Abstract
Profilin 1 (PFN1) is a regulator of the microfilament system and is involved in various signaling pathways. It interacts with many cytoplasmic and nuclear ligands. The importance of PFN1 for human tissue differentiation has been demonstrated by the findings that human cancer cells, expressing conspicuously low PFN1 levels, adopt a nontumorigenic phenotype upon raising their PFN1 level. In the present study, we characterize the ligand binding site crucial for profilin's tumor suppressor activity. Starting with CAL51, a human breast cancer cell line highly tumorigenic in nude mice, we established stable clones that express PFN1 mutants differentially defective in ligand binding. Clones expressing PFN1 mutants with reduced binding to either poly-proline-stretch ligands or phosphatidyl-inositol-4,5-bisphosphate, but with a functional actin binding site, were normal in growth, adhesion, and anchorage dependence, with only a weak tendency to elicit tumors in nude mice, similar to controls expressing wild-type PFN1. In contrast, clones expressing a mutant with severely reduced capacity to bind actin still behaved like the parental CAL51 and were highly tumorigenic. We conclude that the actin binding site on profilin is instrumental for normal differentiation of human epithelia and the tumor suppressor function of PFN1.
INTRODUCTION
Profilins are small (14- to 17-kDa) proteins found in vertebrates and invertebrates, protozoa, fungi, plants, and certain viruses. Most eukaryotes contain more than one profilin gene, and splicing may generate further isoforms. The importance of profilins for normal cell proliferation and differentiation has been documented in genetic studies, showing that profilin gene disruption leads to grossly impaired growth, motility, and cytokinesis in single cells (Haugwitz et al., 1994; Magdolen et al., 1988) and embryonic lethality in multicellular organisms such as insects (Verheyen and Cooley, 1994) and mice (Witke et al., 1993, 2001). A minimum level of profilin was found critical for differentiation of human epithelial cells, whereas a profilin level below this mark correlates with the tumorigenic state of breast cancer cells and tissue, classifying profilin as a tumor suppressor protein (Janke et al., 2000).
Immunofluorescence and fluorescence microscopy with green fluorescent protein (GFP)-profilin–transfected cells revealed profilin as a constituent of highly dynamic microfilament structures associated with cellular membranes, such as the cortical web of lamellipodia (Buss et al., 1992), nascent focal adhesions (Mayboroda et al., 1997), and surface ruffles (Wittenmayer et al., 2000), and also as a component of intracellular vesicles (Dong et al., 2000). Furthermore, profilin was localized in intranuclear bodies, such as speckles, Cajal bodies, and gems (Giesemann et al., 1999; Skare et al., 2003).
Biochemical analysis showed that profilins interact with a plethora of ligands, in particular with actin (Carlsson et al., 1977), the actin-related protein (Arp)2 (Machesky, 1997), gephyrin (Giesemann et al., 2003), the acidic phospholipid phosphatidyl-inositol-4,5-bisphosphate (PIP2; Lassing and Lindberg, 1985; Sohn et al., 1995; Lambrechts et al., 2002; Skare and Karlsson, 2002), and a large catalog of proteins comprising a poly-proline stretch. Due to the functional diversity of the latter, there is no unifying hypothesis on the biological meaning of such interactions. In contrast, the consequences of profilin's interaction with actin and PIP2 in mammalian cells are much better understood (cf. Lassing and Lindberg, 1985; Goldschmidt-Clermont et al., 1990; Aderem, 1992; Goldschmidt-Clermont et al., 1992; Pantaloni and Carlier, 1993; Perelroizen et al., 1996; and for review, Schlüter et al., 1997). Under physiological conditions, profilins are potent regulators of actin filament dynamics, by promoting the exchange of ADP to ATP on actin and by the affinity of profilin–actin complexes for actin filament ends. Regional actin polymerization may be ensured by high local concentrations of profilin–actin complexes, bound to the poly-proline-stretch proteins vasodilator stimulated phosphoprotein (VASP; Reinhard et al., 1995), p140mDia (Watanabe et al., 1997; Krebs et al., 2001), or neuronal Wiscott-Aldrich-Syndrome protein (Suetsugu et al., 1998), which are actin- and plasma membrane-associated proteins (Holt and Koffer, 2001), and by profilin's binding to PIP2. Because a major PIP2 binding site on profilin overlaps with the actin binding site, fluctuations in the PIP2 level, caused by external stimuli, may cause profilin to shuttle between a membrane-bound to an actin-bound form, performing as a potent mediator in the signal cascade leading from external signals to microfilaments (Machesky and Pollard, 1993).
Although this concept of profilin's function is attractive, many details remain unclear. In particular, there is no information on the consequence of profilin's binding to specific ligands for cell proliferation and differentiation in higher eukaryotes. In this context, we challenged the role of three major binding sites on profilin, for actin, PIP2, and the poly-proline-stretch, in human epithelial differentiation. Previously, we had reported that the cell line CAL51 and other human breast cancer cell lines displayed a conspicuously low level of profilin. When the profilin level was raised in CAL51, either by transfer of the chromosomal fragment harboring the profilin 1 gene, or by transfecting the cells with cDNA coding for profilin 1, the transformed phenotype and tumorigenicity of CAL51 cells was markedly reduced (Theile et al., 1995; Janke et al., 2000).
We now transfected CAL51 cells with profilin 1 mutants defective in binding to actin, poly-proline, or PIP2 and generated cell clones stably expressing the mutant profilins. These cells were analyzed with respect to tumor-related phenomena, such as independence of substrate adhesion, inability to differentiate, and the generation of tumors in nude mice. Our results indicate that a minimal level of a functional actin binding site on profilin is critical and indispensable for suppression of tumorigenicity in human epithelia.
MATERIALS AND METHODS
Preparation of Expression Constructs
The mutants of human profilin 1 (PFN1), PFN1/Y59A, PFN1/H133S, and PFN1/R88L were generated by site-directed mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene, La Jolla, CA). For the expression in human cells or bacteria, mutated and wild-type PFN1 cDNAs were cloned either into modified pcDNA3 or pET21c vectors (Novagen, Madison, WI), respectively. Both vectors harbored the BiPro-tag sequence (Rüdiger et al., 1997).
Expression, Purification, and Characterization of Recombinant Proteins
Wild-type, unmodified, and BiPro-tagged human profilin 1 was expressed in Escherichia coli BL21 (DE3) essentially as described previously (Wittenmayer et al., 2000). All mutants were expressed in and purified from E. coli as described previously (Schlüter et al., 1998), and purified from bacterial lysates by affinity chromatography on poly-(l-proline)-columns (Tanaka and Shibata, 1985; Lindberg et al., 1988; Wittenmayer et al., 2000). The mutant PFN1/H133S is not retained by such columns but trails in a separate peak. Collected fractions were analyzed by SDS-PAGE, and purity of the preparation was >90%, as judged from Coomassie-stained gels. Protein concentration was quantified by the Bradford assay, using bovine serum albumin (BSA) as a standard. Native configuration of all proteins was determined by CD spectra. Their respective defect in ligand binding was characterized with untagged proteins in biochemical assays. The affinity for actin was determined with pyrene-labeled skeletal muscle actin at steady state (Schlüter et al., 1998), by using gelsolincapped actin filaments, as described previously (Bjorkegren-Sjogren et al., 1997; Lambrechts et al., 2002). Pig plasma gelsolin was a generous gift of Dr. H. Hinssen (University of Bielefeld, Bielefeld, Germany) and, additionally, was purchased from Sigma Chemie (Taufkirchen, Germany). Retardation on poly-proline-Sepharose columns was used as a criterion for binding to polyproline-stretch ligands. Binding to PIP2 was monitored with two different assays. Dot overlay assays were performed with 0.02–1 μg of PIP2 micelles (prepared as in Hüttelmaier et al., 1998), spotted onto polyvinylidene difluoride membranes (Roth, Karlsruhe, Germany), and sequentially incubated with 3% BSA for 1 h, 30 μM profilin, and 0.3% milk powder in phosphate-buffered saline. Bound profilin was detected with the monoclonal anti-profilin as described for immunoblotting. Microfiltration assays were performed essentially as described previously (Lambrechts et al., 1997; Skare and Karlsson, 2002). Increasing amounts of freshly prepared PIP2 micelles were incubated with 0.5 nmol of profilin. Free and PIP2-bound profilins were separated by centrifugation through PLTK filters (Millipore, Eschborn, Germany). The flow-through, containing unbound profilin, was either concentrated, subjected to SDS-PAGE, and Coomassie Blue staining, or used directly in SDS-PAGE and immunoblotting with anti-profilin. The dot overlay and both variants of the microfiltration assays gave identical results. The data for ligand binding obtained for all recombinant proteins are shown in Table 1.
Table 1.
Ligand binding capacity of the point mutants of human profilin 1 used in this study
| Protein | Binding to actin: Kd valuesa | Binding to poly-proline: binding to poly-proline-Sepharose column | Binding to PIP2 in dot overlay and microfiltration assays |
|---|---|---|---|
| Wild-type PFN1 | 0.71 × 10-6 M | 6-8 M urea needed for elution | 100% |
| PFN1/Y59A | 2.11 × 10-5 M | 6-8 M urea needed for elution | Like wild-type PFN1 |
| PFN1/H133S | 1.09 × 10-6 M | No binding | Like wild-type PFN1 |
| PFN1/R88L | 2.03 × 10-6 M | 6-8 M urea needed for elution | Less than 30% of wild-type value |
Data were obtained with recombinant proteins purified and analysed as described in MATERIALS AND METHODS.
Determined at steady state with gelsolin-capped filaments. The critical concentration of the actin used was 0.87 × 10-6 M
Cell Culture, Transfection, and Isolation of Stable Clones
The mammary epithelial MCF-10A cell line was grown in DMEM/Ham's F12 (1:1) medium supplemented with 20 ng/ml epidermal growth factor, 0.01 mg/ml insulin, 500 ng/ml hydrocortisone, and 5% horse serum. The cell line CAL51 and its nontumorigenic microcell hybrid CAL/17-5 (Theile et al., 1995) were grown in DMEM supplemented with 10% fetal calf serum. CAL51 cells were transfected with plasmid DNAs (pcDNA3, pcDNA-PFN1, pcDNA-PFN1/Y59A, pcDNA-PFN1/H133S, and pcDNA-PFN1/R88L) by using Fu-GENE 6 transfection reagent (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions. Selection with 800 μg/ml G418 started 48 h after transfection and G418-resistant clones were isolated 2–3 wk later.
Preparation of Cell Lysates and Immunoblotting
Cells were harvested and resuspended in ice-cold lysis buffer (50 mM Tris/HCl, pH 7.4, 10 mM NaCl, 5 mM EDTA, 0.3% Triton X-100 containing a protease inhibitor mix [10 μM leupeptin, 20 U/ml aprotinin, 1 μM pepstatin A, and 100 μM pefabloc SC]). After 15-min incubation on ice, cell fragments were homogenized by passing the suspension through a syringe. Analogous procedures were used to obtain total protein extracts from tumor tissue (see below). The resulting suspension was centrifuged for 5 min at 13,000 rpm, and protein concentration of the supernatant was determined by Bradford assay, by using BSA as a standard. Equal amounts of proteins (in the range of 5–15 μg) were separated by SDS-PAGE on tricine-containing gels (Schägger and Jagow, 1987; Wittenmayer et al., 2000) and transferred onto nitrocellulose membrane. Membranes were blocked with 5% nonfat milk in phosphate-buffered saline. Endogenous and transfected BiPro-tagged profilin, and endogenous tubulin were monitored with the following antibodies: monoclonal anti-profilin (Mayboroda et al., 1997), monoclonal antibody against the BiPro tag (Rüdiger et al., 1997) or monoclonal anti-tubulin (Sigma Chemie), respectively. Immunoreactive bands were detected using a horseradish peroxidase-conjugated goat anti-mouse IgG (Sigma Chemie) for a second antibody and by enhanced chemiluminescence (Amersham Biosciences, Freiburg, Germany). The intensity of the signals was determined by densitometry (EASY-win; Herolab, Wiesloch, Germany). For quantitative analysis, standard curves were obtained with recombinant profilin 1 (5–50 ng) and BiPro-tagged profilin 1 (5–30 ng), respectively. The levels of profilin expression were determined for at least five independent experiments and compared by variance analysis (Student-Newman-Keuls; p < 0.05), by using Statview 5.0 (SAS Institute, Cary, NC).
Growth Assays
Colony-forming ability in soft agar of the various cell lines was analyzed essentially as described previously (Theile et al., 1995), seeding 2.5 × 104 cells in 60-mm dishes. After 3 wk at 37°C and in a humidified atmosphere containing 10% CO2, the numbers of colonies were counted. Three independent experiments were performed, and standard deviations were determined. Growth dependence on adhesion to extracellular matrix was investigated by seeding 5 × 104 cells per well on top of a layer of matrigel (BD Biosciences, Palo Alto, CA). Cells were incubated at 37°C in a humidified atmosphere containing 10% CO2. After 48 h to 6 d, colonies were photographed using an Axiovert 100 microscope (Carl Zeiss, Oberkochen, Germany).
Analysis of Cell Spreading
Spreading of CAL51, PFN1-, PFN1/Y59A-, PFN1/H133S-, PFN1/R88L-transfected CAL51 clones, and MCF-10A cells was monitored after seeding them onto glass coverslips, essentially as described previously (Janke et al., 2000). At 30 min after seeding, cells were fixed with 4% formaldehyde for 15 min, permeabilized with 0.2% Triton X-100, and stained with phalloidin-fluorescein isothiocyanate (Sigma Chemie). The cells were divided into two groups according to gross differences in surface area and microfilament organization: flat cells already well spread, with well developed microfilament bundles (group 1) and small, compact cells that displayed most of their actin filaments at their periphery (group 2). Per coverslip, 500 cells were counted in three independent experiments and analyzed by variance analysis (Student-Newman-Keuls; p < 0.05), by using Statview 5.0.
Tumorigenicity Test, Histopathology, and Preparation of Tumor Tissue Extracts
From each clone, 106 or 105 cells, respectively, were suspended in 0.1 ml of serum-free medium and injected subcutaneously into 5- to 6-wk-old female immunodeficient nude mice (BlnA:NMRI-nu/nu). Appearance and size of tumors were examined weekly. After 11 wk, the mice were killed, and subcutaneous tumors were removed and measured. Then, 5-μm sections were obtained from formalin-fixed, paraffin-embedded tumor tissue blocks, stained with hematoxylin/eosin, and examined in a light microscope (BX-50; Olympus, Tokyo, Japan). To quantify the amount of profilin expressed in the tumor tissue, aliquots were homogenized in liquid nitrogen and lysis buffer (20 mM Tris/HCl, pH 8.0, 100 mM NaCl, 500 mM EDTA, 50 mM dithioerythritol, and protease inhibitor mix). The suspension was sheared several times through a 26G gauge needle. Cell debris was removed by centrifugation at 13,000 × g at 4°C for 15 min, and the protein concentration in the supernatant was determined by Bradford assay.
RESULTS
Point Mutations in Profilin1 Yield Proteins Defective in Binding to Specific Ligands
To reduce specific ligand binding in human PFN1, we used three point mutations, created by site-directed mutagenesis. Two of these, PFN1/H133S and PFN1/R88L, had been previously designed to create mutants specifically deficient in polyproline- or PIP2 binding, respectively (Bjorkegren et al., 1993; Sohn et al., 1995). The third one, PFN1/Y59A, was designed according to our previous study on bovine profilin 1, where an analogous mutation (F59A) had displayed a greatly reduced actin binding (Schlüter et al., 1998). The resulting recombinant proteins were expressed in and purified from E. coli and tested for the respective defect in biochemical assays. As can be seen from Table 1, all three mutants showed the expected defect in binding the respective ligand: compared with wild-type PFN1, actin binding of the PFN1/Y59A mutant was decreased by a factor of 30, PIP2 binding of PFN1/R88L was less than one-third of the wild-type value, and PFN1/H133S was not retained on poly-proline columns. In contrast, binding to the other ligands was not or only moderately affected: PIP2 binding of PFN1/Y59A and PFN1/H133S, and poly-proline binding of PFN1/Y59A and PFN1/R88L was as good as seen for wild-type PFN1. PFN1/R88L, however, the mutant substantially defective in PIP2 binding, displayed also a slightly reduced binding to actin.
Expression of Mutated Profilins in CAL51 Cells Yields Clones with Moderate Overall Profilin Levels
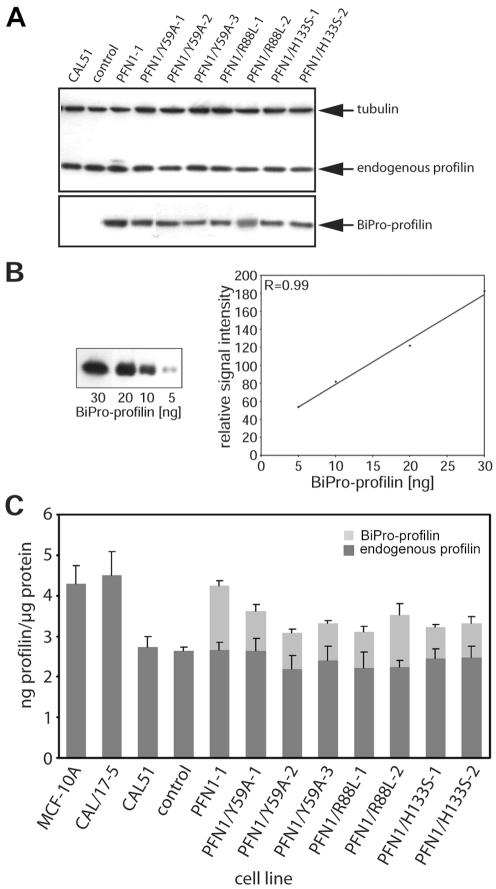
CAL51 cells were transfected with vectors coding for PFN1 wild-type or PFN1 point mutations equipped with the BiPro tag (Rüdiger et al., 1997). Stable clones were selected and the level of endogenous and transfected profilins was determined in quantitative immunoblots. Figure 1 shows the results obtained for the various clones. Tubulin was monitored to ensure equal loading of total proteins (Figure 1A), and the amount of the transfected profilin was determined from standard curves obtained with the tag antibody (Figure 1B), whereas the endogenous profilin was monitored in the same manner with a monoclonal anti-profilin reacting with human profilin 1 (Mayboroda et al., 1997). Figure 1C shows the results obtained for the various clones. The epithelial control cell line MCF-10A contained ∼4.5 ng profilin/μg protein, and a similar value was measured for the nontumorigenic CAL/17-5 microcell hybrid that had been obtained after chromosomal transfer (Theile et al., 1995). The parental tumor cell line CAL51 and a stable clone mock-transfected with the empty vector expressed <3 ng profilin/μg protein. Similar levels were found for a number of other human cancer cell lines (Janke et al., 2000). Transfecting CAL51 with wild-type PFN1 or the three different PFN1 point mutants resulted in clones, expressing the exogenous protein to varying degrees, between 30 and 50% of the endogenous profilin. The difference in total profilin content between CAL51 and mock-transfected cells on one hand and the lines expressing exogenous profilin was statistically significant.
Figure 1.
Expression of endogenous and transfected profilins in derivatives of CAL51. (A) Example of immunoblots of extracts obtained from CAL51 and the following stable clones: one clone transfected with the empty vector (control), one clone transfected with wild-type profilin 1 (PFN1), three clones transfected with the profilin mutant defective in actin binding (PFN1/Y59A), two clones transfected with the profilin mutant defective in PIP2 binding (PFN1/R88L), and two clones transfected with the profilin mutant defective in polyproline-binding (PFN1/H133S). All transfected proteins were equipped with the BiPro tag (see MATERIALS AND METHODS). The blot was reacted with monoclonal antibodies against the BiPro tag, against endogenous profilin, and against tubulin, respectively, to confirm equal loading of total protein. (B) Standard curve obtained with purified BiPro-tagged profilin, to demonstrate that the quantitative data shown in C were obtained within the linear range of anti-BiPro reactivity. (C) Quantitation of the amounts of endogenous and transfected, BiPro-tagged profilin obtained from immunoblots as shown in A. The bars reflect values and SEs obtained from five to 10 independent experiments, compared by variance analysis (see MATERIALS AND METHODS). Values obtained for the CAL/17-5 microcell hybrid and for the MCF-10A normal epithelial line are also shown (left). Note that in all transfected lines the total amount of profilin does not exceed the profilin level of the nontumorigenic lines CAL/17-5 and MCF-10A.
PFN1/Y59A Expression Cannot Restore Anchorage Dependence in CAL51 Cells
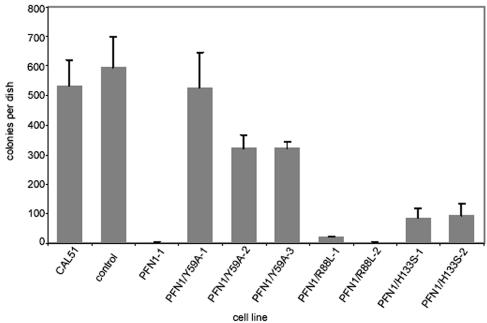
To determine which of the profilin mutants might interfere with the tumorigenic state of CAL51, we first analyzed anchorage independence of growth, a criterion frequently used in the characterization of tumorigenic cells. Previously, we had already shown that CAL51 cells grow well in soft agar, whereas the expression of wild-type profilin 1 induced by either cDNA transfection or chromosomal transfer leads to loss of this ability (Janke et al., 2000). We now tested the various clones stably expressing the respective profilin mutants in this assay. Figure 2 shows that a defect in polyproline binding or in phospholipid binding does not allow for growth in soft agar. Expression of these profilin mutants suppresses this ability drastically, similar to the situation found for clones synthesizing wild-type profilin. In contrast, expression of the profilin mutant with a grossly defective actin binding site (PFN1/Y59A) does not interfere with anchorage-independent growth (Figure 2).
Figure 2.
Growth of CAL51 and derivatives in soft agar. CAL51 cells and the derivative clones indicated were plated in soft agar as described in MATERIALS AND METHODS and examined for colony formation after 3 wk. For explanation of different lines, see Figure 1. CAL51 cells and mock-transfected cells (control) established numerous colonies, whereas a line supplemented with wild-type profilin (PFN1-1) did not grow under these conditions. Note that among the lines expressing mutant profilins, PFN1/R88L and PFN1/H133S are reduced in colony growth, similar to the PFN1 clone, whereas PFN1/Y59A still retains the ability to grow in soft agar. The SD from the mean is given for three independent experiments.
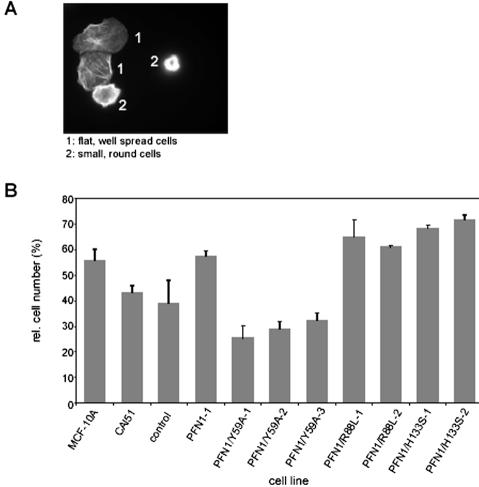
PFN1/Y59A Expression Cannot Normalize Cell Spreading and Microfilament Organization
CAL51, as many tumor lines, displays a delay in adherence and spreading on solid substrates, combined with a not very well organized microfilament system. Elevation of the profilin level in CAL51, however, normalizes these phenomena: CAL51 cells expressing exogenous wild-type profilin 1 adhere and spread faster and show a microfilament organization more reminiscent of epithelial cells (Janke et al., 2000). To elucidate the importance of the profilin-actin binding site for cell spreading and adherence, we analyzed the various clones shortly after seeding them onto glass. Figure 3 demonstrates the data obtained for cells fixed and stained with rhodamine-phalloidin at 30 min after seeding. Based on their morphology and microfilament organization, the cells could be classified into two groups: well spread; flat cells with distinct microfilament bundles and focal contacts; and round, compact cells that had not spread within 30–60 min after seeding (Figure 3A). Figure 3B shows that CAL51 cells expressing wild-type profilin 1 or any of the mutant profilins defective in poly-proline or phospholipid binding displayed a high proportion of cells that were well spread at 30 min after seeding, similar to cells of the normal epithelial line MCF-10A. The cells synthesizing PFN1/R88L or PFN1/H133S were even slightly better in their capacity to adhere and spread shortly after seeding. In contrast, CAL51 parental and CAL51 mock-transfected cells and the clones expressing the profilin mutant defective in actin binding were delayed in adherence and spreading. The observed differences between the CAL51 cells, the mock-transfected line and all three lines expressing PFN1/Y59A on one hand and the MCF-10A epithelial line, the wild-type PFN1 expressing CAL51 line and the five lines either expressing PFN1/R88L or PFN1/H133S on the other hand were statistically significant, emphasizing the importance of an intact actin binding site on profilin for normal epithelioid adherence and microfilament organization.
Figure 3.
Spreading of epithelial, tumor, and profilin-reconstituted cell lines in tissue culture. Cells were seeded on glass coverslips and allowed to adhere for 30 min before fixation and staining with rhodamine-phalloidin to reveal the microfilament system. (A) Fluorescence images to show the criteria used to divide cells into two groups: those that have already spread and display a flat morphology with microfilament bundles (group 1), and those that are still compact and globular (group 2). (B) Quantitation of cells classified in group 1. For explanation of different lines, see Figure 1. Bars reflect the results obtained in three independent experiments, with SEs. Note that the epithelial line MCF-10A, the profilin-reconstituted line PFN1–1 and the lines expressing profilin defective in either PIP2 or poly-proline binding have a high proportion of cells already well spread, whereas for CAL51, the mock-transfected cell line and the lines expressing profilin defective in actin-binding, the proportion of well spread cells is significantly lower (see MATERIALS AND METHODS for statistical analysis).
PFN1/Y59A Expression Does Not Permit an Epithelioid Phenotype on Matrigel
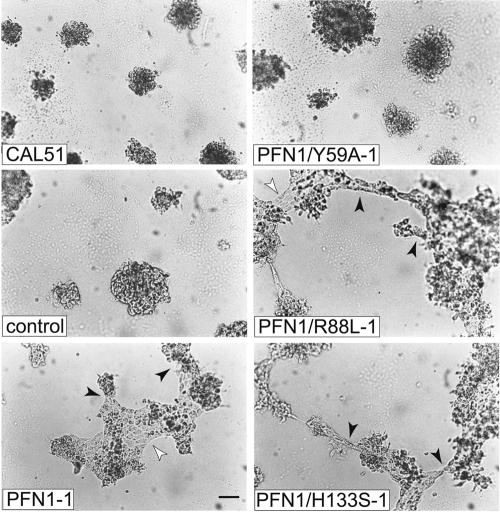
We next tested the various cell clones under experimental conditions that were more similar to the in vivo situation. When embedded in or seeded onto Matrigel, normal epithelial cells have the capacity to organize into tissue-like structures and develop tubular elements reminiscent of epithelial ducts, whereas malignant breast tumor cells have lost this ability and grow in compact, three-dimensional lumps (Petersen et al., 1992). This difference is also observed for MCF-10A normal epithelial cells and CAL51, and expression of wild-type profilin in CAL51 shifts CAL51 toward a more epithelial organization (Janke et al., 2000). To determine the ability of the profilin mutants in this respect, we seeded aliquots of the relevant cell clones onto matrigel and examined them over a period of 6 d. Representative images of the results seen are shown in Figure 4. The CAL51 clones expressing profilin mutants were again seen to fall into two classes: at 6 d after seeding, clones synthesizing profilins defective in either poly-proline or phospholipid binding had formed flat, epitheloid sheets, connected by tubular elements. These were very similar to structures obtained with the normal epithelial line MCF-10A under analogous conditions (Janke et al., 2000). In contrast, the clones expressing profilin defective in actin binding were unable to show such tissue-like organization but grew in small compact foci, like the parental CAL51 line (Figure 4).
Figure 4.
Growth dependence and morphological differentiation of CAL51 parental cells and various derivatives on matrigel. Pictures were taken 6 d after seeding. For explanation of different lines, see Figure 1. Note that the lines reconstituted with either wild-type profilin or profilin mutants defective in PIP2- or poly-proline binding have developed flat sheets of epithelioid cells (white arrowheads) connected by epithelioid tubes (black arrowheads). In contrast, the parental tumor line CAL51, the mock-transfected CAL51 cells and the clone expressing PFN1/Y59A, the mutant severely defective in actin-binding, display compact, undifferentiated colonies. Bar, 100 μm.
PFN1/Y59A Expression Cannot Prevent Tumor Growth in Nude Mice
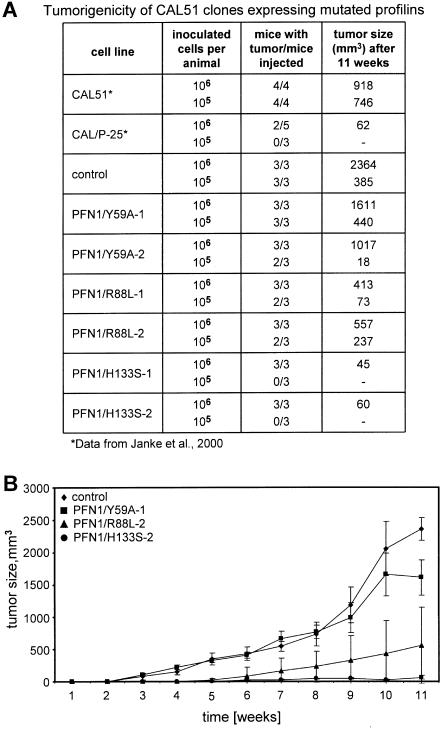
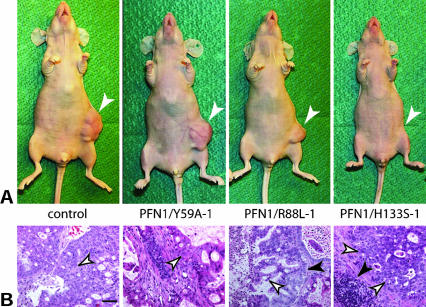
Finally, we challenged the importance of an intact actin binding site on profilin for suppression of tumorigenicity in nude mice. Our previous work had shown that elevation of the profilin level in CAL51 cells by transfer of wild-type profilin correlates with tumor suppression (Janke et al., 2000). Figure 5 summarizes the results obtained for the different cell lines, expressing profilin wild type or the corresponding mutants. Figure 5A compiles the data obtained in relation to numbers of injected cells, injected animals, and for tumor growth. Figure 5B demonstrates the kinetics of tumor development. CAL51 parental and mock-transfected cells elicited rapidly growing tumors after subcutaneous injection of either 105 or 106 cells, and CAL51 cells expressing the mutant PFN1/Y59A were also highly tumorigenic, yielding fast-growing tumors in all mice subcutaneously injected with either 105 or 106 cells. In contrast, tumorigenicity was greatly reduced in animals injected with cells expressing the profilin mutant primarily defective in polyproline binding. PFN1/H133S-containing cells elicited only slow-growing tumors when injected at a high cell number (106), and no tumors in animals injected with a 10-fold lower dosis, comparable with that of CAL51 expressing wild-type profilin (CAL/P-25 in Figure 5A). An intermediate capacity in tumorigenesis was seen in the PFN1/R88L-expressing cells: injection of high and lower cell numbers elicited tumors, but with a considerable delay in growth. Because the profilin mutant PFN1/Y59A is severely, and PFN1/R88L slightly defective in actin binding (Table 1), these data again underline the importance of an intact actin binding site in profilin for tumor suppression. Figure 6 shows the gross morphology and histopathology of the tumors developed in representative animals 11 wk after injection. Mice inoculated with 106 CAL51 parental or PFN1/Y59A-expressing cells displayed large, well vascularized tumors at the site of injection. In contrast, injection of cells expressing the PFN1/R88L profilin mutant resulted in much smaller lumps with less obvious vascularization. Mice injected with PFN1/H133S-expressing cells did not show obvious tumors (Figure 6A). Histopathological examination of tissue samples taken from mice injected with PFN1/Y59A- or PFN1/R88L-expressing cells revealed a high density of typical tumor cells, but tissue from the site of injection of PFN1/H133S-expressing cells still contained some islands of cells different in morphology from the surrounding mouse tissue, possibly human tumor cells (Figure 6B).
Figure 5.
Tumors obtained after injection of the different cell lines in nude mice. For explanation of different lines, see Figure 1. (A) Tumors obtained in relation to the number of cells injected, the number of animals inoculated, and the size of the various tumors obtained after 10 or 11 wk, respectively. For comparison, data obtained from previous experiments are included (Janke et al., 2000) and marked by an asterisk. CAL/P-25: a stable line expressing wild-type profilin 1. (B) Development of tumors over time. The mean tumor size and standard deviations are shown. Note that there is not only a gross difference in the final size of tumors obtained after 11 wk but also in the onset and velocity of tumor growth, and no tumor resorption. Although mock-transfected CAL51 (control) and the lines expressing profilin severely defective in actin-binding (PFN1/Y59A) are highly tumorigenic, the lines expressing profilin defective in PIP2 binding (PFN1/R88L) are much less aggressive in eliciting tumors, and the lines expressing profilin defective in poly-proline binding (PFN1/H133S) are not capable of stimulating substantial tumor growth.
Figure 6.
Pathology of tumors as developed in nude mice. For explanation of different lines, see Figure 1. (A) Gross morphology of the animals, showing the sites of cell injection after 11 wk (arrows). (B) Histopathology of biopsies. Tissue looking abnormal in these stained paraffin sections is marked by white arrows, mouse connective tissue is indicated by black arrows. Note that the large lumps caused by control CAL51 cells and cells expressing the profilin mutant defective in actin-binding (PFN1/Y59A) are heavily vascularized and are primarily composed of tumor cells (white arrows), whereas the smaller, less well vascularized lumps generated by cells expressing the mutant primarily defective in PIP2-binding (PFN1/R88L) contain cells that were histopathologically also identified as tumor cells. Tissue taken from the site where cells defective in poly-proline-binding (PFN1/H133S) had been injected was not vascularized. It contained abnormal looking cells not unequivocally identifiable. They formed only small islands embedded in normal mouse tissue or fibrous material and failed to outgrow. Bar, 50 μm.
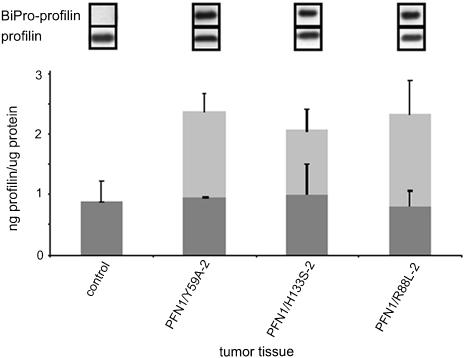
The persistence of the expression of the profilin mutant originally introduced into the various lines was monitored by immunoblotting: aliquots of tissue taken from the injection site were homogenized and prepared for SDS-PAGE and quantitative immunoblotting, with standard curves for the BiPro-tagged profilin and for purified human profilin 1, as described in Figure 1. The results are shown in Figure 7. All tissues of the mice depicted in Figure 6A still expressed the transfected, exogenous profilin mutant, in addition to the endogenous profilin, the level of which was not altered by the transfected variant.
Figure 7.
Expression level of endogenous and transfected profilins, as seen in immunoblots obtained from tumor tissue extracts. For explanation of different lines, see Fig. 1. Top, immunoblots obtained with the tag antibody (BiPro antibody) or anti-profilin, to reveal the transfected and endogenous profilin, respectively. Bottom, diagram revealing the absolute amounts of endogenous profilin (gray columns) and the transfected mutants (white columns), obtained by quantitating immunoblots with antibodies against profilin or the BiPro tag, respectively. The bars reflect the SD from the mean for at least three independent analyses. Note that the tumor tissue derived from all cell clones still express the mutated profilin and that the level of endogenous profilin is not altered in these lines compared with the parental CAL51.
DISCUSSION
In a previous study, we reported tumor suppression by elevating the PFN1 level in various human breast cancer cell lines that display a rather low level of PFN1 mRNA and protein compared with normal breast epithelial cells (Janke et al., 2000). In an attempt to unravel the particular contribution of binding to different ligands in the tumor suppressor activity of PFN1, we used point mutants defective in three major binding sites for ligand proteins. Our results emphasize the particular importance of the profilin–actin interaction for tumor suppression, especially for events related to normal cell spreading, adhesion, and differentiation of human epithelial cells.
Mutants were chosen on the basis of a specific and selective binding defect in one ligand binding site, as seen in biochemical and biophysical assays. As expected from our previous work with bovine profilin 1 (Schlüter et al., 1998), the human profilin mutant PFN1/Y59A was greatly reduced in actin binding but was not affected in binding to polyproline or PIP2. The mutant PFN1/R88L was normal in poly-proline binding, showed a slight reduction in actin binding and a substantial decrease in PIP2 binding. These results confirm previous reports and support the notion that arginine 88, located within the actin binding site and the N-terminal PIP2 binding site on profilin probably contributes somewhat to actin binding, but is definitely very important for PIP2 binding (Sohn et al., 1995; Lambrechts et al., 2002). Analysis of the third mutant, PFN1/H133S, showed the expected failure in binding to poly-proline with well conserved binding to actin and to PIP2. Although there is good evidence for mutual cross talk between the binding sites for actin, PIP2 and poly-proline stretches, extending beyond neighboring binding sites (Lambrechts et al., 2002), the drastic reduction in specific ligand binding by the three mutants chosen, compiled in Table 1, seemed a good basis to investigate the biological function of each binding site with respect to tumor suppression. Equipping the mutants with a tag enabled their identification in transfected cells and discrimination from the endogenous protein.
To ensure that the stably transfected clones were representative, more than one clone was generated for each mutant, and two or three were used for the analyses shown. We found that in all stable clones obtained, the expression of endogenous profilin was unaltered, i.e., still in the range determined for the parental CAL51 line. Remarkably, although we used a promoter capable of driving a strong expression of recombinant proteins (cytomegalovirus promoter), all clones that were easy to propagate, robust for handling, and stable in synthesizing the exogenous protein over several months expressed the transfected profilin mutant to only moderate levels, so that the overall profilin concentration did not exceed the level found for normal human breast epithelial cell lines. The previously determined high levels of profilin overexpression in the nontumorigenic CAL/17-5 line (Janke et al., 2000) were not retained with time, but had also decreased to the level indicative for breast epithelial cells, like MCF-10A. In this context, it is noteworthy that PFN1 expression in most human tissues is within a rather narrow range, not exceeding a threefold difference from the mean. Exceptions from this rule are spleen and tissues with a relatively high expression of other profilin isoforms, i.e., brain and testis (Su et al., 2002). This indicates that human tissue differentiation, in particular epithelial differentiation, requires a minimum level of profilin 1. Consistent with this hypothesis is the finding that PFN1 gene expression in gastric carcinoma is reduced compared with normal stomach (Oien et al., 2003). In contrast, PFN1 gene expression in different mouse tissues is much more variable than seen for humans (Su et al., 2002), and a 50% reduction of profilin 1 levels in heterozygous PFN1 +/- animals does apparently not lead to tumor growth (Witke et al., 2001). Hence, the role of profilin 1 as a tumor suppressor in epithelia may be correlated with an organism-specific tolerance against deviations from the mean.
In standard assays generally accepted to discriminate between a transformed/tumorigenic and normal epithelial state, we confirmed our previous findings that the elevation of the overall profilin 1 level in human breast carcinoma cells was capable to revert the tumorigenic state (Janke et al., 2000). In the analysis reported here, we found prominent differences between mutants defective in the three major ligand binding sites, with respect to anchorage dependence, adhesion and cell morphology, cellular organization on Matrigel, and tumorigenicity in the nude mouse. The results of these assays show that expressing PFN1 mutants with a reduced binding capacity for PIP2 or poly-proline in CAL51 cells can lead to a reversion of several tumor cell-related phenomena, comparable with that found for wild-type profilin. In contrast, a defective actin binding site on profilin is incompatible with a rescue to normal epithelial differentiation. The finding that cells expressing the R88L mutant, which displays a slight reduction in actin binding in addition to the defect in PIP2 binding, possess a reduced capacity to elicit tumors, is consistent with this model.
Although our data stress the importance of the actin binding site on profilin for normal epithelial differentiation, they do not allow for an unequivocal answer to the question of which of the numerous profilin ligand interactions may be involved. However, they permit a qualitative comparison of the importance of the three major binding sites in this process. Regarding PFN1/R88L, quantitative data on the affinity of the profilin–PIP2 complex and the decrease in this mutant are not available. The residual PIP2 binding seen for this mutant may reflect PIP2 binding to the second binding site which is located close to the poly-proline binding site (Lambrechts et al., 2002; Skare and Karlsson, 2002). Apparently, synthesis of this mutant protein, together with the endogenous profilin 1, must be sufficient for epithelial differentiation. Alternatively, reduced PIP2 binding by profilin might be compensated by other components of the microfilament system that also bind to PIP2 and can act as tumor suppressor proteins, like alpha actinin, vinculin, or gelsolin (Vandekerckhove et al., 1990; Rodriguez Fernandez et al., 1992; Glück et al., 1993; Asch et al., 1996; Ben Ze'ev, 1997). Unfortunately, the significance of PIP2 binding of these proteins for normal cell differentiation has not been analyzed.
PFN1/H133S was chosen because of its specific deficiency in binding to poly-proline. In an elaborate and elegant study with fission yeast, by using mutants with a null profilin background, it was seen that only profilin mutants with a residual poly-proline binding of >10% of the wild-type value can restore viability (Lu and Pollard, 2001). Such quantitative data cannot be expected for the system used here, because human breast cancer cells (Janke et al., 2000) as well as human gastric carcinoma cells (Oien et al., 2003) still synthesize some endogenous profilin. For CAL51, we could therefore only analyze the contribution of poly-proline binding activity for epithelial differentiation, rather than viability. CAL51 variants expressing PFN1/H133S, which cannot bind at all to poly-proline in vitro and thus presumably also not to poly-proline-stretch ligands in a cellular environment, were seen to adhere and spread at least as good as CAL51 reconstituted with wild-type profilin, developed epithelioid structures on Matrigel and were very poor in eliciting tumors. Considering the complexity of ligands interacting with profilin via a poly-proline-stretch in mammalian cells, this seems noteworthy. Further analysis concerning the function of specific poly-proline-stretch ligands for profilin in signal transduction, nuclear cytoplasmic traffic, or microfilament organization may unravel more subtle differences that were not seen in our assays.
In striking contrast to the effects seen with PFN1/R88L and PFN1/H133S were the consequences of introducing PFN1/Y59A into CAL51 cells. The stable clones expressing this construct had overall profilin levels well comparable with nontumorigenic lines either reconstituted with wild-type profilin (CAL/17-5) or expressing equivalent levels of endogenous profilin (MCF-10A); yet, they displayed properties comparable with the parental or mock-transfected CAL51 tumorigenic lines. A critical level of profilin with actin binding ability for normal cell viability and proliferation was also found in the fission yeast (Lu and Pollard, 2001), and our data obtained with PFN1/Y59A in CAL51 cells illuminate the importance of profilin's actin binding site for cell adhesion and differentiation of human epithelial cells.
It is tempting to speculate that it is indeed the binding of profilin 1 to G-actin that is critical in this respect. There is a wealth of data revealing the importance of profilin for actin polymerization and dynamics as required for microfilament organization of adherent and motile vertebrate cells (cf. Schlüter et al., 1997; Holt and Koffer, 2001; Pollard and Borisy, 2003). In addition, there is recent evidence that profilin–actin interaction is necessary for the export of nuclear actin (Stüven et al., 2003). However, the actin binding site on profilin is not stringently specific for binding to actin. The same binding site is involved in interacting with the actin-related protein Arp2 (Machesky, 1997; Mullins et al., 1998) which, as part of the Arp2/3 complex, is essential for the initiation and regulation of actin polymerization in the cortical region of motile cells (Pollard et al., 2001; Pollard and Borisy, 2003). Another ligand for the same binding site is gephyrin, a protein essential for molybdenum cofactor synthesis (Stallmeyer et al., 1999; Giesemann et al., 2003). Consequently, lack of a functional actin binding site in profilin may interfere with several processes, like microfilament organization, cell adhesion, nuclear-cytoplasmic transport, and metabolism which are all critical for epithelial differentiation. Furthermore, considering that profilin is not the only actin binding protein classified as a tumor suppressor protein (see above), it is conceivable that profilin has to act in concert with other microfilament proteins in epithelial differentiation. Hence, although we demonstrate that the actin binding site in profilin 1 plays an important role in human epithelial differentiation, further studies will be needed to provide an explanation for this phenomenon on a molecular level.
Acknowledgments
We thank Renate Frege (MDC Berlin-Buch), Eva Saxinger, Nicola Brandt, and Kathrin Schloen (Technical University of Braunschweig) for expert technical help and Dr. S. Seitz (Max-Delbrück-Center, Berlin-Buch) for help with photographic art work. Horst Hinssen (University of Bielefeld) is gratefully acknowledged for a gift of pig plasma gelsolin, and Wolfgang Ziegler (Technical University of Braunschweig) for helpful discussions concerning statistical data evaluation. This work was financially supported by the Deutsche Krebshilfe/Mildred Scheel Stiftung (10-2048-Ja1) and the Fonds der Chemischen Industrie (to B.M.J.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–12–0873. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–12–0873.
Abbreviations used: Arp, actin-related protein; PFN1, profilin 1; PIP2; phospho-inositol-4,5-bisphosphate.
References
- Aderem, A. (1992). Signal transduction and the actin cytoskeleton: the roles of MARCKS and profilin. Trends Biochem. Sci. 17, 438-443. [DOI] [PubMed] [Google Scholar]
- Asch, H.L., Head, K., Dong, Y., Natoli, F., Winston, J.S., Connolly, J.L., and Asch, B.B. (1996). Widespread loss of gelsolin in breast cancers of humans, mice, and rats. Cancer Res. 56, 4841-4845. [PubMed] [Google Scholar]
- Ben Ze'ev, A. (1997). Cytoskeletal and adhesion proteins as tumor suppressors. Curr. Opin. Cell Biol. 9, 99-108. [DOI] [PubMed] [Google Scholar]
- Bjorkegren, C., Rozycki, M., Schutt, C.E., Lindberg, U., and Karlsson, R. (1993). Mutagenesis of human profilin locates its poly(L-proline)-binding site to a hydrophobic patch of aromatic amino acids. FEBS Lett. 333, 123-126. [DOI] [PubMed] [Google Scholar]
- Bjorkegren-Sjogren, C., Korenbaum, E., Nordberg, P., Lindberg, U., and Karlsson, R. (1997). Isolation and characterization of two mutants of human profilin I that do not bind poly(L-proline). FEBS Lett. 418, 258-64. [DOI] [PubMed] [Google Scholar]
- Buss, F., Temm-Grove, C., Henning, S., and Jockusch, B.M. (1992). Distribution of profilin in fibroblasts correlates with the presence of highly dynamic actin filaments. Cell Motil. Cytoskeleton 22, 51-61. [DOI] [PubMed] [Google Scholar]
- Carlsson, L., Nyström, N.E., Sundkvist, I., Markey, F., and Lindberg, U. (1977). Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J. Mol. Biol. 115, 465-483. [DOI] [PubMed] [Google Scholar]
- Dong, J., Radau, B., Otto, A., Muller, E., Lindschau, C., and Westermann, P. (2000). Profilin I attached to the Golgi is required for the formation of constitutive transport vesicles at the trans-Golgi network. Biochim. Biophys. Acta 1497, 253-260. [DOI] [PubMed] [Google Scholar]
- Giesemann, T., Rathke-Hartlieb, S., Rothkegel, M., Bartsch, J.W., Buchmeier, S., Jockusch, B.M., and Jockusch, H. (1999). A role for polyproline motifs in the spinal muscular atrophy protein SMN: profilins bind to and colocalize with SMN in nuclear gems. J. Biol. Chem. 274, 37908-37914. [DOI] [PubMed] [Google Scholar]
- Giesemann, T., Schwarz, G., Nawrotzki, R., Berhörster, K., Rothkegel, M., Schlüter, K., Schrader, N., Schindelin, H., Mendel, R.R., and Jockusch, B.M. (2003). Complex formation between the postsynaptic scaffolding protein gephyrin, profilin and Mena: a possible link to the microfilament system. J. Neurosci. 23, 8330-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glück, U., Kwiatkowski, D.J., and Ben-Ze'ev, A. (1993). Suppression of tumorigenicity in simian virus 40-transformed 3T3 cells transfected with alpha-actinin cDNA. Proc. Natl. Acad. Sci. USA 90, 383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, P.J., Furman, M.I., Wachsstock, D., Safer, D., Nachmias, V.T., and Pollard, T.D. (1992). The control of actin nucleotide exchange by thymosin beta 4 and profilin. A potential regulatory mechanism for actin polymerization in cells. Mol. Biol. Cell 3, 1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, P.J., Machesky, L.M., Baldassare, J.J., and Pollard, T.D. (1990). The actin-binding protein profilin binds to PIP2 and inhibits its hydrolysis by phospholipase C. Science 247, 1575-1578. [DOI] [PubMed] [Google Scholar]
- Haugwitz, M., Noegel, A.A., Karakesisoglou, J., and Schleicher, M. (1994). Dictyostelium amoebae that lack G-actin-sequestering profilins show defects in F-actin content, cytokinesis, and development. Cell 79, 303-314. [DOI] [PubMed] [Google Scholar]
- Holt, M.R., and Koffer, A. (2001). Cell motility: proline-rich proteins promote protrusions. Trends Cell Biol. 11, 38-46. [DOI] [PubMed] [Google Scholar]
- Hüttelmaier, S., Mayboroda, O., Harbeck, B., Jarchau, T., Jockusch, B.M., and Rüdiger, M. (1998). The interaction of the cell-contact proteins VASP and vinculin is regulated by phosphatidylinositol-4,5-bisphosphate. Curr. Biol. 8, 479-488. [DOI] [PubMed] [Google Scholar]
- Janke, J., Schluter, K., Jandrig, B., Theile, M., Kolble, K., Arnold, W., Grinstein, E., Schwartz, A., Estevez-Schwarz, L., Schlag, P. M. et al. (2000). Suppression of tumorigenicity in breast cancer cells by the microfilament protein profilin 1. J. Exp. Med. 191, 1675-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs, A., Rothkegel, M., Klar, M., and Jockusch, B.M. (2001). Characterization of functional domains of mDia1, a link between the small GTPase Rho and the actin cytoskeleton. J. Cell Sci. 114, 3663-3672. [DOI] [PubMed] [Google Scholar]
- Lambrechts, A., Jonckheere, V., Dewitte, D., Vandekerckhove, J., and Ampe, C. (2002). Mutational analysis of human profilin I reveals a second PIP2 binding site neighbouring the poly(L-proline)binding site. BMC Biochem. 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts, A., Verschelde, J.L., Jonckheere, V., Goethals, M., Vandekerckhove, J., and Ampe, C. (1997). The mammalian profilin isoforms display complementary affinities for PIP2 and proline-rich sequences. EMBO J. 16, 484-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassing,I.,andLindberg,U.(1985).Specificinteractionbetweenphosphatidylinositol 4,5-bisphosphate and profilactin. Nature 314, 472-474. [DOI] [PubMed] [Google Scholar]
- Lindberg, U., Schutt, C.E., Hellsten, E., Tjader, A.C., and Hult, T. (1988). The use of poly(L-proline)-Sepharose in the isolation of profilin and profilactin complexes. Biochim. Biophys. Acta 967, 391-400. [DOI] [PubMed] [Google Scholar]
- Lu, J., and Pollard, T.D. (2001). Profilin binding to poly-L-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol. Biol. Cell 12, 1161-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky, L.M. (1997). Cell motility: complex dynamics at the leading edge. Curr. Biol. 7, R164-R167. [DOI] [PubMed] [Google Scholar]
- Machesky, L.M., and Pollard, P.D. (1993). Profilin as a potential mediator of membrane-cytoskeleton communication. Trends Cell Biol. 3, 381-385. [DOI] [PubMed] [Google Scholar]
- Magdolen, V., Oechsner, U., Muller, G., and Bandlow, W. (1988). The intron-containing gene for yeast profilin (PFY) encodes a vital function. Mol. Cell. Biol. 8, 5108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayboroda, O., Schlüter, K., and Jockusch, B.M. (1997). Differential colocalization of profilin with microfilaments in PtK2 cells. Cell Motil. Cytoskeleton 37, 166-177. [DOI] [PubMed] [Google Scholar]
- Mullins, R.D., Heuser, J.A., and Pollard, T.D. (1998). The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA 95, 6181-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oien, K.A., Vass, J.K., Downie, I., Fullarton, G., and Keith, W.N. (2003). Profiling, comparison and validation of gene expression in gastric carcinoma and normal stomach. Oncogene 22, 4287-4300. [DOI] [PubMed] [Google Scholar]
- Pantaloni, D., and Carlier, M.F. (1993). How profilin promotes actin filament assembly in the presence of thymosin beta 4. Cell 75, 1007-1014. [DOI] [PubMed] [Google Scholar]
- Perelroizen, I., Didry, D., Christensen, H., Chua, N.H., and Carlier, M.F. (1996). Role of nucleotide exchange and hydrolysis in the function of profilin in actin assembly. J. Biol. Chem. 271, 12302-12309. [DOI] [PubMed] [Google Scholar]
- Petersen, O.W., Ronnov-Jessen, L., Howlett, A.R., and Bissell, M.J. (1992). Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc. Natl. Acad. Sci. USA 89, 9064-9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T.D., Blanchoin, L., and Mullins, R.D. (2001). Actin dynamics. J. Cell Sci. 114, 3-4. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D., and Borisy, G.G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 113, 549. [DOI] [PubMed] [Google Scholar]
- Reinhard, M., Giehl, K., Abel, K., Haffner, C., Jarchau, T., Hoppe, V., Jockusch, B.M., and Walter, U. (1995). The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 14, 1583-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Fernandez, J.L., Geiger, B., Salomon, D., Sabanay, I., Zoller, M. and Ben-Ze'ev, A. (1992). Suppression of tumorigenicity in transformed cells after transfection with vinculin cDNA. J. Cell Biol. 119, 427-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdiger, M., Jockusch, B.M., and Rothkegel, M. (1997). An epitope tag-antibody combination useful for the detection of protein expression in procaryotic and eukaryotic cells. Biotechniques 23, 96-97. [DOI] [PubMed] [Google Scholar]
- Schägger, H., and Jagow, G.V. (1987). Tricine sodium dodecylsulfate polyacrylamide gel electrophoresis for the separation of proteins in the range from 1–100 kDa. Anal. Biochem. 166, 368-379. [DOI] [PubMed] [Google Scholar]
- Schlüter, K., Jockusch, B.M., and Rothkegel, M. (1997). Profilins as regulators of actin dynamics. Biochim. Biophys. Acta 27, 97-109. [DOI] [PubMed] [Google Scholar]
- Schlüter, K., Schleicher, M., and Jockusch, B.M. (1998). Effects of single amino acid substitutions in the actin-binding site on the biological activity of bovine profilin I. J. Cell Sci. 111, 3261-3273. [DOI] [PubMed] [Google Scholar]
- Skare, P., and Karlsson, R. (2002). Evidence for two interaction regions for phosphatidylinositol (4,5)-bisphosphate on mammalian profilin I. FEBS Lett. 522, 119-124. [DOI] [PubMed] [Google Scholar]
- Skare, P., Kreivi, J.P., Bergstrom, A., and Karlsson, R. (2003). Profilin I colocalizes with speckles and Cajal bodies: a possible role in pre-mRNA splicing. Exp. Cell Res. 286, 12-21. [DOI] [PubMed] [Google Scholar]
- Sohn, R.H., Chen, J., Koblan, K.S., Bray, P.F., and Goldschmidtclermont, P.J. (1995). Localization of a binding site for phosphatidylinositol 4,5-bisphosphate on human profilin. J. Biol. Chem. 270, 21114-21120. [DOI] [PubMed] [Google Scholar]
- Stallmeyer, B., Schwarz, G., Schulze, J., Nerlich, A., Reiss, J., Kirsch, J., and Mendel, R.R. (1999). The neurotransmitter receptor-anchoring protein gephyrin reconstitutes molybdenum cofactor biosynthesis in bacteria, plants, and mammalian cells. Proc. Natl. Acad. Sci. USA 96, 1333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stüven, T., Hartmann, E., and Görlich, D. (2003). Exportin 6, a novel nuclear export receptor that is specific for profilin-actin complexes. EMBO J. 22, 5928-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, A.I., et al. (2002). Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. USA 99, 4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu, S., Miki, H., and Takenawa, T. (1998). The essential role of profilin in the assembly of actin for microspike formation. EMBO J. 17, 6516-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, M., and Shibata, H. (1985). Poly(L-proline)-binding proteins from chick embryos are a profilin and a profilactin. Eur. J. Biochem. 151, 291-297. [DOI] [PubMed] [Google Scholar]
- Theile, M., Hartmann, S., Scherthan, H., Arnold, W., Deppert, W., Frege, R., Glaab, F., Haensch, W., and Scherneck, S. (1995). Suppression of tumorigenicity of breast cancer cells by transfer of human chromosome 17 does not require transferred BRCA1 and p53 genes. Oncogene 10, 439-447. [PubMed] [Google Scholar]
- Vandekerckhove, J., Bauw, G., Vancampernolle, K., Honore, B., and Celis, J. (1990). Comparative two-dimensional gel analysis and microsequencing identifies gelsolin as one of the most prominent downregulated markers of transformed human fibroblast and epithelial cells. J. Cell Biol. 111, 95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen, E.M., and Cooley, L. (1994). Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development 120, 717-728. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., Madaule, P., Reid, T., Ishizaki, T., Watanabe, G., Kakizuka, A., Saito, Y., Nakao, K., Jockusch, B.M., and Narumiya, S. (1997). p140mDia, a mammalian homologue of Drosophila diaphanous, is a target protein for rho small GTPases and is a ligand for profilin. EMBO J. 16, 3044-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witke, W., Sharpe, A.H., and Kwiatkowski, D.J. (1993). Profilin deficient mice are not viable. Mol. Biol. Cell 4, 149a. [Google Scholar]
- Witke, W., Sutherland, J.D., Sharpe, A., Arai, M., and Kwiatkowski, D.J. (2001). Profilin I is essential for cell survival and cell division in early mouse development. Proc. Natl. Acad. Sci. USA 98, 3832-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenmayer, N., Rothkegel, M., Jockusch, B.M., and Schlüter, K. (2000). Functional characterization of green fluorescent protein-profilin fusion proteins. Eur. J. Biochem. 267, 5247-5256. [DOI] [PubMed] [Google Scholar]