Abstract
We explored how transcriptional noise propagates in gene-regulatory pathways by studying the induction of two downstream genes by transcription factors c-fos and c-jun. They are produced for a brief period following serum stimulation of cells and then activate the promoters of their target genes by binding to them as heterodimers. We found that, even though they are coordinately expressed at the population level, in individual cells the expression of c-fos and c-jun is noisy and uncorrelated with each other. The expression of the downstream genes is also noisy, but there is little or no effect of the noise in the upstream genes on the expression of the downstream genes. The noise is not transmitted, because the number of heterodimers present in single cells is relatively invariant, and the induction of downstream genes is insensitive to the number of heterodimers in individual cells. Sequestration of promoters of the downstream genes within compact chromatin is a likely cause of this insensitivity. These barriers to the propagation and amplification of noise are likely to be commonplace in higher eukaryotes.
Keywords: cellular heterogeneity, noise in gene expression, transcription
Introduction
Over the last decade, it has become apparent that gene expression in individual cells deviates substantially from the average behavior of cell populations (Raj and van Oudenaarden, 2008; Eldar and Elowitz, 2010). Often referred to, and analyzed as ‘noise’, this heterogeneity among genetically identical cells has been documented in bacteria, yeast, and higher eukaryotes. Instances have been found in which organisms exploit it for adaptation and development (Losick and Desplan, 2008; Boettiger and Levine, 2009). Underscoring its importance to medicine, cellular heterogeneity in gene expression may explain why only a fraction of cells become induced pluripotent cells during cellular reprogramming and why a fraction of tumor cells often exhibit drug resistance (Sharma et al, 2010; Buganim et al, 2012).
Heterogeneity in gene expression arises both from global factors that impact all genes, such as the differential abundance of RNA polymerase, or of ribosomes, in different cells; and gene-specific factors, such as probabilistic interactions of promoters with the gene activators. Referred to as extrinsic and intrinsic noise, respectively, the nature and the relative contributions of these components to the total noise differ between bacteria, yeast, and higher eukaryotes (Raj and van Oudenaarden, 2008).
Transcriptional heterogeneity can be particularly acute in higher eukaryotes (Raj et al, 2006). It varies from gene to gene and is regulated in certain cases (Raj and van Oudenaarden, 2008; Zenklusen et al, 2008; Boettiger and Levine, 2009; Gandhi et al, 2011; Munsky et al, 2012; Muramoto et al, 2012). The extent of heterogeneity has been recorded for a large number of genes on the genomics scale (Shalek et al, 2013). The heterogeneity arises because mRNAs are produced in bursts of synthesis that start and end randomly and then decay rapidly (Chubb et al, 2006; Raj et al, 2006; Boettiger and Levine, 2009; Suter et al, 2011). One hypothesis that explains the origins of RNA synthesis bursts in eukaryotes is that the promoters are not readily accessible to transcription factors, because they are tightly sequestered within insulating chromatin. Random chromatin remodeling events permit their initial binding, followed by the recruitment of the chromatin decondensation and transcription apparatus, which results in many rounds of concerted mRNA synthesis (Raser and O'Shea, 2004; Raj et al, 2006; Boeger et al, 2008). Observations that genes in which RNA polymerase is prepositioned on the promoters yield relatively less stochastic mRNA synthesis are consistent with this idea (Boettiger and Levine, 2009; Muramoto et al, 2012). Among other hypotheses invoked to explain this heterogeneity is DNA looping between promoter and the 3′-end of the gene which facilitates recycling of RNA polymerase on the same gene (Hebenstreit, 2013).
If transcriptional heterogeneity is so common and unavoidable, then how do the fluctuations in the expression of upstream genes propagate into the expression of the downstream genes that they control (Raj and van Oudenaarden, 2008; Eldar and Elowitz, 2010; Balazsi et al, 2011; Munsky et al, 2012)? Studies of synthetic circuits in bacteria and yeast provided evidence of amplified propagation of noise in those systems (Blake et al, 2003; Pedraza and van Oudenaarden, 2005). If the acute transcriptional noise observed in higher eukaryotes (Raj et al, 2006) was to be similarly amplified in gene-regulatory pathways, then those pathways will be rendered unstable. Although instances where gene-regulatory circuits have redundant pathways to mitigate the effects of noise have been found (Raj et al, 2010), whether and to what extent the noise propagates in gene-regulatory circuits in higher eukaryotes have not been studied so far.
We examined the propagation of noise in an archetypical gene-regulatory pathway in HeLa cells in which transiently expressed transcription factors c-fos and c-jun induce the expression of a set of downstream genes. We found that even though the expression of mRNAs of these transcription factors is noisy, their noise does not propagate into the downstream genes. The transmission of noise is impeded at two different steps: the heterodimers of c-fos and c-jun proteins exhibit a lesser variability than the mRNAs that encode the individual proteins (likely due to the higher stability of the former) and the chromatin context of the downstream genes and the mechanism of their induction is such that it insulates them from the variations in the heterodimers. The existence of these noise buffers explains how cells are able to minimize the propagation of noise in gene-regulatory pathways and maintain their constant phenotypes.
Results
Expression of c-fos and c-jun mRNAs is noisy and is not correlated within individual cells
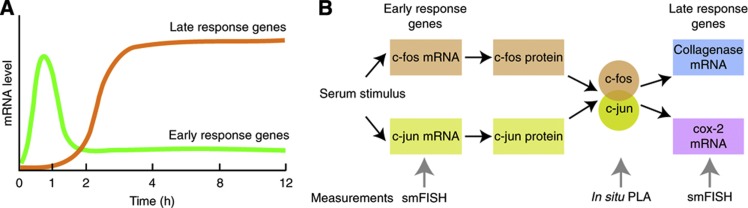
When serum is added to serum-starved cells, a group of genes are expressed immediately. Some of these ‘early response genes’ encode for transcription factors that turn on ‘late response genes’ (Figure 1A) (Iyer et al, 1999). The products of early response genes include a family of transcription factors called AP-1 factors (Figure 1B). They dimerize in a combinatorial manner and then activate the expression of a set of delayed response genes (Figure 1B). c-fos and c-jun are the most well-characterized prototypical members of the AP-1 family. Soon after their syntheses, c-fos and c-jun migrate to the nucleus, heterodimerize, bind to the promoters of their target genes, and then turn them on. Post-translational modifications, particularly phosphorylation of c-jun by the jun amino-terminal kinase (JNK), are important for this gene activation (Derijard et al, 1994). Their syntheses, however, occur only transiently, due to a feedback mechanism that turns off the c-fos gene soon after its induction (Sassone-Corsi et al, 1988; Schonthal et al, 1988). The c-fos c-jun pathway not only allows us to study how noise in gene expression propagates in gene-regulatory pathways in higher eukaryotes, it enables an investigation of a related question as to how heterogeneity in gene expression of components affects the levels of noise in multi-subunit protein complexes.
Figure 1.
(A) The addition of serum to serum-starved cells leads to the immediate induction of expression of a number of genes. Some of these ‘immediate early response genes’ encode transcription factors such as c-fos and c-jun that turn on ‘late response genes.’ The expression of immediate early genes decreases after an initial rise, but the late response genes are expressed in a sustained manner over time. This figure is derived from Lodish et al (2000). (B) Diagram showing the relationships among members of the c-fos/c-jun pathway that were investigated. Species whose levels were measured in single cells are indicated by gray arrows below the blocks. We measured the heterodimers and downstream mRNAs in the same cells, rather than upstream and downstream mRNAs, because by the time downstream mRNAs begin to get expressed, the c-fos and c-jun mRNAs disappear from the cells.
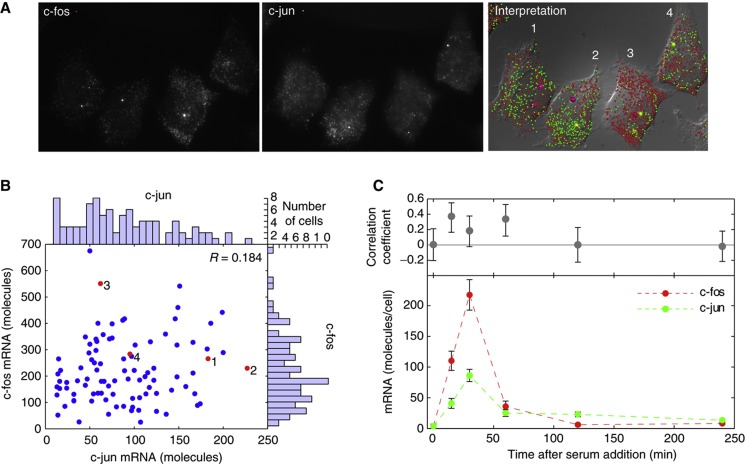
Because they need to act together, the c-fos and c-jun mRNAs are expressed in a coordinated manner during a brief window of time after serum stimulation (Martens et al, 2003). To determine whether the expression of these two genes is also coordinated in individual cells, we investigated whether there is any cell-to-cell heterogeneity in the expression of these mRNAs. To accomplish this, we counted the number of molecules of each mRNA species in individual cells by single-molecule fluorescence in situ hybridization (smFISH) (Raj et al, 2008) at different times after the addition of serum to HeLa cell cultures that were starved of serum (Figure 2). We found that, on average, the kinetics of induction in individual cells mirrors the kinetics of induction observed in earlier studies from ensembles of cells (Martens et al, 2003). However, at the peak of c-fos and c-jun expression at 30 min after serum addition, there was a remarkable lack of correlation between their expressions in individual cells (Figure 2A and B). Similar lack of correlation was observed at the later time points (Figure 2C).
Figure 2.
The numbers of c-fos and c-jun mRNA molecules do not correlate with each other in individual cells. (A) Single-molecule FISH images of representative cells 30 min after the addition of serum to cells that were starved of serum for 48 h. The two left-hand panels are merged z-stacks, indicating the probes used; and the right-hand panel displays the identified mRNA molecules (small balls) and active gene loci (larger balls) on a diffraction interference contrast image. (B) The number of c-fos and c-jun mRNAs in 100 individual cells 30 min after serum stimulation. The red dots correspond to the data obtained from the four cells shown in the images. Marginal histograms show the distribution of each mRNA among the 100 cells that were examined. (C) Number of c-fos and c-jun mRNA molecules per cell as a function of time after the addition of serum and the correlation coefficients between them at each time point. Error bars represent the 95% confidence interval obtained from measurements made on 50–100 cells. Source data for this figure is available on the online supplementary information page.
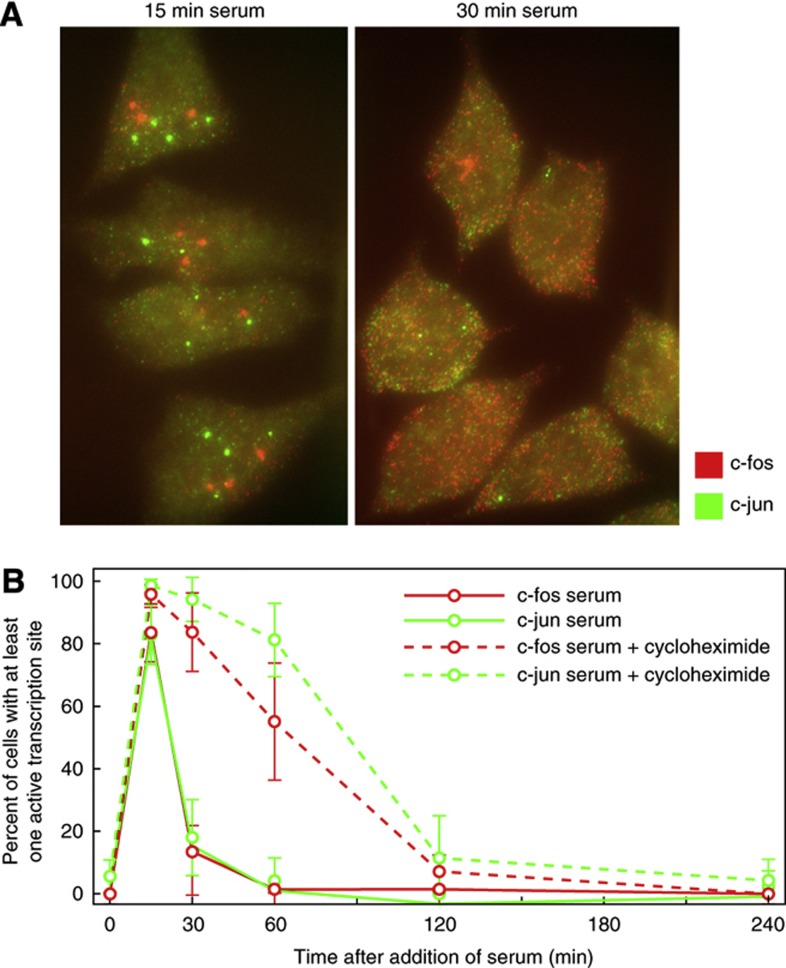
Could a differential onset of mRNA synthesis account for this lack of correlation? One of the manifestations of mRNA synthesis occurring in random bursts is that, during the bursts of synthesis, many mRNA molecules accumulate at the gene locus before they have a chance to disperse into the nucleoplasm (Chubb et al, 2006; Raj et al, 2006). Accordingly, clusters of both c-fos and c-jun mRNAs were visible in the nuclei, marking the onset of gene expression. An analysis of individual cells for the presence of these RNA clusters indicated that by 15 min, at least one of the alleles of each gene is turned on in 83% of the cells (Figure 3), indicating a concordant onset of expression from the two genes. However, by 30 min, as new RNA synthesis comes to a stop due to a negative feedback mechanism in which c-fos protein mediates the suppression of synthesis of its own mRNA (Sassone-Corsi et al, 1988; Schonthal et al, 1988), the clusters of mRNAs at gene loci dissipate due to the dispersal of the mRNA molecules (Figure 3B).
Figure 3.
The c-fos and c-jun genes are turned on in a large fraction of cells within 15 min from the addition of serum. (A) Images showing clusters of nascent mRNA molecules sequestered at gene loci, and their subsequent dispersal into the cell volume 15 and 30 min after serum stimulation. The images are merged z-stacks, color coded as indicated. (B) Proportion of examined cells that displayed at least one active gene locus for the indicated mRNA, as a function of the time elapsed after the addition of serum. The cells displayed up to three active gene loci for both c-fos and c-jun. This likely reflects the known trisomy for chromosomes 14 and 1 in HeLa cells, where c-fos and c-jun are located respectively (Macville et al, 1999).
Evidence that c-fos protein mediates the suppression of mRNA synthesis became apparent when we added the protein synthesis inhibitor cycloheximide to the culture at the same time that we added the serum, which caused the gene loci to remain active for much longer (Figure 3A). These observations indicate that the expression of the two mRNAs begins in a rapid and coordinated manner, but becomes uncorrelated thereafter because different cells yield bursts of different sizes and durations. These observations also suggest that each gene is able to fire off just one burst of expression before the negative feedback mechanism turns it off permanently.
At the peak of their expression (∼30 min after activation), we not only observed the lack of correlation between the syntheses of the two mRNAs, but also observed a large cell-to-cell variation in the number of molecules of each mRNA in individual cells (Figure 2B, marginal histograms). The cell-to-cell variation, or noise strength, is often quantified by either of two parameters—Fano factor (square of the standard deviation (σ) divided by the mean (μ)) or coefficient of variation (σ/μ). The Fano factor provides a measure of how far a population departs from a Poisson distribution, which would occur if mRNAs were to be produced and degraded steadily with equal rates in different cells (Ozbudak et al, 2002; Taniguchi et al, 2010). The Fano factor for the Poisson distribution is one. However, Fano factor is useful only when integer counts for the molecules are available, or when the units of the measurements being compared are the same. Being a unit-less quantity, the coefficient of variation (σ/μ), on the other hand, permits comparison between measurements in different units, but it does not provide a convenient comparison with the Poisson’s distribution. We will use both parameters here.
Using the Fano factor as a measure of noise strength, we found that for c-fos and c-jun the Fano factors were 71 and 30, respectively, deviating significantly from the Poisson behavior. These strikingly high levels of cell-to-cell variations raise a question as to how different cells are able to express the downstream genes controlled by c-fos and c-jun at relatively similar levels to ensure constant phenotypes.
Cell-to-cell variation in c-fos and c-jun proteins
The noise in mRNA expression is often higher than the noise in the corresponding proteins, because the half-life of mRNAs is usually shorter than the half-life of the proteins that they encode (Raj et al, 2006; Taniguchi et al, 2010; Schwanhausser et al, 2011). In the case of c-fos and c-jun, mRNAs degrade rapidly with a half-life of 9 and 11 min, respectively (27), whereas the corresponding proteins are considerably more stable (Rahmsdorf et al, 1987; Kovary and Bravo, 1991). The c-fos protein displays a biphasic stability curve, which has an initial phase of relatively fast decay with 45 min half-life and then a second phase of slower decay with a half-life of 90–120 min (Rahmsdorf et al, 1987; Kovary and Bravo, 1991). A second study estimated an average half-life of 162 min for c-fos protein (Bossis et al, 2003). c-jun protein half-life has been estimated to be 90–120 min (Rahmsdorf et al, 1987; Kovary and Bravo, 1991). On the other hand, the median half-life of most of the mRNAs is 9 h and the median half-life of most of the proteins is 43 h in mammalian cells (Schwanhausser et al, 2011).
To explore whether the relatively longer half-life of c-fos and c-jun proteins buffers them against fluctuations in the synthesis of their mRNAs, we estimated the cell-to-cell variations in their levels by immunofluorescence (IF). We used a pair of antibodies specific for c-fos and c-jun simultaneously to determine the levels of both from the same cells. The analysis was restricted to the nucleus where most of the signals were localized (Supplementary Figure S1A). Although induction of both proteins by serum could be clearly detected by this technique, their mean levels varied to a great extent between different time points after induction (Supplementary Figure S1B). We also estimated the coefficients of variation at each time point and are presenting the data for 6 h in Supplementary Figure S2. This analysis reveals that the coefficient of variation of c-fos protein at 6 h is similar to that of its own mRNA (at 30 min), whereas, the coefficient of variation of c-jun protein at 6 h is lower than its own mRNA (Supplementary Figure S2). During the period of 15 min to 12 h after induction, the coefficients of variation of proteins ranged from 0.26 to 0.49 for c-jun and from 0.42 to 0.58 for c-fos.
Imaging c-fos and c-jun heterodimers
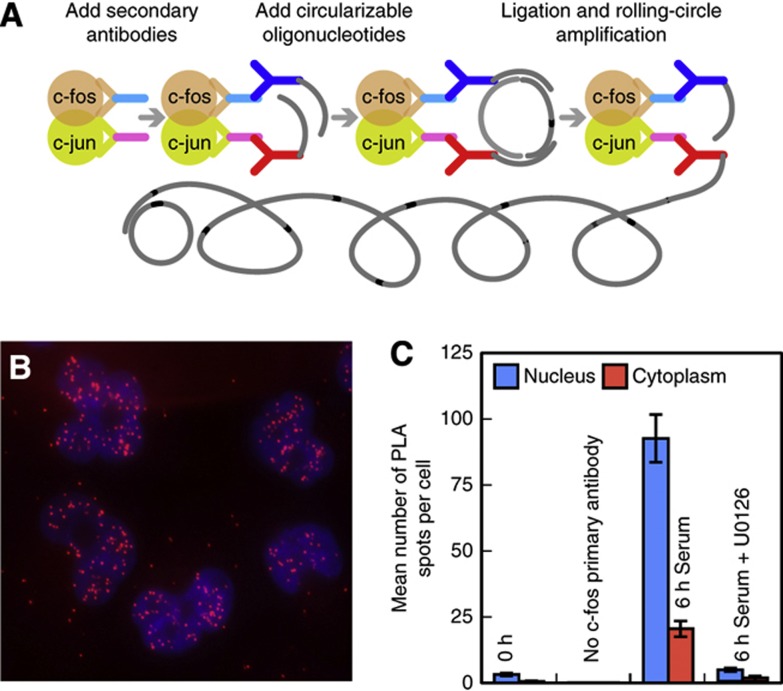
The functional unit responsible for the induction of downstream genes is the heterodimer formed by the c-fos and c-jun proteins, rather than the individual proteins. We therefore developed an approach to obtain a measure of the numbers of these heterodimers in individual cells. In this approach, we utilized an in situ proximity ligation assay (PLA), which enables the specific detection of two protein molecules that exist in close proximity to each other in a complex (Soderberg et al, 2006). During PLA, each of the two proteins in the complex is first probed with a pair of specific primary antibodies that are isolated from two different species. In a second step, species-specific secondary antibodies are bound to each primary antibody. Each of those secondary antibodies is linked to a different oligonucleotide, and if the two different oligonucleotides are in close proximity to each other (by virtue of the two proteins being present in the same complex), then the oligonucleotides are able to serve as templates for the creation of a circular DNA. This circular DNA then serves as a template for an enzymatic ‘rolling-circle’ amplification, generating a tandemly repeated single-stranded DNA sequence that remains tethered to the complex, which is then made visible as an intensely fluorescent spot by the hybridization of labeled probes against the repeated sequences (Figure 4A).
Figure 4.
Detecting c-fos/c-jun heterodimers in situ. (A) Schematic representation of the steps of an in situ proximity ligation assay (PLA). (B) Intracellular distribution of spots generated in an in situ PLA for c-fos/c-jun heterodimers 6 h after serum stimulation. The image is a merged z-stack overlaid on DAPI-stained nuclei. (C) Demonstration of the specificity of detection of c-fos/c-jun heterodimers. U0126 is an inhibitor of mitogen-activated protein kinases that prevent the induction of c-fos.
The detection of the c-fos/c-jun heterodimer was quite specific, because PLA spots appeared only after serum stimulation, the spots were largely restricted to the nucleus, and the spots were not seen when serum induction was accompanied by the addition of U0126, a small-molecule inhibitor of mitogen-activated protein kinases, which specifically induce c-fos expression (Favata et al, 1998) (Figure 4B and C). Additional evidence of the specificity of PLA for the detection of c-fos/c-jun heterodimers has been described by Baan et al (2010).
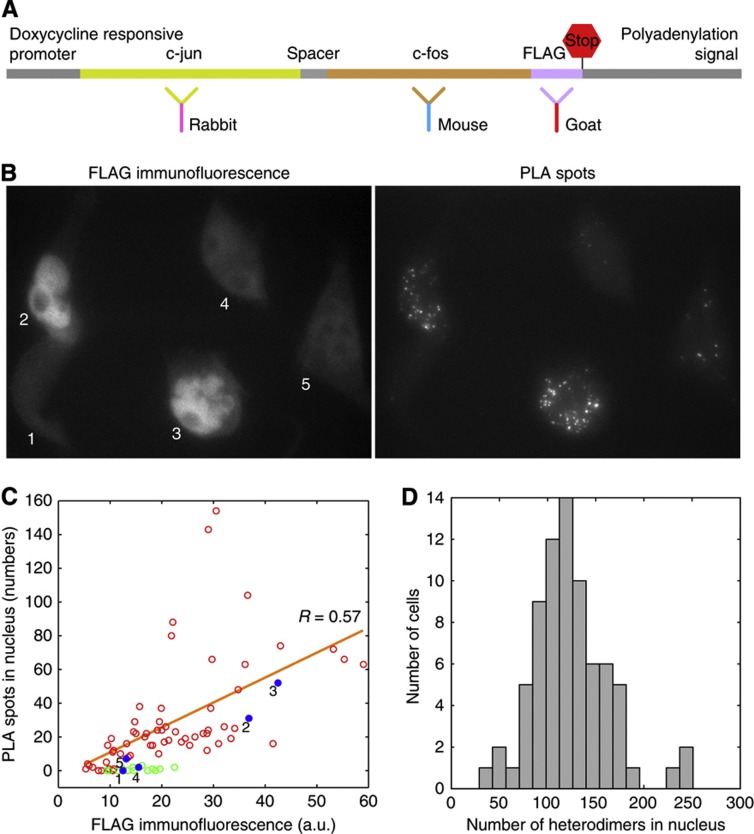
To show that the number of spots quantitatively reflects the number of heterodimers present in individual cells, we engineered a protein in which the c-fos coding sequence was fused with the c-jun coding sequence following an approach described earlier by Bakiri et al (2002). In addition, at the C-terminal of this construct we introduced an artificial sequence, called a FLAG tag, which can serve as a marker for the fused protein. This construct provided us an opportunity to measure the same protein by PLA and IF in the same cells (Figure 5A).
Figure 5.
A demonstration of the proportionality of number of PLA spots with the amount of component proteins and the distribution of PLA spots in HeLa cells. (A) Schematic illustration of an engineered construct in which a c-fos coding sequence was fused to a c-jun coding sequence and a FLAG tag was added at the C-terminal. This fusion protein was detected by PLA (using indicated c-fos and c-jun specific antibodies) and the FLAG tag was detected by direct immunofluorescence (IF) with the help of the indicated third antibody. (B) FLAG IF and PLA signals from the same field of cells expressing the reporter gene integrated into their genomes. (C) Number of PLA spots versus integrated fluorescence intensity in the nuclei of single cells. Red circles, cells in the absence of doxycycline (expressing the reporter), blue spots, data from cells indicated in the image in (B), and green circles, cells in the presence of doxycycline (no expression from the reporter). (D) Distribution of c-fos/c-jun heterodimers from the endogenous gene in unmodified HeLa cells (not the clone discussed above) in the nuclei of 75 cells 6 h after serum induction. Source data for this figure is available on the online supplementary information page.
A gene encoding the fusion protein was placed under the control of a doxycycline promoter and introduced into the genome of a HeLa cell line that also expresses Tet A transactivator and a stable cell line was isolated (Figure 5A). This cell line expresses the reporter in the absence of doxycycline but not in its presence. When c-fos and c-jun mRNAs were imaged in the cells expressing this construct the spots from the two co-localized indicating expression of an mRNA in which both coding sequences are present. This behavior is distinct from cells that express the natural c-fos and c-jun mRNAs whose spots never co-localize (Figure 2A). The fused protein was present largely in the nucleus as indicated by staining with antibodies specific to c-fos, c-jun, and FLAG (Figure 5B). Most importantly, we could detect c-fos and c-jun specific PLA spots and FLAG-specific IF in the same cells (Figure 5B).
Consistent with the previous observations that doxycycline control genes undergo stochastic expression (Raj et al, 2006), there was a large difference in the number of PLA spots and FLAG IF signals in individual cells for this construct. However, when the FLAG signal was large, the number of spots was also large (Figure 5B and C). The number of PLA spots was generally proportional to the integrated IF signal in the nucleus, and in the absence of induction, no PLA spots were detected (Figure 5B and C). These results obtained with an artificial construct can be extrapolated to the natural situation to suggest that the number of PLA spots is proportional to the number of actual c-fos/c-jun heterodimers in the cell.
Number of c-fos/c-jun heterodimers is relatively invariant
After establishing this proportionality with the reporter cell line, we analyzed normal HeLa cells for the number of PLA spots in individual cells after 6 h of serum addition. This analysis revealed that the cell-to-cell variation in the levels of heterodimers (Fano factor 11) is considerably less compared to the variation observed for c-fos and c-jun mRNAs (Figure 5D). How does the cell-to-cell variation in heterodimer compare with the variations in individual proteins? For this comparison, the Fano factor cannot be used, because the units of measurements for the two are different. Instead, we used coefficient of variation which showed that the coefficient of variation of c-fos protein was higher than the coefficient of variation of heterodimers, whereas the coefficient of variation of c-jun protein was about the same as the coefficient of variation of heterodimers (Supplementary Figure S2).
When one component displays higher variability than the other, how will the variation in the product be decided? Since the minority component decides how many heterodimers will be produced, it is likely that the variation in the minority component will dominate the variation in the heterodimers, provided that it remains in the minority over time. However, since the absolute levels of c-fos and c-jun proteins in individual cells are not known presently, we cannot say whether it is c-fos or c-jun that decides the variability of heterodimers.
Nevertheless, the higher stability of individual proteins is likely to contribute to a reduction in the variation. In addition, the proteins are likely to become more stable upon heterodimerization. Earlier observations that c-fos protein exhibits biphasic stability with an early stage of faster decay and a later stage of slower decay (Kovary and Bravo, 1991), implies that the c-fos/c-jun heterodimers are more stable than the individual proteins. This is supported by our own observations (data not shown) that the half-life of the heterodimers is >4 h, which is considerably longer than the reported half-life of the individual proteins. In addition to the higher stability of heterodimers, other factors that are more completely described in the discussion may also contribute to the lower heterodimer variation.
Noise in the expression of downstream genes
Do the relatively constant levels of c-fos/c-jun heterodimers in different cells lead to noise-free expression of downstream genes? To explore this issue, we selected two downstream genes, collagenase 1 (also known as matrix metalloproteinase-1 (MMP-1)) and cyclooxygenase-2 (cox-2) (also known as prostaglandin-endoperoxide synthase 2) that are specifically induced by c-fos/c-jun heterodimers, and are not induced by other members of the AP-1 transcription factor family. Diverse and strong evidence exists for this specificity: (a) c-fos is needed for the expression of collagenase 1 and antisense RNA complementary to c-fos mRNA abolishes this expression (Schonthal et al, 1988); (b) in cell lines derived from mice in which the c-fos gene was knocked out, collagenase 1 gene is not induced by serum activation (Hu et al, 1994); (c) targeted chromatin immunoprecipitation experiments indicate that the collagenase 1 promoter is occupied by both c-fos and c-jun after serum activation (Martens et al, 2003); and (d) among the fused genes constructs obtained by tethering c-jun with AP-1 factors ATF2, c-jun, Fra1, Fra2, or c-fos, only the c-jun/c-fos combination binds to, and induces expression from, the collagenase 1 promoter (Bakiri et al, 2002; Wisniewska et al, 2007). Similarly, the promoter of the cox-2 gene is occupied by both c-fos and c-jun, and is cooperatively induced in their presence (Chen et al, 2005).
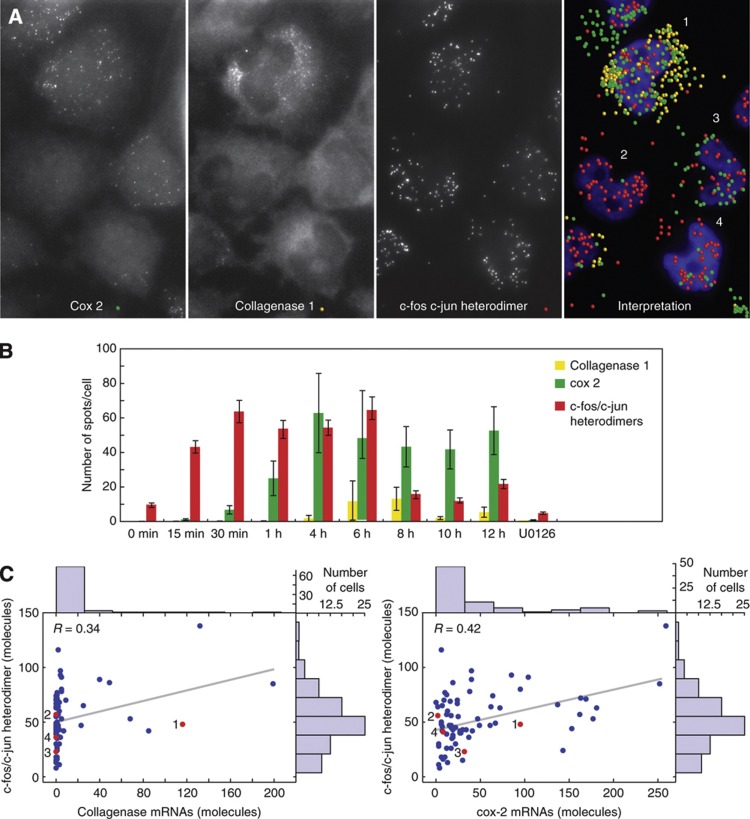
The mRNAs of collagenase 1 and cox-2 were imaged, along with the heterodimers of c-fos and c-jun, in the same cells as a function of time elapsed after the addition of serum. Consistent with previous observations (Martens et al, 2003), the heterodimers became detectable soon after serum induction, yet the two downstream mRNAs appeared after a lag period that was different for each mRNA (Figure 6B). Because of this lag, and because the c-fos and c-jun mRNAs disappear from cells by 60 min (Figure 2C), the upstream and downstream mRNAs do not coexist at the same time. However, the c-fos/c-jun heterodimers do coexist with the two downstream mRNAs (Figure 6A and B). The synthesis of the downstream mRNAs was dependent upon the presence of c-fos/c-jun heterodimers, as the addition of serum in the presence of the c-fos inhibitor U0126 did not result in the synthesis of heterodimers by 4h, and the synthesis of downstream mRNAs was not observed (Figure 6B).
Figure 6.
Noisy expressions of collagenase 1 and cox-2 mRNAs, in spite of relatively constant levels of c-fos/c-jun heterodimers. (A) Triplex detection of cox-2 and collagenase 1 mRNAs and c-fos/c-jun heterodimers 6 h after serum induction. The three left-hand panels are raw merged z-stacks showing the indicated targets, and the right-hand panel shows the identified molecules overlaid on an image of cells stained for their nuclei. (B) Time course of the appearance of nuclear c-fos/c-jun heterodimers and two downstream mRNAs after serum induction in the whole cells. (C) Distributions of nuclear c-fos/c-jun heterodimers and whole cell collagenase 1 and cox-2 mRNAs in 75 single cells 6 h after serum induction. Red dots depict the data obtained from the cells shown in (A). Correlation between collagenase 1 and cox-2 mRNAs is presented in Supplementary Figure S3. Source data for this figure is available on the online supplementary information page.
Strikingly, at the time of the peak synthesis of the two downstream mRNAs (at 6 h), when the number of heterodimers was very similar in different cells, the expression of the two downstream mRNAs was highly stochastic, as indicated by the marginal histograms in Figure 6C. Straight lines fitted to the heterodimer versus collagenase 1 and cox-2 mRNA distributions at 6 h after induction originated at the middle of the heterodimer distributions, and exhibited shallow slopes and low correlation coefficients (Figure 6C). This was also true at other time points, with the slopes of the lines ranging from 0.02 to 0.37 for collagenase 1 and 0.02 to 0.13 for cox-2, and their correlation coefficients ranging from 0.16 to 0.38 for collagenase 1 and 0.10 to 0.42 for cox-2. These observations indicate that although the heterodimers are needed for the expression of downstream mRNAs, the likelihood of induction does not increase substantially with the number of heterodimers present in the cells.
Even though most cells contained similar levels of heterodimers, very few cells expressed collagenase 1 mRNA. However, those that did produce the mRNAs, contained many mRNA molecules, resulting in a ‘long-tailed’ population distribution. Akin to wealth distribution in society, these distributions signify a high cell-to-cell variation, due to the synthesis of mRNA occurring in random bursts (Figure 6C) (Raj et al, 2006). The probability of finding cells expressing cox-2 mRNA was relatively higher; however, its mRNA distribution was also long-tailed (Figure 6C). The noisy synthesis of both mRNAs was reflected in the very high Fano factors associated with their expression (Figure 6C).
Since both downstream genes are induced by the same heterodimers, is their expression correlated? We plotted the number of collagenase 1 mRNAs against the number of cox2 mRNAs in the same cells (Supplementary Figure S3). Only a weak correlation (R=0.47) was observed between them, indicating that the induction of these two gene loci is relatively independent of each other in single cells, even though they both respond to the same transactivator. This is consistent with the previous results where two genes with identical promoters, responding to the same transactivator, but situated at distant genomic loci, displayed uncorrelated noise (Raj et al, 2006).
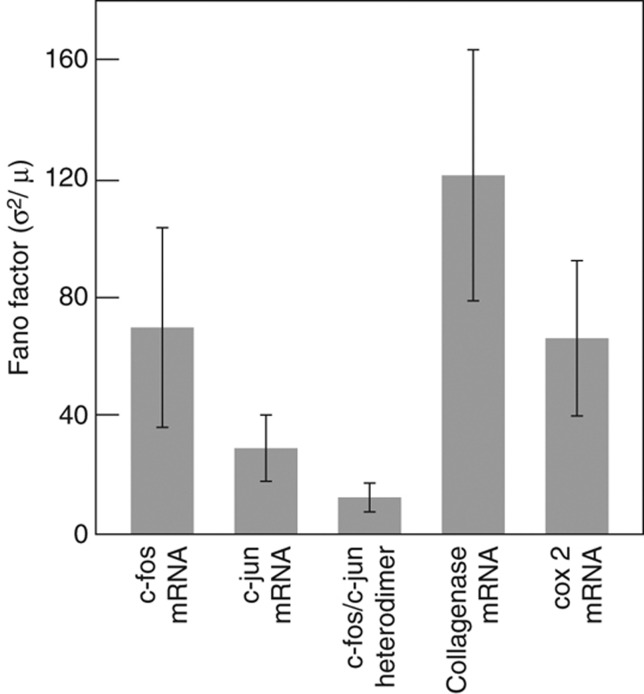
As summarized in Figure 7, we observed that noise levels, denoted by Fano factors, are high in the mRNAs of upstream genes, low in the heterodimers, and again high in the downstream mRNAs. Even though noise in the transcription factor heterodimers is buffered, the expression of the downstream genes is still noisy.
Figure 7.
Cell-to-cell variation (Fano factor, standard deviation2/mean) in mRNAs for the upstream genes c-fos and c-jun, c-fos/c-jun heterodimers, and mRNAs for the downstream genes collagenase 1 and cox-2. The data for c-fos and c-jun mRNAs are from cells at 30 min after serum addition and for the other species it is 6 h after serum addition. The coefficient of variations in these species along with c-fos and c-jun proteins is shown in Supplementary Figure S2. The error bars represent 95% CI. Source data for this figure is available on the online supplementary information page.
Discussion
Different activation mechanisms and noise characteristics of upstream and downstream genes
Our results point to a distinction in the mechanisms of activation of the upstream and downstream genes in this pathway. While the c-fos and c-jun genes are turned on rapidly and concurrently in almost all cells and produce a single burst of mRNA synthesis, the downstream genes are expressed in random bursts in a few cells and after a lengthy lag period. Significantly, earlier studies have shown that RNA polymerase II is prepositioned on the promoters of the c-fos and c-jun genes (Ramirez-Carrozzi et al, 2009), but is recruited de novo to the promoters of the downstream genes before their induction (Martens et al, 2003) (Supplementary Figure S4). It is likely that the prepositioned RNA polymerase II is a part of the mechanism that has evolved to ensure that the mRNA synthesis begins immediately upon receipt of the serum stimulus and occurs synchronously in all cells. A de novo recruitment of the polymerase II to the promoter, on the other hand, leads to a slower and more stochastic onset of the genes as we found in the case of the two downstream genes. A similar observation was made previously by Boettiger and Levine (2009) who found that genes that have RNA Pol II prepositioned on their promoters display synchronous activation in the mesoderm of Drosophila embryo and genes that need to recruit Pol II de novo are expressed in a stochastic manner. Since many genes have prepositioned RNA Pol II, it is likely that they will exhibit a comparatively less cell-to-cell variation in gene expression.
A ‘Noise Bottleneck’ in the c-fos c-jun gene-regulatory pathway
We found that although the expression of both upstream and the downstream genes in the c-fos c-jun pathway is noisy, the noise from upstream genes is not transmitted into the downstream genes. This conclusion is based on two observations each pointing to a separate step. First, the noise in the key intermediate, c-fos/c-jun heterodimer, is lower than either the upstream or the downstream mRNAs, and second, the number of heterodimers does not correlate with the frequency of induction of downstream genes.
Given the high variation in the mRNAs of both c-fos and c-jun, how is low variation in the heterodimers achieved? As we discussed before, that both c-fos and c-jun proteins are more stable than their parent mRNAs, in addition, the heterodimer is likely to be even more stable than the individual proteins. Since higher stability buffers against temporal fluctuations (Raj et al, 2006), this higher stability is expected to be a key reason why heterodimers have lower variation than the parent mRNAs.
It is likely that other factors also contribute to lower the variations in heterodimers. In addition to forming heterodimers with each other, c-fos and c-jun proteins associate with other members of the AP-1 family of transcription factors (Kovary and Bravo, 1991) whose variation will impact the c-fos/c-jun heterodimer variation. A priori, the c-fos/c-jun heterodimer variation will be largely set by the variation of the least abundant component (either c-fos or c-jun), with influences from the noise in other interacting AP-1 transcription factors. Theoretical studies suggest that the dynamics of complex formation can automatically reduce noise in multi-subunit protein complexes (Konkoli, 2010). In addition to this network of transcription factors, other cellular factors influence the activity of c-fos and c-jun. For example, phosphorylation of c-jun by JNKs is necessary for its ability to activate gene (Derijard et al, 1994). Therefore, how many copies of active heterodimers are present in a given cell will also depend upon the levels of JNKs in that cell.
The second barrier that prevents propagation of noise in this gene-regulatory pathway stems from the structure of chromatin and relates to how genes are induced. Our data show that although the variation in heterodimers is low, it is significant. To the extent heterodimers vary from cell-to-cell, this variability is not reflected in the probability with which the cells make the downstream mRNAs (Figure 6C). Although the downstream genes do need heterodimers for their expression, the likelihood of their expression in individual cells is rather insensitive to the number of heterodimers present in the cell. A similar observation was made earlier by Raj et al (2006) who found that in the case of tetracycline controlled genes, the tet transactivator was needed for the activation of its target genes but that activation was rather insensitive to the amount of tet transactivator. This suggests that the sequestration of gene-regulatory regions within chromatin is simultaneously responsible for generating the intrinsic stochasticity of gene expression and for insulating it from fluctuations in the amounts of upstream gene products.
Differences from prokaryotes
In contrast with the observations detailed above, two previous studies, one performed in bacteria (Pedraza and van Oudenaarden, 2005) and the other performed in yeast (Blake et al, 2003), documented amplified propagation of noise into downstream genes. The noise was amplified in those studies because the promoters of the downstream genes in both cases were readily accessible to the transactivator proteins and could thus experience and respond to fluctuations in levels of upstream transcription factors. In E. coli, the subject of the first study, there are no nucleosomes to impede the access of transactivator to the promoter; and in S. cerevisiae, the subject of the second study, the promoters were depleted of nucleosomes relative to the coding sequences (Lee et al, 2007). Furthermore, one of the promoters in the latter study, GAL1-GAL10, harbors a prepositioned accessory transcription factor, RSC, which likely facilitates the entry of transactivator (GAL4p) (Floer et al, 2010). Although the density of nucleosomes in eukaryotic promoters is generally lower than it is in coding regions, they are nonetheless sequestered from transcription factors (as is the case for collagenase 1) (Martens et al, 2003). Furthermore, the other barrier to the transmission of noise in c-fos/c-jun pathway, the heterodimerization-mediated buffering was also absent in the previous studies because the transcription factors functioned as monomers.
Given the prevalence of di- and multi-meric transcription factor complexes and their de novo recruitment to promoters sequestered within tight chromatin, the buffering mechanisms that we describe must be commonplace in higher eukaryotes and likely have an important role in maintaining their constant phenotypes. However, this is likely to be just one out of many mechanisms. For example, Raj et al (2010) discovered that there are redundant arms in some regulatory circuits, where each arm serves to circumvent the noise stemming from the other arm.
Materials and methods
Cell culture
HeLa cells were plated over 0.17 mm thick glass coverslips coated with gelatin and cultured in the α modification of Eagle’s Minimum Essential Medium (Sigma, St Louis, MO) supplemented with 10% fetal bovine serum (Clontech, Mountain View, CA). After culturing the cells for 24 h, the cells were washed and cultured in serum-free medium for 48 h. Thereafter, this medium was replaced with pre-warmed medium containing 20% serum and 200 μM tetradecanoyl phorbol acetate (TPA) (Sigma). The coverslips were withdrawn after indicated periods of time, rapidly washed with phosphate-buffered saline (PBS), and fixed using 4% formaldehyde in PBS solution for 10 min at room temperature. In experiments that included the addition of an inhibitor of mitogen-activated protein kinases, 10 μM U0126 (Promega, Sunnyvale, CA) was added to the serum containing medium. In experiments in which new protein synthesis was inhibited, cycloheximide (Sigma) at a final concentration of 50 μg/ml was added to the culture medium.
Cloning a c-fos/c-jun fusion protein
To fuse the coding sequences of c-fos and c-jun, we followed the approach previously described by Bakiri et al (2002). An important difference was that we used a previously constructed c-fos clone (Vargas et al, 2011) as the source of its coding sequence. In this clone, a full-length human c-fos gene, including its introns, was placed under the control of a doxycycline responsive promoter in vector pTRE2Hyg (Clontech). In our scheme, we first amplified the c-jun coding sequence from genomic DNA, using a forward primer, GTGTCCCCCGCTTGCCACAG, located in the 5′-UTR, and a reverse primer, TCAGCCCCCGACGGTCTCTC, located in the 3′-UTR; and we then cloned it into a pCR-4 TOPO cloning vector (Life Technologies, Carlsbad, CA). Since the c-jun gene harbors no introns, this was equivalent to cloning its cDNA. The resulting clone was modified to introduce a BamH1 site at the 5′ end of the c-jun coding sequence, and to introduce an Mlu1 site at the 3′ end of the c-jun coding sequence, using a QuickChange site-directed mutagenesis kit (Stratagene, San Diego, CA). The BamH1–Mlu1 fragment, containing the c-jun coding sequence, was then excised from this clone and inserted at the 5′ end of the c-fos genomic sequence within pTRE2Hyg-c-fos, using the same pair of restriction enzymes to linearize the host plasmid. The resulting clone was further modified to introduce an 8-amino acid-long linker (Bakiri et al, 2002) between the coding sequence of the two genes. The introduction of this linker also removed the start codon from the c-fos sequence, creating a fused coding frame for the two genes. A DNA fragment (5′-GACTACAAGGACGACGACGACAAG-3′) encoding the FLAG sequence was inserted just before the stop codon of c-fos by site-directed mutagenesis. This construct was linearized with restriction enzyme Fsp1, transfected into HeLa-tet-off cells (Clontech), and then several stable cell lines expressing the continuous gene under doxycycline control were isolated.
Single-molecule fluorescence in situ hybridization
We imaged individual mRNA molecules by hybridizing a set of 48 oligonucleotide probes to each mRNA species. Each oligonucleotide in a set was labeled with one fluorescent dye moiety. The dyes used to label each set were Alexa 594, Cy5, or tetramethylrhodamine, each of which fluoresces in a different color. When so many probes bind simultaneously to the same mRNA, each molecule becomes so intensely fluorescent that it can be seen as a fine fluorescent spot in a fluorescence microscope; whereas the background appears as a low-level, diffused fluorescence (Raj et al, 2008). Evidence for the single-molecule sensitivity and near absolute specificity of this system is reviewed in Supplementary Information provided by Batish et al (2012).
The Ensemble IDs of the sequences of the transcripts that we probed are c-fos, ENST00000303562; c-jun, ENST00000371222; collagenase 1, ENST00000315274; and cox-2, ENST00000367468. The 48 probes in each set, each 20 nucleotides long, were selected for these transcripts using the Stellaris probe designer program available online at http://www.biosearchtech.com/stellarisdesigner/. The probe sequences will be provided upon request. Oligonucleotide sets corresponding to the designed probes, with each probe having a 3′-amino group, were obtained from the Biosearch Technologies (Novato, CA), pooled in equimolar amounts, and coupled to one of the three fluorescent dyes mentioned above, using their respective succinimidyl esters for linkage to their 3′-amino groups, as described previously (Raj et al, 2008). The oligonucleotides coupled to the dye were then purified, using high-pressure liquid chromatography.
Fixed cells attached to coverslips were permeabilized by incubation with 70% alcohol for 1 h, equilibrated briefly with 10% formamide in 2X saline sodium citrate (SSC) buffer (Ambion, Austin, TX), and then hybridized overnight at 37°C in a humid chamber with the probes. Each hybridization reaction (50 μl) contained 10% dextran sulfate (Sigma), 1 μg/μl Escherichia coli tRNA (Sigma), 2 mM ribonucleoside-vanadyl complex (New England Biolabs, Ipswich, MA), 0.02% RNase-free bovine serum albumin (Ambion), 10% formamide, and 1 ng/μl of each probe set. After hybridization, the coverslips were washed twice with 10% formamide in 2X SSC at room temperature, and then mounted in oxygen-depleted mounting medium (Raj et al, 2008).
PLA combined with smFISH
We performed PLA first and then fixed the cells a second time, followed by smFISH for the downstream mRNAs. For PLA, we utilized reagents from a Duolink PLA Kit (Olink Biosciences, Uppsala, Sweden). To protect the cellular RNA during PLA, we included RNase inhibitor from human placenta (New England Biolabs) in the incubation steps, included ribonucleoside-vanadyl complex in washing buffers, and used diethylpyrocarbonate-treated water for dilution of the reagents. For each incubation step, the coverslips were placed over a 50-μl solution (cell-side facing down) on a strip of Parafilm that was stretched over glass and placed in a humid chamber. The humid chamber was pre-equilibrated at the appropriate temperature. For the washing steps, the coverslips were bathed in 1 ml of solution in a 12-well plate.
Our protocol for PLA was composed of the following steps, which refer to reagents included in the Duolink PLA Kit. (1) Fixed cells were permeabilized by bathing the coverslips in 70% ethanol for 1 h. (2) The cells were blocked using 1X blocking solution supplemented with 1 unit/μl of RNase inhibitor for 1 h at room temperature. (3) Primary antibodies against human c-fos (mouse antibody, SC-8047; Santa Cruz Biotechnologies, Santa Cruz, CA) and c-jun (rabbit antibody, SC-1694; Santa Cruz Biotechnologies) were added at 1–100 dilution in the blocking solution, and the coverslips were then incubated overnight at 4°C in a humid chamber. (4) Excess primary antibodies were removed by washing the coverslips twice with wash buffer A (0.01 M Tris–HCl pH 7.4, 0.15 M NaCl, 0.05% Tween-20 (Sigma), and 2 mM ribonucleoside-vanadyl complex). (5) Anti-rabbit plus secondary antibody and anti-mouse minus secondary antibody were incubated in 1X antibody dilution solution supplemented with 1 unit/μl of RNase inhibitor for 1 h at 37°C. (6) Excess secondary antibodies were removed by two washes with buffer A. (7) The cells were incubated with 1X ligation buffer, including ligase, for 30 min at 37°C, followed by two washes in buffer A. (8) The cells were then incubated with 1X rolling-circle amplification mixture, including the DNA polymerase and the detection probes, for 1 h at 37°C, followed by two washes in buffer A. (9) The cells were fixed for a second time with 3.7% formaldehyde in 1X PBS for 10 min, followed by equilibration in 10% formamide in 2X SSC. (10) In situ hybridization was performed as above. The specificity of the primary antibodies was confirmed by preliminary direct IF staining.
The number of spots that are obtained in PLA depends upon the abundance of the complexes, the degree to which the epitopes that are recognized by the primary antibodies are exposed, the distance between the target proteins, and their disposition relative to each other. Proteins that are as far as 30–40 nm from each other in the complex can reliably be detected (http://www.olink.com/node/180). We obtain ∼100 PLA spots per cell for the c-fos/c-jun heterodimer, which is likely to be a small fraction of the total number of heterodimer complexes present in each cell. However, the number of spots detected in a cell is proportional to the number of heterodimers present, and the average number of PLA spots/cell as a function of time follows the expected kinetics (Figure 5).
PLA combined with direct IF for FLAG
The same protocol as above was generally followed with an exception of the RNAse inhibitors, which were excluded. During the step of primary antibody addition an FLAG tag-specific goat antibody A190-101A (Bethyl Laboratories, Montgomery, TX) was included in the incubation mixture in addition to the antibodies for c-fos and c-jun mentioned above. A secondary antibody labeled with fluorescein and specific for the goat constant region was included in the reaction mixture during the DNA replication step. The detection label for PLA was Cy5 rather than fluorescein used in the protocol above.
Imaging and image analysis
An inverted wide-field microscope (Axiovert 200M, Zeiss, Oberkochen, Germany) equipped with a × 100 oil immersion objective with a 1.3 numerical aperture, and a CoolSNAP HQ camera (Photometrics, Tucson, AZ) cooled to −30 °C was used to acquire images. We acquired 20–30 z-sections, separated from each other by 0.2 μm (0.4 μm when PLA and IF were performed) in each channel. The stacks of images were analyzed using custom image analysis programs written in a MATLAB environment, as described previously (Raj et al, 2008). Since PLA spots are larger and more intense than mRNA spots, the size limits used for the detection of mRNA spots were relaxed for the detection of PLA spots. Cell and nuclear boundaries were specified by manually drawing regions of interest over diffraction contrast or DAPI images acquired from a central plane.
To determine the integrated fluorescence intensity in the IF image stacks, we drew regions of interest around the nuclei and in regions that were devoid of cells using the DIC image as a guide. The sum of total fluorescence intensities from the nuclear areas over all the layers was determined for each cell and then divided by the area of region of interest to yield a ‘fluorescence density’. The same was done for the regions of interest from the cell-free blank areas. The average fluorescence density of the blank regions was subtracted from the fluorescence density of each nucleus. The same regions of interest were used to calculate the fluorescence density for IF and number of spots for PLA.
Statistical analysis
The error bars represent 95% confidence intervals (CIs) obtained from measurements made on 50–100 cells. For the data shown in Figure 7 and Supplementary Figure S2 for c-fos RNA, c-jun RNA, and their respective proteins, we obtained 95% CI in standard deviations and means by bootstrapping of data from a single experiment, whereas for heterodimers, collagenase 1, and cox 2 mRNAs these parameters were obtained from three independent experiments. The reported error bars for Fano factors in Figure 7 represent two times the 95% CI in standard deviation, plus 95% CI in mean. The reported error bars for coefficient of variations in Supplementary Figure S2 represent the 95% CI in standard deviations plus 95% CI in means.
Supplementary Material
Supplementary Figures S1-S4
Quantitative data related to figures 2, 5, 6, 7, S2 and S3
Acknowledgments
This research was funded by grants from the National Institute of Mental Health (MH079197), the National Institute of Allergy and Infectious Diseases (AI106036) and royalties received from licensing of Molecular Beacons Technology.
Author contributions: KS and ST designed and performed the research and analyzed the results. KS and ST wrote and edited the manuscript. ST supervised the study.
Footnotes
The authors declare that they have no conflict of interest.
References
- Baan B, Pardali E, ten Dijke P, van Dam H (2010) In situ proximity ligation detection of c-Jun/AP-1 dimers reveals increased levels of c-Jun/Fra1 complexes in aggressive breast cancer cell lines in vitro and in vivo. Mol Cell Proteomics 9: 1982–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakiri L, Matsuo K, Wisniewska M, Wagner EF, Yaniv M (2002) Promoter specificity and biological activity of tethered AP-1 dimers. Mol Cell Biol 22: 4952–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazsi G, van Oudenaarden A, Collins JJ (2011) Cellular decision making and biological noise: from microbes to mammals. Cell 144: 910–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batish M, van den Bogaard P, Kramer FR, Tyagi S (2012) Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci USA 109: 4645–4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake WJ, KAErn M, Cantor CR, Collins JJ (2003) Noise in eukaryotic gene expression. Nature 422: 633–637 [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Kornberg RD (2008) Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell 133: 716–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Levine M (2009) Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325: 471–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossis G, Ferrara P, Acquaviva C, Jariel-Encontre I, Piechaczyk M (2003) c-Fos proto-oncoprotein is degraded by the proteasome independently of its own ubiquitinylation in vivo. Mol Cell Biol 23: 7425–7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R (2012) Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150: 1209–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LC, Chen BK, Chang WC (2005) Activating protein 1-mediated cyclooxygenase-2 expression is independent of N-terminal phosphorylation of c-Jun. Mol Pharmacol 67: 2057–2069 [DOI] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH (2006) Transcriptional pulsing of a developmental gene. Curr Biol 16: 1018–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davis RJ (1994) JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76: 1025–1037 [DOI] [PubMed] [Google Scholar]
- Eldar A, Elowitz MB (2010) Functional roles for noise in genetic circuits. Nature 467: 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, Copeland RA, Magolda RL, Scherle PA, Trzaskos JM (1998) Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem 273: 18623–18632 [DOI] [PubMed] [Google Scholar]
- Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, Hicks J, Bryant GO, Ptashne M (2010) A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141: 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi SJ, Zenklusen D, Lionnet T, Singer RH (2011) Transcription of functionally related constitutive genes is not coordinated. Nat Struct Mol Biol 18: 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebenstreit D (2013) Are gene loops the cause of transcriptional noise? Trends Genet 29: 333–338 [DOI] [PubMed] [Google Scholar]
- Hu E, Mueller E, Oliviero S, Papaioannou VE, Johnson R, Spiegelman BM (1994) Targeted disruption of the c-fos gene demonstrates c-fos-dependent and -independent pathways for gene expression stimulated by growth factors or oncogenes. EMBO J 13: 3094–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J Jr, Boguski MS, Lashkari D, Shalon D, Botstein D, Brown PO (1999) The transcriptional program in the response of human fibroblasts to serum. Science 283: 83–87 [DOI] [PubMed] [Google Scholar]
- Konkoli Z (2010) Exact equilibrium-state solution of an intracellular complex formation model: kA←P reaction in a small volume. Phys Rev E Stat Nonlin Soft Matter Phys 82: 041922. [DOI] [PubMed] [Google Scholar]
- Kovary K, Bravo R (1991) Expression of different Jun and Fos proteins during the G0-to-G1 transition in mouse fibroblasts: in vitro and in vivo associations. Mol Cell Biol 11: 2451–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C (2007) A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Lodish H, Berk A, Lawrence Zipursky S, Matsudaira P, Baltimore D, Darnell J (2000) Molecular Cell Biology 4th edn. W.H. Freeman: New York, [Google Scholar]
- Losick R, Desplan C (2008) Stochasticity and cell fate. Science 320: 65–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macville M, Schrock E, Padilla-Nash H, Keck C, Ghadimi BM, Zimonjic D, Popescu N, Ried T (1999) Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res 59: 141–150 [PubMed] [Google Scholar]
- Martens JH, Verlaan M, Kalkhoven E, Zantema A (2003) Cascade of distinct histone modifications during collagenase gene activation. Mol Cell Biol 23: 1808–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsky B, Neuert G, van Oudenaarden A (2012) Using gene expression noise to understand gene regulation. Science 336: 183–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto T, Cannon D, Gierlinski M, Corrigan A, Barton GJ, Chubb JR (2012) Live imaging of nascent RNA dynamics reveals distinct types of transcriptional pulse regulation. Proc Natl Acad Sci USA 109: 7350–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A (2002) Regulation of noise in the expression of a single gene. Nat Genet 31: 69–73 [DOI] [PubMed] [Google Scholar]
- Pedraza JM, van Oudenaarden A (2005) Noise propagation in gene networks. Science 307: 1965–1969 [DOI] [PubMed] [Google Scholar]
- Rahmsdorf HJ, Schonthal A, Angel P, Litfin M, Ruther U, Herrlich P (1987) Posttranscriptional regulation of c-fos mRNA expression. Nucleic Acids Res 15: 1643–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S (2006) Stochastic mRNA synthesis in mammalian cells. PLoS Biol 4: e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Rifkin SA, Andersen E, van Oudenaarden A (2010) Variability in gene expression underlies incomplete penetrance. Nature 463: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S (2008) Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5: 877–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A (2008) Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135: 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST (2009) A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138: 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK (2004) Control of stochasticity in eukaryotic gene expression. Science 304: 1811–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P, Sisson JC, Verma IM (1988) Transcriptional autoregulation of the proto-oncogene fos. Nature 334: 314–319 [DOI] [PubMed] [Google Scholar]
- Schonthal A, Herrlich P, Rahmsdorf HJ, Ponta H (1988) Requirement for fos gene expression in the transcriptional activation of collagenase by other oncogenes and phorbol esters. Cell 54: 325–334 [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, Trombetta JT, Gennert D, Gnirke A, Goren A, Hacohen N, Levin JZ, Park H, Regev A (2013) Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498: 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, Wong KK, Brandstetter K, Wittner B, Ramaswamy S, Classon M, Settleman J (2010) A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3: 995–1000 [DOI] [PubMed] [Google Scholar]
- Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F (2011) Mammalian genes are transcribed with widely different bursting kinetics. Science 332: 472–474 [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS (2010) Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329: 533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas DY, Shah K, Sinha S, Marras SAE, Schedl P, Tyagi S (2011) Single-Molecule Imaging of Tanrscriptionally Coupled and Uncoupled Splicing. Cell 147: 1054–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska MB, Ameyar-Zazoua M, Bakiri L, Kaminska B, Yaniv M, Weitzman JB (2007) Dimer composition and promoter context contribute to functional cooperation between AP-1 and NFAT. J Mol Biol 371: 569–576 [DOI] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH (2008) Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-S4
Quantitative data related to figures 2, 5, 6, 7, S2 and S3