Abstract
Human leukocyte antigen (HLA) class I and class II alleles are implicated as genetic risk factors for many autoimmune diseases. However, the role of the HLA loci in human systemic lupus erythematosus (SLE) remains unclear. Using a dense map of polymorphic microsatellites across the HLA region in a large collection of families with SLE, we identified three distinct haplotypes that encompassed the class II region and exhibited transmission distortion. DRB1 and DQB1 typing of founders showed that the three haplotypes contained DRB1*1501/ DQB1*0602, DRB1*0801/ DQB1*0402, and DRB1*0301/DQB1*0201 alleles, respectively. By visualizing ancestral recombinants, we narrowed the disease-associated haplotypes containing DRB1*1501 and DRB1*0801 to an ∼500-kb region. We conclude that HLA class II haplotypes containing DRB1 and DQB1 alleles are strong risk factors for human SLE.
Introduction
The human leukocyte antigen (HLA) region on human chromosome 6p21.3 spans ∼3.6 Mb of DNA and contains >200 genes, many of which have important roles in the immune system (Aguado et al. 1999; Dawkins et al. 1999; Rhodes and Trowsdale 1999). The HLA region can be divided roughly into thirds. The telomeric class I region contains the polymorphic HLA A, B, and C genes, which are expressed ubiquitously and function to present antigenic peptides to CD8+ cytotoxic T cells. The centromeric class II region encodes the polymorphic DR, DQ, and DP genes, which are expressed on “professional” antigen-presenting cells and serve to display peptides to CD4+ helper T cells. The class II region also contains genes important for antigen processing, including the TAP, LMP, and DM genes. The class III region lies between the class I and II regions and contains genes important for the innate immune system, including the cytokine tumor necrosis factor α and the complement components C2 and C4. There are also many genes within the HLA region with no obvious function in the immune system.
In addition to their role in determining transplantation tolerance (Snell 1948), HLA class I and II alleles show genetic associations with certain inflammatory and autoimmune disorders (reviewed by Vyse and Todd [1996]). Examples include the association of class I HLA B27 with ankylosing spondylitis and of HLA Cw6 with psoriasis, as well as the association of class II DRB1*0401 (DR4) with rheumatoid arthritis, DRB1*1501 (DR2) with multiple sclerosis, and both DRB1*0301 (DR3) and DRB1*0401 (DR4) with type I diabetes. Because of extensive linkage disequilibrium across the HLA region, it has often been difficult to determine whether the observed associations reflect the class I or class II alleles themselves, linked genes, or combinations of genes carried on a particular haplotype. This difficulty is exemplified by recent data indicating that the long-standing association of psoriasis with HLA-Cw6 is due to the effect of a gene closely linked with, but separate from, the HLA-C loci (Nair et al. 2000).
Systemic lupus erythematosus (SLE [MIM 152700]) is a chronic and severe inflammatory autoimmune disease that affects mostly women (∼10:1 female-to-male ratio). A prominent feature of SLE is the production of pathogenic autoantibodies with specificity for nuclear and other tissue antigens. These autoantibodies lead to tissue damage by depositing in blood vessels as immune complexes and by initiating inflammatory responses after binding to self-antigens. SLE is clinically very heterogeneous and has the potential to affect many different organ systems. There is compelling epidemiological evidence to suggest an important genetic contribution to human SLE (Hochberg 1997), and recent genetic linkage studies in human families with lupus have identified candidate susceptibility loci at 1q23, 1q41-2, 2q37, 4p15, 6p11, and 16q12 (reviewed by Wakeland et al. [2001] and Gaffney et al. [2002]).
Previous case/control association studies have examined the potential role of HLA class I and II alleles in genetic susceptibility to SLE (Harley et al. 1998; Tan and Arnett 1998). The most consistent findings have been modest associations of DRB1*1501 (DR2) and DRB1*0301 (DR3) alleles in white individuals with SLE. However, these associations have, on balance, been much less convincing than those observed in rheumatoid arthritis (DR4) or type I diabetes (DR3). Furthermore, SLE class II associations in nonwhite ethnic groups have shown little consensus.
Here, we present the results of dense HLA microsatellite mapping in a collection of 334 families with SLE. Three risk haplotypes, bearing the class II alleles DRB1*1501/DQB1*0602, DRB1*0301/DQB1*0201, and DRB1*0801/DQB1*0402, were found to exhibit significant transmission distortion in families with SLE and were enriched in individuals with SLE, compared with control individuals. Through visualization of ancestral recombinants, the risk region on each haplotype was localized to defined intervals. The identification of SLE-associated HLA haplotypes and the localization of the effect to discrete risk intervals should facilitate the search for the genes and polymorphisms responsible for mediating susceptibility to SLE.
Material and Methods
Collection of Families with SLE
The demographic and clinical features of the 576 affected individuals (566 female and 10 male) from 211 affected sib-pair and 123 simplex families are described in detail elsewhere (Gaffney et al. 1998, 2000; Graham et al. 2001). All patients fulfilled American College of Rheumatology criteria for SLE. There were 283 white, 22 African American, and 15 Hispanic families, together with 14 families of other/mixed ethnicity. This study was approved by the Human Subjects Review Board at the University of Minnesota.
Samples and Genotyping
Genotyping was performed as described by Gaffney et al. (2000) and Graham et al. (2001). A total of 106 microsatellite markers spanning chromosome 6 were typed in 187 sib-pair families, and 52 HLA markers were typed in the combined 334-family collection (1,121 total samples). Of the markers used to map the HLA, 31 have been described elsewhere (Nair et al. 2000). Fifteen new markers were developed from primary sequence during the course of this project (see supplementary material at the Behrens Lab Web site, for detailed marker information, as well as Jawaheer et al. 2002 [in this issue]). Marker order and intermarker distances were determined from the Human Genome Project Working Draft (UCSC Genome Bioinformatics Web site) and the Current Consensus MHC sequence (Sanger Institute: Human Chromosome 6 Home Web site). HLA-DRB1 and HLA-DQB1 alleles were typed in 208 founders carrying one or more of the risk haplotypes; typing was performed as described by Erlich et al. (1991). After confirming that the D6S2666/2665/2446 haplotype was essentially 100% accurate in predicting the underlying DRB1 and DQB1 risk alleles, we made DR and DQ assignments on the basis of the three-marker microsatellite haplotype.
Data Analysis
All genotyping data were analyzed by PEDCHECK to identify any genotyping/Mendelian errors (O'Connell and Weeks 1998). Linkage analysis was performed using the “maximum likelihood estimate of IBD sharing” function of GENEHUNTER PLUS (Kong and Cox 1997). Association analysis was performed using either the pedigree disequilibrium test (PDT) (PDT version 2.0 [Martin et al. 2000], modified to analyze dyads when triads are unavailable) or the transmission/disequilibrium test (TDT) (TDT/S-TDT version 1.1 [Spielman et al. 1993; Spielman and Ewens 1998]). When the TDT was used, a single affected individual (the index case individual) per family was analyzed in families with multiple affected individuals and/or multiple generations. For association studies, haplotype frequencies were determined from the index case individual of each white family with SLE and from a collection of 174 unrelated female white control individuals. Control individuals were recruited in the New York metropolitan area as part of an ongoing longitudinal cohort study, the New York Cancer Project, administered by the Academic Medicine Development Corporation (AMDeC). The ethnic background of each control individual was established by obtaining information on the origin of all four grandparents. The genotypic relative risk (GRR) was calculated as described by Risch and Merikangas (1996).
Haplotypes across the HLA were generated by GENEHUNTER PLUS for the 275 families that had at least one typed parent. Short exact-match haplotypes were assigned a unique identifying number, and pedigree files were generated for PDT or TDT analyses. Inferred haplotypes (constructed for “missing” parents as determined by transmission data) were not used for association analyses but were used to assist in the determination of ancestral haplotypes and ancestral recombinant breakpoints. Extended haplotype analysis was conducted by first identifying the consensus extended haplotype for each of the three class II risk haplotypes identified by the PDT. This was determined on the basis of allele frequencies of neighboring markers. The consensus microsatellite allele for each founder haplotype was then color-coded maroon. Gold denotes nonconsensus alleles, and gray represents missing data or a genotype with ambiguous phase. The risk founder haplotypes were then sorted on the basis of the number of contiguous consensus alleles (allowing for one marker deviation) radiating from the marker used to select the founder haplotypes in either the telomeric or centromeric direction. Risk haplotypes were then subdivided on the basis of length and composition of consensus alleles and were analyzed using the TDT and PDT. For additional details, see the supplementary material at the Behrens Lab Web site.
Results
Our recent gene mapping in 187 sib-pair families with SLE identified a LOD score of 4.19 on the short arm of human chromosome 6 (Gaffney et al. 1998, 2000). After genotyping of an additional 31 microsatellite markers across 6p (with an average intermarker distance of 2 Mb), including 6 markers within the HLA, the strongest evidence for allele sharing by affected siblings was within the HLA (data not shown).
Single-Marker Family-Based Association Analysis
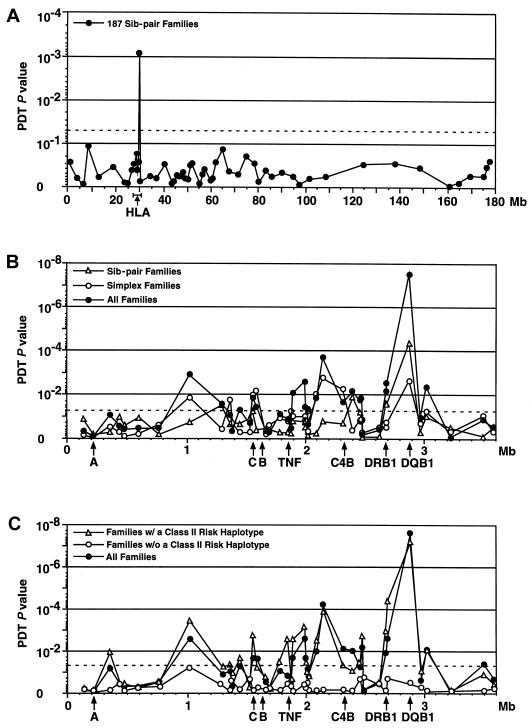
Using the PDT, a multiallelic test robust for complex pedigrees (Martin et al. 2000), we found that marker D6S2446 in the class II region showed the strongest evidence for association (P=9×10-4; fig. 1A). Interestingly, this marker is located in the 75-kb interval between the highly polymorphic DRB1 and DQB1 genes (see online-only figureonline-only figure). To extend these findings, we genotyped the original cohort of 187 families, including an additional 34 sib-pair families, with a dense panel of microsatellite markers across the HLA region (with an average intermarker distance 62 kb; see online-only figureonline-only figure). When the PDT was used, the strongest evidence for association was again found in the class II region at D6S2446 (P=4×10-5; fig. 1B).
Figure 1.
Association between the HLA class II region and SLE. A, PDT global association results for 55 markers across the length of chromosome 6 in 187 sib-pair families with SLE. B, PDT results for 41 HLA markers in 211 multiplex families (unfilled triangles), 123 simplex families (unfilled circles), and all 334 families (filled circles). The heterozygosity index for each marker was >0.70. C, PDT association results in 275 families (filled circles), where at least one parent was available and where phase could be determined unambiguously. Of these, 191 families carried at least one of the three risk haplotypes (unfilled circles), whereas 84 families lacked a risk haplotype (unfilled triangles). The position of selected HLA genes are indicated by the arrows. Dashed lines indicate P=.05.
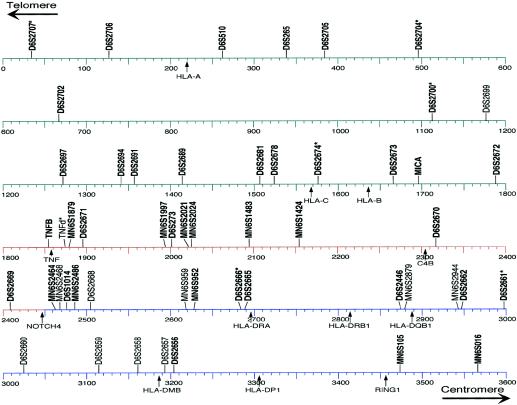
Online-Only Figure .
Physical map of the HLA complex, showing the 52 genotyped microsatellite markers. This map is based on the current consensus 3,673,778-bp sequence available from the Sanger Centre. The positions of the markers indicate the centromeric end of the amplimer, and the positions of genes indicate the translation start codon. The 16 markers with designations beginning with “MN6S” were developed during the course of the present study and have been submitted to the Genome Database. The 11 microsatellite markers with heterozygosities <0.70 are given in nonboldface type. The asterisks (*) indicate that the given primers were modified to amplify the indicated repetitive element.
The same marker panel was then applied to an independent sample of 123 simplex families (single patient with SLE and both parents). Again, the best evidence for association in the simplex collection was at D6S2446 (P=2×10-3; fig. 1B). The results of the PDT at D6S2446 for the combined collection of sib-pair and simplex pedigrees (a total of 334 families) was highly significant (P=3×10-8; fig. 1B). Other regions showed some evidence for association (e.g., ∼1.0 and 2.1 Mb); however, these signals are likely the result of linkage disequilibrium, since the evidence for association at these markers was not observed independent of the class II risk haplotypes (figs. 2 and 3, and data not shown).
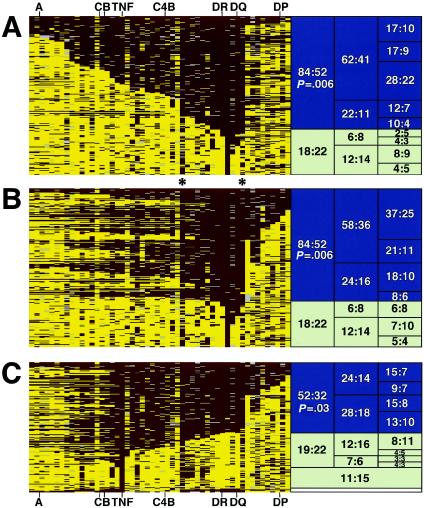
Figure 2.
Visualization of founder risk haplotypes carrying DRB1*1501 (DR2). Shown are all founder haplotypes carrying allele 6 at marker D6S2446 (D6S2446-6). The consensus ancestral haplotype containing D6S2446-6 was determined, and individual alleles were color-coded: marker alleles identical to consensus are shown in maroon, alleles different from consensus are shown in gold, and missing data or alleles with ambiguous phase are shown in gray. After sorting by length, groups of founder haplotypes were tested by TDT, with the numbers shown referring to the ratio of T:NT haplotypes. Blue boxes indicate significant transmission distortion (P<.05) for the larger groups; white boxes indicate nonsignificant TDTs. A, Founder haplotypes (N=176) were sorted by telomeric (left) length, and TDTs were performed. B, Founder haplotypes (N=176) were sorted, first by centromeric (right), and then by telomeric (left) length, to define the centromeric “breakpoint” by TDT. C, Founder haplotypes (N=155) carrying the TNF-α allele found on the DRB1*1501 extended haplotype were sorted on the basis of centromeric (right) length. Only those haplotypes extending to DRB1/DQB1 were significant by TDT. At the bottom of panel C are four “double recombinant” haplotypes, showing consensus markers for the TNF and class II regions but showing nonconsensus in-between; T:NT ratio is 3:1. Asterisks (*) represent the approximate boundaries of the haplotype length showing convincing evidence for transmission distortion. Haplotype groupings that failed to show significant transmission disequilibrium (white boxes) did not contain DRB1*1501, despite carrying D6S2446-6.
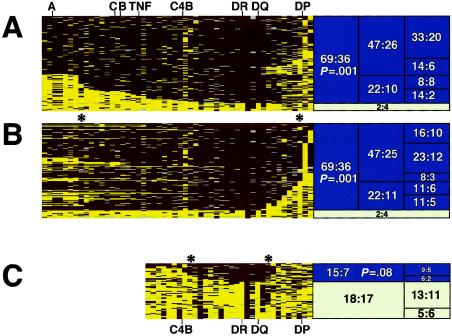
Figure 3.
Haplotypes containing DRB1*0301 (DR3) and DRB1*0801 (DR8) show significant transmission disequilibrium in families with SLE. Founder haplotypes were sorted and analyzed as described in figure 2. A and B, Founder haplotypes (N=110) carrying D6S2446-8 (DRB1*0301 linked) were sorted on the basis of telomeric (left) length (A), and centromeric (right) length (B). C, Founder haplotypes (N=68) carrying D6S2446-5 (DRB1*0801 linked) sorted on the basis of overall length. Asterisks (*) represent the approximate boundaries of the haplotypes showing convincing evidence for transmission distortion. Haplotypes that failed to show significant transmission disequilibrium (white boxes) did not contain DRB1*0301 or DRB1*0801, despite carrying D6S2446-8 (A and B) or D6S2446-5 (C), respectively.
Identification of Risk Haplotypes
To determine whether the observed effect was due to a single or multiple risk haplotypes, the individual alleles of D6S2446 and short haplotypes containing this marker were examined for association in the combined data set. Three individual alleles of D6S2446 (alleles 6, 8, and 5) showed significant transmission disequilibrium, and multimarker haplotypes containing these alleles demonstrated even higher levels of disequilibrium (table 1). We next stratified the families with SLE by ethnicity. The risk haplotypes were found primarily in the white families and were rare in the nonwhite families (table 1). None of the haplotypes common to nonwhite families demonstrated significant disease association (data not shown); however, the current family collection has limited power to address this issue.
Table 1.
Transmission Disequilibrium of Class II Marker Haplotypes in One- and Two-Parent Families with SLE[Note]
|
All (n=273) |
White (n=235) TDT |
Nonwhite (n=38) TDT |
|||||||||||
|
Allele at Markera |
TDT |
||||||||||||
| D6S2666 | D6S2665 | DRB1 | D6S2446 | MN6S2879 | DQB1 | MN6S2944 | T:NT (Ratio) | P Value | PDT P Value | T:NT (Ratio) | P Value | T:NT (Ratio) | P Value |
| … | … | … | 6 | … | … | … | 87:51 (1.7) | .0022 | .0009 | 75:45 (1.66) | .0062 | 12:6 (2.0) | NS (.157) |
| … | 1 | 1501 | 6 | 4 | … | … | 68:33 (2.1) | .0005 | .0005 | 64:31 (2.06) | .0007 | 4:2 (2.0) | NS (.414) |
| 1 | 1 | 1501 | 6 | 4 | 0602 | 3 | 64:32 (2.0) | .0011 | .0013 | 60:30 (2.00) | .0016 | 4:2 (2.0) | NS (.414) |
| … | … | … | 8 | … | … | … | 69:35 (2.0) | .0009 | .0003 | 66:30 (2.2) | .0002 | 3:5 (.6) | NS (.479) |
| … | 8 | 0301 | 8 | 3 | … | … | 51:26 (2.0) | .0044 | .0004 | 50:24 (2.08) | .0025 | 1:2 (.5) | NS (.564) |
| 4 | 8 | 0301 | 8 | 3 | 0201 | 3 | 49:23 (2.1) | .0022 | .0032 | 48:21 (2.28) | .0012 | 1:2 (.5) | NS (.564) |
| … | … | … | 5 | … | … | … | 37:25 (1.5) | NS (.128) | NS (.054) | 27:20 (1.35) | NS (.307) | 10:5 (2.0) | NS (.197) |
| … | 2 | 0801 | 5 | 3 | … | … | 16:6 (2.7) | .0330 | .0260 | 14:5 (2.80) | .0390 | 2:1 (2.0) | NS (.564) |
| 1 | 2 | 0801 | 5 | 3 | 0402 | 3 | 16:4 (4.0) | .0073 | .0055 | 14:3 (4.66) | .0076 | 2:1 (2.0) | NS (.564) |
Note.— Ratio = T/NT. NS = not significant.
Numbers shown refer to arbitrarily assigned allele numbers for each microsatellite marker (see supplementary data).
DRB1 and DQB1 PCR/SSO (sequence-specific oligonucleotide) typing of founders carrying one of the risk alleles at D6S2446 revealed that the three risk haplotypes contained DRB1*1501/DQB1*0602 (DR2/DQ6), DRB1*0301/DQB1*0201 (DR3/DQ2), and DRB1*0801/DQB1*0402 (DR8/DQ4) (table 1). DNA sequencing of genes tightly linked to DRB1 confirmed that the microsatellite haplotypes faithfully identified ancestral fragments of DNA that were identical between unrelated founders bearing the haplotype (R.R.G., P.K.G., and T.W.B., unpublished data).
Of interest, families carrying one or more of the identified risk haplotypes accounted for nearly all of the transmission disequilibrium observed in the class II region across the entire family collection (D6S2446; PDT P=1×10-7) (fig. 1C). Conversely, the 84 families lacking any of the identified class II risk haplotypes showed no disequilibrium at D6S2446 by PDT analysis (P=.50). The sib-pair pedigrees within this group (n=47) also showed no evidence for linkage in the HLA region (LOD=0.17 at D6S2662). We conclude that there are three HLA risk haplotypes in our family collection, with the strongest evidence for transmission disequilibrium in the class II region at D6S2446.
Visualization of Ancestral Recombinant Class II Risk Haplotypes
To visualize the recombinant class II–containing risk haplotypes, we first examined all founder haplotypes containing allele 6 at D6S2446 (D6S2446-6; fig. 2). The consensus allele for each marker on the extended haplotype was determined (see the “Material and Methods” section), and founder haplotypes were color-coded, with maroon indicating an exact match with consensus and with gold denoting a difference from consensus. Missing data or alleles with ambiguous phase are highlighted in gray.
The D6S2446-6 founder haplotypes were first sorted on the basis of the number of contiguous marker alleles identical to the consensus extended haplotype (fig. 2A). This revealed multiple ancestral recombinant DRB1*1501-containing haplotypes (with the size of the recombinant haplotypes decreasing from top to bottom in fig. 2A). Haplotypes were then grouped on the basis of length, and each group was tested for association, using the TDT (Spielman et al. 1993; Spielman and Ewens 1998) and PDT (data not shown). Haplotype clusters showing overall significant transmission distortion are highlighted in blue (right side of the panel). The group of short recombinant haplotypes having a 22:11 transmitted:not transmitted (T:NT) ratio suggested a telomeric ancestral recombinant “breakpoint” at approximately marker MN6S2468 (marked by the left asterisk in fig. 2). To determine the location of the centromeric ancestral “breakpoint,” the founder haplotypes were again sorted, first by identity centromeric to D6S2446 and then by extent of telomeric identity (fig. 2B). This demonstrated that founder haplotypes extending centromeric to marker MN6S2944 (right asterisk in fig. 2) had a level of transmission distortion similar to that of haplotypes that extended beyond. Thus, the estimated centromeric “breakpoint” on this haplotype is ∼60 kb centromeric of DQB1 (∼66 kb centromeric of D6S2446), near marker MN6S2944, a known recombination hotspot within the HLA region (Jeffreys et al. 2001).
Recent studies have suggested that certain tumor-necrosis factor (TNF)–α alleles may contribute to SLE (Rood et al. 2000). To determine whether TNF alleles on the DRB1*1501 haplotype were important for the observed effect, we selected all founder haplotypes that contained the consensus TNF allele from the DRB1*1501 extended haplotype and then sorted and analyzed the data. Strikingly, founder haplotypes carrying the consensus TNF region showed disequilibrium only if they also included DRB1*1501 and DQB1*0602 of the extended haplotype (fig. 2C). Recombinant founder haplotypes that excluded the DRB1*1501 class II region failed to show risk. We suggest that the region of interest on the extended DRB1*1501 haplotype contains both the DRB1 and DQB1 genes and effectively excludes the class I and III regions, including TNF-α.
Founder haplotypes that contained allele 8 at D6S2446 (in strong linkage disequilibrium with DRB1*0301) were then analyzed using the same strategy (fig. 3A and 3B). On the basis of the visualized ancestral haplotypes, it was clear that the level of linkage disequilibrium of markers on this haplotype was much higher than was observed for the DRB1*1501 haplotype (compare with fig. 2). HLA A, B, and C typing confirmed that this haplotype corresponds to the HLA A1-B8-DR3 haplotype, which is known to have unusually extensive disequilibrium (Dawkins et al. 1999). The microsatellites used could distinguish the northern European B8-DR3 haplotype from the southern European B18-DR3 haplotype, which was found in only 1.3% of the founders in the study and showed no association with disease (data not shown). Because of the relative paucity of ancestral recombinants, the risk region within DRB1*0301-containing haplotypes could not be narrowed beyond an ∼1 Mb region encompassing most of the class III and class II regions.
Consensus alleles for the extended ancestral haplotype carrying DRB1*0801 could be identified up to only the class I/class III boundary (fig. 3C). Haplotypes containing DRB1*0801 were less common in our family collection, and ancestral recombinants allowed the risk interval to be narrowed to region of ∼500 kb between markers D6S2669 and D6S2662.
Haplotype Frequencies in SLE Case and Control Individuals
The three SLE class II risk haplotypes were highly enriched in white families. Of the 254 families carrying one or more of the risk haplotypes, 229 (90%) were white or had known white admixture. To explore this further, we compared allele frequencies of 280 female white SLE index case individuals from the family collection and 174 female white control individuals for the identified D6S2666/2665/2446 risk haplotypes (tables 2 and 3). As shown in table 2, the risk haplotypes were more common on the chromosomes of patients with SLE than in control individuals, with an approximately twofold increase in risk haplotype frequency. GRR calculations indicated a doubling of risk for homozygotes (table 2), consistent with an additive model for genetic risk of these haplotypes (Risch and Merikangas 1996). A similar picture emerged when the status of heterozygosity and homozygosity for the various risk haplotypes was examined (table 3). In these analyses, DRB1*0301-containing haplotypes conferred the highest degree of risk, followed by DRB1*1501-containing haplotypes and then DRB1*0801-containing haplotypes. The case/control data for the DRB1*0801 haplotypes trended toward, but did not reach, significance with the current sample size. Given that the TDT results for DRB1*0801-containing haplotypes were significant (table 1), this suggests an improved ability of the TDT to detect association for the relatively rare DRB1*0801 class II haplotype, compared with the case/control approach. Finally, it was striking that nearly two-thirds (63.9%) of the patients carried at least one of the risk haplotypes. We conclude that the identified class II risk haplotypes are enriched in SLE patients and show strong association in a case-control study design.
Table 2.
Frequency of Class II Risk Haplotypes in White Patients with SLE and Control Individuals
|
Frequency(%) |
GRRc |
||||
| Risk Haplotypea | Patient(N=560chromosomes)d | Control(N=348chromosomes)e | P Valueb | Heterozygote | Homozygote |
| DRB1*1501/DQB1*0602 | 20.7 | 12.3 | .0012 | 1.9 | 3.5 |
| DRB1*0301/DQB1*0201 | 17.3 | 7.5 | 2.7×10−5 | 2.6 | 6.7 |
| DRB1*0801/DQB1*0402 | 3.6 | 1.7 | NS (.09) | 2.2 | 4.7 |
| Any of the risk haplotypes | 41.6 | 21.5 | 4.7×10−10 | 2.6 | 6.8 |
Haplotypes containing the indicated DRB1/DQB1 molecules were determined by microsatellite genotypes for markers D6S2666/2665/2446.
Determined by χ2 analysis. NS = not significant.
Calculated as described by Risch and Merikangas (1996).
Index case individuals from white families with SLE.
Unrelated white female control individuals.
Table 3.
Dose-Dependent Risk from Class II Susceptibility Haplotypes
|
Frequency in(%) |
||||
| Risk Haplotypea | Patientsd(N=280) | ControlIndividualse(N=174) | P Valueb | Relative Riskc |
| DRB1*1501/DQB1*0602: | ||||
| Heterozygote | 33.6 | 22.4 | .0110 | 1.5 |
| Homozygote | 3.9 | 1.1 | NS (.084) | 3.5 |
| DRB1*0301/DQB1*0201: | ||||
| Heterozygote | 29.6 | 12.6 | 3.0×10−5 | 2.3 |
| Homozygote | 2.5 | 1.1 | NS (.32) | 2.3 |
| DRB1*0801/DQB1*0402: | ||||
| Heterozygote | 6.4 | 3.4 | NS (.17) | 1.9 |
| Homozygote | .4 | .0 | NS (.43) | … |
| Heterozygote for 1 of 3 risk haplotypes | 45.0 | 33.9 | .0190 | 1.3 |
| Compound heterozygote | 12.1 | 2.3 | 2.3×10−4 | 5.2 |
| Simple homozygote | 6.8 | 2.3 | .0340 | 3.0 |
| Any haplotype combination | 63.9 | 38.5 | 1.3×10−7 | 1.7 |
Haplotypes containing the indicated DRB1/DQB1 molecules were determined by microsatellite genotypes for markers D6S2666/2665/2446
Determined by χ2 analysis. NS = not significant.
Calculated as frequency in patients with SLE/frequency in control individuals.
Index case individuals from white families with SLE.
Unrelated white female control individuals.
Discussion
In the present article, we describe a haplotype-based approach to identify and narrow the HLA risk intervals in a collection of 334 sib-pair and simplex families with SLE. Related approaches have recently been used to dissect the role of HLA alleles in several other autoimmune disorders, including type 1 diabetes (Koeleman et al. 2000; Zavattari et al. 2001), psoriasis (Oka et al. 1999; Nair et al. 2000), Behcet disease (Ota et al. 1999), multiple sclerosis (Marrosu et al. 2001), and rheumatoid arthritis (Jawaheer et al. 2002 [in this issue]). The dense map of microsatellite markers that were typed across the HLA allowed us to reconstruct and then visualize the ancestral DNA haplotypes carrying these risk alleles. For this approach to be successful, it was important that most of the markers maintained stable repeat lengths over long periods of human population history and therefore could be used to identify the underlying ancestral DNA haplotypes with fidelity. This was indeed the case. Sequencing of unrelated individuals carrying the various marker haplotypes showed essentially no sequence diversity at the nucleotide level (R.R.G., P.K.G., and T.W.B., unpublished data). These data show that it is possible to type patients for certain class II DRB1 and DQB1 alleles solely on the basis of their D6S2666/2665/2446 marker haplotype.
Through visualization of recombinations on the ancestral haplotypes carrying the alleles DRB1*1501 and DRB1*0801, the risk regions on these haplotypes could be narrowed to an ∼500-kb region containing both DRB1 and DQB1. The only other genes in this interval are two genes of unknown function—chromosome 6 ORF 10 (C6orf10, formerly known as “testis specific basic protein”) and butyrophilin-like family member II (BTNL2)—together with DRA (invariant), DRB3 and DRB5 (additional β-chain genes present on the DRB1*0301 and DRB1*1501 haplotypes, respectively), and DQA (a different allele for each of the risk haplotypes). Sequencing of these genes on the various risk haplotypes should allow us to determine whether the genetic effect in this region is limited to the class II DRB1, DQA1, and/or DQB1 genes, or whether it includes other tightly linked genes, possibly acting in epistasis. In the future, we will attempt to further narrow the regions of interest by identifying additional families with SLE that carry short recombinant risk haplotypes.
The extensive disequilibrium of the DRB1*0301 haplotype severely limits the ability to localize the genetic effect on this haplotype. It is important to note that the TNF-α and C4 “null” alleles, which have previously been suggested as risk factors for SLE (Harley et al. 1998; Tan and Arnett 1998), are both carried on the extended A1/B8/DR3 haplotype. These data suggest that caution should be exercised before assigning risk to any individual gene on the DRB1*0301 haplotype, given the extensive disequilibrium observed. On the basis of the finding that the DR2- and DR8-containing risk haplotypes exclude most, if not all, of the class I and III regions, we hypothesize that the major genetic effect on the DR3 haplotype may be similarly localized to the class II region. However, we cannot rule out the possibility that other genes on the extended haplotype may also be contributing to risk.
The hypothesis that class II–containing haplotypes are major genetic risk factors in human SLE is supported by studies in the mouse (Morel et al. 1994; Vyse and Kotzin 1998) and has important implications for understanding the pathogenesis of SLE. Previous studies have suggested that the autoimmune response in SLE is driven by antigen, with extensive somatic hypermutation observed in B cells from mice and humans with lupus (Diamond et al. 1992). Somatic hypermutation requires CD4+ helper T cells, which are activated by peptides presented on HLA class II molecules. A recent study showed that serum from some pediatric patients with SLE can induce the generation of class II–bearing dendritic cells from monocytes, and these cells have a greatly enhanced ability to present foreign antigen to CD4+ T cells (Blanco et al. 2001). Given recent progress in understanding the role that apoptotic cells may have in initiating autoantibody responses in SLE (Casciola-Rosen et al. 1994; Mevorach et al. 1998), it will be important to test whether peptides derived from apoptotic cells and presented on the molecules encoded by the identified class II risk alleles contribute to SLE pathogenesis.
Acknowledgments
We thank all the patients, family members, and physicians, for their interest and support. We also thank Therese Ockenden, Michelle Frauenshuh, Sherine Gabriel, Ronald Messner, and Grainne Kearns for their efforts in recruiting families early in the project. We are grateful to the AMDeC and the New York Cancer Project, for providing access to control DNA samples. This work was supported by National Institutes of Health grants R01-AR43274 and P01-AR45231-SCOR in SLE (to T.W.B.) and R01-AR44222 and N01-AR72232 (to P.K.G.), as well as by Lupus Foundation of Minnesota grants (to T.W.B.).
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- Behrens Lab, http://www.behrenslab.org/graham/list.html (for supplementary material)
- Genome Database, The, http://www.gdb.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SLE [MIM 152700]) [PubMed]
- Sanger Institute: Human Chromosome 6 Home, http://www.sanger.ac.uk/HGP/Chr6/MHC.shtml (for current consensus MHC sequence)
- UCSC Genome Bioinformatics Home, http://genome.ucsc.edu/ (for the Human Genome Project working draft)
References
- Aguado B, Bahram S, Beck S, Campbell RD, Forbes SA, Geraghty D, Guillaudeux T, et al (1999) Complete sequence and gene map of a human major histocompatibility complex. Nature 401:921–923 [DOI] [PubMed] [Google Scholar]
- Blanco P, Palucka AK, Gill M, Pascual V, Banchereau J (2001) Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science 294:1540–1543 [DOI] [PubMed] [Google Scholar]
- Casciola-Rosen LA, Anhalt G, Rosen A (1994) Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med 179:1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R, Leelayuwat C, Gaudieri S, Tay G, Hui J, Cattley S, Martinez P, Kulski J (1999) Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol Rev 167:275–304 [DOI] [PubMed] [Google Scholar]
- Diamond B, Katz JB, Paul E, Aranow C, Lustgarten D, Scharff MD (1992) The role of somatic mutation in the pathogenic anti-DNA response. Annu Rev Immunol 10:731–757 [DOI] [PubMed] [Google Scholar]
- Erlich H, Bugawan T, Begovich AB, Scharf S, Griffith R, Saiki R, Higuchi R, Walsh PS (1991) HLA-DR, DQ and DP typing using PCR amplification and immobilized probes. Eur J Immunogenet 18:33–55 [DOI] [PubMed] [Google Scholar]
- Gaffney PM, Kearns GM, Shark KB, Ortmann WA, Selby SA, Malmgren ML, Rohlf KE, Ockenden TC, Messner RP, King RA, Rich SS, Behrens TW (1998) A genome-wide search for susceptibility genes in human systemic lupus erythematosus sib-pair families. Proc Natl Acad Sci USA 95:14875–14879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney PM, Moser KL, Graham RR, Behrens TW (2002) Recent advances in the genetics of systemic lupus erythematosus. Rheum Dis Clin North Am 28:111–126 [DOI] [PubMed] [Google Scholar]
- Gaffney PM, Ortmann WA, Selby SA, Shark KB, Ockenden TC, Rohlf KE, Walgrave NL, Boyum WP, Malmgren ML, Miller ME, Kearns GM, Messner RP, King RA, Rich SS, Behrens TW (2000) Genome screening in human systemic lupus erythematosus: results from a second Minnesota cohort and combined analyses of 187 sib-pair families. Am J Hum Genet 66:547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RR, Langefeld CD, Gaffney PM, Ortmann WA, Selby SA, Baechler EC, Shark KB, Ockenden TC, Rohlf KE, Moser KL, Brown WM, Gabriel SE, Messner RP, King RA, Horak P, Elder JT, Stuart PE, Rich SS, Behrens TW (2001) Genetic linkage and transmission disequilibrium of marker haplotypes at chromosome 1q41 in human systemic lupus erythematosus. Arthritis Res 3:299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley JB, Moser KL, Gaffney PM, Behrens TW (1998) The genetics of human systemic lupus erythematosus. Curr Opin Immunol 10:690–696 [DOI] [PubMed] [Google Scholar]
- Hochberg MC (1997) The epidemiology of systemic lupus erythematosus. In: Wallace DJ, Hahn BH (eds) Dubois' lupus erythematosus. Williams and Wilkins, Baltimore, pp 49–65 [Google Scholar]
- Jawaheer D, Li W, Graham R, Chen W, Damle A, Xiao X, Monteiro J, Khalili H, Lee A, Lundsten R, Begovich A, Bugawan T, Erlich H, Elder J, Criswell L, Seldin M, Amos C, Behrens T, Gregersen P (2002) Dissecting the genetic complexity of the HLA association with rheumatoid arthritis. Am J Hum Genet 71:585–594 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R (2001) Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet 29:217–222 [DOI] [PubMed] [Google Scholar]
- Koeleman BP, Herr MH, Zavattari P, Dudbridge F, March R, Campbell D, Barnett AH, Bain SC, Mulargia AP, Loddo M, Amos W, Cucca F, Todd JA (2000) Conditional ETDT analysis of the human leukocyte antigen region in type 1 diabetes. Ann Hum Genet 64:215–221 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrosu MG, Murru R, Murru MR, Costa G, Zavattari P, Whalen M, Cocco E, Mancosu C, Schirru L, Solla E, Fadda E, Melis C, Porru I, Rolesu M, Cucca F (2001) Dissection of the HLA association with multiple sclerosis in the founder isolated population of Sardinia. Hum Mol Genet 10:2907–2916 [DOI] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL (2000) A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet 67:146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevorach D, Zhou JL, Song X, Elkon KB (1998) Systemic exposure to irradiated apoptotic cells induces autoantibody production. J Exp Med 188:387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel L, Rudofsky UH, Longmate JA, Schiffenbauer J, Wakeland EK (1994) Polygenic control of susceptibility to murine systemic lupus erythematosus. Immunity 1:219–229 [DOI] [PubMed] [Google Scholar]
- Nair RP, Stuart P, Henseler T, Jenisch S, Chia NV, Westphal E, Schork NJ, Kim J, Lim HW, Christophers E, Voorhees JJ, Elder JT (2000) Localization of psoriasis-susceptibility locus PSORS1 to a 60-kb interval telomeric to HLA-C. Am J Hum Genet 66:1833–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A, Tamiya G, Tomizawa M, Ota M, Katsuyama Y, Makino S, Shiina T, Yoshitome M, Iizuka M, Sasao Y, Iwashita K, Kawakubo Y, Sugai J, Ozawa A, Ohkido M, Kimura M, Bahram S, Inoko H (1999) Association analysis using refined microsatellite markers localizes a susceptibility locus for psoriasis vulgaris within a 111 kb segment telomeric to the HLA-C gene. Hum Mol Genet 8:2165–2170 [DOI] [PubMed] [Google Scholar]
- Ota M, Mizuki N, Katsuyama Y, Tamiya G, Shiina T, Oka A, Ando H, Kimura M, Goto K, Ohno S, Inoko H (1999) The critical region for Behcet disease in the human major histocompatibility complex is reduced to a 46-kb segment centromeric of HLA-B, by association analysis using refined microsatellite mapping. Am J Hum Genet 64:1406–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DA, Trowsdale J (1999) Genetics and molecular genetics of the MHC. Rev Immunogenet 1:21–31 [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Rood MJ, van Krugten MV, Zanelli E, van der Linden MW, Keijsers V, Schreuder GM, Verduyn W, Westendorp RG, de Vries RR, Breedveld FC, Verweij CL, Huizinga TW (2000) TNF-308A and HLA-DR3 alleles contribute independently to susceptibility to systemic lupus erythematosus. Arthritis Rheum 43:129–134 [DOI] [PubMed] [Google Scholar]
- Snell GD (1948) Methods for the study of histocompatibility genes. J Genet 49:87 [DOI] [PubMed] [Google Scholar]
- Spielman RS, Ewens WJ (1998) A sibship test for linkage in the presence of association: the sib transmission/disequilibrium test. Am J Hum Genet 62:450–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52:506–516 [PMC free article] [PubMed] [Google Scholar]
- Tan FK, Arnett FC (1998) The genetics of lupus. Curr Opin Rheumatol 10:399–408 [DOI] [PubMed] [Google Scholar]
- Vyse TJ, Kotzin BL (1998) Genetic susceptibility to systemic lupus erythematosus. Annu Rev Immunol 16:261–292 [DOI] [PubMed] [Google Scholar]
- Vyse TJ, Todd JA (1996) Genetic analysis of autoimmune disease. Cell 85:311–318 [DOI] [PubMed] [Google Scholar]
- Wakeland EK, Liu K, Graham RR, Behrens TW (2001) Delineating the genetic basis of systemic lupus erythematosus. Immunity 15:397–408 [DOI] [PubMed] [Google Scholar]
- Zavattari P, Lampis R, Motzo C, Loddo M, Mulargia A, Whalen M, Maioli M, Angius E, Todd JA, Cucca F (2001) Conditional linkage disequilibrium analysis of a complex disease superlocus, IDDM1 in the HLA region, reveals the presence of independent modifying gene effects influencing the type 1 diabetes risk encoded by the major HLA-DQB1, -DRB1 disease loci. Hum Mol Genet 10:881–889 [DOI] [PubMed] [Google Scholar]