Abstract
Objective
DNA methylation has long been implicated in developmental β-globin gene regulation. However, the mechanism underlying this regulation is unclear, especially since these genes do not contain CpG islands. This has led us to propose and test the hypothesis that, just as for histone modifications, developmentally-specific changes in human β-like globin gene expression are associated with long-range changes in DNA methylation.
Methods
Bisulfite sequencing was used to determine the methylation state of individual CpG dinucleotides across the β-globin locus in uncultured primary human erythroblasts from fetal liver and bone marrow, and in primitive-like erythroid cells derived from human embryonic stem cells.
Results
β-globin locus CpGs are generally highly methylated but domains of DNA hypomethylation spanning thousands of base pairs are established around the most highly expressed genes during each developmental stage. These large domains of DNA hypomethylation are found within domains of histone modifications associated with gene expression. We also find hypomethylation of a small proportion of γ-globin promoters in adult erythroid cells, suggesting a mechanism by which adult erythroid cells produce fetal hemoglobin.
Conclusion
This is one of the first reports to show that changes in DNA methylation patterns across large domains around non-CpG island genes correspond with changes in developmentally-regulated histone modifications and gene expression. This data supports a new model in which extended domains of DNA hypomethylation and active histone marks are coordinately established to achieve developmentally-specific gene expression of non-CpG island genes.
Keywords: Methylation, Beta globin, Gene Expression, Red Blood Cells, Chromatin
Introduction
The human β-globin gene locus is an important, clinically-relevant model of tissue-specific, developmentally-regulated gene expression. The five β-like genes are positioned within the locus in the order of their expression (Figure 1) and are activated and repressed at specific time points during development in a process referred to as β-globin “switching”. ε-globin is the predominant gene expressed by primitive erythroid cells generated in the yolk sac. As the site of hematopoiesis moves to fetal liver (FL) where definitive erythrocytes are initially generated, expression switches to the γG- and γA-globin genes, with β-globin expressed at lower but clearly discernible levels. Near the time of birth, the site of hematopoiesis moves to the bone marrow (BM) where β-globin expression predominates with much lower levels of δ-globin and sporadic γ-globin expression (Reviewed in [1]). Understanding the regulation of these genes is a high priority since reactivation of fetal globin gene expression has the potential to produce significant clinical benefits for people with P-thalassemia and sickle cell disease (Reviewed in [2]).
Figure 1. Domains of hypomethylation occur at the highly expressed globin genes.
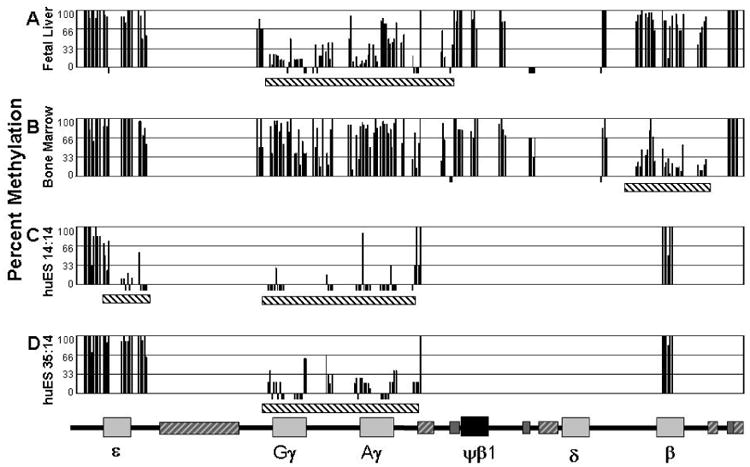
The percentage of methylation (Y axis) at individual CpG's across the human globin locus. Each CpG analyzed is represented by a black bar. CpGs analyzed that are 0% methylated are represented with a bar below 0%. The map of the human globin locus is shown at the bottom is to scale and provides the location of the analyzed CpGs (other than the slightly expanded areas around the γ genes due to the high numbers of CpGs). Striped bars in the map are repetitive elements, dark gray striped bars have been analyzed; light grey striped bars have not been analyzed. A) FL cells sorted for glycophorin A expressing primarily γ-globin. B) BM cells sorted for glycophorin A expressing primarily β-globin. C) huES14:14 cells expressing ε- and γ-globin. D) huES35:14 cells expressing primarily γ-globin. Black and white striped bars under the data sections indicate hypomethylation domains.
DNA methylation is among the mechanisms thought to be critical for appropriate β-globin locus regulation [3, 4]. Evidence supporting a role for methylation in this process dates to studies using methylation-sensitive restriction endonucleases that identified correlations between changes in methylation of promoter CpGs and developmental gene expression [5-7]. These results led to clinical trials demonstrating the ability of the DNA methyltransferase inhibitors 5-azacytidine (5-Aza) and decitabine to induce hypomethylation of a specific γ-globin promoter CpG and to increase γ-globin gene expression [8-11]. More recently, research has shown that developmental changes in γ-globin promoter methylation are transient and inducible by 5-Aza stimulation in adult erythroid cells [12, 13], and that pharmacological stimulation of erythroblasts which results in increased active histone marks, correlates with increased γ-globin expression [3, 14]. While these experiments strongly support a role for DNA methylation in β-like globin gene regulation, the precise mechanisms linking DNA hypomethylation and globin gene expression is still not clear (Reviewed in [4]).
Studies of DNA methylation and gene expression have primarily focused on genes with CpG islands, 200-500bp regions of high CpG content found in the promoters of 72% of human genes, including many ubiquitously expressed housekeeping genes [15-18]. Methylation of CpG islands is associated with gene silencing [19-21], a situation that has been extensively studied in cancer cells [22-26]. The role DNA methylation plays in the mechanism of CpG island gene silencing is well characterized [4, 27], while the role CpG methylation plays in the regulation of non-CpG island genes, like those of the β-globin locus is less clear. In mammals roughly 80% of CpG dinucleotides are methylated [19, 21, 28]. CpGs in non-CpG island promoters are usually methylated, and transcription generally occurs at these genes whether methylation is present or absent [18, 29, 30]. As discussed above, methylation plays a role in γ-globin regulation, but these promoters do not contain CpG islands, which suggests that a more complex mechanism is used to regulate globin gene expression.
Studies of histone modification across the β-globin locus in human primary cells have demonstrated extended domains of transcription-associated histone modifications [31] (Hsu et al, accompanying manuscript). Recently, we have demonstrated that large domains of DNA hypomethylation are present in the mouse β-globin locus [32], and that developmental changes in γ- and β-globin gene methylation extend beyond the promoter cores [12]. These factors led us to consider whether DNA methylation may influence β-globin locus gene expression through a combinatorial mechanism involving domains of DNA hypomethylation and domains of histone modifications. To determine whether extended domains of DNA hypomethylation occur in the human β-globin locus, we have evaluated DNA methylation in uncultured primary human FL and BM erythroid cells and in cells expressing embryonic globin genes derived from human embryonic stem (ES) cells. We have found that developmentally-specific domains of DNA hypomethylation are present. This led us to propose a new model that integrates long-range changes in DNA methylation and histone modification to explain developmental globin gene regulation.
Methods
All human samples were obtained using Institutional Review Board approved protocols. All donors gave informed consent. Adult bone marrow samples were obtained from normal donors at Dartmouth-Hitchcock Medical Center. FL samples were provided by the Birth Defects Research Laboratory of the University of Washington Medical School. Nucleated erythroid cells were purified from FL and BM by ficol gradient separation followed by separation with anti-glycophorin A antibody conjugated to magnetic beads (Miltenyi Biotech, Auburn CA) as previously described [12]. Five BM samples and seven FL samples were used to assay the locus. Human ES cells were co-cultured with FH-B-hTERT feeder cells, a fetal hepatocyte cell line, for 14 days followed by 14 days of liquid culture (huES14:14), or for 35 days in co-culture with FH-B-hTERT cells followed by 14 days in liquid culture (huES35:14) as described [33]. Hematopoietic progenitors produced in these co-cultures are then enriched by selection for CD34 expression and placed in a multi-step serum free liquid culture system for 14 days where they differentiate into mature erythroid cells. β-globin locus chromatin structure was evaluated in these cells without further purification. Data from Figure 3 is combined with data from other studies that used the same methods to purify the erythroblasts.
Figure 3. Domains of DNA hypomethylation are found within extended domains of histone acetylation and marks genes with extremely high transcription levels.
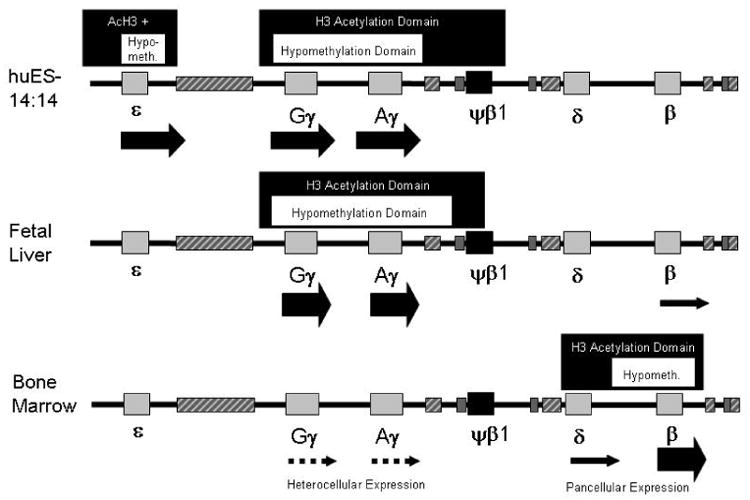
DNA hypomethylation domains are indicated by dark grey boxes above the maps. Domains of active histone acetylation (AcH3) are represented by pale grey boxes above the maps. Transcription level is approximated by the size of the arrows below the genes in the maps, solid arrows indicate pancellular expression, intermittent arrows represent heterocellular expression. Boxes in the maps are as in Figure 1.
Bisulfite conversion of DNA isolated from erythroid cells was performed as previously described [12], using the CpGenometm Fast DNA Modification Kit (Chemicon International, Temecula, CA, USA). Polymerase chain reaction (PCR) of each amplicon used 33ng converted DNA, nested or semi-nested PCR primers and Platinum Taq (Invitrogen, Carlsbad, CA, USA). The web-based program NetPrimer (Premier Biosoft International, Palo Alto, CA, USA) was used to design primers. PCR conditions for primer pairs were individually optimized. The complete primer list is presented in Supplemental Table 1. Purification of PCR amplified DNA was performed with Gel Extraction Kit (Qiagen, Valencia, CA, USA). PCR product was ligated into pGEM®-T Easy Vector System I (Promega, Madison, WI, USA). Inserts were sequenced by the Dartmouth Molecular Biology Core.
Results
To characterize the developmentally-specific changes in long-range DNA methylation of the β-globin locus the state of CpG methylation for 31 PCR amplicons which span the locus was assessed in primary erythroblasts from BM, FL, and human ES cell derived primitive-like erythroid cells. The total region examined spanned 50kb, from 2kb 5′ of the ε-globin gene to 3.3kb 3′ of the β-globin gene coding region. Excluding repetitive elements larger than 700bp, this region contains 252 CpGs. Of these, a total of 158 CpGs (63%) were evaluated in FL and BM cells and 77 CpGs (31%) were evaluated in ES-derived erythroid cells. For each PCR amplicon a median of 8 individual sequences were analyzed (with a range of 3-72 sequences for a given amplicon). FL and BM analysis included analysis of multiple tissue samples for amplicons which were differentially methylated between developmental stages. Data in Figure 1 represents the methylation percentage of each CpG which was analyzed in each tissue. Levels of methylation were divided into three categories for our discussion: hypermethylated (66-100% methylation of a specific CpG), moderately methylated (33-65%) and hypomethylated (0-32%).
β-globin locus DNA methylation in primary definitive erythroid cells
We first evaluated β-globin locus methylation in FL and BM erythroid cells. Analysis of FL reveals an approximately 9kb domain of mixed moderate methylation and hypomethylation that encompasses both γ-globin genes and their flanking regions (Figure 1A). The γG-globin gene is hypomethylated at sites in the promoter region, within the gene and 3′ of the transcribed region. The γA-globin gene is hypomethylated at the promoter and into the intergenic region, while the more 3′ region of the gene shows levels of methylation higher than the corresponding γG-globin regions, but not as high as seen in BM erythroid cells (Figure 1B). This developmentally-specific area of relative hypomethylation extends to the region upstream of a repetitive element 5′ of the Ψβ-globin gene. A region 5′ of each gamma gene promoter is hypermethylated. Outside of the domain surrounding the γ-globin genes, the remainder of the locus, including the β-globin gene, was hypermethylated.
β-globin locus DNA derived from adult BM cells has a very different DNA methylation pattern (Figure 1B). The region around the highly expressed β-globin gene is hypomethylated over a region of approximately 5.5kb in these cells. However, within the hypomethylation domain, and upstream to the β-globin promoter a group of three adjacent CpGs are hypermethylated. The region of the γ-globin genes is generally highly methylated in adult BM cells, although a small number of CpGs remain hypomethylated. In both FL and BM cells the region of the s-globin gene is hypermethylated, consistent with its lack of expression in these cells.
DNA methylation in primitive-like erythroid cells cultured from human ES cells
Based on our identification of domains of differentiation-specific DNA methylation patterns in FL and BM erythroid cells, we next wanted to determine whether characteristic changes in DNA methylation were also present in primitive human erythroid cells. Human ES cells differentiated along the hematopoietic lineage for either 14 days (huES14:14) or 35 days (huES35:14) reflect progression in erythroid development at the early and mid-gestation stages, respectively. The huES14:14 cells express s-globin (40%), γ-globin (60%) and little or no β-globin mRNA, while huES35:14 cells express predominantly γG- and γA-globin mRNA (98%) with low s-globin (2%) and negligible β-globin [33, 34].
In huES14:14 cells, domains of hypomethylation include the s-, γG- and γA-globin genes and surrounding DNA (Figure 1C). The promoter region of the ε gene is moderately methylated, while the gene body and area 3′ to the coding region are hypomethylated. Because we are unable to evaluate methylation of the 13.6kb LINE element between the ε and γG-globin genes, we cannot determine whether this represents a single large hypomethylated domain or two separate domains. The hypomethylated region does not extend 5′ of ε-globin into the region between the locus control region (LCR) and epsilon, indicating that hypomethylation is not “spreading” from the LCR. The promoter of the β-globin gene is hypermethylated in these cells.
In huES35:14 cells, where the ε-globin expression is silenced the region around the gene has become hypermethylated (Figure 1D). The hypomethylation region around the γ-globin genes is maintained. This roughly 9kb region is similar in all γ-globin expressing cells.
γ-Globin promoter DNA methylation patterns are heterogeneous in bone marrow cells
Transcription of the γ-globin genes in BM erythroblasts is heterocellular with most mature cells containing no fetal hemoglobin (HbF) and a few percent of cells, known as F-cells [35], that contain relatively high levels of HbF (14-28% of total cellular hemoglobin) [36, 37]. One possible explanation for this observation is that DNA methylation of the γ-globin promoters is also heterogeneous. By combining data from this study and a previous paper [12], we have analyzed 96 bisulfite converted γ-globin promoter sequences from the BM of five different normal adult donors. Analysis of clones from γG- and γA-globin promoter regions reveals that 5% of alleles are hypomethylated (corresponding to methylation of 0, 1 or 2 of the 7 CpGs between -250 and +50 of the γ-globin promoters) (Figure 2). This degree of hypomethylation is similar to the methylation pattern of the active γ-globin promoters in primary FL cells demonstrated here and reported previously [12]. The remaining 95% of promoter sequences were moderate to hypermethylated.
Figure 2. Heterogeneous methylation of the γ-globin promoters potentially identifies F-cells in bone marrow.
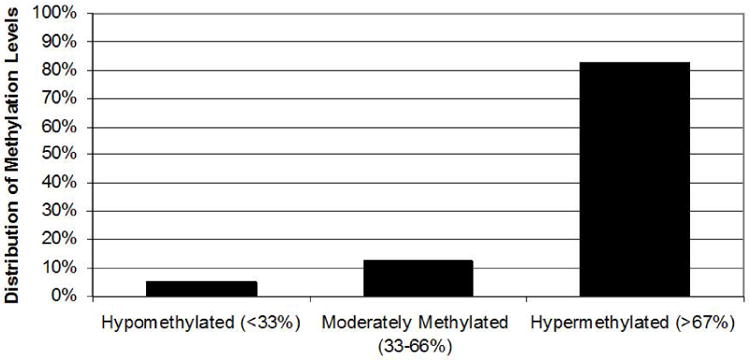
Data is the percentage of γ-globin promoters in bone marrow that are hypomethylated (<33% of promoter CpGs methylated), moderately methylated (33-66% methylated) or hypermethylated (>66% methylated).
Other features of globin locus methylation
There are several repetitive elements in the β-globin locus including the large LINE element upstream of the γ-globin genes. Because of the high copy number of these elements throughout the human genome, it was not possible to determine the methylation status of this element. However, three smaller repetitive elements were included in the assay of FL and BM erythroblasts (Figures 1A and B). The CpGs within the repetitive elements 5′ of Ψβ-globin and 3′ of β-globin are consistently hypermethylated, while CpGs in non-repetitive regions close to these repeats are differentially methylated (hypomethylated 5′ of Ψβ, hypermethylated 3′ of β). All of the CpGs of the small repetitive fragment 5′ of δ-globin were completely unmethylated in FL and hypermethylated in BM. This element is adjacent to the intragenic promoter identified by Gribnau et al [38]. We also found that there are domains of relatively closely spaced CpGs that are constitutively hypermethylated (Figure 1). These include the CpGs upstream of the s-globin gene and downstream of the β-globin gene. Each globin gene has a region ~1kb 5′ of the promoter that is moderate to hypermethylated at all developmental stages, irrespective of the transcription level of the genes. Additional features include several sites that are consistently hypermethylated or hypomethylated regardless of developmental stage and other CpGs that standout as being in the opposite methylation state compared to several near-by CpGs.
Discussion
This study has determined the differentiation-specific long-range methylation patterns of the human β-globin gene locus in primary cells. This work was prompted by recognizing that although the genes do not fit the common model of DNA methylation dependant regulation since they lack CpG island promoters, there is evidence implicating DNA methylation as an important regulatory mechanism for the locus (reviewed in [4]).
The locus has large domains of DNA hypomethylation that change in association with changes in gene expression and changes in coincident histone modifications. While very few examples of extended domains of DNA hypomethylation in any situation have been reported, the IL-4 gene, also a non-CpG island gene, exhibits a differentiation-specific domain similar to the β-globin locus that develops as T cells go from low to high levels of IL-4 expression [29, 39]. The formation of extended domains of DNA hypomethylation may be an important component of gene regulation for these and other highly-expressed, tissue specific genes lacking CpG islands. Possible mechanistic functions of the large hypomethylated domain include permitting the binding of regulatory factors or facilitating chromatin looping for long range chromatin interactions [40].
We have characterized the histone modifications of the β-globin locus in primary erythroid cells (accompanying paper). Combining the data from these two studies reveals the presence of complex chromatin domains that encompass the slightly smaller hypomethylation domains (Figure 3). These data (summarized in Table I) suggest a model of how composite chromatin patterns of DNA methylation and histone modifications correlate with specific changes in gene expression. Highly expressed genes (such as γ-globin in FL or β-globin in BM cells) are surrounded by domains of combined DNA hypomethylation and active histone marks and an absence of any negative histone marks. In contrast, completely silent genes (ε-globin in BM cells and β-globin in primitive cells) are characterized by extended domains of DNA hypermethylation and silent histone marks, with a lack of any active marks. Between these two extremes of expression is the lower-level pancellular expression of β-globin in FL cells [41, 42] and low-level heterocellular expression of γ-globin in BM cells [35, 37]. The pancellular expression of β-globin in FL correlates with a mix of active and silent histone marks at the promoter while the surrounding DNA is hypermethylated. The heterocellular expression of γ-globin in BM erythroid cells exhibits either a silent or an active pattern of DNA methylation marks at the promoter and the absence of active histone marks. A small proportion of γ-globin promoters are hypomethylated, similar to those in FL and it is possible that these hypomethylated alleles are the γ expressing alleles in F cells. Overall, high-level expression is associated with extended domains of DNA hypomethylation and active histone marks and silent genes have the opposite, while low levels of expression are associated with a combination of active and silencing marks which are not found in extended domains. These data support but do not prove a functional role for these chromatin patterns in globin gene regulation.
Table 1.
Composite chromatin patterns across a large region correlate with specific changes in gene expression.
| Expression Level gene (cells) * | Active Histone Marks | Silent Histone Marks | DNA Methylation |
|---|---|---|---|
| High | + | − | Hypomethylated |
| ε-globin (huES14:14) γ-globin (FL) β-globin (BM) | Extended Domain | Extended Domain | |
| Low-Heterocellular | − | + | Heterogeneous |
| γ-globin (BM) | Localized to Promoter/gene | ||
| Low-Pancellular | + | + | Hypermethylated |
| β-globin (FL) | Localized to Promoter/gene | Localized to Promoter/gene | |
| Silent | − | + | Hypermethylated |
| ε-globin (BM) β-globin (huES14:14) | Extended Domain |
Examples of each expression level category are indicated below as ‘expressed gene (cell type)’
: presence of the modification, the extent of the mark is described
: absence of the modification
This study focuses on the domains of hypomethylation, but also observes variation of methylation within individual amplicons. Site-specific CpG methylation has been shown to affect transcription factor binding [43, 44] and increase the presence of methyl-C binding proteins [19, 45-47], leading to gene silencing. An interesting pattern observed in this extended methylation analysis is that within each extended domain of hypomethylation are groups of CpGs roughly 1kb 5′ of the transcriptional start site that are hypermethylated. This is similar to a pattern of many CpG island genes that have hypermethylation in the 5′ region of the promoter [48]. Observing the detailed methylation patterns over greater expanses of DNA may identify individual CpGs which are functionally significant though distant from the promoters.
Our study of β-globin locus methylation also illuminates the previous observation that in fetal erythroid cells there is a 3:1 ratio of γG-globin to γA-globin mRNA [49]. The γG-globin gene is hypomethylated at a greater number of sites compared to γA-globin in fetal liver (Figure 1). Lorincz et al have previously shown that intragenic methylation can inhibit transcription elongation [50], and this mechanism could explain the differential levels of γA-globin compared to γG-globin expression. Experimental induction of site specific DNA hypomethylation at the Aγ gene of human β-YAC transgenic mice led to a 20 fold increase of Aγ transcription in adult mice [51], demonstrating a functional role for demethylation of these CpGs in regulation of γ-globin gene expression levels in this model system.
Our results raise further questions concerning the regulation of the β -like globin genes. How are extended domains of DNA hypomethylation created in a developmentally-regulated fashion? How can individual CpGs that are close to each other have very different methylation states and what is the significance of these differences when they occur outside of known regulatory elements? Although it is now nearly 30 years since methylation of globin genes was first described [5], the exact mechanisms of how this chromatin modification regulates globin gene expression is still poorly understood. Our results suggest that at least part of this complex mechanism involves extensive domains of DNA methylation and histone modification.
Supplementary Material
Four areas (A-D) are expanded to illustrate the PCR fragments amplified for bisulfite sequence analysis, indicated by a black bar below the map, see supplemental table one for the complete list of primers and locations. The number of CpGs assayed in each amplicon is indicated below each bar. Exons are represented by light grey boxes, introns by dark grey boxes. Untranscribed regions are represented in black, white boxes indicate repetitive elements.
Acknowledgments
We wish to acknowledge the University of Washington Birth Defects Laboratory for providing fetal liver samples. Primary funding was provided by NIH grants HL73442 (CL) and HL73431 (SF). Additional funding was provided by NIH grants T32 AI07363, GM075037 (EEB) and HL088467 (EEB) and by the Knights of the York Cross of Honour (CL). No financial interest/relationships with financial interest relating to the topic of this article have been declared.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Experimental Hematology. 2005;33:259–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fathallah H, Atweh GF. Induction of fetal hemoglobin in the treatment of sickle cell disease. Hematology / the Education Program of the American Society of Hematology American Society of Hematology. 2006:58–62. doi: 10.1182/asheducation-2006.1.58. [DOI] [PubMed] [Google Scholar]
- 3.Fathallah H, Weinberg RS, Galperin Y, Sutton M, Atweh GF. Role of epigenetic modifications in normal globin gene regulation and butyrate-mediated induction of fetal hemoglobin. Blood. 2007;110:3391–3397. doi: 10.1182/blood-2007-02-076091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginder GD, Gnanapragasam MN, Mian OY. The role of the epigenetic signal, DNA methylation, in gene regulation during erythroid development. Current topics in developmental biology. 2008;82:85–116. doi: 10.1016/S0070-2153(07)00004-X. [DOI] [PubMed] [Google Scholar]
- 5.van der Ploeg LH, Flavell RA. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980;19:947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]
- 6.Ley TJ, Anagnou NP, Noguchi CT, et al. DNA methylation and globin gene expression in patients treated with 5-azacytidine. Progress in clinical and biological research. 1983;134:457–474. [PubMed] [Google Scholar]
- 7.Mavilio F, Giampaolo A, Care A, et al. Molecular mechanisms of human hemoglobin switching: selective undermethylation and expression of globin genes in embryonic, fetal, and adult erythroblasts. Proc Natl Acad Sci USA. 1983;80:6907–6911. doi: 10.1073/pnas.80.22.6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dover GJ, Charache SH, Boyer SH, Talbot CC, Jr, Smith KD. 5-Azacytidine increases fetal hemoglobin production in a patient with sickle cell disease. Progress in clinical and biological research. 1983;134:475–488. [PubMed] [Google Scholar]
- 9.Ley T, DeSimone J, Anagnou N, et al. 5-azacytidine selectively increases gamma-globin synthesis in a patiaent with ß+thalassemia. New England Journal of Medicine. 1982;301:1469. doi: 10.1056/NEJM198212093072401. [DOI] [PubMed] [Google Scholar]
- 10.Lowrey C, Nienhius A. Brief report: treatment with azacytidine of patients with end-stage thalassemia. N Engl J Med. 1993;329:845–848. doi: 10.1056/NEJM199309163291205. [DOI] [PubMed] [Google Scholar]
- 11.Saunthararajah Y, Hillery CA, Lavelle D, et al. Effects of 5-aza-2′-deoxycytidine on fetal hemoglobin levels, red cell adhesion, and hematopoietic differentiation in patients with sickle cell disease. Blood. 2003;102:3865–3870. doi: 10.1182/blood-2003-05-1738. [DOI] [PubMed] [Google Scholar]
- 12.Mabaera R, Richardson CA, Johnson K, Hsu M, Fiering S, Lowrey CH. Developmental- and differentiation-specific patterns of human gamma- and beta-globin promoter DNA methylation. Blood. 2007;110:1343–1352. doi: 10.1182/blood-2007-01-068635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mabaera R, Greene MR, Richardson CA, Conine SJ, Kozul CD, Lowrey CH. Neither DNA hypomethylation nor changes in the kinetics of erythroid differentiation explain 5-azacytidine's ability to induce human fetal hemoglobin. Blood. 2008;111:411–420. doi: 10.1182/blood-2007-06-093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavelle D, Chin J, Vaitkus K, et al. Oral decitabine reactivates expression of the methylated gamma-globin gene in Papio anubis. American Journal of Hematology. 2007;82:981–985. doi: 10.1002/ajh.21020. [DOI] [PubMed] [Google Scholar]
- 15.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature Genetics. 2007;39:457–466. doi: 10.1038/ng1990. see comment. [DOI] [PubMed] [Google Scholar]
- 19.Bird A, Wolffe A. Methylation-induced repression- belts, braces and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 20.Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO Journal. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klose R, Bird A. Genomic DNA methylation:the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 22.Ehrlich M, Jiang G, Fiala E, et al. Hypomethylation and hypermethylation of DNA in Wilms tumors. Oncogene. 2002;21:6694–6702. doi: 10.1038/sj.onc.1205890. [DOI] [PubMed] [Google Scholar]
- 23.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature Reviews Genetics. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 24.Kanai Y, Hirohashi S. Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a precancerous to a malignant state. Carcinogenesis. 2007;28:2434–2442. doi: 10.1093/carcin/bgm206. [DOI] [PubMed] [Google Scholar]
- 25.Sato N, Maitra A, Fukushima N, et al. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Research. 2003;63:4158–4166. [PubMed] [Google Scholar]
- 26.Sato N, Fukushima N, Matsubayashi H, Goggins M. Identification of maspin and S100P as novel hypomethylation targets in pancreatic cancer using global gene expression profiling. Oncogene. 2004;23:1531–1538. doi: 10.1038/sj.onc.1207269. [DOI] [PubMed] [Google Scholar]
- 27.Ballestar E, Paz MF, Valle L, et al. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. The EMBO journal. 2003;22:6335–6345. doi: 10.1093/emboj/cdg604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber M, Schubeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Current Opinion in Cell Biology. 2007;19:273–280. doi: 10.1016/j.ceb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 29.Reiner SL. Epigenetic control in the immune response. Human Molecular Genetics. 2005;14 Spec No 1:R41–46. doi: 10.1093/hmg/ddi115. [DOI] [PubMed] [Google Scholar]
- 30.Shen HM, Nakamura A, Sugimoto J, et al. Tissue specificity of methylation and expression of human genes coding for neuropeptides and their receptors, and of a human endogenous retrovirus K family. Journal of Human Genetics. 2006;51:440–450. doi: 10.1007/s10038-006-0382-9. [DOI] [PubMed] [Google Scholar]
- 31.Yin W, Barkess G, Fang X, et al. Histone acetylation at the human beta-globin locus changes with developmental age. Blood. 2007;110:4101–4107. doi: 10.1182/blood-2007-05-091256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu M, Mabaera R, Lowrey CH, Martin DI, Fiering S. CpG hypomethylation in a large domain encompassing the embryonic {beta}-like globin genes in primitive erythrocytes. Mol Cell Biol. 2007;27:5047–5054. doi: 10.1128/MCB.02234-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu C, Olivier EN, Velho M, Bouhassira EE. Globin switches in yolk sac-like primitive and fetal-like definitive red blood cells produced from human embryonic stem cells. Blood. 2008;111:2400–2408. doi: 10.1182/blood-2007-07-102087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu C, Hanson E, Olivier E, et al. Differentiation of human embryonic stem cells into hematopoietic cells by coculture with human fetal liver cells recapitulates the globin switch that occurs early in development. Experimental Hematology. 2005;33:1450–1458. doi: 10.1016/j.exphem.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 35.Rochette J, Craig JE, Thein SL. Fetal hemoglobin levels in adults. Blood Rev. 1994;8:213–224. doi: 10.1016/0268-960x(94)90109-0. [DOI] [PubMed] [Google Scholar]
- 36.Boyer SH, Belding TK, Margolet L, Noyes AN. Fetal hemoglobin restriction to a few erythrocytes (F cells) in normal human adults. Science (New York, NY. 1975;188:361–363. doi: 10.1126/science.804182. [DOI] [PubMed] [Google Scholar]
- 37.Wood W, Stamatoyannopolous G, Lim G, Nute P. F-cells in the adult: normal values and levels in individuals with hereditary and acquired elevations of HbF. Blood. 1975;46:671. [PubMed] [Google Scholar]
- 38.Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Molecular Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- 39.Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16:649–660. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- 40.Bulger M, Groudine M. Looping vs. linking: toward a model for long-distance gene activation. Genes and Development. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 41.Papayannopoulou T, Shepard TH, Stamatoyannopoulos G. Studies of hemoglobin expression in erythroid cells of early human fetuses using anti-gamma- and anti-beta-globin chain fluorescent antibodies. Progress in clinical and biological research. 1983;134:421–430. [PubMed] [Google Scholar]
- 42.Stamatoyannopoulos G, grosveld G. Chapter 5: Red cells: hemoglobin switching. In: Stamatoyannopoulos G, majerus PW, Perlmutter RM, varmus h, editors. The Molecular Basis of Blood Disease. 3. WB saunders Co; 2001. pp. 135–182. [Google Scholar]
- 43.Jane SM, Gumucio DL, Ney PA, Cunningham JM, Nienhuis AW. Methylation-enhanced binding of Sp1 to the stage selector element of the human gamma-globin gene promoter may regulate development specificity of expression. Mol Cell Biol. 1993;13:3272–3281. doi: 10.1128/mcb.13.6.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sengupta PK, Lavelle D, DeSimone J. Increased binding of Sp1 to the gamma-globin gene promoter upon site-specific cytosine methylation. Am J Hematol. 1994;46:169–172. doi: 10.1002/ajh.2830460302. [DOI] [PubMed] [Google Scholar]
- 45.Rupon JW, Wang SZ, Gaensler K, Lloyd J, Ginder GD. Methyl binding domain protein 2 mediates gamma-globin gene silencing in adult human betaYAC transgenic mice. Proc Natl Acad Sci U S A. 2006;103:6617–6622. doi: 10.1073/pnas.0509322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singal R, Ferris R, Little J, Wang S, Ginder G. Methylation of the minimal promoter of an embryonic globin gene sliences transcription in primary erythroid cells. Proc Natl Acad Sci USA. 1997;94:13724–13729. doi: 10.1073/pnas.94.25.13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singal R, Wang SZ, Sargent T, Zhu SZ, Ginder GD. Methylation of promoter proximal-transcribed sequences of an embryonic globin gene inhibits transcription in primary erythroid cells and promotes formation of a cell type-specific methyl cytosine binding complex. The Journal of biological chemistry. 2002;277:1897–1905. doi: 10.1074/jbc.M105580200. [DOI] [PubMed] [Google Scholar]
- 48.Brinkman AB, Pennings SW, Braliou GG, Rietveld LE, Stunnenberg HG. DNA methylation immediately adjacent to active histone marking does not silence transcription. Nucleic Acids Research. 2007;35:801–811. doi: 10.1093/nar/gkl1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nute PE, Pataryas HA, Stamatoyannopoulos G. The G and A hemoglobin chains during human fetal development. American Journal of Human Genetics. 1973;25:271–276. [PMC free article] [PubMed] [Google Scholar]
- 50.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 51.Goren A, Simchen G, Fibach E, et al. Fine tuning of globin gene expression by DNA methylation. PLoS ONE. 2006;1:e46. doi: 10.1371/journal.pone.0000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Four areas (A-D) are expanded to illustrate the PCR fragments amplified for bisulfite sequence analysis, indicated by a black bar below the map, see supplemental table one for the complete list of primers and locations. The number of CpGs assayed in each amplicon is indicated below each bar. Exons are represented by light grey boxes, introns by dark grey boxes. Untranscribed regions are represented in black, white boxes indicate repetitive elements.


