Abstract
The relative contribution of immunological dysregulation and impaired epithelial barrier function to allergic diseases is still a matter of debate. Here we describe a new syndrome featuring severe dermatitis, multiple allergies and metabolic wasting (SAM syndrome) caused by homozygous mutations in DSG1. DSG1 encodes desmoglein 1, a major constituent of desmosomes, which connect the cell surface to the keratin cytoskeleton and play a crucial role in maintaining epidermal integrity and barrier function. SAM syndrome-causing mutations resulted in lack of membrane expression of DSG1, leading to loss of cell-cell adhesion. In addition, DSG1 deficiency was associated with increased expression of a number of genes encoding allergy-related cytokines. The deciphering of the pathogenesis of SAM syndrome substantiates the notion that allergy may result from a primary structural epidermal defect.
The epidermis is a stratified squamous epithelium that undergoes a tightly regulated terminal differentiation program culminating in the formation of a functional barrier against environmental agents1. Epidermal barrier disruption is thought to play a critical role in the pathogenesis of various allergic disorders2. Epidermal cell differentiation and barrier formation are critically dependent upon the proper temporal and spatial organization of several intercellular structures3. Among these elements, desmosomes are transmembranal structures that connect the cell surface to the intermediate filament cytoskeleton4. They consist of heterodimers of desmosomal cadherins, desmogleins (DSG1-4) and desmocollins (DSC1-3), which interact within the intercellular space. The intracytoplasmic part of the desmosomal plaque contains a number of associated proteins such as plakoglobin and plakophilins that associate with desmoplakin and thereby link to the keratin cytoskeleton. DSG1 plays a central role in the pathogenesis of three dermatological conditions5: pemphigus foliaceus, an autoimmune blistering disorder caused by autoantibodies directed against DSG1; bullous impetigo and staphylococcal scalded skin syndrome associated with bacterial production of an exfoliative toxin which specifically targets DSG1; and striate palmoplantar keratoderma (PPKS; MIM148700), a rare autosomal dominant disorder featuring hyperkeratotic plaques along the fingers, palms and soles, and caused by heterozygous mutations in the DSG1 gene.
In the present study, we delineate the molecular basis for a syndrome featuring severe allergic dermatitis and resulting from DSG1 dysfunction, suggesting a role for this molecule in maintaining the integrity of the epidermal barrier. More specifically, we studied three individuals who were referred for investigation because of severe skin dermatitis, multiple allergies and metabolic wasting (SAM) (Fig. 1 and Table 1). The first two affected females were born to healthy first degree cousins of Arab Muslim descent (Fig. 2a; family A, II-1 and II-2). Family history was unremarkable. Perinatal course was complicated by severe hypernatremia. The two subjects displayed congenital erythroderma (reminiscent of congenital ichthyosiform erythroderma6), yellowish papules and plaques arranged at the periphery of the palms, along the fingers and over weight-bearing areas of the feet, skin erosions and scaling and hypotrichosis (Fig. 1a,b). In addition, since infancy, they both exhibited severe food allergies, markedly elevated immunoglobulin E (IgE) levels and recurrent infections with severe metabolic wasting. Patient II-1 displayed eosinophilic esophagitis, while patient II-2 had severe esophageal reflux and ventricular septal defect. The third affected individual was a 9 month old female, born to healthy first degree cousins of Druze descent (Fig. 2a; family B, IV-10) with congenital erythroderma, severe dermatitis (Fig. 1c), hypotrichosis (Fig. 1d), recurrent skin and respiratory infections, growth retardation and multiple food allergies. Her sister (family B, individual IV-7) with similar skin and systemic manifestations, elevated IgE levels, microcephaly and a minor cardiac defect (mild pulmonic stenosis) had died at two years of age of sepsis. Two additional family members (IV-1 and IV-2) were reported to have succumbed at 2.5 years of age to a similar disorder.
Figure 1.
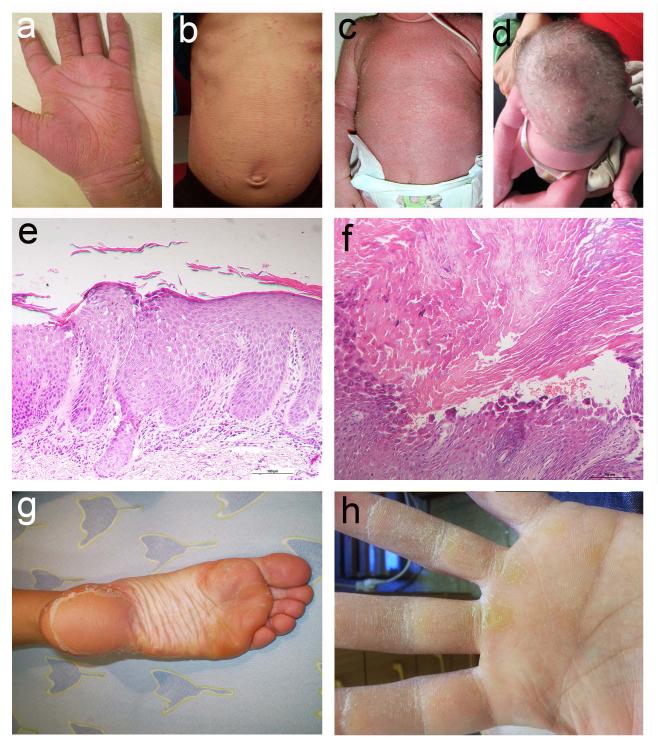
Clinical and pathological features. (a) Individual II-2 of family A displays diffusely red and fissured palms covered with hyperkeratotic yellowish papules and plaques, which are arranged linearly over the fingers. (b,c) Body skin is reddish and covered with scales and erosions in individual II-1 of family A (b) and in individual IV-10 of family B (c). (d) Hypotrichosis is evident in individual IV-10 of family B. (e,f) Skin biopsies obtained from the popliteal area in individual IV-10 of family B (e) and from the palms of individual II-1 of family A (f) demonstrate subcorneal separation associated with acantholysis within the spinous and granular layers (hematoxylin and eosin; bars = 100 μm). (g,h) Heterozygous carriers of DSG1 mutations c.49-1G>A (individual I-2, family A) (g) and c.1861delG (individual III-3, family B) (h) display diffuse plantar and focal palmar keratoderma, respectively.
Table 1. Clinical manifestations in SAM syndrome.
| Family A | Family B | ||
|---|---|---|---|
| II-1 | II-2 | IV-10 | |
| Sex | female | female | female |
| Age | 7y | 3y | 9m |
| Congenital ichthyosis | + | + | + |
| PPKS | + | + | − |
| Malabsorption | + | +a | + |
| Hypoalbuminemia | + | + | + |
| Eosinophilic esophagitis | + | − | − |
| Esophageal reflux | − | + | − |
| Failure to thrive | + | + | + |
| Microcephaly | + | + | − |
| Cardiac defects | − | +b | − |
| GH deficiency | + | − | − |
| Multiple food allergiesc | + | + | + |
| Recurrentinfectionsd | + | + | + |
| IgE (IU/ml)e | 10,900 | 266 | 1,438 |
| Developmental delay | +/− | + | + |
PPKS, palmoplantar keratoderma striata; GH, growth hormone; IgE, immunoglobulin E.
Fed through gastrostomy.
Muscular ventricular-septal defects.
Anaphylactoid reaction to chocolate, allergic rash to eggs, potatoes and peanuts.
Skin infections, severe pneumonia leading to pulmonary insufficiency and intubation.
Normal levels <100 IU/ml.
Figure 2.
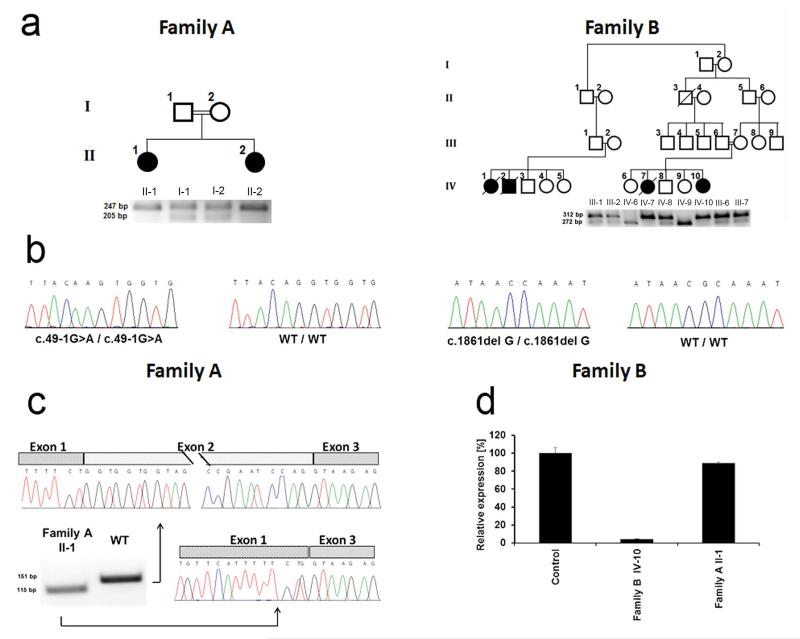
Molecular and immunohistochemical analysis. (a) Family pedigrees are presented in the upper panels. Black symbols denote affected individuals. PCR-RFLP assays (as described in the Online Methods) were used in each family to confirm co-segregation of the mutation with the disease phenotype (lower panels). Mutation c.49-1G>A is associated with the presence of a 247 bp fragment in family A. while mutation c.1861delG results in a 312bp fragment in family B. (b) Direct sequencing of DSG1 identified a homozygous G>A transition at position c.49-1 of the cDNA sequence in patients of family A and a homozygous single nucleotide G deletion at cDNA position c.1861 in family B. The wild type sequences (WT/WT) are given for comparison. (c) cDNA was reverse transcribed from an RNA sample extracted from patient II-1 (family A) skin and from a healthy individual. PCR amplification of a cDNA fragment spanning exons 1-3 demonstrates in the patient skin the presence of an aberrant PCR product shorter than the expected 151-bp fragment. Direct sequencing of both fragment indicated that c.49-1G>A results in skipping of exon 2. (d) Quantitative real-time PCR analysis for gene expression of DSG1 was performed using cDNA derived from skin biopsies of patients II-1 (family A) and IV-10 (family B) as well as a healthy individual. Results are provided as percentage of expression relative to gene expression in control + standard error normalized to ACTB RNA levels.
Histopathological examination of patient skin biopsies showed a psoriasiform dermatitis with alternating para- and ortho-keratosis, hypo- and hyper-granulosis and widespread acantholysis (loss of adhesion between keratinocytes) within the spinous and granular layers, leading to subcorneal and intragranular separation (Fig. 1e,f). Hair microscopy did not disclose any specific abnormality (not shown).
All affected and healthy family members or their legal guardian provided written and informed consent according to a protocol approved by our institutional review boards. The combined occurrence of congenital erythroderma, hypotrichosis, recurrent infections and multiple allergies initially suggested a diagnosis of Netherton syndrome (NTS; MIM 256500). However, no pathogenic mutation could be found in affected individuals of either family in the coding sequence of SPINK5 (encoding LEKTI), and expression of both LEKTI and filaggrin (FLG) protein was detected in all biopsies, although it was more diffuse than in control skin (Supplementary Fig. 1). To identify the causative mutations underlying SAM syndrome in the two families, we used DNA of individuals II-1 and II-2 from family A and of individuals IV-7, IV-8, III-2 and IV-10 from family B for whole-exome capture and NextGen sequencing (see Online Methods and Supplementary Tables 1 and 2). We identified two different homozygous mutations in DSG1 in each kindred (Fig. 2b): c.49-1G>A in family A and c.1861delG in family B. Both mutations were found to co-segregate by PCR-RFLP with the disease phenotype in each family (Fig. 2a). Of note, all heterozygous carriers of the mutations in both families displayed well-demarcated palmoplantar hyperkeratotic papules and plaques most often not arranged in a striate fashion (Fig. 1g,h), in agreement with previous data showing that heterozygous mutations in DSG1 can cause either striate, focal or diffuse PPK7-9, associated with keratinocyte disadhesion (but not frank acantholysis; Supplementary Fig. 2)10,11.
Mutations c.49-1G>A and c.1861delG were excluded from a panel of 170 and 50 population-matched healthy control individuals, respectively, and were absent from the 1000 Genomes Project and the NHLBI Grand Opportunity Exome Sequencing Project databases, including more than 8,200 chromosomes.
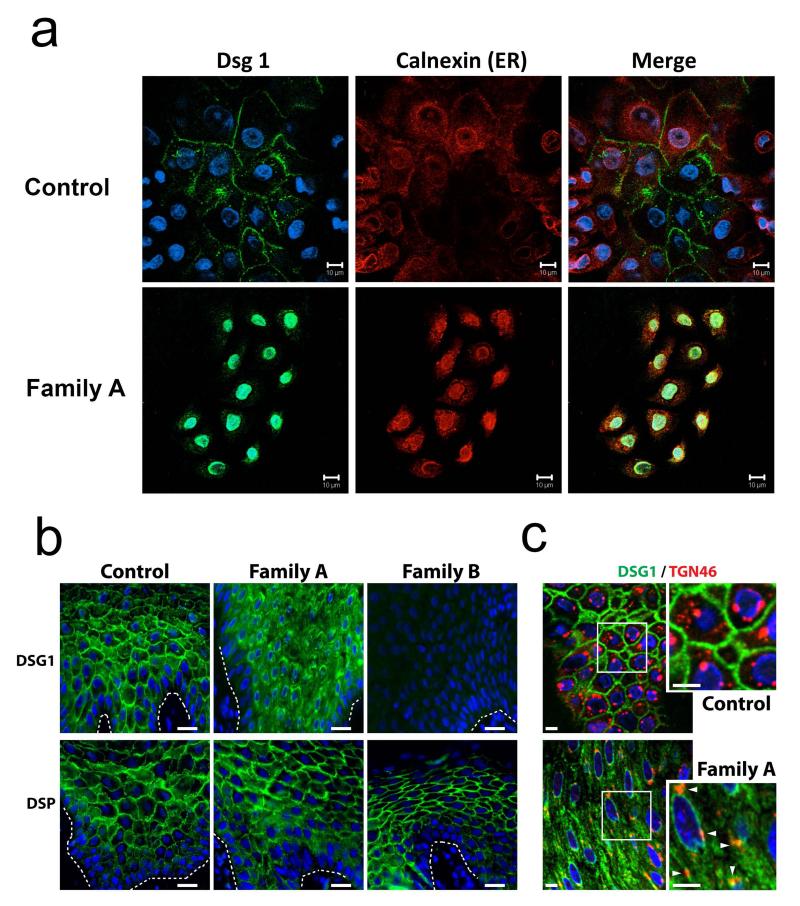
Both mutations are likely to result in loss of function. Indeed, c.49-1G>A is predicted (BDGP) to result in in-frame skipping of exon 2, while c.1861delG is expected to result in frame-shift and premature termination of protein translation (p.Ala612Glnfs.X3). To confirm these predictions, we extracted RNA from skin biopsies obtained from individual II-1 (Family A), individual IV-10 (Family B) and a healthy individual. cDNA direct sequencing confirmed the absence of exon 2 in patient II-1 carrying c.49-1G>A (Fig. 2c), which is predicted to lead to a 13 amino acid deletion and to disrupt the DSG1 signal peptide. This signal peptide sequence has been shown to be required for the correct post-translational maturation of DSG3 (ref. 12). To ascertain the effect of the c.49-1G>A mutation at the cellular level, we established keratinocyte cell cultures from patient II-1, family A. In contrast with control keratinocytes, which demonstrated a continuous membrane staining for DSG1, DSG1 was found to accumulate in the cell cytoplasm of patient cells, particularly around the nucleus (Fig. 3a). Immunostaining of patient skin biopsies also demonstrated cytoplasmic mislocalization of DSG1 as compared with control biopsies (Fig. 3b). Double staining with the endoplasmic reticulum marker calnexin in cultured cells (Fig. 3a) or Golgi/endosome marker TGN46 in tissue sections (Fig. 3c) showed partial co-localization of cytoplasmic DSG1 with these endomembrane compartments in the patient cells.
Figure 3.
DSG1 and DSP expression in patient skin. (a) Keratinocytes were harvested from skin biopsies obtained from a healthy individual (Normal KC) and from individual I-1, family A (Family A). Healthy individual-derived keratinocytes demonstrate normal membrane localization of DSG1 (upper panels). There is no co-localization of DSG1 with the endoplasmic reticulum (ER) chaperone protein calnexin; in contrast, patient keratinocytes (Family A) display no membrane staining of DSG1; instead nuclear and cytoplasmic punctuate DSG1 staining is observed (lower panels). DSG1 occasionally co-localizes with calnexin (scale bar = 10 μm); (b) Immunofluorescence analysis of skin sections from control and affected individuals shows cytoplasmic accumulation of DSG1 and loss of DSG1 staining in patients carrying mutations c.49-1G>A (Family A) and c.1861delG (Family B), respectively, compared with the plasma membrane localization in control samples (upper panels) (scale bars = 20 μm). DSP staining is retained in both c.49-1G>A and c.1861delG-carrying individuals (lower panels). (c) Double label immunofluorescence analysis of skin sections obtained from a healthy individual (Control) and from an individual carrying c.49-1G>A (Family A) showed partial co-localization of cytoplasmic DSG1 with TGN46, a Golgi and endosome marker, in the patient skin (scale bars = 5 μm).
In contrast with the mutation detected in family A, mutation c.1861delG was found to be associated with almost absent DSG1 mRNA expression in the skin (Fig. 2d), probably reflecting mRNA decay, as previously shown for DSG1 heterozygous mutations causing PPKS10. Consistent with this observation, DSG1 staining of a patient (IV-10, family B) skin biopsy was largely negative (Fig. 3b). Staining for desmoplakin (DSP) was retained in affected individuals of both families (Fig. 3b). Thus, both mutations result in loss of expression of DSG1 at the cell membrane. Of interest, mortality was only recorded in subjects carrying c.1861delG, which results in total absence of DSG1 expression (Fig. 3b).
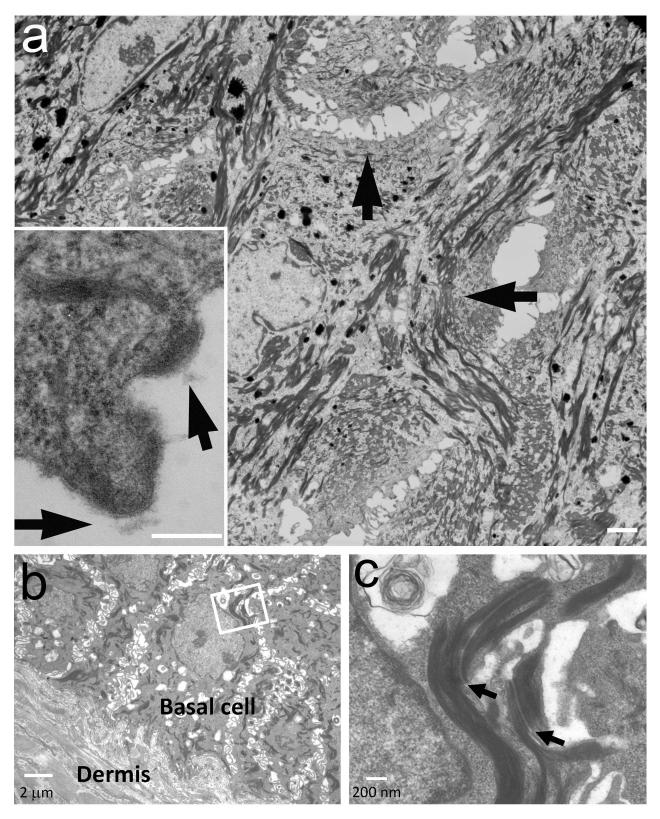
Given the essential role played by DSG1 in the formation of desmosomes in the upper epidermis and the presence of acantholysis in all patients (Fig. 1e,f), we examined desmosome ultrastructure in patient skin. Electron microscopic examination of skin samples obtained from patients II-1 (family A) and IV-10 (family B) demonstrated uneven distribution of desmosomes in the upper epidermis (Fig. 4a). In contrast, desmosome morphology and distribution appeared normal in the basal and lower spinous layers (Fig. 4b,c).
Figure 4.
Electron microscopy. (a) Electron microscopy demonstrates large areas lacking mature desmosomes in the upper spinous and granular layers with retracted keratin filaments (arrows) (bar = 4 μm). In these areas, half split desmosomes can be seen (insert, arrows, bar = 200 nm). (b) Acantholysis is absent in the basal cell layers (bar = 2μm); (c) Desmosomes (arrows) are normal in the basal and lower spinous cells (bar = 200 nm).
Taken together, the above results indicate that SAM syndrome is caused by loss-of-function mutations in DSG1, resulting in impaired desmosome formation in the upper epidermal layers, abnormal differentiation and acantholysis. Although allergic diseases have been traditionally considered as primarily resulting from immunologic dysregulation13, recent data have suggested that impaired epidermal barrier function may also contribute to the development of this group of disorders2. Mutations in FLG, encoding filaggrin, a major constituent of the epidermal barrier, have been shown to predispose to atopic diathesis14 and other Th2-mediated diseases15. In contrast with semi-dominant mutations in FLG, which are only partially penetrant16, recessive mutations in SPINK5, encoding the serine protease inhibitor LEKTI, and in CDSN, encoding corneodesmosin, are fully penetrant and have been shown to cause NTS17 and peeling skin syndrome type B (PSS-B; MIM 270300)18, respectively. Interestingly, NTS, PSS-B and SAM syndrome share numerous clinical features (Supplementary Fig. 3). In addition, both NTS and PSS-B are characterized by superficial intra-epidermal detachment similar to that seen in individuals with SAM syndrome (although frank acantholysis is not observed in either disease) as well as by a heightened Th2 immune response17,18. DSG1 and corneodesmosin are major components of corneodesmosomes, which are responsible for ensuring cohesiveness within the stratum corneum19. Degradation of these two proteins is pivotal for epidermal desquamation, a process that is tightly regulated by epidermal proteases and protease inhibitors such as LEKTI20. These data may explain why these three disorders share common phenotypic features as decreased LEKTI activity or decreased corneodesmosin/DSG1 expression are similarly expected to result in accelerated desquamation21,22. Loss of DSG1 is also likely to interfere with epidermal differentiation by sustaining Erk1/2 signaling suprabasally23.
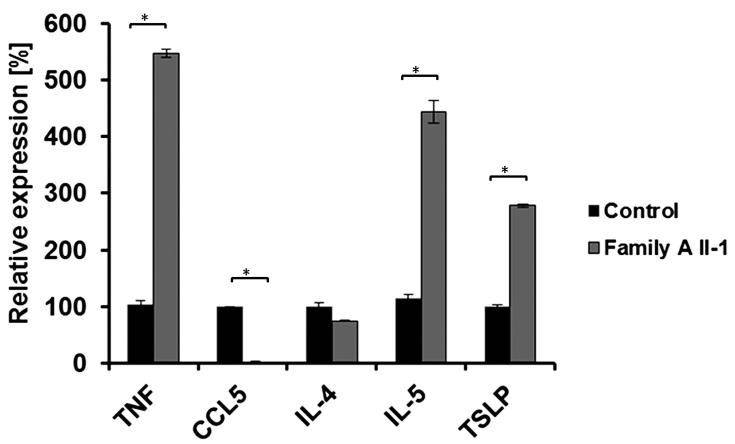
Thus, this report and these previous data demarcate a new group of disorders characterized by aberrant cell-cell adhesion in the upper epidermal layers, resulting in compromised barrier function2, which in turn may expose the immune system to abnormal stimulation, leading to multiple allergies, as barrier dysfunction enhances systemic Th2 responses24. Alternatively, we cannot exclude a role for DSG1 deficiency in stimulating Th2 cytokine production, as previously shown for other elements of the epidermal barrier25. In this regard, it is of interest that the expression of TSLP, IL5 and TNF, encoding mediators involved in skin allergy26,27, was elevated in patient keratinocytes (Fig. 5).
Figure 5.
Cytokine gene expression. Cytokine gene expression in keratinocytes isolated from patient II-1 (Family A) or from a healthy control was measured using qRT-PCR. Results are expressed as percentage of expression relative to gene expression in control cells + standard error (two sided t-test; *P < 0.01). Results are normalized to ACTB RNA levels.
With regard to other manifestations of SAM syndrome, minor cardiac developmental defects were noted in two patients, which is reminiscent of an association between desmosomal diseases and cardiac conditions28. The mechanisms underlying metabolic wasting in SAM syndrome remain to be fully delineated. Although defective epidermal barrier is usually blamed for impaired growth and caloric losses in ichthyoses29,30, malabsorption and enteropathy have been reported in Netherton syndrome31, which is pathogenetically related to SAM syndrome (Supplementary Fig. 3). Given the fact that we observed esophageal involvement in two subjects (Table 1), which may be related to the strong DSG1 expression in the esophagus (Supplementary Fig. 4), the fact that TSLP was up-regulated in patient cells and the fact that TSLP has been shown to induce allergic manifestations in the gastrointestinal tract32, a direct role for DSG1 deficiency in mediating metabolic wasting in SAM syndrome cannot be excluded.
Taken together, the present data substantiate the notion that allergy may result from a primary epidermal defect.
ONLINE METHODS
Subjects
All affected and healthy family members or their legal guardian provided written and informed consent according to a protocol approved by our institutional review board and by the Israel National Committee for Human Genetic Studies in adherence to the Helsinki guidelines (Supplementary Note).
Exome sequencing
EDTA-treated blood samples were obtained from affected individuals and their healthy family members. Genomic DNA was extracted from peripheral blood leukocytes using the 5 Prime ArchivePure DNA Blood Kit (5 Prime Inc.). Exome sequencing of family A patients was performed at King’s College London. Whole-exome capture was done by in-solution hybridization with the SureSelect All Exon 50 Mb Version 4.0 (Agilent) followed by massively parallel sequencing (Illumina HiSeq2000) with 100 bp paired-end reads. Duplicate reads, resulting from PCR clonality or optical duplicates, and reads mapping to multiple locations were excluded from downstream analysis. Single-nucleotide substitutions and small insertion deletions were identified and quality filtered with the SamTools software package and in-house software. Variants were annotated with respect to genes and transcripts with the Annovar tool. Identified variants were cross-referenced with publicly available variant data (dbSNP135 and 1000 Genomes Project). Supplementary Table 1 summarizes the exome sequencing details of family A.
Exome sequencing in family B was performed by BGI Tech Solutions Ltd. Whole-exome capture was done by SeqCap EZ Human Exome Library v3.0 (NimbleGen) followed by massively parallel sequencing (Illumina HiSeq2000) with 90 bp paired-end reads. Reads were aligned to the Genome Reference Consortium Human Build 37 (GRCh37/hg19) using Burrows-Wheeler Aligner (BWA)33. Duplicate reads, resulting from PCR clonality or optical duplicates, and reads mapping to multiple locations were excluded from downstream analysis. Reads mapping to a region of known or detected insertions or deletions were re-aligned to minimize alignment errors. Single-nucleotide substitutions and small insertion deletions were identified and quality filtered using the Genome Analysis Tool Kit (GATK)34. Rare variants were identified by filtering using the data from dbSNP135, the 1000 Genomes Project, the Exome Variant Server (see URLs) and an in-house database of sequenced individuals. Variants were classified by predicted protein effects using Polyphen2 (ref. 35) and SIFT36. Supplementary Table 2 summarizes exome sequencing details of family B.
Mutation analysis
Genomic DNA was PCR-amplified using oligonucleotide primer pairs spanning the entire coding sequence as well as intron–exon boundaries of DSG1 (Supplementary Table 3) and ReddyMix PCR Master Mix (Thermo Scientific). Cycling conditions were as follows: 95 °C, 4 min; 95 °C, 30 sec; 60 °C, 45 sec; 72 °C, 90 sec, for 34 cycles (for exons 1, 6, 12, 15_1, 15_2, 15_3) and 95 °C, 4 min; 95 °C, 30 sec; 52 °C, 45 sec; 72 °C, 90 sec, for 34 cycles (for exons 2, 3, 4, 5, 7, 8, 9, 10, 11, 13, 14, 15_4). Gel-purified (QIAquick gel extraction kit, QIAGEN) amplicons were subjected to bidirectional DNA sequencing with the BigDye terminator system on an ABI Prism 3100 sequencer (Applied Biosystems).
PCR-RFLP
To screen for the c.49-1G>A mutation, we PCR-amplified a 247 bp fragment (for primer sequences, see Supplementary Table 3). The mutation abolishes a recognition site for endonuclease EcoNI (New England Biolabs). To screen for the c.1861delG mutation, we PCR-amplified a 312 bp fragment (for primer sequences, see Supplementary Table 3). The mutation abolishes a recognition site for endonuclease BstUI (New England Biolabs). In both cases, after incubation at 37°C for 8 h, digested PCR products were electrophoresed in ethidium bromide-stained 3% agarose gels.
Cell cultures and reagents
Keratinocyte cell cultures were established from punch biopsies obtained from patient II-1 from family A and healthy controls after written informed consent was obtained, and were maintained in Keratinocyte Growth Medium (KGM) supplemented with 0.4% bovine pituitary extract, 0.1% human epidermal growth factor (hEGF), 0.1% insulin, 0.1% hydrocortisone and 0.1% gentamicin/amphotericin B (all were purchased from Lonza).
Quantitative RT-PCR
For quantitative real-time PCR, cDNA was synthesized from 500 ng of total RNA using qScript kit (Quanta Biosciences). cDNA PCR amplification was carried out with the PerfeCTa SYBR Green FastMix (Quanta Biosciences) on a StepOnePlus system (Applied Biosystems) with gene-specific intron-crossing oligonucleotide pairs (Supplementary Table 3). Cycling conditions were as follows: 95 °C, 30 sec; 95 °C, 4 sec; 60 °C, 30 sec for 40 cycles. Each sample was analyzed in triplicates. For quantification, standard curves were obtained with serially diluted cDNA amplified in the same real-time PCR run. Results were normalized to ACTB mRNA levels.
Immunohistochemistry
For LEKTI and filaggrin staining, formaldehyde-fixed 5-μm paraffin-embedded sections were mounted on SuperFrost®Plus glass (Menzel-Glazer) and processed by using an automated immunostainer (Benchmark-XT, Ventana Medical System). Visualization of the bound primary antibodies was performed using the I-View DAB detection kit (Ventana Medical System). The sections were then counterstained with Gill’s hematoxylin, dehydrated and mounted for microscopic examination. The primary antibodies used were a monoclonal mouse anti-human LEKTI (diluted 1:25) (Santa Cruz Biotechnology) and monoclonal mouse anti-human Filaggrin (diluted 1:40) (Novocastra).
Immunostaining of keratinocytes and skin biopsies
Keratinocytes that were harvested from punch biopsies were grown on glass coverslips and fixed with 4% paraformaldehyde. For immunofluorescence analysis of skin biopsies, 3-5 μm paraffin-embedded sections were baked overnight at 60 °C and de-paraffinized using xylene/ethanol. Following permeabilization with 0.5% Triton/PBS, antigen retrieval was accomplished using 0.01 M citrate buffer, pH 6.0. Sections were blocked in 1% BSA, 2% normal goat serum for 1 h at 37 °C. Primary antibodies were diluted in 1% BSA, 2% normal goat serum and incubated overnight at 4 °C. Primary antibodies used were 4B2 (desmoglein 1) (diluted 1:100)37, 11-5F (desmoplakin; gift from David Garrod, University of Manchester, Manchester, UK) (diluted 1:100), TGN46 (diluted 1:100) (Abnova). Secondary antibody staining was carried out for 30 min at 37 °C using Alexa Fluor anti-mouse or anti-rabbit secondary antibodies (Life Technologies/Invitrogen). Coverslips were mounted in polyvinyl alcohol for microscopical observation using either a DMR Leica microscope (40XNA 1.0 Plan-Fluotar Plan-Apochromat objective) and a charge-coupled device camera (Orca 100 model C47 42-95; Hamamatusu Photonics) for fluorescence image acquisition. For double label immunofluorescence co-localization with TGN46, optical sections were obtained using a 63XNA 1.4 Plan-Apochromat objective on an AxioVision Z1 (Carl Zeiss, Inc,) fitted with an Apotome slide module and a digital camera (AxioCam MRm; Carl Zeiss, Inc).
Electron microscopy
Skin biopsies were fixed in half-strength Karnowski’s fixative and 1% osmium tetroxide. The tissue blocks were dehydrated with ethanol, stained en bloc using 1% uranyl acetate in 50% ethanol, and embedded in Epon resin (TAAB, Aldermaton). Ultrathin sections were stained with 1.5% uranyl acetate in methanol and Reynold’s lead citrate and examined with a JEM-1010 (Jeol Ltd.) transmission electron microscope.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Getsios for the use of the Zeiss Apotome. We thank Kristina Stone (King’s College London) for technical assistance in whole-exome sequencing. This work was supported by a generous donation of the Ram family to E.S.; NIH RO1 AR041836 and the Joseph L. Mayberry Senior Endowment to K.J.G.; the JSPS Kakenhi Grant Number 24591620 and a grant from the Ministry of Health, Labor and Welfare of Japan to A.I.-Y.; DebRA UK to J.A.M.; the Wellcome Trust (Programme grant 092530/Z/10/Z to W.H.I.M. and A.D.I.; Bioresources grant 090066/B/09/Z to A.D.I. and W.H.I.M.); the National Children’s Research Centre to A.D.I.; and the Pachyonychia Congenita Project to F.J.D.S. Some control sections were obtained from the Pathology Core of the Northwestern University Skin Disease Research Center (NIH P30AR057216). The Centre for Dermatology and Genetic Medicine, University of Dundee is supported by a Wellcome Trust Strategic Award (098439/Z/12/Z to W.H.I.M.). L.S. is recipient of an Excellence Award from the Tel Aviv Sourasky Medical Center.
Footnotes
URLs. NHLBI Grand Opportunity Exome Sequencing Project, http://evs.gs. washington.edu/EVS/; 1000 Genomes Project, http://pilotbrowser.1000 genomes.org/index.html; BDGP, http://www.fruitfly.org/seq_tools/splice.html; NCBI, http://www.ncbi.nlm.nih.gov/guide/; DSG1 RefSeq accession number NM_001942, http://www.ncbi.nlm.nih.gov/nuccore/NM_001942
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
EdSumm (same for AOP and issue): Eli Sprecher, Kathleen Green and colleagues show that biallelic mutations in DSG1 cause a syndrome featuring severe dermatitis, multiple allergies and metabolic wasting. The mutations abolish expression of desmoglein 1, resulting in loss of cell adhesion accompanied by increased expression of several allergy-related cytokines.
REFERENCES
- 1.Menon GK, Cleary GW, Lane ME. The structure and function of the stratum corneum. Int. J. Pharm. 2012;435:3–9. doi: 10.1016/j.ijpharm.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Kubo A, Nagao K, Amagai M. Epidermal barrier dysfunction and cutaneous sensitization in atopic diseases. J. Clin. Invest. 2012;122:440–447. doi: 10.1172/JCI57416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simpson CL, Patel DM, Green KJ. Deconstructing the skin: cytoarchitectural determinants of epidermal morphogenesis. Nat. Rev. Mol. Cell. Biol. 2011;12:565–580. doi: 10.1038/nrm3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desai BV, Harmon RM, Green KJ. Desmosomes at a glance. J. Cell Sci. 2009;122:4401–4407. doi: 10.1242/jcs.037457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amagai M, Stanley JR. Desmoglein as a target in skin disease and beyond. J. Invest. Dermatol. 2012;132:776–784. doi: 10.1038/jid.2011.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oji V, et al. Revised nomenclature and classification of inherited ichthyoses: results of the First Ichthyosis Consensus Conference in Soreze 2009. J. Am. Acad. Dermatol. 2010;63:607–641. doi: 10.1016/j.jaad.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 7.Milingou M, Wood P, Masouye I, McLean WH, Borradori L. Focal palmoplantar keratoderma caused by an autosomal dominant inherited mutation in the desmoglein 1 gene. Dermatology. 2006;212:117–122. doi: 10.1159/000090651. [DOI] [PubMed] [Google Scholar]
- 8.Rickman L, et al. N-terminal deletion in a desmosomal cadherin causes the autosomal dominant skin disease striate palmoplantar keratoderma. Hum. Mol. Genet. 1999;8:971–976. doi: 10.1093/hmg/8.6.971. [DOI] [PubMed] [Google Scholar]
- 9.Keren H, Bergman R, Mizrachi M, Kashi Y, Sprecher E. Diffuse nonepidermolytic palmoplantar keratoderma caused by a recurrent nonsense mutation in DSG1. Arch. Dermatol. 2005;141:625–628. doi: 10.1001/archderm.141.5.625. [DOI] [PubMed] [Google Scholar]
- 10.Hershkovitz D, Lugassy J, Indelman M, Bergman R, Sprecher E. Novel mutations in DSG1 causing striate palmoplantar keratoderma. Clin. Exp. Dermatol. 2009;34:224–228. doi: 10.1111/j.1365-2230.2008.02733.x. [DOI] [PubMed] [Google Scholar]
- 11.Bergman R, et al. Disadhesion of epidermal keratinocytes: a histologic clue to palmoplantar keratodermas caused by DSG1 mutations. J. Am. Acad. Dermatol. 2010;62:107–113. doi: 10.1016/j.jaad.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 12.Amagai M, Ishii K, Takayanagi A, Nishikawa T, Shimizu N. Transport to endoplasmic reticulum by signal peptide, but not proteolytic processing, is required for formation of conformational epitopes of pemphigus vulgaris antigen (Dsg3) J. Invest. Dermatol. 1996;107:539–542. doi: 10.1111/1523-1747.ep12582796. [DOI] [PubMed] [Google Scholar]
- 13.Hanifin JM, Butler JM, Chan SC. Immunopharmacology of the atopic diseases. J. Invest. Dermatol. 1985;85:161s–164s. doi: 10.1111/1523-1747.ep12276359. [DOI] [PubMed] [Google Scholar]
- 14.Sandilands A, et al. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat. Genet. 2007;39:650–654. doi: 10.1038/ng2020. [DOI] [PubMed] [Google Scholar]
- 15.Schuttelaar ML, et al. Filaggrin mutations in the onset of eczema, sensitization, asthma, hay fever and the interaction with cat exposure. Allergy. 2009;64:1758–1765. doi: 10.1111/j.1398-9995.2009.02080.x. [DOI] [PubMed] [Google Scholar]
- 16.Brown SJ, et al. Filaggrin haploinsufficiency is highly penetrant and is associated with increased severity of eczema: further delineation of the skin phenotype in a prospective epidemiological study of 792 school children. Br. J. Dermatol. 2009;161:884–889. doi: 10.1111/j.1365-2133.2009.09339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chavanas S, et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat. Genet. 2000;25:141–142. doi: 10.1038/75977. [DOI] [PubMed] [Google Scholar]
- 18.Oji V, et al. Loss of corneodesmosin leads to severe skin barrier defect, pruritus, and atopy: unraveling the peeling skin disease. Am. J. Hum. Genet. 2010;87:274–281. doi: 10.1016/j.ajhg.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishida-Yamamoto A, Igawa S, Kishibe M. Order and disorder in corneocyte adhesion. J. Dermatol. 2011;38:645–654. doi: 10.1111/j.1346-8138.2011.01227.x. [DOI] [PubMed] [Google Scholar]
- 20.Hovnanian A. Netherton syndrome: skin inflammation and allergy by loss of protease inhibition. Cell Tissue Res. 2013;351:289–300. doi: 10.1007/s00441-013-1558-1. [DOI] [PubMed] [Google Scholar]
- 21.Descargues P, et al. Corneodesmosomal cadherins are preferential targets of stratum corneum trypsin- and chymotrypsin-like hyperactivity in Netherton syndrome. J. Invest. Dermatol. 2006;126:1622–1632. doi: 10.1038/sj.jid.5700284. [DOI] [PubMed] [Google Scholar]
- 22.Descargues P, et al. Spink5-deficient mice mimic Netherton syndrome through degradation of desmoglein 1 by epidermal protease hyperactivity. Nat. Genet. 2005;37:56–65. doi: 10.1038/ng1493. [DOI] [PubMed] [Google Scholar]
- 23.Getsios S, et al. Desmoglein 1-dependent suppression of EGFR signaling promotes epidermal differentiation and morphogenesis. J. Cell Biol. 2009;185:1243–1258. doi: 10.1083/jcb.200809044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strid J, Sobolev O, Zafirova B, Polic B, Hayday A. The intraepithelial T cell response to NKG2D-ligands links lymphoid stress surveillance to atopy. Science. 2011;334:1293–1297. doi: 10.1126/science.1211250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonnart C, et al. Elastase 2 is expressed in human and mouse epidermis and impairs skin barrier function in Netherton syndrome through filaggrin and lipid misprocessing. J. Clin. Invest. 2010;120:871–882. doi: 10.1172/JCI41440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Incorvaia C, et al. Allergy and the skin. Clin. Exp. Immunol. 2008;153(Suppl 1):27–29. doi: 10.1111/j.1365-2249.2008.03718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leyva-Castillo JM, Hener P, Jiang H, Li M. TSLP produced by keratinocytes promotes allergen sensitization through skin and thereby triggers atopic march in mice. J. Invest. Dermatol. 2013;133:154–163. doi: 10.1038/jid.2012.239. [DOI] [PubMed] [Google Scholar]
- 28.Petrof G, Mellerio JE, McGrath JA. Desmosomal genodermatoses. Br. J. Dermatol. 2012;166:36–45. doi: 10.1111/j.1365-2133.2011.10640.x. [DOI] [PubMed] [Google Scholar]
- 29.Moskowitz DG, et al. Pathophysiologic basis for growth failure in children with ichthyosis: an evaluation of cutaneous ultrastructure, epidermal permeability barrier function, and energy expenditure. J. Pediatr. 2004;145:82–92. doi: 10.1016/j.jpeds.2004.03.052. [DOI] [PubMed] [Google Scholar]
- 30.Fowler AJ, et al. Nutritional status and gastrointestinal structure and function in children with ichthyosis and growth failure. J. Pediatr. Gastroenterol. Nutr. 2004;38:164–169. doi: 10.1097/00005176-200402000-00012. [DOI] [PubMed] [Google Scholar]
- 31.Sprecher E, et al. The spectrum of pathogenic mutations in SPINK5 in 19 families with Netherton syndrome: implications for mutation detection and first case of prenatal diagnosis. J. Invest. Dermatol. 2001;117:179–187. doi: 10.1046/j.1523-1747.2001.01389.x. [DOI] [PubMed] [Google Scholar]
- 32.Omori-Miyake M, Ziegler SF. Mouse models of allergic diseases: TSLP and its functional roles. Allergol. Int. 2012;61:27–34. doi: 10.2332/allergolint.11-RAI-0374. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 37.Dusek RL, et al. The differentiation-dependent desmosomal cadherin desmoglein 1 is a novel caspase-3 target that regulates apoptosis in keratinocytes. J. Biol. Chem. 2006;281:3614–3624. doi: 10.1074/jbc.M508258200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.