Abstract
Two tribal groups from southern India—the Chenchus and Koyas—were analyzed for variation in mitochondrial DNA (mtDNA), the Y chromosome, and one autosomal locus and were compared with six caste groups from different parts of India, as well as with western and central Asians. In mtDNA phylogenetic analyses, the Chenchus and Koyas coalesce at Indian-specific branches of haplogroups M and N that cover populations of different social rank from all over the subcontinent. Coalescence times suggest early late Pleistocene settlement of southern Asia and suggest that there has not been total replacement of these settlers by later migrations. H, L, and R2 are the major Indian Y-chromosomal haplogroups that occur both in castes and in tribal populations and are rarely found outside the subcontinent. Haplogroup R1a, previously associated with the putative Indo-Aryan invasion, was found at its highest frequency in Punjab but also at a relatively high frequency (26%) in the Chenchu tribe. This finding, together with the higher R1a-associated short tandem repeat diversity in India and Iran compared with Europe and central Asia, suggests that southern and western Asia might be the source of this haplogroup. Haplotype frequencies of the MX1 locus of chromosome 21 distinguish Koyas and Chenchus, along with Indian caste groups, from European and eastern Asian populations. Taken together, these results show that Indian tribal and caste populations derive largely from the same genetic heritage of Pleistocene southern and western Asians and have received limited gene flow from external regions since the Holocene. The phylogeography of the primal mtDNA and Y-chromosome founders suggests that these southern Asian Pleistocene coastal settlers from Africa would have provided the inocula for the subsequent differentiation of the distinctive eastern and western Eurasian gene pools.
Introduction
The origins of the culturally and genetically diverse populations of India have been subject to numerous anthropological and genetic studies (reviewed by Walter et al. 1991; Cavalli-Sforza et al. 1994; Papiha 1996). It remains unsettled whether the genetic diversity seen between different Indian populations primarily reflects their local long-term differentiation or is due to relatively recent migrations from abroad. More than 300 tribal groups are recognized in India, and they are densest in the central and southern provinces. Most of them currently speak Austro-Asiatic, Tibeto-Burman, or Dravidic languages and are often considered representative of the people that preceded the arrival of Aryan populations, whose language is dominant among Indian caste populations now. Historically, it is known that many groups have entered India during the last millennia, as immigrants or as invaders. The magnitude of the genetic contribution of the recent migrations to the Indian subcontinent (assuming, of course, that one can discern the “old and native” genetic contributions) and whether, specifically, the linguistic relatedness of Indo-European speakers is expressed in the genetic landscape (Passarino et al. 1996; Kivisild et al. 1999a, 1999b; Bamshad et al. 2001; Passarino et al. 2001; Quintana-Murci et al. 2001; Shouse 2001; Wells et al. 2001) remains open to debate.
Studies based on mtDNA have shown that, among Indians, the basic clustering of lineages is not language- or caste-specific (Mountain et al. 1995; Kivisild et al. 1999a; Bamshad et al. 2001), although a low number of shared haplotypes indicates that recent gene flow across linguistic and caste borders has been limited (Bamshad et al. 1998; Bhattacharyya et al. 1999; Roychoudhury et al. 2001). More than 60% of Indians have their maternal roots in Indian-specific branches of haplogroup M. Because of its great time depth and virtual absence in western Eurasians, it has been suggested that haplogroup M was brought to Asia from East Africa, along the southern route, by the earliest migration wave of anatomically modern humans, ∼60,000 years ago (Kivisild et al. 1999a, 1999b, 2000; Quintana-Murci et al. 1999). Another deep late Pleistocene link through haplogroup U was found to connect western Eurasian and Indian populations. Less than 10% of the maternal lineages of the caste populations had an ancestor outside India in the past 12,000 years (Kivisild et al. 1999a, 1999b). mtDNA profiles from a larger set of populations all over the subcontinent have bolstered the view of fundamental genomic unity of Indians (Roychoudhury et al. 2001). In contrast, the Y-chromosome genetic distance estimates showed that the chromosomes of Indian caste populations were more closely related to Europeans than to eastern Asians (Bamshad et al. 2001). The tendency of higher caste status to associate with increasing affinities to European (specifically to eastern European) populations hinted at a recent male-mediated introduction of western Eurasian genes into the Indian castes' gene pool. The similarities with Europeans were specifically expressed in substantial frequencies of clades J and R1a (according to Y Chromosome Consortium [YCC] 2002 nomenclature) in India. The exact location of the origin of these haplogroups is still uncertain, as is the timing of their spread (Zerjal et al. 1999; Bamshad et al. 2001; Passarino et al. 2001; Quintana-Murci et al. 2001; Wells et al. 2001).
To address the question of the origin of Indian maternal and paternal lineages further, we analyzed variation in mtDNA, the Y chromosome, and one autosomal locus (Jin et al. 1999) in two southern Indian tribal groups from Andhra Pradesh and compared them with Indian caste groups and populations from Iran, the Middle East, Europe, and central Asia.
Material and Methods
Subjects
Chenchus were first described as shy hunter-gatherers by the Mohammedan army in 1694. They reside in the ranges of Amrabad Plateau, Andhra Pradesh, and have a population size of ∼17,000. Their society is patriarchal and patrilineal, with marriage occurring mostly between clans (kulam) of equal status. Chenchus are described as an australoid population, when physical anthropological features are used as criteria (Bhowmick 1992; Singh 1997; Thurin 1999). The Chenchu language belongs to the Dravidian language family.
More than 300,000 Koyas live in the plains and forests on both sides of the Godavari River in Andhra Pradesh. Their language is related to the Gondi, which connects a large group of Dravidian languages in southern India. They are primarily farmers and live in villages. Exogamous patrilocal clans make up their social structure, as they do for the Chenchus (Singh 1997).
After informed consent was obtained, 180 blood samples were collected from healthy and maternally unrelated volunteers belonging to Chenchu and Koya tribes from Andhra Pradesh, 106 West Bengalis of different caste ranks, 58 Konkanastha Brahmins from Bombay, and 53 Gujaratis; in addition, 132 samples were collected from Sri Lanka (including 40 Sinhalese), 112 from Punjabis of different caste rank, and 139 samples from Uttar Pradesh (including those described by Kivisild et al. 1999a). The Lambadi (n=86) sample from Andhra Pradesh, the Boksas (n=18) from Uttar Pradesh, and the Lobanas (n=62) from Punjab are described by Kivisild et al. (1999a) and were further analyzed here for Y-chromosomal and additional mtDNA markers. The Y-chromosome sample size of each population is shown in figure 3. In addition, 388 Turks from central Turkey (Cappadocia), 202 Kuwaitis, 202 Saudi Arabians, and 440 Iranians were used in mtDNA haplogroup frequency comparisons. In addition, Y-chromosome STR data from six loci were used for comparing intra- and interhaplogroup variances in selected haplogroups and populations. These included 88 central Asians (Altais, Kirghiz, Uzbek, and Tajik) belonging to haplogroup R1a, and Y chromosomes from haplogroups I (12 Estonians and 9 Czechs), J (6 Czechs), and R1 (39 Estonians and 30 Czechs). Further details about these populations will be published elsewhere. DNA was extracted using standard phenol-chloroform methods (Sambrook et al. 1989).
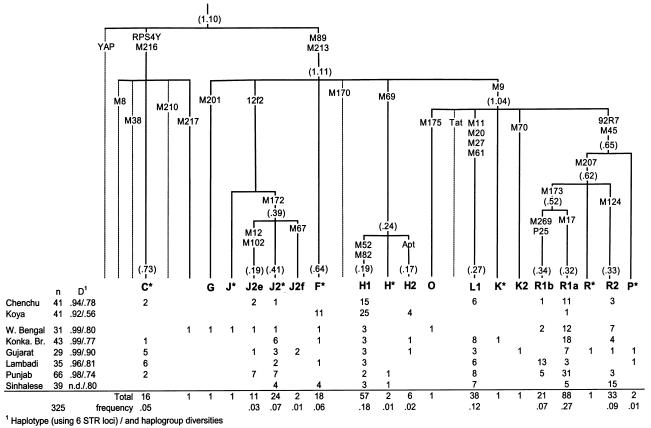
Figure 3.
Y-chromosomal SNP tree and haplogroup frequencies in 8 Indian populations. Haplogroup defining markers (and their background average variances of 6 STR loci) are shown along the branches of the tree.
mtDNA Analyses
Hypervariable segments (HVS) I (nucleotide positions [nps] 16024–16400) and II (nps 16520–300) of the control region were sequenced in 96 Chenchu and 81 Koya samples. In addition, three segments of the coding region (nps 1674–1880, 4761–5260, and 8250–8710) were sequenced, and informative RFLP positions (Macaulay et al. 1999; Quintana-Murci et al. 1999) were checked (table 1) in selected individuals from different haplotypes, to define haplogroup affiliations. Published HVS-I sequence data used for haplotype comparisons included 250 Telugus from Andhra Pradesh (Kivisild et al. 1999a; Bamshad et al. 2001), 48 Haviks, 43 Mukris, and 7 Kadars from Kerala and Karnataka (Mountain et al. 1995).
Table 1.
mtDNA Haplotypes in Chenchu and Koya Populations[Note]
| Type | Group | HVS-I (minus 16000) | 16519 | HVS-II | 447 | 663e | 1674–1880;4761–5260;8250–8710 | 3537a | 4577q | 5584a | 5823a | 7025a | 7598f | 8249b | 9820g | 10237y | 10364e | 10394c | 10397a | 10871z | 11465u | 12308g | 12403z | 12406o | 12704w | 13259o | 14766u | 15606a | 15925i | 15954j | Chenchu | Koya | Matcha |
| H | ref | T | ref | C | − | ref | + | + | + | + | − | + | − | − | − | + | − | − | + | + | − | + | + | + | + | − | − | + | − | ||||
| 1 | M | 223 | C | 73 199 263 | C | 4769 8701 | + | + | + | + | + | − | + | + | − | + | 11 | ** | |||||||||||||||
| 2 | M | 223 | C | 73 200 263 | + | + | + | 1 | ** | ||||||||||||||||||||||||
| 3 | M | 223 | C | 55+T 59del 60del 65+T 73 153 263 | + | + | + | 1 | ** | ||||||||||||||||||||||||
| 4 | M | 223 352 353 | C | 55+T 59del 60del 65+T 73 207 263 | C | 1811 4769 4928 8679 8701 8704 | + | + | − | + | + | + | − | + | + | − | + | 25 | |||||||||||||||
| 5 | M | 223 352 353 | C | 55+T 59del 60del 65+T 73 152 207 263 | + | + | + | 1 | |||||||||||||||||||||||||
| 6 | M | 93 223 | C | 73 146 199 263 | C | 4769 8701 | + | + | − | + | + | 1 | ** | ||||||||||||||||||||
| 7 | M | 183C 189 223 | C | 73 146 153 173 207 246 263 | C | 4769 5027 8701 | − | + | + | + | − | + | 1 | ||||||||||||||||||||
| 8 | M | 129 223 311 | C | 73 152 263 | C | 4769 8701 | + | + | + | − | 1 | ** | |||||||||||||||||||||
| 9 | M | 129 144A 145T 223 362 | C | 73 234 263 | C | 4769 8701 | + | + | + | − | 2 | ||||||||||||||||||||||
| 10 | M | 129 144A 223 362 | C | 73 234 263 | C | 1681 4769 8701 | + | + | + | − | + | 6 | ** | ||||||||||||||||||||
| 11 | M | 129 140 189 243 362 | C | 73 146 152 228 234 263 | C | 4769 9bpdel 8573 8584 8701 | + | + | + | + | + | + | − | + | − | + | 17 | ||||||||||||||||
| 12 | M | 179 223 294 | T | 73 146 152 189 195 249del 263 | C | 4769 8701 | + | + | + | − | + | + | + | 3 | |||||||||||||||||||
| 13 | M | 223 318T 325 | C | 73 246 263 | C | 4769 8277 8279 8701 | + | + | − | + | + | + | − | + | − | + | − | 4 | ** | ||||||||||||||
| 14 | M | 223 327 330 | C | 73 263 | C | 4769 8701 | + | + | + | + | + | − | + | + | − | + | 2 | ||||||||||||||||
| 15 | M | 86 218 223 270 362 | T | 73 185 195 199 204 228 262 263 | C | 4769 8701 | + | + | + | − | + | + | 1 | ||||||||||||||||||||
| 16 | M | 184 223 256G 311 362 | C | 37 73 146 152 263 | C | − | 4769 9bpdel 8701 | + | + | + | − | + | + | + | − | + | − | + | 3 | ||||||||||||||
| 17 | M | 169 172 223 | C | 73 200 263 | C | 4769 8562 8701 | + | + | − | + | + | + | − | + | + | − | 8 | * | |||||||||||||||
| 18 | M | 169 172 176 223 | C | 73 200 263 | C | 4769 8562 8701 | + | + | + | − | + | + | 3 | ||||||||||||||||||||
| 19 | M | 93 169 172 185 186 189 223 278 | C | 73 150 263 | C | 4769 5124A 8562 8701 | + | + | + | − | + | 1 | |||||||||||||||||||||
| 20 | M2a | 110 223 270 274 292 319 352 | C | 73 152 204 263 | G | 1780 4769 5252 8396 8502 8701 | + | + | + | − | 1 | ||||||||||||||||||||||
| 21 | M2a | 187 223 270 274 319 352 | C | 73 200 204 262 263 | G | 1780 4769 5252 8396 8502 8701 | + | + | − | + | + | − | + | − | 8 | ||||||||||||||||||
| 22 | M2a | 223 239 270 319 352 | C | 73 195 203 204 263 | G | + | + | + | − | 5 | |||||||||||||||||||||||
| 23 | M2a | 223 256 270 274 292 319 352 | C | 73 152 204 263 | G | 1780 4769 5252 8396 8502 8701 | + | + | + | − | 1 | ||||||||||||||||||||||
| 24 | M2a | 223 270 274 292 319 352 | C | 73 152 204 263 | G | 1780 4769 5082 5252 8396 8502 8701 | + | + | + | − | 6 | * | |||||||||||||||||||||
| 25 | M2a | 223 270 274 292 319 352 | C | 73 204 263 | + | + | 1 | * | |||||||||||||||||||||||||
| 26 | M2a | 223 270 274 319 352 | C | 73 204 263 | G | 1780 4769 5252 8396 8502 8701 | + | + | + | − | 2 | ** | |||||||||||||||||||||
| 27 | M2a | 223 270 274 319 352 | C | 73 203 204 263 | + | + | 1 | ** | |||||||||||||||||||||||||
| 28 | M2a | 93 223 243 248 270 319 352 | C | 73 195 204 263 | G | 1780 4769 5252 8396 8502 8701 | + | + | + | − | 2 | ||||||||||||||||||||||
| 29 | M2a | 93 223 243 270 319 352 | C | 73 195 204 263 | G | 1780 4769 5252 8396 8502 8701 | + | + | + | − | 2 | ||||||||||||||||||||||
| 30 | M2b | 167+C 183C 189 223 274 295 319 320 | T | G | 1780 4769 8502 8701 | + | + | − | − | 1 | |||||||||||||||||||||||
| 31 | M2b | 183C 189 223 274 295 316 319 320 | C | 73 182 195 263 | G | 1780 4769 8502 8701 | + | + | − | 1 | |||||||||||||||||||||||
| 32 | M2b | 223 274 301 319 | C | 73 146 263 | G | 1780 4769 8502 8701 | − | + | + | + | − | 1 | |||||||||||||||||||||
| 33 | M3 | 126 192 223 | C | 73 131 152 263 | C | 4769 8701 | + | + | + | + | + | + | − | + | − | 2 | ** | ||||||||||||||||
| 34 | M3 | 126 192 223 | C | 73 131 152 228 263 | + | + | 1 | ** | |||||||||||||||||||||||||
| 35 | M3 | 126 223 | C | 73 131 152 263 | C | 4769 4935 8701 | + | + | + | + | − | 1 | ** | ||||||||||||||||||||
| 36 | M3 | 126 223 362 | C | 73 131 152 263 | C | + | + | + | + | − | 1 | ||||||||||||||||||||||
| 37 | M3 | 93 126 145 223 | T | 73 195 263 | C | 4769 4907 8701 | + | + | + | + | − | 1 | * | ||||||||||||||||||||
| 38 | M6 | 188 223 231 362 | C | 73 146 263 | C | 4769 8701 | − | − | + | + | − | + | + | + | − | + | − | + | + | 18 | ** | ||||||||||||
| 39 | N | 111 144 223 256 311 | C | − | 1719 4769 | + | + | + | − | − | + | − | − | + | + | − | + | 2 | |||||||||||||||
| 40 | R | 129 266 318 320 362 | C | 73 228 263 | C | 4769 8511 8584 8650 | + | + | + | + | − | − | + | + | − | + | + | + | − | 5 | |||||||||||||
| 41 | R | 129 362 | C | 73 228 263 | C | 4769 | + | + | + | + | − | − | + | + | − | + | + | + | 1 | ** | |||||||||||||
| 42 | R | 153 266 | C | 16524 16560 73 93 146 200 263 | C | 4769 8594 | + | + | + | + | − | − | + | + | − | + | + | + | − | 2 | |||||||||||||
| 43 | R | 153 302 | C | 73 195 263 | C | 4769 8646 | + | + | + | + | − | − | + | − | + | + | + | − | 3 | ||||||||||||||
| 44 | R | 190+A 291 324 | C | 4769 | + | + | + | − | + | + | + | + | − | 1 | |||||||||||||||||||
| 45 | R | 260 261 311 319 362 | C | 73 146 152 263 | C | 1804 4769 | + | + | + | + | − | − | + | + | − | + | + | + | − | 7 | |||||||||||||
| 46 | R | 260 261 311 319 362 | C | 73 146 152 199 263 | − | − | 2 | ||||||||||||||||||||||||||
| 47 | R | 324 | C | 73 195 198 263 | C | 4769 8598 | + | + | + | − | + | + | − | + | + | + | − | 3 | |||||||||||||||
| 48 | R | 93 179 227 245 266 278 362 | C | 73 195 246 263 | C | 4769 4991 5147 | + | + | + | + | − | − | + | + | − | + | + | + | − | 1 | |||||||||||||
| 49 | R | ref | C | 73 185 189 195 263 | C | 4769 | + | + | + | + | − | − | + | + | − | + | + | + | 1 | ** | |||||||||||||
| 50 | U2c | 51 169 234 278 | C | 73 152 263 | C | 1811 4769 | + | + | + | − | − | + | − | + | + | + | 1 |
** | |||||||||||||||
| Total | 96 | 81 | |||||||||||||||||||||||||||||||
| D | .87 | .94 |
Note.— Sequence differences are numbered according the reference sequence (ref) (Anderson et al. 1981; Andrews et al. 1999). Transversions and length variants (del = deletion; + = insertion) are specified. Restriction enzymes are coded as follows: a = AluI; b = AvaII; c = DdeI; e = HaeIII; f = HhaI; g = HinfI; i = MspI; j = MboI; o = HincII; q = NlaIII; u = MseI; w = MboII; y = HphI; z = MnlI. The presence or absence of a restriction site is indicated by a plus sign (+) or minus sign (−), respectively. “9bpdel” refers to a deletion between nps 8281 and 8289.
Matching HVS-I haplotypes with other Indian populations. One asterisk (*) indicates a match within the state of Andhra Pradesh; two asterisks (**) indicate a match with any Indian population (listed in the “Material and Methods” section).
Y-Chromosome Analyses
Y-chromosomal haplogroups were determined by RFLP and denaturing high-performance liquid chromatography (dHPLC) methods, using 35 biallelic markers (Rosser et al. 2000; Underhill et al. 2000, 2001b) that are shown in hierarchical relation to one another in figure 3. Length variation at six STR loci (DYS19, DYS388, DYS390, DYS391, DYS392, and DYS393) was typed using Cy5-labeled primers, and amplification products were subjected to electrophoresis on ALF Express (Pharmacia-Amersham). Scoring of repeat lengths was standardized by use of controls sequenced by P. de Knijff.
MX1 Locus
A 246-bp segment of the MX1 locus of chromosome 21, containing eight polymorphic sites in humans, was sequenced, and a nearby StuI recognition site polymorphism was determined according to Jin et al. (1999) in 42 Chenchus, 28 Koyas, 35 West Bengalis, 34 Punjabis, and 35 Turks from Cappadocia.
Sequencing
Preparation of sequencing templates was performed according to methods described by Kaessmann et al. (1999). Purified products were sequenced with the DYEnamic ET terminator cycle sequencing kit (Amersham Pharmacia Biotech) and were analyzed on an ABI 377 DNA Sequencer. Sequences were aligned and analyzed with the Wisconsin Package (GCG).
Data Analysis
Median networks (Bandelt et al. 1995, 1999) were constructed using the Network 2.0 program (A. Röhl; Shareware Phylogenetic Network Software Web site) with default settings. Cluster ages were calculated using ρ, the averaged distance to a specified founder haplotype, according to Forster et al. (1996), as well as standard errors as described by Saillard et al. (2000a). In mtDNA coalescent calculations, using the estimator ρ, we use a mutation rate of one transition in the segment between nps 16090 and 16365 per 20,180 years, calibrated with the inference that links Eskimo-Aleutian haplogroup A diversity to their post–Younger Dryas population expansion (Forster et al. 1996).
To calculate the 95% credible regions (CR) from the posterior distribution of the proportion of a group of lineages in the population, we used the program SAMPLING, kindly provided by V. Macaulay.
Haplotype diversity was estimated as
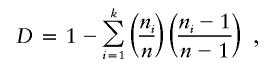 |
where n is the number of sequences, k is the number of distinct haplotypes, and ni is the number of sequences with one distinct haplotype.
A multidimensional scaling (MDS) analysis was performed on the Fst values between populations obtained from Arlequin version 2.0. The Fst scores were entered as distance measures, and a two-dimensional solution was calculated and plotted using SPSS 11.0
Admixture Estimation
Although some admixture models and programs exist and are used in population genetics (Bertorelle and Excoffier 1998; Helgason et al. 2000b; Chikhi et al. 2001, 2002; Qamar et al. 2002), they are not always adequate and realistic estimators of gene flow between populations. This is particularly the case when markers are used that do not have enough restrictive power to determine the source populations (Hammer et al. 2000) or when there are more than two parental populations. In that case, a simplistic model using two parental populations would show a bias towards overestimating admixture. The estimate of 23%–40% Greek admixture in the Pakistani Kalash population (Qamar et al. 2002), for example, is unrealistic and is likely also driven by the low marker resolution that pooled southern and western Asian–specific Y-chromosome haplogroup H together with European-specific haplogroup I, into an uninformative polyphyletic cluster 2.
To avoid these potential artifacts, we used the ratio of the sum of i discriminating haplogroup frequencies in the hybrid (ph) and source (ps) populations to estimate the admixture proportions relative to the closest outgroup population to the hybrid (p0):
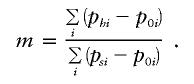 |
The discriminative haplogroups were determined by comparing the 95% confidence regions of the frequency portions of the source and outgroup (the closest known sister population with minimal impact from the source) populations. This method expands upon that of Shriver et al. (1997), by revealing population-specific alleles distinguishing the source populations.
Results
mtDNA Variation
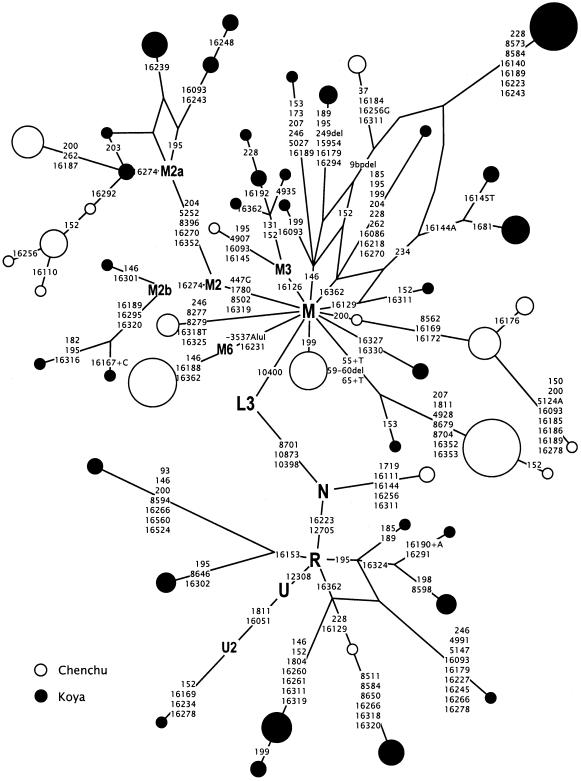
The analysis of mtDNA variation revealed the presence of 18 and 32 different haplotypes among 96 Chenchus and 81 Koyas, respectively (table 1). Although 26 haplotypes occurred in two or more individuals, there was no haplotype sharing between the two regionally close populations. However, an extended haplogroup-based search in the available HVS-I database of 1,093 Indian samples (Mountain et al. 1995; Kivisild et al. 1999a; Bamshad et al. 2001; present study) revealed that 20 haplotypes observed in Chenchus and Koyas are also found in caste populations, and 16 of them were present outside Andhra Pradesh in different populations of India. The unique HVS-I haplotypes of the Chenchus and Koyas had either one- or two-mutational-step neighbors in the total Indian data set, except for one Chenchu haplogroup M sequence, which differed from its closest phylogenetic neighbor by five transitions. In contrast, a search of a western Asian database consisting of 1,232 samples revealed only two unspecific HVS-I matches with Chenchu and Koya HVS-I haplotypes: one in haplogroup M, where 1 Iranian, 12 Chenchus, and 1 Koya shared the root motif (16223T), and in haplogroup R, where 3 Iranians shared the consensus HVS-I type (CRS) with 1 Koya. None of the derived haplotypes of the Chenchus and Koyas yielded a match with any western Asian mtDNA sequence.
All mtDNA sequences of Chenchus and Koyas (table 1; fig. 1) could be allotted to general Eurasian haplogroups M and N (including R) (Quintana-Murci et al. 1999; Kivisild et al. 2002). Asian-specific haplogroup M was the most frequent lineage cluster among both tribal groups, accounting for all but three lineages among Chenchus. Haplotype diversity in Chenchus (0.87; table 1) was small and comparable to values observed among the European outlier population the Saami (Helgason et al. 2000a). The coalescence time estimate 61,800±12,600 years for Chenchu and Koya haplogroup M HVS-I sequences is slightly older than, but within the error range of, previous estimates of haplogroup M age in Indian populations (Kivisild et al. 1999b; Quintana-Murci et al. 1999; Roychoudhury et al. 2001). All of the non-M sequences (31% of Koya mtDNAs) belonged to unclassified branches of haplogroup R, with the exception of one Indian-specific U2 (Kivisild et al. 1999a) sequence. When compared with Indian caste populations, Chenchus and Koyas are characterized by the rarity of haplogroup U and, like tribal groups from West Bengal and Tamil Nadu, by the lack of western Eurasian lineage clusters HV, TJ, N1, and X. These four clades combined cover ∼60% of the western Asian mtDNAs in India. Their frequency is the highest in Punjab, ∼20%, and diminishes threefold, to an average of 7%, in the rest of the caste groups in India (table 2).
Figure 1.
A network relating Chenchu and Koya mtDNA haplotypes. Node areas are proportional to haplotypes frequencies. Variant bases are numbered (Anderson et al. 1981) and shown along links between haplotypes. Character change is specified only for transversions. Insertions and deletions are indicated by “+” and “del,” respectively. Variation at hypervariable positions 16183 and 16517 is not shown.
Table 2.
Major mtDNA Lineage Clusters in India and Western Asia
|
No. of Lineages in Cluster (95% CR for Proportion)a |
||||||||||||
| Population (n) | Db | L1–L3 | M | M2 | M3 | M5 | MΔ9bp | U2i, U7 | U1, U3–U6, U* | HVc, TJ, N1, X | B, Fd | R* |
| Tribal: | ||||||||||||
| Chenchus (96)e | .87 | 0 (.00–.03) | 93 (.91–.99) | 17 (.11–.27) | 1 (.00–.06) | 18 (.12–.28) | 3 (.01–.09) | 0 (.00–.03) | 0 (.00–.03) | 0 (.00–.03) | 0 (.00–.03) | 1 (.00–.06) |
| Koyas (81)e | .94 | 0 (.00–.04) | 56 (.58–.78) | 15 (.12–.28) | 5 (.03–.14) | 0 (.00–.04) | 17 (.14–.31) | 1 (.00–.07) | 0 (.00–.04) | 0 (.00–.04) | 0 (.00–.04) | 25 (.22–.42) |
| Tamil Nadu (49)f | .96 | 0 (.00–.06) | 35 (.58–.82) | 1 (.01–.11) | 12 (.15–.38) | 0 (.00–.06) | 0 (.00–.06) | 6 (.06–.24) | 2 (.01–.14) | 0 (.00–.06) | 0 (.00–.06) | 6 (.06–.24) |
| Western Bengal (34)f | .99 | 0 (.00–.08) | 22 (.48–.79) | 2 (.02–.19) | 3 (.03–.23) | 0 (.00–.08) | 0 (.00–.08) | 7 (.10–.37) | 0 (.00–.08) | 0 (.00–.08) | 0 (.00–.08) | 5 (.07–.30) |
| Caste: | ||||||||||||
| Western Bengalis (106)e | .97 | 0 (.00–.03) | 76 (.63–.79) | 4 (.02–.09) | 7 (.03–.13) | 6 (.03–.12) | 0 (.00–.03) | 10 (.05–.17) | 1 (.01–.05) | 6 (.03–.12) | 0 (.00–.03) | 12 (.07–.19) |
| Gujaratis and Konkanastha Br. (111)e | .99 | 0 (.00–.03) | 53 (.39–.57) | 5 (.02–.10) | 7 (.03–.13) | 0 (.00–.03) | 0 (.00–.03) | 20 (.12–.26) | 5 (.02–.10) | 11 (.06–.17) | 5 (.02–.10) | 12 (.06–.18) |
| Kerala/Karnataka (99)g | .96 | 0 (.00–.03) | 63 (.54–.72) | 15 (.09–.24) | 6 (.03–.13) | 15 (.09–.24) | 0 (.00–.03) | 20 (.14–.29) | 1 (.01–.05) | … | 0 (.00–.03) | 9 (.05–.16) |
| Lambadis (86)h | .99 | 0 (.00–.03) | 55 (.53–.73) | 9 (.06–.19) | 4 (.02–.11) | 9 (.06–.19) | 0 (.00–.03) | 9 (.06–.19) | 2 (.01–.08) | 7 (.04–.16) | 0 (.00–.03) | 11 (.07–.22) |
| Lobanas (62)h | .98 | 0 (.00–.05) | 34 (.43–.67) | 3 (.02–.13) | 3 (.02–.13) | 5 (.04–.18) | 0 (.00–.05) | 2 (.01–.11) | 1 (.00–.09) | 5 (.04–.18) | 0 (.00–.05) | 11 (.10–.29) |
| Punjabis (112)e | .99 | 0 (.00–.03) | 46 (.32–.50) | 1 (.00–.05) | 5 (.02–.10) | 1 (.00–.05) | 0 (.00–.03) | 15 (.08–.21) | 8 (.04–.14) | 21 (.13–.27) | 6 (.03–.11) | 11 (.06–.17) |
| Sri Lanka (132)e | .99 | 0 (.00–.02) | 77 (.50–.66) | 9 (.04–.13) | 6 (.02–.10) | 2 (.01–.05) | 0 (.00–.02) | 19 (.09–.21) | 5 (.02–.09) | 11 (.05–.14) | 2 (.01–.05) | 18 (.09–.21) |
| Telugu, upper (59)i | .99 | 0 (.00–.05) | 36 (.48–.72) | 3 (.02–.14) | 11 (.11–.30) | 0 (.00–.05) | 0 (.00–.05) | 10 (.10–.29) | 1 (.00–.09) | 2 (.01–.11) | 0 (.00–.05) | 9 (.08–.27) |
| Telugu, middle (114)i | .99 | 0 (.00–.03) | 73 (.55–.72) | 7 (.03–.12) | 4 (.01–.09) | 5 (.02–.10) | 0 (.00–.03) | 10 (.05–.15) | 1 (.01–.05) | 6 (.03–.11) | 0 (.00–.03) | 24 (.15–.29) |
| Telugu, lower (70)i | .99 | 0 (.00–.04) | 50 (.60–.81) | 7 (.05–.19) | 1 (.00–.08) | 3 (.02–.12) | 0 (.00–.04) | 5 (.03–.16) | 0 (.00–.04) | 1 (.00–.08) | 0 (.00–.04) | 15 (.14–.32) |
| Uttar Pradesh (139)e,h | .99 | 0 (.00–.02) | 79 (.49–.65) | 4 (.01–.07) | 14 (.06–.16) | 0 (.00–.02) | 0 (.00–.02) | 21 (.10–.22) | 3 (.01–.06) | 9 (.04–.12) | 2 (.00–.05) | 20 (.10–.21) |
| West Asian: | ||||||||||||
| Iranians (440)e | .99 | 2 (.00–.02) | 24 (.04–.08) | 0 (.00–.01) | 5 (.01–.03) | 0 (.00–.01) | 0 (.00–.02) | 41j (.07–.12) | 90 (.17–.25) | 245 (.51–.60) | 2 (.00–.02) | 17 (.02–.06) |
| Turks, Cappadocia (388)e | .99 | 1 (.00–.01) | 16 (.03–.07) | 0 (.00–.01) | 0 (.00–.01) | 0 (.00–.01) | 0 (.00–.01) | 7 (.01–.04) | 93 (.20–29) | 244 (.58–.68) | 1 (.00–.01) | 7 (.01–.04) |
| Middle East (406)e,k | .99 | 41 (.08–.13) | 30 (.05–.10) | 2 (.00–.02) | 1 (.00–.01) | 0 (.00–.01) | 0 (.00–.03) | 15 (.02–.06) | 40 (.07–.13) | 269 (.62–.71) | 1 (.00–.01) | 3 (.00–.02) |
U2i excludes variants of U2 with 16129C; U* = other derivatives of haplogroup U; R* = derivatives of haplogroup R that do not belong to HV, TJ, U, B, and F.
HVS-I haplotype diversity.
Including pre-HV, as defined by Saillard et al. 2000b.
None of the Indian samples belonged to haplogroup B.
From the present study.
From Roychoudhury et al. 2001.
From Moutain et al. 1995.
From Kivisild et al. 1999.
From Bamshad et al. 2001.
Forty Iranians belonged to U7 and one belonged to U2i.
The Middle East sample includes 202 Kuwaitis and 204 Saudi Arabians.
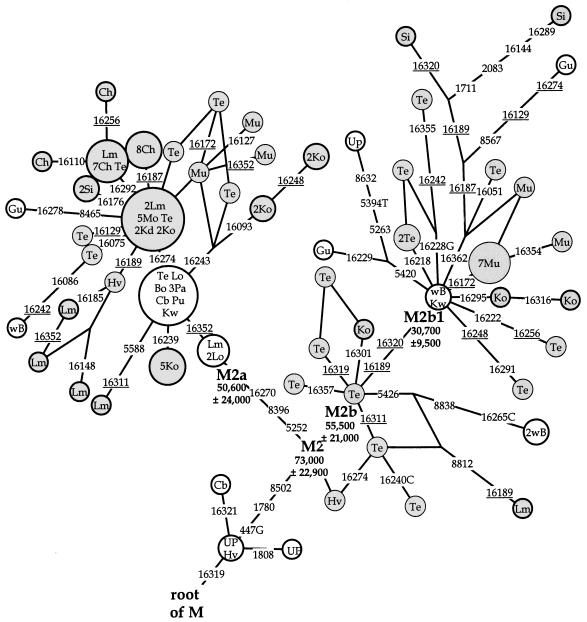
The most frequent M subcluster in Chenchus and Koyas was M2, which was also frequently found in caste groups of southern India but was virtually absent away from India (table 2). Phylogenetic analysis of HVS-I sequence variation in M2 showed that it is composed of two subtrees: M2a and M2b (fig. 2). Additional coding-region sequencing identified three polymorphisms (447G, 1780, and 8502) that define M2. Furthermore, two mutations were found to be specific to subcluster M2a (fig. 2). The calculated cluster ages showed that haplogroup M2 is an ancient haplogroup with a coalescence time of 73,000±22,900 years. Both widely spread subtrees also showed deep coalescence times, consistent with their divergence in India. In addition to M2, two other major clades, M3 and M6 (Bamshad et al. 2001), were found in Chenchus and Koyas in common with the caste groups (table 2). Furthermore, 32% of the Koya M* HVS-I sequences shared an A at hypervariable np 16129, which is characteristic of a likely polyphyletic HVS-I clade M5 (Bamshad et al. 2001). The loss of 12403 MnlI, one of the four defining markers of African M1 cluster (Maca-Meyer et al. 2001), was not found in either tribal sample (table 1).
Figure 2.
A network of haplogroup M2 haplotypes. Circle areas are proportional to haplotypes frequencies. Variant bases are numbered as in figure 1. Bold circles represent haplotypes for which indicated coding region markers were determined. Recurrent mutations are underlined. Haplotypes restricted to South India, including Andhra Pradesh, Kerala, Karnataka, and Sri Lanka are shaded. Hv = Havik, Kd = Kadar, Mu = Mukri (Mountain et al. 1995); Bo = Boksa, Lm = Lambadi, Lo = Lobana, Up = Uttar Pradesh (Kivisild et al. 1999a); Te = Telugu (Bamshad et al. 2001); Cb = Konkanastha Brahmin, Ch= Chenchu, Gu = Gujarat, Ko = Koya, Kw = Kuwait, Mo = Moor, Pa = Parsi, Pu = Punjab, Si = Sinhalese, wB = west Bengal (present study). The coalescence times of the clusters are shown below cluster labels.
A 9-bp deletion between COII/tRNALys occurs in high frequency in eastern Asian and some African populations, because of its independent origins at different phylogenetic backgrounds (Soodyall et al. 1996). It was shown recently that some Indian populations also harbor the 9-bp deletion while clustering separately from Asian and African deleted lineages (Watkins et al. 1999). We found that 21% of Koyas and 3% of Chenchus harbored the deletion at the haplogroup M background. The Chenchu type (16184-16223-16256G-16362) has been previously observed at notable frequencies (44%) among Irulas, another tribe from Andhra Pradesh with australoid anthropological features (Watkins et al. 1999). The presence of the 9-bp deletion at the haplogroup M background was also observed among Kadars of Tamil Nadu and Kerala (Edwin et al. 2002). The HVS-I motif associated with the 9-bp deletion in Koyas has not been observed in previously published studies. Whether the Koya and Chenchu 9-bp deletion types stem from the same deletion event is difficult to judge. They differ by seven HVS-I mutations, suggesting either an ancient common root or independent origins of the deletion.
The haplogroup R lineages of the Koyas (31%) and Chenchus (1%) did not further subdivide into western Eurasian–specific (HV, U, TJ, and R1; Macaulay et al. 1999) or eastern Eurasian–specific branches (B and R9; Kivisild et al. 2002) and showed a coalescence time of 73,000±20,900 years, which overlaps with the age estimate of haplogroup M. Indian-specific derivatives of haplogroup N (other than R) are rare. Only two Chenchus showing no affiliation to known N subbranches stemmed from the haplogroup consensus by four HVS-I mutations (fig. 1; table 1).
Y-Chromosomal Variation
A Y-chromosomal haplogroup tree with 35 biallelic markers in 325 Indian caste and tribal samples is shown with haplogroup frequencies and variances in figure 3. At this resolution, 19 different haplogroups can be distinguished in India, 9 of which occur in four or more different populations and each of which constitutes >5% of the total Y-chromosomal variation in India. There is no distinction, in the presence or absence of these major clades, between tribal and caste groups. When compared with European and Middle Eastern populations (Semino et al. 2000), Indians (i) share with them clades J2 and M173 derived sister groups R1b and R1a, the latter of which is particularly frequent in India; and (ii) lack or show a marginal frequency of clades E, G, I, J*, and J2f. In common with eastern and central Asian populations (Underhill et al. 2000; Karafet et al. 2001; Redd et al. 2002), most Indian populations have clade C lineages. Only one West Bengali sample belonged to clade O (M175), which is the most frequent Y-chromosome clade in east Asia. No YAP+ chromosomes (clades D and E) were detected in either the caste or tribal populations examined here. Clade E has been previously reported in Siddis but attributed to recent gene flow from Africa (Thangaraj et al. 1999; Ramana et al. 2001). Altogether, three clades—H, L, and R2—account for more than one-third of Indian Y chromosomes. They are also found in decreasing frequencies in central Asians to the north and in Middle Eastern populations to the west (table 3). Unclassified derivatives of the general Eurasian clade F were observed most frequently (27%) in the Koyas.
Table 3.
Major Y-Chromosomal Haplogroups in India Compared with Western Eurasia
|
Frequency (95% CR for Proportion) |
||||
| Population (n) | H1 (M52) | L (M11) | R2 (M124) | Reference |
| India: | ||||
| Punjab (66) | .03 (.01–.10) | .12 (.06–.22) | .05 (.02–.13) | Present study |
| West Bengal (31) | .10 (.04–.25) | .00 (.00–.09) | .23 (.12–.40) | Present study |
| AP tribes (82) | .49 (.43–.64) | .07 (.04–.15) | .04 (.01–.10) | Present study |
| AP tribes (67) | .10 (.05–.20) | .04 (.02–.12) | … | Ramana et al. 2001 |
| AP castes (125) | .10 (.06–.16) | .08 (.04–.14) | … | Ramana et al. 2001 |
| Tamil Nadu (259) | .17 (.13–.22) | .29 (.24–.35) | .07 (.05–.11) | Wells et al. 2001 |
| Tadjikistan (168) | .01 (.00–.04) | .11 (.07–.16) | .06 (.03–.11) | Wells et al. 2001 |
| Uzbeks (366) | .02 (.01–.04) | .03 (.02–.05) | .02 (.01–.04) | Wells et al. 2001 |
| Kyrgyzstan (92) | .00 (.00–.03) | .00 (.00–.03) | .02 (.01–.08) | Wells et al. 2001 |
| Kazakstan (95) | .01 (.00–.06) | .01 (.00–.06) | .01 (.00–.06) | Wells et al. 2001 |
| Iran (52) | .00 (.00–.06) | .04 (.01–.13) | .02 (.01–.10) | Wells et al. 2001 |
| Near East (101) | .01 (.00–.05) | .02 (.01–.07) | .00 (.00–.03) | Semino et al. 2000 |
| Caucasus (147) | .00 (.00–.02) | .01 (.00–.05) | .00 (.00–.02) | Wells et al. 2001 |
| Europe (839) | .00 (.00–.01) | .01 (.00–.01) | .00 (.00–.01) | Semino et al. 2000 |
In comparison with caste groups (see fig. 3 and table 3), both tribal populations showed significantly (P<.01) higher frequencies of haplogroup H1. The characteristic M52 A→C transversion has also been described at relatively high frequencies in populations of Tamil Nadu, in southern India (Wells et al. 2001). Among the caste groups, its frequency is the lowest among Punjabis in the northwest. Interestingly, more than one-third of Andhra Pradesh middle and lower caste Y chromosomes were defined as clade 1R in a previous study (Bamshad et al. 2001), which resolves into clades G, F*, and H in the present study. Hence, it seems likely that M52 is not a “tribal”-specific marker but that its frequency is concentrated regionally around Andhra Pradesh. The geographical association is further supported by the similar M52 frequencies found by Ramana et al. (2001) in six Andhra Pradesh caste and tribal populations. Beyond India, this marker has been found occasionally in neighboring populations (table 3) and in the European Roma (Gypsy) population (Kalaydjieva et al. 2001), the major STR haplotype of which matches the common type shared by 3 Chenchus and 11 Koyas (table 4). Interestingly, given its deep phylogenetic position relative to the M89 clade, the STR variance of group H is relatively low compared with other Y-chromosomal haplogroups found in Indians (fig. 3).
Table 4.
Compound Y-Chromosomal Haplotypes in Chenchus and Koyas
|
No. of Repeats at Locus |
||||||||||
| Haplotype | Cladea | DYS019 | DYS388 | DYS390 | DYS391 | DYS392 | DYS393 | Chenchu | Koya | Matchb |
| 1 | C | 15 | 13 | 23 | 10 | 11 | 12 | 1 | ||
| 2 | C | 16 | 13 | 24 | 10 | 11 | 12 | 1 | ||
| 3 | F | 15 | 13 | 21 | 11 | 11 | 14 | 1 | ||
| 4 | F | 15 | 14 | 21 | 11 | 11 | 14 | 1 | ||
| 5 | F | 16 | 13 | 21 | 10 | 12 | 14 | 1 | ||
| 6 | F | 16 | 13 | 21 | 11 | 10 | 14 | 1 | ||
| 7 | F | 16 | 13 | 21 | 11 | 11 | 13 | 1 | ||
| 8 | F | 16 | 13 | 21 | 11 | 11 | 14 | 2 | ||
| 9 | F | 16 | 14 | 21 | 11 | 11 | 14 | 1 | ||
| 10 | F | 17 | 13 | 21 | 11 | 10 | 14 | 2 | ||
| 11 | F | 17 | 13 | 21 | 11 | 11 | 13 | 1 | ||
| 12 | H1 | 13 | 12 | 23 | 11 | 11 | 12 | 1 | ||
| 13 | H1 | 14 | 12 | 21 | 9 | 11 | 12 | 1 | ||
| 14 | H1 | 14 | 12 | 22 | 10 | 11 | 12 | 2 | ||
| 15 | H1 | 14 | 13 | 22 | 10 | 11 | 13 | 1 | ||
| 16 | H1 | 14 | 13 | 22 | 11 | 11 | 12 | 1 | ||
| 17 | H1 | 15 | 12 | 21 | 9 | 11 | 12 | 7 | ||
| 18 | H1 | 15 | 12 | 21 | 10 | 11 | 12 | 2 | ||
| 19 | H1 | 15 | 12 | 21 | 11 | 11 | 12 | 1 | ||
| 20 | H1 | 15 | 12 | 22 | 10 | 11 | 12 | 3 | 11 | * |
| 21 | H1 | 15 | 12 | 22 | 10 | 11 | 13 | 3 | * | |
| 22 | H1 | 15 | 13 | 21 | 11 | 11 | 12 | 1 | ||
| 23 | H1 | 15 | 13 | 22 | 10 | 11 | 13 | 2 | ||
| 24 | H1 | 15 | 13 | 22 | 11 | 11 | 12 | 1 | ||
| 25 | H1 | 16 | 12 | 22 | 10 | 11 | 13 | 1 | ||
| 26 | H1 | 16 | 13 | 22 | 10 | 11 | 12 | 2 | ||
| 27 | H2 | 16 | 12 | 21 | 10 | 11 | 13 | 4 | * | |
| 28 | J2* | 14 | 15 | 23 | 10 | 11 | 13 | 1 | ||
| 29 | J2e | 15 | 14 | 24 | 11 | 11 | 12 | 1 | * | |
| 30 | J2e | 15 | 15 | 24 | 10 | 11 | 12 | 1 | * | |
| 31 | L | 14 | 12 | 22 | 10 | 13 | 11 | 2 | ||
| 32 | L | 14 | 12 | 22 | 10 | 14 | 11 | 4 | * | |
| 33 | R1a | 15 | 12 | 24 | 11 | 11 | 12 | 1 | ||
| 34 | R1a | 15 | 12 | 25 | 10 | 11 | 13 | 1 | * | |
| 35 | R1a | 15 | 12 | 26 | 10 | 11 | 13 | 1 | * | |
| 36 | R1a | 16 | 12 | 24 | 10 | 11 | 13 | 1 | * | |
| 37 | R1a | 17 | 12 | 24 | 11 | 11 | 13 | 1 | * | |
| 38 | R1a | 16 | 12 | 24 | 11 | 11 | 13 | 7 | * | |
| 39 | R1b | 14 | 13 | 24 | 11 | 13 | 13 | 1 | * | |
| 40 | R2 | 14 | 12 | 22 | 10 | 10 | 14 | 1 | ||
| 41 | R2 | 14 | 12 | 23 | 10 | 10 | 14 | 1 | ||
| 42 | R2 | 14 | 13 | 22 | 10 | 10 | 14 | 1 |
||
| Total | 41 | 41 | ||||||||
According to YCC nomenclature.
Compared to 239 Indian caste samples.
The spread of clade L is confined mostly to southern, central, and western Asia (table 3). Being virtually absent in Europe, it is also found irregularly and at low frequencies in populations of the Middle East and southern Caucasus (Nebel et al. 2001; Scozzari et al. 2001; Weale et al. 2001; Wilson et al. 2001). It occurs in Pakistan at a frequency of 13.5% (Qamar et al. 2002). In Indians, all Y chromosomes that had the derived allele at M11, M20, and M61 also shared M27 specific to its subclade L1. When a resolution of six STR loci was used, four Chenchus shared a widespread modal haplotype (14-12-22-10-14-11) with Lambadis, Punjabis, and Iranians. This type differs, however, by three steps from the modal haplotype (15-12-23-10-13-11) commonly found in Armenian M20 chromosomes (Weale et al. 2001).
Clades Q and R share a common phylogenetic node P in the Y-chromosomal tree defined by markers M45 and 92R7 (YCC 2002). The P*(xM207) chromosomes are widespread—although found at low frequencies—over central and eastern Asia (Underhill et al. 2000) and were also found only in two Indian samples (fig. 3). In contrast, their sister branch R, defined by M207, accounts for more than one-third of Indian Y chromosomes and is the most common clade throughout northwestern Eurasia. Its daughter clades R1 and R2 are both found in tribal and caste groups. Clade R1 splits into R1a and R1b, which are similarly variable in Indians (fig. 3) and western Asians but are less so in Estonians, Czechs, and central Asians (table 5). R2 (previously misidentified as “P1” [YCC 2002]) has a more specific spread, being confined to Indian, Iranian, and central Asian populations (table 3).
Table 5.
Y Chromosome Haplogroup Variances when Six STR Loci Are Used
|
Variance of Haplogroup |
|||||||||
| Population | C | H1 | I | J | L | R1a | R1b | R2 | All |
| India | .73 | .19 | .41 | .27 | .32 | .34 | .33 | 1.1 | |
| Pakistana | .47 | .37 | |||||||
| Irana | .57 | .38 | |||||||
| Czechs | .41 | .45 | .14 | .12 | .75 | ||||
| Estonians | .4 | .19 | .17 | .88 | |||||
| Central Asia | .19 | ||||||||
From Quintana-Murci et al. 2001.
Clade J accounts for 13% of Indian Y chromosomes, almost exclusively because of its subcluster J2, defined by M172. Further, one quarter of the J2 chromosomes share an M12 mutation that shows relatively low background STR diversity over all of India (fig. 3). Interestingly, this marker has a wide geographic distribution and has also been found in polymorphic frequencies in Europe, even as far north as Kola-Saamis (Underhill et al. 1997, 2000; Raitio et al. 2001; Scozzari et al. 2001). Only two samples from Gujarat harbored the M67 mutation that is a relatively common marker at the M172 background, from the Middle East through Pakistan (Underhill et al. 2000).
Clade C is widespread in central and eastern Asia, Oceania, and Australia but is rarely found or absent in western Asia and Europe (Bergen et al. 1999; Karafet et al. 1999; Underhill et al. 2000, 2001a; Ke et al. 2001; Wells et al. 2001). In Indians, the defining RPS4Y marker persists at an average frequency of 5%, consistent with its previous accounts in the Andhra Pradesh and Tamil Nadu caste groups of different ranks and tribes (Bamshad et al. 2001). Most eastern Asians and virtually all central Asians with the RPS4Y711 T allele belong to clade C3, carrying a C at the M217 locus (Karafet et al. 2001; Underhill et al. 2001b). In contrast, all but one Indian clade C chromosome harbored the ancestral A allele at M217 and also lacked other SNP markers characterized so far on the RPS4Y711T background. Neither did we find the deletion at the DYS390 locus, typically associated with Australian, Melanesian, and Polynesian populations (Kayser et al. 2000, 2001; Redd et al. 2002).
Using six STR loci, we defined 42 compound haplotypes in 41 Chenchu and 41 Koya Y chromosomes (table 4). Similar to the mitochondrial results, only one shared haplotype was found between the two tribes. However, 12 matches with Indian caste populations were found, mainly in the J and R1 clusters. Whereas the haplotype diversity of Koya (0.92) Y-chromosomal combined haplotypes was similar to that of the Chenchus (0.94), the haplogroup diversity in Koyas was considerably lower (0.56) compared with that in the Chenchus (0.8), who displayed the presence of eight different Y-chromosomal haplogroups. Both populations showed lower levels of diversity than did caste populations (fig. 3). Table 5 compares the STR variances of different Y-chromosomal haplogroups in southern Asia, western Asia, central Asia, and Europe. The variances following the branching order of the Y SNP tree in India are also shown in figure 3. The lowest variance estimates (0.12–0.19), in the predominantly southern Indian cluster H1, were comparable to the equally low variances in the European and central Asian R1a and R1b lineages. Intermediate variances (0.27–0.38) were found in the southern and western Asian L and R clusters, for European specific cluster I (0.41–0.57), and for the cluster J2 in general. The highest variance was observed in haplogroup C, which showed about two-thirds of the variation seen in all other groups together.
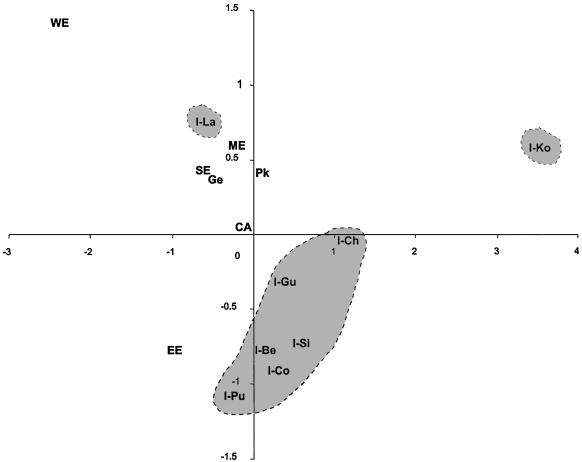
An MDS plot, shown in figure 4, compares eight Indian populations with eight populations from Europe, western Asia, and central Asia, through use of 16 Y-chromosomal haplogroups. Chenchus form a distinctive Indian cluster with five caste populations, whereas the Koyas appear as an outlier, probably because of their low haplogroup diversity. The high frequency of M17 among eastern European and central Asian populations places them next to the Indian cluster. Furthermore, the high frequency of M269 in the Lambadis positions them away from Indians and between the southern and western European populations, among whom this marker is more commonly found (Cruciani et al. 2002).
Figure 4.
Multidimensional scaling plot of eight Indian and seven western Eurasian populations, using Fst distances calculated for 16 Y-chromosomal SNP haplogroups. From India: I-Ch = Chenchus, I-Ko = Koyas, I-Gu = Gujaratis, I-Be = Western Bengalis, I-Si = Singalese, I-Co = Konkanastha Brahmins, I-La = Lambadis. Western Eurasia: CA = Central Asia; Pk = Pakistanis (Underhill et al. 2000); WE = western Europe, including Dutch, French, and German samples; EE = eastern Europe, including Poles, Czechs, and Ukrainians; SE = southern Europe, including Greeks and Macedonians; Ge = Georgians; ME = Middle East, including Turks, Lebanese, and Syrians from Semino et al. (2000).
MX1 Locus in Chromosome 21
Differences between human continental populations were defined by Jin et al. (1999) in a short stretch of sequence of chromosome 21.Table 6 reports the MX1 haplotype profiles in the Chenchus and Koyas, compared with caste groups from Punjab and West Bengal, in the background of data from other world populations. We did not find new Indian-specific mutations, consistent with the idea that the observed variation in the locus has largely arisen in Africa. Haplotype 5, which is common in eastern Asia and Africa but virtually absent in Europe, was found in intermediate frequencies in all Indian populations considered here. Similarly, haplotype 8, which is common in Europe but absent in eastern Asia, was found in India at low frequencies. As is the case in other Eurasian and African populations, haplotypes 3 and 4, which are specific to Australian and Papuan populations, were not found in India. In contrast to the significant differences of haplotype frequencies that were observed between Indian and other world populations, none of the differences in haplotype frequencies was significant within India between caste and tribal groups.
Table 6.
MX1 Haplotypes of Chromosome 21 in Indian Populations, Compared with Continental Groups of the World
|
No. of Lineages (95% CR for Proportion) |
||||||||||
| Population | Ht1 | Ht2 | Ht3 | Ht4 | Ht5 | Ht6 | Ht7 | Ht8 | Ht9 | Ht10 |
| Chenchus (84)a | 0 (.00–.04) | 11 (.08–.22) | 0 (.00–.04) | 0 (.00–.04) | 16 (.12–.29) | 0 (.00–.04) | 40 (.37–.58) | 17 (.13–.30) | 0 (.00–.04) | 0 (.00–.04) |
| Koyas (56)a | 0 (.00–.05) | 4 (.03–.17) | 0 (.00–.05) | 0 (.00–.05) | 15 (.17–.40) | 0 (.00–.05) | 16 (.18–.42) | 21 (.26–.51) | 0 (.00–.05) | 0 (.00–.05) |
| Punjab (68)a | 0 (.00–.04) | 10 (.08–.25) | 0 (.00–.04) | 0 (.00–.04) | 7 (.05–.20) | 0 (.00–.04) | 31 (.34–.57) | 20 (.20–.41) | 0 (.00–.04) | 0 (.00–.04) |
| West Bengal (70)a | 1 (.00–.08) | 2 (.01–.10) | 0 (.00–.04) | 0 (.00–.04) | 16 (.15–.34) | 0 (.00–.04) | 34 (.37–.60) | 17 (.16–.36) | 0 (.00–.04) | 0 (.00–.04) |
| Pakistan (72)b | 0 (.00–.04) | 5 (.03–.15) | 0 (.00–.04) | 0 (.00–.04) | 6 (.04–.17) | 0 (.00–.04) | 27 (.27–.49) | 34 (.36–.59) | 0 (.00–.04) | 0 (.00–.04) |
| Anatolia (70)a | 2 (.01–.10) | 7 (.05–.19) | 0 (.00–.04) | 0 (.00–.04) | 3 (.02–.12) | 1 (.00–.08) | 29 (.31–.53) | 28 (.29–.52) | 0 (.00–.04) | 0 (.00–.04) |
| Europe (192)b | 4 (.01–.05) | 17 (.06–.14) | 0 (.00–.02) | 0 (.00–.02) | 3 (.01–.05) | 0 (.00–.02) | 88 (.39–.53) | 79 (.34–.48) | 1 (.00–.03) | 0 (.00–.02) |
| East Asia (118)b | 0 (.00–.03) | 3 (.01–.07) | 0 (.00–.03) | 0 (.00–.03) | 59 (.41–.59) | 11 (.05–.16) | 45 (.30–.47) | 0 (.00–.03) | 0 (.00–.03) | 0 (.00–.03) |
| Sub-Saharan Africa (102)b | 9 (.05–.16) | 3 (.01–.08) | 0 (.00–.03) | 0 (.00–.03) | 22 (.15–.31) | 23 (.16–.32) | 33 (.24–.42) | 9 (.05–.16) | 0 (.00–.03) | 3 (.01–.08) |
| Amerinds (120)b | 0 (.00–.02) | 14 (.07–.19) | 0 (.00–.02) | 0 (.00–.02) | 78 (.56–73) | 0 (.00–.02) | 2 (.01–.06) | 26 (.15–.30) | 0 (.00–.02) | 0 (.00–.02) |
| Australia and PNG (76)b | 2 (.01–.09) | 10 (.07–.23) | 16 (.13–.32) | 2 (.01–.09) | 27 (.26–47) | 0 (.00–.04) | 15 (.12–.30) | 4 (.02–.13) | 0 (.00–.04) | 0 (.00–.04) |
From the present study.
From Jin et al. 1999.
Discussion
India as an Incubator of Early Genetic Differentiation of Modern Humans Moving out of Africa
Phylogeographic patterns of the Y chromosome and mtDNA support the concept that the Indian subcontinent played a pivotal role in the late Pleistocene genetic differentiation of the western and eastern Eurasian gene pools. All non-Africans, including Indian populations, have inherited a subset of African mtDNA haplogroup L3 lineages, differentiated into groups M and N. Although the frequency of haplogroup M and its diversity are highest in India (Majumder 2001; Edwin et al. 2002), there is no phylogenetic evidence yet from the mtDNA coding region demonstrating that its presence in Africa is due to a back migration. Also, the lack of L3 lineages other than M and N in India and among non-African mitochondria in general (Ingman et al. 2000; Herrnstadt et al. 2002; Kivisild et al. 2002) suggests that the earliest migration(s) of modern humans already carried these two mtDNA ancestors, via a departure route over the horn of Africa (i.e., the southern route migration [Nei and Roychoudhury 1993; Quintana-Murci et al. 1999; Stringer 2000]). More specifically, the ubiquity in India of diverse branches sharing the characteristic 12705T and 16223C transitions (table 2), suggests that the N branch had already given rise to its daughter clade R, which later, in eastern Asians, differentiated into clusters B and R9 (Kivisild et al. 2002) and in western Asia gave rise to haplogroups HV, TJ, and U (Macaulay et al. 1999). The coalescence time of major M subclusters in the Indian subcontinent, which are comparable in diversity and even older than most eastern Asian and Papuan haplogroup M clusters (Forster et al. 2001), suggests that the Indian subcontinent was settled soon after the African exodus (Kivisild et al. 1999b, 2000) and that there has been no complete extinction or replacement of the initial settlers.
In a similar way, Indians show the presence of diverse lineages of the three major Eurasian Y-chromosomal haplogroups C, F, and K, although they have obviously lost the fourth potential founder, D. The presence of several subclusters of F and K (H, L, R2, and F*) that are largely restricted to the Indian subcontinent is consistent with the scenario that the coastal (southern route) migration(s) from Africa carried the ancestral Eurasian lineages first to the coast of Indian subcontinent (or that some of them originated there). Next, the reduction of this general package of three mtDNA (M, N, and R) and four Y-chromosomal (C, D, F, and K) founders to two mtDNA (N and R) and two Y-chromosomal (F and K) founders occurred during the westward migration to western Asia and Europe. After this initial settlement process, each continental region (including the Indian subcontinent) developed its region-specific branches of these founders, some of which (e.g., the western Asian HV and TJ lineages) have, via continuous or episodic low-level gene flow, reached back to India. Western Asia and Europe have thereafter received an additional wave of genes from Africa, likely via the Levantine corridor, bringing forth lineages of Y-chromosomal haplogroup E, for example (Underhill et al. 2001b), which is absent in India.
Gene Flow from Eastern Asia
Although both Indian and eastern Asian populations share, at the interior phylogenetic level, two major trunks of the mtDNA tree (haplogroups M and N), their subsequent branching into boughs and limbs is different (Bamshad et al. 2001; Kivisild et al. 2002): <2% of Indians, whether with tribal or caste affiliation, can trace their maternal ancestry back to eastern Asian–specific (Kivisild et al. 2002) branches (Kivisild et al. 1999a; Bamshad et al. 2001). Analogously, the subclades of the Y-chromosomal clusters C, F, and K do not overlap in southern and eastern Asia. The major continental eastern Asian clade O was virtually absent both in tribal and caste populations, although one particular O subcluster, defined by M95, has been reported in three other tribes of Andhra Pradesh (Ramana et al. 2001) and in castes and tribes of Tamil Nadu (Wells et al. 2001). The frequency of M95 is highest in Austro-Asiatic speakers, Burmese-Lolo, and the Karen of Yunnan, China (Su et al. 1999, 2000) and is virtually absent (1/984) in central Asia (Wells et al. 2001). Its irregular distribution from India to Yunnan might possibly be related to the equally uneven spread of the Austro-Asiatic speakers.
Indian RPS4Y711T chromosomes (clade C), like their Indonesian counterparts (Underhill et al. 2001a), cannot be apportioned between clusters specific to eastern Asian (/M217) and Oceanic populations (/M38, /M210). Given the high hierarchical position of the C clade in the Y binary tree, its wide distribution in the eastern hemisphere, and its high STR variability in India (fig. 3), it seems plausible that the original spread of C was associated with the southern route migration. Although haplogroup C displays idiosyncratic occurrences in Europe (Semino et al. 2000), its presence at 5% in India (perhaps its most reliable westernmost distribution) suggests that the RPS4Y mutation originated in or arrived with the earliest immigrants. Invoking back migrations to India as an explanation is unwarranted, since the absence of derivative RPS4Y lineages common in eastern Asia and Oceania suggests that these differentiations happened after RPS4Y lineages had already transited the subcontinent. Furthermore, the MX1 data distinguish the Indians from the Oceanian population, in which RPS4Y occurs frequently.
Gene Flow from Western Asia, Europe, and Central Asia
Indians virtually lack the HIV-1–protective Δccr5 allele (Majumder and Dey 2001) that is frequent in Europe, western Asia, and central Asia, implying either that this allele arose very recently in Europe or that there has not been substantial gene flow to India from the northwest. Western Eurasian–specific mtDNA haplogroups occur at low frequencies in Indian caste populations (Kivisild et al. 1999a; Bamshad et al. 2001) and are virtually absent among the tribes (Roychoudhury et al. 2001; present study). Southern and western Asian–specific U2i and U7 lineages, which are rare or absent in Europe, however, are found occasionally in the tribes. The copresence of most haplogroup U subclusters (U1–U8) in populations around the Middle East (Macaulay et al. 1999) suggests that the differentiation of haplogroup U occurred mostly west of India. If the ancestor of haplogroup U was brought to the Middle East via northern Africa by the northern route migration—a hypothesis still awaiting support from genetic data—then the presence of haplogroup U in India would be due to an early, Upper Palaeolithic migration from western Asia. Alternatively, one might consider the scenario that all western Eurasian mtDNA variation stems from the coastal southern route migration and that U had already differentiated from R in southern Asia, where it survived only in U2i (and perhaps U7) descendants. Interestingly, mtDNA haplogroup U7 (Richards et al. 2000), like Y-chromosomal clade L (Underhill et al. 2000), is also found, though at low frequencies, in western Asia and occasionally in Mediterranean Europe. The differences in STR modal haplotypes of the L clade between the Caucasus (Weale et al. 2001) and India point to their independent expansions from two distinct founder populations. Given the deep time depth of U7 (Kivisild et al. 1999a), it is possible that this east-to-west link predates the Last Glacial Maximum.
The lack of western Asian and European-specific mtDNA lineages among the tribes and their low frequency in castes of southern and eastern India indicates that the spread of these lineages in India might have been communicated by the caste populations of northwestern India and that there has been limited maternal gene flow from castes to tribes thereafter.
The most common Y-chromosomal lineage among Indians, R1a, also occurs away from India in populations of diverse linguistic and geographic affiliation. It is widespread in central Asian Turkic-speaking populations and in eastern European Finno-Ugric and Slavic speakers and has also been found less frequently in populations of the Caucasus and the Middle East and in Sino-Tibetan populations of northern China (Rosser et al. 2000; Underhill et al. 2000; Karafet et al. 2001; Nebel et al. 2001; Weale et al. 2001). No clear consensus yet exists about the place and time of its origins. From one side, it has been regarded as a genetic marker linked with the recent spread of Kurgan culture that supposedly originated in southern Russia/Ukraine and extended subsequently to Europe, central Asia, and India during the period 3,000–1,000 b.c. (Passarino et al. 2001; Quintana-Murci et al. 2001; Wells et al. 2001). Alternatively, an Asian source (Zerjal et al. 1999) or a deeper Palaeolithic time depth of ∼15,000 years before present for the defining M17 mutation has been suggested (Semino et al. 2000; Wells et al. 2001). Interestingly, the high frequency of the M17 mutation seems to be concentrated around the elevated terrain of central and western Asia. In central Asia, its frequency is highest (>50%) in the highlands among Tajiks, Kyrgyz, and Altais and drops down to <10% in the plains among the Turkmenians and Kazakhs (Wells et al. 2001; Zerjal et al. 2002). Our low STR diversity estimate of haplogroup R1a in central Asians is also consistent with the low diversities found by Zerjal et al. (2002) and suggests a recent founder effect or drift being the reason for the high frequency of M17 in southeastern central Asia. In Pakistan, except for the Hazara, who are supposedly recent immigrants in the region, the frequency of M17 was similarly high in the upper and lower courses of the Indus River valley (Qamar et al. 2002). The frequency of R1a drops from ∼30% in eastern provinces to <10% in the western parts of Iran (Quintana-Murci et al. 2001). Both Pakistanis and Iranians showed STR variances as high as those of Indians, when compared with the lower values in European and central Asian populations. Unexpectedly, both southern Indian tribal groups examined in this study carried M17. The presence of different STR haplotypes and the relatively high frequency of R1a in Dravidian-speaking Chenchus (26%) make M17 less likely to be the marker associated with male “Indo-Aryan” intruders in the area. Moreover, in two previous studies involving southern Indian tribal groups such as the Valmiki from Andhra Pradesh (Ramana et al. 2001) and the Kallar from Tamil and Nadu (Wells et al. 2001), the presence of M17 was also observed, suggesting that M17 is widespread in tribal southern Indians. Given the geographic spread and STR diversities of sister clades R1 and R2, the latter of which is restricted to India, Pakistan, Iran, and southern central Asia, it is possible that southern and western Asia were the source for R1 and R1a differentiation.
Compared with western Asian populations, Indians show lower STR diversities at the haplogroup J background (Quintana-Murci et al. 2001; Nebel et al. 2002) and virtually lack J*, which seems to have higher frequencies in the Middle East and East Africa (Eu10 [Nebel et al. 2001]; Ht25 [Semino et al. 2002]) and is common also in Europe (Underhill et al. 2001b). Therefore, J2 could have been introduced to northwestern India from a western Asian source relatively recently and, subsequently, after comingling in Punjab with R1a, spread to other parts of India, perhaps associated with the spread of the Neolithic and the development of the Indus Valley civilization. This spread could then have also taken with it mtDNA lineages of haplogroup U, which are more abundant in the northwest of India, and the western Eurasian lineages of haplogroups H, J, and T.
The Caste and Tribe Distinction
The example of phylogenetic reconstruction of mtDNA haplogroup M2 showed that individuals from populations of different geographic origin and social status in India share the same branches of the tree. Similarly, since there is no grouping according to language families among the caste groups (Bamshad et al. 2001), no clusters of considerable time depth seem to be rank-specific to Indian tribal or caste groups. Phenomena like the upward social mobility of caste women (Bamshad et al. 1998) could have introduced some tribal genes to the castes more recently, but, given the relatively low proportion of the tribal population size today, recent unidirectional gene flow can be assumed to be a minor modifying force in the formation of the genetic profile of the caste population.
“Gothra” is an identity carried by male lineage in India from time immemorial. The lack of clear distinction between Indian castes and tribes was shown by Ramana et al. (2001), using a two-dimensional PC plot of Y-chromosome haplogroups. The close clustering of Chenchus with the caste groups in our MDS analysis (fig. 4) supports this finding. However, substantial heterogeneity observed in the haplogroup frequencies of the tribes and their generally lower haplotype and haplogroup diversity (e.g., the wide range in frequencies of major clades C*, J, F*, O, and R1a in tribal groups of this study) (Ramana et al. 2001; Wells et al. 2001) suggests that conclusions about Indian prehistory cannot be based on the examination of one or a few groups.
Although, on a general scale, we can argue for largely the same prehistoric genetic inheritance in Indian tribal and caste populations, this does not refute the existence of genetic footprints laid down by known historical events. This would include invasions by the Huns, Greeks, Kushans, Moghuls, Muslims, English, and others. The political influence of Seleucid and Bactrian dynastic Greeks over northwest India, for example, persisted for several centuries after the invasion of the army of Alexander the Great (Tarn 1951). However, we have not found, in Punjab or anywhere else in India, Y chromosomes with the M170 or M35 mutations that together account for >30% in Greeks and Macedonians today (Semino et al. 2000). Given the sample size of 325 Indian Y chromosomes examined, however, it can be said that the Greek homeland (or European, more generally, where these markers are spread) contribution has been 0%–3% for the total population or 0%–15% for Punjab in particular. Such broad estimates are preliminary, at best. It will take larger sample sizes, more populations, and increased molecular resolution to determine the likely modest impact of historic gene flows to India on its pre-existing large populations.
Acknowledgments
We thank Joanna Mountain, for critical advice; Li Jin, for helpful information; Vincent Macaulay, for the program for calculating the credible regions of haplogroup frequencies; and Jaan Lind and Ille Hilpus, for technical assistance. This work was supported by Estonian basic research grant 514 and European Commission Directorates General Research grant ICA1CT20070006 (to R.V.) and by National Institutes of Health grants GM28428 and GM55273 (to L.L.C-S).
Electronic-Database Information
The URL for data presented herein is as follows:
- Shareware Phylogenetic Network Software Web site, http://www.fluxus-engineering.com/sharenet.htm (for Network 2.2 software)
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, et al (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 [DOI] [PubMed] [Google Scholar]
- Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, Naidu JM, Prasad BVR, Reddy PG, Rasanayagam A, Papiha SS, Villems R, Redd AJ, Hammer MF, Nguyen SV, Carroll ML, Batzer MA, Jorde LB (2001) Genetic evidence on the origins of indian caste populations. Genome Res 11:994–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Watkins WS, Dixon ME, Jorde LB, Rao BB, Naidu JM, Prasad BV, Rasanayagam A, Hammer MF (1998) Female gene flow stratifies Hindu castes. Nature 395:651–652 [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen AW, Wang CY, Tsai J, Jefferson K, Dey C, Smith KD, Park SC, Tsai S-J, Goldman D (1999) An Asian–Native American paternal lineage identified by RPS4Y resequencing and by microsatellite haplotyping. Ann Hum Genet 63:63–80 [DOI] [PubMed] [Google Scholar]
- Bertorelle G, Excoffier L (1998) Inferring admixture proportions from molecular data. Mol Biol Evol 15:1298–1311 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya NP, Basu P, Das M, Pramanik S, Banerjee R, Roy B, Roychoudhury S, Majumder PP (1999) Negligible male gene flow across ethnic boundaries in India, revealed by analysis of Y-chromosomal DNA polymorphisms. Genome Res 9:711–719 [PubMed] [Google Scholar]
- Bhowmick PK (1992) The Chenchus of the forests and plateaux: Calcutta. Institute of Anthropology, Calcutta [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton [Google Scholar]
- Chikhi L, Bruford MW, Beaumont MA (2001) Estimation of admixture proportions: a likelihood-based approach using Markov chain Monte Carlo. Genetics 158:1347–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi L, Nichols RA, Barbujani G, Beaumont MA (2002) Y genetic data support the Neolithic demic diffusion model. Proc Natl Acad Sci USA 99:11008–11013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, Santolamazza P, Shen P, Macaulay V, Moral P, Olckers A, Modiano D, Holmes S, Destro-Bisol G, Coia V, Wallace DC, Oefner PJ, Torroni A, Cavalli-Sforza LL, Scozzari R, Underhill PA (2002) A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet 70:1197–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwin D, Vishwanathan H, Roy S, Usha Rani M, Majumder P (2002) Mitochondrial DNA diversity among five tribal populations of southern India. Curr Sci 83:158–163 [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt H-J (1996) Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet 59:935–945 [PMC free article] [PubMed] [Google Scholar]
- Forster P, Torroni A, Renfrew C, Röhl A (2001) Phylogenetic star contraction applied to Asian and Papuan mtDNA evolution. Mol Biol Evol 18:1864–1881 [DOI] [PubMed] [Google Scholar]
- Hammer MF, Redd AJ, Wood ET, Bonner MR, Jarjanazi H, Karafet T, Santachiara-Benerecetti S, Oppenheim A, Jobling MA, Jenkins T, Ostrer H, Bonne-Tamir B (2000) Jewish and Middle Eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc Natl Acad Sci USA 97:6769–6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Sigurðadóttir S, Gulcher J, Ward R, Stefanson K (2000a) mtDNA and the origins of the Icelanders: deciphering signals of recent population history. Am J Hum Genet 66:999–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Sigurðardóttir S, Nicholson J, Sykes B, Hill EW, Bradley DG, Bosnes V, Gulcher JR, Ward R, Stefánsson K (2000b) Estimating Scandinavian and Gaelic ancestry in the male settlers of Iceland. Am J Hum Genet 67:697–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, Howell N (2002) Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet 70:1152–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U (2000) Mitochondrial genome variation and the origin of modern humans. Nature 408:708–713 [DOI] [PubMed] [Google Scholar]
- Jin L, Underhill PA, Doctor V, Davis RW, Shen P, Cavalli-Sforza LL, Oefner PJ (1999) Distribution of haplotypes from a chromosome 21 region distinguishes multiple prehistoric human migrations. Proc Natl Acad Sci USA 96:3796–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Heissig F, von Haeseler A, Pääbo S (1999) DNA sequence variation in a non-coding region of low recombination on the human X chromosome. Nat Genet 22:78–81 [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L, Calafell F, Jobling MA, Angelicheva D, de Knijff P, Rosser ZH, Hurles ME, Underhill P, Tournev I, Marushiakova E, Popov V (2001) Patterns of inter- and intra-group genetic diversity in the Vlax Roma as revealed by Y chromosome and mitochondrial DNA lineages. Eur J Hum Genet 9:97–104 [DOI] [PubMed] [Google Scholar]
- Karafet T, Xu L, Du R, Wang W, Feng S, Wells RS, Redd AJ, Zegura SL, Hammer MF (2001) Paternal population history of East Asia: sources, patterns, and microevolutionary processes. Am J Hum Genet 69:615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet TM, Zegura SL, Posukh O, Osipova L, Bergen A, Long J, Goldman D, Klitz W, Harihara S, deKnijff P, Wiebe V, Griffiths RC, Templeton AR, Hammer MF (1999) Ancestral Asian source(s) of new world Y-chromosome founder haplotypes. Am J Hum Genet 64:817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Brauer S, Weiss G, Schiefenhovel W, Underhill PA, Stoneking M (2001) Independent histories of human Y chromosomes from Melanesia and Australia. Am J Hum Genet 68:173–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Brauer S, Weiss G, Underhill PA, Roewer L, Schiefenhovel W, Stoneking M (2000) Melanesian origin of Polynesian Y chromosomes. Curr Biol 10:1237–1246 [DOI] [PubMed] [Google Scholar]
- Ke Y, Su B, Song X, Lu D, Chen L, Li H, Qi C, Marzuki S, Deka R, Underhill P, Xiao C, Shriver M, Lell J, Wallace D, Wells RS, Seielstad M, Oefner P, Zhu D, Jin J, Huang W, Chakraborty R, Chen Z, Jin L (2001) African origin of modern humans in East Asia: a tale of 12,000 Y chromosomes. Science 292:1151–1153 [DOI] [PubMed] [Google Scholar]
- Kivisild T, Bamshad MJ, Kaldma K, Metspalu M, Metspalu E, Reidla M, Laos S, Parik J, Watkins WS, Dixon ME, Papiha SS, Mastana SS, Mir MR, Ferak V, Villems R (1999a) Deep common ancestry of Indian and western-Eurasian mitochondrial DNA lineages. Curr Biol 9:1331–1334 [DOI] [PubMed] [Google Scholar]
- Kivisild T, Kaldma K, Metspalu M, Parik J, Papiha SS, Villems R (1999b) The place of the Indian mitochondrial DNA variants in the global network of maternal lineages and the peopling of the Old World. In: Deka R, Papiha SS (eds) Genomic diversity. Kluwer/Academic/Plenum Publishers, New York, pp 135–152 [Google Scholar]
- Kivisild T, Papiha SS, Rootsi S, Parik J, Kaldma K, Reidla M, Laos S, Metspalu M, Pielberg G, Adojaan M, Metspalu E, Mastana SS, Wang Y, Gölge M, Demirtas H, Schnakenberg E, De Stefano GF, Geberhiwot T, Claustres M, Villems R (2000) An Indian ancestry: a key for understanding human diversity in Europe and beyond. In: Renfrew C, Boyle K (eds) Archaeogenetics: DNA and the population prehistory of Europe. McDonald Institute for Archaeological Research, University of Cambridge, Cambridge, pp 267–279 [Google Scholar]
- Kivisild T, Tolk H-V, Parik J, Wang Y, Papiha SS, Bandelt H-J, Villems R (2002) The emerging limbs and twigs of the east Asian mtDNA tree. Mol Biol Evol 19:1737–1751 [DOI] [PubMed] [Google Scholar]
- Maca-Meyer N, Gonzalez AM, Larruga JM, Flores C, Cabrera VM (2001) Major genomic mitochondrial lineages delineate early human expansions. BMC Genet 2:13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay VA, Richards MB, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonné-Tamir B, Sykes B, Torroni A (1999) The emerging tree of west Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet 64:232–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder PP (2001) Indian caste origins: genomic insights and future outlook. Genome Res 11:931–932 [DOI] [PubMed] [Google Scholar]
- Majumder P, Dey B (2001) Absence of the HIV-1 protective Delta ccr5 allele in most ethnic populations of India. Eur J Hum Genet 9:794–796 [DOI] [PubMed] [Google Scholar]
- Mountain JL, Hebert JM, Bhattacharyya S, Underhill PA, Ottolenghi C, Gadgil M, Cavalli-Sforza LL (1995) Demographic history of India and mtDNA-sequence diversity. Am J Hum Genet 56:979–992 [PMC free article] [PubMed] [Google Scholar]
- Nebel A, Filon D, Brinkmann B, Majumder PP, Faerman M, Oppenheim A (2001) The Y chromosome pool of Jews as part of the genetic landscape of the Middle East. Am J Hum Genet 69:1095–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebel A, Landau-Tasseron E, Filon D, Oppenheim A, Faerman M (2002) Genetic evidence for the expansion of Arabian tribes into the Southern Levant and North Africa. Am J Hum Genet 70:1594–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Roychoudhury A (1993) Evolutionary relationships of human populations on a global scale. Mol Biol Evol 10:927–943 [DOI] [PubMed] [Google Scholar]
- Papiha SS (1996) Genetic variation in India. Hum Biol 68:607–628 [PubMed] [Google Scholar]
- Passarino G, Semino O, Magri C, Al-Zahery N, Benuzzi G, Quintana-Murci L, Andellnovic S, Bullc-Jakus F, Liu A, Arslan A, Santachiara-Benerecetti AS (2001) The 49a,f haplotype 11 is a new marker of the EU19 lineage that traces migrations from northern regions of the Black Sea. Hum Immunol 62:922–932 [DOI] [PubMed] [Google Scholar]
- Passarino G, Semino O, Modiano G, Bernini LF, Santachiara Benerecetti AS (1996) mtDNA provides the first known marker distinguishing proto-Indians from the other Caucasoids; it probably predates the diversification between Indians and Orientals. Ann Hum Biol 23:121–126 [DOI] [PubMed] [Google Scholar]
- Qamar R, Ayub Q, Mohyuddin A, Helgason A, Mazhar K, Mansoor A, Zerjal T, Tyler-Smith C, Mehdi SQ (2002) Y-chromosomal DNA variation in Pakistan. Am J Hum Genet 70:1107–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Krausz C, Zerjal T, Sayar SH, Hammer MF, Mehdi SQ, Ayub Q, Qamar R, Mohyuddin A, Radhakrishna U, Jobling MA, Tyler-Smith C, McElreavey K (2001) Y-chromosome lineages trace diffusion of people and languages in southwestern Asia. Am J Hum Genet 68:537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt H-J, Passarino G, McElreavey K, Santachiara-Benerecetti AS (1999) Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23:437–441 [DOI] [PubMed] [Google Scholar]
- Raitio M, Lindroos K, Laukkanen M, Pastinen T, Sistonen P, Sajantila A, Syvanen A (2001) Y-chromosomal SNPs in Finno-Ugric-speaking populations analyzed by minisequencing on microarrays. Genome Res 11:471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramana GV, Su B, Jin L, Singh L, Wang N, Underhill P, Chakraborty R (2001) Y-chromosome SNP haplotypes suggest evidence of gene flow among caste, tribe, and the migrant Siddi populations of Andhra Pradesh, South India. Eur J Hum Genet 9:695–700 [DOI] [PubMed] [Google Scholar]
- Redd A, Roberts-Thomson J, Karafet T, Bamshad M, Jorde L, Naidu J, Walsh B, Hammer MF (2002) Gene flow from the Indian subcontinent to Australia: evidence from the Y chromosome. Curr Biol 12:673–677 [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, et al (2000) Tracing European founder lineages in the near eastern mtDNA pool. Am J Hum Genet 67:1251–1276 [PMC free article] [PubMed] [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, et al (2000) Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 67:1526–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhury S, Roy S, Basu A, Banerjee R, Vishwanathan H, Usha Rani MV, Sil SK, Mitra M, Majumder PP (2001) Genomic structures and population histories of linguistically distinct tribal groups of India. Hum Genet 109:339–350 [DOI] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt H-J, Nørby S (2000a) mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 67:718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saillard J, Magalhaes P, Schwartz M, Rosenberg T, Nørby S (2000b) Mitochondrial DNA variant 11719G is a marker for the mtDNA haplogroup cluster HV. Hum Biol 72:1065–1068 [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Scozzari R, Cruciani F, Pangrazio A, Santolamazza P, Vona G, Moral P, Latini V, Varesi L, Memmi MM, Romano V, De Leo G, Gennarelli M, Jaruzelska J, Villems R, Parik J, Macaulay V, Torroni A (2001) Human Y-chromosome variation in the western Mediterranean area: implications for the peopling of the region. Hum Immunol 62:871–884 [DOI] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiæ M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA (2000) The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 290:1155–1159 [DOI] [PubMed] [Google Scholar]
- Semino O, Santachiara-Benerecetti A, Falaschi F, Cavalli-Sforza L, Underhill P (2002) Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am J Hum Genet 70:265–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shouse B (2001) Archaeology: spreading the word, scattering the seeds. Science 294:988–989 [DOI] [PubMed] [Google Scholar]
- Shriver MD, Smith MW, Jin L, Marcini A, Akey JM, Deka R, Ferrell RE (1997) Ethnic-affiliation estimation by use of population-specific DNA markers. Am J Hum Genet 60:957–964 [PMC free article] [PubMed] [Google Scholar]
- Singh KS (1997) The scheduled tribes. In: Singh KS (ed) People of India. Vol III. Oxford University Press, Oxford [Google Scholar]
- Soodyall H, Vigilant L, Hill AV, Stoneking M, Jenkins T (1996) mtDNA control-region sequence variation suggests multiple independent origins of an “Asian-specific” 9-bp deletion in sub-Saharan Africans. Am J Hum Genet 58:595–608 [PMC free article] [PubMed] [Google Scholar]
- Stringer C (2000) Coasting out of Africa. Nature 405:24–25 [DOI] [PubMed] [Google Scholar]
- Su B, Xiao C, Deka R, Seielstad MT, Kangwanpong D, Xiao J, Lu D, Underhill P, Cavalli-Sforza LL, Chakraborty R, Jin L (2000) Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum Genet 107:582–590 [DOI] [PubMed] [Google Scholar]
- Su B, Xiao J, Underhill P, Deka R, Zhang W, Akey J, Huang W, Shen D, Lu D, Luo J, Chu J, Tan J, Shen P, Davis R, Cavalli-Sforza L, Chakraborty R, Xiong M, Du R, Oefner P, Chen Z, Jin L (1999) Y-chromosome evidence for a northward migration of modern humans into Eastern Asia during the last ice age. Am J Hum Genet 65:1718–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn W (1951) The Greeks in Bactria & India. Cambridge University Press, Cambridge [Google Scholar]
- Thangaraj K, Ramana GV, Singh L (1999) Y-chromosome and mitochondrial DNA polymorphisms in Indian populations. Electrophoresis 20:1743–1747 [DOI] [PubMed] [Google Scholar]
- Thurin M (1999) The Chenchu of the Indian Deccan. In: Lee RB, Daily R (eds) The Cambridge encyclopedia of hunters and gatherers. Cambridge University Press, Cambridge, pp 252–256 [Google Scholar]
- Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ (1997) Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res 7:996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Marzuki S, Oefner PJ, Cavalli-Sforza LL, Chambers GK (2001a) Maori origins, Y-chromosome haplotypes and implications for human history in the Pacific. Hum Mutat 17:271–280 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley R, Oefner PJ, Cavalli-Sforza LL (2001a) The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonné-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 [DOI] [PubMed] [Google Scholar]
- Walter H, Danker-Hopfe H, Bhasin MK (1991) Anthropologie Indiens. Gustav Fischer Verlag, Stuttgart [Google Scholar]
- Watkins WS, Bamshad M, Dixon ME, Bhaskara Rao B, Naidu JM, Reddy PG, Prasad BV, Das PK, Reddy PC, Gai PB, Bhanu A, Kusuma YS, Lum JK, Fischer P, Jorde LB (1999) Multiple origins of the mtDNA 9-bp deletion in populations of South India. Am J Phys Anthropol 109:147–158 [DOI] [PubMed] [Google Scholar]
- Weale ME, Yepiskoposyan L, Jager RF, Hovhannisyan N, Khudoyan A, Burbage-Hall O, Bradman N, Thomas MG (2001) Armenian Y chromosome haplotypes reveal strong regional structure within a single ethno-national group. Hum Genet 109:659–674 [DOI] [PubMed] [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, et al (2001) The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 98:10244–10249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JF, Weiss DA, Richards M, Thomas MG, Bradman N, Goldstein DB (2001) Genetic evidence for different male and female roles during cultural transitions in the British Isles. Proc Natl Acad Sci USA 98:5078–5083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y Chromosome Consortium , The (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal T, Pandya A, Santos FR, Adhikari R, Tarazona E, Kayser M, Evgafov O, Singh L, Thangaray K, Destro-Bisol G, Thomas MG, Qamar R, Mehdi SQ, Rosser ZH, Hurles ME, Jobling MA, Tyler-Smith C (1999) The use of Y-chromosomal DNA variation to investigate population history: recent male spread in Asia and Europe. In: Papiha S, Deka R, Chakraborty R (eds) Genomic diversity: applications in human population genetics. Kluwer Academic/Plenum Publishers, New York, pp 91–101 [Google Scholar]
- Zerjal T, Wells R, Yuldasheva N, Ruzibakiev R, Tyler-Smith C (2002) A genetic landscape reshaped by recent events: Y-chromosomal insights into central Asia. Am J Hum Genet 71:466–482 [DOI] [PMC free article] [PubMed] [Google Scholar]