins, protease inhibitors, cellulose-binding elicitor lectin (CBEL)-like proteins with CBM, and expanded families of cell wall degrading enzymes. Overall, depending on the species, 63–78% of the predicted secreted proteins in Pythium species surveyed have expression support (Table S8).
To augment our functional annotation, we annotated the predicted secretome for PFAM domains using InterProScan [66] and associated Gene Ontology (GO) [67] terms using Blast2GO [68]. We also compared the frequency of these annotations with the non-secreted proteome using Chi-square tests with Bonferroni correction [69]. Pathogenesis (GO:0009405), proteolysis (GO:0006508), and carbohydrate metabolic processes (GO:0005975) were highly enriched in the core Pythium secretome (P<0.001) relative to the rest of the predicted proteome from Pythium (Table S9). The protein domains showing the highest enrichment in the core Pythium secretome are elicitin (IPR002200), glycoside hydrolases (IPR000322), glycosyl transferase (IPR001296), peptidase C1A (IPR000668), EGF-like domain (IPR006209), and pectate lyase (IPR004898) (P<0.001) (Table S10).
To document the protein family domains and biological functions enriched in the Pythium species-specific secretome, we compared the frequency of occurrence of PFAM domains and GO terms in the species-specific secretomes relative to the rest of the proteome from that particular species using Chi-square tests. The Py. aphanidermatum-specific secretomes were highly enriched for hydrolase activity (GO:0004553) including cutin hydrolase activity (GO:0050525) and carbohydrate metabolic process (GO:0005975) (P<0.05) (Table S9). Similarly, Py. arrhenomanes-specific secretomes were highly enriched for cellulose catabolic process (GO:0030245), hydrolase activity (GO:0004553), and pectin lyase activity (GO:0047490). The Py. iwayamai-specific secretomes were highly enriched for peptidase activity (GO:0008233), transmembrane transport (GO:0055085), and nucleic acid binding (GO:0003676). The most represented GO terms in Py. ultimum var. sporangiiferum-specific secretome were cysteine-type peptidase activity (GO:0008234), cellulose binding (GO:0030248), and isomerase activity (GO:0016853). The transmembrane transport (GO:0055085) as well as sugar binding and sugar modification activities (GO:0005529, GO:0016787, GO:0004650) were most enriched in Py. vexans-specific secretome while pectate lyase (GO:0030570), proteolysis (GO0006508), and glycosyl bonds hydrolase activity (GO:0016798) were the most enriched GO terms in the Py. irregulare-specific secretome. Enrichment of hydrolase, pectate lyase activity and cell wall modification process in species-specific secretome indicates that degrading host cell wall is one of the major functions of Pythium secretome as illustrated for other oomycete pathogens [55], [60].
Similarly, the protein family domains including cutinase (IPR000675) and glycoside hydrolase (IPR000743) that hydrolyze glycosidic bonds, and peptidase A1 (IPR001461) were highly enriched (P<0.05) in the species-specific secretomes of Py. aphanidermatum (Table S10). In addition, cutinase (IPR000675), glycoside hydrolases (IPR002594, IPR001137), and peptidase inhibitors (IPR011497, IPR013201) domains were enriched in Py. arrhenomanes-specific secretomes relative to their proteomes. The enrichment and their expression upon infection in plant pathogenic oomycetes have already been shown for different families of hydrolases and lyases [55], [60]. The NPP1 domain (IPR008701) that is present in necrosis-inducing proteins was highly enriched in the Py. aphanidermatum and Py. irregulare-specific secretomes. The necrosis-inducing proteins are known for their ability to trigger numerous plant defense responses, necrosis, and cell death in dicotyledonous plants [70]. Several transporter-related domains (IPR005828, IPR003439, IPR011547) along with peptidase S8/S53 (IPR000209) were highly enriched in Py. vexans-specific secretome. The membrane transporter (e.g. ABC transporter) may play important role in counteracting the physiological impact of host defense compounds [71]. Domains containing leucine-rich repeat (IPR001611) were highly enriched in Py. iwayamai along with several peptidase domains (IPR001506, IPR001394, IPR001461). The Py. irregulare-specific secretome was highly enriched for peptidase A1 (IPR001461), pectinesterase (IPR000070), NPP1 domain (IPR008701), serine protease inhibitor (kazal-type (IPR011497)). The protein domains specifically enriched in different Pythium species (e.g., certain transporter families, peptidase, and domains related to metabolism of carbohydrates) highlight the differences between these groups of plant pathogens in terms of their pathogenicity and host preference.
RxLR Effectors
The genomes of the three Phytophthora species encode large numbers (350 to 563) of potential effector proteins that are implicated in pathogenesis [9], [72], [73]. These proteins contain a conserved amino-terminal cell entry domain with the motifs RxLR and dEER [6], [7], which mediate their entry into host cells [74], [75], [76]. RxLR-dEER effectors are hypothesized, and in a few cases experimentally shown, to suppress host defense responses [77]. However, some of these effectors can be recognized by plant immune receptors resulting in programmed cell death and disease resistance [78], [79]. Although no RxLR effectors are present in the Py. ultimum var. ultimum genome [53], evolution under diverse environmental conditions and co-evolution with diverse hosts could lead to inter-specific variation in RxLR effectors among Pythium species.
We used four different bioinformatics approaches to ascertain if RxLR effector genes occurred within the genomes of the six Pythium species sequenced in this study. Consistent with previous analyses of the Py. ultimum var. ultimum genome [53] in which no RxLR effectors were detected, we failed to identify any candidate effectors in any of the six Pythium species sequenced. Our results suggest that in all seven of the Pythium species surveyed, RxLR effectors are absent signifying substantial differences in virulence and the interaction of Pythium species with plant hosts as compared to Phytophthora and Hyaloperonospora species (Table S7). Since Phytophthora genomes have 350–563 RxLR effector candidates [6], [7], the absence of these effectors in Pythium genomes indicates that the effectors are not required for virulence of Pythium species. As compared to hemibiotrophic Phytophthora species, Pythium species are adapted to necrotrophic lifestyle and may not require RxLR effectors for successful colonization and establishment of the infection structure. Other effectors such as secreted hydrolytic enzymes and necrotizing toxins may play important role during necrotrophy.
YxSL[RK] Candidate Effectors
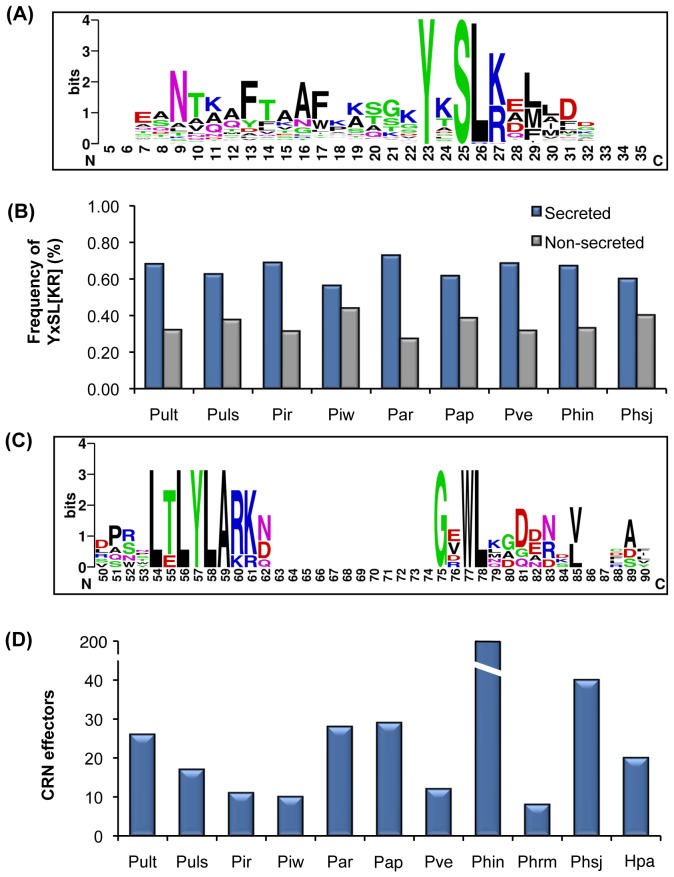
The YxSL[RK] class of putative effectors have been found in many pathogenic oomycetes including Py. ultimum var. ultimum [53], [80], [81]. Interestingly, the YxSL[RK] motif shares similarity in sequence and position with the canonical RxLR motif and appears to be a signature for a novel family of secreted proteins that may function as effectors [81] in Pythium and Phytophthora species. We computationally screened our newly sequenced six Pythium genomes for candidate YxSL[RK] effectors using a HMM profile of a putative YxSL[RK] motif constructed using 57 genes containing the corresponding motif from Py. ultimum var. ultimum [53], three Phytophthora species, and Aphanomyces eutieches [82]. Proteins with the YxSL[RK] motif situated within 30 to 150 amino acids positions after the initial methionine were considered for further analyses. Using the YxSL[RK] motifs previously identified in Py. ultimum var. ultimum as a positive control [53], we identified an initial set of 123 proteins containing the YxSL[RK] motif in the six Pythium species. After searching against the HMM profile and multiple sequence alignment of the 123 proteins, we removed three proteins with an YxSL[RK] motif positioned outside the 30 to 150 amino acid position range. Using the same HMM profile, we were able to identify 21 additional proteins containing the YxSL[RK] motif from Ph. infestans and Ph. sojae. Alignment of the core set of 141 YxSL[RK] effectors from Pythium and Phytophthora show a modular organization with a conserved amino-terminal region, containing four conserved motifs, followed by a highly variable carboxy-terminal region as reported for other oomycete effectors [9] (Figure 4A). The YxSL[RK] candidates are significantly enriched (P≤0.05) within the secretome of Ph. infestans, Ph. sojae, and all seven Pythium species (Figure 4B), nearly two-fold higher in the secreted proteome as compared to the rest of the proteome.
Figure 4. Candidate effector proteins from Pythium.
(A) The typical architecture of a YxSL[RK] effector candidate inferred from 141 sequences from seven Pythium species, Phytophthora infestans, and Phytophthora sojae. The consensus sequence pattern of the YxSL[RK] motif was calculated using WebLogo [103]. The bigger the letter, the more conserved the amino acid site. Please note that the numbers in the sequence logo refer to the corresponding positions in the alignment and thus differ from the average position of the motifs. (B) The YxSL[RK] motif distribution in the proteomes of Pythium species, Phytophthora infestans and Phytophthora sojae is shown. The YxSL[RK] sequence is over-represented in the secretome of Pythium and Phytophthora species relative to the non-secreted proteome (P≤0.05). The YxSL[RK] motifs were counted only if they were within the first 30 to 150 residues from the signal peptide. The frequency was calculated as percentage of either all secreted proteins or all non-secreted proteins. Pult, Pythium ultimum var. ultimum; Puls, Pythium ultimum var. sporangiiferum; Pir, Pythium irregulare; Piw, Pythium iwayamai; Par, Pythium arrhenomanes; Pap, Pythium aphanidermatum; Pve, Pythium vexans; Phin, Phytophthora infestans; Phrm, Phytophthora ramorum; Phsj, Phytophthora sojae; Hpa, Hyaloperonospora arabidopsidis. (C) The typical architecture of an LxLYLAR/K effector motif inferred from 129 sequences from 7 Pythium species. The consensus sequence pattern of the LxLYLAR/K motif was calculated using WebLogo [103]. The bigger the letter, the more conserved the amino acid site. Please note that the numbers in the sequence logo are referring to the corresponding positions in the alignment and thus differ from the average position of the motifs. (D) Number of CRN effector proteins in oomycetes. The number of candidate CRN effectors estimated by Hidden Markov Model (HMM) searches in combination with two other computational methods is shown. The number of CRN effectors from Pythium ultimum var. ultimum, Phytophthora species and H. arabidopsidis were taken from published genome datasets [6], [7], [52], [53]. Pult, Pythium ultimum var. ultimum; Puls, Pythium ultimum var. sporangiiferum; Pir, Pythium irregulare; Piw, Pythium iwayamai; Par, Pythium arrhenomanes; Pap, Pythium aphanidermatum; Pve, Pythium vexans; Phin, Phytophthora infestans; Phrm, Phytophthora ramorum; Phsj, Phytophthora sojae; Hpa, Hyaloperonospora arabidopsidis.
CRN Effectors
The genomes of many oomycete pathogens harbor a large repertoire of a class of candidate effectors termed “Crinklers” (CRN) that are presumed to enter host cytoplasm [6], [83], [84] and elicit necrosis in planta [83]. First identified in Phytophthora based on their ability to elicit plant cell death and defense responses [83], these effectors have been identified in all phytopathogenic oomycete genomes sequenced to date [6], [7], [52], [53], [85]. Similar to the RxLR effectors, CRN effectors contain a conserved motif, LFLAK, following the signal peptide [6], [84]. Contrary to RxLR effectors, CRN effectors are present in all oomycete plant pathogen genomes suggesting that these are an evolutionarily conserved set of effectors in phytopathogenic oomycetes, including Py. ultimum var. ultimum [53]. Through BLASTP [86] searches using 21 well-defined amino-terminal domains from Ph. infestans and Py. ultimum var. ultimum, we identified 45 predicted CRN proteins in the six newly sequenced Pythium species. We aligned all predicted Pythium CRN sequences with the CRN proteins from Ph. infestans and Py. ultimum var. ultimum to build an HMM profile and using HMMer we identified 53 additional candidate effectors with an LFLAK-like domain in other Pythium species. Using the same HMM profile built from alignment of Pythium and Phytophthora CRN sequences, we were able to identify 14 of the 20 candidate CRN effector proteins from the H. arabidopsidis genome [52]. Further string searches of the Pythium proteomes using LxLFLAK and LxLYLAR/K, a conserved motif that is shared amongst Py. ultimum var. ultimum CRN proteins [53], resulted in identification of 5 additional predicted proteins with LxLFLAK-like domains from the six Pythium species. Examination of a set of 129 predicted effector proteins (including 26 from Py. ultimum var. ultimum) from all Pythium species revealed a conserved LxLYLAR/K motif followed by conserved WL motif (Figure 4C) that is shared amongst CRN proteins. Consistent with previous results, the LxLYLAR/K motif was located between 40 and 65 amino acids after the initial methionine, followed by an adjacent diversified WL domain reflecting the modular design of CRN proteins in the oomycetes [6]. Given the important functions of effectors in oomycete pathogenicity, we compared the number of CRN effector classes across oomycete species for which genome sequences are available. Surveys of genome sequences showed that every examined species, including Albugo laibachii, and H. arabidopsidis, have candidate CRN genes [6], [7], [52], [53], [85] indicating that these effectors are ubiquitous in plant pathogenic oomycetes [6], [7], [52], [53], [85]. Comparison of the number of these effectors shows expansion in Ph. infestans [6] and high intraspecific variation in Pythium similar to those found in Phytophthora species (Figure 4D). The intraspecific variation in number of CRN effector indicates that the Pythium species may have adopted species-specific strategies for infection and these strategies could be important during their interaction with different hosts.
Comparative Genomics
Shared gene clusters of oomycetes
The Straminipila includes phytopathogenic oomycetes and autotrophic diatoms. A phylogenetic tree constructed using the Bayesian analyses of nuclear large subunit of rDNA from 14 stramenopiles shows five broad clades: the Phytophthora species with Hyaloperonospora and Py. vexans, the Pythium species with globose sporangia, the Pythium species with filamentous sporangia, the diatom group with T. pseudonana and P. tricornutum, and a separate clade of S. parasitica (Figure 1).
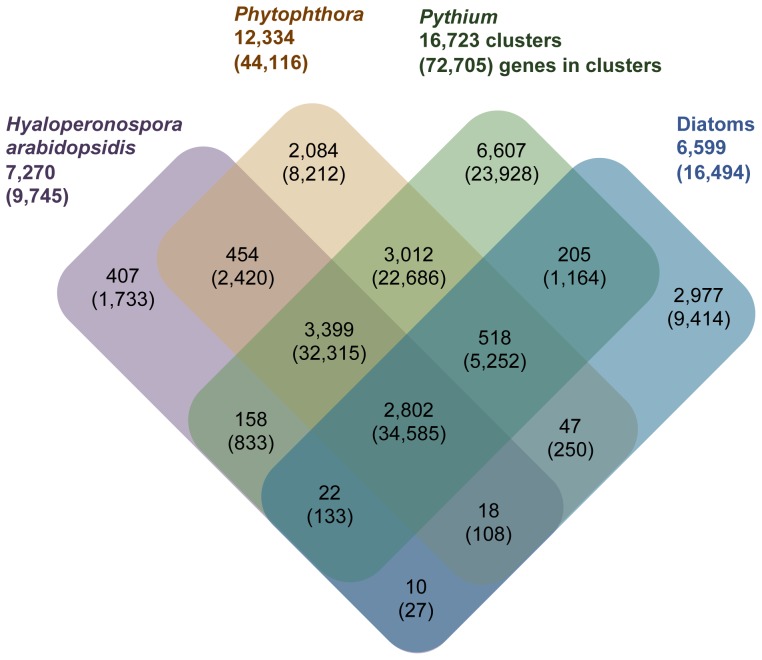
To identify common features in all oomycete genomes, we compared the gene family content of the 11 phytopathogenic oomycetes and two autotrophic diatoms using OrthoMCL [59] using the predicted protein-coding genes from these 13 species. A total of 182,894 protein-coding genes from Pythium (7 species), Phytophthora (3 species), H. arabidopsidis, and diatoms (2 species) were clustered revealing the stramenopile-core (clusters shared by all 13 taxa), Pythium/Phytophthora-specific (including clusters specific to Phytophthora and Pythium), Hpa-specific (clusters specific to H. arabidopsidis) and diatom-specific clusters (clusters specific to two diatom species, Figure 5). A total of 143,060 genes were clustered into 22,720 gene families (clusters) while 39,834 genes were singletons. The core stramenopile cluster of 2,802 gene families contained 34,585 genes. The stramenopile-core genes showed over-representation of major facilitator superfamily (IPR011701) and amino acid transporter, transmembrane (IPR013057) domains, with significant under-representation of glycoside hydrolases (IPR000743, IPR001139), protease inhibitors (Kazal-type (IPR011497)), and necrosis inducing (IPR008701) domains (Bonferroni-corrected P<0.05) relative to the non-core genes (genes other than core genes) from stramenopiles (Table S11). Comparatively higher numbers of diatom-specific genes (2,977 clusters, 9,414 genes) is consistent with the specialized adaptation of diatoms to a phototrophic lifestyle compared to phytopathogenic oomycetes. Kinase-encoding domains (IPR000719, IPR001245) were highly represented in diatom-specific genes while domains including HECT ubiquitin ligase (IPR000569), peptidase M16, C-terminal (IPR007863), proteinase inhibitor I29 (cathespin propeptide (IPR013201)), and glycoside hydrolase, family 30 (IPR001139) were depleted. The Pythium-Phytophthora-specific gene sets (3,012 clusters, 22,686 genes) were highly enriched with protein domains possibly related to pathogenesis including necrosis inducing (IPR008701), elicitin (IPR002200), glycoside hydrolases (IPR000743, IPR011583), and peptidases (IPR001577, IPR001563) (P<0.05) as compared to the rest of the genes from Pythium and Phytophthora. For several groups of over-represented domains in the Pythium-Phytophthora-specific genes, a direct or indirect role in host-pathogen interaction and/or plant pathogen lifestyle has already been hypothesized or demonstrated [6], [55], [60], [87]. Several secreted protease inhibitors (Kazal-type (IPR011497)), peptidase M8 (leishmanolysin (IPR001577)), and peptidase S10 (serine carboxypeptidase (IPR001563)) domains are significantly over-represented in Pythium-Phytophthora-specific clusters, suggestive of a role in protection of the pathogen against host-encoded defense-related proteases as shown in other oomycete pathogens [55]. In comparison to the rest of the genes from H. arabidopsidis, the H. arabidopsidis-specific genes (407 clusters, 1,733 genes) showed under-representation of domains related to transport (IPR011547), host-targeted degradative enzyme (IPR021067), elicitin (IPR002200), and necrosis-inducing protein (IPR008701) (P<0.05) (Table S11). It is possible that in evolving a biotrophic lifestyle, H. arabidopsidis lost many secreted hydrolytic enzymes [52].
Figure 5. Orthologous gene families of Pythium species, Phytophthora species, Hyaloperonospora arabidopsidis and two diatom species.
Protein-coding genes from seven Pythium, three Phytophthora, two diatom species and Hyaloperonospora arabidopsidis were clustered using OrthoMCL [59]. The number of gene families (clusters) and the total number of clustered genes (in parentheses) are indicated for each taxon and their interactions in the Venn diagram. The numbers outside the Venn diagram indicate the total number of orthologous clusters and number of genes (in parentheses) in the clusters for each taxon.
Syntenic Relationships among Oomycetes
The availability of several Pythium genome sequences permits the first detailed investigation of genome evolution within the genus and comparison with that of other straminopiles. By comparison with Py. ultimum var. ultimum, we identified syntenic regions across stramenopiles and analyzed rearrangements in gene order. Previous analyses of synteny between selected regions of Py. ultimum var. ultimum and Phytophthora species showed very similar ortholog content in broad regions but a high level of rearrangement in local gene order [53]. Here, we expanded the syntenic analyses to six other Pythium species, H. arabidopsidis and the diatom T. pseudonana. Due to the fragmented nature of the assemblies in all of the genomes, we identified contigs or scaffolds with a minimum of five genes and identified syntenic blocks in comparison to the Py. ultimum var. ultimum genome using MCscan [88]. A contig or scaffold was considered syntenic if at least three genes in a five-gene block was co-linear with Py. ultimum var. ultimum genome. The comparison of Pythium genomes shows a varying degree of synteny between Py. ultimum var. ultimum and six other Pythium species. Among all Pythium species, Py. ultimum var. ultimum was most syntenic with Py. ultimum var. sporangiiferum and least syntenic with Py. arrhenomanes and Py. iwayamai followed by Py. aphanidermatum (Table S12).
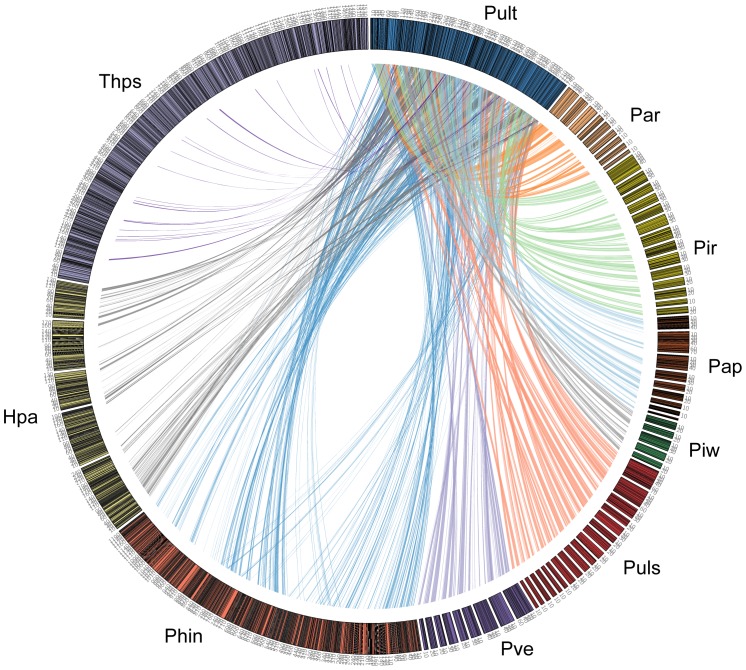
In order to examine the conservation of gene order across three oomycete genera, we compared Py. ultimum var. ultimum not only with the other six Pythium species but also with Ph. infestans, H. arabidopsidis, and T. pseudonana. Figure 6 shows the conservation of gene order between one of the largest scaffolds from Py. ultimum var. ultimum (scf1117875581354) with Ph. infestans (supercontig 1.2), H. arabidopsidis (scaffolds 5,6,7,8 and 9) and T. pseudonana (chromosome 3). As expected, the gene order was highly conserved among Pythium species. The level of synteny revealed by our analyses extends the previously reported synteny between Py. ultimum var. ultimum and Phytophthora species [53] unveiling conservation of a portion of gene order not only within seven Pythium species but also between other stramenopiles. Within oomycetes, the conservation of synteny between species recapitulates the phylogeny shown in Figure 1. The overall degree of conservation is high, being highest among the most closely related species (as shown by the larger spans with fewer breaks in synteny between Pythium species) than with distantly related species (e.g. H. arabidopsidis and T. pseudonana).
Figure 6. Syntenic analyses of stramenopile genomes.
The circle is a graphical representation of the selected regions from Pythium arrhenomanes (contigs 8, 17, 26, 41, 68, 131, 170, 285, 707) Pythium irregulare (contigs 28, 92, 103, 106, 119, 123, 129, 132, 140, 163, 195, 226, 372, 396), Pythium aphanidermatum (scaffolds 4, 6, 23, 80, 88, 96, 115, 150, 327), Pythium iwayamai (contigs 18, 28, 29, 61, 235), Pythium ultimum var. sporangiiferum (contigs 4, 6, 34, 106, 121, 134, 150, 173, 181, 222, 231, 257, 319, 404, 437, 458, 533, 726), Pythium vexans (contigs 9, 31,42, 94, 151, 160, 209, 220, 347), Phytophthora infestans (supercontig 1.2), Hyaloperonospora arabidopsidis (scaffolds 5, 6, 7, 8, 9) and Thalassiosira pseudonana (chromosome 3). Numbers along each ideogram are sequence lengths in kbp. Syntenic regions were identified through reciprocal best matches between gene models and block identification using MCscan [88]. Each line radiating from Py. ultimum var. ultimum (scf1117875581354) links a syntenic gene pair. Each species is represented by a genus-species abbreviation and colored as Pythium ultimum var. ultimum (Pult) in blue, Pythium arrhenomanes (Par) in orange, Pythium irregulare (Pir) in yellow, Pythium aphanidermatum (Pap) in dark brown, Pythium iwayamai (Piw) in green, Pythium ultimum var. sporangiiferum (Puls) in dark red, Pythium vexans (Pve) in purple, Phytophthora infestans (Phin) in brick red, Hyaloperonospora arabidopsidis (Hpa) in olive green, and Thalassiosira pseudonana (Thaps) in light purple.
Conclusions
The genome sequences of 13 stramenopiles enabled genome-wide comparison of gene repertoires within and between phytopathogenic oomycetes and non-pathogenic diatoms. Our comparative analyses of stramenopiles indicate that developmental innovations in oomycete pathogens involve secretion of a large number of effector molecules, proteolytic enzymes, and cell wall hydrolyzing enzymes. However, expansion of a suite of genes encoding effectors and proteolytic enzymes reflect specific adaptations to trophic lifestyle.
These comparative analyses revealed some of the genetic mechanisms underlying necrotrophic and biotrophic lifestyle. The hemibiotrophic Phytophthora species show expansion and diversification of protein families associated with plant infection such as some glycoside hydrolases, ABC transporters and in particular, oomycete pathogenesis related genes. In contrast to the biotrophic H. arabidopsidis, which exhibits dramatic reductions in genes encoding RxLR effectors and other secreted pathogenicity proteins, cell wall hydrolytic enzymes and transporters, the non-biotrophic group (Phytophthora and Pythium) seems to have a large suite of pathogenicity related genes, as a result of expansion of effector families in Phytophthora and proteolytic enzymes in Pythium (Table S7). These differences in rich repertoires of candidate effectors could underlie the coevolution and adaptation of these pathogens to the plant immune system and set them apart from the non-pathogenic autotrophic stramenopiles (e.g. diatoms). A deeper understanding of the complex array of factors, including secreted proteins and proteolytic enzymes identified in this study, which affect host-pathogen interactions and coevolution, could enable efficient targeting of pathogen-control measures in agricultural ecosystems.
Materials and Methods
Illumina Sequencing and Assembly
Approximately 1 g of freshly harvested mycelium from Pythium species was ground in a FastPrep® (BIO 101) machine (Savant, Inc. Holbrook, NY) and the genomic DNA was extracted following a protocol modified from Moller et al. [89]. Libraries were constructed from genomic DNA of Py. arrhenomanes (ATCC 12531 = CBS 324.62), Py. irregulare (DAOM BR486 = CBS 250.28), Py. iwayamai (DAOM 242034 = CBS 132417), Py. ultimum var. sporangiiferum (DAOM BR650 = CBS 219.65), and Py. vexans (DAOM BR484 = CBS 119.80) using the Illumina Genomic DNA Sample kit (Illumina, San Diego, CA). The libraries were paired-end sequenced (76 or 120 bp) on the Illumina Genome Analyzer II/IIx sequencer. The purity-filtered (PF) reads were first trimmed and then quality filtered using custom Perl scripts to remove reads with low quality bases (<Q20) (Table S1).
The trimmed and quality filtered reads were then assembled using Velvet v0.7.63 [90] in conjunction with the VelvetOptimiser tool v2.10 [91], a multi-threaded Perl script for automatically optimizing the parameter options for the Velvet assembler. VelvetOpimiser was run with a k-mer range of 21 to 41 for each assembly the final assembly parameters for each assembly is in Table S2.
Pyrosequencing and Assembly
For Py. aphanidermatum (DAOM BR444 = CBS 132490), a single-end and a paired-end (3 Kb) library was created and sequenced using the Genome Sequencer FLX instrument following the manufacturer’s protocol (Roche Applied Science, Mannheim, Germany). The paired-end library was sequenced using the Titanium chemistry. The single-end library yielded 1,299,108 reads (507 Mb) with an average length of 390 bp. The paired-end library yielded 380,566 reads (137.5 Mb) with an average length of 361 bp. 256,098 of these reads were indentified as a member of a valid read pair. The reads were assembled with the Newbler assembler v2.3 [92] with the large genome mode and paired-end mode flags set. The final assembly statistics is summarized in Table 2 and Table S3.
Genome Annotation
The assembled genomes were annotated using the MAKER v2.03 [93] annotation pipeline. A Pythium specific repeat library constructed previously [53] was supplied to MAKER for the repeat masking step. Gene calls were generated using FGENESH [94] using the Phytophthora matrix and SNAP [95], which was trained using the transcripts from the Pythium ultimum Genome Database (http://pythium.plantbiology.msu.edu/). All the oomycete ESTs in dbEST [96] and all the oomycete proteins in GenBank were provided to MAKER as evidence to refine the annotation. The final annotation set produced by MAKER is summarized in Table 2 and Table S3.
Phylogenetic Analysis
The phylogeny of 14 stramenopiles was created by using 123 ITS rDNA sequences obtained from GenBank. Nucleotide sequences were aligned by ClustalW [97]. Phylogenetic analyses were performed using the MrBayes program for Bayesian analysis [98] using Markov Chain Monte Carlo (MCMC) with the general time reversible (GTR) model. The program was run for 1,000,000 generations and sampled every 100 generations. Phylogenetic tree was visualized in Mega5 [99].
Identification of Orthologous Groups
Orthologous and close paralogous genes were identified using OrthoMCL v1.4 [59] with default parameters. Protein domains were predicted by InterProScan [100]. For each genome or group specific proteins, the total number of proteins with each type of domain was computed.
Identification of Putative Secreted Proteins
Signal peptides were predicted using SignalP v3.0 [64] and transmembrane domains predicted with TMHMM [65]. Proteins showing (i) SignalP3.0 NN Ymax Score ≥0.5 and (ii) SignalP3.0 NN D-score ≥0.5 and (iii) SignalP3.0 HMM S probability ≥0.9 and (iv) predicted localization “Secreted” (S) and (v) no TMHMM predicted transmembrane domain after signal peptide cleavage site were considered to be within the Pythium secretome. Sequences that were predicted to contain transmembrane domains or organelle-targeting signals were omitted from the secretome. The clustering of secreted protein was done using OrthoMCL v1.4 [59].
Enrichment Analyses
InterProScan [100] with default parameters were used to complement the annotation of the secreted proteins. GO terms were assigned using Blast2GO [68] with default parameters. Enrichment frequencies in the core Pythium, stramenopile-core and taxa-specific gene families were calculated as the number of occurrences over the total number of protein domain or GO hits among secreted versus non-secreted proteins. Significance of enrichment/depletion is assessed by a Chi-square test with Bonferroni correction for multiple testing. Only protein domains with enrichment p-value≤0.05 and GO with enrichment p-value≤0.05 were considered.
Carbohydrate-active Enzyme Analyses
The carbohydrate-active enzyme coding genes of Pythium, Phytophthora, H. arabidopsidis and diatom genomes were automatically annotated using the CAZymes Analysis Toolkit – CAT [62] according to the CAZy database classification [61]. Enzyme annotation was done using two approaches. First, a bi-directional BLAST search was performed against the entire non-redundant sequences of the CAZy database. Second, a link or correspondence between the CAZy families and protein family domains was analyzed. A manual scan was also performed based on the PFAM domain information.
Identification of Candidate Effectors
The candidate RxLR effectors were identified using the approach described by Win et al. [101]. We used four different bioinformatics approaches to identify the predicted set of effectors. First, we translated all six frames of the Pythium genome sequences to identify proteins with an amino-terminal signal peptide based on SignalP prediction using SignalP v3.0 [64] with a SignalP HMM score cutoff of ≥0.9. The transmembrane domains were predicted with TMHMM [65] and sequences that were predicted to contain transmembrane domains or organelle-targeting signals were omitted. Candidate RxLR effectors were selected from these secreted translations using custom Perl scripts. Secreted translations with RxLR position between 30 and 150 residues from signal peptide, RxLR position downstream of the signal peptide cleavage site and SignalP v3.0 NN predicted cleavage site of less than 30 amino acids were selected as candidate RxLR effectors. The six frame translation of H. arabidopsidis genome which is reported to have 134 candidate RxLR effectors was used as a positive control. Second, an HMM profile of the RxLR domain was constructed by manually aligning the RxLR domains of the 53 RxLR effectors from Phytophthora species and H. parasitica. The resulting alignment was fed to the hmmbuild program (HMMer software) [102] to generate the HMM profile. The HMM profile was used to search the translations for candidate effectors using the hmmsearch program [102]. To validate our computational approach, the same HMM profile was used to search the six frame translation of H. arabidopsidis genome. Furthermore, the whole Pythium proteome was also searched with the HMM. Third, a comprehensive database of RxLR effector proteins from Phytophthora species [6], [7], H. arabidopsidis [52], and A. laibachii [85] was created. Putative homologs in predicted proteomes of Pythium were identified by BLASTP search against the RxLR effector database at E-value cutoff of 10−5. Fourth, a string searches was performed for the RxLR, RxLx and Rx[LMFY][HKR] motif within the amino terminus of each six frame translation of Pythium genomes, 30 to 150 residues from the signal peptide.
We computationally screened the six Pythium genomes for candidate YxSL[RK] effectors using a HMM profile of the putative YxSL[RK] motif, a novel effector motif identified first in Py. ultimum var. ultimum, constructed using 57 genes containing YxSL[RK] motif from three Phytophthora species and Aphanomyces eutieches. Using the YxSL[RK] motifs from Py. ultimum var. ultimum as a control, we identified an initial set of 123 proteins containing the YxSL[RK] motif in 7 Pythium species. Using the same HMM profile we were able to identify 21 additional proteins with the YxSL[RK] motif in the Ph. infestans and Ph. sojae genomes. After searching against the HMM profile and multiple alignment of the proteins we selected a set of 141 proteins, which includes 120 candidate effectors from seven Pythium species and 21 from two Phytophthora species, with YxSL[RK] motif situated between 30 to 150 amino acids positions.
For the CRN effectors, a BLASTP search against 21 well-defined amino-terminal domains from Ph. infestans and Py. ultimum var. ultimum CRN sequences was performed to identify proteins with putative LFLAK-like domains. The candidate CRN sequences from Ph. infestans and Pythium species were used to construct an HMM profile and the CRN sequences from Py. ultimum var. ultimum were used as a control. Two criteria were used to identify candidate LxLYLAR/K proteins. First, the conserved motif should be preceded by a signal peptide and followed by WL motif. Second, the conserved motif should be located between 40 to 65 amino acids after first methionine. Using the HMM profile, we identified additional candidate effectors with an LxLYLAR/K domain. To validate our computational methods, the same HMM profile was used to identify the CRN effectors from H. arabidopsidis genome which is reported to have 20 candidate CRN effectors [52]. Multiple alignments were conducted using the programs ClustalW and ClustalX [97]. Sequence alignments were submitted to the WebLogo server [103] to generate a sequence logo for the consensus.
Transcriptome Sequencing
One pooled cDNA library was constructed for each of four Pythium species (Py. arrhenomanes, Py. irregulare, Py. iwayamai, and Py. vexans). Initially, plugs of 10% V8 agar containing Pythium species were incubated for 1 day in yeast extract broth (YEB; 30 g/l sucrose, 1 g/l KH2PO4, 0.5 g/l MgSO4·7 H2O, 0.5 g/l KCl, 10 mg/l FeSO4·7 H2O, 1 g/l yeast extract) at 25°C with shaking (200 rpm). Approximately 50 mg of hyphae growing out of the plugs were then transferred to flasks containing media for the various expression assays, with the exception of Py. iwayamai, mycelium was grown under the following conditions: 1, nutrient-rich YEB medium for 3 days at 25°C with shaking (200 rpm); 2, and nutrient-starved Plich medium (S Kamoun, unpublished) for 10 days at 25°C in standing culture, as previously described [104]; 3, YEB medium for 2 days at 25°C with shaking (200 rpm) followed by the addition of 1 and 100 µl/l of the fungicide mefenoxam (Subdue MAXX™, Novartis Crop Production, Greensboro, NC, USA) and subsequent incubation for an additional 3 hours at the same temperature and with agitation; 4, YEB medium for 2 days at 25°C with shaking (200 rpm) followed by a cold stress of 0°C with shaking (200 rpm) for 6 hours; 5, YEB medium for 2 days at 25°C with shaking (200 rpm) followed by a heat stress of 35°C for 6 hours. Py. iwayamai was isolated from a cool temperature site and therefore incubated at 10°C and was not exposed to above mentioned conditions. Condition 5 was not included, because Py. iwayamai did not resist the heat stress at 35°C. For each condition listed above, mycelium was harvested, macerated in liquid nitrogen and RNA was extracted as described [104]. RNA was treated with DNAse (Promega RQ1 RNase-Free DNase, Madison, WI, USA) and subsequently pooled in an equimolar manner with the RNA from other conditions. From each pooled library 10 µg RNA was used to construct cDNA using the mRNA-Seq Sample Prep Kit (Illumina, San Diego, CA, USA), which was sequenced with Illumina Genome Analyzer (GA) II using version 3 sequencing reagents for 36 cycles. Base calling was carried out using the Illumina GA pipeline v1.4.
For each library, the purity filtered reads from the Illumina Genome Analyzer II pipeline were mapped using Tophat v1.1.4 [105], which works in conjunction with Bowtie, a short read aligner v0.12.7 [106]. The minimum and maximum intron sizes were 5 bp and 15 kbp, respectively, for each Tophat run. The final annotation GFF3 file was provided to Tophat and expression values (FPKM) were calculated using the Cufflinks package v0.9.3 [107]. A gene was considered expressed if the FPKM value and FPKM 95% confidence interval lower boundary was greater than 0.001 and zero, respectively.
Synteny Analyses
All protein coding genes from the seven Pythium genomes, Ph. infestans, H. arabidopsidis, and T. pseudonana were compared to each other via an all-by-all BLASTP [108] to generate the appropriate input for the MCscan algorithm [88] (BLASTP, E-value≤1e-10). A python script contained in the MCscan package was used to filter the BLASTP output to remove self-matches and to reorder the list of gene pairs. MCscan v0.8 was used to calculate synteny between all combinations of genomes using the pooled BLASTP output and the genomic coordinates. A minimum of 3 genes within a 5 gene block was required to constitute a syntenic block (default MCscan value is 5). The size of the search window was calculated by MCscan based on the average intergenic distance in the genomes being compared. Default values were used for all other parameters. Each syntenic block is assigned an E-value by MCscan. Custom PERL scripts were used to parse the MCscan output and calculate the total number of syntenic blocks for each genome combination. MCscan output was parsed to create files appropriately formatted for input to Circos [109] for visualization. Figure 6 shows an example spanning selected syntenic regions of Pythium, Ph. infestans, H. arabidopsidis, and T. pseudonana genomes. Each syntenic block is represented as a link whose ends represent the syntenic regions from other species.
Data Access
The whole genome shotgun projects have been deposited at DDBJ/EMBL/GenBank under the accession numbers: AKXX00000000 for Py. aphanidermatum, AKXY00000000 for Py. arrhenomanes, AKXZ00000000 for Py. irregulare, AKYA00000000 for Py. iwayamai, AKYB00000000 for Py. ultimum var. sporangiiferum, and AKYC00000000 for Py. vexans. The genome assemblies, transcript sequences, and protein sequences are also available for download and BLAST searching at Pythium Genome Database (PGD) website (http://pythium.plantbiology.msu.edu/, see download and BLAST pages). Also available for download at the PGD are the annotation files in GFF3 format and the functional annotation of the gene models (http://pythium.plantbiology.msu.edu/download.shtml). The genome assembly and annotation files are also available for download from the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.h748p). The WGS reads are available in the NCBI Short Read Archive (SRA) under the accession SRP006957. The RNA-Seq Reads are available the NCBI SRA under the accession SRP006964.
Supporting Information
Shared clusters of secreted proteins in Pythium . The Venn diagram shows the distribution of secreted protein clusters among Pythium species. The putative secreted proteins from seven Pythium species were predicted by using SignalP v3.0 [64] and clustered using OrthoMCL [59]. The number of gene families (clusters) and the total number of clustered genes (numbers in parentheses) are indicated.
(TIF)
Summary of sequence reads generated for five Pythium species. Sequencing was done using the Illumina Genome Analyzer (GA) II or IIx platform. PF, purity-filtered; Par, Pythium arrhenomanes; Pir, Pythium irregulare; Piw, Pythium iwayamai; Puls, Pythium ultimum var. sporangiiferum; Pve, Pythium vexans.
(XLS)
Assembly parameters for five Pythium species. Sequenced reads were assembled using Velvet v0.7.63 [90] in conjunction with the VelvetOptimiser tool v2.10 [91]. Par, Pythium arrhenomanes; Pir, Pythium irregulare; Piw, Pythium iwayamai; Puls, Pythium ultimum var. sporangiiferum; Pve, Pythium vexans.
(XLS)
Assembly and annotation statistics of thirteen stramenopile genomes. Assembly and annotation statistics were compared to data published by Levesque et al. for Pythium ultimum var. ultimum [53], Haas et al. for Phytophthora infestans [6], Tyler et al. for Phytophthora ramorum and Phytophthora sojae [7], Baxter et al. for Hyaloperonospora arabidopsidis [52], Armbrust et al. for Thalassiosira pseudonana [58] and Bowler et al. for Phaeodactylum tricornutum [57]. Pap, Pythium aphanidermatum; Par, Pythium arrhenomanes; Pir, Pythium irregulare; Piw, Pythium iwayamai; Puls, Pythium ultimum var. sporangiiferum; Pult, Pythium ultimum var. ultimum; Pve, Pythium vexans; Phin, Phytophthora infestans; Phrm, Phytophthora ramorum; Phsj, Phytophthora sojae; Hpa, Hyaloperonospora arabidopsidis; Thps, Thalassiosira pseudonana; Phtr, Phaeodactylum tricornutum.
(XLS)
Whole transcriptome sequencing data for four Pythium species. Sequence reads from each species were mapped to the respective genome using TopHat v1.1.4 [105]. Fragments per kilobase pair of exon model per million fragments mapped (FPKM) values were calculated using Cufflinks v0.9.3 [107] and genome annotation. Genes are considered expressed if FPKM value and FPKM 95% confidence interval lower boundary was greater than 0.001 and zero, respectively.
(XLS)
Protein domains enriched or depleted in core Pythium and Pythium species-specific clusters. Fold change numbers are color coded as black for enriched and red for depleted domains. Only protein domains significantly enriched or depleted are shown.
(XLS)
Sizes of carbohydrate-active enzyme (CAZyme) classes in the stramenopile genomes. The CAZymes coding genes were automatically annotated using the CAZymes Analysis Toolkit – CAT [62] according to the CAZy [61] database classification in combination with protein family domain assignment. Bold text indicate total. Pap, Pythium aphanidermatum; Par, Pythium arrhenomanes; Pir, Pythium irregulare; Piw, Pythium iwayamai; Puls, Pythium ultimum var. sporangiiferum; Pult, Pythium ultimum var. ultimum; Pve, Pythium vexans; Phin, Phytophthora infestans; Phrm, Phytophthora ramorum; Phsj, Phytophthora sojae; Hpa, Hyaloperonospora arabidopsidis; Thps, Thalassiosira pseudonana; Phtr, Phaeodactylum tricornutum.
(XLS)
Quantitative comparison of pathogenicity-related proteins in stramenopiles. Genes were predicted for all datasets using PFAM v23.0 prediction and BLASTP searches. Results were compared to published genome datasets for Pythium ultimum var. ultimum [53], Phytophthora species [6], [7], Hyaloperonospora arabidopsidis [52] and Thalassiosira pseudonana [58]. The numbers in the denominator show the number of genes with expression support. Genes are considered expressed if FPKM value and FPKM 95% confidence interval lower boundary was greater than 0.001 and zero respectively. The expression data for Pythium ultimum var. ultimum were taken from Lévesque et al. [53]. Pap, Pythium aphanidermatum; Par, Pythium arrhemonanes; Pir, Pythium irregulare; Piw, Pythium iwayamai; Puls, Pythium ultimum var. sporangiiferum; Pult, Pythium ultimum var. ultimum; Pve, Pythium vexans; Phin, Phytophthora infestans; Phrm, Phytophthora ramorum; Phsj, Phytophthora sojae; Hpa, Hyaloperonospora arabidopsidis; Thps, Thalassiosira pseudonana.
(XLS)
Number and percentage of secreted proteins from stramenopiles. The table shows the number of secreted protein and the percentage of proteins to be secreted from 13 stramenopiles. Also shown is the number and percentage of secreted proteins with transcript support. The secreted proteins were identified by SignalP v3.0 [64] and transmembrane domains were predicted with TMHMM [65]. Protein coding genes are considered expressed if the FPKM value and FPKM 95% confidence interval lower boundary was greater than 0.001 and zero, respectively. The expression data for Pythium ultimum var. ultimum were taken from Lévesque et al. [53].
(XLS)
Gene ontology (GO) molecular function and biological process categories enriched in core Pythium and species-specific secretome. The table shows the enrichment fold in core Pythium and specific-specific secretome as compared to the non-secretome. Only GO terms significantly enriched in secretome are shown.
(XLS)
Protein domains enriched in core Pythium and species-specific secretome. The table shows the enrichment fold in core Pythium and species-specific secretome as compared to the non-secretome. Only domains significantly enriched in secretome are shown.
(XLS)
Protein domains enriched or depleted in stramenopile-core, diatom, Hpa and Pythium / Phytophthora -specific gene families. The table shows the enriched or depleted protein domains in taxa-specific and stramenopile-core gene families as compared to rest of the genomes. Fold change numbers are color coded as black for enriched and red for depleted domains. Only Protein domains significantly enriched or depleted are shown. Hpa, Hyaloperonospora arabidopsidis.
(XLS)
Number of genes syntenic to Pythium ultimum var. ultimum . Syntenic genes were identified through reciprocal best matches between gene models and block identification using MCscan [88].
(XLS)
Acknowledgments
We acknowledge the assistance of Brieanne Vaillancourt for genome and transcriptome sequencing. We want to thank Tara Rintoul of Agriculture and Agri-Food Canada (AAFC) for culturing and DNA extraction of the strains used for genome sequencing. We are grateful to CBS-KNAW fungal biodiversity center and Carolyn Babcock of AAFC for providing strains for this project.
Funding Statement
Funding for the work was provided by the United States Department of Agriculture National Institute of Food and Agriculture Microbial Genome Sequencing Program to CRB and NT (2007-35600-17774 and 2007-35600-18886). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Beakes GW, Glockling SL, Sekimoto S (2012) The evolutionary phylogeny of the oomycete “fungi”. Protoplasma 249: 3–19. [DOI] [PubMed] [Google Scholar]
- 2. Cavalier-Smith T, Chao E (2006) Phylogeny and megasystematics of phagotrophic heterokonts (Kingdom Chromista). J Mol Evol 62: 388–420. [DOI] [PubMed] [Google Scholar]
- 3.Dick MW (2001) Straminipilous Fungi: systematics of the Peronosporomycetes including accounts of the marine straminipilous protists, the Plasmodiophorids and similar organisms. Dordrecht: Kluwer Academic Publishers.
- 4. Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF (2000) A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290: 972–977. [DOI] [PubMed] [Google Scholar]
- 5. Yoon HS, Hackett JD, Pinto G, Bhattacharya D (2002) The single, ancient origin of chromist plastids. Proc Natl Acad Sci U S A 99: 15507–15512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature 461: 393–398. [DOI] [PubMed] [Google Scholar]
- 7. Tyler BM, Tripathy S, Zhang XM, Dehal P, Jiang RHY, et al. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313: 1261–1266. [DOI] [PubMed] [Google Scholar]
- 8. Baurain D, Brinkmann H, Petersen J, Rodriguez-Ezpeleta N, Stechmann A, et al. (2010) Phyologenomic Evidence for Separate Acquisition of Plastids in Cryptophytes, Haptophytes, and Stramenopiles. Mol Biol Evol 27: 1698–1709. [DOI] [PubMed] [Google Scholar]
- 9. Kamoun S (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol 44: 41–60. [DOI] [PubMed] [Google Scholar]
- 10. Martin FN, Loper JE (1999) Soilborne plant diseases caused by Pythium spp: Ecology, epidemiology, and prospects for biological control. Crit Rev Plant Sci 18: 111–181. [Google Scholar]
- 11. Mendoza L, Ajello L, McGinnis MR (1996) Infections caused by the Oomycetous pathogen Pythium insidiosum . J Mycol Med 6: 151–164. [Google Scholar]
- 12. Saunders GA, Washburn JO, Egerter DE, Anderson JR (1988) Pathogenicity of fungi isolated from field collected larvae of the western treehole mosquito, Aedes sierrensis (Diptera, Culicidae). J Invertebr Pathol 52: 360–363. [DOI] [PubMed] [Google Scholar]
- 13.Van Der Plaats-Niterink AJ (1981) Monograph of the genus Pythium. Stud Mycol: 1–242.
- 14.Erwin DC, Ribeiro OK (1996) Phytophthora diseases worldwide: A.P.S. Press. 562 p. [Google Scholar]
- 15. Cook RJ, Sitton JW, Haglund WA (1987) Influence of soil treatments on growth and yield of wheat and implications for control of Pythium root rot. Phytopathology 77: 1192–1198. [Google Scholar]
- 16. Paulitz TC, Adams K (2003) Composition and distribution of Pythium communities in wheat fields in eastern Washington state. Phytopathology 93: 867–873. [DOI] [PubMed] [Google Scholar]
- 17. Harvey P, Lawrence L (2008) Managing Pythium root disease complexes to improve productivity of crop rotations. Outlooks Pest Manag 19: 127–129. [Google Scholar]
- 18. Dieguez-Uribeondo J, Garcia MA, Cerenius L, Kozubikova E, Ballesteros I, et al. (2009) Phylogenetic relationships among plant and animal parasites, and saprotrophs in Aphanomyces (Oomycetes). Fungal Genet Biol 46: 365–376. [DOI] [PubMed] [Google Scholar]
- 19. Cooke DEL, Drenth A, Duncan JM, Wagels G, Brasier CM (2000) A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet Biol 30: 17–32. [DOI] [PubMed] [Google Scholar]
- 20. Bouwmeester K, Pieter MJAvP, Govers F (2009) Genome biology cracks enigmas of oomycetes plant pathogens. Annu Plant Rev 34: 102–133. [Google Scholar]
- 21. Goker M, Voglmayer H, Riethmuller A, Oberwinkler F (2006) How do obigate parasites evolve? A multi-gene phylgenetic analysis of downy mildews. Fungal Genet Biol 44: 105–122. [DOI] [PubMed] [Google Scholar]
- 22. Runge F, Choi Y-J, Thines M (2011) Phylogenetic investigations in the genus Pseudoperonospora reveal overlooked species and cryptic diversity in the Pseudoperonospora cubensis species cluster. Eur J Plant Pathol 129: 135–146. [Google Scholar]
- 23. Levesque CA, De Cock A (2004) Molecular phylogeny and taxonomy of the genus Pythium . Mycol Res 108: 1363–1383. [DOI] [PubMed] [Google Scholar]
- 24. Bala K, Robideau GP, Desaulniers N, de Cock AWAM, Levesque CA (2010) Taxonomy, DNA barcoding and phylogeny of three new species of Pythium from Canada. Persoonia 25: 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robideau GP, de Cock AWAM, Coffey MD, Voglmayr H, Brouwer H, et al. (2011) DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol Ecol Resour 11: 1002–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cook RJ, Sitton JW, Waldher JT (1980) Evidence for Pythium as a pathogen of direct-drilled wheat in the Pacific Northwest. Plant Dis 64: 102–103. [Google Scholar]
- 27. Larkin RP, English JT, Mihail JD (1995) Effects of infection by Pythium spp. on root-system morphology of alfalfa seedlings. Phytopathology 85: 430–435. [Google Scholar]
- 28.Snowdon AL (1990) A colour atlas of post-harvest diseases and disorders of fruits and vegetables. Vol. 1. General introduction and fruits: 302.
- 29. Sumner DR, Gascho GJ, Johnson AW, Hook JE, Threadgill ED (1990) Root diseases, populations of soil fungi, and yield decline in continuous double-crop corn. Plant Dis 74: 704–710. [Google Scholar]
- 30. Adams PB (1990) The potential of mycoparasites for biological control of plant diseases. Annu Rev Phytopathol 28: 59–72. [DOI] [PubMed] [Google Scholar]
- 31. Martin FN, Hancock JG (1987) The use of Pythium oligandrum for biological control of pre-emergence damping-off caused by Pythium ultimum . Phytopathology 77: 1013–1020. [Google Scholar]
- 32. Takenaka S, Sekiguchi H, Nakaho K, Tojo M, Masunaka A, et al. (2008) Colonization of Pythium oligandrum in the Tomato Rhizosphere for Biological Control of Bacterial Wilt Disease Analyzed by Real-Time PCR and Confocal Laser-Scanning Microscopy. Biol Control 98: 187–195. [DOI] [PubMed] [Google Scholar]
- 33. Takahashi M, Ichitani T, Sasaki M (1977) Pythium porphyrae Takahashi et Sasaki, sp. nov. causing red rot of marine red algae Porphyra spp. Trans Mycol Soc Jpn 18: 279–285. [Google Scholar]
- 34. Brasier CM, Hansen EM (1992) Evolutionary biology of Phytophthora 2. Phylogeny, Speciation and Population Structure. Annu Rev Phytopathol 30: 173–200. [Google Scholar]
- 35.Martin FN (2009) Pythium Genetics. In: Lamour K, Kamoun S, editors. Oomycete Genetics and Genomics: Diversity, Plant and Animal Interactions, and Toolbox. Hoboken: John Willey & Sons. 574.
- 36. Martin FN (2000) Phylogenetic relationships among some Pythium species inferred from sequence analysis of the mitochondrially encoded cytochrome oxidase II gene. Mycologia 92: 711–727. [PubMed] [Google Scholar]
- 37.Koike ST, Gladders P, Paulus AO (2006) Vegetable diseases: a color handbook. Vegetable diseases: a color handbook.
- 38. Gold SE, Stanghellini ME (1985) Effects of temperature on Pythium root rot of spinach Spinacia oleracea grown under hydroponic conditions. Phytopathology 75: 333–337. [Google Scholar]
- 39. Harvey PR, Butterworth PJ, Hawke BG, Pankhurst CE (2000) Genetic variation among populations of Pythium irregulare in southern Australia. Plant Pathol 49: 619–627. [Google Scholar]
- 40. Harvey PR, Butterworth PJ, Hawke BG, Pankhurst CE (2001) Genetic and pathogenic variation among cereal, medic and sub-clover isolates of Pythium irregulare . Mycol Res 105: 85–93. [Google Scholar]
- 41. Spies CF, Mazzola M, Botha WJ, Langenhoven S, Mostert L, et al. (2011) Molecular analyses of Pythium irregulare isolates from grapevines in South Africa suggest that this species complex may be a single variable species. Fungal Biol 115: 1210–1224. [DOI] [PubMed] [Google Scholar]
- 42. Vawdrey LL, Langdon P, Martin T (2005) Incidence and pathogenicity of Phytophthora palmivora and Pythium vexans associated with durian decline in far northern Queensland. Australian Plant Pathology 34: 127–128. [Google Scholar]
- 43. Zeng HC, Ho HH, Zheng FC (2005) Pythium vexans causing patch canker of rubber trees on Hainan Island, China. Mycopathologia 159: 601–606. [DOI] [PubMed] [Google Scholar]
- 44. Spies CFJ, Mazzola M, Botha WJ, Van der Rijst M, Mostert L, et al. (2011) Oogonial biometry and phylogenetic analyses of the Pythium vexans species group from woody agricultural hosts in South Africa reveal distinct groups within this taxon. Fungal Biol 115: 157–168. [DOI] [PubMed] [Google Scholar]
- 45.Quinn E (2012) Resolving the Pythium ultimum species complex. Ottawa: Carleton University. 85 p. [Google Scholar]
- 46. Bridge PD, Newsham KK, Denton GJ (2008) Snow mould caused by a Pythium sp.: a potential vascular plant pathogen in the maritime Antarctic. Plant Pathol 57: 1066–1072. [Google Scholar]
- 47.Iwayama S (1933) On a new snow-rot disease of cereal plants caused by Pythium sp. Pamphlet of the Agricultural Experimental Station Toyama-Ken, Japan.
- 48. McLeod A, Botha WJ, Meitz JC, Spies CFJ, Tewoldemedhin YT, et al. (2009) Morphological and phylogenetic analyses of Pythium species in South Africa. Mycol Res 113: 933–951. [DOI] [PubMed] [Google Scholar]
- 49. Capra JA, Carbone L, Riesenfeld SJ, Wall JD (2010) Genomics through the lens of next-generation sequencing. Genome Biol 11: 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shendure J, Ji HL (2008) Next-generation DNA sequencing. Nat Biotechnol 26: 1135–1145. [DOI] [PubMed] [Google Scholar]
- 51. Soanes DM, Richards TA, Talbot NJ (2007) Insights from sequencing fungal and oomycete genomes: What can we learn about plant disease and the evolution of pathogenicity? Plant Cell 19: 3318–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baxter L, Tripathy S, Ishaque N, Boot N, Cabral A, et al. (2010) Signatures of Adaptation to Obligate Biotrophy in the Hyaloperonospora arabidopsidis Genome. Science 330: 1549–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, et al. (2010) Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol 11. [DOI] [PMC free article] [PubMed]
- 54.Martens C, Van de Peer Y (2010) The hidden duplication past of the plant pathogen Phytophthora and its consequences for infection. BMC Genomics 11. [DOI] [PMC free article] [PubMed]
- 55. Seidl MF, Van den Ackerveken G, Govers F, Snel B (2011) A Domain-Centric Analysis of Oomycete Plant Pathogen Genomes Reveals Unique Protein Organization. Plant Physiol 155: 628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Seidl MF, Van den Ackerveken G, Govers F, Snel B (2012) Reconstruction of Oomycete Genome Evolution Identifies Differences in Evolutionary Trajectories Leading to Present-Day Large Gene Families. Genome Biol Evol 4: 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, et al. (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456: 239–244. [DOI] [PubMed] [Google Scholar]
- 58. Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, et al. (2004) The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 306: 79–86. [DOI] [PubMed] [Google Scholar]
- 59. Li L, Stoeckert CJ, Roos DS (2003) OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raffaele S, Win J, Cano LM, Kamoun S (2010) Analyses of genome architecture and gene expression reveal novel candidate virulence factors in the secretome of Phytophthora infestans. BMC Genomics 11. [DOI] [PMC free article] [PubMed]
- 61. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, et al. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37: D233–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Park BH, Karpinets TV, Syed MH, Leuze MR, Uberbacher EC (2010) CAZymes Analysis Toolkit (CAT): Web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology 20: 1574–1584. [DOI] [PubMed] [Google Scholar]
- 63.Ospina-Giraldo MD, Griffith JG, Laird EW, Mingora C (2010) The CAZyome of Phytophthora spp.: a comprehensive analysis of the gene complement coding for carbohydrate-active enzymes in species of the genus Phytophthora. BMC Genomics 11. [DOI] [PMC free article] [PubMed]
- 64. Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795. [DOI] [PubMed] [Google Scholar]
- 65. Krogh A, Larsson B, von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol 305: 567–580. [DOI] [PubMed] [Google Scholar]
- 66. Zdobnov EM, Apweiler R (2001) InterProScan - an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848. [DOI] [PubMed] [Google Scholar]
- 67. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 69.Abdi H (2007) Bonferroni and Sidak corrections for multiple comparisons. In: Salkind NJ, editor. Encyclopedia of Measurement and Statistics. Thousand Oaks, CA: Sage.
- 70.Gijzen M, Nuernberger T (2006) Nep1-like proteins from plant pathogens: Recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 67. [DOI] [PubMed]
- 71.Stefanato FL, Abou-Mansour E, Buchala A, Kretschmer M, Mosbach A, et al. (2009) The ABC transporter BcatrB from Botrytis cinerea exports camalexin and is a virulence factor on Arabidopsis thaliana. Plant J 58. [DOI] [PubMed]
- 72. Schornack S, van Damme M, Bozkurt TO, Cano LM, Smoker M, et al. (2010) Ancient class of translocated oomycete effectors targets the host nucleus. Proc Natl Acad Sci U S A 107: 17421–17426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stassen JHM, Van den Ackerveken G (2011) How do oomycete effectors interfere with plant life? Curr Opin Plant Biol 14: 407–414. [DOI] [PubMed] [Google Scholar]
- 74. Dou DL, Kale SD, Wang XL, Chen YB, Wang QQ, et al. (2008) Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell 20: 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kale SD (2011) Oomycete and fungal effector entry, a microbial Trojan horse. New Phytol 193: 874–881. [DOI] [PubMed] [Google Scholar]
- 76. Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, et al. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450: 115–118. [DOI] [PubMed] [Google Scholar]
- 77. Sohn KH, Lei R, Nemri A, Jones JDG (2007) The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana . Plant Cell 19: 4077–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bos JIB, Armstrong MR, Gilroy EM, Boevink PC, Hein I, et al. (2010) Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc Natl Acad Sci U S A 107: 9909–9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dou D, Kale SD, Wang X, Chen Y, Wang Q, et al. (2008) Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell 20: 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Horner NR, Grenville-Briggs LJ, van West P (2012) The oomycete Pythium oligandrum expresses putative effectors during mycoparasitism of Phytophthora infestans and is amenable to transformation. Fungal Biol 116: 24–41. [DOI] [PubMed] [Google Scholar]
- 81.van Damme M, Cano LM, Oliva R, Schornack S, Segretin ME, et al.. (2011) Evolutionary and Functional Dynamics of Oomycete Effector Genes. In: Martin F, Kamoun S, editors. Effectors in Plant-Microbe Interactions. Oxford, UK: Wiley-Blackwell. 101–120.
- 82.Gaulin E, Madoui M-A, Bottin A, Jacquet C, Mathe C, et al. (2008) Transcriptome of Aphanomyces euteiches: New Oomycete Putative Pathogenicity Factors and Metabolic Pathways. PLoS ONE 3. [DOI] [PMC free article] [PubMed]
- 83. Torto TA, Li SA, Styer A, Huitema E, Testa A, et al. (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora . Genome Res 13: 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Win J, Morgan W, Bos J, Krasileva KV, Cano LM, et al. (2007) Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell 19: 2349–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kemen E, Gardiner A, Schultz-Larsen T, Kemen AC, Balmuth AL, et al. (2011) Gene Gain and Loss during Evolution of Obligate Parasitism in the White Rust Pathogen of Arabidopsis thaliana. PLoS Biol 9. [DOI] [PMC free article] [PubMed]
- 86. Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, et al. (2005) The genome sequence of the rice blast fungus Magnaporthe grisea . Nature 434: 980–986. [DOI] [PubMed] [Google Scholar]
- 88. Tang H, Wang X, Bowers JE, Ming R, Alam M, et al. (2008) Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res 18: 1944–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moller EM, Bahnweg G, Sandermann H, Geiger HH (1992) A simple and efficient protocol for isolation of high-molecular-weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res 20. [DOI] [PMC free article] [PubMed]
- 90.Zerbino DR, Birney E (2008) Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18. [DOI] [PMC free article] [PubMed]
- 91.VelvetOptimiser website: http://www.bioinformatics.net.au/software.velvetoptimiser.shtml.Accessed 2013 Aug 21.
- 92.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, et al.. (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437. [DOI] [PMC free article] [PubMed]
- 93. Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, et al. (2008) MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18: 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Salamov AA, Solovyev VV (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10. [DOI] [PMC free article] [PubMed]
- 95.Korf I (2004) Gene finding in novel genomes. BMC Bioinformatics 5. [DOI] [PMC free article] [PubMed]
- 96.Boguski MS, Lowe TMJ, Tolstoshev CM (1993) dbEST - Database for “expressed sequence tags”. Nat Genet 4. [DOI] [PubMed]
- 97. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 98. Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 99. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mulder N, Apweiler R (2007) InterPro and InterProScan - Tools for protein sequence classification and comparison. Methods Mol Biol: 59–70. [DOI] [PubMed]
- 101. Bradley A (2007) Encyclopedia of measurement and statistics. Reference & User Services Quarterly 47: 133–133. [Google Scholar]
- 102. Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14: 755–763. [DOI] [PubMed] [Google Scholar]
- 103.WebLogo3 Website: http://weblogo.berkeley.edu.Accessed 2013 Aug 21.
- 104.Cheung F, Win J, Lang JM, Hamilton J, Hue V, et al. (2008) Analysis of the Pythium ultimum transcriptome using Sanger and Pyrosequencing approaches. BMC Genomics 9. [DOI] [PMC free article] [PubMed]
- 105. Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10. [DOI] [PMC free article] [PubMed]
- 107. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 109. Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, et al. (2009) Circos: An information aesthetic for comparative genomics. Genome Res 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, et al.. (2012) MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst Biol 61. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shared clusters of secreted proteins in Pythium . The Venn diagram shows the distribution of secreted protein clusters among Pythium species. The putative secreted proteins from seven Pythium species were predicted by using SignalP v3.0 [64] and clustered using OrthoMCL [59]. The number of gene families (clusters) and the total number of clustered genes (numbers in parentheses) are indicated.
(TIF)
Summary of sequence reads generated for five Pythium species. Sequencing was done using the Illumina Genome Analyzer (GA) II or IIx platform. PF, purity-filtered; Par, Pythium arrhenomanes; Pir, Pythium irregulare; Piw, Pythium iwayamai; Puls, Pythium ultimum var. sporangiiferum; Pve, Pythium vexans.
(XLS)
Assembly parameters for five Pythium species. Sequenced reads were assembled using Velvet v0.7.63 [90] in conjunction with the VelvetOptimiser tool v2.10 [91]. Par, Pythium arrhenomanes; Pir, Pythium irregulare; Piw, Pythium iwayamai; Puls, Pythium ultimum var. sporangiiferum; Pve, Pythium vexans.
(XLS)
Assembly and annotation statistics of thirteen stramenopile genomes. Assembly and annotation statistics were compared to data published by Levesque et al. for Pythium ultimum var. ultimum [53], Haas et al. for Phytophthora infestans [6], Tyler et al. for Phytophthora ramorum and Phytophthora sojae [7], Baxter et al. for Hyaloperonospora arabidopsidis [52], Armbrust et al. for Thalassiosira pseudonana [58] and Bowler et al. for Phaeodactylum tricornutum [57]. Pap, Pythium aphanidermatum; Par, Pythium arrhenomanes; Pir, Pythium irregulare; Piw, Pythium iwayamai; Puls, Pythium ultimum var. sporangiiferum; Pult, Pythium ultimum var. ultimum; Pve, Pythium vexans; Phin, Phytophthora infestans; Phrm, Phytophthora ramorum; Phsj, Phytophthora sojae; Hpa, Hyaloperonospora arabidopsidis; Thps, Thalassiosira pseudonana; Phtr, Phaeodactylum tricornutum.
(XLS)
Whole transcriptome sequencing data for four Pythium species. Sequence reads from each species were mapped to the respective genome using TopHat v1.1.4 [105]. Fragments per kilobase pair of exon model per million fragments mapped (FPKM) values were calculated using Cufflinks v0.9.3 [107] and genome annotation. Genes are considered expressed if FPKM value and FPKM 95% confidence interval lower boundary was greater than 0.001 and zero, respectively.
(XLS)
Protein domains enriched or depleted in core Pythium and Pythium species-specific clusters. Fold change numbers are color coded as black for enriched and red for depleted domains. Only protein domains significantly enriched or depleted are shown.
(XLS)
Sizes of carbohydrate-active enzyme (CAZyme) classes in the stramenopile genomes. The CAZymes coding genes were automatically annotated using the CAZymes Analysis Toolkit – CAT [62] according to the CAZy [61] database classification in combination with protein family domain assignment. Bold text indicate total. Pap, Pythium aphanidermatum; Par, Pythium arrhenomanes; Pir, Pythium irregulare; Piw, Pythium iwayamai; Puls, Pythium ultimum var. sporangiiferum; Pult, Pythium ultimum var. ultimum; Pve, Pythium vexans; Phin, Phytophthora infestans; Phrm, Phytophthora ramorum; Phsj, Phytophthora sojae; Hpa, Hyaloperonospora arabidopsidis; Thps, Thalassiosira pseudonana; Phtr, Phaeodactylum tricornutum.
(XLS)
Quantitative comparison of pathogenicity-related proteins in stramenopiles. Genes were predicted for all datasets using PFAM v23.0 prediction and BLASTP searches. Results were compared to published genome datasets for Pythium ultimum var. ultimum [53], Phytophthora species [6], [7], Hyaloperonospora arabidopsidis [52] and Thalassiosira pseudonana [58]. The numbers in the denominator show the number of genes with expression support. Genes are considered expressed if FPKM value and FPKM 95% confidence interval lower boundary was greater than 0.001 and zero respectively. The expression data for Pythium ultimum var. ultimum were taken from Lévesque et al. [53]. Pap, Pythium aphanidermatum; Par, Pythium arrhemonanes; Pir, Pythium irregulare; Piw, Pythium iwayamai; Puls, Pythium ultimum var. sporangiiferum; Pult, Pythium ultimum var. ultimum; Pve, Pythium vexans; Phin, Phytophthora infestans; Phrm, Phytophthora ramorum; Phsj, Phytophthora sojae; Hpa, Hyaloperonospora arabidopsidis; Thps, Thalassiosira pseudonana.
(XLS)
Number and percentage of secreted proteins from stramenopiles. The table shows the number of secreted protein and the percentage of proteins to be secreted from 13 stramenopiles. Also shown is the number and percentage of secreted proteins with transcript support. The secreted proteins were identified by SignalP v3.0 [64] and transmembrane domains were predicted with TMHMM [65]. Protein coding genes are considered expressed if the FPKM value and FPKM 95% confidence interval lower boundary was greater than 0.001 and zero, respectively. The expression data for Pythium ultimum var. ultimum were taken from Lévesque et al. [53].
(XLS)
Gene ontology (GO) molecular function and biological process categories enriched in core Pythium and species-specific secretome. The table shows the enrichment fold in core Pythium and specific-specific secretome as compared to the non-secretome. Only GO terms significantly enriched in secretome are shown.
(XLS)
Protein domains enriched in core Pythium and species-specific secretome. The table shows the enrichment fold in core Pythium and species-specific secretome as compared to the non-secretome. Only domains significantly enriched in secretome are shown.
(XLS)
Protein domains enriched or depleted in stramenopile-core, diatom, Hpa and Pythium / Phytophthora -specific gene families. The table shows the enriched or depleted protein domains in taxa-specific and stramenopile-core gene families as compared to rest of the genomes. Fold change numbers are color coded as black for enriched and red for depleted domains. Only Protein domains significantly enriched or depleted are shown. Hpa, Hyaloperonospora arabidopsidis.
(XLS)
Number of genes syntenic to Pythium ultimum var. ultimum . Syntenic genes were identified through reciprocal best matches between gene models and block identification using MCscan [88].
(XLS)