Abstract
To investigate the paternal population history of New Guinea, 183 individuals from 11 regional populations of West New Guinea (WNG) and 131 individuals from Papua New Guinea (PNG) were analyzed at 26 binary markers and seven short-tandem-repeat loci from the nonrecombining part of the human Y chromosome and were compared with 14 populations of eastern and southeastern Asia, Polynesia, and Australia. Y-chromosomal diversity was low in WNG compared with PNG and with most other populations from Asia/Oceania; a single haplogroup (M-M4) accounts for 75% of WNG Y chromosomes, and many WNG populations have just one Y haplogroup. Four Y-chromosomal lineages (haplogroups M-M4, C-M208, C-M38, and K-M230) account for 94% of WNG Y chromosomes and 78% of all Melanesian Y chromosomes and were identified to have most likely arisen in Melanesia. Haplogroup C-M208, which in WNG is restricted to the Dani and Lani, two linguistically closely related populations from the central and western highlands of WNG, was identified as the major Polynesian Y-chromosome lineage. A network analysis of associated Y-chromosomal short-tandem-repeat haplotypes suggests two distinct population expansions involving C-M208—one in New Guinea and one in Polynesia. The observed low levels of Y-chromosome diversity in WNG contrast with high levels of mtDNA diversity reported for the same populations. This most likely reflects extreme patrilocality and/or biased male reproductive success (polygyny). Our data further provide evidence for primarily female-mediated gene flow within the highlands of New Guinea but primarily male-mediated gene flow between highland and lowland/coastal regions.
Introduction
The western half of the island of New Guinea, (officially called “Papua,” formerly “Irian Jaya,” also known as “West Papua”), hereafter called “West New Guinea” (WNG), comprises an area of ∼420,000 km2 and contains ∼2,200,000 people belonging to 256 mostly Papuan-speaking language groups (Grimes 2000; Ethnologue Web site). In contrast to many regions in the eastern half of New Guinea (i.e., Papua New Guinea [PNG]), the traditional lifestyle has been maintained in WNG until very recently, and, indeed, many WNG areas remain virtually untouched by outside influence, especially in the highlands and the southern lowlands/coastal areas (Gardner and Heider 1969; Heider 1970; Mitton 1983; van Enk and de Vries 1997). In addition, the Austronesian expansion, a major population movement of eastern Asian Austronesian-speaking people, which, on the basis of linguistic and archaeological evidence, reached the northern, eastern, and southeastern coast of the New Guinea mainland ∼3,500 years ago, appears to have had no influence on the highlands, southern lowlands, and southern coast of WNG (Bellwood 1978; Bellwood 1989). This makes WNG, especially the central and southern regions, a suitable area for investigation of the early human population history of New Guinea, which, according to archaeological evidence, goes back at least 30,000–40,000 years (Groube et al. 1986; Allen et al. 1988; Wickler and Spriggs 1988; Pavlides and Gosden 1994).
Recently, a detailed study of mtDNA variation was performed on ∼200 individuals from 12 regional populations (six language families) of WNG (Tommaseo-Ponzetta et al. 2002). This study revealed high genetic heterogeneity with respect to mtDNA hypervariable region 1 (HV1) sequences, as well as phylogenetic evidence for the great antiquity and long-term isolation of WNG populations. The intergenic COII/tRNALys 9-bp deletion, which is an mtDNA marker for the Austronesian expansion (Redd et al. 1995), was not observed in the entire WNG sample.
In the same way that mtDNA is informative for investigating maternal population history, the analysis of genetic markers from the nonrecombining part of the human Y chromosome enables characterization of paternal lineages. Compared with mtDNA, in general, Y-chromosomal markers tend to exhibit a higher degree of population specificity (Seielstad et al. 1998) and, hence, may be more informative for tracing population history. Recent Y-chromosome analyses performed by us and by others identified paternal lineages that appear to be restricted to Melanesian/eastern Indonesian populations, although all such studies to date include population samples from mainland/island PNG and do not consider populations from WNG, except for a single population sample from the Bird's Head area (Underhill et al. 1997, 2000; Hurles et al. 1998, 2002; Su et al. 2000; Capelli et al. 2001; Kayser et al. 2001a).
We therefore conducted a combined Y-chromosome analysis of 26 binary markers, selected to be informative in Asia/Oceania, as well as seven STR loci, in 183 males from 11 regional populations from the highlands and the southern lowlands/southern coast of WNG. These data were compared with those from 131 males from PNG and 479 additional samples from 14 populations of east and southeastern Asia, Polynesia, and Australia. Most of the WNG samples have previously been sequenced at the HV1 of mtDNA (Tommaseo-Ponzetta et al. 2002), which enables a comparison of paternal and maternal lineages. Also, since the regional WNG populations studied belong to different language families, live at different altitudes, and have different life styles (i.e., horticulture, sago gathering, sea and river fishing, or hunting), the present study allows an investigation of different environmental and cultural influences on the genetic structure of WNG populations.
Samples and Methods
Samples
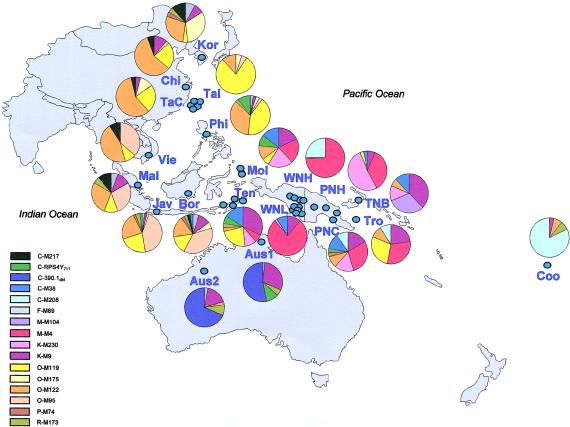
Samples from 183 males from the following 11 WNG populations were analyzed (sample sizes are given in parentheses): Dani or Grant Valley Dani (12), Lani or Western Dani (12), Yali or Northern Ngalik (5), Una (46), Ketengban (19), Awyu (10), Kombai/Korowai (13), Muyu or Kati (8), Mappi (10), Asmat (20), and Citak (28). Samples were collected by M.T.P. in 1996, with informed consent, and are partly identical to those recently studied for mtDNA variation (Tommaseo-Ponzetta et al. 2002). All populations speak Papuan languages that belong to six language families within the Trans New Guinea language phylum: Dani-Kwerba (Dani, Lani, and Yali), Mek (Una and Ketengban), Awyu-Dumut (Awyu, Kombai, and Korowai), Ok (Muyu), Yaqay (Mappi), and Asmat-Kamoro (Asmat and Citak). Populations living in the WNG highlands (central and western highlands: Dani, Lani, and Yali; eastern highlands: Una and Ketengban) practice horticulture (extensive gardening), whereas those living in the southern lowlands (Awyu, Citak, Mappi, Muyu, Kombai, and Korowai) and on the southern coast (Asmat) are sago gatherers and occasional hunters or fishers. Further description of the WNG population samples can be found elsewhere (Tommaseo-Ponzetta et al. 2002). An additional 610 male samples from 18 populations across eastern Asia, mainland and island southeastern Asia, mainland and island Melanesia, Polynesia, and Australia that were analyzed previously at a subset of the markers used here (Kayser et al. 2000a, 2001a) were also included in the present study (fig. 1).
Figure 1.
Y-chromosome haplogroups and their frequency distribution in WNG and an additional 18 populations from Asia/Oceania. Population abbreviations are as follows: Kor = Korea, Chi = China, TaC = Taiwan Chinese, Tai = Taiwan Aborigines, Phi = Philippines, Vie = Vietnam, Mal = Malaysia, Jav = Java, Bor = southern Borneo, Mol = Moluccas, Ten = Nusa Tenggara, TNB = Tolai New Britain, Tro = Trobriand Islands, PNC = PNG coast, PNH = PNG highlands, WNL = WNG lowlands/coast, WNH = WNG highlands, Coo = Cook Islands, Aus1 = Australia Arnhem Land, Aus2 = Australia Sandy Desert. For haplogroup relationships see figure 2.
Genetic Analyses
DNA for the WNG samples was extracted from dried mouth swabs by a standard salting-out procedure (Miller et al. 1988). A total of 26 binary markers (21 SNPs, two short insertion/deletion polymorphisms [in/dels], one Alu insertion polymorphism [YAP], and two specific in/dels at the Y STR locus DYS390]) and seven STR loci from the nonrecombining part of the human Y chromosome (NRY) were studied. The Y STR loci DYS19 (or DYS394), DYS389I, DYS389II, DYS390, DYS391, DYS392, and DYS393 and the binary markers M4, M5, M16, M9, M119, M122, M175, and RPS4Y711 were analyzed, and the DYS390 locus was sequenced, as described elsewhere (Kayser et al. 2000a, 2001a). M89 was analyzed via PCR-RFLP, as described elsewhere (Ke et al. 2001). For the binary markers SRY10831 and synonym SRY-1532 (Whitfield et al. 1995; Rosser et al. 2000); M38, M74, M83, M95, M104, and M173 (Underhill et al. 2000); M186, M189, M208, M210, M216, and M217 (Underhill et al. 2001b); YAP (Hammer 1994); and M230 (P.S. and P.O., unpublished data), standard PCR conditions were used as described elsewhere (Kayser et al. 2001a), with additional details given in tables 1 and 2. Genotyping was performed for some loci via PCR-RFLP (table 1), with subsequent separation of the PCR products in a 3% NuSieve agarose gel (Bio Whittaker Molecular Applications) and visualization via ethidium bromide staining. Alternatively (table 2), genotyping was performed via a primer extension reaction, using the ABI PRISM SNaPshot ddNTP Primer Extension Kit (Applied Biosystems), and subsequent detection was performed with an ABI PRISM 377 DNA Sequencer (Applied Biosystems). SNaPshot reactions were performed according to the instructions of the manufacturer but using 1–2 μl purified PCR product, 0.5 pmol primer, and 5 μl SNaPshot Ready Reaction Mix, in a total volume of 10 μl. The 1-bp insertion/deletion polymorphism M186 was typed via fragment-length analysis with an ABI PRISM 377 DNA Sequencer, using PCR primers 5′-FAM-TCAAAAAGAAATAGGAAACCA-3′ and 5′-CAAGGAGTTAGGAGAACAGAGA-3′ at an annealing temperature of 51°C; resulting fragments were 91 bp (ancestral) and 90 bp (mutant). YAP was typed as described elsewhere (Hammer and Horai 1995). Binary markers were partly analyzed with a hierarchical strategy devised according to the information given by Underhill et al. (2001b), so that more basal markers, such as M9 or RPS4Y711, were typed in all samples, whereas markers associated with, for example, the M9G or the RPS4Y711T mutation were typed only in samples in which the derived state of M9 or RPS4Y711, respectively, was observed. Control DNA samples for the ancestral and mutant state of each binary marker were always included, to aid in accurate typing.
Table 1.
Primers and Parameters for PCR/RFLP/Fragment-Length Typing of Y-Chromosomal Binary Markers
| PCR |
PCR-RFLP |
|||||
| Primer Sequence(5′→3′) |
Fragment(s)(bp) |
|||||
| Marker/Mutation | Forward | Reverse | AnnealingTemperature(°C) | Enzyme | Ancestral | Mutant |
| SRY10831 A→G | CCTCTTGTATCTGACTTTTTCAC | CCACATAGGTGAACCTTGAAAAT | 57 | DraIII | 56 | 30 + 26 |
| M38 T→G | GTATGGCAATGGTATGTAGGCA | AACTTTTTCATACTTCCACAATCTTTTA | 56 | Bst4CI | 37 + 90 | 37 + 23 + 67 |
| M74 G→A | AACTAGGAAAGTCTGAAAAATAATCAGA | GCTGCTGTTGTCTTTTAAGTAACTTACT | 56 | RsaI | 151 | 47 + 104 |
| M83 C→T | AAAGAGAGGCAGCAATGAGAA | CCAAAGTGCTGGGATTACAAG | 64 | HaeIII | 59 + 41 | 100 |
| M189 G→T | TCCTTTCCTTCTTAGGCTTT | ATTAGCACTTACCTATGTGGG | 51 | HindIII | 6 + 96 + 96 | 96 + 106 |
| M208 C→T | CTGTTATTTGACTCACGAAAATT | ACATGAAGTCAGAGAAAGGC | 51 | TaqI | 64 + 97 | 161 |
| M216 C→T | TCAACCAGTTTTTATGAAGCTAG | GGATTATATGAGAGTAGCAAAAGA | 51 | SspI | 158 | 53 + 105 |
| M230 T→A | GATTTTAACAATATATACATGGCCA | ACATTATTAGTATGTAAATCTTCATTGC | 53 | Tsp509I | 79 + 85 | 164 |
Table 2.
Primers and Parameters for PCR/SNaPshot Typing of Y-Chromosomal Binary Markers
| PCR |
SNaPshot Reaction |
|||||
| Primer Sequence(5′→3′) |
||||||
| Marker/Mutation | Forward | Reverse | AnnealingTemperature(°C) | Length(bp) | Primer Sequence(5′→3′) | AnnealingTemperature(°C) |
| M95 C→T | GCAATAGTGTTGCACCTTCT | GACTCTCCTAAGCCTACAGGTT | 52 | 111 | GGATAAGGAAAGACTACCATATTAGTG | 54 |
| M104 A/G→A/A | TGTCAGAGCTGTCAGCCTTC | GGACCTGAGGTCAAGACTTTTAG | 55 | 162 | CTCAGTAGAGGAAAATGCTACAGTC | 55 |
| M173 A→C | CCTAGAAAATTGGAAATAAAGTAA | ACTGGCTTATCATTTCTGAATAT | 50 | 98 | TACAATTCAAGGGCATTTAGAAC | 54 |
| M210 A→T | CAGAAGCCGAGTAGGAAGC | AGGTGATTTTGTATTTATCTTCCC | 54 | 133 | GCTATCTATGACTTTTTAAAACTCTG | 51 |
| M217 A→C | TCTTTAACTTGTGAAGGAGAATGA | GCATTTGATAAAGCTGCTGTG | 54 | 107 | GAATGAAAAAGTTGGGTGACAC | 55 |
Terminology and Nomenclature
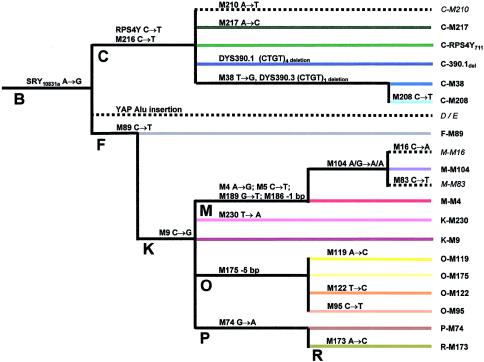
The terminology and nomenclature used here were proposed recently by The Y Chromosome Consortium (YCC), to standardize the use of Y-chromosome markers (YCC 2002). The terms “haplogroup” and “haplotype” are used according to de Knijff (2000): “haplogroups” are NRY lineages defined by binary polymorphisms, and “haplotype” refers to all sublineages of haplogroups that are defined by Y STR variation. Binary NRY haplogroups are designated by a capital letter identifying the major clade from the YCC haplogroup tree (YCC 2002) and the name of the most distal mutated binary marker.
Statistical Analyses
The software package Arlequin, version 2.000 (Schneider et al. 2000; Arlequin's Home on the Web) was used to calculate several population genetic parameters, including diversity of haplogroups and haplotypes and the associated SD, mean number of pairwise haplotype differences (MPD), pairwise FST values (for haplogroups) and RST values (for haplotypes) and associated P values based on 10,000 permutations, Fu's Fs test for population expansion, and the Mantel test for matrix correlation. Multidimensional scaling (MDS) analysis of pairwise FST values was performed by means of the software package STATISTICA (Statsoft), which was also used to calculate Spearman rank correlation coefficients. Nonparametric Mann-Whitney U tests to compare diversity values were calculated with the SPSS (SPSS Inc.) software package. Neighbor-joining trees based on pairwise FST values were constructed using the relevant program in the software package PHYLIP (Felsenstein 1993; PHYLIP Home Page) and were viewed with TreeView software (Page 1996; TreeView Web site). Median-joining network analysis (Bandelt et al. 1999) of haplogroup-associated haplotypes was performed using the software NETWORK2.0b (Shareware Phylogenetic Network Software Web site). For network calculation, each Y STR locus was weighted according to its estimated mutation rate (Kayser et al. 2000b), so that loci with the highest mutation rates were given the lowest weights (ratio of weights for DYS393:DYS392:DYS19:DYS389I:DYS389II:DYS391:DYS390 = 10:10:5:5:2:2:1). Baysian-based coalescence analysis of haplogroup-associated haplotypes was performed using the program BATWING (Mathematical Sciences at Aberdeen Web site), as described elsewhere (Kayser et al. 2001a).
Results
To investigate the Y-chromosome history of WNG, a total of 33 Y-chromosomal markers were analyzed in 183 males from 11 regional groups of WNG and in 610 males from 18 other Asian and Oceanian populations (table 3). In total, 488 different Y chromosomes (based on NRY binary markers and Y STR markers) were characterized among those 793 individuals belonging to 16 haplogroups (based entirely on binary markers) and 464 haplotypes (based entirely on Y STR markers). Only 6 of these 16 haplogroups were observed in WNG: M-M4, C-M208, C-M38, K-M230, O-M122, and K-M9 (tables 3 and 4; figs. 1 and 2); we describe these in more detail.
Table 3.
Y-Chromosome Haplogroup Frequencies (%) and Associated Haplotype Diversity in WNG and 18 Other Populations from Asia/Oceania[Note]
|
Frequency of Haplogroup(%) |
||||||||||||||||
| Population (No. of Individuals) | F-M89 | C-RPS4Y711 | C-M217 | C-M38 | C-M208 | C-390.1del | K-M9 | K-M230 | M-M4 | M-M104 | O-M175 | O-M95 | O-M119 | O-M122 | P-M74 | R-M173 |
| Korea (25) | 8.0 | 0 | 12.0 | 0 | 0 | 0 | 8.0 | 0 | 0 | 0 | 32.0 | 0 | 4.0 | 28.0 | 4.0 | 4.0 |
| China (36) | 0 | 0 | 5.6 | 0 | 0 | 0 | 11.1 | 0 | 0 | 0 | 0 | 2.8 | 22.2 | 58.3 | 0 | 0 |
| Taiwan Chinese (26) | 0 | 0 | 3.8 | 0 | 0 | 0 | 3.8 | 0 | 0 | 0 | 11.5 | 0 | 23.1 | 57.7 | 0 | 0 |
| Taiwan Aborigines (43) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.7 | 4.7 | 79.1 | 11.6 | 0 | 0 |
| Philippines (39) | 2.6 | 10.3 | 0 | 0 | 0 | 0 | 2.6 | 0 | 0 | 0 | 2.6 | 2.6 | 41.0 | 35.9 | 0 | 2.6 |
| Vietnam (11) | 0 | 0 | 9.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 36.4 | 9.1 | 45.5 | 0 | 0 |
| Malaysia (18) | 5.6 | 0 | 11.1 | 0 | 0 | 0 | 11.1 | 0 | 0 | 0 | 0 | 27.8 | 11.1 | 27.8 | 0 | 5.6 |
| Java (53) | 1.9 | 1.9 | 0 | 0 | 0 | 0 | 1.9 | 0 | 0 | 0 | 1.9 | 41.5 | 22.6 | 22.6 | 1.9 | 3.8 |
| Southern Borneo (40) | 5.0 | 0 | 2.5 | 2.5 | 0 | 0 | 10.0 | 0 | 0 | 0 | 5.0 | 37.5 | 15.0 | 17.5 | 2.5 | 2.5 |
| Moluccas (34) | 0 | 8.8 | 0 | 14.7 | 0 | 0 | 17.6 | 20.6 | 20.6 | 0 | 0 | 0 | 5.9 | 11.8 | 0 | 0 |
| Nusa Tenggara (31) | 0 | 6.5 | 0 | 16.1 | 0 | 0 | 32.3 | 9.7 | 6.5 | 0 | 0 | 0 | 22.6 | 3.2 | 0 | 3.2 |
| Tolai New Britain (16) | 0 | 0 | 0 | 12.5 | 0 | 0 | 37.5 | 12.5 | 0 | 31.2 | 0 | 0 | 0 | 6.3 | 0 | 0 |
| Trobriand Islands (53) | 0 | 0 | 0 | 0 | 9.4 | 0 | 22.6 | 0 | 30.2 | 0 | 0 | 0 | 28.3 | 9.4 | 0 | 0 |
| PNG coast (31) | 0 | 3.2 | 0 | 12.9 | 9.7 | 0 | 16.1 | 16.1 | 29.0 | 0 | 0 | 0 | 0 | 9.7 | 3.2 | 0 |
| PNG highlands (31) | 0 | 0 | 0 | 3.2 | 0 | 0 | 6.5 | 51.6 | 35.5 | 0 | 0 | 0 | 3.2 | 0 | 0 | 0 |
| WNG lowlands/coast (89) | 0 | 0 | 0 | 9.0 | 0 | 0 | 11.2 | 2.2 | 77.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| WNG highlands (94) | 0 | 0 | 0 | 0 | 24.5 | 0 | 0 | 0 | 74.5 | 0 | 0 | 0 | 0 | 1.1 | 0 | 0 |
| Cook Islands (28) | 0 | 0 | 0 | 0 | 82.1 | 0 | 3.6 | 0 | 0 | 0 | 0 | 0 | 0 | 7.1 | 0 | 7.1 |
| Australia Arnhem (60) | 1.7 | 10.0 | 0 | 0 | 0 | 53.3 | 30.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.0 |
| Australia Desert (35) | 2.9 | 0 | 0 | 0 | 0 | 68.7 | 17.1 | 0 | 0 | 0 | 0 | 0 | 0 | 3.0 | 0 | 8.6 |
| Overall (793): | ||||||||||||||||
| No. of individuals | 9 | 17 | 10 | 26 | 54 | 56 | 91 | 35 | 184 | 5 | 17 | 50 | 111 | 109 | 4 | 15 |
| No. of haplotypes | 9 | 13 | 10 | 19 | 20 | 31 | 73 | 25 | 75 | 5 | 16 | 37 | 59 | 81 | 4 | 12 |
| Haplotype diversity ± SE | 1.000±.052 | .963±.033 | 1.000±.045 | .966±.022 | .915±.020 | .932±.024 | .991±.004 | .970±.017 | .965±.007 | 1.000±.127 | .993±.023 | .981±.010 | .977±.006 | .983±.007 | 1.000±.177 | .962±.040 |
Note.— WNG data are highlighted in boldface italic type.
Table 4.
Y-Chromosome Haplogroup Frequencies (%) in Regional Populations from New Guinea
|
Frequency of Haplogroup(%) |
||||||||||
| Population | No. of Individuals | C-RPS4Y711 | C-M38 | C-M208 | K-M9 | K-M230 | M-M4 | O-M119 | O-M122 | P-M74 |
| WNG: | ||||||||||
| Dani | 12 | 0 | 0 | 91.7 | 0 | 0 | 0 | 0 | 8.3 | 0 |
| Lani | 12 | 0 | 0 | 100.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Yali | 5 | 0 | 0 | 0 | 0 | 0 | 100.0 | 0 | 0 | 0 |
| Una | 46 | 0 | 0 | 0 | 0 | 0 | 100.0 | 0 | 0 | 0 |
| Ketengban | 19 | 0 | 0 | 0 | 0 | 0 | 100.0 | 0 | 0 | 0 |
| Awyu | 10 | 0 | 0 | 0 | 0 | 0 | 100.0 | 0 | 0 | 0 |
| Kombai/Korowai | 13 | 0 | 0 | 0 | 53.8 | 0 | 46.2 | 0 | 0 | 0 |
| Muyu | 8 | 0 | 12.5 | 0 | 0 | 0 | 87.5 | 0 | 0 | 0 |
| Mappi | 10 | 0 | 10.0 | 0 | 10.0 | 10.0 | 70.0 | 0 | 0 | 0 |
| Asmat | 20 | 0 | 20.0 | 0 | 5.0 | 0 | 75.0 | 0 | 0 | 0 |
| Citak | 28 | 0 | 7.1 | 0 | 3.6 | 3.6 | 85.7 | 0 | 0 | 0 |
| PNGa: | ||||||||||
| Southern highlands | 14 | 0 | 0 | 0 | 7.1 | 57.1 | 35.7 | 0 | 0 | 0 |
| Eastern highlands | 17 | 0 | 5.9 | 0 | 5.9 | 47.0 | 35.3 | 5.9 | 0 | 0 |
| Northern coast | 16 | 0 | 18.8 | 18.8 | 25.0 | 18.8 | 12.5 | 0 | 6.3 | 0 |
| Southern coast | 15 | 6.7 | 6.7 | 0 | 6.7 | 13.3 | 46.7 | 0 | 13.3 | 6.7 |
For other population data from PNG (Tolai, Trobriand) see table 3.
Figure 2.
Relationship of 16 Y-chromosome haplogroups based on 26 binary markers. Color code is as in figure 1. Dashed lines indicate haplogroups not observed in this data set. Nomenclature is according to The YCC (2002).
Haplogroup M-M4
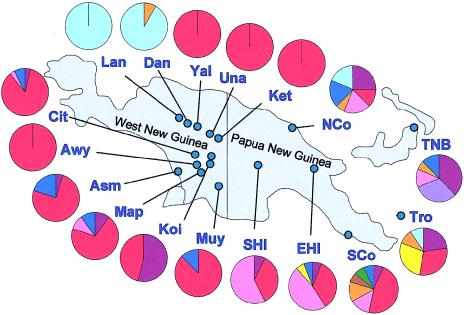
The majority of males analyzed from WNG (77.5% from the lowlands/coast and 74.5% from the highlands) carried the M-M4 haplogroup (tables 3 and 4; figs. 1 and 3), which is additionally characterized by the mutant state of the markers M5, M186, and M189 (fig. 2). Haplogroup M-M4 was previously identified as the predominant Y-chromosome type in Melanesia, on the basis of data from mainland and island PNG (Kayser et al. 2001a), with a frequency of 35.5% in highland PNG, 29% in coastal PNG, 30.2% in the Trobriand Islanders, and 6.5%–20.6% in eastern Indonesia (table 3; fig. 1). It has also been observed in other population samples from Melanesia and eastern Indonesia (Su et al. 2000; Capelli et al. 2001; Hurles et al. 2002). M-M4 has a very high frequency overall in WNG, with some groups completely fixed for this haplogroup, including the Yali, the Una, and the Ketengban from the eastern highlands of WNG, as well as the Awyu from the WNG lowlands (table 4; fig. 3). The coalescence time of chromosomes carrying the M4G mutation was previously estimated as ∼9,700 years on the basis of associated Y STR diversity (Kayser et al. 2001a). Considering the much larger data set from the present study (184 males with 75 haplotypes, compared with 51 males with 41 haplotypes in the previous study), it was only slightly younger (∼8,200 years), with evidence for population expansion ∼4,400 years ago (table 5).
Figure 3.
Y-chromosome haplogroups and their frequency distribution in regional populations from New Guinea. Population abbreviations are as follows: Dan = Dani, Lan = Lani, Yal = Yali, Una = Una, Ket = Ketengban, Awy = Awyu, Koi = Kombai/Korowai, Muy = Muyu, Map = Mappi, Asm = Asmat, Cit = Citak, SHI = PNG southern highlands, EHI = PNG eastern highlands, NCo = PNG northern coast, and SCo = PNG southern coast. Color code is as in figures 1 and 2.
Table 5.
Coalescence-Based Demographic Inference for Haplogroup-Defining Y-SNP Mutations, Based on Associated Y STR Haplotype Diversity[Note]
|
Median (95% Equal-Tailed Interval) |
||||
| Haplogroup (Mutation)a | Initial EffectivePopulation Size/1,000 Individuals | Population Growth Rate/Generation(×10−3) | Start of Population Expansion(×10−3 years) | TMRCA(×10−3 years) |
| M-M4 (M4G) | .21 (.07–.65) | 12.8 (5.2–29.5) | 4.4 (1.8–10.3) | 8.2 (3.8–20.6) |
| C-M208 (M208T) | .21 (.07–.58) | 10.1 (.5–40.0) | 2.2 (.2–9.2) | 6.9 (2.8–19.7) |
| C-M38 (M38G) | .35 (.01–1.06) | 4.6 (.1–22.2) | 4.7 (.1–19.8) | 10.6 (4.5–30.3) |
| K-M230 (M230A) | .23 (.07–.70) | 13.7 (2.9–39.9) | 3.5 (1.2–10.4) | 8.1 (3.4–23.2) |
| O-M95 (M95T) | .27 (.07–.67) | 16.3 (5.3–41.8) | 4.4 (1.9–11.0) | 8.8 (3.9–23.2) |
| O-M119 (M119C)b | .25 (.08–.74) | 18.3 (7.1–39.9) | 3.9 (1.8–9.5) | 12.4 (5.0–34.0) |
| O-M122 (M122C)b | .25 (.07–.78) | 14.5 (5.9–33.2) | 6.0 (2.7–14.5) | 11.1 (5.1–28.3) |
Haplogroup C-M208
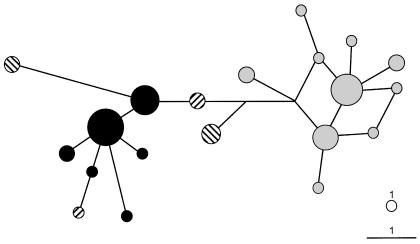
Haplogroup C-M208, a sublineage of haplogroup C-M38 that is also characterized by the DYS390.3 deletion on the RPS4Y711T background (fig. 2), was observed in 24.5% of the WNG highland samples (table 3; fig. 1). However, with respect to regional WNG populations, C-M208 Y chromosomes are completely restricted to the linguistically closely related Dani and Lani from the central/western highlands. In the Lani this haplogroup is completely fixed, and in the Dani it is present in all but one male (table 4; fig. 3). Outside WNG, the C-M208 haplogroup was found at lower frequency in the northern coast of PNG (18.8%) and the Trobriand Islands (9.4%). Interestingly, haplogroup C-M208 is also the major Y-chromosome haplogroup (82%) in the Cook Islanders from Polynesia (table 3; fig. 1). These haplogroup C-M208 Y chromosomes from the Cook Islands were previously shown to carry the DYS390.3 deletion on the RPS4Y711T background (Kayser et al. 2000a, 2001a), so, at the haplogroup level, C-M208 Y chromosomes from Polynesia are completely identical to C-M208 Y chromosomes in WNG and PNG. To further investigate the relationships between C-M208 Y chromosomes in New Guinea and Polynesia, a median-joining network of the 20 different Y STR haplotypes observed in all 54 C-M208 chromosomes was constructed (fig. 4). This network exhibited two distinct clusters of closely related haplotypes, with considerable haplotype sharing within but not between clusters (fig. 4). One cluster consists exclusively of all of the Dani/Lani haplotypes from WNG, as well as two more distantly related haplotypes from coastal PNG and the Trobriand Islands. The other cluster contains exclusively Polynesian haplotypes. Haplotypes from coastal PNG and the Trobriands represent the connection between the two clusters (fig. 4). Y STR haplotype diversity associated with C-M208 was 0.696 in the Dani and Lani (n=23), 0.821 in the Trobriands/coastal PNG (n=8), and 0.824 in the pooled Melanesians (n=31), whereas it was 0.842 in the Polynesians from the Cook Islands (n=23). No haplotype sharing was observed between WNG, PNG, and Polynesia, and differences between the three groups, based on RST values, were highly significant (P<.0001). The Y STR–based coalescence time of haplogroup C-M208 chromosomes was estimated to be ∼6,900 years, with evidence for a population expansion starting ∼2,200 years ago (table 5). When treated separately, the WNG haplotypes had a time back to the most recent common ancestor (TMRCA) of ∼2,400 years, with expansion starting ∼900 years ago; the Polynesian haplotypes had a TMRCA of ∼3,500 years, with expansion starting ∼1,600 years ago; and all Melanesian haplotypes together had a TMRCA of ∼4,800 years, with expansion starting ∼1,500 years ago.
Figure 4.
Median-joining network of 20 Y STR–based haplotypes observed in 54 haplogroup C-M208 chromosomes. Circles denote the haplotype, with the area of the circle proportional to the number of individuals carrying the particular Y STR haplotype. A circle indicating the size that represents one individual is given in the lower right of the figure. Lines denote Y STR mutation steps (one-step distance is indicated in the lower right of the figure); parallel lines represent identical mutations. Network is weighted according to Y STR mutation rates. Population affiliation of haplotypes is as follows: black circles = WNG highlands, striped circles (upward-sloping to the right) = PNG coast, striped circles (downward-sloping to the right) = Trobriand Islands, gray circles = Cook Islands (Polynesia).
Haplogroup C-M38
Haplogroup C-M38 Y chromosomes, which also carry the DYS390.3 deletion on the RPS4Y711T background (fig. 2) but lack the M208T mutation, occurred in 9% of the WNG lowland/coastal samples. This haplogroup was restricted to the Muyu, Mappi, Citak, and Asmat and was not found in the WNG highlands (tables 3 and 4; figs. 1 and 3). Outside WNG, haplogroup C-M38 was found in all other Melanesian groups from PNG except the Trobriand Islanders (table 3; fig. 1). C-M38 Y chromosomes were also observed in the Moluccas (14.7%) and Nusa Tenggara islands (16.1%) of eastern Indonesia, as well as in a single individual from southern Borneo (2.5%), but nowhere else in eastern and southeastern Asia or Australia. The TMRCA of all haplogroup C-M38 chromosomes was estimated to be ∼10,600 years, with a signal of population expansion beginning ∼4,700 years ago (table 5), although this was the smallest signal of population growth (∼0.005/generation) detected for any Y-chromosome mutation analyzed with this model in this or previous studies (Kayser et al. 2000a, 2001a, 2001b). Y chromosomes carrying the M38G mutation have been found in other populations from Indonesia, Micronesia, Melanesia, and Polynesia (Underhill et al. 2000, 2001a; Redd et al. 2002), but, since the M208 marker was not typed in these studies, it is impossible to tell if these are haplogroup C-M38 or haplogroup C-M208 chromosomes.
Haplogroup K-M230
Another haplogroup, which appears to be restricted to Melanesia and eastern Indonesia, is K-M230. The M230 mutation was originally identified in New Guinean samples on the M9G background (P.U., P.S., and P.O., unpublished data), and our results show that haplogroup K-M230 characterizes most of the Melanesian and some of the eastern Indonesian M9G chromosomes, which, in our previous studies (Kayser et al. 2000a, 2001a), could not be further differentiated, because of the lack of appropriate markers. In WNG, haplogroup K-M230 was observed only in a single Mappi and a single Citak (tables 3 and 4; figs. 1 and 3). However, this haplogroup is much more frequent in PNG. In fact, in the PNG highlands, K-M230 is the major haplogroup (51.6%), and it also occurs in the coast of PNG (16%) and in New Britain (12.5%). Outside Melanesia, K-M230 was found only in eastern Indonesia, with a higher frequency in the Moluccas (20.6%) than in the Nusa Tenggara islands (9.7%). The TMCRA for M230A was estimated to be ∼8,200 years, with a signal of population expansion beginning ∼3,500 years ago.
Haplogroup O-M122
Haplogroup O-M122, a sublineage of O-M175 (fig. 2), was, in WNG, found only in a single Dani (tables 3 and 4; figs. 1 and 3). This haplogroup is the major eastern Asian Y-chromosome haplogroup and also occurs in Melanesia and Polynesia (Su et al. 1999, 2000; Kayser et al. 2000a, 2001a; Capelli et al. 2001). We previously suggested that haplogroup O-M122 represents Y-chromosome evidence for the Austronesian expansion from eastern Asia into Melanesia and Polynesia (Kayser et al. 2000a, 2001a). However, since Austronesian languages are completely absent from the region of WNG studied here, the observation of haplogroup O-M122 in a single Dani may be more likely to be explained by recent admixture. Indeed, the Y STR haplotype of this particular Dani individual is shared by four Philippinos, five Javanese, two Trobriand Islanders, and one coastal PNG; such widespread sharing of this haplotype makes recent admixture a more probable explanation than more ancient Austronesian contact.
Haplogroup K-M9
Haplogroup K-M9 most likely represents the common ancestor of the majority of non-African Y chromosomes, and many Y-SNP markers are known on the M9G background (Underhill et al. 1997, 2000, 2001b). Haplogroups carrying the M9G mutation (and additional markers that define sublineages of M9G) are widespread in Asia and account for 78.4% of all Y chromosomes in this study (table 3; fig. 2). For WNG, the proportion of Y chromosomes carrying only M9G and no derived markers (haplogroup K-M9) is small (∼6% of the entire sample), and usually they were found in only single individuals from some populations. An exception is the Korowai/Kombai population, in which haplogroup K-M9 occurs in ∼54% of the samples (table 3 and 4; figs. 1 and 3).
Other Haplogroups not Observed in WNG
Further analyses of additional NRY binary markers in the previously studied non-WNG samples included here (Kayser et al. 2000a, 2001a) revealed a number of haplogroups that were not observed in WNG; we describe these here.
Haplogroup M-M104 was found in all previously characterized M4G Y chromosomes in the Tolai from New Britain (31.2%) (Kayser et al. 2001a), whereas this haplogroup was not observed outside New Britain in the present study (table 3; figs. 1 and 2). The relevant M104 mutation was originally identified in samples from the Bougainville islands of PNG (Underhill et al. 2000) and has also been found on Fiji and on some Polynesian islands (M.K. and M.S., unpublished data). Additional sublineages of M-M104, such as M-M16 or M-M83 (Underhill et al. 2000, 2001b) were not observed in this study (fig. 2).
Haplogroup O-M95, a sublineage of O-M175 (fig. 2), appears to be the predominant southeastern Asian haplogroup, with a high frequency in Java (41.5%), Borneo (37.5%), Malaysia (27.8), and Vietnam (36.4%). It was rarely observed in China (2.8%), Taiwan (4.7%), or the Philippines (2.6%), and it was completely absent from eastern Indonesia, Melanesia, Australia, and Polynesia (table 3; fig. 1). O-M95 Y chromosomes were also previously observed in central Asia/Siberia (Su et al. 1999; Underhill et al. 2000; Wells et al. 2001) and in very low frequencies in coastal PNG and Western Samoa (Capelli et al. 2001). Given this distribution, haplogroup O-M95 is most likely of southeastern Asian origin; its TMRCA was estimated to be ∼8,800 years, with a start of population expansion ∼4,400 years ago (table 5).
Haplogroup O-M175 Y chromosomes, which carry the M175 deletion but no derived markers (fig. 2), were found in high frequency in Korea (32%) but were observed only sporadically in populations from elsewhere in eastern/southeastern Asia (table 3; fig. 1). This haplogroup was completely absent from eastern Indonesia, Melanesia, Australia, and Polynesia but has been reported from central Asia/Siberia (Underhill et al. 2000; Wells et al. 2001).
Y chromosomes carrying the RPS4Y711T mutation together with the M216T mutation represent, after the M9 lineages, the second major paternal lineage in Asia, and this lineage also spread into the Americas (Karafet et al. 1999; Underhill et al. 2000, 2001b; Wells et al. 2001). In the present study, 20.5% of all samples carry RPS4Y711T/M216T and frequently carry additional derived markers. Y chromosomes carrying only RPS4Y711T/M216T and no derived markers—and, thus, belonging to haplogroup C-RPS4Y711—were observed in the present study only at low frequency in the Philippines, Java, eastern Indonesia, coastal PNG, and Australia (table 3; fig. 1).
Haplogroup C-217, a sublineage of C-RPS4Y711 (fig. 2), was observed in Korea (12%), China (4%–6%), Vietnam (9%), Malaysia (11%), and Borneo (2.5%) but not in eastern Indonesia, Melanesia, Polynesia, or Australia (table 3; fig. 1). This haplogroup is reported to be widespread through central and eastern Asia, and it also occurs in North America (Karafet et al. 2001; Underhill et al. 2001b; Redd et al. 2002).
Haplogroup R-M173 is thought to be an ancient Euroasiatic marker, representing the earliest expansion into Europe ∼30,000 years ago (Semino et al. 2000). In addition to its high frequency in Europe it is also found at lower frequencies in central Asian populations (Semino et al. 2000; Underhill et al. 2000; Karafet et al. 2001; Wells et al. 2001), where it may have originated (Wells et al. 2001). Therefore, haplogroup R-M173 Y chromosomes, which were observed here in single individuals from eastern and southeastern Asian populations and more often in Polynesia and Australia (table 3; fig. 1), may reflect an original central Asian contribution. An alternative explanation is recent European admixture, especially in Polynesia and/or Australia, where European influence is well known.
Finally, nine Y chromosomes, which, in our previous study, could not be assigned to any Y-chromosomal haplogroup and thus were referred to as “complete ancestral” (Kayser et al. 2001a), were found here to belong to haplogroup F-M89 (table 3; fig. 1), which occurs in Europe and Asia (Underhill et al. 2000).
Y-Chromosome Diversity in WNG
Y-chromosome diversity in WNG was low when compared with the 18 other populations of Asia/Oceania, including PNG. The lowland/coastal WNG sample and the highland WNG sample showed the third and fourth lowest haplogroup diversity value (table 6; fig. 1), higher only than Polynesians and the aboriginal Taiwanese. For haplotype diversity, the highland WNG had the lowest value and lowland/coastal WNG had the fifth lowest value (together with the Philippines), higher only than that of the Polynesians, the Moluccas, the WNG highlanders, and the Australians from Arnhem Land (table 6).
Table 6.
Y-Chromosome Diversity Characteristics of WNG and 18 Other Populations from Asia/Oceania[Note]
| Population | No. ofIndividuals | No. ofHaplogroups | HaplogroupDiversity ± SD | No. ofHaplotypes | HaplotypeDiversity ± SD |
| Korea | 25 | 8 | .820 ± .050 | 24 | .997 ± .013 |
| China | 36 | 5 | .611 ± .073 | 34 | .997 ± .008 |
| Taiwan (Chinese) | 26 | 5 | .622 ± .085 | 24 | .994 ± .013 |
| Taiwan (Aborigines) | 43 | 4 | .365 ± .088 | 30 | .980 ± .010 |
| Philippines | 39 | 8 | .707 ± .048 | 26 | .964 ± .017 |
| Vietnam | 11 | 4 | .709 ± .099 | 11 | 1.000 ± .039 |
| Malaysia | 18 | 7 | .850 ± .053 | 18 | 1.000 ± .019 |
| Java | 53 | 9 | .736 ± .038 | 38 | .980 ± .009 |
| Southern Borneo | 40 | 10 | .809 ± .045 | 39 | .999 ± .006 |
| Moluccas | 34 | 7 | .863 ± .022 | 21 | .961 ± .017 |
| Nusa Tenggara | 31 | 8 | .826 ± .041 | 30 | .998 ± .009 |
| Tolai New Britain | 16 | 5 | .775 ± .068 | 14 | .983 ± .028 |
| Trobriand Islands | 53 | 5 | .774 ± .024 | 38 | .983 ± .008 |
| PNG coast | 31 | 8 | .854 ± .034 | 27 | .989 ± .012 |
| PNG highlands | 31 | 5 | .622 ± .059 | 26 | .985 ± .013 |
| WNG lowlands/coast | 89 | 4 | .382 ± .061 | 42 | .964 ± .009 |
| WNG highlands | 94 | 3 | .390 ± .046 | 23 | .884 ± .021 |
| Cook Islands | 28 | 4 | .325 ± .110 | 15 | .894 ± .043 |
| Australia Arnhem Land | 60 | 5 | .623 ± .046 | 42 | .962 ± .017 |
| Australia Sandy Desert | 35 |
5 | .506 ± .090 | 25 | .966 ± .020 |
| Overall | 793 | 16 | 464 |
Note.— WNG data are highlighted in boldface italic type.
The reduced Y-chromosome diversity in WNG is more striking in the different regional/linguistic groups (table 7; fig. 3). In a number of WNG groups, only a single haplogroup was observed, even with reasonably large sample sizes (19–46). Reduced haplogroup diversity was observed in nearly all groups from the highlands of WNG (Dani/Lani, Yali, Una, and Ketengban), whereas, in groups from the lowlands and coast, the diversity was usually slightly higher, except for the Awyu (table 7). This is also reflected by the haplotype diversity data based on Y STRs (table 7). The average diversity within WNG highland groups was significantly smaller than within WNG lowland/coastal groups, based on all three Y-chromosome diversity measures (for haplogroups, 0.033 vs. 0.43, Mann-Whitney U test: Z=-2.44, P<.05; for haplotypes, 0.656 vs. 0.890, Z=-2.52, P<.05; for MPD, 1.89 vs. 4.88, Z=-2.84, P<.01). Moreover, WNG groups were, on average, significantly less diverse than PNG groups, on the basis of haplogroup (0.265 vs. 0.685, Mann-Whitney U test: Z=-2.46, P<.05) and haplotype data (0.792 vs. 0.956, Mann-Whitney U test: Z=-2.06, P<.05), but not significantly so on the basis of MPD of haplotypes (3.63 vs. 5.32, Mann-Whitney U test: Z=-1.69, P>.05). Within-PNG diversity values were, on average, lower in highland groups than in the coastal groups for all three measures, although not significantly so (for haplogroups, 0.593 vs. 0.824, Mann-Whitney U test: Z=-1.73, P>.05; for haplotypes, 0.943 vs. 0.977, Z=0, P>.05; MPD for haplotypes, 4.39 vs. 6.72, Z=-1.73, P>.05).
Table 7.
Genetic Diversity Based on Y-Chromosome and mtDNA Analysis of Regional Populations from New Guinea[Note]
| Population | No. of IndividualsY/mtDNA | No. of YHaplogroups | Y-HaplogroupDiversity ± SE | No. of YHaplotypes | Y-HaplotypeDiversity ± SE | Y MPD ± SE | No. of mtDNATypesa | mtDNADiversity ± SEa | mtDNAMPD ± SEa |
| WNG: | |||||||||
| Dani | 12/21 | 2 | .167 ± .134 | 4 | .455 ± .170 | 2.00 ± 3.72 | 17 | .98 ± .02 | 6.00 ± 3.00 |
| Lani | 12/NA | 1 | 0 | 5 | .667 ± .141 | 1.03 ± .84 | NA | NA | NA |
| Yali | 5/NA | 1 | 0 | 3 | .800 ± .164 | 2.20 ± 2.04 | NA | NA | NA |
| Una | 46/50 | 1 | 0 | 11 | .749 ± .061 | 2.80 ± 2.25 | 28 | .96 ± .01 | 7.90 ± 3.72 |
| Ketengban | 19/22 | 1 | 0 | 7 | .608 ± .127 | 1.40 ± 1.40 | 9 | .81 ± .07 | 5.90 ± 2.93 |
| Awyu | 10/12 | 1 | 0 | 6 | .889 ± .075 | 2.98 ± 1.57 | 9 | .89 ± .08 | 8.99 ± 4.45 |
| Kombai/Korowai | 13/NA | 2 | .539 ± .060 | 6 | .718 ± .128 | 2.99 ± 2.17 | NA | NA | NA |
| Muyu | 8/9 | 2 | .250 ± .180 | 7 | .964 ± .077 | 5.39 ± 3.81 | 8 | .97 ± .06 | 5.72 ± 3.03 |
| Mappi | 10/19 | 4 | .533 ± .180 | 6 | .844 ± .103 | 5.42 ± 3.91 | 14 | .92 ± .05 | 8.47 ± 4.10 |
| Asmat | 20/25 | 3 | .416 ± .116 | 14 | .963 ± .025 | 6.52 ± 4.39 | 17 | .95 ± .03 | 4.18 ± 2.15 |
| Citak | 28/39 | 4 | .267 ± .107 | 14 | .860 ± .056 | 4.84 ± 3.57 | 23 | .92 ± .03 | 7.59 ± 3.62 |
| PNGb: | |||||||||
| Southern highlands | 14/7 | 3 | .582 ± .092 | 12 | .967 ± .044 | 4.14 ± 1.85 | 7 | 1.00 ± .08 | 8.76 ± 4.61 |
| Eastern highlands | 17/8 | 5 | .684 ± .081 | 14 | .971 ± .032 | 5.54 ± 3.20 | 7 | .96 ± .08 | 9.39 ± 4.83 |
| Northern coast | 16/13 | 6 | .867 ± .043 | 15 | .992 ± .025 | 7.73 ± 2.89 | 12 | .99 ± .04 | 8.38 ± 4.15 |
| Southern coast | 15/18 | 7 | .781 ± .102 | 12 | .962 ± .040 | 5.71 ± 2.56 | 8 | .70 ± .12 | 2.64 ± 1.48 |
Note.— NA = mtDNA data not available.
mtDNA data are from Tommaseo-Ponzetta et al. ( 2002).
Population Relationship Analyses
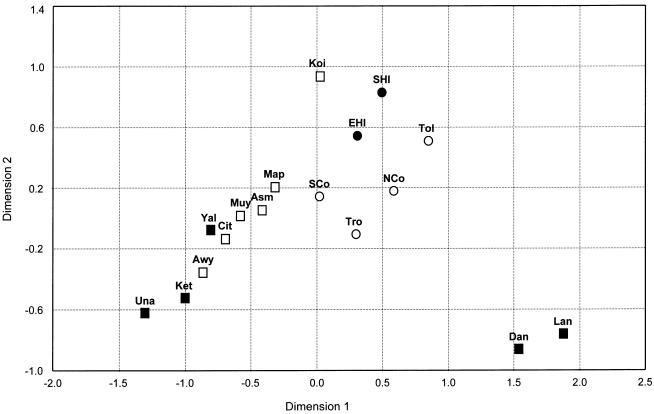
To investigate population relationships, MDS analyses were performed on the basis of pairwise FST values for Y haplogroups. The MDS plot of regional groups from New Guinea (fig. 5) reveals that WNG groups cluster together, except for the Dani/Lani and the Kombai/Korowai, which appear fairly separated from all other WNG populations. The MDS plot also shows that WNG highland groups are closer to WNG lowland/coast groups than to PNG highland groups, and, similarly, PNG highland groups are closer to PNG coastal/island groups than to WNG highland groups. A neighbor-joining tree based on Y haplogroup–based FST values reveals an identical clustering of New Guinea populations (tree not shown). To further investigate the patterns, we examined differentiation of New Guinean populations (excluding the Dani/Lani, who are clear outliers) when grouped according to geography and altitude. Pairwise FST values for Y haplogroups (table 8) revealed that, although always statistically significant, differences between groups within the same geographic region but from different altitudes were, on average, generally smaller than differences between groups from different geographic regions but within the same altitude. That is, the smallest FST values were observed between groups from the PNG highlands and PNG coast and between groups from the WNG highlands and WNG lowlands/coast.
Figure 5.
Two-dimensional MDS plot of regional populations from New Guinea, from pairwise FST values based on Y-chromosome haplogroups. The MDS stress value equals 0.075. Population abbreviations are as in figure 3. Regional code: ▪ = WNG highlands; □ = WNG lowlands/coast; ● = PNG highlands; ○ = PNG coast/islands.
Table 8.
Differentiation of New Guinean Populations, Grouped According to Altitude[Note]
| Population | WNG Highlands | WNG Lowlands/Coast | PNG Highlands | PNG Coast |
| WNG highlandsa | … | .1978 | .0789 | .3315 |
| WNG lowlands/coastb | .1572 | … | .0957 | .3690 |
| PNG highlandsc | .6148 | .3122 | … | .2623 |
| PNG coastd | .4945 | .2026 | .0851 | … |
Note.— Data are average pairwise FST values from Y-chromosome haplogroup analyses (below the diagonal) and pairwise FST values from mtDNA sequence analyses (above the diagonal), based on seven populations in which mtDNA and Y-chromosome data were both available (Una, Ketengban, Awuy, Muyu, Mappi, Asmat, and Citak; the Dani were not included, since they do not share any haplogroups with any WNG highland/lowland or PNG highland group). Samples for mtDNA sequencing from PNG were selected on the basis of the 9-bp deletion (Redd et al. 1995) and/or particular SSO types (Redd and Stoneking 1999) and, hence, are not a random sample.
Includes the Una and Ketengban populations.
Includes the Awyu, Muyu, Mappi, Asmat, and Citak populations.
Includes the southern highland and eastern highland populations.
Includes the northern coast and southern coast populations.
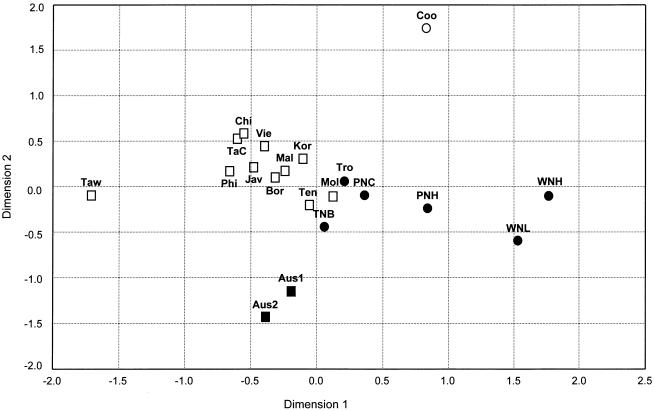
In an MDS plot of all Asian/Oceanian populations studied here, those from WNG and (to a lesser extent) highland PNG are separated from coastal and island Melanesians (fig. 6). The latter form a group with the eastern Indonesian populations and are somewhat separated from a cluster of the remaining eastern and southeastern Asian populations. The Polynesian sample, the two Australian samples together, and the Taiwan aborigine sample are clearly separated. A nearly identical population clustering was observed in a neighbor-joining tree (tree not shown).
Figure 6.
Two-dimensional MDS-plot of populations from Asia/Oceania from pairwise FST values based on Y-chromosome haplogroups. The MDS stress value equals 0.138. Population abbreviations are as in figure 1. Regional code: □ = eastern/southeastern Asia; ▪ = Australia; ● = Melanesia; ○ = Polynesia.
Comparison of Y-Chromosome with mtDNA Data
Sequences of the mtDNA HV1 from eight of the WNG groups analyzed here for Y-chromosome variation (Tommaseo-Ponzetta et al. 2002) were used for comparative analyses of paternal and maternal relationships (the Korowai are omitted because of low sample size in the mtDNA data [n=2]). As seen in table 7, the low Y-haplogroup diversity of WNG populations contrasts sharply with the high mtDNA diversity. Groups such as the Una, Ketengban, Awyu, and Dani, for which the Y-chromosome haplogroup diversity is zero or almost zero, have mtDNA haplotype diversities >0.81 (mostly >0.90). Even for the generally more variable Y STR haplotypes, diversity was always lower than for mtDNA (except for the Asmat), especially in the Dani, in which there were just two Y haplogroups and four Y STR haplotypes in 12 individuals but 17 mtDNA types in 21 individuals (table 7). mtDNA diversity in WNG highland groups is, on average, slightly lower than in WNG lowlands/coastal groups, but the difference is not statistically significant (diversity 0.917 vs. 0.93, Mann-Whitney U test: Z = −0.45, P>.05; MPD 6.60 vs. 6.99, Z=-0.15, P>.05). This is in contrast to the findings for the Y chromosome, where diversity is, on average, significantly lower in WNG highland groups than in lowland/coastal groups, for all three diversity measures (see above). However, if one considers for Y-chromosome analyses only the eight WNG populations for which mtDNA data are available (see footnote “a” of table 9), Y haplogroup diversity is, on average, lower in WNG highland groups (0.056) than in lowland/coastal groups (0.293), although not significantly so (Mann-Whitney U test: Z=-1.68, P>.05), whereas it is, on average, significantly lower for Y haplotypes (diversity: 0.604 vs. 0.904, Z=-2.24, P<.05; MPD: 2.07 vs. 5.03, Z=-2.24, P<.05). Overall, correlation analyses of diversity values from Y-chromosomal haplogroup/haplotype versus mtDNA data revealed no significant relationships (Spearman rank correlation: diversity, mtDNA/Y haplotypes: R=0.19, P>.05; diversity, mtDNA/Y haplogroups: R=0.17, P>.05; MPD, mtDNA/Y haplotypes: R=-0.17, P>.05).
Table 9.
AMOVA Based on Y-Chromosomal and mtDNA Data from WNG Populations[Note]
|
Variance Components (%) |
|||||||||
| Y Haplogroups |
Y Haplotypes |
mtDNA |
|||||||
| Grouping | BetweenGroups | WithinGroups | WithinPopulations | BetweenGroups | WithinGroups | WithinPopulations | BetweenGroups | WithinGroups | WithinPopulations |
| No grouping (8 populations)a | … | 49.38 | 50.62 | … | 32.99 | 67.01 | … | 13.22 | 86.78 |
| Altitude/geography (4 groups) | −16.77 | 62.8 | 53.97 | −1.01 | 33.73 | 67.28 | 1.63 | 11.87 | 86.5 |
| Language (6 groups) | 53.26 | .11 | 46.62 | 23.56 | 11.77 | 64.67 | −2.27 | 15.24 | 87.03 |
| Lowland only (4 populations) | … | −1.33 | 101.33 | … | 9.40 | 90.60 | … | 3.53 | 96.47 |
Note.— Nonsignificant values are shown in boldface italic type.
Includes those eight populations in which mtDNA and Y-chromosome data were available (Dani, Una, Ketengban, Awuy, Muyu, Mappi, Asmat, and Citak). For group definitions, see the “Samples and Methods” section.
Differences between Y chromosomes and mtDNA are also seen in the pairwise FST analysis when New Guinea groups are analyzed according to geography and altitude (table 8). For mtDNA, the average FST value was smallest between WNG highland groups and PNG highland groups, whereas for the Y chromosome the largest average FST value was observed between these same groups. Conversely, the smallest average FST value for the Y chromosome was between PNG highland and PNG coastal groups, whereas the average FST value based on mtDNA was much larger between PNG highland and coastal groups. Mantel tests of the FST matrices derived from Y-chromosome haplogroup and mtDNA data for those eight populations for which both Y chromosomes and mtDNA were available (see footnote “a” of table 9) revealed no significant correlation, either within WNG alone (r=-0.05, P>.05) or when PNG is also included with WNG (r=0.33, P>.05). On the basis of mtDNA, grouping according to either language or altitude does not describe the genetic structure of WNG, since for both comparisons the between-group component is small and not significantly different from 0 (table 9). Overall, we observe a much bigger between-group component for the Y chromosome than for mtDNA. This is especially evident when grouping is performed on the basis of language. On the basis of Y-chromosome data, the between-population component is almost entirely between the language groups, but for mtDNA it is mostly between populations within groups. The four WNG lowland populations (Mappi, Muyu, Citak, and Awyu) are not significantly differentiated from one another with respect to mtDNA or Y haplogroups, although the Y haplotypes do show significant differences among them (table 9).
Discussion
Low Y-Chromosome but High mtDNA Diversity in WNG
WNG, especially the highlands and southern lowlands/southern coast, is considered to be one of the most isolated inhabited regions in the world. This is reflected by the Y-chromosome data presented here. Only 6 Y haplogroups were found, out of 16 so far observed to be present in the entire Asia/Oceania region. In addition, Y STR haplotype diversity was low in WNG compared with that in other populations from Asia/Oceania. This reduced Y-chromosome diversity in WNG was especially striking when individual groups were considered. In 5 of 11 WNG groups, only a single Y haplogroup was observed. Y STR haplotype diversity of WNG groups ranged from 0.455 to 0.963 and was < 0.90 in 9 of the 11 groups. This compares to data from globally dispersed human populations recently obtained for the same Y STR loci used here, where Y haplotype diversity was 0.86–0.997 but was <0.90 in only one (coastal PNG) population and was >0.98 in 13 of the 20 populations analyzed (Kayser et al. 2001c).
Although WNG groups do show reduced genetic diversity of paternal lineages, they do not show reduced diversity of maternal lineages. mtDNA diversity in WNG populations was 0.81–0.98 and was >0.90 in six of the eight populations (Tommaseo-Ponzetta et al. 2002). Moreover, the MPD was 4.18–8.99 (table 7), which is comparable to MPD values for populations from around the world (Oota et al. 2002).
One explanation for observing low levels of Y-chromosome diversity but high levels of mtDNA diversity in the same populations could be extreme patrilocality and/or biased reproductive success in males (e.g., only a few males father most of the offspring). All Papuan-speaking populations in New Guinea are strictly patrilocal, and patrilineal clans are the central units of social, economic, and political organization. In patrilineal clans, all children belong to the father's clan and marriage is exogamous, with wives coming from a different clan. In patrilocal societies, male offspring stay in the family/village where they are born, whereas female offspring do not. Consequently, women in patrilocal societies are brought into the families/villages from outside. The genetic consequences of patrilocal social structure in reducing Y-DNA diversity but maintaining mtDNA diversity (and vice versa for matrilocal societies) has recently been demonstrated for hill tribes from Thailand (Oota et al. 2001). In four of the five highland WNG groups studied here, we observed even less Y STR diversity than in the least diverse patrilocal group studied by Oota et al. (2001). Furthermore, most (if not all) traditional New Guinean populations practice (or practiced until the influence of Christianity) polygyny (Pospisil 1963; Heider 1970; Trenkenschuh and Hoogebrugge 1970; Tommaseo-Ponzetta 1986; Schiefenhövel 1988; Hays 1991; de Vries 1993; van Enk and de Vries 1997), where some men have more than one wife, and others have none. Heider (1970) reports from a Dani community of the Dugum neighborhood, highland WNG, that 29% of the men had more than one wife (range 2–9) and that 38% of the men were not married. Very similar values are reported for a Citak community from the Brazza river in the southern lowlands of WNG, with 27% of the men living in polygynous families and 38% not married (Tommaseo-Ponzetta 1986). In an Eipo community from the upper Eipomek valley in the eastern highlands of WNG, 12% of the men were polygynous and 27% were not married (Schiefenhövel 1988). Polygyny is also reported to be frequent in many other Papuan-speaking groups from WNG (Pospisil 1963; Trenkenschuh and Hoogebrugge 1970; Hays 1991; de Vries 1993; van Enk and de Vries 1997).
Another potential factor that might reduce paternal (but not maternal) genetic diversity is warfare, which predominantly affects males and was common until the second half of the 20th century in most Papuan societies of New Guinea (Brown 1978; Feil 1987; Heider 1997). The number of men involved and killed in warfare varied, but cases are known in which warfare led to the extinction of all adult males in a community, whereas killing of women in warfare is reported to be highly immoral in WNG societies and is thus rare (Pospisil 1963). For example, genealogical studies revealed that 28.5% of the men in a Dani community, but only 2.4% of the women, were killed in warfare (Heider 1997). Also, for many highland PNG societies, the victims of warfare include a much higher proportion of men than women (Feil 1987). Thus, a stronger reduction in Y-chromosome diversity but not in mtDNA diversity would be the consequence of warfare, and this is exactly what we observed in the present study.
An additional explanation for the fairly homogeneous Y-chromosome gene pool in WNG might be lifestyle. Only the highland WNG groups in this study (Dani/Lani, Yali, Una, and Ketengban) practice horticulture (extensive gardening, mostly of sweet potato), whereas the remaining groups from the lowlands and southern coast are hunter/fisher/sago gatherers (Tommaseo-Ponzetta et al. 2002). In all but one of these horticultural highland groups, only a single Y haplogroup was found (in the Dani, one individual carried an additional haplogroup), whereas only one of the seven hunter-gatherer groups (Awyu) showed a single Y haplogroup. Moreover, Y-chromosome diversity in the horticultural groups is, on average, lower than in the nonhorticultural groups, for all three diversity measures estimated (for Y haplogroups, 0.033 vs. 0.43, Mann-Whitney U test: Z=-2.44, P<.05; for Y haplotypes, 0.656 vs. 0.890, Z=-2.52, P<.05; for Y haplotype MPD, 1.89 vs. 4.88, Z=-2.84, P<.01). This severe reduction in Y-chromosome diversity in horticultural groups might be the result of a more recent population expansion caused by food production, in connection with founder effects and isolation. A median-joining network of the C-M208–associated Y STR haplotypes shows a starlike picture for all Dani/Lani (WNG) haplotypes, with most of them being identical or one-step derivatives (fig. 4), and a similar picture was observed for the haplotypes from the Cook Islands of Polynesia. Thus, two different expansions can be assumed, involving C-M208 chromosomes—one in WNG, including the linguistically closely related Dani and Lani from WNG, that began (on the basis of coalescence analysis; table 5) ∼900 years ago, and another in Polynesia, starting ∼1,600 years ago, although only small signals of population growth were detected (rates of 6×10-3 and 9×10-3 per generation, for WNG and Polynesia, respectively). The current population size of the Dani/Lani is ∼270,000 (Grimes 2000), which is the largest observed in WNG (and New Guinea in general), suggesting a dramatic population expansion for this group.
If the reduced Y-chromosome diversity is indeed a result of population expansion associated with horticulture, this should also be detectable in the mtDNA data. Indeed, for two of the highland WNG populations that practice horticulture and show strongly reduced Y-chromosome diversity (Dani and Una; mtDNA data are not available for the Lani), Fu's Fs test for the mtDNA data from Tommaseo-Ponzetta et al. (2002) revealed a statistically significant indication of population expansion (Dani: Fs=-4.90, P<.05, Una: Fs=-6.71, P<.05). However, for the Ketengban, who also practice horticulture and carry only a single Y haplogroup, the mtDNA data do not reveal a significant indication of population growth on the basis of this test (Fs=0.81, P>.05). Moreover, one nonhorticultural WNG group, the Asmat, also shows a significant departure from neutrality in the direction of population expansions (Fs=-5.67, P<.05). mtDNA data reveal an average lower diversity in the horticultural than in the nonhorticultural populations, consistent with the Y-DNA results, but the differences were not significant (diversity: 0.917 vs. 0.93, Mann-Whitney U test: Z=-0.45, P>.05; MPD: 6.60 vs. 6.99, Z=-0.15, P>.05). That there might be reasons for population expansion other than horticulture is indicated by the mtDNA data of the Asmat, a coastal fishing and sago gathering group of WNG, which also show evidence of population expansion from Fu's Fs test and from a large current population size of ∼50,000 (Grimes 2000), which is higher than in all other WNG groups studied here except the Dani/Lani.
Melanesian Y-Chromosome Lineages
On the basis of haplogroup frequency distributions, haplogroup-associated Y STR diversity, and resulting coalescent time estimates, we suggest that at least four Y-chromosomal lineages—namely, haplogroups M-M4, K-M230, C-M38, and C-M208—most likely arose in Melanesia, prior to the Austronesian expansion. Our data for eastern Indonesia provides further evidence for this, with a higher frequency of the M-M4 and K-M230 haplogroups in the Papuan-speaking groups from the Moluccas than in the Austronesian-speaking groups from the Nusa Tenggaras islands. Evidence for a Melanesian, rather than an eastern Indonesian, origin of three of these haplogroups (C-M208 was not found in eastern Indonesia) comes from haplogroup-associated Y STR haplotype diversity data. For haplogroups M-M4 and C-M38, haplotype diversity and MPD are always larger in Melanesia compared with eastern Indonesia (M-M4: haplotype diversity 0.96 vs. 0.94 and MPD 4.4 vs. 3.6; C-M38: diversity 0.96 vs. 0.87 and MPD 5.3 vs. 4.0), and both groups show significant differentiation on the basis of RST (P<.001). For haplogroup K-M230, the MPD in Melanesia is higher than in eastern Indonesia (4.1 vs. 3.0), although haplotype diversity is not (0.96 vs. 0.98), and both groups are not statistically different from each other on the basis of RST (P=.77). A Melanesian origin for these haplogroups is also supported by linguistic evidence for a mainland New Guinea origin of the Papuan languages found in eastern Indonesia (Ross, in press).
Genetic Differences between WNG and PNG
Although the four Melanesian Y haplogroups are present in both PNG and WNG, their frequencies differ between the two regions. This observation is surprising, in that there is no biological or cultural reason to expect differentiation between the eastern and western part of New Guinea, since the central cordillera of New Guinea runs through the entire island from the east to the west. Also, on the basis of linguistic evidence, there is no such differentiation, since all languages spoken in the New Guinea highlands belong to the Trans New Guinea Phylum (Ross 1995, in press; Osmond et al., in press). Nonetheless, there is a clear east-west differentiation, based on haplogroup K-M230, between the WNG highland populations (where K-M230 is completely absent) and the PNG highland populations (where the frequency of K-M230 is 43%–71%). Given the overall rugged topography of the New Guinea highlands, it would seem more reasonable to expect differentiation to occur on the scale of local settlements—for example, between valleys—rather than between WNG and PNG. More-detailed sampling of highland populations, especially from PNG, may help to address this issue.
Furthermore, Y-haplotype diversity associated with these four Y haplogroups differs between WNG and PNG and is always smaller in WNG. For the major Melanesian haplogroup M-M4, of 71 different haplotypes of 175 M-M4 Y chromosomes from New Guinea, only 3 are shared between PNG and WNG; the haplotype diversity of PNG (0.983, based on 36 individuals) is larger than that of WNG (0.942, based on 139 individuals), and, on the basis of RST values, there is statistically significant differentiation between WNG and PNG.
Some of the difference between PNG and WNG can also be explained by the presence of the haplogroups O-M122 and O-M119 in PNG but not in WNG. Both haplogroups are of Asian origin and were most likely brought to New Guinea by the Austronesian expansion (Kayser et al. 2000a, 2001a). In keeping with this hypothesis, both haplogroups are not only essentially absent from WNG, but they also have higher frequencies in coastal PNG than in highland PNG (figs. 1 and 3; tables 3 and 4). This fits well with the assumed route of the Austronesian expansion and the present-day distribution of Austronesian languages along the northern coast of WNG and PNG and the east coast of PNG but not into the highlands of PNG or the highlands and southern lowlands/coastal areas of WNG (Bellwood 1978, 1989). Although the existence of Austronesian haplogroups does enhance diversity in PNG and does contribute to the differences between PNG and WNG, this effect is not significant. After removing haplogroups O-M119 and O-M122 from the analysis, the average diversity in PNG (0.660) is still significantly higher than in WNG (0.301) (Mann-Whitney U test: Z=-2.48, P<.05), and the FST value between PNG and WNG is statistically significant (FST=0.230, P<.001). Moreover, average diversity in highland PNG (0.579) is still lower than in coastal PNG (0.783), but not significantly so (Mann-Whitney U test, Z=-1.73, P=.08), although the FST value between highland and coastal PNG is statistically significant (FST=0.077, P<.05).
When only the four Melanesian haplogroups (M-M4, K-M230, C-M38, and C-M208) are analyzed, the average diversity within WNG groups is still significantly lower (0.112) than in PNG groups (0.565) (Mann-Whitney U test: Z=-3.02, P<.01), and differentiation between WNG and PNG based on FST is still significant (FST=0.287, P<.001). Within WNG, the highland groups are, on average, significantly less diverse (0.000) than the lowland/coastal groups (0.205) (Mann-Whitney U test: Z=-2.12, P<.05), and both regions are significantly different from each other (FST=0.114, P<.001). However, diversity in the PNG highland groups is, on average, lower (0.498) than in PNG coastal/island groups (0.665), but not significantly so (Mann-Whitney U test: Z = −0.577, P>.05), and the differentiation based on FST is not quite statistically significant (FST=0.078, P=.055). Thus, the differences observed in the entire data set between highland and coastal groups from PNG seem to be mainly caused by haplogroups other than the four Melanesian Y haplogroups, whereas in WNG those differences are caused by the four Melanesian haplogroups.
Sex-Specific Migration in New Guinea
On the basis of Y-chromosome data, we observed that WNG groups from the highlands are genetically more similar to WNG lowland/coastal groups than the WNG highland groups are to PNG highland groups or the WNG lowland/coastal groups are to PNG coastal/island groups. Also, PNG highland groups are more similar to PNG coastal/island groups than they are to WNG highland or WNG lowland/coastal groups. In other words, on the basis of New Guinean Y-chromosome data, genetic differences between groups within the same geographic region but at different altitudes were generally smaller than those between groups from the same altitude but different geographic regions. In contrast, on the basis of mtDNA data, WNG groups from the highlands are genetically more similar to the PNG highland groups than to the WNG lowland/coastal groups, and, similarly, the PNG highland groups are more similar to the WNG highland groups than to PNG coastal/island groups. However, the inferences based on the PNG mtDNA sequences may be influenced by the nonrandom ascertainment of the samples selected for sequencing; mtDNA sequence diversity in the coastal PNG samples may be artificially low, since most of these samples were selected on the basis of the 9-bp deletion (Redd et al. 1995), whereas diversity in the PNG highlands may be overestimated, since samples with different SSO types were chosen for sequencing (Redd and Stoneking 1999).
Nevertheless, our results imply that gene flow within the highlands of New Guinea, between PNG and WNG, is primarily female-mediated, whereas gene flow between highland and lowland/coastal regions, within both WNG and PNG, is primarily male-mediated. Male-mediated contact between highland and lowland/coastal regions of New Guinea is also supported by the presence (albeit at very low frequency) of Austronesian Y chromosomes in the highlands of WNG and PNG (such as haplogroups O-M119 and O-M122), which could also reflect recent admixture. In contrast, the mtDNA 9-bp deletion, which is associated with the Austronesian expansion, has so far never been observed in the PNG highlands (Hertzberg et al. 1989; Melton et al. 1995; Redd et al. 1995; Sykes et al. 1995; Redd and Stoneking 1999) nor in WNG (Tommaseo-Ponzetta et al. 2002).
Correlation of Language and Genes in New Guinea
Haplogroups O-M119 and O-M122, which are of Asian origin and in New Guinea are found mainly—but not exclusively—in Austronesian-speaking coastal and island populations and in Austronesian-speaking southeastern Asians and Polynesians but are nearly absent from non-Austronesian-speaking populations of Melanesia, are though to have been brought to New Guinea by the Austronesian expansion (Kayser et al. 2000a, 2001a). Thus, at least for large language categories like the Austronesian and Papuan languages, a general language-gene correlation exists with respect to the Y chromosome. Within WNG, population structure analysis by means of AMOVA from Y-chromosome haplogroup and haplotype data revealed that grouping according to language is supported somewhat more by the Y-chromosome data than by mtDNA data. However, we also identified instances in WNG in which Y chromosomes and language are not correlated. For example, linguistically, the Yali are more closely related to their western neighbors, the Dani/Lani, all belonging to the Dani-Kwerba or Dani language family (Foley 1986). In contrast, our Y-chromosome data show that the Yali are different from the Dani/Lani and identical to their eastern neighbors, the Una and Ketengban, which belong to a different language family, the Mek. This is also reflected in the location of the Yali in the MDS plot (fig. 5), which is close to the Una/Ketengban but distant from the Dani/Lani. Additional genetic evidence for the Y-chromosomal relationship of the Yali with their eastern Mek-speaking, but not their western Dani-Kwerba–speaking, neighbors comes from the Y STR haplotype analysis. For the three haplotypes observed among the five Yali, two are shared with the Una and one with the Ketengban, but none is shared with the Dani/Lani. Linguistic data also indicate contact between the Yali and the Una/Ketengban, since extensive word borrowing is reported between the Yali and Mek languages (Heeschen 1998). Another example in which language and Y-chromosome genetics are not correlated is the Awy-Dumut language family, to which the Kombai, Korowai, and Awyu belong. The Kombai/Korowai show a Y-haplogroup composition different from that of the linguistically related Awyu (table 4; fig. 3), which explains their separate placement in the MDS plot (fig. 5). This difference comes from the large proportion of haplogroup K-M9 in the Kombai/Korowai, whereas K-M9 is completely absent from the Awyu. Thus, in general, there does not seem to be a significant correspondence between language and the Y chromosome in WNG.
Conclusions
We find that genetic variation in WNG is characterized by a reduced diversity of Y-chromosome DNA but not of mtDNA. This seems to reflect cultural features of these Papuan societies, such as their patrilocal residence, their patrilineal and exogamous social clan system, and the high frequency of polygyny. In addition, warfare, which existed until recently in WNG groups and mainly affected men but not women, may have contributed to a reduction of paternal but not maternal genetic lineages. Our data further provide evidence for primarily female-mediated gene flow within the highlands of New Guinea but primarily male-mediated gene flow between highland and lowland/coastal regions, both in WNG and PNG. We also find little correspondence between population relationships based on linguistic versus Y-chromosome data. Further sampling of highland populations, to address the apparent genetic differences between WNG and PNG highlanders, and of northern coast Austronesian and Papuan-speaking groups from WNG and PNG, to address the magnitude of genetic differentiation between these language families, will greatly aid our understanding of the genetic prehistory of New Guinea.
Acknowledgments
We are deeply grateful to the WNG communities, for their participation in this study; to all volunteers, for contributing cheek swab samples; and to the following colleagues, for providing DNA samples: N. Saha, A. G. Soemantri, A. S. M. Sofro, K. Bhatia, J. Kuhl, N. Kretchmer, D. Bugawan, E. Hagelberg, S. Ulijaszek, K. Katayama, J. Martinson, B. Budowle, and C. Tyler-Smith. R. Kittler is acknowledged, for DNA extraction of the WNG samples. We thank B. Voorhoeve, V. Heeschen, G. Reesink, and M. Ross, for useful comments on the linguistics and ethnology of WNG populations. M.T.-P. thanks C. Saccone and M. Attimonelli for helpful collaboration. This study was made possible through financial support from the Max Planck Society (Germany), the Italian Ministry of University and Scientific Research (MURST 60%, P.R.I.N. 2001–2003), and the Ligabue Study and Research Centre, Venice (Italy).
Electronic-Database Information
URLs for data presented herein are as follows:
- Arlequin's Home on the Web, http://lgb.unige.ch/arlequin/ (for Arlequin version 2.000 software)
- Ethnologue, Languages of the World, http://www.ethnologue.com/ (for Ethnologue, 14th edition)
- Mathematical Sciences at Aberdeen, http://www.maths.abdn.ac.uk/ (for BATWING software)
- PHYLIP Home Page, http://evolution.genetics.washington.edu/phylip.html (for PHYLIP version 3.5 software)
- Shareware Phylogenetic Network Software, http://www.fluxus-engineering.com/sharenet.htm (for NETWORK 2.0b software)
- TreeView, http://taxonomy.zoology.gla.ac.uk/rod/treeview.html (for TreeView version 1.6 software)
References
- Allen J, Flannery T, Gosden C, Jones R, White JP (1988) Pleistocene dates for the human occupation of New Ireland, Northern Melanesia. Nature 331:707–709 [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 [DOI] [PubMed] [Google Scholar]
- Bellwood PS (1978) Man's conquest of the Pacific: the prehistory of Southeast Asia and Oceania. Oxford University Press, Oxford [Google Scholar]
- ——— (1989) The colonization of the Pacific: some current hypotheses. In: Hill AVS, Serjeantson SW (eds) The colonization of the Pacific: a genetic trail. Oxford University Press, Oxford, pp 1–59 [Google Scholar]
- Brown P (1978) Highland peoples of New Guinea. Cambridge University Press, Cambridge [Google Scholar]
- Capelli C, Wilson JF, Richards M, Stumpf MPH, Gratrix F, Oppenheimer S, Underhill P, Pascali VL, Ko T-M, Goldstein DB (2001) A predominantly indigenous paternal heritage for the Austronesian-speaking people of insular southeast Asia and Oceania. Am J Hum Genet 68:432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Knijff P (2000) Messages through bottlenecks: on the combined use of slow and fast evolving polymorphic markers on the human Y chromosome. Am J Hum Genet 67:1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries L (1993) Forms and functions in Kombai, an Awyu language of Irian Jaya. Vol B-108 in: Dutton TE, Pawley AK, Ross MD, Tyron DT (eds) Pacific linguistics. Australian National University, Canberra [Google Scholar]
- Feil DK (1987) The evolution of highland Papua New Guinea societies. Cambridge University Press, Cambridge [Google Scholar]
- Felsenstein J (1993) PHYLIP (Phylogeny Inference Package) version 3.5c. Department of Genetics, University of Washington, Seattle [Google Scholar]
- Foley WA (1986) The Papuan languages of New Guinea. Cambridge University Press, Cambridge [Google Scholar]
- Gardner R, Heider KG (1969) Gardens of war: life and death in the New Guinea stone age. Andre Deutsch, London [Google Scholar]
- Grimes BF (2000) (ed) Ethnologue: languages of the world. 14th ed. Summer Institute of Linguistics, Dallas [Google Scholar]
- Groube LM, Chappell J, Muke J, Price D (1986) A 40,000 year-old human occupation site at Huon Peninsula, Papua New Guinea. Nature 324:453–455 [DOI] [PubMed] [Google Scholar]
- Hammer MF (1994) A recent insertion of an Alu element on the Y chromosome is a useful marker for human population studies. Mol Biol Evol 11:749–761 [DOI] [PubMed] [Google Scholar]
- Hammer MF, Horai S (1995) Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 56:951–962 [PMC free article] [PubMed] [Google Scholar]
- Hays TE (1991) Oceania. Vol 2 in: Levinson D (ed) Encyclopedia of world cultures. G.K. Hall, Boston [Google Scholar]
- Heeschen V (1998) An ethnographic grammar of the Eipo language. Vol 23 in: Helfrich K, Jacobshagen V, Koch G, Krieger K, Schiefenhövel W, Schultz W (eds) Mensch, Kultur und Umwelt im zentralen Bergland von West-Neuguinea. Dietrich Reimer Verlag, Berlin [Google Scholar]
- Heider KG (1970) The Dugum Dani: a Papuan culture in the highlands of West New Guinea. Aldine, Chicago [Google Scholar]
- ——— (1997) Grand valley Dani: peaceful warriors. Unnumbered vol in: Spindler G, Spindler L (eds) Case studies in cultural anthropology. Harcourt Brace College Publishers, Forth Worth, Texas [Google Scholar]
- Hertzberg M, Mickleson KNP, Serjeantson SW, Prior JF, Trent RJ (1989) An Asian-specific 9-bp deletion of mitochondrial DNA is frequently found in Polynesians. Am J Hum Genet 44:504–510 [PMC free article] [PubMed] [Google Scholar]
- Hurles ME, Irven C, Nicholson J, Taylor PG, Santos FR, Loughlin J, Jobling MA, Sykes BC (1998) European Y-chromosomal lineages in Polynesia: a contrast to the population structure revealed by mtDNA. Am J Hum Genet 63:1793–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles ME, Nicholson J, Bosch E, Renfrew C, Sykes B, Jobling MA (2002) Y chromosomal evidence of the origins of Oceanic-speaking peoples. Genetics 160:289–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet T, Xu L, Du R, Wang W, Feng S, Wells RS, Redd AJ, Zegura SL, Hammer MF (2001) Paternal population history of East Asia: source, patterns, and microevolutionary processes. Am J Hum Genet 69:615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet TM, Zegura SL, Posukh O, Osipova L, Bergen A, Long J, Goldman D, Klitz W, Harihara S, de Knijff P, Wiebe V, Griffiths RC, Templeton AR, Hammer MF (1999) Ancestral Asian source(s) of New World Y chromosome founder haplotypes. Am J Hum Genet 64:817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Brauer S, Weiss G, Schiefenhövel W, Underhill P, Stoneking M (2001a) Independent histories of human Y chromosomes from Melanesia and Australia. Am J Hum Genet 68:173–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Brauer S, Weiss G, Underhill P, Roewer L, Schiefenhövel W, Stoneking M (2000a) Melanesian origin of Polynesian Y chromosomes. Curr Biol 10:1237–1246 [DOI] [PubMed] [Google Scholar]
- ——— (2001b) Correction: Melanesian origin of Polynesian Y chromosomes. Curr Biol 11:I–II [DOI] [PubMed] [Google Scholar]
- Kayser M, Krawczak M, Excoffier L, Dieltjes P, Corach D, Cagliá A, Gehrig C, Bernini LF, Jespersen J, Bakker E, Roewer L, de Knijff P (2001c) Extensive analysis of chromosome Y microsatellite haplotypes in globally dispersed human populations. Am J Hum Genet 68:990–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Roewer L, Hedman M, Henke L, Henke J, Brauer S, Krüger C, Krawczak M, Nagy M, Dobosz T, Szibor R, de Knijff P, Stoneking M, Sajantila A (2000b) Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am J Hum Genet 66:1580–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Su B, Song X, Lu D, Chen L, Li H, Qi C, Marzuki S, Deka R, Underhill P, Xiao C, Shriver M, Lell J, Wallace D, Wells RS, Seielstad M, Oefner P, Zhu D, Jin J, Huang W, Chakraborty R, Chen Z, Jin L (2001) African origin of modern humans in east Asia: a tale of 12,000 Y chromosomes. Science 292:1151–1152 [DOI] [PubMed] [Google Scholar]
- Melton T, Peterson R, Redd AJ, Saha N, Sofro ASM, Martinson J, Stoneking M (1995) Polynesian genetic affinities with Southeast Asian populations as identified by mtDNA analysis. Am J Hum Genet 57:403–414 [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton R (1983) The lost world of Irian Jaya. Oxford University Press, Melbourne [Google Scholar]
- Oota H, Kitano T, Jin F, Yuasa I, Wang L, Ueda S, Saitou N, Stoneking M (2002) Extreme mtDNA homogeneity in continental Asian populations. Am J Phys Anthropol 118:146–153 [DOI] [PubMed] [Google Scholar]
- Oota H, Settheetham-Ishida W, Tiwawech D, Ishida T, Stoneking M (2001) Human mtDNA and Y-chromosome variation is correlated with matrilocal versus patrilocal residence. Nat Genet 29:20–21 [DOI] [PubMed] [Google Scholar]
- Osmond M, Pawley A, Ross M. Membership and subgrouping of the Trans New Guinea phylum. In: Pawley A, Ross M, Osmond M (eds) Trans New Guinea studies, vol 1. Reconstructing the early history of the Trans New Guinea family. Pacific Linguistics, Canberra (in press) [Google Scholar]
- Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358 [DOI] [PubMed] [Google Scholar]
- Pavlides C, Gosden C (1994) 35,000-year-old sites in the rainforests of West New Britain, Papua New Guinea. Antiquity 68:604–610 [Google Scholar]
- Pospisil L (1963) The Kapauku Papuans of West New Guinea. Unnumbered vol in: Spindler G, Spindler L (eds) Case studies in cultural anthropology. Holt, Rinehart and Winston, New York [Google Scholar]
- Redd AJ, Roberts-Thomson J, Karafet T, Bamshad M, Jorde LB, Naidu JM, Walsh B, Hammer MF (2002) Gene flow from the Indian subcontinent to Australia: evidence from the Y chromosome. Curr Biol 12:673–677 [DOI] [PubMed] [Google Scholar]
- Redd AJ, Stoneking M (1999) Peopling of Sahul: mtDNA variation in aboriginal Australian and Papua New Guinea populations. Am J Hum Genet 65:808–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd AJ, Takezaki N, Sherry ST, McGarvey ST, Sofro ASM, Stoneking M (1995) Evolutionary history of the COII/tRNA-lys intergenic 9 base pair deletion in human mitochondrial DNAs from the Pacific. Mol Biol Evol 12:604–615 [DOI] [PubMed] [Google Scholar]
- Ross M (1995) The great Papuan pronoun hunt: recalibrating our sights. In: Baak C, Bakker M, van der Meij D (eds) Tales from a concave world: liber amicorum Bert Voorhoeve. Leiden University Department of Languages and Cultures of South-East Asia and Oceania, Leiden, pp 139–168 [Google Scholar]
- ——— Defining the Trans New Guinea family: preliminary evidence from pronouns. In: Pawley A, Ross M, Osmond M (eds) Trans New Guinea studies, Vol 1. Reconstructing the early history of the Trans New Guinea family. Pacific Linguistics, Canberra (in press) [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, et al (2000) Y-chromosomal diversity within Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 67:1526–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefenhövel W (1988) Geburtsverhalten und reproduktive Strategien der Eipo. Vol 16 in: Helfrich K, Jacobshagen V, Koch G, Krieger K, Schiefenhövel W, Schultz W (eds) Mensch, Kultur und Umwelt im zentralen Bergland von West-Neuguinea. Dietrich Reimer Verlag, Berlin [Google Scholar]
- Schneider S, Roessli D, Excoffier L (2000) Arlequin ver 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland [Google Scholar]
- Seielstad M, Minch E, Cavalli-Sforza LL (1998) Genetic evidence for a higher female migration rate in humans. Nat Genet 20:278–280 [DOI] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, L.E. B, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA (2000) The genetic legacy of Palaeolithic Homo sapiens sapiens in extant Europeans: a Y-chromosome perspective. Science 290:1155–1159 [DOI] [PubMed] [Google Scholar]
- Su B, Jin L, Underhill P, Martinson J, Saha N, McGarvey ST, Shriver MD, Chu J, Oefner P, Chakraborty R, Deka R (2000) Polynesian origins: insights from the Y chromosome. Proc Natl Acad Sci USA 97:8225–8228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B, Xiao J, Underhill P, Deka R, Zhang W, Akey J, Huang W, Shen D, Lu D, Luo J, Chu J, Tan J, Shen P, Davis R, Cavalli-Sforza L, Chakraborty R, Xiong M, Du R, Oefner P, Chen Z, Jin L (1999) Y-chromosome evidence for a northward migration of modern humans into eastern Asia during the last Ice Age. Am J Hum Genet 65:1718–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes B, Leiboff A, Low-Beer J, Tetzner S, Richards M (1995) The origins of the Polynesians: an interpretation from mitochondrial lineage analysis. Am J Hum Genet 57:1463–1475 [PMC free article] [PubMed] [Google Scholar]
- Tommaseo-Ponzetta M (1986) Gli Asmat della Nuova Guinea. In: Centro studi e ricerche ligabue (ed) Indonesia, la grande deriva etnica. Erizzo, Venice, pp 125–134 [Google Scholar]
- Tommaseo-Ponzetta M, Attimonelli M, De Robertis M, Tanzariello F, Saccone C (2002) Mitochondrial DNA variability of West New Guinea populations. Am J Phys Anthropol 117:49–67 [DOI] [PubMed] [Google Scholar]
- Trenkenschuh FA, Hoogebrugge J (1970) Social structure of Asmat. In: Trenkenschuh FA (ed) Asmat sketch book 1. The Asmat Museum of Culture and Progress and Crosier Missions, Hastings, NE, pp 4–33 [Google Scholar]
- Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ (1997) Detection of numerous Y-chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res 7:996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill P, Passarino G, Lin AA, Marzuki S, Cavalli-Sforza LL, Chambers G (2001a) Maori origins, Y-chromosome haplotypes and implications for human history in the Pacific. Hum Mut 17:271–280 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley RA, Oeffner PJ, Cavalli-Sforza LL (2001b) The phylogeography of Y-chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y-chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 [DOI] [PubMed] [Google Scholar]
- van Enk GJ, de Vries L (1997) The Korowai of Irian Jaya. Vol 9 in: Bright W (ed) Oxford studies in anthropological linguistics. Oxford University Press, New York [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Lin J, Su B, Pitchappan R, Shanmugalakshmi S, Balakrishnan K, Read M, Pearson NM, Zerjal T, Webster MT, Zholoshvili I, Jamarjashvili E, Gambarov S, Nikbin B, Dostiev A, Aknazarov O, Zalloua P, Tsoy I, Kitaev M, Mirrakhimov M, Cariev A, Bodmer WF (2001) The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 98:10244–10249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield LS, Sulston JE, Goodfellow PN (1995) Sequence variation of the human Y chromosome. Nature 378:379–380 [DOI] [PubMed] [Google Scholar]
- Wickler S, Spriggs M (1988) Pleistocene human occupation of the Solomon Islands, Melanesia. Antiquity 62:703–706 [Google Scholar]
- Y Chromosome Consortium, The (2002) A nomenclature system of the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]