Abstract
Huntington’s disease (HD) causes severe motor dysfunction, behavioral abnormalities, cognitive impairment and death. Investigations into its molecular pathology have primarily relied on murine tissues; however, the recent discovery of induced pluripotent stem cells (iPSCs) has opened new possibilities to model neurodegenerative disease using cells derived directly from patients, and therefore may provide a human-cell-based platform for unique insights into the pathogenesis of HD. Here, we will examine the practical implementation of iPSCs to study HD, such as approaches to differentiate embryonic stem cells (ESCs) or iPSCs into medium spiny neurons, the cell type most susceptible in HD. We will explore the HD-related phenotypes identified in iPSCs and ESCs and review how brain development and neurogenesis may actually be altered early, before the onset of HD symptoms, which could inform the search for drugs that delay disease onset. Finally, we will speculate on the exciting possibility that ESCs or iPSCs might be used as therapeutics to restore or replace dying neurons in HD brains.
Keywords: Huntington’s disease, induced pluripotent stem cells, IPSC, MSN, stem cell models, Neurodegenerative disease
Introduction
Huntington’s disease (HD) is a devastating and incurable neurological disorder caused by a CAG expansion in the gene encoding the protein huntingtin (Htt). HD is inherited in an autosomal-dominant fashion and leads to sleep disturbances (Arnulf et al. 2008) motor dysfunction, cognitive impairment and psychiatric abnormalities (Vonsattel et al. 1998) and ultimately premature death (DiFiglia et al. 1995). The age of onset (typically 30–50 years) and severity correlate with the number of CAG-encoded glutamine repeats in mutant Htt (mHtt) (Snell et al. 1993, Andrew et al. 1993), whereby normal, unaffected individuals have 35 or fewer CAG repeats while affected individuals have more than 36 (Langbehn et al. 2004). Individuals with longer expansions have an earlier onset of disease (Langbehn et al. 2004). Cellular pathologies associated with HD result in the death of striatal medium spiny neurons (MSNs) in the basal ganglia (Augood et al. 1997), the executive center for orchestrating motor memory, learning and function in the brain (Packard et al. 2002). At later stages mHtt also affects other cell types, such as cortical neurons and pyramidal projection neurons (Han et al. 2010). Although the mutation that causes HD is known, no therapies that delay or slow the disease progression currently exist (Mestre et al. 2009).
Htt participates in numerous biological processes, such as regulating apoptosis (Hickey et al. 2003), regulating transcription and enhancing microtubule-based transport or scaffolding of cytoskeletal molecules at synapses (Cattaneo et al. 2005). Hallmarks of mHtt-affected cells include abnormal gene expression (Thomas 2006, Cha 2007), aberrant protein folding, degradation and clearance (Finkbeiner 2011), and the formation of large protein aggregates called inclusion bodies (DiFiglia et al. 1997).
It has been hypothesized that the polyglutamine expansion in mHtt alters its conformation, leading to aggregation and abnormal protein-protein interactions (Miller et al. 2010) and may also alter Htt’s normal function (Kratter et al. 2010). In turn, misfolded mHtt may disrupt cellular homeostasis by overwhelming the cell’s proteostasis machinery by interfering with protein-clearance pathways (Finkbeiner 2011). The accumulation of misfolded forms of mHtt may also stress protein homeostasis pathways so that unrelated proteins cannot fold or function properly, thus compromising cell health. As a consequence, the cell sequesters mHtt through the formation of IBs that help mitigate the stress on the proteostasis network by aggregating mHtt into forms that are more inert than misfolded monomers (Arrasate et al. 2004). In addition, mHtt is known to interact promiscuously with other molecules, and thereby may impede their expression or change their function (Li et al. 2004). For example, mHtt interferes with CREB activation of its gene targets (Sugars et al. 2004). Thus, mHtt disrupts multiple fundamental cellular functions, making it challenging to conceive of simple therapeutic strategies.
As with other neurodegenerative diseases, we have learned a great deal about HD from transgenic animals and rodent primary neuronal cultures. However, as diseased human cells are not easily obtainable, much of the work to understand the molecular insults of mHtt has been done in murine tissues. Despite successful modeling of several aspects of HD in vitro and in vivo, therapies that appeared beneficial in these models have been disappointing in human clinical trials.
Until recently, studies in human tissue have been restricted to immortalized or transformed cells, fixed post-mortem tissues and blood samples. While valuable in some respects, immortalized cells have fundamentally different biological properties than brain cells and fixed tissues are subject to the degradation of DNA and RNA. Blood samples have been valuable for detecting gene-expression changes that can be used as biomarkers of disease progression (Lovrecic et al. 2009, Borovecki et al. 2005, Long et al. 2012) but these changes only recapitulate some key molecular characteristics. Further, the availability of such tissues is limited. The dearth of robust human tissue– based systems derived directly from HD patients has slowed the validation of promising therapeutics.
The discovery of induced pluripotent stem cell (iPSC) technology provides exciting new opportunities to develop in vitro HD models that more accurately reflect the human disease. Cells obtained directly from HD patients can be propagated indefinitely and differentiated into the susceptible neuronal subtypes. Although HD is a late-onset disease, recent evidence suggests that these comparatively young HD iPSCs can accurately recapitulate several aspects of the human disease (An et al. 2012, The HD iPSC 2012). Here, we will examine the practical implementation of iPSCs to study HD, including methods to differentiate embryonic stem cells (ESCs) or iPSCs into MSNs, the cell type most susceptible to the pathogenic effects of mHtt. We will explore the HD-related phenotypes identified in iPSCs and ESCs, and review how brain development and neurogenesis may be altered early, before the onset of HD symptoms. Finally, we will explore the exciting possibility that ESCs or iPSCs might be used as therapeutics to restore or replace dying neurons in HD brains. The development of human HD models using iPSCs could lead to critical advances in our understanding of HD progression at its earliest stages, and in turn, could lead to the development of relevant small-molecule or cell-based therapeutics that delay disease onset and progression.
Modeling Human Disease in Rodents
While transgenic animals and rodent primary neuronal cultures have been valuable systems for studying HD, therapies developed using these models have been disappointing in human clinical trials. Clearly, rodents are not humans, having diverged ~96 million years ago (Nei et al. 2001), and the cumulative effect of small and infrequent base-pair changes over time might affect how a putative small-molecule therapeutic interacts with that cell’s proteome. Thus, an effective drug in rodent cells may not work as well or as specifically in humans. Furthermore, many single amino-acid polymorphisms alter molecular stability and global protein-protein interactions (Ferrer-Costa et al. 2002). For example, two single-nucleotide polymorphisms in APOE4 increase the risk of Alzheimer’s disease and significantly alter APOE structure and protein function (Mahley et al. 2006). Further, these polymorphisms yield different functional consequences of protein toxicity in the human versus the rodent (Vance et al. 2010).
In addition to functional differences between human and rodent proteins, the complexity of the human brain far exceeds that of the rodent at the cellular and systems levels. Thus, there are many examples in which murine models do not recapitulate the physiology or anatomy of the human brain (Dolmetsch et al. 2011). For example, anatomically the human brain is simply bigger and far more complex in shape than that of a mouse. There are significant differences in the development and patterning of the cerebral cortex and subventricular zones (Dolmetsch et al. 2011, Clowry et al. 2010) (Table 1). The human brain contains more glial subtypes and a higher ratio of glia to neurons than rodents (Oberheim et al. 2006) while some human neuronal subtypes appear to be completely lacking in rodents altogether (Table 1).
Table 1.
Fundamental differences between rodents and humans
| Event | Examples | Reference |
|---|---|---|
| Patterning and development of the subventricular zone (SVZ) | The SVZ in humans is larger, develops faster and has structures not observed in rodents. | (Clowry et al., 2010) |
| Patterning and development of the cortex | Cortical structures in the human brain, such as the subplate, develop more connections and become more elaborate compared to other mammals. | (Clowry et al., 2010) |
| Neuronal types | Von Economo neurons have only been found in humans, whales and primates but are absent in mice and rats. | (Nimchinsky et al., 1999, Allman et al., 2011) |
| Diversity of cell types | Astrocytes in the human brain are much more abundant, larger and more complex than in mice. | (Oberheim et al., 2006) |
| Gene expression | Regulation of gene-expression regulation varies widely between mice and humans. There is also a divergence of gene function, alternate splicing events and differences in copy number variants between rodents and humans. | (Ginis et al., 2004, Odom et al., 2007, Gharib and Robinson-Rechavi, 2011) |
| Neurogenesis | In the human brain, inhibitory interneurons are generated in cortical progenitor zones; in mice they are formed outside of the cortex and migrate in. | (Clowry et al., 2010) |
| Immunity | There are fundamental differences in the innate and adaptive immune systems between mice and humans, including differences in the concentration and balance of lymphocytes, neutrophils, B and T cells, cytokines and defensins. | (Mestas and Hughes, 2004) |
| Inflammatory Response | Gene expression changes in response to stresses such as burns, injuries, endotoxins and infection vary drastically between murine models and humans. | (Seok et al., 2013) |
In terms of gene-expression profiles in specific cell types, humans and mice can have global differences in the expression of genes involved in pluripotency, cell-cycle regulation, and apoptosis (Ginis et al. 2004) as well as in innate and adaptive immunity (Mestas et al. 2004) and response to inflammatory stress (Zermeno et al. 2009). While some reports show a high conservation in expression of orthologous genes (Zheng-Bradley et al. 2010) there are many examples of divergence of gene function, alternate splicing events and difference in copy number variants between mice an humans (Gharib et al. 2011)(Table 1). In addition, binding sites for highly conserved transcription factors can vary significantly between rodents and humans (Odom et al. 2007). Thus, it is not surprising that so many drugs fail once they reach clinical trials given the heavy reliance on model systems that are so different from human tissues.
Does the HD mutation affect brain development?
Historically, HD has been thought of as a late-onset disorder, with the initiation of the disease marked by the first appearance of symptoms. However, detectable physiological changes may occur much earlier than previously thought. For example, behavioral disturbances such as depression are often evident and can predate the onset of motor dysfunction, (Julien et al. 2007, Duff et al. 2007), suggesting that there are molecular changes that take place in HD brains long before clinical onset of disease symptoms. The longitudinal study PREDICT HD (Nopoulos et al. 2011), investigated intracranial volume (ICV) in HD carriers, which is a measure of early childhood brain development, (Giedd 2004, Rushton et al. 1995). Male HD carriers who were not yet symptomatic had smaller ICVs than non-expanded controls (Nopoulos et al. 2011). Further, another study found that children with 39 or more CAG repeats had smaller head circumference, weight and body mass index than non-affected children (Lee et al. 2012). Thus, mHtt appears to affect brain development, but the mechanisms by which this occurs are unclear.
In mice, null deletion of Htt results in gastrulation defects and embryonic lethality (Zeitlin et al. 1995, Duyao et al. 1995). shRNA-mediated knockdown of Htt expression in the central nervous system (CNS) of late-stage embryos (i.e., embryonic (E) day 12.5) leads to a near-complete loss of neuronal migration from the ventricular zone (VZ) to the cortical plate (CP), while reducing cellular proliferation and increasing cell death in the developing cortex (Tong et al. 2011). To identify Htt’s specific role in maturing tissues, Hdh−/− (Htt knockout) ESCs were injected into the blastocysts of normal mice. Although many cells survived until adulthood, they were mostly found in the cerebellum and hindbrain, and rarely in brain regions most affected in HD, such as the cortex and basal ganglia (Reiner et al. 2001), suggesting that Htt is important in the maturation and development of these cell types. Interestingly, Htt is also a key player in regulating mitosis and neuronal fate (Godin et al. 2010). During mitosis, Htt localizes to the spindle poles during mitosis and aids in the proper orientation of cell division and localization of microtubule-based motors within cells. Loss of Htt expression results in the loss of cortical progenitors in the developing neocortex of the mouse brain (Godin et al. 2010), which directly ties Htt to neurogenesis (see below). Further, in ES cells subjected to a neuronal differentiation paradigm, loss of Htt expression results in cultures that contain fewer mature neurons, but, interestingly, more glial-like cells, compared to WT ES cultures (Conforti et al. 2013).
How might mHtt alter brain development? Misfolded mHtt could induce developmental abnormalities in the striatum, predisposing those neurons to later degeneration. In one study of Hdh-Q111 mice, which contain 111 polyglutamine repeats, distinct differences during embryonic maturation were observed in the expression and localization of several markers in the striatum, including the phosphoprotein DARPP-32, mGluR1 and the neuronal markers NeuN, β-tubulin and Islet-1 (Molero et al. 2009). Furthermore, striatal progenitors from Hdh-Q111 mice were less likely to mature into specific cell types and more likely to maintain their proliferative potential at later stages of development (Molero et al. 2009). A selective delay in neuronal maturation limited to the striatum could have profound implications for the establishment of brain circuitry and function, which could affect brain processes from a very early stage. Recently, in vivo diffusion kurtosis imaging was used to see how a truncated mHTT fragment (51 CAG repeats) affected development in rat pups over time. This methodology can elucidate the structural components of brain tissue in living samples by tracking the distribution of water diffusion (Basser et al. 1996). The study found significant differences in grey and white matter distribution, suggesting that mHTT causes defects in myelination as a result of developmental abnormalities (Blockx et al. 2012). Thus, mHtt expression might compromise oligodendrocyte function during development and alter neuronal maturation in the developing brain, each of which could profoundly affect brain architecture and function before the point in time classically associated with disease onset.
Normal Striatal Development
Much of what we have learned about the process of differentiation and maturation of the brain comes from studying chick or murine tissues during neural development. This begins with induction of the neural plate during gastrulation that, in turn, gives rise to the neural tube. The neural tube is lined with instructive signals from which delineate the anterior and posterior axes of the animal and instruct the subdivision of the three main structures of the brain, namely the 1) forebrain (prosencephalon), which consists of the telencephalon and the diencephalon, 2) midbrain (mesencephalon) and 3) hindbrain (rhombencephalon) (Rubenstein et al. 1998).
The striatum develops from within the telencephalon. The telencephalon is first made up of two distinct domains, the pallium (dorsal telencephalon) and subpallium (ventral telencephalon), which give rise to cortical structures and the basal ganglia respectively (Jain et al. 2001). The subpallium is comprised of the medial ganglionic eminence (MGE), which is established first, followed by subdivision in to the lateral ganglionic eminence (LGE), and the caudal ganglionic eminence (CGE) (Sousa et al. 2010). Neural cells born in the subventricular zone (SVZ) and ventricular zone (VZ) of the ganglionic eminences subsequently migrate into the developing striatum, cortex and other forebrain regions, where they integrate into neural circuits (Marin et al. 2001). MSNs are derived from the LGE (Olsson et al. 1998). In mice, striatal neurogenesis begins around E12 and peaks around E14-E15 (Fentress et al. 1981). Once fully developed, the adult mouse striatum will contain almost 1,400,000 neurons (Fentress et al. 1981) of which ~95% are MSNs (Gerfen 1992a)
What are the factors that govern MSN development in vitro?
An array of morphogens and transcription factors act within the telencephalon to drive LGE patterning and MSN differentiation. For a more detailed summary on normal striatal development, see (Evans et al. 2012). To direct MSN differentiation from iPSCs and ESCs, some of the key factors are reviewed below (An et al. 2012, The HD iPSC 2012, Aubry et al. 2008).
Brain-Derived Neurotrophic Factor (BDNF)
BDNF is a secreted protein that has long been known for its role in promoting survival, axon guidance, synapse formation and neuronal plasticity (Huang et al. 2001). BDNF binds Trk receptors that, in turn, activate downstream signaling events in the CNS. But until recently, BDNF’s role in the developing striatum was unclear. Exposure to BDNF activates BDNF-dependent signaling pathways in embryonic striatal cultures, suggesting that it may be important for striatal differentiation (Ciccolini et al. 2001). In transgenic mice carrying a CNS-specific conditional knockout of the BDNF gene, striatal neurons were much smaller, had fewer branching dendritic segments and reduced spine density (Rauskolb et al. 2010), suggesting a prominent role for BDNF in striatal neuron maturation. During embryonic development, selective removal of the BDNF receptor TrkB from the LGE led to a massive loss of striatopallidal MSNs and a large increase in apoptosis in the SVZ and VZ, suggesting a requirement for BDNF in cell survival pathways as well (Baydyuk et al. 2011). In adult mice, ectopic expression of BDNF in the VZ significantly increased neurogenesis in the striatum, and in particular DARPP-32-positive MSNs (Benraiss et al. 2001). Thus, BDNF is key to striatal neuron differentiation.
Fibroblast Growth Factors (FGFs)
FGFs are a family of ligands that act through their cognate receptors (FGFRs) to promote cell proliferation and differentiation (Mason 2007). They participate in many stages of patterning, starting with neuronal induction, gastrulation and finally telencephalic specification, where they influence patterning of both the dorsal and ventral regions of the telencephalon, and later, the basal ganglia (Mason 2007, Hoch et al. 2009). During telencephalon formation, FGF signaling induces proliferation, promotes survival, and activates ventral gene expression (Hoch et al. 2009, Shinya et al. 2001). There are five FGFs thought to guide telencephalic formation, some of which probably act redundantly, as loss of a single FGF does not terminate development of the telencephalon. However, simultaneous knockout of Fgfr1, Fgfr2, Fgfr3 in mice results in abnormal gene expression and severe defects in telencephalic development (Paek et al. 2009).
FGF-2, has typically been used to generate neural progenitors from ESCs that can give rise to neurons in vitro (Sanalkumar et al. 2010). However, the use of FGFs to generate neural progenitors is complicated, as there appears to be different instructive roles for different FGFs (Sterneckert et al. 2010). Some studies show that they inhibit differentiation of the neural lineage, whereas others show that they are required for neural patterning, and their use in ESC culture remains controversial (Sterneckert et al. 2010). We use FGF-2 along with EGF to generate and maintain a neural progenitor cell (NPC) population of iPSCs (The HD iPSC 2012).
Sonic Hedgehog Homolog (SHH)
Shh, one of the best-studied CNS morphogens, acts early in development to establish a dorsal-ventral axis in the embryo, and at later stages participates in cell proliferation, axon guidance, neurogenesis and neuronal specification (Sousa and Fishell, 2010). During early patterning, Shh is expressed in the ventral telencephalon and activates Gli transcription factors that are critical for establishing the LGE and MGE (Sousa and Fishell, 2010). Genetic studies revealed that the loss of Shh expression leads to a loss of ventral cell types as well as expression of the LGE markers Dlx2 and Gsh2 (Rallu et al. 2002), whereas ectopic Shh expression in the telencephalon induces the expression of LGE and striatal markers, such as GAD67 and Dlx2 (Kohtz et al. 1998).
This patterning effect persists in culture when differentiating stem cells into forebrain neurons. In ES cultures, inhibition of SHH signaling promotes the differentiation of dorsal telencephalic structures that give rise to cortical neurons (Gaspard et al. 2008, Eiraku et al. 2008, Eiraku et al. 2012), whereas exposure to SHH stimulates the expression of subpallial LGE markers (Ma et al. 2012, Danjo et al. 2011). Addition of the small-molecule agonist purmorphamine to human neural stem cell cultures also significantly increased the percentage of DARPP-32-positive neurons after differentiation (Ma et al. 2012, El-Akabawy et al. 2011). Further, exposure of ESC-derived primitive neuroepithelial cells to SHH triggered ventralization and, in a subset of cells, promoted differentiation into DARPP-32-expressing GABAergic neurons (Li et al. 2009). Nevertheless, the timing of exposure and concentration of Shh seems to be critical for the patterning of cells towards the LGE fate. In studies aimed at defining the optimal Shh exposure conditions that promote LGE differentiation in vitro, it has been shown that exposure of mouse ESCs to exogenous Shh during striatal differentiation is critical, as is the eventual removal of exogenous Shh to induce terminal maturation into GABAergic DARPP-32-positive cells (Danjo et al. 2011).
Wnt Inhibitors
Wnts are a group of secreted palmitoylated signaling proteins that bind to Frizzled receptors which, in turn, signal beta-catenin translocation to the nucleus. Once there, beta-catenin activates the transcription of many target genes involved in cell fate, differentiation, proliferation and cell migration (Logan et al. 2004). In early telencephalic development, Wnts induce the expression of PAX-6 and other genes that stimulate dorsal development and patterning ((Li et al. 2009, Gunhaga et al. 2003) that will give rise to cortical neurons.
Wnts are aquired for neurogenesis and proliferation of the MGE (Gulacsi et al. 2008). During subdivision of the MGE and LGE, inhibiting Wnt signalling in dorsal structures is key to promoting LGE patterning. Specifically, inhibition of Wnt signaling in the pallium results in the ectopic expression of ventrally associated genes such as Gsh2, Mash1, Dlx2 (Backman et al. 2005), whereas inducing Wnt activity in ventral LGE-derived explants can stimulate the formation of dorsal telencephalic structures in vitro (Gunhaga et al. 2003). Finally, inhibiting Wnt activity during the differentiation of ESC cultures seems key to promoting the MSN fate, as exposure to the Wnt inhibitor Dkk-1 along with SHH can induce dorsal progenitor cells to adopt a ventral fate and eventually give rise to GABAergic neurons (Aubry et al. 2008, Li et al. 2009, Watanabe et al. 2005).
Other compounds that induce striatal development include the anticonvulsant and mood-stabilizing drug valproic acid, which when ectopically expressed increases the neurogenesis of several cell types in the brain (Zhang et al. 2010a). Valproic acid was shown to increase the production of GABAergic neurons from cultured primoridal stem cells more than five fold (Laeng et al. 2004). The morphogen retinoic acid (RA) stimulates the expression of Nolz1, a key transcription factor that promotes striatal differentiation in LGE-patterned cells (Urban et al. 2010). Further, RA activates the expression of GAD67, an enzyme involved in GABA neurotransmitter synthesis and GABAergic neuron differentiation in vitro (Chatzi et al. 2011). Many ESC differentiation methods include retinoic acid to increase GABAergic neuronal differentiation (Ma et al. 2012, Shin et al. 2011).
MSN Differentiation in Culture
To successfully model HD in culture, the ability to differentiate iPSCs or ESCs into inhibitory GABAergic MSNs—the most susceptible cell type in HD pathogenesis—along with other affected cell types, including cortical neurons, astrocytes and possibly oligodendrocytes, will be key. Protocols for differentiating these cell types have been described (Shi et al. 2012, Krencik et al. 2011, Czepiel et al. 2011) and are being further refined as iPSC technology becomes more widely used. Protocols to differentiate ESCs into GABAergic neurons generally follow a multi-step process of 1) induction of the neural lineage, 2) regional patterning and differentiation of neuronal progenitor cells and 3) specialization of mature neurons (Aubry et al. 2008, Carri et al. 2013). Differentiated GABAergic neurons may be generated by several methods including the embryoid body (EB), monolayer, five-stage methods (Shin et al. 2011) and a cell-clustering stepwise differentiation (The HD iPSC 2012).
The EB method fosters the formation of aggregates or clusters of cells that recapitulate the 3D structure and intracellular signaling processes that occur during native development to produce all three germ layers (Itskovitz-Eldor et al. 2000). After EB generation, morphogens such as retinoic acid are added to induce neural patterning (Bain et al. 1995), after which cells are plated to induce GABAergic specification (Strubing et al. 1995). Similar to the EB method, the monolayer protocol utilizes patterning morphogens, such as FGF, in defined concentrations and for specific periods of time to induce GABAergic specification (Westmoreland et al. 2001). This methodology is postulated to generate neural cells more efficiently than other methods as the media and conditions are more defined and likely more closely recapitulate neural development (Abranches et al. 2009). Further these cells tend to be less aggregated and more uniform making them easy to characterize. The five-stage method involves EB formation that is followed by dissociation and monolayer formation. This protocol also utilizes instructive cues such as FGF to direct neuronal patterning, followed by FGF deprivation to induce GABAergic specification (Okabe et al. 1996). Other GABAergic differentiation protocols incorporate feeder layers for neuronal induction (Aubry et al. 2008); however, these methods can introduce unknown factors that limit differentiation potential and restrict the resulting neural cell types to a midbrain or hindbrain fate (Pankratz et al. 2007). All of these methods have been used with different iterations of varying morphogens and media.
Although all of these methods produce GABAergic neurons, the first protocol designed to specifically direct the differentiation of ESCs towards GABAergic MSNs was established using ESCs co-cultured with murine MS5 stromal cells for ~3 months (Aubry et al. 2008). The protocol is divided into several stages, beginning with the generation of neural rosettes, then the patterning towards telencephalic progenitors, and a final stage of induction of specialized, mature striatal cells using morphogens known to drive striatal neuron patterning, such as SHH, BDNF, valproic acid and Dkk-1. Cultures differentiated in this way are composed of ~20% mature MAP-2 positive neurons that stain for GABA, calbindin and calretinin, with only about ~10% of these being DARPP-32-positive. Given that the vast majority of striatal neurons are GABAergic MSNs (Gerfen 1992b), the proportion of DARPP-32-positive cells produced with this differentiation method is quite low. Further, high levels of Nestin- and PAX-6-positive cells continue to propagate, even after 3 months in culture, suggesting that many cells in these cultures are still dividing and differentiating. Indeed, when injected into the rat striatum, younger, less differentiated populations of cells never became DARPP-32-positive and instead formed teratomas, whereas cells injected at later stages of maturation produced no teratomas and differentiated into DARPP-32-positive cells in vivo (Aubry et al. 2008). Still, massive overgrowth and proliferation eventually occurred even with the more mature cells, demonstrating that they do not receive, or are unreceptive to, the appropriate signals that induce functional integration in the brain. In another study, a slightly different version of this protocol (that subjected the cells to EB stage), was used to differentiate HD iPSCs and a similar-sized subset (~10%) became DARPP-32-positive (Zhang et al. 2010b).
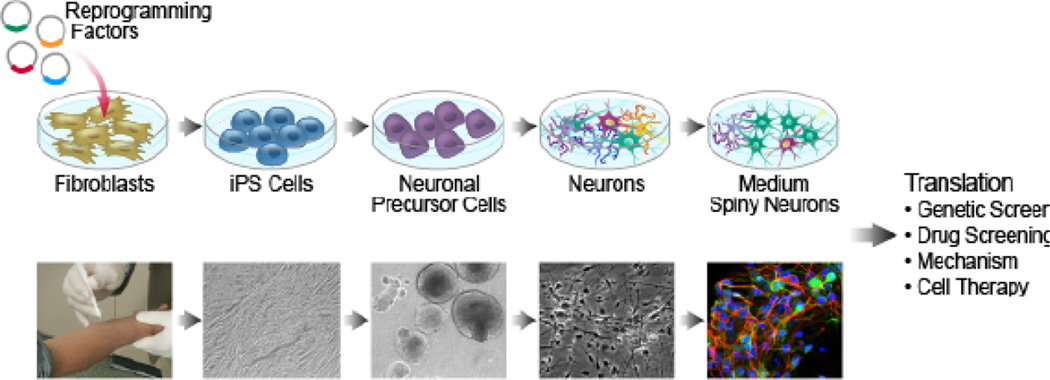
The HD iPS consortium used the protocol developed by Aubry et al. (2008), but did not use a stromal cell line for the neural induction of ES cells; rather, iPSCs were first differentiated into a population of intermediate renewable neural stem cells (NSCs) that were then exposed to striatal patterning morphogens (The HD iPSC 2012) (Figure 1). Differentiating MSNs from NSCs makes the production of striatal neurons much more feasible, as it is less time consuming to maintain NSCs in culture than ESCs/iPSCs. Using this method, the HD iPS consortium reported the generation of cultures containing ~5% DARPP-32-positive neurons after ~2.5 months.
Figure 1.
Modeling HD in culture using IPS cells; from patients to cultured Medium Spiny-Like neurons.
a. Skin fibroblasts are reprogrammed by expressing "reprogramming factors" that covert the differentiated cells into iPS that are capable of becoming any cell type. To make striatal-like cultures, iPS are exposed to growth factors that induce patterning of neural lineage. Addition of growth factors and neurotropins induce development of neurons and eventual maturation of cultures that contain MSNs.
b. Skin cells are safely removed from a patient (far left) and transformed into self renewing iPS cells (micrograph second panel from left). NSCs (middle) are converted into neurons (micrograph second panel from right) and matured into DARPP-32 positive cells (right). Confocal image of fixed and stained iPSs differentiated for 60 days in culture (The Hd Ipse, 2012). Blue= DAPI, Green=DARPP-32, red= MAP-2.
Other methods have also been reported for generating MSNs from ESCs. A monolayer method was used to differentiate mouse ESCs into GABAergic neurons, of which a subset (~30%) became DARPP-32-positive after just three weeks (Shin et al. 2011). Another protocol using hESCs resulted in a significantly increased differentiation efficiency, yielding cultures comprised of 85% DARPP-32-positive cells that were capable of functionally integrating and restoring motor function in the QA-lesion-induced HD model (Ma et al. 2012). While still time consuming, this differentiation protocol is not as lengthy (~1.5 months) as the Aubry et al., (2008) method.
Most recently, a very elegant study relied on the expression of markers found in the developing human brain to gauge the maturation of striatal MSNs from hESCs differentiated using a monolayer method (Carri et al. 2013). Although the protocol is quite long (~2.5 months) and the efficiency of generation of DARPP-32-positive cells is low (~20%), the characterization of the cells generated is by far the most thorough to date. The group performed extensive co-staining experiments to define striatal identity, comprehensive electrophysiology that revealed a subset of cells containing MSN-like properties, and determined that these cells were able to integrate and form synaptic connections upon transplantation into the rat brain (Carri et al. 2013).
Although these different protocols underscore the potential of this technique for generating MSNs, they are fairly time consuming and resource intensive, both of which make the generation of these neurons highly susceptible to failure. These obstacles need to be addressed in order to develop these cells as an effective platform for genetic or drug screening.
What markers do MSNs express once they mature?
To monitor the differentiation success of ESCs or iPSCs into striatal neurons, it is important to define the gene-expression patterns indicative of mature MSNs. For details on the transcription factors that influence LGE patterning and genes that are expressed during MSN development see (Jain et al. 2001, Evans et al. 2012, Carri et al. 2013).
The most commonly used marker of mature MSNs of either type is DARPP-32, which is expressed in ~95% of all MSNs (Ouimet et al. 1984, Ouimet et al. 1998). Other general MSN markers include Rhes, GPR88, GAD-65/67, CALBINDIN and CTIP-2 (Table 2). Expression of many of these genes is downregulated in mouse models of HD as well as in human HD brains (Table 2), but alterations in the expression of these genes in MSNs differentiated from HD iPSCs have not yet been detected (An et al. 2012, The HD iPSC 2012, Zhang et al. 2010b). Antibodies against most of these proteins are available; however, some are better than others. We generally rely on immunohistochemical staining of DARPP-32 and CTIP-2 to assess the differentiation of striatal-like neurons in iPSC cultures (The HD iPSC 2012). Because many of these proteins, such as DARPP-32, calbindin, and CTIP2, are not solely expressed in MSNs but are also found in other neuronal cell types (Ouimet et al. 1984, Liu et al. 1992, Rajput et al. 2009, Arlotta et al. 2008), co-staining of multiple striatal markers is ideal. A recent study examined both the simultaneous expression of CTIP2 with DARPP-32 to better confirm MSN identity (Carri et al. 2013, Rajput et al. 2009). Further, to ascertain that LGE patterning is occurring during differentiation, the examination of proteins expressed in striatal precursors, such as FOXP1 (Carri et al. 2013, Tamura et al. 2004), FOXP2 (Carri et al. 2013, Takahashi et al. 2003), and GSX2 (El-Akabawy et al. 2011, Waclaw et al. 2009) would be optimal.
Table 2.
Genes enriched in Mature MSNs
| Gene | Protein | Expression | Expression Change in HD |
Reference |
|---|---|---|---|---|
| Adora2a | Adenosine A2a receptor | Enriched in striatopallidal MSNs but is also expressed in the olfactory tubule, cortex and hippocampus and peripheral tissues | mHtt reduced expression of the core promoter of the A2a receptor in primary striatal neurons invitro | (Lobo et al. 2006, Heiman et al. 2008, Chiang et al. 2005, Blum et al. 2003; Wei et al., 2011) |
| CALB | Calbindin-D28k, Calcium Binding Protein | Enriched in spiny neurons of the striatal matrix but also widely expressed in the brain and nervous system and peripheral tissues | Expression increases in both HD mouse models and HD brains | (Ferrante et al. 1991, Sun et al. 2005, Huang et al. 1995, Pickel et al. 1996; Schwaller et al., 2002) |
| CTIP-2 | COUP TF1–interacting factor 2 | Expressed in early postmitotic migratory MSNs during development; expression increases and is maintained in the adult. Also expressed in cortex, hippocampus and olfactory bulb | Expression decreases in mouse models of HD | (Arlotta et al. 2008, Desplats et al. 2008) |
| DARPP-32 | Dopamine and adenosine 3, 5-cyclic monophosphate (cAMP)-regulated phosphoprotein, 32 kDa | Highest in mature, post mitotic MSNs, but also expressed in other cell types in the cerebellum, cortex. In addition, some expression is found in glial cells | Expression decreases in HD mouse models | (Ouimet et al. 1984, Ouimet et al. 1998, Bibb et al. 2000, Luthi-Carter et al. 2000) |
| DRD1 | Dopamine receptor 1 | Highly enriched in striatonigral MSNs but expressed in other cell types in the cortex and basal ganglia | Expression decreases in HD brains | (Augood et al. 1997, Gerfen et al. 1990, Lobo et al. 2006, Heiman et al. 2008, Meador-Woodruff et al. 1996, Hodges et al. 2006) |
| DRD2 | Dopamine receptor 2 | Highly enriched in striatopallidal MSNs but also expressed in other parts of the basal ganglia | Expression decreases in HD brains | (Augood et al. 1997, Gerfen et al. 1990, Lobo et al. 2006, Heiman et al. 2008, Meador-Woodruff et al. 1996) |
| Enkephalin (Penk) | Neuropeptide | Highly enriched in striatopallidal MSNs | Expression decreases in mouse models of HD | (Lobo et al. 2006, Heiman et al. 2008, Hodges et al. 2006, Menalled et al. 2000) |
| GAD65/67 | Glutamic acid decarboxylase | Enriched in Striatal neurons of the basal ganglia but expresses in all GABAergic neurons | Expression does not change in the striatum of mouse models of HD but is altered in other parts of the brain | (Menalled et al. 2000, Pickel et al. 1980, Mercugliano et al. 1992, Gourfinkel-An et al. 2003) |
| GPR6 | G protein–coupled receptor 6 | Enriched in striatopallidal MSNs | Expression decreases in HD brains | (Hodges et al. 2006, Lobo et al. 2007) |
| GPCR88 | G protein–coupled receptor 88 | Enriched in the caudate and putamen but also expressed in the olfactory tubercule, nucleus accumbens and inferior olive nucleus | Expression decreases in HD brains | (Hodges et al. 2006, Mizushima et al. 2000, Van Waes et al. 2011) |
| Rhes | Rhes homolog enriched in the striatum | Enriched in adult striatum but also expressed in many other parts of the brain including the cortex, hippocampus and cerebellum | Unknown | (Vargiu et al. 2001) |
Mature MSNs are functionally classified as either striatopallidal or striatonigral MSNs, which make up the direct or indirect pathways of the striatum, respectively (Gerfen et al. 1990). There is evidence that there is a differential susceptibility of these two MSN populations to the effects of mHtt, whereby striatopallidal neurons are lost first while striatonigral neurons are spared until later in the disease course (Albin et al. 1992). Many genes are selectively enriched in one of these two neuronal populations (Lobo et al. 2006, Heiman et al. 2008) (Table 2). It will be intriguing to probe this potential differential susceptibility using neuronal cultures derived from HD versus WT iPSCs.
Does the HD mutation affect neurogenesis?
Changes in the architecture of synapses and circuitry during neurogenesis could predispose the HD-affected brain to cell dyshomeostasis and eventual death, so a thorough understanding of whether mHtt affects these processes is critical for the development of HD iPS cell models. There is increasing evidence from both murine and human studies that neurogenesis is altered in both maturing and adult HD brains. However, there seems to be a discrepancy between the sorts of changes that occur in humans versus mice.
Among the first rodent models of HD were the R6/1 and R6/2 transgenic mouse lines. R6/1 mice express a variant of human mHtt that contains ~115 repeats within exon 1, and show HD-like symptoms around 15–21 weeks of age (Mangiarini et al. 1996). R6/2 mice express the same mHtt fragment but with 28 more CAG repeats, which results in a more severe phenotype (Mangiarini et al. 1996). Neurosphere assays, which show how many neural stem cells (NSCs) are present in tissues, revealed that the number of hippocampal precursor stem cells was significantly more in R6/1 mice compared to WT mice, while no change in the number of SVZ stem cells was observed between WT and R6/1 mice (Walker et al. 2011). However, curiously in vivo bromodeoxyuridine (BRDU) labeling, revealed that neurogenesis in the hippocampus was significantly decreased compared to WT animals suggesting that although cellular proliferation is altered, a latent stem cell population in the hippocampus is maintained as a response to mHtt (Walker et al. 2011). The same reduction of cell proliferation in the hippocampus of R6/1 was observed by another independent study (Lazic et al. 2004).
Another group studying cell proliferation in the hippocampus of R6/2 mice found that as early as 3.5 weeks of age, R6/2 mice had fewer proliferating cells in the dentate gyrus compared to WT animals. Most of the proliferating cells stained for NeuN, confirming that they were neuronal precursors (Gil et al. 2005). The reduction of proliferating cells resulted in significantly fewer neuroblasts in the dentate gyrus at ten weeks. However, similar to the study by Walker and colleagues, there were no differences in the number of proliferating cells in the SVZ in R6/2 or WT mice (Gil et al. 2005). Likewise, direct comparison of proliferating BRDU-positive and differentiating DCX-positive cells in the R6/2 showed similar numbers of newly born cells in the SVZ (Phillips et al. 2005).
In mice, mHtt may also affect neurogenesis in parts of the brain other than the SVZ and hippocampus. For example, impaired neurogenesis was observed in the piriform cortex, which is required for odor perception and olfactory memory (Lazic et al., 2007), in both R6/2 (Phillips et al. 2006) and R6/1 mice (Lazic et al. 2007). This finding is consistent with the observation that HD patients have defects in olfactory discrimination (Lazic et al. 2007, Nordin et al. 1995).
In contrast, a study looking at neurogenesis in the forebrain subependymal layers in the SVZ of R6/2 mice contradict the preceding studies but are consistent with observations in human tissues. They showed a significant increase in the number of proliferating NSCs in the neurosphere assay compared to WT animals, which directly corresponded to the onset of symptoms. No neurosphere differences were observed at 4 weeks of age, when symptoms are subtle, but by 8 weeks—at about the time when motor symptoms start to become apparent—these changes became significant (Batista et al., 2006). Interestingly, NSCs that normally migrate to the olfactory bulb were redirected to the striatum (Batista et al. 2006), suggesting that the striatum can recruit proliferating cells, possibly in reaction to the cellular insults caused by mHtt. In agreement with this, after injection of quinolinic acid–(QA)–a potent neurotoxic substance that creates lesions in the brain to model HD—neuroblast proliferation increased and the migration of these cells appeared to be redirected to the striatum from the SVZ (Tattersfield et al. 2004).
Indeed, studies in human tissues suggest proliferative rather than inhibitory changes in neurogenesis in response to mHtt expression. Before the HTT gene was mapped, Ferrante et al. (1991) examined the subcellular changes in postmortem HD brains at different disease stages. Single-section Golgi staining of the striatum of HD patients revealed two surprising morphological changes compared to controls: (1) a substantial increase in spine density in a subset of dendrites from “Grade 2”–stage HD MSNs, and (2) an increase in the size of growth-cone-like processes with filipodia on the tips of distal dendrites, suggesting proliferation rather than degeneration in neurons from HD brains (Ferrante et al. 1991). Although, in the more severe grades of HD, the majority of structural changes observed in MSNs were those associated with degeneration such as spine loss, abnormal recurving of distal dendritic segments and aberrant branching of dendrites (Ferrante et al. 1991).
The first study to show that neurogenesis takes place in the brain as a direct consequence of cell death was in postmortem brain tissue from HD patients. Curtis et al. (2003) observed a significant increase in cell proliferation in the subependymal layer (SEL) or (SVZ) adjacent to the caudate in HD brains compared to unaffected controls (Curtis et al. 2003), and this increase was positively correlated with the number of CAG repeats. Further, these proliferating cells gave rise to both astrocytes and newborn neurons. The extent of neurogenesis was highly significant, such that HD brains exhibited a 2.6-fold increase in the number of newborn neurons in the SVZ compared to controls (Curtis et al. 2005). Interestingly, a study examined the number of proliferating cells in the hippocampal subgranular zone (SGZ) of HD human postmortem brains, and found no differences in cellular proliferation compared to controls. (Low et al. 2011). This contrasts with what has been observed in murine studies of hippocampal neurogenesis in the presence of mHtt.
In summary, the majority of murine-based HD models report alterations of neurogenesis in the hippocampus while SVZ neurogenesis is spared. Whereas, in the brains of humans with HD, changes in neurogenesis are found in the SVZ but not in the hippocampus. The reason for these discrepancies between murine and human neurogenesis in the context of HD is unclear. One possibility is that because symptoms of disease appear only after decades of exposure to endogenous levels of mHtt in humans, whereas transgenic HD models experience a much shorter time course of disease progression and typically have higher levels of transgenic mHtt, different signaling pathways and coping mechanisms are triggered. A finding consistent with this possibility was observed in the knock-in Hdh-Q111 model, in which mHtt expression is under the control of the endogenous promoter. In these mice, neurogenesis within the striatal SVZ was shown to be lower in young animals (E11.5), but at later stages (E15.5) became higher compared to WT (Molero et al. 2009), suggesting that rodent mHtt-expressing models may better recapitulate the human disease state when mHtt is expressed at near-physiological levels.
However, another study suggests that this explanation still falls short. Using a mouse model in which full-length Htt carrying 128 CAG repeats is expressed under the control of the human promoter (YAC-128 model), leading to reduced mHtt expression compared to R6 mice, YAC-128 mice exhibited a marked reduction in neural precursor proliferation and differentiation in the hippocampus but no changes in the SVZ were observed (Simpson et al. 2011).
The differences observed may alternatively be due to CAG repeat length, as most rodent models have upwards of ~100 CAG repeats, whereas in humans CAG repeats generally range from 40–50. It would be of interest to look at neurogenesis in juvenile cases of HD where extreme CAG repeat lengths are observed. Finally, it is likely that the lack of access to early stage (i.e., young to mid-adult) post-mortem human tissue makes resolving these discrepancies difficult, given that increases in human neurogenesis may take place only very late in the disease course, and therefore, a comparable time frame may be challenging to witness in mice. Regardless, these discrepancies illustrate the challenges with using rodents to model human disease, which in our view may be circumvented by the development of in vitro models that rely on human stem cells from HD patients.
STEM CELLS and HD
ESCs are self-renewing pluripotent cells derived from embryos that are capable of giving rise to all cell types from the three germ layers. They have been used extensively to model development, differentiation and some disease states in vitro (Ben-David et al. 2012). The first stem cells made from HD patients were part of an effort to make ESC lines from individuals with different genetic abnormalities (Verlinsky et al. 2005). Since then, other HD lines have been made from donated embryos containing 40–51 repeats (Niclis et al. 2009, Bradley et al. 2011, Mateizel et al. 2006). One line was generated from blastocysts that were fertilized in vitro and propagated on mouse feeder layers (Mateizel et al. 2006), and since then several of these lines have been successfully differentiated into neurons (Niclis et al. 2009, Bradley et al. 2011), confirming the potential of stem cells to generate neurons carrying mHtt. Importantly, some of the HD ESC lines retain stable CAG repeats over multiple passages and following neuronal differentiation (Seriola et al. 2011). Although many human lines have been generated to date, no HD-related phenotypes have been reported with these human ESC lines.
Murine ESC-based models can reveal HD-related phenotypes. ESCs generated and propagated in vitro from mice with either 150 or 77 CAG repeats (Hdh CAG150 and Hdh CAG77, respectively), showed similar neural proliferative properties to human SVZ cells in HD brains (Curtis et al. 2003, Curtis et al. 2005). Specifically, at the ESC stage, the HD cell lines showed no differences in cell-cycle progression compared to cells from a WT Hdh CAG7 line. However, following neuronal differentiation, more cells became positive for Sox-3 and β-tubulin in the Hdh CAG150 and CAG77 lines than in WT cells, suggesting that the HD lines transitioned faster to the neural progenitor and neuron stages (Lorincz et al. 2009). NSCs isolated from the SVZ of mice at postnatal week 20 exhibited similar differentiation properties, indicating that these phenotypes were not specific to ESCs. After subjecting the ES cells to only ~1 week of a neuronal differentiation paradigm, increased cell death was observed in the CAG150 line at suggesting that the polyglutamine expansion becomes toxic as soon as the cells take on a neuronal fate. Finally, transcriptional profiling revealed many gene-expression changes in pathways regulating axonal growth and neuronal differentiation, suggesting that the increase in neurogenesis observed in mutant cells may be a result of mHtt-induced transcriptional dysregulation. As changes in gene expression are a hallmark of HD in human brains (Cha 2007), these findings demonstrate that ESCs can recapitulate key aspects of the disease.
In another study, a mES cell line carrying 150 CAG repeats in HTT (HdhQ150) was used to confirm the activity of a compound identified in a screen for molecules capable of enhancing mHtt degradation. This compound induced the expression of heat-shock protein 70 (HSP70) and reduced mHtt expression in HdhQ150 ESCs differentiated into neurons (Baldo et al. 2012).
Recently, Jacobsen et al. created a series of isogenic mouse ES–cell lines carrying 7– 111 CAG repeats, as well as a line in which Htt was completely knocked out (Jacobsen et al. 2011). With these lines, they uncovered gene-expression changes attributable specifically to CAG-repeat size and not loss of Htt function. In doing so, they were able to parse out how the of loss of Htt expression versus varying CAG-repeat lengths alter biological pathways in distinct or overlapping ways (Jacobsen et al. 2011). This allelic series was subsequently used to investigate the effects of mHtt during neuronal differentiation (Conforti et al. 2013). Changes in cell death, measured by caspase activation and transcription, were observed in mHtt cells compared to WT, although no relationship unique to CAG length was detected. However, both high CAG-repeat length and loss of Htt expression resulted in similarly decreased proportion of mature neurons, where a loss of mHtt resulted exclusively in increased numbers of glial cells (Conforti et al. 2013). This allelic series will be useful for exploring other CAG-dependent phenotypes, and efforts are underway to establish similar isogenic lines in human iPSCs (unpublished data).
Although still a relatively new undertaking, these results suggest that ESCs can be used to explore the effects of mHtt in cells that are renewable and fully expandable, which could be extremely valuable for drug screening efforts. Given that ESCs from a single genetic background can be differentiated into multiple cell types, we can tease out the mechanisms by which mHtt exerts its cell-type-selective effects in a controlled genetic background. Further, ESC-based models will to allow for the study of mHtt’s effects on cells during early differentiation and development, which may reveal novel insight into HD pathogenesis.
iPSCs and HD
The revolutionary discovery by Yamanaka et al., (2006) that fibroblasts could be reprogrammed into self-renewing pluripotent cells displaying many of the properties of ESCs has transformed our ability to study disease in human cells. The reprogramming of fibroblasts into iPSCs depends on the expression of 4–6 factors that silence genes involved in cell differentiation while activating genes that promote pluripotency (Takahashi et al. 2007, Yu et al. 2007). Like human ESCs (hESCs), iPSCs can self-renew for long periods of time and differentiate into nearly all cell lineages (Takahashi et al. 2007, Yu et al. 2007).
Importantly, iPSC technology enables models to be created of both genetically inherited and sporadic diseases in vitro (Ebert et al. 2009, Park et al. 2008a). Since iPSCs can be derived directly from patients with a given disease, they carry all of the associated genetic components that make certain cell types susceptible to a disease state, thereby representing the most genetically precise model of a disease (Figure 1). Despite the promise of this technology for modeling complex neurodegenerative diseases like HD, several considerations must be addressed. The first is: can we observe disease-relevant phenotypic differences in patient-derived iPSCs compared with healthy controls? There are many limitations of cell culture that might interfere with this possibility. Cells growing in a dish lack the complexity of neural cells that have developed in vivo. The complex 3D environment of the brain and the plethora of intrinsic and extrinsic cues may be important to fully understand how the insults of mHtt alter cellular function. Second, there are many other cell types within the brain other than neurons that may contribute to the neurodegenerative disease process. The stoichiometry and concentration of these different cell types may be important for detecting HD-related pathology and these features may be hard to recapitulate in culture. Finally, a third important consideration is the role of aging in neurodegeneration. Since the symptoms of HD typically emerge in late adulthood, years rather than weeks of mHtt expression may be necessary to fully recapitulate disease. It is possible that the effects of mHtt may immediately disrupt cellular function, but an organism’s ability to deal with the insults of mHtt may be offset more readily in young versus aged cells; in this way, symptoms can be staved off until late age. Thus, how easily will we be able to capture the phenotypes associated with the initiation of late-onset disease symptoms in a short-term culture? So far, the reports of using iPSCs been generated from patients with various neurodegenerative disorders indeed display robust phenotypes that replicate many features of their respective diseases (An et al. 2012, The HD iPSC 2012, Ebert et al. 2009, Park et al. 2008b, Marchetto et al. 2010, Lee et al. 2009, Egawa et al. 2012, Brennand et al. 2011, Bilican et al. 2012). We will review the progress made to model HD using this exciting new technology.
Fibroblasts from HD patients have shown HD-related phenotypes, such as alterations in proteasome activity (Seo et al. 2004) and altered gene-expression profiles (Valenza et al. 2005), that conceivably could hinder the successful generation of iPSCs. However, proof of principle that iPSCs can successfully be made from individuals with severe neurodegenerative disorders has been demonstrated, including pluripotent HD lines that exhibit no apparent difference from control iPSCs (Park et al. 2008a). It was not until these HD iPSCs were subjected to a neuronal differentiation paradigm that differences between WT and HD iPSCs emerged (An et al. 2012, Jeon et al. 2012, Chae et al. 2012). Proteomic analysis revealed hundreds of distinct changes in protein expression in the HD iPSCs subjected to a short neuronal induction protocol (Chae et al. 2012). Further, neuronal differentiation efficiency and neurite length was reduced in HD iPSCs (Chae et al. 2012).
An independent group developed iPSC-based models from the HD lines originally developed by Daley and colleagues (Park et al. 2008b). These lines were subjected to a modified differentiation protocol to generate MSNs (Aubry et al. 2008) that express DARPP-32 (Zhang et al. 2010b). As mHtt specifically affects striatal neurons, differentiated HD cultures might be expected to contain fewer DARPP-32-positive neurons if these cells are more susceptible than other cell types in culture. Although their analysis was limited by technical concerns (i.e., formation of dense cell clusters, making it difficult to identify individually labeled cells), no difference in the percentage of DARPP-32-positive neurons could be detected between HD and WT cells. We use a slightly different version of the protocol by Aubry et al. (2008) to generate striatal neurons (The HD iPSC 2012), and observed that the cells cluster into tightly bound 3D structures rather than a monolayer of neurons. Indeed, these structures make it quite difficult to accurately estimate the percentage of DARPP-32-positive cells within defined areas of each well (The HD iPSC 2012).
However, Ellerby and colleagues (An et al. 2012, Zhang et al. 2010b), found other HD-related phenotypes in the lines made by Daley and colleagues (Park et al. 2008a). Removing growth factors at an early NSC stage resulted in more caspase 3/7 activity in the HD compared to WT Daley iPSCs, suggesting that cells with mHtt are more susceptible to stress even at a very early stage of differentiation (Zhang et al. 2010b). In a subsequent study, additional HD- related phenotypes were reported and the CAG expansion of the Daley HD-iPSC line was corrected to WT lengths through homologous recombination. Restoration of the CAG repeat to WT length was able to restore distinct transcriptional changes in the E-cadherin and TGF-beta signaling pathways to WT levels, prevented the cell death, altered BDNF transcription, mitochondrial dysfunction and elevated caspase 3/7 activity observed in the HD-iPSC lines (An et al. 2012), suggesting these alterations were directly induced by CAG expansion and were not caused by inter-cell-line variability.
Other HD iPSCs have been generated and examined, including lines generated from two individuals homozygous for mHtt (repeat sizes between 39–44 CAGs) and a heterozygote with 44 repeats (Camnasio et al. 2012). Surprisingly, this group reported few differences between these HD and WT lines, as neuronal differentiation, growth rate and caspase activation were indistinguishable. However, lysosomal activity was higher in HD iPSCs, suggesting that cellular coping mechanisms, including the protein-clearance machinery, may be abnormal in HD.
Scientists from the HD and iPSC communities have united to form a consortium under the auspices of the National Institutes of Neurological Disorders and Stroke (NINDS). The so-called HD iPS consortium has generated a large panel of HD iPSC lines and HD fibroblasts that are being reprogrammed into iPSCs, and made these lines available to the public through the Coriell Institute for Medical Research, which maintains a cell repository (http://ccr.coriell.org/) (The HD iPSC 2012). This collaborative effort discovered numerous differences between HD and WT iPSCs, and most importantly, uncovered HD-related phenotypes in several HD iPSC lines. Whole-transcript expression profiling and analysis of genes involved in signaling, neuronal development and axon guidance confirmed previously described aberrant changes in HD tissues compared to WT, but new and interesting changes were also observed that were CAG expansion-dependent. Further, decreased ATP/ADP levels were detected in the HD-derived NSCs, revealing that changes in energy metabolism may be an early component of HD. The group further differentiated the NSCs to produce mature neurons, and found alterations in their ability to fire both spontaneous and evoked action potentials. In cultures differentiated for longer periods of time to yield DARPP-32-positive neurons, HD-derived lines exhibited properties seen in cells harboring disease-associated CAG expansions, including increased cell death, increased susceptibility to cell stressors such as glutamate and trophic factor removal, and calcium dyshomeostasis (The HD iPSC 2012). These findings reveal that HD iPSCs can exhibit many of the biological properties found in the human HD brain.
These previous studies report phenotypes in cells differentiated towards a neuronal lineage. However, other cell types are affected by mHtt expression, such as astrocytes (Bradford et al. 2010). iPSCs made from a juvenile onset (109 repeats) and an adult-onset (50 CAG repeats) HD patient and differentiated towards an astrocytic lineage displayed elevated cytoplasmic vacuolation that increased over time (Juopperi et al. 2012). Differentiating HD iPSCs into different cellular lineages may reveal novel insights into the cell-specific affects of mHtt.
One of the most widely documented phenotypes in human HD brains and HD mouse models is the formation of mHtt aggregates (DiFiglia et al. 1997, Arrasate et al. 2012). However, in human HD iPSCs, the spontaneous formation of aggregates has not been observed. Recently, the addition of a stressor, the proteasome inhibitor MG132, was shown to induce aggregation in iPSCs containing 72 repeats, as revealed by immunostaining using the mHtt-specific antibody EM48 (Gutekunst et al. 1999) (Jeon et al. 2012). It may be that the effects of endogenous levels of mHtt—or the exposure time to mHtt during typical iPSC culture—are not sufficient to disrupt cell proteostasis and promote aggregation in the absence of stress. Interestingly, the same study observed EM48-positive aggregates after transplantation of HD iPSCs into mouse brains after ~9 months in vivo (Jeon et al. 2012), suggesting that cellular age may be a key factor in identifying aggregates in human HD iPSCs. Curiously, aggregates in iPS cells generated in HD models from other species have been observed in culture. For example, iPSC lines have been generated from skin cells obtained from transgenic rhesus monkeys expressing Htt exon 1 (Chan et al. 2010) and from R6/2 mouse tissues (Castiglioni et al. 2011). Interestingly, in both models, mHtt-specific aggregates were not detectable in pluripotent cells, but were seen only after the cells differentiated into neurons.
Finally, other phenotypes in iPS cells from mouse models have been observed. In the R6/2 iPSC lines, transcriptional dysregulation—a hallmark of HD—was observed for genes involved in the cholesterol and lysosomal pathways (Castiglioni et al. 2011). Curiously, the expression of several genes previously linked to HD pathogenesis, such as BDNF, were unaffected in these cells (Castiglioni et al. 2011, Zuccato et al. 2001, Zuccato et al. 2005).
Applications for Elucidating Mechanism and Drug Discovery
iPSC technology allows cells obtained directly from patients with HD to be propagated indefinitely and differentiate into the susceptible neuronal subtypes (An et al. 2012, The HD iPSC 2012), offering the promise of discovering unique, human-specific effects of polyglutamine-expanded Htt that cannot be revealed by rodent models. A human model may eventually reveal how mHtt affects the molecular changes that lead to alterations in behavioral, cognitive, and motor impairments characteristic of patients with HD. iPSCs carrying mHtt may also form the basis for improved high-throughput drug screening, bioinformatics, and global gene-expression analyses. They have been proposed as potentially powerful diagnostic tools for patient stratification in clinical trials and personalized medicine approaches. For example, a potential therapy might be tested first in iPSCs from a patient to determine whether they respond to the compound and should therefore be included in a clinical trial. If the therapy becomes an approved drug, the same approach could be used to determine who should receive it, thereby avoiding potentially harmful side effects in patients who don’t stand to benefit.
The next challenge will be to employ HD iPSCs to screen for drugs that attenuate mHtt-induced pathology. Exposure to IGF-1 and gentamicin has been shown to restore normal glutamatergic synapse numbers in iPSCs derived from Rett’s syndrome patients (Marchetto et al. 2010), and high-dose BDNF can increase cell-survival rates in HD iPSCs (The HD iPSC 2012). Altered RNA metabolism, a hallmark of ALS iPSCs, can be attenuated by exposure to the transcriptional inhibitor anacardiac acid in differentiated motor neurons generated from ALS patients (Egawa et al. 2012). Exposure of iPSCs from patients with familial dysautonomia (FD) to kinetin led to reduced expression of the aberrantly spliced form of the IκB kinase complex–associated protein (Lee et al. 2009). These studies suggest that iPSCs can be used to find small molecules that are active in primary human cells and can reverse phenotypes relevant to a given disease process.
The critical unanswered question is whether drugs discovered this way will be more successful in clinical trials than the preclinical model systems that have been used previously. Importantly, the next steps en route to developing iPSCs as a screening platform will necessarily involve establishing the best methodologies to differentiate large numbers of cells with minimal variability, and to devise sensitive, high-throughput approaches to detect relevant phenotypic changes. iPSCs may be well-suited to identify reversible phenotypes that appear early in the course of a disease and at a time when pharmacological intervention might be feasible and effective, long before their devastating late-stage consequences take hold.
The Next Steps: Early Phenotypes in HD iPSCs
For future efforts involving iPS cell–based systems to model HD, it may be useful to focus on changes in HD iPSCs that occur in patients well before cell death or the onset of symptoms. For example, alterations in gene transcription are present early on before symptoms of HD begin (Cha 2007), and some are thought to predate symptoms. Postmortem in situ hybridization studies of presymptomatic or early-HD human tissue helped to establish the Vonsattel grading system, in which grade 0 denotes no detectable abnormality, grade 1 suggests only microscopic changes such as gliosis of the caudate and putamen, while grade 3 indicates more severe neuropathology (Vonsattel et al. 1998). At early stages, there was a significant reduction in the expression levels of enkephalin and substance-P mRNAs. Similarly, D1 and D2 receptor expression was significantly decreased in early-stage HD tissue without degeneration or apparent cell loss (Augood et al. 1997). In a comprehensive gene-expression study, principal-component analysis of microarray data from blood samples of both early and late presymptomatic individuals revealed distinct gene- and HD-specific clustering patterns, suggesting that significant transcriptional changes were present early on. Further, the group identified a cohort of 12 biomarker genes that could distinguish the stages of disease progression from very early to advanced HD (Borovecki et al. 2005).
In R6/2 mice, postnatal MSNs had abnormally large NMDA currents and current densities just two weeks after birth, suggesting the presence of aberrant synaptic function early in development (Starling et al. 2005) that could lead to excitotoxicity, mitochondrial damage and cell death (Bano et al. 2011). There is also evidence for bioenergetic changes caused by mHtt before HD symptoms start. For example, asymptomatic R6/2 mice as young as 4 weeks old have been shown to display increased levels of phosphocreatine and creatine that precede a decrease in ATP levels in the striatum and cortex (Mochel et al. 2012). Interestingly, decreased glucose uptake was observed in the cortex and striatum of presymptomatic HD patients, suggesting that altered metabolic changes take place in the brain long before symptoms begin (Ciarmiello et al. 2006). This study also reported a substantial loss in the volume of white-matter in the brains of presymptomatic HD carriers compared to controls (Ciarmiello et al. 2006) supporting the idea that cell loss and alterations in brain structure are occurring well before disease onset. Other reports revealed changes in cellular trafficking as well as significantly altered staining patterns for dynamin and pascin1 in the cortex of presymptomatic HD brains, suggesting that axonal transport is impeded (DiProspero et al. 2004). This already long list of molecular changes is growing. Uncovering such phenotypes in iPSCs, and adapting these cells for use in assays to test therapeutics, may be key to finding drugs that can offset HD at its earliest stages.
In addition, it may be that improved methods to generate bona fide MSNs will be needed to identify phenotypes associated with the early-disease stage of HD. Thus far, the investigation of HD-related phenotypes in human HD iPS cells have used methodologies that produce DARPP-32-positive cells or cell types which exhibit action potentials with features characteristic of immature cells (The HD iPSC 2012), and it is not clear how similar these iPSC-derived cells are to native MSNs in the human brain. Investigations of HD phenotypes should be performed in cultures where comparison studies of human fetal or newborn MSN phenotypes and their electrophysiological properties were used to gauge the extent of development and maturation of iPSC-derived MSNs, as recently reported in ES cells (Carri et al. 2013).
Stem Cells as Therapeutics: Transplantation
iPSCs and ESCs are not just invaluable tools for modeling HD in vitro, but also hold promise as cell-replacement therapies for treating HD. Some of the first human cell-implantation studies used fetal neural tissues that were grafted into the striatum of HD patients (Sramka et al. 1992, Bachoud-Levi et al. 2000). These efforts showed modest results: about half of the patients showed some clinical improvement for ~2 years post-surgery but deterioration resumed thereafter (Bachoud-Levi et al. 2006). Although, the grafts seemed to integrate, they did not protect the surrounding tissues from degeneration (Strubing et al. 1995). Although, fetal striatal neurons have proved to be the most efficacious cell type for use in transplantation studies (Evans et al. 2012) the most intractable problem associated with fetal stem cells is their source: in addition to the controversy that surrounds the use of brain tissues from aborted fetuses, the availability of high-quality material is limited (Evans et al. 2012, Dunnett et al. 2007). Thus, a more reliable source of pluripotent cells will be required to replace dying or damaged striatal tissues in humans.
Studies in rodent HD models have yielded modest but promising results using NPCs or ESCs (Dunnett et al. 2007, Kim et al. 2008) that although are less mature than fetal striatal tissues are more easily obtainable. Adult NPCs isolated from the SVZ and injected into QA-lesioned rats not only survive but can migrate and successfully differentiate in the striatum, and most importantly, can significantly improve motor impairment (Vazey et al. 2006). A similar study was performed using hESCs differentiated into NPCs and injected into the striatum of QA-lesioned animals. Like NPCs, these hESCs differentiated into mature cell types that rescued behavioral deficits (Song et al. 2007). However, some of the most exciting studies have employed stem cells genetically engineered to deliver trophic factors to support the survival of existing cells and to induce new cell growth. An impressive study engineered mouse NPCs (mNPCs) to express glial-derived neurotrophic factor (GDNF). When these GDNF-expressing mNPCs were injected into the striatum of presymptomatic mice containing 82 polyglutamine repeats (N171-82Q), the GDNF-producing cells significantly reduced weight gain, latency to fall in the rotarod test and neurodegeneration within the striatum (Ebert et al. 2010).
Injecting ESCs directly into patients could be potentially harmful since these cells have the ability to self-renew, proliferate, migrate and become tumorigenic (Kulbatski 2010). It is well-established that ESCs can also accumulate chromosomal abnormalities in culture over time, raising the risk of eventual transformation (Ross et al. 2011). Further, a common measure of pluripotency of an ESC or iPSC line is the formation of teratomas after injection into immunocompromised mice (Takahashi et al. 2007, Ross et al. 2011). This potential for unregulated growth must be addressed prior to their application as cell-based therapies in humans.
One way to circumvent this is to differentiate ESCs and iPSCs into a mature non-dividing state prior to transplantation. Noggin-treated hESCs injected into the QA-lesioned rat striatum were found to migrate, mature into MAP-2 positive cells and survive for up to 8 weeks post-transplantation, but did not form tumors; in contrast, noggin-untreated hESCs were hyperplasic and migrated outside of the transplantation site (Vazey et al. 2010). However, a different study revealed that ESCs, which were differentiated into mature DARPP-32-positive cells, eventually formed highly proliferative teratomas, but only 3–5 months after injection (Aubry et al. 2008). Thus, following the graft site throughout the life of injected animals is crucial to determine the long-term consequences of stem cell–based therapies. A recent study used a different striatal protocol to generate DARPP-32-positive GABAergic neurons that were then transplanted into the striatum of QA-lesioned mice (Ma et al. 2012). The grafts did not overgrow or form tumors and rescued motor deficits in the animals; however, it was unclear how long the grafts remained intact after transplantation. Although ESCs hold great promise as clinical HD treatments, better differentiation protocols are needed that will select for the cell type least likely to exhibit hyperplastic overgrowth and tumorigenesis, while relieving the symptoms of disease.
Another promising cell type is the human multipotent stromal cell/mesenchymal stem cell (MSC) derived from bone marrow, thought to promote repair by releasing trophic cytokines and chemokines. Injection of hMSCs into a mouse model containing 82 polyglutamine repeats (N171-82Q) increased neurogenesis in the striatum and prevented striatal loss (Snyder et al. 2010), in part by inducing expression of the neurotrophins FGF-2 and VEGF in the striatum. Interestingly, the majority of injected hMSCs did not survive for more than 5 days within the striatum, whereas the neuroprotective changes were observed up to 30 days post-injection; thus, these cells probably exerted their beneficial effects mainly by stimulating surrounding striatal cells rather than by replacing dying neurons (Snyder et al. 2010). In a separate study, MSCs engineered to express BDNF were injected into the striatum of YAC-128 mice, which led to marked improvements in motor function and a reduction in striatal cell loss (Dey et al. 2010). These effects may be mediated by MSC-induced secretion of trophic factors and cytokines that induce angiogenesis and prevent apoptosis by promoting the generation of new neurons (Joyce et al. 2010). MSC-based therapies may circumvent overgrowth or abnormal proliferation in the brain caused by injection of neuronal progenitors. Another interesting approach has been to employ MSCs to deliver shRNAs that can stimulate the degradation of mHtt mRNAs (Olson et al. 2012). Although this approach has not yet been used in HD mouse models in vivo, it has been used to successfully knock down the expression of mHtt in neuron-like cell lines (Olson et al. 2012). Even if MSCs are not able to replace dead or dying cells, their ability to release factors that can support neurons and/or glia or block the effects of potentially harmful proteins is tantalizing. It will be interesting to see how these gene-deliverable, cell-based therapies develop.
Concluding Remarks
Modeling HD using iPSCs is well underway. Because iPSCs come directly from affected patients, they are likely to represent the most molecularly and genetically precise cell model of the disease. Thus, using iPSCs may bridge the gaps in knowledge gleaned from rodent models. Furthermore, this technology holds great promise for elucidating the mechanisms of disease progression, in light of the growing body of evidence showing that many molecular changes take place in cells well before the onset of clinical symptoms, such as during development and neurogenesis. iPSCs may therefore be key to helping unravel the underlying pathogenic mechanisms of mHtt early in its course that eventually culminate in the symptoms of HD. Finally, these cells offer great hope as a potentially invaluable drug-screening platform to uncover and devise therapies targeting the disease course well before cell death and clinical symptoms emerge, which could lead to greater success in clinical trials.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Julia A. Kaye, Gladstone Institute of Neurological Disease, 1650 Owens Street, San Francisco, CA 94158, julia.kaye@gladstone.ucsf.edu (415) 734-2535.
Steven Finkbeiner, Gladstone Institute of Neurological Disease, 1650 Owens Street, San Francisco, CA 94158, sfinkbeiner@gladstone.ucsf.edu, (415) 734-2508 Professor, Departments of Neurology and Physiology, University of California, San Francisco, Senior Investigator & Associate Director, Gladstone Institute of Neurological Disease Director, Taube-Koret Center for Neurodegenerative Disease Research and the Hellman Family Foundation Program in Alzheimer’s Disease Research.
References Cited
- 1.Arnulf I, Nielsen J, Lohmann E, Schiefer J, Wild E, Jennum P, Konofal E, Walker M, Oudiette D, Tabrizi S, Durr A. Rapid eye movement sleep disturbances in Huntington disease. Arch Neurol. 2008;65:482–488. doi: 10.1001/archneur.65.4.482. [DOI] [PubMed] [Google Scholar]
- 2.Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 3.DiFiglia M, Sapp E, Chase K, Schwarz C, Meloni A, Young C, Martin E, Vonsattel JP, Carraway R, Reeves SA. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14:1075–1081. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- 4.Snell RG, MacMillan JC, Cheadle JP, Fenton I, Lazarou LP, Davies P, MacDonald ME, Gusella JF, Harper PS, Shaw DJ. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington's disease. Nat Genet. 1993;4:393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- 5.Andrew SE, Goldberg YP, Kremer B, Telenius H, Theilmann J, Adam S, Starr E, Squitieri F, Lin B, Kalchman MA, et al. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- 6.Langbehn DR, Brinkman RR, Falush D, Paulsen JS, Hayden MR. A new model for prediction of the age of onset and penetrance for Huntington's disease based on CAG length. Clinical genetics. 2004;65:267–277. doi: 10.1111/j.1399-0004.2004.00241.x. [DOI] [PubMed] [Google Scholar]
- 7.Augood SJ, Faull RL, Emson PC. Dopamine D1 and D2 receptor gene expression in the striatum in Huntington's disease. Ann Neurol. 1997;42:215–221. doi: 10.1002/ana.410420213. [DOI] [PubMed] [Google Scholar]
- 8.Packard MG, Knowlton BJ. Learning and memory functions of the Basal Ganglia. Annu Rev Neurosci. 2002;25:563–593. doi: 10.1146/annurev.neuro.25.112701.142937. [DOI] [PubMed] [Google Scholar]
- 9.Han I, You Y, Kordower JH, Brady ST, Morfini GA. Differential vulnerability of neurons in Huntington's disease: the role of cell type-specific features. Journal of neurochemistry. 2010;113:1073–1091. doi: 10.1111/j.1471-4159.2010.06672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mestre T, Ferreira J, Coelho MM, Rosa M, Sampaio C. Therapeutic interventions for disease progression in Huntington's disease. Cochrane Database Syst Rev. 2009. CD006455. [DOI] [PMC free article] [PubMed]
- 11.Hickey MA, Chesselet MF. Apoptosis in Huntington's disease. Prog Neuropsychopharmacol Biol Psychiatry. 2003;27:255–265. doi: 10.1016/S0278-5846(03)00021-6. [DOI] [PubMed] [Google Scholar]
- 12.Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington's disease. Nat Rev Neurosci. 2005;6:919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- 13.Thomas EA. Striatal specificity of gene expression dysregulation in Huntington's disease. J Neurosci Res. 2006;84:1151–1164. doi: 10.1002/jnr.21046. [DOI] [PubMed] [Google Scholar]
- 14.Cha JH. Transcriptional signatures in Huntington's disease. Prog Neurobiol. 2007;83:228–248. doi: 10.1016/j.pneurobio.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finkbeiner S. Huntington's Disease. Cold Spring Harbor perspectives in biology. 2011. p. 3. [DOI] [PMC free article] [PubMed]
- 16.DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 17.Miller J, Arrasate M, Shaby BA, Mitra S, Masliah E, Finkbeiner S. Quantitative relationships between huntingtin levels, polyglutamine length, inclusion body formation, and neuronal death provide novel insight into huntington's disease molecular pathogenesis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:10541–10550. doi: 10.1523/JNEUROSCI.0146-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kratter IH, Finkbeiner S. PolyQ disease: too many Qs, too much function? Neuron. 2010;67:897–899. doi: 10.1016/j.neuron.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 20.Li SH, Li XJ. Huntingtin-protein interactions and the pathogenesis of Huntington's disease. Trends Genet. 2004;20:146–154. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Sugars KL, Brown R, Cook LJ, Swartz J, Rubinsztein DC. Decreased cAMP response element-mediated transcription: an early event in exon 1 and full-length cell models of Huntington's disease that contributes to polyglutamine pathogenesis. J Biol Chem. 2004;279:4988–4999. doi: 10.1074/jbc.M310226200. [DOI] [PubMed] [Google Scholar]
- 22.Lovrecic L, Kastrin A, Kobal J, Pirtosek Z, Krainc D, Peterlin B. Gene expression changes in blood as a putative biomarker for Huntington's disease. Movement disorders : official journal of the Movement Disorder Society. 2009;24:2277–2281. doi: 10.1002/mds.22477. [DOI] [PubMed] [Google Scholar]
- 23.Borovecki F, Lovrecic L, Zhou J, Jeong H, Then F, Rosas HD, Hersch SM, Hogarth P, Bouzou B, Jensen RV, Krainc D. Genome-wide expression profiling of human blood reveals biomarkers for Huntington's disease. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long JD, Matson WR, Juhl AR, Leavitt BR, Paulsen JS. 8OHdG as a marker for Huntington disease progression. Neurobiology of disease. 2012;46:625–634. doi: 10.1016/j.nbd.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An MC, Zhang N, Scott G, Montoro D, Wittkop T, Mooney S, Melov S, Ellerby LM. Genetic Correction of Huntington's Disease Phenotypes in Induced Pluripotent Stem Cells. Cell stem cell. 2012. [DOI] [PMC free article] [PubMed]
- 26.The HD iPSC, C. Induced Pluripotent Stem Cells from Patients with Huntington's Disease Show CAG-Repeat-Expansion-Associated Phenotypes. Cell stem cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nei M, Xu P, Glazko G. Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms. Proc Natl Acad Sci U S A. 2001;98:2497–2502. doi: 10.1073/pnas.051611498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrer-Costa C, Orozco M, de la Cruz X. Characterization of diseaseassociated single amino acid polymorphisms in terms of sequence and structure properties. J Mol Biol. 2002;315:771–786. doi: 10.1006/jmbi.2001.5255. [DOI] [PubMed] [Google Scholar]
- 29.Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer's disease. Proc Natl Acad Sci U S A. 2006;103:5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vance JE, Hayashi H. Formation and function of apolipoprotein E-containing lipoproteins in the nervous system. Biochimica et biophysica acta. 2010;1801:806–818. doi: 10.1016/j.bbalip.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 31.Dolmetsch R, Geschwind DH. The human brain in a dish: the promise of iPSC-derived neurons. Cell. 2011;145:831–834. doi: 10.1016/j.cell.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clowry G, Molnar Z, Rakic P. Renewed focus on the developing human neocortex. Journal of anatomy. 2010;217:276–288. doi: 10.1111/j.1469-7580.2010.01281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oberheim NA, Wang X, Goldman S, Nedergaard M. Astrocytic complexity distinguishes the human brain. Trends in neurosciences. 2006;29:547–553. doi: 10.1016/j.tins.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Ginis I, Luo Y, Miura T, Thies S, Brandenberger R, Gerecht-Nir S, Amit M, Hoke A, Carpenter MK, Itskovitz-Eldor J, Rao MS. Differences between human and mouse embryonic stem cells. Developmental biology. 2004;269:360–380. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 35.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. Journal of immunology. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 36.Zermeno V, Espindola S, Mendoza E, Hernandez-Echeagaray E. Differential expression of neurotrophins in postnatal C57BL/6 mice striatum. Int J Biol Sci. 2009;5:118–127. doi: 10.7150/ijbs.5.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng-Bradley X, Rung J, Parkinson H, Brazma A. Large scale comparison of global gene expression patterns in human and mouse. Genome biology. 2010;11:R124. doi: 10.1186/gb-2010-11-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gharib WH, Robinson-Rechavi M. When orthologs diverge between human and mouse. Brief Bioinform. 2011;12:436–441. doi: 10.1093/bib/bbr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nature genetics. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Julien CL, Thompson JC, Wild S, Yardumian P, Snowden JS, Turner G, Craufurd D. Psychiatric disorders in preclinical Huntington's disease. Journal of neurology, neurosurgery, and psychiatry. 2007;78:939–943. doi: 10.1136/jnnp.2006.103309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duff K, Paulsen JS, Beglinger LJ, Langbehn DR, Stout JC. Psychiatric symptoms in Huntington's disease before diagnosis: the predict-HD study. Biological psychiatry. 2007;62:1341–1346. doi: 10.1016/j.biopsych.2006.11.034. [DOI] [PubMed] [Google Scholar]
- 42.Nopoulos PC, Aylward EH, Ross CA, Mills JA, Langbehn DR, Johnson HJ, Magnotta VA, Pierson RK, Beglinger LJ, Nance MA, Barker RA, Paulsen JS. Smaller intracranial volume in prodromal Huntington's disease: evidence for abnormal neurodevelopment. Brain : a journal of neurology. 2011;134:137–142. doi: 10.1093/brain/awq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giedd JN. Structural magnetic resonance imaging of the adolescent brain. Annals of the New York Academy of Sciences. 2004;1021:77–85. doi: 10.1196/annals.1308.009. [DOI] [PubMed] [Google Scholar]
- 44.Rushton JP, Ankney CD. Brain size matters: a reply to Peters. Canadian journal of experimental psychology = Revue canadienne de psychologie experimentale. 1995;49:562–569. doi: 10.1037/1196-1961.49.4.562. discussion 570–6. [DOI] [PubMed] [Google Scholar]
- 45.Lee JK, Mathews K, Schlaggar B, Perlmutter J, Paulsen JS, Epping E, Burmeister L, Nopoulos P. Measures of growth in children at risk for Huntington disease. Neurology. 2012. [DOI] [PMC free article] [PubMed]
- 46.Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nature genetics. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 47.Duyao MP, Auerbach AB, Ryan A, Persichetti F, Barnes GT, McNeil SM, Ge P, Vonsattel JP, Gusella JF, Joyner AL, et al. Inactivation of the mouse Huntington's disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- 48.Tong Y, Ha TJ, Liu L, Nishimoto A, Reiner A, Goldowitz D. Spatial and temporal requirements for huntingtin (Htt) in neuronal migration and survival during brain development. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:14794–14799. doi: 10.1523/JNEUROSCI.2774-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reiner A, Del Mar N, Meade CA, Yang H, Dragatsis I, Zeitlin S, Goldowitz D. Neurons lacking huntingtin differentially colonize brain and survive in chimeric mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:7608–7619. doi: 10.1523/JNEUROSCI.21-19-07608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Godin JD, Colombo K, Molina-Calavita M, Keryer G, Zala D, Charrin BC, Dietrich P, Volvert ML, Guillemot F, Dragatsis I, Bellaiche Y, Saudou F, Nguyen L, Humbert S. Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron. 2010;67:392–406. doi: 10.1016/j.neuron.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 51.Conforti P, Camnasio S, Mutti C, Valenza M, Thompson M, Fossale E, Zeitlin S, Macdonald ME, Zuccato C, Cattaneo E. Lack of huntingtin promotes neural stem cells differentiation into glial cells while neurons expressing huntingtin with expanded polyglutamine tracts undergo cell death. Neurobiology of disease. 2013;50:160–170. doi: 10.1016/j.nbd.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 52.Molero AE, Gokhan S, Gonzalez S, Feig JL, Alexandre LC, Mehler MF. Impairment of developmental stem cell-mediated striatal neurogenesis and pluripotency genes in a knock-in model of Huntington's disease. Proc Natl Acad Sci U S A. 2009;106:21900–21905. doi: 10.1073/pnas.0912171106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. Journal of magnetic resonance. Series B. 1996;111:209–219. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- 54.Blockx I, De Groof G, Verhoye M, Van Audekerke J, Raber K, Poot D, Sijbers J, Osmand AP, Von Horsten S, Van der Linden A. Microstructural changes observed with DKI in a transgenic Huntington rat model: evidence for abnormal neurodevelopment. NeuroImage. 2012;59:957–967. doi: 10.1016/j.neuroimage.2011.08.062. [DOI] [PubMed] [Google Scholar]
- 55.Rubenstein JL, Shimamura K, Martinez S, Puelles L. Regionalization of the prosencephalic neural plate. Annual review of neuroscience. 1998;21:445–477. doi: 10.1146/annurev.neuro.21.1.445. [DOI] [PubMed] [Google Scholar]
- 56.Jain M, Armstrong RJ, Barker RA, Rosser AE. Cellular and molecular aspects of striatal development. Brain research bulletin. 2001;55:533–540. doi: 10.1016/s0361-9230(01)00555-x. [DOI] [PubMed] [Google Scholar]
- 57.Sousa VH, Fishell G. Sonic hedgehog functions through dynamic changes in temporal competence in the developing forebrain. Curr Opin Genet Dev. 2010;20:391–399. doi: 10.1016/j.gde.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marin O, Rubenstein JL. A long, remarkable journey: tangential migration in the telencephalon. Nat Rev Neurosci. 2001;2:780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- 59.Olsson M, Bjorklund A, Campbell K. Early specification of striatal projection neurons and interneuronal subtypes in the lateral and medial ganglionic eminence. Neuroscience. 1998;84:867–876. doi: 10.1016/s0306-4522(97)00532-0. [DOI] [PubMed] [Google Scholar]
- 60.Fentress JC, Stanfield BB, Cowan WM. Observation on the development of the striatum in mice and rats. Anatomy and embryology. 1981;163:275–298. doi: 10.1007/BF00315705. [DOI] [PubMed] [Google Scholar]
- 61.Gerfen CR. The neostriatal mosaic: multiple levels of compartmental organization. Journal of neural transmission Supplementum. 1992a;36:43–59. doi: 10.1007/978-3-7091-9211-5_4. [DOI] [PubMed] [Google Scholar]
- 62.Evans AE, Kelly CM, Precious SV, Rosser AE. Molecular regulation of striatal development: a review. Anatomy research international. 2012. p. 106529. [DOI] [PMC free article] [PubMed]
- 63.Aubry L, Bugi A, Lefort N, Rousseau F, Peschanski M, Perrier AL. Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. Proc Natl Acad Sci U S A. 2008;105:16707–16712. doi: 10.1073/pnas.0808488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annual review of neuroscience. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ciccolini F, Svendsen CN. Neurotrophin responsiveness is differentially regulated in neurons and precursors isolated from the developing striatum. Journal of molecular neuroscience : MN. 2001;17:25–33. doi: 10.1385/JMN:17:1:25. [DOI] [PubMed] [Google Scholar]
- 66.Rauskolb S, Zagrebelsky M, Dreznjak A, Deogracias R, Matsumoto T, Wiese S, Erne B, Sendtner M, Schaeren-Wiemers N, Korte M, Barde YA. Global deprivation of brain-derived neurotrophic factor in the CNS reveals an area-specific requirement for dendritic growth. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:1739–1749. doi: 10.1523/JNEUROSCI.5100-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baydyuk M, Russell T, Liao GY, Zang K, An JJ, Reichardt LF, Xu B. TrkB receptor controls striatal formation by regulating the number of newborn striatal neurons. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1669–1674. doi: 10.1073/pnas.1004744108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benraiss A, Chmielnicki E, Lerner K, Roh D, Goldman SA. Adenoviral brain-derived neurotrophic factor induces both neostriatal and olfactory neuronal recruitment from endogenous progenitor cells in the adult forebrain. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:6718–6731. doi: 10.1523/JNEUROSCI.21-17-06718.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mason I. Initiation to end point: the multiple roles of fibroblast growth factors in neural development. Nature reviews Neuroscience. 2007;8:583–596. doi: 10.1038/nrn2189. [DOI] [PubMed] [Google Scholar]
- 70.Hoch RV, Rubenstein JL, Pleasure S. Genes and signaling events that establish regional patterning of the mammalian forebrain. Seminars in cell & developmental biology. 2009;20:378–386. doi: 10.1016/j.semcdb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 71.Shinya M, Koshida S, Sawada A, Kuroiwa A, Takeda H. Fgf signalling through MAPK cascade is required for development of the subpallial telencephalon in zebrafish embryos. Development. 2001;128:4153–4164. doi: 10.1242/dev.128.21.4153. [DOI] [PubMed] [Google Scholar]
- 72.Paek H, Gutin G, Hebert JM. FGF signaling is strictly required to maintain early telencephalic precursor cell survival. Development. 2009;136:2457–2465. doi: 10.1242/dev.032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanalkumar R, Vidyanand S, Lalitha Indulekha C, James J. Neuronal vs. glial fate of embryonic stem cell-derived neural progenitors (ES-NPs) is determined by FGF2/EGF during proliferation. Journal of molecular neuroscience : MN. 2010;42:17–27. doi: 10.1007/s12031-010-9335-z. [DOI] [PubMed] [Google Scholar]
- 74.Sterneckert J, Stehling M, Bernemann C, Arauzo-Bravo MJ, Greber B, Gentile L, Ortmeier C, Sinn M, Wu G, Ruau D, Zenke M, Brintrup R, Klein DC, Ko K, Scholer HR. Neural induction intermediates exhibit distinct roles of Fgf signaling. Stem cells. 2010;28:1772–1781. doi: 10.1002/stem.498. [DOI] [PubMed] [Google Scholar]
- 75.Rallu M, Machold R, Gaiano N, Corbin JG, McMahon AP, Fishell G. Dorsoventral patterning is established in the telencephalon of mutants lacking both Gli3 and Hedgehog signaling. Development. 2002;129:4963–4974. doi: 10.1242/dev.129.21.4963. [DOI] [PubMed] [Google Scholar]
- 76.Kohtz JD, Baker DP, Corte G, Fishell G. Regionalization within the mammalian telencephalon is mediated by changes in responsiveness to Sonic Hedgehog. Development. 1998;125:5079–5089. doi: 10.1242/dev.125.24.5079. [DOI] [PubMed] [Google Scholar]
- 77.Gaspard N, Bouschet T, Hourez R, Dimidschstein J, Naeije G, van den Ameele J, Espuny-Camacho I, Herpoel A, Passante L, Schiffmann SN, Gaillard A, Vanderhaeghen P. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- 78.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell stem cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 79.Eiraku M, Sasai Y. Mouse embryonic stem cell culture for generation of three-dimensional retinal and cortical tissues. Nature protocols. 2012;7:69–79. doi: 10.1038/nprot.2011.429. [DOI] [PubMed] [Google Scholar]
- 80.Ma L, Hu B, Liu Y, Vermilyea SC, Liu H, Gao L, Sun Y, Zhang X, Zhang SC. Human Embryonic Stem Cell-Derived GABA Neurons Correct Locomotion Deficits in Quinolinic Acid-Lesioned Mice. Cell stem cell. 2012. [DOI] [PMC free article] [PubMed]
- 81.Danjo T, Eiraku M, Muguruma K, Watanabe K, Kawada M, Yanagawa Y, Rubenstein JL, Sasai Y. Subregional specification of embryonic stem cell-derived ventral telencephalic tissues by timed and combinatory treatment with extrinsic signals. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31:1919–1933. doi: 10.1523/JNEUROSCI.5128-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.El-Akabawy G, Medina LM, Jeffries A, Price J, Modo M. Purmorphamine increases DARPP-32 differentiation in human striatal neural stem cells through the Hedgehog pathway. Stem cells and development. 2011;20:1873–1887. doi: 10.1089/scd.2010.0282. [DOI] [PubMed] [Google Scholar]
- 83.Li XJ, Zhang X, Johnson MA, Wang ZB, Lavaute T, Zhang SC. Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development. 2009;136:4055–4063. doi: 10.1242/dev.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annual review of cell and developmental biology. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 85.Gunhaga L, Marklund M, Sjodal M, Hsieh JC, Jessell TM, Edlund T. Specification of dorsal telencephalic character by sequential Wnt and FGF signaling. Nature neuroscience. 2003;6:701–707. doi: 10.1038/nn1068. [DOI] [PubMed] [Google Scholar]
- 86.Gulacsi AA, Anderson SA. Beta-catenin-mediated Wnt signaling regulates neurogenesis in the ventral telencephalon. Nature neuroscience. 2008;11:1383–1391. doi: 10.1038/nn.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Backman M, Machon O, Mygland L, van den Bout CJ, Zhong W, Taketo MM, Krauss S. Effects of canonical Wnt signaling on dorso-ventral specification of the mouse telencephalon. Developmental biology. 2005;279:155–168. doi: 10.1016/j.ydbio.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 88.Watanabe K, Kamiya D, Nishiyama A, Katayama T, Nozaki S, Kawasaki H, Watanabe Y, Mizuseki K, Sasai Y. Directed differentiation of telencephalic precursors from embryonic stem cells. Nature neuroscience. 2005;8:288–296. doi: 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- 89.Zhang XZ, Li XJ, Zhang HY. Valproic acid as a promising agent to combat Alzheimer's disease. Brain research bulletin. 2010a;81:3–6. doi: 10.1016/j.brainresbull.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 90.Laeng P, Pitts RL, Lemire AL, Drabik CE, Weiner A, Tang H, Thyagarajan R, Mallon BS, Altar CA. The mood stabilizer valproic acid stimulates GABA neurogenesis from rat forebrain stem cells. Journal of neurochemistry. 2004;91:238–251. doi: 10.1111/j.1471-4159.2004.02725.x. [DOI] [PubMed] [Google Scholar]
- 91.Urban N, Martin-Ibanez R, Herranz C, Esgleas M, Crespo E, Pardo M, Crespo-Enriquez I, Mendez-Gomez HR, Waclaw R, Chatzi C, Alvarez S, Alvarez R, Duester G, Campbell K, de Lera AR, Vicario-Abejon C, Martinez S, Alberch J, Canals JM. Nolz1 promotes striatal neurogenesis through the regulation of retinoic acid signaling. Neural development. 2010;5:21. doi: 10.1186/1749-8104-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chatzi C, Brade T, Duester G. Retinoic acid functions as a key GABAergic differentiation signal in the basal ganglia. PLoS biology. 2011;9:e1000609. doi: 10.1371/journal.pbio.1000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shin E, Palmer MJ, Li M, Fricker RA. GABAergic Neurons from Mouse Embryonic Stem Cells Possess Functional Properties of Striatal Neurons In Vitro, and Develop into Striatal Neurons In Vivo in a Mouse Model of Huntington's Disease. Stem cell reviews. 2011. [DOI] [PubMed]
- 94.Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nature neuroscience. 2012;15:477–486. doi: 10.1038/nn.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krencik R, Weick JP, Liu Y, Zhang ZJ, Zhang SC. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nature biotechnology. 2011;29:528–534. doi: 10.1038/nbt.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Czepiel M, Balasubramaniyan V, Schaafsma W, Stancic M, Mikkers H, Huisman C, Boddeke E, Copray S. Differentiation of induced pluripotent stem cells into functional oligodendrocytes. Glia. 2011;59:882–892. doi: 10.1002/glia.21159. [DOI] [PubMed] [Google Scholar]
- 97.Carri AD, Onorati M, Lelos MJ, Castiglioni V, Faedo A, Menon R, Camnasio S, Vuono R, Spaiardi P, Talpo F, Toselli M, Martino G, Barker RA, Dunnett SB, Biella G, Cattaneo E. Developmentally coordinated extrinsic signals drive human pluripotent stem cell differentiation toward authentic DARPP-32+ medium-sized spiny neurons. Development. 2013;140:301–312. doi: 10.1242/dev.084608. [DOI] [PubMed] [Google Scholar]
- 98.Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, Amit M, Soreq H, Benvenisty N. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Molecular medicine. 2000;6:88–95. [PMC free article] [PubMed] [Google Scholar]
- 99.Bain G, Kitchens D, Yao M, Huettner JE, Gottlieb DI. Embryonic stem cells express neuronal properties in vitro. Dev Biol. 1995;168:342–357. doi: 10.1006/dbio.1995.1085. [DOI] [PubMed] [Google Scholar]
- 100.Strubing C, Ahnert-Hilger G, Shan J, Wiedenmann B, Hescheler J, Wobus AM. Differentiation of pluripotent embryonic stem cells into the neuronal lineage in vitro gives rise to mature inhibitory and excitatory neurons. Mechanisms of development. 1995;53:275–287. doi: 10.1016/0925-4773(95)00446-8. [DOI] [PubMed] [Google Scholar]
- 101.Westmoreland JJ, Hancock CR, Condie BG. Neuronal development of embryonic stem cells: a model of GABAergic neuron differentiation. Biochemical and biophysical research communications. 2001;284:674–680. doi: 10.1006/bbrc.2001.5031. [DOI] [PubMed] [Google Scholar]
- 102.Abranches E, Silva M, Pradier L, Schulz H, Hummel O, Henrique D, Bekman E. Neural differentiation of embryonic stem cells in vitro: a road map to neurogenesis in the embryo. PloS one. 2009;4:e6286. doi: 10.1371/journal.pone.0006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Okabe S, Forsberg-Nilsson K, Spiro AC, Segal M, McKay RD. Development of neuronal precursor cells and functional postmitotic neurons from embryonic stem cells in vitro. Mechanisms of development. 1996;59:89–102. doi: 10.1016/0925-4773(96)00572-2. [DOI] [PubMed] [Google Scholar]
- 104.Pankratz MT, Zhang S-C. Embryoid bodies and neuroepithelial development. In: Loring JF, Wesselschmidt RL, Schwartz PH, editors. Human Stem Cell Manual. Elsevier, New York: 2007. pp. 185–198. A Laboratory Guide. [Google Scholar]
- 105.Gerfen CR. The neostriatal mosaic: multiple levels of compartmental organization in the basal ganglia. Annual review of neuroscience. 1992b;15:285–320. doi: 10.1146/annurev.ne.15.030192.001441. [DOI] [PubMed] [Google Scholar]
- 106.Zhang N, An MC, Montoro D, Ellerby LM. Characterization of Human Huntington's Disease Cell Model from Induced Pluripotent Stem Cells. PLoS Curr. 2010b;2:RRN1193. doi: 10.1371/currents.RRN1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ouimet CC, Miller PE, Hemmings HC, Jr, Walaas SI, Greengard P. DARPP-32, a dopamine- and adenosine 3':5'-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. III. Immunocytochemical localization. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1984;4:111–124. doi: 10.1523/JNEUROSCI.04-01-00111.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ouimet CC, Langley-Gullion KC, Greengard P. Quantitative immunocytochemistry of DARPP-32-expressing neurons in the rat caudatoputamen. Brain research. 1998;808:8–12. doi: 10.1016/s0006-8993(98)00724-0. [DOI] [PubMed] [Google Scholar]
- 109.Liu FC, Graybiel AM. Transient calbindin-D28k-positive systems in the telencephalon: ganglionic eminence, developing striatum and cerebral cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1992;12:674–690. doi: 10.1523/JNEUROSCI.12-02-00674.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rajput PS, Kharmate G, Somvanshi RK, Kumar U. Colocalization of dopamine receptor subtypes with dopamine and cAMP-regulated phosphoprotein (DARPP-32) in rat brain. Neuroscience research. 2009;65:53–63. doi: 10.1016/j.neures.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 111.Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tamura S, Morikawa Y, Iwanishi H, Hisaoka T, Senba E. Foxp1 gene expression in projection neurons of the mouse striatum. Neuroscience. 2004;124:261–267. doi: 10.1016/j.neuroscience.2003.11.036. [DOI] [PubMed] [Google Scholar]
- 113.Takahashi K, Liu FC, Hirokawa K, Takahashi H. Expression of Foxp2, a gene involved in speech and language, in the developing and adult striatum. Journal of neuroscience research. 2003;73:61–72. doi: 10.1002/jnr.10638. [DOI] [PubMed] [Google Scholar]
- 114.Waclaw RR, Wang B, Pei Z, Ehrman LA, Campbell K. Distinct temporal requirements for the homeobox gene Gsx2 in specifying striatal and olfactory bulb neuronal fates. Neuron. 2009;63:451–465. doi: 10.1016/j.neuron.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gerfen CR, Engber TM, Mahan LC, Susel Z, Chase TN, Monsma FJ, Jr, Sibley DR. D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science. 1990;250:1429–1432. doi: 10.1126/science.2147780. [DOI] [PubMed] [Google Scholar]
- 116.Albin RL, Reiner A, Anderson KD, Dure LS, Handelin B, Balfour R, Whetsell WO, Jr, Penney JB, Young AB. Preferential loss of striato-external pallidal projection neurons in presymptomatic Huntington’s disease. Ann. Neurol. 1992;31:425–430. doi: 10.1002/ana.410310412. [DOI] [PubMed] [Google Scholar]
- 117.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nature neuroscience. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 118.Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 120.Walker TL, Turnbull GW, Mackay EW, Hannan AJ, Bartlett PF. The latent stem cell population is retained in the hippocampus of transgenic Huntington's disease mice but not wild-type mice. PloS one. 2011;6:e18153. doi: 10.1371/journal.pone.0018153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lazic SE, Grote H, Armstrong RJ, Blakemore C, Hannan AJ, van Dellen A, Barker RA. Decreased hippocampal cell proliferation in R6/1 Huntington's mice. Neuroreport. 2004;15:811–813. doi: 10.1097/00001756-200404090-00014. [DOI] [PubMed] [Google Scholar]
- 122.Gil JM, Mohapel P, Araujo IM, Popovic N, Li JY, Brundin P, Petersen A. Reduced hippocampal neurogenesis in R6/2 transgenic Huntington's disease mice. Neurobiology of disease. 2005;20:744–751. doi: 10.1016/j.nbd.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 123.Phillips W, Morton AJ, Barker RA. Abnormalities of neurogenesis in the R6/2 mouse model of Huntington's disease are attributable to the in vivo microenvironment. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:11564–11576. doi: 10.1523/JNEUROSCI.3796-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Phillips W, Morton AJ, Barker RA. Limbic neurogenesis/plasticity in the R6/2 mouse model of Huntington's disease. Neuroreport. 2006;17:1623–1627. doi: 10.1097/01.wnr.0000236855.85962.f6. [DOI] [PubMed] [Google Scholar]
- 125.Lazic SE, Goodman AO, Grote HE, Blakemore C, Morton AJ, Hannan AJ, van Dellen A, Barker RA. Olfactory abnormalities in Huntington's disease: decreased plasticity in the primary olfactory cortex of R6/1 transgenic mice and reduced olfactory discrimination in patients. Brain research. 2007;1151:219–226. doi: 10.1016/j.brainres.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 126.Nordin S, Paulsen JS, Murphy C. Sensory- and memory-mediated olfactory dysfunction in Huntington's disease. Journal of the International Neuropsychological Society : JINS. 1995;1:281–290. doi: 10.1017/s1355617700000278. [DOI] [PubMed] [Google Scholar]
- 127.Batista CM, Kippin TE, Willaime-Morawek S, Shimabukuro MK, Akamatsu W, van der Kooy D. A progressive and cell non-autonomous increase in striatal neural stem cells in the Huntington's disease R6/2 mouse. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:10452–10460. doi: 10.1523/JNEUROSCI.2850-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tattersfield AS, Croon RJ, Liu YW, Kells AP, Faull RL, Connor B. Neurogenesis in the striatum of the quinolinic acid lesion model of Huntington's disease. Neuroscience. 2004;127:319–332. doi: 10.1016/j.neuroscience.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 129.Ferrante RJ, Kowall NW, Richardson EP., Jr Proliferative and degenerative changes in striatal spiny neurons in Huntington's disease: a combined study using the section-Golgi method and calbindin D28k immunocytochemistry. J Neurosci. 1991;11:3877–3887. doi: 10.1523/JNEUROSCI.11-12-03877.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Curtis MA, Penney EB, Pearson AG, van Roon-Mom WM, Butterworth NJ, Dragunow M, Connor B, Faull RL. Increased cell proliferation and neurogenesis in the adult human Huntington's disease brain. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9023–9027. doi: 10.1073/pnas.1532244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Curtis MA, Penney EB, Pearson J, Dragunow M, Connor B, Faull RL. The distribution of progenitor cells in the subependymal layer of the lateral ventricle in the normal and Huntington's disease human brain. Neuroscience. 2005;132:777–788. doi: 10.1016/j.neuroscience.2004.12.051. [DOI] [PubMed] [Google Scholar]
- 132.Low VF, Dragunow M, Tippett LJ, Faull RL, Curtis MA. No change in progenitor cell proliferation in the hippocampus in Huntington's disease. Neuroscience. 2011;199:577–588. doi: 10.1016/j.neuroscience.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 133.Simpson JM, Gil-Mohapel J, Pouladi MA, Ghilan M, Xie Y, Hayden MR, Christie BR. Altered adult hippocampal neurogenesis in the YAC128 transgenic mouse model of Huntington disease. Neurobiology of disease. 2011;41:249–260. doi: 10.1016/j.nbd.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 134.Ben-David U, Kopper O, Benvenisty N. Expanding the boundaries of embryonic stem cells. Cell Stem Cell. 2012;10:666–677. doi: 10.1016/j.stem.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 135.Verlinsky Y, Strelchenko N, Kukharenko V, Rechitsky S, Verlinsky O, Galat V, Kuliev A. Human embryonic stem cell lines with genetic disorders. Reproductive biomedicine online. 2005;10:105–110. doi: 10.1016/s1472-6483(10)60810-3. [DOI] [PubMed] [Google Scholar]
- 136.Niclis JC, Trounson AO, Dottori M, Ellisdon AM, Bottomley SP, Verlinsky Y, Cram DS. Human embryonic stem cell models of Huntington disease. Reproductive biomedicine online. 2009;19:106–113. doi: 10.1016/s1472-6483(10)60053-3. [DOI] [PubMed] [Google Scholar]
- 137.Bradley CK, Scott HA, Chami O, Peura TT, Dumevska B, Schmidt U, Stojanov T. Derivation of Huntington's disease-affected human embryonic stem cell lines. Stem cells and development. 2011;20:495–502. doi: 10.1089/scd.2010.0120. [DOI] [PubMed] [Google Scholar]
- 138.Mateizel I, De Temmerman N, Ullmann U, Cauffman G, Sermon K, Van de Velde H, De Rycke M, Degreef E, Devroey P, Liebaers I, Van Steirteghem A. Derivation of human embryonic stem cell lines from embryos obtained after IVF and after PGD for monogenic disorders. Human reproduction. 2006;21:503–511. doi: 10.1093/humrep/dei345. [DOI] [PubMed] [Google Scholar]
- 139.Seriola A, Spits C, Simard JP, Hilven P, Haentjens P, Pearson CE, Sermon K. Huntington's and myotonic dystrophy hESCs: down-regulated trinucleotide repeat instability and mismatch repair machinery expression upon differentiation. Human molecular genetics. 2011;20:176–185. doi: 10.1093/hmg/ddq456. [DOI] [PubMed] [Google Scholar]
- 140.Lorincz MT, Zawistowski VA. Expanded CAG repeats in the murine Huntington's disease gene increases neuronal differentiation of embryonic and neural stem cells. Mol Cell Neurosci. 2009;40:1–13. doi: 10.1016/j.mcn.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Baldo B, Weiss A, Parker CN, Bibel M, Paganetti P, Kaupmann K. A screen for enhancers of clearance identifies huntingtin as a heat shock protein 90 (Hsp90) client protein. The Journal of biological chemistry. 2012;287:1406–1414. doi: 10.1074/jbc.M111.294801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jacobsen JC, Gregory GC, Woda JM, Thompson MN, Coser KR, Murthy V, Kohane IS, Gusella JF, Seong IS, MacDonald ME, Shioda T, Lee JM. HD CAG-correlated gene expression changes support a simple dominant gain of function. Human molecular genetics. 2011;20:2846–2860. doi: 10.1093/hmg/ddr195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 144.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 145.Ebert AD, Yu J, Rose FF, Jr, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008a;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Generation of human-induced pluripotent stem cells. Nat Protoc. 2008b;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- 148.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, Ganat YM, Menon J, Shimizu F, Viale A, Tabar V, Sadelain M, Studer L. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Egawa N, Kitaoka S, Tsukita K, Naitoh M, Takahashi K, Yamamoto T, Adachi F, Kondo T, Okita K, Asaka I, Aoi T, Watanabe A, Yamada Y, Morizane A, Takahashi J, Ayaki T, Ito H, Yoshikawa K, Yamawaki S, Suzuki S, Watanabe D, Hioki H, Kaneko T, Makioka K, Okamoto K, Takuma H, Tamaoka A, Hasegawa K, Nonaka T, Hasegawa M, Kawata A, Yoshida M, Nakahata T, Takahashi R, Marchetto MC, Gage FH, Yamanaka S, Inoue H. Drug Screening for ALS Using Patient-Specific Induced Pluripotent Stem Cells. Science translational medicine. 2012;4 doi: 10.1126/scitranslmed.3004052. 145ra104. [DOI] [PubMed] [Google Scholar]
- 151.Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, McCarthy S, Sebat J, Gage FH. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bilican B, Serio A, Barmada SJ, Nishimura AL, Sullivan GJ, Carrasco M, Phatnani HP, Puddifoot CA, Story D, Fletcher J, Park IH, Friedman BA, Daley GQ, Wyllie DJ, Hardingham GE, Wilmut I, Finkbeiner S, Maniatis T, Shaw CE, Chandran S. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proceedings of the National Academy of Sciences of the United States of America. 2012. [DOI] [PMC free article] [PubMed]
- 153.Seo H, Sonntag KC, Isacson O. Generalized brain and skin proteasome inhibition in Huntington's disease. Annals of neurology. 2004;56:319–328. doi: 10.1002/ana.20207. [DOI] [PubMed] [Google Scholar]
- 154.Valenza M, Rigamonti D, Goffredo D, Zuccato C, Fenu S, Jamot L, Strand A, Tarditi A, Woodman B, Racchi M, Mariotti C, Di Donato S, Corsini A, Bates G, Pruss R, Olson JM, Sipione S, Tartari M, Cattaneo E. Dysfunction of the cholesterol biosynthetic pathway in Huntington's disease. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:9932–9939. doi: 10.1523/JNEUROSCI.3355-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jeon I, Lee N, Li JY, Park IH, Park KS, Moon J, Shim SH, Choi C, Chang DJ, Kwon J, Oh SH, Shin DA, Kim HS, Do JT, Lee DR, Kim M, Kang KS, Daley GQ, Brundin P, Song J. Neuronal properties, in vivo effects, and pathology of a Huntington's disease patient-derived induced pluripotent stem cells. Stem cells. 2012;30:2054–2062. doi: 10.1002/stem.1135. [DOI] [PubMed] [Google Scholar]
- 156.Chae JI, Kim DW, Lee N, Jeon YJ, Jeon I, Kwon J, Kim J, Soh Y, Lee DS, Seo KS, Choi NJ, Park BC, Kang SH, Ryu J, Oh SH, Shin DA, Lee DR, Do JT, Park IH, Daley GQ, Song J. Quantitative proteomic analysis of induced pluripotent stem cells derived from a human Huntington's disease patient. The Biochemical journal. 2012;446:359–371. doi: 10.1042/BJ20111495. [DOI] [PubMed] [Google Scholar]
- 157.Camnasio S, Carri AD, Lombardo A, Grad I, Mariotti C, Castucci A, Rozell B, Riso PL, Castiglioni V, Zuccato C, Rochon C, Takashima Y, Diaferia G, Biunno I, Gellera C, Jaconi M, Smith A, Hovatta O, Naldini L, Di Donato S, Feki A, Cattaneo E. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington's disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiology of disease. 2012;46:41–51. doi: 10.1016/j.nbd.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 158.Bradford J, Shin JY, Roberts M, Wang CE, Sheng G, Li S, Li XJ. Mutant huntingtin in glial cells exacerbates neurological symptoms of Huntington disease mice. The Journal of biological chemistry. 2010;285:10653–10661. doi: 10.1074/jbc.M109.083287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Juopperi TA, Kim WR, Chiang CH, Yu H, Margolis RL, Ross CA, Ming GL, Song H. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington's disease patient cells. Molecular brain. 2012;5:17. doi: 10.1186/1756-6606-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Arrasate M, Finkbeiner S. Protein aggregates in Huntington's disease. Experimental neurology. 2012;238:1–11. doi: 10.1016/j.expneurol.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gutekunst C-A, Li S-H, Yi H, Mulroy JS, Kuemmerle S, Jones R, Rye D, Ferrante RJ, Hersch SM, Li X-J. Nuclear and neuropil aggregates in Huntington’s disease: Relationship to neuropathology. J. Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chan AW, Cheng PH, Neumann A, Yang JJ. Reprogramming Huntington monkey skin cells into pluripotent stem cells. Cellular reprogramming. 2010;12:509–517. doi: 10.1089/cell.2010.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Castiglioni V, Onorati M, Rochon C, Cattaneo E. Induced pluripotent stem cell lines from Huntington's disease mice undergo neuronal differentiation while showing alterations in the lysosomal pathway. Neurobiology of disease. 2011. [DOI] [PubMed]
- 164.Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, Timmusk T, Sipione S, Cattaneo E. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 165.Zuccato C, Liber D, Ramos C, Tarditi A, Rigamonti D, Tartari M, Valenza M, Cattaneo E. Progressive loss of BDNF in a mouse model of Huntington's disease and rescue by BDNF delivery. Pharmacological research : the official journal of the Italian Pharmacological Society. 2005;52:133–139. doi: 10.1016/j.phrs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 166.Starling AJ, Andre VM, Cepeda C, de Lima M, Chandler SH, Levine MS. Alterations in N-methyl-D-aspartate receptor sensitivity and magnesium blockade occur early in development in the R6/2 mouse model of Huntington's disease. Journal of neuroscience research. 2005;82:377–386. doi: 10.1002/jnr.20651. [DOI] [PubMed] [Google Scholar]
- 167.Bano D, Zanetti F, Mende Y, Nicotera P. Neurodegenerative processes in Huntington's disease. Cell death & disease. 2011;2:e228. doi: 10.1038/cddis.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mochel F, Durant B, Meng X, O'Callaghan J, Yu H, Brouillet E, Wheeler VC, Humbert S, Schiffmann R, Durr A. Early alterations of brain cellular energy homeostasis in Huntington disease models. The Journal of biological chemistry. 2012;287:1361–1370. doi: 10.1074/jbc.M111.309849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ciarmiello A, Cannella M, Lastoria S, Simonelli M, Frati L, Rubinsztein DC, Squitieri F. Brain white-matter volume loss and glucose hypometabolism precede the clinical symptoms of Huntington's disease. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2006;47:215–222. [PubMed] [Google Scholar]
- 170.DiProspero NA, Chen EY, Charles V, Plomann M, Kordower JH, Tagle DA. Early changes in Huntington's disease patient brains involve alterations in cytoskeletal and synaptic elements. Journal of neurocytology. 2004;33:517–533. doi: 10.1007/s11068-004-0514-8. [DOI] [PubMed] [Google Scholar]
- 171.Sramka M, Rattaj M, Molina H, Vojtassak J, Belan V, Ruzicky E. Stereotactic technique and pathophysiological mechanisms of neurotransplantation in Huntington's chorea. Stereotactic and functional neurosurgery. 1992;58:79–83. doi: 10.1159/000098976. [DOI] [PubMed] [Google Scholar]
- 172.Bachoud-Levi AC, Remy P, Nguyen JP, Brugieres P, Lefaucheur JP, Bourdet C, Baudic S, Gaura V, Maison P, Haddad B, Boisse MF, Grandmougin T, Jeny R, Bartolomeo P, Dalla Barba G, Degos JD, Lisovoski F, Ergis AM, Pailhous E, Cesaro P, Hantraye P, Peschanski M. Motor and cognitive improvements in patients with Huntington's disease after neural transplantation. Lancet. 2000;356:1975–1979. doi: 10.1016/s0140-6736(00)03310-9. [DOI] [PubMed] [Google Scholar]
- 173.Bachoud-Levi AC, Gaura V, Brugieres P, Lefaucheur JP, Boisse MF, Maison P, Baudic S, Ribeiro MJ, Bourdet C, Remy P, Cesaro P, Hantraye P, Peschanski M. Effect of fetal neural transplants in patients with Huntington's disease 6 years after surgery: a long-term follow-up study. Lancet Neurol. 2006;5:303–309. doi: 10.1016/S1474-4422(06)70381-7. [DOI] [PubMed] [Google Scholar]
- 174.Dunnett SB, Rosser AE. Stem cell transplantation for Huntington's disease. Experimental neurology. 2007;203:279–292. doi: 10.1016/j.expneurol.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 175.Kim M, Lee ST, Chu K, Kim SU. Stem cell-based cell therapy for Huntington disease: a review. Neuropathology : official journal of the Japanese Society of Neuropathology. 2008;28:1–9. doi: 10.1111/j.1440-1789.2007.00858.x. [DOI] [PubMed] [Google Scholar]
- 176.Vazey EM, Chen K, Hughes SM, Connor B. Transplanted adult neural progenitor cells survive, differentiate and reduce motor function impairment in a rodent model of Huntington's disease. Experimental neurology. 2006;199:384–396. doi: 10.1016/j.expneurol.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 177.Song J, Lee ST, Kang W, Park JE, Chu K, Lee SE, Hwang T, Chung H, Kim M. Human embryonic stem cell-derived neural precursor transplants attenuate apomorphine-induced rotational behavior in rats with unilateral quinolinic acid lesions. Neuroscience letters. 2007;423:58–61. doi: 10.1016/j.neulet.2007.05.066. [DOI] [PubMed] [Google Scholar]
- 178.Ebert AD, Barber AE, Heins BM, Svendsen CN. Ex vivo delivery of GDNF maintains motor function and prevents neuronal loss in a transgenic mouse model of Huntington's disease. Experimental neurology. 2010;224:155–162. doi: 10.1016/j.expneurol.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 179.Kulbatski I. Stem/precursor cell-based CNS therapy: the importance of circumventing immune suppression by transplanting autologous cells. Stem cell reviews. 2010;6:405–410. doi: 10.1007/s12015-010-9141-6. [DOI] [PubMed] [Google Scholar]
- 180.Ross AL, Leder DE, Weiss J, Izakovic J, Grichnik JM. Genomic instability in cultured stem cells: associated risks and underlying mechanisms. Regenerative medicine. 2011;6:653–662. doi: 10.2217/rme.11.44. [DOI] [PubMed] [Google Scholar]
- 181.Vazey EM, Dottori M, Jamshidi P, Tomas D, Pera MF, Horne M, Connor B. Comparison of transplant efficiency between spontaneously derived and noggin-primed human embryonic stem cell neural precursors in the quinolinic acid rat model of Huntington's disease. Cell transplantation. 2010;19:1055–1062. doi: 10.3727/096368910X494632. [DOI] [PubMed] [Google Scholar]
- 182.Snyder BR, Chiu AM, Prockop DJ, Chan AW. Human multipotent stromal cells (MSCs) increase neurogenesis and decrease atrophy of the striatum in a transgenic mouse model for Huntington's disease. PloS one. 2010;5:e9347. doi: 10.1371/journal.pone.0009347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Dey ND, Bombard MC, Roland BP, Davidson S, Lu M, Rossignol J, Sandstrom MI, Skeel RL, Lescaudron L, Dunbar GL. Genetically engineered mesenchymal stem cells reduce behavioral deficits in the YAC 128 mouse model of Huntington's disease. Behavioural brain research. 2010;214:193–200. doi: 10.1016/j.bbr.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 184.Joyce N, Annett G, Wirthlin L, Olson S, Bauer G, Nolta JA. Mesenchymal stem cells for the treatment of neurodegenerative disease. Regenerative medicine. 2010;5:933–946. doi: 10.2217/rme.10.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Olson SD, Kambal A, Pollock K, Mitchell GM, Stewart H, Kalomoiris S, Cary W, Nacey C, Pepper K, Nolta JA. Examination of mesenchymal stem cell-mediated RNAi transfer to Huntington's disease affected neuronal cells for reduction of huntingtin. Molecular and cellular neurosciences. 2012;49:271–281. doi: 10.1016/j.mcn.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chiang MC, Lee YC, Huang CL, Chern Y. cAMP-response element-binding protein contributes to suppression of the A2A adenosine receptor promoter by mutant Huntingtin with expanded polyglutamine residues. The Journal of biological chemistry. 2005;280:14331–14340. doi: 10.1074/jbc.M413279200. [DOI] [PubMed] [Google Scholar]
- 187.Blum D, Hourez R, Galas MC, Popoli P, Schiffmann SN. Adenosine receptors and Huntington's disease: implications for pathogenesis and therapeutics. Lancet neurology. 2003;2:366–374. doi: 10.1016/s1474-4422(03)00411-3. [DOI] [PubMed] [Google Scholar]
- 188.Sun Z, Wang HB, Deng YP, Lei WL, Xie JP, Meade CA, Del Mar N, Goldowitz D, Reiner A. Increased calbindin-D28k immunoreactivity in striatal projection neurons of R6/2 Huntington's disease transgenic mice. Neurobiology of disease. 2005;20:907–917. doi: 10.1016/j.nbd.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 189.Huang Q, Zhou D, Sapp E, Aizawa H, Ge P, Bird ED, Vonsattel JP, DiFiglia M. Quinolinic acid-induced increases in calbindin D28k immunoreactivity in rat striatal neurons in vivo and in vitro mimic the pattern seen in Huntington's disease. Neuroscience. 1995;65:397–407. doi: 10.1016/0306-4522(94)00494-p. [DOI] [PubMed] [Google Scholar]
- 190.Pickel VM, Heras A. Ultrastructural localization of calbindin-D28k and GABA in the matrix compartment of the rat caudate-putamen nuclei. Neuroscience. 1996;71:167–178. doi: 10.1016/0306-4522(95)00441-6. [DOI] [PubMed] [Google Scholar]
- 191.Desplats PA, Lambert JR, Thomas EA. Functional roles for the striatal-enriched transcription factor, Bcl11b, in the control of striatal gene expression and transcriptional dysregulation in Huntington's disease. Neurobiology of disease. 2008;31:298–308. doi: 10.1016/j.nbd.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bibb JA, Yan Z, Svenningsson P, Snyder GL, Pieribone VA, Horiuchi A, Nairn AC, Messer A, Greengard P. Severe deficiencies in dopamine signaling in presymptomatic Huntington's disease mice. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6809–6814. doi: 10.1073/pnas.120166397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Luthi-Carter R, Strand A, Peters NL, Solano SM, Hollingsworth ZR, Menon AS, Frey AS, Spektor BS, Penney EB, Schilling G, Ross CA, Borchelt DR, Tapscott SJ, Young AB, Cha JH, Olson JM. Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Human molecular genetics. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- 194.Meador-Woodruff JH, Damask SP, Wang J, Haroutunian V, Davis KL, Watson SJ. Dopamine receptor mRNA expression in human striatum and neocortex. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 1996;15:17–29. doi: 10.1016/0893-133X(95)00150-C. [DOI] [PubMed] [Google Scholar]
- 195.Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, Elliston LA, Hartog C, Goldstein DR, Thu D, Hollingsworth ZR, Collin F, Synek B, Holmans PA, Young AB, Wexler NS, Delorenzi M, Kooperberg C, Augood SJ, Faull RL, Olson JM, Jones L, Luthi-Carter R. Regional and cellular gene expression changes in human Huntington's disease brain. Hum Mol Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 196.Pickel VM, Sumal KK, Reis DJ, Miller RJ, Hervonen A. Immunocytochemical localization of enkephalin and substance P in the dorsal tegmental nuclei in human fetal brain. The Journal of comparative neurology. 1980;193:805–814. doi: 10.1002/cne.901930315. [DOI] [PubMed] [Google Scholar]
- 197.Menalled L, Zanjani H, MacKenzie L, Koppel A, Carpenter E, Zeitlin S, Chesselet MF. Decrease in striatal enkephalin mRNA in mouse models of Huntington's disease. Experimental neurology. 2000;162:328–342. doi: 10.1006/exnr.1999.7327. [DOI] [PubMed] [Google Scholar]
- 198.Mercugliano M, Soghomonian JJ, Qin Y, Nguyen HQ, Feldblum S, Erlander MG, Tobin AJ, Chesselet MF. Comparative distribution of messenger RNAs encoding glutamic acid decarboxylases (Mr 65,000 and Mr 67,000) in the basal ganglia of the rat. The Journal of comparative neurology. 1992;318:245–254. doi: 10.1002/cne.903180302. [DOI] [PubMed] [Google Scholar]
- 199.Gourfinkel-An I, Parain K, Hartmann A, Mangiarini L, Brice A, Bates G, Hirsch EC. Changes in GAD67 mRNA expression evidenced by in situ hybridization in the brain of R6/2 transgenic mice. Journal of neurochemistry. 2003;86:1369–1378. doi: 10.1046/j.1471-4159.2003.01916.x. [DOI] [PubMed] [Google Scholar]
- 200.Lobo MK, Cui Y, Ostlund SB, Balleine BW, Yang XW. Genetic control of instrumental conditioning by striatopallidal neuron-specific S1P receptor Gpr6. Nature neuroscience. 2007;10:1395–1397. doi: 10.1038/nn1987. [DOI] [PubMed] [Google Scholar]
- 201.Mizushima K, Miyamoto Y, Tsukahara F, Hirai M, Sakaki Y, Ito T. A novel G-protein-coupled receptor gene expressed in striatum. Genomics. 2000;69:314–321. doi: 10.1006/geno.2000.6340. [DOI] [PubMed] [Google Scholar]
- 202.Van Waes V, Tseng KY, Steiner H. GPR88 - a putative signaling molecule predominantly expressed in the striatum: Cellular localization and developmental regulation. Basal ganglia. 2011;1:83–89. doi: 10.1016/j.baga.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Vargiu P, Morte B, Manzano J, Perez J, de Abajo R, Gregor Sutcliffe J, Bernal J. Thyroid hormone regulation of rhes, a novel Ras homolog gene expressed in the striatum. Brain research. Molecular brain research. 2001;94:1–8. doi: 10.1016/s0169-328x(01)00140-1. [DOI] [PubMed] [Google Scholar]