Abstract
Mammalian TopBP1 is a BRCT domain–containing protein whose function in mitotic cells is linked to replication and DNA damage checkpoint. Here, we study its possible role during meiosis in mice. TopBP1 foci are abundant during early prophase I and localize mainly to histone γ-H2AX–positive domains, where DNA double–strand breaks (required to initiate recombination) occur. Strikingly, TopBP1 showed a pattern almost identical to that of ATR, a PI3K-like kinase involved in mitotic DNA damage checkpoint. In the synapsis-defective Fkbp6-/- mouse, TopBP1 heavily stains unsynapsed regions of chromosomes. We also tested whether Schizosaccharomyces pombe Cut5 (the TopBP1 homologue) plays a role in the meiotic recombination checkpoint, like spRad3, the ATR homologue. Indeed, we found that a cut5 mutation suppresses the checkpoint-dependent meiotic delay of a meiotic recombination defective mutant, indicating a direct role of the Cut5 protein in the meiotic checkpoint. Our findings suggest that ATR and TopBP1 monitor meiotic recombination and are required for activation of the meiotic recombination checkpoint.
INTRODUCTION
Meiosis generates haploid gametes from diploid parental cells in a process that involves two consecutive nuclear divisions without an intervening S phase. A process unique to meiosis is the recombination that occurs between homologous chromosomes during prophase I. Meiotic recombination initiates with the generation of DNA double-strand breaks (DSBs) by the Spo11 endonuclease (Keeney et al., 1997), which are repaired via homologous recombination with sequences on a nonsister chromatid. A set of proteins monitor recombination and activate a checkpoint during late prophase I (pachytene) if the recombination is not finished. This checkpoint, known as the “pachytene checkpoint” or “meiotic recombination checkpoint” (Roeder and Bailis, 2000), prevents segregation of homologous chromosomes until recombination is completed and ensures proper distribution of the genetic material to the gametes. Many of the proteins involved in meiotic recombination and in pachytene checkpoint can be visualized, forming foci, associated with the “axial core,” a proteinaceous structure that holds sister chromatids together at early stages of prophase I, and/or to the “synaptonemal complex” (SC), a protein scaffold that maintains homologous chromosomes together along their entire length during the pachytene stage of prophase I.
Key proteins during the recombination process are Rad51, a bacterial RecA homologue that is also involved in homologous strand exchange during recombinational repair of DSBs in somatic cells (Baumann et al., 1996), and the meiotic Rad51-homologue protein, Dmc1, which promotes recombination between nonsister chromatids (Schwacha and Kleckner, 1997). Rad51 together with Dmc1 forms foci associated with the SC (Barlow et al., 1997; Tarsounas et al., 1999). There are many other proteins that play a role in meiotic recombination and are also associated with the SC, for example, RPA, MLH1, and BLM (Moens et al., 2002).
Mammalian ATR protein belongs to a family of protein kinases termed phosphatidyl-inositol 3-kinase-like protein kinases (PIKKs) and plays an important role during the somatic DNA-damage activated checkpoint (Abraham, 2001). ATR is proposed to sense the damage and send an inhibitory signal to the cell cycle progression machinery. During meiosis, ATR forms foci associated with the SC at prophase I (Keegan et al., 1996; Moens et al., 1999). Both yeast ATR homologues mutants (Schizosaccharomyces pombe rad3 and Saccharomyces cerevisiae mec1) show a defect in the pachytene checkpoint when activated by a recombination-defective mutation (Lydall et al., 1996; Shimada et al., 2002), suggesting that ATR might be playing the same role during mammalian meiosis. Together with spRad3 and scMec1, other proteins are required for the pachytene checkpoint in yeast (Lydall et al., 1996; San-Segundo and Roeder, 1999, 2000; Roeder and Bailis, 2000; Shimada et al., 2002; Perez-Hidalgo et al., 2003); many of them are also involved in the mitotic DNA damage checkpoint. In mammals, apart from ATR, other checkpoint proteins, such as Chk1 and Rad1, have been located associated with the SC (Flaggs et al., 1997; Freire et al., 1998).
Human BRCT domain–containing protein TopBP1 was first identified through its association with topoisomerase IIβ (Yamane et al., 1997), and its function seems to be linked to replication and DNA damage checkpoint processes (Mäkiniemi et al., 2001; Yamane et al., 2003). Indeed, TopBP1 localizes to the sites of DNA damage in response to DSBs and replication blocks (Honda et al., 2002; Yamane et al., 2002). Both TopBP1 protein homologues in yeast (S. pombe Rad4/Cut5 and S. cerevisiae Dpb11) are required for DNA replication and for the DNA damage and DNA replication checkpoints and act upstream of effector kinases spChk1, spCds1, and scRad53 (Saka and Yanagida, 1993; Araki et al., 1995; McFarlane et al., 1997; Wang and Elledge, 1999; Harris et al., 2003). Recently, studies in Xenopus indicate that the TopBP1 homologue, xCut5, is required for ATR binding to the sites of DNA damage (Parrilla-Castellar and Karnitz, 2003).
In this report, we show that TopBP1 and ATR colocalize in meiotic prophase I chromosomes, and also, that they are associated with DSBs sites. Moreover, in a synapsis-deficient mouse (Fkbp6-/-), TopBP1 heavily stains unsynapsed chromosomes at late prophase I. We also show that a S. pombe cut5 mutant is defective in the meiotic recombination checkpoint. We, therefore, suggest that TopBP1 and ATR are part of the machinery that monitors recombination during prophase I and activates the pachytene checkpoint.
MATERIALS AND METHODS
Reagents and Secondary Antibodies
All reagents were purchased from Sigma (St. Louis, MO) unless otherwise stated. Fluorescein isothiocyanate (FITC) and rhodamine-conjugated secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA), and immunogold-conjugated secondary antibodies were obtained from Cedarlane Laboratories (Hornby, Ontario, Canada).
Primary Antibodies
We used the polyclonal rabbit anti-TopBP1 antibody previously described (Mäkiniemi et al., 2001). ATR antiserum was raised in sheep against a recombinant protein spanning amino acids 2341–2641 of the ATR protein. Mouse anti-Dmc1, anti-Cor1, and anti-Syn1 antibodies have been described previously (Dobson et al., 1994; Tarsounas et al., 1999). Cor1/Scp3 is the main component of the axial core and forms the so called lateral element of the SC (Dobson et al., 1994; Pelttari et al., 2001). Syn1/Scp1 protein is one of the components of the SC (Meuwissen et al., 1992; Dobson et al., 1994). Dmc1 and Rad51 proteins share epitopes and our mouse anti-Dmc1 serum recognizes both antigens unless immunodepleted for Rad51 cross-reacting antibodies. With such immunodepleted antibodies, it was described that Dmc1 and Rad51 colocalize at foci using immunofluorescence and electron microscopy (Tarsounas et al., 1999). We therefore used the mouse anti-Dmc1 to detect the Rad51/Dmc1 foci. Colocalization of Rad51 and Dmc1 was reported earlier in S. cerevisiae (Bishop, 1994). Antibodies against rat testis-specific histone 1 (H1t) were generated in rabbit with Escherichia coli–expressed H1t protein (Moens, 1995). The CREST anticentromere serum was obtained from a patient with CREST syndrome (Dobson et al., 1994). Mouse monoclonal anti-γ-H2AX and rabbit polyclonal anti-γ-H2AX were purchased from Upstate Biotechnology (Lake Placid, NY).
Mouse Testis Immunocytology
The nuclear contents of whole-mount spermatocytes were displayed by surface spreading of a testicular cell suspension on a hypotonic liquid surface. The intact nuclei became attached to plastic-coated glass slides and were fixed briefly in 2% paraformaldehyde with 0.03% SDS. After washing in PhotoFlo 200 (Eastman Kodak, Rochester, NY) and Triton X-100 in PBS, cells were blocked with goat serum (we used donkey serum for sheep anti-ATR antibody) and incubated overnight with primary antibodies at room temperature. TopBP1 and ATR sera were used at 1:300 and 1:500 dilutions, respectively. After washes, slides were incubated with secondary antibodies for1hat37°C. Cells were mounted in ProLong Antifade (Molecular Probes, Eugene, OR) with 4 μg/ml 4′,6-diamidino-2-phenylindole (DAPI), and viewed with a 200 W fluorescence microscope. To avoid seeing bleeding from one fluorescence channel into another, we performed controls that did not include each of the primary antibodies but included the secondary antibodies. Images were recorded on a CCD camera and digitally enhanced with Adobe Photoshop 6.0 (San Jose, CA).
For electron microscopy, cells were treated briefly with DNase I after fixation. Secondary antibodies were conjugated with 5- or 10-nm gold particles. After washes, the plastic was floated off and transferred to nickel grids. The electron microscopy images were recorded at 10,000× or 20,000× magnification on 35-mm film.
Fission Yeast Strains and Genetic Methods
The following S. pombe diploid pat1 strains were used for the meiotic recombination checkpoint studies: S964 (wild type), h-/h- pat1-114/pat1-114 leu1-32/leu1-32 ade6-M210/ade6-M216; S1329 (rad4-116), h-/h- pat1-114/pat1-114 leu1-32/leu1-32 ade6-M210/ade6-M216 rad4-116/rad4-116; S1295 (meu13), h-/h- pat1-114/pat1-114 leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-M210/ade6-M216 meu13::ura4+/meu13::ura4+; S1330 (rad4-116 meu13), h-/h- pat1-114/pat1-114 leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-M210/ade6-M216 rad4-116/rad4-116 meu13::ura4+/meu13::ura4+; and S1331 (rec12), h-/h- pat1-114/pat1-114 ade6-M210/ade6-M216 mek1-HA::kanMX6/mek1-HA::kanMX6 leu1-32/leu1-32 rec12-152::LEU2/rec12-152::LEU2. The following S. pombe haploid strains were used for analysis of meiotic recombination: PN22 (wild type), h- leu1-32; S1285 (wild type), h+ his5–303; S1382 (rad4-116), h- leu1-32 rad4-116; and S1383 (rad4-116), h+ his5–303 rad4-116. The rad4-116 mutation (McFarlane et al., 1997) was introduced by genetic crosses using the strain S1304 (h- ura4-D18 leu1-32 rad4-116) obtained from A. Carr (Genome Damage and Stability Center, United Kingdom). The response to methyl methanesulfonate (MMS) treatment at different temperatures was used to follow the rad4-116 mutation in the segregants from crosses. Media and growth conditions were as described (Moreno et al., 1991). Synchronous meiosis and analyses of meiotic progression and premeiotic DNA replication were carried out as described (Perez-Hidalgo et al., 2003), except that the temperature of 32°C was used to inactivate the Pat1 kinase and induce meiosis. Spore viability was determined by plating 2000 randomly isolated spores from each strain onto YES plates and counting the number of colonies formed after incubation at 25°C for 5 days. Meiotic intergenic recombination was analyzed as described (Perez-Hidalgo et al., 2003), except that crosses were carried out at two different temperatures.
Fission Yeast Cytology and Immunoblot Analysis
S. pombe nuclear spreads and immunofluorescence were performed essentially as described for S. cerevisiae (Sym et al., 1993; San-Segundo and Roeder, 1999), except that fission yeast cells were spheroplasted by treatment with 0.5 mg/ml 20T zymolyase, 2 mg/ml Novozyme, 10 mM DTT in PEMS buffer (100 mM PIPES, 1 mM EGTA, 1 mM MgSO4, 1 M sorbitol) for 30 min at 30°C. Rabbit polyclonal anti-γ-H2AX was used at 1:250 dilution. Affinity-purified rabbit anti-Cut5 antibodies were used at 1:100 dilution. The anti-Cut5 crude serum was kindly provided by M. Yanagida (Kyoto University, Japan). To ensure that the signal was specific of the Cut5 antibody, we performed controls that contained a nonrelated primary antibody but included the same secondary antibody. Goat anti-rabbit antibodies conjugated to CY3 (Jackson ImmunoResearch) were used at 1:200 dilution as secondary antibodies. Immunoblot analysis was performed as described (Perez-Hidalgo et al., 2003). Rabbit polyclonal anti-γ-Cdc2(Tyr15) (Cell Signaling Technology, Beverly, MA) and rabbit polyclonal anti-Cdc2 antibodies were used at 1:1000 and 1:200 dilutions, respectively.
RESULTS
TopBP1 Associates with Chromosome Cores at Early Prophase I
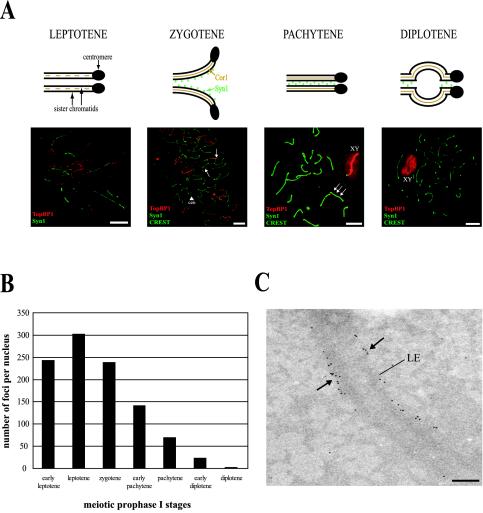
We have found that, during meiosis in mice, the TopBP1 protein associates with the chromosome cores forming foci (Reini et al., 2004). To gain more information about the expression and location of TopBP1 on meiotic chromosomes, we carried out a more detailed study by immunofluorescence, and we counted TopBP1 foci at different stages of meiotic prophase I (Figure 1). The staging of meiotic prophase I nuclei was based on morphology of chromosome cores (detected with mouse anti-Cor1 antibody), SCs (visualized with mouse anti-Syn1 antibody), number of centromeres (detected with human CREST anticentromere serum), and appearance of histone H1t at midpachytene (detected with rabbit anti-H1t; see MATERIALS AND METHODS). Figure 1A shows an schematic view of the different stages of prophase I together with the observed staining with TopBP1 (red) and Syn1 (green). Figure 1B shows the average number of TopBP1 foci per nucleus found at the different stages (at least 3 different nuclei per stage were analyzed). At early leptotene we observed ∼245 bright TopBP1 foci along the chromosome cores (Figure 1, A and B). The highest number of TopBP1 foci (∼300) is reached at late leptotene, when more chromosome cores appear. The number of foci decreases to ∼250 during the synaptic stage (zygotene), when chromosome cores are completely assembled and homologous chromosomes attach to each other, forming the SCs. At this stage, TopBP1 is located mainly on yet unsynapsed chromosome cores (Figure 1A, zygotene). During pachytene, the number of TopBP1 foci declines to ∼150, but they are mainly concentrated on nonsynapsed chromosomes and, also, the X-Y pair becomes strongly stained (Figure 1A, pachytene). At diplotene, when the SC begins to break down and chromosomes move apart, remaining associated only at the cross-over sites (“chiasmata”), TopBP1 foci were no longer detected (Figure 1A, diplotene). As shown in the pachytene and diplotene images, the bright TopBP1 staining on the X-Y pair is not circumscribed to the chromosomal cores, but is also visible on the surrounding area containing the condensed chromatin.
Figure 1.
Localization of TopBP1 protein during meiotic prophase I in mouse testis. (A) The four stages of meiotic prophase I (top). Fluorescence microscopy of mouse meiotic chromosomes stained with rabbit anti-TopBP1 antibody (red), mouse anti-Syn1 and human CREST anticentromere serum (green; bottom). In leptotene, TopBP1 is associated with newly formed chromosome cores. In zygotene, TopBP1 is mainly located to chromosome cores that are not completely synapsed (arrows). The arrowhead indicates two single centromeres (cen). In the fully synapsed pachytene stage there are fewer TopBP1 foci on autosomes but the X-Y pair is brightly stained (XY). The green and red images are slightly offset to show the association of TopBP1 foci with the chromosome cores (arrows). In diplotene, TopBP1 is staining only the X-Y pair (XY). Bars, 20 μm. (B) Average number of TopBP1-containing foci during the different stages of the meiotic prophase I. Thirty-two nuclei in total (at least 3 nuclei per stage) stained with rabbit anti-TopBP1 antibody were analyzed. (C) Electron micrograph of a full pachytene SC with many 10-nm gold grains marking TopBP1 sites (arrows) at the lateral elements. LE, lateral element. Magnification, 20.000×. Bar, 200 nm.
To characterize in more detail the location of TopBP1 in meiotic chromosomes, we performed immunoelectron microscopy using 10-nm gold grain-conjugated secondary antibodies. Figure 1C shows a pachytene SC with many TopBP1 foci located along the lateral elements of the SC.
TopBP1 Does Not Fully Associate with Dmc1 Foci
To assess the possible role of TopBP1 during mammalian meiosis, we investigated its association with other known proteins that participate in this process. For this purpose, we first analyzed the spatial and temporal distribution of TopBP1 relative to the Dmc1 protein. Rad51 and Dmc1 are RecA-type recombinases that form mixed complexes associated with mouse meiotic chromosome cores and SCs (Tarsounas et al., 1999). These Rad51/Dmc1 foci are the sites of single-strand DNA tails that are active in homology search and strand exchange (Bishop, 1994). The distribution of Rad51/Dmc1 foci is similar to the one described here for TopBP1 protein (abundant foci at leptotene and zygotene and less foci in later stages; Moens et al., 2000).
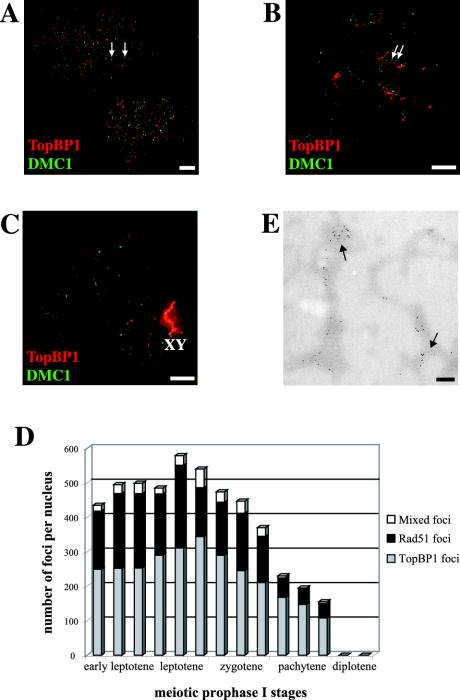
Immunofluorescence observations at successive stages of meiotic prophase I on spermatocyte nuclei that are doubly labeled with rabbit anti-TopBP1 (red) and mouse anti-Dmc1 (green) antibodies show that these two proteins partially colocalize. Figure 2A shows two leptotene nuclei with numerous red and green bright foci, but only some of them (∼10%, see Figure 2D for quantification) colocalize (yellow foci, arrows). Figure 2B shows one zygotene nucleus with many bright foci associated with the chromosome cores. At this stage, there is a higher degree of association (∼20%, Figure 2D) of the two types of foci (arrows). A pachytene nucleus is shown in Figure 2C. There are only a few weak red foci on the autosomes, but the X-Y pair is strongly labeled by the anti-TopBP1 antibody. On the contrary, there is almost no Dmc1 staining in this nucleus.
Figure 2.
Coimmunostaining of TopBP1 and Dmc1 proteins in different stages of mouse meiotic prophase I. A, B, and C are stained with rabbit anti-TopBP1 (red) and mouse anti-Dmc1 (green). (A) Two leptotene nuclei showing numerous bright TopBP1 and Dmc1 foci, ∼10% of them colocalizing (yellow foci, arrows). (B) One zygotene nucleus showing bright TopBP1 staining on yet unsynapsed chromosome cores. The number of Dmc1 foci is lower than in leptotene, but there is a higher degree of colocalization (20%) of the two types of foci (arrows). The red and green images are slightly offset to better show this association. (C) One pachytene nucleus showing very bright TopBP1 staining on the X-Y pair (XY) and a few foci on the autosomes. Dmc1 protein is coming off the chromosome cores at this stage, as shown by the weak green signal. Bars, 20 μm. (D) Numbers of Rad51/Dmc1, TopBP1, and mixed foci per nucleus that are associated with chromosome cores/SCs at successive developmental stages, based on 14 completely analyzed nuclei. A small proportion of the foci contain all three proteins represented by the white segments of the bars. (E) Immunoelectron micrograph of chromosome cores stained with 5- and 10-nm gold particles marking Dmc1 and TopBP1 sites, respectively. Bar, 200 nm.
We also used immunoelectron microscopy for a more detailed study of the possible association of TopBP1 with Dmc1. In this case, we used 5-nm gold grains for Dmc1 and 10 nm for TopBP1. Figure 2E shows chromosome core fragments from an early prophase nucleus with many Dmc1 and TopBP1 foci, some of them colocalizing (arrows). In a somewhat later nucleus, we observed almost no colocalization of these two types of foci, in agreement with what we observed using immunofluorescence microscopy (unpublished data).
TopBP1 and ATR Colocalize at the Chromosome Cores and Are Associated with γ-H2AX Positive Domains
Apart from Dmc1, the number and distribution of TopBP1 foci during mouse spermatogenesis is similar to that of ATR (Keegan et al., 1996; Moens et al., 1999). Therefore, we decided to study whether these two proteins colocalize. For this purpose, we raised a sheep polyclonal antibody against the C-terminus of the human ATR protein (see MATERIALS AND METHODS) and analyzed surface-spread spermatocytes immunostained with this antibody together with our rabbit anti-TopBP1 antibody.
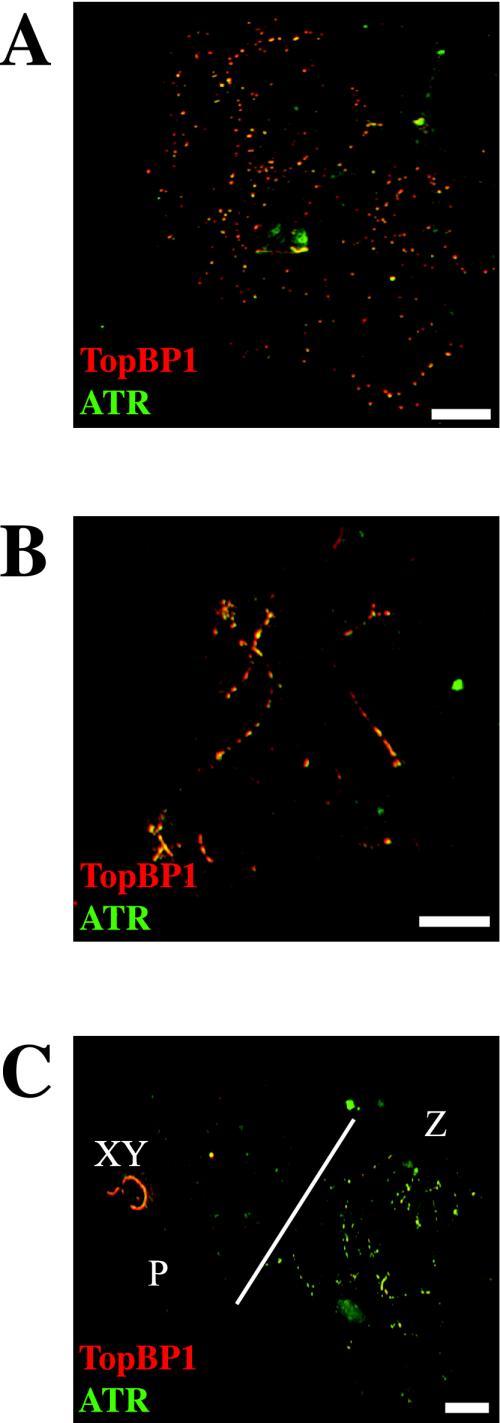
Strikingly, we observed an almost complete colocalization of TopBP1 and ATR using immunofluorescence. Figure 3A shows one leptotene nucleus with numerous bright yellow foci, indicating that many TopBP1 (red) and ATR (green) foci do colocalize at this stage. The number of foci is almost the same for the two proteins, and they are associated with the chromosome cores. In Figure 3B, there is a late zygotene nucleus with intense yellow staining on the chromosomes that have not completed synapsis. Figure 3C shows two nuclei, one in zygotene (right) and the other in pachytene (left). The zygotene nucleus shows the same distribution of foci as the one in Figure 3B, with extensive labeling of yet unsynapsed regions. The pachytene nucleus shows bright ATR and TopBP1 staining only on the X-Y pair, but there are some weak TopBP1 foci also on the autosomes. These findings correlate well with previous data using a different ATR antibody (Moens et al., 1999); ATR foci were not detected on late pachytene nuclei.
Figure 3.
Coimmunostaining of TopBP1 and ATR proteins in different stages of mouse meiotic prophase I. A, B, and C are stained with rabbit anti-TopBP1 (red) and sheep anti-ATR (green) antibodies. (A) One leptotene nucleus showing many yellow bright foci, demonstrating that TopBP1 and ATR colocalize in most, if not all, foci. (B) One zygotene nucleus showing an intense yellow staining of yet unsynapsed regions. (C) Two different nuclei are shown. The one on the right (Z) is a zygotene nucleus showing the same pattern as in B. The one on the left (P) is a pachytene nucleus showing TopBP1 on the X-Y pair (XY) and a few weak foci on the autosomes. In this nucleus, ATR is only present on the X-Y pair. Bars, 20 μm.
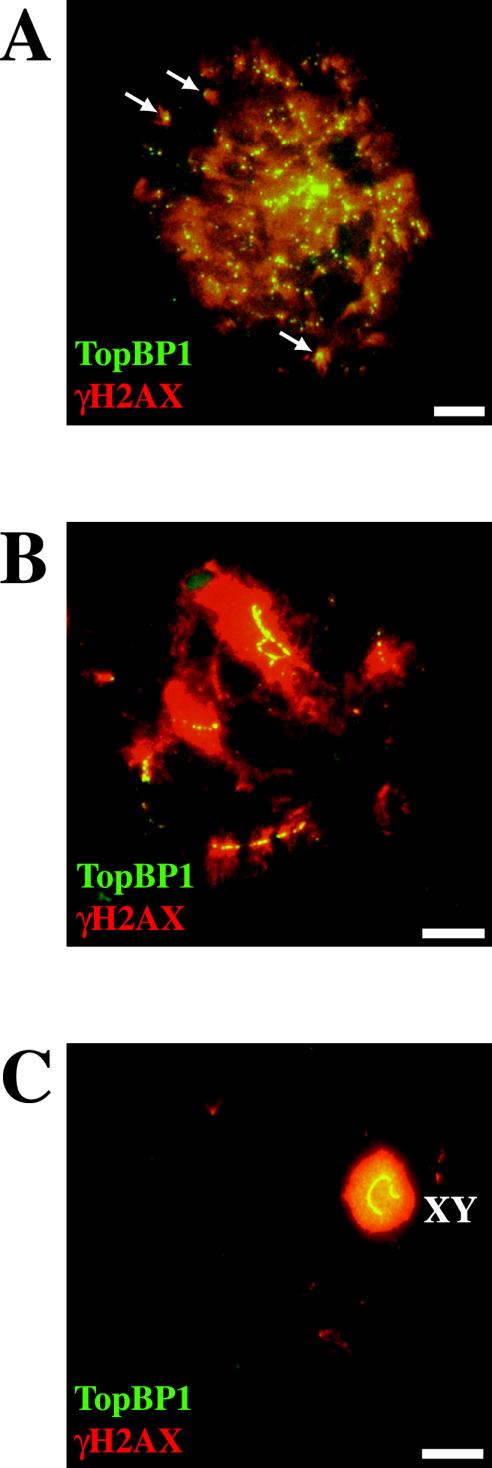
The mitotic checkpoint function of ATR homologous proteins scMec1 and spRad3 has been related to their physical localization to DSBs (Kondo et al., 2001; Melo et al., 2001). Therefore, we decided to test whether ATR-TopBP1 foci localize to DSBs. In response to the generation of DSBs, histone H2AX becomes phosphorylated at serine 139 (Rogakou et al., 1998); H2AX is also phosphorylated during meiotic prophase I (Mahadevaiah et al., 2001). Figure 4 shows the coimmunostaining of mouse surface-spread spermatocytes with anti-TopBP1 and anti-γ-H2AX antibodies. All TopBP1 foci were located within γ-H2AX-positive domains in early stages of meiotic prophase I (Figure 4A), although it is difficult to observe a precise colocalization because of the diffuse staining with the anti–γ-H2AX antibody. Particularly interesting are isolated γ-H2AX domains (Figure 4A, arrows), where a colocalization with TopBP1 is observed. As described earlier (Mahadevaiah et al., 2001), during the synaptic stage γ-H2AX domains were less abundant and were concentrated over the last stretches of axial elements to synapse. At zygotene, all TopBP1 foci (mainly located to yet unsynapsed regions) colocalized with γ-H2AX–positive domains (Figure 4B). At the end of prophase I, TopBP1 and γ-H2AX are concentrated solely over the chromatin of the sex chromosomes, as shown in Figure 4C.
Figure 4.
Coimmunostaining of TopBP1 and γ-H2AX proteins in different stages of mouse meiotic prophase I. A, B, and C are stained with rabbit anti-TopBP1 (green) and mouse anti-γ-H2AX (red). (A) One late leptotene nucleus showing the colocalization of the two proteins (yellow). All TopBP1 foci colocalize with γ-H2AX, but there are many γ-H2AX–positive domains that do not contain TopBP1 foci. Arrows indicate isolated γ-H2AX domains containing TopBP1 foci. (B) One zygotene nucleus showing TopBP1 staining on yet unsynapsed regions that are also positive for γ-H2AX staining. (C) A late pachytene nucleus showing TopBP1 and γ-H2AX uniformly coating the sex chromosomes. Bars, 20 μm.
TopBP1 Staining Is Restricted to Unsynapsed Regions in a Fkbp6-/- Mouse
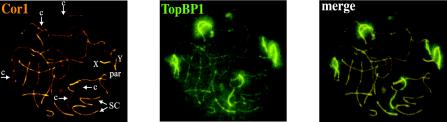
Next, we decided to study the localization of TopBP1 in a genetic background with a recombination defect. Fkbp6 protein (FK506 Binding Protein 6) is a component of the synaptonemal complex. The Fkbp6-/- mouse was recently reported to show several alterations during homologous chromosome pairing in meiosis (Crackower et al., 2003). During early meiotic prophase I, the Fkbp6-/- mouse carries out normal sister chromatid pairing, and normal Rad51/Dmc1 staining was observed, indicating an initiation of recombination. At later stages (late zygotene-pachytene), homologous chromosome pairing is deficient and abnormal pairing, nonhomologous partner switches and autosynapsis of X chromosome cores occur. Accumulation of Rad51/Dmc1 foci is also observed (Crackower et al., 2003). Therefore, this mouse cannot fully accomplish the recombination process and activates the pachytene checkpoint, resulting in apoptosis and absence of germinal cells. We found that, in the Fkbp6-/- mouse, TopBP1 showed the previously described pattern during leptotene and early zygotene (unpublished data). However, at later stages, TopBP1 displays a very intense staining in unsynapsed chromosomes. Figure 5 shows an early pachytene nucleus doubly labeled with anti-Cor1 and anti-TopBP1 antibodies, where the strongest TopBP1 signal is restricted to unsynapsed regions (Figure 5, Cor1, c), whereas synapsed chromosomes show much less staining (Figure 5, Cor1, SC).
Figure 5.
Coimmunostaining of TopBP1 and Cor1 proteins in a Fkbp6-/- mouse pachytene nucleus. Fluorescence microscopy of mouse meiotic chromosomes from a pachytene nucleus stained with mouse anti-Cor1 (red, left) and rabbit anti-TopBP1 (green, middle). Right panel: merged image. TopBP1 is mainly located to chromosome cores that are not completely synapsed. The arrowheads indicate chromosome cores (c) and synaptonemal complexes (SC). The X and Y chromosomes and the pseudoautosomal region (PAR) are also showed.
S. pombe Cut5 Is Involved in the Meiotic Recombination Checkpoint
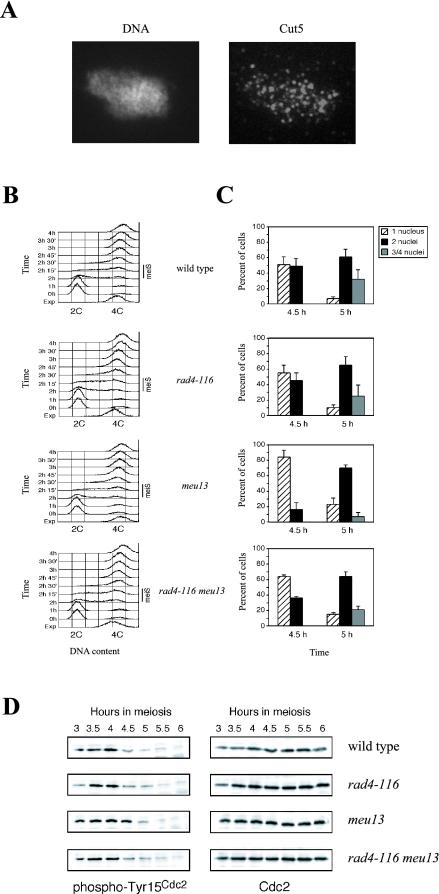
Because ATR and TopBP1 colocalize and the fission yeast ATR homologue, spRad3, has been recently described to be involved in the pachytene checkpoint (Shimada et al., 2002), we decided to study the meiotic role of Cut5, the TopBP1 homologue in S. pombe. First, using a purified antibody against Cut5 (Saka et al., 1994), we found that Cut5 localizes to multiple foci in S. pombe nuclear spreads during meiotic prophase I, in agreement with the data obtained in mammals (Figure 6A).
Figure 6.
The S. pombe Cut5/Rad4 protein is required for the meiotic recombination checkpoint. (A) Spread nuclei of S. pombe strain S964 (wild type) prepared after 3 h of induction of the meiotic program (corresponding to meiotic prophase I; see Perez-Hidalgo et al., 2003) stained with DAPI (left) and anti-Cut5 antibody (right). (B) Flow cytometry analysis of DNA content during pat1-induced synchronous meiosis at 32°C of strains S964 (wild type; top panel), S1329 (rad4-116; second panel from the top), S1295 (meu13; second panel from the bottom), and S1330 (rad4-116 meu13; bottom panel). Note that the timing of premeiotic DNA replication (meiS) is the same in all strains. A representative experiment is shown, but it has been repeated three times obtaining similar results. (C) The rad4-116 mutation abolishes a checkpoint-dependent delay of entry into meiosis I. Kinetics of meiotic progression at 32°C of strains S964 (wild type; top panel), S1329 (rad4-116; second panel from the top), S1295 (meu13; second panel from the bottom) and S1330 (rad4-116 meu13; bottom panel) was followed by DAPI staining of nuclei. The whole time course of meiosis (from 0 to 9 h) was analyzed (unpublished data; see Perez-Hidalgo et al., 2003 for details) but, for clarity, only the data corresponding to the relevant time points (4.5 and 5 h) representing the onset of the first meiotic division are presented. The percent of uninucleated cells (striped bars) that have not yet undergone meiosis I, binucleated cells (black bars) that have undergone meiosis I, and tri- or tetranucleated cells (gray bars) that have undergone meiosis II is shown. The average values (including SD) of three independent experiments are presented. At least 400 cells were scored for each strain at every time point in each experiment. (D) The Cut5-dependent meiotic recombination checkpoint regulates phosphorylation of Cdc2 on tyrosine 15. Western blot analysis of meiotic time courses of strains S964 (wild type), S1329 (rad4-116), S1295 (meu13), and S1330 (rad4-116 meu13), using antiphospho-Cdc2(Tyr15) and anti-Cdc2 antibodies. Note that phosphorylation of Cdc2 on Tyr15 persists longer in the meu13 mutant compared with the rad4-116 meu13 double mutant (see the 4.5-h time point).
The cut5+ gene (also known as rad4+) is essential in S. pombe; therefore, for our studies of the meiotic recombination checkpoint, we used the rad4-116 allele, which at the semipermissive temperature of 32°C is viable because retains the normal replication function of Cut5, but is defective in the checkpoint response to DNA damage and replication blocks in vegetative cells (McFarlane et al., 1997 and unpublished data). First, we examined premeiotic DNA replication in the rad4-116 mutant by flow cytometry using pat1 diploid strains to induce synchronous meiosis (Perez-Hidalgo et al., 2003). Although at 34°C the rad4-116 mutant displays a ∼30 min delay in DNA replication (unpublished data), at 32°C premeiotic DNA replication occurs with similar kinetics in both wild type and rad4-116 (Figure 6B). Thus, we used 32°C in our analyses of meiotic recombination checkpoint function in the rad4-116 mutant.
To trigger the meiotic recombination checkpoint in fission yeast, the meu13 mutant was used. Like its S. cerevisiae homologue (Hop2), the meiosis-specific S. pombe Meu13 protein is required for homologous pairing and meiotic recombination (Nabeshima et al., 2001); in the meu13 mutant, DSBs persist longer than in wild type, resulting in a checkpoint-dependent delay of entry into meiosis I (Shimada et al., 2002; Perez-Hidalgo et al., 2003). We carefully examined kinetics of meiotic progression during pat1-induced synchronous meiosis of wild type, rad4-116, meu13, and rad4-116 meu13 diploid strains (Figure 6C and unpublished data). The rad4-116 single mutant behaves essentially like the wild type. The meu13 mutant displayed a ∼30 min delay of entry into meiosis I, as observed earlier. Strikingly, the rad4-116 mutation partially abolishes the meu13 meiotic delay; the rad4-116 meu13 double mutant proceeds to meiosis I with faster kinetics than the meu13 single mutant. For example, at the 4.5-h time point, only 16% of meu13 cells have completed meiosis I, compared with 49% in wild type and 36% in the rad4-116 meu13 double mutant.
It has been previously described that the meiotic recombination checkpoint in fission yeast delays entry into meiosis I by maintaining inhibitory phosphorylation of the cyclin-dependent kinase Cdc2 on tyrosine 15 (Shimada et al., 2002; Perez-Hidalgo et al., 2003). To confirm that the faster meiotic progression in the rad4-116 meu13 double mutant is a consequence of a defective checkpoint response, we examined the status of Tyr15 phosphorylation during synchronous meiosis (Figure 6D). As reported, phosphorylation of Cdc2 on Tyr15 persists longer when the checkpoint is triggered by the meu13 mutation. In contrast, dephosphorylation of Tyr15 occurs earlier in the rad4-116 meu13 double mutant (compare the 4.5-h time point in Figure 6D), correlating with faster entry into the first meiotic division (Figure 6C). Moreover, spore viability in the rad4-116 meu13 double mutant is significantly reduced compared with the meu13 or the rad4-116 single mutants (Table 1).
Table 1.
Analysis of spore viability
| Relevant genotype | Total spores | Viable spores | Spore viability (%) | Fold decrease |
|---|---|---|---|---|
| Wild type | 2000 | 994 | 49.7 | 1 |
| rad4-116 | 2000 | 1088 | 54.4 | 0.9 |
| meu13 | 2000 | 824 | 41.2 | 1.2 |
| rad4-116 meu13 | 2000 | 231 | 11.5 | 4.3 |
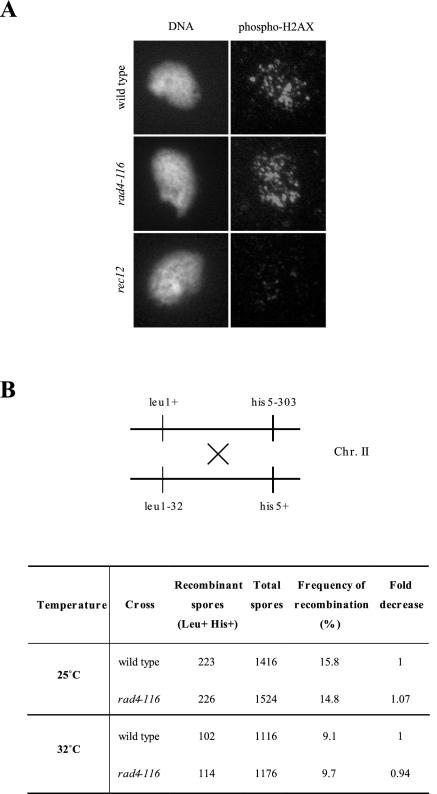
To rule out the possibility that rad4-116 may be defective in DSBs formation, bypassing the meu13 delay, as occurs in a rec12 meu13 double mutant (Shimada et al., 2002), we examined formation of DSBs in spread nuclei, prepared after 3 h in meiosis, of wild type, rec12, and rad4-116 cells, using an anti-γ-H2AX antibody (Rogakou et al., 1998). No difference in γ-H2AX staining could be observed between wild type and rad4-116 during meiosis at 32°C (Figure 7A). In contrast, very little γ-H2AX signal could be detected in the S. pombe rec12 mutant (the SPO11 homologue), which lacks the endonuclease responsible for generation of meiotic DSBs (Figure 7A). In addition, we measured intergenic recombination in the leu1-his5 interval on chromosome II; there is not a recombination defect in the rad4-116 mutant (Figure 7B). Thus, the faster progression of rad4-116 meu13 is likely caused by a defective meiotic recombination checkpoint.
Figure 7.
The rad4-116 allele does not alter meiotic recombination. (A) Spread nuclei of S. pombe strains S964 (wild type), S1329 (rad4-116), and S1331 (rec12) prepared after 3 h of induction of the meiotic program (corresponding to meiotic prophase I; see Perez-Hidalgo et al., 2003) stained with DAPI (left) and anti-γ-H2AX antibody (right). Representative images are shown, but at least 20 nuclei from each strain were examined. (B) Frequency of intergenic meiotic recombination measured on the leu1-his5 interval on chromosome II by random spore analysis of crosses between wild type (PN22 × S1285) and rad4-116 (S1382 × S1383) strains. Crosses were performed at the permissive (25°C) and semipermissive (32°C) temperature for the rad4-116 allele.
DISCUSSION
In this article we report a detailed characterization of the number and distribution of TopBP1 foci during the different stages of male mouse meiotic prophase I. The observed pattern is very defined: the number of foci reaches its maximum at leptotene and then decreases gradually when homologous chromosomes begin to synapse. During early pachytene, most of TopBP1 is localized along unsynapsed chromosomes but almost none is present at the aligned chromosomes, where the SC is fully formed. Interestingly, TopBP1 strongly stains the XY pair until late diplotene. Altogether, this pattern suggests that TopBP1 plays a role during early prophase I.
TopBP1 colocalization with ATR was almost total; in contrast, colocalization with Dmc1, which marks early recombination nodules, was variable depending on the stage (10–35% of colocalization during early prophase I and <10% in later stages, Figure 2D). TopBP1 foci colocalize with γ-H2AX–positive domains that are thought to mark DSBs. Similarly, Dmc1 foci also colocalize to γ-H2AX–positive domains (Mahadevaiah et al., 2001). The reason why the two proteins do not colocalize with each other but are located within the γ-H2AX–positive domains could be due to the fact that the γ-H2AX domains mark extensive chromatin regions (up to a megabase away from the DSB; Rogakou et al., 1999). In mitotic cells, upon DSB formation, TopBP1 localizes to nuclear foci that also show a γ-H2AX–positive staining (Honda et al., 2002; Yamane et al., 2002). Moreover, ATR and its homologues in different organisms have been located physically to the regions of DSBs (Kondo et al., 2001; Melo et al., 2001; Zou et al., 2002). Taking into account the data presented in this article and the data in the literature obtained from studies in mitotic cells, we speculate that TopBP1 and ATR could be present at DSBs sites in meiotic chromosomes. At the particular time after a DSB is formed and resected, Rad51/Dmc1, TopBP1, and ATR could colocalize. In fact, the higher colocalization values are obtained during leptotene and early zygotene (Figure 2D), when DSBs are reported to occur. Later on, the Rad51/Dmc1 complexes could move along the chromosomes carrying out the process of strand exchange, whereas the TopBP1–ATR foci are located in a different position.
Strikingly, TopBP1 localization pattern changes in meiotic spreads of mice with an activated pachytene checkpoint. Male Fkbp6-/- mice display pachytene arrest and spermatocytes undergo apoptosis, resulting in a sterility phenotype (Crackower et al., 2003). Fkbp6 is a component of the synaptonemal complex and the knockout mice initiate recombination but are not able to carry out the full process because of deficiencies in synapsis. In mice, both DSB formation and synapsis among homologues are required for proper recombination (Romanienko and Camerini-Otero, 2000). The staining pattern of TopBP1 in Fkbp6-/- spermatocytes may reflect its function during meiosis. The fact that the heavy stain (up to levels equivalent to XY staining in a normal pachytene) is observed only in unsynapsed regions may reflect a signaling in checkpoint response, because it is precisely in those chromosome arms where, presumably, recombination has started but has not finished. In agreement with the TopBP1 localization pattern in the Fkbp6-/- mouse, ATR also shows a heavy staining in unsynapsed chromosomes in ATM-deficient mice, reported to display pairing defects (Barlow et al., 1998; Moens et al., 1999).
Remarkably, the functional data indicating that TopBP1 may indeed play a role in the meiotic recombination checkpoint come from our studies of its S. pombe homologue Rad4/Cut5. As expected, when we monitored kinetics of meiosis in the recombination-defective meu13 mutant, we observed a delay in the onset of the first meiotic division (Figure 6; Shimada et al., 2002; Perez-Hidalgo et al., 2003); however, this meiotic delay was partially suppressed in the rad4-116 meu13 double mutant (Figure 6C). This observation was also confirmed by monitoring the phosphorylation status of spCdc2 on Tyr 15 (Figure 6D). The relief of the meu13 delay by the rad4-116 mutation is probably due to the lack of checkpoint function and not a consequence of a defect in the initiation of meiotic recombination (i.e., generation of DSBs), because formation of DSBs, as assessed by γ-H2AX staining at 3 h in meiosis, is essentially normal in the rad4-116 mutant at 32°C (Figure 7A). The possibility that the rad4-116 mutation alters another aspect of meiotic recombination different from DSB formation, leading to a faster meiotic progression in the rad4-116 meu13 double mutant, cannot be excluded. However, we do not favor this possibility because spore viability and recombination frequency are similar in wild type and rad4-116 strains (Table 1, Figure 7B), indicating that homologous recombination is not altered in the rad4-116 mutant at 32°C. Importantly, the existence of this Cut5-dependent meiotic delay is crucial to ensure proper distribution of the genetic material to meiotic progeny because the spore viability, which reflects the fidelity in chromosome segregation, is severely compromised in the rad4-116 meu13 double mutant compared with the meu13 single mutant, in which the meiotic checkpoint is active (Table 1). Nevertheless, the meiotic checkpoint is not fully abolished by the rad4-116 mutation at 32°C, because meiotic progression in the rad4-116 meu13 double mutant is still slower than that of wild type or the rad4 single mutant. This observation may indicate that spCut5 plays a partial role in the meiotic checkpoint and there could be several redundant pathways that contribute to it. Alternatively, this effect could be due to the presence of a partially active Cut5 protein at that temperature. Favoring this hypothesis, we found that at 32°C the rad4-116 strain displays a fully active meiotic DNA replication checkpoint when it is triggered with hydroxyurea (unpublished data) and that it does not show any replication deficiency (Figure 6B). It is interesting to note that at that temperature we do find a defective meiotic recombination checkpoint, indicating that there might be regions in the protein that are important for the meiotic replication checkpoint and others important for the meiotic recombination checkpoint.
Studies from yeast to mammals have shown that a group of proteins conserved along evolution are involved in the checkpoint activated upon DNA damage in mitotically dividing cells (Caspari and Carr, 2002; Melo and Toczyski, 2002; Rouse and Jackson, 2002). These proteins are thought to act in a signal transduction pathway in a hierarchy consisting of a group of proteins that initially sense DNA damage and then, in conjunction with intermediate adaptor proteins, signal this DNA damage to various effector kinases (Melo and Toczyski, 2002). These effector kinases finally send the inhibitory signal to the cell cycle progression machinery and induce other DNA damage responses, such as DNA repair, apoptosis, etc. Data in the literature indicate that ATR and homologues are placed in the first group of proteins very close to the recognition of DNA damage (Kondo et al., 2001; Melo et al., 2001; Zou et al., 2002). In addition, studies in Xenopus showed that, in xCut5-depleted extracts, xATR was not able to be loaded onto regions of DNA damage, whereas xCut5 required xRPA, but not xATR, to be loaded onto the damaged DNA (Parrilla-Castellar and Karnitz, 2003). Studies in yeast have shown the existence of adaptor proteins (scRad9, spCrb2/Rhp9) that constitute a communication link between the ATR-like kinase and downstream effector kinases (named Chk1 and Chk2 in mammals; Saka et al., 1997; Emili, 1998; Sun et al., 1998; Vialard et al., 1998). Moreover, the budding yeast TopBP1 homologue, scDpb11, is required for proper activation of the effector kinase scRad53 (Wang and Elledge, 1999), and the fission yeast TopBP1 homologue, spCut5, interacts with spChk1 protein (Saka et al., 1997) and is required for activation of spChk1 and spCds1 (Harris et al., 2003). Interestingly, a common feature to scRad9 and spCrb2/Rhp9 is that, like TopBP1, they contain BRCT domains. For this reason, it is tempting to speculate that TopBP1 could function in the pachytene checkpoint as an adaptor protein that interacts with ATR and, perhaps, with other proteins downstream in this pathway.
Our localization and functional studies of Cut5 in fission yeast and the colocalization of TopBP1 with ATR in mouse spermatocytes, together with the conservation of checkpoint mechanisms through evolution, strongly suggest that TopBP1 functions in the mammalian pachytene checkpoint. However, we do not exclude a role of TopBP1 during meiotic recombination as well; in fact, mutations in ATR homologues in Drosophila (mei41) and S. cerevisiae (mec1) have been described to affect the recombination process (Grushcow et al., 1999; McKim et al., 2000). In the future, it will be interesting to determine where in the pachytene checkpoint pathway is TopBP1/Cut5 functioning. Molecular and genetic studies in S. pombe will be used to study whether spCut5 might be an adaptor protein between spRad3 (ATR homologue) and spMek1 (a meiosis-specific FHA domain-containing scRad53/hChk2 homologue), both involved in the meiotic recombination checkpoint (Shimada et al., 2002; Perez-Hidalgo et al., 2003), similarly to the situation in the S. cerevisiae DNA damage checkpoint, where the BRCT domain-containing protein scRad9 is an adaptor between the PIKK scMec1 and the FHA effector kinase scRad53 (Emili, 1998; Sun et al., 1998; Vialard et al., 1998).
In mammals, it will be of great interest to study the effects of a conditional knockout of TopBP1 in testis. Moreover, a Fkbp6-/- TopBP1-/- double knockout mouse would be useful to determine whether the deficiency in TopBP1 can suppress the pachytene arrest and if TopBP1 plays a role in triggering the apoptosis observed in the Fkbp6-/- mouse. It will be also interesting to perform colocalization studies with other checkpoint proteins already described to form foci at meiotic spermatocytes, like Chk1 and Rad1 (Flaggs et al., 1997; Freire et al., 1998). Meiotic distribution of those different proteins could reflect a temporal and spatial pathway between them and much research on that field remains to be done.
Acknowledgments
We thank Tony Carr and Marius Poitelea for the fission yeast rad4-116 strain. We are grateful to Eduardo Salido for comments and to Julie Bailis for advice in S. pombe spreads. We thank Michael Crackower and Josef Penninger for providing the Fkbp6-/- mice. We are grateful to Satoru Mochida and Mitsuhiro Yanagida for the Cut5 antibody. L.P.-H. and P.A.S.-S. thank Sergio Moreno for encouragement and support. D.P. is supported by a predoctoral fellowship from the Spanish Ministry of Education and Culture. L.P.-H. is a recipient of a predoctoral fellowship from Ministry of Science and Technology of Spain. R.F. is currently supported by a FIS contract. J.E.S. is supported by grants from the Academy of Finland and from the Cancer Society of Finland. This work was supported by grants from Fondo de Investigaciones Sanitarias (FIS) (01/1624) andFUNCIS (PI 45/00) to R.F. and Grant BMC2002–00121 from the Ministry of Science and Technology of Spain to P.A.S.-S.
Abbreviations used: SC, synaptonemal complex.; DSBs, DNA double-strand breaks.; PIKK, phosphatidyl-inositol 3-kinase-like protein kinase.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–06–0444. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-06-0444.
References
- Abraham, R.T. (2001). Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15, 2177-2196. [DOI] [PubMed] [Google Scholar]
- Araki, H., Leem, S.H., Phongdara, A., and Sugino, A. (1995). Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl. Acad. Sci. USA 92, 11791-11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow, A.L., Benson, F.E., West, S.C., and Hultén, M.A. (1997). Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J. 16, 5207-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow, C. et al. (1998). Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development 125, 4007-4017. [DOI] [PubMed] [Google Scholar]
- Baumann, P., Benson, F.E., and West, S.C. (1996). Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell 87, 757-766. [DOI] [PubMed] [Google Scholar]
- Bishop, D. (1994). RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79, 1081-1092. [DOI] [PubMed] [Google Scholar]
- Caspari, T., and Carr, A.M. (2002). Checkpoints: how to flag up double-strand breaks. Curr. Biol. 12, R105-R107. [DOI] [PubMed] [Google Scholar]
- Crackower, M.A. et al. (2003). Essential role of Fkbp6 in male fertility and homologous chromosome pairing in meiosis. Science 300, 1291-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson, M.J., Pearlman, R.E., Karaiskakis, A., Spyropoulos, B., and Moens, P.B. (1994). Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J. Cell Sci. 107(Pt 10), 2749-2760. [DOI] [PubMed] [Google Scholar]
- Emili, A. (1998). MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell 2, 183-189. [DOI] [PubMed] [Google Scholar]
- Flaggs, G. et al. (1997). Atm-dependent interactions of a mammalian Chk1 homolog with meiotic chromosomes. Curr. Biol. 7, 977-986. [DOI] [PubMed] [Google Scholar]
- Freire, R., Murguia, J.R., Tarsounas, M., Lowndes, N.F., Moens, P.B., and Jackson, S.P. (1998). Human and mouse homologs of Schizosaccharomyces pombe rad1(+) and Saccharomyces cerevisiae RAD 17, linkage to checkpoint control and mammalian meiosis. Genes Dev. 12, 2560-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grushcow, J.M., Holzen, T.M., Park, K.J., Weinert, T., Lichten, M., and Bishop, D.K. (1999). Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics 153, 607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S., Kemplen, C., Caspari, T., Chan, C., Lindsay, H.D., Poitelea, M., Carr, A.M., and Price, C. (2003). Delineating the position of rad4+/cut5+ within the DNA-structure checkpoint pathways in Schizosaccharomyces pombe. J. Cell Sci. 116, 3519-3529. [DOI] [PubMed] [Google Scholar]
- Herold, S. et al. (2002). Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell 10, 509-521. [DOI] [PubMed] [Google Scholar]
- Honda, Y., Tojo, M., Matsuzaki, K., Anan, T., Matsumoto, M., Ando, M., Saya, H., and Nakao, M. (2002). Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response. J. Biol. Chem. 277, 3599-3605. [DOI] [PubMed] [Google Scholar]
- Keegan, K., Holtzman, D., Plug, A., Christenson, E., Brainerd, E., Flaggs, G., Bently, N., and Taylor, E. (1996). The ATR and ATM protein kinases associate with different sites along meiotically paired chromosomes. Genes Dev. 10, 2423-2437. [DOI] [PubMed] [Google Scholar]
- Keeney, S., Giroux, C.N., and Kleckner, N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375-384. [DOI] [PubMed] [Google Scholar]
- Kondo, T., Wakayama, T., Naiki, T., Matsumoto, K., and Sugimoto, K. (2001). Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294, 867-870. [DOI] [PubMed] [Google Scholar]
- Lydall, D., Nikolsky, Y., Bishop, D.K., and Weinert, T. (1996). A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383, 840-843. [DOI] [PubMed] [Google Scholar]
- Mahadevaiah, S.K. et al. (2001). Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27, 271-276. [DOI] [PubMed] [Google Scholar]
- Mäkiniemi, M. et al. (2001). BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem. 276, 30399-30406. [DOI] [PubMed] [Google Scholar]
- McFarlane, R.J., Carr, A.M., and Price, C. (1997). Characterisation of the Schizosaccharomyces pombe rad4/cut5 mutant phenotypes: dissection of DNA replication and G2 checkpoint control function. Mol. Gen. Genet. 255, 332-340. [DOI] [PubMed] [Google Scholar]
- McKim, K.S., Jang, J.K., Sekelsky, J.J., Laurencon, A., and Hawley, R.S. (2000). mei-41 is required for precocious anaphase in Drosophila females. Chromosoma 109, 44-49. [DOI] [PubMed] [Google Scholar]
- Melo, J., and Toczyski, D. (2002). A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14, 237-245. [DOI] [PubMed] [Google Scholar]
- Melo, J.A., Cohen, J., and Toczyski, D.P. (2001). Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15, 2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen, R.L., Offenberg, H.H., Dietrich, A.J., Riesewijk, A., van Iersel, M., and Heyting, C. (1992). A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J. 11, 5091-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens, P.B. (1995). Histones H1 and H4 of surface-spread meiotic chromosomes. Chromosoma 104, 169-174. [DOI] [PubMed] [Google Scholar]
- Moens, P.B., Tarsounas, M., Morita, T., Habu, T., Rottinghaus, S.T., Freire, R., Jackson, S.P., Barlow, C., and Wynshaw-Boris, A. (1999). The association of ATR protein with mouse meiotic chromosome cores. Chromosoma 108, 95-102. [DOI] [PubMed] [Google Scholar]
- Moens, P.B., Freire, R., Tarsounas, M., Spyropoulos, B., and Jackson, S.P. (2000). Expression and nuclear localization of BLM, a chromosome stability protein mutated in Bloom's syndrome, suggest a role in recombination during meiotic prophase. J. Cell Sci. 113, 663-672. [DOI] [PubMed] [Google Scholar]
- Moens, P.B., Kolas, N.K., Tarsounas, M., Marcon, E., Cohen, P.E., and Spyropoulos, B. (2002). The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 115, 1611-1622. [DOI] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. [DOI] [PubMed] [Google Scholar]
- Nabeshima, K., Kakihara, Y., Hiraoka, Y., and Nojima, H. (2001). A novel meiosis-specific protein of fission yeast, Meu13p, promotes homologous pairing independently of homologous recombination. EMBO J. 20, 3871-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrilla-Castellar, E.R., and Karnitz, L.M. (2003). Cut5 is required for binding of Atr and DNA polymerase alpha to genotoxin-damaged chromatin. J. Biol. Chem. Published on-line as Manuscript C300418200. [DOI] [PubMed]
- Pelttari, J. et al. (2001). A meiotic chromosomal core consisting of cohesin complex proteins recruits DNA recombination proteins and promotes synapsis in the absence of an axial element in mammalian meiotic cells. Mol. Cell. Biol. 21, 5667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Hidalgo, L., Moreno, S., and San-Segundo, P.A. (2003). Regulation of meiotic progression by the meiosis-specific checkpoint kinase Mek1 in fission yeast. J. Cell Sci. 116, 259-271. [DOI] [PubMed] [Google Scholar]
- Reini, K., Uitto, L., Perera, D., Moens, P.B., Freire, R., Syväoja, J.E. (2004). TopBP1 localises to centrosomes in mitosis and to chromosome cores in meiosis. Chromosoma. (in press). [DOI] [PubMed]
- Roeder, G.S. (1997). Meiotic chromosomes: it takes two to tango. Genes Dev. 11, 2600-2621. [DOI] [PubMed] [Google Scholar]
- Roeder, G.S., and Bailis, J.M. (2000). The pachytene checkpoint. Trends Genet. 16, 395-403. [DOI] [PubMed] [Google Scholar]
- Rogakou, E.P., Boon, C., Redon, C., and Bonner, W.M. (1999). Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146, 905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou, E.P., Pilch, D.R., Orr, A.H., Ivanova, V.S., and Bonner, W.M. (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858-5868. [DOI] [PubMed] [Google Scholar]
- Romanienko, P.J., and Camerini-Otero, R.D. (2000). The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6, 975-987. [DOI] [PubMed] [Google Scholar]
- Rouse, J., and Jackson, S.P. (2002). Interfaces between the detection, signaling, and repair of DNA damage. Science 297, 547-551. [DOI] [PubMed] [Google Scholar]
- Saka, Y., Esashi, F., Matsusaka, T., Mochida, S., and Yanagida, M. (1997). Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 11, 3387-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka, Y., Fantes, P., Sutani, T., McInerny, C., Creanor, J., and Yanagida, M. (1994). Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 13, 5319-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka, Y., and Yanagida, M. (1993). Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+. Cell 74, 383-393. [DOI] [PubMed] [Google Scholar]
- San-Segundo, P.A., and Roeder, G.S. (1999). Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97, 313-324. [DOI] [PubMed] [Google Scholar]
- San-Segundo, P.A., and Roeder, G.S. (2000). Role for the silencing protein Dot1 in meiotic checkpoint control. Mol. Biol. Cell 11, 3601-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha, A., and Kleckner, N. (1997). Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90, 1123-1135. [DOI] [PubMed] [Google Scholar]
- Shimada, M., Nabeshima, K., Tougan, T., and Nojima, H. (2002). The meiotic recombination checkpoint is regulated by checkpoint rad+ genes in fission yeast. EMBO J. 21, 2807-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z., Hsiao, J., Fay, D.S., and Stern, D.F. (1998). Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281, 272-274. [DOI] [PubMed] [Google Scholar]
- Sym, M., Engebrecht, J.A., and Roeder, G.S. (1993). ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72, 365-378. [DOI] [PubMed] [Google Scholar]
- Tarsounas, M., Morita, T., Pearlman, R.E., and Moens, P.B. (1999). RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J. Cell Biol. 147, 207-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialard, J.E., Gilbert, C.S., Green, C.M., and Lowndes, N.F. (1998). The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17, 5679-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., and Elledge, S.J. (1999). DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96, 3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane, K., Chen, J., and Kinsella, T.J. (2003). Both DNA topoisomerase II-binding protein 1 and BRCA1 regulate the G2-M cell cycle checkpoint. Cancer Res. 63, 3049-3053. [PubMed] [Google Scholar]
- Yamane, K., Kawabata, M., and Tsuruo, T. (1997). A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur. J. Biochem. 250, 794-799. [DOI] [PubMed] [Google Scholar]
- Yamane, K., Wu, X., and Chen, J. (2002). A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol. Cell. Biol. 22, 555-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, L., Cortez, D., and Elledge, S.J. (2002). Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16, 198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]