Abstract
BACKGROUND AND PURPOSE
Endocannabinoid signalling has been shown to have a role in the control of epidermal physiology, whereby anandamide is able to regulate the expression of skin differentiation genes through DNA methylation. Here, we investigated the possible epigenetic regulation of these genes by several phytocannabinoids, plant-derived cannabinoids that have the potential to be novel therapeutics for various human diseases.
EXPERIMENTAL APPROACH
The effects of cannabidiol, cannabigerol and cannabidivarin on the expression of skin differentiation genes keratins 1 and 10, involucrin and transglutaminase 5, as well as on DNA methylation of keratin 10 gene, were investigated in human keratinocytes (HaCaT cells). The effects of these phytocannabinoids on global DNA methylation and the activity and expression of four major DNA methyltransferases (DNMT1, 3a, 3b and 3L) were also examined.
KEY RESULTS
Cannabidiol and cannabigerol significantly reduced the expression of all the genes tested in differentiated HaCaT cells, by increasing DNA methylation of keratin 10 gene, but cannabidivarin was ineffective. Remarkably, cannabidiol reduced keratin 10 mRNA through a type-1 cannabinoid (CB1) receptor-dependent mechanism, whereas cannabigerol did not affect either CB1 or CB2 receptors of HaCaT cells. In addition, cannabidiol, but not cannabigerol, increased global DNA methylation levels by selectively enhancing DNMT1 expression, without affecting DNMT 3a, 3b or 3L.
CONCLUSIONS AND IMPLICATIONS
These findings show that the phytocannabinoids cannabidiol and cannabigerol are transcriptional repressors that can control cell proliferation and differentiation. This indicates that they (especially cannabidiol) have the potential to be lead compounds for the development of novel therapeutics for skin diseases.
Keywords: phytocannabinoids, endocannabinoid system, gene expression, DNA methylation, skin
Introduction
Endocannabinoids (eCBs) are lipid mediators derived from membrane precursors and are involved in multiple regulatory functions, both in health and disease (Di Marzo and Petrosino, 2007). The two most important eCBs are N-arachidonylethanolamine (‘anandamide’, AEA) and 2-arachidonoylglycerol (2-AG) that elicit their activity via at least two G-protein–coupled cannabinoid receptors (CB1 and CB2), both widely distributed throughout the body (Rodríguez de Fonseca et al., 2005). AEA and 2-AG can also activate non-CB1/non-CB2 receptors and/or a purported ‘CB3’ (or GPR55) receptor (Baker et al., 2006); yet, there is controversy about the actual involvement of GPR55 in eCBs signalling (Pertwee et al., 2010). Furthermore AEA, but not 2-AG, behaves as a ligand to type-1 vanilloid receptor (transient receptor potential vanilloid 1, TRPV1) channels (Pertwee et al., 2010). Several enzymes are involved in eCBs synthesis and degradation: AEA is synthesized mainly by N-acyl-phosphatidylethanolamines-specific phospholipase, and is degraded by fatty acid amide hydrolase (FAAH); 2-AG is mainly synthesized by an sn-1-specific diacylglycerol lipase, and is degraded by a specific monoacylglycerol lipase (Ahn et al., 2008; Di Marzo, 2008; Ueda et al., 2011). Within the CNS and in the peripheral tissues, eCBs, their target receptors and metabolic enzymes, along with the proteins responsible for their transport and intracellular trafficking, form the endocannabinoid system (ECS) (Maccarrone et al., 2010).
Recently, the ECS has been reported to have a role in the control of skin physiology (Bíró et al., 2009; Pasquariello et al., 2009), and it has been suggested that constituents of the ECS have the potential to be exploited as new targets for future therapies in dermatology (Paus et al., 2006; Karsak et al., 2007; Kupczyk et al., 2009; Petrosino et al., 2010).
The epidermis is the outer layer of the skin serving as a physical and chemical barrier to the environment, provided by terminally differentiated keratinocytes (Nemes and Steinert, 1999; Kalinin et al., 2001). Epidermal differentiation begins with the migration of keratinocytes from a basal layer, composed of proliferating cells, and ends with the formation of the cornified cell envelope, an insoluble protein structure found in differentiated keratinocytes (Candi et al., 2005).
All major ECS components have been found to be active in human epidermis, where CB1 cannabinoid receptor expression is higher in more differentiated (i.e. granular and spinous) layers of skin (Casanova et al., 2003; Stander et al., 2005). Also immortalized and normal epidermal keratinocytes have a fully functional ECS (Berdyshev et al., 2000; Maccarrone et al., 2003; Oddi et al., 2005). In these cells, AEA mediates transcriptional effects associated with epidermal differentiation and skin development, through a CB1-dependent mechanism (Maccarrone et al., 2003). In line with this, in spontaneously immortalized human keratinocytes (HaCaT cells) and in normal human epidermal keratinocytes (NHEK cells) induced to differentiate in vitro by 12-O-tetradecanoylphorbol 13-acetate (TPA) plus calcium, AEA levels were reduced due to enhanced degradation by FAAH (Maccarrone et al., 2003). Moreover, in HaCaT cells exposed to AEA, there is a reduction in the formation of cornified envelopes (Maccarrone et al., 2003) and a reduction in the expression of keratins 1 (K1) and 10 (K10), involucrin and transglutaminase 5 (TGase5) genes, which are all up-regulated during cornification (Paradisi et al., 2008).
Gene expression is controlled by epigenetic mechanisms that cause heritable but potentially reversible changes in DNA methylation, histone modification and RNA-associated silencing (Jaenisch and Bird, 2003). Epigenetics is thus the study of molecular mechanisms by which the environment controls gene activity independently of DNA sequence. It is well established that complex diseases are generally caused by both genetic and environmental factors, but even though the role of genetic abnormalities in the pathogenesis of many skin diseases has been thoroughly investigated (for review see Zhang, 2012), there have been few studies on the importance of epigenetics in altering the course of these diseases (Chen et al., 2008; Millington, 2008; Lopez et al., 2009).
Variations in global DNA methylation have been reported between differentiated and undifferentiated cells (Lyon et al., 1987; Ehrlich, 2003), and in particular a hypomethylation in differentiated versus undifferentiated keratinocytes has been documented (Veres et al., 1989). Moreover, inhibition of DNA methylation and histone deacetylation has been shown to promote keratinocyte differentiation (Rosl et al., 1988; Schmidt et al., 1989; Staiano-Coico et al., 1989), and an inverse correlation between DNA methylation and the expression of differentiating genes has been demonstrated in human keratinocytes (Engelkamp et al., 1993; Elder and Zhao, 2002). It has also been suggested that inhibition of differentiation by AEA occurs through changes in chromatin methylation patterns (Paradisi et al., 2008; Pasquariello et al., 2009), and that AEA induces DNA methylation of keratinocyte-differentiating genes by increasing DNA methyltransferase (DNMT) activity via a CB1-dependent involvement of p38 and p42/p44 MAPK (Paradisi et al., 2008).
Based on these findings, in the present study we investigated the possible epigenetic regulation of skin differentiation genes by selected phytocannabinoids that are plant-derived cannabinoids, which mimic the natural eCBs, and thus have potential as novel therapeutics for human diseases (Hill et al., 2012a).
Phytocannabinoids are known to have anti-inflammatory properties (Klein, 2005) and to inhibit growth of proliferating carcinogenic cells (Kogan, 2005). These compounds are lipophilic, and hence are readily absorbed through the skin. In particular, it has been documented that cannabidiol (CBD) accumulates only in the stratum corneum, without penetrating into the deeper layers (Lodzki et al., 2003). However, the therapeutic potential of cannabinoid-based preparations for skin diseases has not yet been investigated. Up to now, just one study has reported the inhibition of human keratinocyte proliferation by phytocannabinoids, suggesting that phytocannabinoids could be beneficial in the treatment of psoriasis (Wilkinson and Williamson, 2007).
In this study, we investigated the effects of three major non-psychoactive components of Cannabis sativa (Izzo et al., 2009): CBD and its precursor cannabigerol (CBG), that are, together with Δ9-tetrahydrocannbinol, the most abundant phytocannabinoids (Hill et al., 2012b); and cannabidivarin (CBDV), a propyl analogue of CBD which like its congener has anticonvulsant properties (Jones et al., 2010; Hill et al., 2012a).
The understanding of the epigenetic regulation of keratinocyte differentiation by phytocannabinoids may pave the way to the development of new drugs for skin diseases, analogous to other human disorders like multiple sclerosis (Rog, 2010), bowel disease (Lal et al., 2011) and cancer (Solinas et al., 2012).
Methods
The nomenclature of all drug/molecular targets used in this study conforms to BJP's Guide to Receptors and Channels (Alexander et al., 2011).
Materials
Chemicals were of the purest analytical grade. Anandamide (AEA) and TPA were purchased from Sigma Chemical Co. (St. Louis, MO, USA). S-Adenosyl-L-[methyl-3H]-methionine was from Amersham Biosciences (Buckinghamshire, UK). CBD, CBG and CBDV were kind gifts of GW Pharma Ltd (Sittingbourne, UK). Capsazepine (N-[2-(4-chlorophenyl) ethyl]-1,3, 4, 5-tetrahydro-7, 8-dihydroxy-2H-2-benzazepine-2-carbothioamide, CPZ) was from Calbiochem (San Diego, CA, USA). N-Piperidino-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-3-pyrazole carboxamide (SR141716) and N-[(1)-endo-1,3,3-trimethy-1-bicyclo [2.2.1]-heptan-2-yl]5-(4-chloro-3-methyl-phenyl)-1-(4-methyl-benzyl)-pyrazole-3-carboxamide (SR144528) were from Sanofi-Aventis Recherche (Montpellier, France). Goat anti-DNMT1 and anti-Lamin A polyclonal antibodies, and rabbit anti-goat antibody conjugated to HRP were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Cell culture and treatment
HaCaT cells were grown in a 1:1 mixture of minimum essential medium and Ham's F-12 medium (Invitrogen, Berlin, Germany), supplemented with 10% fetal calf serum and 1% non-essential amino acids, at 37°C in a 5% CO2 humidified atmosphere. Cell differentiation was induced by treating HaCaT cells with TPA (10 ng·mL−1) plus CaCl2 (1.2 mM) for 5 days (Candi et al., 2001).
AEA, CBD, CBG and CBDV were dissolved in methanol; SR141716, SR144528 and CPZ were dissolved in DMSO; these compounds were added at the indicated concentrations directly to the serum-free culture medium, at the same time as TPA plus calcium (Paradisi et al., 2008). Culture medium containing vehicles alone was added to controls under the same conditions. After each treatment, cell viability was determined by Trypan Blue dye exclusion, as described previously (Paradisi et al., 2008).
NHEK (Lonza Group Ltd, Basel, Switzerland) were grown at 37°C in a humidified 5% CO2 atmosphere in KGM-Gold™ growth medium (Lonza Group Ltd), according to the manufacturer's instructions. NHEKs were treated for 5 days with AEA, CBD and CBG at the indicated concentrations, as described above for HaCaT cells.
Quantitative real-time (qRT)-PCR
RNA was extracted using RNeasy extraction kit (Qiagen, Crawley, UK) from proliferating and differentiated HaCaT cells, following the manufacturer's instructions. RT-PCR reactions were performed using the QuantiTect Reverse Transcription Kit (Qiagen). The relative abundance of each mRNA species was assessed by qRT-PCR, using QuantiFast Multiplex PCR Kit (Qiagen) on a DNA Engine Opticon 2 Continuous Fluorescence Detection System (MJ Research, Waltham, MA, USA). The primers used for PCR amplification are shown in Table 1. Actin was used as a housekeeping gene for quantity normalization (D'Addario et al., 2008). One microlitre of the first strand cDNA product was used for amplification in triplicate in 20 μL reaction solution, containing 10 μL of QuantiFast Multiplex PCR Kit (Qiagen) and 10 pmol of each primer. The following PCR programme was used: 95°C for 10 min, followed by 50 amplification cycles of 95°C for 10 s and 60°C for 30 s.
Table 1.
Primer sequences used for reverse transcription–PCR
| Human gene | Forward (5′ → 3′) | Reverse (3′ → 5′) |
|---|---|---|
| K10 | ACGAGGAGGAAATGAAAGAC | GGACTGTAGTTCTATCTCCAG |
| K1 | AGAAAGCAGGATGTCTGG | AAACAAACTTCACGCTGG |
| Involucrin | CTCTGCCTCAGCCTTACT | GCTGCTGATCCCTTTGTG |
| TGase5 | TCAGCACAAAGAGCATCCAG | TTCAGGGAGACTTGCACCAC |
| β-actin | TGACCCAGATCATGTTTGAG | TTAATGTCACGCACGATTTCC |
| DNMT1 | CCCCTGAGCCCTACCGAAT | CTCGCTGGAGTGGACTTGTG |
| DNMT3a | TATTGATGAGCGCACAAGAGAGC | GGGTGTTCCAGGGTAACATTGAG |
| DNMT3b | GGCAAGTTCTCCGAGGTCTCTG | TGGTACATGGCTTTTCGATAGGA |
| DNMT3L | GGCTCTGGTTTCGGAAGAA | TCTCTTAGGGGGAGAAAGCA |
| GAPDH | CAGCCTCAAGATCATCAGCA | TGTGGTCATGAGTCCTTCCA |
| M K10 | AGTTTTCGTTTTCGTAGTCGTC | CGAATATAACCTCACCCCG |
| U K10 | GGAGTTTTTGTTTTTGTAGTTGTT | AACCAAATATAACCTCACCCCA |
| myoD | CCAACTCCAAATCCCCTCTCTAT | TGATTAATTTAGATTGGGTTTAGAGAAGGA |
Genomic methylation level
A modification of the methyl-accepting assay (Broday et al., 1999) was used to determine the methylation level of DNA isolated from HaCaT cells. DNA (200 ng) was incubated with four units of SssI methylases (New England Biolabs, Ipswich, MA, USA) in the presence of 1.5 mM S-adenosyl-L-methyl-[3H]-methionine and 1.5 mM nonradioactive S-adenosylmethionine (New England Biolabs). The reaction mixtures (20 μL) were incubated at 37°C for 4 h in the manufacturer's buffer containing 0.1 μg of RNase A. The reactions were terminated by adding 300 μL of stop solution (1% sodium dodecyl sulfate (SDS), 2 mM EDTA, 5% 2-propyl alcohol, 125 mM NaCl, 1 mg of proteinase K mL−1, 0.25 mg of carrier DNA mL−1) for 1 h at 37°C. DNA was extracted with phenol-chloroform and was ethanol-precipitated. The DNA recovered was resuspended in 30 μL of 0.3 M NaOH and incubated for 30 min at 37°C. DNA was spotted on Whatman GF/C filter discs, dried and then washed five times with 5% (w v−1) trichloroacetic acid followed by 70% (v v−1) ethanol. Filters were placed in scintillation vials and incubated for 1 h at 60°C with 500 μL of 0.5 M perchloric acid. Then, 5 mL of scintillation mixture was added, and tritium incorporation was determined in a Tri-Carb 2810 TR liquid scintillation analyser (Perkin Elmer, Waltham, MA, USA). Higher levels of 3H methyl group incorporated into DNA were indicative of lower levels of genomic DNA methylation (Paradisi et al., 2008).
Assay of DNA methyltransferase activity
Cell extracts were prepared in ice-cold lysis buffer containing 50 mM Tris-HCl, pH 7.8, 1 mM EDTA, 10% glycerol, 0.01% sodium azide, 10% Tween-80, 100 μg·mL−1 RNase A and 0.5 mM PMSF. De novo methyltransferase activity was measured in cell extracts (30 μg proteins per test), that were incubated in the presence of 3 μg double-stranded oligonucleotides and 2.4 μCi of S-adenosyl-L-methyl-[3H]-methionine (Amersham Biosciences), at 37°C for 1 h. The reaction was terminated by adding 90 μL of stop solution [1% SDS, 2 mM EDTA, 3% (wv−1) 4-amino salicylate, 5% butanol, 0.25 mg·mL−1 calf thymus DNA, and 1 mg·mL−1 proteinase K], followed by incubation at 37°C for 45 min. The reaction mixture was then spotted on a Whatman GF/C filter paper disc (Sigma Chemical Co.). Filters were washed twice with 5% trichloroacetic acid, rinsed in 70% ethanol and dried at 56°C for 20 min. Finally, filters were submerged in UltimaGold (Packard, Meriden, CT, USA) scintillation solution and radioactivity was measured in a Tri-Carb 2810 TR liquid scintillation analyzer (Perkin Elmer). A blank control reaction was done simultaneously using cell extracts that were heated to 80°C for 15 min to inactivate DNMT. The results were expressed as counts per minute and were corrected by background subtraction.
Analysis of DNA methylation by methylation-specific primer RT-PCR
Genomic DNA was isolated from HaCaT cells using DNeasy kit (Qiagen). After DNA extraction, DNA (2 μg) was treated with bisulfite, using the Methyl Detector Bisulfite Modification Kit (Active Motif, Carlsbad, CA, USA) according to the manufacturer's protocol. The relative abundance of each mRNA species was assessed by qRT-PCR, using QuantiFast Multiplex PCR Kit (Qiagen) on a DNA Engine Opticon 2 Continuous Fluorescence Detection System (MJ Research). The amplification programme was as follow: 95°C for 2 min, 50 cycles at 95°C for 10 s and 60°C for 30 s. PCR was also performed for the non-CpG-containing region of myoD, that served as a control gene (D'Addario et al., 2012). One microlitre of bisulfite-treated DNA was used for amplification in triplicate in a 20 μL reaction solution containing 10 μL of QuantiFast SYBR Green PCR (Qiagen) and 10 pmol of each primer. The DNA methylation level was calculated as (1/1 + 2−ΔCt), where ΔCt = CtU-CtM (Lu et al., 2007). The data are presented as fold induction over proliferating cells (Prol = 1). The primers used for PCR amplification for both gene expression and K10 DNA methylation levels are shown in Table 1.
Immunochemical analysis
The protein content of the nuclear extracts was determined by the Bio-Rad Protein Assay Kit (Bio-Rad, Hercules, CA, USA). For Western blotting, equal amounts of protein (25 μg per lane) were loaded onto 8% SDS–PAGs, and were electroblotted onto PVDF sheets (GE-Healthcare, Pollards Woods, UK). Membranes were blocked with 3% BSA for 2 h, and then were incubated with anti-DNMT1 (1:500), or Lamin A (1:1000) antibodies. Then, membranes were rinsed and incubated with HRP-conjugated secondary antibody (diluted 1:1000) in blocking solution. Membranes were washed with TBS-T buffer and incubated with a 1:1000 dilution of HRP–conjugated secondary antibodies (Sigma Chemical Co.), for 1 h at room temperature. After being washed with TBS-T buffer, proteins ware visualized using the HRP substrate ECL Prime (GE-Healthcare).
Statistical analysis
The data are presented as mean ± SEM of at least three independent determinations, each performed in triplicate. Statistical analysis was performed by Student's unpaired t-test or one-way anova, as appropriate. Post hoc comparisons between pairs of groups were performed by Bonferroni test, using GraphPad Software for Science (San Diego, CA, USA).
Results
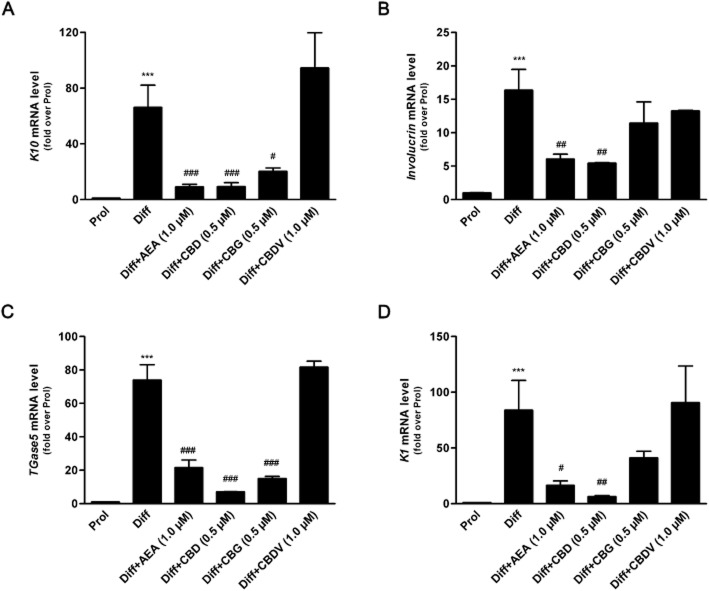
The effects of the three major phytocannabinoids CBD, CBG and CBDV on proliferating and differentiated HaCaT cells were tested and were compared to those of the endogenous cannabinoid AEA as a control (Maccarrone et al., 2003; Paradisi et al., 2008). In a preliminary set of dose-response experiments on K10 gene expression levels (Figure 1), the lowest effective dose of CBD (P < 0.001) and CBG (P < 0.05) was found to be 0.5 μM, whereas CBDV was ineffective up to 1.0 μM, previously found to be the lowest effective dose of AEA (Paradisi et al., 2008). Therefore, in all subsequent experiments CBD and CBG were used at 0.5 μM, and CBDV and AEA at 1.0 μM. By using qRT-PCR analysis, we showed a significant reduction in the expression of K10 and TGase5 genes after treatment of differentiated HaCaT cells with 0.5 μM CBD (P < 0.001) or CBG (P < 0.05 for K10; P < 0.001 for TGase5), as well as with 1.0 μM AEA (P < 0.001) (Figure 2). Also the expression of involucrin and K1 genes was significantly inhibited by CBD but not by CBG, under the same experimental conditions, once again resembling the effect of AEA. In contrast, 1.0 μM CBDV did not change the expression of any gene tested (Figure 2). Based on these findings, we chose to perform further analyses on K10 only, as done previously to test the effects of AEA (Paradisi et al., 2008). We also extended our study by investigating the most relevant effects of AEA, CBD and CBG on primary cells, NHEKs. Analysis of the results showed that all the treatments induced a consistent down-regulation of K10 gene expression (see Supporting Information Table S1). Unfortunately, difficulties in growing NHEKs prevented any further extension of our analyses in these primary cells. We investigated the molecular mechanism by which CBD and CBG affect K10 gene expression, and found that the effect of 0.5 μM CBD was reversed by 0.05 μM SR141716 (P < 0.05; Figure 3), a selective CB1 receptor antagonist (Pertwee et al., 2010), but not by 0.05 μM SR144528, a selective antagonist of CB2 receptors (Pertwee et al., 2010). In addition, 0.5 μM CPZ, a selective antagonist of TRPV1 receptors (De Petrocellis and Di Marzo, 2010), was ineffective in keeping with the absence of TRPV1 receptors in HaCaT cells (Maccarrone et al., 2003) (Figure 3). Collectively, these data suggest that CBD triggered a CB1-dependent mechanism that resembled that already observed for AEA (Paradisi et al., 2008). In contrst, the effect of CBG on K10 mRNA levels was not inhibited by any of the three selective receptor antagonists, supporting the involvement of a distinct transduction pathway (Figure 3). Incidentally, SR141716 and SR144528 were used at concentrations previously shown to block their specific targets in HaCaT cells (Maccarrone et al., 2003; Paradisi et al., 2008). Next, we assessed the methylation status of K10 gene, using a bisulfite-based methylation specific PCR assay. Indeed, it is known that gene expression is regulated by epigenetic mechanisms such as DNA methylation. As shown in Figure 4, the methylation level of K10 significantly decreased (P < 0.001) upon differentiation of proliferating HaCaT cells with TPA plus calcium. Interestingly, both CBD and CBG significantly increased (P < 0.001 for CBD; P < 0.05 for CBG) K10 gene methylation in differentiated cells (Figure 4), thus resembling the effect of AEA (Paradisi et al., 2008). The effect of CBD was due to a CB1-dependent mechanism, because it was prevented by SR141716 (P < 0.05; Figure 4). In contrast, CBG did not trigger CB1 signalling, because SR141716 did not counteract the effect of this phytocannabinoid on K10 gene (Figure 4). In addition, the overall methylation levels were measured in human keratinocytes by using an SssI methylase assay. Firstly, differentiation of HaCaT cells led to a significant reduction (P < 0.05, Figure 5A) in global DNA methylation; secondly, AEA (P < 0.01) and CBD (P < 0.05), but not CBG, increased DNA methylation levels in differentiated cells, up to those of proliferating cells (Figure 5A). Once again, the effect of CBD was reversed by SR141716 (P < 0.05), indicating a CB1-dependent mechanism. In contrast, the effect of CBG was not dependent on this receptor (Figure 5A). We also tested whether CBD and CBG could affect genomic DNA methylation through the regulation of DNMT activity. Similarly to AEA, CBD induced a slight increase (P = 0.4156) in DNMT activity in differentiating cells, whereas CBG induced a small (yet not significant; P = 0.1043) decrease in the activity of this enzyme in the same cells (Figure 5B). Finally, we demonstrated selective alterations in the gene expression of the various DNMTs in differentiated HaCaT cells, either untreated or after exposure to AEA, CBG and CBD (Table 2). In particular, we found that DNMT1 gene expression was significantly reduced (P = 0.0039) in differentiated cells and was up-regulated by AEA (P = 0.1014), CBD (P = 0.3290) and CBG (P = 0.0520), although this did not reach statistical significance. Consistently, densitometric analysis of DNMT1 levels revealed a reduction in the enzyme protein in differentiated cells, and a recovery towards that of proliferating cells after any treatment (Table 2). The levels of gene expression of all the other DNMTs analysed (DNMT3a, DNMT3b, DNMT3L) were not affected by any of the compounds tested under the same experimental conditions (Table 2).
Figure 1.
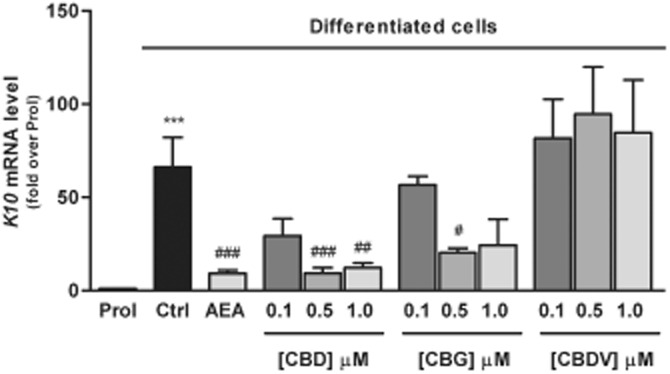
Expression of K10 gene in HaCaT cells. Keratinocytes were induced to differentiate by treatment with TPA plus calcium for 5 days. Differentiated HaCaT cells were treated with 1 μM AEA and different amounts (0.1–0.5–1.0 μM) of CBD, CBG and CBDV. K10 was detected by quantitative RT-PCR, under different conditions and with primers, as described in the Methods section. For the quantification of gene expression, β-actin was used as a housekeeping gene. The results are shown as fold induction over proliferating cells of three independent experiments. Prol, proliferating cells; Ctrl, differentiated cells. ***P < 0.001 versus Prol; ###P < 0.001 versus Ctrl; ##P < 0.01 versus Ctrl; #P < 0.05 versus Ctrl.
Figure 2.
Expression of epidermal differentiation-related genes in HaCaT cells. Differentiated HaCaT cells were treated with 1 μM AEA, 0.5 μM CBD, 0.5 μM CBG or 1.0 μM CBDV. K10 (A), involucrin (B), TGase5 (C) and K1 (D) were detected by quantitative RT-PCR, under condition and with primers described in the Methods section. The results are shown as fold induction over proliferating cells of three independent experiments. Prol, proliferating cells; Diff, differentiated cells. ***P < 0.001 versus Prol; ###P < 0.001 versus Diff; ##P < 0.01 versus Diff; #P < 0.05 versus Diff.
Figure 3.
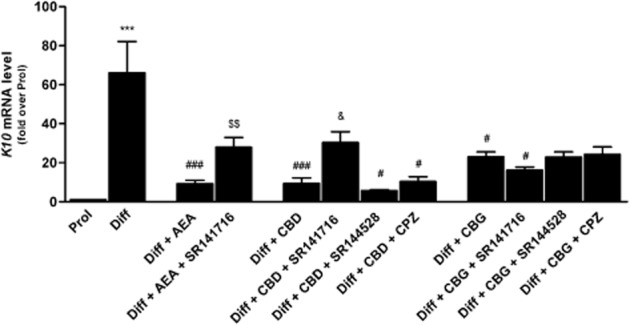
Effect of AEA (1.0 μM), CBD and CBG (both used at 0.5 μM), alone or in the presence of 0.05 μM SR141716, 0.05 μM SR144528 or 0.5 μM capsazepine (CPZ), on K10 gene expression in HaCaT cells. SR141716, SR144528 and CPZ were ineffective when used alone. Prol, proliferating cells; Diff, differentiated cells. ***P < 0.001 versus Prol; ###P < 0.001 versus Diff; #P < 0.05 versus Diff; $$P < 0.01 versus Diff + AEA; &P < 0.05 versus Diff + CBD.
Figure 4.
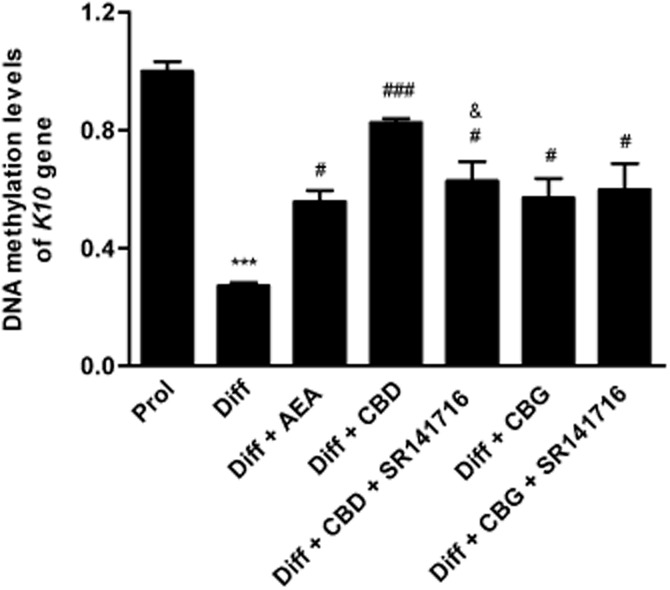
Methylation-specific primed PCR. DNA methylation levels of K10 gene in differentiated HaCaT cells treated with CBD and CBG (both used at 0.5 μM), alone or in the presence of SR141716 (0.05 μM). SR141716 was ineffective when used alone. The methylation status of the K10 gene was analysed as described in the Methods section. Prol, proliferating cells; Diff, differentiated cells. ***P < 0.001 versus Prol; ###P < 0.001 versus Diff; #P < 0.05 versus Diff; &P < 0.05 versus Diff + CBD.
Figure 5.
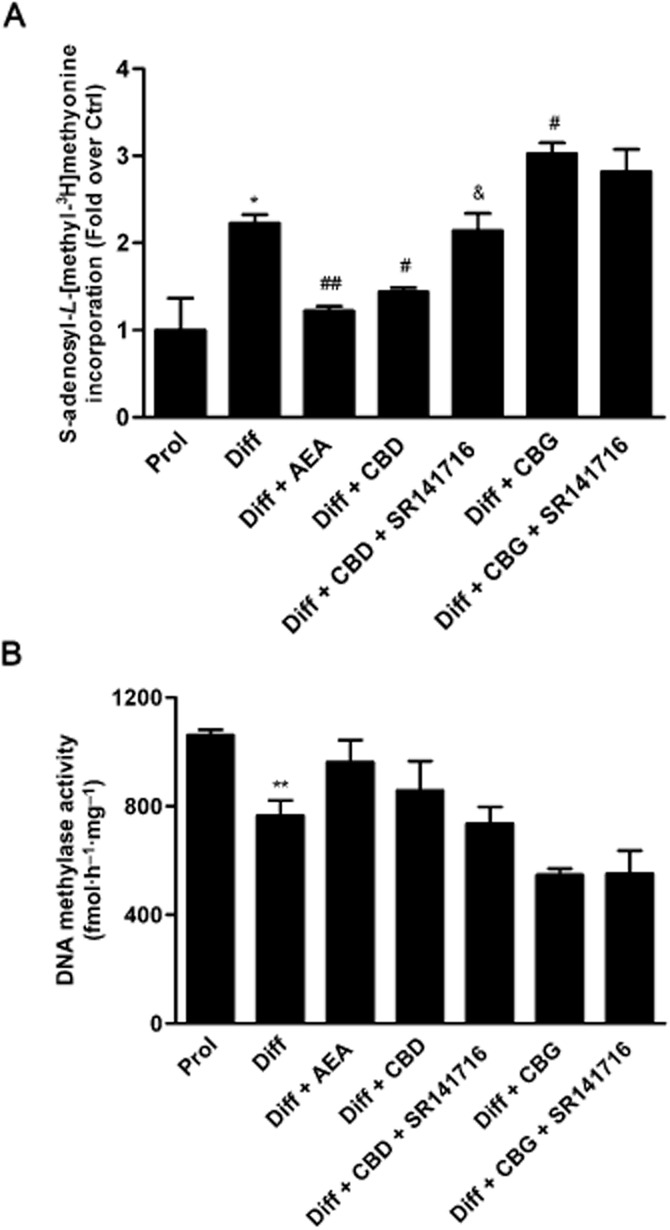
(A) Methylation levels of genomic DNA were measured by methyl-accepting assay with CpG methylase SssI, in the presence of S-adenosyl-L-[methyl-3H]-methyonine (see Methods for details). Higher levels of [3H]methyl group incorporated into DNA indicated lower level of genomic DNA methylation. Prol, proliferating cells; Diff, differentiated cells. *P < 0.05 versus Prol; ##P < 0.01 versus Diff; #P < 0.05 versus Diff; &P < 0.05 versus Diff + CBD. (B) Proliferating and differentiated keratinocytes treated with 1 μM AEA, 0.5 μM CBD or 0.5 μM CBG were lysed, and DNA methyltransferase activity was measured as described in the Methods section. Prol, proliferating cells; Diff, differentiated cells. **P < 0.01 versus Prol.
Table 2.
Effect of AEA, CBD and CBG on DNMTs gene expression and on DNMT1 protein levels
| mRNA level | Prola | Diffb | Diff + AEA | Diff + CBD | Diff + CBG |
|---|---|---|---|---|---|
| DNMT1 | 0.99 ± 0.11 | 0.44 ± 0.06c | 1.92 ± 0.76 | 1.24 ± 0.55 | 1.46 ± 0.47 |
| DNMT3a | 1.03 ± 0.20 | 1.40 ± 0.40 | 2.45 ± 1.12 | 1.77 ± 0.61 | 1.25 ± 0.35 |
| DNMT3b | 1.10 ± 0.32 | 1.07 ± 0.24 | 2.91 ± 1.45 | 2.65 ± 1.00 | 0.85 ± 0.27 |
| DNMT3L | 1.10 ± 0.28 | 1.82 ± 0.31 | 2.94 ± 1.23 | 2.48 ± 0.50 | 1.01 ± 0.28 |
| DNMT1 (protein level)d | 1.00 ± 0.11 | 0.74 ± 0.08 | 0.96 ± 0.12 | 1.13 ± 0.11 | 1.30 ± 0.15 |
Prol, proliferating keratinocytes.
Diff, differentiated keratinocytes.
P < 0.01 versus Prol.
Protein levels were quantified by densitometric analysis of three independent Western blots (see Supporting Information Figure S1 for a representative blot).
Discussion and conclusions
In this study, we showed that the expression of epidermal differentiation genes (i.e. keratins, involucrin and transglutaminase) is regulated by the phytocannabinoids CBD and CBG, but not CBDV, through distinct mechanisms. Indeed, the effect of CBD was dependent on CB1 cannabinoid receptors, similar that previously reported for AEA (Paradisi et al., 2008), whereas CBG did not affect either this or the other AEA-binding receptor subtype, the CB2 cannabinoid receptor. Moreover, CBG did not affect the transcription of involucrin and K1, but it down-regulated that of K10 and TGase5. In this context, it should be recalled that CBD and CBG also dose-dependently inhibit keratinocyte proliferation (Wilkinson and Williamson, 2007), although at an effective dose (>1 μM) higher than the optimal dose (0.5 μM) found here to reduce the differentiation markers K10 and TGase5. Additionally, we suggest that inhibition of epidermal differentiation elicited by CBD shares the same CB1-dependent mechanism of action as that of AEA. This seems remarkable, because CBD is generally reported to have a very low affinity (in the micromolar range) for CB1 and CB2 cannabinoid receptors, although independent investigations have recently shown that it can also behave as an inverse agonist or antagonist at the same receptors (Thomas et al., 2007; Castillo et al., 2010). Moreover, it should be recalled that CBD, like AEA, might enhance the biological activity of endogenous cannabinoids by increasing their release and/or by inhibiting their degradation. Such an ‘entourage effect’ (Ben-Shabat et al., 1998; Ligresti et al., 2006) may represent an additional indirect mechanism by which CBD might modulate CB1/CB2 signalling. On the other hand, the effects of CBG on K10 gene expression were not mediated by CB1 or CB2 cannabinoid receptors. In vitro studies have shown that CBG is also an α2-adrenoceptor agonist, and an antagonist of 6-HT1A (Cascio et al., 2010) and TRPV1 (De Petrocellis et al., 2008; De Petrocellis et al., 2011) receptors. Moreover, the possibility that these receptors as well as other eCBs targets like peroxisome PPAR-γ, which play a role in skin biology (Ellis et al., 2000; Bhagavathula et al., 2004; Kuenzli and Saurat, 2004), or GPR55 might be triggered by CBG remains to be explored. In this context, it should be mentioned that SR141716 also behaves as an agonist at GPR55 (Kapur et al., 2009), although there are no data on a possible involvement of this receptor in the epidermis. It should also be noted that recent findings have shown that the barrier recovery is delayed in CB1 knock-out mice, while it is accelerated in CB2 knock-out mice (Roelandt et al., 2012). Additionally, CB1 activation in human keratinocytes by high doses (2.5 and 10 μM) of arachidonoylcyclopropylamide for 24 h increased the mRNA level of K10 at high Ca2+ concentrations, while reducing K10 protein level under the same conditions (Roelandt et al., 2012). On the one hand, it can be proposed that knock-out animals might have developed different compensatory mechanisms that do not fully reflect the physiology of normal (wild-type) keratinocytes. On the other hand, the opposite effects of arachidonoylcyclopropylamide on human keratinocytes (so called ‘cannabinoid paradox’) at doses well above those used here might be due to complex mechanisms, which may be related to eCBs signalling mechanisms that inhibit mRNA translation (Roelandt et al., 2012), as well as to reduced cell viability and proliferation induced by eCBs and phytocannabinoids at concentrations >1 μM (Siegmund et al., 2006; Wilkinson and Williamson, 2007; Pucci et al., 2011; Tóth et al., 2011). At any rate, consistent with our previous findings with AEA (Paradisi et al., 2008), here we show that changes in K10 gene expression induced by CBD, but not CBG, are due to increased methylation of genomic DNA. It is noteworthy that an inverse correlation between DNA methylation and the expression of differentiating genes has already been identified in human keratinocytes (Engelkamp et al., 1993; Elder and Zhao, 2002), although a role for phytocannabinoids in this process is unprecedented.
We also observed an overall reduction in DNA methylation in differentiating keratinocytes, in agreement with an early study showing that DNA methylation in human keratinocytes varies depending on the differentiation state, whereby there is a lower methylcytosine content in the DNA of differentiated versus undifferentiated cells (Veres et al., 1989). CBD was able to reverse these changes and to increase global DNA methylation in differentiated cells, thus suggesting a broader effect, not restricted to the K10 gene alone.
Finally, we evaluated the effect of CBD and CBG, as well as of AEA, on the expression of DNMTs, the enzymes that catalyse DNA methylation (Baylin and Herman, 2000), in order to better dissect the role of DNA methylation on the modulation of epidermal differentiation by phytocannabinoids. We observed that the induction of HaCaT cell differentiation for 5 days determined a selective and significant reduction of DNMT1 gene expression. Consistently, DNMT1 was previously found to be down-regulated during epidermal differentiation (Sen et al., 2010). CBG and CBD, like AEA, were able to reverse these changes, and thus to induce an up-regulation, even if not in a significant manner, of DNMT1 in line with the observed increase in DNA methylation and reduction in mRNA levels. It is important to point out that these changes were selective, since we did not observe any alteration of DNMT3a, DNMT3b and DNMT3L gene expression whatever the treatment, or upon cell differentiation alone. Consistently, DNMT3a and DNMT3b are known to mediate methylation-independent gene repression (Bachman et al., 2001). Overall, our data confirm that DNA methylation is altered during cell differentiation, and that DNMT1 is required to maintain a progenitor state. In addition, our results also suggest changes in cellular maintenance but not de novo methyltransferase activity, because DNMT3a and 3b can methylate unmethylated DNA, and are thus referred to as de novo DNMTs. In contrast, DNMT1 primarily functions to maintain DNA methylation by preferentially methylating hemimethylated DNA (Dodge et al., 2005).
Taken together, the present data clearly identify the phytocannabinoids CBD and CBG as transcriptional repressors, further suggesting a role for eCBs signalling in the control of cell proliferation and differentiation (Maccarrone et al., 2003; Aguado et al., 2006; Galve-Roperh et al., 2006; Laezza et al., 2006; Matias et al., 2006; Ofek et al., 2006; Cavuoto et al., 2007; Telek et al., 2007).
In conclusion, understanding the nature of genetic and epigenetic interactions in the regulation of epidermal differentiation, and clarifying how phytocannabinoids could possibly modulate these effects represent a major challenge in the skin biology arena. Our data might pave the way to the development of preventive strategies, for example aimed at reducing allergic inflammation, or to the design of new and more effective therapeutics for the management of skin cancer. Plant-derived cannabinoids that are devoid of psychoactive effects appear to be good candidates for these purposes. In general, our findings also suggest that phytocannabinoids might act through epigenetic mechanisms in other human diseases (e.g. multiple sclerosis), where their administration has been proven to be beneficial (Rog, 2010). Yet, major differences in signalling mechanisms triggered by different phytocannabinoids that might act through CB1-dependent (CBD), CB1-independent (CBG), or have no effect at all (CBDV), call for a careful investigation into their activity before any exploration of their therapeutic potential.
Finally, we believe that the importance of our findings goes beyond the role of phytocannabinoids in keratinocyte differentiation that we have shown here. In fact, DNA methylation is an epigenetic mechanism involved in the regulation of different cellular processes, including embryonic development, transcription, chromatin structure, X chromosome inactivation, genomic imprinting and chromosome stability. A reduction in DNA methylation has been demonstrated in different human diseases, most notably cancer (Robertson, 2005). Therefore, natural compounds that act as DNA methyltransferase enhancers, like phytocannabinoids, may well be exploited for elucidating mechanisms beyond those involved in skin biology.
Acknowledgments
We are grateful to GW Pharmaceuticals for financial support.
Glossary
- AEA
anandamide
- 2-AG
2-arachidonoylglycerol
- CB1
cannabinoid receptor type I
- CB2
cannabinoid receptor type II
- CBD
cannabidiol
- CBDV
cannabidivarin
- CBG
cannabigerol
- CPZ
capsazepine
- DAGL
diacylglycerol lipase
- DNMT
DNA methyltransferase
- eCBs
endocannabinoids
- ECS
endocannabinoid system
- FAAH
fatty acid amide hydrolase
- K1
keratin 1
- K10
keratin 10
- NAPE-PLD
N-acyl-phosphatidylethanolamines-specific phospholipase D
- NHEK
normal human epidermal keratinocytes
- MAGL
monoacylglycerol lipase
- SR141716
N-piperidino-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-3-pyrazole carboxamide
- SR144528
N-[(1)-endo-1,3,3-trimethy-1-bicyclo [2.2.1]-heptan-2-yl]5-(4-chloro-3-methyl-phenyl)-1-(4-methyl-benzyl)-pyrazole-3-carboxamide
- TGase 5
transglutaminase 5
- THC
Δ9-tetrahydrocannbinol
- TPA
12-O-tetradecanoylphorbol 13-acetate
- TRPV1
transient receptor potential vanilloid 1
Conflict of interest
The authors declare that there are no conflicts of interest.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Figure S1 Western blot analysis of DNMT1 and Lamin A in proliferating and differentiated HaCaT cells treated with AEA, CBD or CBG.
Table S1 Effect of CBD and CBG on K10 gene expression in NHEK cells.
References
- Aguado T, Palazuelos J, Monory K, Stella N, Cravatt B, Lutz B, et al. The endocannabinoid system promotes astroglial differentiation by acting on neural progenitor cells. J Neurosci. 2006;26:1551–1561. doi: 10.1523/JNEUROSCI.3101-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn K, McKinney MK, Cravatt BF. Enzymatic pathways that regulate endocannabinoid signaling in the nervous system. Chem Rev. 2008;108:1687–1707. doi: 10.1021/cr0782067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Mathie A, Peters JA. Guide to receptors and channels (GRAC), 5th edition. Br J Pharmacol. 2011;164(Suppl. 1):S1–S324. doi: 10.1111/j.1476-5381.2011.01649_1.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman KE, Rountree MR, Baylin SB. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- Baker D, Pryce G, Davies WL, Hiley CR. In silico patent searching reveals a new cannabinoid receptor. Trends Pharmacol Sci. 2006;27:1–4. doi: 10.1016/j.tips.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- Ben-Shabat S, Fride E, Sheskin T, Tamiri T, Rhee MH, Vogel Z, et al. An entourage effect: inactive endogenous fatty acid glycerol esters enhance 2-arachidonoyl-glycerol cannabinoid activity. Eur J Pharmacol. 1998;353:23–31. doi: 10.1016/s0014-2999(98)00392-6. [DOI] [PubMed] [Google Scholar]
- Berdyshev EV, Schmid PC, Dong Z, Schmid HH. Stress-induced generation of N-acylethanolamines in mouse epidermal JB6 P+ cells. Biochem J. 2000;346:369–374. [PMC free article] [PubMed] [Google Scholar]
- Bhagavathula N, Nerusu KC, Lal A, Ellis CN, Chittiboyina A, Avery MA, et al. Rosiglitazone inhibits proliferation, motility, and matrix metalloproteinase production in keratinocytes. J Invest Dermatol. 2004;122:130–139. doi: 10.1046/j.0022-202X.2003.22111.x. [DOI] [PubMed] [Google Scholar]
- Bíró T, Tóth BI, Haskó G, Paus R, Pacher P. The endocannabinoid system of the skin in health and disease: novel perspectives and therapeutic opportunities. Trends Pharmacol Sci. 2009;30:411–420. doi: 10.1016/j.tips.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broday L, Lee YW, Costa M. 5-azacytidine induces transgene silencing by DNA methylation in Chinese hamster cells. Mol Cell Biol. 1999;19:3198–3204. doi: 10.1128/mcb.19.4.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Oddi S, Terrinoni A, Paradisi A, Ranalli M, Finazzi-Agro A, et al. Transglutaminase 5 cross-links loricrin, involucrin, and small proline-rich proteins in vitro. J Biol Chem. 2001;276:35014–35023. doi: 10.1074/jbc.M010157200. [DOI] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–340. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- Casanova ML, Blázquez C, Martínez-Palacio J, Villanueva C, Fernández-Aceñero MJ, Huffman JW, et al. Inhibition of skin tumor growth and angiogenesis in vivo by activation of cannabinoid receptors. J Clin Invest. 2003;111:43–50. doi: 10.1172/JCI16116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascio MG, Gauson LA, Stevenson LA, Ross RA, Pertwee RG. Evidence that the plant cannabinoid cannabigerol is a highly potent alpha2-adrenoceptor agonist and moderately potent 5HT1A receptor antagonist. Br J Pharmacol. 2010;159:129–141. doi: 10.1111/j.1476-5381.2009.00515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo A, Tolón MR, Fernández-Ruiz J, Romero J, Martinez-Orgado J. The neuroprotective effect of cannabidiol in an in vitro model of newborn hypoxic-ischemic brain damage in mice is mediated by CB(2) and adenosine receptors. Neurobiol Dis. 2010;37:434–440. doi: 10.1016/j.nbd.2009.10.023. [DOI] [PubMed] [Google Scholar]
- Cavuoto P, McAinch AJ, Hatzinikolas G, Cameron-Smith D, Wittert GA. Effects of cannabinoid receptors on skeletal muscle oxidative pathways. Mol Cell Endocrinol. 2007;267:63–66. doi: 10.1016/j.mce.2006.12.038. [DOI] [PubMed] [Google Scholar]
- Chen M, Chen ZQ, Cui PG, Yao X, Li YM, Li AS, et al. The methylation pattern of p16INK4a gene promoter in psoriatic epidermis and its clinical significance. Br J Dermatol. 2008;158:987–993. doi: 10.1111/j.1365-2133.2008.08505.x. [DOI] [PubMed] [Google Scholar]
- D'Addario C, Ming Y, Ogren SO, Terenius L. The role of acetaldehyde in mediating effects of alcohol on expression of endogenous opioid system genes in a neuroblastoma cell line. FASEB J. 2008;22:662–670. doi: 10.1096/fj.07-8346com. [DOI] [PubMed] [Google Scholar]
- D'Addario C, Di Francesco A, Arosio B, Gussago C, Dell'Osso B, Bari M, et al. Epigenetic regulation of fatty acid amide hydrolase in Alzheimer disease. PLoS ONE. 2012;7:e39186. doi: 10.1371/journal.pone.0039186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Petrocellis L, Di Marzo V. Non-CB1, non-CB2 receptors for endocannabinoids, plant cannabinoids, and synthetic cannabimimetics: focus on G-protein-coupled receptors and transient receptor potential channels. J Neuroimmu Pharmacol. 2010;5:103–121. doi: 10.1007/s11481-009-9177-z. [DOI] [PubMed] [Google Scholar]
- De Petrocellis L, Vellani V, Schiano-Moriello A, Marini P, Magherini PC, Orlando P, et al. Plant-derived cannabinoids modulate the activity of transient receptor potential channels of ankyrin type-1 and melastatin type-8. J Pharmacol Exp Ther. 2008;325:1007–1015. doi: 10.1124/jpet.107.134809. [DOI] [PubMed] [Google Scholar]
- De Petrocellis L, Ligresti A, Moriello AS, Allarà M, Bisogno T, Petrosino S, et al. Effects of cannabinoids and cannabinoid-enriched Cannabis extracts on TRP channels and endocannabinoid metabolic enzymes. Br J Pharmacol. 2011;163:1479–1494. doi: 10.1111/j.1476-5381.2010.01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marzo V. Endocannabinoids: synthesis and degradation. Rev Physiol Biochem Pharmacol. 2008;160:1–24. doi: 10.1007/112_0505. [DOI] [PubMed] [Google Scholar]
- Di Marzo V, Petrosino S. Endocannabinoids and the regulation of their levels in health and disease. Curr Opin Lipidol. 2007;18:129–140. doi: 10.1097/MOL.0b013e32803dbdec. [DOI] [PubMed] [Google Scholar]
- Dodge JE, Okano M, Dick F, Tsujimoto N, Chen T, Wang S, et al. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J Biol Chem. 2005;280:17986–17991. doi: 10.1074/jbc.M413246200. [DOI] [PubMed] [Google Scholar]
- Ehrlich M. Expression of various genes is controlled by DNA methylation during mammalian development. J Cell Biochem. 2003;88:899–910. doi: 10.1002/jcb.10464. [DOI] [PubMed] [Google Scholar]
- Elder JT, Zhao X. Evidence for local control of gene expression in the epidermal differentiation complex. Exp Dermatol. 2002;11:406–412. doi: 10.1034/j.1600-0625.2002.110503.x. [DOI] [PubMed] [Google Scholar]
- Ellis CN, Varani J, Fisher GJ, Zeigler ME, Pershadsingh HA, Benson SC, et al. Troglitazone improves psoriasis and normalizes models of proliferative skin disease: ligands for peroxisome proliferator-activated receptor-gamma inhibit keratinocyte proliferation. Arch Dermatol. 2000;136:609–616. doi: 10.1001/archderm.136.5.609. [DOI] [PubMed] [Google Scholar]
- Engelkamp D, Schafer BW, Mattei MG, Erne P, Heizmann CW. Six S100 genes are clustered on human chromosome 1q21: identification of two genes coding for the two previously unreported calcium-binding proteins S100D and S100E. Proc Natl Acad Sci U S A. 1993;90:6547–6551. doi: 10.1073/pnas.90.14.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galve-Roperh I, Aguado T, Rueda D, Velasco G, Guzman M. Endocannabinoids: a new family of lipid mediators involved in the regulation of neural cell development. Curr Pharm Des. 2006;12:2319–2325. doi: 10.2174/138161206777585139. [DOI] [PubMed] [Google Scholar]
- Hill AJ, Williams CM, Whalley BJ, Stephens GJ. Phytocannabinoids as novel therapeutic agents in CNS disorders. Pharmacol Ther. 2012a;133:79–97. doi: 10.1016/j.pharmthera.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Hill AJ, Mercier MS, Hill TD, Glyn SE, Jones NA, Yamasaki Y, et al. Cannabidivarin is anticonvulsant in mouse and rat. Br J Pharmacol. 2012b;167:1629–1642. doi: 10.1111/j.1476-5381.2012.02207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo AA, Borrelli F, Capasso R, Di Marzo V, Mechoulam R. Non-psychotropic plant cannabinoids: new therapeutic opportunities from an ancient herb. Trends Pharmacol Sci. 2009;30:515–527. doi: 10.1016/j.tips.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jones NA, Hill AJ, Smith I, Bevan SA, Williams CM, Whalley BJ, et al. Cannabidiol displays antiepileptiform and antiseizure properties in vitro and in vivo. J Pharmacol Exp Ther. 2010;332:569–577. doi: 10.1124/jpet.109.159145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinin A, Marekov LN, Steinert PM. Assembly of the epidermal cornified cell envelope. J Cell Sci. 2001;114:3069–3070. doi: 10.1242/jcs.114.17.3069. [DOI] [PubMed] [Google Scholar]
- Kapur A, Zhao P, Sharir H, Bai Y, Caron MG, Barak LS, et al. Atypical responsiveness of the orphan receptor GPR55 to cannabinoid ligands. J Biol Chem. 2009;284:29817–29827. doi: 10.1074/jbc.M109.050187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsak M, Gaffal G, Date R, Wang-Eckhardt L, Rehnelt J, Petrosino S, et al. Attenuation of allergic contact dermatitis through the endocannabinoid system. Science. 2007;316:1494–1497. doi: 10.1126/science.1142265. [DOI] [PubMed] [Google Scholar]
- Klein TW. Cannabinoid-based drugs as anti-inflammatory therapeutics. Nat Rev Immunol. 2005;5:400–411. doi: 10.1038/nri1602. [DOI] [PubMed] [Google Scholar]
- Kogan NW. Cannabinoids and cancer. Mini Rev Med Chem. 2005;5:941–952. doi: 10.2174/138955705774329555. [DOI] [PubMed] [Google Scholar]
- Kuenzli SS, Saurat JH. Peroxisome proliferator-activated receptors as new molecular targets in psoriasis. Curr Drug Targets Inflamm Allergy. 2004;3:205–211. doi: 10.2174/1568010043343976. [DOI] [PubMed] [Google Scholar]
- Kupczyk P, Reich A, Szepietowski JC. Cannabinoid system in the skin – a possible target for future therapies in dermatology. Exp Dermatol. 2009;18:669–679. doi: 10.1111/j.1600-0625.2009.00923.x. [DOI] [PubMed] [Google Scholar]
- Laezza C, Pisanti S, Crescenzi E, Bifulco M. Anandamide inhibits Cdk2 and activates Chk1 leading to cell cycle arrest in human breast cancer cells. FEBS Lett. 2006;580:6076–6082. doi: 10.1016/j.febslet.2006.09.074. [DOI] [PubMed] [Google Scholar]
- Lal S, Prasad N, Ryan M, Tangri S, Silverberg MS, Gordon A, et al. Cannabis use amongst patients with inflammatory bowel disease. Eur J Gastroenterol Hepatol. 2011;23:891–896. doi: 10.1097/MEG.0b013e328349bb4c. [DOI] [PubMed] [Google Scholar]
- Ligresti A, Moriello AS, Starowicz K, Matias I, Pisanti S, De Petrocellis L, et al. Antitumor activity of plant cannabinoids with emphasis on the effect of cannabidiol on human breast carcinoma. J Pharmacol Exp Ther. 2006;318:1375–1387. doi: 10.1124/jpet.106.105247. [DOI] [PubMed] [Google Scholar]
- Lodzki M, Godin B, Rakou L, Mechoulam R, Gallily R, Touitou E. Cannabidiol-transdermal delivery and anti-inflammatory effect in a murine model. J Control Release. 2003;93:377–387. doi: 10.1016/j.jconrel.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Lopez J, Percharde M, Coley HM, Webb A, Crook T. The context and potential of epigenetics in oncology. Br J Cancer. 2009;100:571–577. doi: 10.1038/sj.bjc.6604930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Katsaros D, de la Longrais IA, Sochirca O, Yu H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007;67:10117–10122. doi: 10.1158/0008-5472.CAN-07-2544. [DOI] [PubMed] [Google Scholar]
- Lyon SB, Buonocore L, Miller M. Naturally occurring methylation inhibitor: DNA hypomethylation and hemoglobin synthesis in human K562 cells. Mol Cell Biol. 1987;7:1759–1763. doi: 10.1128/mcb.7.5.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccarrone M, Di Rienzo M, Battista N, Gasperi V, Guerrieri P, Rossi A, et al. The endocannabinoid system in human keratinocytes. Evidence that anandamide inhibits epidermal differentiation through CB1 receptor-dependent inhibition of protein kinase C, activation protein-1, and transglutaminase. J Biol Chem. 2003;278:33896–33903. doi: 10.1074/jbc.M303994200. [DOI] [PubMed] [Google Scholar]
- Maccarrone M, Dainese E, Oddi S. Intracellular trafficking of anandamide: new concepts for signaling. Trends Biochem Sci. 2010;35:301–308. doi: 10.1016/j.tibs.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Matias I, Gonthier MP, Orlando P, Martiadis V, De Petrocellis L, Cervino C, et al. Regulation, function, and dysregulation of endocannabinoids in models of adipose and beta-pancreatic cells and in obesity and hyperglycemia. J Clin Endocrinol Metab. 2006;91:3171–3180. doi: 10.1210/jc.2005-2679. [DOI] [PubMed] [Google Scholar]
- Millington GW. Epigenetics and dermatological disease. Pharmacogenomics. 2008;9:1835–1850. doi: 10.2217/14622416.9.12.1835. [DOI] [PubMed] [Google Scholar]
- Nemes Z, Steinert PM. Bricks and mortar of the epidermal barrier. Exp Mol Med. 1999;31:5–19. doi: 10.1038/emm.1999.2. [DOI] [PubMed] [Google Scholar]
- Oddi S, Bari M, Battista N, Barsacchi D, Cozzani I, Maccarrone M. Confocal microscopy and biochemical analysis reveal spatial and functional separation between anandamide uptake and hydrolysis in human keratinocytes. Cell Mol Life Sci. 2005;62:386–395. doi: 10.1007/s00018-004-4446-8. [DOI] [PubMed] [Google Scholar]
- Ofek O, Karsak M, Leclerc N, Fogel M, Frenkel B, Wright K, et al. Peripheral cannabinoid receptor, CB2, regulates bone mass. Proc Natl Acad Sci U S A. 2006;103:696–701. doi: 10.1073/pnas.0504187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradisi A, Pasquariello N, Barcaroli D, Maccarrone M. Anandamide regulates keratinocyte differentiation by inducing DNA methylation in a CB1 receptor-dependent manner. J Biol Chem. 2008;283:6005–6012. doi: 10.1074/jbc.M707964200. [DOI] [PubMed] [Google Scholar]
- Pasquariello N, Oddi S, Malaponti M, Maccarrone M. Regulation of gene transcription and keratinocyte differentiation by anandamide. Vitam Horm. 2009;81:441–446. doi: 10.1016/S0083-6729(09)81017-0. [DOI] [PubMed] [Google Scholar]
- Paus R, Schmelz M, Bíró T, Steinhoff M. Frontiers in pruritus research: scratching the brain for more effective itch therapy. J Clin Invest. 2006;116:1174–1186. doi: 10.1172/JCI28553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertwee RG, Howlett AC, Abood ME, Alexander SP, Di Marzo V, et al. International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid receptors and their ligands: beyond CB1 and CB2. Pharmacol Rev. 2010;62:588–631. doi: 10.1124/pr.110.003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosino S, Cristino L, Karsak M, Gaffal E, Ueda N, Tüting T, et al. Protective role of palmitoylethanolamide in contact allergic dermatitis. Allergy. 2010;65:698–711. doi: 10.1111/j.1398-9995.2009.02254.x. [DOI] [PubMed] [Google Scholar]
- Pucci M, Pirazzi V, Pasquariello N, Maccarrone M. Endocannabinoid signaling and epidermal differentiation. Eur J Dermatol. 2011;2:29–34. doi: 10.1684/ejd.2011.1266. [DOI] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Rodríguez de Fonseca F, Del Arco I, Bermudez-Silva FJ, Bilbao A, Cippitelli A, Navarro M. The endocannabinoid system: physiology and pharmacology. Alcohol Alcohol. 2005;40:2–14. doi: 10.1093/alcalc/agh110. [DOI] [PubMed] [Google Scholar]
- Roelandt T, Heughebaert C, Bredif S, Giddelo C, Baudouin C, Msika P, et al. Cannabinoid receptors 1 and 2 oppositely regulate epidermal permeability barrier status and differentiation. Exp Dermatol. 2012;21:688–693. doi: 10.1111/j.1600-0625.2012.01561.x. [DOI] [PubMed] [Google Scholar]
- Rog DJ. Cannabis-based medicines in multiple sclerosis – a review of clinical studies. Immunobiology. 2010;215:658–672. doi: 10.1016/j.imbio.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Rosl F, Durst M, Zur Hausen H. Selective suppression of human papillomavirus transcription in non-tumorigenic cells by 5-azacytidine. EMBO J. 1988;7:1321–1328. doi: 10.1002/j.1460-2075.1988.tb02947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Cathelineau C, Cavey MT, Dionisius V, Michel S, Shroot B, et al. Sodium butyrate selectively antagonizes the inhibitory effect of retinoids on cornified envelope formation in cultured human keratinocytes. J Cell Physiol. 1989;140:281–287. doi: 10.1002/jcp.1041400213. [DOI] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund SV, Seki E, Osawa Y, Uchinami H, Cravatt BF, Schwabe RF. Fatty acid amide hydrolase determines anandamide-induced cell death in the liver. J Biol Chem. 2006;281:10431–10438. doi: 10.1074/jbc.M509706200. [DOI] [PubMed] [Google Scholar]
- Solinas M, Massi P, Cantelmo A, Cattaneo M, Cammarota R, Bartolini D, et al. Cannabidiol inhibits angiogenesis by multiple mechanisms. Br J Pharmacol. 2012;167:1218–1231. doi: 10.1111/j.1476-5381.2012.02050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiano-Coico L, Helm RE, McMahon CK, Pagan-Charry I, LaBruna A, Piraino V, et al. Sodium-N-butyrate induces cytoskeletal rearrangements and formation of cornified envelopes in cultured adult human keratinocytes. Cell Tissue Kinet. 1989;22:361–375. doi: 10.1111/j.1365-2184.1989.tb00221.x. [DOI] [PubMed] [Google Scholar]
- Stander S, Schmelz M, Metze D, Luger T, Rukwied R. Distribution of cannabinoid receptor 1 (CB1) and 2 (CB2) on sensory nerve fibers and adnexal structures in human skin. J Dermatol Sci. 2005;38:177–188. doi: 10.1016/j.jdermsci.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Telek A, Biro T, Bodo E, Toth BI, Borbiro I, Kunos G, et al. Inhibition of human hair follicle growth by endo- and exocannabinoids. FASEB J. 2007;21:3534–3541. doi: 10.1096/fj.06-7689com. [DOI] [PubMed] [Google Scholar]
- Thomas A, Baillie GL, Phillips AM, Razdan RK, Ross RA, Pertwee RG. Cannabidiol displays unexpectedly high potency as an antagonist of CB1 and CB2 receptor agonist. Br J Pharmacol. 2007;150:613–623. doi: 10.1038/sj.bjp.0707133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth BI, Dobrosi N, Dajnoki A, Czifra G, Oláh A, Szöllosi AG, et al. Endocannabinoids modulate human epidermal keratinocyte proliferation and survival via the sequential engagement of cannabinoid receptor-1 and transient receptor potential vanilloid-1. J Invest Dermatol. 2011;131:1095–1104. doi: 10.1038/jid.2010.421. [DOI] [PubMed] [Google Scholar]
- Ueda N, Tsuboi K, Uyama T, Ohnishi T. Biosynthesis and degradation of the endocannabinoid 2-arachidonoylglycerol. Biofactors. 2011;37:1–7. doi: 10.1002/biof.131. [DOI] [PubMed] [Google Scholar]
- Veres DA, Wilkins L, Coble DW, Lyon SB. DNA methylation and differentiation of human keratinocytes. J Invest Dermatol. 1989;93:687–690. doi: 10.1111/1523-1747.ep12319883. [DOI] [PubMed] [Google Scholar]
- Wilkinson JD, Williamson EM. Cannabinoids inhibit human keratinocyte proliferation through a non-CB1/CB2 mechanism and have a potential therapeutic value in the treatment of psoriasis. J Dermatol Sci. 2007;45:87–92. doi: 10.1016/j.jdermsci.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Zhang X. Genome-wide association study of skin complex diseases. J Dermatol Sci. 2012;6:89–97. doi: 10.1016/j.jdermsci.2012.02.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.