Significance
In cyanobacteria, the photosystem II (PSII) core subunit, D1, is synthesized as a precursor and must be processed by a C-terminal peptidase before functional PSII can be assembled. Our work shows that in land plants, D1 maturation is not only required for the assembly of a functional PSII, but also is essential for the formation of supercomplexes—structures that are absent in cyanobacteria. This unexpected link opens an avenue for exploring the mechanism for the association of light harvesting complexes with the PSII core complexes—a final step in the development of the naturally occurring form of PSII in land plants.
Keywords: photosynthesis, photoinhibition
Abstract
Photosystem II (PSII) reaction center protein D1 is synthesized as a precursor (pD1) with a short C-terminal extension. The pD1 is processed to mature D1 by carboxyl-terminal peptidase A to remove the C-terminal extension and form active protein. Here we report functional characterization of the Arabidopsis gene encoding D1 C-terminal processing enzyme (AtCtpA) in the chloroplast thylakoid lumen. Recombinant AtCtpA converted pD1 to mature D1 and a mutant lacking AtCtpA retained all D1 in precursor form, confirming that AtCtpA is solely responsible for processing. As with cyanobacterial ctpa, a knockout Arabidopsis atctpa mutant was lethal under normal growth conditions but was viable with sucrose under low-light conditions. Viable plants, however, showed deficiencies in PSII and thylakoid stacking. Surprisingly, unlike its cyanobacterial counterpart, the Arabidopsis mutant retained both monomer and dimer forms of the PSII complexes that, although nonfunctional, contained both the core and extrinsic subunits. This mutant was also essentially devoid of PSII supercomplexes, providing an unexpected link between D1 maturation and supercomplex assembly. A knock-down mutant expressing about 2% wild-type level of AtCtpA showed normal growth under low light but was stunted and accumulated pD1 under high light, indicative of delayed C-terminal processing. Although demonstrating the functional significance of C-terminal D1 processing in PSII biogenesis, our study reveals an unsuspected link between D1 maturation and PSII supercomplex assembly in land plants, opening an avenue for exploring the mechanism for the association of light-harvesting complexes with the PSII core complexes.
Photosystem II (PSII) consists of more than 20 subunits. Assembly of this photosystem is a multistep process that functions in a highly coordinated fashion (1–3). The process starts with PSII initiation complexes (D2, PsbE, PsbF, and PsbI), and then D1 and CP47 are sequentially recruited to form CP47-RC complexes, followed by addition of PsbH, PsbM, PsbTc, and PsbR subunits. Next, CP43 and other subunits are added to generate PSII monomers, which develop into PSII dimers. Finally, light-harvesting complex (LHC) II is attached to form PSII supercomplexes. The D1 protein of PSII is prone to photodamage under excessive light conditions (4). To sustain photosynthesis, damaged D1 protein is degraded and replaced with a newly synthesized copy via PSII repair—a highly complex and critical process whose mechanism remains unclear (3, 4).
In most oxygen-evolving photosynthetic organisms, D1 protein is synthesized as a precursor (pD1) with a C-terminal tail. The pD1 protein is integrated into the thylakoid membrane and forms the initial PSII reaction center combined with other PSII subunits. The C-terminal tail of pD1 must be cleaved by an endopeptidase named the carboxyl terminal peptidase (Ctp) to produce mature D1, the functional form (5). In the cyanobacterium Synechocystis PCC 6803, there are three Ctp homologs (CtpA, CtpB, and CtpC), but only one, CtpA, is responsible for cleavage of the pD1 C-terminal extension (5). Disruption of CtpA leads to a loss of PSII activity and oxygen evolution from failure to form the manganese cluster (4, 6). The processing of pD1 is also critical for the association of extrinsic proteins on the luminal side to stabilize the PSII complexes (6, 7).
In contrast to cyanobacteria, our knowledge of the significance of Ctp enzymes and D1 C-terminal processing is limited in land plants. Previous researchers reported the purification of CtpA-like protein from pea (8) and spinach (9). The spinach study further showed that the recombinant Ctp protein expressed from Escherichia coli displays activity against pD1 (9). However, because we lack a genetic approach, the functional significance of CtpA and C-terminal processing remains unknown in those and other land plants. In this study, we applied genetics to identify a gene (At4g17740) encoding a CtpA enzyme in Arabidopsis and showed that it is required for PSII function and chloroplast development. We found that Arabidopsis CtpA is essential for assembling functional PSII core complexes, dimers, and PSII supercomplexes. The enzyme is also critical for the PSII damage–repair cycle during the photoinhibition process.
Results and Discussion
CtpA Is Conserved in Higher Plants and Cyanobacteria.
In Arabidopsis thaliana, three genes (At4g17740, At3g57680, and At5g46390) are predicted to encode Ctp homologs according to amino acid sequence homology analysis (5, 10). Initially, we compared the cyanobacterial CtpA with these three Arabidopsis Ctp proteins and found that it shared 42%, 36%, and 31% amino acid identity with At4g17740, At3g57680, and At5g46390, respectively. Furthermore, the five amino acids required for activity of the cyanobacterial enzyme (11) were conserved in all three putative Arabidopsis homologs (Fig. S1A). We further compared the three putative Ctp proteins of Arabidopsis with spinach CtpA (9) and found that the product of gene At4g17740 showed 77% amino acid identity, whereas the products of other two genes shared less than 40% identity (Fig. S1B). We thus concluded that At4g17740 is likely to encode the D1 C-terminal processing protease CtpA in Arabidopsis. This protein is thus referred to as putative AtCtpA.
The Putative AtCtpA Is Located in the Chloroplast Thylakoid Lumen.
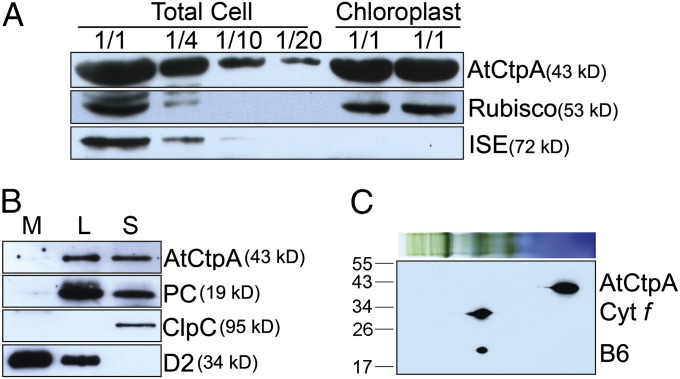
In cyanobacteria and the bacterium Pseudomonas aeruginosa, CtpA was found to be associated, respectively, with the plasma membrane and periplasmic space (12, 13). Proteomic analysis suggested, by contrast, that the Arabidopsis At4g17740 gene product was located in the chloroplast lumen in Arabidopsis (14). To confirm this observation, we performed an immunoblot analysis of proteins isolated from whole leaves, purified chloroplasts, and subchloroplast compartments. The AtCtpA protein was detected in the chloroplast fraction and total cell extract, similar to the blotting pattern for Rubisco (Fig. 1A). By contrast, ISE1, a mitochondria-specific RNA helicase (15), was not detected in the chloroplast fraction. Further experiments confirmed that, like plastocyanin (PC), the putative AtCtpA is a luminal protein (Fig. 1B). Because thylakoid breakage is unavoidable during chloroplast fractionation, stromal fraction is typically contaminated by thylakoid lumen proteins. Therefore, we observed that both PC—a well-known luminal protein—and AtCtpA were detected in the stromal fraction (Fig. 1B). Further, Blue Native (BN) gel followed by a 2D SDS/PAGE immunoblot revealed that At4g17740 protein was in the soluble fraction (Fig. 1C). However, this does not exclude the possibility that it may be loosely or transiently associated with membrane complexes and may fall off the membrane after the solubilization procedure used for Blue Native gel.
Fig. 1.
AtCtpA (At4g17740) localization. (A) AtCtpA is present in chloroplasts. Protein samples (corresponding to 4 µg chlorophyll or fractions of this amount—1/2, 1/4, 1/10, or 1/20) from total leaf extract or chloroplasts were separated by 12% SDS/PAGE and probed with antibodies against AtCtpA, Rubisco (a marker for chloroplast proteins), or ISE1 (a marker for mitochondria proteins). (B) AtCtpA is located in the thylakoid lumen. L, thylakoid lumen fraction; M, thylakoid membrane fraction; S, stromal fraction. Immunoblot analysis was conducted using antibodies against ClpC (a stromal protein), D2, and PC. (C) AtCtpA is not associated with high molecular PSII complexes. BN-PAGE and 2D SDS/PAGE immunoblot with an antibody against AtCtpA. Cytf, cytochrome f; B6, cytochrome b6. Auto-peroxidase activity of heme group in Cytf and Cytb6 produced signals in this assay.
Putative AtCtpA Is Essential for Autotrophic Growth and Chloroplast Development.
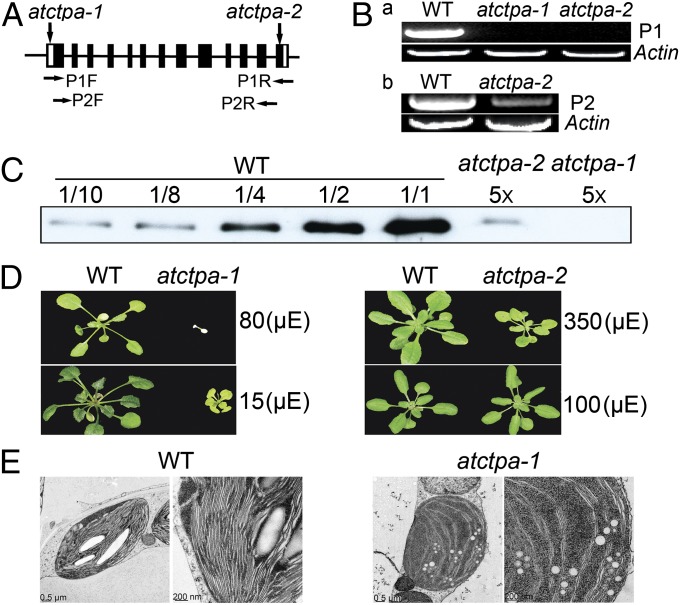
The function of putative AtCtpA was explored using two transfer DNA (T-DNA) insertion mutant alleles of the gene. The first allele, referred to as atctpa-1 (SALK_056011), contained a T-DNA insert in the 5′ UTR that would presumably block the initiation of transcription. The second allele, atctpa-2 (SALK_070529), had a T-DNA insert in the last exon of the coding region (Fig. 2A) that would result in a protein in which the C-terminal 18 aa of the wild-type (WT) protein was replaced by a new 11-aa stretch (Fig. S2). The full-length coding region of the At4g17740 transcript was not detected by RT-PCR in either mutant (Fig. 2 B, a). The atctpa-2 mutant plants showed a significantly reduced level of transcript that corresponded to the region before the T-DNA insertion site (P2 product) (Fig. 2 B, b). This result suggested that although neither mutant could synthesize a full-length WT protein, the atctpa-2 mutant might produce a mutated protein lacking the C-terminal 18 aa. Indeed, immunoblot analysis showed that the atctpa-1 mutant was deficient in AtCtpA protein and that the atctpa-2 mutant contained about 2% of the WT protein (Fig. 2C). Studies on cyanobacteria and a eukaryotic alga (Scenedesmus obliquus) identified several amino acids essential for CtpA activity (Fig. S1A) (11, 16). None of these was affected by the T-DNA insertion in atctpa-2. It seemed likely, therefore, that the mutated atctpa-2 protein still had enzyme activity.
Fig. 2.
Characterization of atctpa mutants. (A) Location of T-DNA insertion sites in the AtCtpA gene of atctpa-1 and atctpa-2 mutants. Solid bars indicate exons that are separated by introns (black lines). The 5′- and 3′-UTRs are indicated by white boxes. The horizontal arrows denote primers used in B. (B) RT-PCR analysis of AtCtpA transcripts in the atctpa mutants. (a) RT-PCR products using forward and reverse primers (P1F and P1R) indicated in A. (b) RT-PCR analysis performed with primers P2F and P2R shown in A. (C) Immunoblot analysis of AtCtpA protein in WT and atctpa mutants. Protein samples equivalent to 1 µg chlorophyll as 1/1 or fractions of 1/2, 1/4, 1/8, or a 5× multiple of this amount were analyzed from WT or mutants. (A–C) atctpa-1 grown under 15 µE in 1/2 MS medium containing 2% sucrose for 5 wk; atctpa-2 and WT grown under 350 µE in soil for 4 wk. (D) Phenotypes of 5-wk-old WT atctpa-1 and 4-wk-old WT atctpa-2 plants. atctpa-1 and its WT control were grown in 1/2 MS medium containing 2% sucrose under 15 or 80 µE light conditions; atctpa-2 and its WT counterpart were grown in soil under 100- or 350-µE light intensity. (E) Electron micrographs of chloroplasts were taken as described elsewhere (34) from 5-wk-old WT and atctpa-1 grown in 1/2 MS medium containing 2% sucrose under 15 µE.
Based on these results, we concluded that atctpa-1 is a null mutant and atctpa-2 represents a weak allele. The null mutant was lethal when grown in soil under all light conditions tested, but was viable in 1/2 Murashige and Skoog (MS) medium with 2% (wt/vol) sucrose under low light (15 µE), conditions used to grow this mutant in further analyses. These plants were, however, much smaller and appeared yellowish compared with WT. Relatively high light (80 µE) proved to be lethal (Fig. 2D). Plants containing the weak allele also showed reduced growth and yellowish leaves compared with WT when grown under high light (350 µE), but showed no significant differences when grown under normal light (100 µE) (Fig. 2D). The atctpa-2 mutants thus appeared to be sensitive to high light stress.
We examined the inner structure of chloroplasts of the WT and null mutant by electron microscopy (Fig. 2E) and found a dramatic difference: chloroplasts in the WT accumulated large starch grains, whereas starch grains were not visible in the mutant. Chloroplasts in WT also displayed well-organized thylakoid stacking—a feature essentially absent in the mutant. These chloroplast defects are consistent with the inability to grow photoautotrophically.
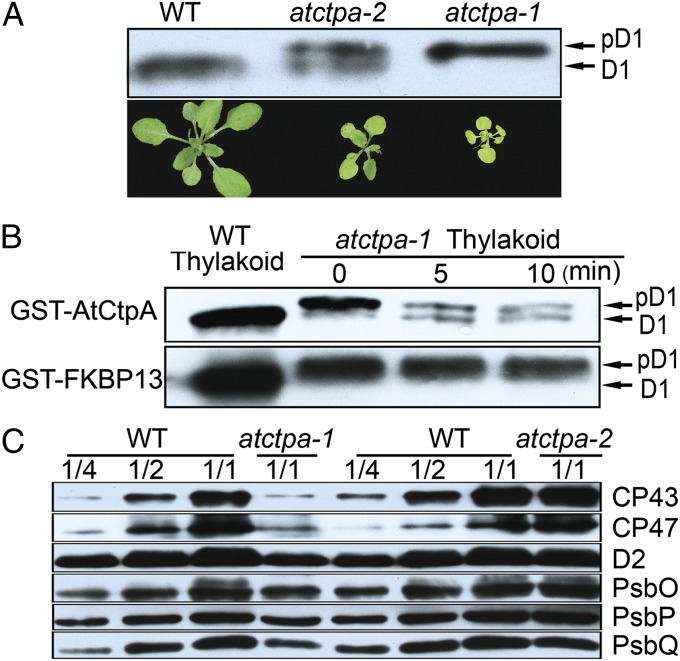
The AtCtpA Null Mutant Lacks Mature D1 Protein and Shows a Reduced Level of PSII Core Proteins.
The putative AtCtpA gene appeared to be essential for autotrophic growth and starch synthesis, consistent with its role in producing mature D1 protein for PSII assembly and function. To test this point, we analyzed D1 protein in thylakoid membranes from both mutants and compared them with WT (Fig. 3A). Strikingly, although observed only in its mature form in the WT plants, D1 was present only in pD1 form in the null mutant and in both pD1 and mature forms (mD1) in the weak allele. This experiment confirmed that At4g17740 encodes the D1 C-terminal processing protease CtpA. That the null mutant failed to produce detectable levels of mD1 indicated that this is the only gene encoding the C-terminal processing enzyme, and that the two other peptidase homologs (At3g57680 and At5g46390) cannot function in this capacity. This agrees with the previous study concluding that At3g57680 is not related to pD1 C-terminal processing (10). The presence of both pD1 and mD1 in the weak allele suggested that the AtCtpA protein (∼2% WT level) found in this mutant had limited D1 C-terminal processing activity. However, this activity was sufficient for growth under normal light (100 µE) (Fig. 2D).
Fig. 3.
AtCtpA is responsible for D1 C-terminal processing. (A) Western blot analysis of D1 protein in WT, atctpa-1, and atctpa-2 mutant plants (Upper). Photographs of WT, atctpa-1, and atctpa-2 plants (Lower). Thylakoid protein samples equivalent to 2 µg of chlorophyll were separated by 10% SDS/PAGE containing 8M urea, blotted and probed with D1 antibody. (B) D1 C-terminal processing activity assay in vitro. WT thylakoid sample was used as a control for mD1 in the left lane. Processing of pD1 into D1 in the presence of 5 µg of recombinant GST-AtCtpA is shown at different times. GST-FKBP13 (FK506-binding protein 13) protein served as a negative control. (C) Western blot analysis of other PSII subunits in WT, atctpa-1, and atctpa-2 plants. Immunoblot analysis was performed as in A with antibodies against the indicated thylakoid membrane proteins. Plant growth conditions: when WT was compared with atctpa-2, both plants were grown in the soil under 350 µE for 4 wk. When WT was compared with atctpa-1, plants were grown in 1/2 MS medium with 2% sucrose under 15 µE for 5 wk.
To demonstrate the role of AtCtpA activity in pD1 cleavage directly, we assayed the C-terminal processing enzyme in vitro. In the assay, we used purified GST-AtCtpA fusion protein expressed in E. coli as the enzyme and pD1 from the thylakoid membrane of atctpa-1 null mutant as the substrate. Before addition of the enzyme, we treated thylakoid membrane with 0.1% n-Dodecyl β-d-maltoside to ensure that pD1 in the thylakoid is accessible to the enzyme. The results showed that the GST-AtCtpA fusion protein rapidly converted pD1 to mD1, whereas a GST fusion control (GST-FKBP13) (17) did not cleave pD1 (Fig. 3B).
In further experiments, we examined other thylakoid membrane proteins of the two atctpa mutants (Fig. 3C and Fig. S3). In atctpa-2 mutant plants, the profile of thylakoid membrane proteins was largely comparable with that of WT. However, in atctpa-1, the PSII core proteins D2, CP43, and CP47 decreased to about 25% of WT. The oxygen evolution complex proteins, PsbO, PsbP, and PsbQ, were reduced to about 50% compared with WT (Fig. 3C). The PSI components, PsaF and PsaD, also decreased to this level (Fig. S3). We suspected that the reduced level of PSI subunits might be a secondary effect of PSII deficiency as described for other PSII mutants (18, 19).
Essential Role of AtCtpA in PSII Function.
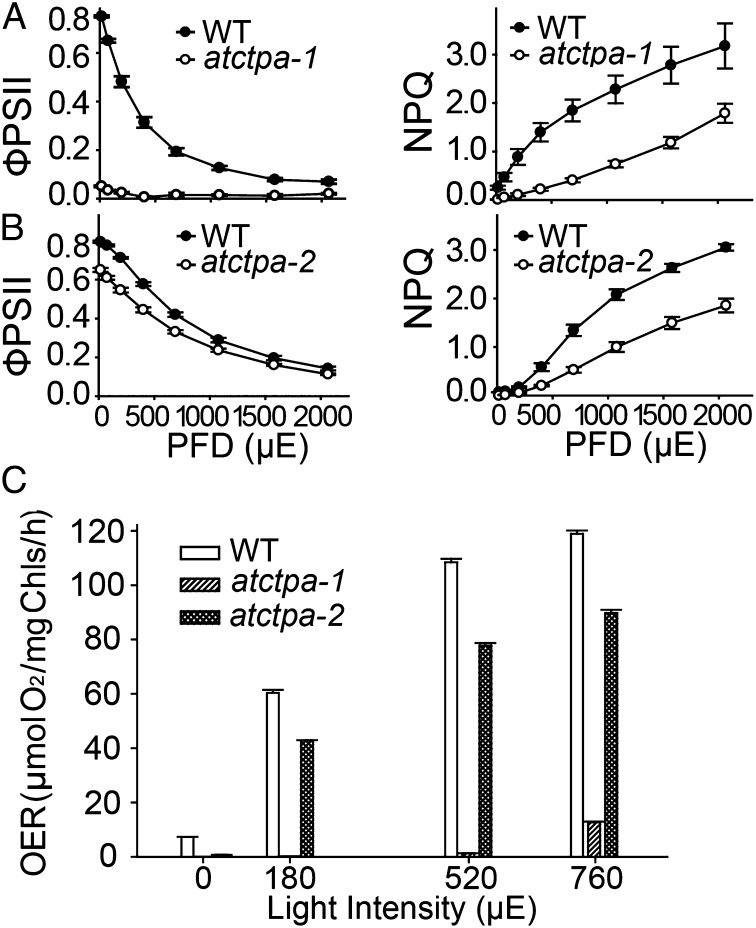
Although C-terminal processing of D1 protein is essential for PSII assembly and function in cyanobacteria, this requirement has not been documented for land plants. The severe growth defects observed in the atctpa null mutant suggested that the C-terminal processing of D1 is essential for photosynthesis in Arabidopsis. To identify the defect, we initially conducted chlorophyll fluorescence analyses using rosette leaves of whole plants. We observed that quantum efficiency of PSII (ФPSII), which represents photochemistry at different photon flux density (20), was below detection in atctpa-1 and was reduced in atctpa-2 (Fig. 4 A and B, Left). A defect was also observed for the nonphotochemical quenching (NPQ), which represents plants’ ability to dissipate excess light energy as heat (21) (Fig. 4 A and B, Right). Rapid NPQ induction and dark relaxation kinetics revealed that total NPQ is lower in atctpa-2 than WT and is rapidly reversible during a dark relaxation phase (Fig. S4), indicating that the decrease of NPQ was due to the rapidly reversible pH/energy-dependent component qE (21).
Fig. 4.
PSII activity in WT and mutant plants. (A) Chlorophyll fluorescence parameters of 5-wk-old WT and atctpa-1 plants grown in 1/2 MS plates with 2% sucrose (15 µE). (B) Chlorophyll fluorescence parameters of 4-wk-old WT and atctpa-2 plants grown in soil (350 µE). Data are presented as means ± SD (n = 4). PFD, photon flux density. WT and mutant data are significantly different (P < 0.05). (C) Oxygen evolution rate (OER) of WT, atctpa-1, and atctpa-2 plants. atctpa-1 was grown under 15 µE in 1/2 MS medium containing 2% sucrose for 5 wk; atctpa-2 and WT were grown under 350 µE in soil for 4 wk. Data shown as means ± SD (n = 4). WT and mutant data are significantly different (P < 0.05).
We also measured oxygen evolution rates in the mutants (Fig. 4C). The results showed that knockout mutant plants lacked oxygen evolution activity and that the knock-down counterparts had a reduced level, which is in agreement with plant growth phenotype. This result is also consistent with the finding that the CtpA-deficient mutants of a green alga (S. obliquus) and a cyanobacterium (Synechocystis PCC 6803) showed reduced oxygen evolution (22, 23). Collectively, the results indicated that PSII activity depends heavily on the function of the C-terminal processing enzyme, thus making mature D1 essential for PSII function in Arabidopsis and possibly land plants in general.
D1 C-Terminal Processing Is Required for the Assembly of PSII Supercomplexes.
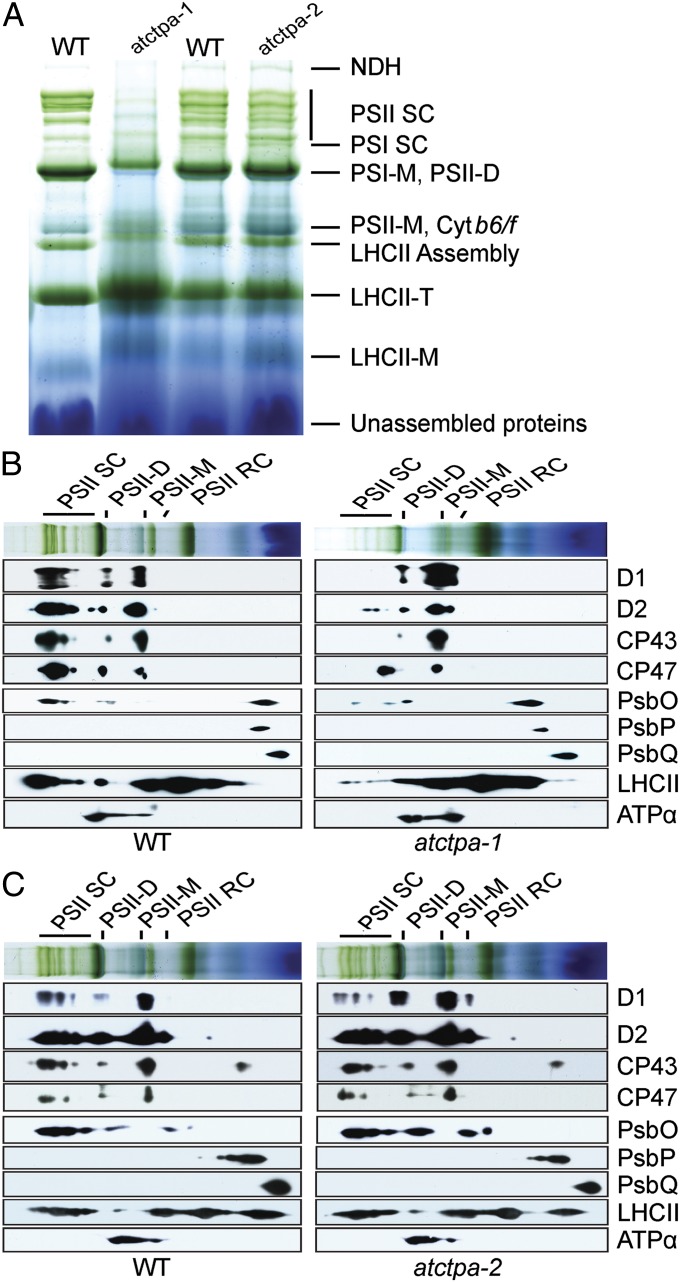
The loss of PSII activity could result from either a lack of, or a defect in, assembly of the complexes. We analyzed the profiles of photosynthetic complexes in the thylakoid membranes of WT and mutant plants. Because of differences in growth phenotype, we grew atctpa-1 under low light (15 µE) in 1/2 MS medium containing sucrose and atctpa-2 under high light (350 µE) in soil. PSII profiles in the atctpa-2 mutant were similar to WT in BN gels. By contrast, the knockout atctpa-1 mutant displayed a severely reduced level of PSII supercomplexes (PSII SC) and possibly PSII dimers (PSII-D) (Fig. 5A). Because supercomplexes are formed by the attachment of LHCII trimers to PSII dimers (24, 25), null mutant plants accumulated higher levels of LHCII trimers because of the deficiency in supercomplexes.
Fig. 5.
Protein complexes in the thylakoid membranes of WT and mutant plants. (A) BN gel analysis of thylakoid membrane protein complexes (7.5 µg chlorophyll) of WT and atctpa-1 grown under 15 µE in 1/2 MS medium with 2% sucrose for 5 wk and WT, atctpa-2 plants grown under 350 µE in soil for 4 wk. Protein complexes were identified as described previously (31). (B) Thylakoid proteins of WT and atctpa-1 separated by BN gel in A were further subjected to SDS/PAGE and immunoblotted with antibodies as indicated. (C) Thylakoid proteins of WT and atctpa-2 separated by BN gel in A were further subjected to SDS/PAGE and immunoblotted with antibodies as indicated. Plant growth conditions were the same as described in Fig. 3. LHCII-M, PSII LHC monomers; LHCII-T: PSII LHC trimers; NDH, NADPH dehydrogenase complexes; PSI-M, PSI monomers; PSI SC, PSI supercomplexes; PSII-M, PSII monomers.
To confirm the PSII pattern and further investigate changes in PSII subunits, we performed a 2D SDS/PAGE immunoblot after BN gel separation using antibodies against various PSII subunits (Fig. 5 B and C). Under the indicated low light (15 µE), PSII core subunits D1, D2, CP43, and CP47 were associated with PSII supercomplexes, PSII dimers, and PSII monomers in WT plants (Fig. 5B). By contrast, the unprocessed D1 protein and other PSII subunits in the atctpa-1 plants were negligible in the supercomplexes but were present in low-molecular-weight forms, including PSII dimers and PSII monomers (Fig. 5B). In the atctpa-1 mutant, a major portion of pD1, D2, and CP43 accumulated in the form of PSII monomers, suggesting that pD1 incorporation into the PSII monomers hinders further assembly of larger forms of PSII complexes. Under high light (350 µE), D2, CP43, and CP47 showed a similar pattern in the atctpa-2 mutant and WT, but the D1 pattern was quite different: the atctpa-2 mutant had less D1 in PSII supercomplexes and more in PSII dimers and monomers (Fig. 5C), consistent with a deficiency in the assembly of PSII into supercomplexes.
In the cyanobacterial ctpa mutant, a key defect lies in the assembly of the Mn cluster, leaving the extrinsic subunits of PSII in the soluble form (6). We thus examined the distribution pattern of three extrinsic subunits (PsbO, PsbP, and PsbQ) in WT and the atctpa-1 mutant using the 2D immunoblot approach. Unlike the situation in cyanobacteria, the three subunits showed a similar pattern in the WT and null mutant (Fig. 5B). This result indicates that although PSII function requires mature D1 in both land plants and cyanobacteria, the defect caused by pD1 may not be the same in the two organisms.
According to the current model of PSII assembly, the D1 C-terminal extension is believed to be cleaved by CtpA after insertion of pD1 into the reaction center core complexes (RC) (3, 26). Our results show that when the C-terminal extension is not cleaved, pD1 is nonetheless recruited to form PSII monomers and dimers (Fig. 5B), suggesting that the presence of the C-terminal extension does not block early steps of PSII assembly. The absence of PSII supercomplexes in the knockout mutant indicated, however, that the presence of pD1 hinders the association of LHCII complexes with PSII dimers.
Defect in C-Terminal Processing Renders Mutant Plants Hypersensitive to Photoinhibition.
Photoinhibition refers to the process in which PSII activity and thus the rate of photosynthesis is impeded by high light intensity (4). At the molecular level, the major target of photodamage is D1 protein that is degraded after damage and replaced with newly synthesized D1 during repair (3, 4, 27).
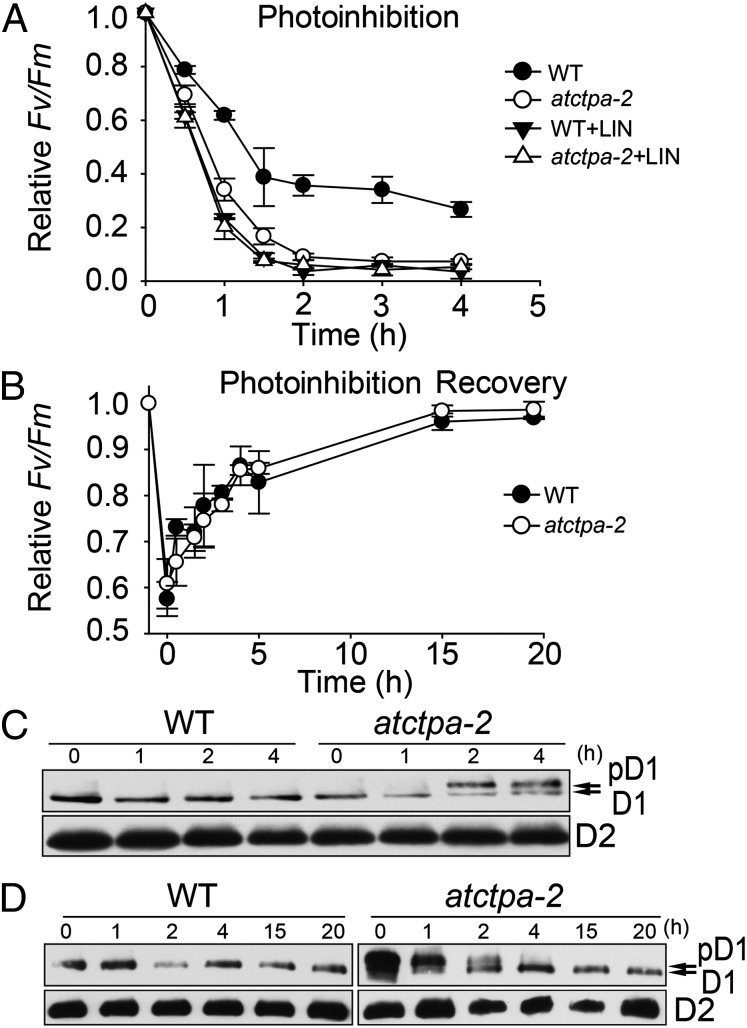
These data show that the atctpa mutants were sensitive to high light (Fig. 2D), raising the possibility that D1 C-terminal processing is important in the photodamage–repair cycle. To study the functional significance of D1 maturation in this cycle, we conducted photoinhibition and recovery assays using atctpa-2 mutant and WT plants, which had similar photosynthesis when grown under 100 µE. Under high light exposure (1,800 µE), atctpa-2 mutant plants displayed more severe photoinhibition than WT (Fig. 6A). When we added lincomycin to inhibit protein biosynthesis in chloroplasts, both WT and mutant were hypersensitive to photoinhibition under high light conditions (Fig. 6A). Without lincomycin, photodamage was effectively repaired in the mutant as well as in the WT under low light (15 µE), indicating that protein biosynthesis was not affected in the mutant (Fig. 6B). To examine the levels of D1 and D2 in parallel with PSII activity, we performed immunoblot analysis using protein extracts from the atctpa-2 and WT plants at different times during the photoinhibition and recovery treatments (Fig. 6 C and D). More pD1 accumulated in the mutant (Fig. 6C), along with a more severe decrease in PSII activity (Fig. 6A), when exposed to high light. In a separate experiment, plants were transferred to low light to allow recovery after high light treatment for 2 h (Fig. 6B). Immunoblot analysis showed that with recovery of PSII activity, the level of pD1 decreased and that of mature D1 increased (Fig. 6D). After low light treatment for 4 h, most pD1 disappeared, in parallel with the recovery of PSII activity (Fig. 6B). Based on this observation, we conclude that the processing of pD1 is light-dependent: under high light, the D1 turnover rate is high and so is pD1 processing; under low light, D1 turnover is slow and pD1 processing also slows down. This is in agreement with previous work (28).
Fig. 6.
PSII activity and D1 processing during photoinhibition and repair. (A) PSII activity (FV/FM) was recorded using detached leaves from 4-wk-old WT and atctpa-2 plants grown under 350 µE and monitored during exposure to high light (1,800 µE) for 4 h in the presence or absence of 1.5 mM lincomycin. Data for WT and atctpa-2 are not significantly different (P > 0.1) in the presence of lincomycin but are significantly different (P < 0.05) without lincomycin. (B) Photoinhibition recovery of WT and atctpa-2. Plants with 50% photoinhibition recovered under low light (15 µE) for up to 20 h. Data represent means ± SD (n = 3). WT and atctpa-2 data are not significantly different (P > 0.1). (C) D1 status during photoinhibition treatment. Samples were taken from plants identical to those used in Fig. 6A. Total proteins (equivalent to 2 µg chlorophyll) were subjected to immunoblotting with antibodies against D1 and D2, respectively. (D) D1 status during photoinhibition recovery. Samples were taken from plants as used in Fig. 6B and subjected to the same assay as that in C.
To confirm that the phenotype of atctpa was due to mutation of the At4g17740 gene, we conducted a complementation experiment. The results showed that the AtCtpA protein was restored to WT level in atctpa-2 mutant plants transformed with a construct containing the At4g17740 coding region under the control of the CaMV 35S promoter. The phenotype of the complemented transgenic plants was indistinguishable from WT (Fig. S5A). Immunoblot analysis showed that complemented plants, such as WT, produced mature D1 but not pD1 (Fig. S5B). These results thus confirmed that the growth defect in atctpa-2 resulted from mutation of the At4g17740 gene that encodes AtCtpA.
Concluding Remarks.
We have identified a gene encoding the D1 C-terminal processing protease CtpA in the chloroplast thylakoid lumen of Arabidopsis. Loss of AtCtpA activity resulted in the recovery of D1 protein in its precursor form. The C-terminal extension of pD1 prevented assembly of functional PSII complexes, thus leading to inability to grow photoautotrophically. Mutant plants producing only 2% of WT level of AtCtpA were hypersensitive to photoinhibition under high light because of their limited ability to satisfy the increased demand of D1 processing and repair. However, the same mutant plants grew well under low light when less photodamage took place; thus, less mature D1 was required. The results confirmed that, although two other genes encoding related proteases are present in the genome, a single gene is exclusively responsible for the production of CtpA activity in Arabidopsis.
The D1 protein is encoded by the plastid genome, making it difficult to conduct genetic analysis of this important element. The mutant plants described in this study will facilitate functional studies on mature D1 in the assembly of PSII complexes in land plants. The present results have confirmed that mature D1 is required for the assembly of functional PSII. As a result of D1 deficiency, the abundance of other PSII core subunits was also reduced—particularly D2, CP43, and CP47—supporting a general theme that deficiency of any core components in PSII complexes affects the abundance of other subunits. The observation that CP43 and CP47 were most dramatically affected by D1 deficiency agrees with earlier work on the sequence of PSII subunit incorporation—i.e., pD1 is incorporated into the D2-cyt559 complex before processing pD1 into mD1, whereas CP43 and CP47 are incorporated into the D1-D2 complex after pD1 processing (26). Our results further show that the majority of the pD1, D2, CP43, and CP47 components was incorporated into the PSII complexes, including both monomer and dimer forms. This observation suggests that, although much preferred, mD1 is not absolutely required for subunit incorporation into PSII even though the photosystem containing pD1 is inactive. Perhaps the most severe effect pD1 has on PSII assembly lies in the formation of supercomplexes. The pD1-containing monomers and dimers were not capable of interacting with LHCII trimers, a prerequisite to the formation of supercomplexes. Because cyanobacterial PSII does not form similar supercomplexes, this finding prompts further functional analysis of mD1 in the interaction between the PSII core complexes and LHCII light-harvesting complexes. The mechanism for this association remains unknown.
The current work demonstrates that C-terminal processing of D1 protein is required for photosynthesis in land plants, as found earlier for cyanobacteria and green algae. The results raise the question of how the formation of PSII supercomplexes is linked to D1 processing and, furthermore, how PSII assembly differs in cyanobacteria and land plants. Further studies using atctpa mutants as a model should address these questions.
Materials and Methods
A. thaliana (Columbia-0) and atctpa T-DNA insertion mutants were grown in the soil or on MS plates under conditions described in the figure legends. Homozygous T-DNA insertion lines were isolated by PCR genotyping of two putative mutant lines (29). The primers used are shown in Table S1. For mutant complementation, the coding region of AtCtpA was amplified by RT-PCR from total RNA and cloned behind the CaMV 35S promoter in the vector pBASTA-35S-FLAG. We transferred the construct into atctpa-2 mutant plants using the floral dip method (30).
Measurement of chlorophyll fluorescence parameters and immunoblot analysis of thylakoid membrane proteins was conducted as described previously (31). BN SDS/PAGE analysis, oxygen evolution measurements, and production of recombinant AtCtpA protein were done as described elsewhere (32). Recombinant GST-fusion protein was used to raise rabbit antibodies by the Cocolico Biotechnical Company.
The in vitro AtCtpA enzyme activity assay was performed according to previous work (33) using pD1 recovered from the detergent-treated (0.1% nN-Dodecyl β-d-maltoside) thylakoid membranes of the atctpa-1 mutant as substrate. The proteolytic reaction was carried out at 25 °C for 0, 5, and 10 min. The “0” time point contained a sample in which we stopped the reaction immediately after adding the enzyme. Therefore, residual amounts of pD1 would be processed into mature D1. Thylakoid membranes were collected and subjected to immunoblot analysis and probed with an antibody against D1 protein.
For photoinhibition assays, detached leaves from plants grown in soil under normal light (100 µE) were first soaked in water or 1.5 mM lincomycin for 3 h at an irradiance of 15 µE, followed by exposure to 1,800 µE. For recovery assays, leaves showing 50% photoinhibition (after 1,800 µE light exposure) were incubated under low light (15 µE) to recover for the indicated times. Samples were dark-adapted for 15 min before measuring Fv/Fm (the ratio of variable fluorescence to maximum fluorescence, a parameter of the maximum PSII activity). All data were subjected to Student t test in statistical analysis.
Supplementary Material
Acknowledgments
We thank Drs. A. Melis, S. Park, R. Malkin, and K. Niyogi for providing antibodies and helpful discussions. This work was supported in part by a fellowship from the Chinese Scholarship Council (to Y.C.) and by Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy Grant DE-FG02-11ER16274 (to S.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313894110/-/DCSupplemental.
References
- 1.Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004;303(5665):1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- 2.Guskov A, et al. Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat Struct Mol Biol. 2009;16(3):334–342. doi: 10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- 3.Baena-González E, Aro EM. Biogenesis, assembly and turnover of photosystem II units. Philos Trans R Soc Lond B Biol Sci. 2002;357(1426):1451–1459, discussion 1459–1460. doi: 10.1098/rstb.2002.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adir N, Zer H, Shochat S, Ohad I. Photoinhibition - a historical perspective. Photosynth Res. 2003;76(1-3):343–370. doi: 10.1023/A:1024969518145. [DOI] [PubMed] [Google Scholar]
- 5.Satoh K, Yamamoto Y. The carboxyl-terminal processing of precursor D1 protein of the photosystem II reaction center. Photosynth Res. 2007;94(2-3):203–215. doi: 10.1007/s11120-007-9191-z. [DOI] [PubMed] [Google Scholar]
- 6.Roose JL, Pakrasi HB. Evidence that D1 processing is required for manganese binding and extrinsic protein assembly into photosystem II. J Biol Chem. 2004;279(44):45417–45422. doi: 10.1074/jbc.M408458200. [DOI] [PubMed] [Google Scholar]
- 7.Roose JL, Wegener KM, Pakrasi HB. The extrinsic proteins of Photosystem II. Photosynth Res. 2007;92(3):369–387. doi: 10.1007/s11120-006-9117-1. [DOI] [PubMed] [Google Scholar]
- 8.Bowyer JR, et al. Carboxyl-terminal processing of the D1 protein and photoactivation of water-splitting in photosystem II. Partial purification and characterization of the processing enzyme from Scenedesmus obliquus and Pisum sativum. J Biol Chem. 1992;267(8):5424–5433. [PubMed] [Google Scholar]
- 9.Fabbri BJ, et al. The carboxyterminal processing protease of D1 protein: Expression, purification and enzymology of the recombinant and native spinach proteins. Pest Manag Sci. 2005;61(7):682–690. doi: 10.1002/ps.1038. [DOI] [PubMed] [Google Scholar]
- 10.Yin S, Sun X, Zhang L. An Arabidopsis ctpA homologue is involved in the repair of photosystem II under high light. Chin Sci Bull. 2008;53(7):1021–1026. [Google Scholar]
- 11.Inagaki N, Maitra R, Satoh K, Pakrasi HB. Amino acid residues that are critical for in vivo catalytic activity of CtpA, the carboxyl-terminal processing protease for the D1 protein of photosystem II. J Biol Chem. 2001;276(32):30099–30105. doi: 10.1074/jbc.M102600200. [DOI] [PubMed] [Google Scholar]
- 12.Zak E, et al. The initial steps of biogenesis of cyanobacterial photosystems occur in plasma membranes. Proc Natl Acad Sci USA. 2001;98(23):13443–13448. doi: 10.1073/pnas.241503898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoge R, Laschinski M, Jaeger KE, Wilhelm S, Rosenau F. The subcellular localization of a C-terminal processing protease in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2011;316(1):23–30. doi: 10.1111/j.1574-6968.2010.02181.x. [DOI] [PubMed] [Google Scholar]
- 14.Schubert M, et al. Proteome map of the chloroplast lumen of Arabidopsis thaliana. J Biol Chem. 2002;277(10):8354–8365. doi: 10.1074/jbc.M108575200. [DOI] [PubMed] [Google Scholar]
- 15.Stonebloom S, et al. Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. Proc Natl Acad Sci USA. 2009;106(40):17229–17234. doi: 10.1073/pnas.0909229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao DI, Qian J, Chisholm DA, Jordan DB, Diner BA. Crystal structures of the photosystem II D1 C-terminal processing protease. Nat Struct Biol. 2000;7(9):749–753. doi: 10.1038/78973. [DOI] [PubMed] [Google Scholar]
- 17.Gupta R, Mould RM, He Z, Luan S. A chloroplast FKBP interacts with and affects the accumulation of Rieske subunit of cytochrome bf complex. Proc Natl Acad Sci USA. 2002;99(24):15806–15811. doi: 10.1073/pnas.222550399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meurer J, Plücken H, Kowallik KV, Westhoff P. A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 1998;17(18):5286–5297. doi: 10.1093/emboj/17.18.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plücken H, Müller B, Grohmann D, Westhoff P, Eichacker LA. The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett. 2002;532(1-2):85–90. doi: 10.1016/s0014-5793(02)03634-7. [DOI] [PubMed] [Google Scholar]
- 20.Maxwell K, Johnson GN. Chlorophyll fluorescence—a practical guide. J Exp Bot. 2000;51(345):659–668. doi: 10.1093/jxb/51.345.659. [DOI] [PubMed] [Google Scholar]
- 21.Müller P, Li XP, Niyogi KK. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 2001;125(4):1558–1566. doi: 10.1104/pp.125.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diner BA, Ries DF, Cohen BN, Metz JG. COOH-terminal processing of polypeptide D1 of the photosystem II reaction center of Scenedesmus obliquus is necessary for the assembly of the oxygen-evolving complex. J Biol Chem. 1988;263(18):8972–8980. [PubMed] [Google Scholar]
- 23.Nixon PJ, Trost JT, Diner BA. Role of the carboxy terminus of polypeptide D1 in the assembly of a functional water-oxidizing manganese cluster in photosystem II of the cyanobacterium Synechocystis sp. PCC 6803: Assembly requires a free carboxyl group at C-terminal position 344. Biochemistry. 1992;31(44):10859–10871. doi: 10.1021/bi00159a029. [DOI] [PubMed] [Google Scholar]
- 24.Minagawa J, Takahashi Y. Structure, function and assembly of Photosystem II and its light-harvesting proteins. Photosynth Res. 2004;82(3):241–263. doi: 10.1007/s11120-004-2079-2. [DOI] [PubMed] [Google Scholar]
- 25.Nelson N, Yocum CF. Structure and function of photosystems I and II. Annu Rev Plant Biol. 2006;57:521–565. doi: 10.1146/annurev.arplant.57.032905.105350. [DOI] [PubMed] [Google Scholar]
- 26.Komenda J, Sobotka R, Nixon PJ. Assembling and maintaining the Photosystem II complex in chloroplasts and cyanobacteria. Curr Opin Plant Biol. 2012;15(3):245–251. doi: 10.1016/j.pbi.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 27.Edelman M, Mattoo AK. D1-protein dynamics in photosystem II: The lingering enigma. Photosynth Res. 2008;98(1-3):609–620. doi: 10.1007/s11120-008-9342-x. [DOI] [PubMed] [Google Scholar]
- 28.Oelmüller R, Herrmann RG, Pakrasi HB. Molecular studies of CtpA, the carboxyl-terminal processing protease for the D1 protein of the photosystem II reaction center in higher plants. J Biol Chem. 1996;271(36):21848–21852. doi: 10.1074/jbc.271.36.21848. [DOI] [PubMed] [Google Scholar]
- 29.Sessions A, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14(12):2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 31.Fu A, et al. A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104(40):15947–15952. doi: 10.1073/pnas.0707851104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lima A, et al. A redox-active FKBP-type immunophilin functions in accumulation of the photosystem II supercomplex in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2006;103(33):12631–12636. doi: 10.1073/pnas.0605452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto Y, Inagaki N, Satoh K. Overexpression and characterization of carboxyl-terminal processing protease for precursor D1 protein: Regulation of enzyme-substrate interaction by molecular environments. J Biol Chem. 2001;276(10):7518–7525. doi: 10.1074/jbc.M008877200. [DOI] [PubMed] [Google Scholar]
- 34.McDonald KL, Webb RI. Freeze substitution in 3 hours or less. J Microsc. 2011;243(3):227–233. doi: 10.1111/j.1365-2818.2011.03526.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.