Abstract
Background
Molecular phylogenetic investigations have revolutionized our understanding of the evolutionary history of ferns—the second-most species-rich major group of vascular plants, and the sister clade to seed plants. The general absence of genomic resources available for this important group of plants, however, has resulted in the strong dependence of these studies on plastid data; nuclear or mitochondrial data have been rarely used. In this study, we utilize transcriptome data to design primers for nuclear markers for use in studies of fern evolutionary biology, and demonstrate the utility of these markers across the largest order of ferns, the Polypodiales.
Principal Findings
We present 20 novel single-copy nuclear regions, across 10 distinct protein-coding genes: ApPEFP_C, cryptochrome 2, cryptochrome 4, DET1, gapCpSh, IBR3, pgiC, SQD1, TPLATE, and transducin. These loci, individually and in combination, show strong resolving power across the Polypodiales phylogeny, and are readily amplified and sequenced from our genomic DNA test set (from 15 diploid Polypodiales species). For each region, we also present transcriptome alignments of the focal locus and related paralogs—curated broadly across ferns—that will allow researchers to develop their own primer sets for fern taxa outside of the Polypodiales. Analyses of sequence data generated from our genomic DNA test set reveal strong effects of partitioning schemes on support levels and, to a much lesser extent, on topology. A model partitioned by codon position is strongly favored, and analyses of the combined data yield a Polypodiales phylogeny that is well-supported and consistent with earlier studies of this group.
Conclusions
The 20 single-copy regions presented here more than triple the single-copy nuclear regions available for use in ferns. They provide a much-needed opportunity to assess plastid-derived hypotheses of relationships within the ferns, and increase our capacity to explore aspects of fern evolution previously unavailable to scientific investigation.
Introduction
Over the past twenty years, molecular phylogenetic approaches have radically altered our understanding of relationships in the fern tree of life. Arguably the most important finding (and among the most contentious) is that ferns, including the horsetails (Equisetum) and whisk ferns (Psilotaceae), form a clade sister to seed plants [1]. Ferns are therefore one of the great vascular plant radiations; only the angiosperm clade has more extant species. Broad studies of fern phylogeny, e.g., [1-18] have increasingly found stronger resolution and support across the majority of the backbone nodes, many of which were unanticipated based on morphological data. This rewriting of fern phylogeny has resulted in a novel emerging consensus of deep fern relationships [19-22]. Similarly, new molecular data have greatly facilitated inquiries into fern relationships at much finer scales, such as within genera, e.g., [23-46].
The vast majority of these studies, however, have been limited to data from the plastid genome; nuclear and mitochondrial data have not been widely used, cf. [4,9,47-49]. This dependence upon plastid data reflects a general absence of genomic resources available for ferns [50-53]—for example, no fern mitochondrial or nuclear genome has been sequenced yet—which has impeded the development of novel markers. To date only seven nuclear regions have been used in fern phylogeny investigations: ITS [13,54-57]; ribosomal 18S [4,58]; LEAFY [59-62]; gapCpSh (gapCp “short”) [36,42,43,49,57,63-67]; gapCpLg (gapCp “long”) [67]; cam [67]; and pgiC [57,59,66,68-75].
This strong reliance on the plastid genome makes fern phylogenetics vulnerable to misleading inferences, such as failures of this linkage group to track the organismal divergences (e.g., due to deep coalescence or reticulation). In addition, plastid data are poorly suited for species-level work in the many fern groups that have reticulate evolutionary histories [76-79]. Polyploidy and hybridization are common in ferns [80], and fully unraveling relationships in these groups will require the development of multiple unlinked markers. Regions from the nucleus are particularly attractive for this purpose because that genome has multiple linkage groups that are expected to have an elevated rate of evolution in ferns, e.g., [49].
The recent sequencing of approximately 1000 green plant transcriptomes by the One Thousand Plants Project (1KP; onekp.com) provides an unprecedented opportunity to facilitate the development of novel low-copy nuclear markers for use in ferns. There is no fern nuclear genome that has been sequenced to date, and only a handful of EST libraries, sequenced plastomes, or transcriptomes, e.g., [81-89]. Included in the 1KP sampling (as of January 2013, when we finished the sampling for this project) are 62 fern accessions, comprising 60 unique species. Our sampling from this time point is particularly rich in members of the leptosporangiate order Polypodiales, especially Pteridaceae (sensu [20,90]) and eupolypods II (sensu [8,16,19,91]), but also includes representatives of each of the major eusporangiate clades (Ophioglossales, Psilotales, Equisetales, Marattiales), as well as each of the leptosporangiate orders, except for the Osmundales [20]. Additional taxa, including representatives of the Osmundales, were sequenced in the 1KP project after we had finished our sampling. The full list—including algae, bryophytes, lycophytes, ferns, and seed plants—is available at http://www.onekp.com/samples/list.php.
Here, we utilize these transcriptome data to design primers for 20 nuclear markers across ten protein-coding genes. Our primary goals are: 1) to provide primers that will amplify single-copy nuclear markers across the majority of the Polypodiales; 2) to demonstrate the relative success of those primers in amplifying the desired region from genomic DNA, using a test set of 15 diploid Polypodiales species, and; 3) to characterize the resulting sequences and their efficacy in inferring relationships at various phylogenetic depths. In addition, we provide transcriptome alignments—curated broadly across ferns—for each of our target loci (including closely related paralogs in the case of gene families) to assist other investigators in designing primers for fern taxa of interest outside of the Polypodiales.
Results
Primer Development
We developed primer pairs for 20 regions across a total of ten distinct single-copy protein-coding genes: ApPEFP_C, cryptochrome 2, cryptochrome 4, DET1, gapCpSh, IBR3, pgiC, SQD1, TPLATE, and transducin (Tables 1, 2, 3; Figure 1). Each primer pair successfully amplifies the majority of taxa in our genomic DNA test set (comprising DNA from 15 diploid Polypodiales species; Table 3; Figure 2; Appendix S1). In general, we only attempted to sequence PCR products that had strong single bands (viewed with agarose gel electrophoresis). Many of the missing sequences are likely to be attainable by applying cloning protocols (e.g., see 64). Those sequences that we did attain by cloning are noted in the Methods section and in Table 3.
Table 1. Summary of the genes for which we designed primers.
| Length (CDS; in bp) |
Arabidopsis
|
||||||
|---|---|---|---|---|---|---|---|
| Abbreviation | Protein Name | Arabid. | Ferns | TAIR Gn# | # of Introns | Chromo. # | |
| 1 | ApPEFP_C | appr-1-p processing enzyme family protein | 1689 | ~1650-1743 | AT1G69340 | 13 | 1 |
| 2 | CRY2 | cryptochrome 2 | 2046 | ~2000 | AT4G08920 | 3 | 4 |
| 3 | CRY4 | cryptochrome 4 | 1839 | ~2100 | AT1G04400 | 3 | 1 |
| 4 | DET1 | Nuclear-localized regulator of plant development | 1632 | ~1600-2700 | AT4G10180 | 9 | 4 |
| 5 | gapCpSh | Plastid-localized GAPDH, short copy | 1266; 1260 | 1315 | AT1G79530; AT1G16300 | 13 | 1 |
| 6 | IBR3 | IBA-Response 3 (acyl-CoA dehydrogenase) | 2475 | ~2445-2490 | AT3G06810 | 16 | 3 |
| 7 | pgiC | glucose-6-phosphate isomerase / sugar isomerase family protein | 1683 | --a | AT5G42740 | 21 | 5 |
| 8 | SQD1 | Sulfoquinovosyldiacylglyerol 1 | 1431 | ~1515-1521 | AT4G33030 | 1 | 4 |
| 9 | TPLATE | a cytokinesis protein targeted to the cell plate | 3531 | --a | AT3G01780 | 6 | 3 |
| 10 | transducin | transducin family protein / WD-40 repeat family protein | 2868 | --a | AT3G21540 | 11 | 3 |
For each gene, we list its length in ferns and in Arabidopsis, provide the TAIR accession number for the Arabidopsis sequence (as well as its number of introns and chromosomal position). The TreeBASE accession number for our “all-in” fern alignments is S14616. Comparisons with Arabidopsis thaliana are based on the most closely related homolog(s). a These loci were trimmed to a focal region prior to completion, so the full length of the coding DNA sequence (CDS) is unknown.
Table 2. Priming details for 20 novel nuclear markers.
|
Primers (Forward, Reverse)
|
||||
|---|---|---|---|---|
| Protein Region | Name | Sequence (5'–3') | PCR Program | |
| ApPEFP_C | 1 | 4218Cf4, 4218Cr12 | GGACCTGGSCTYGCTGARGAGTG, GCAACRTGAGCAGCYGGTTCRCGRGG | 6512035 |
| ApPEFP_C | 1a | 4218Cf4, 4218Cr3 | GGACCTGGSCTYGCTGARGAGTG, TCGTAAGCRTTYGTTACTTTDGCC | 5506035 |
| ApPEFP_C | 1b | 4218Cf6, 4218Cr6 | AAAGTTATACATACTGTTGGTCC, GCAACATGAGCAGCTGGTTCACGAGG | 5506035 |
| ApPEFP_C | 2 | 4218f25, 4218r7 | AATGCTCTRAGTCAYTGYTAYMGATC, TTGTAAATCTCTGTRTCRGATGYYGT | 5509035 |
| ApPEFP_C | 3 | 4218f26, 4218r13 | CAAAGGCGCAARGAACARTGGGARAGRGTTGC, TCAAGACAYCGTAGCAGRAARTGBGCYCC | 6512035 |
| CRY2 | 1 | CRY2F3289_Pt, CRY2R3838_Pt | AGGATGARYTGGAGAAAGGYAGCAATG, GTRTCCCAGAAATAYTTCATACCCC | 5209035 |
| CRY4 | 1 | CRY2F3289_Pt, CRY2R3838_Pt | AGGATGARYTGGAGAAAGGYAGCAATG, GTRTCCCAGAAATAYTTCATACCCC | 5209035 |
| DET1 | 1 | det1-335all, det1-906all | TATGAYGTGGARTGCCCAGAT, TCTCTGCAGAAHKGYCCAA | 5506035 |
| gapCpSh | 1 | gapCpShF1, gapCpShR2 | TGCACMACHAACTGCCTTGCRCCBCTTGC, CCATTYARCTCTGGRAGCACCTTTCC | 6512035 |
| IBR3 | 1 | 4321F2, 4321R2 | TCTGCMCATGCMATTGAAAGAGAG, CCCARKGTYGAAAGYTCCCAATC | 6312035 |
| IBR3 | 2 | 4321F5, 4321R6 | ATGACYGAACCAGATGTKGCDTCVTCRGATGC, TGRTGGAGYCTKCCTGGGCCTA | 6512035 |
| pgiC | 1 | pgic_1156F, pgiC_1900R | GGYCTYYTRAGYGTYTGGAATGT, GGTGAAATYGAYTTYGGDGARC | 5812035 |
| SQD1 | 1 | EMSQD1E1F6, EMSQD1E1R2 | GCAAGGGTACHAAGGTHATGATCATAGG, CCTTTDCCRTARACTGTAAGAGGATG | 5512035 |
| SQD1 | 1a | EMSQD1E1F6, EMSQD1E1R4 | GCAAGGGTACHAAGGTHATGATCATAGG, GCGTGARTCRTGCACTTTGCTRAGATG | 5512035 |
| SQD1 | 2 | EMSQD1E2F4, EMSQD1E2R8 | CGHGTRTTYAATCARTTYACAGAAC, GTCACTGTHACAGGTTTYACDCCAGC | 5512035 |
| TPLATE | 1 | 6560_1630F, 6560_2329R | TGCYTAGTSGARAGYTGYTTTCA, AATGTAGCAACTAACAGGCTTCAAGA | 5812035 |
| TPLATE | 2 | 6560_3136F, 6560_3686R | AAYCTYCARCATCTYCAGTCTC, GCAACKGCHGCDGTBGAAAG | 5812035 |
| transducin | 1 | 6928_850F, 6928_1357R | TTRCGBGGRCAYARAGATCA, GGAWCSTTARTSGGYTGCCAA | 5812035 |
| transducin | 2 | 6928_1955F, 6928_2816R | AAGGCDGGRAARCTNGAGAT, ATGGAYATYTCCWCYGATGC | 5812035 |
| transducin | 3 | 6928_3406F, 6928_3802R | TCBATTCGRMGATGGGAGCG, CAAACYCARGARWCYSTGAC | 5812035 |
The first two digits of the PCR program is the annealing temperature, followed by a three-digit elongation time (in seconds), followed by the number of cycles.
Table 3. Sequence characteristics for the single-copy regions developed in this study.
|
# of Differences
|
Length of Amplified Region
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein Region | Cys. pair | Poly. pair | Cya. | Lin. | Sac. | Adi. | Che. | Cry. | Den. | Dry. | P.am. | P.gl. | Ath. | C.bu. | C.pr. | The. | Woo. | |
| ApPEFP_C | 1 | >38; >25a | 50; 25b | ? | ? | >751 | >690 | >829 | 721 | 931 | 761 | 846; >871c | 843; 837c | >849 | >866 | 932; 934c | 687 | 939 |
| ApPEFP_C | 1a | >13; >12a | 10; 9b | 145 | ? | 258d | 321d | 281 | 300d | 293 | 267 | ~257 | ~255 | 201d | 245 | 161 | >225 | 255 |
| ApPEFP_C | 1b | 6; 6a | 11; 3b | ? | ? | ? | 224 | 209d | 223d | 269 | 223 | 224; 223d | 216; 223d | 224 | 214d | 223d | 223 | 222 |
| ApPEFP_C | 2 | 5 | ? | 382 | 334 | 360 | 360 | 340 | 357 | 406 | 359 | ? | ? | 360 | 359 | 358 | 359 | 357 |
| ApPEFP_C | 3 | 14 | 11; 25 | ? | 377 | ? | >506 | >776 | 770 | 802 | 454 | 385 | 378; 384c | 490 | 482 | 461c | 489 | 457 |
| CRY2 | 1 | 9 | 14 | 516 | 516 | 516 | 516 | 516 | 516 | ? | 516 | 516 | 516 | 516 | 516 | 516 | 516 | 516 |
| CRY4 | 1 | 2 | ? | ? | 516 | 516 | 516 | 516 | ? | 516 | 516 | ? | 516 | 516 | 516 | 516 | 516 | ? |
| DET1 | 1 | ~5 | 6 | ? | ? | ? | ~630 | ? | ? | ? | 667 | 668 | 668 | ~670 | 665 | 664 | 669 | 669 |
| gapCpSh | 1 | ? | 14 | ? | 455 | 459 | ? | ? | 482 | 476 | 522c | 531 | 525 | ? | ? | 466 | 517 | 592 |
| IBR3 | 1 | >16 | 29; 31 | ? | ? | 870 | 817 | >700 | 827 | 836 | 819; 828c | 843c | 844 | 815 | >819 | 840 | >910 | 821 |
| IBR3 | 2 | ~6 | ~21 | ~600 | >766 | 611 | ~574 | 581 | 568 | ~1196 | ? | ~590 | 595 | 586 | ~580 | ~582 | 579 | 588 |
| pgiC | 1 | >32 | >19 | 674 | ? | ? | ? | ? | ? | ? | 625 | >615 | 678 | >474 | >664 | >581 | 619 | 620 |
| SQD1 | 1 | 10 | >8e | ? | 700 | 668 | 700 | 700 | 700 | 700 | 700 | ? | 700 | 700 | 700 | 700 | 700 | 685 |
| SQD1 | 1a | 8 | 8 | 530 | 530f | 530f | 530f | 530f | 530f | 530f | 530f | 529 | 530f | 530f | 530f | 530f | 530f | 530f |
| SQD1 | 2 | 1 | 3 | 264 | 263 | 264 | 264 | 256 | 264 | 263 | 264 | 264 | 264 | 264 | 264 | 233 | 264 | ? |
| TPLATE | 1 | 12 | 14 | 719 | >657 | ? | 646 | >561 | >529 | 627 | 711 | 696 | 698 | >638 | 710 | >662 | >692 | 687 |
| TPLATE | 2 | 5 | 10 | >327 | ? | ? | ? | 551 | ? | 529 | 512 | 497 | 493 | 302 | 427 | 424 | 722 | 541 |
| transducin | 1 | 11 | ? | ? | ? | ? | 402 | 397 | 426 | 416 | ? | >381 | ? | 436 | 437 | 435 | 420 | 421 |
| transducin | 2 | 11 | >7 | >521 | ? | ? | ? | >426 | ? | 539 | 534 | 529 | >445 | 518 | 525 | 517 | 514 | 504 |
| transducin | 3 | 6 | 7 | ? | ? | ? | 227 | 308 | 231 | ? | 268 | 251 | 251 | 242 | 244 | 244 | 243 | 242 |
Cys: Cystopteris, Poly: Polypodium, Cya: Cyatheales, Lin.: Lindsaea, Sac.: Saccoloma, Adi: Adiantum, Che: Cheilanthes, Cry: Cryptogramma, Den: Dennstaedtia, P.am.: Polypodium amorphum, P.gly.: Polypodium glycyrrhiza, Ath.: Athyrium, C.bu.: Cystopteris bulbifera, C.pr.: Cystopteris protrusa, The: Thelypteris, Woo: Woodsia. a The two values come from comparing the single incomplete Cystopteris bulbifera sequence against two sequences cloned from C. protrusa. b This locus has a duplication in Polypodium; these values are the number of bp changes between each of the ortholog pairs. c Required cloning. d These lengths are derived from the corresponding portion of the APPEFP_C Region 1 alignment (we did not attempt to amplify Region 1a or 1b for all taxa). e For P. amorphum we were not able to amplify Region 1 for this locus, only Region 1a. f These lengths are derived from the corresponding portion of the SQD1 Region 1a alignment (Region 1a for all taxa).
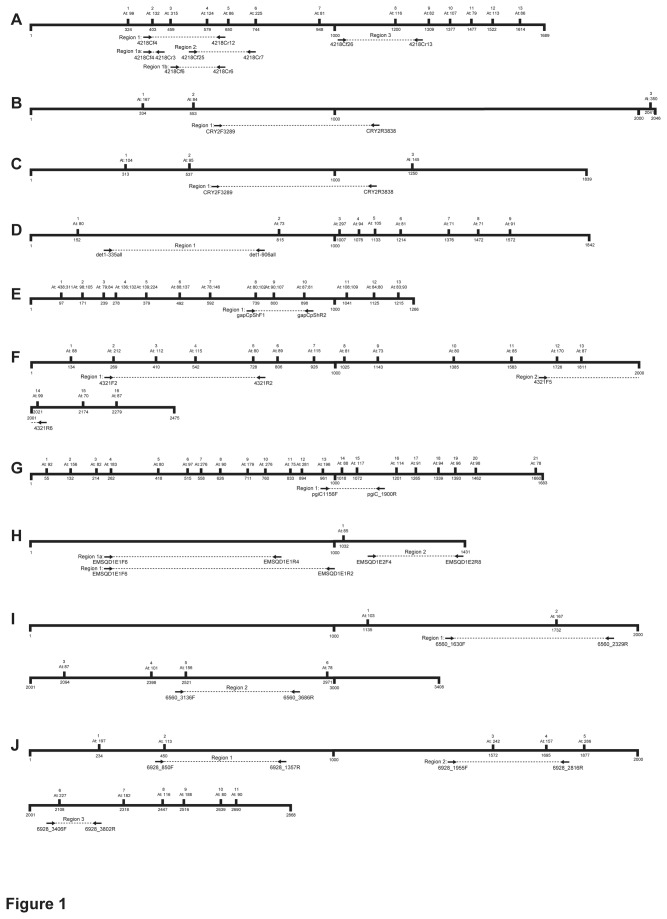
Figure 1. Schematic diagrams of the ten nuclear genes for which we developed fern-specific primers.
(A) ApPEFP_C; (B) CRY2; (C) CRY4; (D) DET1; (E) gapCpSh; (F) IBR3; (G) pgiC; (H) SQD1; (I) TPLATE; (J) transducin. Each subset of the figure represents one protein-coding locus, using the most closely related Arabidopsis thaliana homolog as the template. The coding sequence is measured (in base pairs) along the bottom of the thickened horizontal line, with each locus wrapping onto a new line every 2000 base pairs, when necessary. Intron location, number, and length (in base pairs in Arabidopsis) are given above the line. Also shown below the line are the priming locations for each of the markers we developed. For gapCpSh, intron locations are based on Arabidopsis gapCp1: the first two exons of Arabidopsis gapCp2 are each one codon shorter than in gapCp1.
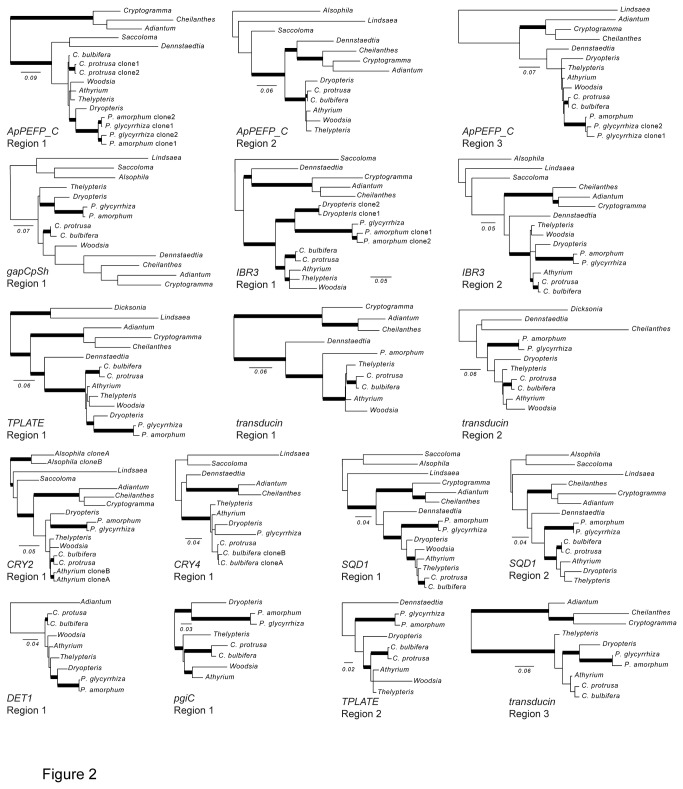
Figure 2. Maximum likelihood phylograms for each region, including only those taxa that were successfully sequenced from our 15-taxon genomic DNA test set.
Bold branches indicate strong support (≥70% bootstrap support). Scale bars are in units of substitutions per site. In the taxon names, “C.” and “P.” refer to Cystopteris and Polypodium, respectively. These phylograms are unrooted, but oriented as if rooted by the Cyatheales (or our best guess, when the Cyatheales accession did not sequence successfully), when space permits.
ApPEFP_C
We developed primers for three regions of ApPEFP_C (Figure 1A), and for one of those regions (Region 1) we designed non-overlapping internal primers that can amplify two smaller subset regions (Figure 1A). Region 1 is approximately 700–1000bp long in ferns (Table 3) and spans introns 2, 3, and 4, exons 3 and 4, and half of exon 5 (Figure 1A). It could be direct-sequenced for most taxa in our genomic DNA test set, although cloning was necessary for Polypodium (due to a hypothesized gene duplication in the Polypodiaceae; see Figure 2) and Cystopteris protrusa. Within Region 1, the additional reverse primer 4218Cr3 allows for the amplification of the subset Region 1a, and the forward primer 4218Cf6 yields Region 1b (Figure 1A). Both these smaller regions are approximately 200–300 bp long. Region 1a is the more variable of the two, whereas Region 1b has good length conservation among taxa and is easy to align across the complete breadth of the Polypodiales (Table 3). Region 2 overlaps with the 3’ end of Region 1; it includes a portion of exon 4, introns 4 and 5, exon 5, and most of exon 6 (Figure 1A). Finally, Region 3 is intermediate in length and includes much of the large exon 8, intron 8, and most of exon 9. In the eupolypods, Region 3 ranges in length from approximately 400 to 500 bp, but is consistently larger in the Pteridaceae and Dennstaedtiaceae (reaching 802 bp in Dennstaedtia). It amplified and direct-sequenced well (Figure 2), but required cloning for P. glycyrrhiza and C. protrusa (see above); however, the P. amorphum Region 3 sequence was clean (not double-peaked when directly sequenced), and did not require cloning.
CRY2 and CRY4
We designed primers to target a 516 bp region in the third exon of CRY2 (Figure 1B). However, these primers also cross-amplify the same region in CRY4 (also 516 bp; Figure 1C), and, less frequently, in CRY3 and CRY5 (see Figure S2). The PCR products thus cannot be directly sequenced. Nevertheless, after cloning, we recovered CRY2 and CRY4 for 14 and 11 (respectively) of the 15 taxa in our genomic DNA test set. These two loci are sufficiently divergent that assigning sequences to the correct copy is straightforward. CRY2 appears to have higher sequence variation, with nine nucleotide differences between the Cystopteris species pair, whereas there are only two nucleotide differences between the same species pair in CRY4 (Cystopteris protrusa and C. bulbifera constituted one of the two pairs of closely related species that we used as a metric for informativeness at shallow phylogenetic depths—see Methods and Table 3).
DET1
We focused on a single region of DET1 (Figure 1D). The primer pair 4321F2-4321R2 amplifies a ~670 bp region that includes most of the second exon (in Arabidopsis; ferns contain an additional intron within this region). All sequences were obtained by direct-sequencing.
GapCpSh
We designed primers for a single region of gapCpSh. The forward primer is situated just before intron 8 and the reverse priming site is just after intron 10 (Figure 1E); this region ranges in length from ~450 to 590 bp in our genomic DNA test set. In general GapCpSh amplified and direct-sequenced well, although we were not able to obtain clean sequences for Alsophila, Adiantum, Cheilanthes, Athyrium, or Cystopteris bulbifera (cloning not attempted) and the Dryopteris sequence required cloning. This region physically overlaps with the gapCp region amplified with the primers of Schuettpelz et al. [64], but differs in that our primers are specific to the gapCp Short (sensu [64]) copy in Polypodiales, and amplify a region slightly shorter than that of Schuettpelz et al. [64].
IBR3
We designed primers for two regions of IBR3. Region 1 spans introns 2–5 and exons 3–5 (Figure 1F); it is approximately 900bp long in the Polypodiales species we examined (Table 3). Region 2, at the 3’ end of the gene, is shorter, at around 600 bp in most species; however, it is much larger (1200 bp) in Dennstaedtia. It spans introns 12–14, exons 13 and 14, and the end of exon 12. Both regions amplified well, and gave clean direct sequences for the majority of taxa in our test set.
PgiC
We developed one novel primer pair for pgiC—a locus already known to work well in fern phylogenetics [57,66,71-75,92]. Our primers are situated in exons 14 and 16, amplifying introns 14, 15, and exon 15 (Figure 1G). The amplified region ranges in length between 600 and 700bp across our test set (Table 3), and the range of variation observed is appropriate for resolving infrageneric relationships (Table 3 and see citations above). It amplified and direct-sequenced well for 10 of the 15 test set taxa; Lindsaea, Saccoloma, Dennstaedtia, Cheilanthes and Adiantum failed amplify and/or direct-sequence cleanly.
SQD1
Primers were designed for two regions of SQD1: a 700 bp region within the first exon and a 264 bp region within the second exon (Figure 1H; Table 3). The Region 1 forward and reverse primers—EMSQDE1F6 and EMSQDE1R2—produced successful amplifications for 13 of the 15 taxa in our test set. An additional reverse primer, EMSQDE1R4, was designed to amplify a 530 bp subset (henceforth designated Region 1a; Figure 1H, Table 3), which resulted in successful amplification and direct-sequencing of the two remaining accessions (Alsophila and Polypodium amorphum). Primers designed for Region 2 resulted in the successful sequencing of all taxa in our test set, except Woodsia.
TPLATE
We designed primers for two regions of TPLATE (Figure 1I). Our primers for Region 1 were highly successful (only Saccoloma failed to amplify). It spans part of exon 2, all of intron 2, and part of exon 3, ranging in length among taxa in our test set from 650-720bp. Region 1 had moderate levels of variation (16 differences for the Cystopteris species pair, and 15 for Polypodium; Table 3). Region 2 is 400-550bp long (Table 3), and slightly less variable than Region 1. Its primers are situated in exon 5 and exon 6 and span intron 5 (Figure 1I). We managed to sequence this region for 11 of the 15 test set taxa (Adiantum, Cryptogramma, Lindsaea, and Saccoloma were unsuccessful). In our phyogenetic analyses of these data, we had to exclude Cheilanthes and Dicksonia because they were too divergent from the other taxa to align confidently.
Transducin
For transducin we designed primers for three regions. Region 1 extends from the 5’ end of intron 2 through to the middle of exon 3 (Figure 1J), and is approximately 400-450 bp long (Table 3). Amplification and sequencing was successful for 10 of our 15 test set species (Lindsaea, Saccoloma, Dicksonia, Dryopteris, and Polypodium glycyrrhiza were unsuccessful). Region 2 is approximately 550bp long, spanning exons 3 to 5 (Figure 1J, Table 3). It was amplified and sequenced successfully for 11 of the test set species (Adiantum, Cryptogramma, Lindsaea, and Saccoloma failed). Region 3 is 250-300 bp long, amplifying intron 6 and portions of exons 6 and 7 (Figure 1J, Table 3). It was successfully amplified and sequenced for all test set taxa except Lindsaea and Saccoloma.
Model selection and the Polypodiales phylogeny
The combined alignment of our 19 newly developed regions (SQD1 Region 1 and Region 1a were merged for these analyses) across our 15-taxon Polypodiales genomic DNA test set (the set of genomic DNAs that we used to test our new primer sets) is 9007 base pairs long; 42 percent of the sites are variable. Twenty-eight percent of the characters in this alignment are missing (i.e., gaps or question marks). We investigated five models for these data (where “model” refers to the product of the partitioning scheme and the substitution model applied to each subset of the data), which ranged from one subset and 37 free parameters to 30 subsets and 238 free parameters (see Methods; Table 4; Appendix S2). In their extremes, these models differed by over 2600 in their log likelihood scores, and by nearly 4500 Bayesian information criterion (BIC) points (Table 4). Model selection based on the BIC favored a relatively simple model for these data: four data subsets corresponding to the three codon positions and the noncoding sites, respectively, with the first two codon positions optimized under a GTR+G model and the two other subsets including an additional proportion invariant parameter (GTR+I+G; Table 4; Appendix S2). No parameters, other than relative branch lengths, were linked across partitions.
Table 4. Model comparison, by the Bayesian Information Criterion (BIC).
| Support Values by Branch (ML bootstrap percent) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | lnL | BIC | Subsets | Param. | A | B | C | D | E | F | G | H | I | J | K | L |
| 1: Pos. & Locus | -40574.4 | 83316.0 | 30 | 238 | 56 | 100 | 53 | 100 | 100 | 100 | 100 | 99 | 100 | 100 | 87 | 58 |
| 2a: Locus (each) | -42767.5 | 86819.0 | 19 | 141 | 49 | 100 | 74 | 100 | 100 | 100 | 100 | 96 | 100 | 100 | 88 | 61 |
| 2b: Locus (scheme) | -42748.0 | 86470.3 | 11 | 107 | 52 | 100 | 72 | 100 | 100 | 100 | 100 | 97 | 100 | 100 | 87 | 67 |
| 3: Pos. | -40875.4 | 82370.0 | 4 | 68 | 55 | 100 | 41 | 100 | 100 | 100 | 100 | 98 | 100 | 100 | 82 | 42 |
| 4: Unpartitioned | -43190.2 | 86717.3 | 1 | 37 | 66 | 100 | 61 | 100 | 100 | 100 | 100 | 95 | 100 | 100 | 80 | 41 |
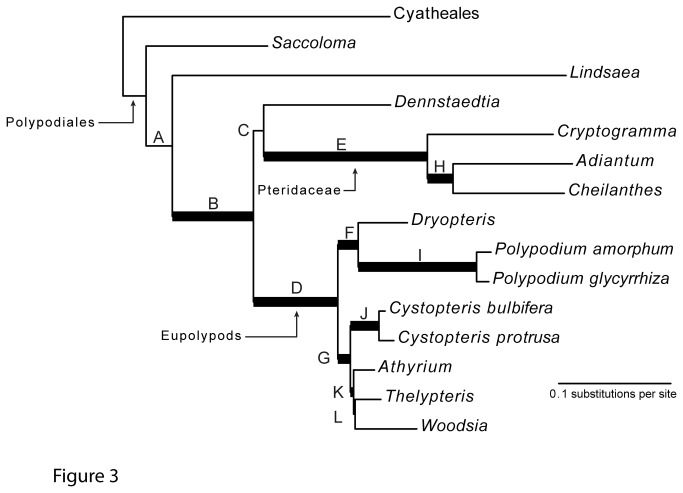
Values in bold face indicate strong support (≥70%). Branch designations (A – L) refer to Figure 3. Model 1 is the best PartitionFinder scheme given each codon position, for each locus, as the data blocks. In model 2a each locus gets its own partition, across codon positions. Model 2b is the best PartitionFinder scheme given the loci as the data blocks. Model 3 is partitioned by codon position, across loci. Model 4 is not partitioned. For substitution model parameterization, see Appendix S2. Subsets = the final number of subsets (“partitions”) for that model. Param. = number of free parameters.
Model parameterization had strong effects on the fit to our data and on our subsequent inference. In general, the models without a codon-position component to their partitioning schemes (the unpartitioned model—model 4, and the two models partitioned by locus—models 2a and 2b) performed very poorly. The addition of codon-position-based partitions dramatically improved model fit (Table 4), such that the subsequent addition of locus-based partitions resulted in a decline in model fit. For example, the BIC favored the simple by-position partitioning scheme (model 3: four subsets, 68 free parameters) over the best by-position-and-locus scheme (model 1: 30 subsets, 238 free parameters; Table 4).
Model choice impacted the ML estimate of topology, but only slightly: model 2a resolved Lindsaea as sister to the rest of the Polypodiales, whereas all other models put Saccoloma in that position. However, model choice had a stronger effect on support values. The most extreme example of this effect was branch C (Figure 3), which ranged from 41 percent ML bootstrap support under model 3 (our best-fitting model) to 74 percent support under model 2a (our worst-fitting model; Table 4).
Figure 3. Combined data maximum likelihood phylogram of our 15-taxon genomic DNA test set.
Analyses were performed under our best-fitting model (model 3, see Table 3). Bold branches indicate strong support (≥70% bootstrap support); internal branches are labeled A – L for ease of discussion.
Despite the high proportion of missing data, ML analyses of this alignment under our best-fitting model yielded a well supported phylogeny, with only three branches lacking strong support: Branch A (the earliest divergence in the Polypodiales), Branch C (the position of Dennstaedtia with respect to Pteridaceae and the eupolypods), and Branch L (the relationships among the three non-Cystopteridaceae eupolypod II accessions; branch labels refer to Figure 3). The transcriptome alignments themselves also contain rich phylogenetic data. The analysis of these data is beyond the scope of this paper, and is the focus of a forthcoming manuscript.
Discussion
Single-copy locus identification, and alignment inference
Our approach to single-copy locus identification and development was highly effective, albeit labor-intensive. Specifically, we combined repeated rounds of sequence-merging and tree-building for each of our candidate loci. This approach allowed us to build compact matrices (long reads for each accession with relatively little missing data) despite the fragmentary nature of the source assemblies. Our hands-on method of alignment development and curation also allowed us to identify novel gene duplication events, to distinguish among paralogs, and to detect both contaminants and misidentifications. We are therefore confident that our final alignments are both of high data quality (data density, taxon representation, and alignment inference quality) and high accuracy (free from contaminants, inter-paralog chimaeras, etc.; see Figures S1-S9). This approach was only possible given the modest amount of data that we worked with (the “moderate data approach”, see 6), and would not scale to large genomic datasets [93,94].
Nuclear genes with newly designed primer sets
ApPEFP_C is poorly characterized and does not appear to have a history as a phylogenetic marker; in Arabidopsis it is described simply as an “appr-1-p processing enzyme family protein” [95]. ApPEFP, formerly thought to be single-copy across much of the green plants (1KP data, Norman Wicket, pers. comm.), appears to have duplicated early within leptosporangiate ferns (Figure S1). We designated the pre-duplication version ApPEFP_A and the two post-duplication copies ApPEFP_B and ApPEFP_C, respectively. The ApPEFP_B/C duplication may have taken place early within leptosporangiate diversification; Dipteris and Lygodium each have both copies. Further sampling (particularly of the Osmundales) will be necessary to refine the timing of this duplication.
There appears to be an additional duplication of ApPEFP_A in the Equisetales, and probably at least one other duplication in the Marattiales (Figure S1). The ApPEFP_B phylogeny is well resolved, with no additional apparent duplications in this paralog. ApPEFP_C was the best represented in our transcriptome sampling, and is the only copy that we pursued for primer generation. Within ApPEFP_C there is an apparent duplication in the Polypodiaceae. This duplication occurred in the ancestry of Polypodium, after the divergence of Phlebodium and Pleopeltis (Figure S1).
CRY2 and CRY4 are members of the cryptochrome family of blue light photoreceptors, a gene family known in both prokaryotes and eukaryotes. In Arabidopsis, cryptochromes are responsible for circadian clock entrainment, flower induction, and de-etiolation [96,97]. Their function in ferns is not entirely clear, although some copies may be involved in inhibition of spore germination under blue light [98]. There are three cryptochrome copies in Arabidopsis [97], and five copies described in Adiantum capillus-veneris [98]. In our data, we recover these five copies (which we denote as CRY1 through CRY5) from the majority of the polypod fern transcriptomes (Figure S2). The gene family appears to have evolved via an initial duplication on the fern stem lineage, producing the ancestral CRY1/2 and CRY3/4 paralogs. CRY5 originated around this time, too, perhaps from duplication of the CRY1/2 paralog. Two additional duplications followed, producing CRY1 and CRY2 on the stem branch of Cyatheales + Polypodiales, and CRY3 and CRY4 on the stem branch of Polypodiales, after the divergence of Cyatheales (Figure S2).
The first intron of fern CRY2 is currently being developed as a phylogenetic marker in Deparia (Li-Yaung Kuo pers. comm.) and Adiantum (Wanyu Zhang pers. comm.). We designed our primer pair to target the third exon instead, and found that it recovers the corresponding region from both CRY2 and CRY4 for most of our test set.
The DET1 protein is an important regulator in the ubiquitin-proteasome system as part of the CDD (COP10-DET1-DDB1) complex. It also has been found to be a transcriptional co-repressor recruited to target genes by specific transcription factors [99]. The gene appears to be single copy in polypod ferns (Figure S3) and is present in other eukaryotes, including humans [100]. Of all the nuclear regions for which we designed primers, DET1 is the most conserved (Table 3).
GapCpSh is a member of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene family and is one of the most frequently used nuclear loci in ferns, following the pioneering work of Ebihara et al. [63] and Schuettpelz et al. [64]. Land plants have four deeply divergent GAPDH genes—gapA, gapB, gapC, and gapCp—each of which is nuclear encoded. The first two are originally of mitochondrial origin, and the latter two were plastid encoded prior to their relocation to the nucleus [101-104]. Although we used only fern gapCp sequences as queries to build our all-in transcriptome alignments, our pool of transcriptome hits included representatives of each of the four main copies, as well as a fifth clade of uncertain identity (Figure S4). This myster copy appears to be a member of the GAPDH family (it is readily alignable to other members of the family and all our sequences in this clade have well-characterized members of the GAPDH family as their closest blast hits), but is deeply divergent from the known copies. It appears to be most closely related to the gapC/gapCp copies, but diverged from their ancestor prior to the gapC/gapCp duplication event (Figure S4a). Our transcriptome hits included a good representation of this mystery copy from across the Polypodiales, with an additional hit in Anemia (Schizaeales). Presumably it has been either lost from other ferns or was transcribed at insufficient levels to be captured in many of our source transcriptomes.
GapA and gapB are very poorly represented in our blast hits, as might be expected, given their phylogenetic distance from our query sequences. GapC sequences, however, are well represented, with a broad sample of sequences from the Ophioglossales and Polypodiales, and sparser representation from the Cyatheales (Culcita) and Salviniales (Azolla and Pilularia). Within the gapC portion of the phylogeny (Figure S4a), species are generally in their expected phylogenetic position, with two main exceptions. The first is the position of Culcita (a member of the Cyatheales) within the Pteridaceae (in the Polypodiales). This Culcita sequence, however, is very short (128 bp) and its position is likely an artifact due to limited data. The second irregularity is more difficult to explain: a clade of three Pteridaceae sequences is effectively sister to the rest of the leptosporangiate sequences and far from the Pteridaceae. The relationship among these three sequences corresponds with their expected species relationships, and each of the three species also has a “good” gapC sequence in the appropriate phylogenetic position. These three anomalous sequences may represent an otherwise uncaptured gapC duplication early in the leptosporangiate fern evolution.
The position of the Equisetales sequences is also ambiguous. Each of the two Equisetum accessions (E. diffusum and E. hyemale) has multiple gapC/Cp type sequences, but they fall together in a clade that is resolved in our maximum likelihood (ML) analyses as sister to the fern + seed plant gapCp clade, rather than in separate gapC and gapCp clades (Figure S4). Based on this result, we tentatively treat them as gapCp copies, with an Equisetum-specific gapCp duplication. Consistent with the results of Schuettpelz et al. [64], we recovered three main gapCp copy types in the ferns: a pre-duplication copy, and a duplication in the leptosporangiates forming gapCp “short” (gapCpSh) and gapCp “long” (gapCpLg). Schuettpelz et al. [64] hypothesized that the gapCpSh/Lg duplication event occurred near the base of the Polypodiales, or possibly more deeply (with subsequent losses, based on their sampling). Our transcriptome data suggest that the duplication very likely occurred at a point after the divergence of the Hymenophyllales, Gleicheniales, and Schizaeales, but prior to the divergence of the Salviniales, from the remaining leptosporangiates (Figure S4). Within the gapCp clade there is one group of sequences that is difficult to reconcile with the organismal phylogeny: a clade of five Cyatheales sequences (three from Thyrsopteris, and one each from Plagiogyria and Culcita) that appear to have diverged before the gapCpSh/Lg duplication (Figure S4b). The three species represented also have “good” gapCpSh and gapCpLg sequences, so it is unclear what paralog this anomalous clade represents.
GapCpLg is represented in our transcriptome sample by a single Salviniales sequence (Pilularia), and by sequences from the majority of our sampled species of Cyatheales and Polypodiales. The phylogeny of these sequences is consistent with the currently accepted fern topology [8,20], and does not show any indication of subsequent duplication. GapCpSh is even better represented, with sequences from both Pilularia and Azolla, plus broad representation across Cyatheales and Polypodiales. As with the Adiantum-specific gapCpSh duplication found by Rothfels and Schuettpelz [49], the two Astrolepis-specific duplications found by Beck et al. [42], and the gapCp duplication documented in the evolution of Arabidopsis [105] our data suggest at least two more duplications of gapCpSh: one in a common ancestor of Culcita and Plagiogyria, and another in the Lindsaeaceae (Figure S4b).
IBR3 has not been previously used as phylogenetic marker. It is related to acyl-CoA dehydrogenases and, while its subcellular location has not been confirmed, it contains a peroxisomal targeting sequence and likely is localized to that organelle [95,106]. IBR3 appears to be present as a single copy throughout the fern tree, and is thought to be single copy across land plants (1KP data; Norman Wicket, pers. comm.). One possible exception in our data is in the Psilotaceae, where there may be a duplication (Figure S5).
PgiC is one of the most extensively used nuclear markers in ferns (e.g., [57,59,66,71-75]). It also has a history in angiosperm phylogenetics, e.g., [107,108], was one of the most frequently used enzymes in allozyme studies, e.g., [109,110], and is single-copy in ferns [71] (Figure S6). The gene codes for phosphoglucose isomerase, an enzyme active in the glycolysis of glucose-6-phosphate isomerase [95]. In our phylogenetic analyses, we excluded the Dicksonia sequence because it was too divergent from the other taxa to align confidently.
The SQD1 gene encodes a protein required for synthesis of sulfoquinovosyldiacylglycerol (SQDG), a well-characterized sulfolipid found in chloroplast membranes, and is widely distributed across land plants, green algae, and cyanobacteria [111]. It is hypothesized that SQD1 permits proper functioning of photosystem II under phosphorous limited conditions [112]. Studies utilizing Southern hybridization demonstrated that SQD1 is single-copy in Arabidopsis thaliana and the chlorophyte algae Chlamydomonas reinhardtii [113,114]. In silico analysis of fully annotated genomes indicated that SQD1 is also present as a single copy in Oryza sativa and Populus trichocarpa, prompting the development of angiosperm-specific primers [115,116]. Additional genomic analyses confirmed single copies of SQD1 in Physcomitrella patens, Selaginella moellendorffii, Vitis vinifera, Zea mays, and Sorghum bicolor [117]. Our study suggests that SQD1 is a single copy gene for the majority of fern taxa (Figure S7). A notable exception is the presence of an apparent duplication in an ancestor of the Marattiaceae. Several other more-recent duplications have occurred in isolated genera or species such as Lindsaea, Culcita macrocarpa and Nephrolepis exaltata. Notably, our Ophioglossum (Ophioglossales) SQD1 sequence is resolved as sister to Lygodium (Schizeaceae), a position incompatible with the current, accepted understanding of fern phylogeny [20]. It is possible that this is an alignment artifact. We do not suspect contamination, because an Ophioglossum + Lygodium clade is not recovered in phylogenies of any of other loci in this study.
TPLATE has been identified as a cytokinesis protein involved in the formation of the cell plate [95,118]. Van Damme et al. [119] have also shown that it is important for the formation of viable pollen. It is a member of the group of putatively single-copy markers identified by the 1KP project (Norman Wicket pers. comm.), and to our knowledge has not previously been used as a phylogenetic marker. It is single-copy in our transcriptome sample except for possible duplications in the Ophioglossaceae (Figure S8).
The function of the transducin protein in plants is not well described. It belongs to the G-protein complex, which is involved in signaling across the cell membrane [120]. In Arabidopsis this complex is thought to be involved in the export and import of mRNA and protein to the nucleus [95]. It is a member of the group of putatively single-copy markers identified by the 1KP project (Norman Wicket pers. comm.), and is single-copy in our sample (Figure S9). To our knowledge it has not previously been used as a phylogenetic marker.
Model selection and the Polypodiales phylogeny
The strong improvement in fit to our data that is provided by selecting a model with codon-position based partitions, and the correspondingly weak (or negative) contribution of locus-based partitions, is consistent with other studies [121-124]. This result both emphasizes the importance of including codon position information in model selection procedures, and suggests that our loci share organismal histories: the absence of strong by-locus effects on model fit suggests congruence among the gene trees. Also notable is the strong effect of model choice on ML bootstrap support levels (Table 4). Each of our five models was the best of its “class,” in the sense that each represented the optimal parameterization for the chosen partitioning scheme (by the Akaike information criterion—AIC), and each was at least moderately parameterized (had a minimum of 37 free parameters; Table 4). One might thus naïvely expect that these models, on the same data, would perform similarly. Instead, they resulted in the inference of quite divergent levels of bootstrap support for some nodes (up to a 33 percentage point difference in support; Table 4). Interestingly, the poorer-fitting models tended to find higher levels of support for both branch C of Figure 3 (a branch that was unsupported or weakly supported in earlier phylogenetic investigations [7,8,16]) and for branch L (a branch that is inconsistent with the strongly supported—74 percent ML bootstrap support and 1.0 posterior probability—results of Rothfels et al. [6]). The importance of adequate partitioning schemes for accurate phylogenetic inference has been long acknowledged [125,126], and our results mirror those of other recent empirical studies that found strong effects—both on inference of topology and support levels—of partitioning methods [122,123,127-129].
The resulting phylogenetic conclusions under our best-fitting model (model 3; see Table 4; Figure 3; Appendix S2) are comfortingly consistent with earlier results (e.g., [7,8,90]). The nine branches that are highly supported in our analyses have been inferred with high support in earlier studies, and the three branches that lack support in our data likewise have historically resisted resolution [7,8,16]. The sole exception to this pattern is the relationship among the three non-Cystopteridaceae members of the eupolypods II, which our data do not support (branch L in Figure 3), but which Rothfels et al. [6] were able to resolve with strong support (using much denser taxon sampling).
Conclusions
The 1KP fern transcriptomes provide a powerful means to generate new single-copy nuclear regions for use by evolutionary biologists. The 20 primer pairs presented here (amplifying regions across 10 protein-coding genes) more than triple the number of such regions available for ferns. Moreover, across most of our Polypodiales genomic DNA test set (Appendix S1), the majority of these primer pairs yield PCR products that can be directly sequenced. Our sample spans the phylogenetic breadth of the Polypodiales, which includes approximately two thirds of extant fern species. Our test set, however, was focused on diploid species; researchers working with polyploids, questions of hybridization, or heterozygous individuals will need to clone their PCR products.
These newly available markers vary in their degree of variation and phylogenetic informativeness at a range of evolutionary depths (see Table 3, Figures 2, 3). In combination they yield the first broad multi-gene nuclear phylogeny for ferns. This phylogeny features strong levels of support, is consistent with the results of earlier studies, and thus provides critical evidence for the general consistency of inferences from these two genomic compartments.
For researchers working on groups outside of the Polypodiales (or those with a narrower focus within the Polypodiales), our new primers may not be directly applicable, but serve instead as a proof of concept. For these researchers, our fern-wide “all-in” alignments (see Figures S1-S9; TreeBASE accession number S14616) will provide an opportunity to design primers for their study group of choice, regardless of the position of that group within the fern phylogeny.
Methods
Extracting transcriptome sequences of interest and creating “all-in” alignments
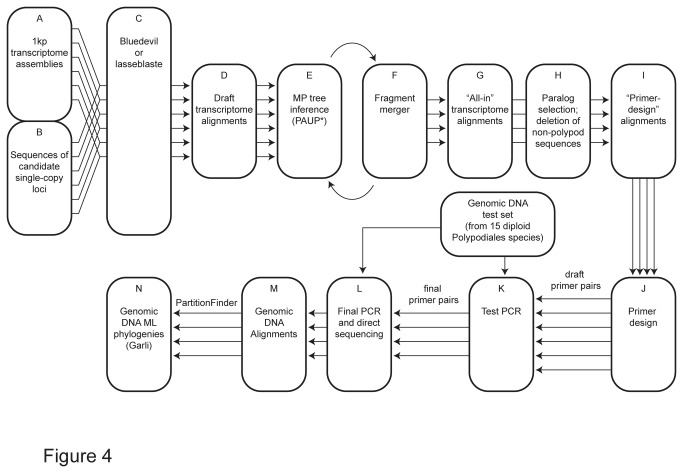
As of January 2013, the 1KP project (www.onekp.com) had sequenced 62 fern transcriptomes, spanning the deepest branches in the fern phylogeny. RNA extraction protocols used here varied [130] although we found that the Spectrum Total Plant RNA Kit (Sigma-Aldrich, St. Louis, Missouri, U.S.A.) was effective for use with ferns. The sequencing was performed on Illumina’s GAIIx (earlier samples) or HiSeq (later samples) sequencing platforms at BGI-Shenzhen, and the 2x75 bp (GAIIx) or 2x90 bp (HiSeq) paired-end reads were assembled with SOAPdenovo (http://soap.genomics.org.cn/soapdenovo.html [131]) and SOAPdenovo-trans (http://soap.genomics.org.cn/SOAPdenovo-Trans.html); for further details on RNA extractions, transcriptome sequencing, and assembly, see Johnson et al. [130]. We took a top-down approach to finding single-copy loci in the transcriptome data. Potential single-copy loci were first selected based on personal interest or from a list of markers (generated by the 1KP project) that are putatively single-copy across a broad sample of land plants (Norman Wickett, pers. comm.). Subsequently, for each of these loci we used a combination blast [132] and tree-searching approach (Figure 4), which allowed us to confirm that the loci were single-copy (in the transcriptome data), and to focus on those with particularly good representation in the transcriptomes available to us.
Figure 4. Flowchart of our transcriptome-mining pipeline.
We inferred fern-wide alignments for our candidate loci using one of two broad approaches (Figure 4). The first utilized the python script Blue Devil v0.6 [133], which detects the longest open reading frames (ORFs) in a series of query sequences, blasts those ORFs against a pool of transcriptome assemblies and provides a MUSCLE-based [134] alignment of the resulting hits. Blue Devil provides the options of using either blastn or tblastx [135], of varying the blast significance cut-off values, and of using CAP3 [136] to re-assemble the blast hits prior to producing the alignment. CAP3 was particularly useful in our pipeline because it allowed the SOAPdenovo and SOAPdenovo-trans assemblies of each transcriptome to be assembled together into one “master” assembly.
Our second main approach to producing transcriptome alignments was based on a nested series of blast searches using lasseblaste [137]. This script takes a series of query sequences as input (we used the entire pool of putatively single-copy markers listed by the 1KP project; Norman Wicket pers. comm.) and blasts each of these sequences against the pool of transcriptomes. It then takes the resulting hits and blasts them back to the full transcriptomes. From this final pool of hits, lasseblaste utilizes MAFFT [138] to produce a separate alignment of the hit sequences obtained for each query sequence and provides an accompanying quality score. The scoring system rewards alignments that have broad representation across the included transcriptomes, indicating good taxon coverage and penalizes alignments that have many hits per transcriptome, suggesting multiple paralogs and/or short read lengths. We selected five of the top 10 best-scoring of these alignments to pursue for primer design.
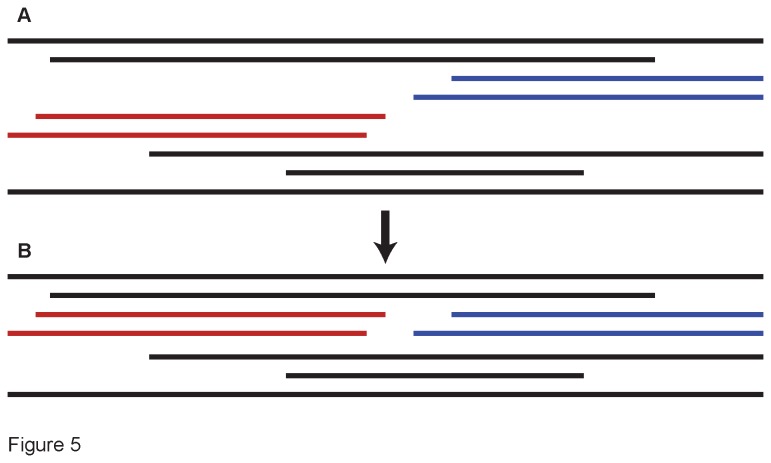
Regardless of whether we used Blue Devil or lasseblaste to infer the initial alignment, we subsequently refined that alignment manually, in an iterative manner. First, we inferred a preliminary phylogenic tree from that alignment using maximum parsimony (MP) in PAUP* v4.0a125 [139]. Groups of discontinuous (or slightly overlapping) sequences from a given accession that appeared closely related in the resulting tree and did not have any conflicts with each other were combined into a single sequence in Mesquite v2.75 [140] (Figure 5). We then repeated the MP analyses on this new alignment. The resulting tree had fewer terminals, and was inferred from longer average sequences, and so provided greater power to place previously uncertain fragments. We continued this “infer-tree, group-sequences” approach until no further fragments met our criteria for merging. This process allowed us to produce an alignment with minimal missing data, and to effectively distinguish among paralogs. The final alignments generated in this way are referred to as our “all-in” alignments (see TreeBASE study number S14616).
Figure 5. Example of our sequence-merging protocol.
(A) In this schematic of a transcriptome alignment, aligned sequence fragments are indicated by the horizontal bars. Included are four fragments (colored) from our focal accession, which group together in the maximum parsimony tree. However, the two fragments from the 5’ end of the protein (in red) have some base pair conflicts with each other, as do the fragments from the 3’ end (in blue). Since the two sets of fragments do not overlap, and they group in the same area of the MP tree, it is not possible to determine which 5’ fragment belongs with which 3’ one. In this case we merged the sequences arbitrarily (B). The resulting alignment retains the full nucleotide data for primer-design purposes, but the relationships at the tips of the tree may be erroneous due to the two potentially chimaeric sequences.
Despite our targeting putatively single-copy genes, some of the transcriptome queries returned a variety of paralogs. In these cases, our sequence pools occasionally included two or more sequence fragments of different paralogs from a single individual taxon, where it was unclear which fragments belonged together. For example, an accession might have two fragments from the 5’ end of the protein that conflict with each other, and two conflicting sequences from the 3’ end, without any indication of which one of the 5’ sequences corresponds to which of the 3’ sequences. In this case, we created two sequences by merging the non-conflicting fragments arbitrarily (Figure 5). All sequence variation is thus preserved for primer generation purposes, but the resulting sequences may be chimeras, and their fine-scale phylogenetic relationships incorrect.
We inferred a final phylogenetic tree from each all-in alignment by ML, using Garli 2.0 [141], under the best-fitting model and partitioning scheme as determined by PartitionFinder v1.0.1 [124]. In each case we designated three data blocks (one for each codon position), and used PartitionFinder to evaluate all partitioning schemes, with the best selected according to the AIC. The subsequent tree searches (in Garli) were each run ten times, independently, from different random addition starting trees (see Figures S1-S9).
Polypod-only alignment and primer design
From each all-in alignment we identified the copy (if multiple paralogs were present) that included the best representation of polypod sequences, and extracted those sequences to produce a new, polypod-only alignment. To this alignment we added the related Arabidopsis thaliana genomic DNA and cDNA sequences based on blast searches of TAIR [95] using Mesquite’s pair-wise alignment tool with a high gap-opening penalty (40). We were able to use the comparison of Arabidopsis genomic and cDNA sequences to estimate the location of exon-intron boundaries in the fern transcriptome sequences. In cases where the exact beginning and end of the Arabidopsis introns were ambiguous, we refined the boundaries to match known exon-intron boundary sequence signatures as closely as possible (e.g., see 142).
The resulting alignments are our “primer-design” alignments—they contain all available information for our taxonomic target (the Polypodiales) for each region of choice. Using the primer-design alignment we searched for conserved sites for primer design. Each primer pair was checked for hairpins, melting point, self-dimers, and hetero-dimers with Integrated DNA Technologies’ OligoAnalyzer v3.1 (http://www.idtdna.com/analyzer/applications/oligoanalyzer/).
Amplification of genomic DNA and sequence characterization
Primer pairs were assayed against the test set of genomic DNA from 15 fern taxa, spanning the major polypod divergences (Appendix S1). PCR conditions followed published protocols [143] with two adjustments: (1) We incorporated one additional microliter of each primer (to compensate for primer degeneracy) and (2) reduced the volume of water by two microliters (to keep reaction size constant). Total reaction size was 21 microliters. The initial PCRs were performed across a temperature gradient, with the final optimal thermocycling conditions listed in Table 2.
For each region that amplified consistently (produced strong single bands for the majority of the test genomic DNAs), we purified and direct sequenced the products following established protocols [6,8]. For high priority targets that gave poor sequencing results, we cloned the PCR products following established protocols [64], and sequenced them as listed in Table 2. For the cryptochrome loci (CRY2 and CRY4), the PCR products were gel-extracted using the QIAquick Gel Extraction Kit (QIAGEN Inc., Gaithersburg, MD) prior to cloning. We aligned the resulting sequences by hand or MAFFT [138] and used Garli v2.0 [141] to infer the best ML phylogenetic tree under a GTR+I+G model. Support was assessed via 1000 bootstrap pseudoreplicates, with each bootstrap tree search performed twice, from different random addition starting trees (Figure 2).
Due to the breadth of our taxon sample, much of the intron data could not be unambiguously aligned and thus were excluded prior to tree-searching, which reduced our ability to assess the utility of these markers at shallower phylogenetic depths. To overcome this weakness, we chose two pairs of closely related species (Cystopteris bulbifera and C. protrusa and Polypodium amorphum and P. glycyrrhiza) to provide metrics for the variability of each region. For each species pair we computed the total number of base differences between the sequences of the two species (with each indel counted as a single “difference” regardless of its length) for each region (Table 3). All newly generated genomic sequences are available in GenBank (Appendix S1).
Polypodiales combined data phylogeny
To demonstrate the utility of our markers across various phylogenetic depths (the earliest divergences in the Polypodiales occurred approximately 190 million years ago [144]) and to attempt to resolve polytomies in the backbone of the Polypodiales phylogeny [8,20] we combined the genomic DNA alignments for our loci and inferred their phylogeny by ML. Some of the locus alignments contained multiple sequences for individual accessions (representing paralogs, or allelic variation; see Figure 2). In these cases, the longest sequence was retained. In the event of a predicted duplication affecting multiple accessions, the copy that had the greatest average length was kept, rather than the longest sequence within each copy. The resulting alignments were combined into a single alignment using abioscripts (available at http://ormbunkar.se/phylogeny/abioscripts/). This script produces a concatenated alignment, inserting blank characters for accessions not represented in a particular locus, while maintaining exclusion set, codon position, and character set information.
We used PartitionFinder v1.0.1 [124] to find the best model for the analysis of these data. We performed three PartitionFinder runs to investigate a spectrum of possible models (Table 4). The first had four predefined data blocks (one for each codon position, and one for the noncoding sequences), the second had 19 data blocks (one for each locus), and the third had 72 data blocks (each codon position/noncoding sequence considered separately, for each locus). For each of these three runs, we set PartitionFinder to find the best partitioning scheme while considering all possible substitution models (with subset-specific substitution models selected by the AIC), testing all possible schemes in the first case, and using a greedy heuristic for the latter two runs. We selected the final model (optimal partitioning scheme with accompanying substitution models for each subset) by fit, as assessed by the BIC. To this set of three models, we added two others (see Table 4). The simplest is an unpartitioned GTR+I+G model, and the more complicated is partitioned by locus, with each locus given its own best-fit substitution model (manually derived from the subset output files from the by-locus PartitionFinder run).
We performed ML tree searches under these five models in Garli 2.0 [141]. For each model we did 10 best-tree searches, from different random-addition sequence starting trees, and assessed support via 1000 bootstrap pseudoreplicates, each from a single random-addition starting tree (Table 4, Figure 3). These bootstrap runs, and other computation-intensive analyses, were run on the Duke Shared Cluster Resource (https://wiki.duke.edu/display/SCSC/DSCR).
Supporting Information
Voucher data and GenBank accession numbers for our Polypodiales genomic DNA test set. Numbers in parenthesis following the species names are Fern Lab Database accession numbers (fernlab.biology.duke.edu); letters in parentheses are acronyms for the herbaria where the vouchers are deposited, from Index Herbariorum [145]. Missing data are indicated by an n-dash (“-”).
(DOCX)
Full description of partitioning schemes and substitution models applied for the five models investigated (1, 2a, 2b, 3, and 4). In the "Subset Contents" field for model 2a, terminal digits refer to codon position: _1= First codon position; _2= Second codon position; _3= Third codon position; _N= Non-coding sequence.
(XLSX)
ApPEFP all-in maximum likelihood transcriptome phylogeny.
(PDF)
CRY all-in maximum likelihood transcriptome phylogeny. a) preduplication CRY3/4, CRY3, and CRY4; b) CRY5, preduplication CRY1/2, and CRY2; c) CRY1, and a cartoon “map” of the entire cryptochrome fern phylogeny.
(PDF)
DET1 all-in maximum likelihood transcriptome phylogeny.
(PDF)
GAP all-in maximum likelihood transcriptome phylogeny. a) gapA, gapB, mystery gap, and gapC; b) gapCp (including Cp Short and Cp Long), and a cartoon map of the GAP family phylogeny.
(PDF)
IBR3 all-in maximum likelihood transcriptome phylogeny.
(PDF)
pgiC all-in maximum likelihood transcriptome phylogeny.
(PDF)
SDQ1 all-in maximum likelihood transcriptome phylogeny.
(PDF)
TPLATE all-in maximum likelihood transcriptome phylogeny.
(PDF)
transducin all-in maximum likelihood transcriptome phylogeny.
(PDF)
Acknowledgments
We thank the many people responsible for the availability of transcriptomes through the 1KP project, particularly Eric Carpenter for his many contributions, the staff at BGI-Shenzhen for the transcriptome sequencing, Norman Wicket for providing the draft list of single-copy loci, and M. Barker, A. Calcedo, L. DeGironimo, M. Deyholos, N. Stewart, T. Shen, and D. Soltis for providing important fern material. The Juniper Level Botanic Garden at Plant Delights Nursery (www.juniperlevelbotanicgarden.org) generously allowed sampling of their collections for RNA extractions, as did the North Carolina Botanical Garden at Chapel Hill, Royal Botanic Gardens at Sydney, and the Duke University Live Plant Collection (http://liveplantcollections.biology.duke.edu). Computations were enabled by facilities at WestGrid, through Compute/Calcul Canada, and at Texas Advanced Computing Center, through iPlant Collaborative. Finally, we thank David Swofford for assistance with analyses, Li-Yaung Kuo for providing unpublished sequence information on fern cryptochromes, and Editor Keith Crandall and two anonymous reviewers for their comments and edits.
Funding Statement
This work was supported by funds from the National Science Foundation (www.nsf.gov) DDIG DEB-1110767 to KMP and CJR (co-authors), DDIG DEB-1110775 to KMP and EMS (co-authors), and DEB-1145614 to KMP and LH; a Natural Science and Engineering Research Council (Canada) Postgraduate Scholarship – Doctoral of NSERC (Natural Sciences and Engineering Research Council) to CJR and Discovery grant to SWG; grants from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas) to PK (2006-429 and 2010-585); GKSW acknowledges the support of Alberta Enterprise and Advanced Education, Genome Alberta, Alberta Innovates Technology Futures iCORE, Musea Ventures, and BGI-Shenzhen. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pryer KM, Schneider H, Smith AR, Cranfill R, Wolf PG et al. (2001) Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature 409: 618–622. doi: 10.1038/35054555. PubMed: 11214320. [DOI] [PubMed] [Google Scholar]
- 2. Hasebe M, Wolf PG, Pryer KM, Ueda K, Ito M et al. (1995) Fern phylogeny based on rbcL nucleotide sequences. Am Fern J 85: 134–181. doi: 10.2307/1547807. [DOI] [Google Scholar]
- 3. Korall P, Pryer KM, Metzgar JS, Schneider H, Conant DS (2006) Tree ferns: Monophyletic groups and their relationships as revealed by four protein-coding plastid loci. Mol Phylogenet Evol 39: 830–845. doi: 10.1016/j.ympev.2006.01.001. PubMed: 16481203. [DOI] [PubMed] [Google Scholar]
- 4. Pryer KM, Schuettpelz E, Wolf PG, Schneider H, Smith AR et al. (2004) Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences. Am J Bot 91: 1582–1598. doi: 10.3732/ajb.91.10.1582. PubMed: 21652310. [DOI] [PubMed] [Google Scholar]
- 5. Pryer KM, Smith AR, Skog JE (1995) Phylogenetic relationships of extant ferns based on evidence from morphology and rbcL sequences. Am Fern J 85: 205–282. doi: 10.2307/1547810. [DOI] [Google Scholar]
- 6. Rothfels CJ, Larsson A, Kuo L-Y, Korall P, Chiou W-L et al. (2012) Overcoming deep roots, fast rates, and short internodes to resolve the ancient rapid radiation of eupolypod II ferns. Syst Biol 61: 490–509. doi: 10.1093/sysbio/sys001. PubMed: 22223449. [DOI] [PubMed] [Google Scholar]
- 7. Schuettpelz E, Korall P, Pryer KM (2006) Plastid atpA data provide improved support for deep relationships among ferns. Taxon 55: 897–906. doi: 10.2307/25065684. [DOI] [Google Scholar]
- 8. Schuettpelz E, Pryer KM (2007) Fern phylogeny inferred from 400 leptosporangiate species and three plastid genes. Taxon 56: 1037–1050. doi: 10.2307/25065903. [DOI] [Google Scholar]
- 9. Wikström N, Pryer KM (2005) Incongruence between primary sequence data and the distribution of a mitochondrial atp1 group II intron among ferns and horsetails. Mol Phylogenet Evol 36: 484–493. doi: 10.1016/j.ympev.2005.04.008. PubMed: 15922630. [DOI] [PubMed] [Google Scholar]
- 10. Hasebe M, Omori T, Nakazawa M, Sano T, Kato M et al. (1994) rbcL gene sequences provide evidence for the evolutionary lineages of leptosporangiate ferns. Proc Natl Acad Sci USA 91: 5730–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sano R, Takamiya M, Ito M, Kurita S, Hasebe M (2000) Phylogeny of the lady fern group, tribe Physematieae (Dryopteridaceae), based on chloroplast rbcL gene sequences. Mol Phylogenet Evol 15: 403–413. doi: 10.1006/mpev.1999.0708. PubMed: 10860649. [DOI] [PubMed] [Google Scholar]
- 12. Wolf PG (1995) Phylogenetic analyses of rbcL and nuclear ribosomal RNA gene sequences in Dennstaedtiaceae. Am Fern J 85: 306–327. doi: 10.2307/1547812. [DOI] [Google Scholar]
- 13. Wolf PG (1996) Pteridophyte phylogenies based on analyses of DNA sequences: A multiple gene approach. In: Camus JM, Gibby M, Johns RJ. Pteridology in perspective. Kew. Royal Botanical Gardens; pp. 203–215. [Google Scholar]
- 14. Wolf PG (1997) Evaluation of atpB nucleotide sequences for phylogenetic studies of ferns and other pteridophytes. Am J Bot 84: 1429–1440. doi: 10.2307/2446141. PubMed: 21708550. [DOI] [PubMed] [Google Scholar]
- 15. Wolf PG, Soltis PS, Soltis DE (1994) Phylogenetic relationships of dennstaedtioid ferns: Evidence from rbcL sequences. Mol Phylogenet Evol 3: 383–392. doi: 10.1006/mpev.1994.1044. PubMed: 7697195. [DOI] [PubMed] [Google Scholar]
- 16. Kuo L-Y, Li F-W, Chiou W-L, Wang C-N (2011) First insights into fern matK phylogeny. Mol Phylogenet Evol 59: 556–566. doi: 10.1016/j.ympev.2011.03.010. PubMed: 21402161. [DOI] [PubMed] [Google Scholar]
- 17. Rai HS, Graham SW (2010) Utility of a large, multigene plastid data set in inferring higher-order relationships in ferns and relatives (monilophytes). Am J Bot 97: 1444–1456. doi: 10.3732/ajb.0900305. PubMed: 21616899. [DOI] [PubMed] [Google Scholar]
- 18. Lehtonen S, Wahlberg N, Christenhusz MJM (2012) Diversification of lindsaeoid ferns and phylogenetic uncertainty of early polypod relationships. Bot J Linn Soc 170: 489–503. doi: 10.1111/j.1095-8339.2012.01312.x. [DOI] [Google Scholar]
- 19. Rothfels CJ, Sundue MA, Kuo L-Y, Larsson A, Kato M et al. (2012) A revised family-level classification for eupolypod II ferns (Polypodiidae: Polypodiales). Taxon 61: 515–533. [Google Scholar]
- 20. Smith AR, Pryer KM, Schuettpelz E, Korall P, Schneider H et al. (2006) A classification for extant ferns. Taxon 55: 705–731. doi: 10.2307/25065646. [DOI] [Google Scholar]
- 21. Christenhusz MJM, Zhang X-C, Schneider H (2011) A linear sequence of extant families and genera of lycophytes and ferns. Phytotaxa 19: 7–54. [Google Scholar]
- 22. Schuettpelz E, Pryer KM (2008) Fern phylogeny. In: Ranker TA, Haufler CH. Biology and evolution of ferns and lycophytes. New York: Cambridge University Press; pp. 395–416. [Google Scholar]
- 23. Haufler CH, Ranker TA (1995) RbcL sequences provide phylogenetic insights among sister species of the fern genus Polypodium . American Fern Journal 85: 361-374. [Google Scholar]
- 24. Nagalingum NS, Schneider H, Pryer KM (2007) Molecular phylogenetic relationships and morphological evolution in the heterosporous fern genus Marsilea . Syst Bot 32: 16–25. doi: 10.1600/036364407780360256. [DOI] [Google Scholar]
- 25. Metzgar J, Skog JE, Zimmer EA, Pryer KM (2008) The paraphily of Osmunda is confirmed by phylogenetic analyses of seven plastid loci. Syst Bot 33: 31–36. doi: 10.1600/036364408783887528. [DOI] [Google Scholar]
- 26. Des Marais DL, Smith AR, Britton DM, Pryer KM (2003) Phylogenetic relationships and evolution of extant horsetails, Equisetum, based on chloroplast DNA sequence data (rbcL and trnL-F). Int J Plant Sci 164: 737–751. doi: 10.1086/376817. [DOI] [Google Scholar]
- 27. Dubuisson JY, Hennequin S, Douzery EJP, Cranfill RB, Smith AR et al. (2003) rbcL phylogeny of the fern genus; Trichomanes (Hymenophyllaceae), with special reference to neotropical taxa. Int J Plant Sci 164: 753–761. doi: 10.1016/S0168-9452(03)00060-8. [DOI] [Google Scholar]
- 28. Rothfels CJ, Windham MD, Grusz AL, Gastony GJ, Pryer KM (2008) Toward a monophyletic Notholaena (Pteridaceae): Resolving patterns of evolutionary convergence in xeric-adapted ferns. Taxon 57: 712–724. [Google Scholar]
- 29. Labiak PH, Rouhan G, Sundue MA (2010) Phylogeny and taxonomy of Leucotrichum (Polypodiaceae): A new genus of grammitid ferns from the Neotropics. Taxon 59: 911–921. [Google Scholar]
- 30. McKeown M, Sundue MA, Barrington DS (2012) Phylogenetic analyses place the Australian monotypic Revwattsia in Dryopteris (Dryopteridaceae). Phytokeys 14: 43–56. doi: 10.3897/phytokeys.14.3446. PubMed: 23170072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sundue MA (2010) A monograph of Ascogrammitis, a new genus of grammitid ferns (Polypodiaceae). Brittonia 62: 357–399. doi: 10.1007/s12228-009-9108-6. [DOI] [Google Scholar]
- 32. Zhang L-B, Zhang L, Dong S-Y, Sessa EB, Gao X-F et al. (2012) Molecular circumscription and major evolutionary lineages of the fern genus Dryopteris (Dryopteridaceae). BMC Evol Biol 12: 180. doi: 10.1186/1471-2148-12-180. PubMed: 22971160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Adjie B, Takamiya M, Ohto M, Ohsawa TA, Watano Y (2008) Molecular phylogeny of the lady fern genus Athyrium in Japan based on chloroplast rbcL and trnL-trnF sequences. Acta Phytotaxonomica Geobotanica 59: 79–95. [Google Scholar]
- 34. Wang M-L, Chen ZD, Zhang X-C, Lu S-G, Zhao G-F (2003) Phylogeny of the Athyriaceae: Evidence from chloroplast trnL-F region sequences. Acta Phytotaxonomica Sin 41: 416–426. [Google Scholar]
- 35. Liu Y-C, Chiou W-L, Kato M (2011) Molecular phylogeny and taxonomy of the fern genus Anisocampium (Athyriaceae). Taxon 60: 824–830. [Google Scholar]
- 36. Li F-W, Pryer KM, Windham MD (2012) Gaga, a new fern genus segregated from Cheilanthes (Pteridaceae). Syst Bot 37: 845–860. doi: 10.1600/036364412X656626. [DOI] [Google Scholar]
- 37. Windham MD, Huiet L, Schuettpelz E, Grusz AL, Rothfels CJ et al. (2009) Using plastid and nuclear DNA sequences to redraw generic boundaries and demystify species complexes in cheilanthoid ferns. Am Fern J 99: 68–72. [Google Scholar]
- 38. Cranfill R, Kato M (2003) Phylogenetics, biogeography, and classification of the woodwardioid ferns (Blechnaceae). In: Chandra S, Srivastava M. Pteridology in the New Millennium. Dordrecht: Kluwer Publishing House Academic Publishers; pp. 25–47. [Google Scholar]
- 39. He L-J, Zhang X-C (2012) Exploring generic delimitation within the fern family Thelypteridaceae. Mol Phylogenet Evol, 65: 1–8. PubMed: 22877644. [DOI] [PubMed] [Google Scholar]
- 40. Smith AR, Cranfill RB (2002) Intrafamilial relationships of the thelypteroid ferns (Thelypteridaceae). Am Fern J 92: 131–149. doi:10.1640/0002-8444(2002)092[0131:IROTTF]2.0.CO;2. [Google Scholar]
- 41. Sigel EM, Windham MD, Huiet L, Yatskievych G, Pryer KM (2011) Species relationships and farina evolution in the cheilanthoid fern genus Argyrochosma (Pteridaceae). Syst Bot 36: 554–564. doi: 10.1600/036364411X583547. [DOI] [Google Scholar]
- 42. Beck JB, Windham MD, Yatskievych G, Pryer KM (2010) A diploids-first approach to species delimitation and interpreting polyploid evolution in the fern genus Astrolepis (Pteridaceae). Syst Bot 35: 223–234. doi: 10.1600/036364410791638388. [DOI] [Google Scholar]
- 43. Grusz AL, Windham MD, Pryer KM (2009) Deciphering the origins of apomictic polyploids in the Cheilanthes yavapensis complex (Pteridaceae). Am J Bot 96: 1636–1645. doi: 10.3732/ajb.0900019. PubMed: 21622350. [DOI] [PubMed] [Google Scholar]
- 44. Kreier H-P, Schneider H (2006) Phylogeny and biogeography of the staghorn fern genus Platycerium (Polypodiaceae, Polypodiidae). Am J Bot 93: 217–225. doi: 10.3732/ajb.93.2.217. PubMed: 21646182. [DOI] [PubMed] [Google Scholar]
- 45. Schneider H, Russell S, Cox C, Bakker F, Henderson S et al. (2004) Chloroplast phylogeny of asplenioid ferns based on rbcL and trnL-F spacer sequences (Polypodiidae, Aspleniaceae) and its implications for biogeography. Syst Bot 29: 260–274. doi: 10.1600/036364404774195476. [DOI] [Google Scholar]
- 46. Hennequin S, Hovenkamp P, Christenhusz MJM, Schneider H (2010) Phylogenetics and biogeography of Nephrolepis–A tale of old settlers and young tramps. Bot J Linn Soc 164: 113–127. doi: 10.1111/j.1095-8339.2010.01076.x. [DOI] [Google Scholar]
- 47. Qiu YL, Cho Y, Cox JC, Palmer JD (1998) The gain of three mitochondrial introns identifies liverworts as the earliest land plants. Nature 394: 671–674. doi: 10.1038/29286. PubMed: 9716129. [DOI] [PubMed] [Google Scholar]
- 48. Vangerow S, Teerkorn T, Knoop V (1999) Phylogenetic information in the mitochondrial nad5 gene of pteridophytes: RNA editing and intron sequences. Plant Biol 1: 235–243. doi: 10.1111/j.1438-8677.1999.tb00249.x. [DOI] [Google Scholar]
- 49. Rothfels CJ, Schuettpelz E (2013) Accelerated rate of molecular evolution for vittarioid ferns is strong and not driven by selection. Syst Biol. In press [DOI] [PubMed] [Google Scholar]
- 50. Pryer KM, Schneider H, Zimmer EA, Banks JA (2002) Deciding among green plants for whole genome studies. Trends Plant Sci 7: 550–554. doi: 10.1016/S1360-1385(02)02375-0. PubMed: 12475497. [DOI] [PubMed] [Google Scholar]
- 51. Barker MS, Wolf PG (2010) Unfurling fern biology in the genomics age. BioScience 60: 177–185. [Google Scholar]
- 52. Nakazato T, Barker MS, Rieseberg LH, Gastony GJ (2008) Evolution of the nuclear genome of ferns and lycophytes. In: Ranker TA, Haufler CH. Biology and Evolution of Ferns and Lycophytes. Cambridge: Cambridge University Press; pp. 175–198. [Google Scholar]
- 53. Zimmer EA, Wen J (2012) Using nuclear gene data for plant phylogenetics: Progress and prospects. Mol Phylogenet Evol 65: 774–785. doi: 10.1016/j.ympev.2012.07.015. PubMed: 22842093. [DOI] [PubMed] [Google Scholar]
- 54. Gastony GJ, Rollo DR (1998) Cheilanthoid ferns (Pteridaceae: Cheilanthoideae) in the southwestern United States and adjacent Mexico--A molecular phylogenetic reassessment of generic lines. Aliso 17: 131–144. [Google Scholar]
- 55. Reid JD, Plunkett GM, Peters GA (2006) Phylogenetic relationships in the heterosporous fern genus Azolla (Azollaceae) based on DNA sequence data from three noncoding regions. Int J Pl Sci 167: 529–538. doi: 10.1086/501071. [DOI] [Google Scholar]
- 56. Maggini F, Marrocco R, Gelati MT, Dominicis RI (1998) Lengths and nucleotide sequences of the internal spacers of nuclear ribosomal DNA in gymnosperms and pteridophytes. Plant Syst Evol 213: 199–205. doi: 10.1007/BF00985200. [DOI] [Google Scholar]
- 57. Schneider H, Navarro-Gomez A, Russell SJ, Ansell SW, Grundmann M et al. (2012) Exploring the utility of three nuclear regions to reconstruct reticulate evolution in the fern genus Asplenium . J Syst Evolution 51: 142–153. [Google Scholar]
- 58. Kranz H, Huss V (1996) Molecular evolution of pteridophytes and their relationship to seed plants: Evidence from complete 18S rRNA gene sequences. Plant Syst Evol 202: 1–11. doi: 10.1007/BF00985814. [DOI] [Google Scholar]
- 59. Wang L, Schneider H, Wu Z, He L, Zhang X et al. (2012) Indehiscent sporangia enable the accumulation of local fern diversity at the Qinghai-Tibetan Plateau. BMC Evol Biol 12: 158. doi: 10.1186/1471-2148-12-158. PubMed: 22929005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shepherd LD, Perrie LR, Brownsey PJ (2008) Low-copy nuclear DNA sequences reveal a predominance of allopolyploids in a New Zealand Asplenium fern complex. Mol Phylogenet Evol 49: 240–248. doi: 10.1016/j.ympev.2008.06.015. PubMed: 18640280. [DOI] [PubMed] [Google Scholar]
- 61. Adjie B, Masuyama S, Ishikawa H, Watano Y (2007) Independent origins of tetraploid cryptic species in the fern Ceratopteris thalictroides . J Plant Res 120: 129–138. doi: 10.1007/s10265-006-0032-5. PubMed: 16955374. [DOI] [PubMed] [Google Scholar]
- 62. Chen C-W, Kuo L-Y, Wang C-N, Chiou W-L (2012) Development of a PCR primer set for intron 1 of the low-copy gene LEAFY in Davalliaceae. Am J Bot, 99: e223–e225. PubMed: 22623608. [DOI] [PubMed] [Google Scholar]
- 63. Ebihara A, Ishikawa H, Matsumoto S, Lin S-J, Iwatsuki K et al. (2005) Nuclear DNA, chloroplast DNA, and ploidy analysis clarified biological complexity of the Vandenboschia radicans complex (Hymenophyllaceae) in Japan and adjacent areas. Am J Bot 92: 1535–1547. doi: 10.3732/ajb.92.9.1535. PubMed: 21646171. [DOI] [PubMed] [Google Scholar]
- 64. Schuettpelz E, Grusz AL, Windham MD, Pryer KM (2008) The utility of nuclear gapCp in resolving polyploid fern origins. Syst Bot 33: 621–629. doi: 10.1600/036364408786500127. [DOI] [Google Scholar]
- 65. Nitta JH, Ebihara A, Ito M (2011) Reticulate evolution in the Crepidomanes minutum species complex (Hymenophyllaceae). Am J Bot, 98: 1–19. PubMed: 22012924. [DOI] [PubMed] [Google Scholar]
- 66. Sessa EB, Zimmer EA, Givnish TJ (2012) Unraveling reticulate evolution in North American Dryopteris (Dryopteridaceae). BMC Evol Biol 12: 104. doi: 10.1186/1471-2148-12-104. PubMed: 22748145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang R, Liu T, Wu W, Li Y, Chao L et al. (2013) Molecular evidence for natural hybridization in the mangrove fern genus Acrostichum . BMC Plant Biol 13: 74. doi: 10.1186/1471-2229-13-74. PubMed: 23634934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee S-J, Park C-W (2013) Relationships and origins of the Dryopteris varia (L.) Kuntze species complex (Dryopteridaceae) in Korea inferred from nuclear and chloroplast DNA sequences. Biochem Syst Ecol 50: 371–382. [Google Scholar]
- 69. Chang Y, Li J, Lu S, Schneider H (2013) Species diversity and reticulate evolution in the Asplenium normale complex (Aspleniaceae) in China and adjacent areas. Taxon. In press [DOI] [PubMed] [Google Scholar]
- 70. Dyer RJ, Savolainen V, Schneider H (2012) Apomixis and reticulate evolution in the Asplenium monanthes fern complex. Ann Bot 110: 1515–1529. doi: 10.1093/aob/mcs202. PubMed: 22984165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ishikawa H, Watano Y, Kano K, Ito M, Kurita S (2002) Development of primer sets for PCR amplification of the PgiC gene in ferns. J Plant Res 115: 65–70. doi: 10.1007/s102650200010. PubMed: 12884051. [DOI] [PubMed] [Google Scholar]
- 72. James KE, Schneider H, Ansell SW, Evers M, Robba L et al. (2008) Diversity arrays technology (DArT) for pan-genomic evolutionary studies of non-model organisms. PLOS ONE 3: e1682. doi: 10.1371/journal.pone.0001682. PubMed: 18301759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chang H-M, Chiou W-L, Wang J-C (2009) Molecular evidence for genetic heterogeneity and the hybrid origin of Acrorumohra subreflexipinna from Taiwan. Am Fern J 99: 61–77. doi: 10.1640/0002-8444-99.2.61. [DOI] [Google Scholar]
- 74. Chao Y-S, Dong S-Y, Chiang Y-C, Liu H-Y, Chiou W-L (2012) Extreme multiple reticulate origins of the Pteris cadieri complex (Pteridaceae). Int J Mol Sci 13: 4523–4544. doi: 10.3390/ijms13044523. PubMed: 22605994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Juslén A, Väre H, Wikström N (2011) Relationships and evolutionary origins of polyploid Dryopteris (Dryopteridaceae) from Europe inferred using nuclear pgiC and plastid trnL-F sequence data. Taxon 60: 1284–1294. [Google Scholar]
- 76. Vogel JC, Russell SJ, Rumsey FJ, Barrett JA, Gibby M (1998) Evidence for the maternal transmission of chloroplast DNA in the genus Asplenium (Aspleniaceae, Pteridophyta). Bot Acta 111: 247–249. [Google Scholar]
- 77. Stein DB, Barrington DS (1990) Recurring hybrid formation in a population of Polystichum x potteri: Evidence from chloroplast DNA comparisons. Ann Mo Bot Gard 77: 334–339. doi: 10.2307/2399548. [DOI] [Google Scholar]
- 78. Guillon JM, Raquin C (2000) Maternal inheritance of chloroplasts in the horsetail Equisetum variegatum (Schleich.). Curr Genet 37: 53–56. doi: 10.1007/s002940050008. PubMed: 10672445. [DOI] [PubMed] [Google Scholar]
- 79. Gastony GJ, Yatskievych G (1992) Maternal inheritance of the chloroplast and mitochondrial genomes in cheilanthoid ferns. Am J Bot 79: 716–722. doi: 10.2307/2444887. [DOI] [Google Scholar]
- 80. Lovis J (1978) Evolutionary patterns and processes in ferns. Adv Bot Res 4: 229–415. doi: 10.1016/S0065-2296(08)60371-7. [DOI] [Google Scholar]
- 81. Der J, Barker M, Wickett NJ, dePamphilis CW, Wolf PG (2011) De novo characterization of the gametophyte transcriptome in bracken fern, Pteridium aquilinum . BMC Genomics 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wolf PG, Karol KG (2012) Plastomes of bryophytes, lycophytes and ferns. Genomics Chloroplasts Mitochondria Advances Photosynth Respiration 35: 89–102. doi: 10.1007/978-94-007-2920-9_4. [DOI] [Google Scholar]
- 83. Karol KG, Arumuganathan K, Boore JL, Duffy AM, Everett KD et al. (2010) Complete plastome sequences of Equisetum arvense and Isoetes flaccida: Implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evol Biol 10: 321. doi: 10.1186/1471-2148-10-321. PubMed: 20969798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wolf PG, Der JP, Duffy AM, Davidson JB, Grusz AL et al. (2010) The evolution of chloroplast genes and genomes in ferns. Plant Mol Biol 76: 251–261. PubMed: 20976559. [DOI] [PubMed] [Google Scholar]
- 85. Salmi ML, Bushart TJ, Stout SC, Roux SJ (2005) Profile and analysis of gene expression changes during early development in germinating spores of Ceratopteris richardii . Plant Physiol 138: 1734–1745. doi: 10.1104/pp.105.062851. PubMed: 15965014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yamauchi D, Sutoh K, Kanegae H, Horiguchi T, Matsuoka K et al. (2005) Analysis of expressed sequence tags in prothallia of Adiantum capillus-veneris. J Plant Res 118: 223–227. doi: 10.1007/s10265-005-0209-3. PubMed: 15940394. [DOI] [PubMed] [Google Scholar]
- 87. Grewe F, Guo W, Gubbels EA, Hansen AK, Mower JP (2013) Complete plastid genomes from Ophioglossum californicum, Psilotum nudum, and Equisetum hyemale reveal an ancestral land plant genome structure and resolve the position of Equisetales among monilophytes. BMC Evol Biol 13: 8-. PubMed: 23311954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Roper JM, Kellon Hansen S, Wolf PG, Karol KG, Mandoli DF et al. (2007) The complete plastid genome sequence of Angiopteris evecta (G Forst.) Hoffm. (Marattiaceae). Am Fern J 97: 95–106. doi:10.1640/0002-8444(2007)97[95:TCPGSO]2.0.CO;2. [Google Scholar]
- 89. Gao L, Wang B, Wang ZW, Zhou Y, Su YJ et al. (2013) Plastome sequences of Lygodium japonicum and Marsilea crenata reveal the genome organization transformation from basal ferns to core leptosporangiates. Genome Biol Evolution 5: 1403–1407. doi: 10.1093/gbe/evt099. PubMed: 23821521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Schuettpelz E, Schneider H, Huiet L, Windham MD, Pryer KM (2007) A molecular phylogeny of the fern family Pteridaceae: Assessing overall relationships and the affinities of previously unsampled genera. Mol Phylogenet Evol 44: 1172–1185. doi: 10.1016/j.ympev.2007.04.011. PubMed: 17570688. [DOI] [PubMed] [Google Scholar]
- 91. Schneider H, Schuettpelz E, Pryer KM, Cranfill R, Magallón S et al. (2004) Ferns diversified in the shadow of angiosperms. Nature 428: 553–557. doi: 10.1038/nature02361. PubMed: 15058303. [DOI] [PubMed] [Google Scholar]
- 92. Larsson A, Windham MD, Korall P (2013) Phylogeny of Woodsia (Woodsiaceae). In prep.
- 93. Wong KM, Suchard MA, Huelsenbeck JP (2008) Alignment uncertainty and genomic analysis. Science 319: 473–476. doi: 10.1126/science.1151532. PubMed: 18218900. [DOI] [PubMed] [Google Scholar]
- 94. Rannala B, Yang Z (2008) Phylogenetic inference using whole genomes. Annu Rev Genomics Hum Genet 9: 217–231. doi: 10.1146/annurev.genom.9.081307.164407. PubMed: 18767964. [DOI] [PubMed] [Google Scholar]
- 95. Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C et al. (2011) The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210. PubMed: 22140109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Möglich A, Yang X, Ayers RA, Moffat K (2010) Structure and function of plant photoreceptors. Annu Rev Plant Biol 61: 21–47. doi: 10.1146/annurev-arplant-042809-112259. PubMed: 20192744. [DOI] [PubMed] [Google Scholar]
- 97. Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T et al. (2011) The cryptochromes: Blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62: 335–364. doi: 10.1146/annurev-arplant-042110-103759. PubMed: 21526969. [DOI] [PubMed] [Google Scholar]
- 98. Imaizumi T, Kanegae T, Wada M (2000) Cryptochrome nucleocytoplasmic distribution and gene expression are regulated by light quality in the fern Adiantum capillus-veneris . Plant Cell 12: 81–96. doi: 10.2307/3871031. PubMed: 10634909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593. doi: 10.1016/j.tplants.2012.05.004. PubMed: 22705257. [DOI] [PubMed] [Google Scholar]
- 100. Wertz IE, O’Rourke KM, Zhang Z, Dornan D, Arnott D et al. (2004) Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303: 1371–1374. doi: 10.1126/science.1093549. PubMed: 14739464. [DOI] [PubMed] [Google Scholar]
- 101. Petersen J, Teich R, Becker B, Cerff R, Brinkmann H (2006) The GapA/B gene duplication marks the origin of Streptophyta (charophytes and land plants). Mol Biol Evol 23: 1109–1118. doi: 10.1093/molbev/msj123. PubMed: 16527864. [DOI] [PubMed] [Google Scholar]
- 102. Martin W, Cerff R (1986) Prokaryotic features of a nucleus-encoded enzyme. cDNA sequences for chloroplast and cytosolic glyceraldehyde-3-phosphate dehydrogenases from mustard (Sinapis alba). Eur J Biochem 159: 323–331. doi: 10.1111/j.1432-1033.1986.tb09871.x. PubMed: 3530755. [DOI] [PubMed] [Google Scholar]
- 103. Brinkmann H, Martinez P, Quigley F, Martin W, Cerff R (1987) Endosymbiotic origin and codon bias of the nuclear gene for chloroplast glyceraldehyde-3-phosphate dehydrogenase from maize. J Mol Evol 26: 320–328. doi: 10.1007/BF02101150. PubMed: 3131533. [DOI] [PubMed] [Google Scholar]
- 104. Petersen J, Brinkmann H, Cerff R (2003) Origin, evolution, and metabolic role of a novel glycolytic GAPDH enzyme recruited by land plant plastids. J Mol Evol 57: 16–26. doi: 10.1007/s00239-002-2441-y. PubMed: 12962302. [DOI] [PubMed] [Google Scholar]
- 105. Muñoz-Bertomeu J, Cascales-Miñana B, Mulet JM, Baroja-Fernández E, Pozueta-Romero J et al. (2009) Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis . Plant Physiol 151: 541–558. doi: 10.1104/pp.109.143701. PubMed: 19675149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zolman BK, Nyberg M, Bartel B (2007) IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol Biol 64: 59–72. doi: 10.1007/s11103-007-9134-2. PubMed: 17277896. [DOI] [PubMed] [Google Scholar]
- 107. Ford VS, Lee J, Baldwin BG, Gottlieb LD (2006) Species divergence and relationships in Stephanomeria (Compositae): PgiC phylogeny compared to prior biosystematic studies. Am J Bot 93: 480–490. doi: 10.3732/ajb.93.3.480. PubMed: 21646207. [DOI] [PubMed] [Google Scholar]
- 108. Kamiya K, Harada K, Tachida H, Ashton PS (2005) Phylogeny of PgiC gene in Shorea and its closely related genera (Dipterocarpaceae), the dominant trees in Southeast Asian tropical rain forests. Am J Bot 92: 775–788. doi: 10.3732/ajb.92.5.775. PubMed: 21652457. [DOI] [PubMed] [Google Scholar]
- 109. Charlesworth D, Yang Z (1998) Allozyme diversity in Leavenworthia populations with different inbreeding levels. Heredity 81(4): 453–461. doi: 10.1046/j.1365-2540.1998.00415.x. PubMed: 9839439. [DOI] [PubMed] [Google Scholar]
- 110. Witter MS (1990) Evolution in the Madiinae: Evidence from enzyme electrophoresis. Ann Mo Bot Gard 77: 110–117. doi: 10.2307/2399630. [DOI] [Google Scholar]
- 111. Sato N (2004) Roles of the acidic lipids sulfoquinovosyl diacylglycerol and phosphatidylglycerol in photosynthesis: Their specificity and evolution. J Plant Res 117: 495–505. doi: 10.1007/s10265-004-0183-1. PubMed: 15538651. [DOI] [PubMed] [Google Scholar]
- 112. Mizusawa N, Wada H (2012) The role of lipids in photosystem II. Biochim Biophys-Bioenerget 1817: 194–208. doi: 10.1016/j.bbabio.2011.04.008. PubMed: 21569758. [DOI] [PubMed] [Google Scholar]
- 113. Essigmann B, Güler S, Narang RA, Linke D, Benning C (1998) Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana . Proc Natl Acad Sci USA 95: 1950–1955. doi: 10.1073/pnas.95.4.1950. PubMed: 9465123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Sato N, Sugimoto K, Meguro A, Tsuzuki M (2003) Identification of a gene for UDP-sulfoquinovose synthase of a green alga, Chlamydomonas reinhardtii, and its phylogeny. DNA Res 10: 229–237. doi: 10.1093/dnares/10.6.229. PubMed: 15029954. [DOI] [PubMed] [Google Scholar]
- 115. Li M, Wunder J, Bissoli G, Scarponi E, Gazzani S et al. (2008) Development of COS genes as universally amplifiable markers for phylogenetic reconstructions of closely related plant species. Cladistics 24: 727–745. doi: 10.1111/j.1096-0031.2008.00207.x. [DOI] [Google Scholar]
- 116. Bruni I, De Mattia F, Galimberti A, Galasso G, Banfi E et al. (2010) Identification of poisonous plants by DNA bar coding approach. Int J Leg Med 124: 595–603. doi: 10.1007/s00414-010-0447-3. PubMed: 20354712. [DOI] [PubMed] [Google Scholar]
- 117. Moummou H, Kallberg Y, Tonfack LB, Persson B, van der Rest Bt (2012) The plant short-chain dehydrogenase(SDR) superfamily: Genome-wide inventory and diversification patterns. BMC Plant Biol 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Van Damme D, Gadeyne A, Vanstraelen M, Inzé D, Van Montagu MCE et al. (2011) Adaptin-like protein TPLATE and clathrin recruitment during plant somatic cytokinesis occurs via two distinct pathways. Proc Natl Acad Sci U S A 108: 615–620. doi: 10.1073/pnas.1017890108. PubMed: 21187379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Van Damme D, Coutuer S, De Rycke R, Bouget FY, Inzé D et al. (2006) Somatic cytokinesis and pollen maturation in Arabidopsis depend on TPLATE, which has domains similar to coat proteins. Plant Cell 18: 3502–3518. doi: 10.1105/tpc.106.040923. PubMed: 17189342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Neves SR, Prahlad TR, Iyengar R (2002) G protein pathways. Science 296: 1636–1639. doi: 10.1126/science.1071550. PubMed: 12040175. [DOI] [PubMed] [Google Scholar]
- 121. Shapiro B, Rambaut A, Drummond AJ (2005) Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol 23: 7–9. doi: 10.1093/molbev/msj021. PubMed: 16177232. [DOI] [PubMed] [Google Scholar]
- 122. Strugnell J, Norman M, Jackson J, Drummond AJ, Cooper A (2005) Molecular phylogeny of coleoid cephalopods (Mollusca: Cephalopoda) using a multigene approach; the effect of data partitioning on resolving phylogenies in a Bayesian framework. Mol Phylogenet Evol 37: 426–441. doi: 10.1016/j.ympev.2005.03.020. PubMed: 15935706. [DOI] [PubMed] [Google Scholar]
- 123. Brandley MC, Schmitz A, Reeder TW (2005) Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Syst Biol 54: 373–390. doi: 10.1080/10635150590946808. PubMed: 16012105. [DOI] [PubMed] [Google Scholar]
- 124. Lanfear R, Calcott B, Ho SYW, Guindon S (2012) PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29: 1695–1701. doi: 10.1093/molbev/mss020. PubMed: 22319168. [DOI] [PubMed] [Google Scholar]
- 125. Bull JJ, Huelsenbeck JP, Cunningham CW, Swofford DL, Waddell PJ (1993) Partitioning and combining data in phylogenetic analysis. Syst Biol 42: 384–397. doi: 10.2307/2992473. [DOI] [Google Scholar]
- 126. Brown JM, Lemmon AR (2007) The importance of data partitioning and the utility of Bayes factors in Bayesian phylogenetics. Syst Biol 56: 643–655. doi: 10.1080/10635150701546249. PubMed: 17661232. [DOI] [PubMed] [Google Scholar]
- 127. Ward PS, Brady SG, Fisher BL, Schultz TR (2010) Phylogeny and biogeography of dolichoderine ants: Effects of data partitioning and relict taxa on historical inference. Syst Biol 59: 342–362. doi: 10.1093/sysbio/syq012. PubMed: 20525640. [DOI] [PubMed] [Google Scholar]
- 128. Li C, Lu G, Ortí G (2008) Optimal data partitioning and a test case for ray-finned fishes (Actinopterygii) based on ten nuclear loci. Syst Biol 57: 519–539. doi: 10.1080/10635150802206883. PubMed: 18622808. [DOI] [PubMed] [Google Scholar]
- 129. McGuire JA, Witt CC, Altshuler DL, Remsen JV, [!(surname)!] (2007) Phylogenetic systematics and biogeography of hummingbirds: Bayesian and maximum likelihood analyses of partitioned data and selection of an appropriate partitioning strategy. Syst Biol 56: 837–856. doi: 10.1080/10635150701656360. PubMed: 17934998. [DOI] [PubMed] [Google Scholar]
- 130. Johnson MTJ, Carpenter EJ, Tian Z, Bruskiewich R, Burris JN et al. (2012) Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PLOS ONE 7: e50226. doi: 10.1371/journal.pone.0050226. PubMed: 23185583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Luo R, Liu B, Xie Y, Li Z, Huang W et al. (2012) SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 1: 18. doi: 10.1186/2047-217X-1-18. PubMed: 23587118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. doi: 10.1016/S0022-2836(05)80360-2. PubMed: 2231712. [DOI] [PubMed] [Google Scholar]
- 133. Li F-W (2013) Blue Devil Blast Utility Seq Extr Devised By Li. Available: http://pryerlab.biology.duke.edu/software.
- 134. Edgar RC (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. doi: 10.1093/nar/gkh340. PubMed: 15034147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J et al. (2009) BLAST+: Architecture and applications. BMC Bioinformatics 10: 421. doi: 10.1186/1471-2105-10-421. PubMed: 20003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Huang X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9: 868–877. doi: 10.1101/gr.9.9.868. PubMed: 10508846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Larsson A (2013) lasseblaste. Available: ormbunkar.se/phylogeny/lasseblaste/.
- 138. Katoh K, Misawa K, Kuma K-i, Miyata T (2002) MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066. doi: 10.1093/nar/gkf436. PubMed: 12136088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Swofford DL (2002) PAUP*: Phylogenetic analysis using parsimony (* and other methods). 4.0 ed Sunderland, MA: Sinauer. [Google Scholar]
- 140. Maddison WP, Maddison DR (2009) Mesquite: A modular system for evolutionary analysis. Version 2.72 ed: http://mesquiteproject.org.
- 141. Zwickl DJ (2006) GARLI. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. [PhD]. Austin: the University of Texas. [Google Scholar]
- 142. Brown JWS, Smith P, Simpson CJ (1996) Arabidopsis consensus intron sequences. Plant Mol Biol 32: 531–535. doi: 10.1007/BF00019105. PubMed: 8980502. [DOI] [PubMed] [Google Scholar]
- 143. Rothfels CJ, Windham MD, Pryer KM (2013) A plastid phylogeny of the cosmopolitan fern family Cystopteridaceae (Polypodiopsida). Syst Bot 38: 295–306. doi: 10.1600/036364413X666787. [DOI] [Google Scholar]
- 144. Schuettpelz E, Pryer KM (2009) Evidence for a Cenozoic radiation of ferns in an angiosperm-dominated canopy. Proc Natl Acad Sci U S A 106: 11200–11205. doi: 10.1073/pnas.0811136106. PubMed: 19567832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Thiers B ([continuously updated]) Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Voucher data and GenBank accession numbers for our Polypodiales genomic DNA test set. Numbers in parenthesis following the species names are Fern Lab Database accession numbers (fernlab.biology.duke.edu); letters in parentheses are acronyms for the herbaria where the vouchers are deposited, from Index Herbariorum [145]. Missing data are indicated by an n-dash (“-”).
(DOCX)
Full description of partitioning schemes and substitution models applied for the five models investigated (1, 2a, 2b, 3, and 4). In the "Subset Contents" field for model 2a, terminal digits refer to codon position: _1= First codon position; _2= Second codon position; _3= Third codon position; _N= Non-coding sequence.
(XLSX)
ApPEFP all-in maximum likelihood transcriptome phylogeny.
(PDF)
CRY all-in maximum likelihood transcriptome phylogeny. a) preduplication CRY3/4, CRY3, and CRY4; b) CRY5, preduplication CRY1/2, and CRY2; c) CRY1, and a cartoon “map” of the entire cryptochrome fern phylogeny.
(PDF)
DET1 all-in maximum likelihood transcriptome phylogeny.
(PDF)
GAP all-in maximum likelihood transcriptome phylogeny. a) gapA, gapB, mystery gap, and gapC; b) gapCp (including Cp Short and Cp Long), and a cartoon map of the GAP family phylogeny.
(PDF)
IBR3 all-in maximum likelihood transcriptome phylogeny.
(PDF)
pgiC all-in maximum likelihood transcriptome phylogeny.
(PDF)
SDQ1 all-in maximum likelihood transcriptome phylogeny.
(PDF)
TPLATE all-in maximum likelihood transcriptome phylogeny.
(PDF)
transducin all-in maximum likelihood transcriptome phylogeny.
(PDF)