Summary
Controlling the position of the nucleus is vital for a number of cellular processes from yeast to humans. In Drosophila nurse cells, nuclear positioning is crucial during dumping, when nurse cells contract and expel their contents into the oocyte. We provide evidence that in nurse cells, continuous filopodia-like actin cables, growing from the plasma membrane and extending to the nucleus, achieve nuclear positioning. These actin cables move nuclei away from ring canals. When nurse cells contract, actin cables associate laterally with the nuclei, in some cases inducing nuclear turning so that actin cables become partially wound around the nuclei. Our data suggest that a perinuclear actin meshwork connects actin cables to nuclei via actin-crosslinking proteins such as the filamin Cheerio. We provide a revised model for how actin structures position nuclei in nurse cells, employing evolutionary conserved machinery.
Graphical Abstract
Highlights
-
•
Actin cables in Drosophila nurse cells are unsegmented filopodia-like structures
-
•
E-cadherin is required for the orientation of actin cables toward the nucleus
-
•
Nuclear positioning is achieved by continuous elongation of actin cables
-
•
Actin cables associate with perinuclear actin-containing crosslinkers like filamin
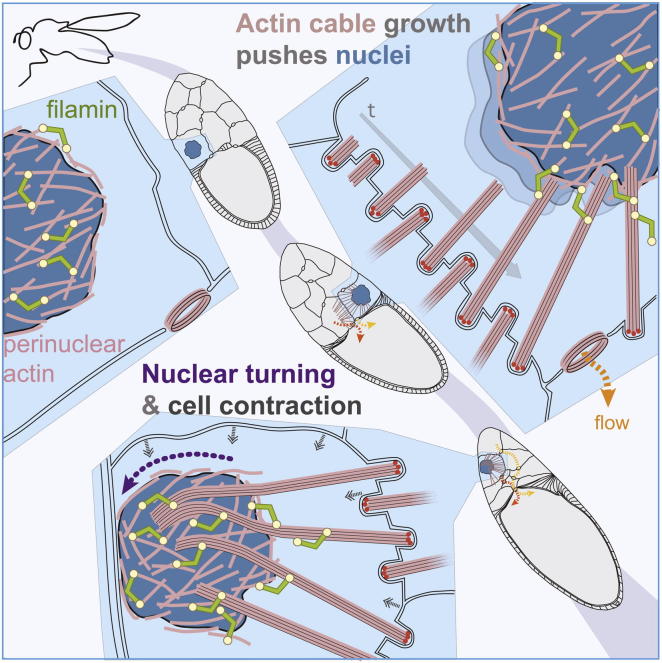
Huelsmann et al. propose a nuclear positioning model by visualizing the interplay between actin structures as Drosophila nurse cells contract to expel their contents. Filopodia-like actin cables grow from the plasma membrane, connect with an actin meshwork surrounding the nucleus, and push nuclei out of the path of cytoplasmic flow.
Introduction
Many cells control the position of their nucleus (e.g., during polarization, cell division, or cell migration). Nuclear mispositioning can lead to chromosomal defects during cell division and impairs cell migration (Burke and Roux, 2009). In humans, defects in nuclear positioning are linked to myopathies (Zhang et al., 2007), brain disorders (Tsai et al., 2007), and cancers (Doherty et al., 2010).
In order to position their nucleus, cells use cytoskeletal structures built of microtubules, actin, intermediate filaments or a combination of them, to generate force on their nucleus, either to push or pull it into the correct position (Dupin and Etienne-Manneville, 2011; Zhao et al., 2012). Recent studies revealed how actin structures and their link to the nuclear envelope can be critical for nuclear positioning (Folker et al., 2011; Luxton et al., 2010). When mouse fibroblasts polarize for migration, transmembrane actin-associated nuclear (TAN) lines link the retrograde flow of dorsal actin cables to nuclear movement in order to position the nucleus. Anchoring of the nucleus to the moving actin cables requires myosin and LINC (linker of nucleoskeleton and cytoskeleton) complexes, in which Nesprin-2G binds with its KASH domain to Sun2, forming a bridge between actin cables in the cytoplasm and A-type lamins inside the nucleus (Folker et al., 2011; Luxton et al., 2010). This connection between actin cables and nuclei is crucial for repolarization and migration of fibroblasts, but it is unclear whether the mechanism described above is common to diverse cell types.
In Drosophila, nurse cells regulate the position of their nuclei via actin cables during late oogenesis, where this process is essential for fertility (Cooley et al., 1992). During oogenesis, each daughter of a germline stem cell undergoes a series of four incomplete cell divisions to produce an oocyte and 15 nurse cells interconnected by ring canals (Spradling, 1993). Products synthesized in nurse cells are transported through the ring canals into the growing oocyte. Toward the end of oogenesis, nurse cells contract to expel their cytoplasmic contents into the oocyte, a process called “dumping” (Spradling, 1993). Just before dumping begins, arrays of cytoplasmic actin cables arise in the nurse cells that extend from the plasma membrane to the nucleus and keep the nucleus away from the ring canals during dumping. Mutations affecting the formation of actin cables result in nuclei that clog the ring canals producing small, “dumpless” eggs (Hudson and Cooley, 2002).
Previous research on the structure of the actin cables (Guild et al., 1997) developed a “fire engine extension ladder” model for the segmented appearance of actin cables, in which the cables are composed of small units of bundled actin filaments. All bundles consist of parallel actin filaments that point with their barbed ends toward the plasma membrane. The bundles of a single cable are thought to connect side by side, so that when nurse cells contract the bundles slide against each other to retract the cable, reminiscent of the retraction of an extension ladder on a fire engine. In electron micrographs, actin cables did not show any contact to the nuclei of nurse cells (Guild et al., 1997), and LINC complexes are not required for dumping (Technau and Roth, 2008; Xie and Fischer, 2008). This led to the view that the actin cables function as a passive obstacle to block nuclear movement toward the ring canals during dumping.
Despite these data, it was unclear how actin cables are generated and oriented toward the nucleus and how actin cables are able to prevent a soft nucleus from squeezing between them and blocking ring canals. Here, we have reexamined the structure and formation of actin cables and found that they are in fact similar to filopodia, i.e., unsegmented bundles of parallel actin filaments. Using live imaging, we observed actin cables making contact with the nucleus, and through continued elongation, they actively moved the nucleus away from the ring canals. As nurse cells contracted, the filopodia-like actin cables packed tightly around the nucleus, sometimes forcing the nucleus to turn, and did not retract in length. Finally, our data show that actin cables associate with a perinuclear actin meshwork containing the filamin Cheerio and other actin crosslinking proteins. Thus, we propose a revised model for nuclear positioning, in which elongating filopodia-like actin cables position the nucleus of a cell in association with perinuclear actin.
Results
Nurse Cell Actin Cables Are Unsegmented, Filopodia-like Structures
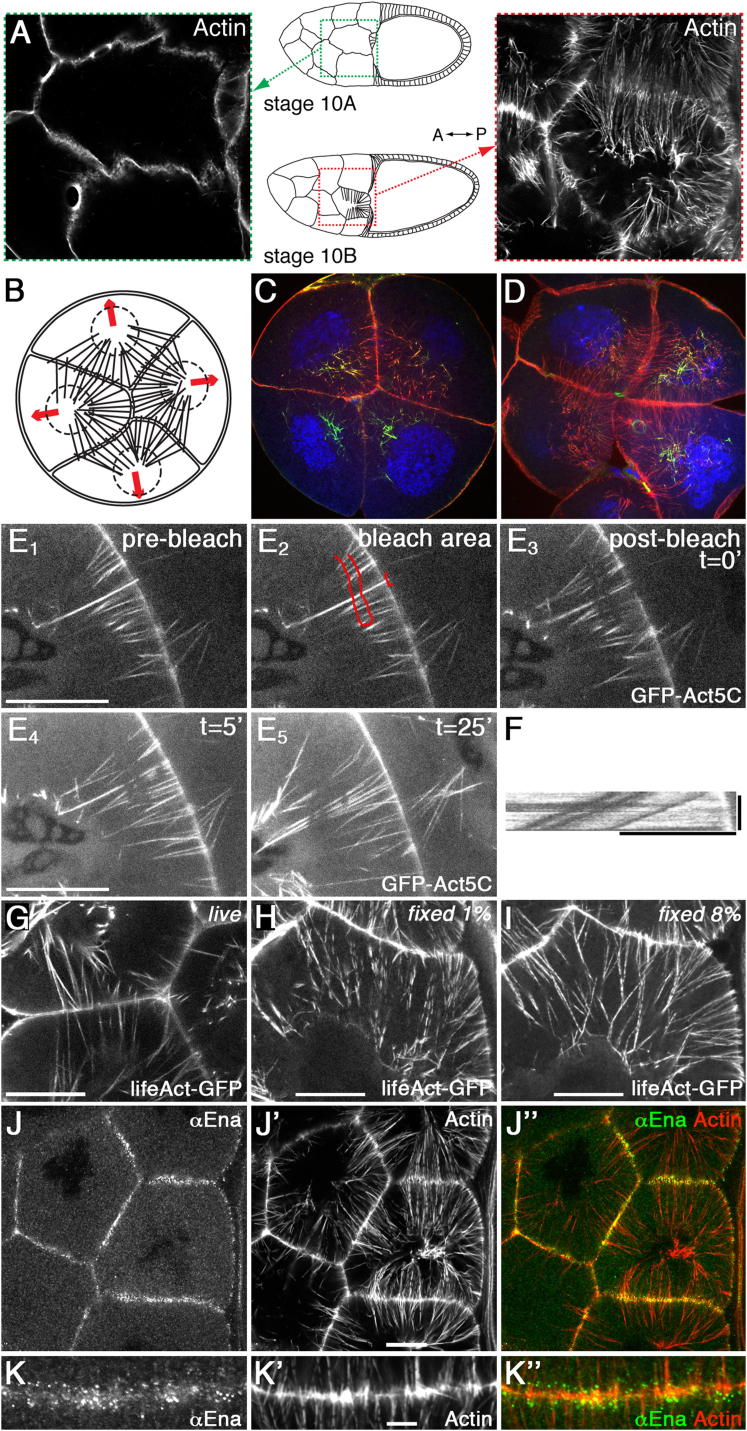
During stage 10B, nurse cells generate actin cables spanning from the membrane toward the nucleus (Figure 1A). Cross-sections revealed that these actin cables formed only at basolateral membranes between nurse cells and between nurse cells and the centripetal follicle cells/oocyte (Figures 1B–1D). Apical membranes of nurse cells facing the outer stretched follicle cells did not generate arrays of actin cables (Figures 1B–1D). In order to reveal the mechanisms generating actin cables and the dynamics of the process, we examined actin cables in live ovaries, using actin-green fluorescent protein (GFP), lifeact-GFP, and fascin-GFP. In all cases, the live-imaged actin cables appeared as continuous, unsegmented actin bundles extending from the membrane to the nucleus (Figures 1E and 1G; Figure S1B available online). However, in fixed samples actin cables appeared segmented (Figures 1H, 1I, and S1A). This might suggest that fixation and phalloidin staining reveal an underlying periodicity in the cable structure. However, changing the concentration of fixative or length of fixation altered the degree of segmentation (Figures 1H, 1I, and S1E–S1I), with both the segments and gaps became smaller in more strongly fixed samples (Figures S1E–S1I). Furthermore, the segment length and/or periodicity was variable, even along single actin cables (Figures S1E–S1I). These finding are inconsistent with an underlying regular segmentation of actin cables. Thus, we conclude that nurse cell actin cables are continuous filopodia-like structures.
Figure 1.
Actin Cables of Nurse Cells Are Filopodia-like Structures
(A) Illustration showing representative areas within the Drosophila egg chambers, from which most pictures were taken for this article; stage 10A is before actin cable formation, stage 10B is after formation of actin cable at the start of dumping.
(B–D) Cross-sections through nurse cell illustrating the membrane areas with actin cables. (B) A schematic drawing of four nurse cells with their nuclei and actin cables, surrounded by stretched follicle cells. (C) A cross-section through nurse cells of a stage 10B egg chamber (single z section). (D) A cross-section through nurse cells of a stage late10B/11 chamber (Venus-Cheerio in green, actin in red, nuclei in blue, projection of four z sections covering 6 μm). Note the size of the nuclei compared to the length of actin cables.
(E) Live confocal microscopy images from Movie S1 showing the parts of two nurse cells expressing GFP-Act5C (stage 10B). E1 shows the actin cables before bleaching, E2 the bleached area depicted in red, and E3 the postbleach image. Note that the smaller bleached area affected the indentation containing the tip complex. E4 and E5 display two time frames of Movie S1 from the bleached cables (each a projection of four z sections covering 3 μm).
(F) Kymograph of bleached points along one of the actin cables in Movie S1. Horizontal bar represents 10 μm, vertical bar represents 20 min.
(G) Live-imaged actin cables marked with Lifeact-GFP.
(H) Actin cables marked with Lifeact-GFP and fixed with 1% formaldehyde (projection of two z sections covering 1 μm).
(I) Actin cables marked with Lifeact-GFP and fixed with 8% formaldehyde (projection of two z sections covering 1 μm).
(J) Enabled localized to the tip complex of each actin cable, which resided in a small membrane indentation extending into the neighboring cell (J, αEnabled; J′, actin; J″, overlay).
(K) Enlargement highlighting the tip complexes in membrane indentations (K, αEnabled; K′, actin; K″, overlay). Scale bar represents 5 μm.
Unless otherwise stated, scale bars represent 20 μm in all figures, and each picture is a single confocal section.
Filopodial actin filaments nucleate from a tip complex and are bundled by specific proteins (Faix et al., 2009). We therefore asked if the distribution of actin nucleators and actin bundling proteins is consistent with actin cables being filopodia-like. As previously shown (Gates et al., 2009), the filopodia tip complex protein Enabled localized in a dot at the membrane-associated end of each actin cable (Figures 1J and 1K). Similarly, Pico, the single ortholog of Riam and Lamellipodin (Lyulcheva et al., 2008), localized to the tip complex when tagged with GFP and expressed under its endogenous promoter (data not shown, Figure 2A), colocalizing with Enabled (Figure S2A). The tip complexes and part of the actin cables were situated in small membrane protrusions with an average length of ∼1 μm (Figure 1K) (Guild et al., 1997; Gutzeit, 1986; Riparbelli and Callaini, 1995). The actin-bundling proteins Fascin (Drosophila Singed) and Villin (Drosophila Quail) were localized along the entire actin cable length, as previously reported (Cant and Cooley, 1996; Mahajan-Miklos and Cooley, 1994; Zanet et al., 2009), and we found that Drosophila Fimbrin/Plastin also localized to actin cables (Figures S1C and S1D). Thus, like other continuous filopodia-like structures, nurse cell actin cables have a tip complex and the actin-bundling proteins Fascin, Villin and Fimbrin.
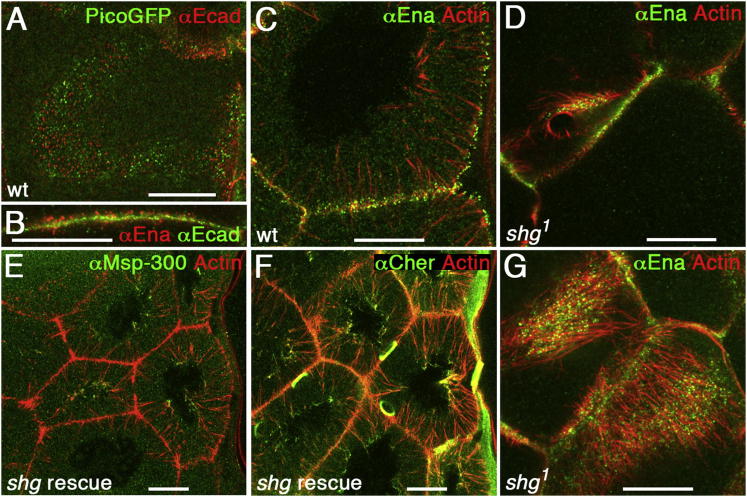
Figure 2.
E-Cadherin Is Required for Actin Cable Orientation
(A) E-cadherin complexes (red) were interspersed between tip complexes of actin cables marked with PicoGFP (green) along the plasma membrane between nurse cells.
(B) Enlargement showing the E-cadherin in green and the tip complexes in red (αEnabled).
(C) In wild-type nurse cells, actin cables extended to the nucleus perpendicular to the cortex. Tip complexes, green (αEnabled); actin, red.
(D) In shg1 germline clones, actin cables formed, had a tip complex, but do not span the cytoplasm but lie on the membrane marked by ring canal and tip complexes (αEnabled, green; actin, red). See also Movies S2 and S3.
(E and F) Egg chambers mutant for shg are rescued by ubiquitous expression of E-cadherin-GFP (genotype: shgR69/shg1; ubi::E-cadherin-GFP). Actin cables are oriented toward the nucleus as in wild-type egg chambers (E and F). Msp-300 (E) and Cheerio (F) localize to the perinuclear ends of actin cables.
(G) Projection of four z sections spanning 3 μm of nurse cells shown in (D) with actin cables lying on the membrane marked by tip complexes (αEnabled, green; actin, red).
In filopodia and microvilli, actin cables grow from a tip complex at the interface with the plasma membrane and protrude outward. We reasoned actin cables may also grow at the membrane from a tip complex, but extend inward. To test this, we marked points along GFP-Actin cables in live samples by bleaching and followed cable growth by time-lapse confocal microscopy. In all cases the bleached points moved away from the plasma membrane (Figures 1E and 1F; Movie S1). The rate of growth of the cables was remarkably steady and proceeded at ∼0.3 μm/min, as shown in the kymograph in Figure 1F. All bleached points remained as discrete points as the cables extended, demonstrating that all actin filaments within each cable elongated at the same rate (Figures 1E and 1F; Movie S1). These data establish actin cables as filopodia-like bundles extending in parallel from the membrane toward the nucleus.
E-Cadherin Is Essential for Orientation of Actin Cables toward the Nucleus
One obvious difference between other filopodia-like structures that protrude outward and actin cables, which only elongate inward into the cell, is that actin cables extend from membranes adjacent to other cells. This raised the idea that cell-cell adhesion could be crucial for inward-growing actin cables (e.g., tight cell-cell adhesion between nurse cells could block actin cables protruding outward). Consistent with this, E-cadherin is required for cell-cell adhesion of nurse cells and dumping (Oda et al., 1997). We found that E-cadherin and β-catenin colocalized in clusters interspersed between tip complexes of actin cables (Figures 2A and 2B) suggesting that adhesion could act locally on actin cables. To test the role of E-cadherin, we generated germline clones of mutations in the gene encoding E-cadherin, shotgun (shg1 and shgR69). However, loss of E-cadherin did not prevent actin cable formation nor did it cause sustained extension of membrane protrusions containing actin cables. Instead, actin cables fell over so that they were lying flat on the plasma membrane rather than standing perpendicular to it (Figures 2C, 2D, 2G, and S2B–S2D; Movies S2 and S3), and in general they were oriented in the same direction (Figure 2G). The tip complex proteins Enabled and Pico still clustered at the membrane at the tip of each actin cable (Figures 2D and 2G and not shown). Consistent with actin cables failing to reach the nuclei, some nuclei blocked ring canals, explaining why shg mutant eggs are dumpless (Figure 5E). We confirmed that these phenotypes were due to a lack of E-cadherin function as GFP-tagged E-cadherin rescued them (Figures 2E, 2F, 5E). Thus, E-cadherin junctional complexes interspersed between the tip complexes of actin cables are essential for the orientation of actin cables and thereby for nuclear positioning.
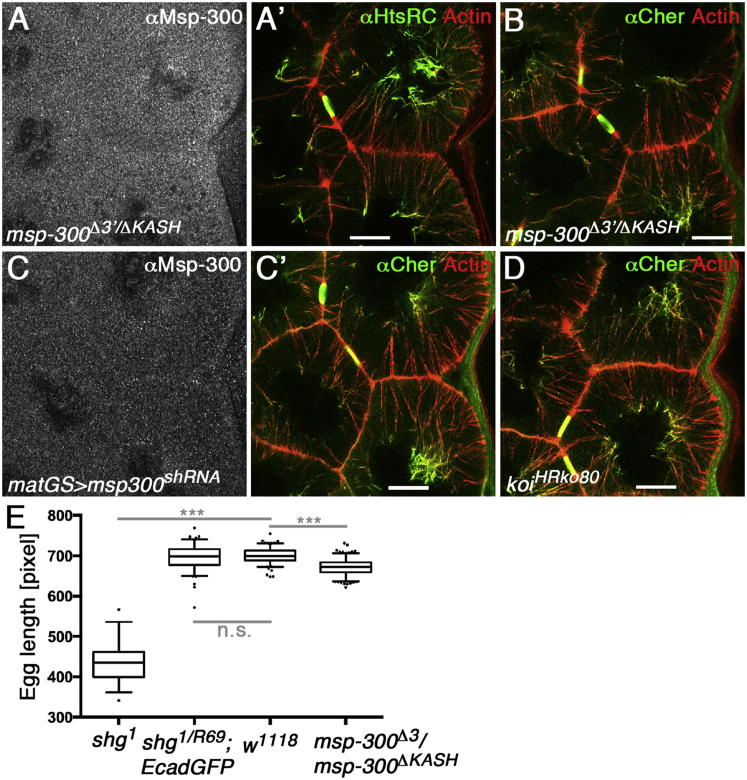
Figure 5.
LINC Complexes Are Not Essential to Link Actin Cables to the Nucleus in Nurse Cells
(A and B) Deletion of the KASH domain eliminates localization of Msp-300 from actin cables but does not affect the actin cables or their contact to the nucleus (A, αMsp-300; A′, overlay of αHtsRC with actin; B, overlay of αCheerio with actin).
(C) Knockdown of Msp-300 eliminates localization of Msp-300 from actin cables but does not affect actin cables nor nuclear positioning (C, αMsp-300; C′, overlay of αCheerio with actin).
(D) Eliminating the SUN protein Koi in koiHRko80 mutants does not affect the localization of Cheerio to the distal ends of actin cables nor nuclear positioning (overlay of αCheerio in green with actin in red).
(E) Box and whisker graph of egg length quantification; boxes are extending from the 25th to the 75th percentile, the line in the middle shows the median, whiskers are extending from the 5th to the 95th percentile, points below or above the whiskers are shown as individual dots (shg1 n = 39, shg1/shgR69;ubi::E-cadherin-GFP n = 105, w1118 n = 142, msp-300ΔKASH/msp-300Δ3′ n = 193; ∗∗∗p < 0.001, Student’s t test, n.s. = not significant).
See also Figure S4.
Nuclear Positioning through Actin Cable Elongation
The fact that actin cables were filopodia-like and not retractable ladder-like structures demanded an alternative mechanism to explain the behavior of actin cables during nurse cell contraction. Our time-lapse movies revealed two further aspects of actin cable behavior during dumping. First, once actin cables have reached the nucleus, actin cable elongation continued and actin cables pushed against the nucleus. This led to the deformation of the nucleus and eventually to its localization away from the ring canals (Figures 3A–3C; Movies S4A and S4B). This suggests that actin cables actively position the nuclei rather than just being a passive barrier preventing nuclei from clogging ring canals.
Figure 3.
Actin Cables Associate Laterally around Turning Nuclei
(A) Six time frames from Movie S4A showing the formation of actin cables and their behavior during dumping. Following the nurse cell at the bottom right (marked with an asterisk, t = 0′), actin cables extended toward the nucleus (t = 10′), pushed against the nucleus (t = 20′), and forced the nucleus toward the side of the nurse cell (t = 30′). Subsequently, the nucleus turned with associated actin cables, which grew over it (t = 40′ and t = 50′). Each frame is a projection of five z sections covering 4 μm; due to the projection, actin cables appear sometimes segmented. Note that the entire length of some actin cables are not in the projected focal planes and so falsely appear unconnected to the plasma membrane.
(B) Actin cables pushing nuclei to the outside in a stage 11 egg chamber; note the deformation of the asterisked nucleus (projection of three z sections covering 2 μm).
(C) During nurse cell contraction actin cables continue growing, bundle at the nucleus, and start to associate laterally with it (projection of three z sections covering 4 μm).
(D) After contraction at stage 14, actin cables localize around the nurse cell nuclei (projection of six z sections covering 5 μm).
See also Movies S4A–S4C.
Second, during further cell contraction, actin cables continued growing and clustered together at the nuclei. The continued growth is accompanied by distortion or turning of the nuclei (Figures 3A and 3C; Movies S4A–S4C). The last frame of Movie S4B shows how actin cables bend during the partial turning. Due to the large size of the nucleus, this partial turn or distortion is sufficient to provide lateral association with most of the length of actin cables (see cross-sections in Figures 1B–1D). At the end of dumping, the remaining actin cables were located in bundles around the nuclei (Figure 3D). These data indicate that as nurse cells contract, the actin cables partially wrap around the nuclei rather than shortening.
Actin-Associated Proteins Bind Differentially along Actin Cables
The live imaging described above suggested that actin cables are actively linked to the nucleus. First, we never observed an example where the ends of the cables that are closest to the nucleus (hereafter called the perinuclear ends of actin cables) became detached from the nucleus. On the contrary, Movie S4B shows that occasionally actin cables detach from the plasma membrane and appear to be pulled toward the nucleus (marked by yellow arrow in Movie S4B). Second, the close proximity of the stiff actin cables with turning nuclei suggests a firm association of actin cables with nuclei.
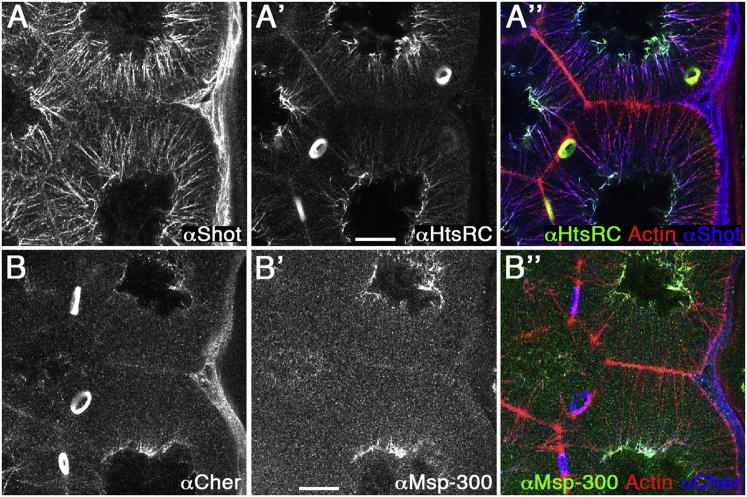
In order to identify mechanisms linking actin cables to nuclei, we examined the distribution of several actin-associated proteins in nurse cells. We found four proteins enriched at the perinuclear end of actin cables, which contains the pointed ends of the actin filaments. The spectraplakin Short stop (Shot), an actin-microtubule crosslinker, and the HtsRC isoform of the adducin-like Hu li tai shao, a potential actin bundling and capping protein, localized in a gradient along actin cables, with their highest levels close to the nucleus (Figures 4A and S3A–S3E). The filamin Cheerio, an actin crosslinker, and the nesprin Msp-300, a KASH protein that binds to the nuclear envelope, were more concentrated at the perinuclear end of actin cables (Figure 4B), with Msp-300 more confined to the perinuclear ends than Cheerio. Before dumping, Msp-300, HtsRC and Cheerio appeared in patches around the nucleus and Shot localized to microtubules around the nucleus, in the cytoplasm and at the cortex (Figure S3F and data not shown). This adds to the previously described localization of Cheerio and HtsRC to ring canals (Petrella et al., 2007; Sokol and Cooley, 1999) and the perinuclear localization of Msp300 in early nurse cells and oocytes (Technau and Roth, 2008; Xie and Fischer, 2008; Yu et al., 2006).
Figure 4.
Actin-Binding Proteins at the Perinuclear Ends of Actin Cables
(A) The spectraplakin Shot and the adducin-like HtsRC localized in gradients along actin cables with their highest expression at the perinuclear end of actin cables (A, αShot; A′, αHtsRC; A″, overlay with actin staining).
(B) The perinuclear end of actin cables is marked by Cheerio and Msp-300 (B, αCheerio; B′, αMsp-300; B″, overlay with actin staining).
See also Figure S3.
Actin Cables Connect to Nuclei Independently of LINC Complexes
We hypothesized that LINC complexes may connect actin cables to nurse cell nuclei, especially because the KASH domain protein Msp-300 localized to the perinuclear end of actin cables (Figure 4B). Quantification of egg lengths revealed that Msp-300 mutant eggs were slightly smaller than wild-type eggs (Figure 5E), showing a minor impairment of dumping. But, as previously reported (Technau and Roth, 2008; Xie and Fischer, 2008), we found that deletion of the Msp-300 KASH domain did not have an obvious effect on actin cables or nuclear positioning, even though Msp-300 was no longer localized to the perinuclear ends of actin cables (Figure 5A). Shot, Cheerio, and HtsRC localized normally in Msp-300 mutants (Figures 5A, 5B, and S4A). Because it was shown that KASH-independent forms of Msp-300 anchor nuclei to the actomyosin compartment in striated muscles (Elhanany-Tamir et al., 2012), we repeated our experiments using RNAi affecting both, KASH-dependent and KASH-independent forms of Msp-300 (Ni et al., 2011). This led to an effective reduction of Msp-300 protein, but normal actin cables (Figures 5C and S4B). Similarly, the deletion of the SUN-protein encoded by klaroid (koi) did not lead to nuclear mispositioning or mislocalization of Cheerio, Shot, or HtsRC (Figures 5D and S4C). These data demonstrate that only a minor part of dumping depends on Msp-300 function and suggest that other mechanisms are involved in connecting actin cables to nuclei.
Alternative Mechanisms Lead to Concentration of Proteins at the Perinuclear End of Actin Cables
To confirm that actin cables contact the nucleus, we examined how the actin-associated proteins Shot, HtsRC, Cheerio, and Msp-300 become enriched at the perinuclear end of actin cables. We envisioned two mechanisms that could explain how proteins localize differentially along actin cables: first, it depends on intrinsic actin cable features (i.e., the actin cable close to the membrane is different from the perinuclear end), or second, the localization to the perinuclear ends depends on the close proximity to the nucleus (i.e., proteins at the nucleus associate with actin cables only after the cables have reached the nucleus).
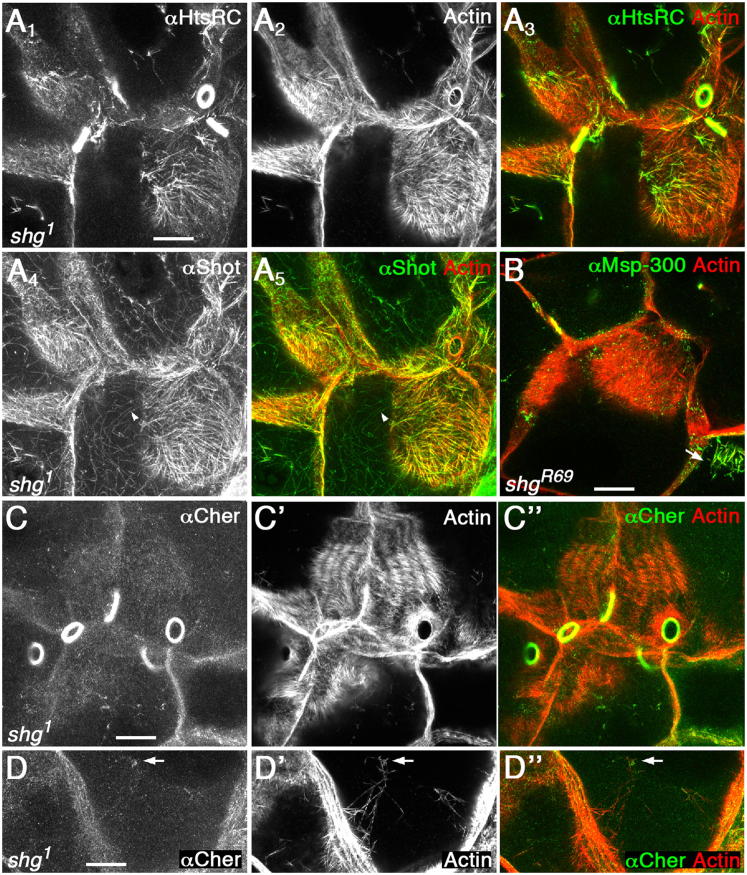
We distinguished between these two mechanisms by examining the distribution of these proteins on actin cables that have not reached the nucleus, using the E-cadherin mutants described above. In shg mutant nurse cells, HtsRC and Shot still localized to actin cables lying on the plasma membrane (Figure 6A), whereas in contrast, Cheerio and Msp-300 did not (Figures 6B–6D). In rare cases when actin cables did reach the nucleus in the absence of E-cadherin, Cheerio and Msp-300 accumulated at their ends (Figures 6B and 6D, arrows). Adding back E-cadherin-GFP in shg mutant egg chambers restored the localization of Msp-300 and Cheerio (Figures 2E and 2F). Thus, our results supported both mechanisms: the enrichment of HtsRC and Shot toward the pointed end of the actin filaments reflects an inherent property of the actin cable itself, whereas Cheerio and Msp-300 require actin cable proximity to the nuclei to accumulate at the perinuclear ends.
Figure 6.
Two Mechanisms of Protein Concentration at the Perinuclear Ends of Actin Cables
(A) In shg mutants, HtsRC and Shot still bound to actin cables with no contact to the nucleus. Note, the actin-microtubule crosslinker Shot also localized to microtubules (arrowhead in A4 and A5).
(B–D) In contrast, Msp-300 (B) and Cheerio (C and D) did not bind actin cables lying on the membrane of shg mutants. They only localized to actin cables that appeared to be in contact with the nucleus (arrows in B and D).
Images of egg chambers derived from germline clones of the E-cadherin mutants shg1 (A, C, D) or shgR69 (B).
The Filamin Cheerio Reveals a Perinuclear Actin Network in Nurse Cells
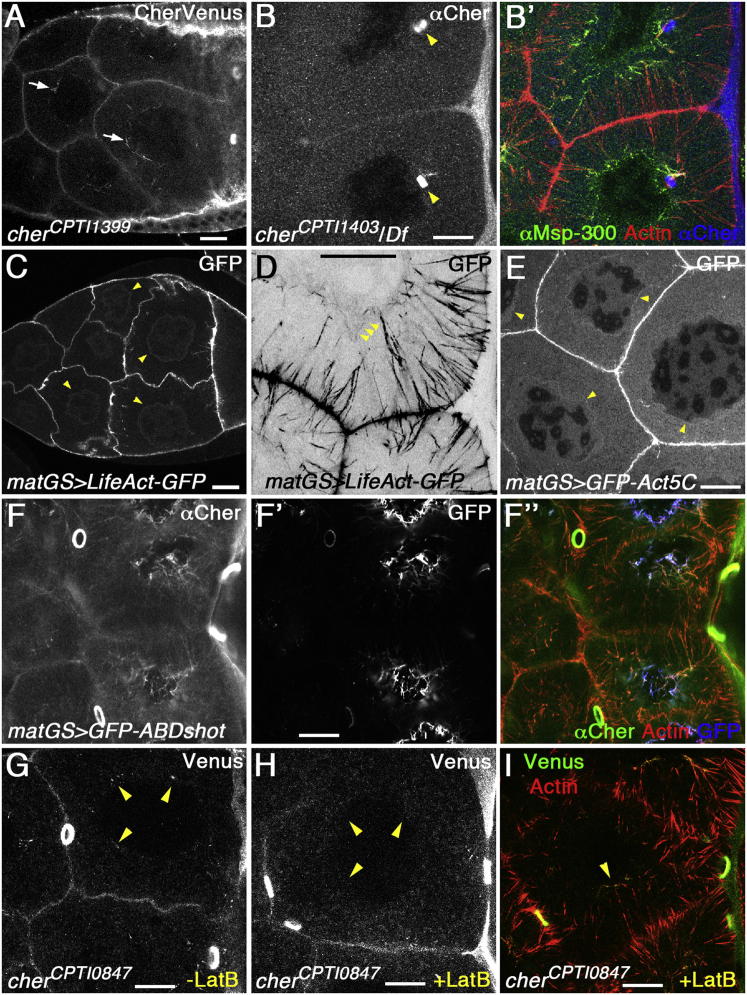
Because Cheerio only accumulated on actin cables that had reached the nucleus, we examined whether the Cheerio localized to the nucleus prior to the formation of actin cables. We used a viable gene trap that inserts Venus into the N-terminal part of filamin repeat 12 after the first hinge (Rees et al., 2011). We found that Cheerio-Venus localized in a weak filamentous pattern around the nucleus long before actin cables formed (Figure 7A). This early perinuclear Cheerio localization was sensitive to extensive detergent treatment; in nurse cells stained with antibodies we detected early perinuclear Cheerio in irregular patches. Msp-300 colocalized with Cheerio in these perinuclear patches (Figure S3F). Another gene trap, in which Venus replaces part of the actin-binding domain of Cheerio, failed to localize around both the nucleus and the perinuclear ends of actin cables (Figure 7B). Even though the mutant Cheerio-Venus localized to ring canals, ring canal development was disrupted and egg chambers were dumpless. Thus, perinuclear localization of Cheerio requires a functional actin-binding domain, suggesting that Cheerio binds to a perinuclear actin structure. To confirm the presence of perinuclear actin we examined the distribution of Lifeact-GFP and GFP-Actin5C and found that both had a perinuclear localization prior to actin cable formation (Figures 7C and 7E). The perinuclear actin was also identified as halos around nuclei in live images of egg chambers expressing GFP-Actin (Movie S4B). During dumping, perinuclear actin was also visible with Lifeact-GFP or Fascin-GFP, in the presence of actin cables (Figures 7D and S1A). The presence of perinuclear actin filaments is further supported by two genetic manipulations that stabilized perinuclear actin and increased the perinuclear accumulation of Msp-300 and Cheerio: the expression of the actin-binding domain of Shot (Figures 7F, S5A, and S5B) and mutation of the ovarian tumor gene (Figures S5C–S5E) (Rodesch et al., 1997).
Figure 7.
The Perinuclear Actin Meshwork of Nurse Cells
(A) Image of Cheerio marked with a viable insertion of YFP into filamin repeat 12 (CPTI-001399) at stage 9 of oogenesis. Cheerio marked a perinuclear filamentous structure (arrows) before the formation of actin cables.
(B) Image of the YFP insertion CPTI-001403 disrupting the actin binding domain of Cheerio trans-heterozygous over the deficiency Df(3R)Exel6176. The insertion abolished the localization of Cheerio to the perinuclear ends of actin cables (B, Cheerio; B′, overlay of Cheerio with actin and Msp-300). The mutant protein still localized to ring canals (yellow arrowheads in B) but ring canal development is severely disrupted.
(C) Lifeact-GFP marked perinuclear actin at stage 9 of oogenesis, before the formation of actin cables. The arrows highlight GFP-positive lines around nuclei.
(D) Lifeact-GFP also marked the perinuclear actin during dumping (highlighted by the arrows), in the presence of actin cables (inverted image).
(E) Actin5C-GFP also marked perinuclear actin before the formation of actin cables. The arrows highlight GFP-positive lines around nuclei.
(F) Overexpression of the actin-binding domain of Shot stabilized the perinuclear actin network (F, Cheerio; F′, GFP; F″, overlay of Cheerio with actin and GFP).
(G) Control egg chamber showed perinuclear accumulations of Cheerio-Venus before the formation of actin cables (stage 10a).
(H) At the same developmental stage, 500 μM LatB abolished perinuclear Cheerio-Venus.
(I) After the formation of actin cables (stage 10b), LatB reduced Cheerio-Venus at the perinuclear ends of actin cables (Cheerio-Venus in green, actin in red). The yellow arrowheads within nuclei point to the border between nuclei and cytoplasm (G-I).
In order to perturb perinuclear actin, we applied different concentrations of Latrunculin B (LatB) and Cytochalasin D (CytD) to egg chambers in culture and assayed perinuclear actin and Cheerio localization. Prior to actin cable formation, LatB caused disassembly of perinuclear actin filaments (Movie S5A) and loss of perinuclear Cheerio (Figures 7G and 7H). This fits with Cheerio being recruited by binding to perinuclear actin. The presence of LatB during dumping did not affect already formed actin cables but reduced Cheerio at the perinuclear ends (Figure 7I; Movie S5D). The application of CytD resulted in the dramatic formation of cytoplasmic actin patches, before and after actin cable formation. Cheerio did not localize to these actin patches (Figures S5F–S5H), even when they were close to the nucleus, indicating that there is specificity to the binding of Cheerio to perinuclear actin. Finally, inhibition of actin polymerization resulted in the immediate block of dumping (Movies S5B and S5C) supporting the model that cortical actin and nonmuscle Myosin II mediate nurse cell contraction (Wheatley et al., 1995) but precluding our assessment of perinuclear actin function during dumping.
The dissection of the specific function of the association of actin cables with perinuclear actin during dumping is difficult because (1) Cheerio, Shot, and HtsRC are essential during early oogenesis (Sokol and Cooley, 1999; Röper and Brown, 2004; Petrella et al., 2007), and (2) LatB not only disrupted perinuclear actin but also blocked nurse cell contraction (Movies S5B and S5C). Therefore, we attempted to knock down Cheerio function after it had performed its ring canal function by RNAi- and GFP-mediated degradation (Caussinus et al., 2012; Ni et al., 2011), but without success: RNAi-mediated knockdown did not sufficiently reduce Cheerio to cause an effect, whereas degradation of Cheerio-GFP caused ring canal defects (data not shown). In summary, all of our data are consistent with a model in which elongating actin cables position nuclei by crosslinking with perinuclear actin filaments, but the robustness and overlapping functions of proteins within the actin structures of the nurse cell have prevented us from specifically disrupting and testing the role of this link between actin cables and perinuclear actin.
Discussion
Here, we propose a revised model for nuclear positioning in Drosophila nurse cells. Filopodia-like actin cables grow from the plasma membrane toward the nucleus just prior to dumping. The orientation of actin cables toward the nucleus requires the function of E-cadherin in the membrane. During dumping, actin cables associate with a perinuclear actin meshwork and position the nucleus away from the ring canals. When nurse cells contract further, actin cables make lateral associations with turning nuclei and form irregular bundles. What are the implications of this revised model?
Our results demonstrate that actin cables in Drosophila nurse cells share many features with filopodia-like structures like filopodia, microvilli, or stereocilia. Actin cables are unsegmented bundles of parallel extending actin filaments, with their barbed ends in a tip complex within a membrane indentation. The presence of a membrane-associated tip complex, from which actin cables elongate, and the binding of bundling proteins are in accordance with their function and distribution on other bundles of parallel actin filaments (Lin et al., 2005). These findings eliminate several issues of the ladder model (e.g., how ladder units are generated or kept aligned). How then do actin cables get segmented? A periodic structure in the actin cables, which is revealed by fixation and staining by breakage and depolymerization at weak “nodes” in the structure, should generate segment/gap lengths that correlate with the extent of fixation (e.g., stronger fixation would shorten gaps and lengthen segments). Instead, stronger fixation reduced both segment and gap length. The variability in the unit length “gap + segment” is inconsistent with this interpretation. We also ruled out that the segmentation reflects regions of the cable that are inaccessible to phalloidin, because we see segments with GFP-actin in fixed samples. We speculate that the actin cables breaking and the new free ends depolymerizing during fixation cause the segmented appearance.
Our live imaging revealed that actin filaments elongate in parallel within cables from the membrane at a rate of 0.3 μm/min. This elongation rate is similar to microvilli (0.21 μm/min) (Loomis et al., 2003) and slow-growing filopodia (0.3–1.0 μm/min) (Mallavarapu and Mitchison, 1999), whereas stereocilia are much slower (up to 0.007 μm/min) (Manor and Kachar, 2008), and fast-growing filopodia can extend much faster (1–60 μm/min) (Mallavarapu and Mitchison, 1999; Medalia et al., 2007). In contrast to actin cables, these bundles generate outward-directed protrusions, rather than cytoplasmic actin cables. Actin cables do protrude a short distance, but it remains unclear what limits their outward protrusion and generates their inward elongation. We have ruled out that the short protrusions arise from the “pushing back” of the nucleus, as we see protrusions in cables that have yet to reach the nucleus. The pointed ends of the actin bundles of filopodia and microvilli become integrated with actin networks at the membrane, cortical actin, or terminal web, respectively, which may brace the outward directed protrusion (Gupton and Gertler, 2007). The actin cables must either lack the mechanism to connect with such a bracing structure at the membrane, or this connection is not strong enough to overcome the resistance of pushing into the adjacent cell. From filopodia, it is also known that they coordinate actin elongation with membrane extension during their outgrowth (Mattila et al., 2007). For actin cables, we found that E-cadherin complexes interspersed between tip complexes of actin cables did not restrict the outgrowth of membrane protrusions harboring the tip complexes in nurse cells. Instead, E-cadherin is required for the orientation of actin cable toward the nucleus. Loss of E-cadherin also leads to cell membrane defects (Oda et al., 1997) and thereby might affect cell contraction as well. Thus, E-cadherin could establish a specific cortical tension within the cell to aid perpendicular growth of actin cables. Alternatively, E-cadherin-dependent struts may link to the actin cables to hold them perpendicular. Actin filaments generated by E-cadherin complexes, as shown in mammalian systems (Kobielak et al., 2004; Kovacs et al., 2002), could orient cable outgrowth. It is also possible that E-cadherin adhesion affects indirectly the orientation of actin cables (e.g., by generating specific membrane domains or by modulating either the signaling to or the expression of actin regulators).
Our live imaging also sheds light on the behavior of actin cables during dumping. It revealed that actin cables actively pushed nuclei away from ring canals and during nurse cell contraction their lateral sides become associated with turning nuclei. Thus, the process of nuclear positioning during cell contraction looks more like curling up a fire hose than retracting a fire ladder. In addition, the observed actin cables lying around the nuclei reveals why multiple cables appear adjacent to each other following nurse cell contraction (Guild et al., 1997). The nuclear positioning by actin cable elongation can also explain how the start of nurse cells contraction close to the oocyte (Gutzeit and Koppa, 1982) does not block the flow of material from more distal nurse cells. These must expel their cytoplasm through already contracted proximal nurse cells. Because actin cables positioned the nucleus to the side of nurse cells, the nuclei of the almost empty nurse cells do not encumber the flow-through of cytoplasm from the more distal nurse cells. Thus, the sustained growth of actin cables and their wrapping around the nucleus could allow the efficient transport of the entire contents of all nurse cells into the oocyte.
Finally, our data reveal a perinuclear actin meshwork and suggest that actin cables associate with the nucleus via the perinuclear actin during nuclear positioning. This connection might enable the stiff actin cables, which nonetheless do bend under pressure (see Movies S4A and S4B), to stay attached to the turning nuclei. We have not been able to test genetically the possible functions of the link between actin cables and the perinuclear actin, because mutations in proteins localizing to the perinuclear ends of actin cables affect early oogenesis or the ring canals that are also needed for dumping. We also suspect that the actin binding proteins function redundantly during the association of actin cables to perinuclear actin. Recently, we also found α-Actinin and α-Spectrin localized to actin cable perinuclear ends (unpublished data) supporting the idea that multiple actin binding proteins ensure a robust association of actin cables with perinuclear actin. The idea of a molecular connection between actin cables and perinuclear actin is supported by the observation that (1) the filamin Cheerio and the nesprin-like Msp-300 only localize to perinuclear ends of actin cables when they are in contact with the nucleus, and (2) disrupting perinuclear actin leads to a loss of perinuclear Cheerio and reduction of Cheerio at actin cable perinuclear ends. Thus, we propose a model in which perinuclear actin crosslinkers like Cheerio connect actin cables to the perinuclear actin once the cables reach the nuclear envelope. An alternative model that we cannot exclude is that these crosslinking proteins travel with or along the elongating actin cables and get stabilized at the pointed ends of actin cables once they reach the perinuclear actin. Nonetheless, both models require linkage between the actin cable ends and the perinuclear actin. This link is further supported by the report that actin cables remain associated with dissected nuclei of nurse cells (Gutzeit, 1986).
Multiple indirect links to the nucleus via a perinuclear actin meshwork might permit actin cables to move nuclei with a weak nucleoskeleton. A relative low stiffness of the nuclear envelope is supported by actin cables distorting nuclei (Figure 3; Movies S4A–S4C) and multilobed nurse cell nuclei in EM micrographs (Guild et al., 1997). With a flexible nuclear envelope, direct links between actin cables and nuclear envelope might be inefficient at withstanding the force generated by growing actin cables and nurse cell contraction, which can be overcome by connecting to an actin meshwork surrounding the nucleus. This could explain why, in contrast to other tissues, LINC complexes have a redundant role, if any, in connecting actin cables to nurse cell nuclei (Elhanany-Tamir et al., 2012; Khatau et al., 2012; Luxton et al., 2010; Technau and Roth, 2008; Xie and Fischer, 2008). Alternatively, other proteins at the nuclear envelope could connect to actin cables. Nuclear pore complexes have been linked to dumping in Drosophila, but their exact role in this process is unknown (Gigliotti et al., 1998; Riparbelli et al., 2007).
Compared to nuclear positioning by bundles of parallel actin filaments via TAN lines in mammalian cells (Luxton et al., 2010), our data show that nurse cells employ diverse mechanisms that differ in the nature of the link between actin cables and nuclei (see above) and in the driving force that positions the nucleus. The driving force in mouse fibroblasts is the Myosin II-dependent retrograde flow of dorsal actin cables (Luxton et al., 2010) whereas in nurse cells it is actin polymerization. Another example of actin bundles exerting force on nuclei are apical actin caps that regulate nuclear shape in cultured mammalian cells (Gay et al., 2011). Here, Myosin II-containing bundles of parallel actin filaments extend over the nucleus and connect with their ends to basal focal adhesions (Khatau et al., 2009). Interestingly, Filamin A is involved in the formation of theses parallel actin bundles. Despite the above-mentioned differences, TAN lines, apical caps, and actin cables involve perinuclear actin (Gay et al., 2011; Luxton et al., 2010) and filamins (Gay et al., 2011). This suggests that the fundamental molecular machinery is conserved but its configuration is adapted to a variety of cellular contexts.
Experimental Procedures
Fly Stocks
Fly stocks used from the Bloomington Stock Center are: w1118, Df(3R)Exel6176, Df(2L)Exel6011, Df(2R)Exel6050; TRiP-HMS00368, TRiP-HMS00632, TRiP-GL00344, TRiP-HMS01501 (Ni et al., 2011). Fly stocks used from Kyoto stock center are: cherCPTI001399, cherCPTI000847, cherCPTI001403, fimCPTI003498 (Rees et al., 2011). Other fly stocks used are: fimCC01493 (Buszczak et al., 2007); FRTG13 shot3 (Röper and Brown, 2004), Msp-300ΔKASH (Xie and Fischer, 2008), Msp-300Δ3′ (Technau and Roth, 2008), koiHRko80 (Kracklauer et al., 2007), UASP-GFP_Actin5C (Röper et al., 2005), UASP-GFP_Fascin (Zanet et al., 2009), FRTG13 shg1 (González-Reyes and St Johnston, 1998), FRT42D shgR69 (Godt and Tepass, 1998), mat-tub-gal4_GeneSwitch (abbreviated as matGS, gift from N. Lowe), UASP-GFP_ABDshot (gift from K. Röper); shgR69;DEFL#23 and shgR69;DEFL#65 are amorphic mutants of E-cadherin rescued with ubi::E-cadherin-GFP (Haruta et al., 2010). Germline clones for shg1 were generated using FRTG13 ovoD/T(1;2)OR64/CyO, clones of shgR69 using FRT42D ubiGFP(S65T)nls/CyO. UASP-Lifeact lines were generated according to Riedl et al. (2008), optimized to Drosophila codon usage, and tagged with C-terminal fluorescent proteins. UASP-dGradFP lines were generated according to Caussinus et al. (2012). All flies were kept at 25°C; heat shocks were induced for 1 hr in a 37°C water bath on 2 consecutive days during larval stages. Before ovary dissections, 1-day-old flies were kept on fresh baker’s yeast for 1–2 days.
Live Imaging and Culture of Egg Chambers
Live imaging of dissected ovaries was performed as described in Cliffe et al. (2007). Ovaries were imaged on an Olympus FluoView1000 inverted confocal. For bleaching experiments, actin cables were imaged, bleached using the line tool of the Olympus software, and then imaged as z stacks over time. Time series images were handled and processed in ImageJ, which was also used to generate kymographs. To mark the nucleus, we added Hoechst 33342 (Invitrogen) to the medium (1.5 μg/ml final concentration). To disrupt the actin cytoskeleton egg chambers were incubated in medium containing different concentrations of Latrunculin B (500–0.5 μM, Invitrogen) or Cytochalasin D (400–5 μg/ml, Invitrogen). In controls the medium contained the equivalent amount of DMSO only.
Immunofluorescence and Microscopy
For antibody stainings, ovaries were dissected in PBS and subsequently fixed with 4% methanol-free formaldehyde (Polyscience) in PBS at room temperature for 10–30 min. After blocking and permeabilization with PBS with 0.5% BSA and 0.3% Triton X-100 (PBT), ovaries were incubated in PBT containing primary or secondary antibodies at 4°C over night. The following primary antibodies were used: antibodies maintained by Developmental Studies Hybridoma Bank: 5G2 αEnabled (1:10), αHtsRC (1:10), DCAD2 αE-Cadherin (1:10); other antibodies: αMsp-300 (1:400; Volk, 1992), αCheerio (1:400, αNtCher recognizing the N terminus of Cheerio; Sokol and Cooley, 2003), and αShot (1:1,000; Strumpf and Volk, 1998). Secondary antibodies coupled to Alex488 or Cy3 were used at 1:200 (Invitrogen) or coupled to Cy5 at 1:100 (Jackson ImmunoResearch Laboratories). We used Rhodamine-phalloidin (1:400) or Alexa647-phalloidin (1:200) from Invitrogen to stain the actin cytoskeleton in fixed tissue. Fluorescent proteins were directly detected. All samples were mounted in Vectashield (Vector Laboratories). For ovary cross-section we adapted a protocol for Drosophila embryos (Narasimha and Brown, 2006). Images were generated on an Olympus FluoView1000 upright confocal and then analyzed and further processed using ImageJ and Photoshop CS4. For egg length measurements eggs were rinsed in water, dried, and mounted in 3S Voltalef oil, and imaged with a 10× objective on a Leica DMR microscope with a MacroFire camera (Optronix). Egg lengths were determined using ImageJ, graphs generated using Prism 6 (GraphPad) and the means of the different genotypes tested for significant differences using Student’s t tests.
Acknowledgments
We would like to thank K. Röper for comments on the manuscript. Thank you also to K. Röper, D. Nashchekin, E. Caussinus, L. Cooley, N. Lowe, H. Oda, S. Plaza, S. Roth, P. Rørth, U. Tepass, the Bloomington and Kyoto Drosophila Stock Centres, and the Developmental Studies Hybridoma Bank (Iowa) for providing reagents. We are indebted to J. Overton for excellent technical assistance and A. Cliffe and the St Johnston laboratory for sharing their expertise in live imaging egg chambers and oogenesis, respectively. We would also like to thank the reviewers for their constructive comments. This work was funded by Wellcome Trust grant 086451 to N.H.B., Academy of Finland grants 135473 and 138327 to J.Y., and Gurdon Institute core funding from the Wellcome Trust (092096) and Cancer Research UK (CRUK) (C6946/A14492).
Published: September 30, 2013
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information includes five figures and five movies and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2013.08.014.
Supplemental Information
Time-lapse movie of bleached actin cables (see Figures 1A and 1B) in nurse cells expressing GFPActin5C under the control of matGS-gal4. Actin cables extended continuously from the membrane to the nucleus (projection of four z sections spanning 3 μm; recorded 1 frame/30 s and playing with 10 frames/s, total length 22 min; genotype: mat-gal4.GS>UASp-GFP_actin5C).
Animated z stack through fixed and stained wild-type nurse cells showing actin cables and tip complexes (1 μm between each section, covering a total of 21 μm; αEnabled in green, actin in red; playing with 10 frames/s; genotype: w1118).
Animated z stack through fixed and stained shg1 mutant nurse cells showing actin cables and tip complexes lying on the membrane (1 μm between each section, covering a total of 11 μm; αEnabled in green, actin in red; playing with 10 frames/s; genotype: shg1).
Egg chambers of the genotype mat-gal4.GS>UASp-GFP_actin5C are shown.
(A) Frames 1–132. Actin cables wrap around turning nuclei during nurse cell contraction. Single frames are shown in Figure 3. The movie was created from z projections of five sections spanning 8 μm, recorded 1 frame/60 s, and playing at 10 frames/s, total length 120 min. The stage position readjusted partway through. Due to the distance between projected sections, some actin cables falsely appear segmented. In addition, the entire length of some actin cables is not in the focal planes, so these cables incorrectly appear unattached from the membrane.
(B) Frames 134–233. Outgrowth of actin cables and nurse cell dumping. As discussed in the text, an actin cable pulled toward the nucleus is marked with a yellow arrow. The movie frames are projections of three sections spanning 2 μm, recorded 1 frame/91.5 s, and playing at 10 frames/s, total length 128 min.
(C) Frames 235–303. Continued growth of actin cables leading to the turning of nuclei during nurse cell contraction. Nuclear turning is clearest in the marked nucleus (actin in grey, overlay of actin in green, and Hoechst in magenta). The movie is a single section, recorded at 1 frame/62 s and playing at 10 frames/s.
(A) Frames 1–66. Latrunculin B leads to the disappearance of perinuclear actin-GFP (while increasing nuclear actin-GFP). Latrunculin B (125 μM) was added at frame “36 min.” Actin is white or green and Hoechst staining of nuclei is magenta. The movie is a single section recorded at 1 frame/60.3 s and playing at 10 frames/s. The genotype was mat-gal4.GS>UASp-GFP_actin42A.
(B) Frames 68–114. Latrunculin B blocks dumping. Addition of 125 μM Latruculin B at frame “12 min” led to a halt of dumping but not to the disassembly of actin cables. Actin is white or green and Hoechst is magenta. The movie is single section recorded at 1 frame/60.2 s and playing at 10 frames/s. The genotype was mat-gal4.GS>UASp-GFP_actin5C.
(C) Frames 118–218. Latrunculin B blocks dumping. Continuation of (B) showing that dumping behavior does not recover within the time of the movie.
(D) Frames 220–266. Cheerio in an egg chamber treated with Latrunculin B. Addition of Latrunculin B at frame “33 min” reduced Cheerio-GFP (white or green) at the perinuclear ends of actin cables; nuclei marked with Hoechst are in magenta. The movie is a single section recorded at 1 frame/118 s and playing at 10 frames/s. The genotype was cherCPTI000847.
References
- Burke B., Roux K.J. Nuclei take a position: managing nuclear location. Dev. Cell. 2009;17:587–597. doi: 10.1016/j.devcel.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Buszczak M., Paterno S., Lighthouse D., Bachman J., Planck J., Owen S., Skora A.D., Nystul T.G., Ohlstein B., Allen A. The Carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cant K., Cooley L. Single amino acid mutations in Drosophila fascin disrupt actin bundling function in vivo. Genetics. 1996;143:249–258. doi: 10.1093/genetics/143.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussinus E., Kanca O., Affolter M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 2012;19:117–121. doi: 10.1038/nsmb.2180. [DOI] [PubMed] [Google Scholar]
- Cliffe A., Poukkula M., Rørth P. Culturing Drosophila egg chambers and imaging border cell migration. Nat. Protoc. 2007 Published online July 23, 2007. [Google Scholar]
- Cooley L., Verheyen E., Ayers K. chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophila oogenesis. Cell. 1992;69:173–184. doi: 10.1016/0092-8674(92)90128-y. [DOI] [PubMed] [Google Scholar]
- Doherty J.A., Rossing M.A., Cushing-Haugen K.L., Chen C., Van Den Berg D.J., Wu A.H., Pike M.C., Ness R.B., Moysich K., Chenevix-Trench G., Australian Ovarian Cancer Study Management Group. Australian Cancer Study (Ovarian Cancer) Ovarian Cancer Association Consortium (OCAC) ESR1/SYNE1 polymorphism and invasive epithelial ovarian cancer risk: an Ovarian Cancer Association Consortium study. Cancer Epidemiol. Biomarkers Prev. 2010;19:245–250. doi: 10.1158/1055-9965.EPI-09-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin I., Etienne-Manneville S. Nuclear positioning: mechanisms and functions. Int. J. Biochem. Cell Biol. 2011;43:1698–1707. doi: 10.1016/j.biocel.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Elhanany-Tamir H., Yu Y.V., Shnayder M., Jain A., Welte M., Volk T. Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J. Cell Biol. 2012;198:833–846. doi: 10.1083/jcb.201204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J., Breitsprecher D., Stradal T.E.B., Rottner K. Filopodia: Complex models for simple rods. Int. J. Biochem. Cell Biol. 2009;41:1656–1664. doi: 10.1016/j.biocel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Folker E.S., Ostlund C., Luxton G.W.G., Worman H.J., Gundersen G.G. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc. Natl. Acad. Sci. USA. 2011;108:131–136. doi: 10.1073/pnas.1000824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates J., Nowotarski S.H., Yin H., Mahaffey J.P., Bridges T., Herrera C., Homem C.C.F., Janody F., Montell D.J., Peifer M. Enabled and Capping protein play important roles in shaping cell behavior during Drosophila oogenesis. Dev. Biol. 2009;333:90–107. doi: 10.1016/j.ydbio.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay O., Gilquin B., Nakamura F., Jenkins Z.A., McCartney R., Krakow D., Deshiere A., Assard N., Hartwig J.H., Robertson S.P., Baudier J. RefilinB (FAM101B) targets filamin A to organize perinuclear actin networks and regulates nuclear shape. Proc. Natl. Acad. Sci. USA. 2011;108:11464–11469. doi: 10.1073/pnas.1104211108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigliotti S., Callaini G., Andone S., Riparbelli M.G., Pernas-Alonso R., Hoffmann G., Graziani F., Malva C. Nup154, a new Drosophila gene essential for male and female gametogenesis is related to the nup155 vertebrate nucleoporin gene. J. Cell Biol. 1998;142:1195–1207. doi: 10.1083/jcb.142.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godt D., Tepass U. Drosophila oocyte localization is mediated by differential cadherin-based adhesion. Nature. 1998;395:387–391. doi: 10.1038/26493. [DOI] [PubMed] [Google Scholar]
- González-Reyes A., St Johnston D. The Drosophila AP axis is polarised by the cadherin-mediated positioning of the oocyte. Development. 1998;125:3635–3644. doi: 10.1242/dev.125.18.3635. [DOI] [PubMed] [Google Scholar]
- Guild G.M., Connelly P.S., Shaw M.K., Tilney L.G. Actin filament cables in Drosophila nurse cells are composed of modules that slide passively past one another during dumping. J. Cell Biol. 1997;138:783–797. doi: 10.1083/jcb.138.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton S.L., Gertler F.B. Filopodia: the fingers that do the walking. Sci. STKE. 2007;2007:re5. doi: 10.1126/stke.4002007re5. [DOI] [PubMed] [Google Scholar]
- Gutzeit H.O. The role of microfilaments in cytoplasmic streaming in Drosophila follicles. J. Cell Sci. 1986;80:159–169. doi: 10.1242/jcs.80.1.159. [DOI] [PubMed] [Google Scholar]
- Gutzeit H., Koppa R. Time-lapse film analysis of cytoplasmic streaming during late oogenesis of Drosophila. J. Embryol. Exp. Morphol. 1982;67:101–111. [Google Scholar]
- Haruta T., Warrior R., Yonemura S., Oda H. The proximal half of the Drosophila E-cadherin extracellular region is dispensable for many cadherin-dependent events but required for ventral furrow formation. Genes Cells. 2010;15:193–208. doi: 10.1111/j.1365-2443.2010.01389.x. [DOI] [PubMed] [Google Scholar]
- Hudson A.M., Cooley L. Understanding the function of actin-binding proteins through genetic analysis of Drosophila oogenesis. Annu. Rev. Genet. 2002;36:455–488. doi: 10.1146/annurev.genet.36.052802.114101. [DOI] [PubMed] [Google Scholar]
- Khatau S.B., Hale C.M., Stewart-Hutchinson P.J., Patel M.S., Stewart C.L., Searson P.C., Hodzic D., Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. USA. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatau S.B., Kusuma S., Hanjaya-Putra D., Mali P., Cheng L., Lee J.S.H., Gerecht S., Wirtz D. The differential formation of the LINC-mediated perinuclear actin cap in pluripotent and somatic cells. PLoS ONE. 2012;7:e36689. doi: 10.1371/journal.pone.0036689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobielak A., Pasolli H.A., Fuchs E. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat. Cell Biol. 2004;6:21–30. doi: 10.1038/ncb1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs E.M., Goodwin M., Ali R.G., Paterson A.D., Yap A.S. Cadherin-directed actin assembly: E-cadherin physically associates with the Arp2/3 complex to direct actin assembly in nascent adhesive contacts. Curr. Biol. 2002;12:379–382. doi: 10.1016/s0960-9822(02)00661-9. [DOI] [PubMed] [Google Scholar]
- Kracklauer M.P., Banks S.M.L., Xie X., Wu Y., Fischer J.A. Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly (Austin) 2007;1:75–85. doi: 10.4161/fly.4254. [DOI] [PubMed] [Google Scholar]
- Lin H.W., Schneider M.E., Kachar B. When size matters: the dynamic regulation of stereocilia lengths. Curr. Opin. Cell Biol. 2005;17:55–61. doi: 10.1016/j.ceb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Loomis P.A., Zheng L., Sekerková G., Changyaleket B., Mugnaini E., Bartles J.R. Espin cross-links cause the elongation of microvillus-type parallel actin bundles in vivo. J. Cell Biol. 2003;163:1045–1055. doi: 10.1083/jcb.200309093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton G.W.G., Gomes E.R., Folker E.S., Vintinner E., Gundersen G.G. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyulcheva E., Taylor E., Michael M., Vehlow A., Tan S., Fletcher A., Krause M., Bennett D. Drosophila pico and its mammalian ortholog lamellipodin activate serum response factor and promote cell proliferation. Dev. Cell. 2008;15:680–690. doi: 10.1016/j.devcel.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S., Cooley L. The villin-like protein encoded by the Drosophila quail gene is required for actin bundle assembly during oogenesis. Cell. 1994;78:291–301. doi: 10.1016/0092-8674(94)90298-4. [DOI] [PubMed] [Google Scholar]
- Mallavarapu A., Mitchison T. Regulated actin cytoskeleton assembly at filopodium tips controls their extension and retraction. J. Cell Biol. 1999;146:1097–1106. doi: 10.1083/jcb.146.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor U., Kachar B. Dynamic length regulation of sensory stereocilia. Semin. Cell Dev. Biol. 2008;19:502–510. doi: 10.1016/j.semcdb.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila P.K., Pykäläinen A., Saarikangas J., Paavilainen V.O., Vihinen H., Jokitalo E., Lappalainen P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J. Cell Biol. 2007;176:953–964. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medalia O., Beck M., Ecke M., Weber I., Neujahr R., Baumeister W., Gerisch G. Organization of actin networks in intact filopodia. Curr. Biol. 2007;17:79–84. doi: 10.1016/j.cub.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Narasimha M., Brown N.H. Confocal microscopy of Drosophila embryos. In: Celis J.E., editor. Cell Biology: A Laboratory Handbook. Academic Press; New York: 2006. pp. 77–86. [Google Scholar]
- Ni J.-Q., Zhou R., Czech B., Liu L.-P., Holderbaum L., Yang-Zhou D., Shim H.-S., Tao R., Handler D., Karpowicz P. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H., Uemura T., Takeichi M. Phenotypic analysis of null mutants for DE-cadherin and Armadillo in Drosophila ovaries reveals distinct aspects of their functions in cell adhesion and cytoskeletal organization. Genes Cells. 1997;2:29–40. doi: 10.1046/j.1365-2443.1997.d01-284.x. [DOI] [PubMed] [Google Scholar]
- Petrella L.N., Smith-Leiker T., Cooley L. The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development. 2007;134:703–712. doi: 10.1242/dev.02766. [DOI] [PubMed] [Google Scholar]
- Rees J.S., Lowe N., Armean I.M., Roote J., Johnson G., Drummond E., Spriggs H., Ryder E., Russell S., St Johnston D. In vivo analysis of proteomes and interactomes using parallel affinity capture (iPAC) coupled to mass spectrometry. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.002386. M110.002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli M.G., Callaini G. Cytoskeleton of the Drosophila egg chamber: new observations on microfilament distribution during oocyte growth. Cell Motil. Cytoskeleton. 1995;31:298–306. doi: 10.1002/cm.970310406. [DOI] [PubMed] [Google Scholar]
- Riparbelli M.G., Gigliotti S., Callaini G. The Drosophila nucleoporin gene nup154 is required for correct microfilament dynamics and cell death during oogenesis. Cell Motil. Cytoskeleton. 2007;64:590–604. doi: 10.1002/cm.20206. [DOI] [PubMed] [Google Scholar]
- Rodesch C., Pettus J., Nagoshi R.N. The Drosophila ovarian tumor gene is required for the organization of actin filaments during multiple stages in oogenesis. Dev. Biol. 1997;190:153–164. doi: 10.1006/dbio.1997.8697. [DOI] [PubMed] [Google Scholar]
- Röper K., Brown N.H. A spectraplakin is enriched on the fusome and organizes microtubules during oocyte specification in Drosophila. Curr. Biol. 2004;14:99–110. [PubMed] [Google Scholar]
- Röper K., Mao Y., Brown N.H. Contribution of sequence variation in Drosophila actins to their incorporation into actin-based structures in vivo. J. Cell Sci. 2005;118:3937–3948. doi: 10.1242/jcs.02517. [DOI] [PubMed] [Google Scholar]
- Sokol N.S., Cooley L. Drosophila filamin encoded by the cheerio locus is a component of ovarian ring canals. Curr. Biol. 1999;9:1221–1230. doi: 10.1016/s0960-9822(99)80502-8. [DOI] [PubMed] [Google Scholar]
- Sokol N.S., Cooley L. Drosophila filamin is required for follicle cell motility during oogenesis. Dev. Biol. 2003;260:260–272. doi: 10.1016/s0012-1606(03)00248-3. [DOI] [PubMed] [Google Scholar]
- Spradling A.C. Developmental genetics of oogenesis. In: Bate M., Martinez Arias A., editors. The Development of Drosophila Melanogaster. Cold Spring Harbor Laboratory Press; New York: 1993. pp. 1–70. [Google Scholar]
- Strumpf D., Volk T. Kakapo, a novel cytoskeletal-associated protein is essential for the restricted localization of the neuregulin-like factor, vein, at the muscle-tendon junction site. J. Cell Biol. 1998;143:1259–1270. doi: 10.1083/jcb.143.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau M., Roth S. The Drosophila KASH domain proteins Msp-300 and Klarsicht and the SUN domain protein Klaroid have no essential function during oogenesis. Fly (Austin) 2008;2:82–91. doi: 10.4161/fly.6288. [DOI] [PubMed] [Google Scholar]
- Tsai J.-W., Bremner K.H., Vallee R.B. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Volk T. A new member of the spectrin superfamily may participate in the formation of embryonic muscle attachments in Drosophila. Development. 1992;116:721–730. doi: 10.1242/dev.116.3.721. [DOI] [PubMed] [Google Scholar]
- Wheatley S., Kulkarni S., Karess R. Drosophila nonmuscle myosin II is required for rapid cytoplasmic transport during oogenesis and for axial nuclear migration in early embryos. Development. 1995;121:1937–1946. doi: 10.1242/dev.121.6.1937. [DOI] [PubMed] [Google Scholar]
- Xie X., Fischer J.A. On the roles of the Drosophila KASH domain proteins Msp-300 and Klarsicht. Fly (Austin) 2008;2:74–81. doi: 10.4161/fly.6108. [DOI] [PubMed] [Google Scholar]
- Yu J., Starr D.A., Wu X., Parkhurst S.M., Zhuang Y., Xu T., Xu R., Han M. The KASH domain protein MSP-300 plays an essential role in nuclear anchoring during Drosophila oogenesis. Dev. Biol. 2006;289:336–345. doi: 10.1016/j.ydbio.2005.10.027. [DOI] [PubMed] [Google Scholar]
- Zanet J., Stramer B., Millard T., Martin P., Payre F., Plaza S. Fascin is required for blood cell migration during Drosophila embryogenesis. Development. 2009;136:2557–2565. doi: 10.1242/dev.036517. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Bethmann C., Worth N.F., Davies J.D., Wasner C., Feuer A., Ragnauth C.D., Yi Q., Mellad J.A., Warren D.T. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum. Mol. Genet. 2007;16:2816–2833. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- Zhao T., Graham O.S., Raposo A., St Johnston D. Growing microtubules push the oocyte nucleus to polarize the Drosophila dorsal-ventral axis. Science. 2012;336:999–1003. doi: 10.1126/science.1219147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapse movie of bleached actin cables (see Figures 1A and 1B) in nurse cells expressing GFPActin5C under the control of matGS-gal4. Actin cables extended continuously from the membrane to the nucleus (projection of four z sections spanning 3 μm; recorded 1 frame/30 s and playing with 10 frames/s, total length 22 min; genotype: mat-gal4.GS>UASp-GFP_actin5C).
Animated z stack through fixed and stained wild-type nurse cells showing actin cables and tip complexes (1 μm between each section, covering a total of 21 μm; αEnabled in green, actin in red; playing with 10 frames/s; genotype: w1118).
Animated z stack through fixed and stained shg1 mutant nurse cells showing actin cables and tip complexes lying on the membrane (1 μm between each section, covering a total of 11 μm; αEnabled in green, actin in red; playing with 10 frames/s; genotype: shg1).
Egg chambers of the genotype mat-gal4.GS>UASp-GFP_actin5C are shown.
(A) Frames 1–132. Actin cables wrap around turning nuclei during nurse cell contraction. Single frames are shown in Figure 3. The movie was created from z projections of five sections spanning 8 μm, recorded 1 frame/60 s, and playing at 10 frames/s, total length 120 min. The stage position readjusted partway through. Due to the distance between projected sections, some actin cables falsely appear segmented. In addition, the entire length of some actin cables is not in the focal planes, so these cables incorrectly appear unattached from the membrane.
(B) Frames 134–233. Outgrowth of actin cables and nurse cell dumping. As discussed in the text, an actin cable pulled toward the nucleus is marked with a yellow arrow. The movie frames are projections of three sections spanning 2 μm, recorded 1 frame/91.5 s, and playing at 10 frames/s, total length 128 min.
(C) Frames 235–303. Continued growth of actin cables leading to the turning of nuclei during nurse cell contraction. Nuclear turning is clearest in the marked nucleus (actin in grey, overlay of actin in green, and Hoechst in magenta). The movie is a single section, recorded at 1 frame/62 s and playing at 10 frames/s.
(A) Frames 1–66. Latrunculin B leads to the disappearance of perinuclear actin-GFP (while increasing nuclear actin-GFP). Latrunculin B (125 μM) was added at frame “36 min.” Actin is white or green and Hoechst staining of nuclei is magenta. The movie is a single section recorded at 1 frame/60.3 s and playing at 10 frames/s. The genotype was mat-gal4.GS>UASp-GFP_actin42A.
(B) Frames 68–114. Latrunculin B blocks dumping. Addition of 125 μM Latruculin B at frame “12 min” led to a halt of dumping but not to the disassembly of actin cables. Actin is white or green and Hoechst is magenta. The movie is single section recorded at 1 frame/60.2 s and playing at 10 frames/s. The genotype was mat-gal4.GS>UASp-GFP_actin5C.
(C) Frames 118–218. Latrunculin B blocks dumping. Continuation of (B) showing that dumping behavior does not recover within the time of the movie.
(D) Frames 220–266. Cheerio in an egg chamber treated with Latrunculin B. Addition of Latrunculin B at frame “33 min” reduced Cheerio-GFP (white or green) at the perinuclear ends of actin cables; nuclei marked with Hoechst are in magenta. The movie is a single section recorded at 1 frame/118 s and playing at 10 frames/s. The genotype was cherCPTI000847.