Significance
The induction of immediate early genes (IEGs) by neural stimuli underlies much of the plasticity of brain function, but regulatory mechanisms have been obscure. Inositol polyphosphate multikinase (IPMK) is a notably pleiotropic enzyme that displays inositol phosphate kinase activity and phosphatidylinositol kinase activity and exhibits physiologically noncatalytic actions such as stabilizing the mammalian target of rapamycin complex 1 complex. We report that IPMK is required for IEG induction by neural activation and neurotrophic stimuli. We have elucidated the molecular mechanisms responsible for IPMK influences; namely, that it enhances the transcriptional coactivation ability of Creb-binding protein (CBP). This epigenetic regulation of IEGs may have both neural and nonneural implications, as IPMK and CBP are broadly expressed in a variety of tissues.
Keywords: inositol phosphates, learning
Abstract
Profound induction of immediate early genes (IEGs) by neural activation is a critical determinant for plasticity in the brain, but intervening molecular signals are not well characterized. We demonstrate that inositol polyphosphate multikinase (IPMK) acts noncatalytically as a transcriptional coactivator to mediate induction of numerous IEGs. IEG induction by electroconvulsive stimulation is virtually abolished in the brains of IPMK-deleted mice, which also display deficits in spatial memory. Neural activity stimulates binding of IPMK to the histone acetyltransferase CBP and enhances its recruitment to IEG promoters. Interestingly, IPMK regulation of CBP recruitment and IEG induction does not require its catalytic activities. Dominant-negative constructs, which prevent IPMK-CBP binding, substantially decrease IEG induction. As IPMK is ubiquitously expressed, its epigenetic regulation of IEGs may influence diverse nonneural and neural biologic processes.
Immediate early genes (IEGs) are a family of rapidly inducible genes that are transcriptionally activated in response to a wide variety of stimuli (1–5). In the brain, IEG induction mediates the plasticity underlying complex processes such as learning, memory, and behavior (6–8). Thus, genetic deletion of these IEGs often leads to deficits in the encoding and consolidation of long-term memory (9–11). Signaling cascades in which neural activity induces IEGs have been elusive.
Inositol polyphosphate multikinase (IPMK) is a multifunctional enzyme that possesses both inositol phosphate kinase (IP3-kinase) (12, 13) and phosphatidylinositol kinase (PI3-kinase) activities (14, 15). In addition, IPMK displays physiologic activities that are independent of its catalytic activity, such as binding and stabilizing the mTOR1 complex (16), as well as enhancing p53-mediated transcription and cell death (17, 18). We show that IPMK is a transcriptional coactivator that is required for induction of a multiplicity of IEGs in the brain by binding to and regulating the recruitment of the histone acetyltransferase CBP.
Results
IPMK Is Required for IEG Induction.
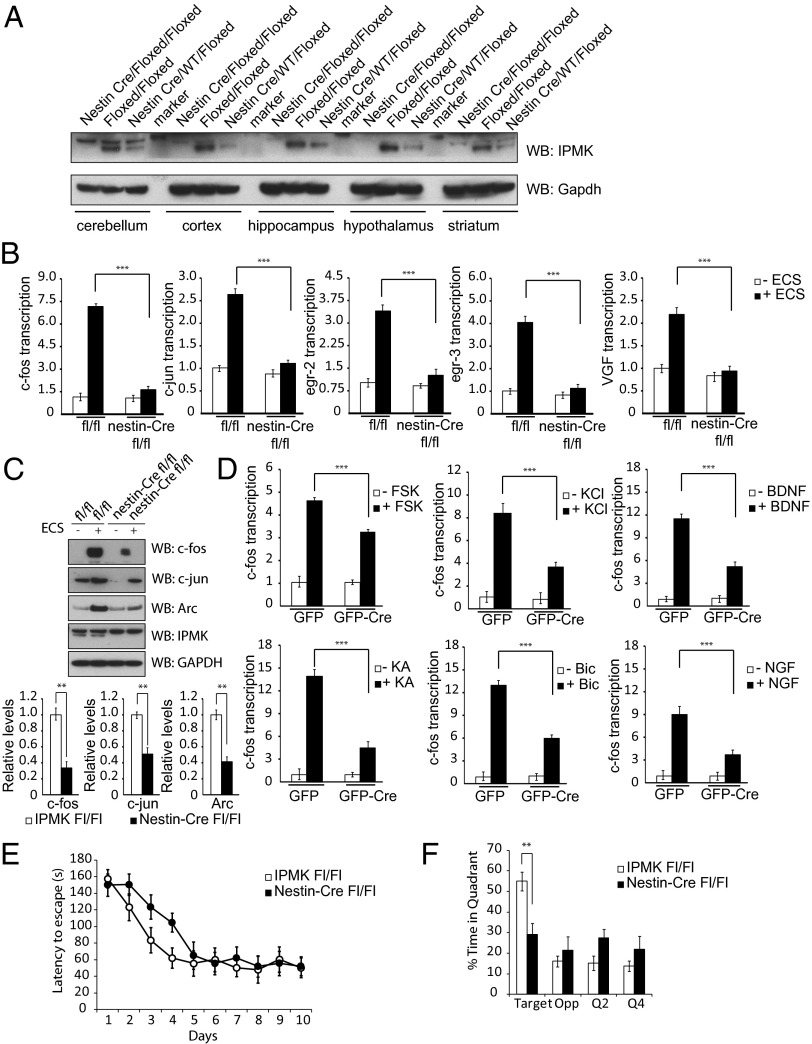
IPMK is essential for mammalian physiology, as conventional IPMK−/− mice perish at embryonic day 9.5 (E9.5) as a result of multiple morphological deficits (19). Accordingly, we created conditional knockout IPMK mice, using the Cre-loxP system. To elucidate neural functions of IPMK, we created brain-specific IPMK knockout mice by mating IPMKfloxed/floxed mice with nestin-Cre+/− mice, which mediate excision of floxed alleles in neuroprogenitor cells before neuronal-astrocyte differentiation (20). These IPMK brain knockout mice are viable, grossly normal, and give rise to offspring in predicted Mendelian frequencies, despite the abolition of IPMK protein in several brain regions (Fig. 1A).
Fig. 1.
IPMK enhances IEG expression. (A) Nestin-Cre IPMKfl/fl mice display uniform depletion of IPMK in cerebellum, cortex, hippocampus, hypothalamus, and striatum of the brain. Nestin-Cre IPMKwt/fl mice exhibit half the levels of IPMK present in each brain region tested, reflecting deletion of only the floxed allele. (B) Levels of IEG expression as assessed via quantitative PCR (qPCR) are increased in the hippocampus of IPMKfl/fl mice after ECS. In nestin-Cre IPMKfl/fl mice, levels of IEG expression are decreased 60%–80%. ***P < 0.001. Data are means ± SEM from four experiments. Data are expressed as fold over baseline, where baseline is defined as IPMKfl/fl mice without ECS treatment. (C) Western blotting of hippocampal tissue in IPMKfl/fl mice after ECS reveals robust enhancement of c-fos, c-jun, and Arc protein levels. In nestin-Cre IPMKfl/fl mice, levels of IEG expression are decreased 50–70%. **P < 0.01. Data are means ± SEM from four experiments. (D) Primary cortical neurons from IPMKfl/fl mice are infected with lentivirus expressing GFP or GFP-Cre and treated with KCl, forskolin (Fsk), NGF, BDNF, bicuculline (Bic), and kainic acid (KA). In all circumstances, neurons with IPMK deletion exhibit impaired c-fos mRNA induction. ***P < 0.001, Student t test. Data are means ± SEM from three experiments. (E) Performance on the Barnes maze task during training for 10 d (two trials per day, 1 h intertraining interval). Nestin-Cre IPMKfl/fl (n = 16) mice exhibit higher latencies to find the exit versus control littermates (n = 18) during the initial phase of training. F1,32 = 4.24, P < 0.05; F1,32 = 5.37, P < 0.05; F1,32 = 7.80, P < 0.01. (F) Nestin-Cre IPMKfl/fl mice show a deficiency in spatial memory during a probe trial performed 1 wk after training. IPMK brain knockout mice are significantly impaired in spatial localization for the correct quadrant. F1,32 = 5.88, P < 0.05.
The most dramatic induction of IEGs in the brain is elicited by electroconvulsive shock (ECS) (6). Within minutes after ECS, we observe four- to eightfold increases in hippocampal mRNA for the FBJ murine osteosarcoma viral oncogene homolog (c-fos), Jun proto-oncogene (c-jun), early growth response protein 2 (egr2), early growth response protein 3 (egr3), and VGF nerve growth factor inducible (VGF), which are substantially decreased in IPMK-deleted mice (Fig. 1B). This decrease is still seen 1 h after ECS treatment (Fig. S1A). Protein levels for c-fos, c-jun, and activity-regulated cytoskeleton-associated protein (Arc/Arg3.1), maximally induced by ECS, are markedly diminished in IPMK mutants (Fig. 1C). This decrease in IEGs reflects neuronal mechanisms, as IPMK is predominantly expressed in neurons. Thus, synapsin-Cre+/− IPMKfl/fl mice in which IPMK deletion occurs at E12.5 in developing neurons display essentially a complete absence of IPMK via Western blot (Fig. S1B), as well as pronounced deficits in IEG induction in the cortex after ECS (Fig. S1C) (21). Synapsin-Cre+/− IPMKfl/fl mice also exhibit decreased c-fos and c-jun protein levels after ECS treatment (Fig. S1D).
As IEGs are responsive to a variety of cellular stimuli, we sought to determine those influenced by IPMK. Induction of c-fos is impaired more than 50% in primary IPMKfl/fl neurons, with lentiviral-mediated deletion of IPMK after treatment with potassium chloride (KCl), forskolin, nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), the GABA antagonist bicuculline, and the glutamate agonist kainic acid (Fig. 1D). To examine regulation by IPMK of IEGs in a continuous cell line, we used the neuronal-like PC12 cells (Fig. S1E). Induction of several IEGs by potassium depolarization (Fig. S1F) or NGF (Fig. S1G) in these cells is reduced about 60% by depletion of IPMK, using RNA interference.
IPMK affects IEG-sensitive behaviors. Thus, nestin-Cre+/− IPMKfl/fl mice display deficient spatial memory, as assessed by the Barnes maze, a test in which mice use visual cues to find a hidden escape box. Nestin-Cre+/− IPMKfl/fl mice exhibit longer escape latencies during days 2–4 of training compared with IPMKfl/fl littermate control mice, although by day 10 this difference is absent (Fig. 1E). However, the IPMK mutants manifest deficits in spatial localization of the target during a probe trial 1 wk after training (Fig. 1F). The spatial memory deficit of nestin-Cre+/− IPMKfl/fl mice is consistent with the impaired memory for context of these mice tested 1 wk after habituation to a novel environment (Fig. S2 A and B). Short-term memory in nestin-Cre+/− IPMKfl/fl mice appears normal, as these mice perform as well as littermate controls in the novel object recognition test 3 h after training (Fig. S2C). However, the IPMK knockout mice are impaired in novel object recognition 24 h after training (Fig. S2D), which is commensurate with a deficit in long-term memory.
IPMK Binds to CBP and Enhances Its Recruitment to the c-fos Promoter.
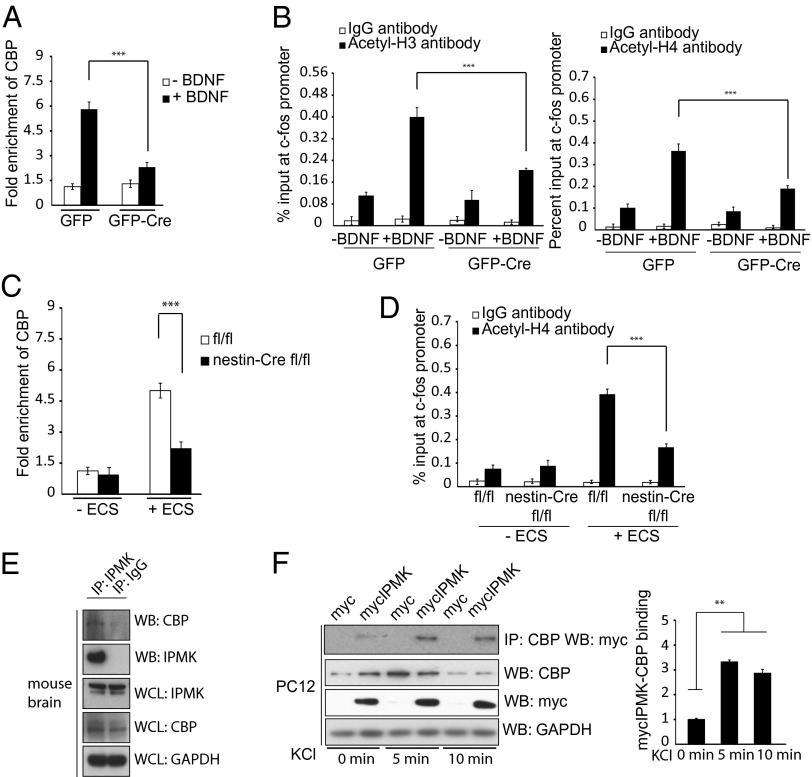
We explored molecular mechanisms underlying the ability of IPMK to mediate induction of IEGs. The histone acetylating enzyme CBP is recruited to IEG promoters immediately after neuronal activity (22). We monitored the recruitment of CBP to the promoter of c-fos, using chromatin immunoprecipitation (ChIP) (Fig. 2A). Recruitment of CBP to the c-fos promoter in response to BDNF stimulation is reduced more than 50% in IPMK-deleted neuronal cultures. Associated histone H3 and H4 acetylation is similarly diminished (Fig. 2B). NGF-associated recruitment of CBP to the c-fos promoter in PC12 cells is also substantially reduced (Fig. S3A) after IPMK depletion by RNA interference with similar diminution of histone acetylation (Fig. S3B). The importance of IPMK in this process is evident in intact mice subjected to ECS, in which CBP recruitment to the c-fos promoter (Fig. 2C) and associated histone acetylation is markedly reduced in IPMK knockout mice (Fig. 2D).
Fig. 2.
IPMK binds to CBP and enhances its role as a transcriptional activator of c-fos. (A) IPMKfl/fl primary cortical neurons are infected with GFP or GFP-Cre expressing lentivirus, treated with BDNF, and assayed via ChIP for CBP recruitment to the c-fos promoter. Deletion of IPMK decreases CBP recruitment to the c-fos promoter by ∼60%. ***P < 0.001. Data are means ± SEM from three experiments. (B) Histone H3 and H4 acetylation is significantly decreased after treatment with BDNF in primary cortical neurons with IPMK deletion. ***P < 0.001. Data are means ± SEM from three experiments. (C) Nestin-Cre IPMKfl/fl mice exhibit ∼60% less CBP recruitment to the c-fos promoter after ECS treatment compared with littermate IPMKfl/fl control mice. ***P < 0.001. Data are means ± SEM from three experiments. (D) Histone H4 acetylation after ECS treatment is decreased by 65% in IPMK knockout mice compared with controls. ***P < 0.001. Data are means ± SEM from three experiments. (E) Endogenous IPMK and CBP interact. Endogenous IPMK is immunoprecipitated with IPMK antibody versus control IgG antibody in wild-type mice. Coimmunoprecipitates are separated via gel electrophoresis, and Western blotting is performed with anti-CBP antibody. (F) PC12 cells are transfected with myc-IPMK and treated with KCl. Overexpressed IPMK elicits a threefold increase in binding to CBP after depolarization. **P < 0.01. Data are means ± SEM from three experiments.
To elucidate how IPMK affects CBP, we examined physical interactions between the two proteins. Endogenous IPMK binds to CBP in mouse brain lysates (Fig. 2E). Overexpressed IPMK and CBP bind robustly in HEK293 cells (Fig. S3C). This binding is regulated by neuronal depolarization, as within 5 min of KCl treatment, binding between the two proteins increases more than threefold (Fig. 2F). Similarly, NGF stimulates IPMK-CBP binding more than twofold in Neuro2A cells (Fig. S3D).
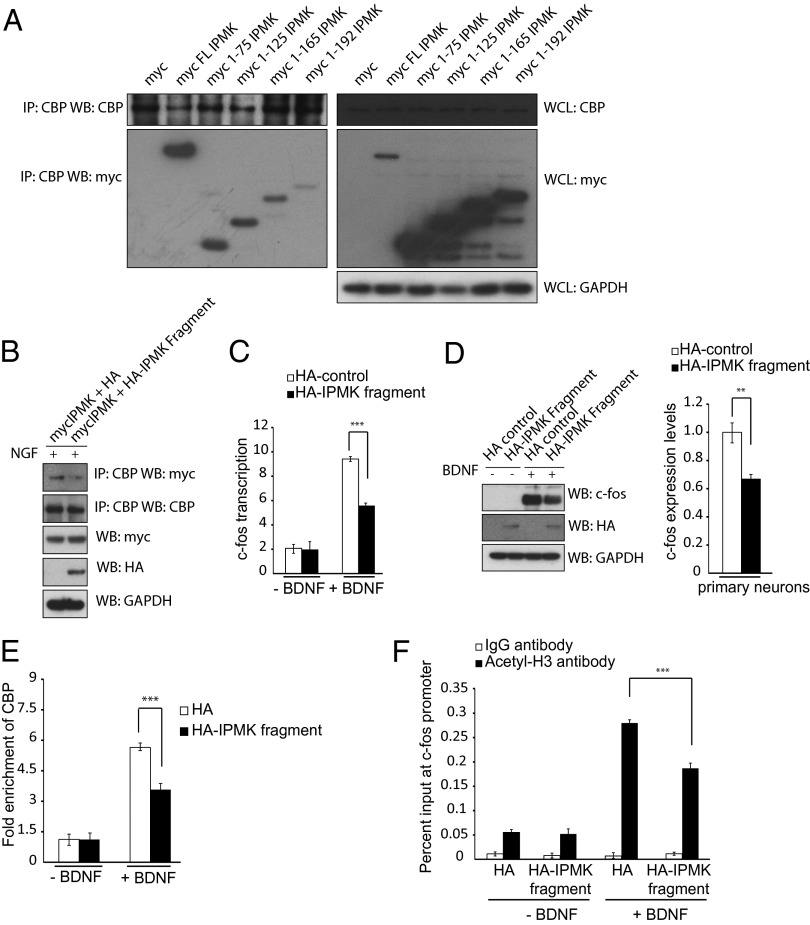
To ascertain whether binding of IPMK to CBP is responsible for regulation of CBP function, we mapped binding interactions between IPMK and CBP. A construct comprising amino acids 1–75 of IPMK binds robustly to CBP (Fig. 3A). This IPMK 1–75 fragment acts as a dominant-negative, preventing binding of IPMK to CBP (Fig. 3B). We monitored influences of the 1–75 fragment on BDNF-elicited transcriptional events. Induction of c-fos mRNA (Fig. 3C) and protein (Fig. 3D) by BDNF is reduced substantially by overexpression of the IPMK dominant-negative fragment. Similarly, recruitment of CBP to the c-fos promoter (Fig. 4E) and associated histone acetylation (Fig. 4F) are both markedly diminished by the 1–75 IPMK fragment.
Fig. 3.
A dominant negative construct of IPMK prevents IPMK-CBP association and CBP-mediated acetylation of the c-fos promoter. (A) C-terminal deletion constructs of myc-IPMK are transfected into PC12 cells. CBP is immunoprecipitated, and the myc tag is probed via Western blot. (B) PC12 cells overexpressing a dominant-negative fragment of HA-IPMK encompassing amino acids 1–75 show decreased levels of IPMK-CBP binding. (C) Real-time PCR is used to assess c-fos transcription levels in primary cortical neurons after BDNF treatment. Neurons expressing a dominant-negative IPMK fragment show decreased transcription levels of c-fos. ***P < 0.001. Data are means ± SEM from three experiments. (D) c-fos protein levels are assayed in primary neurons expressing control HA-tag or HA-tagged IPMK dominant negative after BDNF treatment using Western blot. Neurons containing an overexpressed dominant-negative IPMK fragment show diminished c-fos protein levels. **P < 0.01. Data are means ± SEM from three experiments. (E) The ChIP assay is used to assess CBP recruitment to the c-fos promoter after BDNF treatment in neurons infected with control HA-tag or HA-tagged dominant negative construct. Cells containing the dominant-negative IPMK fragment show decreased recruitment to the promoter. ***P < 0.001. Data are means ± SEM from three experiments. (F) The ChIP assay is used to assess histone H3 acetylation at the c-fos promoter after BDNF treatment. Cells containing the dominant-negative IPMK fragment demonstrate decreased levels of histone acetylation. ***P < 0.001. Data are means ± SEM from three experiments.
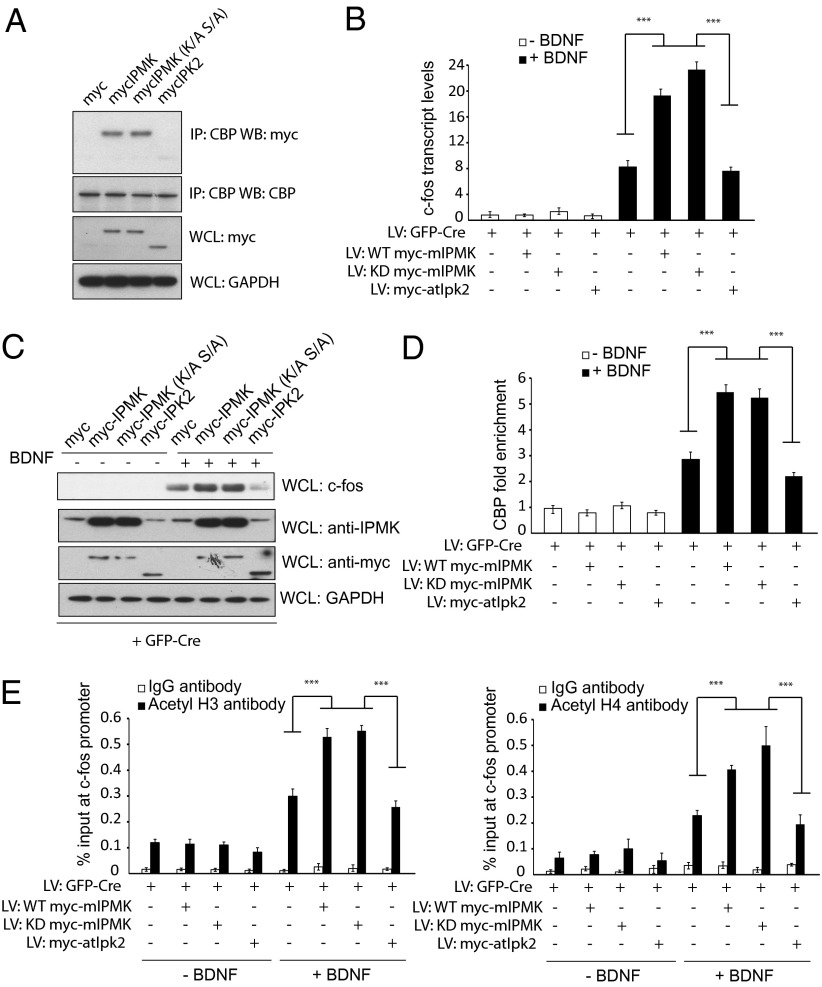
Fig. 4.
IPMK kinase activity is not required for binding to CBP. (A) Determinants of the IPMK-CBP interaction. Myc-tagged wild-type mIPMK, kinase-dead mIPMK, and atIpk2 are transfected into PC12 cells. Binding between various constructs of IPMK and CBP is assessed via coimmunoprecipitation experiments. Although wild-type mIPMK and kinase-dead mIPMK coimmunoprecipitates with CBP, atIpk2 does not. (B) Wild-type and kinase-dead IPMK, but not atIpk2, can complement IPMK deficiency in c-fos transcription. Endogenous IPMK is depleted in IPMKfl/fl neurons by infection with lentiviral particles expressing GFP-Cre. Control myc peptide, wild-type mIPMK, kinase-dead mIPMK, and atIpk2 are coexpressed using lentivirus. Neurons are stimulated with BDNF, and mRNA of c-fos was assessed via qPCR. (C) Western blotting reveals an increase in c-fos protein levels in IPMK-null neurons complemented with wild-type and kinase-dead IPMK, but not atIpk2. (D) ChIP using anti-CBP antibody reveals increased CBP recruitment to the c-fos promoter on complementation of IPMK-deleted neurons with wild-type or kinase-dead, but not atIpk2, IPMK. P < 0.001. Data are means ± SEM from three experiments. (E) Infection of IPMK-deleted neurons with wild-type or kinase-dead IPMK enhances histone H3 and H4 acetylation at the c-fos promoter after treatment with BDNF. Such an increase is not seen with expression of atIpk2 IPMK in IPMK-null neurons. P < 0.001. Data are means ± SEM from three experiments.
IEG Induction Does Not Require IPMK Catalytic Ability.
IPMK acts catalytically as both an IP3-kinase phosphorylating inositol phosphates (12, 13) and a PI3-kinase phosphorylating phosphatidylinositol 4,5-bisphosphate (14, 23). Noncatalytically, IPMK binds and stabilizes the mTORC1 complex (16). We explored properties of IPMK that may mediate its stimulation of IEGs. In primary cortical cultures, we deleted IPMK by lentivirus GFP-Cre and attempted rescue by infection with lentiviruses encoding wild-type IPMK, kinase-dead IPMK (K129A, S235A), or Arabidopsis thaliana IPMK (atIpk2). AtIpk2 is notable for lacking PI3-kinase activity and possessing only IP3-kinase activity, thus allowing selective complementation of pathways that require soluble inositol polyphosphates (24, 25). The divergence in structure of atIpk2 from mammalian IPMK (24) is evident in the failure of atIpk2 to bind CBP (Fig. 4A). The decrease in induction of c-fos RNA (Fig. 4B) and protein (Fig. 4C) in IPMK-deleted primary neurons is rescued both by wild-type and kinase dead mammalian IPMK, but not by atIpk2.
We substantiated these findings in experiments using PC12 cells that were stably expressing wild-type mouse IPMK, kinase-dead IPMK, or IPMK (atIpk2) and that were transiently transfected with shRNAs to knock down endogenous rat IPMK. Depolarization with KCl increases c-fos mRNA more than twofold in cells overexpressing wild-type IPMK and kinase-dead IPMK, but not atIpk2 (Fig. S4A). Both wild-type and kinase-dead IPMK, but not atIpk2, increase c-fos protein levels (Fig. S4B). These experiments confirm that catalytic activity of IPMK is not required for gene activation and that inositol phosphate repletion is not sufficient to rescue c-fos transcription.
In primary IPMK knockout cortical cultures, recruitment of CBP to the c-fos promoter (Fig. 4D) and concomitant histone H3 acetylation (Fig. 4E) are rescued by both wild-type and kinase dead IPMK, but not by atIpk2. Thus, regulation of IEGs by IPMK is independent of catalytic function but displays certain structural requirements, as atIpk2 fails to elicit these actions.
Discussion
In sum, we show that IPMK is critical for the induction of IEGs by diverse stimuli including neuronal depolarization and various growth factors. IPMK regulates IEGs by binding to CBP to augment its recruitment to promoters of the IEGs. Because of the importance of CBP function and IEGs for plasticity of neuronal function (22, 26–32), we suggest that IPMK is a novel determinant of mechanisms underlying learning and memory. Such a possibility is supported by the impaired performance of nestin-Cre+/− IPMKfl/fl mice in contextual habituation and the Barnes maze test.
A notably pleiotropic protein, IPMK possesses two distinct kinase functions as well as noncatalytic functions. In the cytosol, IPMK is a physiologic PI3-kinase that enhances signaling downstream of Akt phosphorylation (14). In addition, IPMK is a crucial cofactor in the mTORC1 complex independent of its catalytic function (16). Within the nucleus, yeast IPMK (also known as Arg82) regulates genes responsive to arginine and phosphate disposition (33). Although considerable debate has centered on the role of IPMK’s catalytic metabolites in yeast transcriptional regulation (34–36), recent complementation experiments reveal that arginine gene transcription does not require IPMK’s catalytic activity (37). This is in contrast with PHO5 gene transcription, which requires IPMK’s IP3-kinase functionality (38). Whereas IPMK’s ability to regulate transcription in yeast is well-established, nuclear roles of mammalian IPMK are less understood. We recently reported that IPMK binds to p53 during cell death and enhances p53-mediated gene transcription independent of IPMK catalytic activity (17, 18). Blind et al. (23) showed that IPMK’s nuclear PI3 kinase activity generating PIP3 is integral to the transcription of steroidogenic factor 1 targets (23).
Recently, transcriptional repressive functions have been implicated for inositol 1,4,5,6-tetrakisphosphate (39). In a crystal structure of histone deacetylase 3 (HDAC3) in complex with the deacetylase activating domain (DAD) domain of nuclear receptor co-repressor/silencing mediator for retinoid or thyroid-hormone receptors (NCoR/SMRT), inositol 1,4,5,6-tetrakisphosphate localizes at the interface of these two proteins and stimulates HDAC catalytic activity. Thus, the soluble inositol phosphate kinase activity of IPMK might repress gene transcription. In contrast, we show that IPMK noncatalytically stimulates histone acetylation by CBP. Conceivably, posttranslational modifications may elicit a switch between IPMK’s catalytic versus noncatalytic states, differentiating transcriptional coactivation versus corepression via recruitment of IPMK into CBP versus HDAC-containing complexes.
Our study has focused on neural roles of IPMK-CBP interactions. Both IPMK and CBP are expressed ubiquitously (31, 40). CBP regulates many transcriptional events other than IEGs. Accordingly, IPMK’s influence on gene transcription may be relevant to a wide range of physiologic events.
Materials and Methods
Mice.
IPMKfl/fl mice were generated at Ozgene, as described previously (14). IPMKfl/fl mice were mated with nestin-Cre mice (Jackson Laboratory, stock no. 003771) or synapsin-Cre mice (Jackson Laboratory, stock no. 003966) to create IPMK brain-knockout mice. Mice were weaned at postnatal day 20 and housed by sex in groups of three to five mice. Male mice were used for experiments at 6–12 wk of age. All mice were housed with a 12-h light-dark schedule and received food and water ad libitum. Animal protocols were performed in accordance with National Institutes of Health guidelines and approved by the Johns Hopkins University Committee on Animal Care.
ECS.
ECS was administered as previously described (41), wherein the electroshock consisted of 0.5-s, 100-Hz, 10-mA stimulus of 0.5-ms square wave pulses delivered using the Ugo Basile ECT unit, Model 7801. At these parameters, both IPMK deletion mutants and control littermates showed similar minor convulsions lasting 1–2 s; 15 min after ECS, animals were decapitated and their brains processed for RNA isolation. For Western blotting, animals were processed 1 h after ECS.
Dissociated Neuron Culture.
Dissociated cortical neurons were prepared from E16/17 mouse pups and maintained in a humidified incubator with 5% CO2 at 37 °C. Cultures were maintained in Neurobasal medium supplemented with B27 (Invitrogen) and glutamine. Neurons were plated at 1,500,000 per well in a six-well plate, or 8,000,000 per 10-cm plate. Plates were coated with poly-d-lysine. For viral transduction, neurons were infected overnight with 1 μL of 1.5 × 108 infectious units per milliliter per 1,000,000 cells. For stimulation experiments, neurons were depolarized for 30 min for RNA and 1 h for Western blots and ChIP. Neurons were treated with KCl (55 mM), forskolin (10 μM), BDNF (50 ng/mL), bicuculline (50 µM/mL), kainate (100 nM), or NGF (100 ng/mL).
ChIP Assay.
ChIP assays were performed as described previously (42). See SI Materials and Methods for detailed protocol.
Barnes Maze Test.
The Barnes Maze was performed as previously described (43). Animals were trained with two trials each day for 10 d, with an intertrial interval of 1 h. After training, animals were tested via probe trial 7 d later. Data were compared with two-way ANOVA followed by Holm-Sidak post hoc tests.
Novel Object Recognition Test.
This task was performed as previously described (26). Mice were tested at 3 and 24 h. Exploration times were recorded and used to calculate the discrimination index (time spent with object A − time spent with object B)/(total time exploring both objects) for training and test sessions. Discrimination indices of 0 indicate equal exploration of both objects. Discrimination indices were compared via one-way ANOVA with Dunnett's post hoc analysis.
Context Habituation.
To examine habituation of activity and memory for a context, mice were allowed to freely explore and open a field chamber for two 5-min sessions on each of 2 consecutive days (day 1 and day 2). Mice were then returned to the same chamber 7 d later for two additional 5-min sessions (day 9). Average movement times were calculated for mice in each group, and the movement times of day 2 and day 9 were compared to assess retention for context via one-way ANOVA.
Supplementary Material
Acknowledgments
We thank A. Snowman, L. Hester, R. Barrow, J. Xu, M. Ma, J. Sbodio, R. Tokhunts, R. Mealer, P. Scherer, and other members of the S.H.S. laboratory for their helpful discussions and B. Ziegler for organizing the manuscript. This work was funded by US Public Health Service Grant MH-18501 and Medical Scientist Training Program T-32 Grant (to R.X. and M.S.V.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315551110/-/DCSupplemental.
References
- 1.Cole AJ, Saffen DW, Baraban JM, Worley PF. Rapid increase of an immediate early gene messenger RNA in hippocampal neurons by synaptic NMDA receptor activation. Nature. 1989;340(6233):474–476. doi: 10.1038/340474a0. [DOI] [PubMed] [Google Scholar]
- 2.Curran T, Morgan JI. Superinduction of c-fos by nerve growth factor in the presence of peripherally active benzodiazepines. Science. 1985;229(4719):1265–1268. doi: 10.1126/science.4035354. [DOI] [PubMed] [Google Scholar]
- 3.Cole RL, Konradi C, Douglass J, Hyman SE. Neuronal adaptation to amphetamine and dopamine: Molecular mechanisms of prodynorphin gene regulation in rat striatum. Neuron. 1995;14(4):813–823. doi: 10.1016/0896-6273(95)90225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hope B, Kosofsky B, Hyman SE, Nestler EJ. Regulation of immediate early gene expression and AP-1 binding in the rat nucleus accumbens by chronic cocaine. Proc Natl Acad Sci USA. 1992;89(13):5764–5768. doi: 10.1073/pnas.89.13.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunt SP, Pini A, Evan G. Induction of c-fos-like protein in spinal cord neurons following sensory stimulation. Nature. 1987;328(6131):632–634. doi: 10.1038/328632a0. [DOI] [PubMed] [Google Scholar]
- 6.Cole AJ, Abu-Shakra S, Saffen DW, Baraban JM, Worley PF. Rapid rise in transcription factor mRNAs in rat brain after electroshock-induced seizures. J Neurochem. 1990;55(6):1920–1927. doi: 10.1111/j.1471-4159.1990.tb05777.x. [DOI] [PubMed] [Google Scholar]
- 7.Bozon B, Davis S, Laroche S. A requirement for the immediate early gene zif268 in reconsolidation of recognition memory after retrieval. Neuron. 2003;40(4):695–701. doi: 10.1016/s0896-6273(03)00674-3. [DOI] [PubMed] [Google Scholar]
- 8.Jones MW, et al. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci. 2001;4(3):289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- 9.Ramamoorthi K, et al. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science. 2011;334(6063):1669–1675. doi: 10.1126/science.1208049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alberi L, et al. Activity-induced Notch signaling in neurons requires Arc/Arg3.1 and is essential for synaptic plasticity in hippocampal networks. Neuron. 2011;69(3):437–444. doi: 10.1016/j.neuron.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plath N, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52(3):437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 12. Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH (1999) Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol 9(22):1323–1326. [DOI] [PubMed]
- 13.Saiardi A, et al. Mammalian inositol polyphosphate multikinase synthesizes inositol 1,4,5-trisphosphate and an inositol pyrophosphate. Proc Natl Acad Sci USA. 2001;98(5):2306–2311. doi: 10.1073/pnas.041614598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maag D, et al. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc Natl Acad Sci USA. 2011;108(4):1391–1396. doi: 10.1073/pnas.1017831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Resnick AC, et al. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc Natl Acad Sci USA. 2005;102(36):12783–12788. doi: 10.1073/pnas.0506184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S, et al. Amino acid signaling to mTOR mediated by inositol polyphosphate multikinase. Cell Metab. 2011;13(2):215–221. doi: 10.1016/j.cmet.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu R, et al. Inositol polyphosphate multikinase is a coactivator of p53-mediated transcription and cell death. Sci Signal. 2013;6(269):ra22. doi: 10.1126/scisignal.2003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu R, Snyder SH. Gene transcription by p53 requires inositol polyphosphate multikinase as a co-activator. Cell Cycle. 2013;12(12):1819–1820. doi: 10.4161/cc.25119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frederick JP, et al. An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc Natl Acad Sci USA. 2005;102(24):8454–8459. doi: 10.1073/pnas.0503706102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23(1):99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Y, et al. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15(7):859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blind RD, Suzawa M, Ingraham HA. Direct modification and activation of a nuclear receptor-PIP₂ complex by the inositol lipid kinase IPMK. Sci Signal. 2012;5(229):ra44. doi: 10.1126/scisignal.2003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Endo-Streeter S, Tsui MK, Odom AR, Block J, York JD. Structural studies and protein engineering of inositol phosphate multikinase. J Biol Chem. 2012;287(42):35360–35369. doi: 10.1074/jbc.M112.365031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevenson-Paulik J, Odom AR, York JD. Molecular and biochemical characterization of two plant inositol polyphosphate 6-/3-/5-kinases. J Biol Chem. 2002;277(45):42711–42718. doi: 10.1074/jbc.M209112200. [DOI] [PubMed] [Google Scholar]
- 26.Alarcón JM, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/- mice: A model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42(6):947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 27.Caccamo A, Maldonado MA, Bokov AF, Majumder S, Oddo S. CBP gene transfer increases BDNF levels and ameliorates learning and memory deficits in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA. 2010;107(52):22687–22692. doi: 10.1073/pnas.1012851108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crump NT, et al. Dynamic acetylation of all lysine-4 trimethylated histone H3 is evolutionarily conserved and mediated by p300/CBP. Proc Natl Acad Sci USA. 2011;108(19):7814–7819. doi: 10.1073/pnas.1100099108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giralt A, et al. Long-term memory deficits in Huntington’s disease are associated with reduced CBP histone acetylase activity. Hum Mol Genet. 2012;21(6):1203–1216. doi: 10.1093/hmg/ddr552. [DOI] [PubMed] [Google Scholar]
- 30.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42(6):961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrij F, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376(6538):348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, et al. Metformin activates an atypical PKC-CBP pathway to promote neurogenesis and enhance spatial memory formation. Cell Stem Cell. 2012;11(1):23–35. doi: 10.1016/j.stem.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 33.Dubois E, Messenguy F. Pleiotropic function of ArgRIIIp (Arg82p), one of the regulators of arginine metabolism in Saccharomyces cerevisiae. Role in expression of cell-type-specific genes. Mol Gen Genet. 1994;243(3):315–324. doi: 10.1007/BF00301067. [DOI] [PubMed] [Google Scholar]
- 34.Dubois E, Dewaste V, Erneux C, Messenguy F. Inositol polyphosphate kinase activity of Arg82/ArgRIII is not required for the regulation of the arginine metabolism in yeast. FEBS Lett. 2000;486(3):300–304. doi: 10.1016/s0014-5793(00)02318-8. [DOI] [PubMed] [Google Scholar]
- 35.El Alami M, Messenguy F, Scherens B, Dubois E. Arg82p is a bifunctional protein whose inositol polyphosphate kinase activity is essential for nitrogen and PHO gene expression but not for Mcm1p chaperoning in yeast. Mol Microbiol. 2003;49(2):457–468. doi: 10.1046/j.1365-2958.2003.03562.x. [DOI] [PubMed] [Google Scholar]
- 36.Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287(5460):2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 37.Bosch D, Saiardi A. Arginine transcriptional response does not require inositol phosphate synthesis. J Biol Chem. 2012;287(45):38347–38355. doi: 10.1074/jbc.M112.384255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steger DJ, Haswell ES, Miller AL, Wente SR, O’Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299(5603):114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watson PJ, Fairall L, Santos GM, Schwabe JW. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481(7381):335–340. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14(13):1553–1577. [PubMed] [Google Scholar]
- 41.Ramanan N, et al. SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat Neurosci. 2005;8(6):759–767. doi: 10.1038/nn1462. [DOI] [PubMed] [Google Scholar]
- 42.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39(5):715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 43. Sunyer B, Patil S, Höger H, Lubec G (October 4, 2007) Barnes maze, a useful task to assess spatial reference memory in the mice. Nature Protocol Exchange, 10.1038/nprot.2007.390.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.