Significance
Here we report that addiction to histone lysine demethylase (KDM)6B expression is shared by a number of cervical carcinoma lines and arises as a direct and immediate consequence of human papillomavirus (HPV)16 E7 oncoprotein expression. We also report that the p16INK4A tumor suppressor is a mediator of KDM6B addiction and that p16 expression is necessary for survival of HPV E7-expressing cells. The addiction of p16INK4A is based on its ability to inhibit CDK4/CDK6 activity. We also present data that KDM6 inhibition may be evaluated as a therapeutic modality for high-risk HPV-associated lesions and cancers.
Keywords: synthetic lethality, apoptosis, biomarker, cancer therapy
Abstract
The tumor suppressor p16INK4A inhibits formation of enzymatically active complexes of cyclin-dependent kinases 4 and 6 (CDK4/6) with D-type cyclins. Oncogenic stress induces p16INK4A expression, which in turn triggers cellular senescence through activation of the retinoblastoma tumor suppressor. Subversion of oncogene-induced senescence is a key step during cancer development, and many tumors have lost p16INK4A activity by mutation or epigenetic silencing. Human papillomavirus (HPV)-associated tumors express high levels of p16INK4A in response to E7 oncoprotein expression. Induction of p16INK4A expression is not a consequence of retinoblastoma tumor suppressor inactivation but is triggered by a cellular senescence response and is mediated by epigenetic derepression through the H3K27-specific demethylase (KDM)6B. HPV E7 expression causes an acute dependence on KDM6B expression for cell survival. The p16INK4A tumor suppressor is a critical KDM6B downstream transcriptional target and its expression is critical for cell survival. This oncogenic p16INK4A activity depends on inhibition of CDK4/CDK6, suggesting that in cervical cancer cells where retinoblastoma tumor suppressor is inactivated, CDK4/CDK6 activity needs to be inhibited in order for cells to survive. Finally, we note that HPV E7 expression creates a unique cellular vulnerability to small-molecule KDM6A/B inhibitors.
Posttranslational histone modifications, including phosphorylation, ubiquitination, acetylation, and methylation, impact both the physical state and the transcriptional competence of chromatin. These modifications are dynamic and regulate a variety of cellular processes, such as stem cell maintenance, cell fate determination and maintenance, cell cycle control, and epigenetic heritability of transcriptional programs (reviewed in refs. 1 and 2). Methylation of lysine residues on histones is modulated by histone lysine methyltransferases (KMTs) and histone lysine demethylases (KDMs). Although histone lysine methylation is involved in both transcriptional activation and repression, histone H3 trimethylation of lysine 27 (H3K27me3) on gene promoters is crucial for epigenetic silencing mediated by polycomb group proteins (3–5). The JmjC-domain containing histone demethylases, KDM6A (UTX) and KDM6B (JMJD3) remove this repressive mark, allowing for transcriptional activation (6–10). Although KDM6A and KDM6B appear identical with regards to catalytic activities and histone substrate specificities, they have a number of nonoverlapping and nonredundant biological activities. KDM6B—but not KDM6A—regulates RAS/RAF-mediated oncogene-induced senescence (OIS) (11, 12). OIS was originally described as a cell-abortive response to ectopic RAS oncogene expression in normal human cells. It is now recognized that OIS represents one of several cell-intrinsic tumor-suppressor responses that function to eliminate aberrantly proliferating, potentially premalignant cells. Evidence for OIS has been detected in premalignant lesions, including prostatic intraepithelial neoplasia, melanoma, colon adenoma, and lung adenoma (13–15). It represents a major barrier to malignant progression, and additional genetic or epigenetic lesions are necessary for progression to invasive cancer. Consistent with this notion, frank tumors contain lower numbers of senescent cells than premalignant lesions (15).
The retinoblastoma tumor suppressor (pRB) is a key mediator of OIS. Activation of pRB in response to OIS occurs through transcriptional induction of the cyclin-dependent kinases 4 and 6 (CDK4/6) inhibitor and tumor suppressor p16INK4A. In normal cells p16INK4A transcription is epigenetically repressed through a polycomb repressive complex-dependent mechanism (16). OIS causes KDM6B-mediated H3K27 demethylation and increased p16INK4A expression, which in turn causes G1 arrest and senescence through activation of the pRB tumor suppressor (11, 12). Presumably to inactivate OIS, p16INK4A is frequently mutated or silenced by CpG methylation in human tumors. However, some human cancers—most notably those caused by infections with high-risk human papillomaviruses (HPVs),—express p16INK4A at high levels. We have previously shown that the HPV16 E7 oncoprotein triggers p16INK4A expression through an OIS-related mechanism that involves KDM6B-mediated H3K27 demethylation. Inactivation of pRB does not signal HPV16 E7 OIS-mediated p16INK4A induction, but is necessary to quench OIS signaling (17, 18). Hence, although high-level p16INK4A expression is a valuable biomarker for high-risk HPV-associated lesions and cancers, given that the OIS response is inactivated by pRB degradation, one would surmise that high-level p16INK4A expression in HPV+ tumors is biologically inconsequential. Surprisingly, however, KDM6B depletion in the HPV16+ CaSki cervical carcinoma cell line caused a dramatic decrease in viability (17). We hypothesize that the observed KDM6B “addiction” of CaSki cells arose as a consequence of HPV16 E7 expression and was caused by KDM6B-mediated alterations in host gene expression.
Here we report that p16INK4A is a key downstream transcriptional mediator of KDM6B addiction. Those cervical carcinoma lines that require KDM6B also require p16INK4A expression for survival. The requirement of KDM6B and p16INK4A expression for cervical carcinoma survival arises as immediate and direct consequences of HPV16 E7 expression, even in cells that do not depend on E7 expression for proliferation and survival. A KDM6 selective small-molecule inhibitor has cytostatic/cytotoxic efficacy in KDM6B/p16INK4A-addicted cervical cancer lines, suggesting that inhibition of these enzymes may be evaluated as a therapeutic modality for HPV-associated lesions and cancers.
Results
Cervical Cancer Cells Are Addicted to KDM6B.
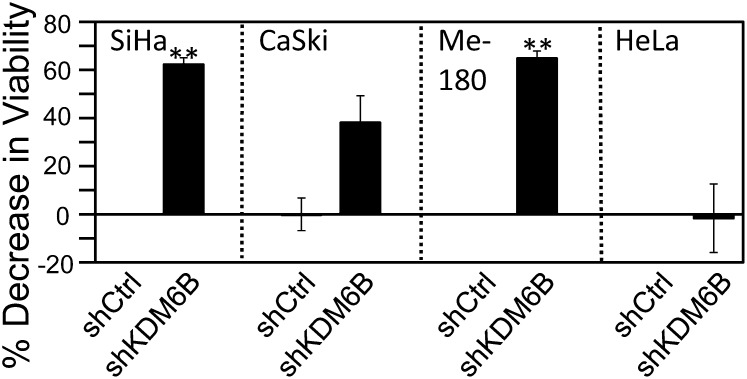
HPV16 E7 induces the expression of the cervical cancer biomarker, p16INK4A, through transcriptional induction of the H3K27-specific histone demethylase KDM6B. HPV16 E7-mediated p16INK4A induction is thus mechanistically similar to RAS-induced OIS activation. We also reported that KDM6B depletion in the HPV16+ cervical cancer line CaSki markedly inhibited cell proliferation/survival (17). To determine whether other HPV+ cervical carcinoma cells showed a similar phenotype, we depleted KDM6B in the HPV16+ cervical cancer line SiHa, the HPV39+ cervical cancer cell line Me-180, and the HPV18+ cervical cancer cell line HeLa by transfection of a KDM6B-specific shRNA. Depletion of KDM6B was verified by quantitative PCR (qPCR). Three days posttransfection, cell viability was assayed by reduction of Alamar Blue, a resazurin-based measure of mitochondrial fitness. Consistent with our previously published results (17), KDM6B depletion caused a significant decrease in cell proliferation/survival of CaSki cells (−38 ± 11%; P = 0.01). Similarly, KDM6B depletion also inhibited cell proliferation/survival of SiHa cells (−62 ± 3%; P < 0.0001), and Me-180 cells (−65 ± 3%; P < 0.0001). Surprisingly, however, KDM6B depletion caused no significant decrease in cell proliferation/survival of HeLa cells (−1.6 ± 14%; P = 0.8535) (Fig. 1). These results indicate that most, but not all, high-risk HPV+ cervical carcinoma lines are addicted to KDM6B expression.
Fig. 1.
KDM6B addiction of cervical cancer lines. KDM6B was depleted in the HPV16+ SiHa and CaSKi cervical carcinoma cell lines, the HPV39+ cervical cancer cell line Me-180, and the HPV18+ cervical cancer cell line HeLa. Cell viability was measured by AlamarBlue assay. Averages and SDs for three independent experiments are shown. Statistically significant changes are indicated, **P < 0.01.
KDM6B Addiction Represents a Direct Consequence of HPV16 E7 Expression.
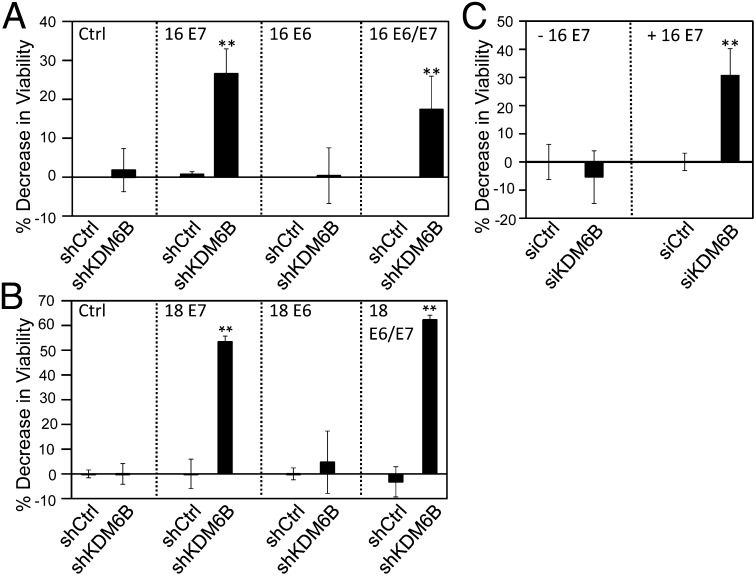
Cervical carcinoma lines contain integrated HPV genomes and consistently express the E6 and E7 oncoproteins. Given that HPV16 E7 causes increased expression of KDM6B, we next determined whether KDM6B addiction of cervical cancer cell lines was caused by HPV E7 oncoprotein expression. To test this hypothesis we engineered primary human foreskin keratinocyte (HFK) populations with ectopic expression of HPV16 or HPV18 E6 and/or E7. Retroviral vectors were used, and HPV16 E7 expression was assessed by Western blotting and HPV18 E7 expression was assessed by qPCR. Because of the absence of appropriate antibodies, HPV16 and HPV18 E6 expression was determined by assessing p53 levels, which are decreased in HPV16 and HPV18 E6-expressing cells because of E6-mediated proteasomal degradation (19). HPV16 E7 levels were similar to those detected in HPV16+ SiHa and CaSki cervical carcinoma lines. KDM6B was depleted by RNAi in donor- and passage-matched primary HFK populations; depletion was verified by qPCR (Fig. S1 A and B), and cell proliferation/survival was assessed by Alamar Blue. Infection efficiency of these cells is low; hence, the decreases in viability that we observed with these cells are less pronounced than with other cell types. We observed a significant (P = 0.0134) 20 ± 5% decrease in cell viability in HPV16 E7-expressing HFKs. Similarly, cell proliferation/survival was also significantly decreased (15 ± 2%; P = 0.0031) in cells that are similar to cervical carcinoma cells that coexpress E6 and E7. In contrast, HPV16 E6-expressing HFKs as well as control-vector–infected HFK populations were not significantly affected (P = 0.9482 and 0.7750, respectively) by KDM6B depletion (Fig. 2A). These experiments were performed in multiple independently derived populations with similar results. Moreover, similar results were obtained when KDM6B was depleted in HPV18 E6- and E7-expressing HFKs (Fig. 2B). These results support the model that KDM6B addiction in cervical carcinoma cells is caused by HPV E7 protein expression.
Fig. 2.
KDM6B addiction is caused by the HPV E7 protein. KDM6B was depleted in: (A) HFKs expressing control vector, HPV16 E7, E6, or E6 and E7; (B) HFKs expressing HPV18 E7, E6, or E6 and E7; and (C) U2OS-tet on cells with doxycycline-inducible expression of HPV16 E7. Cell viability was measured by AlamarBlue assay. Averages and SDs for three independent experiments are shown. Statistically significant changes are indicated, **P < 0.01.
KDM6B Addiction Arises as an Immediate Consequence of HPV16 E7 Expression.
To determine whether KDM6B addiction develops as an immediate consequence of HPV16 E7 expression or whether it is acquired after long-term E7 expression, we performed similar experiments in U2OS osteosarcoma cells with doxycycline-inducible expression of HPV16 E7. Upon doxycycline treatment, these cells express HPV16 E7, KDM6B and p16INK4A are induced, and the H3K27me3 mark is decreased. This process is reversed when doxycycline is removed (17). Depletion of KDM6B did not significantly inhibit the proliferation/survival of these cells before HPV16 E7 induction (P = 0.5197). In contrast, KDM6B depletion caused a 31 ± 9% (P < 0.0001) decrease in viability after HPV16 E7 expression was induced by 72 h of doxycycline treatment. Depletion of KDM6B was verified by qPCR (Fig. S1C). This result shows that KDM6B addiction arises as a direct and immediate consequence of HPV16 E7 expression. This finding is particularly remarkable because, unlike HPV-expressing cervical cancer cells, HPV E7 expression is not necessary for proliferation/survival of U2OS cells (Fig. 2C).
Cervical Cancer Cells Are Addicted to p16INK4A.
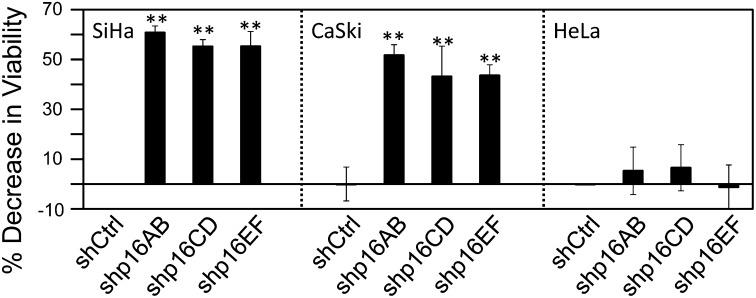
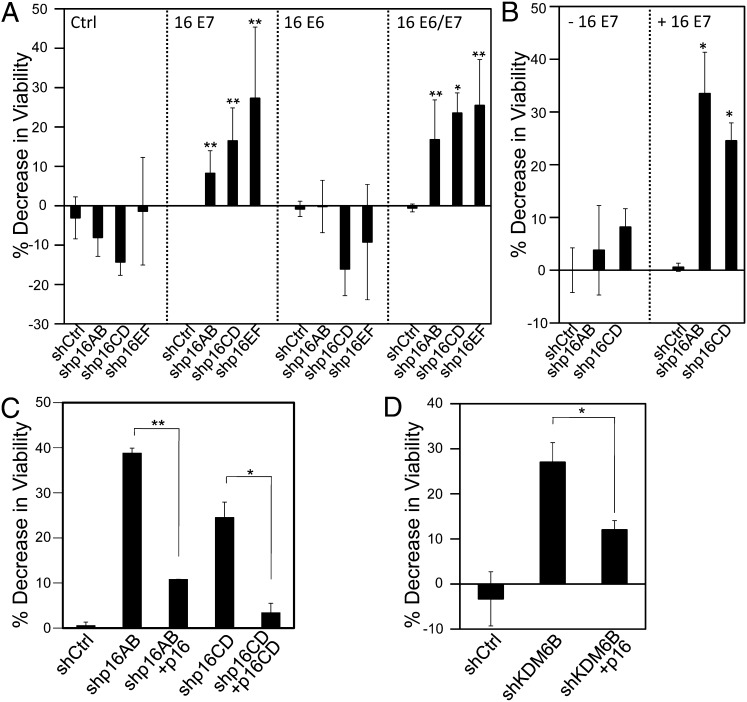
We next wanted to determine the mediators of the observed KDM6B addiction. KDM6B modulates expression of multiple target genes by altering the physical state of chromatin. In addition, KDM6B may also have additional biological activities that are independent of its histone demethylase activity (6). The best-known epigenetic KDM6B target gene in HPV-associated cervical carcinoma cells is p16INK4A (11, 12, 17). To determine if the addiction to KDM6B was dependent on its ability to regulate p16INK4A expression, we used three independent p16INK4A shRNAs to specifically deplete p16INK4A without affecting expression of the overlapping p14ARF gene (20) in the HPV16+ SiHa and CaSki and the HPV18+ HeLa cervical cancer cell lines, and cell proliferation/survival was assessed by Alamar Blue. Depletion of p16INK4A was verified by qPCR (Fig. S2). SiHa cells displayed significant decreases in cell viability upon p16INK4A depletion with each of the three shRNAs (shp16AB: 61 ± 3% P < 0.0001; shp16CD: 55 ± 3% P < 0.0001; shp16EF: 55 ± 5% P < 0.0001). Similarly, CaSki cells also displayed a significant decrease in cell viability (shp16AB: 52 ± 4% P = 0.0007; shp16CD: 43 ± 12% P = 0.0073; shp16EF: 44 ± 4% P = 0.0013). On the other hand, similar to what we observed when KDM6B was depleted, proliferation/survival of HeLa cells was not significantly affected by p16INK4A depletion (shp16AB: P = 0.6977, shp16CD: P = 0.4181, shp16EF: P = 0.5848) (Fig. 3).
Fig. 3.
Cervical cancer cell addiction to p16INK4A. p16INK4A was depleted in the HPV16+ SiHa and CaSki cervical carcinoma cell lines, and the HPV18+ HeLa cervical cancer line. Three independent p16 shRNA constructs (shp16AB, shp16CD, shp16EF) were used. Cell viability was measured by AlamarBlue assay. Averages and SDs for three independent experiments are shown. Statistically significant changes are indicated, **P < 0.01.
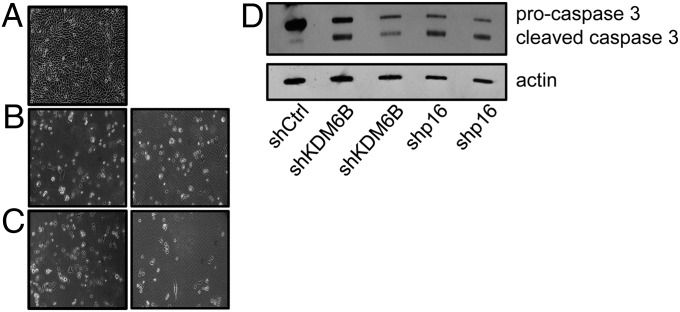
To determine whether KDM6B and p16INK4A depletion caused growth arrest or cell death, we analyzed cell numbers and procaspase 3 cleavage in SiHa cells. Depletion of KDM6B or p16INK4A (Fig. S3) caused a marked decrease in cell numbers and a decrease in procaspase 3 levels with a concomitant increase in cleaved caspase 3 levels (Fig. 4). This finding suggests that KDM6B or p16INK4A depletion in SiHa cells caused cell death, presumably by apoptosis.
Fig. 4.
SiHa cervical cancer cells show evidence of cell death and caspase 3 cleavage upon KDM6B and p16INK4A depletion. p16INK4A and KDM6B were depleted in the HPV16+ cervical carcinoma cell line SiHa. (A) SiHa cells with control shRNA (magnification, 10×); (B) SiHa cells with KDM6B shRNA (magnification, 10×); (C) SiHa cells with p16INK4A shRNA (magnification, 10×). (D) Western blot analysis of p16INK4A, procaspase 3, and cleaved caspase 3 levels. Lysates were separated by SDS/PAGE, transferred, and probed for p16INK4A and procaspase 3. An actin blot is included as a loading control.
Addiction to p16INK4A Arises as a Direct and Immediate Consequence of HPV E7 Expression.
To determine whether p16INK4A addiction was caused by HPV E7 oncoprotein expression, we next depleted p16INK4A by RNAi in donor- and passage-matched primary HFK populations with ectopic expression of the HPV16 E6 and/or E7 oncoproteins, and assessed cell proliferation/survival by Alamar Blue. Depletion of p16INK4A was verified by qPCR (Fig. S4A). We observed a significant decrease in cell viability in HPV16 E7 expressing HFKs (shp16AB: 15 ± 3%, P = 0.0015; shp16CD: 25 ± 6%, P = 0.0019; shp16EF: 33 ± 11%, P = 0.0055). Similarly, cell proliferation/survival was also significantly decreased in cells that coexpress HPV16 E6 and E7 (shp16AB: 17 ± 10%, P = 0.0156; shp16CD: 24 ± 5%, P = 0.0034; shp16EF: 25 ± 12%, P = 0.0402). In contrast, HPV16 E6-expressing HFKs as well as control-vector–infected HFK populations were not significantly affected by p16INK4A depletion (Fig. 5A). Similar results were obtained with HPV18 E6- and E7-expressing HFKs (Fig. S5). To determine whether p16INK4A addiction arises as an immediate consequence of HPV16 E7 expression or whether it develops after long-term E7 expression, we performed similar experiments in U2OS osteosarcoma cells with doxycycline-inducible expression of HPV16 E7. Depletion of p16INK4A was assessed by qPCR (Fig. S4B) and did not significantly inhibit the proliferation/survival of these cells before HPV16 E7 induction (shp16AB: 0.9439; shp16CD: P = 0.1632). In contrast, p16INK4A depletion caused a statistically significant decrease in viability after HPV16 E7 expression was induced by 72 h of doxycycline treatment (shp16AB: 33 ± 8%, P = 0.0274; shp16CD: 25 ± 3%, P = 0.0105) (Fig. 5B).
Fig. 5.
HPV16 E7 induces p16INK4A addiction. p16INK4A was depleted in (A) HFKs expressing control vector, HPV16 E7, E6, or E6 and E7; and (B) U2OS-tet on cells with doxycycline-inducible expression of HPV16 E7. Three independent p16 shRNA constructs (shp16AB, shp16CD, shp16EF) were used in A and two in B. (C) p16INK4A was depleted in U2OS-tet on cells with doxycycline-inducible expression of HPV16 E7. Two independent p16 shRNA constructs (shp16AB, shp16CD) and their respective rescue constructs (p16, 16CD) were used. (D) KDM6B was depleted in U2OS-tet on cells with doxycycline-inducible expression of HPV16 E7 and rescued with p16INK4A. Cell viability was measured by AlamarBlue assay. Averages and SDs for three independent experiments are shown. Statistically significant changes are indicated: *P < 0.05, **P < 0.01.
To rule out off-target effects of shRNA, we determined whether the loss of cell proliferation/survival as a consequence of p16INK4A depletion was rescued by ectopic p16INK4A expression. The p16AB shRNA targets sequences corresponding to amino acids 6 through 12 and, because we used a p16INK4A cDNA expression vector that is lacking the first eight amino acids, it is not targeted by the p16AB construct and could be used directly for rescue experiments. For the p16CD shRNA, we generated a p16INK4A rescue construct (p16CD) harboring silent mutations in the targeting sequences of the shRNA. Rescue experiments were performed in the U2OS cells with inducible HPV6 E7 expression. Twenty-four hours after we cotransfected the p16INK4A shRNA construct together with its corresponding rescue construct, HPV16 E7 expression was induced by the addition of doxycycline, and cell viability was measured 72 h after the addition of doxycycline. Reexpression of p16INK4A together with p16INK4A shRNA rescued the decrease in viability observed with depletion of p16INK4A. This finding confirms that the decrease in viability is a result of p16INK4A depletion and is not caused by off-target effects of these shRNAs (Fig. 5C). Reexpression of p16INK4A together with KDM6B shRNA rescued the decrease in viability observed with depletion of KDM6B, functionally linking KDM6B and p16INK4A (Fig. 5D).
Cell Death in Response to p16INK4A Depletion is CDK4- and CDK6-Dependent.
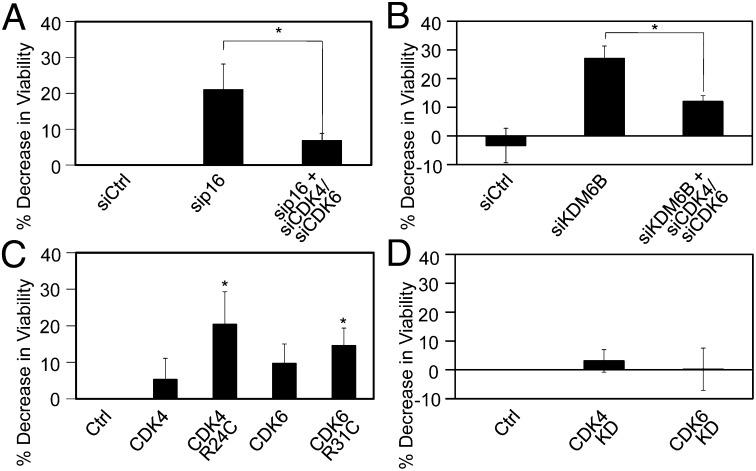
The p16INK4A tumor suppressor acts as an inhibitor of CDK4/6-cyclin D complexes. To determine if the addiction to p16INK4A was dependent on p16’s ability to inhibit CDK4/6 activity, we codepleted CDK4, CDK6, and p16INK4A in U2OS cells and induced the expression of HPV16 E7 by the addition of doxycycline. Cell viability was assessed 72 h after the induction of HPV16 E7 expression. Depletion of p16INK4A, CDK4 and CDK6 was assessed by qPCR (Fig. S6) and rescued the loss of viability that is observed upon p16INK4A or KDM6B depletion (Fig. 6 A and B).
Fig. 6.
Cell death caused by p16INK4A depletion in p16INK4A-addicted cells is dependent on CDK4 and CDK6. (A) p16INK4A or p16INK4A, together with CDK4 and CDK6, were depleted in U2OS-tet on cells with doxycycline-inducible expression of HPV16 E7 by transfection with p16INK4A, CDK4-, or CDK6-specific siRNA duplexes. Transfection of the nontargeting siRNA pool was used as a control (siCtrl). (B) KDM6B or KDM6B in combination with CDK4 and CDK6 were depleted in U2OS-tet on cells with doxycycline-inducible expression of HPV16 E7 by transfection with KDM6B, CDK4-, or CDK6-specific siRNA duplexes. Transfection of a nontargeting siRNA pool was used as a control (siCtrl). (C) U2OS-tet on cells with doxycycline-induced HPV16 E7 expression were transfected with CDK4, CDK6, and oncogenic CDK4 or CDK6 mutants that cannot be inhibited by p16INK4A (R24C and R31C, respectively). Cell viability was measured by AlamarBlue assay. Averages and SDs for three independent experiments are shown. (D) Kinase-defective CDK4 or CDK6 mutants were transfected into U2OS-tet on cells with doxycycline-induced HPV16 E7 expression and did do not affect viability. Averages and SDs for three independent experiments are shown. Statistically significant changes are indicated: *P < 0.05.
To determine whether the observed cytotoxicity of p16INK4A depletion was caused by CDK4 or CDK6 hyperactivity, we transiently transfected U2OS cells with an oncogenic CDK4 arginine 24 to cysteine (R24C) mutant derived from a human melanoma (21) that is insensitive to inhibition by p16INK4A and the corresponding CDK6 arginine 31 to cysteine (R31C) mutant. Doxycycline was added 24 h after transfection and cell viability was assessed 72 h after doxycycline addition. Expression of these p16INK4A insensitive CDK4 and CDK6 mutants caused a decrease in cell viability in the doxycycline-treated cells, but not in the vehicle-treated cells, which is similar to that observed with p16INK4A depletion (Fig. 6C and Fig. S7). In contrast, transfection of kinase defective CDK4 (CDK4 KD) or CDK6 (CDK6 KD) mutants did not affect cell viability (Fig. 6D and Fig. S7B). As expected, HeLa cell viability was not affected by CDK4/6 depletion or expression of the p16INK4A insensitive, oncogenic CDK4 R24C and CDK6 R31C mutants (Fig. S8).
Cervical Cancer Cells Are Sensitive to Treatment with the KDM6A/6B Inhibitor GSK-J4.
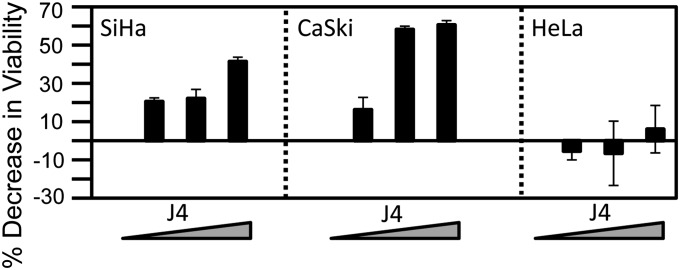
In contrast to mutations, epigenetic alterations are reversible by modulating the activities of the corresponding epigenetic enzymes. Given our finding that some cervical cancer cells are “addicted” to KDM6B and KDM6A, we wanted to determine whether a recently developed KDM6-selective small-molecule inhibitor, GSK-J4, may show some efficacy in inhibiting proliferation and/or survival cervical carcinoma cells. Hence, we treated SiHa and CaSki cervical cancer cells, which were sensitive to KDM6B and p16INK4A depletion, as well as HeLa cells, which show no dependence on these molecules, with GSK-J4. GSK-J4 treatment of SiHa and CaSki cells resulted in a marked decrease in cell viability, whereas GSK-J4 treatment of HeLa cervical cancer cells, which were not sensitive to KDM6B and p16INK4A depletion, did not affect cell viability (Fig. 7).
Fig. 7.
Effect of GSK-J4 on cervical cancer cells. Cervical cancer cell lines SiHa, CaSki, and HeLa were treated with 0–30 μM GSK-J4 (CaSki) or 25–100 μM GSK-J4 (SiHa and HeLa) for 72 h. Cell viability was measured by AlamarBlue assay. Averages and SDs for three independent experiments are shown.
Discussion
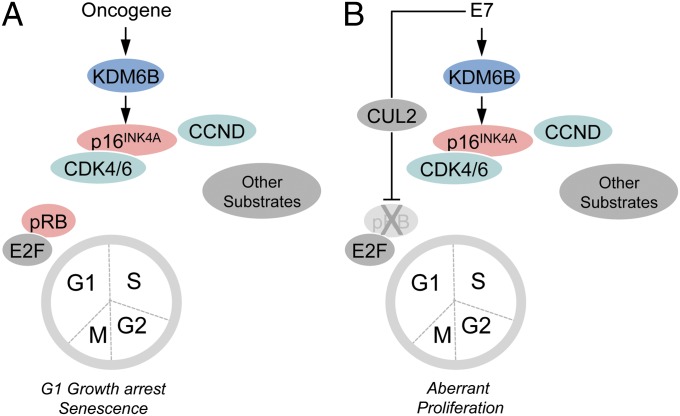
OIS is a cell-intrinsic tumor-suppressive mechanism that protects cells from unrestrained proliferation following oncogenic insults, such as RAS/RAF activation or expression of a high-risk HPV E7 protein (reviewed in ref. 22). Evidence of OIS has been detected in premalignant lesions, but much less so in frank cancers, suggesting that OIS must be evaded or bypassed by additional mutations in order for premalignant lesions to progress (15). Indeed, HPV16 E7 both triggers OIS and quenches this response by targeting the pRB tumor suppressor for proteasomal degradation (17). In addition to proliferative arrest, OIS involves the activation of a variety of signaling pathways, and senescent cells are characterized by increased size, activation of β-galactosidase, chromatin aggregates enriched for H3K9me3, activated DNA damage markers, and extracellular matrix-degrading enzymes (reviewed in ref. 23).
Because KDM6B, which mediates RAS and HPV E7 OIS, has targets other than p16INK4A, OIS activation causes a range of epigenetic alterations. These alterations are biologically inconsequential as long as p16INK4A expression is induced and the cells undergo senescence, but may contribute to carcinogenesis once the OIS response has been evaded. KDM6B may have both tumor-suppressive and oncogenic activities in different cancer types. KDM6B is up-regulated in prostate cancer and its expression is higher in metastatic prostate cancer (24); it has been shown to promote epithelial-to-mesenchymal transition and cancer cell invasion (25). On the other hand, KDM6B has been reported to be down-regulated in certain human cancers, and the KDM6B gene is located at 5q31, an area that is frequently lost in a number of malignancies, including myeloid leukemias (26).
As evidenced by aberrant homeobox gene expression, HPV16 E7-expressing cells undergo profound epigenetic alterations as a consequence of enhanced KDM6A and KDM6B expression (17, 27). Indeed, several homeobox genes (HOX) have been identified as bona fide oncogenes. For example, HOXC10 overexpression is associated with increased invasion and the transition of cervical high-grade squamous intraepithelial lesions to cervical squamous cell carcinoma (28). HOXB7 mediates aspects of epithelial-to-mesenchymal transition in breast cancer, promotes proliferation in oral cancer, and is associated with progression and development of metastases in lung cancer (29–31). In addition, germ-line HOXB13 mutations have been linked to an increased prostate cancer risk (32).
Hence, the molecular basis for the observed dependence of cervical carcinoma cells on KDM6A and KDM6B expression was expected to be complex. Our finding that p16INK4A expression is a key downstream mediator of KDM6B addiction of cervical carcinoma cells, therefore, is quite surprising. After all, p16INK4A is a well-established and frequently mutated or epigenetically silenced tumor suppressor and key regulator of pRB activity. Because HPV E7 inactivates pRB by targeting it for proteasomal degradation, one would predict that enhanced expression of the p16INK4A tumor suppressor by E7, while providing a valuable surrogate for E7 expression (33), would have only minimal biological consequences. The finding that KDM6B-triggered p16INK4A expression is required for cell viability that arises as a direct and immediate consequence of HPV16 E7 oncoprotein expression suggests that the biological activity of p16INK4A in HPV-associated cancers is more akin to that of an oncogene, as opposed its well-established tumor-suppressor activity in most other human tumor types. Because this pro-oncogenic activity of p16INK4A is dependent on its ability to inhibit CDK4/6 activity, our results suggest that the dogmatic tumor-suppressive activity of p16INK4A, and by extension the entire pRB tumor-suppressor pathway, is strictly dependent on cellular context. The finding that ectopic expression of a human melanoma-derived, oncogenic CDK4 mutant causes cell death, and thus has tumor-suppressive activity in HPV E7-expressing cells (Fig. 6), further supports this contention. It is particularly remarkable that observed reversals in oncogenic and tumor-suppressive activities within the pRB pathway arise as an immediate consequence of HPV16 E7 expression, even in U2OS cells that are not intrinsically dependent on E7 expression for continued growth/survival.
The pRB tumor suppressor is a rate-limiting substrate for CDK4/6 control of G1/S transition and senescence. Our results, however, suggest that there are other relevant CDK4/CDK6 substrates that need to be retained in an unphosphorylated state when pRB is inactivated, and that continued p16INK4A expression is required to quench their aberrant phosphorylation in such cells. An unbiased screen for CDK4 and CDK6 substrates yielded 68 potential phosphorylation targets, in addition to pRB, p107, and p130 (34). Several of these putative substrates, including FOXM1 and SMAD3, have been implicated in senescence signaling (34–36). Thus, the tumor-suppressive activities of CDK4/6 in pRB/p107/p130-defective cells may be mediated through one or several of these, or involve as yet unidentified pathways (Fig. 8).
Fig. 8.
Induction and abrogation of the OIS tumor suppressor response by HPV16 E7. (A) Oncogenic stimuli such as RAS/RAF signaling or HPV16 E7 expression causes KDM6B expression and derepression of p16INK4A expression, which inhibits CDK4/6 activity and pRB phosphorylation, causing G1 cell cycle arrest and senescence (OIS). (B) HPV16 E7 has evolved to evade OIS by targeting pRB for ubiquitin-mediated proteasomal degradation.
Unlike the HPV16+ SiHa and CaSki cervical cancer lines, the HPV18+ HeLa cervical carcinoma line is insensitive to KDM6B or p16INK4A depletion and KDM6 inhibition by the GSK-J4 small-molecule inhibitor. This result is not because of an inability of HPV18 E7 to induce KDM6B and p16INK4A expression, nor is it because of inherent differences in the expression levels of KDM6B, p16INK4A, CDK4, or CDK6 in HeLa cells (Fig. S9). Rather, we hypothesize that HeLa cells acquired a downstream secondary mutation that renders these cells no longer dependent on the oncogenic activities of p16INK4A. Elucidating the mechanisms of resistance may thus aid in identifying molecules that, when CDK4/CDK6 phosphorylated, cause cell death or growth arrest in cancers with mutant pRB. However, we did not observe emergence of resistant SiHa or CaSki cell populations upon stable depletion of p16INK4A or KDM6B.
Other cancers, including high-grade serous ovarian carcinoma, a subset of lung cancers, and highly aggressive breast and prostate cancers, also express high levels of p16INK4A (24, 37–41). Such high-level p16INK4A-expressing cancers generally also have pRB mutations (reviewed in ref. 42). It will be interesting to determine whether p16INK4A expression in these tumors is also mechanistically linked to KDM6B expression and whether they may also be susceptible to KDM6 inhibitors.
Finally, it is interesting to note that epigenetic alterations other than H3K27 methylation/demethylation also contribute to senescence regulation. H3K4 demethylation, for example, may also play a role in OIS, and the H3K4 demethylases KDM5A and KDM5B (JARID1A and JARID1B) contribute to the repressive chromatin environment of senescent cells (43). Conversely, depletion of KDM2B (NDY1) results in premature senescence (44, 45), whereas overexpression of KDM2B results in immortalization of primary mouse embryo fibroblasts (44–46). It will be interesting to determine the cellular targets of these enzymes and whether any of these may contribute to the observed resistance to KDM6 inhibition or p16INK4A depletion.
Materials and Methods
Cells.
Primary HFKs were isolated and cultured as previously described (5). HFKs were transduced by recombinant retroviruses carrying either the control vector (LXSN) or vectors encoding HPV16 E6 or E7 or both oncogenes (47). U2OS-tet on, CaSki, SiHa, and HeLa cells were maintained as described in SI Materials and Methods.
Cell Viability Assay.
Three days after transfection, media was removed and 100-μL AlamarBlue reagent (Invitrogen; diluted 10-fold in growth medium) was added to each well. The plates were incubated for 1–3 h at 37 °C and then read at 570 and 600 nm on a microtiter well plate reader (Biotek).
Quantitative RT-PCR.
Lysates were prepared and quantitative RT-PCR was performed using the Cells-to-CT kit as per the manufacturer’s instructions (Applied Biosystems). Predesigned Taqman qPCR assays for KDM6B, p16INK4A, CDK4, and CDK6 were supplied by Applied Biosystems as a 20× premix containing both primers and FAM-nonfluorescent quencher probe. Analysis was performed using a StepOnePlus Real-Time PCR System (Applied Biosystems).
RNAi.
For RNAi, 1 × 104 cells were seeded onto 96-well plates 1 d before transfection with 100 ng of DNA using Lipofectamine 2000 (Invitrogen). The cells were transfected with the shRNA, siRNA and rescue constructs described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank our colleagues for their gifts of reagents, Miranda Grace for technical assistance, and Ed Harlow, Nick Dyson, James Rocco, James DeCaprio, and the two anonymous reviewers for discussions and suggestions. This work was supported by Public Health Service Grants K01CA143010 (to M.E.M.-D.) and R01CA066980 (to K.M.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310432110/-/DCSupplemental.
References
- 1.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8(1):9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 2.Tolhuis B, et al. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet. 2006;38(6):694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- 3.Lund AH, van Lohuizen M. Epigenetics and cancer. Genes Dev. 2004;18(19):2315–2335. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- 4.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300(5616):131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 5.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136(21):3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 6.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 7.De Santa F, et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130(6):1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Jepsen K, et al. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450(7168):415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- 9.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449(7163):689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 10.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318(5849):447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 11.Agger K, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23(10):1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barradas M, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23(10):1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michaloglou C, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 14.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436(7051):725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collado M, et al. Tumour biology: Senescence in premalignant tumours. Nature. 2005;436(7051):642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 16.Popov N, Gil J. Epigenetic regulation of the INK4b-ARF-INK4a locus: In sickness and in health. Epigenetics. 2010;5(8):685–690. doi: 10.4161/epi.5.8.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLaughlin-Drubin ME, Crum CP, Münger K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci USA. 2011;108(5):2130–2135. doi: 10.1073/pnas.1009933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez SL, Stremlau M, He X, Basile JR, Münger K. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J Virol. 2001;75(16):7583–7591. doi: 10.1128/JVI.75.16.7583-7591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63(6):1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 20.Shin JJ, Katayama T, Michaud WA, Rocco JW. Short hairpin RNA system to inhibit human p16 in squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2004;130(1):68–73. doi: 10.1001/archotol.130.1.68. [DOI] [PubMed] [Google Scholar]
- 21.Wölfel T, et al. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269(5228):1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 22.Campisi J, d’Adda di Fagagna F. Cellular senescence: When bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8(9):729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 23.Bianchi-Smiraglia A, Nikiforov MA. Controversial aspects of oncogene-induced senescence. Cell Cycle. 2012;11(22):4147–4151. doi: 10.4161/cc.22589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarrard DF, et al. Alterations in the p16/pRb cell cycle checkpoint occur commonly in primary and metastatic human prostate cancer. Cancer Lett. 2002;185(2):191–199. doi: 10.1016/s0304-3835(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 25.Ramadoss S, Chen X, Wang CY. Histone demethylase KDM6B promotes epithelial-mesenchymal transition. J Biol Chem. 2012;287(53):44508–44517. doi: 10.1074/jbc.M112.424903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Z, et al. A novel nuclear protein, 5qNCA (LOC51780) is a candidate for the myeloid leukemia tumor suppressor gene on chromosome 5 band q31. Oncogene. 2001;20(47):6946–6954. doi: 10.1038/sj.onc.1204850. [DOI] [PubMed] [Google Scholar]
- 27.Hyland PL, et al. Evidence for alteration of EZH2, BMI1, and KDM6A and epigenetic reprogramming in human papillomavirus type 16 E6/E7-expressing keratinocytes. J Virol. 2011;85(21):10999–11006. doi: 10.1128/JVI.00160-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhai Y, et al. Gene expression analysis of preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer Res. 2007;67(21):10163–10172. doi: 10.1158/0008-5472.CAN-07-2056. [DOI] [PubMed] [Google Scholar]
- 29.Wu X, et al. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 2006;66(19):9527–9534. doi: 10.1158/0008-5472.CAN-05-4470. [DOI] [PubMed] [Google Scholar]
- 30.De Souza Setubal Destro MF, et al. Overexpression of HOXB7 homeobox gene in oral cancer induces cellular proliferation and is associated with poor prognosis. Int J Oncol. 2010;36(1):141–149. [PubMed] [Google Scholar]
- 31.Chen H, et al. Hoxb7 inhibits transgenic HER-2/neu-induced mouse mammary tumor onset but promotes progression and lung metastasis. Cancer Res. 2008;68(10):3637–3644. doi: 10.1158/0008-5472.CAN-07-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ewing CM, et al. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366(2):141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sano T, Oyama T, Kashiwabara K, Fukuda T, Nakajima T. Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am J Pathol. 1998;153(6):1741–1748. doi: 10.1016/S0002-9440(10)65689-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders L, et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell. 2011;20(5):620–634. doi: 10.1016/j.ccr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu F, Matsuura I. Inhibition of Smad antiproliferative function by CDK phosphorylation. Cell Cycle. 2005;4(1):63–66. doi: 10.4161/cc.4.1.1366. [DOI] [PubMed] [Google Scholar]
- 36.Matsuura I, et al. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430(6996):226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 37.Milde-Langosch K, Riethdorf S. Role of cell-cycle regulatory proteins in gynecological cancer. J Cell Physiol. 2003;196(2):224–244. doi: 10.1002/jcp.10286. [DOI] [PubMed] [Google Scholar]
- 38.Herschkowitz JI, He X, Fan C, Perou CM. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res. 2008;10(5):R75. doi: 10.1186/bcr2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kommoss S, et al. AGO-OVAR Independent prognostic significance of cell cycle regulator proteins p16(INK4a) and pRb in advanced-stage ovarian carcinoma including optimally debulked patients: A translational research subprotocol of a randomised study of the Arbeitsgemeinschaft Gynaekologische Onkologie Ovarian Cancer Study Group. Br J Cancer. 2007;96(2):306–313. doi: 10.1038/sj.bjc.6603531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andujar P, et al. p16INK4A inactivation mechanisms in non-small-cell lung cancer patients occupationally exposed to asbestos. Lung Cancer. 2010;67(1):23–30. doi: 10.1016/j.lungcan.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 41.Milde-Langosch K, Hagen M, Bamberger AM, Löning T. Expression and prognostic value of the cell-cycle regulatory proteins, Rb, p16MTS1, p21WAF1, p27KIP1, cyclin E, and cyclin D2, in ovarian cancer. Int J Gynecol Pathol. 2003;22(2):168–174. doi: 10.1097/00004347-200304000-00009. [DOI] [PubMed] [Google Scholar]
- 42.Malumbres M, Barbacid M. To cycle or not to cycle: A critical decision in cancer. Nat Rev Cancer. 2001;1(3):222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- 43.Chicas A, et al. H3K4 demethylation by Jarid1a and Jarid1b contributes to retinoblastoma-mediated gene silencing during cellular senescence. Proc Natl Acad Sci USA. 2012;109(23):8971–8976. doi: 10.1073/pnas.1119836109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfau R, et al. Members of a family of JmjC domain-containing oncoproteins immortalize embryonic fibroblasts via a JmjC domain-dependent process. Proc Natl Acad Sci USA. 2008;105(6):1907–1912. doi: 10.1073/pnas.0711865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tzatsos A, Pfau R, Kampranis SC, Tsichlis PN. Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus. Proc Natl Acad Sci USA. 2009;106(8):2641–2646. doi: 10.1073/pnas.0813139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15(11):1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halbert CL, Demers GW, Galloway DA. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65(1):473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.